Abstract
Objectives:
The skin microbiome modulates immunity by interacting with keratinocytes to combat pathogens. Allergic disorders are classified by IgE sensitivity and aberrant Th2 responses, and an increasing number of studies describe associations with skin microbiome fluctuations. In this review, we discuss commensal-epidermal homeostasis and its influence on allergic disease.
Data sources:
All included references were obtained from PubMed.
Study Selections:
Studies addressing relevant aspects of commensal-epidermal homeostasis, skin microbiome dysbiosis, and microbiome-targeted therapeutics and prevention in allergy were included.
Results:
Homeostasis between the commensal microbiome and the epidermis is important in protecting against allergic disease. Commensals promote anti-allergic Th1 and Th17 immunophenotypes within the skin and induce keratinocytes to secrete antimicrobial peptides and alarmins that enhance barrier function and antagonize pro-allergic organisms. Perturbations in this homeostasis, however, is associated with allergic disease development. Atopic dermatitis is associated with decreases in skin commensals and increases in the pathogen, Staphylococcus aureus. Fluctuations in the skin microbiome contribute to decreased barrier dysfunction, allergic sensitization, and Th2 cytokine secretion. Little is known about how the skin microbiome impacts food allergy, allergic rhinitis, and asthma, and it is poorly understood how cutaneous inflammation influences systemic allergic responses. Therapies are targeted towards maintenance of the skin barrier, replacement of healthy commensals, and anti-Th2 biologic therapy.
Conclusions:
Although the impacts of commensal-epidermal homeostasis on allergy within the skin are becoming increasingly clear, future studies are necessary to assess its impacts on extracutaneous allergic disorders and to explore potential therapeutics targeting the skin microbiome.
Keywords: Skin Microbiome, Allergy, Skin, Atopic Dermatitis, Asthma, Allergic Rhinitis, Food Allergy
Introduction
Allergic disorders, classified by IgE sensitivity, are steadily rising in prevalence worldwide and significantly impact quality of life in all age groups1. A growing body of literature is revealing that the skin microbiome, the community of bacteria, viruses and fungi present on the skin, plays a significant role in modulating allergic disorders. The microbiome is vital for immune system development and homeostasis. Changes in microbial composition and function, termed dysbiosis, have been linked to alterations in immune responses and to the development of allergic diseases. Here we provide a comprehensive review on the impact of the skin microbiome on allergic disease, including atopic dermatitis (AD), allergic asthma, food allergy, and allergic rhinitis (AR). We discuss the modulation of allergic disease by commensal-epidermal homeostatic interactions, and the role of the skin microbiome in major allergic diseases. Lastly, we discuss the implications of the skin microbiome on allergic disease diagnosis, therapeutics, and prevention.
Mechanisms of Commensal-Epidermal Homeostasis:
The human skin microbiome is a highly variable community of bacteria, viruses, and fungi (Table 1) on the skin that modulates host immunity and provides protection from pathogens. The composition of the microbiome is site-specific, varying with the physiologic environments at each body site2. Body sites differ by pH, fatty acid composition, and ability of the skin to retain water, but can be influenced by gender, antibiotic treatment, and cosmetic use3. These factors in turn affect microbial ecology3. Site-specific microbial communities (Table 1) develop around three months of age4, and early-life colonization is needed for the development of commensal tolerance and decreased risk of allergic diseases like asthma5. In the skin, microbial ecology also varies by depth within the epidermis6. There is a growing body of literature regarding the human skin microbiome composition under healthy and pathogenic states and the homeostatic interactions between normal skin commensals and the epidermis. As discussed below, healthy commensal-epidermal homeostasis predominantly protects against allergic disease by promoting barrier function, deterring colonization of pro-allergic pathogens including Staphylococcus aureus (S. aureus), and maintaining an anti-allergic Th1/Th17 immunophenotype.
Table 1.
Commensal skin organisms and skin pathogens in allergic disease
| Common commensal organisms of the healthy skin microbiome | ||||
|---|---|---|---|---|
| Microbiome Component | Phyla | Genera | Species | Source |
| Bacterial Skin Microbiome | Actinobacteria | Corynebacteria | 33, 4, 105, 106 | |
| Cutibacteria * | C. acnes | |||
| Firmicutes | Staphylococci | S. epidermidis | ||
| Proteobacteria | Roseomonas | R. mucosa | ||
| Skin Mycobiome | Basidiomycota | Malassezia | M. furfur, M. restrica, M. globosa, M. sympodialis | 56 |
| Ascomycota | Candida | C.albicans | ||
| Taxa | ||||
| Skin Virome | Phages: | Pseudomonas phage, Bacillus phage, Staphylococcus phage, Propionibacterium phage, Planktothrix phage, Streptococcus phage, Burkholderia Phage, Mycobacterium Phage | 59, 60 | |
| Other: | Human Papillomavirus | |||
| Pathogenic organisms of the skin microbiome in allergic disease | ||||
| Microbiome Component | Phyla | Genera | Species | Source |
| Bacterial Skin Microbiome | Actinobacteria | Corynebacteria | C. striatum | 65, 72, 113 |
| Firmicutes | Staphylococci | S. aureus | ||
| Streptococci | S. pyogenes | |||
| Skin Mycobiome | Basidiomycota | Malassezia |
M. dermatis, M. sloofiae,
M. sympodialis |
74, 75 |
| Ascomycota | Candida | C. albicans | ||
| Taxa | ||||
| Skin Virome | Herpesviridae | Herpes Simplex Virus | 113 | |
| Poxviridae | Molluscum contagiosum, Vaccinica Virus | |||
Formerly Propionobacteria
Commensal-Epidermal Homeostasis: Epidermal Barrier-mediated Immunity
The skin consists of the dermis and epidermis (Figure 1), with the latter subdivided into the stratum basale, stratum spinosum, stratum granulosum (SG)7 and stratum corneum (SC). Barrier integrity is primarily maintained by the SC layer and tight junctions (TJs) within the SG. The SC is composed of a cornified envelope, in which keratinocytes containing cross-linked structural proteins like filaggrin (FLG) are connected by corneodesmosomes, and a lipid-rich lamellar layer produced and secreted by keratinocytes8,9. TJs are multimeric protein complexes that physically link keratinocytes within the SG10. Together the SC and TJs form a physical barrier impermeable to microbes, including skin commensals9. However, as discussed below, disruptions in epidermal-microbiome homeostasis can compromise SC or TJ formation and function, thereby disrupting the skin barrier and increasing susceptibility to allergic disease.
Figure 1. Properties of the epidermal barrier.
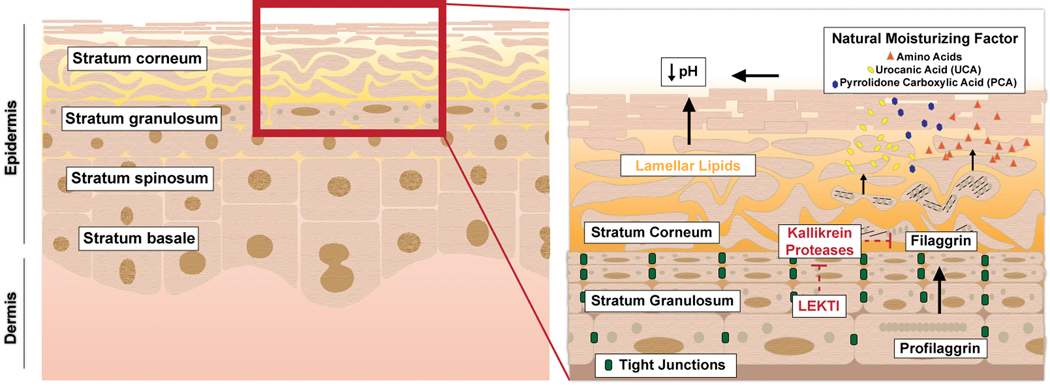
The skin consists of a dermis and epidermis, which is further subdivided into the stratum basale, stratum spinosum, stratum granulosum, and stratum corneum. The epidermal barrier is formed primarily by (a) tight junctions within the stratum granulosum and (b) the stratum corneum. The stratum corneum consists of lamellar lipids (e.g. ceramides and long-chain fatty acids) and a cornified envelope. Natural moisturizing factors (byproducts of filaggrin catabolism) and fatty acids of the lamellar layer create an acidic pH. Kallikrein proteases (KLKs) promote desquamation by degrading filaggrin and corneodesmosomes. The KLKs are inhibited by LEKTI.
