Abstract
Investigation of Caenorhabditis elegans act-5 gene function revealed that intestinal microvillus formation requires a specific actin isoform. ACT-5 is the most diverged of the five C. elegans actins, sharing only 93% identity with the other four. Green fluorescent protein reporter and immunofluorescence analysis indicated that act-5 gene expression is limited to microvillus-containing cells within the intestine and excretory systems and that ACT-5 is apically localized within intestinal cells. Animals heterozygous for a dominant act-5 mutation looked clear and thin and grew slowly. Animals homozygous for either the dominant act-5 mutation, or a recessive loss of function mutant, exhibited normal morphology and intestinal cell polarity, but died during the first larval stage. Ultrastructural analysis revealed a complete loss of intestinal microvilli in homozygous act-5 mutants. Forced expression of ACT-1 under the control of the act-5 promoter did not rescue the lethality of the act-5 mutant. Together with immuno-electron microscopy experiments that indicated ACT-5 is enriched within microvilli themselves, these results suggest a microvillus-specific function for act-5, and further, they raise the possibility that specific actins may be specialized for building microvilli and related structures.
INTRODUCTION
Actin proteins are fundamental structural components of eukaryotic cells that mediate a wide array of cellular processes (Pollard and Cooper, 1986; Herman, 1993). Actin is required for cell polarization and division, organelle transport and cell migration in addition to more specialized cellular phenomena such as muscle contraction and ring canal formation (Tilney et al., 1996). Most eukaryotes express multiple actin proteins, each encoded by a distinct gene, but these actin isoforms themselves share remarkable sequence conservation, suggesting that common functional properties have constrained their divergence during evolution. On the other hand, many organisms express more than one actin protein within the same cell, or differentially between cell types, raising the possibility that nearly identical actins evolved for specialized functions.
Gene rescue experiments in both Drosophila and mouse, in addition to vertebrate overexpression studies, have indicated that functional differences do indeed exist between at least some actins within an organism (Schevzov et al., 1992; Kumar et al., 1997; Fyrberg et al., 1998). However, the extent to which the residues that distinguish isoforms confer functional differences to the three-dimensional actin molecule, and similarly, whether specific cellular structures use specific actin isoforms, is poorly understood.
The C. elegans genome encodes five actin genes. Whereas ACT-1, -2, -3, and -4 all share 99% protein sequence identity, ACT-5, the most divergent, shares 93% identity with the others. Prior phenotypic analysis of animals that carry dominant actin mutations affecting the act-1,2,3 gene cluster suggest that these genes act primarily in muscle and myofilament-containing cells (Waterston et al., 1984) and may exert their dominance because of an altered rate or extent of polymerization (Frieden et al., 2000). Moreover, molecular analysis of revertants of the dominant actin mutations indicate functional redundancy within this set of actins (Landel et al., 1984). Although no dominant or recessive mutations have been identified for act-4, transgene reporter studies indicate that it, too, is predominantly expressed in muscle cells (Stone and Shaw, 1993), although preliminary in situ hybridization analysis with act-1, act-2, and act-4 probes reveals more widespread expression of actin genes 1–4 (Shin-i and Kohara, 1998).
Here, we report a functional analysis of a single C. elegans actin isoform, ACT-5. Genetic analysis led us to discover that act-5 does not serve a muscle cell function nor does it encode a general cytoplasmic actin. Instead, act-5 expression is limited to a subset of C. elegans somatic cells, all of which contain microvilli or microvilli-like structures, and act-5 function is essential for the stable morphogenesis of intestinal microvilli. Microvilli are actin-based cellular structures that form plasma membrane projections into the extracellular space and whose specialized shape provides increased cellular surface area. Microvilli are a crucial component of the terminal differentiation process for many epithelial cell types, usually ones that play an absorptive or filtering role. Although its function has long been recognized and appreciated (Hirokawa and Heuser, 1981), the molecular mechanisms that generate and stabilize a microvillus are poorly understood. Actin filaments are known to be the core building blocks of microvilli, but whether a specific actin isoform has evolved to be functionally specialized for this role has not been demonstrated (but see Discussion; Sawtell et al., 1988). The results presented here suggest that microvilli are built, at least in part, by a specialized actin protein and that in C. elegans, the ACT-5 isoform is uniquely adapted for this function.
MATERIALS AND METHODS
Worm Strains and Plasmids
All strain backgrounds are Bristol N2. The following mutations were used: IN700 dtEx418[pCKE1 (act-5::gfp), pRF4]; IN2054 dtIs419[pJAW70, pRF4]I; IN2052 dtIs419(I);act-5 (dt2017)(III): IN2050 dtIs419[pJAW70, pRF4](I) act-5 (dt2019 dt2017)(III); IN2500 dtEx68[pRF4,pJJB8]; IN2501 dtEx69[pRF4,pJJB8]; IN2506 act-5(dt2017)III;dtEx68[pRF4,pJJB8]; IN2507 act-5(dt2017)III;dtEx69 [pRF4,pJJB8]; IN2502 dtEx70[pRF4,pJJB10]; IN2503 dtEx71[pRF4,pJJB10]; IN2505 dtEx75[pRF4,pJJB10].
A wild-type act-5(+) transgene, pJAW70, was made by amplification of C. elegans genomic DNA with oligonucleotides 5′-TGTTATTTTTAACTAGTGTAGAG-3′ and 5′-GTCCCAAGAATTGATCAATGACG-3′. The amplification product was digested with SpeI and BclI and cloned into pJAW72. pJAW72 is a variant of the cloning vector pSL1180 in which the ApaI site was deleted.
Generation of the act-5 promoter-gfp transcriptional reporter pCKE1 used two intermediate constructs, pNP2 and pNP4. pNP2 is identical to pJAW70 except the act-5 stop codon is mutated to an ApaI site. pNP4 is identical to pNP2 except that a gfp cassette (derived from pPD95.69; kindly provided by Andy Fire, Stanford University School of Medicine, Stanford, CA) was cloned into the unique ApaI site of pNP2 to create a full-length act-5::gfp translational fusion. pNP2 was modified by strand overlap extension (Ho et al., 1989) with the following four primers: 5′-TGTTATTTTTAACTAGTGTAGAG-3′ paired with 5′-AATTTGAAAAAAAATCAGCGGGCCCGAAGCACTTTCGGTGAAC-3′ and 5′-GTCCCAAGAATTGATCAATGACG-3′ paired with 5′-GTTCACCGAAAGTGCTTCGGGCCCGCTGATTTTTTTTCAAATT-3′. The amplification products of the first round reaction were then used as template for a second round of amplification in combination with the outermost primers used in the first round reactions. The resulting product was digested with SpeI and BclI and cloned into pJAW72 to create pNP2. To make pNP4, An ApaI gfp coding region cassette was produced by amplification of pPD95.69 (kindly provided by A. Fire) with primers 5′ GATCGGGCCCAGTAAAGGAGAAGAACTTTTC-3′ and 5′-GATCGGGCCCTTATTTGTATAGTTCATCCATGCC-3′, digested with ApaI, and ligated into the unique ApaI site of pNP2. pCKE1 was derived from the full-length act-5::gfp translational fusion by first linearizing pNP4 at the unique SalI site in the act-5 coding region and then priming in opposite directions away from the SalI site with primers 5′-ACGTAGAGCTCAGTAAAGGAGAAGAACTTTTCACTG-3′ and 5′-CTAGCGAGCTCCATCTGAAATTAATAAAAGGTTGAAG-3′. The primers create a synthetic SacI site (two amino acids) in place of all but the initiator ATG of act-5 and maintain the reading frame with downstream gfp cassette. The methylation-defective strain SCS110 (Stratagene, La Jolla, CA), was used for all cloning procedures.
Gateway-based technology (Invitrogen) was used to drive expression of either ACT-1 or ACT-5 cDNA open reading frames under the control of the act-5 promotor. A Gateway destination vector was constructed using the Gateway Vector Conversion System. Briefly, sequential QuikChange (Stratagene) reactions were carried out to add 5′ and 3′ SnaBI sites for blunt cutting and ligation of the Gateway Reading Frame Cassette B into pCKE1 (replacing GFP). The mutagenic primers used were 5′-ACTCGTACATCACTTAATAACGTACGTATTTTCTATAAACTCGACTTCTTCAACC-3′ and its complement and 5′-GATGAACTATACAAATAGGGGCTACGTACCGCTGATTTTTTTTCAAATTTTTTTTTTC-3′ and its complement. After confirmation of QuikChange reactions, Gateway Reading Frame Cassette B was ligated into the vector backbone, replacing the sequence between the two constructed SnaBI sites; this vector was named pJJB3.
pJJB8 (ACT-5 cDNA) and pJJB10 (ACT-1 cDNA), each flanked by act-5 regulatory regions were constructed by PCR amplification and recombination into pJJB3. The ACT-5 open reading frame was amplified using primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTTTTCTATAAACTCGACTTCTTCAACCTTTTATTAATTTCAGATGGAAGAAGAAATCGC-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGCTTAGAAGCACTTTCGGTGAACAATCG-3′ and the ACT-5 cDNA clone cm19a6 as a template (Waterston et al., 1992). The ACT-1 open reading frame was amplified by reverse transcription (RT)-PCR (Titan One Tube RT-PCR; Roche Diagnostics, Indianapolis, IN) by using the primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCAGGTACATTAAAAACTAATCAAAATGTGTGACGACGAGGTTGC-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGTGCATTTAGAAGCACTTGCGGTGAACGATG-3′ and total worm RNA as template. Each PCR fragment was recombined into the intermediate entry clone pDONR201 by using the Gateway BP clonase enzyme mix. The entry clones were then recombined with pJJB3 by using the Gateway LR clonase enzyme mix and sequenced to confirm that there were no unpredicted mutations.
