Abstract
The bacterial cell-division protein FtsZ has been a promising antibiotic target for over a decade now, but there is still a need for more work in this area. So far there are no FtsZ targeting drugs commercially available. We have analyzed a wide variety of prospective drugs and their interactions with multiple FtsZ species using both free and directed docking simulations. Our goal is to present a standardized computational screening method for potential drug compounds targeting FtsZ. Our work is an example of a way to compare many proposed drugs and FtsZ species combinations relatively quickly. A common method for comparison can yield new results that individual studies and varying methods might not show, as we demonstrate here. To our knowledge this is one of the first, if not the first, computational docking study on the new E. coli FtsZ structures obtained in 2020.
MSC: 0000, 1111
Keywords: FtsZ, Antibiotics, Molecular docking
Graphical abstract
Highlights
-
•
Computational docking study of FtsZ and many proposed drugs
-
•
Includes the new E. coli FtsZ structures obtained in 2020
-
•
Favorable binding observed at many sites on FtsZ, drugs compete with the nucleotides
1. Introduction
After Sir Alexander Fleming received his Nobel prize for the discovery of penicillin, he hypothesized that antibiotic resistance would become a problem as use of penicillin and similar drugs increased. It took 9 years for resistance to develop to tetracycline (introduced in 1950), but only one year for ceftaroline (introduced in 2010) [1]. There are now several infections (MRSA, VRSA, and others) that cannot be adequately treated with current antibiotics, and we are approaching a time when we will have no reliable treatments [2]. In 2019 the World Health Organization listed antimicrobial resistance as one of their top 10 threats to global health [3]. This shows the need for new antibiotic strategies is urgent, and the FtsZ protein is one potential solution.
Filamentous Temperature-sensitive protein Z (FtsZ) is a bacterial protein present in most species (both Gram-positives and Gram-negatives) and is a key component in cell division. By assembling into long protofilaments [4], FtsZ with other associated proteins forms the Z-ring, which then constricts to divide cells [5], [6], [7], [8], [9]. It is hoped that by interfering with FtsZ function and preventing cell division it will be much harder for bacteria to evolve resistance, or to pass any obtained resistance down to daughter cells [10], [11], [12]. Additionally, the homologous eukaryotic protein tubulin has been a successful cancer drug target, typically requiring small doses, which suggests that FtsZ could be drugable likewise [13], [14], [15], [16]. Most current drugs target the interdomain cleft, which is an allosteric site [17], [18], [19], or a discovered PC190273 site near the H7 helix [2], [19], [20], [21], [22], [23]. Any compound that binds in this area could affect the to R state transition and therefore affect the cell division process [24], [25], [26]. Some compounds have also been designed to out-compete the nucleotide, GTP or GDP, to bind at the nucleotide site [27], [28], [29], [30], see Fig. 1. The nucleotide site is occupied in the cell, and the nucleotide binding stabilizes the FtsZ protein, so this binding site is difficult to target. There have also been two cryptic sites discovered [31] that are transient in nature and bind very small compounds.
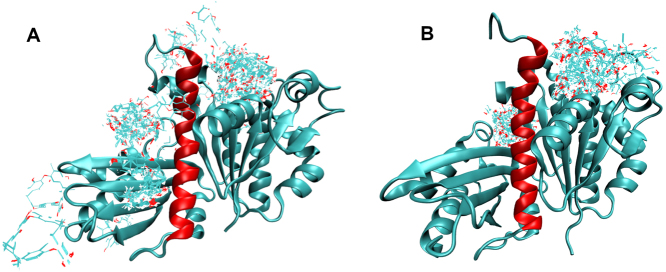
Fig. 1.
View of (A/C) 3wgn and (B/D) 6umk structures showing the H7 helix (red), the T7 loop (dark blue), the flexible loop containing amino acids 71-73 (purple), and the nucleotide binding site (orange) with nucleotide bound (surface representation). Panels C and D are rotated views of A and B, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The current study aims to do several things. First, to use one computational method to investigate many compounds and species of FtsZ. It is difficult to directly compare the results of previous studies due to varying methodologies and a focus on dosage, kinetics, and polymerization results in the literature. In vitro controls are limited due to the inability to grow bacterial cells without FtsZ present, therefore in silico studies are an important addition [32], [33]. Second, we aim to include the newly discovered structures of E. coli FtsZ [34], [35] which were not previously available and provide an important comparison point. Finally, to compare the results of free vs. directed docking simulations. Many past docking studies chose specific sites that were known previously [36], but allowing drug compounds to search the entire protein has yielded surprising results.
Our study includes several controls, including: re-binding GTP and GDP to the nucleotide site, PC190723 binding to the site identified in crystal structures [20], [22] as our positive controls, and Cilagicin-BP, which is an antibiotic compound not associated with FtsZ [37] and has an unfavorable binding energy, as our negative control. We have found that for many drugs and species of FtsZ the preferred binding site from free docking is the nucleotide site. Additionally, most drugs, excluding our negative control and GTP/GDP, show only modest negative G values for directed docking, indicating mildly favorable binding to the selected sites. Taken together, this may be why many current drug families, while showing some promising experimental results [19], [20], [21], [23], [30], [38], [39], [40], do not always have the same effects for in vivo results and/or require very large doses to work. A promising result of our study is that most compounds are capable of favorable binding at multiple sites on the FtsZ protein. This is a good indicator of low resistance potential as it would require multiple mutations to combat binding in bacterial cells.
2. Materials and methods
We ran free and restricted/directed SwissDock [41] docking simulations of 15 proposed drugs with a wide range of formulations against the FtsZ structures from four different bacterial species, both Gram-positive and Gram-negative, see Table 1. Note that we have classified M. tuberculosis as Gram-negative for the purposes of this study, though this classification is debatable [42], [43]. MarvinSketch [44] and UCSF-Chimera [45] were used to draw and prepare the drug compounds for docking. Key structural features of the FtsZ protein are given in Table 2. Free docking allowed the compounds to bind anywhere on the surface of the protein, and the binding energy results are shown in Table 3. Similarly, the key binding location results for free docking are shown in Table 4. Restricted docking sites are identified by the amino acid used as the center of the box, see Table 5. Restricted docking was completed for the most populated cluster and/or the cluster that overlapped with key binding sites from literature and POCASA [46] results. The restricted docking was used to obtain an average G value for that binding location alone, as compared to free docking which gives an average for all results regardless of binding site.
Table 1.
