Abstract
Standard first-line therapy for patients with metastatic non-small cell lung cancer (mNSCLC) without identified actionable mutations consists of regimens comprising immune checkpoint inhibitors (ICIs), alone or in combination with platinum-based chemotherapy (CTx). However, approximately 20–30% of patients with mNSCLC (including some patients with high tumor programmed cell death ligand-1 expression) display primary resistance to ICIs, either alone or in combination with CTx. Mutations in tumor suppressor genes serine/threonine kinase 11 (STK11), and Kelch-like ECH-associated protein 1 (KEAP1) often detected in patients with Kirsten rat sarcoma virus mutations, are associated with an aggressive disease phenotype and resistance to standard ICI regimens. Consequently, there is an important need for effective treatments for patients with NSCLC with STK11 or KEAP1 mutations. In this article, we describe new data on the prevalence of STK11 and KEAP1 mutations in a large clinical population, consider practicalities around the detection of these mutations using available biomarker testing methodologies, and describe experiences of managing some of these difficult-to-treat patients in our clinical practice.
Keywords: protein serine-threonine kinases, kelch-like ECH-associated protein 1, c proto-oncogene proteins p21(ras), carcinoma, non-small-cell lung, immunotherapy
Introduction
Globally, lung cancer is the second most commonly diagnosed malignancy, but the leading cause of cancer mortality (1). Among the more than 2 million cases of lung cancer diagnosed each year (1), most are diagnosed at an advanced stage, leading to a poor prognosis for these patients (2). The most common lung cancer subtype, non-small cell lung cancer (NSCLC) accounts for 85% of cases (3), but can be further divided by histological subtype, with adenocarcinoma being the most common (4, 5). NSCLC is driven by an array of genomic events and molecular mechanisms which lead to disparities in outcomes (5, 6). As our understanding of the molecular pathology of NSCLC has advanced, so has our ability to distinguish between specific molecular subtypes, to reach more accurate judgments regarding the prognoses of the patients in our care, and to tailor treatment regimens that target the specific etiology of each individual’s disease.
Advances in molecular subtyping have facilitated the development of personalized treatments for NSCLC (7), and organizations such as the National Comprehensive Cancer Network® (NCCN®), the American Society of Clinical Oncology (ASCO), and the European Society for Medical Oncology (ESMO) now advocate the use of biomarker testing at the time of diagnosis of advanced/metastatic NSCLC, to ensure that patients with actionable molecular alterations are treated with the appropriate targeted therapies (8–10). For those without actionable mutations, the addition of an anti-programmed cell death ligand-1 (PD-L1)/programmed cell death-1 (PD-1) agent to chemotherapy (CTx) provides a survival benefit compared with chemotherapy alone (11–18). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) recommend the use of chemoimmunotherapy (CIT) regimens as options in this broad subset of patients, irrespective of PD-(L)1 expression status (9). However, in some patients with mutations in certain tumor suppressor genes, including the serine/threonine kinase 11(STK11) and Kelch-like ECH-associated protein (KEAP1) genes, the benefit of CIT appears to be less clear (19–24). Understanding the molecular and cellular basis of resistance to PD-(L)1 inhibition may allow us to better refine treatment strategies for patients with STK11 and KEAP1 mutation-positive NSCLC. In this article we consider the latest information and provide our perspectives on the treatment of patients with NSCLC tumors harboring STK11 and KEAP1 mutations.
Therapeutic options for patients with advanced NSCLC and KRAS mutations
The Kirsten rat sarcoma virus (KRAS) mutation is the most frequently occurring genomic abnormality in NSCLC, being present in up to 30% of tumors (19). The KRAS inhibitors sotorasib and adagrasib were approved by the US Food and Drug Administration (FDA) for second-line treatment of KRAS G12C-mutated NSCLC (25, 26) after demonstrating acceptable objective response rates in these patients after failure of standard therapies, including CIT (27, 28). More information is required on the effectiveness of KRAS inhibitors for first-line treatment of metastatic (m)NSCLC; the results of several ongoing trials of KRAS inhibitors in combination with PD-(L)1 inhibitors are awaited.
In the absence of data on first-line therapy for tumors with KRAS mutations (of any type), the NCCN Guidelines® continue to recommend the use of CIT as a treatment option (9). Although immunotherapy (IO), with or without CTx, has demonstrated efficacy in patients with a KRAS mutation (24, 29), there are notable variabilities in reported outcomes among these patients, some of which may be explained by genetic heterogeneity. Firstly, while KRAS mutations arise most commonly in the G12 codon (30, 31), increasing evidence suggests that there are important differences between the three most common G12 mutations: G12C (40% of KRAS mutations in lung), G12V (22%), and G12D (16%) (32). G12C and G12V mutations both result from a smoking-induced point mutation, whereas G12D mostly occurs in low/never-smokers (33, 34). G12C and G12V appear to be induced by chronic exposure to tobacco carcinogens, which also increases tumor mutational burden (TMB) and PD-1/PD-L1 expression, both of which are predictors of good response to immunotherapy (35–39). In contrast, neither PD-L1 expression nor TMB are enriched in tumors with KRAS G12D mutations. Rather, such tumors have an immunologically ‘cold’ immune microenvironment, with relatively low expression of tumor neoantigens and little T-cell infiltration (39). Consequently, compared with the favorable outcomes observed in patients with tumors bearing KRAS G12C or G12V mutations, those with G12D mutations have worse outcomes on any line of anti-PD-(L)1 therapy (33, 40).
Other forms of genomic diversity exist among patients with NSCLC tumors harboring KRAS mutations. As described in the following sections, recent advances in our understanding of the influence of this genomic diversity are beginning to inform our clinical practice.
The influence of STK11 and KEAP1 mutations on outcomes of advanced NSCLC
Around half of patients with identified KRAS mutations have been found to harbor additional cancer-associated mutations, most frequently in tumor suppressor genes, specifically STK11 and KEAP1 (41–43). These genes normally act to inhibit cell proliferation and tumor development, so their loss or inactivation removes this inhibition of cell proliferation and contributes to abnormal proliferation of the tumor cells (42–44). Tumors bearing such mutations tend to have an aggressive disease biology, due to alterations in metabolic pathways that contribute to an immunosuppressive tumor environment (24, 45).
As mentioned, the results of several analyses suggest that patients with KRAS mutations benefit from standard-of-care CIT (24, 29, 46); however, the presence of concurrent STK11 and KEAP1 may lead to relatively poor outcomes (19, 20, 22, 47–49). In some real-world studies, overall survival (OS) outcomes in patients with advanced NSCLC treated with CTx, IO, or CIT were found to be worse among patients with KRAS-mutated NSCLC with co-mutations in STK11 or KEAP1, compared with tumors with KRAS mutations alone ( Table 1A ).