PCR-based Screen and Identification of act-5 Deletion Mutants
A PCR-based screening strategy was used to isolate the dt2017 allele from a library of mutagenized C. elegans genomes, by using the following nested primers: outer, CAGTTCTCCTTACCGAGGCTCC and AGTATCTCATGGAATTTGTGTC; and inner, AGTATCTCATGGAATTTGTGTC and GTGTTTCGCCTAGAAATGGAAG. dt2017 and dt2019 alleles contain a deletion between the coordinates 2535 and 2042 of cosmid T25C8; this deletion is flanked directly by (5′) TGTCACTCACACCGTTCCAA, (3′) ACGTCGCCCACGACTTCGAG on the act-5 coding strand. dt2019 was derived from the dt2017 allele and represents the sole mutation isolated among ∼250,000 mutagenized haploid genomes that could suppress the starved appearance and slow growth phenotype of IN2052 [pJAW70, pRf4] act-5 (dt2017) animals. In addition to the deletion described above, dt2019 contains a nonsense mutation (G→ A on the coding strand) at coordinate 2792 of cosmid T25C8.
Animals carrying dt2017 or dt2019 were identified by PCR-based molecular typing with the following primers: CAGTTCTCCTTACCGAGGCTCC and GTGTTTCGCCTAGAAATGGAAG (these primers amplify a 1268-base pair product in wild type, and a shorter 774-base pair product if the deletion allele is present). dt2017 heterozygous animals also could be identified based on their relatively small, starved appearance. To identify the homozygote, heterozygotes were allowed to lay eggs over a period of ∼8 h; all F1 eggs were counted and monitored throughout their development. After 2 d, all F1 progeny were lysed and individually assessed by PCR for the deletion or wild-type act-5 sequence. Animals carrying act-5(dt2017) were backcrossed more than 5 times before phenotypic analysis.
RNA Interference (RNAi)
Oligos 5′-GTTAGTCTAGAACATGTGCCTTCCATTTCTAGGCG-3′ and 5′-TCGGACTGCAGGAGAAAATGAAGTATCTCATGGAATTTG-3′ were used to amplify a region from the 3′ untranslated region (UTR) of the act-5 mRNA and flank it with synthetic XbaI and PstI sites. The 94-base pair PCR product was cut with XbaI-PstI and cloned into the C. elegans feeding RNAi vector pPD129.36 and transformed into HT115 cells (Timmons et al., 2001). RNAi was performed as described previously (Kamath et al., 2001).
Mutant Rescue Assay
Because act-5(dt2107)/+ animals grow slowly and act-5(dt2017) homozygotes are inviable, transgenes can be used to test whether other worm actins can substitute for ACT-5 in vivo. Rescue assays for ACT-5 or ACT-1 driven by the act-5 regulatory regions were performed by first transforming wild-type animals with either pJJB8 (ACT-5) or pJJB10 (ACT-1) and pRF4 as a coinjection marker (Mello et al., 1991) to establish extrachromosomal array lines. Independent transgenic lines were isolated by identification of F1 Rol progeny (N = 2 for pJJB8, N = 3 for pJJB10). The transgenic lines were then crossed to dt2017 heterozygotes to create dt2017/+;Ex[pJJB8,pRF4] or dt2017/+; Ex[pJJB10,pRF4] strains. The genotypes were confirmed by PCR genotyping (see below) to identify dt2017 heterozygotes and Rol progeny to identify the presence of transgene. dt2107/+ animals carrying the transgene were then transferred individually to plates and allowed to reproduce at three intervals for a total of 22 h to generate three relatively synchronized broods of F1 progeny. F1 embryos were counted and monitored for ∼48 h to determine those that arrest relative to those that continue development beyond the L1 larval stage. Worms were then collected for genotyping, and their larval stage recorded. Animals carrying dt2017 were identified by PCR-based molecular typing with the following primers: CAGTTCTCCTTACCGAGGCTCC and CTCATGTACTGAGAAATGTGAAGC (these primers amplify a 1027-base pair product in wild type, and a shorter 533-base pair product if the deletion allele is present). In all lines, we attempted to isolate homozygous dt2017 animals carrying the array as an indicator of rescue.
To confirm that ACT-1 cDNA array-carrying lines were expressing ACT-1 at the level of mRNA, we isolated total RNA from each independent transgenic line. RT-PCR (Titan One Tube RT-PCR; Roche Diagnostics) with the following primers in all combinations was performed to confirm that the lines carrying ACT-1 cDNA exhibited a new mRNA, corresponding to ACT-1 coding and ACT-5 3′ UTR, that was not present in nonarray-carrying lines. Primers used were ACT-1 coding 5′-CTGATCGTATGCAGAAGGAAATC-3′, ACT-1 3′ UTR 5′-GAAATAGAAAGCTGGTGGTGACGATGG-3′, ACT-5 coding 5′-GATCGTATGCAAAAGGAGATTCAAC-3′, and ACT-5 3′ UTR 5′-GAAGTATCTCATGGAATTTGTGTC-3′.
Antibody Purification
Rabbit antisera raised against a synthetic ACT-5 peptide fragment (VAHDFESELAAA) was used to generate anti-ACT-5–enriched antibodies. The synthetic peptide contained an artificial N-terminal cysteine and was coupled to KLH as a carrier. Two cyanogen bromide-Sepharose columns (Sigma-Aldrich, St. Louis, MO) were the basis of our affinity purification procedure: one column contained coupled GST::ACT-5 (amino acids 167–322), whereas the other column contained coupled GST::ACT-1 (amino acids 168–323). Glutathione S-transferase (GST) fusions were expressed in Escherichia coli, isolated from the insoluble fraction by using glutathione agarose, and visualized on a Coomassie-stained protein gel (Smith and Johnson, 1988). ACT-1 and ACT-5 protein fusions (11.6 and 10.6 mg, respectively) were coupled to 3.5 ml of resin. For purification, R7400 anti-ACT-5 antisera was passed through a 0.22-μm filter and then loaded onto the GST::ACT-1 column. Flowthrough was then loaded onto the GST::ACT-5 column. After several washes, bound proteins were eluted using 0.1 M glycine, pH 2.5, and immediately neutralized with 1/10 volume of 1 M Tris, pH 8. Purified antibodies strongly recognized ACT-5::GST but only weakly bound to ACT-1::GST on Western blots (our unpublished data).
Immunofluorescence and Light Microscopy
Wild-type embyros were collected in M9 buffer (22 mM KH2PO4, 22 mM Na2HPO4, 85 mM NaCl, and 1 mM MgSO4) and bleached (Lewis and Fleming, 1995) for 5 min with intermittent inversion. Samples were washed with M9, followed by fixation in 3% formaldehyde (in 0.1 M sodium phosphate buffer, pH 7.0, 0.1 mM EDTA) for 5 min. Embryos were transferred into –20°C methanol and stored at –20°C for up to 3 wk. For staining, samples were flash frozen (in tubes) in liquid nitrogen and then thawed at room temperature, twice. Samples were washed in 1× phosphate-buffered saline (PBS) + 0.001% Tween 20 (PBT) and then blocked for 2 h at room temperature in 10% normal goat serum (Invitrogen) and 1% bovine serum albumin (BSA) (fraction V; Roche Diagnostics) in PBT. After labeling (see below) samples were stained with 0.1 μg/ml 4,6-diamidino-2-phenylindole (DAPI) for 5 min and mounted in 5% N-propyl gallate (Sigma-Aldrich) in 90% glycerol. Images obtained through this staining procedure are suboptimal for labeling antigens present in the embryonic intestine; however, the preparation conditions that optimize for intestinal staining result in a loss of muscle actin antigens. The method used maintains high levels of muscle actin C4 reactivity, while allowing some antibodies to reach the intestine, providing a specificity comparison between anti-ACT-5 polyclonal antibodies and the pan-reactive monoclonal antibody (mAb) C4.