Filaggrin:
Mutations in FLG, or disrupted FLG expression or processing alters epidermal-microbiome homeostasis which can predispose microbial invasion11, altered microbiome composition (i.e. reduced diversity)12,13, increased susceptibility to viral and bacterial skin infections14, and increased risk of allergic sensitization15. Reduced FLG levels and/or processing are major risk factors for allergic sensitization and allergic disease16, and FLG expression is reduced by pro-allergic Th2 cytokines1,11,17. Reduced FLG expression and commensal dysbiosis may thus create a positive feedback loop culminating in a dysfunctional barrier and predisposition to allergic disease.
Epidermal Proteases and Antiproteases:
Keratinocyte desquamation is regulated by the balance between kallikrein proteases (KLKs) that cleave FLG and corneodesmosomes, and antiproteases, namely lympho‐epithelial Kazal-type-related inhibitor (LEKTI), encoded by SPINK518. Reduced corneodesmosomes, reduced LEKTI, or unchecked KLK activity are associated with AD and other atopic diseases19–21. Indeed, individuals with Netherton Syndrome (NS), caused by SPINK5 mutations, often exhibit AD, elevated IgE and varying degrees of asthma, food allergy and allergic rhinitis19,20. Individuals with NS are also susceptible to recurrent skin infections and dysbiosis, including infection by S. aureus which can further disrupt barrier function22. Finally, S. aureus phenol-soluble modulins (PSMs) enhance epidermal KLK activity and promote barrier dysfunction23.
Stratum Corneum Lipids:
The lipids of the lamellar layer include cholesterols, fatty acids (FAs) and ceramides8,24 that may influence skin microbiome composition, skin barrier function and allergic disease susceptibility. FAs contribute to the acidic epidermal pH that favors commensals over pro-allergic bacteria25. Skin abundant in ceramides and long-chain FAs is associated with increased abundance of normal skin commensals, such as Corynebacterium and Cutibacterium26. Additionally, long-chain FAs exert antimicrobial activity against bacteria, fungi and enveloped viruses27. Perturbations in the lipidome, however, can induce epidermal barrier defects and are associated with AD8,24,25. For example, reduced ceramides, long-chain FAs, and expression of FA elongases (which synthesize long-chain FAs) have been observed in the epidermis of individuals with AD28, and Th2 cytokines can suppress FA elongase expression in human keratinocytes28.
Tight Junctions:
The importance of TJs as an epidermal barrier component highlighted by its frequent targeting for destruction by allergens and pro-allergic pathogens29,30. S. aureus, for example, prevents TJ complex membrane localization, thereby reducing intercellular connectivity and increasing epidermal permeability31. TJ perturbations can further disrupt the SC by reducing FLG and lamellar lipid synthesis, and are associated with AD32. Contrarily, commensal Staphylococcus epidermidis (S. epidermidis) promotes TJ formation both directly and indirectly by inducing keratinocyte antimicrobial peptide (AMP) secretion (discussed below)29.
Skin pH:
The acidic 4–6 pH range of the healthy epidermis favors commensal over pro-allergic bacteria25,33. Contributors to this acidic pH are natural moisturizing factors (NMFs, which are byproducts of FLG catabolism) and FAs produced by keratinocytes or as metabolites of commensal bacteria (e.g. Cutibacterium acnes)25,33. This acidity antagonizes pathogens by several means, including reducing S. aureus keratinocyte adhesion and altering the conformation and activity of S. aureus virulence factors25. However, the inability to maintain acidic skin (e.g. defective FLG catabolism) alters microbial diversity and allows S. aureus keratinocyte adhesion25,33, demonstrating the importance of skin pH in commensal-epidermal homeostasis.
Commensal-Epidermal Homeostasis: Keratinocyte-mediated Immunity
Keratinocytes release a multitude of proteins that can impact commensal-epidermal homeostasis and thus influence allergic disease (Figure 2). Percutaneous allergens activate type 2 dendritic cells (DCs) and trigger the secretion of innate type 2 cytokines (thymic stromal lymphopoietin [TSLP], IL-33 and IL-25) from keratinocytes, which in turn stimulate canonical type 2 cytokine (e.g. IL-4) release from type 2 innate lymphoid cells (ILC2s). Together, allergen-activated DCs and ILC2-derived IL-4 co-stimulate the differentiation of naïve cells (Th0) into pro-allergic T-helper type 2 (Th2) cells34–36. The immune microenvironment of the skin is an important mediator of allergic disease susceptibility, and it is impacted by the commensal skin microbiome. It is beyond the scope of this review to comprehensively review skin immunity. A few key keratinocyte-derived factors are discussed here. Chief among them are AMPs and the alarmins S100A8 and S100A9, which are predominantly anti-allergic, and the innate type 2 cytokines, which are pro-allergic.
Figure 2. Adaptive Immunity in Commensal-Epidermal Homeostasis.
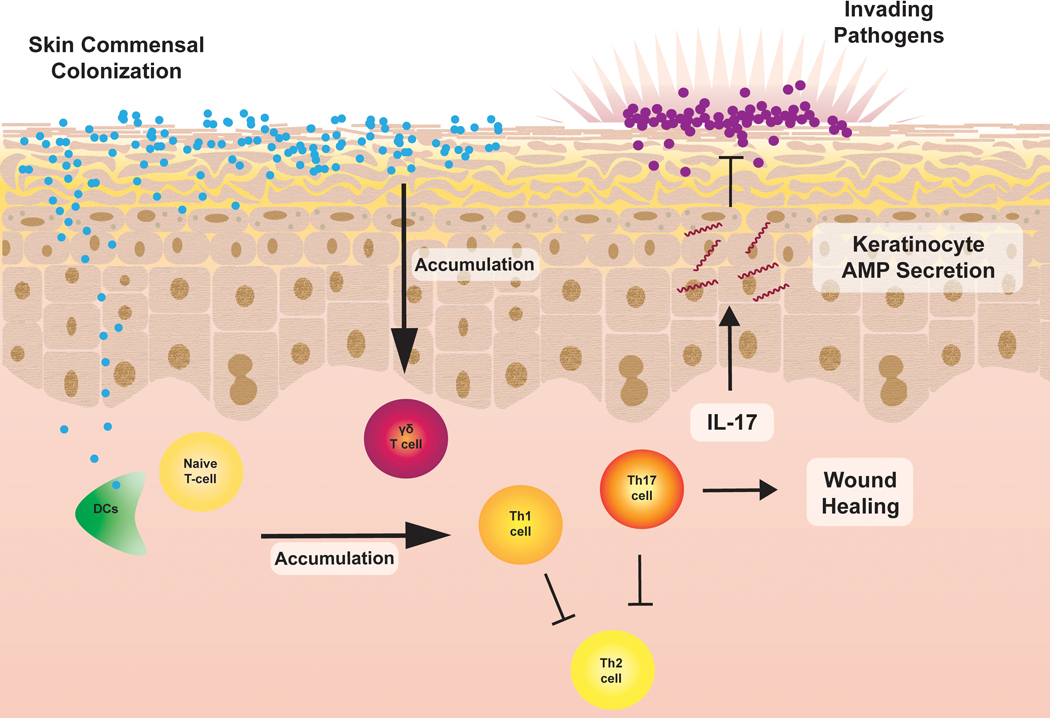
Commensals induce the accumulation of γδ-T-cells and commensal-specific type 1 and type 17 T-cells. These cells protect against allergic disease by suppressing type 2 immune responses, promoting wound healing, and inhibiting pathogens by secreting IL-17 which induces keratinocyte antimicrobial peptide secretion.
Antimicrobial Peptides:
Human beta-defensins (hBDs) and cathelicidins (e.g. LL-37) are AMPs secreted by keratinocytes constitutively or in response to inflammatory or infectious stimuli37–39. These AMPs are active against many bacteria, fungi, and viruses38,39 and primarily kill via membrane permeabilization, though other mechanisms have been described37,39. Bacterial and fungal skin commensals stimulate hBD or LL-37 secretion from keratinocytes which can synergize with antimicrobial compounds secreted from commensals to attenuate pathogen colonization40,41. hBDs and LL-37 can promote type 1 over type 2 immunity38,39, though conflicting studies have observed pro-allergic activity in some contexts39. Finally, AMPs can increase epidermal barrier integrity by inducing TJs and promoting wound healing38,39.