FtsZ species investigated.
| PDB code | Ligand bound | Bacterial species | Classification |
|---|---|---|---|
| 3wgn | GTP | S. aureus | Gram-positive |
| 3vo8 | GDP | S. aureus | Gram-positive |
| 2vam | N/A | B. subtilis | Gram-positive |
| 2vxy | citric acid | B. subtilis | Gram-positive |
| 6unx | GTP | E. coli | Gram-negative |
| 6umk | GDP | E. coli | Gram-negative |
| 5zue | GTP | M. tuberculosis | Gram-negative |
Table 2.
Key structural features of FtsZ with amino acid locations by species. For E. coli the loop containing amino acids 71-73 is shifted and does not interact with GTP/GDP, see Fig. 1.
| Bacterial species | H7 Helix |
|---|---|
| S. aureus | 179-203: MMEAFKEADNVLRQGVQGISDLIAV |
| B. subtilis | 179-203: MLEAFREADNVLRQGVQGISDLIAT |
| E. coli | 178-202: ELDAFGAANDVLKGAVQGIAELITR |
| M. tuberculosis | 176-201: LMDAFRSADEVLLNGVQGITDLITTP |
| Bacterial species | T7 Loop |
| S. aureus | 204-210: SGEVNLD |
| B. subtilis | 204-210: PGLINLD |
| E. coli | 203–209: PGLMNVD |
| M. tuberculosis | 202–207: GLINVD |
| Bacterial species | Nucleotide binding pocket |
| S. aureus | 22-26: GGNNA, 71-73: AGA, |
| 104-110: GMGGGTG, 134-136: RPF, H7 Helix | |
| B. subtilis | 22-26: GGNNA, 71-73: AGA, |
| 104-110: GMGGGTG, 134-136: RPF, H7 Helix | |
| E. coli | 20-25: GGGNA, 70-72: AGA, |
| 104-110: (G)MGGGTGT, 133-135: KPF, H7 Helix | |
| M. tuberculosis | 18-22: GGGVN, 68-70: AGA, |
| 101-106: GEGGGT, 131-133: RPF, H7 Helix | |
Table 3.
FtsZ free docking binding data. Only one cluster was found for the 3wgn/Cilagicin-BP system, so the calculations for the average G for the top cluster and the average G for all results are the same. Only two clusters were found for the 6umk/Cilagicin-BP system, row 1 is the top cluster, row 2 is the second cluster, column 5 is the average of all results.
| FtsZ Code, | Compound | Average full fitness, | Average G, | Average G, |
|---|---|---|---|---|
| Monomers | Top cluster | Top cluster | All results | |
| (kcal/mol) | (kcal/mol) | (kcal/mol) | ||
| 3wgn | GTP | −2231.5 | −17.7 | −13.8 |
| 6umk | GTP | −2389.2 | −15.0 | −12.8 |
| 3wgn | GDP | −2063.0 | −14.7 | −12.1 |
| 6umk | GDP | −2244.6 | −12.8 | −10.7 |
| 3wgn | Cilagicin-BP [37] | 2951.0 | 162.3 | 162.3 |
| 6umk | Cilagicin-BP | −1723.8 | −9.0 | 120.1 |
| 2811.3 | 249.1 | |||
| 3wgn | PC190723 [20] | −1674.1 | −7.7 | −6.9 |
| 5zue | Stokes Compound 1 [38] | −1555.1 | −8.1 | −7.5 |
| 2vam | Stokes Compound 1 | −1556.9 | −8.1 | −7.4 |
| 3vo8 | Stokes Compound 1 | −1593.5 | −8.2 | −7.6 |
| 5zue | Stokes Compound 2 [38] | −1602.6 | −8.7 | −7.9 |
| 2vam | Stokes Compound 2 | −1559.8 | −8.4 | −7.2 |
| 3vo8 | Stokes Compound 2 | −1640.4 | −11.0 | −8.6 |
| 3wgn | TXA709 [55] | −1716.0 | −8.2 | −7.7 |
| 6umk | TXA709 | −1893.6 | −8.5 | −7.9 |
| 3wgn | TXA6101 [56] | −1644.1 | −8.0 | −7.3 |
| 6umk | TXA6101 | −1829.7 | −8.0 | −7.5 |
| 3wgn | TXY6129 [39] | −1636.0 | −8.9 | −7.7 |
| 6umk | TXY6129 | −1816.2 | −8.0 | −7.6 |
| 3wgn | Chrysophaentin A [50] | −1651.7 | −6.7 | −6.7 |
| 6umk | Chrysophaentin A | −1854.6 | −7.3 | −7.1 |
| 6unx | Chrysophaentin A | −1820.5 | −6.6 | −7.0 |
| 5zue | Scopoletin [52] | −1586.3 | −6.2 | −6.0 |
| 5zue | Plumbagin [51] | −1599.1 | −6.4 | −5.9 |
| 6umk | TXY436 [40] | −1940.3 | −8.2 | −7.5 |
| 2vxy | TXY436 | −1655.9 | −8.4 | −8.1 |
| 2vam | TXY436 | −1681.0 | −8.6 | −7.2 |
| 3wgn | Resveratrol [57] | −1718.9 | −7.5 | −6.2 |
| 3wgn | Compound 12 [29] | −1695.3 | −7.1 | −7.0 |
Table 4.
FtsZ free docking binding location(s). Some ligands were able to dock through the flexible loop at the nucleotide binding site of 6umk due to the gap in the crystal structure, which would not be possible in the cell.
