Abstract
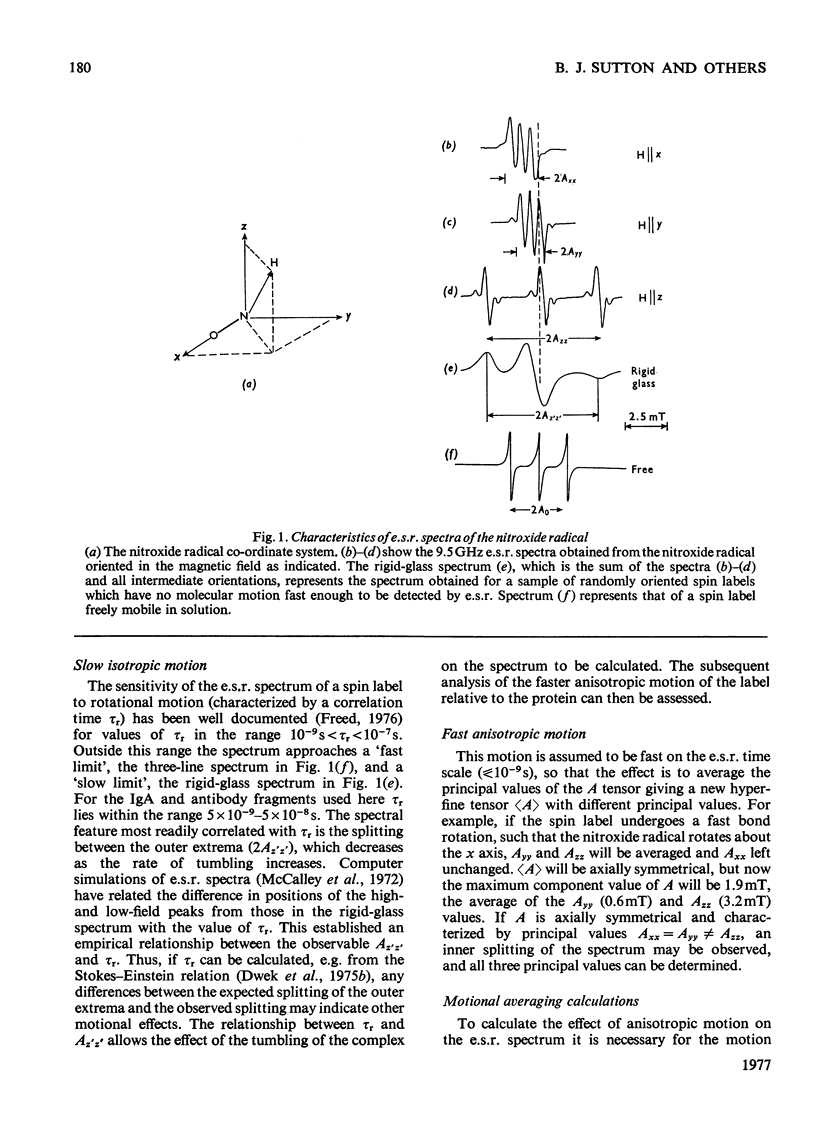
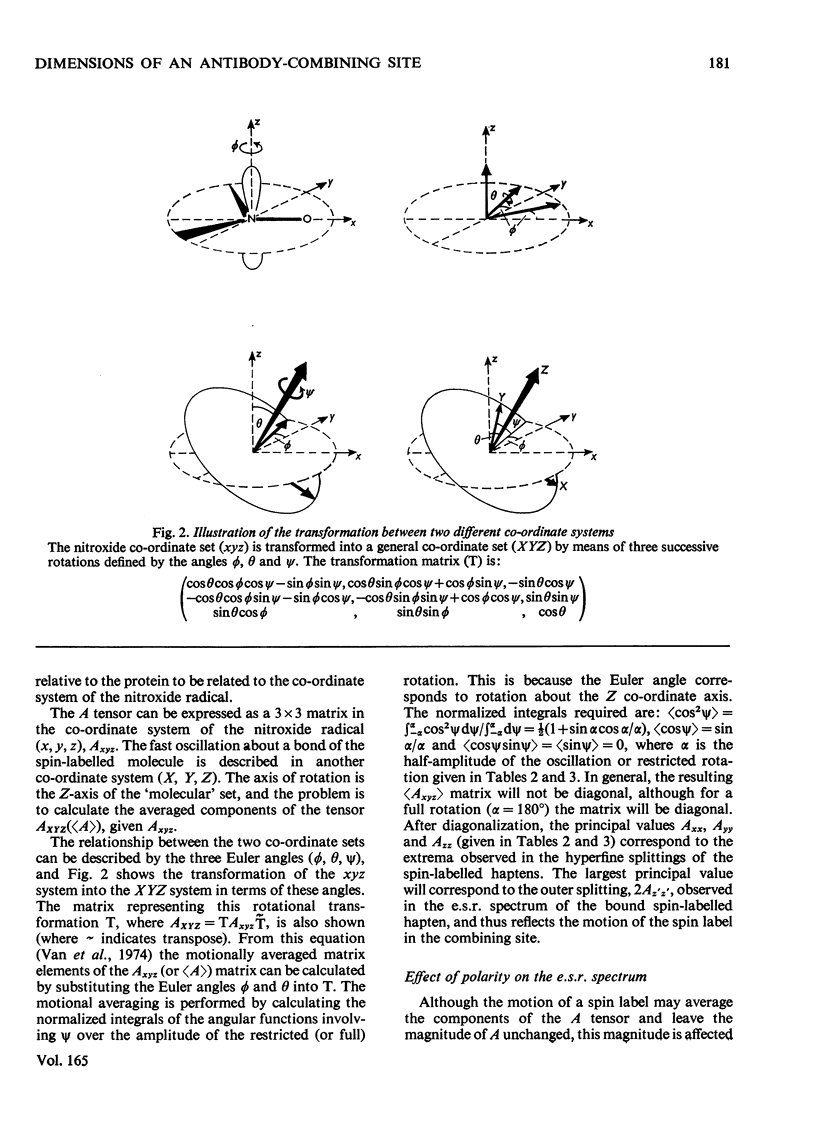
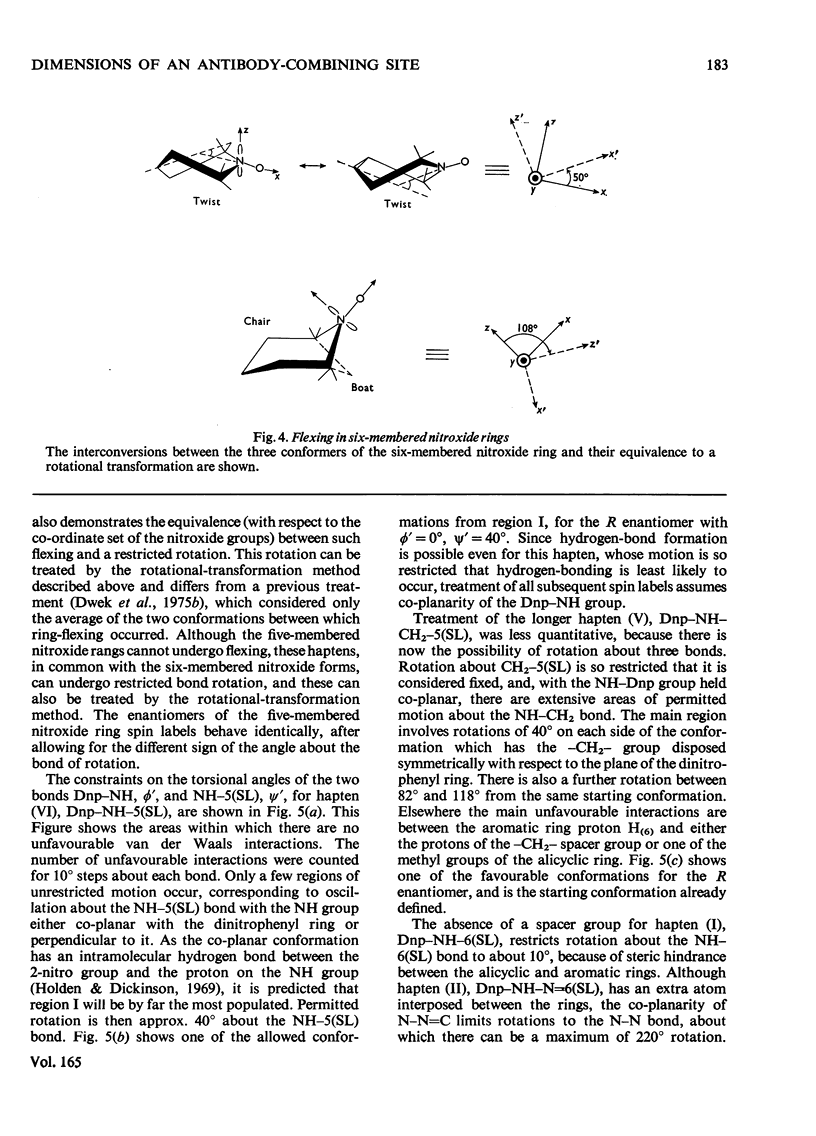
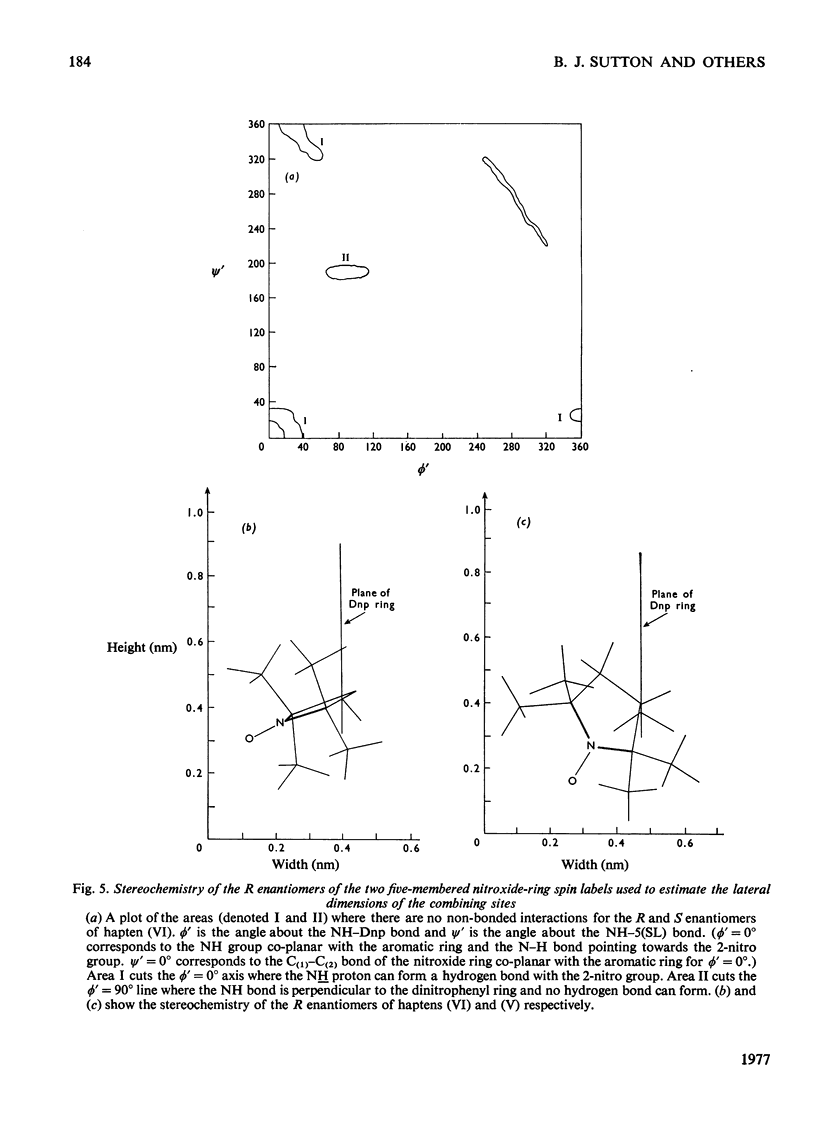
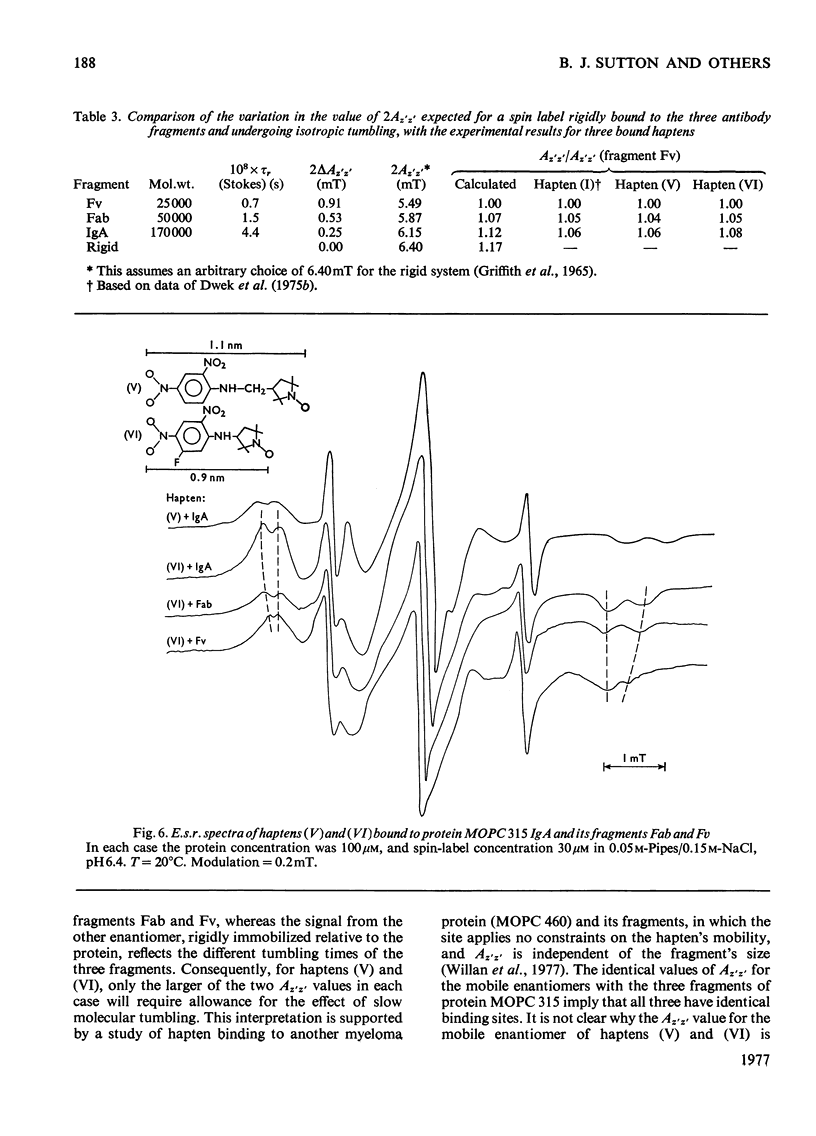
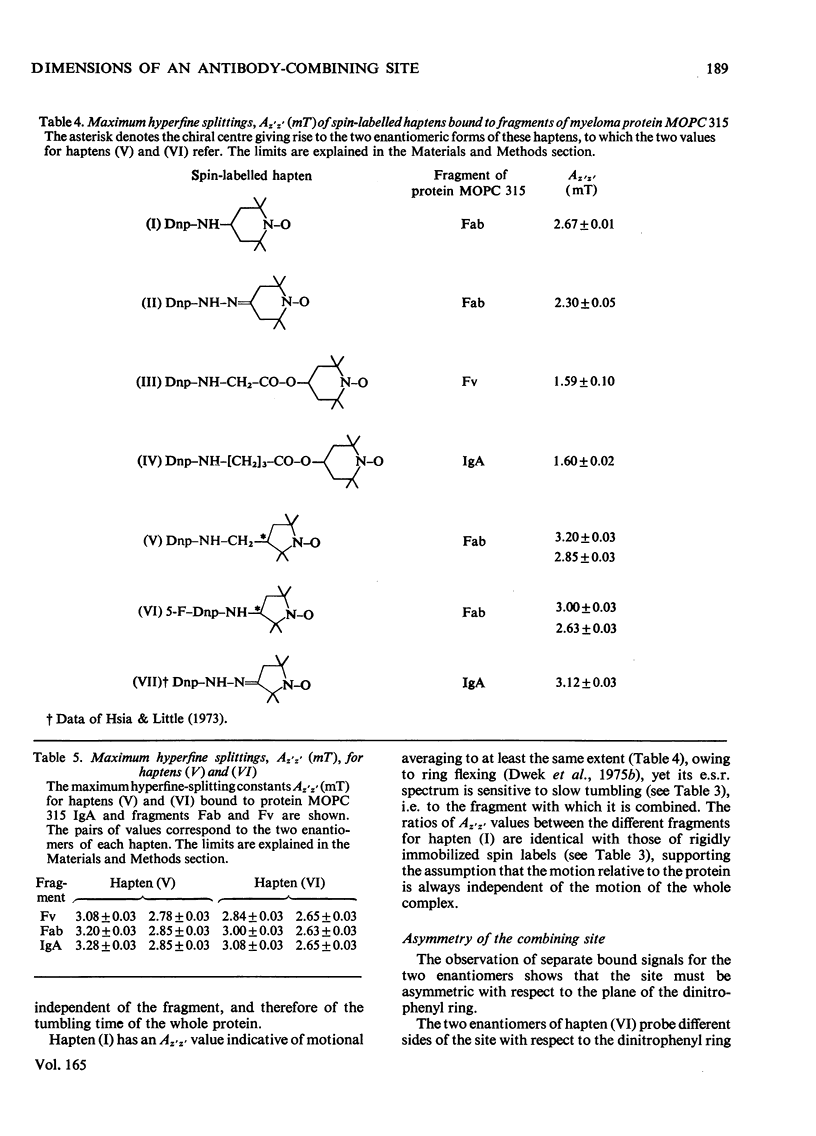
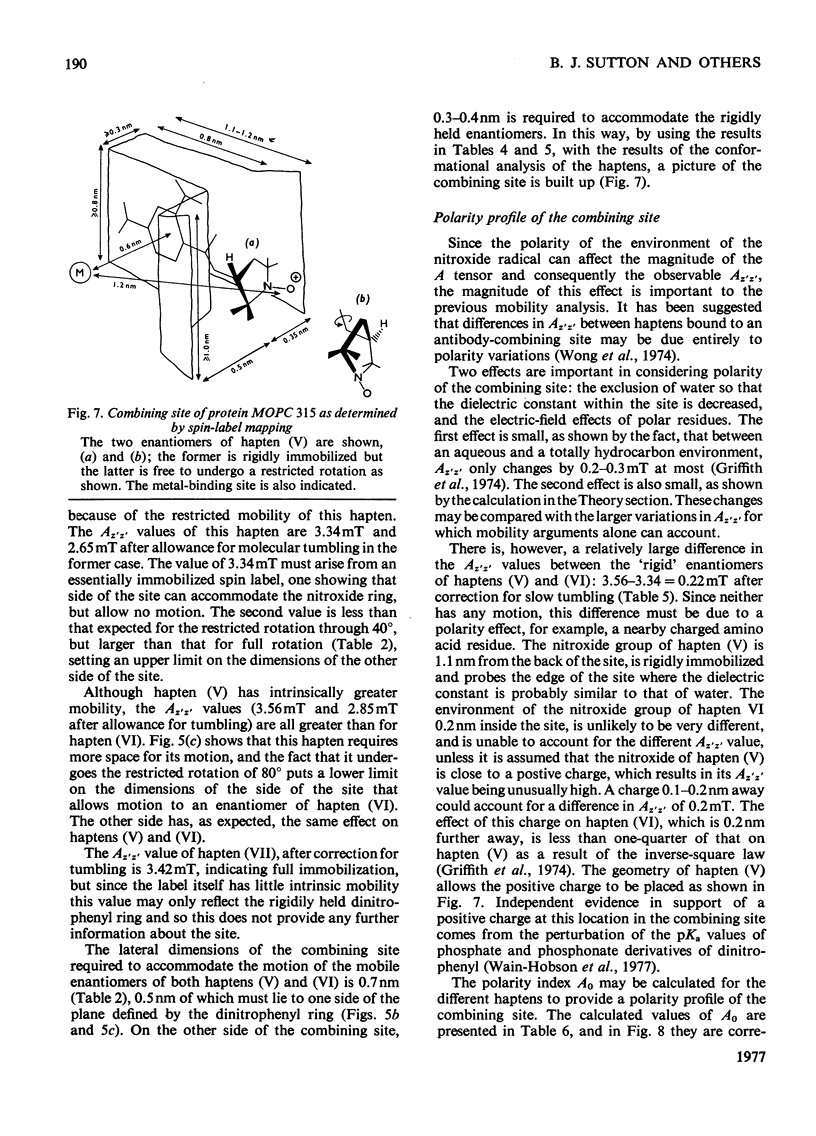
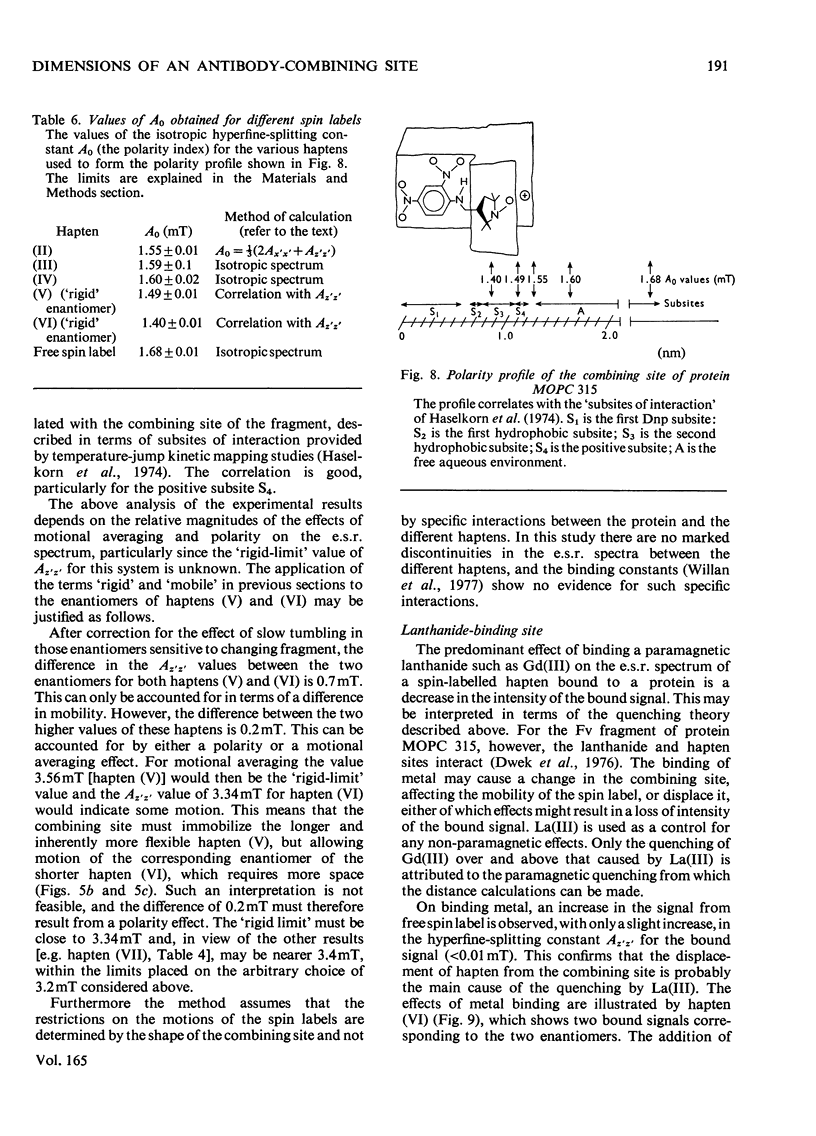
1. A series of Dnp (dinitrophenyl) nitroxide spin labels was used to map the dimensions of the combining site of the Dnp-binding immunoglobulin A myeloma protein MOPC 315. The method compares the observed e.s.r. (electron-spin-resonance) hyperfine splittings with those calculated on the basis of different postulated motions for the spin label. The analysis is complicated by the sensitivity of the e.s.r. hyperfine splitting to the overall `tumbling' time of the antibody–hapten complex and the polarity of the spin-label's environment. When these effects are considered quantitatively, it is then possible to determine the degree of mobility of each