Abstract
Homologous recombination (HR)-based gene therapy using adeno-associated viruses (AAV-HR) without nucleases has several advantages over classic gene therapy, especially the potential for permanent transgene expression. However, the low efficiency of AAV-HR remains a major limitation. Here, we tested a series of small-molecule compounds and found that ribonucleotide reductase (RNR) inhibitors substantially enhance AAV-HR efficiency in mouse and human liver cell lines approximately three-fold. Short-term administration of the RNR inhibitor fludarabine increased the in vivo efficiency of both non-nuclease- and CRISPR/Cas9-mediated AAV-HR two- to sevenfold in the murine liver, without causing overt toxicity. Fludarabine administration induced transient DNA damage signaling in both proliferating and quiescent hepatocytes. Notably, the majority of AAV-HR events occurred in non-proliferating hepatocytes in both fludarabine-treated and control mice, suggesting that the induction of transient DNA repair signaling in non-dividing hepatocytes was responsible for enhancing AAV-HR efficiency in mice. These results suggest that use of a clinically approved RNR inhibitor can potentiate AAV-HR-based genome-editing therapeutics.
Main
Recombinant AAVs are highly popular gene transfer vectors1,2. Despite their widespread use, classic gene replacement approaches are not suitable to treat newborn or pediatric populations for some target organs. Specifically, in the liver, hepatocyte proliferation and turnover dilutes vector episomes, eliminating therapeutic expression during growth and development3–5. To overcome the challenge of treating pediatric populations, integrative vectors may achieve lifelong therapeutic gene expression. Correcting a disease early in a individual’s life may prevent disease progression, secondary complications or death.
As an alternative to gene replacement, we have developed a promoterless recombinant AAV (rAAV)-based gene-targeting strategy referred to as ‘GeneRide’ or AAV-HR6. This gene-editing technology targets the 3′ coding region of a highly expressed gene, such as albumin (Alb), using homology arms surrounding a self-cleaving peptide (P2A) and a transgene. Successful gene targeting via HR results in conversion of the native mRNA to a fusion mRNA (Alb-P2A-transgene) and translation of the endogenous and transgene proteins. This technology has shown permanent correction in animal models of several human diseases6,7.
Despite its effectiveness as an experimental therapy, the overall rate of AAV-HR remains low. Furthermore, the molecular mechanism of such gene targeting is not fully understood. Here, we identify a small-molecule drug that can be administered transiently and safely to enhance gene editing in developing somatic mammalian tissue.
Results
RNR inhibition enhances gene targeting in cultured cells.
Because increased vector copy numbers might improve AAV-HR, we first evaluated compounds previously reported to enhance standard rAAV transduction, confirming their transduction-improving properties8–16 (Supplementary Table 1 and Supplementary Fig. 1a). We then used a promoterless vector targeting the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) locus for HR, designated rAAVDJ-GAPDH-P2A-GFP, to evaluate if these drugs could also increase AAV-HR efficiency. This vector expresses green fluorescent protein (GFP) following on-target HR, allowing fluorescence-activated cell sorting (FACS) quantification of HR events17 (Fig. 1a). As rAAVDJ-GAPDH-P2A-GFP lacks a promoter and start codon at the N terminus of GFP, there is little functional expression from episomal rAAV genomes; as such, detection of GFP+ cells following transduction at a high multiplicity of infection (20,000 viral genomes per cell) in vitro was near the limit of detection (Extended Data Fig. 1a). However, when treated with hydroxyurea (HU), cells showed a three- to fourfold increase in GFP+ cells (Fig. 1b), whereas most of other compounds failed to increase the GFP+ fraction to the extent of HU (Extended Data Fig. 1a).
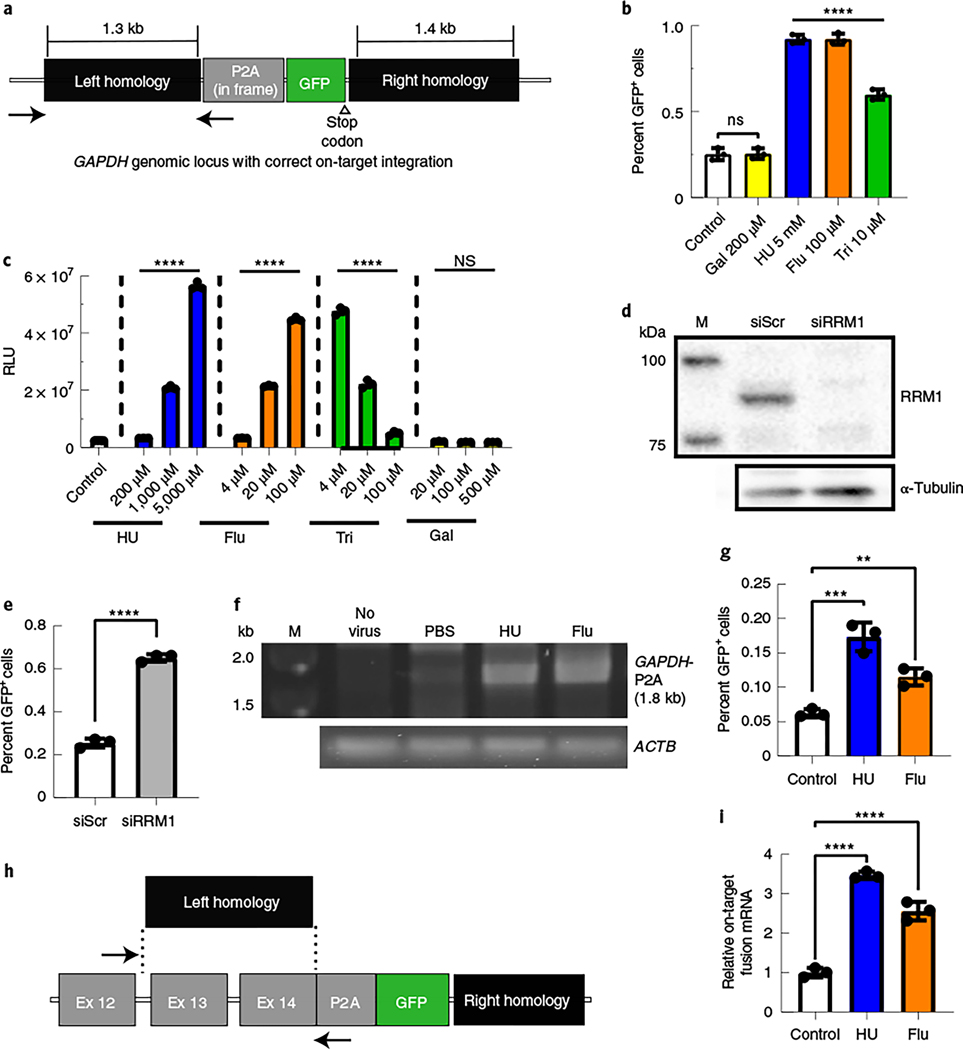
Fig. 1 |. Inhibition of RNR increased the efficiency of gene targeting in human and mouse cell lines.
a, The GAPDH genomic locus after on-target HR with the rAAVDJ-GAPDH-P2A-GFP gene-targeting vector. The positions of in and out PCR primers, to detect site-specific integration, are indicated by black arrows. b, Huh7 cells were treated with RNR inhibitors and transduced with the gene-targeting vector rAAVDJ-GAPDH-P2A-GFP. Flow cytometry analysis of GFP+ fractions was performed after 14 d; Flu, fludarabine; Tri, triapine; Gal, gallium nitrate. Bars represent the group mean, and error bars represent s.d.; n = 3 biological replicate wells. Significance was determined by one-way analysis of variance (ANOVA) with Dunnett’s test for multiple comparisons. P values of all groups were <0.0001, except Gal, which was 0.9998. c, Huh7 cells were treated with RNR inhibitors and transduced with an rAAVDJ vector expressing Firefly luciferase (Fluc) from the CAG promoter. Luciferase activity was measured 24 h later; RLU, relative light units. Data from b and c are representative of two independent experiments. Values are displayed as the group means, with error bars representing s.d.; n = 3 biological replicate wells. Significance was determined by one-way ANOVA analysis with Dunnett’s test for multiple comparisons. d,e, Huh7 cells were transfected with RRM1 small interfering RNA (siRNA; siRRM1) and transduced with rAAVDJ-GAPDH-P2A-GFP; siScr, scrambled siRNA control. Western blotting of RRM1 was performed 2 d after siRNA transfection (d). The GFP+ fraction was analyzed by flow cytometry 3 d after transduction (e). Bars represent the group mean, and error bars represent s.d.; n = 3 biological replicate wells. Data are representative of two independent experiments. Significance was determined using a two-tailed t-test. f, A junction capture PCR was used to detect on-target integration at the GAPDH locus using gDNA extracted from Huh7 cells treated with the indicated RNR inhibitors 14 d after AAV transduction. Control reactions included amplification of the ACTB locus; M, size marker. g, Murine Hepa1–6 cells were treated with RNR inhibitors and transduced with the rAAVDJ-Alb-P2A-GFP targeting vector. Flow cytometry analysis of GFP+ fractions at 14 d after AAV transduction is shown. Bars represent the group mean, and error bars represent s.d.; n = 3 biological replicate wells. Data are from two independent experiments. The HU group P values were 0.0002 and 0.0076 for the fludarabine group. h, The mouse Alb locus after HR with the gene-targeting rAAVDJ-Alb-P2A-GFP vector is shown. The positions of quantitative PCR (qPCR) primers to detect on-target integrated fusion mRNA are indicated; Ex, exon. i, RNA was extracted from transduced Hepa1–6 cells, and qPCR was performed to quantify expression levels of on-target Alb-P2A-GFP fusion mRNA. Actb mRNA was used for normalization, and data are shown as relative expression to control. Bars represent the group mean, and error bars represent s.d.; n = 3 biological replicate wells. Data are from two independent experiments. Significance was determined by one-way ANOVA with Dunnett’s test for multiple comparisons for all data, unless otherwise indicated; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; NS, not significant.
A well-defined target of HU is the RNR enzyme, which generates deoxyribonucleotides from ribonucleotides18. Therefore, we examined the effects of other RNR inhibitors (fludarabine, triapine and gallium nitrate) on transduction and found that these drugs also enhanced promoter-driven expression, except gallium nitrate18 (Fig. 1c). In the flow cytometry HR assay, we observed an increase in GFP+ Huh7 cells pretreated with fludarabine and triapine (Fig. 1b). To explore RNR inhibition as a mechanism for increased HR, we used siRNA knockdown of RRM1, the main subunit of the RNR complex18. Knockdown of RRM1 also increased GFP+ fractions in rAAVDJ-GAPDH-P2A-GFP-transduced cells (Fig. 1d,e). These data supported that suppressing RNR can increase gene targeting in vitro. To confirm on-target HR, we used PCR to amplify genomic DNA (gDNA) at the left integration junction in Huh7 cells after infection. One primer binds to the upstream genomic sequence, past the homology arms, and the second primer binds to the P2A sequence (Fig. 1a). As such, correctly integrated genomic sequences with the expected size were amplified only in transduced cells (Fig. 1f).
We also examined HU or fludarabine treatment in the mouse hepatoma cell line Hepa1–6 using a vector targeting the mouse Alb locus, named rAAVDJ-Alb-P2A-GFP (Fig. 1g). As before, HU or fludarabine pretreatment increased the GFP+ fractions in rAAVDJ-Alb-P2A-GFP-infected Hepa1–6 cells (Fig. 1g). To further confirm targeted integration, we analyzed on-target Alb-P2A-GFP fusion mRNA levels by qPCR (Fig. 1h) and observed 3.5- or 2.6-fold higher levels in HU- or fludarabine-treated cells, respectively (Fig. 1i). These data showed that RNR inhibitors increased rAAV gene targeting in vitro, independent of species and target locus.
Fludarabine increases AAV-mediated gene targeting in vivo.
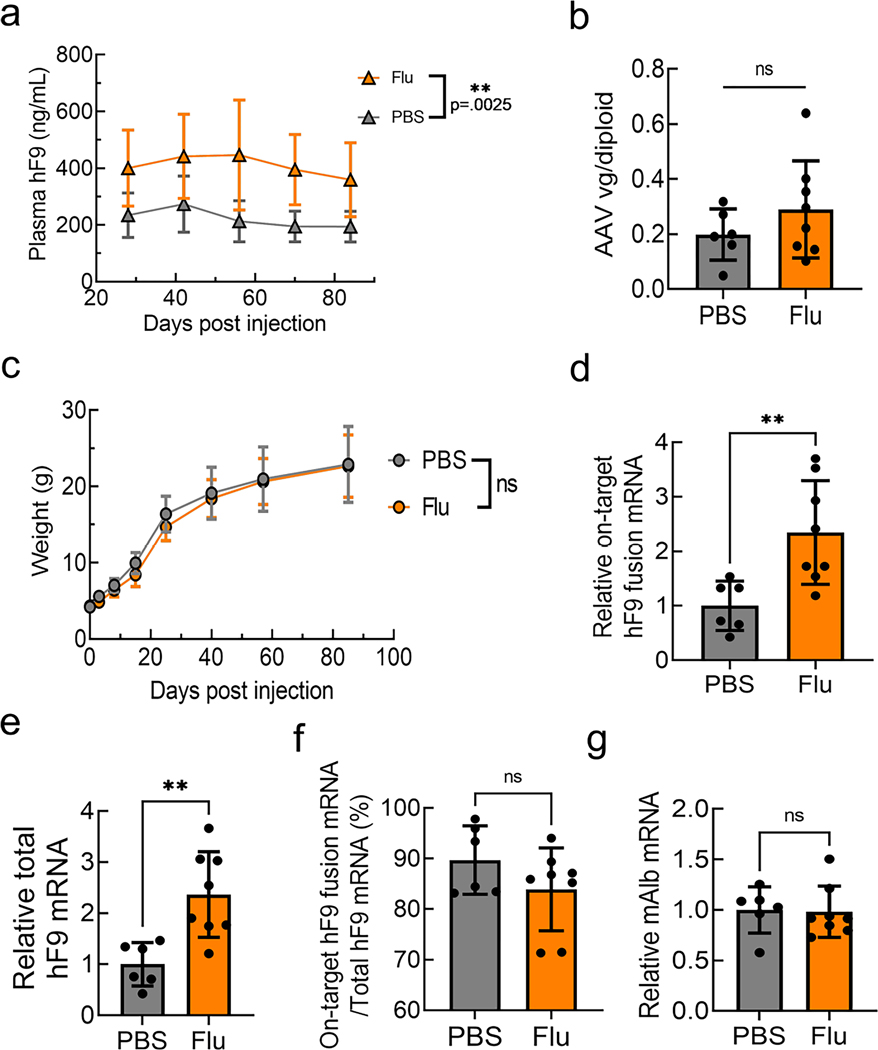
Next, we injected a gene-targeting vector named rAAV8-Alb-P2A-hF9, containing human coagulation factor 9 (hF9; gene name F9) within homology arms targeting the mouse Alb locus, with or without administration of HU or fludarabine into 4-week-old mice6 (Fig. 2a). Strikingly, we observed a sustained 2.7- to 4.6-fold increase of plasma hF9 levels in mice treated with fludarabine, but not HU, compared to control mice (Fig. 2b). Importantly, fludarabine administration did not alter body or liver weights, and no toxicity was observed based on complete blood counts, blood biochemical examination, alanine transaminase levels and liver histopathological analysis by a trained veterinarian pathologist (Supplementary Table 2 and Extended Data Fig. 1b–e). Furthermore, rAAV genomes per diploid hepatocyte were unchanged by either drug treatment, showing that improved hF9 expression was not the result of increased vector delivery into the liver (Fig. 2c).