| FtsZ code | Compound | Key binding location (s) |
|---|---|---|
| From most to least significant clusters | ||
| 3wgn | GTP | Nucleotide binding site (NBS) |
| 6umk | GTP | NBS |
| 3wgn | GDP | NBS |
| 6umk | GDP | NBS |
| 3wgn | Cilagicin-BP | Pocket formed by amino acids 20, 70, 107, 142 |
| 6umk | Cilagicin-BP | Pocket formed by amino acids 20, 70, 107, 142 |
| 3wgn | PC190723 | NBS; top of H7 helix; |
| PC190723 site from crystal structure | ||
| 5zue | Stokes Compound 1 | NBS; between T7 loop and N-terminus |
| 2vam | Stokes Compound 1 | NBS; interdomain cleft; PC190723 site |
| 3vo8 | Stokes Compound 1 | Left of interdomain cleft; NBS; PC190723 site |
| 3wgn | Stokes Compound 2 | Left of interdomain cleft; NBS; PC190723 site |
| 5zue | Stokes Compound 2 | NBS; between H7 helix, T7 loop, and the N-terminus |
| 2vam | Stokes Compound 2 | NBS; interdomain cleft |
| 3vo8 | Stokes Compound 2 | Left of interdomain cleft; NBS; PC190723 site |
| 3wgn | TXA709 | NBS; pocket formed by amino acids |
| 75, 112-116, 146-157 | ||
| 6umk | TXA709 | NBS; behind interdomain cleft near the C-term |
| 3wgn | TXA6101 | NBS; pocket formed by amino acids |
| 75, 112-116, 146-157 | ||
| 6umk | TXA6101 | NBS; behind interdomain cleft near the C-term |
| 3wgn | TXY6129 | NBS; pocket formed by amino acids |
| 75, 112-116, 146-157 | ||
| 6umk | TXY6129 | NBS; behind interdomain cleft near the C-term |
| 3wgn | Chrysophaentin A | NBS; left of interdomain cleft; PC190723 site |
| 6umk | Chrysophaentin A | NBS; behind interdomain cleft near the C-term |
| 6unx | Chrysophaentin A | NBS |
| 5zue | Scopoletin | NBS; between H7 helix and N-terminus |
| 5zue | Plumbagin | Between H7 helix and N-terminus; NBS; |
| near the T7 loop | ||
| 6umk | TXY436 | NBS; behind interdomain cleft near the C-term |
| 2vxy | TXY436 | H7 helix and interdomain cleft plus |
| pocket formed by amino acids 171, 230, 248; | ||
| pocket formed by amino acids 75, 112-116, 146-157 | ||
| 2vam | TXY436 | NBS; interdomain cleft plus pocket formed by |
| amino acids 171, 230, 248 | ||
| 3wgn | Resveratrol | NBS; pocket formed by amino acids |
| 75, 112-116, 146-157; pocket formed by | ||
| amino acids 171, 230, 248; PC190723 site | ||
| 3wgn | Compound 12 | NBS; PC190723 site; left of interdomain cleft |
Table 5.
FtsZ restricted docking data.
| FtsZ code, | Average full fitness, | Average G, | Average G, |
| Compound | Top cluster | Top cluster | All results |
| (kcal/mol) | (kcal/mol) | (kcal/mol) | |
| 3wgn, Resveratrol | −1713.1 | −6.8 | −5.9 |
| 5zue, Scopoletin | −1581.8 | −6.5 | −5.9 |
| 5zue, Plumbagin | −1596.7 | −6.1 | −5.5 |
| 3wgn, Chrysophaentin A | −1653.2 | −7.1 | −6.5 |
| 3wgn, Compound 12 | −1682.5 | −7.3 | −6.1 |
| FtsZ code, | Box center | Box size | |
| Compound | Amino acid | (X, Y, Z) (Å) | |
| 3wgn, Resveratrol | Val132 | 15, 15, 15 | |
| 5zue, Scopoletin | Phe180 | 10.5, 10.5, 10.5 | |
| 5zue, Plumbagin | Phe180 | 10, 10, 10 | |
| 3wgn, Chrysophaentin A | Gly22 | 15, 15, 15 | |
| 3wgn, Compound 12 | Gly22 | 15, 15, 15 | |
The negative control, Cilagicin-BP, is also an antibiotic compound, but one which is not expected to interact with FtsZ based on the proposed method of action (MOA) [37]. This negative control was done to test the SwissDock algorithm; we expected to see poor binding and unfavorable binding energies for a compound that has no ties to FtsZ. This is confirmed by the strong positive average G value, see Table 3. The binding location is the same for both species of FtsZ with species adjusted residue numbering. Of the two clusters identified (already a poor result compared to the typical approximately 30 clusters for all other compounds), one cluster is negative for 6umk, but the binding location is the same for both clusters and the overall average energy is positive. All of this indicates a poor binding partner, and therefore a suitable negative control.
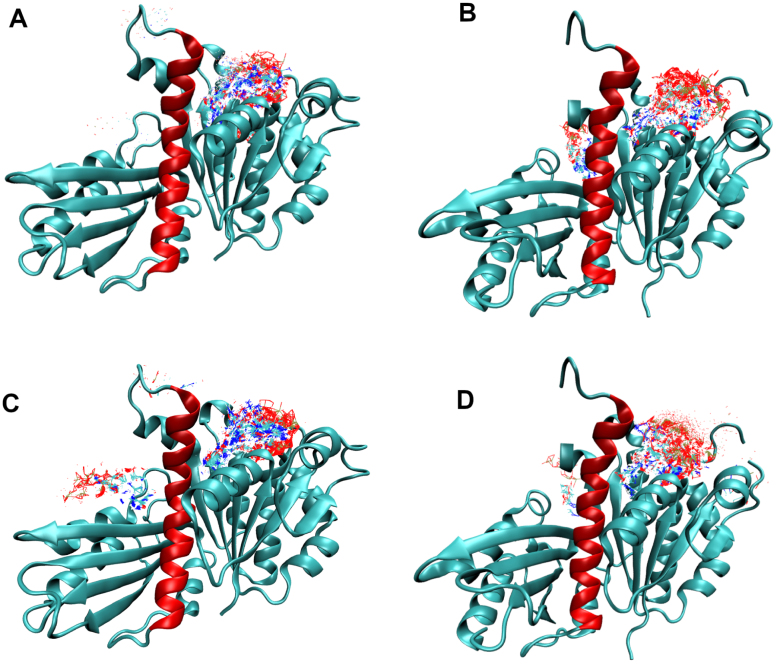
Our main positive controls are the nucleotides GTP and GDP, which are known to bind FtsZ, and indeed appear in many of the crystal structures used here. SwissDock was able to dock both nucleotides nearly exclusively to the nucleotide binding site in free docking simulations. Fig. 2 shows that the vast majority of docked structures are in the correct location, which indicates a suitable positive control and speaks to the reliability of the simulations. If SwissDock was a poor model for this protein we would have expected the nucleotide to bind in multiple locations, as observed for the drug compounds. Our data also show more favorable binding for the nucleotides than for any drug tested, see Table 6 and additional discussion in the results section. Further, we see that GTP has a more favorable binding energy than GDP for both FtsZ structures tested, consistent with experimental results [47].
Fig. 2.
View of the free docking results for the nucleotide structures (positive controls). (A) 3wgn with GDP, (B) 6umk with GDP, (C) 3wgn with GTP, and (D) 6umk with GTP. The clusters are depicted as a composite of all docked results to illustrate that nearly all are found at the nucleotide binding site. The H7 helix is shown in red. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 6.
