Abstract
In recent years, many neuroimaging studies have applied artificial intelligence (AI) to facilitate existing challenges in Parkinson's disease (PD) diagnosis, prognosis, and intervention. The aim of this systematic review was to provide an overview of neuroimaging‐based AI studies and to assess their methodological quality. A PubMed search yielded 810 studies, of which 244 that investigated the utility of neuroimaging‐based AI for PD diagnosis, prognosis, or intervention were included. We systematically categorized studies by outcomes and rated them with respect to five minimal quality criteria (MQC) pertaining to data splitting, data leakage, model complexity, performance reporting, and indication of biological plausibility. We found that the majority of studies aimed to distinguish PD patients from healthy controls (54%) or atypical parkinsonian syndromes (25%), whereas prognostic or interventional studies were sparse. Only 20% of evaluated studies passed all five MQC, with data leakage, non‐minimal model complexity, and reporting of biological plausibility as the primary factors for quality loss. Data leakage was associated with a significant inflation of accuracies. Very few studies employed external test sets (8%), where accuracy was significantly lower, and 19% of studies did not account for data imbalance. Adherence to MQC was low across all observed years and journal impact factors. This review outlines that AI has been applied to a wide variety of research questions pertaining to PD; however, the number of studies failing to pass the MQC is alarming. Therefore, we provide recommendations to enhance the interpretability, generalizability, and clinical utility of future AI applications using neuroimaging in PD. © 2024 The Author(s). Movement Disorders published by Wiley Periodicals LLC on behalf of International Parkinson and Movement Disorder Society.
Keywords: artificial intelligence, quality control, Parkinson's disease, neuroimaging

Parkinson's disease (PD) is an age‐related neurodegenerative disease associated with debilitating motor deficits but also accompanied by nonmotor symptoms. 1 Approximately 1% of the elderly population develops PD, hallmarked by a gradual loss of dopaminergic neurons in the substantia nigra. 2 Diagnosis relies on clinically detectable motor symptoms, with the Movement Disorder Society‐Unified Parkinson's Disease Rating Scale (MDS‐UPDRS) serving as a clinical tool to assess motor (II–IV) and nonmotor (I) symptoms. 3 Despite the high prevalence of PD, efficient early or differential diagnoses, prognoses, and treatment strategies remain a major clinical challenge. The heterogeneous disease phenotype and overlapping symptoms with atypical parkinsonian syndromes (APS) contribute to high misdiagnosis rates 4 , 5 and hamper accurate prognoses. Moreover, by the time of a PD diagnosis, up to 70% of dopaminergic neurons are already degenerated, 2 , 6 significantly reducing the potential effectiveness of intervention therapies. For this reason, various neuroimaging modalities were proposed and validated to enhance diagnostic accuracy. 7 , 8 , 9 Unfortunately, interpreting neuroimaging data, especially when considering additional risk factors for understanding individual PD trajectories, can be a highly complex task and may require advanced automated solutions.
“Artificial intelligence” (AI) is an umbrella term for powerful tools (eg, from machine or deep learning) capable of integrating and interpreting the information of diverse clinical datasets. AI can be defined as the ability of computers to learn and perform tasks without specific instructions based on data patterns. 10 In recent years, AI has garnered increasing attention in the field of neuroimaging in PD, particularly with increasing availability of data and advancing computational resources. With their strong potential for detecting complex associations within multidimensional (eg, imaging) and/or multimodal data (eg, incorporating imaging, genetic, and demographic data), AI can aid diagnosis, estimate disease course trajectories, or contribute to treatment planning. This can pave the way for more patient‐tailored (eg, precision medicine) rather than uniform approaches in PD. 11
However, with the ever‐increasing number of studies using neuroimaging‐based AI techniques in PD, maintaining an overview of potentially helpful applications for daily practice is becoming more challenging. It has been repeatedly recommended that quality standards be ensured among medical AI studies 12 , 13 , 14 to achieve generalizable results and interpretable decision‐making. Without proper regulation, overfitting can cause issues of generalizability. Overfitting occurs when AI algorithms capture noise that is specific to the training data, resulting in strong performance during training but poor performance at the validation stage and real‐world settings. 15 Another common problem is data leakage, which happens when information from data used for validation is introduced into the training process. 16 , 17 , 18 Additionally, overly complex approaches that are trained on a relatively large number of features are prone to overfitting as they are likely to fit noise in the training data rather than disease‐specific patterns. 19 , 20 Incomplete reporting of model performance further prevents interpretability of a model's strengths and pitfalls. 20 Together, these (mal)practices have likely impeded the implementation of AI into clinical routines. Although these issues are well known, the quality of neuroimaging‐based AI studies in PD has not yet been systematically analyzed.