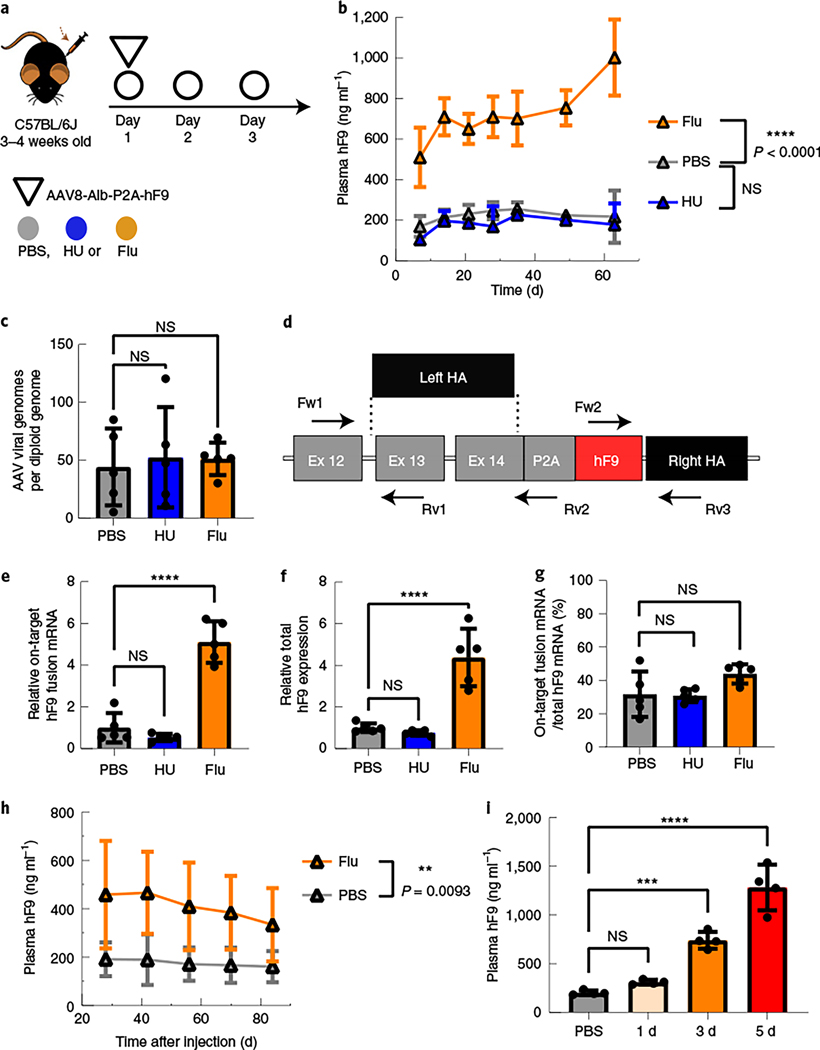
Fig. 2 |. Fludarabine administration increased the efficiency of gene targeting in mouse hepatocytes.
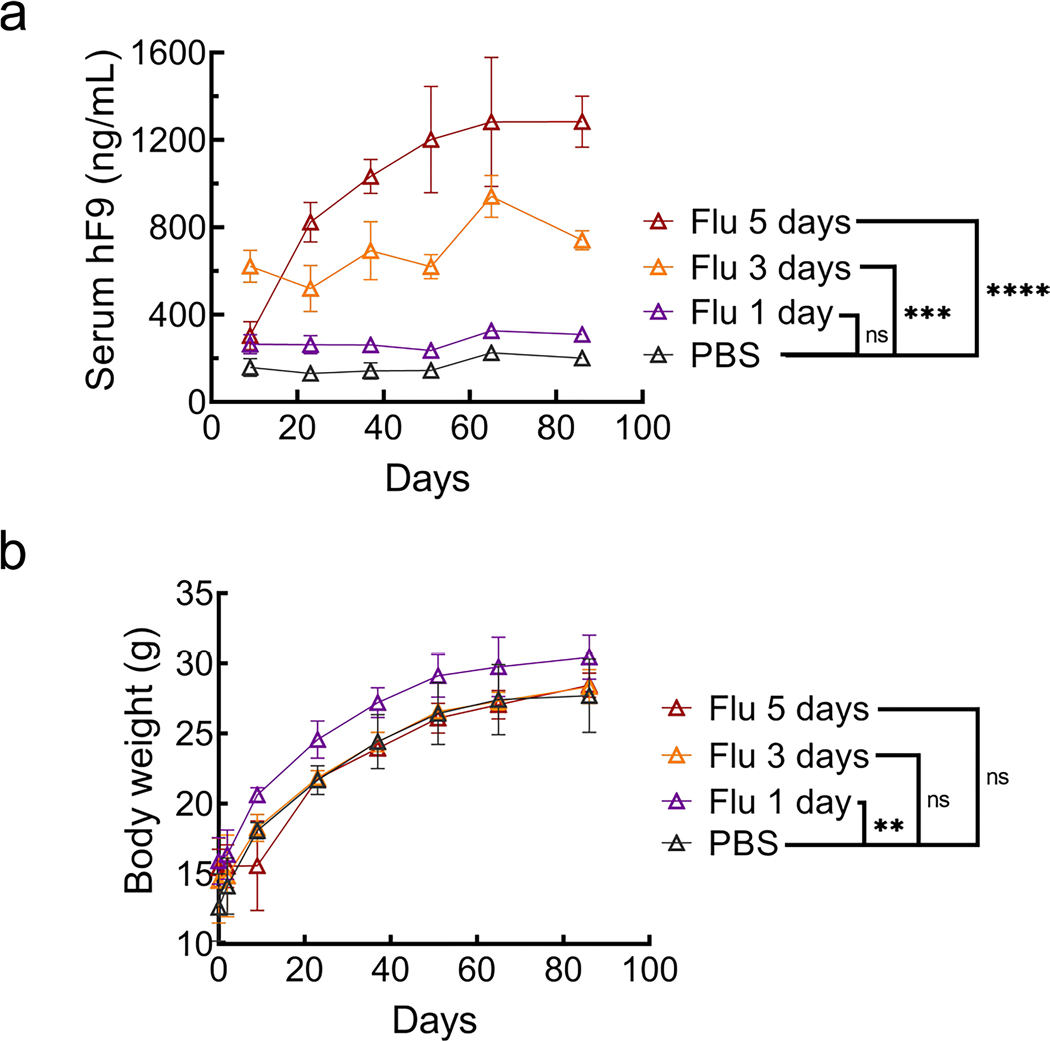
a, Four-week-old mice were administered intraperitoneal (i.p.) injections of HU (300 mg kg–1) once per day or fludarabine (125 mg kg–1) three times per day through day 1 to day 3. Mice were also administered intravenous (i.v.) injections with the rAAV8-Alb-P2A-hF9 targeting vector (1.0 × 1011 viral genomes per mouse) on day 1 immediately after the HU or second fludarabine injection. b, hF9 protein levels in mouse plasma were determined using an enzyme-linked immunosorbent assay (ELISA) following rAAV8-Alb-P2A-hF9 targeting vector injection with or without drug treatment over a 65-d period. Values are displayed as the group mean with error bars representing s.d.; n = 5 mice per group. Significance testing was performed by two-way ANOVA analysis. c, gDNA was extracted from liver tissues 65 d after rAAV8-Alb-P2A-hF9 injection, and qPCR was performed to quantify total AAV genomes. Actb primers were used to quantify mouse diploid genomes. Each point represents data from one mouse. Bars represent the group mean, and error bars represent the s.d.; n = 5 mice per group. Significance testing was performed using a one-way ANOVA analysis followed by Dunnett’s t-test. d, The mouse Alb locus after HR with the gene-targeting AAV-Alb-P2A-hF9 vector is shown. Exon–intron structure and the positions of qPCR primer pairs used for e–g are indicated; HA, homology arm; Fw, forward; Rv, reverse. e–g, Total RNA was extracted from mouse liver tissues in Fig. 2b. qPCR assays quantified the expression levels of on-target integration-derived Alb-P2A-hF9 fusion mRNA (primers Fw1 and Rv2; arrows) (e), total hF9 mRNA (Fw2 and Rv3) (f) and the fraction of hF9 fusion mRNA derived from on-target HR out of the total amount of hF9 mRNA (g). Actb mRNA was used for normalization, and data are shown as relative expression to the PBS-treated group. Bars represent the group mean, and error bars represent the s.d.; n = 5 mice per group. Significance was determined using a one-way ANOVA analysis followed by Dunnett’s t-test, unless otherwise indicated. h, One-week-old male neonatal mice were injected i.p. with PBS or fludarabine (375 mg kg–1) and 4 h later with rAAV8-Alb-P2A-hF9 (2.5 × 1013 viral genomes per kilogram of body weight). A second fludarabine dose was given 1 d after vector injection. Four weeks later, plasma was drawn, and hF9 levels were measured by ELISA; n = 6 PBS-treated mice and n = 8 fludarabine-treated mice. Values are displayed as the group mean, with error bars representing s.d. Significance testing was performed by two-way ANOVA. i, Fludarabine dosing regimens were tested by i.p. administration (125 mg kg–1) three times per day for one, three or five sequential days. Four-week-old mice were injected i.v. at day 1 with the rAAV8-Alb-P2A-hF9 targeting vector (1.0 × 1011 viral genomes per mouse) immediately after the second fludarabine administration injection. Blood was collected 2 months later, and hF9 protein levels were determined via ELISA. Values are displayed as the group mean with error bars representing s.d.; n = 4 mice per group (see also Extended Data Fig. 6). Significance was determined using a one-way ANOVA followed by Dunnett’s t-test; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To confirm HR in liver gDNA, we amplified the right-sided integration junction using a primer binding in the mouse genome and a primer in hF9 (Supplementary Fig. 2a). An on-target HR-derived amplicon (expected size of 1.6 kilobases (kb)) appeared in rAAV-injected mouse samples only (Supplementary Fig. 2b). A 1.5-kb nested PCR amplicon (primers Fw3 and Rv2) was TOPO cloned, and five clones (one from each mouse) were Sanger sequenced, confirming correct on-target integration without any indels (Supplementary Fig. 2c,d). We next quantified the amount of fusion hF9 mRNA derived from on-target integration (Fig. 2d). Fusion mRNA levels were significantly higher in the fludarabine-treated animals (5.1-fold increase; Fig. 2e). qPCR of total hF9 mRNA with transgene-internal primers, which detects mRNA from targeted integration, random integration or episomal-derived expression, showed a 4.4-fold increase in fludarabine-treated versus control mice (Fig. 2f). To determine the level of expression from sources other than HR, we compared the abundance of the on-target fusion mRNA to the total amount of hF9 mRNA (Fig. 2g). hF9 fusion mRNA from on-target HR represented ~30–40% of the total hF9 mRNA; importantly, this ratio was unchanged by fludarabine treatment. Endogenous Alb expression also remained unchanged (Extended Data Fig. 1f). These data demonstrated that fludarabine increased hF9 expression through enhanced AAV-HR efficiency.