FtsZ binding energy comparisons: Z-test statistical results.
| Nucleotide pocket restricted | ||
| Drug docking compared to GTP/GDP | ||
| Data Set 1 | Data Set 2 | Calculated P-Value |
| 3wgn-Resveratrol | 3wgn-GTP | 0.0274 |
| 3wgn-Resveratrol | 3wgn-GDP | 0.01 |
| 3wgn-Chrysophaentin A | 3wgn-GTP | 0.01 |
| 3wgn-Chrysophaentin A | 3wgn-GDP | 0.01 |
| 6umk-Chrysophaentin A | 6umk-GTP | 0.01 |
| 6umk-Chrysophaentin A | 6umk-GDP | 0.01 |
| 3wgn-Compound 12 | 3wgn-GTP | 0.01 |
| 3wgn-Compound 12 | 3wgn-GDP | 0.01 |
| 6umk-TXY436 | 6umk-GTP | 0.01 |
| 6umk-TXY436 | 6umk-GDP | 0.01 |
| Restricted drug docking | ||
| Species comparison | ||
| Data Set 1 | Data Set 2 | Calculated P-Value |
| 2vxy-TXY436 | 6umk-TXY436 | 0.01 |
| 3wgn-Chrysophaentin A | 6umk-Chrysophaentin A | 0.2978 |
| Same species nucleotide | ||
| Binding comparison | ||
| Data Set 1 | Data Set 2 | Calculated P-Value |
| 3wgn-GTP | 3wgn-GDP | 0.2183 |
| 6umk-GTP | 6umk-GDP | 0.0268 |
| Different species | ||
| Comparison of nucleotides | ||
| Data Set 1 | Data Set 2 | Calculated P-Value |
| 3wgn-GTP | 6umk-GTP | 0.01 |
| 3wgn-GDP | 6umk-GDP | 0.01 |
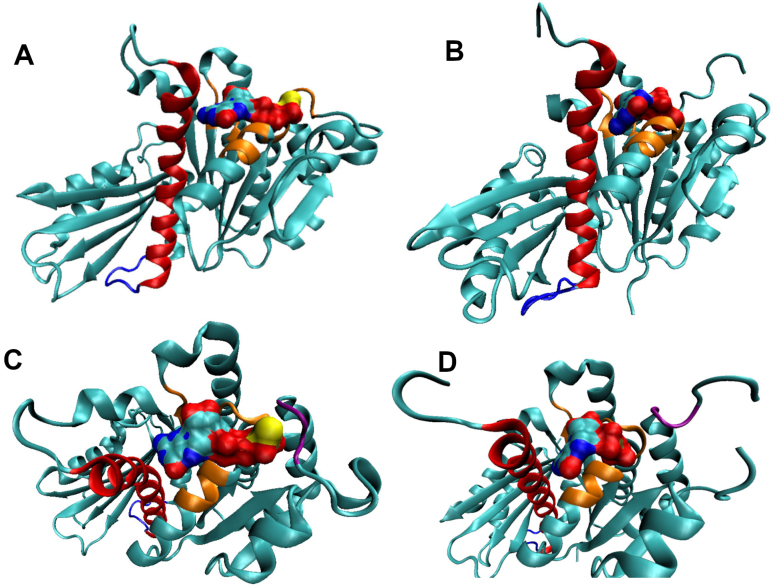
An additional positive control, PC190723, shows clusters near the H7 helix at the site identified by crystal structures [20], [22], see the figure in the SI. The largest cluster is at the GTP/GDP binding site, which is not accessible under the conditions used to obtain the crystal structure. SwissDock does not take small molecules into account during docking, only protein atoms, which is why PC190723 was able to bind at this site during our simulations. Previous simulations [2], [23] also found favorable binding energies for PC190723, on the order of −40 to −50 kcal/mol, similar to our results. Experimentally determined K values of a similar drug, S2727 [23] as well as PC190723 [19] were converted to G values using G = RT ln(K) at 298K; the rough values are G = −8.48 kcal/mol and G = −9.54 kcal/mol, respectively. These values are not strongly favorable, similar to our results of approximately −7 kcal/mol for PC190723.
For PDB structure files that contained more than one FtsZ chain, only one was chosen to run in SwissDock, so all FtsZ structures listed are monomers. When FtsZ is assembled into protofilaments it is usually considered to be in the Tense (T) state. However, when existing as a monomer FtsZ is in the Relaxed (R) state. The structures used here are bound to GTP, for 3wgn, 6unx, and 5zue, or to GDP for 6umk, and 3vo8 as examples [24]. In the T-state the interdomain cleft is open, while in the R-state it is less accessible. The type of docking done here does not allow the protein to be flexible, so the structures are submitted as is from the PDB and do not change during the simulations.
The top cluster is “cluster 0” determined by SwissDock, and contains around 3–15 members depending on the data set. The average G and average full fitness reported for these top clusters are in Table 3. The average G for all results includes the values for all clusters calculated by SwissDock (typically 250 individual results), see exceptions in Table 3.
The Z-test was run on the restricted docking binding energies to compare different species of FtsZ and different drug compounds to the nucleotide binding energies. The full results are given in Table 6, and the full process for obtaining the P-values is given in the SI. Figures and docking location analysis used the Visual Molecular Dynamics (VMD) software [48].
3. Results
All the free docking results show similar negative (favorable) binding energies, and are the same/similar order of magnitude, except for Cilagicin-BP (negative control). The binding energies are also similar for some drugs binding Gram-positive vs. Gram-negative FtsZ, in agreement with previous results [49] that drug effectiveness differences may not be due to the drugs binding FtsZ, but rather to how difficult it is for drugs to enter Gram-negative bacterial cells. The binding energies between the same drug for 3wgn and 6umk are significant in one case but not the other; Chrysophaentin A binds with statistically the same energy to both species (S. aureus vs. E. coli) but TXY436 has a statistically different binding energy between the same two species, see Table 6. This suggests that some of the current drugs might be better candidates for broad-spectrum use than others, if the binding energies could be improved.
