Abstract
The intestinal cells of Caenorhabditis elegans embryos contain prominent, birefringent gut granules that we show are lysosome-related organelles. Gut granules are labeled by lysosomal markers, and their formation is disrupted in embryos depleted of AP-3 subunits, VPS-16, and VPS-41. We define a class of gut granule loss (glo) mutants that are defective in gut granule biogenesis. We show that the glo-1 gene encodes a predicted Rab GTPase that localizes to lysosome-related gut granules in the intestine and that glo-4 encodes a possible GLO-1 guanine nucleotide exchange factor. These and other glo genes are homologous to genes implicated in the biogenesis of specialized, lysosome-related organelles such as melanosomes in mammals and pigment granules in Drosophila. The glo mutants thus provide a simple model system for the analysis of lysosome-related organelle biogenesis in animal cells.
INTRODUCTION
Lysosomes are ubiquitous membrane-bound organelles that function as major degradative sites within eukaryotic cells (Tappel, 1969). Lysosomes contain an assortment of acid-dependent hydrolases that function in the breakdown of proteins, lipids, nucleic acids, and oligosaccharides. Lysosomes receive exogenous material through the endocytic pathway and are characterized as being the terminal compartment of the endocytic pathway. Lysosomes also receive material via the secretory pathway and directly from the cytoplasm (Kornfeld and Mellman, 1989; Mullins and Bonifacino, 2001; Luzio et al., 2003). Lysosomes function in diverse and important cellular processes including cell surface receptor turnover, destruction of pathogens, antigen processing, digestion, starvation responses, tissue remodeling, ion storage, autophagy, and plasma membrane repair.
The yeast vacuole shares several characteristics with the lysosomes of higher animals. Genetic screens have led to the identification of >150 genes necessary for the transport and sorting of newly synthesized proteins to the yeast vacuole (Jones, 1977; Bankaitis et al., 1986; Rothman and Stevens, 1986; Bonangelino et al., 2002). These genes control two pathways of Golgi-to-vacuole transport, the carboxypeptidase Y (CPY) and alkaline phosphatase (ALP) sorting pathways (Burd et al., 1998; Conibear and Stevens, 1998; Mullins and Bonifacino, 2001). Proteins trafficked via the CPY pathway transit an endosomal prevacuolar compartment en route to the vacuole. The ALP pathway mediates transport to the vacuole independent of the prevacuolar compartment.
Many of the genes involved in transport to the yeast vacuole have homologues in higher animals (Lemmon and Traub, 2000; Mullins and Bonifacino, 2001; Bonangelino et al., 2002). For example, the HOPS complex proteins (Vps11p, Vps16p, Vps18p, and Vps33p) regulate membrane fusion events necessary for lysosomal delivery within yeast (Rieder and Emr, 1997; Peterson and Emr, 2001), Drosophila melanogaster (Sevrioukov et al., 1999; Sriram et al., 2003), and mammalian (Poupon et al., 2003; Richardson et al., 2004) endosomal systems. Similarly, the proteins composing the AP-3 complex control the formation of transport vesicles for lysosomal cargo in each of these species (Odorizzi et al., 1998; Bonifacino and Traub, 2003; Luzio et al., 2003).
Although the yeast vacuole has proven to be a useful model for studying basic features of lysosomes, studies in Drosophila, mammals, and Caenorhabditis elegans have identified genes necessary for the formation of lysosomes that are not conserved in yeast (Lloyd et al., 1998; Spritz, 1999; Marks and Seabra, 2001; Piper and Luzio, 2004; Treusch et al., 2004). For example, the Hook family of proteins are present in Drosophila (Kramer and Phistry, 1996), C. elegans (Malone et al., 2003), and mammals but not in yeast (Walenta et al., 2001). The Drosophila Hook and human Hook1 proteins likely function in transport to lysosomes by mediating interactions between endosomal membranes and microtubules (Kramer and Phistry, 1996; Sunio et al., 1999; Richardson et al., 2004). In addition, animal cells can contain specialized lysosome-related organelles that are not found in yeast (Dell'Angelica et al., 2000b). Examples include melanosomes in vertebrate melanocytes and pigment granules in Drosophila retinal pigment cells (Lloyd et al., 1998; Marks and Seabra, 2001; Raposo and Marks, 2002). Mammalian cytotoxic T lymphocytes, basophils, and platelets each have secreted lysosomes whose contents initiate and modulate immune and clotting responses (Blott and Griffiths, 2002; King and Reed, 2002; Stinchcombe et al., 2004). Several human diseases are associated with defects in the formation or function of these specialized lysosomes, such as Hermansky-Pudlak syndrome (HPS), Chediak-Higashi syndrome, and Griscelli syndrome (Stinchcombe et al., 2004). HPS in mammals is a genetic disorder characterized by partial albinism and prolonged bleeding resulting from defects in melanosome and platelet-dense granule formation, respectively (Huizing et al., 2002; Li et al., 2004).
The Rhabditis genus of nematodes contains intestine-specific birefringent material sometimes called rhabditin granules, or gut granules in C. elegans (Chitwood and Chitwood, 1974; Laufer et al., 1980). The gut granules have been hypothesized to correspond to intestine-specific lysosomes (Clokey and Jacobson, 1986; Kostich et al., 2000; Hersh et al., 2002) and thus represent a potential system for studying lysosome biogenesis. The birefringence of gut granules in C. elegans allows the granules to be scored easily by polarization microscopy in living animals (Laufer et al., 1980). Moreover, the formation of gut granules is highly robust; embryos that are cleavage blocked after only one or two divisions can develop gut granules in the precursors of the intestinal lineage (Laufer et al., 1980).
In this report, we provide evidence that the birefringent gut granules are lysosome-related organelles; the gut granules stain with dyes that mark lysosomes, and previously identified genes involved in lysosomal biogenesis alter the formation of gut granules. We describe results from a screen to identify new genes involved in gut granule development and present a phenotypic and molecular characterization of one of these mutants, glo-1.
MATERIALS AND METHODS
Strains and Alleles
N2 was used as the wild-type strain. Mutant alleles used are listed by chromosome: linkage group I (LGI): apt-6(ok429), arIs37[pmyo3::ssGFP] (Fares and Greenwald, 2001b), dpy-5(e61), glo-2(zu455), rme-8(b1023), unc-13(e450), vps-34/let-512(h797).
LGII: cad-1(j1), rrf-3(pk1426), unc-52(e444), ypt-7/rab-7(ok511).
LGIII: cup-5(ar465), cup-5(n3194), daf-4(e1364), flu-3(e1001), unc-36(e251), mtm-6(ok330), vps-16(ok719), vps-29(tm1320).
LGIV: vps-27/pqn-9(ok579), vps-30/bec-1(ok700).
LGV: apt-2(tm935), flu-1(e1002), rme-1(b1045), glo-4(ok623), vac-1/eea-1(tm933), vps-54(tm584).
LGX: apt-7(tm920), bIs1[vit-2::gfp] (Grant and Hirsh, 1999), dpy-8(e130), dpy-23(e840), flu-2(e1003), flu-4(e1004), glo-1(kx21), glo-1(kx92), glo-1(zu391), glo-1(zu430), glo-1(zu437), glo-1(zu442), glo-3(zu446), glo-3(kx91), glo-3(kx92), glo-3(gm125), lin-2(e1309), unc-6(e78), vps-5/snx-1(tm847), vps-41(ep402), vps-45(tm246).
LG unknown: bIs33[rme-8::gfp] (Zhang et al., 2001), pwIs50[lmp-1::gfp] (Treusch et al., 2004), pwIs87[vha-6::rme-1::gfp] (this work), pwIs72[vha-6::rab-5::gfp] (this work). References for strains used are provided at Wormbase (www.worm-base.org). The glo-3(gm125) allele was provided by Gian Garriga (University of California, Berkeley, Berkeley, CA). C. elegans were cultured as described previously (Brenner, 1974). All strains were grown at 22°C unless otherwise noted.
Genetics
Glo alleles were isolated using the method described by Priess et al. (1987) with the following modifications for a nonclonal, F2 screen. Individual F1 progeny of ethylmethane sulfonate or ethyl nitroso urea-mutagenized hermaphrodites were not separated, F2 embryos within the carcass of their mothers were screened using polarization or differential interference contrast (DIC) microscopy, and carcasses containing 1/4 glo(–) embryos were recovered. zu455 was identified in a nonclonal, F3 screen (Priess et al., 1987). Genetic complementation tests placed the glo mutants into three complementation groups as follows: glo-1: kx21, kx92, zu391, zu430, zu437, zu442; glo-2: zu455; and glo-3: zu446, kx91, kx94, gm125. glo-1 mapped to the aex-2 unc-115 genetic interval. glo-2 mapped to the unc-57 dpy-5 genetic interval. glo-3 mapped to the unc-115 egl-15 genetic interval. Standard genetic techniques were used to determine that all of the glo alleles were recessive, that glo-1 and glo-3 alleles were zygotically expressed, and that glo-2 could be maternally or paternally rescued. All glo alleles used for phenotypic analysis were backcrossed at least three to six times to N2.
For analysis of brood size, embryonic lethality, and larval lethality, five fourth larval (L4)-stage animals from each strain were placed on individual plates and grown at 22°C. Once egg laying commenced, adults were moved to new plates every 8 h, and the number of embryos on each plate was scored. Twenty-four hours after removing an adult, the plate was scored again for the number of unhatched embryos. Wild-type embryos failed to hatch (embryonic lethality) only 1% of the time (n = 809), whereas 24% (n = 838) of glo-1(zu437) embryos failed to hatch, 13% (n = 766) of glo-2(zu455) embryos failed to hatch, and 30% (n = 841) of glo-3(zu446) embryos failed to hatch. Dead Glo embryos seemed to elongate but failed to hatch. Ninety-six hours after removing an adult, the plate was scored for the number of L4/adult stage progeny. Wild-type embryos always progressed to adulthood (n = 799), whereas only 62% (n = 640) of glo-1(zu437) larvae, 63% (n = 669) of glo-2(zu455) larvae, and 43% (n = 589) of glo-3(zu446) larvae progressed to adulthood. In nearly all cases, dead larvae did not progress beyond the L1 stage. Fertility was not affected by the Glo mutations: average brood size ± SEM for wild type was 202 ± 20, for glo-1(zu437) was 210 ± 11;=, for glo-2(zu455) was 192 ± 21, and for glo-3(zu446) was 210 ± 10.
The apt-6(ok429) allele is predicted to be null causing a Val 366-stop in APT-6. The glo phenotypes of apt-6(ok429) were rescued in eight of eight transgenic lines carrying cosmid ZK1043 that contained the wild-type apt-6 gene. apt-6/R11A5.1 feeding RNA interference (RNAi) by using the rrf-3(pk1426) strain resulted in embryonic and adult Glo phenotypes. The apt-7(tm920) allele is predicted to be null, encoding an in frame deletion of APT-7 from Gly 234-Leu332. The glo phenotypes of apt-7(tm920) were rescued in eight of eight transgenic lines carrying cosmid F53H8 that contained the wild-type apt-7 gene. apt-7/F52H8.1 feeding RNAi using the rrf-3(pk1426) strain resulted in embryonic and adult Glo phenotypes. The glo-4(ok623) allele is predicted to cause a Met 428-stop in GLO-4. The glo phenotypes of glo-4(ok623) were rescued in three of three transgenic lines carrying fosmid H22C01 that contained the wild-type glo-4 gene. glo-4/F07C3.4 feeding RNAi using the rrf-3(pk1426) strain resulted in an adult Glo phenotype. Genetic analysis revealed that apt-6 (ok429), apt-7(tm920), and glo-4(ok623) alleles were maternally rescued for the embryonic glo phenotype and that the adult glo phenotype was zygotically expressed.