Table 1.
Impact of STK11, KEAP1, and KRAS mutations or co-mutations on survival outcomes with standard therapies (A, B), and on survival outcomes with IO + CTx combinations vs. CTx alone (C).
| A. Patients with KRAS mutations: HRs for PFS and OS between subgroups with/without STK11 and/or KEAP1 co-mutations | |||
|---|---|---|---|
| Analysis | Therapy type | PFS HR (95% CI)a |
OS HR (95% CI)a |
| HRs for KRASm + STK11m vs. KRASm + STK11wt | |||
| Julianb ( 49) | IO ± CTx, or CTx (1L) | – | 1.00 (0.67, 1.51) P=0.99 |
| Julianb ( 49) | IO ± CTx, or CTx (2L) | – | 1.02 (0.56, 1.84) P=0.96 |
| Peters ( 48) | IO ± CTx, or IO + CTx + VEGFi |
– | 1.63 (1.12, 2.39) P=0.0136 |
| HRs for KRAS G12Cm + KEAP1m vs. KRAS G12Cm + KEAP1wt | |||
| Julian ( 49) | IO ± CTx, or CTx (1L) | – | 1.57 (0.95, 2.60) P=0.08 |
| Julian ( 49) | IO ± CTx, or CTx (2L) | – | 1.63 (0.77, 3.45) P=0.20 |
| HRs for KRAS G12Cm + STK11m + KEAP1m vs. KRAS G12Cm + STK11wt + KEAP1wt | |||
| Julian ( 49) | IO ± CTx, or CTx (1L) | – | 1.93 (1.35, 2.75) P<0.001 |
| Julian ( 49) | IO ± CTx, or CTx (2L) | – | 2.20 (1.27, 3.81) P=0.005 |
| B. HRs for PFS and OS between subgroups with/without STK11, KEAP1 and STK11 + KEAP1 mutations | |||
|---|---|---|---|
| Analysis | Therapy type | PFS HR (95% CI)c |
OS HR (95% CI)c |
| HRs for STK11m vs. STK11wt | |||
| Papillon-Cavanagh ( 22) | CTx | 1.32 (1.04, 1.68) P=0.01–0.05 | 1.19 (0.89, 1.6)d |
| Papillon-Cavanagh ( 22) | Anti PD-(L)1 | 1.33 (0.93, 1.9)d | 1.43 (0.91, 2.26)d |
| Cordeiro de Lima ( 51) | Anti PD-(L)1 | 1.31 (1.12, 1.88) P=0.02 | 1.33 (1.13, 2.21) P=0.001 |
| Julian ( 49) | IO ± CTx, or CTx (1L) | - | 1.27 (1.00, 1.62) P=0.05 |
| Julian ( 49) | IO ± CTx, or CTx (2L) | – | 1.32 (0.95, 1.83) P=0.10 |
| Peters ( 48) | IO ± CTx, or IO + CTx + VEGFie |
– | 1.46 (1.12, 1.92) P=0.0070 |
| HRs for KEAP1m vs. KEAP1wt | |||
| Papillon-Cavanagh ( 22) | CTx | 1.53 (1.22, 1.93) P ≤ 0.001 | 1.49 (1.14, 1.95) P=0.001–0.01 |
| Papillon-Cavanagh ( 22) | Anti PD-(L)1 | 1.71 (1.2, 2.45) P=0.001–0.01 | 1.71 (1.04, 2.81) P=0.01–0.05 |
| Cordeiro de Lima ( 51) | Anti PD-(L)1 | 1.27 (0.97, 1.29) P=0.32 | 1.19 (1.18, 1.31) P=0.04 |
| Julian ( 49) | IO ± CTx, or CTx (1L) | – | 1.21 (1.00, 1.48) P=0.06 |
| Julian ( 49) | IO ± CTx, or CTx (2L) | – | 1.20 (0.93, 1.55) P=0.17 |
| Peters ( 48) | IO ± CTx, or IO + CTx + VEGFie |
– | 1.26 (0.95, 1.68) P=0.1172 |
| HRs for STK11m + KEAP1m vs. STK11wt + KEAP1wt | |||
| Julian ( 49) | IO ± CTx, or CTx (1L) | – | 1.81 (1.44, 2.26) P<0.001 |
| Julian ( 49) | IO ± CTx, or CTx (2L) | – | 1.83 (1.35, 2.49) P<0.001 |
| C. HRs for PFS and OS between investigational therapy and CTx (for subgroups with/without KRAS, STK11 and KEAP1 mutations) | |||||
|---|---|---|---|---|---|
| Analysis | Comparison | PFS HR (95% CI)f |
OS HR (95% CI)f |
||
| KRAS subgroups | KRASm | KRASwt | KRASm | KRASwt | |
| POSEIDONg | Anti-PD-L1 vs. CTx | 0.82 (0.53, 1.29) (57) | NR | 0.74 (0.50, 1.09) (70) | 0.87 (0.68, 1.12) (70) |
| CheckMate 227 ( 54) | Anti-PD-1 + Anti-CTLA-4 vs. CTx | – | 0.79 (0.55, 1.12) | 0.73 (0.56, 0.95) | |
| CheckMate 9LA ( 55) | Anti-PD-1 + Anti-CTLA-4 + CTx vs. CTx | 0.74 (0.50, 1.10) | 0.73 (0.52, 1.02) | 0.72 (0.48, 1.08) | 1.0 (0.72, 1.39) |
| POSEIDONg | Anti-PD-1 + Anti-CTLA-4 + CTx vs. CTx | 0.57 (0.35, 0.92) (57) | NR | 0.55 (0.36, 0.83) (70) | 0.78 (0.61, 1.00) (70) |
| STK11 subgroups | STK11m | STK11wt | STK11m | STK11wt | |
| POSEIDONg | Anti-PD-L1 vs. CTx | 1.02 (0.55, 1.93) (57) | NR | 1.02 (0.59, 1.80) (70) | 0.79 (0.63, 1.00) (70) |
| CheckMate 227 ( 54) | Anti-PD-1 + Anti-CTLA-4 vs. CTx | – | – | 0.78 (0.48, 1.27) | 0.75 (0.59, 0.94) |
| CheckMate 9LA ( 55) | Anti-PD-1 + Anti-CTLA-4 + CTx vs. CTx | 0.61 (0.37, 1.00) | 0.77 (0.57, 1.04) | 0.79 (0.48, 1.28) | 0.90 (0.67, 1.22) |
| POSEIDONg | Anti-PD-1 + Anti-CTLA-4 + CTx vs. CTx | 0.47 (0.23, 0.93) | NR | 0.57 (0.32, 1.04) (70) | 0.71 (0.56, 0.90) (70) |
| KEAP1 subgroups | KEAP1m | KEAP1wt | KEAP1m | KEAP1wt | |
| POSEIDONg | Anti-PD-L1 vs. CTx | 1.51 (0.55, 5.25) (57) | NR | 0.77 (0.31, 2.15) (70) | 0.83 (0.70, 0.98) (70) |
| CheckMate 227 ( 54) | Anti-PD-1 + Anti-CTLA-4 vs. CTx | – | – | 0.31 (0.14, 0.70) | 0.80 (0.65, 1.00) |
| CheckMate 9LA ( 55) | Anti-PD-1 + Anti-CTLA-4 + CTx vs. CTx | 0.34 (0.14, 0.83) | 0.79 (0.60,1.03) | 0.51 (0.24, 1.08) | 0.94 (0.71, 1.23) |
| POSEIDONg | Anti-PD-1 + Anti-CTLA-4 + CTx vs. CTx | 0.94 (0.33, 3.35) (57) | NR | 0.43 (0.16, 1.25) (70) | 0.76 (0.64, 0.90) (70) |
aHR >1.0 indicates that the comparison favors the subgroup without STK11/KEAP1 co-mutations; b KRAS G12Cm only; cHR >1.0 indicates that the comparison favors the subgroup without STK11/KEAP1 mutations; dP value not available; ePatients received pembrolizumab (n=94), pembrolizumab + chemotherapy, NSQ (n=462), pembrolizumab + chemotherapy, SQ (n=122), or atezolizumab + bevacizumab + chemotherapy (n=4); fHR <1.0 indicates that the comparison favors the investigational therapy; gAnalysis performed in patients with NSQ histology only.