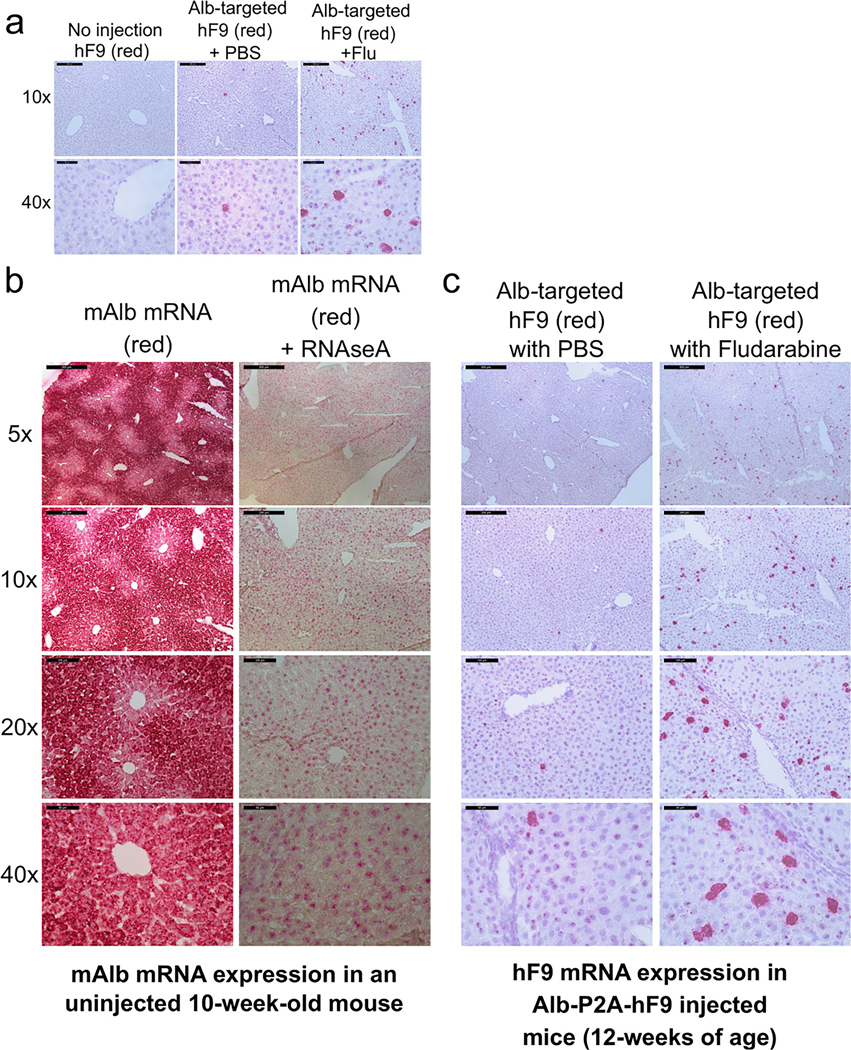
We next used RNAScope in situ hybridization to visualize hF9 and mouse Alb mRNAs on a single-cell level in mouse liver. Mouse Alb mRNA was immensely abundant in all hepatocytes, while hF9 mRNA was expressed in relatively few hepatocytes, consistent with the low frequency of gene targeting and a paucity of episomal expression7,19 (Extended Data Fig. 2a–c). These infrequent hF9+ cells displayed strong intracellular staining and were readily visible in fludarabine-treated animals, supporting that fludarabine enhanced AAV-HR efficiency.
Our following experiment was to confirm that transgene- expressing hepatocytes contained on-target HR using the targeting vector rAAV8-Alb-P2A-GFP. After pretreatment with PBS or fludarabine and injection with rAAV8-Alb-P2A-GFP in 4-week-old mice, we isolated hepatocytes and sorted the GFP+ hepatocyte population by FACS; the GFP+ hepatocyte population was increased by fludarabine (~fivefold), as seen before (Supplementary Fig. 3). gDNA was then extracted, and droplet digital PCR (ddPCR) reactions were used to amplify an untargeted region of Alb to quantify the number of endogenous unedited Alb copies (Supplementary Fig. 4a). In the same reactions, primers and a probe for on-target HR were also included (Supplementary Fig. 4b). We found that in PBS-treated mice, approximately ~1.8% of Alb alleles contained on-target HR; in fludarabine-treated animals, ~3.5% of Alb alleles contained on-target HR (Supplementary Fig. 4c,d). This characterization confirmed that some GFP+ hepatocytes possessed on-target HR and that fludarabine did not decrease the ratio of on-target-to-off-target integration.
Next, we examined the use of fludarabine in neonatal mice as a test of efficacy in a stringent model of vector dilution, organ development and cell growth. As such, we injected 7-d-old mice with rAAV-Alb-hF9 in combination with a 2-d administration of PBS or fludarabine. We then monitored hF9 plasma levels for 84 d, which showed a significant increase in expression in the fludarabine-treated groups, despite no difference in body weight gain or remaining viral genomes in the mouse livers at the end of the experiment (Fig. 2h and Extended Data Fig. 3a–c). qPCR data showed a greater than twofold increase in both on-target fusion mRNA and total hF9 mRNA in fludarabine-treated mice (Extended Data Fig. 3d,e). On-target fusion mRNA comprised ~80% of total hF9 mRNA in both treatment groups, and Alb mRNA remained unchanged (Extended Data Fig. 3f,g). Separately, hematoxylin and eosin (H&E) sections of livers from mice treated as neonates with rAAV and PBS or fludarabine were examined by a pathologist 157 d after treatment (Supplementary Fig. 5a). Although minor inflammation was noted in some animals from both treatment groups, there were no signs that fludarabine caused liver damage, with no group-dependent signs of neoplasms, bilirubin retention, microcytosis or fatty livers. Liver weights remained unchanged by fludarabine treatment (Supplementary Fig. 5b). In all neonatally treated mice, no visable tumor growth was observed at the time of liver collection. Thus, fludarabine did not cause overt harm to the liver of neonatally treated mice but successfully increased AAV-HR.
We also examined a fludarabine dose response in 4-week-old mice. Five days of fludarabine injections resulted in the highest plasma hF9 levels (6.4-fold versus PBS) and did not significantly decrease body weight (Fig. 2i and Extended Data Fig. 4). Delayed fludarabine administration, 4 weeks after vector injection, did not alter hF9 expression, suggesting that the enhancing effect of fludarabine requires temporal proximity to rAAV transduction (Supplementary Fig. 6a,b). Additionally, fludarabine increased gene targeting to the Apoe locus, demonstrating that its effect is not restricted to a single locus (Supplementary Fig. 6c). After pretreatment with fludarabine and injection of a promoter-bearing vector expressing human α−1-antitrypsin (SERPINA1; also known as hAAT), rAAV8-HLP-hAAT, there was no sustained increase in serum hAAT levels (Supplementary Fig. 7a). At 11 weeks (end of experiment), there was no difference in SERPINA1 mRNA levels (Supplementary Fig. 7b). We also determined that neither HU nor fludarabine changed the amount of total nuclear or double-stranded rAAV genomes in Huh7 cells (Supplementary Fig. 7c,d). Thus, fludarabine may alter the dynamics of rAAV genome processing without altering overall entry efficiency.
Fludarabine transiently suppresses hepatocyte proliferation.
HU induces cell cycle arrest and can synchronize cultured cells in S phase; moreover, HR is most active in the S/G2 phases of the cell cycle20–23. Such an effect could increase HR, so we investigated the cycling status of liver cells via bromodeoxyuridine (BrdU) labeling of DNA synthesis. Mice were injected with BrdU for 3 d with or without fludarabine, and liver sections were stained with anti-BrdU (Fig. 3a and Supplementary Fig. 8a,b). BrdU incorporation averaged ~50 positive nuclei per field of view in control mice, while BrdU-labeled nuclei were nearly absent in fludarabine-treated mice, demonstrating a strong inhibitory effect of fludarabine on hepatocyte DNA synthesis (Fig. 3b,c). Following fludarabine removal, BrdU incorporation began to recover, suggesting that BrdU+ hepatocytes progressed through S phase after drug washout (Fig. 3b,c). These data indicate that fludarabine treatment causes a transient and substantial inhibition of cell proliferation/DNA synthesis in mouse livers. Thus, increased hepatocyte proliferation is not the cause of increased gene targeting with fludarabine treatment.
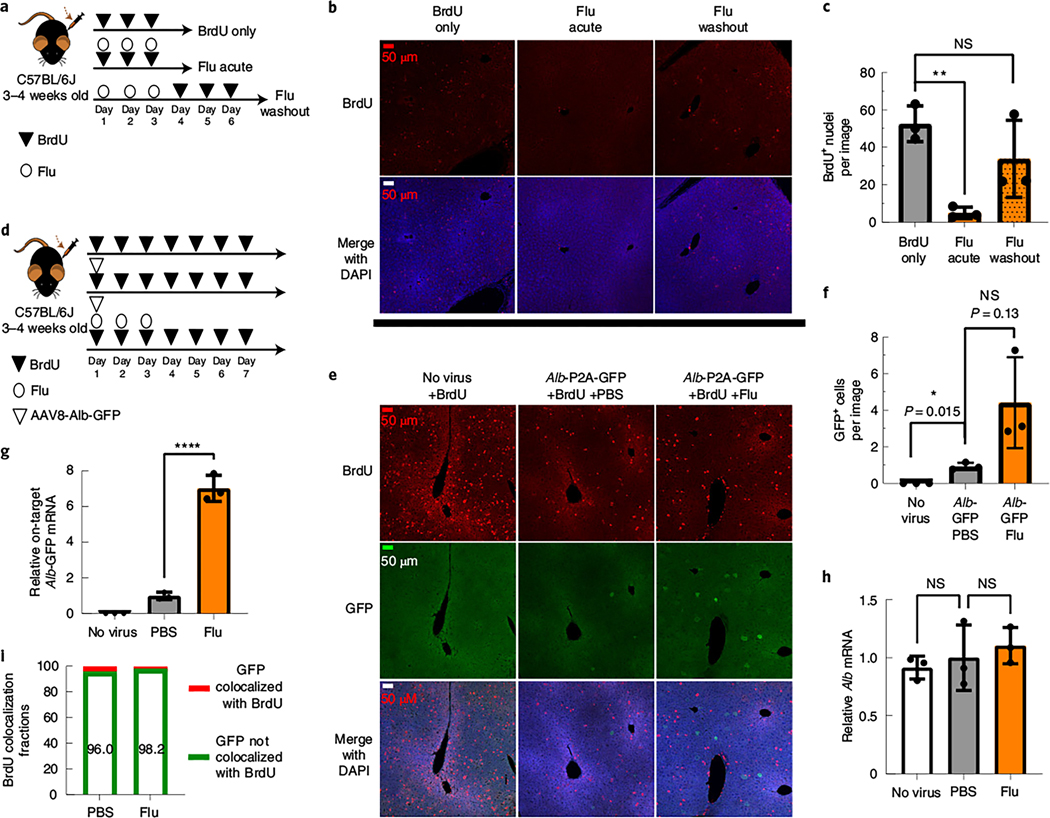
Fig. 3. Fludarabine transiently inhibits S phase progression, and rAAV gene targeting occurs in hepatocytes that have not progressed through S phase.
3 |a, Mice were injected i.p. with BrdU (200 mg kg–1) once per day for 3 d to label proliferating hepatocytes. Some mice were simultaneously injected with fludarabine (125 mg kg–1 three times per day for 3 d), while the final group was treated with fludarabine before the 3 d of BrdU injection; n = 3 mice per group. b, Six hours after the last injection, mice were killed, and livers were collected for immunostaining with anti-BrdU. Representative images from each group are shown with BrdU-labeled nuclei (red) and a DAPI counterstain (blue). All images were taken with a ×20 objective with identical exposure and settings. c, Images of BrdU-labeled nuclei were quantified from each group as the number of BrdU+ nuclei per field of view. Images used for quantification are from two or more slides per mouse, three mice per group and two or more independent strains. Significance was determined using a Shapiro–Wilk test for normal distribution and two-tailed Student’s t-test for normally distributed data or non-parametric Mann–Whitney U-test for non-normally distributed data. Each point represents average data from one mouse; n = 3 per group. Bars represent the group mean, and error bars represent the s.d. The P value of the fludarabine acute group is 0.0012, and the P value for the fludarabine washout group is 0.4. d, To label proliferating cells in the liver, mice were i.p. injected with BrdU once per day for 7 d. On day 1 of BrdU injections, two groups of mice were i.v. injected with the rAAV8-Alb-P2A-GFP targeting vector (1 × 1011 viral genome per mouse). One group was also treated with fludarabine (125 mg kg–1, three times per day) for the first 3 d. e, On day 7, mice were killed, and livers were collected for immunohistochemistry staining of BrdU (red) and GFP (green) with a DAPI nuclear stain (blue). Representative images are shown. f, GFP+ cells were quantified as the number of positive cells per 20× field of view. An F-test was used to determine variance between groups of normally distributed data, and a two-tailed Student’s t-test or t-test with Welch’s correction was used to test for significance. Images used for quantification are from two or more slides per mouse with three mice per group and two or more independent strains. Each point represents average data from one mouse; n = 3 per group. Bars represent the group mean, and error bars represent s.d. g,h, Total RNA was extracted from liver tissue, and qPCR was used to determine the levels of on-target integration-derived Alb-P2A–GFP mRNA and endogenous Alb mRNA, as described previously. Each point represents average data from one mouse; n = 3 per group. Bars represent the group mean, and error bars represent s.d. Statistical testing was performed using a one-way ANOVA followed by Dunnett’s t-test; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. i, The number of GFP+ cells that colocalized with BrdU (that is, the number of cells that have undergone S phase DNA synthesis) were quantified and displayed as the percentage of total GFP+ cells per group.
To further study the relationship between cell cycling and HR, we injected rAAV8-Alb-P2A-GFP simultaneously with BrdU and co-stained hepatocytes, which underwent HR (GFP signal) and DNA synthesis (BrdU) (Fig. 3d). As before, fludarabine increased GFP+ cells ~4.7-fold (Fig. 3e,f). On-target fusion GFP mRNA significantly increased six- to sevenfold in fludarabine- versus PBS-treated animals, whereas Alb mRNA expression remained unchanged (Fig. 3g,h). Surprisingly, we found that BrdU (S phase marker) colocalized with GFP (HR marker) infrequently at 4.0% of all GFP+ cells in PBS-treated mice and 1.8% in fludarabine-treated mice (Fig. 3i). Thus, most GFP+ cells had not progressed through S phase during the timeframe of the experiment, regardless of drug treatment. Overall, progression through S phase may not be a primary determinant of rAAV-HR in vivo.
Fludarabine induces a transient DNA damage response.