S100A8 and S100A9:
The alarmins S100A8 and S100A9 are inflammatory mediators within the skin that function as either homodimers or as the heterodimer calprotectin42. Their expression is influenced by the skin microbiome; for example, commensal S. epidermidis upregulates keratinocyte s100a8 and s100a9 expression by stimulating IL-17A secretion from commensal-specific Tc17 cells in mice43. Moreover, elevated calprotectin is associated with Th1 and Th17 inflammatory phenotypes44. S100A8 and S100A9 exert antimicrobial activity (e.g against S. aureus) by several means including the chelation of the essential metal ions Zn2+ and Mn2+ by calprotectin and S100A9-mediated neutrophil recruitment/activity42. Calprotectin also promotes skin barrier integrity by stimulating keratinocyte proliferation and differentiation42.
Epithelial-derived Innate Type 2 Cytokines:
The innate type 2 cytokines TSLP, IL-25 and IL-33 promote pro-allergic type 2 immunity through various mechanisms, including Th2 polarization, type 2 cell activation/expansion and type 2 cytokine induction1,11,17. These cytokines also induce barrier dysfunction directly by suppressing FLG expression11,17. Their influence on allergic disease is illustrated by their association with an AD-like phenotype if increased within the skin11,17,45 and, for TSLP specifically, an asthma-like phenotype if it enters systemic circulation46,47. Various pro-allergic stimuli induce TSLP, IL-25 and IL-33 production, including type 2 cytokines, allergens, and mechanical damage1,11,17. The skin microbiome also can influence their synthesis; for example, notch-deficient mice exhibit skin inflammation and systemic atopy, and have higher epidermal TSLP mRNA and serum TSLP levels in the absence versus presence of normal commensals48. Pro-allergic S. aureus, on the other hand, induces TSLP and IL-33 expression by keratinocytes49,50.
Commensal-Epidermal Homeostasis: Adaptive Immunity
Bacterial Microbiome:
Just as in the gut, recent studies have demonstrated that commensal bacteria protect against allergic disease by priming the adaptive immune system. In addition to directly inducing AMPs from keratinocytes, commensal S. epidermidis in mice primes epidermal accumulation of commensal-specific type 1 and type 17 helper and cytotoxic T-cells (Th1, Th17, Tc1 and Tc17)43,51,52. Commensals induce these cells to secrete IL-17A which promotes keratinocytes to produce anti-allergic peptides such as s100a8 and s100a943. Similarly, commensal Corynebacteria accolens can induce epidermal IL-17A+ γδ-T-cell influx, thereby promoting a type 17 immunophenotype53. Commensal-specific Tc17 T-cells can promote barrier integrity in mice directly by expressing tissue-repair genes and accelerating wound healing51. Interestingly, S. epidermidis can suppress S. aureus colonization when the skin barrier is intact, but when disrupted (e.g. tape-stripping) it instead promotes S. aureus colonization54. This phenomenon may be explained by a “poised” Th2 transcriptional profile identified within S. epidermidis-specific Tc17/Th17 cells which, upon exposure to pro-allergic innate type 2 cytokines, is translated, thereby superimposing or substituting the type 17 profile55.
Mycobiome:
Information regarding the mycobiome’s influence on allergic disease development is limited, and available studies draw contradicting conclusions. For example, many Malassezia species induce Malassezia-specific Th17 cells56 and keratinocyte hBD production57, which protects from allergy (see below). Contrarily, Malassezia globosa and restricta can promote pro-allergic TSLP release from keratinocytes57, and Malassezia-specific IgE and Th2-polarized T-cells have been reported in AD patients58. Further study is warranted to determine how fungal commensals influence allergic disease and if these effects are species- and/or context-dependent.
Virome:
The typical skin virome is not well characterized, nor is its role in allergic disease. In an analysis of double-stranded DNA viruses, bacteriophages were prevalent on human skin59 (Table 1) and topical phage therapy reduced AD severity in one study, though the mechanism is unclear60. More controlled and mechanistic studies are nevertheless required to address the role of the skin virome in allergic disease.
The Skin Biome in Allergic Disease
Atopic Dermatitis
AD affects 10–20% of children worldwide, often in industrialized countries61, with onset before one year of age in up to 60% of children62. Healthy and AD skin exhibit many structural and immunological differences. AD is also characterized by skin microbiome dysbiosis. However, whether these microbiome disruptions precede63,64 or cause13,26 skin disease or are a consequence of disease remains unclear. Studies suggest that commensals may play a role in preventing AD since commensal staphylococcal colonization at 2 months of age is associated with decreased AD incidence at one year63 and commensals are positively correlated with the levels of long-chain FAs12. The loss of commensal abundance is correlated with individuals that have FLG deficiency12. Decreased commensal abundance is also observed during AD flares, but can be reversed with bleach baths, emollients, or topical steroids65. Birth cohorts suggest that infants who develop AD have a decrease in the heterogeneous commensal population 66, which is accompanied by increased abundance of the pathogen S. aureus. Infants with S. aureus colonization or lacking S. aureus-inhibiting commensals67 at 3 months are more likely to develop AD compared to those who are not colonized64.
Various S. aureus virulence factors have been associated with AD68,69. The S. aureus cell wall component lipoteichoic acid (LTA) upregulates over 300 genes in keratinocytes including Th2-polarizing genes such as TSLP and IL-4. In addition, LTA reduces FLG expression70. S. aureus also secretes enterotoxins capable of inducing inflammation by acting as superantigens and activating T cells via major histocompatibility complex II. Individuals with AD have significantly more specific-IgE to the S. aureus alpha, delta, and Toxic Shock Syndrome Toxin-1 toxins than healthy controls. S. aureus-derived PSMs can also increases keratinocyte KLK activity23, which may promote S. aureus skin penetration by increasing desquamation and reducing barrier integrity (Figure 3).
Figure 3. Effect of the skin microbiome on allergic disease.
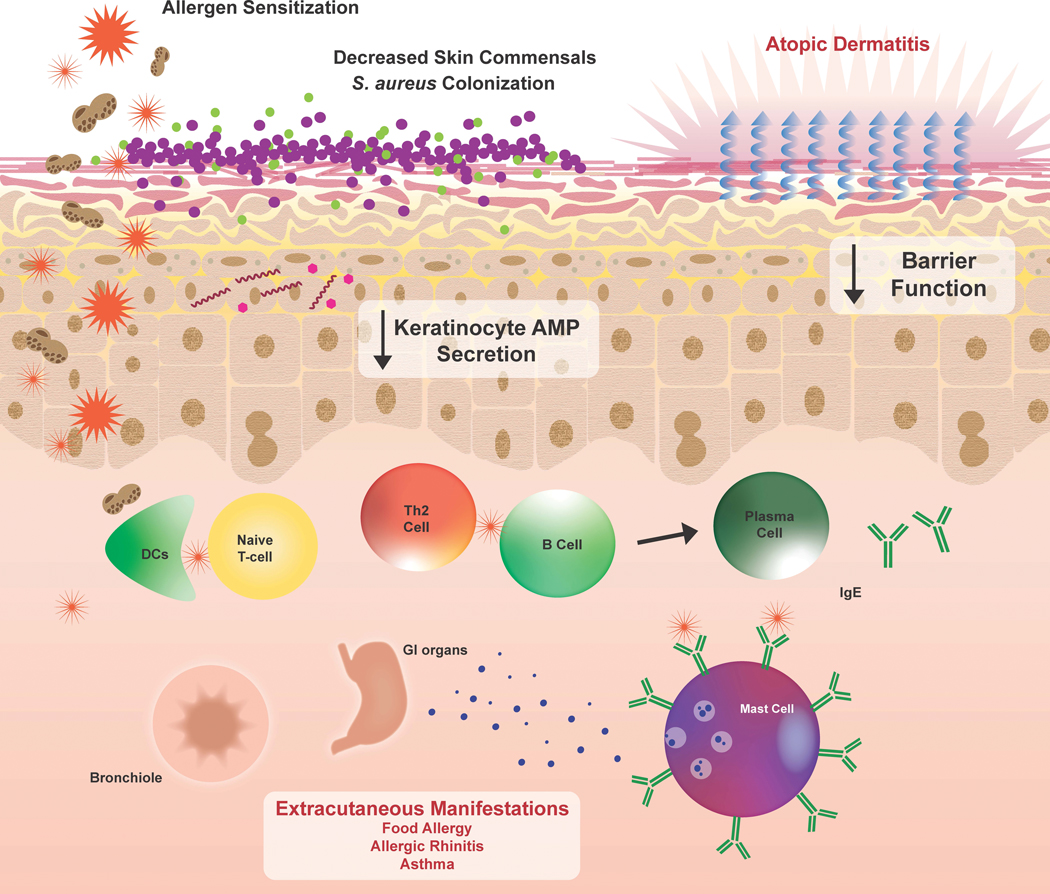
A decrease in skin commensals and the presence of pathogens contributes to skin microbiome dysbiosis that can occur in allergic disease. Decreases in keratinocyte AMPs also prevent clearance of these pathogens. In particular, S. aureus is shown to decrease barrier function, induce IgE production, and induce secretion of Th2 cytokines, which can have systemic consequences and exacerbate extracutaneous manifestations of allergy.