The loop containing amino acids 71-73 is part of the GTP/GDP binding pocket for S. aureus, M. tuberculosis and B. subtilis, but not for E. coli FtsZ. In the 6umk structure the loop has moved outward, see Fig. 1. This could account for the statistical species difference observed for binding energies of the nucleotides between species, as SwissDock treats the protein as a static structure. It is not a guarantee that this is true in the cell, due to the flexibility of proteins in general. It is possible that this loop is flexible and has an undefined range of motion between the two extremes shown by the crystal structures of 3wgn and 6umk. The protein could also exhibit a conformational change at this loop from one extreme to the other, as these motions are not often captured by crystal structures. Additionally, there is a small gap in the 3wgn crystal structure, which could contribute to this result. The binding energies between the nucleotides GTP and GDP for the same species are not significantly different to the same threshold of the compounds.
All of the drug compounds are statistically different from the binding energies of GTP/GDP at a threshold -value of 0.01, except for Resveratrol which is statistically different at a threshold -value of 0.05, see Table 6. This indicates that the binding energies of all the drugs tested are not enough to out-compete the nucleotide compounds in order to bind in the pocket. As indicated by our free docking simulations, this is the preferred binding location for most of the compounds. Previous work has found binding of many drugs at the nucleotide site, including Chrysophaentin A determined by NMR [50], and plumbagin [51] and Compound 12 [29] with E. coli FtsZ determined by simulations.
Plumbagin was also found to bind near the H7 helix for B. subtilis FtsZ [51], another cluster location observed here when binding to M. tuberculosis FtsZ. Scopoletin was found by simulations to bind near the T7 loop with a G of −5.06 kcal/mol [52], which agrees with our results. The study of scopoletin also appears to show a correlation between binding energy and the GTPase and/or polymerization disruption effects of the drug [52], in line with our hypothesis. Taken together these results could explain why many in vivo tests have required large dosages of drugs [21], [53], and why only one has shown enough potential to make it to clinical trials yet [39], [54].
4. Discussion
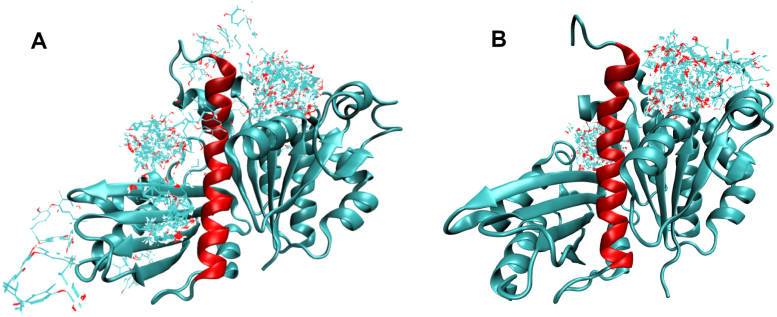
Our results provide evidence for why FtsZ targeting drugs may have proved elusive so far. Many past docking simulations were targeted to known or hypothesized sites, but here we have allowed the compounds to dock freely first and then followed up with restricted docking. In the cell, FtsZ is bound to either GTP or GDP, which is why so many crystal structures feature one or the other bound to the protein. Therefore, a drug hoping to bind to the nucleotide binding pocket would be required to compete with the nucleotides during the swap of a GDP for a new GTP. We have shown that the most common preferred binding location with the largest clusters (i.e. the clusters with the most individual results) for all tested compounds is in fact the nucleotide binding site, see Fig. 3, and further that none of the tested drugs have the binding energy (G) necessary to out-compete the nucleotides. In the cell, the drug compounds would need to bind at one of the other cluster locations, of which several have been identified in Table 4. We also see some clusters at the interdomain cleft, for 3vo8, 2vxy, and 3wgn, in agreement with previous research. More figures of the additional binding sites identified by free docking can be found in the SI.
Fig. 3.
View of (A) 3wgn and (B) 6umk docked with the drug compound Chrysophaentin A. The H7 helix is shown in red. The largest cluster is at the nucleotide binding site for both. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Together this may explain previous observations of large dosage requirements, and the failure to advance to clinical trails for almost all FtsZ targeting drugs so far. One exception is TXA709, developed by Taxis Pharmaceuticals, Inc., which planned to start clinical trials according to a 2022 news release [39]. Our results for this drug do not show a more favorable binding energy for this drug compared to others, so more comparisons will be needed later when the clinical trial results are released. A few runs of the homologous protein tubulin (PDB code 1jff) were run with the compounds TXY436 and PC190723, which resulted in similar G values (−7.56 and −7.22 kcal/mol, respectively) but with fewer binding sites (two sites for each compound). While not conclusive, these results suggest that off-target binding may be possible with some of these compounds. This result is not unexpected due to the homologous nature of FtsZ and tubulin, and is something for future research to consider when looking at drug compounds targeting FtsZ.
One promising result is that most drug compounds do show variable binding sites, which is one way to avoid evolved drug resistance [10], [18]. To prevent all binding interactions with the drug, a bacterium would need to mutate FtsZ at all possible binding sites, which is more difficult than a single point mutation. The drugs all also show overall favorable binding energies. For a few examples, the average G was also calculated for all clusters except the top cluster, to show any variation in binding energies, with no major differences observed. For 2vxy with TXY436 the average of all results without the top cluster was −6.85 kcal/mol, and for 6umk with TXY436 it was −7.13 kcal/mol, both of which are similar to the top cluster average (see Table 3). For 3wgn with Chrysophaentin A the G for all but the top cluster is −6.46 kcal/mol, and for 6umk with the same compound it is −6.23 kcal/mol, again similar to the results shown in Table 3, Table 5. This indicates that the compounds can bind favorably in all identified cluster locations, not only at the top cluster location.
This compares well with previous experiments, including plasma binding AUC assays for Compound 1 [38], TXA709 [55], and PC190723 [58], all of which show over 90% of the drug molecules binding to FtsZ. A new strategy for FtsZ targeting drugs could be to attempt much larger compounds that decorate the surface of the protein and interfere with protofilament formation. These compounds would not be required to target a specific area, in fact larger binding areas would be ideal. Larger binding areas would also avoid nucleotide competition given the size of the binding pocket. Such compounds might require larger doses but would retain the advantage of multiple biding sites. Larger and more non-polar compounds have also been suggested by previous research [10], [37], [56].
5. Conclusions
Our current work is not meant to be a fully exhaustive survey but rather an example of a way to compare many proposed drugs and FtsZ species combinations relatively quickly and cheaply. Other simulations can provide additional information, but take longer to run and require more resources (funding and/or cluster resources). The existing literature has far more studies showing dosage and the kinetic/polymerization effects than binding energy data, and most binding energy data available has been obtained computationally using various types of simulations. This makes direct comparisons between research labs difficult, and highlights the importance of computational work in this area. A single computational method for comparison can yield new results that individual studies and varying methods might not show, as we demonstrated here by using SwissDock to compare the binding locations and energies of proposed drugs to the GDP/GTP nucleotides already known to bind FtsZ.