1L, first-line; 2L, second-line; CI, confidence interval; CTLA-4, cytotoxic T−lymphocyte-associated antigen 4; CTx, chemotherapy; HR, hazard ratio; IO, immunotherapy; KEAP1(m/wt), Kelch-like ECH-associated protein 1 (mutation-positive/wild-type); KRAS(m/wt), Kirsten rat sarcoma virus (mutation-positive/wild-type); NR, not reached; NSQ, non-squamous; OS, overall survival; PD-(L)1; programmed cell death (ligand)-1; PFS, progression-free survival; SQ, squamous; STK11(m/wt), serine/threonine kinase 11 (mutation-positive/wild-type); VEGF, vascular endothelial growth factor.
The presence of STK11 and/or KEAP1 mutations (without concurrent KRAS mutations) also appears to predict for poor therapeutic outcomes in patients with advanced NSCLC. In some studies, OS and progression-free survival (PFS) were worse among patients with STK11 and/or KEAP1 mutations treated with CTx, IO, or CIT, compared with patients with STK11 or KEAP1 wild-type tumors ( Table 1B ). KEAP1 mutations have also been shown to confer resistance to radiotherapy (50). Although it is currently recommended that patients with STK11 or KEAP mutation-positive NSCLC should receive standard-of-care CIT (8–10), the introduction of IO has not improved the outlook for these patients (22, 51), who continue to have relatively poor outcomes, irrespective of the treatment regimen given ( Table 1B ). There is no firm consensus on whether the presence of these mutations is negatively prognostic or predicts poor survival outcomes with specific therapies.
There is a clear unmet need for effective treatments for patients with STK11 and KEAP1 mutations (with or without concurrent KRAS mutations). Select treatment regimens comprising PD-(L)1 inhibitors in combination with CTx, and specific combinations incorporating anti-cytotoxic T−lymphocyte-associated antigen 4 (CTLA-4) agents are NCCN-recommended and FDA-approved first-line treatment options for patients with mNSCLC with performance status (PS) 0–1 and without driver mutations (9, 52, 53). In three Phase 3 studies (CheckMate 227 [NCT02477826], POSEIDON [NCT03164616] and CheckMate 9LA [NCT03215706]), conducted in treatment-naïve patients with EGFR and ALK wild-type mNSCLC, dual PD-(L)1 and CTLA-4 inhibition with CTx led to improvements in OS and PFS compared with CTx alone (54–56). Exploratory analyses suggest that such regimens may also be beneficial for patient subgroups with historically poor outcomes (specifically, patients with STK11 and/or KEAP1 mutations). In each of these studies, patients treated with these regimens achieved improvements in OS compared to those treated with CTx, in subsets both with and without STK11, KEAP1, or KRAS mutations ( Table 1C ) (54, 55, 57). However, considering the small numbers in these subsets, prospective analyses based on larger sample sizes are required to adequately evaluate the effects of these regimens in these difficult-to-treat patients, and to fully establish the value of STK11, KEAP1, and KRAS mutations as biomarkers to inform routine clinical practice.
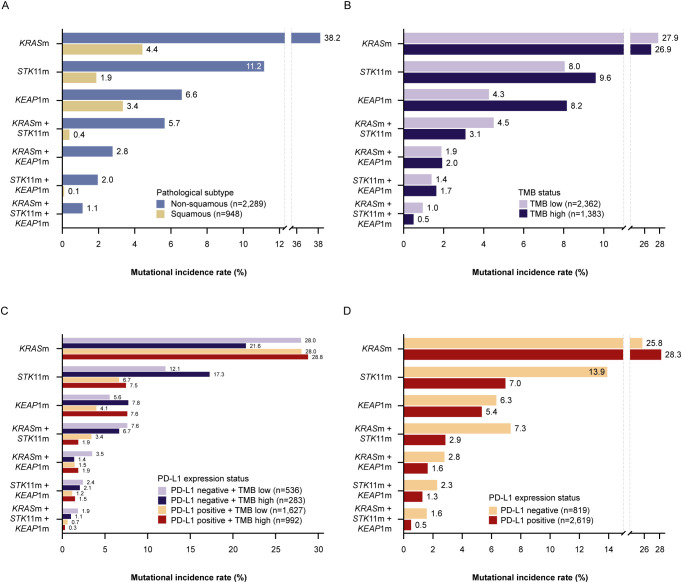
Prevalence of STK11 and KEAP1 mutations in NSCLC: pathological subtyping of NSCLC in a large clinical population
At Baylor University Medical Center, Dallas, Texas patients with NSCLC undergo routine pathological subtyping using the OncoKB platform. Subsequently, biomarker testing is conducted at PathGroup [RA] using the Endeavor test, a broad panel designed to detect variants in more than 500 cancer genes, with full exon coverage, as well as TMB.