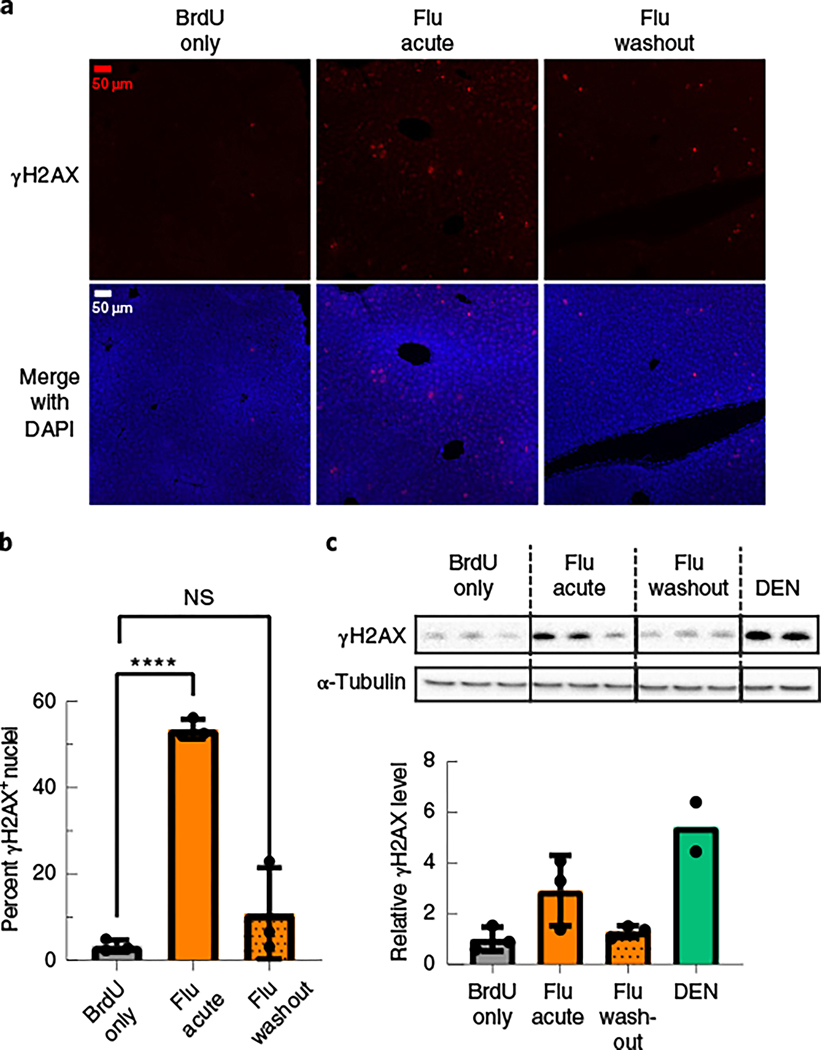
Fludarabine has numerous effects on cells; it is incorporated into DNA during replication or repair24–26, reduces the dNTP pool by RNR inhibition and causes premature transcription termination26–28. Any of these events can trigger DNA repair pathways, which can be linked to increased HR efficiency29–31. To assay for a DNA damage response, we stained for phosphorylated Ser 139 H2AX (γH2AX) in liver sections from the mice studied in Fig. 3a32. Minimal levels of γH2AX were detected in untreated or BrdU-only-treated livers (Fig. 4a,b and Supplementary Fig. 8c,d). Mice treated with diethylnitrosamine (DEN), a potent mutagen, showed intense γH2AX staining (Supplementary Fig. 8c)33. During fludarabine treatment, mouse livers possessed widespread pan-nuclear γH2AX foci (Fig. 4a,b). The number of γH2AX+ nuclei lessened after fludarabine removal (Fig. 4a,b). Western blotting for γH2AX using liver lysates from the same animals provided similar and hence consistent results (Fig. 4c). We also found that siRNA-mediated knockdown of RRM1 or fludarabine treatment strongly increased γH2AX levels in Huh7 cells (Supplementary Fig. 9). Thus, fludarabine temporarily activated DNA repair signaling, which, in part, explains the mechanism of increased gene-targeting efficiency.
Fig. 4 |. Fludarabine induces a transient DNA damage response in mice.
a, Liver tissue sections from the same mice in Fig. 3a–c were also stained for the DNA damage response marker γH2AX. Representative images are shown with γH2AX (red) and DAPI (blue). b, Images of γH2AX nuclei were quantified from each group and are displayed as the percentage of γH2AX+ nuclei out of all nuclei. Images used for quantification are from two or more slides per mouse, three mice per group and two or more independent strains. An F-test was used to determine variance between groups of normally distributed data, and a two-tailed Student’s t-test or t-test with Welch’s correction was used to test for significance. Each point represents average data from one mouse; n = 3 per group. Bars represent group means, and error bars represent s.d. c, Liver tissue lysates from the same mice were used for western blotting of γH2AX, and α-tubulin was used as a loading control (top). Image analysis quantification of the γH2AX band intensity in the western blot was normalized to α-tubulin (bottom). Each lane (top) and point (bottom) represents average data from one mouse; n = 3 mice per group, except the DEN data, which are two technical replicates from one mouse. Error bars represent s.d.
DEN can increase AAV-HR but also off-target expression.
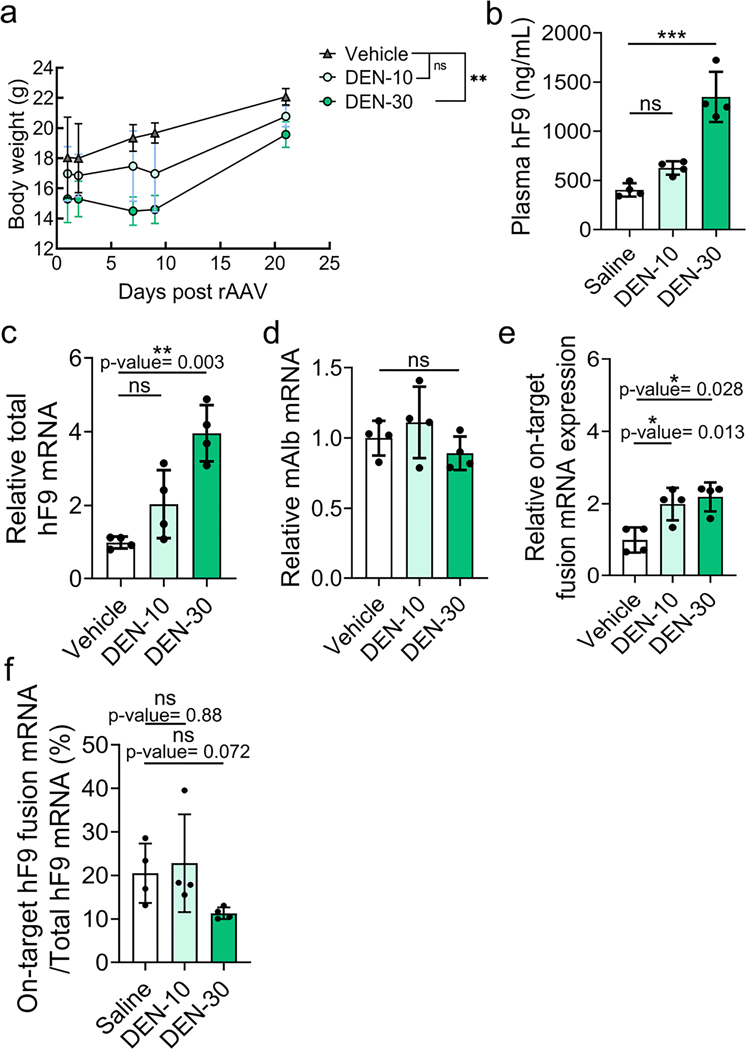
Treatments that induce DNA damage can increase gene targeting rates in cultured cells34 (Extended Data Fig. 1a). To determine if genotoxicity is also a governing principle for rAAV-HR in vivo, we used DEN and rAAV8-Alb-P2A-hF9 co-injections in mice. DEN administration significantly decreased body weight in a dose-dependent manner, indicative of toxicity (Extended Data Fig. 5a). DEN at a dose of 30 mg kg–1 increased hF9 plasma levels ~3.6-fold (Extended Data Fig. 5b), whereas a lower dose (10 mg kg–1) did not significantly change hF9 levels. Total hF9 mRNA expression was only significantly increased by high-dose DEN administration (two- and fourfold increase at low and high doses, respectively), whereas Alb mRNA expression was unchanged (Extended Data Fig. 5c,d). On-target HR-derived fusion hF9 mRNA was ~twofold higher at both doses (Extended Data Fig. 5e). Comparing the on-target fusion mRNA to total hF9 mRNA showed a trend toward decreased on-target-derived mRNA with the high dose of DEN (Extended Data Fig. 5f). Thus, low-dose DEN did not significantly increase plasma hF9 or total hF9 mRNA levels; high-dose DEN showed some limited enhancement of hF9 expression, however, this treatment resulted in toxicity and increased off-target effects. In total, DEN could not safely enhance AAV-HR to the extent that we observed with fludarabine.
Fludarabine increases CRISPR/Cas9 gene editing efficiency.
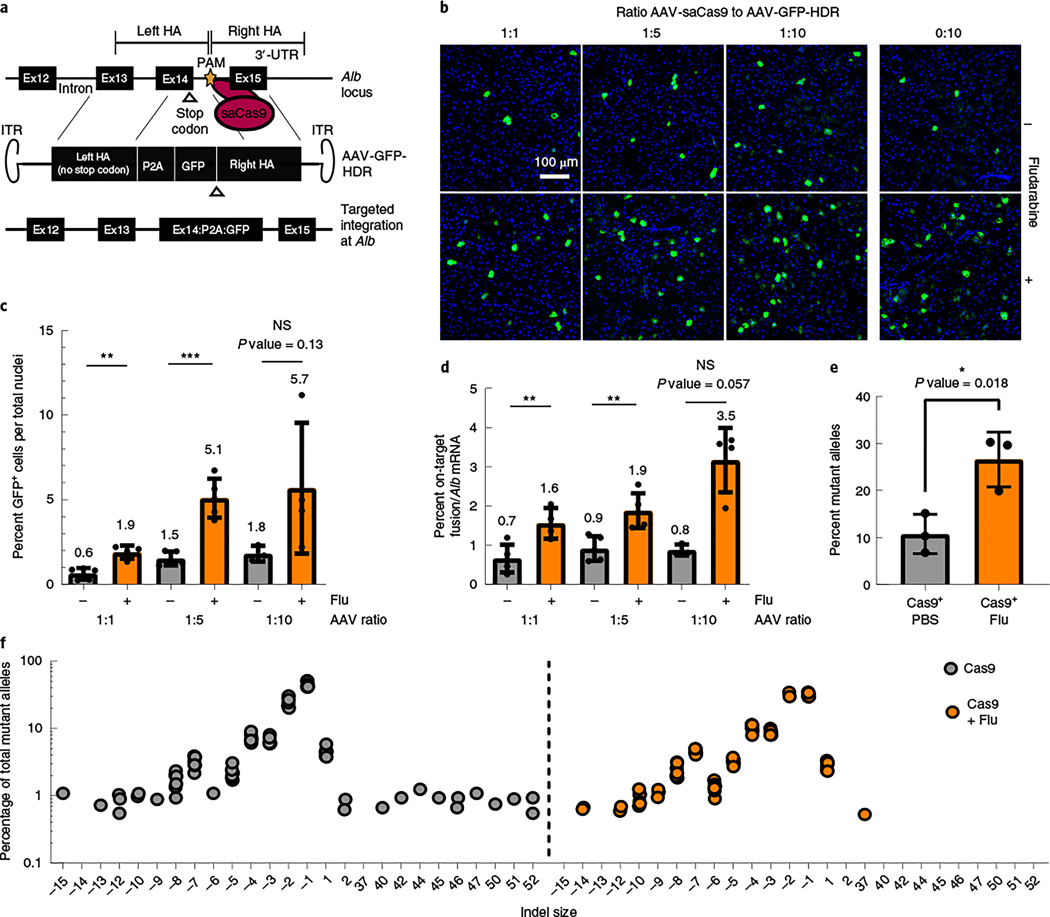
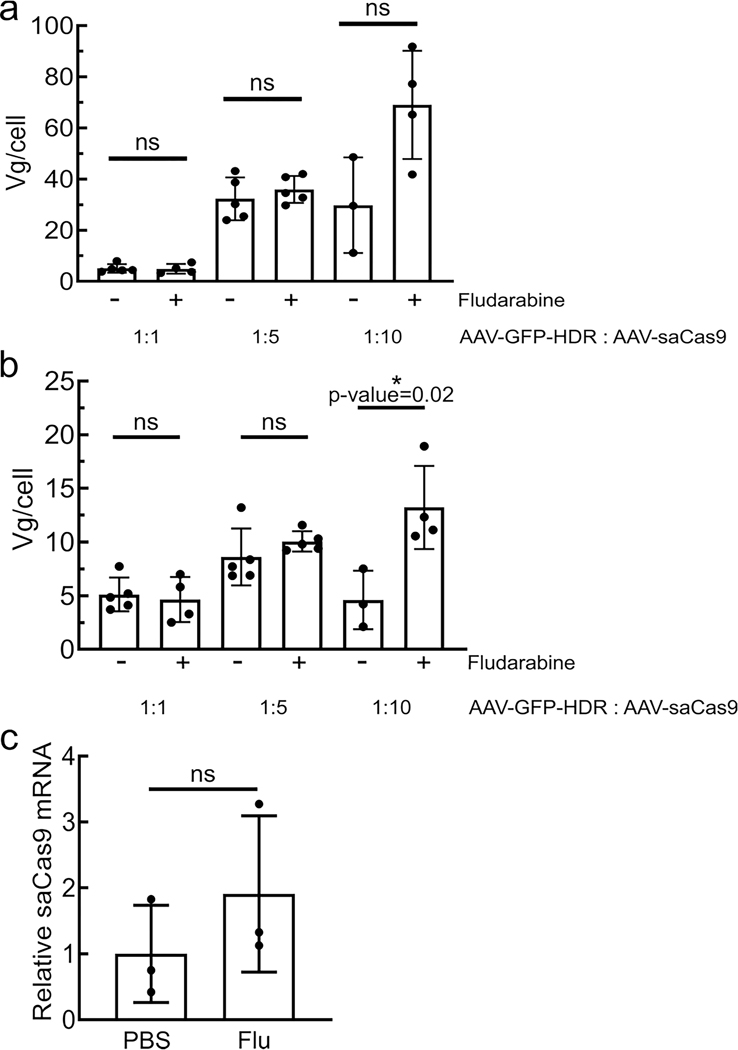
Programmable nucleases, such as CRISPR/Cas9, can also increase rAAV-HR efficiency through DNA damage19,35. To investigate if CRISPR/Cas9-mediated HR is influenced by fludarabine, mice were injected with two rAAVs: one delivered Staphylococcus aureus Cas9 (saCas9) plus a gRNA targeting the intron after exon 14 in mouse Alb (AAV-Cas9) and a second AAV vector encoding a promoterless GFP sequence flanked by homology arms (AAV-GFP-HDR; Fig. 5a). We examined various ratios of AAV-Cas9 to AAV-GFP-HDR (1:1, 1:5 and 1:10) co-injections, with or without fludarabine. We found that fludarabine treatment increased the abundance of GFP+ hepatocytes at all ratios (Fig. 5b,c). On-target fusion mRNA also showed an increase of 2- to 3.6-fold with fludarabine treatment (Fig. 5d). Viral genome copies showed no significant differences between treatments, except in the 1:10 group (Extended Data Fig. 6a,b). To determine if fludarabine influenced DNA repair at the target site, we performed targeted deep sequencing using liver gDNA from mice injected with AAV-Cas9 alone36. There was a two- to threefold increase in insertions/deletions (indels) present in fludarabine-treated mice (Fig. 5e). Additionally, analysis of the top 12 indel-containing alleles showed an absence of larger insertions from fludarabine-treated animals (Fig. 5f). saCas9 mRNA levels were unchanged by fludarabine treatment (Extended Data Fig. 6c). In total, fludarabine also enhanced CRISPR/Cas9-mediated gene editing in vivo, likely through stimulation of DNA repair and/or altered repair characteristics at the target site.