To construct double mutants, unmarked Glo mutants were mated into genetically marked (unc-27 or dpy-11) Glo mutants to generate transheterozygous animals, which in all cases were wild type. We then isolated Glo nonUnc or Glo nonDpy adult progeny of the transheterozygotes. These were homozygous for the unmarked Glo mutation and two of three were predicted to be heterozygous for the marked Glo mutation. We isolated homozygous double mutants by picking Unc or Dpy progeny. At least two independent double mutant lines were isolated, and in all cases they showed the same phenotype.
Microscopy
All microscopic analysis was performed with a Zeiss Axioskop II plus microscope equipped with DIC, polarization, and fluorescence optics. Digital images were captured using an Insight Spot QE 4.1 camera with Spot Basic software (Diagnostic Instruments, Sterling Heights, MI). Standard polarization optics was used to detect birefringent intestinal material. Intestinal autofluorescence was detected using the Zeiss 09 (Ex:BP450-490; Em:LP515) or 15 (Ex:BP586/12; Em:LP590) fluorescence filters. To label acidic compartments with acridine orange, L4/young adults were placed in a drop of 1× M9 containing 0.1 mg/ml acridine orange and incubated in the dark for 6–20 h (Kostich et al., 2000). Scored adults were allowed to recover for 1 h on an OP50 seeded NGM plate before viewing, or they were immediately cut to release embryos for analysis. To label acidic compartments with LysoTracker Red (Molecular Probes, Eugene, OR), L4/young adults were placed onto an OP50 seeded NGM plate containing 2 μM LysoTracker Red. Animals were grown on these plates for 12–48 h without exposure to light (Hersh et al., 2002). Adults, larvae, and embryos were removed from LysoTracker Red plates and directly observed. For analysis of colocalization of LysoTracker Red and autofluorescence, the Zeiss 15 (Ex:BP586/12; Em:LP590) and (Ex: BP480/20; Em:BP530/20) filters were used. Endocytosis into the intestine was analyzed by feeding animals FM4-64 (Molecular Probes) (Griffiths et al., 2001). L4/young adults were incubated in a drop of 32 mM FM4-64 containing 0.2% dimethyl sulfoxide in the dark for 90 min. Animals were allowed to recover for 30 min on an OP50 seeded NGM plate before viewing. Fluid phase endocytosisintotheintestinewasanalyzedbyfeedinganimalstetramethylrhodamine B isothiocyanate (TRITC)-bovine serum albumin (Sigma-Aldrich) (Clokey and Jacobson, 1986). L4/young adults were incubated in a drop of 1× M9 containing 5 mg/ml TRITC-bovine serum albumin for 15 h without exposure to light. Animals scored were allowed to recover for 4 h on an OP50 seeded NGM plate before viewing. The presence of dyes that label lysosomes was scored using a Zeiss 15 fluorescent filter. To analyze receptor mediated endocytosis in glo-1(–) oocytes, glo-1(zu391) bIs1[vit-2::gfp] strains were constructed and evaluated for the localization of VIT-2/YP170::GFP as described previously (Grant and Hirsh, 1999). To determine whether glo-1(–) animals have defects in fluid phase endocytosis into coelomocytes, arIs37[pmyo3::ssGFP]; glo-1(zu437) strains were constructed and analyzed for the presence of green fluorescent protein (GFP) within coelomocytes as described previously (Fares and Greenwald, 2001b). Larvae and adults were immobilized for photography by mounting them in 1× M9 containing 10 mM levamisole. Embryos greater than 1.5-fold in length were immobilized for photography by chilling embryo-containing slides to 4°C for 2–5 min or by subjecting them to a hypoxic environment. At least 20 L4/young adult stage and at least 40 embryos were scored in each experiment reported.
For the time-course analysis of embryo elongation and localization of birefringent intestinal material, individual 1.5-fold stage embryos (420 min from first cell cleavage) were raised at 22°C and analyzed every 45 min by using DIC and polarization optics.
For immunostaining with affinity-purified rabbit GLO-1 P176–192, FUS-1 (K. Kontani and J. Rothman, University of California, Santa Barbara, Santa Barbara, CA), VHA-11 (M. Futai, Osaka University, Osaka, Japan), and mouse 3E6 GFP antisera (Qbiogene, Carlsbad, CA), embryos were fixed with methanol and stained as described previously (Leung et al., 1999). Standard C. elegans immunofluorescence protocols result in the loss of birefringent and autofluorescent gut granules.
Molecular Identification of glo-1
Standard three factor genetic crosses with visible markers were used to map glo-1(zu391) to the aex-2 unc-115 interval of LGX. Further mapping relative to DNA polymorphisms placed glo-1(zu391) in the pkP643 pkP5126 interval. Mapping data are available from Wormbase (www.wormbase.org). The mapping positioned glo-1(zu391) to a region spanned by three cosmids: R07B1, F21G4, and C17G1. Two of eight glo-1(zu391); R07B1 stable transmitting lines were GLO(+). R07B1 was digested with AgeI and religated leaving only two intact genes, R07B1.8 and R07B1.9. Ten of 17 R07B1-AgeI dropout containing glo-1(zu391) lines were GLO(+). The sequence of glo-1 mutants was obtained by PCR amplifying (Expand high fidelity; Roche Diagnostics, Indianapolis, IN) the glo-1 genomic region from glo-1 homozygous worms and analyzing the DNA sequence. Amplifications and sequencing were performed in duplicate. DNA sequencing was performed at the Fred Hutchinson Cancer Research Center Genomics Core Facility (Seattle, WA) or the Oregon Health and Science University Vollum Institute (Portland, OR). The glo-1(zu442) mutant did not contain any changes from wild-type in glo-1 exons, introns, or 1-kb 5′ or 3′ flanking regions. To obtain the glo-1 cDNA, total RNA from an adult N2 population was isolated using a TRIzol (Invitrogen, Carlsbad, CA)/chloroform extraction. cDNA was generated using a d(T)30 primed reaction with Superscript II (Invitrogen). Primers specific for the 5′ and 3′ end of the predicted glo-1 gene were used to PCR amplify the glo-1 cDNA, which was TA cloned into pCR2.1 (Invitrogen). The 5′ end of the glo-1 cDNA was amplified using 5′ RACE (Invitrogen). glo-1 cDNAs were sequenced and found to be SL1 trans-spliced and differ substantially from the Genefinder predictions for gene R07B1.8. Searches of the public sequence databases were performed at National Center for Biotechnology Information (Bethesda, MD) by using BLAST. Alignments were performed using ClustalW and displayed using BOXSHADE.
GLO-1 Antibodies
Affinity-purified antibodies recognizing GLO-1 were generated by Bethyl Laboratories (Montgomery, TX). A GLO-1–specific peptide was synthesized (176cys-VISTEQGGQYDVPFMNR192), coupled to KLH, and used in immunization. Antibodies binding the peptide were affinity purified using an agarose linked GLO-1 P176-192 peptide. The specificity of the antisera was tested using glo-1(zu437) embryos that contain a Trp106-stop mutation.
Generation of glo-1::gfp Reporters
A pVHA-6.GLO plasmid was generated by cloning the entire coding region of glo-1, PCR amplified from a full-length cDNA, into a vha-6 promoter-driven GFP vector [vha-6-GFP-GtwyB(EcoRI) derived from pPD117.01; gift of Andrew Fire, Stanford University, Stanford, CA] by using the Gateway recombination cloning system (Invitrogen). The complete plasmid sequence is available upon request. glo-1 was amplified using primers ggggacaagtttgtacaaaa aagcaggcttggcagcactcacaa (glo1_gtwyF) and ggggaccactttgtacaagaaagctggg tttagcaacatttcgagtcgt (glo1_gtwyR).
To generate the glo-1 promoter-gfp and glo-1 promoter-glo-1::gfp reporters, we used a PCR fusion technique (Hobart, 2002). A 2.1-kb sequence 5′ to the start of glo-1 was amplified and fused to a 1.7-kb gfp-containing sequence from pDP95.67 or a 2-kb gfp-glo-1–containing sequence from pVHA-6.GLO. glo-1 promoter-gfp fusions were injected into N2 at 10 ng/μl with the dominant marker pRF4 at 100 ng/μl. Cellular expression was identical in five independent transmitting lines. glo-1 promoter-glo-1::gfp fusions were injected into glo-1(zu437) animals at 0.3–1.3 ng/μl with the dominant marker pRF4 at 100 ng/μl. The subcellular expression pattern was identical in four independent transmitting lines, and each line was rescued for embryonic and adult Glo phenotypes.
RESULTS
Birefringent Gut Granules Are Lysosome-related Organelles
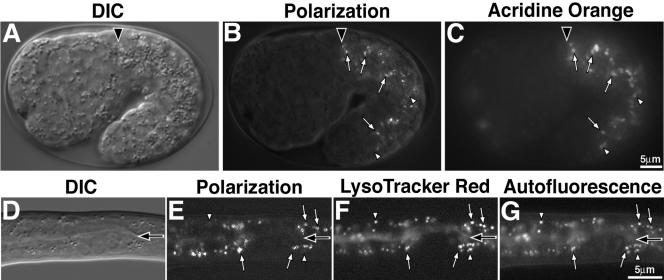
C. elegans embryos undergo rapid cell proliferation, followed by distinctive morphogenesis stages named for the shape or length of the body, such as “bean,” “1.5-fold,” and “pretzel” stages. In normal development, birefringent gut granules are only seen in intestinal cells. The granules are first observed at the bean stage and persist in intestinal cells throughout larval and adult development (Figures 1, 2, and 4). We examined embryos after staining with the lysosomal marker acridine orange, a fluorescent acidophilic dye that stains lysosomal compartments in C. elegans (Clokey and Jacobson, 1986; Kostich et al., 2000; Oka and Futai, 2000; Hersh et al., 2002). Before the bean stage, acridine orange stained structures throughout all embryonic cells. However, from the bean stage onward, the most prominent staining was in intestinal cells, and much of the staining coincided with the birefringent gut granules (Figure 1, B and C). Similarly, staining with the fluorescent lysosomal dye LysoTracker Red (Hersh et al., 2002) often marked birefringent gut granules in late-stage pretzel embryos (our unpublished data) and newly hatched larvae (Figure 1, E and F). Examples of sites stained with acridine orange or LysoTracker Red that did not show birefringence could represent different types of lysosomes or lysosomes at different stages of development. By comparing images collected with polarization optics and fluorescence microscopy, we found that the birefringent gut granules in larvae stained with LysoTracker Red were identical to autofluorescent intestinal granules (Figure 1, E–G) that have been previously shown to be acidified and terminal endocytic lysosomes (Clokey and Jacobson, 1986). Thus, intestinal cells contain birefringent, autofluorescent gut granules that stain with LysoTracker Red and acridine orange, strongly suggesting that the birefringent granules are components of lysosome-related organelles.
Figure 1.
Birefringent gut granules are located within acidic compartments. Birefringent gut granules (B) in a wild-type 1.5-fold stage embryo colocalized with acidified, acridine orange-stained (C) compartments (white arrows). Some acidic intestinal compartments stained by acridine orange did not contain detectable birefringent material (white arrowheads). In A–C, a black arrowhead marks the anterior of the intestinal primordium. A newly hatched L1-stage larvae (D–G) contained birefringent gut granules within acidified and autofluorescent intestinal organelles. Birefringent gut granules colocalized with autofluorescent and acidic LysoTracker Red-stained compartments (white arrows). Some acidic intestinal compartments stained by LysoTracker Red colocalized with autofluorescent, but not birefringent, gut granules (white arrowheads). In D–G, the intestinal lumen is marked with a black arrow.