The authors [MS and RA] analyzed tumor samples collected from 3,745 patients with NSCLC of any stage, including patients with actionable oncogenic alterations. Patients with mixed histology and unknown PD-L1 status were excluded from relevant subgroup analyses. In addition to routine biomarker testing using the Endeavor test, the PGDx elio™ tissue complete assay was used to evaluate TMB; the results were cross-validated with FoundationOne® CDx and MSK-IMPACT® assays. TMB-high status was defined as ≥16.0 mutations/megabase (mut/Mb), TMB-low status was defined as <16.0 mut/Mb. PD-L1 testing was performed using the PD-L1 IHC 22C3 pharmDx assay; a tumor proportion score (TPS) ≥1% was deemed to be positive and TPS <1% negative (indeterminate results were excluded from the analysis).
The current analysis revealed that KRAS mutations were present in 27.5% of NSCLC tumors, similar to previous reports that indicate the overall prevalence to be ~30% (19). In this analysis, we report the prevalence of pathogenetic variants only. Without this curation, our dataset could include ‘passenger’ mutations that would unknowingly be interpreted as false positives for oncogenicity. Other studies may not have curated their prevalence data in the same way. Accordingly, in the current analysis, STK11 mutations and KEAP1 mutations were identified relatively infrequently (in 8.6% and 5.7% of patients, respectively), compared with the previously reported prevalence ranges of 18–25% for STK11 (24, 58, 59) and 10–15% for KEAP1 (24, 59). KRAS was co-mutated with STK11 in 4.0% of tumors, and with KEAP1 in 1.9% of tumors. Co-mutations in KEAP1 and STK11 occurred in 1.5%, while 0.8% harbored triple mutations in KRAS, STK11, and KEAP1.
Relationships between mutation frequencies and tumor histology, TMB, and PD-L1 status
All the mutations and co-mutations evaluated (in STK11, KEAP1, and KRAS) were markedly more frequent in tumors with non-squamous (n=2,289) than squamous histology (n=948) ( Figure 1A ). Moreover, no squamous cell tumors harbored KRAS and KEAP1 co-mutations or triple mutations (in KRAS, STK11 and KEAP1). The marked differences between patients with non-squamous and squamous histology were also apparent in subgroups defined both by histology and either TMB ( Supplementary Figure S1A ) or PD-L1 expression status ( Supplementary Figure S1B ).
Figure 1.
Incidence of KRAS, STK11, and KEAP1 (co-)mutations by pathological subtype (A), TMBa status (B), TMBa and PD-L1 expressionb status (C), and PD-L1 expressionb status (D). aThe PGDx elio™ tissue complete assay was used to determine TMB high (≥16.0 mut/Mb) vs. TMB low (<16.0 mut/Mb) status; bThe PD-L1 IHC 22C3 pharmDx assay was used to determine PD-L1 positive (TPS ≥1%) vs. PD-L1 negative (TPS <1%) status. KEAP1(m), Kelch-like ECH-associated protein 1 (mutation-positive); KRAS(m), Kirsten rat sarcoma virus (mutation-positive); PD-L1, programmed cell death ligand-1; STK11(m), serine/threonine kinase 11 (mutation-positive); TMB, tumor mutational burden; TPS, tumor proportion score.
The curation of prevalence data to identify only pathogenic variants of KRAS, STK11, and KEAP1 is especially pertinent to comparisons between the TMB-high and -low subsets, as non-oncogenic passenger mutations are more likely to occur in the TMB-high subset than the TMB-low subset. By reporting pathogenic variants specifically, our dataset is more likely to represent the clinically relevant phenotype. In our analysis, KEAP1 mutations were identified in almost twice the proportion of patients with TMB-high than TMB-low tumors ( Figure 1B ). This difference between patients with TMB-high and TMB-low status was also apparent in the subgroups with either PD-L1-positive or PD-L1 negative tumors ( Figure 1C ), and in the non-squamous but not the squamous subgroup ( Supplementary Figure S1A ).
The rate of STK11 mutations (occurring alone and concurrently with KRAS mutations) was substantially higher in the subset with PD-L1 negative tumors, almost twice the rate in the subset with PD-L1 positive tumors ( Figure 1D ). This difference between patients with PD-L1 negative and PD-L1 positive tumors was also apparent in the subgroups with either TMB-high or TMB-low status ( Figure 1C ) and in the non-squamous but not the squamous subgroup ( Supplementary Figure S1B ).
Perspectives on the use of STK11 and KEAP1 biomarker testing in current clinical practice
Currently, biomarker testing is routinely used at diagnosis of NSCLC, to determine the patient’s eligibility for one of the currently available targeted therapies. When feasible, testing for additional markers (other than actionable abnormalities) could provide more detailed and specific insights into the molecular pathology that drives the disease in individual patients. We believe that integrating STK11 and KEAP1 biomarker testing into routine practice can be valuable for clinicians making treatment decisions for patients with mNSCLC. Understanding the potential impact of these biomarkers on prognosis provides some foresight into how the patient’s disease would be likely to develop during planned treatment. Thus, broader genomic testing may lead to improvements in selection and sequencing of treatment.
At the Sarah Cannon Research Institute, Nashville, a 65-year-old male presented to MJ with mNSCLC and several metastatic brain lesions. In line with our usual biomarker testing practice, next-generation sequencing (NGS) and immunohistochemistry testing for PD-L1 in tissue specimens were performed. The patient’s tumor had an elevated PD-L1 expression of 75%, together with mutations in both STK11 and KRAS G12D. TMB reported as part of NGS was 12 mut/Mb. The patient was treated with whole-brain radiation and an initial course of steroids, followed by pembrolizumab 200 mg every 3 weeks. Although his PS was robust at diagnosis, by the time the course of radiotherapy had been completed, he began to show signs of disease progression and systemic decline. We expected his condition to improve once IO was started, but unfortunately, he continued to weaken, until he became too frail to receive additional therapy. Nowadays, we would still use radiotherapy to treat this type of patient, as this can achieve clinically valuable improvements when added to CTx or IO. However, knowing the negative prognostic impact of STK11 and KEAP1 mutations, we would also consider combining radiotherapy with concurrent or sequential CIT.
Obtaining information on STK11 and KEAP1 mutation status ‘up-front’ may further inform the choice of therapeutic regimen offered to our patients. Given that the incorporation of an additional IO agent can impose a greater financial burden on patients than PD-(L)1 inhibition alone, and that many oncologists are cautious of immune-mediated adverse events (IMAEs) that may arise with dual ICIs, being able to select patients who are characterized by good response may help us to refine our treatment strategy.