L1 animals were collected in M9 buffer (act-5 mutants were picked between 20 and 36 h after egg laying, and wild-type L1 animals were collected by allowing embryos to hatch overnight without food) before processing. For MH33 and anti-actin staining, worms were fixed as described previously (Ruvkun and Giusto, 1989) with modifications. In brief, worms were shaken at 250 rpm in 0.3 ml of heptane, 0.1 ml of 90% methanol; 10% 0.1 M EGTA, 0.5 ml of 1× PBS, 0.08 M HEPES, 1.6 mM MgSO4, 0.8 mM EGTA, 3% formaldehyde for 30′ at room temperature and then rapid-frozen in liquid nitrogen followed by additional shaking for 1 h. Preparations were incubated for 12 h in 5% 2-mercaptoethanol, 1% Triton X-100, 0.1 M Tris, pH 7.4, at 37°C, and then they were rinsed three times in PBT, followed by a 1.75-h incubation at 37°C in 1 U/μl collagenase VII (C0773; Sigma-Aldrich). After five washes in PBT, samples were blocked for 1 h in 1% BSA (in PBT). For MH27 staining, L1 animals were processed as described previously (Finney and Ruvkun, 1990). After labeling, samples were stained with 1 μg/ml DAPI for 10 min and then rinsed several times in PBT before mounting in 60% glycerol.
Primary antibody incubations were performed at room temperature overnight; secondary antibody incubations were performed for 2 to 3 h at room temperature. Antibodies and (dilutions) used were anti-ACT-5 (1:10); anti-actin C4 (ICN) (1:100); MH33 (1:100); MH27 (1:100); Alexa 488 goat anti-mouse and Alexa 568 goat anti-rabbit (Molecular Probes, Eugene, OR) (1:300). MH33 and MH27 antisera were kindly provided by R. Frances and R. Waterston (Washington University School of Medicine, St. Louis, MO).
Immunofluorescence preparations were examined using an Olympus BMAX 60F microscope equipped with a Princeton Instruments KAF-1400 charge-coupled device camera, by using either a 60× (1.4 numerical aperture [NA]) or 100× (1.35 NA) objective. Embryonic anti-actin, and L1 MH27 images were obtained using a Zeiss 510 META confocal microscope. Images in Figures 2 and 6 were processed by a blind deconvolution algorithm, by using the AutoDeblur software from AutoQuant.
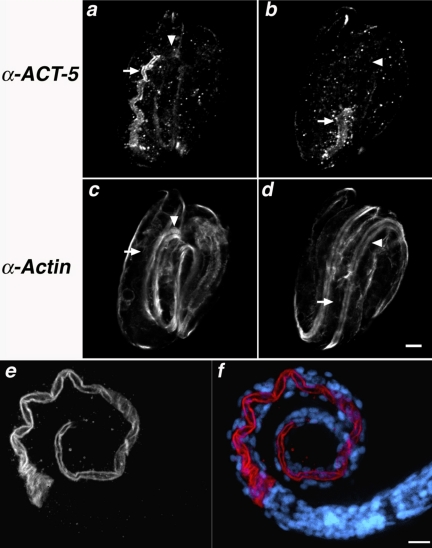
Figure 2.
anti-ACT-5 specifically labels the apical intestine. (a–d) Wild-type embryos costained with purified anti-ACT-5 antibodies (a and b) and C4, a pan-reactive anti-actin mAb (c and d). Images represent single confocal optical sections from the middle third (a and c) and top third (b and d) of the same threefold embryo. Arrows indicate the posterior (a and c) and anterior (b and d) of the intestine, where robust anti-ACT-5 label, but very little C4 staining is present (a vs. c); arrowheads point to a single longitudinal body wall muscle quadrant, which is not detected with anti-ACT-5 but readily labeled with C4 (b vs. d). Bar, 5 μm (a–d). (e and f) Wild-type L1 animals stained with anti-ACT-5, in white (e) or red (f); DAPI-stained nuclei are shown in blue (f). ACT-5 label decorates the lumenal surface of the intestine, showing as a tube along most of the length of this L1 animal. Images in e and f are projections of serial image slices that encompass approximately one-half the volume of an intestine and derive from the central portion of the intestine; this processing reveals the unlabeled lumenal area at the center of the intestine. Bar, 10 μm (e and f).
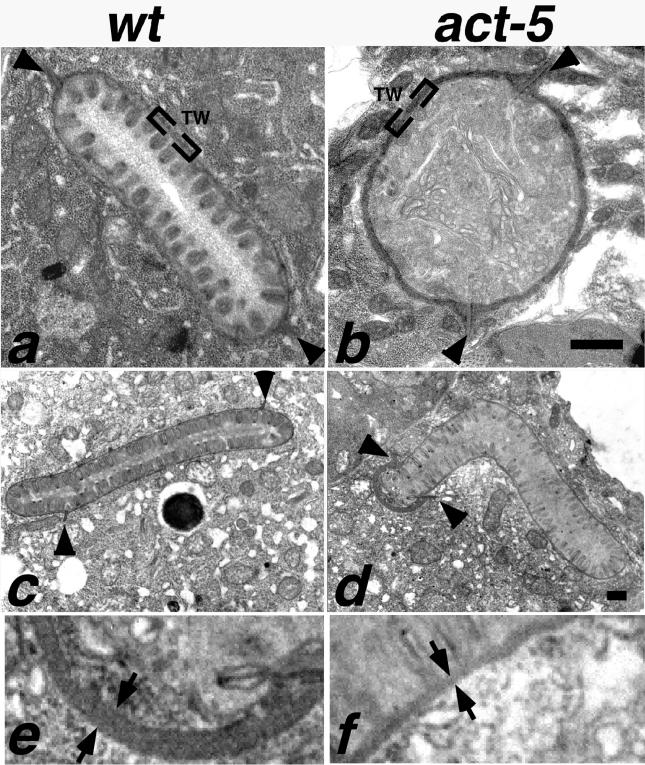
Figure 6.
Intestinal microvilli are defective or absent from ACT-5–depleted animals. In wild-type ultrastructural preparations (a and c), intestinal cross sections reveal tightly packed, finger-like projections that correspond to microvilli projecting from the lumenal surface. At the base of these projections is an electron-dense region called the terminal web (TW). A small segment of the terminal web is enclosed by brackets in a and b. Two finger-like projections pointing toward the cytoplasmic side of the intestinal cells are the apical junctions (arrowheads) that mark the boundary between apical regions of two intestinal cells. Microvilli are absent from intestinal cross sections of act-5 (dt2017) animals (b). act-5(RNAi) animals frequently exhibited one shortened apical region (distance between apical junctions) on one of a pair of intestinal cells; this shortened apical surface was typically associated with an abnormally thick terminal web (d). Equivalent higher magnification views of a portion of the thick (e) and normal terminal web width (f) from the micrograph shown in d reveal the difference in thickness as the gap between the arrows (e and f). Control RNAi cross section from an L2–L3 stage animal shown in c. Bar for a and b in b; for c and d in d, 500 nm.
Electron Microscopy
Animals grown for 2 d on act-5(RNAi) or control RNAi plates were collected and processed for electron microscopy by high-pressure freezing followed by freeze substitution (McDonald, 1999). A paste of diluted baker's yeast was used to gather ∼100 animals into one of each of five, 200-μm specimen carriers, and specimens were cryofixed in a high-pressure freezer (Leica EMPact). Freeze substitution was carried out using a freeze substitution machine (Leica AFS) as follows: samples were held in 2% osmium tetroxide, 0.1% uranyl acetate, and 8% dimethoxypropane in acetone at –78°C for 2 d; warmed to –20°C >12 h; held at –20°C 12 h; and then warmed to 20°C >6 h. Samples were rinsed six times in 100% acetone and infiltrated with Epon resin as follows: 2:1 acetone:resin for 3 h, 1:1 acetone:resin overnight, 1:2 acetone: resin 3 h followed by two changes in pure resin overnight. After embedding, 80-nm sections were cut onto Formvar grids and poststained in uranyl acetate and lead citrate. Cross sections from 10 individuals were examined for act-5 and control RNAi animals.
L1 worms were collected in M9 buffer (see below) and prepared by chemical/microwave fixation as described previously (Hall, 1995) with minor modifications. Worms were placed in a prechilled ceramic dish on ice, with prechilled fix (2.5% glutaraldehyde, 0.2 M sucrose, 1 mM MgCl2, 1% paraformaldehyde, and 0.05 M cacodylate buffer). Fixations were carried out on ice with three, 2-min pulses in a temperature-controlled microwave (Pelco 3451 laboratory microwave system; Ted Pella, Redding, CA). Animals were rinsed three times in 0.2 M cacodylate buffer and postfixed for 1 h in 1% osmium tetroxide. After rinsing again in cocodylate buffer, animals were mounted in 2.5% agarose blocks and held at 4°C overnight. Blocks were stained with 1% uranyl acetate, dehydrated with an increasing series of ethanol:water washes, rinsed twice in propylene oxide, and then infiltrated with Epon resin gradually over the course of 2 d.
