Abstract
Monoclonal antibody (mAb) technology has significantly contributed to basic research and clinical settings for various purposes, including protective and therapeutic drugs. However, a rapid and convenient method to generate high-affinity antigen-specific mAbs has not yet been reported. Here, we developed a rapid, easy, and low-cost protocol for antigen-specific mAb production from single memory B cells. Using this method, high-affinity IgG1 mAbs specific to the hapten 4-hydroxy-3-nitrophenylacetyl (NP) were established from NP-CGG immunized C57BL/6 mice within 6 days. Our mAb production system allows flexible switching of IgG1 to any other isotype with the same paratope, enabling the absolute quantification of antigen-specific serum antibody titers and affinity maturation. Additionally, we established a protocol for the production of IgM and IgA, retaining their functional pentamer and dimer structures. This method is also effective against human antigens and pathogens, making it a powerful tool for mAb development in both research and clinical settings.
Keywords: Monoclonal antibody, Memory B cells, B cell receptor, Single cell, Affinity maturation, IgM, IgG, IgE, IgA, J chain, Secretory component, 4-Hydroxy-3-nitrophenylacetyl (NP)
1. Motivation
The use of monoclonal antibodies is not only indispensable for basic research but also in clinical settings such as prophylactic, therapeutic, and diagnostic applications. Over the past few decades, advances in genetic engineering technologies have led to the development of methods to produce monoclonal antibodies from single cells, however, existing protocols do not allow to produce antigen-specific mAbs at the laboratory level rapidly, easily, and inexpensively. In this study, we developed a novel protocol that overcomes all these issues and created a standard mAbs that enables absolute quantification of antigen-specific serum antibody titers.
2. Introduction
Antibodies play an important role in biological defense through a variety of mechanisms such as the neutralization of viruses, bacterial toxins, and antibody-dependent cellular cytotoxicity [[1], [2], [3]]. Monoclonal antibodies (mAbs) that specifically bind to pathogen molecules are extremely useful for the treatment and diagnosis of infectious diseases, therefore, it is of great significance to establish technology for the rapid production of mAbs against emerging infectious diseases [[4], [5], [6], [7]]. FDA-approved antiviral mAbs were limited to respiratory syncytial virus, human immunodeficiency virus, Ebola virus, and rabies virus until 2019 when the number nearly doubled with the approval of several antibodies against COVID-19 [8]. While most developed mAbs are IgG subclass, it has been reported that airway mucosal protection by IgA prevents infectious diseases of influenza and SARS-CoV-2 viruses [9,10]. Thus, establishing a convenient protocol to switch virus-neutralizing IgG to other subclasses such as IgA is urgently required for research and development of therapeutic drugs against infectious diseases associated with mucosal immunity.
Hybridoma technology to produce mAbs was originally discovered by Kohler and Milstein in 1975, which has been widely applied in research and therapy [11,12]. For the generation of mAbs by hybridoma technology, mice are injected with a specific antigen that provokes an immune response. Then, cells producing antibodies are harvested from the mouse and fused with immortal myeloma cells to obtain hybridoma cells which have the antibody-producing ability and immortality allowing unlimited antibody secretion. A disadvantage of the hybridoma method is marked by long screening processes and periods (6–9 months), suboptimal selection of specific mAb-producing cells, no guarantee that mAbs are virus-free, and not applicable to human mAbs [13]. On the other hand, the phage display technology produces antibodies with affinity to target molecules using a library in which the variable regions of antibodies are displayed on phages. Phage display is more efficient than hybridoma technology, but it has the disadvantages of producing antibodies whose heavy and light chains do not naturally pair and of limited antibody repertoire making it difficult to isolate effective antibodies.
The single-cell technologies for mAb production have been widely utilized in recent years allowing the efficient establishment of mAbs specific for the antigen of interest by cloning the sequence encoding the B cell receptor (BCR) from antigen-specific B cells [14,15]. These methods include two major approaches: one is PCR amplification of the reverse transcribed BCR genes from FACS-sorted single cells, and the other is direct synthesis of the BCR sequences obtained by high-throughput single-cell sequencing. Problems with the former method include that 1) a limited number of alleles that can be covered, resulting in a low success rate of amplification by PCR, 2) additional PCR is required for sequencing and cloning of variable portions, 3) it takes several weeks in large scale systems to produce the amount of antibody required for evaluation, and 4) switching to various constant regions is not facilitated. The latter strategy could solve some of these problems [[16], [17], [18]]. While the single-cell RNA-seq method has proven to be highly accurate and allows for a deep analysis of the antibody repertoire [18], the costs of high-throughput library preparation and sequencing are substantial, and the sequencing cores can take weeks to months to process. Furthermore, most laboratories are not familiar with high-throughput sequencing techniques, which require expertise in RNA-seq library preparation and computational analysis.
Here, we describe a rapid, simple, and low-cost method to generate antigen-specific mAbs derived from mouse single-memory B cells. Using this method, hapten 4-hydroxy-3-nitrophenylacetyl (NP)-specific high-affinity IgG1 mAbs were obtained from NP-CGG immunized C57BL/6 mouse within 6 days. Our mAb production system applies to a variety of antigens including viral antigens and human antigens and allows flexible conversion of IgG1 into any other immunoglobulin isotypes that exert identical epitope-binding properties. The developed isotype-converted mAbs enabled us the absolute quantification of antigen-specific serum antibody titers.
3. Result
3.1. Designing multiplexed PCR primers for single-cell ig gene amplification
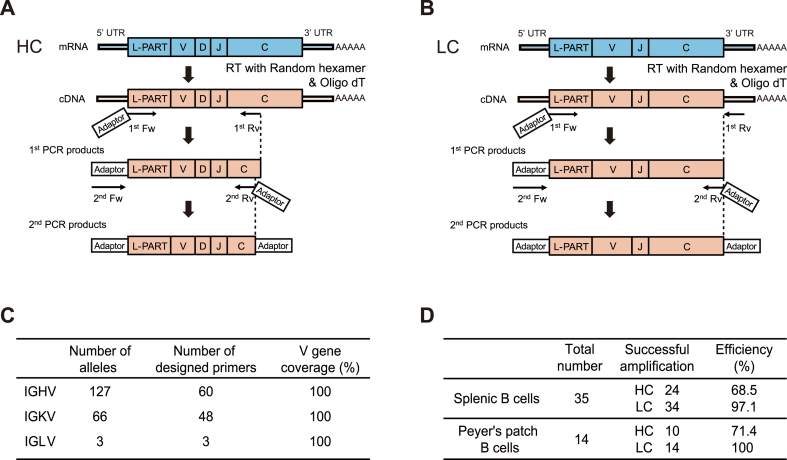
To establish a rapid, simple, and low-cost single-cell-based method for antigen-specific mAbs production, first, we attempted to develop PCR primers amplifying immunoglobulin (Ig) heavy and light chain genes. A schematic diagram of the strategy for amplifying heavy chain (HC) and light chain (LC) genes and cloning them into expression vectors is shown (Fig. 1A–B). Single cell-derived cDNAs synthesized using random hexamers and oligo dT primers were subjected to semi-nested RT-PCR amplification of HC and LC chain genes. The first PCR was performed using the single-cell cDNAs with multiplexed forward primers for the leader part and the reverse primers for the CH1 of IgH constant region or 3′ UTR of Igκ and Igλ genes. The second PCR was performed between 5′ and 3′ adapter sequences using the first PCR product as a template.
Fig. 1.
Designing multiplexed PCR primers for single-cell Ig gene amplification
(A-B) Schematic representation of HC (A) and LC (B) amplification by PCR. L-PART, leader part; UTR, untranslated region; Fw, forward primer; Rv, reverse primer
(C) The number and coverage of designed forward primers. The number of alleles representing the V gene sequences retrieved from the IMGT database. V gene coverage indicates the percentage of V genes covered by the primers.
(D) Efficiency of paired HC and LC amplification using designed primers. The total number represents the number of non-specific B220+CD19+IgG1+CD38+ live single memory B cells sorted from the spleen or Peyer's patch. Successful amplification means the number of amplified samples of HC and LC. The agarose gel electrophoresis of spleen-derived samples is shown in Supplementary Fig. 1. HC, heavy chain; LC, light chain.
The multiplexed forward primers for the first PCR were designed to cover all Ig variable gene alleles by annealing under specific conditions. The number of IGHV, IGKV, and IGLV alleles are 127, 66, and 3, respectively in C57BL/6 mice (Fig. 1C). We designed the first forward primers in the leader sequences to minimize the number of primers by excluding identical sequences and employing degenerate bases. Finally, 60, 48, and 3 forward primers were pooled as the multiplexed primer sets for IGHV, IGKV, and IGLV, respectively (Fig. 1C and Table S1). To confirm the successful amplification using designed primer sets, we performed dry sorting of single CD19+B220+IgG1+CD38+ memory B cells from C57BL/6 mouse spleen and Peyer's patch. Sorted single cells were reverse transcribed to cDNAs, then divided into two parts for independent HC and LC PCRs. Even though sorted B cells carried diverse V genes, the successful amplification rate of HC and LC was 68.5–71.4 % and 97.1–100 %, respectively, which was lower in HC compared to LC (Fig. 1D and Fig. S1).