At UC San Diego (UCSD) Health, a patient with a history of moderate smoking presented to SPP with non-squamous mNSCLC. Plasma was sent to a vendor for cell-free DNA (cfDNA) NGS testing, given minimal tissue availability and the need for rapid diagnosis. The patient was found to have a KRAS G12V mutation with co-mutations in STK11 and KEAP1. The PD-L1 expression score was 0 and TMB was approximately 9 mut/Mb. As is typical for patients with these disease characteristics treated at UCSD Health, the patient began combination treatment with CTx, a PD-1 inhibitor, and an anti-CTLA-4 agent. The patient tolerated treatment well and remains in remission over a year into their therapy.
In our experience, IMAEs in patients receiving an additional CTLA-4 inhibitor with CIT can be managed by the tumor board, and this can prolong the benefits achieved with IO. In this case, the patient had an IMAE (Grade 2 immune colitis with no blood). This was managed by a tumor board which included an immunologist who recommended treatment with steroids. The patient responded rapidly to prednisone (1 mg/kg oral starting dose), and the dose was tapered over four weeks. When immune colitis recurred, treatment with vedolizumab (a selective biologic drug that decreases gut inflammation, with only limited system-wide immunosuppression) was initiated. CIT was continued successfully without the CTLA-4 agent for two years.
Further considerations on biomarker testing
Comprehensive genomic profiling (CGP) is recommended by ASCO, ESMO, and NCCN Guidelines, and is specified in the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology guidelines (8, 9, 60, 61). Guidelines suggest broad biomarker testing may be used to support access to new treatments or clinical trials (8). Most often, CGP is carried out using an NGS-based platform; many commercially-available assays ( Supplementary Table S1 ) include STK11 and KEAP1 mutations in their gene panel. Although tissue sampling remains the gold-standard for diagnostic testing of mNSCLC, in our experience, plasma (‘liquid biopsies’) may also be suitable for analysis of STK11 and KEAP1 mutations, particularly for patients with advanced disease, where the rate of detection tends to be higher. In the case of tissue testing, the need to screen for an increasingly broad range of markers will require a sufficient quantity of high-quality specimens that may not be available to clinicians in some clinical settings (62). Furthermore, as patients with STK11 and KEAP1 mutations often show rapid clinical deterioration, there is a limited therapeutic window. Consequently, the relatively long turnaround time required for tissue biopsy and testing may prove to be unfeasible in some circumstances.
Conclusions
We believe that all patients with NSCLC should receive NGS-based CGP at diagnosis, using tumor tissue or cfDNA (particularly when tissue is limited or unavailable). As shown in our analysis, the distinct patterns of mutational prevalence between PD-L1 and TMB subgroups only further highlight the complex relationship between these two biomarkers. In light of recent guidelines that TMB should not be used as a sole indicator for ICIs (63), there is a need to identify new confounding factors that may influence treatment outcomes. This could include improving TMB scoring through more robust filtering of ancestral bias, assessment of the mutational status of KEAP1 and STK11, and integrating PD-L1 positivity into a comprehensive score predictive of response. Emerging evidence on the prevalence of these mutations in specific racial/ethnic groups may also help to shed light on the heterogeneous nature of such tumors, and may allow us to tailor our clinical practice accordingly (51).
Given their poor prognosis when receiving standard CIT, patients with STK11 and KEAP1 mutations should be offered clinical trials with novel agents that specifically target on these genomic mechanisms. Increasing the ease of NGS testing for all patients with NSCLC, including in the first-line setting, will be vital for identifying these patients before they initiate less effective treatments. Future prospective studies will have a critical role in evaluating STK11 and KEAP1 mutations as predictors of resistance to anti-PD-1 (only)-directed strategies, and in determining the efficacy of combinatorial strategies (including combinations with anti-CTLA-4). Other emerging biomarkers of potential interest include TP53 and SMARCA4, both being associated with aggressive disease biology (64–67), and LRP1B, which is potentially predictive of favorable outcomes with IO (68, 69).
Acknowledgments
Support for third-party writing assistance under the direction of the authors for this manuscript was provided by Lara Higham, BSc, of Ashfield MedComms and was funded by AstraZeneca.
Funding Statement
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Data availability statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.
Ethics statement
PathGroup (Nashville, TN, United States) did not require the study to be reviewed or approved by an ethics committee because the samples analyzed were a by-product of routine care.
Author contributions
MS: Writing – original draft, Writing – review & editing. MJ: Writing – original draft, Writing – review & editing. RA: Writing – original draft, Writing – review & editing. SP: Writing – original draft, Writing – review & editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2024.1459737/full#supplementary-material
References
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. (2018) 68:7–30. doi: 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 3. Sher T, Dy GK, Adjei AA. Small cell lung cancer. Mayo Clin Proc. (2008) 83:355–67. doi: 10.4065/83.3.355 [DOI] [PubMed] [Google Scholar]
- 4. Zappa C, Mousa SA. Non-small cell lung cancer: current treatment and future advances. Transl Lung Cancer Res. (2016) 5:288–300. doi: 10.21037/tlcr.2016.06.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang M, Herbst RS, Boshoff C. Toward personalized treatment approaches for non-small-cell lung cancer. Nat Med. (2021) 27:1345–56. doi: 10.1038/s41591-021-01450-2 [DOI] [PubMed] [Google Scholar]
- 6. Paliogiannis P, Colombino M, Sini MC, Manca A, Casula M, Palomba G, et al. Global prognostic impact of driver genetic alterations in patients with lung adenocarcinoma: a real-life study. BMC Pulm Med. (2022) 22:32. doi: 10.1186/s12890-021-01803-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mascaux C, Tomasini P, Greillier L, Barlesi F. Personalised medicine for nonsmall cell lung cancer. Eur Respir Rev. (2017) 26:170066. doi: 10.1183/16000617.0066-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hendriks LE, Kerr KM, Menis J, Mok TS, Nestle U, Passaro A, et al. Oncogene-addicted metastatic non-small-cell lung cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol. (2023) 34:339–57. doi: 10.1016/j.annonc.2022.12.009 [DOI] [PubMed] [Google Scholar]
- 9.. Referenced with permission from the NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) for Non-Small Cell Lung Cancer V.11.2024. © National Comprehensive Cancer Network, Inc. 2024. All rights reserved. Accessed October 15, 2024. To view the most recent and complete version of the guideline, go online to NCCN.org. NCCN makes no warranties of any kind whatsoever regarding their content, use or or application and disclaims any responsibility for their application or use in any way.
