Abstract
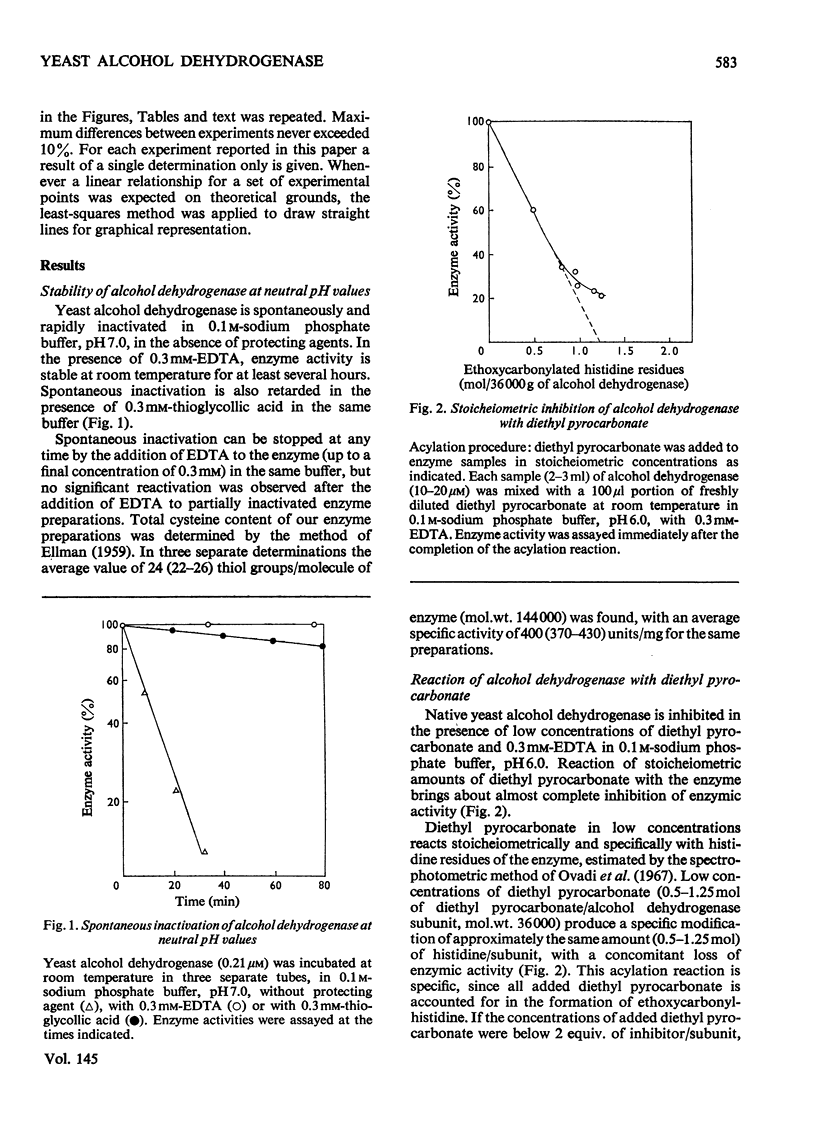
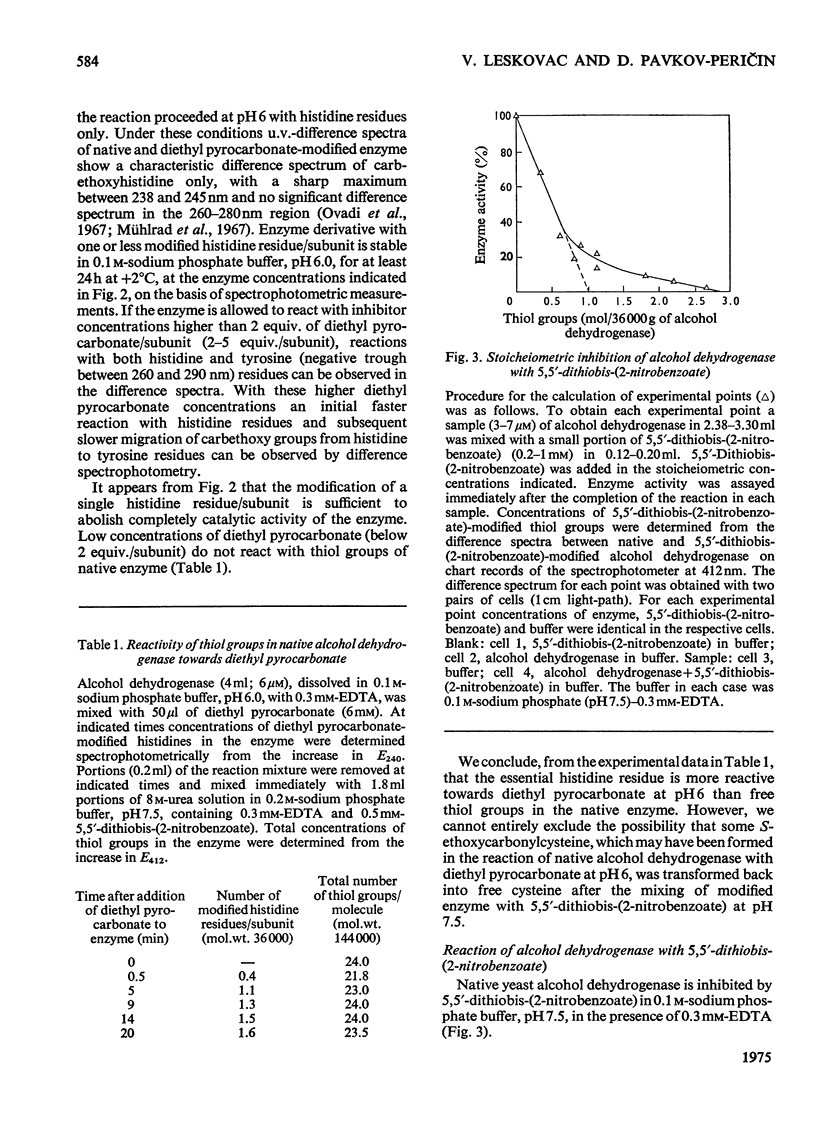
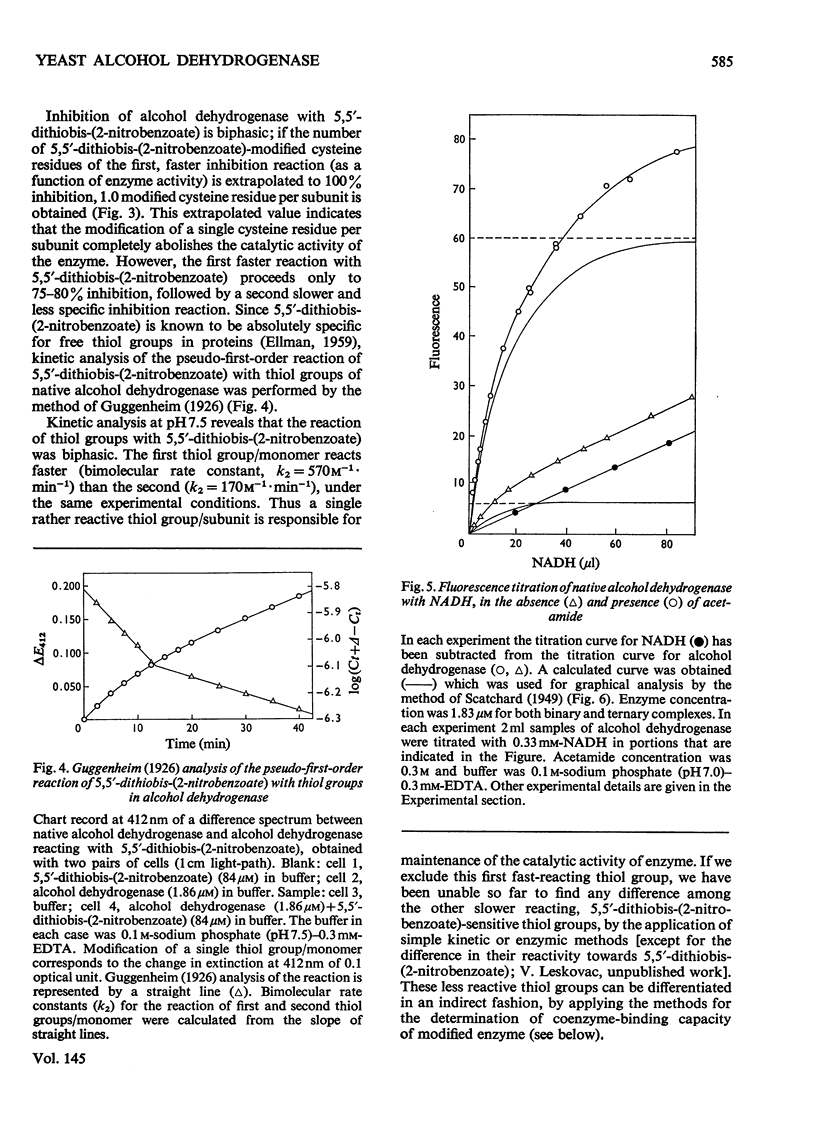
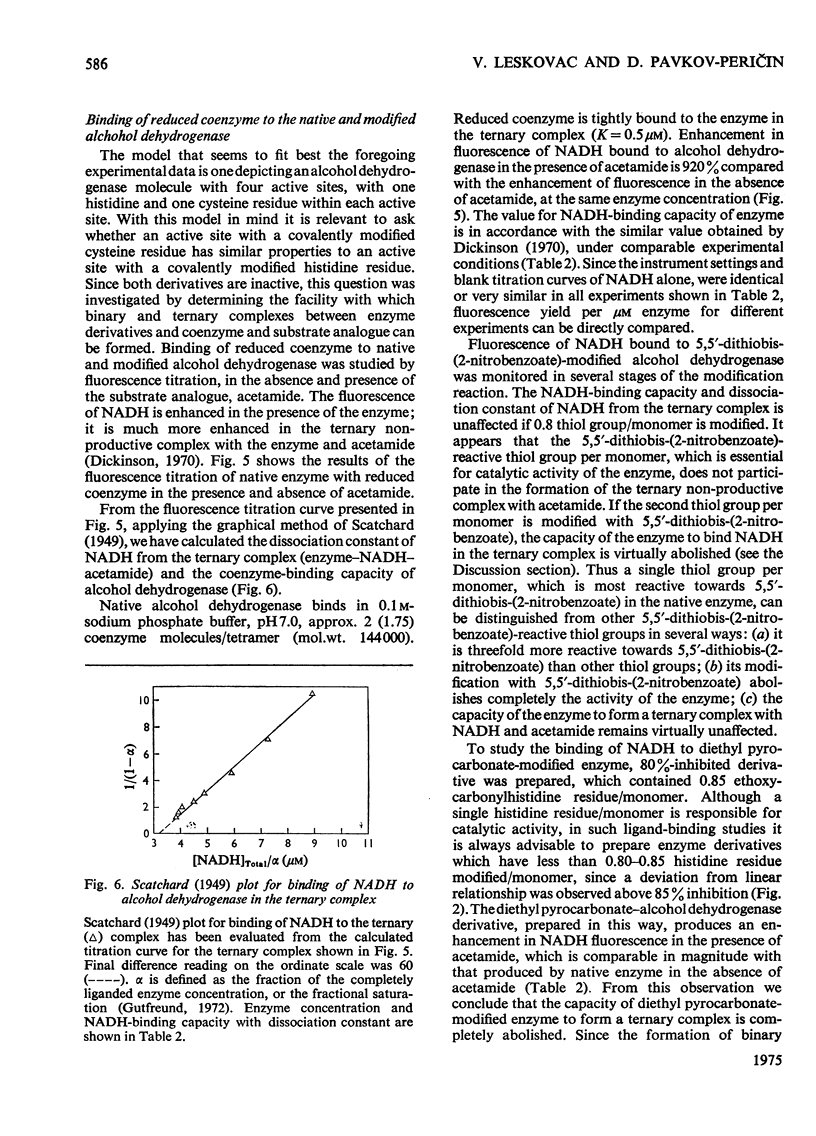
1. Yeast alcohol dehydrogenase (EC 1.1.1.1) is inhibited by stoicheiometric concentrations of diethyl pyrocarbonate. The inhibition is due to the acylation of a single histidine residue/monomer (mol.wt. 36000). 2. Alcohol dehydrogenase is also inhibited by stoicheiometric amounts of 5,5'-dithiobis-(2-nitrobenzoate), owing to the modification of a single cysteine residue/monomer. 3. Native alcohol dehydrogenase binds two molecules of reduced coenzyme/molecule of enzyme (mol.wt. 144000). 4. Modification of a single histidine residue/monomer by treatment with diethyl pyrocarbonate prevents the binding of acetamide in the ternary complex, enzyme-NADH-acetamede, but does not prevent the binding of NADH to the enzyme. 5. Modification of a single cysteine residue/monomer does not prevent the binding of acetamide to the ternary complex. After the modification of two thiol groups/monomer by treatment with 5,5'-dithiobis-(2-nitrobenzoate), the capacity of enzyme to bind coenzyme in the ternary complex was virtually abolished. 6. From the results presented in this paper we conclude that at least one histidine and one cysteine residue are closely associated in the substrate-binding site of alcohol dehydrogenase.