Therefore, the aims of this systematic review were threefold: (1) to compile a comprehensive overview of AI applications based on neuroimaging in PD, (2) to evaluate the quality of existing AI research studies using minimal quality criteria (MQC), and (3) to suggest actionable recommendations to enhance the quality of medical research utilizing AI.
Methods
All decisions were performed by two independent raters; in cases of disagreement consensus was reached by discussion. This review was not registered.
PubMed Search
A PubMed search was conducted on January 1, 2024, with the search term shown in Figure 1 (full search term available in the Supplementary Materials, M1), yielding 810 articles. The search term was deliberately broad to capture an inclusive range of literature. We established the following exclusion criteria based on consultations with experts in the fields of PD/neurology (T.E.) and AI (E.D.): articles had to be original research articles (eg, no reviews or case reports), written in English, and not be retracted. Moreover, the study had to be done in humans, and PD had to be a stand‐alone diagnosis (ie, PD could not be grouped with other diseases as “parkinsonian syndromes”). Similarly, neuroimaging had to be the primary input modality for the AI model, which needed to adhere to our definition of AI and serve a clinically relevant research aim. Consequently, studies on AI‐based preprocessing or symptom classification and those lacking a defined outcome were excluded.
FIG. 1.
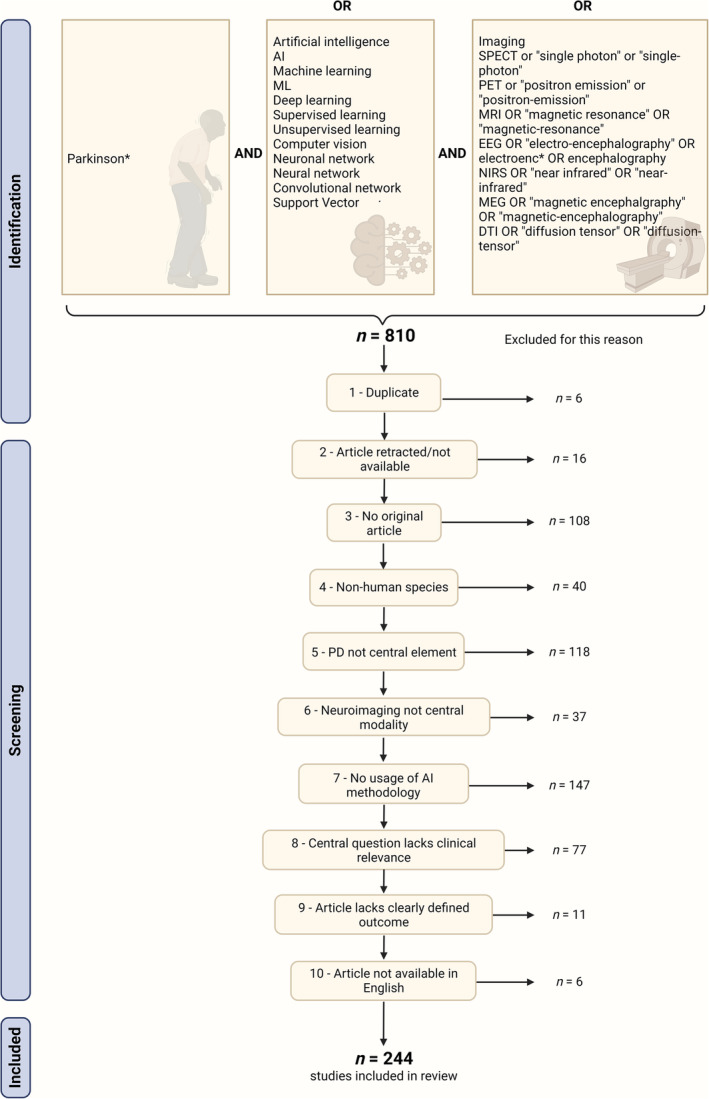
Visualization of PubMed search term and the respective exclusion criteria for current systematic review. Top: the search term incorporated three main domains with the respective possible expressions: (1) the clinical condition of interest (Parkinson's disease [PD]), (2) the techniques of interest (artificial intelligence [AI]), and (3) the neuroimaging modality (eg, MRI [magnetic resonance imaging]). Bottom: exclusion criteria and number of studies failing the criteria. [Color figure can be viewed at wileyonlinelibrary.com]
Categorization of Research
According to their aims, papers were categorized into diagnosis, prognosis, and intervention, with some papers fitting into multiple categories.