Corynebacterium species are another common skin commensal (Table 1). Although C. accolens may protect from allergy by promoting IL-17A+ γδ-T-cell aggregation53, Corynebacterium may be involved in allergic disease via the induction of type 2 responses71. C. striatum can increase expression of genes associated with S. aureus carriage in the nares, suggesting C. striatum promotes a shift towards S. aureus commensalism72. Several studies have demonstrated Corynebacterium presence in individuals with AD73.
Malassezia species are common fungi on skin (Table 1) and healthy individuals have Malassezia-specific T-cells and antibodies56. However, AD subjects are characterized by the presence of M. dermatis and M. sloofiae74, and several studies reveal the presence of Malassezia-specific IgE58 in individuals with AD. Moreover, the presence of Malassezia-specific IgE has been correlated with increasing severity of AD in adults. Malassezia antigens can induce autoreactive T-cells, which induce further inflammation independent of the fungal antigens75. Malassezia exposure led to stratum corneum colonization, myelocytic infiltration, and IL-17 induction in mice56. This IL-17 induction was crucial to controlling fungal burden; however, Malassezia presence on the disrupted skin barrier induced inflammation, and IL-17-deficient mice showed a diminished inflammatory response Malassezia overgrowth56. These studies demonstrate that Malassezia-driven inflammation promotes AD-like disease in mice56. Studies are needed to characterize the differences, transition, or expansion of Malassezia-specific memory T-cells in healthy adults versus those with AD.
The skin microbiome also extends protection against viral pathogens76. Children with AD are prone to viral infections, though these are less common than bacterial infections. Viral infections are known as complications of AD rather than a direct contributor to its pathogenesis. The most common viral infection in children with AD is Herpes Simplex Virus (HSV) which causes eczematicum herpeticum (EH). EH manifestations range from fever and malaise to encephalitis or septic shock and is recurrent in up to 25% of cases77. Viral infections are more likely in those with severe AD and often occur concurrently with S. aureus infections77. In fact, S. aureus alpha-toxin aids in HSV infection by promoting viral binding to keratinocyte receptors78. T-cell lines generated from individuals with EH showed increased secretion of IL-4+ when compared to T-cell lines generated from healthy controls, suggesting virus-specific T cells generate a type-2 response in EH79. Treatment of AD with topical corticosteroids did not prevent subsequent EH, nor did interferon-γ therapy result in symptom improvement77.
Molluscum contagiosum is a common childhood viral infection caused by a DNA virus of the poxviridae family. While molluscum is usually self-limiting, those with AD experience prolonged infection with a more widespread distribution. Skin barrier defects, such as FLG mutations, can predispose to molluscum infection, while AD-induced pruritus promotes autoinoculation80,81.
AD patients who had previously received a smallpox vaccination can develop eczema vaccinatum, characterized by disseminated vaccinia virus which, if systemic, is lethal82. FLG seems to protect from vaccinia, as FLG-deficient mice suffer from disseminated vaccinia infection83. As such, alternatives to the current smallpox vaccine are needed in those with AD. Imvamune, a vaccine using the modified Vaccinia Ankara virus, is safe in patients with mild-moderate AD and elicits a specific immune response84.
Food Allergy
Food allergy affects approximately 1 in 13 children in the US and is defined as type-1 hypersensitivity to a given food antigen85. The LEAP/LEAP-ON study (n=640) observed that skin and nasal S. aureus colonization in children is associated with concurrent eczema. Total and specific IgE, particularly to egg white and peanut, were significantly associated S. aureus colonization of the skin but not the nares, suggesting that IgE sensitization may be promoted by skin biome interactions. However, this study is limited by use of less sensitive culture techniques to collect S. aureus , thus future studies should test these associations with sequencing-based approaches86. Further studies of the impact of the skin microbiome on food allergy are needed.
Allergic Rhinitis
Allergic rhinitis (AR) is characterized by seasonal allergens that trigger upper airway inflammation inducing rhinorrhea, sneezing, and nasal congestion. AR affects up to 25% of the global population and steadily continues to rise87. The nasal cavity harbors its own community of organisms to defend against environmental insults88. A study sampling S. aureus from both children and adults demonstrated that 85% of subjects were colonized with S. aureus in the nares, and approximately 77% of AD subjects colonized by S. aureus in the nares were also colonized on the skin89. These findings suggest S. aureus proliferates in the nasal cavity and a connection exists between the biomes at these 2 surfaces.
Asthma
Allergic asthma affects up to 18% of the global population and is defined by airway hyperresponsiveness and airway inflammation90. A major risk factor for asthma development is allergic sensitization61. The absence of skin commensals and antimicrobial responses91 predispose to microbiome dysbiosis, associated with inflammation and Th2 responses.
Organisms in household dust can be transferred to the skin via surfaces92 and those sharing a household have been reported to have similar skin commensals93, highlighting the influence of the environment on the skin microbiome. S. aureus persists in dust94 and is highly abundant in the microbiome of purified house dust mite bodies95. Interestingly, Staphylococcus, Haemophilus, and Corynebacterium species in house dust are associated with increased asthma risk, while house dust levels of cockroach, cat, and mouse allergens are inversely related to asthma development in an early life cohort96. IgE sensitization to house dust mite 97 and S. aureus is common in those with asthma and/or AD95, and percutaneous exposure to S. aureus via HDM exposure could be another potential route for sensitization to S. aureus. Bacterial antigens in house dust that encounter the skin may reduce barrier integrity. The possibility that bacterial antigens are taken up through the skin in addition to the airways may provide insight into the progression of atopic comorbidities.
Therapeutic interventions
Understanding of the role the skin microbiome in allergic disease and the underlying mechanisms has revealed unique opportunities for targeted therapy (Figure 4).
Figure 4. Interventions influencing the skin microbiome.
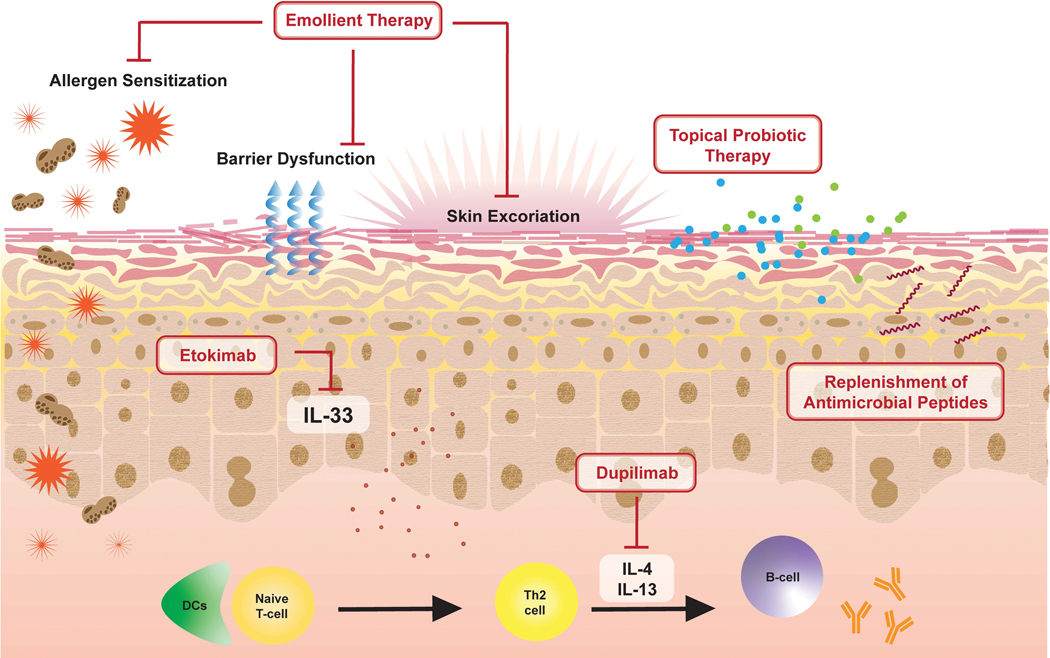
Emollient therapy aids in repairing a damaged skin barrier preventing inflammation and sensitization. The replacement of skin commensals and antimicrobial peptides are also being studied as potential therapeutics. Biologic therapeutics, which act by blocking cytokines implicated in allergic disease such as IL-33, IL-4 and IL-13, may also have an impact on skin microbiome dysbiosis.