- 10. Singh N, Temin S, Baker S, Blanchard E, Brahmer JR, Celano P, et al. Therapy for stage IV non-small-cell lung cancer with driver alterations: ASCO living guideline. J Clin Oncol. (2022) 40:3310–22. doi: 10.1200/jco.22.00824 [DOI] [PubMed] [Google Scholar]
- 11. Gandhi L, Rodríguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F, et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med. (2018) 378:2078–92. doi: 10.1056/NEJMoa1801005 [DOI] [PubMed] [Google Scholar]
- 12. Paz-Ares L, Luft A, Vicente D, Tafreshi A, Gümüş M, Mazières J, et al. Pembrolizumab plus chemotherapy for squamous non-small-cell lung cancer. N Engl J Med. (2018) 379:2040–51. doi: 10.1056/NEJMoa1810865 [DOI] [PubMed] [Google Scholar]
- 13. Mok TSK, Wu Y-L, Kudaba I, Kowalski DM, Cho BC, Turna HZ, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet. (2019) 393:1819–30. doi: 10.1016/s0140-6736(18)32409-7 [DOI] [PubMed] [Google Scholar]
- 14. Gogishvili M, Melkadze T, Makharadze T, Giorgadze D, Dvorkin M, Penkov K, et al. Cemiplimab plus chemotherapy versus chemotherapy alone in non-small cell lung cancer: a randomized, controlled, double-blind phase 3 trial. Nat Med. (2022) 28:2374–80. doi: 10.1038/s41591-022-01977-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sezer A, Kilickap S, Gümüş M, Bondarenko I, Özgüroğlu M, Gogishvili M, et al. Cemiplimab monotherapy for first-line treatment of advanced non-small-cell lung cancer with PD-L1 of at least 50%: a multicentre, open-label, global, phase 3, randomised, controlled trial. Lancet. (2021) 397:592–604. doi: 10.1016/S0140-6736(21)00228-2 [DOI] [PubMed] [Google Scholar]
- 16. Herbst RS, Giaccone G, de Marinis F, Reinmuth N, Vergnenegre A, Barrios CH, et al. Atezolizumab for first-line treatment of PD-L1-selected patients with NSCLC. N Engl J Med. (2020) 383:1328–39. doi: 10.1056/NEJMoa1917346 [DOI] [PubMed] [Google Scholar]
- 17. Socinski MA, Jotte RM, Cappuzzo F, Orlandi F, Stroyakovskiy D, Nogami N, et al. Atezolizumab for first-line treatment of metastatic nonsquamous NSCLC. N Engl J Med. (2018) 378:2288–301. doi: 10.1056/NEJMoa1716948 [DOI] [PubMed] [Google Scholar]
- 18. West H, McCleod M, Hussein M, Morabito A, Rittmeyer A, Conter HJ, et al. Atezolizumab in combination with carboplatin plus nab-paclitaxel chemotherapy compared with chemotherapy alone as first-line treatment for metastatic non-squamous non-small-cell lung cancer (IMpower130): a multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. (2019) 20:924–37. doi: 10.1016/s1470-2045(19)30167-6 [DOI] [PubMed] [Google Scholar]
- 19. Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer. (2019) 19:495–509. doi: 10.1038/s41568-019-0179-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discovery. (2018) 8:822–35. doi: 10.1158/2159-8290.cd-18-0099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Arbour KC, Rizvi H, Plodkowski AJ, Hellmann MD, Knezevic A, Heller G, et al. Treatment outcomes and clinical characteristics of patients with KRAS-G12C-mutant non–small cell lung cancer. Clin Cancer Res. (2021) 27:2209–15. doi: 10.1158/1078-0432.ccr-20-4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Papillon-Cavanagh S, Doshi P, Dobrin R, Szustakowski J, Walsh AM. STK11 and KEAP1 mutations as prognostic biomarkers in an observational real-world lung adenocarcinoma cohort. ESMO Open. (2020) 5:e000706. doi: 10.1136/esmoopen-2020-000706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shire NJ, Klein AB, Golozar A, Collins JM, Fraeman KH, Nordstrom BL, et al. STK11 (LKB1) mutations in metastatic NSCLC: prognostic value in the real world. PloS One. (2020) 15:e0238358. doi: 10.1371/journal.pone.0238358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tanaka I, Koyama J, Itoigawa H, Hayai S, Morise M. Metabolic barriers in non-small cell lung cancer with LKB1 and/or KEAP1 mutations for immunotherapeutic strategies. Front Oncol. (2023) 13:1249237. doi: 10.3389/fonc.2023.1249237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. XALKORI® (crizotinib) [prescribing information]. New York City, NY: Pfizer Inc; (2022). [Google Scholar]
- 26. LUMAKRAS® (sotorasib) [prescribing information]. Thousand Oaks, CA: Amgen Inc; (2021). [Google Scholar]
- 27. Jänne PA, Riely GJ, Gadgeel SM, Heist RS, Ou S-HI, Pacheco JM, et al. Adagrasib in non-small-cell lung cancer harboring a KRAS G12C mutation. N Engl J Med. (2022) 387:120–31. doi: 10.1056/nejmoa2204619 [DOI] [PubMed] [Google Scholar]
- 28. Skoulidis F, Li BT, Dy GK, Price TJ, Falchook GS, Wolf J, et al. Sotorasib for lung cancers with KRAS p.G12C mutation. N Engl J Med. (2021) 384:2371–81. doi: 10.1056/nejmoa2103695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aggarwal C, Nimeiri H, Chen J, Huerga I, Horn L, Trunova N, et al. (2022). 1403 Outcomes following first-line immunotherapy with or without chemotherapy stratified by KRAS mutational status – A real world analysis in patients with advanced NSCLC. In: Romero PJ, editor, in: Society for Immunotherapy of Cancer Annual Meeting, 2022 Nov 8–12. [Google Scholar]
- 30. Prior IA, Hood FE, Hartley JL. The frequency of ras mutations in cancer. Cancer Res. (2020) 80:2969–74. doi: 10.1158/0008-5472.Can-19-3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Haigis KM. KRAS alleles: the devil is in the detail. Trends Cancer. (2017) 3:686–97. doi: 10.1016/j.trecan.2017.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jacobs F, Cani M, Malapelle U, Novello S, Napoli VM, Bironzo P. Targeting KRAS in NSCLC: old failures and new options for “non-G12c” patients. Cancers. (2021) 13:6332. doi: 10.3390/cancers13246332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ricciuti B, Alessi JV, Elkrief A, Wang X, Cortellini A, Li YY, et al. Dissecting the clinicopathologic, genomic, and immunophenotypic correlates of KRASG12D-mutated non-small-cell lung cancer. Ann Oncol. (2022) 33:1029–40. doi: 10.1016/j.annonc.2022.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Riely GJ, Kris MG, Rosenbaum D, Marks J, Li A, Chitale DA, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. (2008) 14:5731–4. doi: 10.1158/1078-0432.ccr-08-0646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Judd J, Abdel Karim N, Khan H, Naqash AR, Baca Y, Xiu J, et al. Characterization of KRAS mutation subtypes in non-small cell lung cancer. Mol Cancer Ther. (2021) 20:2577–84. doi: 10.1158/1535-7163.mct-21-0201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Falk AT, Yazbeck N, Thon L, Guibert N, Hofman V, Zahaf K, et al. Impact of Kras mutant subtypes on PD-L1 expression in lung adenocarcinoma. In: Soria J-C, editor. European Society for Medical Oncology Congress, 2016 Oct 7–11. Elsevier, Copenhagen, Denmark: Amsterdam: (2016). p. vi28. [Google Scholar]