Future work in our lab will expand to further PDB available FtsZ structures, larger and more non-polar compounds as explained above, and any new proposed drugs made by other labs. We invite anyone working on new drug compounds who would like to have them tested in the same manner as this study to contact our lab.
CRediT authorship contribution statement
Ileini N. Espino: Writing – review & editing, Investigation, Formal analysis. Julia Drolet: Investigation, Data curation. Ty-niquia Jones: Investigation, Data curation. Antonette Uwechue: Investigation, Data curation. Brittany Koehler: Investigation, Data curation. Raquel Beaird: Investigation, Data curation. Sanni Maione: Investigation, Data curation. Christine Darrah: Investigation, Data curation. Rana Hijazi: Investigation, Data curation. Christopher James: Investigation, Data curation. Annabelle Dupre: Investigation, Data curation. Ewa Koscinski: Investigation, Data curation. Leilani Creft: Investigation, Data curation. Michael Giampaolo: Investigation, Data curation. Alexandre Bernier: Investigation, Data curation. Kelly E. Theisen: Writing – review & editing, Writing – original draft, Visualization, Supervision, Resources, Project administration, Methodology, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We wish to acknowledge SUNY-Plattsburgh and the Department of Chemistry and Biochemistry for start-up research funds, lab space, and support throughout this project. We also acknowledge the work of Samara Acevedo, Shaniel Fox, Jheaneel Rose, and Taylor VanWeort, undergraduate researchers, and Adavita Jain, a summer high school student researcher. Additionally, we would like to thank the MDH Cures Community for their resources on SwissDock and associated programs that were extremely helpful.
Footnotes
Supplementary material related to this article can be found online at https://doi.org/10.1016/j.bbrep.2024.101796.
Supplementary information can be found in the file ‘SI_FtsZ_Docking_Theisen.pdf’.
Appendix A. Supplementary data
The following is the Supplementary material related to this article.
Supplementary information can be found in the file ‘SI_FtsZ_Docking_Theisen.pdf’.
Contains statistics calculations, more figures, additional data.
References
- 1.Ventola C. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Therap. 2015;40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 2.Ma Y., Zhang S., Zhou L., Zhang L., Zhang P., Ma S. Exploration of the inhibitory mechanism of PC190723 on FtsZ protein by molecular dynamics simulation. J. Molec. Graph. Model. 2022;114 doi: 10.1016/j.jmgm.2022.108189. [DOI] [PubMed] [Google Scholar]
- 3.. WHO, 2019.
- 4.Löwe J., Amos L. Tubulin-like protofilaments in -induced FtsZ sheets. EMBO J. 1999;18:2364–2371. doi: 10.1093/emboj/18.9.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.den Blaauwen T., Hamoen L., Levin P. The divisome at 25: The road ahead. Curr. Opin. Microbiol. 2017;36:85–94. doi: 10.1016/j.mib.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corbin L., Erickson H. A unified model for treadmilling and nucleation of single-stranded FtsZ protofilaments. Biophys. J. 2020;119:792–805. doi: 10.1016/j.bpj.2020.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huecas S., Ramírez-Aportela E., Vergoñós A., Nùñez-Ramírez R., Llorca O., Díaz J., Juan-Rodr’iguez D., Oliva M., Castellen P., Andreu J. Self-organization of FtsZ polymers in solution reveals spacer role of the disordered C-terminal tail. Biophys. J. 2017;113:1831–1844. doi: 10.1016/j.bpj.2017.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X., Ma S. Design, synthesis and antibacterial activity of cinnamaldehyde derivatives as inhibitors of the bacterial cell division protein FtsZ. Eur. J. Med. Chem. 2015;95:1–15. doi: 10.1016/j.ejmech.2015.04.048. [DOI] [PubMed] [Google Scholar]
- 9.Lock R., Harry E. Cell-division inhibitors: New insights for future antibiotics. Nat. Rev. 2008;7:324–338. doi: 10.1038/nrd2510. [DOI] [PubMed] [Google Scholar]
- 10.Kusuma K., Payne M., Ung A., Bottomley A., Harry E. Ftsz as an antibacterial target: Status and guidelines for progressing this avenue. ACS Infect. Dis. 2019;5:1279–1294. doi: 10.1021/acsinfecdis.9b00055. [DOI] [PubMed] [Google Scholar]
- 11.Silber N., de Opitz C., Mayer C., Sass P. Cell division protein FtsZ: From structure and mechanism to antibiotic target. Fut. Microbiol. 2020;15:801–831. doi: 10.2217/fmb-2019-0348. [DOI] [PubMed] [Google Scholar]
- 12.Tripathy S., Sahu S. Ftsz inhibitors as a new genera of antibacterial agents. Bioorg. Chem. 2019;91 doi: 10.1016/j.bioorg.2019.103169. [DOI] [PubMed] [Google Scholar]
- 13.Nogales E., Downing K., Amos L., Löwe J. Tubulin and FtsZ form a distinct family of GTPases. Nat. Struct. Biol. 1998;5:451–458. doi: 10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]
- 14.Mahendran R., Jenifer F., Palanimuthu M., Subasri S. Sequence and structural analysis of FtsZ homologs and comparison of bacterial FTsZ with eukaryotic tubulins. Ind. J. Sci. Tech. 2011;4:141–146. [Google Scholar]
- 15.den Blaauwen T., Andreu J., Monasterio O. Bacterial cell division proteins as antibiotic targets. Bioorg. Chem. 2014;55:27–38. doi: 10.1016/j.bioorg.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Hurley K., Santos T., Nepomuceno G., Huynh V., Shaw J., Weibel D. Targeting the bacterial division protein FtsZ. J. Med. Chem. 2016;59:6975–6998. doi: 10.1021/acs.jmedchem.5b01098. [DOI] [PubMed] [Google Scholar]