3.2. Amplification of paired immunoglobulin genes from antigen-specific single memory B cells
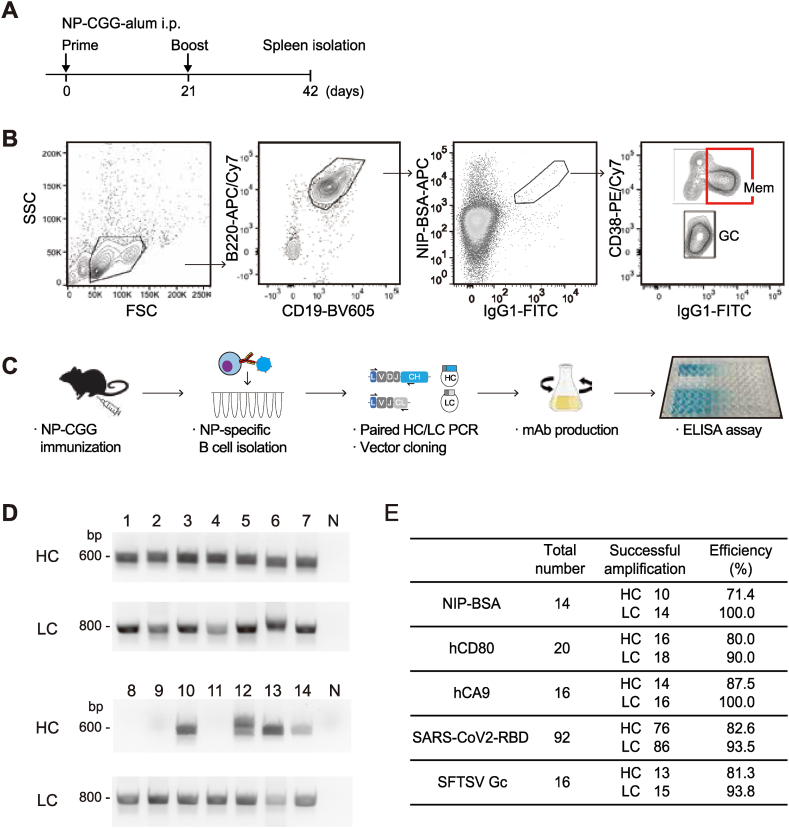
To test whether newly designed PCR primers are potent for the amplification of HC and LC genes from antigen-specific single memory B cells, we immunized C57BL/6 mice with 100 μg of alum-precipitated NP18-CGG. NP-specific B cells were detected using flow cytometry with hapten 4-hydroxy-5-iodo-3-nitrophenylacetyl (NIP) coupled to BSA because anti-NP antibodies bind the cross-reacting hapten NIP with higher affinity than the homologous hapten NP [19]. Immunization was performed twice in total at 3-week intervals and collected spleen on day 42 from the initial immunization (Fig. 2A). The NP-specific IgG1+ memory B cells (CD19+B220+IgG1+CD38+NIP+) were detected and single-cell sorted by flow cytometry as the gating strategy shows (Fig. 2B). The experimental workflow indicates the generation and evaluation of mAbs derived from antigen-specific single memory B cells (Fig. 2C). From the NP-specific IgG1+ memory B cells, we could amplify the HC and LC genes with success rates of 71.4 % and 100 %, respectively (Fig. 2D–E). Furthermore, we took this approach to other antigens and confirmed that our protocol efficiently works to obtain not only NP hapten but also various antigen-specific mAbs such as CD80 and CA9 human membrane proteins as well as SARS-CoV-2 spike receptor-binding domain (RBD) and SFTSV Gc viral antigens, in which the successful amplification rates were similar to randomly isolated B cells and NP-specific B cells (Fig. 1, Fig. 2E). These results imply that the established protocol works effectively to isolate paired HC and LC genes from various B cells as far as we have tested.
Fig. 2.
Amplification of paired immunoglobulin genes from antigen-specific single memory B cells
(A) Schematic diagram of experiment. C57BL/6 mouse was immunized twice at 3-week intervals. Each immunization was performed using 100 μg/mouse NP-CGG with alum adjuvant. The mouse was analyzed on day 42.
(B) Gating strategy for FACS sorting of antigen-specific memory B cells. Lymphocytes were gated by forward and side scatters. B220+CD19+IgG1+CD38+ live NP-specific memory B cells were sorted.
(C) Schematic workflow of mAb production and ELISA. Step 1: a mouse was immunized twice with NP-CGG. Step 2: NP-specific memory B cells were single-cell sorted (Day 1). Step 3: RT-PCR amplification of paired HC and LC genes. PCR products were cloned in the expression vectors (Day 1–2). Step 4: Expression vector plasmids were co-transfected into Expi293F cells and cultured for 4 days for mAb production (Day 2–6). mAbs were purified from the supernatant (Day 6). Step 5: Binding affinity to NP2-BSA and NP21-BSA was determined by ELISA (Day 6).
(D) Agarose gel electrophoresis of second PCR products. IgG1+NIP+ memory B cells were sorted and cDNAs were synthesized by reverse transcription. The HC and LC genes were amplified separately. The expected PCR products of HC and LC were approximately 600 and 800 base pairs (bp), respectively.
(E) Efficiency of successful amplification of HC and LC genes against various antigens. Total number represents the number of antigen-specific B220+CD19+IgG1+CD38+ live single memory B cells sorted from the spleen. Successful amplification means the number of amplified samples of HC and LC. hCD80, human CD80; hCA9, human CA9; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; RBD, receptor-binding domain; SFTSV, severe fever with thrombocytopenia syndrome virus; Gc, SFSTV viral glycoprotein Gc.
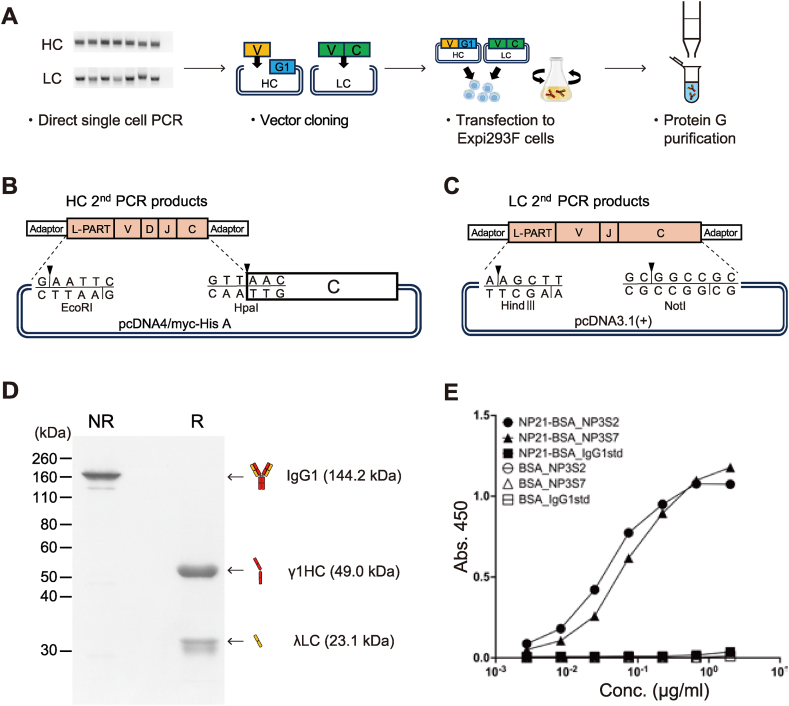
3.3. Production of antigen-specific mAbs from single B cells
The workflow from PCR to IgG1 antibody purification is shown (Fig. 3A). Expression vectors were designed for direct cloning of the second PCR products of HC and LC (Fig. 3B–C). The second PCR products were cloned into expression vectors and co-transfected into Expi293F cells and antibodies were purified from supernatants after the culture. For efficient mAb production, a series of HC and LC transfection ratios were compared. SDS-PAGE analysis showed that antibody production is higher at a 1:2 (HC:LC) plasmid weight ratio (Fig. S2A). After the Protein G purification, NP3S2 antibody was treated under non-reducing or reducing conditions for SDS-PAGE analysis that revealed a full-length IgG1 of approximately 150 kDa in the non-reducing condition and approximately 50 kDa of γ1HC and 25 kDa of λLC in the reducing condition (Fig. 3D). ELISA evaluation against BSA and NP21-BSA confirmed that IgG1 mAb clones NP3S2 and NP3S7 specifically bind to NP haptens by 0.054 μg/ml and 0.056 μg/ml of IC50 values, respectively (Fig. 3E).
Fig. 3.
Development of antigen-specific mAbs from single B cells
(A) Schematic workflow of mAb purification from single memory B cells. Step 1: HC and LC gene amplification by RT-PCR. Step 2: PCR products were cloned into expression vectors. Step 3: HC and LC expression plasmids were co-transfected into Expi293F cells. Step 4: mAbs were purified from culture supernatant by Protein G affinity purification.
(B) Cloning of variable domain into IgG1 HC expression vector containing IgG1 CH1 (from 41Asn) to CHS domain.
(C) Cloning of full-length LC gene into the expression vector.
(D) γ1HC and λLC expression plasmids encoding NP3S2 IgG1 were transfected into Expi293F cells. 3 μg of purified NP3S2 IgG1 antibody per well was separated on 10 % SDS-PAGE either non-reducing (NR) or reducing (R) conditions.
(E) mAbs NP3S2 and NP3S7 were produced in a combination of HC with paired LC obtained from single cells. NP3S2 and NP3S7 correspond to #2 and #7 in Fig. 2D. The NP-binding activity was measured by ELISA against NP21-BSA or BSA. IgG1std, mouse IgG1 standard.
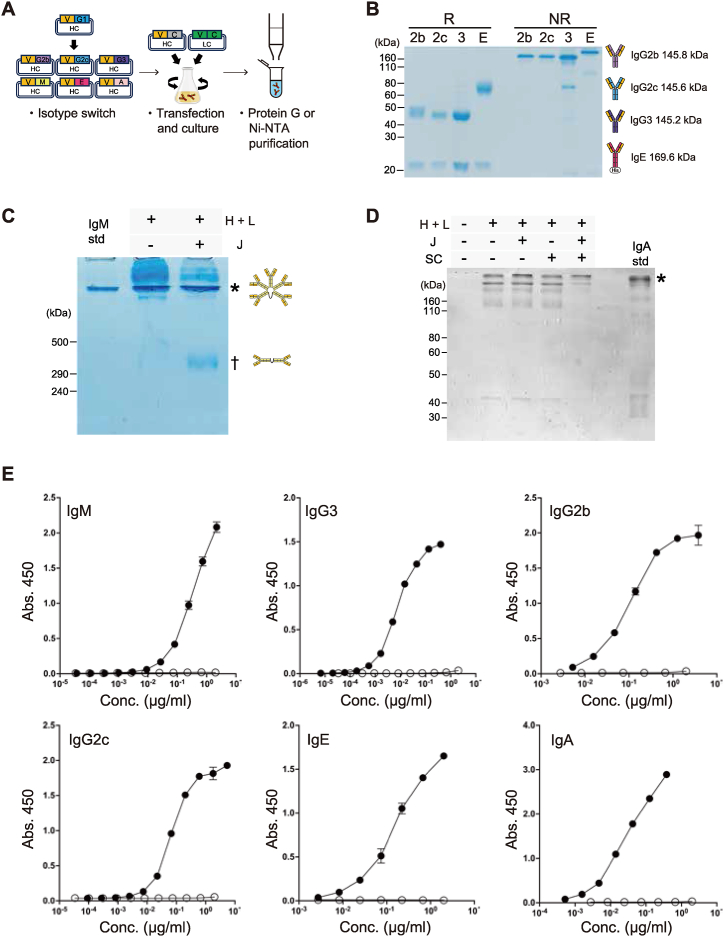
3.4. Isotype conversion of NP-specific antibodies with identical epitope binding properties
To produce a series of isotype-converted mAbs, the VDJ part of the NP3S2 γ1HC was cloned into the HC expression vectors of different isotypes and co-transfected with the original λLC into Expi293F (Fig. 4A). The IgG2b, IgG2c, and IgG3 mAbs were purified by affinity column purification using Protein G, and the IgE antibody was purified by Histidine-tag purification. Then, SDS-PAGE was performed for purified mAbs under the reducing or nonreducing conditions. We found HC2-LC2 bands at the expected size under the nonreducing condition and HC and LC bands at the expected size under the reducing condition (Fig. 4B). ELISA against NP-BSA confirmed that NP-specific IgG2b, IgG2c, IgG3, and IgE were successfully produced and purified (Fig. 4E).
Fig. 4.