Fig. 5 |. Fludarabine increases CRISPR/Cas9 gene editing efficiency in vivo.
a, saCas9 is targeted to the intron downstream of exon 14 in the mouse Alb locus, delivered by AAV-saCas9. Homology-directed repair is accomplished using an AAV-encoded repair template, AAV-GFP-HDR, which contains a P2A-GFP transgene flanked by sequences homologous to the gRNA target site. The PAM site of AAV-GFP-HDR was mutated to avoid self-targeting; 3′-UTR, 3′-untranslated region. ITR, inverted terminal repeats. b, Mice were treated with PBS or fludarabine, as previously described, and co-injected with AAV-saCas9 and AAV-GFP-HDR. Three ratios of AAV-saCas9 to AAV-GFP-HDR vector were used (1:1, 1:5 and 1:10), with each part representing 6.0 × 1012 viral genomes per kilogram of body weight. Two weeks later, mice were sacrificed, and livers were imaged for native GFP expression. Representative images are shown; n = 3 to 5 mice per group. c, GFP+ cells were quantified from multiple images per mouse. A Shapiro–Wilk test was used to test for normal distribution, an F-test determined variation between groups, and a two-tailed Student’s t-test or t-test with Welch’s correction tested for significance. Each point represents data from an individual mouse; n = 3 to 5 mice per group. Bars represent the group mean, with error bars representing s.d. The P value was 0.0012 for the 1:1 group, 0.0002 for the 1:5 group and 0.138 for the 1:10 group. d, Total RNA was extracted from mouse livers and used for quantification of the on-target HR-derived fusion mRNA or Alb mRNA by qPCR. mRNA levels were normalized to Actb mRNA. Data are displayed as a percentage of fusion mRNA out of all Alb mRNA. Each point represents data from an individual mouse; n = 3 to 5 mice per group. Bars represent the group mean, with error bars representing s.d. Significance testing was performed as in c. The P value was 0.0084 for the 1:1 group, 0.004 for the 1:5 group and 0.057 for the 1:10 group. e, Separately, mice were injected with 6.0 × 1012 viral AAV-saCas9 genomes per kilgram of body weight with or without fludarabine treatment. Two weeks later, gDNA was extracted from livers, and targeted deep sequencing of the gRNA target site was performed. Data are normalized to sequencing data from a non-injected control mouse and are displayed as a percentage of alleles containing indels; n = 3 mice per group. Bars represent the group mean, and error bars represent s.d. Significance testing was performed with a two-tailed t-test after testing for distribution and variance. f, The top 12 mutant alleles from targeted deep sequencing were analyzed for all mice and technical replicates. Indels were graphed as the percentage of reads from all mutant alleles based on size (x axis).
Discussion
In this study, we showed that short-term treatment with fludarabine, a Food and Drug Administration (FDA)-approved drug currently in clinical use for pediatric populations37, significantly enhanced HR gene editing without causing overt toxicity in juvenile and neonatal mice. To date, no other compounds have been proven to increase gene editing efficiencies in somatic mammalian tissue; heretofore, edit-enhancing drugs have been applied only in cultured cells or zygotes38–44. We provide functional and molecular evidence that fludarabine significantly enhances both AAV-HR and CRISPR/Cas9-mediated HR efficiencies. Thus, fludarabine can increase editing efficiencies across numerous contexts, editing technologies and genomic targets.
While our data implicated that some expression from episomes or off-target integration of the rAAV vector may occur, it is important to note that treatment with fludarabine did not alter the ratio of on-target-to-off-target expression. Future studies will focus on examining the off-target-mediated expression and integration of the promoterless rAAV vectors. While increasing the frequency of random rAAV off-target integration may confer some risk of hepatocellular carcinoma, we emphasize that our vectors lack exogenous promoter sequences, which are known as the key drivers of oncogenesis after rAAV or lentiviral insertion near proto-oncogenes45,46. In this regard, promoterless rAAVs were shown to be far safer than classical promoter-bearing vectors in a mouse model of methylmalonic acidemia, and AAV integration has not been established as a cause of hepatocellular carcinoma in humans47. As such, increased integration of promoterless vectors, due to fludarabine treatment, is unlikely to be oncogenic; this theory was supported by the overt lack of any tumor generation in our long-term experiments.
Certain HR pathway factors have been identified as important components for rAAV-HR41. In this regard, canonical HR is predominantly active during the S/G2 phases of the cell cycle21–23. Contrary to this S phase-centric principle, our present data showed that ~96% of hepatocytes undergoing AAV-HR were not coimmunostained with BrdU, suggesting that S phase progression is not a major determining factor in AAV-HR in vivo. It should be noted that most in vivo hepatocytes are in G0, which is considerably different from cultured cells and could contribute to the discordance with previous studies conducted in cell culture48. Additionally, others have reported that AAV-mediated HR can occur independent of S phase in cardiomyocytes49. Another consideration is the possibility for gene expression to occur following non-HR events when using promoterless vectors, although successful transcription and translation is likely to be rare due to the lack of an N-terminal start codon and promoter.
The lack of S phase progression in AAV-HR+ hepatocytes suggested an effect of fludarabine in non-proliferating hepatocytes. Fludarabine inhibits the catalytic subunit of the RNR complex RRM1, which functions throughout the cell cycle18. Thus, fludarabine can antagonize basal DNA repair levels in non-dividing cells due to decreased intracellular dNTPs24. Moreover, fludarabine, a purine analog, is incorporated into replicating DNA but also into nascent RNA, leading to premature transcription termination50. Reduction of intracellular dNTP pools, replication stalling and premature transcription termination can all activate DNA repair pathways, which could explain the enhancing effect of fludarabine in non-proliferating hepatocytes. By examining γH2AX levels, we confirmed widespread transient activation of DNA repair signaling in fludarabine-treated mouse livers. γH2AX is a hallmark of various DNA repair pathways and facilitates the recruitment and accessibility of repair factors to damaged DNA, which could directly or indirectly promote HR51,52. Lastly, the interaction with DNA repair by fludarabine was also observed by the altered repair characteristics at the targeted locus in mice injected with AAV-Cas9. Although γH2AX and DNA damage could be associated with apoptosis, H&E examination of murine livers showed no signs of cell death or necrosis 12 h after treatment with fludarabine. Furthermore, the induction of DNA repair signaling likely influences transduction and perhaps recombinogenic forms of rAAV53. Indeed, we noted transient effects on vector transduction with fludarabine use in vivo. Therefore, fludarabine treatment likely improved AAV-HR through increased activation of DNA repair, through vector capture at fludarabine-promoted breaks or by altering intracellular vector processing. Most importantly, fludarabine did not reduce the ratio of on-target integration, as measured by ddPCR, or the ratio of on-target-to-off-target expression, as measured by qPCR, but rather increased the likelihood for on-target HR to occur, resulting in greater numbers of transgene-expressing hepatocytes. By contrast, DEN, a potent mutagen, induced higher levels of γH2AX and increased total hF9 mRNA fourfold at a high dose, yet mediated a lesser enhancement of HR-derived on-target mRNA than fludarabine (twofold or less) while showing signs of toxicity. Thus, it is still unclear what forms of DNA damage are required for AAV-HR, as increased vector capture would be expected by the more genotoxic DEN treatment, unless cells were lost due to DEN toxicity. It is likely that the magnitude and type of DNA damage, in addition to other factors such as vector processing, are the important determinants for AAV-HR efficiency54. Because the precise process of rAAV-mediated gene targeting is not fully understood, particularly in vivo, unveiling the details of such mechanisms will be the focus of our future studies.
Overall, these findings showed that transient treatment with fludarabine can safely enhance rAAV gene editing efficiencies in a clinically relevant manner. In the future, formulations using established liver-targeting drug delivery technology, such as PEGylation or lipid nanoparticles, can specifically deliver fludarabine to the liver and reduce its systemic effects, further facilitating successful clinical applications. These refined approaches, coupled with testing in other mouse strains and large animal models, will advance this HR-enhancing small molecule to the clinic and permit lifelong therapeutic gene modification after a single administration of rAAV vectors.
online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41587–022-01240–2.
Methods
Cell culture and compound treatment.
Huh7 cells were purchased from JCRB, 293T and Hepa1–6 cells were purchased from ATCC, and all lines were cultured in DMEM with 10% fetal bovine serum (FBS) and 2 mM glutamine. For HU (Sigma-Aldrich), fludarabine phosphate (C10H13FN5O7P, Fisher Scientific), triapine (Fisher Scientific) and gallium(III) nitrate hydrate (Fisher Scientific), cells were pretreated with each compound for 16 h at the indicated concentration, and the culture medium was replaced with fresh medium without compounds before rAAV transduction. For Torin-1 (10 μM; Fisher Scientific), FK228 (100 nM; Fisher Scientific), trichostatin A (200 nM; Sigma-Aldrich), MG132 (1 μM; Sigma-Aldrich) and teniposide (100 nM; Abcam), cells were treated with each compound for 24 h, and rAAV was transduced 1 h after compound addition. All compounds were dissolved in DMSO for in vitro assay.
Vectors.
The AAV vectors containing ITR sequences used in this study are based on AAV type 2 backbone. The cloning of CAG-Fluc(Addgene Cat# 83281)55 and Alb-P2A-hF9 vectors were prepared as described previously6,56. The HLP-hAAT vector was derived from the CAG-Fluc vector by replacing the CAG promoter with the HLP promoter (a kind gift from A. Nathwani, University College London, UK) and the Fluc gene with the hAAT gene. The Alb-P2A-GFP vector was generated by replacing hF9 coding sequence of the Alb-P2A-hF9 vector with GFP coding sequence using an In-Fusion HD Cloning kit (Takara). SaCas9-sgRNA8 vector and Alb-P2A-GFP vectors used for Fig. 5 were prepared as described previously19. For construction of the GAPDH-P2A-GFP vector, human genomic GAPDH segments were PCR amplified using 5′-GACTGTACAGGGCTGCTCACATATTCTGG-3′ (Fw) and 5′-CTGTGTACAGAGTGTATGTGGCTGTGGCCC-3′ (Rv; both containing BsRG1 sites for cloning) and inserted between AAV2 ITRs into BsrGI restriction sites in a modified pTRUF backbone56. The genomic segment spans approximately 1.7 kb upstream and 1.7 kb downstream to the GAPDH stop codon. We then synthesized a 1,359-base pair (bp) fragment spanning the region at the end of the GAPDH locus between the two SexA1 sites to be cloned in the vector. In this fragment, the GAPDH stop codon was removed, and it was inserted into an optimized P2A coding sequence preceded by a linker coding sequence (Gly-Ser-Gly) and followed by the GFP sequence (without the start codon). For Apoe-P2A-hF9 vector construction, a genomic fragment containing sequences used for both homology arms was amplified from mouse gDNA. Primers 5′-TCCACACCTGCCTAGTCTCG-3′ and 5′-GTGCCAGAGGCAGTTGAGTT-3′ were used to amplify a 2.9-kb fragment, and the PCR product was directly cloned into the pCR Blunt II TOPO vector using the Zero Blunt TOPO PCR cloning kit (Invitrogen) and was sequence verified. The left homology arm was amplified from the cloned Apoe genomic fragment using primers 5′-atatcatcgatcgcgatgcattaattaagcggccgAAGACTGTAGGTCCTGACCC-3′ and 5′-ggtggcgccgcttccTTGATTCTCCTGGGCCAC-3′, the middle part containing hF9 sequence was amplified from Alb vector using primers 5′-gcccaggagaatcaaGGAAGCGGCGCCACCAAT-3′ and 5′-ggagaagga tactcaTGTCAGCTTGGTCTTTTCTTTGATCC-3′, and the right homology arm was amplified from the cloned Apoe genomic fragment using primers 5′-aagaccaagctgacaTGAGTATCCTTCTCCTGTCCTGC-3′ and 5′-acgtaacagatctgatatcacgcgtgtacactagtGCCCTGCTGAGTCCCTGAG-3′. Lowercase letters indicate the overlapping sequences. Phusion Hot Start Flex (NEB) was used for all amplifications. Amplicons were assembled using the NEBuilder HiFi DNA Assembly Master Mix (NEB) according to instructions.
Animals.
All animal work was performed in accordance to the guidelines for animal care at Stanford University or approved by the International Centre for Genetic Engineering and Biotechnology review board, with full respect to the European Union Directive 2010/63/EU for animal experimentation and by the Italian Health Minister (authorization 996/2017-PR). Wild-type C57BL/6 (B6) mice were purchased from Jackson Laboratory. We used 3- to 4-week-old juvenile male mice to test the effect of RNR small-molecule inhibitors on gene targeting efficiency, as these animals are still undergoing development and growth, including in the liver. Neonatal mice were 1-week-old females and males at the time of vector and drug injection. Both vector and drug were injected i.p. in neonatal mice. Mice were housed at 18.3–23.9 °C, with 40–60% humidity on a 12-h light/12-h dark cycle. Food and water were given ad libitum.