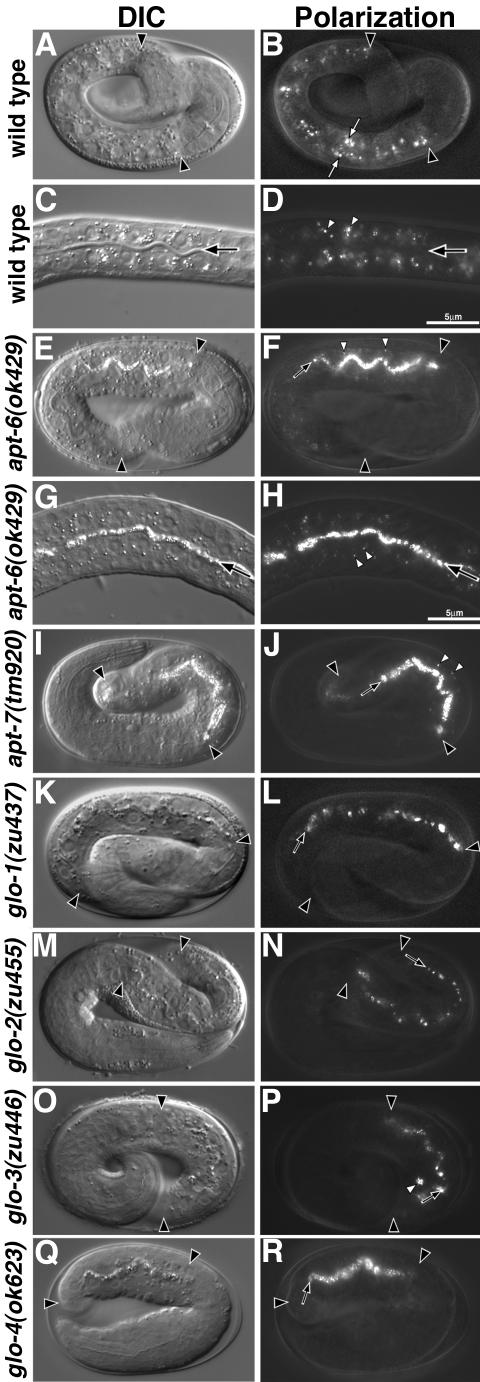
Figure 2.
Birefringent gut granule mislocalization in Glo mutants. Wild-type pretzel-stage embryos (A and B) and L1-stage larvae (C and D) contained birefringent gut granules (white arrows in B and white arrowheads in D) within intestinal cells. The intestinal lumen is marked with a black arrow in C and D. apt-6 (E and F) pretzel-stage embryos contained gut granules within the intestinal lumen (black arrow) and a reduced number of gut granules (white arrowheads) within intestinal cells. (G and H) The presence of birefringent material within the intestinal lumen (black arrow) and intestinal cells (white arrowheads) is shown at higher magnification in a newly hatched apt-6 larva. apt-7 (I and J) pretzel-stage embryos contained gut granules within the intestinal lumen (black arrow) and a reduced number of gut granules (white arrowheads) within intestinal cells. glo-1 (K and L), glo-2 (M and N), and glo-4 (Q and R) pretzel-stage embryos lacked gut granules within the intestinal cell cytoplasm and instead gut granules (black arrows) were localized in the intestinal lumen. glo-3 (O and P) pretzel stage embryos mislocalized birefringent granules to the intestinal lumen (black arrow) and typically contained between one and five birefringent gut granules (white arrowhead) within intestinal cells. In A, B, E, and F, and I-R intestinal cells are located between the black arrowheads. C. elegans embryos are ∼50 μm in length.
Figure 4.
Embryos require glo-1 for the assembly of acidified gut granules within the intestine. Intestinal cells in wild-type bean (A and B), 1.5-fold (C and D), and pretzel stage (E and F) embryos contained many acidic compartments stained by acridine orange. In wild type, the majority of gut granules were present within acidified compartments. (G–L) glo-1 intestinal cells at the same embryonic stages lacked birefringent gut granules and acidic structures stained by acridine orange. In all panels, the intestinal primordium is located between the black arrowheads. Acridine orange-stained apoptotic corpses anterior of the intestinal primordium in wild type (B and D) and glo-1 (H and J) embryos.
Biogenesis of Gut Granules Requires the Activity of Genes Implicated in Transport to Lysosomes
We next asked whether mutations in genes implicated in trafficking to lysosomes would alter the morphology or distribution of gut granules. Late Golgi-to-vacuole transport in yeast involves the ALP and CPY pathways (Bryant and Stevens, 1998). ALP trafficking requires the adaptor protein complex AP-3, a heterotetrameric complex composed of δ, β3, μ3, and σ3 chains (Boehm and Bonifacino, 2002). C. elegans apt-6 and apt-7 encode the β3 and μ3 subunits of AP-3, respectively (Boehm and Bonifacino, 2001). We found that 62% of apt-6(ok429) and 78% of apt-7(tm920) embryos had relatively few gut granules in their intestinal cells (Table 1 and Figure 2, E–J). Interestingly, these embryos accumulated birefringent material extracellularly in the intestinal lumen (Figure 2, E–J). Previous studies have shown that defects in the trafficking to yeast vacuoles and mammalian lysosomes can cause lysosomal components to be secreted (Kornfeld and Mellman, 1989; Bryant and Stevens, 1998; Mullins and Bonifacino, 2001). Birefringent material in the intestinal lumen was expelled as newly hatched apt-6(–) and apt-7(–) larvae defecated, and larvae and adult animals had few autofluorescent and acidified gut granules (Figure 3, D–I). We define this embryonic and larval phenotype as the Glo phenotype.
Table 1.
Birefringent and autofluorescent gut granules
| Genotype | % >2-fold stage embryos with Glo phenotype (n) | % L4/young adults with Glo phenotype (n) |
|---|---|---|
| Wild type | 0 (>200) | 0 (>200) |
| AP-3 subunit mutants | ||
| apt-6(ok429) | 62 (90) | 100 (50) |
| apt-7(tm920) | 78 (72) | 100 (36) |
| AP-1 / AP-2 subunit mutants | ||
| apt-2(tm935)a | 0 (84) | nd |
| dpy-23(e840) | 2 (138) | 0 (49) |
| CPY sorting pathway homologue mutantsb | ||
| vps-5/snx-1(tm847) | 0 (41) | 0 (43) |
| vps-16(ok719)c | 0 (35) | 100 (38) |
| vps-27/pqn-9(ok579)d | 0 (150) | nd |
| vps-29(tm1320) | 0 (52) | 0 (39) |
| vps-30/bec-1(ok700)e | 0 (20) | nd |
| vps-34/let-512(h797)f | 11 (122) | nd |
| vps-41(ep402)g | 0 (59) | 100 (23) |
| vps-45(tm246) at 25°Ch | 0 (21) | 0 (7) |
| vps-54(tm584) | 0 (58) | 0 (50) |
| vac-1/eea-1(tm933) | 0 (100) | 0 (38) |
| ypt-7/rab-7(ok511)i | 0 (31) | 0 (21) |
| flu mutants | ||
| flu-1(e1002) | 0 (157) | 0 (57) |
| flu-2(e1003) | 0 (161) | 0 (50) |
| flu-3(e1001) | 0 (147) | 0 (60) |
| flu-4(e1004) | 0 (124) | 0 (51) |
| Mutants with reduced aspartyl protease activity | ||
| daf-4(e1364) at 15°C | 0 (123) | 0 (42) |
| unc-52(e444) | 0 (100) | 0 (67) |
| cad-1(j1)j | 8 (60) | 0 (32) |
| Endocytosis mutants | ||
| cup-5(ar465) | 0 (40) | 0 (30) |
| cup-5(n3194)1k | 0 (104) | 0 (39) |
| mtm-6(ok330) | 0 (108) | 0 (27) |
| rme-1(b1045) | 0 (53) | 0 (47)l |
| rme-8(b1023) at 15°C | 0 (42) | 0 (32) |
| rme-8(b1023) at 25°Cm | 0 (44) | 0 (15) |
| glo mutants | ||
| glo-1(zu437) | 100 (>200) | 100 (>200) |
| glo-2(zu455) | 100 (>200) | 100 (>200) |
| glo-3(zu446) | 100 (>200) | 0 (>200) |
| glo-4(ok623) | 100 (139) | 100 (54) |
| Double mutantsn | ||
| apt-6(ok429); unc-27(e155) glo-1(zu437) | 100 (46) | 100 (61) |
| glo-4(ok623); unc-27(e155) glo-1(zu437) | 100 (109) | 100 (77) |
| dpy-11(e244) glo-4(ok623); apt-6(ok429) | 100 (98) | 100 (67) |
All strains were grown at 22°C unless otherwise noted. Embryos were analyzed using polarization microscopy and were scored as Glo when birefringentgut granules were lacking from intestinal cells or present within the intestinal lumen. L4/young adults were analyzed using fluorescence microscopy with a standard fluorescein isothiocyanate filter and were scored as Glo when <100 autofluorescent gut granules were present within the intestine. nd, not done because the mutation resulted in embryonic or larval lethality.
Embryos scored were second generation progeny of apt-2(tm935)/+ adults. Because apt-2(—) is necessary to progress beyond the 2-fold stage of embryonic development, 17% of the embryos scored (bean to 2-fold stage) were expected to be apt-2(—)/apt-2(—).
CPY pathway alleles predicted to be null: vps-5, vps-16, vps-27, vps-29, vps-30, vps-45, vps-54, vac-1, rab-7.
Scored progeny of vps-16(ok719)/hT2[qIs48(GFP)]. vps-16(—)/vps-16(—) animals were identified as lacking GFP expression.
Embryos scored were the progeny of vps-27(ok579)/nT1 adults. Nonaneuploid embryos were scored, 17% were predicted to be vps-27(—)/vps-27(—). Because vps-27(—) mutants do not develop into adults, L1-to-L3 stage progeny of vps-27(ok579)/nT1 animals were scored for the presence of autofluorescent gut granules. Ninety-four of 94 larvae were wild type, 17% were expected to be vps-27(—)/vps-27(—).
Embryos scored were the progeny of vps-30(ok700)/nT1[qIs51(GFP)] adults. vps-30(—)/vps-30(—) embryos were identified as lacking GFP expression. Because vps-30(—) mutants do not develop into adults, L2-to-L4 stage progeny of vps-30(ok700)/nT1[lsqb]qIs51(GFP)] animals lacking GFP expression were scored. Thirty-three of 33 vps-30(—)/vps-30(—) larvae had wild-type autofluorescent gut granules.
Embryos scored were the progeny of dpy-5(e61) vps-34(h797) unc-13(e450); sDp2 adults. 60% of the embryos were expected to have lost sDp2 (McKim and Rose, 1990) and be vps-34(—). All Glo embryos were 2- to 3-fold stage and lacked gut granules. dpy-5(—) and unc-13(—) embryos were wild type.
Embryos scored were the progeny of vps-41(ep402)/dpy-8(e130) unc-6(e78) adults. Twenty-five percent of the embryos were predicted to be vps-41(—)/vps-41(—). Adult vps-41(—)/vps-41(—) progeny were identified based on an increase in germline apoptosis (Costa, personal communication).
Animals scored were the progeny of vps-45(—)/vps-45(—) adults grown at 25°C.