Isotype conversion of NP-specific antibodies with identical epitope binding properties
(A) Schematic workflow for the isotype conversion. Step 1: The variable domain of the HC was cloned into each isotype expression vectors. Step 2: HC and LC expression plasmids were co-transfected into Expi293F cells. For IgM expression, J chain expression plasmid was added to HC and LC. For secretory IgA expression, J chain and SC expression plasmids were added to HC and LC. Step 3: mAbs were purified from the culture supernatant. IgG2b, IgG2c, and IgG3 mAbs were purified by Protein G affinity purification. Histidine-tagged IgE, IgM, and IgA antibodies were purified by Ni- NTA affinity purification.
(B) NP3S2-derived γ2bHC, γ2cHC, γ3HC, or εHC were transfected into Expi293F cells with λLC expression plasmid. 3 μg of purified mAb per well was separated on 10 % SDS-PAGE either non-reducing (NR) or reducing (R) conditions.
(C) IgM multimer formation in the absence or presence of the J chain. NP3S2-derived μHC and λLC were transfected into Expi293F cells with or without J chain expression plasmid and purified using Ni-NTA agarose. 3 μg of purified mAb per well was separated on 6 % SDS-PAGE under the NR condition. The molecular weights of IgM pentamer and dimer are 890.7 kDa and 365.7 kDa (μHC 64.4 kDa, λLC 23.1 kDa, J chain 15.7 kDa), respectively. Asterisk, pentamer; dagger indicates dimer; IgM std, mouse IgM standard; H + L, HC and LC; J, J chain.
(D) IgA dimer formation in the presence of J chain and SC. The molecular weight of sIgA is 381.8 kDa (αHC 51.1 kDa, λLC 23.1 kDa, J chain 15.7 kDa, SC 69.3 kDa). IgA std, mouse IgA standard; SC, secretory component.
(E) ELISA analysis of isotype switched NP3S2 mAbs. For IgM, IgG3, IgG2b, IgG2c, and IgA, NP-binding activity was measured by ELISA using plates coated with NP21-BSA. Since the IgE standard used in this study was an anti-TNP antibody that binds to NP21-BSA but not to NP2-BSA, NP2-BSA was used for the detection of NP3S2-IgE. Closed circles are NP3S2-derived mAbs. Open circles are each isotype standard.
3.5. Production of secretory IgM and IgA holding functional structures
The VDJ part of NP3S2 was cloned into the μHC expression vector and co-transfected with the original λLC into Expi293F resulting in a high occupancy of multimeric antibodies exceeding pentameric size (Fig. 4C, no J chain). Since IgM antibodies form stable pentamers via the J chain in vivo [20], we predicted that co-expression of the J chain would be necessary in this Expi293F cell system, unlike hybridoma. Simultaneous transfection of the J chain expression vector dramatically increased the occupancy of pentamers (Fig. 4C and Fig. S2B).
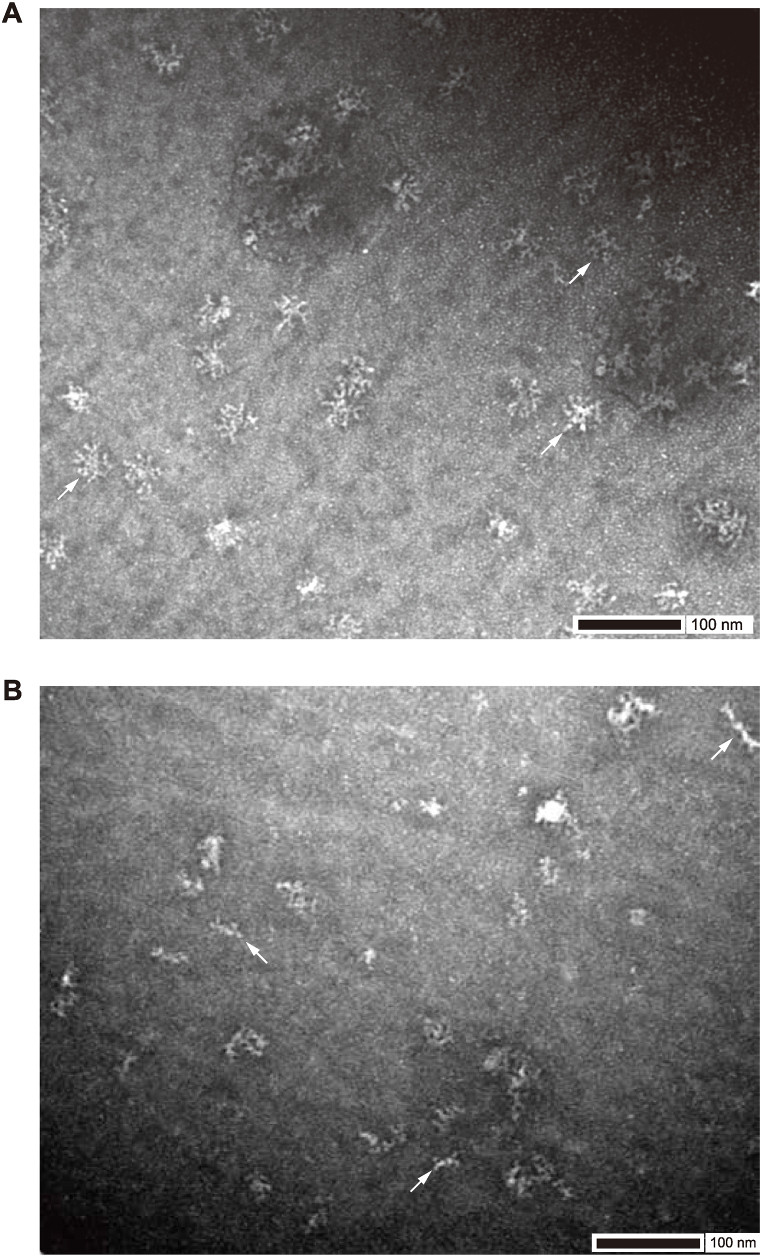
Intracellularly, IgA antibodies form a dimer where J chains are present in intermediate positions. Furthermore, a membrane protein called poly Ig receptor (pIgR), which is expressed in mucosal tissues, wraps around the IgA dimer and forms a secretory IgA (sIgA) by detaching the extracellular domain to form a state called secretory component (SC), which is secreted out of the cell as a stable dimer [21,22]. Dimer formation was examined by co-transfection of the J chain and SC expression constructs together with αHC and λLC. About 90 % of the compounds formed a dimer under conditions in which both J chain and SC were added, whereas the proportion of dimers was only about half when either J chain or SC was used separately (Fig. 4D). After the purification of IgA, we noticed that the position of the purified IgA band was higher than that of hybridoma-derived IgA under non-reducing conditions, which was caused by the presence of SC in our system (Fig. 4D and Fig. S2C). We confirmed that purified IgM and IgA mAbs bind to NP similar to IgG and IgE mAbs (Fig. 4E). Furthermore, we performed electron microscopy analysis of purified IgM and IgA to confirm whether the secretary IgM and IgA retained functional pentamer and dimer structures. The electron microscopy images indicated that purified IgM and IgA by our protocol formed IgM pentamers and IgA dimers as expected (Fig. 5A–B).
Fig. 5.
Electron microscopy revealed IgM pentamer and IgA dimer structures
(A) Purified NP3S2-IgM antibodies were observed using TEM. Typical IgM pentamers are indicated by arrows.
(B) Purified NP3S2-IgA antibodies were observed using TEM. Typical IgA dimers are indicated by arrows.
3.6. Affinity quantification of mAbs to specific antigens
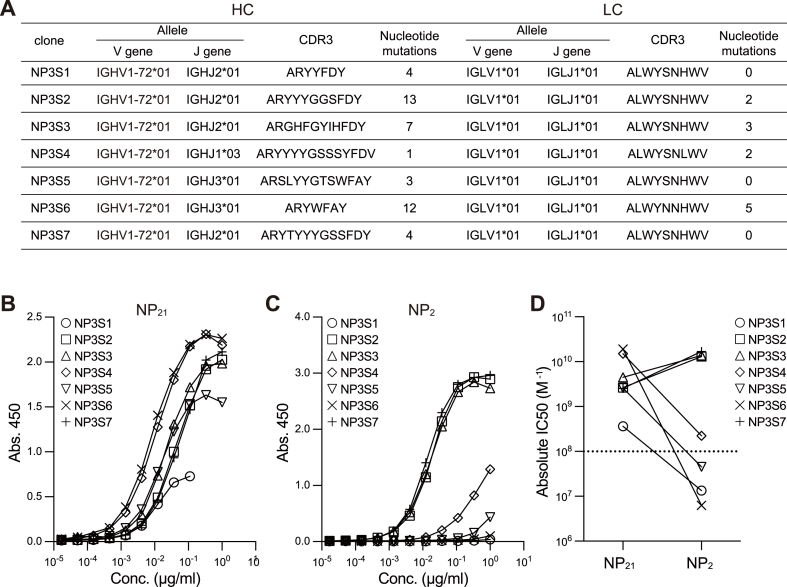
Sanger sequencing of the second PCR products confirmed that the NP-specific antibody genes were amplified as independent clones containing variable number of nucleotide mutations, suggesting that mAbs were derived from B cells that had undergone somatic hypermutation during the germinal center reaction (Fig. 6A). As expected, most clones consisted of IGHV1-72∗01 (also known as Vh186.2) HC genes and Igλ LC genes. Binding affinity against NP2-BSA or NP21-BSA was determined by ELISA for isolated mAbs, NP3S1, NP3S2, NP3S3, NP3S4, NP3S5, NP3S6, and NP3S7. These seven mAbs showed strong binding to NP21-BSA (Fig. 6B). Out of seven mAbs, four mAbs showed decreased affinity to NP2-BSA, whereas three mAbs showed increased affinity to NP2-BSA indicating varying affinities between high- and low-valent NPs (Fig. 6C–D). Thus, the established protocol is useful to determine the binding specificity and affinity levels of BCR encoded by antigen-specific memory B cells.
Fig. 6.
Affinity quantification of antigen-specific mAbs to antigens
(A) VH and VL genes, CDR3 amino acid sequence, and the number of nucleotide mutations of each NP-specific mAb clone are shown.
(B–C) ELISA analysis of NP-specific mAbs (clone: NP3S1-NP3S7). The NP-binding activity was measured by ELISA using plates coated with NP21-BSA (B) or NP2-BSA (C).
(D) The absolute IC50 was calculated using a nonlinear regression curve fit on Prism. The dotted line indicates the detection limit.
3.7. Absolute quantification of serum antigen-specific antibody titers
Quantification of antigen-specific antibody concentration in serum is often problematic because each polyclonal antibody has a different affinity to the specific antigen, therefore, antigen-specific antibody titer in serum is usually indicated by the relative titers. As a result, the concentration of antigen-specific antibodies, especially of all isotypes in serum has been hardly compared between different isotypes. Developing standard mAbs of all isotypes recognizing identical antigen by uniform affinity can solve this issue. Therefore, we established the method for absolute quantification of serum antigen-specific antibody titers by utilizing the NP-specific mAbs of all isotypes with identical paratopes.