- 37. Liu C, Zheng S, Jin R, Wang X, Wang F, Zang R, et al. The superior efficacy of anti-PD-1/PD-L1 immunotherapy in KRAS-mutant non-small cell lung cancer that correlates with an inflammatory phenotype and increased immunogenicity. Cancer Lett. (2020) 470:95–105. doi: 10.1016/j.canlet.2019.10.027 [DOI] [PubMed] [Google Scholar]
- 38. Pan LN, Ma YF, Li Z, Hu JA, Xu ZH. KRAS G12V mutation upregulates PD-L1 expression via TGF-β/EMT signaling pathway in human non-small-cell lung cancer. Cell Biol Int. (2021) 45:795–803. doi: 10.1002/cbin.11524 [DOI] [PubMed] [Google Scholar]
- 39. Watterson A, Coelho MA. Cancer immune evasion through KRAS and PD-L1 and potential therapeutic interventions. Cell Commun Signal. (2023) 21:45. doi: 10.1186/s12964-023-01063-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu C, Zheng S, Wang Z, Wang S, Wang X, Yang L, et al. KRAS-G12D mutation drives immune suppression and the primary resistance of anti-PD-1/PD-L1 immunotherapy in non-small cell lung cancer. Cancer Commun (Lond). (2022) 42:828–47. doi: 10.1002/cac2.12327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Scheffler M, Ihle MA, Hein R, Merkelbach-Bruse S, Scheel AH, Siemanowski J, et al. K-ras mutation subtypes in NSCLC and associated co-occuring mutations in other oncogenic pathways. J Thorac Oncol. (2019) 14:606–16. doi: 10.1016/j.jtho.2018.12.013 [DOI] [PubMed] [Google Scholar]
- 42. Murray CW, Brady JJ, Tsai MK, Li C, Winters IP, Tang R, et al. An LKB1–SIK axis suppresses lung tumor growth and controls differentiation. Cancer Discovery. (2019) 9:1590–605. doi: 10.1158/2159-8290.CD-18-1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Romero R, Sayin VI, Davidson SM, Bauer MR, Singh SX, LeBoeuf SE, et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat Med. (2017) 23:1362–8. doi: 10.1038/nm.4407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 45. Gillette MA, Satpathy S, Cao S, Dhanasekaran SM, Vasaikar SV, Krug K, et al. Proteogenomic characterization reveals therapeutic vulnerabilities in lung adenocarcinoma. Cell. (2020) 182:200–25.e35. doi: 10.1016/j.cell.2020.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nakajima EC, Ren Y, Vallejo JJ, Akinboro O, Mishra-Kalyani PS, Larkins EA, et al. (2022). Outcomes of first-line immune checkpoint inhibitors with or without chemotherapy according to KRAS mutational status and PD-L1 expression in patients with advanced NSCLC: FDA pooled analysis. In: Friedberg JW, editor, in: American Society of Clinical Oncology Annual Meeting, 2022 Jun 3–7. p. 9001. Chicago, IL: Philadelphia: Lippincott Williams & Wilkins. [Google Scholar]
- 47. Sitthideatphaiboon P, Galan-Cobo A, Negrao MV, Qu X, Poteete A, Zhang F, et al. STK11/LKB1 mutations in NSCLC are associated with KEAP1/NRF2-dependent radiotherapy resistance targetable by glutaminase inhibition. Clin Cancer Res. (2021) 27:1720–33. doi: 10.1158/1078-0432.ccr-20-2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peters S, Salomonsen RJB, Skoulidis F, Perez ID, Wang A, Cooper M, et al. (2023). 68P Real-world (rw) outcomes in patients (pts) with metastatic (m) NSCLC and STK11, KEAP1 and/or KRAS mutations (mut) receiving PD-(L)1-based treatment (tx): CORRELATE. In: Haanen JB, editor, in: European Society for Medical Oncology Immuno-Oncology Congress, 2023 Dec 6–8. p. 100540. Geneva, Switzerland: Amsterdam: Elsevier. [Google Scholar]
- 49. Julian C, Pal N, Gershon A, Evangelista M, Purkey H, Lambert P, et al. Overall survival in patients with advanced non-small cell lung cancer with KRAS G12C mutation with or without STK11 and/or KEAP1 mutations in a real-world setting. BMC Cancer. (2023) 23:352. doi: 10.1186/s12885-023-10778-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jeong Y, Hoang NT, Lovejoy A, Stehr H, Newman AM, Gentles AJ, et al. Role of KEAP1/NRF2 and TP53 mutations in lung squamous cell carcinoma development and radiation resistance. Cancer Discovery. (2017) 7:86–101. doi: 10.1158/2159-8290.cd-16-0127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cordeiro De Lima VC, Corassa M, Saldanha E, Freitas H, Arrieta O, Raez L, et al. STK11 and KEAP1 mutations in non-small cell lung cancer patients: descriptive analysis and prognostic value among Hispanics (STRIKE registry-CLICaP). Lung Cancer. (2022) 170:114–21. doi: 10.1016/j.lungcan.2022.06.010 [DOI] [PubMed] [Google Scholar]
- 52. Olivares-Hernández A, González Del Portillo E, Tamayo-Velasco Á, Figuero-Pérez L, Zhilina-Zhilina S, Fonseca-Sánchez E, et al. Immune checkpoint inhibitors in non-small cell lung cancer: from current perspectives to future treatments—a systematic review. Ann Transl Med. (2023) 11:354. doi: 10.21037/atm-22-4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Olivares-Hernández A, Del Barco-Morillo E, Miramontes-González JP. New developments and updates in non-small cell lung cancer and immunotherapy: adjuvant, neoadjuvant and advanced stages. Ann Transl Med. (2023) 11:423. doi: 10.21037/atm-23-1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ramalingam SS, Balli D, Ciuleanu TE, Pluzanski A, Lee JS, Schenker M, et al. (2021). 4O Nivolumab (NIVO) + ipilimumab (IPI) versus chemotherapy (chemo) as first-line (1L) treatment for advanced NSCLC (aNSCLC) in CheckMate 227 part 1: efficacy by KRAS, STK11, and KEAP1 mutation status. In: Friedberg JW, editor, in: American Society of Clinical Oncology Annual Meeting, 2021 Jun 4–8. pp. S1375–S6. Chicago, IL: Philadelphia: Lippincott Williams & Wilkins. [Google Scholar]