Mouse AAV injection, drug dosing, bleeding and tissue sampling.
B6 mice received tail vain injections of rAAV8 packaging each vector at the designated dose and were bled at indicated time points into heparinized capillary tubes (Thermo Fisher). Mouse body weight was measured using a Scout Pro portable scale (Ohaus) at the indicated time points. Plasma samples were obtained by centrifugation at 9,600g for 10 min and used for ELISA assays of hF9 or hAAT, alanine aminotransferase (ALT) enzyme measurements and complete blood count tests. HU and fludarabine were resuspended in PBS and injected i.p. with the indicated dose/regimen. For our studies examining the in vivo effects of RNR inhibitors, we chose to use HU and fludarabine due to their FDA approval and well-established safety profiles. The drug dosing was based on information from several preclinical studies.
Briefly, an initial dose of 1,000 mg kg–1 injection of HU showed severe toxicity, leading us to reduce the dose to 300 mg kg–1 per day. For fludarabine, a 10% lethal dose (LD10) for a single dose or for five daily i.v. infusions was about 980 and 400 mg kg–1 per dose, respectively, in male mice according to the product monograph of fludarabine. Additionally, 5-d doses of half the LD10 or ~200 mg kg–1 per dose was considered safe, and administration of a smaller dose (125 mg kg–1) three times a day over 3 d (1,125 mg kg–1 total dose) produced the greatest activity in a leukemia tumor xenograft model. Using this information, we decided to use a dose of three 125 mg kg–1 injections per day (375 mg kg–1 d–1) of fludarabine for 3 d. All drug administrations were i.p. For BrdU labeling of proliferating mouse hepatocytes, BrdU was resuspended in PBS and i.p. injected at 200 mg kg–1 per day for 3 d or 7 d. DEN solution was prepared using saline. We determined an appropriate dose of DEN after finding that administration at 100 mg kg–1 resulted in severe weight loss requiring euthanasia. Dosing at 10 or 30 mg kg–1 as a single injection per day for 3 sequential days was well tolerated and used to examine the drug’s genotoxic effect on gene targeting. At the end of each experiment, mice were anesthetized with isoflurane and perfused transcardially with PBS, and liver tissues were quickly collected and cut into several pieces. The tissues for mRNA extractions were immediately submerged in RNAlater solution (Sigma-Aldrich) and stored at 4 °C until use. For gDNA or protein extraction, tissues were snap-frozen in liquid nitrogen and stored at −80 °C until use.
Hematology and liver pathology.
Complete blood count and liver histopathological analyses were performed in mice injected with three 125 mg kg–1 injections of fludarabine per day for 3 d. On the day of the last injection and 4 weeks after that, mice were submitted to the Veterinary Service Center at Stanford University for standard blood paneling tests and blinded analysis of liver pathology by a skilled veterinary pathologist.
AAV production.
rAAV vectors were produced as previously described using a triple transfection protocol with Ca3(PO4)2 or PEI 25K, followed by purification by CsCl gradient57 or using an AAVpro Purification kit (all serotypes; Takara). Purified rAAVs were stored at –80 °C until use. rAAV genomes were extracted and purified using a QIAamp MinElute Virus Spin kit (Qiagen) and were titered by qPCR. The sequence information of primers is shown in Supplementary Table 3.
Firefly luciferase assay.
Luciferase assays were performed using the ONE-Glo Luciferase Assay System (Promega) following the manufacturer’s instruction. Briefly, at indicated time points after rAAV (CAG-Fluc) transduction, an equal volume of the reconstituted substrate to the cultured medium was added to the cells grown in 96-well plates and incubated for 10 min with gentle shaking. Luminescent activity was measured using a plate reader and Tecan i-control Microplate Reader Software.
siRNA transfection.
ON-TARGETplus siRNA against human RRM1 and POLR2A as well as scramble control siRNA (Dharmacon) were transfected into Huh7 cells using RNAiMAX (Life Technologies) according to the manufacturer’s instructions. The final concentration of siRNA was 20 nM. AAVDJ packaging GAPDH-P2A-GFP vectors were added into the cultured medium at a multiplicity of infection of 20,000 vector copies per cell at 48 h after siRNA transfection. Flow cytometry was performed for detection of GFP+ fractions at the indicated time points.
Flow cytometry.
Huh7 or Hepa1–6 cells were collected and washed with cold PBS and resuspended in cold PBS containing 3% FBS. Cells were kept on ice and protected from light until analyzed. Singlet cells were determined based on forward scatter/side scatter (FSC/SSC) plot, and GFP+ fractions were gated based on a negative control, which was non-transduced cells. The number of GFP-expressing cells was evaluated using the BD FACSCalibur instrument with CellQuest software, and data were analyzed using the FlowJo software package. GFP+ hepatocytes were sorted by FACS at the Stanford Shared FACS Facility core. Singlet cells were determined based on FSC/SSC plot, and GFP+ fractions were gated based on a negative control, which was hepatocytes isolated from non-injected mice.
RNA extraction and cDNA preparation.
Total RNA was extracted using an RNeasy micro plus kit (Qiagen) according to the manufacturer’s protocol with DNase treatment. Liver tissue samples stabilized in RNAlater solution (~100 mg) were homogenized in RINO 1.5-ml Screw-Cap tubes filled with stainless steel beads and 600 μl of RLT buffer (including β-mercaptoethanol) using a bead homogenizer (Next Advance Bullet Blender Storm). Total RNA was extracted from the tissue lysates using an RNeasy plus mini kit (Qiagen) with additional on-column DNase treatment. cDNA was synthesized from 200–500 ng of total RNA using a High-Capacity RNA-to-cDNA kit (Life Technologies) according to the manufacturer’s instructions.
gDNA extraction.
Cultured cells were collected by trypsinization and washed with PBS. Total gDNA was extracted using a QIAamp DNA Mini kit (Qiagen) according to the manufacturer’s protocol with RNase A treatment. Snap-frozen liver tissue (~100 mg) was homogenized in RINO 1.5-ml Screw-Cap tubes filled with stainless steel beads and 600 μl of AL buffer using a bead homogenizer. Total RNA was extracted from the tissue lysates using a DNeasy Blood & Tissue kit (Qiagen).
PCR, qPCR and qPCR with reverse transcription.
PCRs to amplify genomic regions where homologous integrations occurred (junction PCR) were performed using Q5 Hot Start High-Fidelity 2× Master Mix (NEB). The following cycling conditions were used in 20-μl reactions: human ACTB (one cycle of 98 °C for 30 s, 28 cycles of 98 °C for 10 s, 60 °C for 15 s and 72 °C for 10 s and one cycle of 72 °C for 2 min), GAPDH-P2A junction (one cycle of 98 °C for 30 s, 35 cycles of 98 °C for 10 s, 62 °C for 15 s and 72 °C for 1 min and one cycle of 72 °C for 2 min), mouse Alb (one cycle of 98 °C for 30 s, 32 cycles of 98 °C for 10 s, 60 °C for 10 s and 72 °C for 2 min and one cycle of 72 °C for 2 min), hF9-Alb junction nested PCR (one cycle of 98 °C for 30 s, 20 cycles using 50 ng of gDNA for the first PCR and 25 cycles using 2 μl of the first PCR product for the second PCR of 98 °C for 10 s, 62 °C for 15 s and 72 °C for 1 min and one cycle of 72 °C for 2 min). Ten microliters of each PCR product was analyzed on 1% agarose gels containing ethidium bromide and visualized using ChemiDoc Imaging Systems (Bio-Rad). Primer sequence information is listed in Supplementary Table 3.
qPCR was performed in duplicate using Apex qPCR Green Master Mix (Genesee Scientific) and a CFX384 Touch Real-Time PCR Detection System (Bio-Rad) using the following cycling conditions: 95 °C for 15 min, 45 cycles of 95 °C for 10 s, 60 °C for 10 s and 72 °C for 10 s and one cycle of 95 °C for 10 s and 65 °C for 1 min and 65–97 °C (5 °C s–1). Standard curves for each primer set were generated using serially diluted linearized plasmid and used for quantification. CFX Maestro Software was used for data analysis, and relative mRNA expression levels were calculated by normalization against Actb. All sequence information of primers is listed in Supplementary Table 3.
Hepatocyte isolation.
Primary hepatocytes were isolated by anesthetizing mice, cannulating a 23-gauge needle into the inferior vena cava, cutting the liver portal vein and performing whole-liver perfusion with Hank’s balanced salt solution (HBSS) supplemented with 0.5 mM EDTA. Subsequently, a 0.25 mg ml–1 solution of collagenase (Sigma-Aldrich) and 5 mM CaCl2 (Thermo Fisher) dissolved in HBBS was used to perfuse and dissolve the liver tissue. Gentle physical agitation was used to fully dissociate the liver tissue, and hepatocytes were isolated by three serial centrifugations (50g) at 4 °C and counted following resuspension. Hepatocytes were kept on ice until FACS.
ddPCR.
Approximately 150 to 300 ng of gDNA from GFP+ hepatocytes or negative-control hepatocytes were digested in 25-μl reactions with SpeI at 37 °C for 1 h. Then, approximately 25 ng of digested gDNA was added to a 25-μl PCR reaction containing ddPCR Supermix for Probes (no dUTP; Bio-Rad), 900 nM target-specific primers and 250 nM amplicon-specific probes. Droplets were generated using 22 μl of PCR reactions and 70 μl of oil (Bio-Rad) according to manufacturer’s instructions using a QX200 droplet generator. Reactions were cycled as follows: 95 °C for 10 min, 50 cycles of 95 °C for 30 s, 60 °C for 30 s and 72 °C for 6 min and one cycle of 95 °C for 10 min, followed by a hold at 4 °C until droplet reading. Droplets were read using absolute quantification on the QX200 system. Primers to amplify a 1.6-kb non-targeted region of endogenous mouse Alb were 5′-CTGCTGTGCACCAGTTGATGTT-3′ and 5′-TGCTTTCTGGGTGTAGCGAACT-3′, combined with a HEX-labeled probe (5′-TCTGGTGCTGAGGACACGTAGCCCAGT-3′). Primers to amplify on-target HR with a 1.4-kb amplicon were 5′-GGGCAAGGCAACGTCATGG-3′ and 5′- CCAGGGTTCTCTTCCACGTC-3′, combined with a FAM-labeled probe (5′-GCCCAAGGCTAC AGCGGAGC-3′). Data were analyzed with Quantasoft software.
Protein extraction and western blotting.
Total cell lysates from cultured cells or mouse liver tissues were prepared using RIPA buffer containing Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher). Liver tissues were homogenized in RINO 1.5-ml screw-cap tubes filled with stainless steel beads and 600 μl of RIPA buffer using a bead homogenizer. Protein concentrations were measured using a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Fisher), and the same amount of protein for each sample was loaded into NuPAGE 4–12% Bis-Tris Protein Gels (Thermo Fisher). An iBlot2 transfer system (Thermo Fisher) was used for western blotting. PVDF membranes were blocked with 5% bovine serum albumin containing TBS plus 0.1% Tween 20 (TBS-T) buffer, and the following first antibodies were used: anti-RRM1 (D12F12; 1:1,000; Cell Signaling Technology), horseradish peroxidase (HRP)-conjugated anti-α-tubulin (11H10; Cell Signaling Technology; 1:2,000) and anti-γH2AX (EP854(2)Y; Abcam; 1:2,000). An anti-rabbit IgG HRP-conjugated antibody (1:5,000) (GE Healthcare) was used, and signals were detected using Pierce ECL Plus Western Blotting Substrate (Thermo Fisher) and ChemiDoc Imaging Systems (Bio-Rad).
Southern blotting.
Nuclear fractions of Huh7 cells were obtained using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher), and gDNA was extracted using a QIAamp DNA Mini kit (Qiagen) and digested overnight with XhoI (NEB) to cut only host gDNA. Digested DNA was run in a 1% TAE agarose gel at room temperature overnight. After electrophoresis, the gel was washed with denaturing buffer (3 M NaCl and 400 mM NaOH) twice for 5 min, and DNA was transferred to an Amersham Hybond-XL membrane (GE Healthcare) using transfer buffer (3 M NaCl and 8 mM NaOH) overnight. Membranes were washed with 2× saline sodium citrate (SSC) buffer for 5 min and blocked with UltraPure Salmon Sperm DNA (Thermo Fisher) in QuikHyb Hybridization Solution (Agilent Technologies) for 1 h at 65 °C. Probes for GFP (574 bp) were generated using gel-purified PCR amplicons containing GFP sequence and a BcaBEST Labeling kit (Takara) and [α−32P]-dCTP (PerkinElmer), and probe hybridization were performed overnight at 65 °C with rotation. The membrane was washed with 2× SSC buffer and with 2× SSC containing 0.1% SDS at 65 °C. Signals were visualized using a Personal Molecular Imager System (Bio-Rad) and analyzed with Quantity One 1-D software (Bio-Rad).
Immunohistochemistry staining of liver sections.
For all in situ hybridization and immunostaining experiments, liver tissue was dissected into 2- to 3-mm pieces and fixed for 24 h in 10% neutral buffered formalin (Sigma-Aldrich) at 4 °C. Tissue was subsequently processed through 10%, 20% and 30% sucrose solutions for 24 h each and frozen embedded into optimal cutting temperature (OCT) medium (Sakura Finetek) with liquid nitrogen and 2-methylbutane (Sigma-Aldrich). Frozen tissue was sectioned into 16-μm-thick sections using a Microm HM550 Microtome (Thermo Scientific). Tissue sections were blocked with antibody diluent comprised of 5.0% normal donkey serum (Jackson Immuno Research) and 0.1% Triton-X 100 (Sigma-Aldrich). GFP was stained with an anti-GFP chicken IgY primary antibody (1:100; Invitrogen), and phosphorylated Ser 139 γH2AX was stained with a rabbit monoclonal antibody (20E3; 1:250; Cell Signaling Technology). Polyclonal secondary detection antibodies consisted of anti-chicken IgY antibody conjugated to Alexa Fluor 488 (1:400; Jackson Immuno Research) and polyclonal anti-rabbit IgG antibody conjugated to Alexa Fluor 594 (1:400; Thermo Scientific).