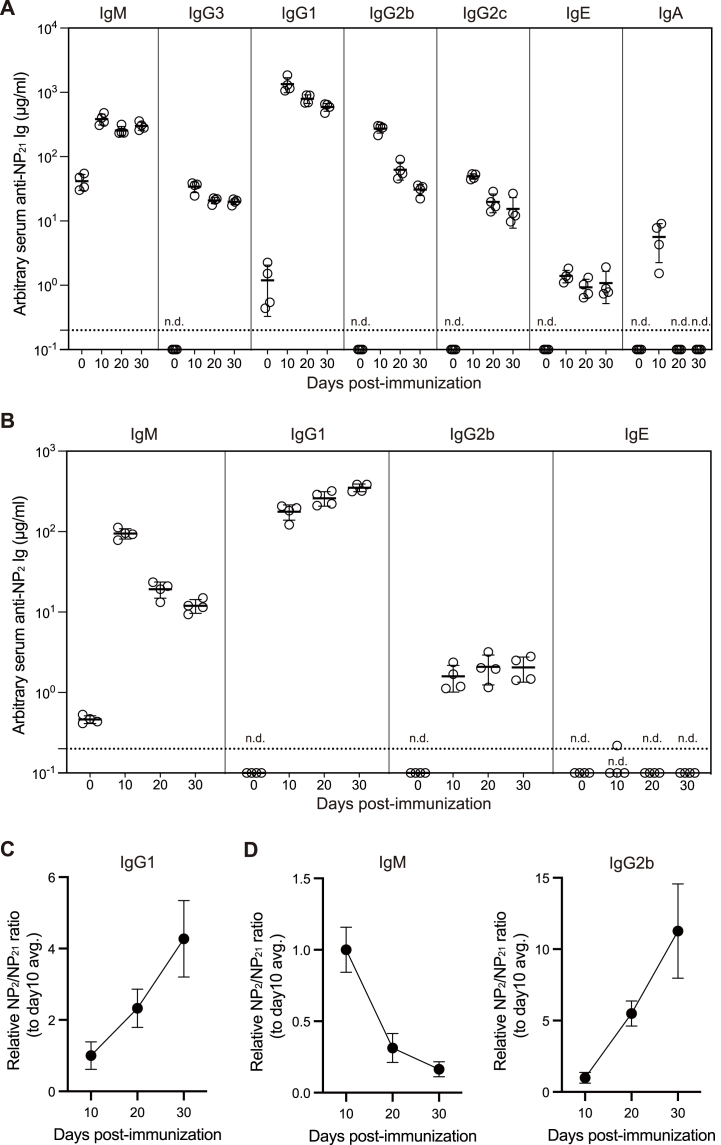
C57BL/6 mice were immunized with 100 μg of alum-precipitated NP18-CGG, and sera were collected every 10 days. The anti-NP antibody titers in serum were measured by ELISA against NP21-BSA using NP-specific standard isotypes retaining the VDJ gene from NP3S2 IgG1. Consistent with previous studies, NP-specific IgG, IgE, and IgA isotypes were virtually absent before immunization whereas IgM antibodies binding to high-valent NPs were present at low concentrations (Fig. 7A) [23]. NP-CGG immunization increased NP-specific antibody concentrations, which peaked on day 10 and then decreased in all isotypes. NP-specific IgE concentrations were 10- to 1000-fold lower than NP-specific IgM and IgG, indicating that NP-CGG does little to increase serum IgE concentrations, as expected from previous studies [24]. In addition, NP-specific IgA concentrations reached higher levels than IgE, however, that dropped to undetectable on days 20 and 30, unlike other isotypes.
Fig. 7.
Absolute quantification of serum antigen-specific antibody titers
(A-B) Absolute quantification of NP-specific antibody concentration in serum. C57BL/6 mice were immunized with the alum-precipitated NP-CGG. ELISA plates were coated with NP21-BSA (A) or NP2-BSA (B). Antibodies bound to the plates were detected using HRP-conjugated anti-mouse isotype-specific antibodies. NP3S2-IgG1 mAb from Fig. 3 or NP3S2-IgM, IgG3, IgG1, IgG2b, IgG2c, IgE, and IgA mAbs developed from NP3S2-IgG1, which are consisted of identical V-domain recognizing NP, were used as the standard. From the four-parameter logistic (4 PL) curve fit of NP3S2-derived mAbs on Prism, NP-specific antibody concentration of each sample was determined. n.d., not detectable.
(C–D) Post-immunization NP2/NP21 ratios in IgG1 (C), IgM (D), and IgG2b (D). The NP2/NP21 ratios were calculated based on the NP2-binding antibody concentration and the NP21-binding antibody concentration. Relative NP2/NP21 ratios were calculated by dividing the ratio at each time point by the average ratio at day 10. avg., average.
It is widely accepted that antibodies that bind to low-valent NPs represent high-affinity antibodies [25]. Therefore, we next determined NP-specific antibody titers against NP2-BSA using NP3S2-derived standard mAbs. Analysis of serum antibody concentrations indicated that IgG1 and IgG2b titers were maintained or even increased, whereas IgM titers decreased after the peak on day 10. IgE antibodies binding to NP2-BSA were undetectable at all time points (Fig. 7B).
Based on these data, the NP2 to NP21 binding ratio relative to day 10 was calculated. The NP2/NP21 ratio of IgG1 is thought to reflect affinity maturation, that increased over time as previously reported (Fig. 7C) [26]. The NP2/NP21 ratio of IgG2b increased over time as in IgG1, whereas that of IgM decreased as previously suggested [23] (Fig. 7D). Taken together, the present method and standard mAbs provide a unique method for absolute quantification of antigen-specific antibody titers and quantitative assessment of antibodies binding to haptens of different valencies.
4. Discussion
Affinity maturation of BCRs occurs in the germinal center (GC) B cells due to the accumulation of nucleotide mutations known as somatic hypermutation. Here, we reported a rapid, simple, and low-cost method to generate antigen-specific mAbs from mouse single-memory B cells. The present study provides a novel protocol to isolate mouse mAbs edited by somatic hypermutations that bind to NP antigen with high affinity.
The affinity of the NP-specific antibodies has been assessed by differential binding to antigens with either low or high haptenation ratios [25]. Tarlinton's group reported that frequencies of the high (>0.8) NP2/NP15 ratio is only 3 % on day 6 post-immunization whereas that reaches 55 % on day 21 post-immunization indicating the importance of germinal center reaction for the production of high-affinity B cells [27]. Using absolute quantification data in this study, we observed that the NP2/NP21 ratio of IgG1 and IgG2b increased from day 10 to day 30 after primary immunization, where NP2/NP21 ratio of IgM decreased consistently with the previous report [23]. Furthermore, a method for absolute quantification of antigen-specific antibody titers of all isotypes using standard mAbs with identical V domain enabled comparison of produced Ig concentrations among isotypes at different time points.
Rajewsky's group demonstrated that somatic antibody mutants become detectable on day 6 after immunization, and most of the somatic mutations accumulating in the memory compartment are introduced until day 14, which is predicted to cease around day 22 after primary immunization [28]. Milstein's group also found that somatic hypermutation is activated during the secondary response using oxazolone-chicken serum albumin as an antigen [29]. In this study, “boost” immunization was performed on day 21, a late phase of the ongoing GC reaction. The mutation frequency of mAbs isolated on day 42 was 2.14 % on average (0.34–4.42 %), which is much higher than that of the primary response (roughly 0.3, 0.6, 0.9, and 1.2 % on days 6, 8, 10, and 14 in the primary response [28]) suggesting those clones were from primary response. Our immunization strategy may contribute to the isolation of high-affinity IgG1 mAbs with a large number of accumulated mutations after the selective pressure in long-term GC reaction. The NP3S2 antibody obtained in this study had the p.W33L mutation, conventionally considered an indicator of high affinity for NPs. In addition, the antibody contained a total of nine amino acid mutations, including p.K38R and p.S66N, which have been observed in previous hybridoma-derived high-affinity antibodies [30], and these mutations are thought to contribute to the high affinity of the present NP3S2 antibody. The mechanism by which each mutation contributes to antigen binding can be clarified by structural analysis in the future.
For efficient recombinant antibody production, there is the optimal ratio of heavy and light chain genes to transfect in CHO cells [31]. We found that IgG1 production is higher at a 1:2 (HC:LC) plasmid weight ratio in Expi293F system. HC is maintained in the endoplasmic reticulum (ER) bound to binding immunoglobulin protein (BiP) and secreted out of the cell in the HC2LC2 form when it associates with LC. Moreover, since LC is known to decrease the degradation of unfolded HC polypeptide [32], a relatively high excess of LC may be useful for the efficient formation and secretion of antibodies.
Compared to the method using hybridomas, the probability of generating antibodies from individual memory B cells was higher, i.e., about 70–80 % in this study vs. about 1 % in the success rate of hybridoma fusion [28]. The present method may be useful for the study of affinity maturation by reducing bias caused by low efficiency in hybridoma generation. Moreover, the present technique is more cost-effective and time-saving than the production of mAbs from synthesizing heavy- and light-chain DNA from single-cell RNA seq [33]. On the other hand, the advantages of the method using single-cell RNA seq include the flexibility of codon optimization adapted to the expressing cell type, the replacement with the most efficient leader sequence, and the absence of bias due to PCR.
The mAbs specific for the antigen of interest can be generated in a short period using this method, allowing rapid preparation of new candidate therapeutic antibodies against emerging pandemics. In addition, epitopes of neutralizing antibodies have been identified for some pre-existing infectious diseases, such as gp41 of HIV-1 [34] and H protein of measles virus [35]. Immunization of mice with such antigens may lead to the discovery of mAbs with higher affinity than conventional ones, which could be used for the development of humanized antibody drugs as therapeutic or prophylactic agents.
By enabling absolute quantification of antibody titer of each isotype against a specific antigen, it can be applied to studies focusing on IgG2b, IgE, IgA, and others. Sandwich ELISA for each isotype antibody or ELISA using pooled serum from mice immunized with the relevant antigen has been used as a standard in the absence of monoclonal antibodies [36]. The former is difficult to achieve adequate absolute quantification because of the different coated antigens and the latter is a polyclonal antibody, which means that the binding mode of each antibody is different. The same problem exists in humans, especially with reference sera, which are difficult to standardize internationally [37]. The present method can produce standard antibodies with a single binding mode in a short time and is expected to partially solve the aforementioned problems.
In general, IgM is produced by the transfection of μHC, LC, and J chains in CHO cells for stable expression or in HEK293 cells for transient expression, however, IgM secreted by these methods includes not only pentamers but also dimers [38]. Therefore, IgM pentamers have been obtained through affinity chromatography purification of IgM or a combination of anion exchange chromatography and polyethylene glycol precipitation [39,40]. In this study, by adding a Histidine-tag to the carboxyl terminus of μHC, high-purity IgM purification was obtained only through Ni-NTA purification with a low proportion of impurities as confirmed by SDS-PAGE.