Emollients
Although their utility in AD prevention has recently been cast into doubt98, emollients are typically first-line therapy for AD, reduce the need for pharmaceutical interventions99, and decrease AD severity100. Emollient administration increases bacterial diversity100 and decreases S. aureus detection in lesional skin101. Coal tar emollients are an established AD therapy as they activate the aryl hydrocarbon receptor and thus induce keratinocyte differentiation, increase FLG expression, reduce Th2 responses, and decrease the abundance of pro-allergic S. aureus102.
Emollients are also an efficient vehicle for other potential therapies, including those that specifically target the skin microbiome. Commensal- and keratinocyte-derived AMPs (e.g. hBDs and LL-37) prevent/reduce pathogen colonization41,67. Therefore, emollients combined with AMPs may be an effect treatment for allergic diseases. In fact, the application of emollients containing bacteria-derived Sh-lantibiotics to individuals with AD reduced detectable S. aureus67. Emollients containing keratinocyte AMPs may also be useful to treat biofilms, since LL-37 was able to eradicate pre-existing MRSA biofilms in a wounded skin model without compromising keratinocyte function103. However, additional in vivo studies are needed to establish the utility and effectiveness of replenishing AMPs through emollients or other means.
Probiotics
An alternative to utilizing AMPs is the transplantation of microbes themselves. Living microorganisms that modulate the existing microbiota are known as probiotics104. The commensal Roseomonas mucosa isolated from healthy individuals inhibited S. aureus growth in vitro, and its application to the skin of an AD mouse model decreased S. aureus colonization and improved barrier function105. Furthermore, the use of topical R. mucosa in individuals with AD demonstrated that the probiotic use was associated with decreased disease severity and S. aureus burden with no adverse events or treatment complications106. Oral probiotics have modest effects in those with moderate-severe AD, but little effect in those with mild disease107. It will be interesting for future studies to assess if S. epidermidis or other commensals would be effective in oral or topical probiotics, and to determine the contribution of individual species or strains to improved AD outcomes.
Biologics
The type 2 cytokines IL-4 and IL-13 impact the skin microbiome by suppressing keratinocyte-derived AMPs and thus weakening microbial defenses. IL-4 and IL-13 also increase the efficacy of S. aureus alpha-toxin, ultimately perpetuating cell death108. The human monoclonal antibody dupilumab blocks the IL-4 and IL-13 receptor and, thus, reduces their effector functions. The AD-LIBERTY EXPLORE study subjected moderate-severe AD patients to either weekly treatments of dupilumab or a control and found that subjects treated with dupilumab showed increased microbial diversity and decreased S. aureus in both nonlesional and lesional skin109. Additional studies are necessary to elucidate the underlying mechanisms and determine the impact on other organisms.
As discussed above, the innate type 2 cytokine IL-33 is associated with both local and systemic allergic responses when released from epidermal keratinocytes in response to damage, allergens, or S. aureus colonization1,11,17. The downstream effects of IL-33 include increased IgE and decreased FLG expression. Etokimab is a humanized monoclonal antibody against IL-33; its efficacy was assessed in 12 adult patients with moderate-severe AD. Etokimab reduced AD severity by over 50% with only minimal adverse effects110. Although the trial did not directly examine etokimab’s impact on S. aureus colonization, anti-IL-33 therapy may be a useful to abrogate dysbiosis-induced inflammatory responses.
Can the Skin Biome Aid Diagnosis and Prevention
AD is a clinical diagnosis lacking biomarkers or diagnostic testing. Skin dysbiosis often occurs prior to the presentation of AD symptoms making the microbiome a potential diagnostic tool. Unfortunately, although the influence of pathogens on atopy has been demonstrated64,66,69,111–113, there are obstacles preventing the efficient, cost-effective, and reliable use of the microbiome for diagnostic purposes114. Currently, metagenomic analysis is expensive, non-quantitative, and difficult to interpret in a clinical laboratory setting, making it a poor diagnostic tool115. Additionally, despite the fact that S. aureus colonization has been associated with increased AD severity in many studies7,116, S. aureus colonization status does not influence current clinical practice62.
Primary prevention of AD is focused on maintenance of the skin barrier. The Barrier Enhancement for Eczema Prevention (BEEP) trial and the Prevention of Eczema By a Barrier Lipid Equilibrium Strategy (PEBBLES) trial are randomized control trials in which infants are subjected to standard skin care regimens or standard skin care in addition to a designated emollient117,118. The PEBBLES trial is distinguished by the use of an emollient based on the ratio of ceramides, cholesterol, and fatty acids118. Although no statistically significant effect was found, a trend suggests that the use of emollients early in life as prevention of AD needs to be explored further. The more recent PREVENTADALL trial found that emollient therapy did not prevent AD by the age of 12 months98. With the understanding that emollients can minimize S. aureus colonization119, it would be interesting to examine if shifts in the skin microbiome occur with emollient therapy. To our knowledge there are no primary preventions targeting the skin microbiome; therefore, future studies are needed to determine the associated benefits and risks of targeting the skin microbiome in both current and developing studies.
Conclusions
In summary, we have outlined recent literature regarding the influence of the skin microbiome on allergic disease. We discussed how commensal-epidermal homeostasis influences allergic pathologic processes in commensal-epidermal homeostasis. We then outlined the contributions of skin microbiome dysbiosis in the development of allergy and progression through the atopic march. Finally, we discussed how our improved understanding of the skin microbiome’s influence of allergy is being leveraged to develop therapeutic interventions to prevent the development and progression of allergy.
Supplementary Material
Funding Source:
This work was supported by National Institutes of Health grants U19 AI070235 (GKH, MGS, ABH), T32 GM063483–17 (SBD and TG), T32 ES010957 (SBD), R01 GM094363 (ABH), and K12 HD028827–27 (MGS).