Embryos scored were the progeny of rab-7(—)/rab-7(—) animals. L4-young adults scored were the progeny of rab-7(ok511)/mIn1[mIs14(GFP) dpy-10(e128)] animals and rab-7(—)/rab-7(—) animals were recognized as lacking GFP expression.
Glo embryos were 2-fold stage and lacked birefringent gut granules.
cup-5(n3194) animals are unc-36(e251). Embryos scored were the progeny of cup-5(—)/cup-5(—) adults and L4-adults scored were the progeny of cup-5(—) unc-36(e251)/qC1 dpy-19(e1259) glp-1(q339) animals.
95% of rme-1(b1045) adults lacked autofluorescent gut granules in the anterior most ring of intestinal cells (int-1).
rme-8(b1023) animals were grown at 25°C starting at the L1 stage. At this temperature, rme-8(—) embryos arrested and were scored at the 1.25-fold stage. Wild-type 1.25-fold stage embryos contain birefringent gut granules (Figure 4A).
The linked unc-27(e155) and dpy-11(e244) markers did not alter the Glo phenotype.
Figure 3.
Analysis of autofluorescent and acidified gut granules in young adult-stage Glo mutants. (A–C) Wild-type animals contained numerous autofluorescent and LysoTracker Red-stained acidified gut granules within intestinal cells, visualized in fluorescein isothiocyanate and rhodamine channels, respectively. Intestinal cells in young adult apt-6 (D–F), apt-7 (G–I), vps-16 (J–L), glo-1 (P–R), glo-2 (S–U), and glo-4 (Y–A′) animals contained few or no autofluorescent and acidified gut granules. vps-41 (M–O) animals contained a reduced number of gut granules that were localized in intestinal cells near the basal membrane. Reduced numbers of gut granules were present within the intestinal cells of glo-3 (V–X) animals. Posterior glo-3 intestinal cells (shown in V–X) contained on average 2 to 3 times the number of autofluorescent and acidic gut granules as anterior intestinal cells. In all panels the intestinal lumen (apical) is marked with a black arrow.
The AP-1 and AP-2 heterotetrameric complexes function in targeting to lysosomes via the Golgi and plasma membrane, respectively (Boehm and Bonifacino, 2002). C. elegans apt-2 encodes the σ1 chain of the AP-1 complex, and dpy-23 encodes the μ2 chain of the AP-2 complex (Boehm and Bonifacino, 2001). We found that neither apt-2(tm935) or dpy-23(e840) embryos nor larvae had obvious gut granule defects (Table 1).
CPY trafficking involves endosomal compartments as intermediates en route to the yeast vacuole (Bryant and Stevens, 1998; Mullins and Bonifacino, 2001; Raiborg et al., 2003; Sannerud et al., 2003). We analyzed strains carrying mutations in C. elegans genes that are homologous to CPY trafficking genes for defects in gut granule biogenesis. Mutations in the C. elegans homologues of the vac-1 and vps-45 genes that function in late Golgi-to-endosome transport, the vps5, vps29, vps30, and vps54 genes that control the recycling of transport machinery from endosomes back to the Golgi, and the vps-27 gene necessary for endosome maturation did not result in Glo phenotypes (Table 1). Mutations in the vps-34 gene (Fares and Greenwald, 2001a; Roggo et al., 2002; Xue et al., 2003) that in yeast regulates trafficking to endosomes, resulted in some 2- to 3-fold stage embryos that were Glo (Table 1). However, the Glo phenotype of vps-34(h797) was transient, because older embryos always contained gut granules (Table 1). The final step of both the CPY and ALP sorting pathways require VPS16, VPS41, and YPT7, a Rab7 homologue (Mullins and Bonifacino, 2001; Luzio et al., 2003). Mutations in the C. elegans rab-7 gene did not result in a Glo phenotype (Table 1). In contrast, all of vps-16(ok719) and vps-41(ep402) adults had Glo phenotypes with a loss or significant reduction in the number of autofluorescent and acidic gut granules (Table 1 and Figure 3, J–O); the embryos of these adults arrested in early development and lacked birefringent gut granules (our unpublished data). We conclude that homologues of some genes involved in the ALP trafficking pathway are required for proper development of gut granules in C. elegans.
We examined several other candidate genes with possible roles in lysosome biogenesis. Briefly, abnormal fluorescence (flu) mutants have abnormal gut granule autofluorescence (Babu, 1974) and are thought to function in tryptophan catabolism (Siddiqui and Babu, 1980). We found that none of the four flu mutants had Glo phenotypes as embryos or adults (Table 1). CUP-5 is a predicted Ca2+ channel localized to lysosomes that functions in the reformation of lysosomes from endo-lysosomal organelles in C. elegans coelomocytes (Fares and Greenwald, 2001b; Hersh et al., 2002; Treusch et al., 2004). cup-5 mutant embryos did not display Glo phenotypes (Table 1). Because yeast and mammalian cells defective in lysosomal trafficking often secrete lysosomal proteases (Kornfeld and Mellman, 1989; Burd et al., 1998; Conibear and Stevens, 1998), we examined daf-4(e1364), unc-52(e444), and cad-1(j1) mutants that have been reported to have significantly reduced levels of aspartyl protease activity (Jacobson et al., 1988). daf-4(–) and unc-52(–) animals did not display embryonic or adult Glo phenotypes (Table 1). In contrast, we found that 8% of cad-1(j1) embryos were Glo (Table 1). However, the Glo phenotypes all occurred earlier than the 2-fold stage, and older cad-1(–) embryos developed gut granules similar to vps-34(h797) mutants (Table 1). From our staining results, and the finding that mutations in several ALP trafficking pathway homologous are defective in the formation of gut granules, we conclude that the gut granules are components of intestine-specific, lysosome-related organelles.
Identification and Characterization of Glo Mutants
To identify additional genes involved in lysosomal biogenesis in C. elegans, we used polarization microscopy to screen mutagenized embryos for defects in birefringent gut granules. Ten mutants were identified with Glo phenotypes. Although our genetic screens should have allowed the recovery of inviable, as well as viable alleles, all of the out-crossed mutants could be maintained as homozygous strains with variable amounts of embryonic and larval lethality (13–30% embryonic and 38–57% larval inviablity; see Materials and Methods for details). Genetic tests showed that these mutants were defective in three genes we call glo-1 (six alleles), glo-2 (one allele), and glo-3 (three alleles). We obtained a candidate Glo mutant, gm125, isolated independently by Gian Garriga, and showed that it was an allele of glo-3.
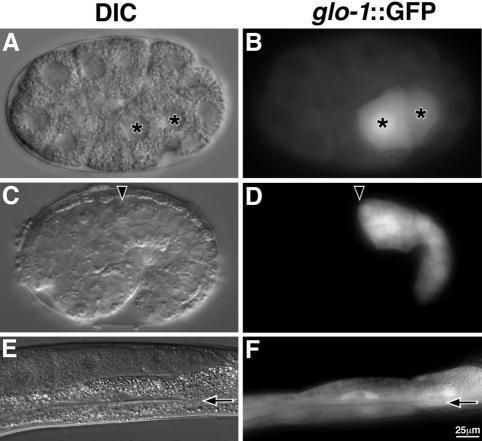
The embryonic Glo phenotype of glo-1, glo-2, and glo-3 mutants is dynamic and changes during development. At the bean stage of embryogenesis, when birefringent material first becomes detectable in wild-type intestinal cells (Figure 4A), the Glo mutants lacked, or had significantly reduced levels of, birefringent gut granules. Birefringent gut granules were not detected in the intestinal cells, or in the intestinal lumen, of glo-1 and glo-2 bean-stage mutants (Figure 4G; our unpublished data). glo-3 bean-stage embryos often contained one to five intracellular birefringent gut granules that persisted throughout embryogenesis (Figure 2, O and P). We used polarization microscopy to monitor individual glo-1(zu437), glo-2(zu455), and glo-3(zu446) embryos at 45-min intervals beginning at the 1.5-fold stage. In all three mutants, significant numbers of birefringent gut granules were not detectable until late in embryogenesis (the 4-fold or pretzel stage), and the birefringent material was located extracellularly in the intestinal lumen (Figure 2, K–P). Similar to apt-6 and apt-7 mutants described above, the birefringent material was expelled from gut lumen when newly hatched larvae defecated.
The intestines of wild-type, L4 stage animals or adults contain hundreds of autofluorescent and acidified gut granules (Clokey and Jacobson, 1986; Figure 3, A–C). Most L4 and adult glo-1 mutant animals completely lacked acidified gut granules (Figure 3, P–R, and Table 1), and glo-2 mutants had only 10–50 acidified gut granules (Figure 3, S–U, and Table 1). L4 and adult glo-3 mutants contained more that 100 acidified gut granules but significantly less than wild-type animals (Figure 3, V–X, and Table 1).
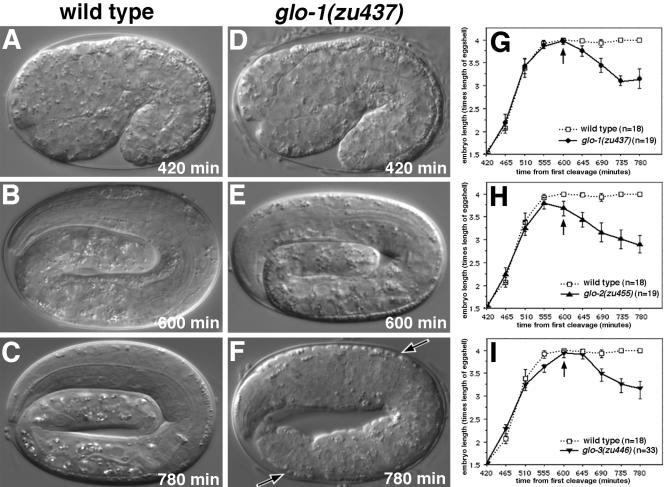
In addition to defects in the formation of birefringent and autofluorescent lysosomes, the Glo mutants have defects in embryonic body morphogenesis. Wild-type embryos divide into an ellipsoidal ball of cells and then elongate fourfold into a worm (Sulston et al., 1983; Priess and Hirsh, 1986) (Figure 5, A–C). At hatching Glo embryos were shorter and fatter than wild-type embryos (our unpublished data), suggesting that they might have a defect in either body elongation or body length maintenance. We found that all three mutants elongated from 1.5- to 4-fold at a rate indistinguishable from wild type (Figure 5, G–I). However, all three Glo mutants retracted to a 3-fold length before hatching (Figure 5, D–I); by the end of the first larval stage, the majority of Glo animals were essentially wild type in length.
Figure 5.
Elongation of wild-type and Glo mutant embryos. Images of wild-type (A–C) and glo-1 (D–F) embryos were captured at the times listed after first cell cleavage. glo-1 embryos elongated normally and reached the fourfold stage (E and G), however soon after they reduced to 3-fold in length (F and G) and displayed ripples in their epidermis/cuticle (F, black arrows). glo-2 (H) and glo-3 (I) embryos similarly elongated to fourfold before reducing to 3-fold length. (G–I) Glo embryos lacked birefringent gut granules before they occurred in the intestinal lumen (arrow denotes average time of occurrence). In G–I, the length of individual embryos and the presence/localization of birefringent material was monitored every 45 min starting at the 1.5-fold stage. Bars represent 95% confidence interval.