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams M. J., Buehner M., Chandrasekhar K., Ford G. C., Hackert M. L., Liljas A., Rossmann M. G., Smiley I. E., Allison W. S., Everse J. Structure-function relationships in lactate dehydrogenase. Proc Natl Acad Sci U S A. 1973 Jul;70(7):1968–1972. doi: 10.1073/pnas.70.7.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury S. L., Jakoby W. B. Ligand interactions with yeast aldehyde dehydrogenase. J Biol Chem. 1971 Nov 25;246(22):6929–6932. [PubMed] [Google Scholar]
- Bühner M., Sund H. Yeast alcohol dehydrogenase: SH groups, disulfide groups, quaternary structure, and reactivation by reductive cleavage of disulfide groups. Eur J Biochem. 1969 Nov;11(1):73–79. doi: 10.1111/j.1432-1033.1969.tb00741.x. [DOI] [PubMed] [Google Scholar]
- Dickinson F. M. Role of the essential thiol groups of yeast alcohol dehydrogenase. Biochem J. 1972 Jan;126(1):133–138. doi: 10.1042/bj1260133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson F. M. The binding of dihydronicotinamide--adenine dinucleotide and pyridine-3-aldehyde--adenine dinucleotide by yeast alcohol dehydrogenase. Biochem J. 1970 Dec;120(4):821–830. doi: 10.1042/bj1200821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson M. Measurements of the concentration of active sites in preparations of yeast alcohol dehydrogenase. Eur J Biochem. 1974 Jan 3;41(1):31–36. doi: 10.1111/j.1432-1033.1974.tb03240.x. [DOI] [PubMed] [Google Scholar]