Abbreviations/Acronyms:
- AD
Atopic Dermatitis
- AMP
Antimicrobial Peptide
- AR
Allergic Rhinitis
- DC
Dendritic Cell
- EH
Eczematicum Herpeticum
- FA
Fatty acid
- FLG
Filaggrin
- hBD
Human Beta-Defensin
- HDM
House dust mite
- HSV
Herpes Simplex Virus
- ILC
Innate Lymphoid Cell
- IgE
Immunoglobulin E
- LEKTI
Lympho‐epithelial Kazal-type-related Inhibitor
- LTA
Lipoteichoic Acid
- NMF
Natural Moisturizing Factor
- NS
Netherton Syndrome
- PSM
Phenol-Soluble Modulin
- SC
Stratum Corneum
- SG
Stratum Granulosum
- Th1
T-helper 1 Cell
- Th2
T-helper 2 Cell
- Th17
T-helper 17 Cell
- TJ
Tight Junctions
- TSLP
Thymic Stromal Lymphopoietin
Footnotes
Trial Registration: Not Applicable
Conflict of Interest Statement: None
References
- 1.Burks AW, Holgate ST, O’Hehir RE, et al. Middleton’s allergy: principles and practice. 2020. [Google Scholar]
- 2.Oh J, Byrd AL, Deming C, et al. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514(7520):59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cundell AM. Microbial Ecology of the Human Skin. Microb Ecol. 2018;76(1):113–120. [DOI] [PubMed] [Google Scholar]
- 4.Capone KA, Dowd SE, Stamatas GN, Nikolovski J. Diversity of the human skin microbiome early in life. J Invest Dermatol. 2011;131(10):2026–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ege MJ, Mayer M, Normand A-C, et al. Exposure to Environmental Microorganisms and Childhood Asthma. New England Journal of Medicine. 2011;364(8):701–709. [DOI] [PubMed] [Google Scholar]
- 6.Stevens ML GT, Schauberger E, Baatyrbek kyzy A, Andersen, SD H, Kalra MK, Martin LJ, Haslam D, Herr AB, Biagini Myers JM, Khurana Hershey GK,. Simultaneous skin biome and keratinocyte genomic capture reveals microbiome differences by depth of sampling Journal of Allergy and Clinical Immunology. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Totte JEE, Pardo LM, Fieten KB, et al. Nasal and skin microbiomes are associated with disease severity in paediatric atopic dermatitis. Br J Dermatol. 2019;181(4):796–804. [DOI] [PubMed] [Google Scholar]
- 8.Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6(4):328–340. [DOI] [PubMed] [Google Scholar]
- 9.Elias PM. The skin barrier as an innate immune element. Seminars in Immunopathology. 2007;29(1):3–14. [DOI] [PubMed] [Google Scholar]
- 10.Brandner JM, Zorn-Kruppa M, Yoshida T, Moll I, Beck LA, De Benedetto A. Epidermal tight junctions in health and disease. Tissue Barriers. 2014;3(1–2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thyssen JP, Kezic S. Causes of epidermal filaggrin reduction and their role in the pathogenesis of atopic dermatitis. Journal of Allergy and Clinical Immunology. 2014;134(4):792–799. [DOI] [PubMed] [Google Scholar]
- 12.Baurecht H, Rühlemann MC, Rodríguez E, et al. Epidermal lipid composition, barrier integrity, and eczematous inflammation are associated with skin microbiome configuration. J Allergy Clin Immunol. 2018;141(5):1668–1676.e1616. [DOI] [PubMed] [Google Scholar]
- 13.van Drongelen V, Haisma EM, Out-Luiting JJ, Nibbering PH, El Ghalbzouri A. Reduced filaggrin expression is accompanied by increased Staphylococcus aureus colonization of epidermal skin models. Clin Exp Allergy. 2014;44(12):1515–1524. [DOI] [PubMed] [Google Scholar]
- 14.McAleer MA, Irvine AD. The multifunctional role of filaggrin in allergic skin disease. J Allergy Clin Immunol. 2013;131(2):280–291. [DOI] [PubMed] [Google Scholar]
- 15.Biagini Myers JM, Sherenian MG, Baatyrbek Kyzy A, et al. Events in Normal Skin Promote Early-life Atopic Dermatitis - the MPAACH Cohort. J Allergy Clin Immunol Pract. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van den Oord RA, Sheikh A. Filaggrin gene defects and risk of developing allergic sensitisation and allergic disorders: systematic review and meta-analysis. Bmj. 2009;339:b2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imai Y, Yasuda K, Sakaguchi Y, et al. Skin-specific expression of IL-33 activates group 2 innate lymphoid cells and elicits atopic dermatitis-like inflammation in mice. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(34):13921–13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakabe J-i, Yamamoto M, Hirakawa S, et al. Kallikrein-related peptidase 5 functions in proteolytic processing of profilaggrin in cultured human keratinocytes. J Biol Chem. 2013;288(24):17179–17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kishibe M Physiological and pathological roles of kallikrein-related peptidases in the epidermis. J Dermatol Sci. 2019;95(2):50–55. [DOI] [PubMed] [Google Scholar]
- 20.Hovnanian A Netherton syndrome: skin inflammation and allergy by loss of protease inhibition. Cell Tissue Res. 2013;351(2):289–300. [DOI] [PubMed] [Google Scholar]
- 21.Oji V, Eckl K-M, Aufenvenne K, et al. Loss of corneodesmosin leads to severe skin barrier defect, pruritus, and atopy: unraveling the peeling skin disease. Am J Hum Genet. 2010;87(2):274–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams MR, Cau L, Wang Y, et al. Interplay of Staphylococcal and Host Proteases Promotes Skin Barrier Disruption in Netherton Syndrome. Cell Reports. 2020;30(9):2923–2933.e2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams MR, Nakatsuji T, Sanford JA, Vrbanac AF, Gallo RL. Staphylococcus aureus Induces Increased Serine Protease Activity in Keratinocytes. J Invest Dermatol. 2017;137(2):377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elias PM. Lipid abnormalities and lipid-based repair strategies in atopic dermatitis. Biochim Biophys Acta. 2014;1841(3):323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rippke F, Schreiner V, Doering T, Maibach HI. Stratum corneum pH in atopic dermatitis: impact on skin barrier function and colonization with Staphylococcus Aureus. American Journal of Clinical Dermatology. 2004;5(4):217–223. [DOI] [PubMed] [Google Scholar]
- 26.Baurecht H, Ruhlemann MC, Rodriguez E, et al. Epidermal lipid composition, barrier integrity, and eczematous inflammation are associated with skin microbiome configuration. J Allergy Clin Immunol. 2018;141(5):1668–1676 e1616. [DOI] [PubMed] [Google Scholar]
- 27.Drake DR, Brogden KA, Dawson DV, Wertz PW. Thematic review series: skin lipids. Antimicrobial lipids at the skin surface. J Lipid Res. 2008;49(1):4–11. [DOI] [PubMed] [Google Scholar]
- 28.Berdyshev E, Goleva E, Bronova I, et al. Lipid abnormalities in atopic skin are driven by type 2 cytokines. JCI Insight. 2018;3(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bäsler K, Bergmann S, Heisig M, Naegel A, Zorn-Kruppa M, Brandner JM. The role of tight junctions in skin barrier function and dermal absorption. Journal of Controlled Release. 2016;242:105–118. [DOI] [PubMed] [Google Scholar]
- 30.Runswick S, Mitchell T, Davies P, Robinson C, Garrod DR. Pollen proteolytic enzymes degrade tight junctions. Respirology. 2007;12(6):834–842. [DOI] [PubMed] [Google Scholar]
- 31.Bäsler K, Brandner JM. Tight junctions in skin inflammation. Pflugers Archiv: European Journal Of Physiology. 2017;469(1):3–14. [DOI] [PubMed] [Google Scholar]
- 32.Bergmann S, von Buenau B, Vidal-y-Sy S, et al. Claudin-1 decrease impacts epidermal barrier function in atopic dermatitis lesions dose-dependently. Scientific Reports. 2020;10(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9(4):244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Na H, Cho M, Chung Y. Regulation of Th2 Cell Immunity by Dendritic Cells. Immune Netw. 2016;16(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stier MT, Peebles RS Jr., Innate lymphoid cells and allergic disease. Ann Allergy Asthma Immunol. 2017;119(6):480–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Benedetto A, Kubo A, Beck LA. Skin barrier disruption: a requirement for allergen sensitization? J Invest Dermatol. 2012;132(3 Pt 2):949–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dürr UHN, Sudheendra US, Ramamoorthy A. LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2006;1758(9):1408–1425. [DOI] [PubMed] [Google Scholar]
- 38.van Harten RM, van Woudenbergh E, van Dijk A, Haagsman HP. Cathelicidins: Immunomodulatory Antimicrobials. Vaccines (Basel). 2018;6(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meade KG, O’Farrelly C. β-Defensins: Farming the Microbiome for Homeostasis and Health. Frontiers in Immunology. 2019;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakatsuji T, Chen TH, Narala S, et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med. 2017;9(378). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ommori R, Ouji N, Mizuno F, Kita E, Ikada Y, Asada H. Selective induction of antimicrobial peptides from keratinocytes by staphylococcal bacteria. Microbial Pathogenesis. 2013;56:35–39. [DOI] [PubMed] [Google Scholar]