- 55. Paz-Ares LG, Ciuleanu T-E, Cobo M, Bennouna J, Schenker M, Cheng Y, et al. First-line nivolumab plus ipilimumab with chemotherapy versus chemotherapy alone for metastatic NSCLC in CheckMate 9LA: 3-year clinical update and outcomes in patients with brain metastases or select somatic mutations. J Thorac Oncol. (2023) 18:204–22. doi: 10.1016/j.jtho.2022.10.014 [DOI] [Google Scholar]
- 56. Johnson ML, Cho BC, Luft A, Alatorre-Alexander J, Geater SL, Laktionov K, et al. Durvalumab with or without tremelimumab in combination with chemotherapy as first-line therapy for metastatic non-small-cell lung cancer: the phase III POSEIDON study. J Clin Oncol. (2023) 41:1213–27. doi: 10.1200/jco.22.00975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Peters S, Cho BC, Luft A, Alatorre-Alexander J, Geater SL, Kim SW, et al. (2022). OA15.04 association between KRAS/STK11/KEAP1 mutations and outcomes in POSEIDON: durvalumab ± tremelimumab + chemotherapy in mNSCLC. In: Adjei AA, editor, in: International Association for the Study of Lung Cancer World Conference on Lung Cancer, 2022 Aug 6–9. pp. S39–41. Vienna, Austria Amsterdam: Elsevier. [Google Scholar]
- 58. Devarakonda S, Pellini B, Verghese L, Park H, Morgensztern D, Govindan R, et al. A phase II study of everolimus in patients with advanced solid malignancies with TSC1, TSC2, NF1, NF2 or STK11 mutations. J Thorac Dis. (2021) 13:4054–62. doi: 10.21037/jtd-21-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dabbous F, Wang C-Y, Simmons D, Huse S, Jassim R. (2023). Prevalence of STK11, KEAP1, and KRAS mutations/co-mutations and associated clinical outcomes for patients newly diagnosed with metastatic non-small cell lung cancer. In: Friedberg JW, editor, in: American Society of Clinical Oncology Annual Meeting, 2023 Jun 2–6. p. e21186. Chicago, IL: Philadelphia: Lippincott Williams & Wilkins. [Google Scholar]
- 60. Kalemkerian GP, Narula N, Kennedy EB, Biermann WA, Donington J, Leighl NB, et al. Molecular testing guideline for the selection of patients with lung cancer for treatment with targeted tyrosine kinase inhibitors: American Society of Clinical Oncology endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology clinical practice guideline update. J Clin Oncol. (2018) 36:911–9. doi: 10.1200/jco.2017.76.7293 [DOI] [PubMed] [Google Scholar]
- 61. Jaiyesimi IA, Leighl NB, Ismaila N, Alluri K, Florez N, Gadgeel S, et al. Therapy for stage IV non–small cell lung cancer with driver alterations: ASCO living guideline. J Clin Oncol. (2024) 42:e1–e22. doi: 10.1200/jco.23.02744 [DOI] [PubMed] [Google Scholar]
- 62. Raez LE, Brice K, Dumais K, Lopez-Cohen A, Wietecha D, Izquierdo PA, et al. Liquid biopsy versus tissue biopsy to determine front line therapy in metastatic non-small cell lung cancer (NSCLC). Clin Lung Cancer. (2023) 24:120–9. doi: 10.1016/j.cllc.2022.11.007 [DOI] [PubMed] [Google Scholar]
- 63. Sholl LM, Awad M, Basu Roy U, Beasley MB, Cartun RW, Hwang DM, et al. Programmed death ligand-1 and tumor mutation burden testing of patients with lung cancer for selection of immune checkpoint inhibitor therapies: guideline from the college of American Pathologists, Association for Molecular Pathology, International Association for the Study of Lung Cancer, Pulmonary Pathology Society, and LUNGevity foundation. Arch Pathol Lab Med. (2024) 148:757–74. doi: 10.5858/arpa.2023-0536-cp [DOI] [PubMed] [Google Scholar]
- 64. Saleh MM, Scheffler M, Merkelbach-Bruse S, Scheel AH, Ulmer B, Wolf J, et al. Comprehensive analysis of TP53 and KEAP1 mutations and their impact on survival in localized- and advanced-stage NSCLC. J Thorac Oncol. (2022) 17:76–88. doi: 10.1016/j.jtho.2021.08.764 [DOI] [PubMed] [Google Scholar]
- 65. Tsao M-S, Aviel-Ronen S, Ding K, Lau D, Liu N, Sakurada A, et al. Prognostic and predictive importance of p53 and RAS for adjuvant chemotherapy in non-small-cell lung cancer. J Clin Oncol. (2007) 25:5240–7. doi: 10.1200/jco.2007.12.6953 [DOI] [PubMed] [Google Scholar]
- 66. Murakami I, Hiyama K, Ishioka S, Yamakido M, Kasagi F, Yokosaki Y. p53 gene mutations are associated with shortened survival in patients with advanced non-small cell lung cancer: an analysis of medically managed patients. Clin Cancer Res. (2000) 6:526–30. [PubMed] [Google Scholar]
- 67. Liang X, Gao X, Wang F, Li S, Zhou Y, Guo P, et al. Clinical characteristics and prognostic analysis of SMARCA4-deficient non-small cell lung cancer. Cancer Med. (2023) 12:14171–82. doi: 10.1002/cam4.6083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chen H, Chong W, Wu Q, Yao Y, Mao M, Wang X. Association of LRP1B mutation with tumor mutation burden and outcomes in melanoma and non-small cell lung cancer patients treated with immune check-point blockades. Front Immunol. (2019) 10:1113. doi: 10.3389/fimmu.2019.01113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Brown LC, Tucker MD, Sedhom R, Schwartz EB, Zhu J, Kao C, et al. LRP1B mutations are associated with favorable outcomes to immune checkpoint inhibitors across multiple cancer types. J Immunother Cancer. (2021) 9:e001792. doi: 10.1136/jitc-2020-001792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Peters S, Cho BC, Luft A, Alexander JAA, Geater SL, Laktionov K, et al. (2023). LBA3 Durvalumab (D) ± tremelimumab (T) + chemotherapy (CT) in first-line (1L) metastatic NSCLC (mNSCLC): 5-year (y) overall survival (OS) update from the POSEIDON study. In: Haanen JB, editor, in: European Society for Medical Oncology Immuno-Oncology Congress, 2023 Dec 6–8. p. 100693. Geneva, Switzerland: Amsterdam: Elsevier. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.