To obtain wild-type L1 animals, gravid adults were treated with bleach and potassium hydroxide according to standard methods (Lewis, 1995) to isolate several hundred eggs, and the eggs were hatched in S-Medium (Johnson et al., 1984) solution overnight. To obtain a sufficient quantity of act-5 mutant L1 animals, 100 act-5(dt2017)/+ gravid animals were placed on each of seven egg collection plates, allowed to lay eggs for 8 h, and then removed. After 2 d, L1 arrested animals were manually picked into M9 buffer and immediately processed as described above. Approximately 5000 wild-type and 500-1000 mutant L1 animals were processed. Cross sections of intestines from eight mutant and eight wild-type animals were examined.
For immuno-electron microscopy, freeze substitution was carried out on high-pressure frozen worm tissue as described in Bossinger et al. (2004), except 0.1% uranyl acetate, instead of safranin O, was added to methanol for the freeze substitution media. Samples were gradually infiltrated with LR white resin over the course of 3 d and then embedded in gelatin capsules. Sections (80 nm) were loaded onto nickel grids and processed for antibody labeling, at room temperature, as follows: sections were rinsed in PBS and then held 15′ in 0.05 M glycine, 1× PBS, rinsed in PBS, and then held for 30′ in blocking buffer (BB) (0.1% cold water fish gelatin, 0.8% BSA, 0.02% Tween 20, and 1× PBS. Samples were incubated in 15 μl of primary antibody (undiluted MH33 or anti-ACT-5; 1:50 anti-C4 actin antibody) for 1.5 h, rinsed one time in BB and then twice in PBS, incubated in secondary antibody (1:50 5-nm gold-conjugated anti-mouse antibody or 10-nm gold-conjugated anti-rabbit antibody; Amersham Biosciences, Piscataway, NJ) for 1.5 h. Samples were rinsed 3 × 5′ in PBS and postfixed in 0.5% glutaraldehyde, 1× PBS for 5′. Grids were rinsed 3 × 5′ in PBS and then twice in distilled water and stained with uranyl acetate. All samples were analyzed using a JEOL 1200 EX electron microscope operating at 80 kV.
Protein Extracts and Western Blotting
Total SDS-soluble nematode proteins were prepared as described previously (Moerman et al., 1988) with the final extracts containing ∼0.18 g of packed volume of worms per milliliter of solubilization buffer. Discontinuous 12% SDS-polyacrylamide gels were used to separate 40 μl of sample per well and then blotted to nitrocellulose (Bio-Rad, Hercules, CA) in 25 mM Tris, pH 8.3, 192 mM glycine, and 20% (vol/vol) methanol. After blotting, the membrane was probed with either a 1:2500 dilution of the C4 mouse anti-actin mAb, or a 1:200 dilution of affinity-purified rabbit anti-ACT-5 in blocking buffer TTBS [0.3 M NaCl, 20 mM Tris-HCl, pH 8.0, 0.1% (vol/vol) Tween 20, and 0.01% NaN3 containing 5% BSA (wt/vol) and 5% (wt/vol) normal goat serum] for 6 h. Blots were washed four times in TTBS and then incubated for 2 h in blocking buffer containing alkaline phosphatase-conjugated goat anti-mouse (C4 blots) or goat anti-rabbit (anti-ACT-5 blots) antibodies from Pierce Chemical (Rockford, IL). After four additional washes in TTBS, the blots were developed for 1–10 min as described previously (Ey and Ashman, 1986). To visualize the ∼38-kDa truncation product in act-5(dt2017)-carrying lines, the blot must be developed for nearly 10 times longer than it takes to visualize the full-length ACT-5 polypeptide (Supplemental Figure 1).
RESULTS
act-5 Is Expressed in Cells That Contain Microvilli
Analysis of a reporter transgene revealed that act-5 expression is restricted to a small subset of cells within the C. elegans alimentary tract and excretory systems. A transgene construct containing GFP (gfp) (Chalfie et al., 1994), flanked upstream and downstream by roughly 1.5 kb of T25C8.2 regulatory sequence (pCKE1; see Materials and Methods), was heritably transformed into wild-type animals. GFP fluorescence was easily detected within a subset of embryonic, larval, and adult cells in transgenic animals, consistent with previous findings on the act-5 expression pattern (Figure 1; Gobel et al., 2004). Within larvae and adults, GFP expression occurred within the 20 cells that comprise the adult intestine, and in two sets of three associated cells: one just anterior and one posterior to the intestine. The anterior set corresponds to the middle of three sets of pharyngeal-intestinal valve cells (vpi 2), whereas the posterior group of associated cells comprises the rectal epithelial cell (rectD, rect_VL, rect_VR) (Sulston et al., 1983). Finally, the single excretory canal cell, which extends processes along the entire length of the worm, also showed GFP expression. The aforementioned act-5-expressing cells derive from three separate embryonic founder cell lineages yet, interestingly, each belongs to the small number of cells reported to contain microvilli or canaliculi (microvilli-like processes that increase the lumenal surface area of the excretory canal cells) in C. elegans (Sulston et al., 1983; Buechner, 2002).
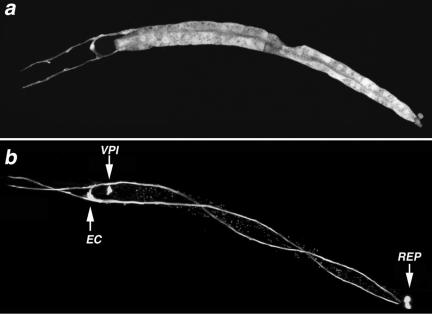
Figure 1.
Excretory and alimentary tract cells express act-5. (a and b) GFP expression, controlled by act-5 promoter sequences, is observed within each of the 20 pairs of intestinal cells that form a tube along most of the length of the animal; the H-shaped excretory cell (EC), pharyngeal-intestinal valve cells (VPI), and rectal epithelial cells (REP) also express the transgene reporter. The act-5::gfp transgene has been lost from intestinal cells of the animal in b, allowing better visualization of the EC, REP, and VPI cells. Anterior is left.
Anti-ACT-5-enriched Antibodies Label the Apical Domain of Polarized Intestinal Cells
The level of shared identity between actin proteins makes immunolocalization of specific isoforms challenging (Herman, 1993). To create antibodies that label ACT-5 with minimal cross-reactivity to other C. elegans actins, we performed a two-step affinity purification procedure on polyclonal antisera, which was generated using a divergent 13-amino acid stretch within ACT-5 (see Materials and Methods). Both initial and purified anti-ACT-5 antisera recognized a major 43-kDa band on protein blots of wild-type animals (Supplemental Figure 1; our unpublished data).
Evidence that the purified antibodies are significantly enriched for anti-ACT-5 activity came from immunolabeling experiments on fixed C. elegans embryos (Figure 2, a–d). Whereas a pan-reactive anti-actin antibody labeled numerous cell types, including prominent staining of the four body wall muscle quadrants that run the length of the worm, anti-ACT-5 enriched antisera showed robust label only in the small subset of embryonic cells that belong to the intestine, accompanied by an extremely low level of staining in muscle cells.
Anti-ACT-5 immunolabeling further indicated that ACT-5 is apically localized within intestinal cells. An apical subcellular localization pattern of ACT-5 within the intestine is consistent with previous studies that exploited a yellow fluorescent protein (YFP)::ACT-5 or GFP::ACT-5 translational fusion protein expressed from a transgene (Bossinger et al., 2004; Gobel et al., 2004). Analysis of larval and adult animals labeled with anti-ACT-5 revealed robust, continuous antibody localization along the length of the intestine immediately adjacent to the lumenal surface, whereas almost no label could be detected in cytoplasmic and basal regions of intestinal cells (Figure 2, e and f; Bossinger et al., 2004; Gobel et al., 2004). In contrast to the act-5::GFP transcriptional reporter, no antibody staining of the excretory cell was observed. The latter could be due to the stringent extraction procedures that are required to optimize immunovisualization of ACT-5 in the gut, or the much lower level of ACT-5 expression in the excretory cell relative to the gut. It is noteworthy that the pan-reactive mAb C4 also does not reveal the actin within the excretory cell using the whole mount fixation methods used for these studies. An apical subcellular localization of ACT-5 within the intestine is intriguing in light of the fact that act-5 expression is limited to cells containing microvilli and suggests the possibility that act-5 might play a role in microvillus formation or function.