- 42.Wang S, Song R, Wang Z, Jing Z, Wang S, Ma J. S100A8/A9 in Inflammation. Frontiers in Immunology. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naik S, Bouladoux N, Linehan JL, et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 2015;520(7545):104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bourgonje AR, von Martels JZH, de Vos P, Faber KN, Dijkstra G. Increased fecal calprotectin levels in Crohn’s disease correlate with elevated serum Th1- and Th17-associated cytokines. PLoS One. 2018;13(2):e0193202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hvid M, Vestergaard C, Kemp K, Christensen GB, Deleuran B, Deleuran M. IL-25 in atopic dermatitis: a possible link between inflammation and skin barrier dysfunction? J Invest Dermatol. 2011;131(1):150–157. [DOI] [PubMed] [Google Scholar]
- 46.Leyva-Castillo JM, Hener P, Jiang H, Li M. TSLP produced by keratinocytes promotes allergen sensitization through skin and thereby triggers atopic march in mice. J Invest Dermatol. 2013;133(1):154–163. [DOI] [PubMed] [Google Scholar]
- 47.Morita H, Arae K, Unno H, et al. IL-25 and IL-33 Contribute to Development of Eosinophilic Airway Inflammation in Epicutaneously Antigen-Sensitized Mice. PLOS ONE. 2015;10(7):e0134226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yockey LJ, Demehri S, Turkoz M, et al. The absence of a microbiota enhances TSLP expression in mice with defective skin barrier but does not affect the severity of their allergic inflammation. J Invest Dermatol. 2013;133(12):2714–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li M Current evidence of epidermal barrier dysfunction and thymic stromal lymphopoietin in the atopic march. European Respiratory Review. 2014;23(133):292–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vu AT, Baba T, Chen X, et al. Staphylococcus aureus membrane and diacylated lipopeptide induce thymic stromal lymphopoietin in keratinocytes through the Toll-like receptor 2-Toll-like receptor 6 pathway. J Allergy Clin Immunol. 2010;126(5):985–993, 993 e981–983. [DOI] [PubMed] [Google Scholar]
- 51.Linehan JL, Harrison OJ, Han S-J, et al. Non-classical immunity controls microbiota impact on skin immunity and tissue repair. Cell. 2018;172(4):784–796.e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tamoutounour S, Han SJ, Deckers J, et al. Keratinocyte-intrinsic MHCII expression controls microbiota-induced Th1 cell responses. Proc Natl Acad Sci U S A. 2019;116(47):23643–23652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ridaura VK, Bouladoux N, Claesen J, et al. Contextual control of skin immunity and inflammation by Corynebacterium. J Exp Med. 2018;215(3):785–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burian M, Bitschar K, Dylus B, Peschel A, Schittek B. The Protective Effect of Microbiota on S. aureus Skin Colonization Depends on the Integrity of the Epithelial Barrier. Journal of Investigative Dermatology. 2017;137(4):976–979. [DOI] [PubMed] [Google Scholar]
- 55.Harrison OJ, Linehan JL, Shih HY, et al. Commensal-specific T cell plasticity promotes rapid tissue adaptation to injury. Science. 2019;363(6422). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sparber F, Gregorio CD, Steckholzer S, et al. The Skin Commensal Yeast Malassezia Triggers a Type 17 Response that Coordinates Anti-fungal Immunity and Exacerbates Skin Inflammation. Cell Host & Microbe. 2019;25(3):389–403.e386. [DOI] [PubMed] [Google Scholar]
- 57.Gaitanis G, Magiatis P, Hantschke M, Bassukas ID, Velegraki A. The Malassezia Genus in Skin and Systemic Diseases. Clinical Microbiology Reviews. 2012;25(1):106–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nowicka D, Nawrot U. Contribution of Malassezia spp. to the development of atopic dermatitis. Mycoses. 2019;62(7):588–596. [DOI] [PubMed] [Google Scholar]
- 59.Hannigan GD, Meisel JS, Tyldsley AS, et al. The human skin double-stranded DNA virome: topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome. mBio. 2015;6(5):e01578–01515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhvania P, Hoyle NS, Nadareishvili L, Nizharadze D, Kutateladze M. Phage Therapy in a 16-Year-Old Boy with Netherton Syndrome. Front Med (Lausanne). 2017;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bantz SK, Zhu Z, Zheng T. The Atopic March: Progression from Atopic Dermatitis to Allergic Rhinitis and Asthma. J Clin Cell Immunol. 2014;5(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weidinger S, Novak N. Atopic dermatitis. Lancet. 2016;387(10023):1109–1122. [DOI] [PubMed] [Google Scholar]
- 63.Kennedy EA, Connolly J, Hourihane JO, et al. Skin microbiome before development of atopic dermatitis: Early colonization with commensal staphylococci at 2 months is associated with a lower risk of atopic dermatitis at 1 year. J Allergy Clin Immunol. 2017;139(1):166–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meylan P, Lang C, Mermoud S, et al. Skin Colonization by Staphylococcus aureus Precedes the Clinical Diagnosis of Atopic Dermatitis in Infancy. J Invest Dermatol. 2017;137(12):2497–2504. [DOI] [PubMed] [Google Scholar]
- 65.Kong HH, Oh J, Deming C, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22(5):850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Byrd AL, Deming C, Cassidy SKB, et al. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci Transl Med. 2017;9(397). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nakatsuji T, Chen TH, Narala S, et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Science Translational Medicine. 2017;9(378). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim J, Kim BE, Ahn K, Leung DYM. Interactions Between Atopic Dermatitis and Staphylococcus aureus Infection: Clinical Implications. Allergy Asthma Immunol Res. 2019;11(5):593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iwamoto K, Moriwaki M, Miyake R, Hide M. Staphylococcus aureus in atopic dermatitis: Strain-specific cell wall proteins and skin immunity. Allergology International. 2019;68(3):309–315. [DOI] [PubMed] [Google Scholar]
- 70.Brauweiler AM, Goleva E, Leung DYM. Staphylococcus aureus Lipoteichoic Acid Initiates a TSLP-Basophil-IL4 Axis in the Skin. J Invest Dermatol. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ege MJ. Exposure to Environmental Microorganisms and Childhood Asthma. n engl j med. 2011:9. [DOI] [PubMed] [Google Scholar]
- 72.Ramsey MM, Freire MO, Gabrilska RA, Rumbaugh KP, Lemon KP. Staphylococcus aureus Shifts toward Commensalism in Response to Corynebacterium Species. Front Microbiol. 2016;7:1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Furue M, Iida K, Imaji M, Nakahara T. Microbiome analysis of forehead skin in patients with atopic dermatitis and healthy subjects: Implication of Staphylococcus and Corynebacterium. J Dermatol. 2018;45(7):876–877. [DOI] [PubMed] [Google Scholar]
- 74.Han SH, Cheon HI, Hur MS, et al. Analysis of the skin mycobiome in adult patients with atopic dermatitis. Exp Dermatol. 2018;27(4):366–373. [DOI] [PubMed] [Google Scholar]
- 75.Glatz M, Buchner M, von Bartenwerffer W, et al. Malassezia spp.-specific immunoglobulin E level is a marker for severity of atopic dermatitis in adults. Acta Derm Venereol. 2015;95(2):191–196. [DOI] [PubMed] [Google Scholar]
- 76.Rowan-Nash AD, Korry BJ, Mylonakis E, Belenky P. Cross-Domain and Viral Interactions in the Microbiome. Microbiol Mol Biol Rev. 2019;83(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Seegraber M, Worm M, Werfel T, et al. Recurrent eczema herpeticum - a retrospective European multicenter study evaluating the clinical characteristics of eczema herpeticum cases in atopic dermatitis patients. J Eur Acad Dermatol Venereol. 2019. [DOI] [PubMed] [Google Scholar]
- 78.Bin L, Kim BE, Brauweiler A, et al. Staphylococcus aureus α-toxin modulates skin host response to viral infection. J Allergy Clin Immunol. 2012;130(3):683–691.e682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Traidl S, Kienlin P, Begemann G, et al. Patients with atopic dermatitis and history of eczema herpeticum elicit herpes simplex virus-specific type 2 immune responses. J Allergy Clin Immunol. 2018;141(3):1144–1147 e1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Damevska K, Emurlai A. Molluscum Contagiosum in a Patient with Atopic Dermatitis. N Engl J Med. 2017;377(21):e30. [DOI] [PubMed] [Google Scholar]