hapten which is allowed by the shape of the combining site. 2. The dinitrophenyl ring is rigidly held, and the depth of the site is 1.1–1.2nm and has lateral dimensions at the entrance to the site ≥0.6nm×0.9nm. The analysis of the results for spin-labelled haptens with chiral centres allows these lateral dimensions to be refined to 0.8nm and 1.1nm, and it is shown that the site is asymmetric with respect to the plane of the dinitrophenyl ring. 3. A polarity profile of the combining site was also obtained and a positively charged amino acid residue, possibly arginine-95L (light chain), was located at the entrance to the site. 4. The binding of Gd(III) to the antibody–hapten complexes results in quenching of the e.s.r. signal of the nitroxide. By using La(III) as a control, the paramagnetic contribution to the quenching is measured. 5. Analysis of the differential quenchings of the enantiomers of two five-membered nitroxide ring spin labels gives two possible locations of the metal-binding site. One of these is equidistant (0.7nm) from each of the three dinitrophenyl aromatic protons, and nuclear-magnetic-resonance relaxation studies, at 270MHz, on solutions of dinitrobenzene, Gd(III) and the Fv fragment (variable region of heavy and light chain) from protein MOPC 315 support this location for the metal site. 6. The e.s.r. and metal-binding data were then compared with the results of a model of the combining site constructed on the basis of framework invariance in immunoglobulins [Padlan, Davies, Pecht, Givol & Wright (1976) Cold Spring Harbor Symp. Quant. Biol. 41, in the press]. The overall agreement is very good. Assignments of possible chelating groups for the metal can be made.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burton D. R., Forsén S., Karlström G., Dwek R. A., McLaughlin A. C., Wain-Hobson S. Difficulties in determining accurate molecular motion parameters from proton relaxation enhancement measurements as illustrated by the immunoglobulin G-Gd(III) system. Eur J Biochem. 