- 17.Pradhan P., Margolin W., Beuria T. Targeting the achilles heel of FtsZ: The interdomain cleft. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.732796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andreu J., Huecas S., Araújo-Bazán L., Vázquez-Villa H., Martín-Fontecha M. The search for antibacterial inhibitors targeting cell division protein FtsZ at its nucleotide and allosteric binding sites. Biomedicines. 2022;10:1825. doi: 10.3390/biomedicines10081825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huecas S., Araújo-Bazán L., Ruiz F., Ruiz-Àvila L., Mart’inez R., Escobar-Peña A., Artola M., Vásquez-Villa H., Martín-Fontecha M., Fernández-Tornero C., López-Rodr’iguez M., Andreu J. Targeting the FtsZ allosteric binding site with a novel fluorescence polarization screen, cytological and structural approaches for antibacterial discovery. J. Med. Chem. 2021;64:5730–5745. doi: 10.1021/acs.jmedchem.0c02207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andreu J., Schaffner-Barbero C., Huecas S., Alonso D., López-Rodríguez M., Ruiz-Avila L., nez Ramírez R.N., Llorca O., Martín-Galiano A. The antibacterial cell division inhibitor PC190723 is an FtsZ polymer-stabilizing agent that induces filament assembly and condensation. J. Biol. Chem. 2010;285:14239–14246. doi: 10.1074/jbc.M109.094722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elsen N., Lu J., Parthasarathy G., Reid J., Sharma S., Soisson S., Lumb K. Mechanism of action of the cell-division inhibitor PC190723: Modulation of FtsZ assembly cooperativity. J. Am. Chem. Soc. 2012;134:12342–12345. doi: 10.1021/ja303564a. [DOI] [PubMed] [Google Scholar]
- 22.Matsui T., Yamane J., Mogi N., Yamaguchi H., Takemoto H., Yao M., Tanaka I. Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus. Acta. Crystallogr. D. Biol. Crystallogr. 2012;D68:1175–1188. doi: 10.1107/S0907444912022640. [DOI] [PubMed] [Google Scholar]
- 23.Du R., Sun N., Fung Y., Zheng Y., Chen Y., Chan P., Wong W., Wong K. Discovery of FtsZ inhibitors by virtual screening as antibacterial agents and study of the inhibition mechanism. RSC Med. Chem. 2022;13:79–89. doi: 10.1039/d1md00249j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsui T., Han X., Yu J., Yao M., Tanaka I. Structural change in FtsZ induced by intermolecular interactions between bound GTP and the T7 loop. J. Biol. Chem. 2014;289:3501–3509. doi: 10.1074/jbc.M113.514901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Battaje R., Bhondwe P., Dhaked H., Panda D. Evidence of conformational switch in Streptococcus pneumoniae FtsZ during polymerization. Prot. Sci. 2021;30:523–530. doi: 10.1002/pro.4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruiz F., Huecas S., Santos-Aledo A., Prim E., Andreu J., Fernández-Tornero C. Ftsz filament structures in different nucleotide states reveal the mechanism of assembly dynamics. PLOS Biol. 2022;3001497:1–22. doi: 10.1371/journal.pbio.3001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Artola M., Ruiz-Avila L., Vergoñós A., Huecas S., Araújo-Bazán L., Martín-Fontecha M., Vázquez-Villa H., Turrado C., Ramírez-Aportela E., Hoegl A., Nodwell M., Barasoain I., Chacóon P., Sieber S., Andreu J., López-Rodríguez M. Effective GTP-replacing FtsZ inhibitors and antibacterial mechanism of action. ACS Chem. Biol. 2014;10:834–843. doi: 10.1021/cb500974d. [DOI] [PubMed] [Google Scholar]
- 28.Läppchen T., Pinas V., Hartog A., Koomen G., Schaffner-Barbero C., Andreu J., Trambaiolo D., Lówe J., Juhem A., Popov A. Probing FtsZ and tubulin with C8-substituted GTP analogs reveals differences in their nucleotide binding sites. Chem. Biol. 2008;15:189–199. doi: 10.1016/j.chembiol.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 29.Chan F., Sun N., Neves M., Lam P., Chung W., Wong L., Chow H., Ma D., Chan P., Leung Y., Chan T., Abagyan R., Wong K. Identification of a new class of FtsZ inhibitors by structrue-based design and in vitro screening. J. Chem. Inform. Model. 2013;53:2131–2140. doi: 10.1021/ci400203f. [DOI] [PubMed] [Google Scholar]
- 30.Chan F., Sun N., Leung Y., Wong K. Antimicrobial activity of a quinuclidine-based FtsZ inhibitor and its synergistic potential with -lactam antibiotics. J. Antibiot. 2015;68:253–258. doi: 10.1038/ja.2014.140. [DOI] [PubMed] [Google Scholar]
- 31.Alnami A., Norton R., Pena H., Haider S., Kozielski F. Conformational flexibility of A highly conserved helix controls cryptic pocket formation in FtsZ. J. Molec. Biol. 2021;433 doi: 10.1016/j.jmb.2021.167061. [DOI] [PubMed] [Google Scholar]
- 32.Anderson A. The process of structure-based drug design. Chem. Biol. 2003;10:787–797. doi: 10.1016/j.chembiol.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Haranahalli K., Tong S., Ojima I. Recent advances in the discovery and development of antibacterial agents targeting the cell-division protein FtsZ. Bioorg. Med. Chem. 2016;24:6354–6369. doi: 10.1016/j.bmc.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schumacher M., Ohashi T., Corbin L., Erickson H. High-resolution crystal structures of Escherichia coli FtsZ bound to GDP and GTP. Acta Cryst. 2020;F76:94–102. doi: 10.1107/S2053230X20001132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshizawa T., Fujita J., Terakado H., Ozawa M., Kuroda N., Tanaka S., Uehara R., Matsumura H. Crystal structures of the cell-division protein FtsZ from Klebsiella pneumoniae and Escherichia coli. Acta Crystallogr. F. Struct. Biol. Commun. 2020;F76:86–93. doi: 10.1107/S2053230X2000076X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Casiraghi A., Suigo L., Valoti E., Straniero V. Targeting bacterial cell division: A binding site-centered approach to the most promising inhibitors of the essential protein FtsZ. Antibiot. 2020;9:69. doi: 10.3390/antibiotics9020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z., Koirala B., Hernandez Y., Zimmerman M., Brady S. Bioinformatic prospecting and synthesis of a bifunctional lipopeptide antibiotic that evades resistance. Science. 2022;376:991–996. doi: 10.1126/science.abn4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stokes N., Baker N., Bennett J., Berry J., Collins I., Czaplewski L., Logan A., Macdonald R., MacLeod L., Peasley H., Mitchell J., Nayal N., Yadav A., Srivastava A., Haydon D. An improved small-molecule inhibitor of FtsZ with superior in vitro potency, drug-like properties, and in vivo efficacy. Antimicrob. Agents. Chemother. 2013;57:317–325. doi: 10.1128/AAC.01580-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosado-Lugo J., Sun Y., Banerjee A., Cao Y., Datta P., Zhang Y., Yuan Y., Parhi A. Evaluation of 2,6-difluoro-3-(oxazol-2-ylmethoxy)benzamide chemotypes as gram-negative FtsZ inhibitors. J. Antibiot. 2022;75:385–395. doi: 10.1038/s41429-022-00531-9. [DOI] [PubMed] [Google Scholar]