Detection of BrdU-incorporated DNA was accomplished with heat denaturing in an antigen retrieval buffer (Advanced Cell Diagnostics), followed by staining with rat monoclonal anti-BrdU (BU1/75 (ICR1); 1:150; Abcam) and secondary Alexa Fluor 594 antibody (1:400; Thermo Scientific). All immunohistochemistry slides were mounted with Prolong Diamond Antifade with DAPI (Thermo Scientific) and imaged on a Zeiss LSM 880 confocal microscope with identical laser intensities and exposure times. Specificity of all staining procedures was ensured with appropriate biological controls and control slides stained with secondary antibody only.
For enhanced GFP (eGFP) experiments with Cas9, specimens were frozen in OCT compound (BioOptica), and 4-μm slices were obtained on a cryostat. The percentage of eGFP+ cells in liver specimens was detected by natural GFP fluorescence, while nuclei were visualized by Hoechst (10 lg ml–1) staining. Slides were mounted in Mowiol 4–88 (Sigma-Aldrich). Images were acquired on a Nikon Eclipse E-800 epifluorescence microscope with a charge-coupled device camera (DMX 1200F; Nikon). Digital images were acquired on a Nikon Ti Eclipse inverted fluorescence microscope equipped with an Intensilight Epi-fluorescence Illuminator, a Perfect Focus 3 system and a ×20 (0.45-NA) objective. Acquisition was performed with a DS-Qi2 16 Mpixel camera (Nikon). NIS Elements microscope imaging software (Nikon) was used to quantify the total number of cells (Hoechst staining) and GFP+ cells. The settings of Automated Spot Detection were set to detect all bright spots with a typical minimum diameter of 8 μm. The function ‘Detect all objects’ was activated, and spot detection output was directly exported to Excel, where data analysis was performed. For each animal, an average of 10,000 nuclei were counted. Measurements (GFP+ cells per total nuclei) were averaged for each animal, and the results were expressed as mean ± s.d. for each treatment.
RNAScope in situ hybridization of hF9.
Liver tissue was processed for RNA in situ hybridization as described above. Fixed frozen tissue was sectioned into 9-μm-thick sections, and RNAScope hybridization was performed according to the manufacturer’s protocol (Advanced Cell Diagnostics). A custom probe was designed to detect codon-optimized hF9 mRNA, while control probes targeted either murine peptidylprolyl isomerase B (PPIB) (positive control for RNA quality) or bacterial 4-hydroxy-tetrahydrodipicolinate reductase (dapB; negative control). RNA specificity was confirmed using RNase digestion of control tissue sections, and slides were counterstained with 50% hematoxylin (Thermo Scientific). Imaging was performed using a Leica DM2000 brightfield microscope.
Image analysis.
All image analysis was performed in a blinded manner. Analysis of BrdU incorporation was performed manually, requiring nuclear colocalization of BrdU signal and greater signal intensity over background to be recorded as a positive nucleus. Signal from overtly non-hepatocyte nuclei directly associated with larger liver structures, such as central veins or bile ducts, were not included in the tally. Analysis of BrdU incorporation and GFP colocalization was performed similarly. Scoring of phosphorylated Ser 139 γH2AX was performed using ImageJ software.
ELISA.
Mouse plasma samples were used to quantify hF9 or hAAT protein expression levels. ELISAs for hF9 were performed as previously described6 with the following antibodies: mouse anti-hF9 IgG (1:1,000; HIX-1, Sigma-Aldrich) and polyclonal goat anti-hF9 peroxidase-conjugated IgG (1:4,000; Enzyme Research). ELISAs for hAAT were performed as previously described56.
Targeted Illumina deep sequencing.
gDNA was extracted from liver tissue using phenol/chloroform extraction following incubation with RNase A and proteinase K. gDNA was submitted to the Genome Engineering and iPSC Center at Washington University in St. Louis for targeted deep sequencing of the gRNA target site on the 2 × 250 Illumina MiSeq platform.
Statistics.
GraphPad Prism was used for statistical analysis. Groups of two were analyzed by unpaired t-test. More than two groups were compared by one-way ANOVA with Bonferroni correction. ELISA and body weight time courses were analyzed by two-way ANOVA. Statistical significance was assumed with a P value of <0.05 (*), <0.01(**) and <0.001 (***). Bars in graphs represent the s.d. for each group.
Image analysis was performed using two-tailed t-tests between groups. Normally distributed data, determined by a D’Agostino Pearson test or Shapiro– Wilk test, were analyzed with a parametric Student’s t-test if groups had equal variance. Groups with unequal variance were analyzed with an unpaired t-test with Welch’s correction. Variance testing was performed with an F-test. Significance in non-normally distributed data was determined using a non-parametric Mann– Whitney U-test.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
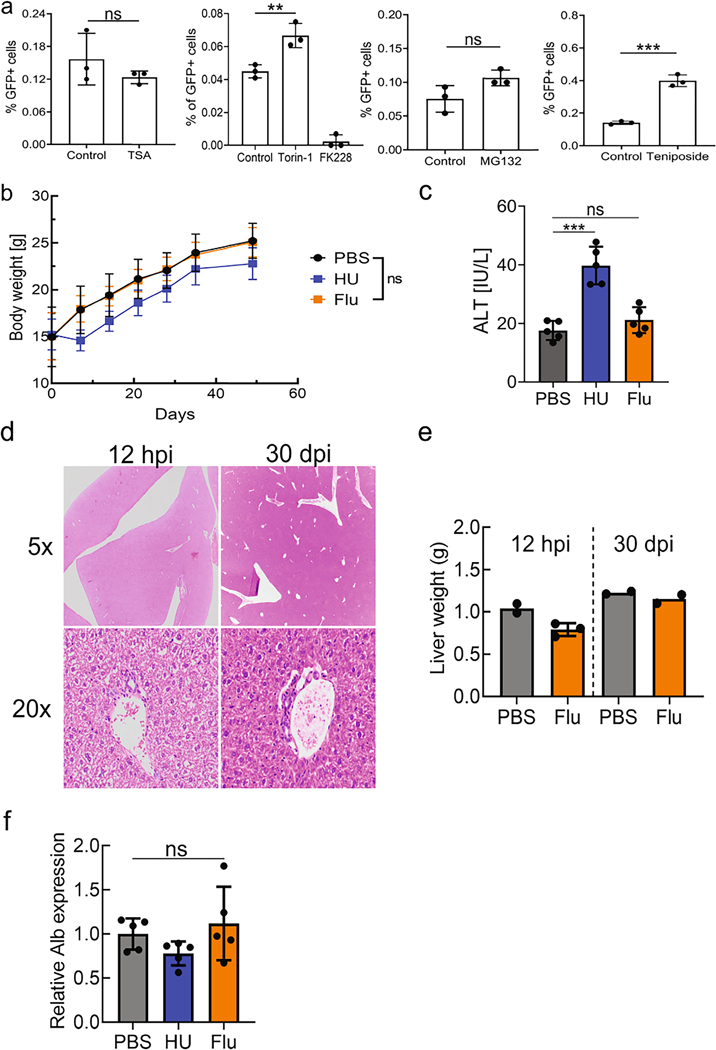
Extended Data Fig. 1 |. rAAV-mediated gene targeting efficiency in vitro with various compounds and the assessment of toxicity from fludarabine or hydroxyurea administration in mice.
a, The effect of each compound on rAAV-mediated gene targeting efficiency was tested in Huh7 cells by treatment with each drug followed by transduction with an AAVDJ gene targeting vector, GAPDH-P2A-GFP. Flow cytometry analysis of GFP positive cells 2 days after treatment is shown. Data is representative of two independent experiments, each with three biological replicates. Data is displayed as the group mean with error bars representing s.d.; n = 3 biological replicate wells. Significance testing was performed by a one-way ANOVA with Dunnett’s multiple comparison test and two-tailed t test. P-value for Torin-1 was .0046, MG132 was .076, TSA was .301, and .0003 for Teniposide. b, Mice were treated with hydroxyurea (HU) (300 mg/kg/day) or fludarabine (Flu) (375 mg/kg/day) for three days. Body weight was monitored over 6 weeks after drug administration. Data is displayed as the group mean with error bars representing s.d.; n = 5 individual mice per group. Significance testing was performed by a two-way ANOVA. c, Serum alanine transaminase (ALT) levels were evaluated 3 days after the last drug administration. Data is displayed as the group mean with error bars representing s.d.; n = 5 mice per group. Significance testing was performed by a one-way ANOVA with Dunnett’s multiple comparison test. P-value for HU-treatment was <0.0001 and .4343 for Flu-treatment. d, Mice were treated with fludarabine (Flu) (375 mg/kg/day) or PBS for three days. Groups of two PBS mice or three fludarabine-treated mice were submitted 12 hours after the final drug injection (hpi). A second cohort of mice was submitted 30 days after injections (dpi). Livers were collected for H&E staining and subsequent blinded analysis by a trained veterinary pathologist. Representative images of H&E-stained liver sections from fludarabine treated mice are shown, at 12 hpi (left column) and 28 dpi (right column). Top panel is images with a 5x objective and bottom panel with a 20x objective. e, Livers were weighed at the time of collection at either 12 hpi or 30 dpi. Hematological analysis of these mice is available in Supplementary Table 2. Each point represents data from one animal with n = 2 or 3 per group from one independent experiment. Error bars represent s.d., where applicable. f, Total RNA was extracted from liver tissue from the animals in Fig. 2. qPCR using primers (Fw1 and Rv1) (Fig. 2d) determined the levels of endogenous albumin mRNA following treatment with PBS, HU, or Flu. Data is shown as the mean and error bars represent s.d; n = 5 animals per group. Statistical testing was performed with a one-way ANOVA analysis followed by Dunnett’s t-test from one independent experiment.
Extended Data Fig. 2 |. RNAScope in situ hybridization of mAlb and hF9 in mouse liver.
a, Detection of hF9 mRNA (red) in liver sections using RNAScope in situ hybridization. Liver sections of mice from non-injected, PBS-treated, and Flu-treated groups at the end of experiment in Fig. 2 (60 days after rAAV transduction, ~12 weeks of age) were used for hybridization and counterstained with hematoxylin. Representative images from each injected group are shown. b. Representative images of RNAScope in situ hybridization are provided. In the first two columns, mAlb mRNA was stained in a non-injected normal mouse to determine the albumin locus expression characteristics. Specificity for RNA was confirmed by digestion of tissue with RNAseA. c, Additional images of hF9 mRNA staining in mice injected with Alb-P2A-hF9 vector, with or without fludarabine treatment are shown at various levels of magnification.
Extended Data Fig. 3 |. Fludarabine treatment increases AAV-HR in neonatal mice.
a, Female neonatal mice were injected with i.p. with PBS or Fludarabine (375 mg/kg) and four hours later with rAAV8-Alb-P2A-hF9 (2.5 × 1013 vg/kg) at one week of age. Fludarabine was administered once more, one day after vector injection. Four weeks later, plasma was drawn and hF9 levels measured at various time points by ELISA. Data shown is from n = 9 mice per group and displayed as the group mean with error bars representing s.d. Significance testing was performed by two-way ANOVA analysis. b, Genomic DNA was extracted from liver tissues of male neonates (from Fig. 2h) 84 days after rAAV8-Alb-P2A-hF9 vector injection and qPCR was performed to quantify the amount of total AAV genomes. Actb primers were used for quantification of the number of diploid genomes. Data is displayed as the group mean with error bars representing s.d.; n = 6 PBS-treated and n = 8 fludarabine-treated mice. Significance in all qPCR data in this figure was determined using two-tailed Student’s T tests, after testing for normal distribution with Shapiro-Wilk test and F tests for variance. For all qPCR data Actb mRNA was used for normalization and data is shown as relative expression to the PBS-treated group. c, Male and female mouse weights were monitored following treatment with PBS or Fludarabine and rAAV8-Alb-P2A-hF9. Data is displayed as the group mean with error bars representing s.d.; n = 17 PBS-treated and n = 18 fludarabine-treated mice. Significance testing was performed by 2-way ANOVA analysis. d, On-target fusion hF9 mRNA was quantified from liver tissues of the male mice at the end of the time course, using primers Fw1 and Rv2 (from Fig. 2e). Data is displayed as the group mean with error bars representing s.d.; n = 6 PBS-treated and n = 8 fludarabine-treated mice. A Shapiro-Wilk test was used to test for normal distribution, and F test determined variation between groups, and two-tailed Student’s t-test was used to test for significance in d-g. p-value was .008. e, Total hF9 mRNA was also quantified from the male mice using primers Fw2 and Rv3 (from Fig. 2f). Data is displayed as the group mean with error bars representing s.d.; n = 6 PBS-treated and n = 8 fludarabine-treated mice. p-value was .0035. f, The fraction of hF9 fusion mRNA derived from on-target HR out of the total amount of hF9 mRNA is shown. Data is displayed as the group mean with error bars representing s.d.; n = 6 PBS-treated and n = 8 fludarabine-treated mice. g, mAlb mRNA was quantified from the male mice using primers Fw1 and Rv1 (from Fig. 2). Data is displayed as the group mean with error bars representing s.d.; n = 6 PBS-treated and n = 8 fludarabine-treated mice.
Extended Data Fig. 4 |. The effect of different Flu dosing regimens on gene targeting efficiency.
a, Four-week-old mice were treated with various Flu doses, differing in the length of administration, as described in Fig. 2i, as well as injected i.v. with the gene targeting Alb-P2A-hF9 vector (1 × 1011 vg/ mouse) on Day 1 of Flu treatment. Plasma hF9 protein levels from the same mice in Fig. 2i were determined via ELISA for a 90-day time course. Data is displayed as the group mean with error bars representing s.d.; n = 4 mice per group. Significance testing was performed by a two-way ANOVA with Dunnett’s multiple comparison test. P-value of the 3-day treatment was .0004 and < .0001 for the 5-day treatment. b, Body weight of mice in each treated group were monitored throughout the time course. Data is displayed as the group mean with error bars representing s.d.; n = 4 mice per group. Statistics were performed by two-way ANOVA. P-value for the 1-day treatment was .001.