- 81.Manti S, Amorini M, Cuppari C, et al. Filaggrin mutations and Molluscum contagiosum skin infection in patients with atopic dermatitis. Ann Allergy Asthma Immunol. 2017;119(5):446–451. [DOI] [PubMed] [Google Scholar]
- 82.von Sonnenburg F, Perona P, Darsow U, et al. Safety and immunogenicity of modified vaccinia Ankara as a smallpox vaccine in people with atopic dermatitis. Vaccine. 2014;32(43):5696–5702. [DOI] [PubMed] [Google Scholar]
- 83.He Y, Sultana I, Takeda K, Reed JL. Cutaneous Deficiency of Filaggrin and STAT3 Exacerbates Vaccinia Disease In Vivo. PLoS One. 2017;12(1):e0170070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Greenberg RN, Hurley MY, Dinh DV, et al. A Multicenter, Open-Label, Controlled Phase II Study to Evaluate Safety and Immunogenicity of MVA Smallpox Vaccine (IMVAMUNE) in 18–40 Year Old Subjects with Diagnosed Atopic Dermatitis. PLoS One. 2015;10(10):e0138348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bird JA, Lack G, Perry TT. Clinical management of food allergy. J Allergy Clin Immunol Pract. 2015;3(1):1–11; quiz 12. [DOI] [PubMed] [Google Scholar]
- 86.Tsilochristou O, du Toit G, Sayre PH, et al. Association of Staphylococcus aureus colonization with food allergy occurs independently of eczema severity. J Allergy Clin Immunol. 2019;144(2):494–503. [DOI] [PubMed] [Google Scholar]
- 87.Cruz AA, Popov T, Pawankar R, et al. Common characteristics of upper and lower airways in rhinitis and asthma: ARIA update, in collaboration with GA(2)LEN. Allergy. 2007;62 Suppl 84:1–41. [DOI] [PubMed] [Google Scholar]
- 88.Kozik AJ, Huang YJ. The microbiome in asthma: Role in pathogenesis, phenotype, and response to treatment. Ann Allergy Asthma Immunol. 2019;122(3):270–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wrobel J, Tomczak H, Jenerowicz D, Czarnecka-Operacz M. Skin and nasal vestibule colonisation by Staphylococcus aureus and its susceptibility to drugs in atopic dermatitis patients. Ann Agric Environ Med. 2018;25(2):334–337. [DOI] [PubMed] [Google Scholar]
- 90.Reddel HK, FitzGerald JM, Bateman ED, et al. GINA 2019: a fundamental change in asthma management: Treatment of asthma with short-acting bronchodilators alone is no longer recommended for adults and adolescents. Eur Respir J. 2019;53(6). [DOI] [PubMed] [Google Scholar]
- 91.Naik S, Bouladoux N, Linehan JL, et al. Commensal–dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 2015;520(7545):104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Prescott SL, Larcombe DL, Logan AC, et al. The skin microbiome: impact of modern environments on skin ecology, barrier integrity, and systemic immune programming. World Allergy Organ J. 2017;10(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Maier RM, Palmer MW, Andersen GL, et al. Environmental determinants of and impact on childhood asthma by the bacterial community in household dust. Appl Environ Microbiol. 2010;76(8):2663–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ludwig S, Jimenez-Bush I, Brigham E, et al. Analysis of home dust for Staphylococcus aureus and staphylococcal enterotoxin genes using quantitative PCR. Sci Total Environ. 2017;581–582:750–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dzoro S, Mittermann I, Resch-Marat Y, et al. House dust mites as potential carriers for IgE sensitization to bacterial antigens. Allergy. 2018;73(1):115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.von Mutius E The microbial environment and its influence on asthma prevention in early life. J Allergy Clin Immunol. 2016;137(3):680–689. [DOI] [PubMed] [Google Scholar]
- 97.Leonel C, Sena IFG, Silva WN, et al. Staphylococcus epidermidis role in the skin microenvironment. J Cell Mol Med. 2019;23(9):5949–5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Skjerven HO, Rehbinder EM, Vettukattil R, et al. Skin emollient and early complementary feeding to prevent infant atopic dermatitis (PreventADALL): a factorial, multicentre, cluster-randomised trial. The Lancet. 2020. [DOI] [PubMed] [Google Scholar]
- 99.Eichenfield LF, Tom WL, Chamlin SL, et al. Guidelines of care for the management of atopic dermatitis: section 1. Diagnosis and assessment of atopic dermatitis. J Am Acad Dermatol. 2014;70(2):338–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Glatz M, Jo JH, Kennedy EA, et al. Emollient use alters skin barrier and microbes in infants at risk for developing atopic dermatitis. PLoS One. 2018;13(2):e0192443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Inoue Y, Nakagawara R, Kambara T, et al. Prevalence of atopic dermatitis in Japanese infants treated with moisturizer since birth and its relation to FLG mutations. Eur J Dermatol. 2013;23(2):288–289. [DOI] [PubMed] [Google Scholar]
- 102.Smits JPH, Ederveen THA, Rikken G, et al. Targeting the Cutaneous Microbiota in Atopic Dermatitis by Coal Tar via AHR-Dependent Induction of Antimicrobial Peptides. J Invest Dermatol. 2019. [DOI] [PubMed] [Google Scholar]
- 103.Haisma EM, de Breij A, Chan H, et al. LL-37-derived peptides eradicate multidrug-resistant Staphylococcus aureus from thermally wounded human skin equivalents. Antimicrob Agents Chemother. 2014;58(8):4411–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chang YS, Trivedi MK, Jha A, Lin YF, Dimaano L, Garcia-Romero MT. Synbiotics for Prevention and Treatment of Atopic Dermatitis: A Meta-analysis of Randomized Clinical Trials. JAMA Pediatr. 2016;170(3):236–242. [DOI] [PubMed] [Google Scholar]
- 105.Myles IA, Williams KW, Reckhow JD, et al. Transplantation of human skin microbiota in models of atopic dermatitis. JCI Insight. 2016;1(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Myles IA, Earland NJ, Anderson ED, et al. First-in-human topical microbiome transplantation with Roseomonas mucosa for atopic dermatitis. JCI Insight. 2018;3(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhao M, Shen C, Ma L. Treatment efficacy of probiotics on atopic dermatitis, zooming in on infants: a systematic review and meta-analysis. Int J Dermatol. 2018;57(6):635–641. [DOI] [PubMed] [Google Scholar]
- 108.Li R, Hadi S, Guttman-Yassky E. Current and emerging biologic and small molecule therapies for atopic dermatitis. Expert Opin Biol Ther. 2019;19(4):367–380. [DOI] [PubMed] [Google Scholar]
- 109.Callewaert C, Nakatsuji T, Knight R, et al. IL-4Ralpha Blockade by Dupilumab Decreases Staphylococcus aureus Colonization and Increases Microbial Diversity in Atopic Dermatitis. J Invest Dermatol. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen YL, Gutowska-Owsiak D, Hardman CS, et al. Proof-of-concept clinical trial of etokimab shows a key role for IL-33 in atopic dermatitis pathogenesis. Sci Transl Med. 2019;11(515). [DOI] [PubMed] [Google Scholar]
- 111.Park KD, Pak SC, Park KK. The Pathogenetic Effect of Natural and Bacterial Toxins on Atopic Dermatitis. Toxins (Basel). 2016;9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Paller AS, Kong HH, Seed P, et al. The microbiome in patients with atopic dermatitis. J Allergy Clin Immunol. 2019;143(1):26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ong PY, Leung DY. Bacterial and Viral Infections in Atopic Dermatitis: a Comprehensive Review. Clin Rev Allergy Immunol. 2016;51(3):329–337. [DOI] [PubMed] [Google Scholar]
- 114.Reiger M, Traidl-Hoffmann C, Neumann AU. The skin microbiome as a clinical biomarker in atopic eczema: Promises, navigation, and pitfalls. J Allergy Clin Immunol. 2020;145(1):93–96. [DOI] [PubMed] [Google Scholar]
- 115.Greninger AL. The challenge of diagnostic metagenomics. Expert Rev Mol Diagn. 2018;18(7):605–615. [DOI] [PubMed] [Google Scholar]
- 116.Wichmann K, Uter W, Weiss J, et al. Isolation of alpha-toxin-producing Staphylococcus aureus from the skin of highly sensitized adult patients with severe atopic dermatitis. Br J Dermatol. 2009;161(2):300–305. [DOI] [PubMed] [Google Scholar]
- 117.Chalmers JR, Haines RH, Bradshaw LE, et al. Daily emollient during infancy for prevention of eczema: the BEEP randomised controlled trial. Lancet. 2020;395(10228):962–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lowe A, Su J, Tang M, et al. PEBBLES study protocol: a randomised controlled trial to prevent atopic dermatitis, food allergy and sensitisation in infants with a family history of allergic disease using a skin barrier improvement strategy. BMJ Open. 2019;9(3):e024594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Seite S, Zelenkova H, Martin R. Clinical efficacy of emollients in atopic dermatitis patients - relationship with the skin microbiota modification. Clin Cosmet Investig Dermatol. 2017;10:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