Previous methods for purification of sIgA include chromatography by mixing purified SC with a solution of IgA dimers purified from culture supernatants of hybridoma [41] or HC, LC, and J chain-expressing CHO cells [42]. These methods require purification by size exclusion chromatography or affinity chromatography for J chain or SC because of the high proportion of monomers. With the method of constant expression of HC, LC, J chain, and SC expression vectors in CHO cells and purification with Protein L [43], a high dimer ratio could be achieved, but the time required to obtain expression lines is a disadvantage. IgA dimerized by the J chain binds to pIgR expressed on the basolateral surface of the small intestine and is transported and secreted as sIgA [44]. Since 18 amino acids in the C terminal domain of αHC are required for binding to the J chain [45], the present expression vector preserves these amino acids and carries an 8xHistidine-tag at the end of the C terminal domain. The presence of the Histidine-tag may partially inhibit the binding to the J chain, and this point remains to be improved. The mouse sIgA purification method in this study is simple and yields higher purity than previous methods.
4.1. Limitations of the study
Although antibody genes containing leader sequences are amplified from single cell-derived cDNAs, expression efficiency can be reduced due to lack of introns [46]. While Expi293F is a human-derived cell line, antibody genes are not codon-optimized, thus, the 1:2 ratio of HC:LC is not always the best. The optimal ratio may differ from clone to clone. Furthermore, Histidine-tag was added to the heavy chains of IgE, IgM, and IgA for purification. We have tested functionality of these mAbs in ELISA, but the in vivo function of mAb has not been tested. This study does not go into the details of monoclonal antibodies against virus-related antigens or cell surface antigens. Therefore, it remains to be clarified whether present method can be applied to a wide range of biological antigens.
5. STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| TruStain FcX™ (anti-mouse CD16/32) Antibody | Biolegend | Cat#101319; RRID: AB_1574973 |
| FITC anti-mouse IgG1 Antibody | Biolegend | Cat#406605; RRID: AB_493292 |
| Brilliant Violet 605™ anti-mouse CD19 Antibody | Biolegend | Cat#115539; RRID: AB_11203538 |
| PE/Cyanine7 anti-mouse CD38 Antibody | Biolegend | Cat#102717; RRID: AB_2072892 |
| APC/Cyanine7 anti-mouse/human CD45R/B220 Antibody | Biolegend | Cat#103223; RRID: AB_313006 |
| Mouse IgG1 UNLB | SouthernBiotech | Cat#0102-01 |
| Mouse IgG2b UNLB | SouthernBiotech | Cat#0104-01 |
| Mouse IgG2c UNLB | SouthernBiotech | Cat#0122-01 |
| Mouse IgG3 UNLB | SouthernBiotech | Cat#0105-01 |
| Mouse IgA UNLB | SouthernBiotech | Cat#0106-01 |
| Mouse IgM UNLB | SouthernBiotech | Cat#0101-01 |
| Purified Mouse IgE, κ Isotype Control Standard | BD Pharmingen | Cat#557080 |
| Goat Anti-Mouse IgG1, Human ads-UNLB | SouthernBiotech | Cat#1070-01 |
| Goat Anti-Mouse IgG2b, Human ads-UNLB | SouthernBiotech | Cat#1090-01 |
| Goat Anti-Mouse IgG2c, Human ads-UNLB | SouthernBiotech | Cat#1079-01 |
| Goat Anti-Mouse IgG3, Human ads-UNLB | SouthernBiotech | Cat#1100-01 |
| Goat Anti-Mouse IgA-UNLB | SouthernBiotech | Cat#1040-01 |
| Goat Anti-Mouse IgM-UNLB | SouthernBiotech | Cat#1021-01 |
| Goat Anti-Mouse IgE-UNLB | SouthernBiotech | Cat#1110-01 |
| Goat Anti-Mouse IgG1, Human ads-HRP | SouthernBiotech | Cat#1070-05 |
| Goat Anti-Mouse IgG2b, Human ads-HRP | SouthernBiotech | Cat#1090-05 |
| Goat Anti-Mouse IgG2c, Human ads-HRP | SouthernBiotech | Cat#1079-05 |
| Goat Anti-Mouse IgG3, Human ads-HRP | SouthernBiotech | Cat#1100-05 |
| Goat Anti-Mouse IgA-HRP | SouthernBiotech | Cat#1040-05 |
| Goat Anti-Mouse IgM-HRP | SouthernBiotech | Cat#1021-05 |
| Goat Anti-Mouse IgE-HRP | SouthernBiotech | Cat#1110-05 |
| Bacterial and virus strains | ||
| DH5α competent cell, Champion™ DH5α high | Smobio | Cat#CC5202 |
| Chemicals, peptides, and recombinant proteins | ||
| Fetal Bovine Serum | Nichirei biosciences | Cat#175012 |
| Chicken gamma globulin fraction | Rockland | Cat#D602-0100 |
| NP-OSu | Biosearch Technologies | Cat#N-1010-100 |
| Critical commercial assays | ||
| Allophycocyanin Labeling Kit - SH | Dojindo | Cat#LK24 |
| NAP™-25 Columns | GE Healthcare | Cat#17085201 |
| TRI Reagent | Molecular Research Center | Cat#TR118 |
| ReverTra Ace® qPCR RT Master Mix | TOYOBO | Cat#FSQ-201 |
| PrimeScript™ II Reverse Transcriptase | Takara Bio | Cat#2690A |
| PrimeSTAR® HS DNA Polymerase | Takara Bio | Cat#R010A |
| Random Primers | Thermo Fisher | Cat#48190011 |
| NEBuilder HiFi DNA Assembly Master Mix | New England Biolabs | Cat#E2621S |
| FastGene™ Gel/PCR Extraction Kit | FastGene | Cat#FG-91302 |
| FastGene™ PlasmidMini | FastGene | Cat#FG-90502 |
| FavorPrep Plasmid Extraction Midi Kit | Favorgen | Cat#FAPDE 002 |
| Protein G Sepharose™ 4 Fast Flow | Cytiva | Cat#17061801 |
| Ni-NTA Agarose | QIAGEN | Cat#30210 |
| SureBlue™ TMB 1-Component Microwell Peroxidase Substrate | SeraCare | Cat#5120-0075 |
| ELISA POD Substrate TMB Kit (Popular) | Nacalai | Cat#05298-80 |
| Experimental models: Cell lines | ||
| Expi293™ Expression System Kit | Thermo Fisher | Cat#A14635 |
| Expi293F | Thermo Fisher | RRID:CVCL_D615 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6JJcl | CLEA Japan | N/A |
| Oligonucleotides | ||
| Primers for single cell PCR, see Table S1 | This paper | N/A |
| Primers for expression vector preparation, see Table S2 | This paper | N/A |
| Software and algorithms | ||
| FlowJo v10 | FlowJo, LLC | https://www.flowjo.com/ |
| CytExpert software v2.4 | Beckman Coulter | https://www.beckman.pt/ |
| GraphPad Prism 10 | GraphPad software, LLC | https://www.graphpad.com/ |
| Other | ||
| pcDNA3.1 (+) | Invitrogen | Addgene Plasmid; V790-20 |
| pcDNA4/myc-His A | Invitrogen | Addgene Plasmid; V86320 |
| Water deionized & sterilized | Nacalai tesque | Cat#06442-95 |
| Falcon® 5 mL Round Bottom Polystyrene Test Tube, with Cell Strainer Snap Cap | Corning | Cat#352235 |
| Clear Flat-Bottom Immuno Nonsterile 96-Well Plates | Thermo Fisher | Cat#442404 |
| Econo-Column® Chromatography Columns | Bio-Rad | Cat##7371012 |
| Superose™ 6 Increase 10/300 GL | Cytiva | Cat# 29091596 |
| 200 mesh Cu grid with a formvar carbon substrate | EM Japan | Cat#U1007 |
| Polyoxymethylene Sorbian Monolaurate | Nacalai tesque | Cat#28353-85 |
| Bovine serum albumin | Sigma | Cat#A9647-50G |
| Sodium Chloride | Nacalai tesque | Cat#31320-05 |
| Disodium Hydrogen phosphate Dodecahydrate | Fujifilm | Cat#196-02835 |
| Potassium dihydrogen phosphate | Sigma | Cat#24-5260-5 |
| Potassium Chloride | Katayama Chemical | Cat#24-4290 |
| Hydrochloric Acid | Fujifilm | Cat#080-01066 |
| Sulfuric Acid | Fujifilm | Cat#190-04675 |
| D-MEM(High Glucose) with L-Glutamine, Phenol Red and Sodium Pyruvate | Fujifilm | Cat#043-30085 |
| Cell Culture Plate VTC-P12 | Violamo | Cat#2-8588-02 |
| Bambanker | GC Lymphotec | Cat#CS-02-002 |
5.1. Mice
C57BL/6JJcl mice were purchased from CLEA Japan. Mice were bred and maintained in a specific pathogen-free facility. Animal care and experiments were conducted in accordance with the international guidelines of the ARRIVE and the Japanese Association for Laboratory Animal Science (JALAS), and the institutional guidelines established by the Ethics Review Committee for Animal Experimentation of Hiroshima University with the approval number: A24-61.
5.2. Protein conjugation
For 4-hydroxy-3-nitrophenylacetyl (NP)-chicken gamma globulin (CGG) conjugation, 67 μl of 50 mg/ml NP-OSu/DMSO was added to 5 ml of 5 mg/ml CGG in 100 mM NaHCO3 and incubated at RT for 60 min. For NP-bovine serum albumin (BSA) conjugation, 200 μl of 1 mg/ml or 50 mg/ml NP-OSu/DMSO was added to 3 ml of 2.5 mg/ml BSA in 100 mM NaHCO3 and incubated at RT for 15 min. For 4-hydroxy-3-iodo-5-nitrophenylacetyl (NIP)-BSA conjugation, 216 μl of 10 mg/ml NIP-OSu/DMSO was added to 2.7 ml of 5.6 mg/ml BSA in 100 mM NaHCO3 and incubated at RT for 30 min. Conjugated NP-CGG, NP-BSA, and NIP-BSA were purified using NAP-25 columns (GE Healthcare). The coupling ratio of NPs or NIPs per protein molecule was determined by measuring NP or NIP at 430 nm absorption. For NIP-BSA-APC, NIP-BSA was labeled with Allophycocyanin (APC) using the Allophycocyanin labeling kit-SH (Dojindo).
5.3. Immunization
Mice were intraperitoneally injected with alum-precipitated hapten-conjugated or recombinant proteins as previously described [47]. The part of mAb production can be started at any time points depends on purpose of study but should be after day 10 from primary immunization to detect antigen specific IgG1 memory B cells. Isolated memory B cells can be frozen at −80 °C until starting mAb production process.
5.4. Preparation of single-cell suspension
Single-cell suspensions were prepared in Dulbecco's modified Eagle's medium (DMEM) supplemented with 1 % fetal calf serum (FCS) and 1 mM EDTA. To isolate mononuclear cells from spleens, tissues were mashed using frosted glass microscope slides. Single cell suspensions from spleen were resuspended in Gey's solution (130 mM NH4Cl, 5.0 mM KCl, 0.8 mM Na2HPO4, 0.18 mM KH2PO4, 5.6 mM Glucose, 0.03 mM Phenol red, 1.0 mM MgCl2, 0.3 mM MgSO4, 1.5 mM CaCl2, 13 mM NaHCO3) for red blood cell lysis. Cell debris was removed using 70 μm nylon mesh. Cell pellets were resuspended in the appropriate buffer. For cryopreservation of single cell suspension, cells were resuspended in Bambanker (GC Lymphotec) according to manufacturer's instructions and stored in a −80 °C freezer.