1976 Dec 11;71(2):519–528. doi: 10.1111/j.1432-1033.1976.tb11140.x. [DOI] [PubMed] [Google Scholar]
- Dower S. K., Wain-Hobson S., Gettins P., Givol D., Jackson W. R., Perkins S. J., Sunderland C. A., Sutton B. J., Wright C. E., Dwek R. A. The combining site of the dinitrophenyl-binding immunoglobulin A myeloma protein MOPC 315. Biochem J. 1977 Aug 1;165(2):207–223. doi: 10.1042/bj1650207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwek R. A., Givol D., Jones R., McLaughlin A. C., Wain-Hobson S., White A. I., Wright C. Interactions of the lanthanide- and hapten-binding sites in the Fv fragment from the myeloma protein MOPC 315. Biochem J. 1976 Apr 1;155(1):37–53. doi: 10.1042/bj1550037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwek R. A., Jones R., Marsh D., McLaughlin A. C., Press E. M., Price N. C., White A. I. Antibody--hapten interactions in solution. Philos Trans R Soc Lond B Biol Sci. 1975 Nov 6;272(915):53–74. doi: 10.1098/rstb.1975.0070. [DOI] [PubMed] [Google Scholar]
- Dwek R. A. Structural studies in solution on the combining site of the myeloma protein MOPC 315. Contemp Top Mol Immunol. 1977;6:1–52. doi: 10.1007/978-1-4684-2841-4_1. [DOI] [PubMed] [Google Scholar]
- Griffith O. H., Dehlinger P. J., Van S. P. Shape of the hydrophobic barrier of phospholipid bilayers (evidence for water penetration in biological membranes). J Membr Biol. 1974;15(2):159–192. doi: 10.1007/BF01870086. [DOI] [PubMed] [Google Scholar]
- Haselkorn D., Friedman S., Givol D., Pecht I. Kinetic mapping of the antibody combining site by chemical relaxation spectrometry. Biochemistry. 1974 May 7;13(10):2210–2222. doi: 10.1021/bi00707a030. [DOI] [PubMed] [Google Scholar]
- Hochman J., Inbar D., Givol D. An active antibody fragment (Fv) composed of the variable portions of heavy and light chains. Biochemistry. 1973 Mar 13;12(6):1130–1135. doi: 10.1021/bi00730a018. [DOI] [PubMed] [Google Scholar]