Formation of Endo-Lysosomal Organelles in glo-1 Mutants
In this report, we describe our phenotypic and molecular analysis of glo-1; detailed studies of glo-2 and glo-3 will be presented elsewhere. We found that glo-1(zu437) L4/adult intestinal cells showed little or no staining with the lysosomal markers LysoTracker Red (Figure 3, P–R) and acridine orange (Figure 6, O and P). Similarly, we found that intestinal cells lacked staining with the lysosomal marker acridine orange throughout embryonic development (Figure 4, G–L).
Figure 6.
glo-1 is necessary for the formation of acidified and terminal endocytic organelles in larvae and adults. In wild type, gut granules were weakly autofluorescent in the rhodamine channel (B). After staining with dyes that label acidic (D) or terminal endocytic (F and H) compartments, gut granules in wild type accumulated the dyes and displayed much stronger fluorescence. In contrast, intestinal cells in glo-1 mutants did not stain with acidic (P) or terminal the endocytic (R) organelle marker TRITC-bovine serum albumin. In 25% (n = 20) of glo-1 animals, the endocytic marker FM4-64 accumulated in organelles localized near the apical surface of intestinal cells (T). In A–H and M–T, the intestinal lumen is marked with a black arrow. glo-1 mutants did not have defects in receptor mediated endocytosis (compare J and V) of YP170::GFP into oocytes (white arrowheads mark the most proximal oocytes). Fluid phase endocytosis of GFP localized within the body cavity by coelomocytes (white circle marks cell periphery) was not disrupted in glo-1 mutants (compare L and X).
In C. elegans, V-ATPase (vacuolar H+-ATPase) activity is necessary for acidification of intestinal lysosomes (Oka and Futai, 2000). VHA-17/FUS-1 and VHA-11 are integral membrane, and peripherally associated, subunits of the V-ATPase (Oka and Futai, 2000; Sambade and Kane, 2004; Kontani et al., 2005). Both subunits are present in punctate structures within intestinal precursors of early embryos when there are only eight cells in the intestinal primordium (the E8 stage) (Figure 10K). At later stages, both subunits seem to localize to organelles similar in distribution and number to gut granules (Figure 7, A and C). In contrast, glo-1(zu437) embryos lacked detectable expression of both VHA-17/FUS-1 and VHA-11 (Figure 7, B and D). These data, together with the lack of staining by lysosomal markers (Figures 3, 4, and 6), suggest that the Glo phenotype of glo-1(–) results from a defect in gut granule biogenesis, rather than a specific defect in the birefringent components of intestinal lysosomes.
Figure 10.
Subcellular localization of GLO-1. Wild-type (A and B) and glo-1(zu437) (C and D) E8 stage embryos stained with anti-GLO-1 antibodies. (E–H) glo-1(zu437) strains carrying an extrachromosomal transgene containing the glo-1 promoter driving the expression of GLO-1::GFP in intestinal precursor cells at the E4 (E and F) and E8 (G and H) stages. glo-1(zu437); GLO-1::GFP embryos at the E8 stage (I–L) and 1.5-fold stage (M-P) after staining with anti-GFP and anti-VHA-17/FUS-1 antibodies. GLO-1::GFP and FUS-1 colocalized to the same intestinal organelles (white arrows). (A–L) Black asterisks marks intestinal nuclei. (M–P) Black arrowheads mark the anterior of the intestine. (R–T) High magnification of the intestine of a 1.5-fold, glo-1(zu437); GLO-1::GFP embryo (boxed region in Q). Most gut granules, pseudocolored red in S and T, are present within vesicles containing GLO-1::GFP (R and T) (white arrows). Some gut granules are not present within GLO-1::GFP containing organelles (white arrowhead).
Figure 7.
Analysis of endo-lysosomal organelles in glo-1 embryos. Antibodies that recognize the V-ATPase subunits VHA-17/FUS-1 (A and B) and VHA-11 (C and D) stained organelles in wild-type 1.5-fold (A) and bean-stage (C) embryos. These organelles were absent glo-1(–) embryos at similar stages (B and D). Anti-GFP antibodies used to stain embryos expressing RME-1 (E and F), RAB-5 (G and H) LMP-1 (I and J), and RME-8 (K and L) GFP protein fusions revealed a similar staining pattern in wild-type and glo-1(–) pretzel-stage embryos, with the exception that LMP-1::GFP structures were slightly enlarged in glo-1 embryos (white arrow in J). In all panels, intestinal cells are located between the black arrowheads.
Wild-type animals that were fed TRITC-bovine serum albumin or FM4-64, markers of terminal endocytic compartments, showed colocalization of these markers with gut granules (Figures 6, E–H). We found that 25% of glo-1(zu437) animals (n = 20) had FM4-64–stained organelles that were localized near the apical intestinal cell surface (Figure 6, S and T), indicating that endocytosis into intestinal cells is not blocked by glo-1(–). However, glo-1(zu437) animals did not accumulate TRITC-bovine serum albumin (Figure 6, Q and R) in intestinal organelles. The absence of staining by TRITC-bovine serum albumin suggests that endocytosis might be affected in glo-1(–) mutants.
Previous studies have provided experimental paradigms for assaying both receptor-mediated and fluid phase endocytosis in C. elegans. YP170::GFP that is synthesized in, and secreted from, the adult intestine is recognized by the RME-2 receptor and endocytosed into oocytes (Grant and Hirsh, 1999) (Figure 6, I and J). We found that glo-1(zu437) oocytes seemed to show wild-type accumulation of YP170::GFP (Figure 6, U and V). In wild-type animals, soluble GFP protein in the body cavity is endocytosed and degraded by specialized cells called coelomocytes (Fares and Greenwald, 2001a) (Figure 6, K and L). Similarly, GFP was effectively removed from the body cavity of glo-1(zu437) mutant animals (Figure 6, W and X).
We next asked whether mutations in genes with known roles in endocytosis would cause a Glo phenotype. The MTM-6 protein is enriched at the apical surface of the intestine and is necessary for an early step in coelomocyte endocytosis (Xue et al., 2003; Dang et al., 2004). We found that null mutations in mtm-6 did not cause an embryonic or adult Glo phenotype (Table 1). RME-1 is required for endocytosis by oocytes, coelomocytes, and from the basolateral intestinal cell membrane (Fares and Greenwald, 2001a; Grant et al., 2001). We found that a null mutation in rme-1 did not cause an embryonic or adult Glo phenotype (Table 1). RME-8 is essential for endocytosis across the apical intestinal membrane (Fares and Greenwald, 2001a). A temperature-sensitive allele of rme-8 did not affect the formation of gut granules at 15 or 25°C (Table 1). We conclude that the Glo phenotype does not seem to result from general defects in endocytotic processes known to function in C. elegans intestinal cells.
To examine the formation of endosomes in glo-1 mutants, we analyzed the localization of endosomal proteins in glo-1(zu437) embryos. RME-1 is localized to early/recycling endosomal membranes in intestinal cells (Grant et al., 2001). We constructed a transgene of rme-1, fused to gfp under the control of the intestinal-specific vha-6 promoter (Oka et al., 2001; Pereira-Leal and Seabra, 2001; Pujol et al., 2001). RME-1::GFP was associated with small punctate structures throughout the cytoplasm of intestinal cells in both wild-type and glo-1 mutant embryos (Figure 7, E and F). In mammalian cells, the GTPase Rab5 is localized to early endosomes (Segev, 2001; Zerial and McBride, 2001). When C. elegans RAB-5::GFP was expressed in wild-type embryos under the control of the vha-6 promoter, fluorescence was observed in punctate structures near the apical surfaces of intestinal cells (Figure 7G); an identical distribution was observed in glo-1 mutants (Figure 7H). The C. elegans lmp-1 gene encodes a homologue of the well characterized LAMP and CD68 lysosomal membrane proteins (Kostich et al., 2000). LMP-1::GFP is localized to lysosomes in coelomocytes (Treusch et al., 2004), but in intestinal cells LMP-1::GFP was localized to unidentified organelles that likely originate from endosomes (our unpublished observations). In wild-type and glo-1 mutant embryonic intestinal cells, LMP-1::GFP was associated with the basolateral membrane and with punctate organelles located near the apical surface (Figure 7I); some of the glo-1 mutant puncta seemed slightly larger than wild-type (Figure 7J). In adult intestinal cells, RME-8::GFP seems to be localized to late endosomal compartments located near the apical cell surface (Zhang et al., 2001). We found RME-8::GFP localized near the apical surface in embryonic intestinal cells (Figure 7K) and that this distribution was not altered in glo-1(zu437) mutants (Figure 7L). We conclude that although the biogenesis of gut granules is disrupted in glo-1(–) embryos, at least some aspects of the endosomal system in glo-1 mutant embryos seem to be organized properly.
glo-1 Encodes a Rab GTPase Homologous to Mammalian Rab38 and Drosophila Rab-RP1/Lightoid
We identified the glo-1 gene by using a positional cloning strategy and cosmid rescue experiments. glo-1(zu391) mapped to a 93-kb interval defined by transposon polymorphisms pkP643 and pkP5126. The Glo phenotypes of embryo and adult glo-1(zu391) mutants were fully rescued by a cosmid, R07B1, that covered part of this interval, by a subclone containing the predicted genes R07B1.8 and R07B1.9, and by an R07B1.8 cDNA expressed under the control of its own promoter. DNA sequencing showed that the glo-1 alleles kx21, kx92, zu391, zu430, and zu437 each had mutations in R07B1.8 (Figure 8A), a gene with no predicted function. However, in isolating and sequencing R07B1.8 cDNAs, we found that the predicted coding sequence for R07B1.8 was in error. The corrected sequence for R07B1.8, hereafter referred to as the glo-1 gene, is predicted to encode a 24-kDa member of the Rab family of small GTPases, hereafter referred to as GLO-1.
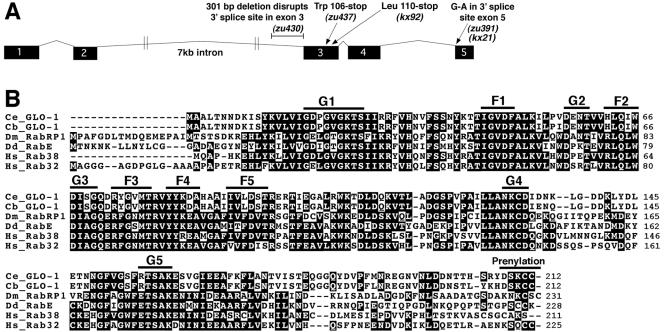
Figure 8.
Predicted structure of the glo-1 gene and encoded protein. (A) The intron-exon structure of glo-1 and location of the glo-1 mutations are shown. (B) The predicted C. elegans GLO-1 protein sequence (AAY42967) is aligned to the C. briggsae GLO-1 (CBP14321 and CBP14320), D. melanogaster RabRP1 (AB035646), Dictyostelium RabE (AF116859), human Rab38 (NP071732), and human Rab32 (Q13637) proteins. Positions of GTPase (G1-G5), Rab family (F1–F5), and prenylation motifs are shown.
GLO-1 contains the five conserved GTPase sequence motifs (G1–G5) (Figure 8B) that are important for guanine nucleotide binding and hydrolysis (Bourne et al., 1991; Valencia et al., 1991). In addition, the GLO-1 sequence displays the five well conserved Rab family motifs (RabF1–5) and a C-terminal Rab prenylation signal (Figure 8B) that distinguish the Rab family from other small GTPases (Pereira-Leal and Seabra, 2000, 2001). The zu437 allele changes the Trp106 codon to a stop codon predicted to result in production of a GLO-1 protein truncated between the G3 and G4 motifs. The truncated protein lacking the G4, G5, and prenylation motifs will not be able to bind guanine nucleotides or associate with cellular membranes. Given that both characteristics are absolutely necessary for the function of Rab GTPases (Seabra, 1998; Collins and Brennwald, 2000), glo-1(zu437) likely represents a null allele.