act-5(RNAi) Causes a Reversible Defect in Intestinal Lumen Morphogenesis
Previous studies implicated a functional role for act-5 in intestinal lumen morphogenesis (Gobel et al., 2004). RNAi directed at the 3′ UTR of act-5 produces low-penetrance lumenal phenotypes that mimic those caused by a mutation in erm-1, a gene that encodes an actin-binding protein that is required for proper intestinal lumen morphogenesis (Gobel et al., 2004). act-5(RNAi) also causes a significant amount of L1 arrest and lethality (Gobel et al., 2004). Using a feeding RNAi strategy (Kamath et al., 2001; Timmons et al., 2001), we observed the same constellation of act-5(RNAi) phenotypes as those described previously (Gobel et al., 2004), with one important addition; act-5(RNAi) is reversible. When several animals, belonging to the most extreme growth-arrested class, were removed to standard bacteria plates after 72 h of development on act-5(RNAi) plates, about one-half of the animals (10/18) grew into reproductive adults within the next 3 d. The reversibility of act-5(RNAi) indicates that the process affected by act-5 depletion is ongoing throughout larval growth, rather than a discrete embryonic developmental event. Together with the expression pattern and subcellular localization of act-5, the phenotypic effects of act-5(RNAi) suggest the possibility that act-5 function is required for proper morphogenesis of the intestinal lumenal surface (Gobel et al., 2004).
act-5 Function Is Dispensable for Embryogenesis but Essential for Larval Growth
To more rigorously address the developmental requirement for act-5 function, we carried out a molecular genetic screen for act-5 mutants. We used a PCR-based strategy to screen a frozen mutant bank representing ∼1 million mutagenized haploid genomes (kindly provided by V. Reinke, Yale University, New Haven, CT). This effort identified act-5 (dt2017), which contains a 494-base pair deletion within T25C8.2. dt2017 is predicted to encode a protein that has replaced 54 central amino acids (residues 166–219 of ACT-5) with a single asparagine residue (Figure 3). The portion deleted from the dt2017 gene product normally spans parts of both subdomains 3 and 4 of the three-dimensional actin molecule, and moreover, includes residues thought to be important for ATP/ADP binding (Pollard and Cooper, 1986; Kabsch and Vandekerckhove, 1992; Herman, 1993). The extent of the lesion, in light of the fact that actins have diverged so little during eukaryotic evolution, indicates that the putative protein product of the dt2017 deletion allele has lost actin function. A second allele, act-5 (dt2019), which is derived from dt2017 (see Materials and Methods), additionally contains a premature stop codon upstream of the dt2017 deletion. The dt2019 allele thus encodes a 78-amino acid peptide fragment of the 375 residue ACT-5 protein and is also almost certainly a null allele for a conventional actin such as ACT-5 (Figure 3).
Figure 3.
Actin protein alignment and sequence changes caused by act-5 mutations. C. elegans actin protein alignment. ClustalW 1.83 alignment (Thompson et al., 1994) between the five C. elegans actins (CeACT-1–5, accession nos. CAB04678, CAB04675, CAB04676, AAB04575, and CAB05817, respectively) and the predicted Caenorhabditis briggsae ACT-5 (CbACT-5, accession no. CAE71353). The protein sequence for C. elegans ACT-1 and ACT-3 are identical and denoted as CeACT-1_3. Identical residues are shaded in black, conserved changes in gray. Sequence coordinates are given for the entire alignment, which is offset 1 residue for CeACT-5 and CbACT-5. The boxed region at residues 220–232 indicates the synthetic peptide used to elicit anti-ACT-5 antibodies. Asterisk indicates the position of the premature stop codon in dt2019; down arrows denote the region deleted in dt2017 and replaced by asparagine.
Observation of the growth of all individuals within a brood from dt2017 heterozygotes revealed a class of progeny that move slowly, exhibit slowed or nonexistent pharyngeal pumping, and die, as early L1 larvae, within 2 to 3 d after hatching. Scoring each animal within this brood for both growth rate and genotype demonstrated that the L1 lethal class corresponds to act-5 homozygotes (Figure 4a). This analysis further revealed that act-5 (dt2017) heterozygotes have a slightly slower growth rate than wild-type animals, accompanied by a smaller size and clear appearance, reminiscent of act-5(RNAi) animals (Figure 4b). dt2017 heterozygotes do, however, reach adulthood and are fertile. The dt2017 heterozygote phenotype raised the possibility that dt2017 has dominant negative activity, or alternatively, that act-5 is a rare haploinsufficient locus in C. elegans.
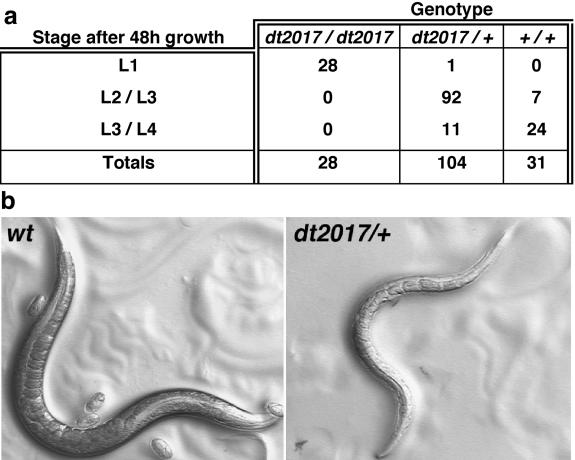
Figure 4.
act-5 is essential for growth. (a) Table presents an analysis of the growth of individual animals in a brood, after 2 d. Animals were assayed by PCR to identify genotype after their phenotype was recorded. act-5 (dt2017) homozygotes correspond to animals that fail to grow beyond the first larval stage (L1), even after 2 d of development. (b) Adults carrying one copy of act-5 (dt2017) are skinny and clear (right), compared with wild-type adults (left). Images were taken with bright field optics.
A transgene rescue experiment confirmed that the L1 lethal phenotype associated with act-5 mutations is indeed a consequence of missing act-5 function. The lethality associated with either act-5 allele is completely suppressed in transgenic worms carrying pJAW70, which contains roughly 3 kb of genomic flanking sequence accompanying a wild-type act-5 gene. However, although the L1 lethality of act-5(dt2017) homozygotes was rescued by pJAW70 carried either as an extrachromosomal or integrated transgene, these animals still looked clear and smaller than wild-type animals, similar to dt2017 heterozygotes. This phenotype is dependent on the dt2017 allele, as pJAW70-carrying animals that are wild-type at the act-5 locus are normal in size and appearance. The failure of act-5 (+) to suppress the phenotypic effects of one copy of the dt2017 allele argues against haploinsufficiency as the basis for dominance and is more consistent with the dt2017 gene product having dominant negative activity. Consistent with this hypothesis, protein extracts from animals that are dt2017 homozygotes but carry a wild-type act-5(+) transgene show accumulation of an ∼38-kDa product that is absent in wild-type worms that carry the integrated act-5(+) transgene (Supplemental Figure 1).
Further support for the notion that dt2017 is a dominant-negative allele comes from the fact the act-5 (dt2019) mutation, which contains a premature stop codon that predicts production of a significantly smaller mutant derivative of actin than the dt2017 product, was isolated as a suppressor of the slow growth and starved appearance of pJAW70-carrying, dt2017 homozygotes. At the level of the dissection microscope, animals homozygous for either dt2017 or dt2019 are indistinguishable and die as young L1 larvae. Moreover, dt2019 is by all apparent criteria, completely recessive.
act-5 Mutant Intestinal Cells Undergo Cell Division, Gastrulation, and Are Properly Polarized
Although actin often functions in basic cellular processes that are essential for cell polarity, cytokinesis, and viability, intestinal cells in act-5 mutants form a grossly normal and intact organ comprised of viable, correctly positioned, and properly polarized cells. First, using Nomarski differential interference contrast microscopy, we did not observe any obvious disruption along the length of act-5 mutant L1 animals that might indicate a developmental defect in intestinal morphogenesis, although, due to the small size and unhealthy appearance of act-5 mutant L1 animals, a completely continuous lumen was difficult to trace by using this method. Immunolocalization of actin, IFB-2 (the gut-specific intermediate filament protein recognized by MH33 mAb; Francis and Waterston, 1991; Bossinger et al., 2004), or the adherens junction marker JAM-1 (recognized by MH27 mAb; Francis and Waterston, 1991; Legouis et al., 2000; Koppen et al., 2001) confirmed that act-5 mutants have completely elaborated intestines with lumens that are continuous along their length (Figure 5; our unpublished data).
Figure 5.
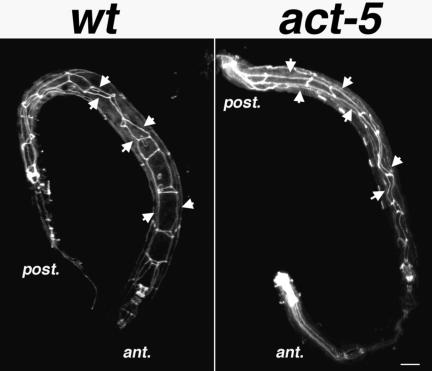
Intestinal cell polarization does not require act-5 function. MH27 staining (white) labels apical regions of polarized cells, giving rise to a web-like pattern along the bodies of the wild-type (left) and act-5(dt2017) mutant (right) worms. Although act-5 mutant intestinal lumens seem slightly deformed relative to control animals, MH27 is properly restricted to apical cell regions in both genotypes. Projections encompass the entire volume of each animal. Anterior (ant.) and posterior (post.) ends of the worm are indicated. Bar, 10 μm.