- Hsia J. C., Little J. R. Structural properties of the ligand binding sites of murine myeloma proteins. FEBS Lett. 1973 Apr 1;31(1):80–84. doi: 10.1016/0014-5793(73)80077-8. [DOI] [PubMed] [Google Scholar]
- Hsia J. C., Piette L. H. Spin-labeling as a general method in studying antibody active site. Arch Biochem Biophys. 1969 Jan;129(1):296–307. doi: 10.1016/0003-9861(69)90179-9. [DOI] [PubMed] [Google Scholar]
- Inbar D., Hochman J., Givol D. Localization of antibody-combining sites within the variable portions of heavy and light chains. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2659–2662. doi: 10.1073/pnas.69.9.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klostergaard J., Grossberg A. L., Krausz L. M., Pressman D. Absence of lysine from the DNP-lysine binding site of protein 315; designation of lysine 52 of the heavy chain as a peripheral residue. Immunochemistry. 1977 Jan;14(1):37–44. doi: 10.1016/0019-2791(77)90331-7. [DOI] [PubMed] [Google Scholar]
- Klostergaard J., Krausz L. M., Grossberg A. L., Pressman D. Arginine as a contact residue in the hapten-binding site of protein 315. Immunochemistry. 1977 Feb;14(2):107–110. doi: 10.1016/0019-2791(77)90288-9. [DOI] [PubMed] [Google Scholar]
- Poljak R. J. X-ray diffraction studies of immunoglobulins. Adv Immunol. 1975;21:1–33. [PubMed] [Google Scholar]
- Shimshick E. J., McConnell H. M. Rotational correlation time of spin-labeled alpha-chymotrypsin. Biochem Biophys Res Commun. 1972 Jan 14;46(1):321–327. doi: 10.1016/0006-291x(72)90665-1. [DOI] [PubMed] [Google Scholar]
- Stryer L., Griffith O. H. A spin-labeled hapten. Proc Natl Acad Sci U S A. 1965 Dec;54(6):1785–1791. doi: 10.1073/pnas.54.6.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. S., Leigh J. S., Jr, Cohn M. Magnetic resonance studies of spin-labeled creatine kinase system and interaction of two paramagnetic probes. Proc Natl Acad Sci U S A. 1969 Sep;64(1):219–226. doi: 10.1073/pnas.64.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wain-Hobson S., Dower S. K., Gettins P., Givol D., McLaughlin A. C., Pecht I., Sunderland C. A., Dwek R. A. Specificity of interactions of hapten side chains with the combining site of the myeloma protein MOPC 315. Biochem J. 1977 Aug 1;165(2):227–235. doi: 10.1042/bj1650227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willan K. J., Marsh D., Sunderland C. A., Sutton B. J., Wain-Hobson S., Dwek R. A., Givol D. Comparison of the dimensions of the combining sites of the dinitrophenyl-binding immunoglobulin A myeloma proteins MOPC 315, MOPC 460 and XRPC 25 by spin-label mapping. Biochem J. 1977 Aug 1;165(2):199–206. doi: 10.1042/bj1650199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L. T., Peitte L. H., Little J. R., Hsia J. C. Stereospecificity of murine myeloma protein-315 to enantiomeric spin labeled dinitrophenyl hapten. Immunochemistry. 1974 Jul;11(7):377–379. doi: 10.1016/0019-2791(74)90191-8. [DOI] [PubMed] [Google Scholar]