- 40.Kaul M., Mark L., Zhang Y., Parhi A., LaVoie E., Pilch D. An FtsZ-targeting prodrug with oral antistaphylococcal efficacy in vivo. Antimicrob. Ag. Chemother. 2013;57:5860–5869. doi: 10.1128/AAC.01016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grosdidier A., Zoete V., Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nuc. Acids Res. 2011;39:270–277. doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu L., Fu-Liu C. Is mycobacterium tuberculosis a closer relative to gram-positive or gram-negative bacterial pathogens? Tuberculosis. 2002;82:85–90. doi: 10.1054/tube.2002.0328. [DOI] [PubMed] [Google Scholar]
- 43.Ferrer-González E., Fujita J., Yoshizawa T., Nelson J., Pilch A., Hillman E., Ozawa M., Kuroda N., Al-Tameemi H., Boyd J., LaVoie E., Matsumura H., Pilch D. Structure-guided design of a fluorescent probe for the visualization of FtsZ in clinically important gram-positive and gram-negative bacterial pathogens. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-56557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marvin E., 21.4.6 J. 2022. Marvin was used for drawing, displaying and characterizing chemical structures, substructures and reactions. ChemAxon. ( http://www.chemaxon.com) [Google Scholar]
- 45.Petersen E., Goddard T., Huang C., Couch G., Greenblatt D., Meng E., Ferrin T. UCSF chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 46.Yu J., Zhou Y., Tanaka I., Yao M. Roll: A new algorithm for the detection of protein pockets and cavities with a rolling probe sphere. Bioinform. 2010;26:46–52. doi: 10.1093/bioinformatics/btp599. [DOI] [PubMed] [Google Scholar]
- 47.Scheffers D., den Blaauwen T., Driessen A. Non-hydrolysable GTP--S stabilizes the FtsZ polymerin a GDP-bound state. Molec. Microbiol. 2000;35:1211–1219. doi: 10.1046/j.1365-2958.2000.01791.x. [DOI] [PubMed] [Google Scholar]
- 48.Humphrey W., Dalke A., Schulten K. VMD – Visual molecular dynamics. J. Molec. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 49.Ruiz-Avila L., Huecas S., Artola M., Vergoñós A., Ramírez-Aportela E., Cercenado E., Barasoain I., Vázquez-Villa H., Martín-Fontecha M., Chacón P., López-Rodríguez M., Andreu J. Synthetic inhibitors of bacterial cell division targeting the GTP-binding site of FtsZ. ACS Chem. Biol. 2013;8:2072–2083. doi: 10.1021/cb400208z. [DOI] [PubMed] [Google Scholar]
- 50.Plaza A., Keffer J., Bifulco G., Lloyd J., Bewley C. Chrysophaentins A-H, antibacterial bisdiarylbutene macrocycles that inhibit the bacterial cell division protein FtsZ. J. Amer. Chem. Soc. 2010;132:9069–9077. doi: 10.1021/ja102100h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhattacharya A., Jindal B., Singh P., Datta A., Panda D. Plumbagin inhibits cytokinesis in bacillus subtilis by inhibiting FtsZ assembly – A mechanistic study of its antibacterial activity. FEBS J. 2013;280:4585–4599. doi: 10.1111/febs.12429. [DOI] [PubMed] [Google Scholar]
- 52.Duggirala S., Nankar R., Rajendran S., Doble M. Phytochemicals as inhibitors of bacterial cell division protein FtsZ: Coumarins are promising candidates. Appl. Biochem. Biotech. 2014;174:283–296. doi: 10.1007/s12010-014-1056-2. [DOI] [PubMed] [Google Scholar]
- 53.Lepak A., Parhi A., Madison M., Marchillo K., VanHecker J., Andes D. In vivo pharmacodynamic evaluation of an FtsZ inhibitor, TXA-709, and its active metabolite, TXA-707, in a murine neutropenic thigh infection model. Antimicrob. Ag. Chemother. 2015;59:6568–6574. doi: 10.1128/AAC.01464-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Butler M., Paterson D. Antibiotics in the clinical pipeline in October 2019. J. Antibiot. 2020;73:329–364. doi: 10.1038/s41429-020-0291-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaul M., Mark L., Zhang Y., Parhi A., Lyu Y., Pawlak J., Saravolatz S., Saravoltaz L., Weinstein M., LaVoie E., Pilch D. TXA709, an FtsZ-targeting benzamide prodrug with improved pharmacokinetics and enhanced in vivo efficacy against methicillin-resistant Staphylococcus aureus. Antimicrob. Ag. Chemother. 2015;59:4845–4855. doi: 10.1128/AAC.00708-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fujita J., Maeda Y., Mizohata E., Inoue T., Kaul M., Parhi A., LaVoie E., Pilch D., Matsumura H. Structural flexibility of an inhibitor overcomes drug resistance mutations in Staphylococcus aureus FtsZ. ACS Chem. Biol. 2017;12:1947–1955. doi: 10.1021/acschembio.7b00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hwang D., Lim Y.-H. Resveratrol antibacterial activity against Escherichia coli is mediated by Z-ring formation inhibition via suppression of FtsZ expression. Sci. Rep. 2015;5:10029. doi: 10.1038/srep10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haydon D., Bennett J., brown D., Collins I., Galbraith G., Lancett P., Macdonald R., Stokes N., Chauhan P., Sutariya J., Nayal N., Srivastava A., Beanland J., Hall R., Henstock V., Noula C., Rockley C., Czaplewski L. Creating an antibacterial with in vivo efficacy: Synthesis and characterization of potent inhibitors of the bacterial cell division protein FtsZ with improved pharmaceutical properties. J. Med. Chem. 2010;53:3927–3936. doi: 10.1021/jm9016366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Contains statistics calculations, more figures, additional data.