Extended Data Fig. 5 |. The effect of DeN administration on the efficiency of gene targeting in mice liver.
a, DEN (10 or 30 mg/kg) was administered through a single i.p. injection per day for three days. Mice were also injected i.v. with rAAV8 packaged Alb-P2A-hF9 gene targeting vector (1.0 × 1011 vg/ mouse) on Day 1. Body weight was measured at the indicated time points. Data is displayed as the group mean with error bars representing s.d.; n = 4 mice per group. Significance testing was performed by two-way ANOVA with Dunnett’s multiple comparison test. P-value for DEN-10 treatment was .1589 and .0065 for DEN-30 treatment. b, Plasma hF9 protein levels in each treatment group was determined by ELISA. Data is displayed as the group mean with error bars representing s.d.; n = 4 mice per group. Significance testing was performed by one-way ANOVA with Dunnett’s multiple comparison test. P-value for DEN-10 treatment was .1301 and < .0001 for DEN-30 treatment. c-f, Total RNA was extracted from liver tissues and qPCR was performed to quantify the expression levels of (c) total hF9 mRNA, (d) endogenous albumin mRNA and (e) on-target integration derived Alb-P2A-hF9 fusion mRNA. (f) The fraction of hF9 fusion mRNA derived from on-target HR out of the total amount of hF9 mRNA is given. Actb mRNA was used for normalization and each data is shown as relative expression to the vehicle (saline)-injected control group. Data is displayed as the group mean with error bars representing s.d.; n = 4 mice per group. Significance was determined using two-tailed unpaired t tests for normally distributed data with equal variance or Welch’s correction for data with significantly different variance. Testing of non-normally distributed data was performed with a non-parametric Mann-Whitney U test.
Extended Data Fig. 6 |. Fludarabine does not increase transduction of CRISPR/Cas9 encoded rAAV vectors or saCas9 mRNA.
a-b, At the conclusion of the two-week experiment in Fig. 5, DNA was extracted from livers of rAAV-injected animals. Viral genomes were quantified by qPCR and normalized to cellular genomic DNA. The donor rAAV repair template AAV-GFP-HDR is presented on the top panel (a), while AAV-Cas9 is displayed in the middle panel (b). Each point represents data from one animal, with n = 3 to 5 mice per group. Error bars represent s.d. Significance testing was performed using two-tailed t tests or Mann-Whitney U test if data was non-normally distributed. c, Mice were injected with SaCas9 (6.0E12 vg/kg) following fludarabine or PBS treatment as in Fig. 5f–g. Two weeks later, total RNA was extracted and saCas9 mRNA levels quantified by qPCR. n = 3 mice per group. Error bars represent s.d. Significance was determined using two-tailed t test.
Supplementary Material
Acknowledgements
This work was supported by grants from the NIH 2R01HL06427418A1 (M.A.K.), the Falk Medical Research Trust (M.A.K.), NIH DK098132 (C.J.S.) and the National Hemophilia Foundation (C.J.S.). A.F.M. was supported by intramural funds. We wish to acknowledge the Stanford Cell Sciences Imaging Facility for use of imaging equipment, the Stanford Genomics Facility for performing next-generation sequencing and the Stanford Department of Comparative Medicine’s Animal Histology Services and Stanford Veterinary Service Center for animal health examinations. We thank M.J. Finegold and J.G. Vilches-Moure for histology analysis. We also thank the Genome Engineering and iPSC Center at Washington University in St. Louis for performing next-generation sequencing and analysis. The microscope was funded by the Stanford Beckman Center. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the various funding bodies or universities involved.
Footnotes
Competing interests
S.T. and M.A.K. are named on patent applications related to this article. M.A.K. has commercial affiliations and stock and/or equity in companies with technology broadly related to this article. S.T. is a current employee of Daiichi-Sankyo Co., Ltd. G.A. is a current employee of Sangamo Therapeutics. The remaining authors declare no competing interests.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41587-022-01240-2.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41587-022-01240-2.
Data availability
The data generated in this manuscript are fully available upon reasonable request made to the corresponding author. Source data are provided with this paper.
References
- 1.Kotterman MA & Schaffer DV Engineering adeno-associated viruses for clinical gene therapy. Nat. Rev. Genet 15, 445–451 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keeler AM & Flotte TR Recombinant adeno-associated virus gene therapy in light of Luxturna (and Zolgensma and Glybera): where are we, and how did we get here? Annu. Rev. Virol 6, 601–621 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L, Wang H, Bell P, Mcmenamin D. & Wilson JM Brief report hepatic gene transfer in neonatal mice by adeno-associated virus serotype 8 vector. Hum. Gene Ther 23, 533–539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L. et al. AAV8-mediated hepatic gene transfer in infant rhesus monkeys (Macaca mulatta). Mol. Ther 19, 2012–2020 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunningham SC, Dane AP, Spinoulas A. & Alexander IE Gene delivery to the juvenile mouse liver using AAV2/8 vectors. Mol. Ther 16, 1081–1088 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Barzel A. et al. Promoterless gene targeting without nucleases ameliorates haemophilia B in mice. Nature 517, 360–364 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Porro F. et al. Promoterless gene targeting without nucleases rescues lethality of a Crigler–Najjar syndrome mouse model. EMBO Mol. Med 9, 1346–1355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hösel M. et al. Autophagy determines efficiency of liver-directed gene therapy with adeno-associated viral vectors. Hepatology 66, 252–265 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schreiber CA et al. An siRNA screen identifies the U2 snRNP spliceosome as a host restriction factor for recombinant adeno-associated viruses. PLoS Pathog. 11, e1005082 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson JS & Samulski RJ Enhancement of adeno-associated virus infection by mobilizing capsids into and out of the nucleolus. J. Virol 83, 2632–2644 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kia A, Yata T, Hajji N. & Hajitou A. Inhibition of histone deacetylation and DNA methylation improves gene expression mediated by the adeno-associated virus/phage in cancer cells. Viruses 5, 2561–2572 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okada T. et al. A histone deacetylase inhibitor enhances recombinant adeno-associated virus-mediated gene expression in tumor cells. Mol. Ther 13, 738–746 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Russell DW, Alexander IE & Miller AD DNA synthesis and topoisomerase inhibitors increase transduction by adeno-associated virus vectors. Proc. Natl Acad. Sci. USA 92, 5719–5723 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicolson SC, Li C, Hirsch ML, Setola V. & Samulski RJ Identification and validation of small molecules that enhance recombinant adeno-associated virus transduction following high-throughput screens. J. Virol 90, 7019–7031 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong L. et al. Heat-shock treatment-mediated increase in transduction by recombinant adeno-associated virus 2 vectors is independent of the cellular heat-shock protein 90. J. Biol. Chem 279, 12714–12723 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcus-Sekura CJ & Carter BJ Chromatin-like structure of adeno-associated virus DNA in infected cells. J. Virol 48, 79–87 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Alencastro G. et al. Improved genome editing through inhibition of FANCM and members of the BTR dissolvase complex. Mol. Ther 29.3, 1016–1027 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aye Y, Li M, Long MJC & Weiss RS Ribonucleotide reductase and cancer: biological mechanisms and targeted therapies. Oncogene 34, 2011–2021 (2015). [DOI] [PubMed] [Google Scholar]
- 19.De Caneva A. et al. Coupling AAV-mediated promoterless gene targeting to SaCas9 nuclease to efficiently correct liver metabolic diseases. JCI Insight 5, e128863. (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maurer-Schultze B, Siebert M. & Bassukas ID An in vivo study on the synchronizing effect of hydroxyurea. Exp. Cell. Res 174, 230–243 (1988). [DOI] [PubMed] [Google Scholar]
- 21.Wongt EA & Capecchi MR Homologous recombination between coinjected DNA sequences peaks in early to mid-S phase. Mol. Cell Biol 7, 2294–2295 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rothkamm K, Krüger I, Thompson LH, Löbrich M. & Biophysik F. Pathways of DNA double-strand break repair during the mammalian cell cycle the induction and repair of individual IR-induced DSBs. Mol. Cell. Biol 23, 5706–5715 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heyer W-D, Ehmsen KT & Liu J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet 44, 113–139 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandoval A, Consoli U, Plunkett W. & Anderson MD Fludarabine- mediated inhibition of nucleotide excision repair induces apoptosis in quiescent human lymphocytes. Clin. Cancer Res 2, 1731–1741 (1996). [PubMed] [Google Scholar]
- 25.Huang P, Chubb S. & Plunketts W. Termination of DNA synthesis by 9-β-d-arabinofuranosyl-2-fluoroadenine. A mechanism for cytotoxicity. J. Biol. Chem 265, 16617–16625 (1990). [PubMed] [Google Scholar]
- 26.Huang P, Sandoval A, Van Den Neste E, Keating MJ & Plunkett W. Inhibition of RNA transcription: a biochemical mechanism of action against chronic lymphocytic leukemia cells by fludarabine. Leukemia 14, 1405–1413 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Pettitt AR Mechanism of action of purine analogues in chronic lymphocytic leukaemia. Br. J. Haematol 121, 692–702 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Tseng WC, Derse D, Cheng YC, Brockman RW & Bennett LL In vitro biological activity of 9-β-d-arabinofuranosyl-2-fluoroadenine and the biochemical actions of its triphosphate on DNA polymerases and ribonucleotide reductase from HeLa cells. Mol. Pharmacol 21, 474–477 (1982). [PubMed] [Google Scholar]
- 29.Lans H, Hoeijmakers JHJ, Vermeulen W. & Marteijn JA The DNA damage response to transcription stress. Nat. Rev. Mol. Cell Biol 20, 766–784 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Yasuhara T. et al. Human Rad52 promotes XPG-mediated R-loop processing to initiate transcription-associated homologous recombination repair. Cell 175, 558–570 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Stoimenov I, Gottipati P, Schultz N. & Helleday T. Transcription inhibition by 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) causes DNA damage and triggers homologous recombination repair in mammalian cells. Mutat. Res 706, 1–6 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Stiff T. et al. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 64, 2390–2396 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Den Engelse L. & Philippus EJ In vivo repair of rat liver DNA damaged by dimethylnitrosamine or diethylnitrosamine. Chem. Biol. Interact 19, 111–124 (1977). [DOI] [PubMed] [Google Scholar]
- 34.Ferrara L, Parekh-Olmedo H. & Kmiec EB Enhanced oligonucleotide-directed gene targeting in mammalian cells following treatment with DNA damaging agents. Exp. Cell. Res 300, 170–179 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Porteus MH, Cathomen T, Weitzman MD & Baltimore D. Efficient gene targeting mediated by adeno-associated virus and DNA double-strand breaks. Mol. Cell. Biol 23, 3558–3565 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sentmanat MF, Peters ST, Florian CP, Connelly JP & Pruett-Miller SM A survey of validation strategies for CRISPR–Cas9 editing. Sci. Rep 8, 888 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ju HY, et al. Pharmacokinetics of fludarabine in pediatric hematopoietic stem cell transplantation. Blood 124, 2466–2466 (2014). [Google Scholar]
- 38.Maruyama T. et al. Increasing the efficiency of precise genome editing with CRISPR–Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol 33, 538–542 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin S, Staahl BT, Alla RK & Doudna JA Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 3, e04766 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu C. et al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell 16.2, 142–147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vasileva A, Linden RM & Jessberger R. Homologous recombination is required for AAV-mediated gene targeting. Nucleic Acids Res. 34, 3345–3360 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song J. et al. RS-1 enhances CRISPR/Cas9-and TALEN-mediated knock-in efficiency. Nat. Commun 7, 10548 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang W. et al. A high-throughput small molecule screen identifies farrerol as a potentiator of CRISPR/Cas9-mediated genome editing. eLife 9, e56008 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao T. et al. Small-molecule compounds boost genome-editing efficiency of cytosine base editor. Nucleic Acids Res. 49, 8974–8986 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chandler RJ et al. Vector design influences hepatic genotoxicity after adeno-associated virus gene therapy. J. Clin. Invest 125, 870–880 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Montini E. et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J. Clin. Invest 119, 964–975 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chandler RJ et al. Promoterless, nuclease-free genome editing confers a growth advantage for corrected hepatocytes in mice with methylmalonic acidemia. Hepatology 73, 2223–2237 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trobridge G, Hirata RK & Russell DW Gene targeting by adeno-associated virus vectors is cell-cycle dependent. Hum. Gene Ther 16, 522–526 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Kohama Y. et al. Adeno-associated virus-mediated gene delivery promotes S-phase entry-independent precise targeted integration in cardiomyocytes. Sci. Rep 10, 15348 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang P. & Plunkett W. Action of 9-β-d-arabinofuranosyl-2-fluoroadenine on RNA metabolism. Mol. Pharmacol 39, 449–455 (1991). [PubMed] [Google Scholar]
- 51.Mailand N. et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131, 887–900 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Ji JH et al. De novo phosphorylation of H2AX by WSTF regulates transcription-coupled homologous recombination repair. Nucleic Acids Res. 47, 6299–6314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mano M, Ippodrino R, Zentilin L, Zacchigna S. & Giacca M. Genome-wide RNAi screening identifies host restriction factors critical for in vivo AAV transduction. Proc. Natl Acad. Sci. USA 112, 11276–11281 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cervelli T. et al. Processing of recombinant AAV genomes occurs in specific nuclear structures that overlap with foci of DNA-damage-response proteins. J. Cell Sci 121, 349–357 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Pekrun K. et al. Using a barcoded AAV capsid library to select for clinically relevant gene therapy vectors. JCI Insight 4, e131610 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu J. et al. A 5′ noncoding exon containing engineered intron enhances transgene expression from recombinant AAV vectors in vivo. Hum. Gene Ther 28, 125–134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grimm D, Pandey K, Nakai H, Storm TA & Kay MA Liver transduction with recombinant adeno-associated virus is primarily restricted by capsid serotype not vector genotype. J. Virol 80, 426–439 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this manuscript are fully available upon reasonable request made to the corresponding author. Source data are provided with this paper.