5.5. Flow cytometry for single-cell sorting
For single-cell sorting, spleen cells were treated with TruStain FcX™ (anti-mouse CD16/32) antibody (BioLegend, #101319) to reduce non-specific labeling of the cells. Cells were stained with anti-IgG1-FITC (BioLegend, #406605) for 30 min on ice. After washing, cells were stained with NIP-BSA-APC, CD19-BV605 (BioLegend, #115539), CD38-PE/Cy7 (BioLegend, #102717), and B220-APC/Cy7 (BioLegend, #103223) for 30 min on ice. The cells were resuspended in FACS buffer (PBS containing 1 % FCS, 1 mM EDTA, and 0.05 % NaN3) supplemented with 0.2 μg/ml propidium iodide (PI) to exclude dead cells. Single-cell sorting was performed on Special Order System BD FACSAria II (BD Biosciences) or FACSAria Fusion (BD Biosciences). Cells were directly sorted into 8-strip PCR tubes. Tubes containing single cells were stored at −80 °C until proceeding to RT-PCR. Flow cytometric data were acquired on CytoFLEX S (Beckman Coulter). Flow cytometric data were analyzed using CytExpert software (v2.4, Beckman Coulter), or FlowJo software (v10.8.1, BD Biosciences).
5.6. Single-cell RT-PCR
C57BL/6 V genes are selected from IMGT (http://www.imgt.org), and alleles with matching first 30 bases are integrated. Melting temperatures from the leader part were adjusted to 58–60 °C. Custom primers designed with the adopter sequences are shown in Table S1.
Each 5 μl of pre-RT mix 1 containing 10 nmol dNTP, 50 pmol random hexamer and 50 pmol oligo dT was added to single-cell sorted PCR tubes. After heating at 65 °C for 5 min, tubes were immediately cooled on ice. 5 μl of the pre-RT mix 2 containing 10 U RNase inhibitor, 100 U PrimeScript™Ⅱ RTase and 1x PrimeScript Buffer (PrimeScript™ II Reverse Transcriptase, Takara Bio) was added to each well. For RT reaction, samples were incubated at 30 °C for 10 min, 42 °C for 40 min followed by heating at 72 °C for 15 min, then cooled on ice.
For PCR amplification of HC and LC genes, PrimeSTAR® HS DNA polymerase (Takara Bio), custom primers (See Table S1), and SimpliAmp Thermal Cycler (Applied Biosystems) were used. 1 μl of RT products added to each PCR tube containing 9 μl of 1st PCR mix includes 0.2 mM dNTP, 0.04 μM Forward primers, 0.2 μM reverse primers, 0.25 U PrimeSTAR HS DNA Polymerase, 1x PrimeSTAR Buffer as the final concentration. For 1st PCR, the initial denaturation at 98 °C for 3 min was followed by 40 cycles of sequential reaction of 98 °C for 10 s, 55 °C for 5 s, and 72 °C for 40 s. The 1st PCR products were diluted 10-fold by adding 90 μl of dH2O, of which 1 μl was added to the second PCR mix including 0.2 mM dNTP, 0.2 μM forward and reverse primers, 0.25 U PrimeSTAR HS DNA Polymerase, 1x PrimeSTAR Buffer as the final concentration. For the second PCR, the initial denaturation at 98 °C for 3 min was followed by 40 cycles of sequential reaction of 98 °C for 10 s, 68 °C for 40 s.
PCR fragments were purified with FastGene™ Gel/PCR Extraction Kit (FastGene) and assembled into a linearized pcDNA vector using NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs) according to the manufacturer's instructions. Final 1 μl of assembled products mixed with 10 μl DH5α competent cell (Smobio) for transformation. The vectors were extracted and purified using FastGene Plasmid Mini Kit (FastGene) or FavorPrep Plasmid Extraction Midi Kit (Favorgen)
HC and LC genes were cloned into each expression vector and sequenced using a CMV-forward primer (5′-CGCAAATGGGCGGTAGGCGTG-3′) located upstream of the cloning part. For the calculation of mutation frequency, the number of nucleotide mutations in each IGHV gene was divided by the length of each V gene. Since the error frequency of PrimeSTAR HS DNA Polymerase is extremely low (1 in 2.08 x 104 bases), we did not consider the polymerase error rate in the calculation.
5.7. Plasmid
The complementary DNA (cDNA) of C57BL/6JJcl spleen and Peyer's patch were prepared using TRIreagent (Cosmo Bio) and ReverTra Ace (TOYOBO) according to the manufacturer's instructions. The IgG1, IgG2b, IgG2c, IgG3, IgE, and IgM Fc domain fragments were amplified from cDNA of spleen using custom primers (See Table S2). The IgA Fc domain, Joining chain (J chain), and secretory component (SC) fragments were amplified from cDNA of Peyer's patch using custom primers (See Table S2). pcDNA4 expression plasmids encoding IgG1/IgG2b/IgG2c/IgG3/IgA/IgE/IgM Fc domain, J chain, and SC were generated by assembly of PCR products using NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs) according to the manufacturer's instructions. Final 1 μl of assembled products mixed with 10 μl DH5α competent cell (Smobio) for transformation. The vectors were extracted and purified using FastGene Plasmid Mini Kit (FastGene) or FavorPrep Plasmid Extraction Midi Kit (Favorgen).
To produce each isotype expression vector with the NP-specific variable domain, the amplified variable domain of NP3S2 using custom primers (Table S2) was assembled into linearized each Fc domain-containing vector using NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs) according to the manufacturer's instructions.
5.8. Monoclonal antibody production
For IgG1/IgG2b/IgG2c/IgG3/IgE preparation, the pcDNA3 (Invitrogen) vectors containing an LC gene and the pcDNA4 (Invitrogen) vectors containing an HC gene were simultaneously transfected into Expi293F cells using Expi293 Expression System Kit (Thermo Fisher Scientific). Expi293F cells were incubated at 37 °C, 8 % CO2 incubator with shaking at 125 rpm (for 25 ml culture in 200 ml flask) or 260 rpm (for 1 ml culture in 12-well plate (Violamo)). For IgM production, the J chain-containing vector was simultaneously transfected with μHC and λLC. For secretory IgA production, the J chain containing vector and the secretory component containing vector were simultaneously transfected with αHC and λLC. Five days after the transfection, the culture supernatants were collected and subjected to ELISA or SDS-PAGE.
For Fig. 4D, the J chain and SC expression vector were transfected with αHC and λLC in a 1 ml culture scale of Expi293F. The amounts of αHC, λLC, J chain, and SC expression vectors were 200 ng, 400 ng, 100 ng, and 300 ng, respectively.
5.9. Antibody purification
For IgG1/IgG2b/IgG2c/IgG3 purification, Protein G Sepharose™ 4 Fast Flow (cytiva) was used according to the manufacturer's instructions. For IgA/IgM/IgE purification from 25 ml culture volume, 500 μl Ni-NTA Agarose (QIAGEN) was mixed with culture supernatants, rotated at 4 °C for 1 overnight, and loaded onto the Econo-Column® Chromatography Columns (Bio-Rad) followed by wash step with 50 ml Wash buffer (50 mM Tris-HCl pH 8.0, 500 mM NaCl, 30 mM Imidazole). Histidine-tagged antibodies were eluted with 500 μl Elution buffer (50 mM Tris-HCl pH 8.0, 500 mM NaCl, 500 mM Imidazole) 5–10 times. For IgA/IgM/IgE purification from 1 ml culture volume, 50 μl Ni-NTA Agarose (QIAGEN) was mixed with culture supernatants and rotated at 4 °C for 1 overnight. After a centrifugation step (5 min, 300g, 4 °C), supernatants were discarded and pellets were recovered by 1 ml Wash buffer and centrifuged (5 min, 300g, 4 °C) followed by discarding supernatants. Histidine-tagged antibodies were eluted with 50 μl Elution buffer (50 mM Tris-HCl pH 8.0, 500 mM NaCl, 500 mM Imidazole) once.
5.10. ELISA
For the measurement of antibody concentration, ELISA plates (Thermo Fisher Scientific) were coated with 2 μg/ml anti-mouse IgM/IgG3/IgG1/IgG2b/IgG2c/IgE/IgA overnight at 4 °C. Plates were washed three times with PBS supplemented with 0.05 % Tween 20 (PBS-T) and coated with (1 % BSA in PBS) for at least 1 h at room temperature and then washed three times with PBS-T. Supernatants containing monoclonal antibodies and 2 μg/ml standard IgM/IgG3/IgG1/IgG2b/IgG2c/IgA (Southern Biotech) or IgE (BD Biosciences) were diluted in PBS-T and three-fold series dilutions were performed in 5–10 steps. After wells were washed with PBS-T three times, horseradish peroxidase (HRP)-conjugated anti-mouse IgM/IgG3/IgG1/IgG2b/IgG2c/IgE/IgA (Southern Biotech) antibodies (1:3000) diluted in PBS-T were added.
For Fig. 3E, ELISA plates were coated with 2 μg/ml BSA or NP21-BSA for 1 overnight. Plates were washed with PBS-T three times and coated with (1 % BSA in PBS) for at least 1 h at room temperature. After wells were washed with PBS-T three times, developed antibodies or standard antibodies (Mouse IgG1 UNLB) were tested at 2 μg/ml in the top well and three-fold series dilutions were performed in 6 steps. After wells were washed with PBS-T three times, HRP-conjugated anti-mouse IgG1 antibody (1:3000) diluted in PBS-T was added.
For Fig. 4E, ELISA plates were coated with 2 μg/ml NP2- or NP21-BSA for 1 overnight. Plates were washed with PBS-T three times and coated with (1 % BSA in PBS) for at least 1 h at room temperature. After wells were washed with PBS-T three times, NP3S2 antibodies or each isotype standard antibody (Mouse IgM/IgG3/IgG2b/IgG2c/IgE/IgA UNLB) tested 2 μg/ml in top well and three-fold series dilutions were performed in 6 steps. After wells were washed with PBS-T three times, HRP-conjugated anti-mouse IgM/IgG3/IgG2b/IgG2c/IgE/IgA antibody (1:3000) diluted in PBS-T was added.
For Fig. 6B–D, ELISA plates were coated with 2 μg/ml NP2- or NP21-BSA for 1 overnight. Plates were washed with PBS-T three times and coated with (1 % BSA in PBS) for at least 1 h at room temperature. After the wells were washed with PBS-T three times, antibodies tested 1 μg/ml in the top well, and three-fold series dilutions were performed in 10 steps. Only NP3S1 was started at 0.1 μg/ml due to low yield. After wells were washed with PBS-T three times, HRP-conjugated anti-mouse IgG1 antibody (1:3000) diluted in PBS-T was added.