- ELLMAN G. L. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959 May;82(1):70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- HAYES J. E., Jr, VELICK S. F. Yeast alcohol dehydrogenase: molecular weight, coenzyme binding, and reaction equilibria. J Biol Chem. 1954 Mar;207(1):225–244. [PubMed] [Google Scholar]
- Holbrook J. J., Ingram V. A. Ionic properties of an essential histidine residue in pig heart lactate dehydrogenase. Biochem J. 1973 Apr;131(4):729–738. doi: 10.1042/bj1310729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook J. J., Pfleiderer G., Mella K., Volz M., Leskowac W., Jeckel R. The importance of SH-groups for enzymic activity. 7. The amino acid sequence around the essential SH-group of pig heart lactate dehydrogenase, isoenzyme I. Eur J Biochem. 1967 Jun;1(4):476–481. doi: 10.1111/j.1432-1033.1967.tb00095.x. [DOI] [PubMed] [Google Scholar]
- Hucho F., Markau U., Sund H. Studies of glutamate dehydrogenase. Characterization of histidine residues involved in the activity and association. Photoactivated labelling with pyridoxal 5'-phosphate. Eur J Biochem. 1973 Jan 3;32(1):69–75. doi: 10.1111/j.1432-1033.1973.tb02580.x. [DOI] [PubMed] [Google Scholar]
- KAGI J. H., VALLEE B. L. The role of zinc in alcohol dehydrogenase. V. The effect of metal-binding agents on thestructure of the yeast alcohol dehydrogenase molecule. J Biol Chem. 1960 Nov;235:3188–3192. [PubMed] [Google Scholar]