Our results add GLO-1 to a previous list of 29 predicted Rab family members in C. elegans (Nonet et al., 1997; Pereira-Leal and Seabra, 2001). GLO-1 is most similar to the vertebrate Rab32 and Rab38, Drosophila Rab-RP1/Lightoid, and Dictyostelium RabE proteins, showing between 45 and 47% identity with each (Figure 8B). Interestingly, the vertebrate and Drosophila proteins homologous to GLO-1 are implicated in the biogenesis of lysosome-related organelles. The mouse Rab32 and Rab38 proteins associate with melanosomes (Loftus et al., 2002; Cohen-Solal et al., 2003); Rab38 is necessary for the biogenesis of melanosomes (rats and mice) and platelet dense granules (rat) (Tschopp and Zucker, 1972; Loftus et al., 2002; Oiso et al., 2004). Drosophila Rab-RP1/Lightoid associates with, and is required for, the formation of pigment granules in retinal pigment cells (Fujikawa et al., 2002; Ma et al., 2004). Human Rab32, in contrast to mouse Rab32, can associate with cAMP-dependent protein kinase A (PKA) on mitochondria where it has been proposed to control mitochondrial fission (Alto et al., 2002). The interaction between human Rab32 and PKA requires an Ala at position 185 and replacement with a Phe abolishes this binding (Alto et al., 2002). The presence of a Phe at the corresponding position in the wild-type GLO-1 protein strongly suggests that GLO-1 cannot interact with PKA and does not control mitochondrial dynamics.
GLO-1 Is Expressed in the Intestine and Is Associated with Gut Granules
We analyzed the expression and localization of GLO-1 by using GFP reporters expressed under the control of the glo-1 promoter; the GLO-1::GFP fusion completely rescued the glo-1(zu437) embryonic and adult Glo phenotypes. In addition, we generated antibodies against GLO-1; the localization patterns described below were not observed in glo-1(zu437) mutant embryos, indicating that the affinity-purified antiserum is specific (Figure 10, C and D). glo-1::gfp expression was first detected in the daughters of the E blastomere (the E2 stage) that produces the entire intestine (Figure 9, A and B). This early expression suggests that glo-1 might be a target of the GATA transcription factor END-1 that specifies the intestinal fate (Zhu et al., 1997). Expression continued in all cells of the embryonic and adult intestines (Figure 9, C–F). After the daughters of the E blastomere divide (the E4 stage), GLO-1::GFP was enriched in punctate organelles within the intestinal precursors (Figure 10, E and F), and a similar localization was observed with the anti-GLO-1 antiserum (our unpublished data). Similar structures were present in later intestinal precursors (Figure 10, A, G, J, and N). We never observed expression of GLO-1::GFP in embryonic or adult stage hypodermal (skin) cells. We conclude that GLO-1 is localized in structures that are consistent with lysosomes and that GLO-1::GFP is an accurate reporter for GLO-1 localization in intestinal cells.
Figure 9.
Expression of glo-1 in embryos and adults. (A–F) Wild-type strains carried an extrachromosomal transgene containing the glo-1 promoter driving the expression of gfp (glo-1::gfp). gfp expression in intestinal precursors at the E2 (A and B) and 1.5-fold (C and D) stages are shown. (A and B) Nuclei of intestinal precursors are marked with asterisks. (C and D) Black arrowheads denote the anterior of the intestinal primordium. (E and F) GFP was expressed within the adult intestine (black arrow marks lumen). Bar (E and F), 25 μm.
We next compared the localizations of GLO-1::GFP and the lysosomal V-ATPase subunit VHA-17/FUS-1. From the E8 stage, when VHA-17/FUS-1 first becomes detectable in the intestine, through embryonic development, most GLO-1::GFP colocalized with VHA-17/FUS-1 (Figure 10, I–P). These observations indicate that GLO-1 is localized to V-ATPase–containing compartments during most of embryogenesis. We also determined whether GLO-1::GFP colocalized with birefringent gut granules. In 1.5-fold stage embryos, the majority of gut granules were present within GLO-1::GFP-containing organelles (Figure 10, Q–T). Together, our localization studies demonstrate that in embryonic intestinal cells GLO-1::GFP is associated with lysosome-related organelles.
Identification of GLO-4, a Putative Guanine Nucleotide Exchange Factor for GLO-1
Drosophila Rab-RP1/Lightoid protein physically interacts with Claret, a putative guanine nucleotide exchange factor (GEF) that contains seven RCC1-like repeats (Ma et al., 2004). Because GLO-1 is homologous to Rab-RP1, we searched the C. elegans genome for a Claret homologue. F07C3.4 showed the highest similarity with Claret, having 30% identity within a region predicted to contain six RCC1-like repeats. We analyzed animals that were homozygous for ok623, an allele of F07C3.4 predicted to truncate the protein before the RCC1-like repeats. ok623 embryos and L4/adults had highly penetrant Glo phenotypes (Table 1), ok623 embryos mislocalized birefringent material into the intestinal lumen (Figure 2, Q and R), and ok623 L4/adult stage animals lacked autofluorescent acidified gut granules (Figure 3, Y–A′). Because glo-2 and glo-3 mutations do not map near the F07C3.4 locus, we hereafter refer to this gene as glo-4. The Glo phenotypes of glo-1(–) and glo-4(–) animals were indistinguishable, and the phenotype of a glo-4(ok623); glo-1(zu437) double mutant was identical to each single mutant (Table 1). These genetic results and sequence homology make GLO-4 a strong candidate for a GLO-1 GEF.
glo-1 and glo-4 Are Epistatic to Mutations in the AP-3 Complex
The intestinal cells of glo-1 and glo-4 mutants lacked embryonic gut granules (Figure 2). In contrast, gut granules were always present within the intestines of apt-6 and apt-7 mutant embryos (Figure 2). We generated double mutants between these groups and analyzed the resulting Glo phenotypes. We found that glo-1 and glo-4 embryonic Glo phenotypes were epistatic to the apt-6 and apt-7 phenotypes; apt-6; glo-1 and glo-4; apt-7 embryonic intestinal cells always lacked birefringent gut granules (Table 1).
DISCUSSION
Evolutionarily Conserved Biogenesis of C. elegans Gut Granules, Drosophila Pigment Granules, and Mammalian Melanosomes
In this report, we have presented evidence that the intestine-specific gut granules of C. elegans are components of lysosome-related organelles and have described eight Glo genes that are required for the normal development of gut granules (Table 1 and Figures 2 and 3). Four of these genes (apt-6, apt-7, vps-16, and vps-41) encode proteins homologous to components of the AP-3–mediated, Golgi-to-vacuole trafficking pathway in yeast (Odorizzi et al., 1998; Mullins and Bonifacino, 2001). glo-1 encodes a predicted Rab GTPase that is expressed only in the intestinal lineage, and glo-4 encodes a possible GLO-1 guanine nucleotide exchange factor. The predicted GLO-1/Rab protein is localized to intestinal lysosome-related organelles in C. elegans (Figure 10) and is most similar to Rabs in other organisms that have been implicated in the biogenesis of specialized, lysosome-related organelles, mammalian melanosomes (Rab32 and Rab38 proteins), and Drosophila pigment granules (Rab-RP1/Lightoid) (Loftus et al., 2002; Cohen-Solal et al., 2003; Ma et al., 2004). Indeed, four other Glo genes are homologous to Drosophila eye pigmentation genes; defects in these genes cause reduced pigmentation (Huizing et al., 2002; Dell'Angelica, 2004). glo-4 is homologous to claret, apt-6 is homologous to ruby, apt-7 is homologous to carmine, and vps-41 is homologous to light. The product of the Drosophila vps-16 gene is part of complex that includes Vps18/Deep orange and Vps41/Carnation, but it has not been shown to be required for pigment granule formation (Luzio et al., 2003).
Defects in mammalian genes related to glo-1 and apt-6 are associated with HPS. HPS is a genetic disorder characterized by partial albinism and prolonged bleeding, resulting from defects in melanosome and platelet-dense granule formation, respectively (Huizing et al., 2002; Li et al., 2004). apt-6 is homologous to the HPS2 gene in humans and the Pearl locus in mice. A null mutation in the rat ruby/glo-1 gene causes a penetrant HPS phenotype (Oiso et al., 2004), and a likely reduction of function mutation in the mouse chocolate/glo-1 gene results in a partial HPS phenotype (Loftus et al., 2002).
Drosophila pigment granules and mammalian melanosomes are specialized lysosomes with cell type-specific characteristics and functions (Dell'Angelica et al., 2000b). For example, pigment granules and melanosomes contain transporters and machinery that result in the formation of pigments within these lysosome-related organelles. C. elegans lysosomal gut granules contain cell type-specific autofluorescent pigments (Clokey and Jacobson, 1986) and birefringent granules. Our findings that the glo genes glo-1, glo-4, apt-6, apt-7, and vps-41 are homologous to genes implicated in the formation of pigment granules and/or melanosomes support the view that gut granules are components of specialized, lysosome-related organelles in C. elegans.
Regulation of Body Length by the Glo Genes
Late-stage embryos and early L1-stage larvae of the Glo mutants that mislocalize birefringent gut granules into the intestinal lumen are Dpy (shorter and fatter than wild type). For glo-1, glo-2, and glo-3, we found that the Dpy phenotype results from retraction after reaching full body length (Figure 5). Body length retraction has been reported for sqt-3(e2117) and dpy-18(e364);phy-2(RNAi) embryos (Priess and Hirsh, 1986; Winter and Page, 2000). We have additionally found that dpy-17(e164) results in the same phenotype (our unpublished observations). These mutations disrupt cuticle collagens secreted by the hypodermis, or cuticle collagen-modifying proteins (Van der Keyl et al., 1994; Johnstone, 2000; Winter and Page, 2000); however, we did not detect GLO-1 expression in the hypodermis. Moreover, we found that Dpy Glo mutants typically return to a wild-type length soon after hatching (our unpublished observations), whereas we know of no evidence that Dpy phenotypes resulting from cuticle defects are reversible. Because C. elegans adults exposed to hyperosmotic stress quickly become Dpy in a process that is rapidly reversible (Lamitina et al., 2004), and yeast mutants lacking lysosomes are osmotically sensitive (Banta et al., 1988), we propose that the Dpy phenotypes of Glo mutants may arise from defects in osmotic regulation. The site of this regulation could be the intestinal cells but also might be the excretory cell that has a well characterized role in osmoregulation (Nelson and Riddle, 1984).
Misrouting of Lysosomal Components in Glo Mutants
Defects in six of the Glo genes cause a mislocalization of birefringent material into the intestinal lumen during embryogenesis. Although embryos that lack either glo-1 or glo-4 activity contain few if any birefringent gut granules in their intestinal cells, embryos defective in any of the AP-3 group of Glo genes contain moderate amounts of birefringent gut granules (Figure 2). In mammalian cells, the AP-3 pathway normally traffics membrane-associated proteins such as LAMP and tyrosinase to lysosomes and melanosomes, respectively (Le Borne et al., 1998; Dell'Angelica et al., 1999; Huizing et al., 2001). Human cells lacking AP-3 activity inappropriately reroute lysosomal proteins to the plasma membrane before their eventual delivery to lysosomes (Le Borne et al., 1998; Dell'Angelica et al., 1999, 2000a; Rous et al., 2002). By analogy, intestinal gut granules in the AP-3 pathway mutants could arise through an aberrant trafficking pathway. Gut granule precursors might be inappropriately targeted to, and secreted from, the plasma membrane. Material secreted from the apical plasma membrane into the gut lumen presumably would be lost as larvae defecate, but material secreted from the basolateral plasma membrane into the body cavity might eventually be reinternalized and incorporated into intestinal lysosomes. Indeed, we occasionally observed birefringent material accumulating in the excretory cell (our unpublished observations), which is located within the body cavity and separate from the intestine. If internalization is an obligatory step toward forming lysosome-related gut granules in the AP-3–defective larvae, it may be interesting to test whether genes involved in endocytic pathways are required for these intestinal gut granules because such genes are not required for the formation of wild-type gut granules (Table 1).