Intestinal cells maintain different molecular and functional microenvironments between apical and basal regions (Legouis et al., 2000). In C. elegans, MH27 mAb highlights this epithelial cell polarity, because it specifically labels junctions between the apical regions of cells (Leung et al., 1999). The polarized distribution of the MH27 antigen within the intestine looks like two narrow linear axes running immediately adjacent to and along the length of the intestinal lumen, bridged at specific locations throughout their lengths by perpendicular-oriented axes (Figure 5). In act-5 mutants, MH27 is clearly restricted to regions along the length of the narrow intestinal lumen, demonstrating that act-5 function is dispensable for establishing apical-basal polarity within intestinal cells. Further support for this conclusion was provided by ultrastructural analysis (see below), because apical junctions were observed exclusively associated with the lumenal side of act-5 mutant intestinal cells. The MH27 distribution pattern was found to be somewhat distorted in act-5 mutants relative to wild-type animals, which may reflect a requirement for act-5 in forming normal apical junction structures. Alternatively, this distortion might reflect the fact that act-5 animals are more vulnerable to damage during the immuno-fixation and processing procedure, because they normally die as L1 animals, shortly after hatching.
act-5 Is Essential for Gut Microvillus Formation
Although the L1 lethality associated with mutations in act-5 could reflect a defect in any of a number of actin-dependent processes, the act-5(RNAi) phenotypes together with the subcellular localization of ACT-5 hinted at a possible role for this gene product in microvillus formation or function. To ask whether act-5 is involved in microvillus morphogenesis, we analyzed the ultrastructure of intestinal cells from wild-type and act-5 mutant L1 animals by using transmission electron microscopy. Several consecutive cross sections of intestines from seven or more wild-type and mutant animals were analyzed. In wild-type cross-sections prepared by chemical/microwave fixation, microvilli looked like broad finger-like projections reaching into an oblong intestinal lumen (Figure 6). Two apical junctions, located roughly equidistant from one another along the circumference of the lumen, were apparent as microvilli-like structures but their orientation was pointed away from the lumen (Figure 6, arrowheads). The terminal web was notable in wild-type sections as a continuous, electron-dense border at the base of microvilli and along the circumference of the intestinal lumen. In contrast to wild type, intestinal cross sections from act-5 mutants contained lumens that were frequently round instead of oblong, and often, but not always, associated with an abnormally thick terminal web structure. The circumference of the lumen itself did not significantly differ between wild-type and mutant animals. Furthermore, the apical junctions that accompany act-5 mutant intestinal cells often seemed abnormally lengthened, although to varying extents from section to section. Most striking, however, was the complete absence of microvilli within the lumens of act-5 mutant intestines (Figure 6B). Out of five cross sections analyzed from each of seven animals, no microvilli were detected in act-5 mutant intestines, whereas microvilli were always readily visible in wild-type intestinal cross sections.
Defects in the ultrastructural appearance of intestines from growth-retarded, act-5(RNAi) animals were less obvious than those for dt2017 animals; however, one-half of analyzed cross sections (n = 20) exhibited unequal apical surface lengths between the two cells comprising a given section of the lumen, accompanied by an abnormally thick terminal web associated with the shorter apical surface (Figure 6D). Distorted and elongated apical junction structures, and/or regions with severely disrupted or missing microvilli also were exhibited by a subset of sections, especially among those sections exhibiting abnormally short apical regions. The relatively mild severity of the defects exhibited by act-5(RNAi) intestinal cross-sections is consistent with the fact that act-5(RNAi) elicits a weaker phenotype compared with that of act-5 loss-of-function genetic mutants.
ACT-5–enriched Antibodies Predominantly Label Microvilli in Thin Sections
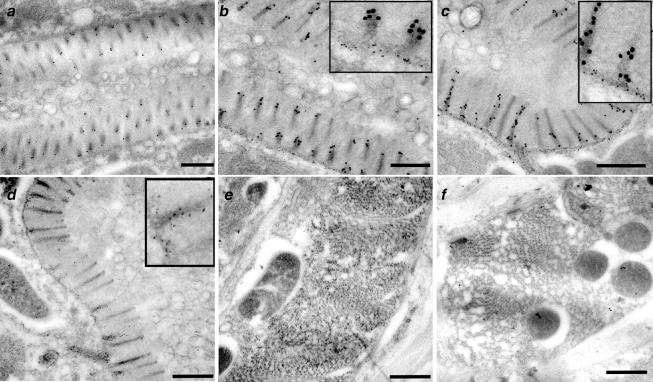
To begin to ask whether a specialized actin isoform could be responsible for building microvilli in the C. elegans intestine, we analyzed the distribution pattern of the ACT-5–enriched antisera on cross sections of wild-type adult worms. We labeled consecutive sections with either MH33 antisera, which detects intermediate filaments, or a pan-reactive anti-actin antibody (C4) to control for the success of our technique. Additional sections were incubated with secondary antibody only, to assess nonspecific background labeling. As has been reported previously (Bossinger et al., 2004), MH33 antisera decorated the terminal web and apical junction structures, with little or no detectable label in other regions of the cross section (Figure 7, b and c). The pan-reactive anti-actin antibody displayed some labeling throughout the terminal web, apical junctions, and along the length of microvilli (Figure 7d). Anti-actin label also decorated other regions of the section, such as areas containing muscle cells (Figure 7e). In contrast to either MH33 or C4 anti-actin antibody, the anti-ACT-5–enriched antibodies exhibited robust label along the lengths of microvilli, sparse labeling at the terminal web, and no above-background label in other regions of the sections, most notably those containing muscle cells (Figure 7, a–c, and f). This distribution pattern is exactly what one would predict for a building block component of microvilli, and as such it provides strong evidence that act-5 encodes a major actin component of microvilli.
Figure 7.
anti-ACT-5 antisera labels microvilli on thin sections. Gold particles (10 nm), detecting anti-ACT-5 antisera, exhibit robust labeling on microvilli of wild-type intestinal cross sections (a–c), whereas exhibiting little or no label in other areas of the section (muscle cells in f). MH33 antisera, marked by 5-nm gold particles, labels the terminal web and apical junction areas (b and c). C4 (marked by 5-nm gold particles), a pan-reactive monoclonal anti-actin antibody, which detects several, if not all, C. elegans actins, labels microvilli (d) in addition to several additional cells within the worm; muscle cell labeling is shown in e. Insets in b, c, and d show enlarged regions to enable better visualization of 5-nm gold particles. Bar, 500 nm.
The ACT-5 Protein Isoform Is Uniquely Adapted for the Essential act-5 Function
The requirement for wild-type act-5(+) gene function for microvillus biogenesis and viability could be at the level of gene expression or protein isoform specialization. Previous work on the Act5C gene of Drosophila showed that the inviability associated with the loss of this essential gene could be rescued by a chimeric transgene in which the Act5C regulatory sequences drive expression of the closely related Drosophila actin, Act42A (Wagner et al., 2002). In this case, the unique temporal and quantitative regulation of actin levels, dictated by the Act5C promoter, served the essential function, rather than the distinct amino acid differences of Act5C relative to Act42A. Considering this precedent, it is possible that in C. elegans, the act-5 regulatory sequences may simply produce gut actin levels that reach a critical temporal or quantitative threshold optimized for the biogenesis of microvilli and that any of the other four highly related worm actins, 99% identical to each other and 93% identical to ACT-5, could serve the essential functions of the ACT-5 protein if expressed under the control of the act-5 regulatory sequences. To address this possibility, we chose ACT-1, which is 100% identical to ACT-3 and 99% identical to ACT-2,4, as a representative of the non-ACT-5 actins, and drove the expression of the ACT-1 cDNA under the control of the act-5 regulatory sequences (Pact-5::ACT-1). Because the Pact-5::ACT-1 construct lacks introns relative to the genomic act-5 construct, we generated the analogous construct with the ACT-5 cDNA (Pact-5::ACT-5) as a positive control for comparison. Suitable transgenic lines were made by microinjection and then these transgenes were crossed with the dominant dt2017/+ animals to obtain animals with genotypes dt2017/+;Pact-5::ACT-1 or dt2017/+;Pact-5::ACT-5. F1 progeny from the transgenic dt2017/+ adults were scored for L1 arrest versus continuous development and to assay whether rescued dt2017 homozygotes could be recovered. Table 1 compares the genotype of the progeny and their developmental stage for the Pact-5::ACT-5 and Pact-5::ACT-1 transgenic lines. The genotyping data clearly indicate that although the Pact-5::ACT-5 transgene rescued 76% of the dt2017 homozygotes, all of the dt2017 homozygotes in the Pact-5::ACT-1 transgenic line arrest as L1 larvae (Table 1). Moreover, the dt2017; Pact-5::ACT-5 animals are easy to establish as stable, heritable strains. These rescue data are most consistent with the ACT-5 isoform having unique properties that are adapted to its specialized function in microvilli or other structures required for viability.