For Fig. 7A–D, ELISA plates were coated with 2 μg/ml NP2- or NP21-BSA for 1 overnight. Plates were washed with PBS-T three times and coated with (1 % BSA in PBS) for at least 1 h at room temperature. After wells were washed with PBS-T three times, 2 μg/ml NP3S2-each isotype antibody and 40- to 400-times diluted NP-CGG immunized mouse serum added in top well followed by three-fold series dilutions in 2–5 steps. After wells were washed with PBS-T three times, HRP-conjugated anti-mouse each isotype antibody (1:3000) diluted in PBS-T was added.
Finally, wells were developed with the SureBlue™ TMB 1-Component Microwell Peroxidase Substrate (SeraCare) and stopped reaction with 1 M HCl, or ELISA POD Substrate TMB Kit (Nacalai) and stopped reaction with 1 M H2SO4. The absorbance at 450 nm was measured using iMark microplate absorbance reader (Bio-Rad). The absolute IC50 was calculated using a nonlinear regression curve fit on Prism (v10.0, GraphPad).
5.11. Transmission electron microscopy (TEM) observation
To evaluate each antibody by TEM, size-exclusion chromatography (SEC) was performed using an NGC Chromatography System (Bio-Rad). After Ni-NTA affinity purification, antibody samples were applied to a SEC column (Superose 6 increase 10/300 GL, Cytiva) equilibrated with PBS, and the elution was detected by absorbance at 280 nm. Three μl of the fraction containing the resulting peak top was adsorbed onto a glow-discharged carbon-coated Cu grid (EM Japan) for 30 s, and the excess solution was absorbed using filter paper. In addition, 3 μl of PBS was adsorbed onto the grid for 30 s, and the grid was washed by absorbing the excess solution with filter paper in the same manner. Finally, 3 μl of staining solution (2 % [w/v] uranyl acetate) was adsorbed onto the grid for 30 s, and the excess solution was removed using filter paper. Staining was performed twice. The sample grids were allowed to air dry and observed using TEM (JEM-1400Plus, JEOL) with a tungsten filament at 80 kV at the Natural Science Center for Basic Research and Development, Hiroshima University. Images were recorded using a CCD camera (2048 × 2048 pixels).
7. Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
CRediT authorship contribution statement
Rin Yoshizato: Writing – original draft, Methodology, Investigation, Conceptualization. Mariko Miura: Methodology, Investigation, Conceptualization. Kiyomi Shitaoka: Methodology. Yuri Matsuoka: Methodology. Akifumi Higashiura: Writing – review & editing, Investigation. Akima Yamamoto: Investigation. Yun Guo: Investigation. Hitoshi Azuma: Investigation. Yohei Kawano: Writing – review & editing, Supervision, Methodology. Shouichi Ohga: Supervision. Tomoharu Yasuda: Writing – review & editing, Writing – original draft, Supervision, Project administration, Conceptualization.
6. Resource availability
6.1. Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Tomoharu Yasuda (yasudat@hiroshima-u.ac.jp)
Data and code availability
-
●
All data reported in this paper will be shared by the lead contact upon request.
-
●
This paper does not report original code.
-
●
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We thank T. Kawaguchi and Y. Hayashi for technical assistance; N. Nishimichi for discussion and technical advice; N. Kikkawa and M. Tawa for administrative assistance; and all lab members for useful discussion and comments. We also thank the staff of the Analysis Center of Life Science, Hiroshima University for the use of their facilities. This work was supported in part by the Natural Science Center for Basic Research and Development (NBARD-00134). This work was supported by the JSPS KAKENHI Grant Number JP21H02751 to T.Y.; JSPS KAKENHI Grant Number JP22K16372 to K.S.; Sumitomo Mitsui Trust Bank-New Corona Vaccine and Therapeutics Development Donation Account to T.Y.; the READYFOR crowdfunding donation to T.Y.; Japan Agency for Medical Research and Development (AMED) Research Grant for COVID-19, JP20fk0108453 to T.Y.; Research Funding for Longevity Sciences from the National Center for Geriatrics and Gerontology (grant 21-2 to T.Y.)
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e40837.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Table S1. Excel file containing the primer sequences used for HC and LC RT-PCR from single B cells
Table S2. Excel file containing the primer sequences used for expression vector preparation and variable domain cloning.
Figure S1. Agarose gel electrophoresis of the second PCR products.
Splenic B220+CD19+IgG1+CD38+ live single memory B cells were sorted and cDNAs were synthesized by reverse transcription. HC and LC genes were separately amplified. The amplification efficiencies were 68.6 % (24/35) in HC and 97.1 % (34/35) in LC. H, Heavy chain; L, Light chain
Figure S2. SDS-PAGE analysis of produced IgG1, IgM, and IgA mAbs.
(A) SDS-PAGE image of monoclonal IgG1 antibodies secreted in the supernatant of Expi293F cells. Each expression vectors were transfected into Expi293F cells by the indicated proportions. For lanes 1 and 2, NP3S2 expression vectors were used, and for lanes 3 and 4, NP3S7 expression vectors were used.
(B) Different amounts of J chain expression plasmids were transfected with μHC and LC expression plasmids into Expi293F cells. Purified IgM antibodies were eluted in 50 μl buffer, then, 9 μl of purified antibody per well was separated on 6 % SDS-PAGE gel. The molecular weight of IgM pentamer is 890.7 kDa (HC 64.4 kDa, LC 23.1 kDa, J chain 15.7 kDa). The asterisk indicates pentamer. IgM std, Mouse IgM-UNLB
(C) SC expression plasmid was transfected with αHC and LC expression plasmids into Expi293F cells. 3 μg of purified antibody per well was separated on 10 % SDS-PAGE either non-reducing (NR) or reducing (R) conditions. IgA std, Mouse IgA-UNLB; dIgA, dimeric IgA; sIgA, secretory IgA (NP3S2-sIgA generated in this study).
References
- 1.Taylor P.C., Adams A.C., Hufford M.M., de la Torre I., Winthrop K., Gottlieb R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021;21:382–393. doi: 10.1038/s41577-021-00542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim A.S., Diamond M.S. A molecular understanding of alphavirus entry and antibody protection. Nat. Rev. Microbiol. 2023;21:396–407. doi: 10.1038/s41579-022-00825-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klein F., Mouquet H., Dosenovic P., Scheid J.F., Scharf L., Nussenzweig M.C. Antibodies in HIV-1 vaccine development and therapy. Science. 2013;341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merkuleva Y.A., Shcherbakov D.N., Ilyichev A.A. Methods to produce monoclonal antibodies for the prevention and treatment of viral infections. Russ. J. Bioorg. Chem. 2022;48:256–272. doi: 10.1134/s1068162022020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caskey M., Klein F., Nussenzweig M.C. Broadly neutralizing anti-HIV-1 monoclonal antibodies in the clinic. Nat. Med. 2019;25:547–553. doi: 10.1038/s41591-019-0412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Ayoubi L.W., Mahmoud O., Zakhour J., Kanj S.S. Recent advances in the treatment of Ebola disease: a brief overview. PLoS Pathog. 2024;20 doi: 10.1371/journal.ppat.1012038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shitaoka K., Higashiura A., Kawano Y., Yamamoto A., Mizoguchi Y., Hashiguchi T., Nishimichi N., Huang S., Ito A., Ohki S., Kanda M., Taniguchi T., Yoshizato R., Azuma H., Kitajima Y., Yokosaki Y., Okada S., Sakaguchi T., Yasuda T. Structural basis of spike RBM-specific human antibodies counteracting broad SARS-CoV-2 variants. Commun. Biol. 2023;6:395. doi: 10.1038/s42003-023-04782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Jong H.K., Grobusch M.P. Monoclonal antibody applications in travel medicine. Trop Dis Travel Med Vaccines. 2024;10:2. doi: 10.1186/s40794-023-00212-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamura S., Funato H., Hirabayashi Y., Kikuta K., Suzuki Y., Nagamine T., Aizawa C., Nakagawa M., Kurata T. Functional role of respiratory tract haemagglutinin-specific IgA antibodies in protection against influenza. Vaccine. 1990;8:479–485. doi: 10.1016/0264-410x(90)90250. [DOI] [PubMed] [Google Scholar]
- 10.Dowell A.C., Waiblinger D., Wright J., Ladhani S.N., Moss P. Nucleocapsid-specific antibodies as a correlate of protection against SARS-CoV-2 reinfection in children. J. Infect. 2023;87:267–269. doi: 10.1016/j.jinf.2023.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Köhler G., Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 12.Rajewsky K. The advent and rise of monoclonal antibodies. Nature. 2019;575:47–49. doi: 10.1038/d41586-019-02840-w. [DOI] [PubMed] [Google Scholar]
- 13.Mitra S., Tomar P.C. Hybridoma technology; advancements, clinical significance, and future aspects. J. Genet. Eng. Biotechnol. 2021;19:159. doi: 10.1186/s43141-021-00264-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Boehmer L., Liu C., Ackerman S., Gitlin A.D., Wang Q., Gazumyan A., Nussenzweig M.C. Sequencing and cloning of antigen-specific antibodies from mouse memory B cells. Nat. Protoc. 2016;11:1908–1923. doi: 10.1038/nprot.2016.102. [DOI] [PubMed] [Google Scholar]
- 15.Jones B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne J.L., Heinz B.A., Foster D., Higgs R.E., Balasubramaniam D., Wang L., Zhang Y., Yang E.S., Bidshahri R., Kraft L., Hwang Y., Žentelis S., Jepson K.R., Goya R., Smith M.A., Collins D.W., Hinshaw S.J., Tycho S.A., Pellacani D., Xiang P., Muthuraman K., Sobhanifar S., Piper M.H., Triana F.J., Hendle J., Pustilnik A., Adams A.C., Berens S.J., Baric R.S., Martinez D.R., Cross R.W., Geisbert T.W., Borisevich V., Abiona O., Belli H.M., de Vries M., Mohamed A., Dittmann M., Samanovic M.I., Mulligan M.J., Goldsmith J.A., Hsieh C.L., Johnson N.V., Wrapp D., McLellan J.S., Barnhart B.C., Graham B.S., Mascola J.R., Hansen C.L., Falconer E. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuniyoshi Y., Maehara K., Iwasaki T., Hayashi M., Semba Y., Fujita M., Sato Y., Kimura H., Harada A., Ohkawa Y. Identification of immunoglobulin gene sequences from a small read number of mRNA-seq using hybridomas. PLoS One. 2016;11 doi: 10.1371/journal.pone.0165473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schanz M., Liechti T., Zagordi O., Miho E., Reddy S.T., Günthard H.F., Trkola A., Huber M. High-throughput sequencing of human immunoglobulin variable regions with subtype identification. PLoS One. 2014;9 doi: 10.1371/journal.pone.0111726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henry K.A. Next-generation DNA sequencing of V(H)/V(L) repertoires: a primer and guide to applications in single-domain antibody discovery. Methods Mol. Biol. 2018;1701:425–446. doi: 10.1007/978-1-4939-7447-4_24. [DOI] [PubMed] [Google Scholar]