Irrespective of whether the intestinal gut granules of AP-3–defective larvae arise from an aberrant pathway, this pathway remains dependent on GLO-1 and GLO-4 functions; double mutant embryos that are defective in an AP-3 component and either glo-1 or glo-4 lack intestinal gut granules (Table 1). Genetic analyses in Drosophila have provided similar evidence that GLO-1/Rab-RP1/Lightoid and GLO-4/Claret function at a step that is distinct from the AP-3 pathway during pigment granule biogenesis (Ma et al., 2004). An attractive model is that GLO-1 and GLO-4 function in a late step in lysosomal targeting, as does the HOPS complex for both CPY and ALP pathways in yeast (Mullins and Bonifacino, 2001).
In conclusion, we have demonstrated here that the Glo phenotype identifies evolutionarily conserved genes that function during gut granule biogenesis in C. elegans. Because Glo phenotypes can be identified readily in inviable embryos (our unpublished data), it should be straightforward to saturate for Glo mutants through standard genetic screens. We propose that the gut granules of C. elegans provide a model system for understanding the formation of specialized, lysosome-related organelles, such as melanosomes and platelet granules in mammals, as well as providing a simple model for HPS in humans.
Acknowledgments
We gratefully acknowledge Ben Leung, Mike Costa, Russell Hill, and Elizabeth Kwan for identification of glo mutants. We thank Erin Currie, Nicole Kaupp, Karen Kelly, Elizabeth Kwan, and Natalie Miller for assistance with genetic characterization of the glo mutants. We thank members of the Hermann, Priess, Lycan, and Binford laboratories for advice, especially Ben Leung, Jeremy Nance, and Barbara Page. We thank Gary Reiness and Deborah Lycan for helpful comments on an earlier draft of this manuscript. We gratefully acknowledge the gift of antisera from Masamitsu Futai, Kenji Kontani, and Joel Rothman. We thank Andy Fire for the C. elegans Vector Toolkit. We thank Gian Garriga for gm125 and Mike Costa/Exelixis for vps-41(ep402). All other nematode strains were provided by the Caenorhabditis Genetics Center, the C. elegans Knockout Consortium, and the National Bioresource Project for C. elegans. Some strains were generated as part of the C. elegans Genetic Toolkit. This work was supported by grants from the National Science Foundation (MCB-0314332) and the M. J. Murdock Charitable Trust (to G.J.H.); the John S. Rogers Summer Research Program (to L.K.S. and C.A.H.); the National Institutes of Health (GM-67237) and March of Dimes (5-FY02-252) (to B.D.G.); and the Howard Hughes Medical Institute (to J.R.P.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–01–0060) on April 20, 2005.
References
- Alto, N. M., Soderling, J., and Scott, J. D. (2002). Rab32 is an A-kinase anchoring protein and participates in mitochondrial dynamics. J. Cell Biol. 158, 659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu, P. (1974). Biochemical genetics of Caenorhabditis elegans. Mol. Gen. Genet. 135, 39–44. [Google Scholar]
- Bankaitis, V. A., Johnson, L. M., and Emr, S. D. (1986). Isolation of yeast mutants defective in protein targeting to the vacuole. Proc. Natl. Acad. Sci. USA 83, 9075–9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banta, L. M., Robinson, J. S., Klionsky, D. J., and Emr, S. D. (1988). Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J. Cell Biol. 107, 1369–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blott, E. J., and Griffiths, G. M. (2002). Secretory lysosomes. Nat. Rev. Mol. Cell. Biol. 3, 122–131. [DOI] [PubMed] [Google Scholar]
- Boehm, M., and Bonifacino, J. S. (2001). Adaptins: the final recount. Mol. Biol. Cell 12, 2907–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm, M., and Bonifacino, J. S. (2002). Genetic analyses of adaptin function from yeast to mammals. Gene 286, 175–186. [DOI] [PubMed] [Google Scholar]
- Bonangelino, C. J., Chavez, E. M., and Bonifacino, J. S. (2002). Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 2486–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino, J. S., and Traub, L. M. (2003). Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72, 395–447. [DOI] [PubMed] [Google Scholar]
- Bourne, H. R., Sanders, D. A., and McCormick, F. (1991). The GTPase superfamily: conserved structure and molecular mechanism. Nature 349, 117–127. [DOI] [PubMed] [Google Scholar]
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant, N. J., and Stevens, T. H. (1998). Vacuole biogenesis in Saccharomyces cerevisiae: protein transport pathways to the yeast vacuole. Microbiol. Mol. Biol. Rev. 62, 230–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd, C. G., Babst, M., and Emr, S. D. (1998). Novel pathways, membrane coats and PI kinase regulation in yeast lysosomal trafficking. Semin. Cell Dev. Biol. 9, 527–533. [DOI] [PubMed] [Google Scholar]
- Chitwood, B. G., and Chitwood, M. B. (1974). Introduction to Nematology, Baltimore, MD: University Park Press.
- Clokey, G. V., and Jacobson, L. A. (1986). The autofluorescent “lipofuscin granules” in the intestinal cells of Caenorhabditis elegans are secondary lysosomes. Mech. Ageing Dev. 35, 79–94. [DOI] [PubMed] [Google Scholar]
- Cohen-Solal, K. A., Sood, R., Marin, Y., Crespo-Carbone, S. M., Sinsimer, D., Martino, J. J., Robbins, C., Makalowska, I., Trent, J., and Chen, S. (2003). Identification and characterization of mouse Rab32 by mRNA and protein expression analysis. Biochem. Biophys. Acta 1651, 68–75. [DOI] [PubMed] [Google Scholar]
- Collins, R. N., and Brennwald, P. (2000). Rab. In: GTPases, ed. A. Hall, New York: Oxford University Press, 137–175.
- Conibear, E., and Stevens, T. H. (1998). Multiple sorting pathways between late Golgi and the vacuole in yeast. Biochim. Biophys. Acta 1404, 211–230. [DOI] [PubMed] [Google Scholar]
- Dang, H., Li, Z., Skolnik, E. Y., and Fares, H. (2004). Disease-related myotubularins function in endocytic traffic in Caenorhabditis elegans. Mol. Biol. Cell 15, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Angelica, E. C. (2004). The building BLOC(k)s of lysosomes and related organelles. Curr. Opin. Cell Biol. 16, 458–464. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica, E. C., Aguilar, R. C., Wolins, N., Hazelwood, S., Gahl, W. A., and Bonifacino, J. S. (2000a). Molecular characterization of the protein encoded by the Hermansky-Pudlak syndrome type 1 gene. J. Biol. Chem. 275, 1300–1306. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica, E. C., Mullins, C., Caplan, S., and Bonifacino, J. S. (2000b). Lysosome-related organelles. FASEB J. 14, 1265–1278. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica, E. C., Shotelersuk, V., Aguilar, R. C., Gahl, W. A., and Bonifacino, J. S. (1999). Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the β3A subunit of the AP-3 adaptor. Mol. Cell 3, 11–21. [DOI] [PubMed] [Google Scholar]
- Fares, H., and Greenwald, I. (2001a). Genetic analysis of endocytosis in Caenorhabditis elegans: coelomocyte uptake defective mutants. Genetics 159, 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares, H., and Greenwald, I. (2001b). Regulation of endocytosis by CUP-5, the Caenorhabditis elegans mucolipin-1 homolog. Nat. Genet. 28, 64–68. [DOI] [PubMed] [Google Scholar]
- Fujikawa, K., Satoh, A. K., Kawamura, S., and Ozaki, K. (2002). Molecular and functional characterization of a unique Rab protein, RABRP1, containing the WDIAGQE sequence in a GTPase motif. Zool. Sci. 19, 981–993. [DOI] [PubMed] [Google Scholar]
- Grant, B., and Hirsh, D. (1999). Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol. Biol. Cell 10, 4311–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, B., Zhang, Y., Paupard, M.-C., Lin, S. X., Hall, D., and Hirsh, D. (2001). Evidence that RME-1, a conserved C. elegans EH domain protein, functions in endocytic recycling. Nat. Cell Biol. 3, 573–579. [DOI] [PubMed] [Google Scholar]
- Hersh, B. M., Hartwig, E., and Horvitz, H. R. (2002). The Caenorhabditis elegans mucolipin-like gene cup-5 is essential for viability and regulates lysosomes in multiple cell types. Proc. Natl. Acad. Sci. USA 99, 4355–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobart, O. (2002). PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32, 728–730. [DOI] [PubMed] [Google Scholar]
- Jacobson, L. A., Jen-Jacobson, L., Hawdon, J. M., Owens, G. P., Bolanowski, M. A., Emmons, S. W., Shah, M. V., Pollock, R. A., and Conklin, D. S. (1988). Identification of a putative structural gene for cathepsin D in Caenorhabditis elegans. Genetics 119, 355–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone, I. L. (2000). Cuticle collagen genes: expression in C. elegans. Trends Genet. 16, 21–27. [DOI] [PubMed] [Google Scholar]
- Jones, E. W. (1977). Proteinase mutants of Saccharomyces cerevisiae. Genetics 85, 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, S. M., and Reed, G. L. (2002). Development of platelet secretory granules. Semin. Cell Dev. Biol. 13, 293–302. [DOI] [PubMed] [Google Scholar]
- Kontani, K., Moskowitz, I.P.G., and Rothman, J. H. (2005). Repression of cell-cell fusion by components of the C. elegans vacuolar ATPase complex. Dev. Cell 8, 787–794. [DOI] [PubMed] [Google Scholar]
- Kornfeld, S., and Mellman, I. (1989). The biogenesis of lysosomes. Annu. Rev. Cell Biol. 5, 483–525. [DOI] [PubMed] [Google Scholar]
- Kostich, M., Fire, A., and Fambrough, D. M. (2000). Identification and molecular-genetic characterization of a LAMP/CD68-like protein from Caenorhabditis elegans. J. Cell Sci. 113, 2595–2606. [DOI] [PubMed] [Google Scholar]
- Kramer, H., and Phistry, M. (1996). Mutations in the Drosophila hook gene inhibit endocytosis of the Boss transmembrane ligands into multivesicular bodies. J. Cell Biol. 133, 1205–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamitina, S. T., Morrison, R., Moeckel, G. W., and Strange, S. (2004). Adaptation of the nematode Caenorhabditis elegans to extreme osmotic stress. Am. J. Physiol. 286, C785–C791. [DOI] [PubMed] [Google Scholar]
- Laufer, J. S., Bazzicalupo, P., and Wood, W. B. (1980). Segregation of developmental potential in early embryos of Caenorhabditis elegans. Cell 19, 569–577. [DOI] [PubMed] [Google Scholar]
- Le Borne, R., Alconada, A., Bauer, U., and Hoflack, B. (1998). The mammalian AP-3 adaptor-like complex mediates the intracellular transport of lysosomal membrane glycoproteins. J. Biol. Chem. 273, 29451–29461. [DOI] [PubMed] [Google Scholar]