Table 1.
Actin isoform-specific rescue of act-5(dt2017)
| Genotype
|
|||||
|---|---|---|---|---|---|
| Transgene | Stage (N) | dt2017/dt2017 | dt2017/+ | +/+ | % Rescuea |
| Pact-5::ACT-5 | L1 (3) | 3 | 0 | 0 | 76 |
| >L2 (92) | 10 | 57 | 25 | ||
| Pact-5::ACT-1 | L1 (33) | 33 | 0 | 0 | 0 |
| >L2 (167) | 0 | 103 | 64 | ||
Shown are data for rescue assays performed on independent lines carrying an extrachromasomal transgene expressing either ACT-5 (2 independent lines) or ACT-1 (3 independent lines) both under the control of act-5 5′ and 3′ regulatory sequences. Rescue assays measured the developmental stage and genotype of multiple individual progeny of dt2017/+, transgene-carrying hermaphrodites ∼48 h after egg laying.
Percentage of rescue is defined as the number of dt2017 homozygotes that grow beyond the L1 stage after ∼48 h.
DISCUSSION
A Functional Link between a Divergent Actin and Microvillus Formation
We provide genetic evidence that implicates a distinct C. elegans actin isoform in microvillus morphogenesis. Furthermore, our immunocytological analysis revealed that ACT-5 is specifically expressed and apically enriched within microvilli-containing cells of the intestine, and ultrastructural analysis indicated that ACT-5 is likely located within microvilli themselves. Together, our findings and previous work on the distribution of YFP-ACT-5 (Bossinger et al., 2004) and the phenotype induced by act-5(RNAi) (Gobel et al., 2004) strongly suggest that ACT-5 may directly serve as a core building block for microvillus formation.
Whereas the function and ultrastructure of microvilli have been appreciated for decades (Hirokawa and Heuser, 1981), the molecular pathways that underlie their initial formation and stabilization have remained elusive. This could, in part, be due to the fact that genes required for microvilli formation also likely serve additional essential functions. Mutations in such loci might thus exhibit severe defects that precede or preclude an ability to examine microvillus formation. eps-8 is an excellent example of a gene required for early embryonic development but that also has later functions in the intestine (Croce et al., 2004). EPS-8 is a C. elegans orthologue of a receptor tyrosine kinase substrate that has barbed-end actin capping activity and is widely expressed and enriched at the apical tip of microvilli (Croce et al., 2004; Disanza et al., 2004). eps-8 is essential for embryonic development but produces two isoforms, EPS-8A and EPS-8B (Croce et al., 2004). An EPS-8A–specific knockout permits normal embryonic development but animals arrest as L1-L2 larvae with major defects in the gut, and the mutant can be rescued by intestine-specific expression of EPS-8A (Croce et al., 2004). Given that EPS-8A is uniquely adapted among EPS-8 isoforms to serve an essential function in the gut, it is reasonable to suggest that specific actin isoforms also might be uniquely adapted to the microvillus-specialized actin-binding proteins.
Larval Lethality and act-5 Function
The experiments reported here show that act-5 loss-of-function mutations result in early larval lethality. An absence of microvilli is expected to result in severely diminished intestinal absorption, but starvation per se is typically not associated with L1 lethality since a “checkpoint” pathway, which arrests the growth of wild-type L1 animals, can be triggered during absolute starvation conditions. Because of this check-point, starvation-arrested L1 larvae can live at least 10 d in the absence of food (Johnson et al., 1984), whereas act-5 mutants lived no longer than 72 h.
Perhaps through its role in building proper microvilli, act-5 is required for execution of the L1 starvation arrest pathway. In particular, one could imagine that proteins on the surface of microvilli themselves might monitor nutrient absorption and/or food availability, and thus serve as an essential component of the initiating signals that trigger starvation arrest. Alternatively, act-5 function might be required for apical surface remodeling that perhaps needs to occur within the intestines of L1 animals for survival during starvation conditions. Finally, a high surface area intestine might be required for efficient elimination of feces or metabolic waste generated by the organism, regardless of food supply status.
Another potential explanation for L1 lethality is that act-5 animals are on one or the other side of a critical threshold of nutrient deprivation that is required for starvation arrest to be elicited. On the one hand, act-5 mutants might absorb enough food to prevent starvation arrest but an insufficient amount to nourish proper growth. On the other hand, perhaps an absence of microvilli leads to irreparable damage during late embryonic stages, before the worm has the capacity to trigger the starvation arrest checkpoint pathway, the latter of which presumably occurs after hatching.
Finally, rather than lethality due to loss of intestinal microvilli, L1 lethality might reflect an essential function for act-5 in the excretory cell, which is thought to control osmoregulation (Nelson and Riddle, 1984). It will be interesting to explore this possibility by assessing whether L1 lethality of act-5 mutants can be rescued by a wild-type act-5 transgene expressed exclusively within the excretory cell, the intestine, or other cell types, even those for which we have not yet detected expression of ACT-5.
ACT-5 Is Uniquely Specialized for Microvilli Formation
Suggestion of the existence of a microvillus-specific actin in vertebrates was provided over a decade ago by analyses of isoform-restricted antibodies on rat intestinal epithelia (Sawtell et al., 1988). Here, we provide the first genetic evidence in support of the notion that a specialized actin functions in microvilli biogenesis. How, at the molecular level, might act-5 be unique among C. elegans actins in participating in microvilli morphogenesis?
One possibility is that act-5 function is required for microvilli formation simply because it is the sole actin expressed in intestinal cells. Two observations in this study strongly argue against this possibility. First, a scenario in which act-5 encodes the only actin in intestinal cells is not compatible with the terminal phenotype of act-5 loss-of-function mutants because embryonic intestinal cells are competent to grow, divide, and terminally differentiate into polarized epithelia in these animals. Second, and more importantly, expression of ACT-1 under the control of the ACT-5 promoter does not rescue the inviability of dt2017 mutants. Based on our data, protein sequence differences between ACT-5 and the other C. elegans actins most likely render ACT-5 functionally distinct and specialized for microvilli formation or an unknown essential function.
Evidence for functional differences among actin isoforms also has been found in Drosophila, where a cytoplasmic actin failed to rescue the flight defect caused by a muscle actin mutation, and in mammals, where a mouse lacking cardiac muscle actin could not be rescued by cardiac expression of an endogenous but noncardiac muscle actin protein (Kumar et al., 1997; Fyrberg et al., 1998). Similarly, overexpression studies in vertebrate cultured cells support the notion that some actin isoforms exhibit distinct functional capacities (Schevzov et al., 1992). Finally, point mutations in the human ACTG1 γ actin isoform have been implicated as the causative defect in autosomal dominant, progressive hearing loss in humans (van Wijk et al., 2003). Given that the stereocilia of the inner ear contain numerous microvilli-like structures whose characteristic staircase organization and length probably play a central role in hearing (Hudspeth and Jacobs, 1979; Howard and Hudspeth, 1987), and the potential for microvilli-specialized actins described here, it is possible that ACTG1 is uniquely adapted for function in the microvilli of the inner ear.
Future exploration of the molecular relationship between act-5 function and microvilli morphogenesis by using C. elegans promises to shed new light on the mechanisms underlying both actin functional diversity and the biogenesis of microvilli in the gut and other tissues that exploit microvilli for specific physiological needs.