- Pfleiderer G., Auricchio F. The DPNH-binding capacity of various dehydrogenases. Biochem Biophys Res Commun. 1964 May 22;16(1):53–59. doi: 10.1016/0006-291x(64)90210-4. [DOI] [PubMed] [Google Scholar]
- Rossmann M. G., Adams M. J., Buehner M., Ford G. C., Hackert M. L., Lentz P. J., Jr, McPherson A., Jr, Schevitz R. W., Smiley I. E. Structural constraints of possible mechanisms of lactate dehydrogenase as shown by high resolution studies of the apoenzyme and a variety of enzyme complexes. Cold Spring Harb Symp Quant Biol. 1972;36:179–191. doi: 10.1101/sqb.1972.036.01.025. [DOI] [PubMed] [Google Scholar]
- Twu J. S., Chin C. C., Wold F. Studies on the active-site sulfhydryyl groups of yeast alcohol dehydrogenase. Biochemistry. 1973 Jul 17;12(15):2856–2862. doi: 10.1021/bi00739a013. [DOI] [PubMed] [Google Scholar]
- Twu J. S., Wold F. Butyl isocyanate, an active-site-specific reagent for yeast alcohol dehydrogenase. Biochemistry. 1973 Jan 30;12(3):381–386. doi: 10.1021/bi00727a003. [DOI] [PubMed] [Google Scholar]
- Woenckhaus C., Berghäuser J., Pfleiderer G. Markierung essentieller Aminosäurereste der Lactat-Dehydrogenase aus Schweineherz mit (Carbonyl-14C)3-(2-Brom-acetyl)-pyridin. Hoppe Seylers Z Physiol Chem. 1969 Apr;350(4):473–483. [PubMed] [Google Scholar]