- Lemmon, S. K., and Traub, L. M. (2000). Sorting in the endosomal system in yeast and animal cells. Curr. Opin. Cell Biol. 12, 457–466. [DOI] [PubMed] [Google Scholar]
- Leung, B., Hermann, G. J., and Priess, J. R. (1999). Organogenesis of the Caenorhabditis elegans intestine. Dev. Biol. 216, 114–134. [DOI] [PubMed] [Google Scholar]
- Li, W., Rusiniak, M. E., Chintala, S., Gautam, R., Novak, E. K., and Swank, R. T. (2004). Murine Hermansky-Pudlak syndrome genes: regulators of lysosome-related organelles. Bioessays 26, 616–628. [DOI] [PubMed] [Google Scholar]
- Lloyd, V., Ramaswami, M., and Kramer, H. (1998). Not just pretty eyes: Drosophila eye color mutations and lysosomal delivery. Trends Cell Biol. 8, 257–259. [DOI] [PubMed] [Google Scholar]
- Loftus, S. K., Larson, D. M., Baxter, L. L., Antonellis, A., Chen, Y., Wu, X., Jiang, Y., Bittner, M., Hammer, J. A., and Pavan, W. J. (2002). Mutation of melanosome protein RAB38 in chocolate mice. Proc. Natl. Acad. Sci. USA 99, 4471–4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone, C. J., Misner, L., Le Bot, N., Tsai, M. C., Campbell, J. M., Ahringer, J., and White, J. G. (2003). The C. elegans Hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell 115, 825–836. [DOI] [PubMed] [Google Scholar]
- Marks, M. S., and Seabra, M. C. (2001). The melanosome: membrane dynamics in black and white. Nat. Rev. Mol. Cell. Biol. 2, 738–748. [DOI] [PubMed] [Google Scholar]
- McKim, K. S., and Rose, A. M. (1990). Chromosome I duplications in Caenorhabditis elegans. Genetics 124, 115–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins, C., and Bonifacino, J. S. (2001). The molecular machinery for lysosome biogenesis. Bioessays 23, 333–343. [DOI] [PubMed] [Google Scholar]
- Nelson, F. K., and Riddle, D. L. (1984). Functional study of the Caenorhabditis elegans secretory-excretory system using laser microsurgery. J. Exp. Zool. 231, 45–56. [DOI] [PubMed] [Google Scholar]
- Nonet, M. L., Staunton, J. E., Kilgard, M. P., Fergestad, T., Hartweig, E., Horvitz, H. R., Jorgensen, E. M., and Meyer, B. J. (1997). Caenorhabditis elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J. Neurosci. 17, 8061–8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi, G., Cowles, C. R., and Emr, S. D. (1998). The AP-3 complex: a coat of many colours. Trends Cell Biol. 8, 282–288. [DOI] [PubMed] [Google Scholar]
- Oiso, N., Riddle, S. R., Serikawa, T., Kuramoto, T., and Spritz, R. A. (2004). The rat Ruby (R) locus is Rab38: identical mutations in Fawn-hooded and Tester-Moriyama rats derived from an ancestral Long Evans rat sub-strain. Mamm. Genome 15, 307–314. [DOI] [PubMed] [Google Scholar]
- Oka, T., and Futai, M. (2000). Requirement of V-ATPase for ovulation and embryogenesis in Caenorhabditis elegans. J. Biol. Chem. 275, 29556–29561. [DOI] [PubMed] [Google Scholar]
- Oka, T., Toyomura, T., Honjo, K., Wada, Y., and Futai, M. (2001). Four subunit a isoforms of Caenorhabditis elegans vacuolar H+-ATPase. J. Biol. Chem. 276, 33079–33085. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal, J. B., and Seabra, M. C. (2000). The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J. Mol. Biol. 301, 1077–1087. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal, J. B., and Seabra, M. C. (2001). Evolution of the Rab family of small GTP-binding proteins. J. Mol. Biol. 313, 889–901. [DOI] [PubMed] [Google Scholar]
- Peterson, M. R., and Emr, S. D. (2001). The class C Vps complex functions at multiple stages of the vacuolar transport pathway. Traffic 2, 476–486. [DOI] [PubMed] [Google Scholar]
- Piper, R. C., and Luzio, J. P. (2004). CUPpling calcium to lysosomal biogenesis. Trends Cell Biol. 14, 471–473. [DOI] [PubMed] [Google Scholar]
- Poupon, V., Stewart, A., Gray, S. R., Piper, R. C., and Luzio, J. P. (2003). The role of mVps18p in clustering, fusion, and intracellular localization of late endocytic organelles. Mol. Biol. Cell 14, 4015–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess, J. R., and Hirsh, D. I. (1986). Caenorhabditis elegans morphogenesis: the role of the cytoskeleton in elongation of the embryo. Dev. Biol. 117, 156–173. [DOI] [PubMed] [Google Scholar]
- Priess, J. R., Schnabel, H., and Schnabel, R. (1987). The glp-1locus and cellular interactions in early C. elegans embryos. Cell 51, 601–611. [DOI] [PubMed] [Google Scholar]
- Pujol, N., Bonnerot, C., Ewbank, J. J., Kohara, Y., and Thierry-Mieg, D. (2001). The Caenorhabditis elegans unc-32gene encodes alternative forms of a vacuolar ATPase a subunit. J. Biol. Chem. 276, 11913–11921. [DOI] [PubMed] [Google Scholar]
- Raiborg, C., Rusten, T. E., and Stenmark, H. (2003). Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 15, 446–455. [DOI] [PubMed] [Google Scholar]
- Raposo, G., and Marks, M. S. (2002). The dark side of lysosome-related organelles: specialization of the endocytic pathway for melanosome biogenesis. Traffic 3, 237–248. [DOI] [PubMed] [Google Scholar]
- Richardson, S.C.W., Winistorfer, S. C., Poupon, V., Luzio, J. P., and Piper, R. C. (2004). Mammalian late vacuole protein sorting orthologues participate in early endosomal fusion and interact with the cytoskeleton. Mol. Biol. Cell 15, 1197–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, S. E., and Emr, S. D. (1997). A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol. Biol. Cell 8, 2307–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roggo, L., Bernard, V., Kovacs, A. L., Rose, A. M., Savoy, F., Zetka, M., Wymann, M. P., and Muller, F. (2002). Membrane transport in Caenorhabditis elegans: an essential role for VPS34 at the nuclear membrane. EMBO J. 21, 1673–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman, J. H., and Stevens, T. H. (1986). Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the secretory pathway. Cell 47, 1041–1051. [DOI] [PubMed] [Google Scholar]
- Rous, B. A., Reaves, B. J., Ihrke, G., Briggs, J.A.G., Gray, S. R., Stephens, D. J., Banting, G., and Luzio, J. P. (2002). Role of adaptor complex AP-3 in targeting wild-type and mutated CD63 to lysosomes. Mol. Biol. Cell 13, 1071–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambade, M., and Kane, P. M. (2004). The yeast vacuolar proton-translocating ATPase contains a subunit homologous to the Manduca sexta and bovine e subunits that is essential for function. J. Biol. Chem. 279, 17361–17365. [DOI] [PubMed] [Google Scholar]
- Sannerud, R., Saraste, J., and Goud, B. (2003). Retrograde traffic in the biosynthetic-secretory route: pathways and machinery. Curr. Opin. Cell Biol. 15, 438–445. [DOI] [PubMed] [Google Scholar]
- Seabra, M. C. (1998). Membrane association and targeting of prenylated Ras-like GTPases. Cell Signal. 10, 167–172. [DOI] [PubMed] [Google Scholar]
- Segev, N. (2001). Ypt and Rab GTPases: insight into functions through novel interactions. Curr. Opin. Cell Biol. 13, 500–511. [DOI] [PubMed] [Google Scholar]
- Sevrioukov, E. A., He, J. P., Moghrabi, N., Sunio, A., and Kramer, H. (1999). A role for the deep orange and carnation eye color genes in lysosomal delivery in Drosophila. Mol. Cell 4, 479–486. [DOI] [PubMed] [Google Scholar]
- Siddiqui, S. S., and Babu, P. (1980). Kynurenine hydroxylase mutants of the nematode Caenorhabditis elegans. Mol. Gen. Genet. 179, 21–24. [DOI] [PubMed] [Google Scholar]
- Spritz, R. A. (1999). Multi-organellar disorders of pigmentation: intracellular traffic jams in mammals, flies and yeast. Trends Genet. 15, 337–340. [DOI] [PubMed] [Google Scholar]
- Sriram, V. S., Krishnan, K. S., and Mayor, S. (2003). Deep-orange and carnation define distinct stages in late endosomal biogenesis in Drosophila melanogaster. J. Cell Biol. 161, 593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe, J., Bossi, G., and Griffiths, G. M. (2004). Linking albinism and immunity: the secrets of secretory lysosomes. Science 305, 55–59. [DOI] [PubMed] [Google Scholar]
- Sulston, J. E., Schierenberg, E., White, J. G., and Thomson, J. N. (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64–119. [DOI] [PubMed] [Google Scholar]
- Sunio, A., Metcalf, A. B., and Kramer, H. (1999). Genetic dissection of endocytic trafficking in Drosophila using a horseradish peroxidase-Bride of Sevenless chimera: hook is required for normal maturation of multivesicular endosomes. Mol. Biol. Cell 10, 847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tappel, A. L. (1969). Lysosomal Enzymes and Other Components, Amsterdam: North-Holland Publishing Co.
- Treusch, S., Knuth, S., Slaugenhaupt, S. A., Goldin, E., Grant, B. D., and Fares, H. (2004). Caenorhabditis elegans functional orthologue of human protein hmucolipin-1 is required for lysosome biogenesis. Proc. Natl. Acad. Sci. USA 13, 4483–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp, T. B., and Zucker, M. B. (1972). Hereditary defect in platelet function in rats. Blood 40, 217–226. [PubMed] [Google Scholar]
- Valencia, A., Chardin, P., Wittinghofer, A., and Sander, C. (1991). The ras protein family: evolutionary tree and role of conserved amino acids. Biochemistry 30, 4637–4648. [DOI] [PubMed] [Google Scholar]
- Van der Keyl, H., Kim, H., Oke, C. V., and Edwards, M. K. (1994). Caenorhabditis elegans sqt-3 mutants have mutations in the col-1collagen gene. Dev. Dyn. 201, 86–94. [DOI] [PubMed] [Google Scholar]
- Walenta, J. H., Didier, A. J., and Kramer, H. (2001). The Golgi-associated Hook3 protein is a member of a novel family of microtubule-binding proteins. J. Cell Biol. 152, 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter, A. D., and Page, A. P. (2000). Prolyl 4-hydroxylase is an essential procollagen-modifying enzyme required for exoskeleton formation and the maintenance of body shape in the nematode Caenorhabditis elegans. Mol. Cell. Biol. 20, 4084–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial, M., and McBride, H. (2001). Rab proteins as membrane organizers. Nat. Rev. Mol. Cell. Biol. 2, 107–117. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Grant, B., and Hirsh, D. (2001). RME-8, a conserved J-domain protein, is required for endocytosis in Caenorhabditis elegans. Mol. Biol. Cell 12, 2011–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., Hill, R. J., Heid, P. J., Fukuyama, M., Sugimoto, A., Priess, J. R., and Rothman, J. H. (1997). end-1 encodes an apparent GATA factor that specifies the endoderm precursor in Caenorhabditis elegans embryos. Genes Dev. 11, 2883–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]