Supplementary Material
Acknowledgments
We thank Bob Waterston in whose laboratory this work was first initiated. We also appreciate the help of the Molecular Cellular Imaging Facility at University of Texas Southwestern for all ultrastructural experiments, and Valerie Reinke for allowing us to screen a deletion library. Confocal imaging was made possible by a shared instrumentation National Institutes of Health S10-RR019406 (to J.A.W.). This work also was supported by a Burroughs Wellcome Fund Career Award and National Institutes of Health Grant R01-AG19983 (to J.A.W.). A.J.M. was supported by a postdoctoral research fellowship from the Helen Hay Whitney Foundation.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–12–1061) on May 4, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Bossinger, O., Fukushige, T., Claeys, M., Borgonie, G., and McGhee, J. D. (2004). The apical disposition of the Caenorhabditis elegans intestinal terminal web is maintained by LET-413. Dev. Biol. 268, 448–456. [DOI] [PubMed] [Google Scholar]
- Buechner, M. (2002). Tubes and the single C. elegans excretory cell. Trends Cell Biol. 12, 479–484. [DOI] [PubMed] [Google Scholar]
- Chalfie, M., Tu, Y., Euskirchen, G., Ward, W. W., and Prasher, D. C. (1994). Green fluorescent protein as a marker for gene expression. Science 263, 802–805. [DOI] [PubMed] [Google Scholar]
- Croce, A., Cassata, G., Disanza, A., Gagliani, M. C., Tacchetti, C., Malabarba, M. G., Carlier, M. F., Scita, G., Baumeister, R., and Di Fiore, P. P. (2004). A novel actin barbed-end-capping activity in EPS-8 regulates apical morphogenesis in intestinal cells of Caenorhabditis elegans. Nat. Cell Biol. 6, 1173–1179. [DOI] [PubMed] [Google Scholar]
- Disanza, A., Carlier, M. F., Stradal, T. E., Didry, D., Frittoli, E., Confalonieri, S., Croce, A., Wehland, J., Di Fiore, P. P., and Scita, G. (2004). Eps8 controls actin-based motility by capping the barbed ends of actin filaments. Nat. Cell Biol. 6, 1180–1188. [DOI] [PubMed] [Google Scholar]
- Ey, P. L., and Ashman, L. K. (1986). The use of ALP-conjugated anti-immunoglobulin with immunoblots for determining the specificity of monoclonal antibodies to protein mixtures. Methods Enzymol. 121, 497–509. [DOI] [PubMed] [Google Scholar]
- Finney, M., and Ruvkun, G. (1990). The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell 63, 895–905. [DOI] [PubMed] [Google Scholar]
- Francis, R., and Waterston, R. H. (1991). Muscle cell attachment in Caenorhabditis elegans. J Cell Biol. 114, 465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieden, C., Du, J., Schriefer, L., and Buzan, J. (2000). Purification and polymerization properties of two lethal yeast actin mutants. Biochem. Biophys. Res. Commun. 271, 464–468. [DOI] [PubMed] [Google Scholar]
- Fyrberg, E. A., Fyrberg, C. C., Biggs, J. R., Saville, D., Beall, C. J., and Ketchum, A. (1998). Functional nonequivalence of Drosophila actin isoforms. Biochem. Genet. 36, 271–287. [DOI] [PubMed] [Google Scholar]
- Gobel, V., Barrett, P. L., Hall, D. H., and Fleming, J. T. (2004). Lumen morphogenesis in C. elegans requires the membrane-cytoskeleton linker erm-1. Dev. Cell 6, 865–873. [DOI] [PubMed] [Google Scholar]
- Hall, D. H. (1995). Electron microscopy and 3D image reconstruction. In: Caenorhabditis elegans: Modern Biological Analysis of an Organism, vol. 48, ed. D.S.H.F. Epstein, New York: Academic Press, 395–436. [Google Scholar]
- Herman, I. M. (1993). Actin isoforms. Curr. Opin. Cell Biol. 5, 48–55. [DOI] [PubMed] [Google Scholar]
- Hirokawa, N., and Heuser, J. E. (1981). Quick-freeze, deep-etch visualization of the cytoskeleton beneath surface differentiations of intestinal epithelial cells. J Cell Biol. 91, 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K., and Pease, L. R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Howard, J., and Hudspeth, A. J. (1987). Mechanical relaxation of the hair bundle mediates adaptation in mechanoelectrical transduction by the bullfrog's saccular hair cell. Proc. Natl. Acad. Sci. USA 84, 3064–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudspeth, A. J., and Jacobs, R. (1979). Stereocilia mediate transduction in vertebrate hair cells (auditory system/cilium/vestibular system). Proc. Natl. Acad. Sci. USA 76, 1506–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, T. E., Mitchell, D. H., Kline, S., Kemal, R., and Foy, J. (1984). Arresting development arrests aging in the nematode Caenorhabditis elegans. Mech. Ageing Dev. 28, 23–40. [DOI] [PubMed] [Google Scholar]
- Kabsch, W., and Vandekerckhove, J. (1992). Structure and function of actin. Annu. Rev. Biophys. Biomol. Struct. 21, 49–76. [DOI] [PubMed] [Google Scholar]
- Kamath, R. S., Martinez-Campos, M., Zipperlen, P., Fraser, A. G., and Ahringer, J. (2001). Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2, RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppen, M., Simske, J. S., Sims, P. A., Firestein, B. L., Hall, D. H., Radice, A. D., Rongo, C., and Hardin, J. D. (2001). Cooperative regulation of AJM-1 controls junctional integrity in Caenorhabditis elegans epithelia. Nat. Cell Biol. 3, 983–991. [DOI] [PubMed] [Google Scholar]
- Kumar, A., et al. (1997). Rescue of cardiac α-actin-deficient mice by enteric smooth muscle γ-actin. Proc. Natl. Acad. Sci. USA 94, 4406–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landel, C. P., Krause, M., Waterston, R. H., and Hirsh, D. (1984). DNA rearrangements of the actin gene cluster in Caenorhabditis elegans accompany reversion of three muscle mutants. J. Mol. Biol. 180, 497–513. [DOI] [PubMed] [Google Scholar]
- Legouis, R., Gansmuller, A., Sookhareea, S., Bosher, J. M., Baillie, D. L., and Labouesse, M. (2000). LET-413 is a basolateral protein required for the assembly of adherens junctions in Caenorhabditis elegans. Nat. Cell Biol. 2, 415–422. [DOI] [PubMed] [Google Scholar]
- Leung, B., Hermann, G. J., and Priess, J. R. (1999). Organogenesis of the Caenorhabditis elegans intestine. Dev. Biol. 216, 114–134. [DOI] [PubMed] [Google Scholar]
- Lewis, J.A.a.F., J.T. (1995). Basic culture methods. In: Methods in Cell Biology, vol. 48, ed. D.S.H.F. Epstein, San Diego: Academic Press, 3–29. [PubMed] [Google Scholar]
- McDonald, K. (1999). High-pressure freezing for preservation of high resolution fine structure and antigenicity for immunolabeling. Methods Mol. Biol. 117, 77–97. [DOI] [PubMed] [Google Scholar]
- Mello, C. C., Kramer, J. M., Stinchcomb, D., and Ambros, V. (1991). Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman, D. G., Benian, G. M., Barstead, R. J., Schriefer, L. A., and Waterston, R. H. (1988). Identification and intracellular localization of the unc-22 gene product of Caenorhabditis elegans. Genes Dev. 2, 93–105. [DOI] [PubMed] [Google Scholar]
- Nelson, F. K., and Riddle, D. L. (1984). Functional study of the Caenorhabditis elegans secretory-excretory system using laser microsurgery. J. Exp. Zool. 231, 45–56. [DOI] [PubMed] [Google Scholar]
- Pollard, T. D., and Cooper, J. A. (1986). Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annu. Rev. Biochem. 55, 987–1035. [DOI] [PubMed] [Google Scholar]
- Ruvkun, G., and Giusto, J. (1989). The Caenorhabditis elegans heterochronic gene lin-14 encodes a nuclear protein that forms a temporal developmental switch. Nature 338, 313–319. [DOI] [PubMed] [Google Scholar]
- Sawtell, N. M., Hartman, A. L., and Lessard, J. L. (1988). Unique isoactins in the brush border of rat intestinal epithelial cells. Cell Motil. Cytoskeleton 11, 318–325. [DOI] [PubMed] [Google Scholar]
- Schevzov, G., Lloyd, C., and Gunning, P. (1992). High level expression of transfected β- and γ-actin genes differentially impacts on myoblast cytoarchitecture. J. Cell Biol. 117, 775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin-i, T., and Kohara, Y. (1998). NEXTDB: the expression pattern map database for C. elegans. Genome Informatics 1998, 224–225. [Google Scholar]
- Smith, D. B., and Johnson, K. S. (1988). Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67, 31–40. [DOI] [PubMed] [Google Scholar]
- Stone, S., and Shaw, J. E. (1993). A Caenorhabditis elegans act-4::lacZ fusion: use as a transformation marker and analysis of tissue-specific expression. Gene 131, 167–173. [DOI] [PubMed] [Google Scholar]
- Sulston, J. E., Schierenberg, E., White, J. G., and Thomson, J. N. (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64–119. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney, L. G., Tilney, M. S., and Guild, G. M. (1996). Formation of actin filament bundles in the ring canals of developing Drosophila follicles. J Cell Biol. 133, 61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons, L., Court, D. L., and Fire, A. (2001). Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103–112. [DOI] [PubMed] [Google Scholar]
- van Wijk, E., Krieger, E., Kemperman, M. H., De Leenheer, E. M., Huygen, P. L., Cremers, C. W., Cremers, F. P., and Kremer, H. (2003). A mutation in the γ actin 1 (ACTG1) gene causes autosomal dominant hearing loss (DFNA20/26). J. Med. Genet. 40, 879–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, C. R., Mahowald, A. P., and Miller, K. G. (2002). One of the two cytoplasmic actin isoforms in Drosophila is essential. Proc. Natl. Acad. Sci. USA 99, 8037–8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston, R., et al. (1992). A survey of expressed genes in Caenorhabditis elegans. Nat. Genet. 1, 114–123. [DOI] [PubMed] [Google Scholar]
- Waterston, R. H., Hirsh, D., and Lane, T. R. (1984). Dominant mutations affecting muscle structure in Caenorhabditis elegans that map near the actin gene cluster. J. Mol. Biol. 180, 473–496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.