- 19.Imanishi T., Mäkelä O. Strain differences in the fine specificity of mouse anti-hapten antibodies. Eur. J. Immunol. 1973;3:323–330. doi: 10.1002/eji.1830030602. [DOI] [PubMed] [Google Scholar]
- 20.Miller F., Metzger H. Characterization of a human macroglobulin. II. Distribution of the disulfide bonds. J. Biol. Chem. 1965;240:4740–4745. [PubMed] [Google Scholar]
- 21.Strugnell R.A., Wijburg O.L. The role of secretory antibodies in infection immunity. Nat. Rev. Microbiol. 2010;8:656–667. doi: 10.1038/nrmicro2384. [DOI] [PubMed] [Google Scholar]
- 22.Woof J.M., Russell M.W. Structure and function relationships in IgA. Mucosal Immunol. 2011;4:590–597. doi: 10.1038/mi.2011.39. [DOI] [PubMed] [Google Scholar]
- 23.Tashiro Y., Murakami A., Hara Y., Shimizu T., Kubo M., Goitsuka R., Kishimoto H., Azuma T. High-affinity IgM(+) memory B cells are defective in differentiation into IgM antibody-secreting cells by re-stimulation with a T cell-dependent antigen. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-32926-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee H.K., Lee H.H., Park Y.M., Park H.J., Lee J.H., Ha T.Y. Anti-IL-4 antibody inhibits antigen specific IgE response but fails to prevent chicken gamma globulin-induced active systemic anaphylaxis: evidence for the involvement of IgG antibodies. J. Korean Med. Sci. 1996;11:111–117. doi: 10.3346/jkms.1996.11.2.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herzenberg L.A., Black S.J., Tokuhisa T., Herzenberg L.A. Memory B cells at successive stages of differentiation. Affinity maturation and the role of IgD receptors. J. Exp. Med. 1980;151:1071–1087. doi: 10.1084/jem.151.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi Y., Dutta P.R., Cerasoli D.M., Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. V. Affinity maturation develops in two stages of clonal selection. J. Exp. Med. 1998;187:885–895. doi: 10.1084/jem.187.6.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith K.G., Light A., Nossal G.J., Tarlinton D.M. The extent of affinity maturation differs between the memory and antibody-forming cell compartments in the primary immune response. EMBO J. 1997;16:2996–3006. doi: 10.1093/emboj/16.11.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiss U., Zoebelein R., Rajewsky K. Accumulation of somatic mutants in the B cell compartment after primary immunization with a T cell-dependent antigen. Eur. J. Immunol. 1992;22:511–517. doi: 10.1002/eji.1830220233. [DOI] [PubMed] [Google Scholar]
- 29.Rada C., Gupta S.K., Gherardi E., Milstein C. Mutation and selection during the secondary response to 2-phenyloxazolone. Proc. Natl. Acad. Sci. U. S. A. 1991;88:5508–5512. doi: 10.1073/pnas.88.13.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen D., Simon T., Sablitzky F., Rajewsky K., Cumano A. Antibody engineering for the analysis of affinity maturation of an anti-hapten response. EMBO J. 1988;7:1995–2001. doi: 10.1002/j.1460-2075.1988.tb03038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlatter S., Stansfield S.H., Dinnis D.M., Racher A.J., Birch J.R., James D.C. On the optimal ratio of heavy to light chain genes for efficient recombinant antibody production by CHO cells. Biotechnol. Prog. 2005;21:122–133. doi: 10.1021/bp049780w. [DOI] [PubMed] [Google Scholar]
- 32.Fagioli C., Mezghrani A., Sitia R. Reduction of interchain disulfide bonds precedes the dislocation of Ig-mu chains from the endoplasmic reticulum to the cytosol for proteasomal degradation. J. Biol. Chem. 2001;276:40962–40967. doi: 10.1074/jbc.M107456200. [DOI] [PubMed] [Google Scholar]
- 33.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., Chai X., He R., Li X., Lv Q., Zhu H., Deng W., Xu Y., Wang Y., Qiao L., Tan Y., Song L., Wang G., Du X., Gao N., Liu J., Xiao J., Su X.D., Du Z., Feng Y., Qin C., Qin C., Jin R., Xie X.S. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J.H., Leaman D.P., Kim A.S., Torrents de la Peña A., Sliepen K., Yasmeen A., Derking R., Ramos A., de Taeye S.W., Ozorowski G., Klein F., Burton D.R., Nussenzweig M.C., Poignard P., Moore J.P., Klasse P.J., Sanders R.W., Zwick M.B., Wilson I.A., Ward A.B. Antibodies to a conformational epitope on gp41 neutralize HIV-1 by destabilizing the Env spike. Nat. Commun. 2015;6:8167. doi: 10.1038/ncomms9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zemella A., Beer K., Ramm F., Wenzel D., Düx A., Merkel K., Calvignac-Spencer S., Stern D., Dorner M.B., Dorner B.G., Widulin N., Schnalke T., Walter C., Wolbert A., Schmid B.G., Mankertz A., Santibanez S. Vaccine-induced neutralizing antibodies bind to the H protein of a historical measles virus. Int. J. Med. Microbiol. 2024;314 doi: 10.1016/j.ijmm.2024.151607. [DOI] [PubMed] [Google Scholar]
- 36.Choi G.S., Huh D.H., Han S.B., Ahn D.H., Kang K.R., Kim J.A., Choi B.M., Kim H.R., Kang J.H. Enzyme-linked immunosorbent assay for detecting anti-pertussis toxin antibody in mouse. Clin Exp Vaccine Res. 2019;8:64–69. doi: 10.7774/cevr.2019.8.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carreño J.M., Singh G., Tcheou J., Srivastava K., Gleason C., Muramatsu H., Desai P., Aberg J.A., Miller R.L., Study Group P., Pardi N., Simon V., Krammer F. mRNA-1273 but not BNT162b2 induces antibodies against polyethylene glycol (PEG) contained in mRNA-based vaccine formulations. Vaccine. 2022;40:6114–6124. doi: 10.1016/j.vaccine.2022.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hennicke J., Schwaigerlehner L., Grünwald-Gruber C., Bally I., Ling W.L., Thielens N., Reiser J.B., Kunert R. Transient pentameric IgM fulfill biological function-Effect of expression host and transfection on IgM properties. PLoS One. 2020;15 doi: 10.1371/journal.pone.0229992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gagnon P., Hensel F., Richieri R. Purification of IgM monoclonal antibodies. BioPharm Int Sup. 2008:26–35. [Google Scholar]
- 40.Tchoudakova A., Hensel F., Murillo A., Eng B., Foley M., Smith L., Schoenen F., Hildebrand A., Kelter A.R., Ilag L.L., Vollmers H.P., Brandlein S., McIninch J., Chon J., Lee G., Cacciuttolo M. High level expression of functional human IgMs in human PER.C6 cells. mAbs. 2009;1:163–171. doi: 10.4161/mabs.1.2.7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corthésy B., Benureau Y., Perrier C., Fourgeux C., Parez N., Greenberg H., Schwartz-Cornil I. Rotavirus anti-VP6 secretory immunoglobulin A contributes to protection via intracellular neutralization but not via immune exclusion. J. Virol. 2006;80:10692–10699. doi: 10.1128/jvi.00927-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shoji K., Takahashi T., Kurohane K., Iwata K., Matsuoka T., Tsuruta S., Sugino T., Miyake M., Suzuki T., Imai Y. Recombinant immunoglobulin A specific for influenza A virus hemagglutinin: production, functional analysis, and formation of secretory immunoglobulin A. Viral Immunol. 2015;28:170–178. doi: 10.1089/vim.2014.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li C., An X., Butt A.M., Zhang B., Zhang Z., Wang X., Huang Y., Zhang W., Zhang B., Mi Z., Tong Y. Construction of a chimeric secretory IgA and its neutralization activity against avian influenza virus H5N1. J Immunol Res. 2014;2014 doi: 10.1155/2014/394127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brandtzaeg P., Prydz H. Direct evidence for an integrated function of J chain and secretory component in epithelial transport of immunoglobulins. Nature. 1984;311:71–73. doi: 10.1038/311071a0. [DOI] [PubMed] [Google Scholar]
- 45.Pasalic D., Weber B., Giannone C., Anelli T., Müller R., Fagioli C., Felkl M., John C., Mossuto M.F., Becker C.F.W., Sitia R., Buchner J. A peptide extension dictates IgM assembly. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E8575–e8584. doi: 10.1073/pnas.1701797114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaul O. How introns enhance gene expression. Int. J. Biochem. Cell Biol. 2017;91:145–155. doi: 10.1016/j.biocel.2017.06.016. [DOI] [PubMed] [Google Scholar]
- 47.Azuma H., Kawano Y., Shitaoka K., Kawahara T., Ito A., Higashiura A., Kitajima Y., Ohki S., Yasuda T. Vaccination with the Omicron spike RBD boosts broadly neutralizing antibody levels and confers sustained protection even after acquiring immunity to the original antigen. Int. Immunol. 2023;35:197–207. doi: 10.1093/intimm/dxac055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Excel file containing the primer sequences used for HC and LC RT-PCR from single B cells
Table S2. Excel file containing the primer sequences used for expression vector preparation and variable domain cloning.
Figure S1. Agarose gel electrophoresis of the second PCR products.
Splenic B220+CD19+IgG1+CD38+ live single memory B cells were sorted and cDNAs were synthesized by reverse transcription. HC and LC genes were separately amplified. The amplification efficiencies were 68.6 % (24/35) in HC and 97.1 % (34/35) in LC. H, Heavy chain; L, Light chain
Figure S2. SDS-PAGE analysis of produced IgG1, IgM, and IgA mAbs.
(A) SDS-PAGE image of monoclonal IgG1 antibodies secreted in the supernatant of Expi293F cells. Each expression vectors were transfected into Expi293F cells by the indicated proportions. For lanes 1 and 2, NP3S2 expression vectors were used, and for lanes 3 and 4, NP3S7 expression vectors were used.
(B) Different amounts of J chain expression plasmids were transfected with μHC and LC expression plasmids into Expi293F cells. Purified IgM antibodies were eluted in 50 μl buffer, then, 9 μl of purified antibody per well was separated on 6 % SDS-PAGE gel. The molecular weight of IgM pentamer is 890.7 kDa (HC 64.4 kDa, LC 23.1 kDa, J chain 15.7 kDa). The asterisk indicates pentamer. IgM std, Mouse IgM-UNLB
(C) SC expression plasmid was transfected with αHC and LC expression plasmids into Expi293F cells. 3 μg of purified antibody per well was separated on 10 % SDS-PAGE either non-reducing (NR) or reducing (R) conditions. IgA std, Mouse IgA-UNLB; dIgA, dimeric IgA; sIgA, secretory IgA (NP3S2-sIgA generated in this study).
Data Availability Statement
-
●
All data reported in this paper will be shared by the lead contact upon request.
-
●
This paper does not report original code.
-
●
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.