The first category was diagnosis, which was further subdivided into six subcategories:
Classification of PD versus healthy controls (HC, ie, a cohort without neurological impairment)
Classification of PD versus APS (where PD was a distinct group)
Identification of PD subtypes (eg, clinically established subtypes like tremor dominant or new biological PD subtypes; biological subtypes needed to be related to clinically relevant explanatory variables like genetic variants)
Disease staging and symptom severity estimation
Classification of cognitive impairment
Classification of freezing of gait (FOG)
The second category was prognosis. Four subcategories were defined:
Prediction of conversion to PD (ie, from prodromal to clinical PD)/monitoring of PD (ie, changes from clinical to biomarker‐confirmed diagnosis)
Prediction of disease and symptom severity
Prediction of symptom occurrence
Conversion to PD dementia
The third category was therapeutic interventions. Two subcategories were formed:
Drug therapy
Deep brain stimulation (DBS)
MQC
We defined MQC and applied them to all included papers. The criteria were based on commonly applied evaluation methods to assess medical AI approaches and cover validity of methods at the implementation level (MQC 1–3) and diligence of reporting at the interpretation level (MQC 4 and 5):
Train/test split: data were split into separate cohorts for training and evaluating the model. This criterion was fulfilled if (at least) a training and a test set existed, or if cross‐validation was applied. This criterion did not apply to unsupervised algorithms.
No data leakage: no information from the test set or target variable was available to train the algorithm or transform the training data (eg, this criterion was not passed when feature selection was based on the whole dataset rather than only on training data). 16 , 17 , 18
Minimal model complexity: the number of features had to be smaller than that of individuals, or samples included in the training set. 19 , 20 This criterion did not apply to deep learning–based models.
Generalizability: test or average cross‐validation performance was reported to infer model performance on unseen data, 16 without selective reporting of favorable results. This criterion did not apply to unsupervised algorithms.
Indication of biological plausibility: outlining how strongly features contributed to the model's performance to ensure that these features can be interpreted in a biologically meaningful context. 20
MQC could be rated as “passed” (✓), “failed” (✗), “not applicable” (NA), or “unclear” (?). Unclear indicated that the raters could not rate the MQC due to the paper lacking adequate information. MQC were evaluated only for algorithms specific to the research aim of the respective study. The full MQC details are presented in the Supplementary Materials (section M3).
Meta‐Data
Next to assessing the quality of included studies, we also recorded the aim (ie, outcome), modality, best‐performing model, most effective features, sample size, and overall performance of each study's best model. For comparability, we reported accuracy (the most common metric), where available, and otherwise area under the curve (AUC) for classification; and r, r2, or the mean absolute error (MAE) for regression approaches. If cross‐validation and test results were available, according to best practices, we reported the test performance of the model that performed best during cross‐validation. Although reporting of performance on external test sets demonstrates generalizability, only a few studies did so. Therefore, we reported performance on test (ie, validation data derived from the same cohort as the training data) and external test set (ie, additional validation data derived from a different cohort than the training data) when available. For comparability, we specifically noted usage of Parkinson's Progression Markers Initiative (PPMI) data.
Statistical Analysis
Statistical analyses were conducted per study (ie, each unique digital object identifier, n = 244), per subcategory entry (ie, counting a study allocated to two subcategories as two unique subcategory entries, n = 284), or per study aim (ie, counting each of a study's aims within or across subcategories, n = 327). For an example, see Supplementary Materials, section M4. Within the “classification of PD vs. HC” subcategory, we analyzed whether model accuracy was different between model types or modalities using a t test weighted by sample size (at the level of study aims). Only studies reporting an accuracy were included in this analysis.
To assess the methodological quality of studies, we evaluated the percentage of studies meeting MQC at the subcategory level (the same study never differed in MQC across study aims within one category). To assess whether recency or journal quality influences MQC adherence, we assessed frequencies of MQC adherence by year and impact factors, respectively, across subcategory entries. Impact factors were retrieved via Clarivate's Journal Citation Reports (https://jcr.clarivate.com/jcr/) for the publication year and arranged into quantiles. Finally, we examined which MQC indicate overfitting by comparing the accuracies of study aims (noting that accuracies might vary across multiple aims within the same subcategory) that passed versus those that did not pass each implementation‐level MQC (MQC 1–3). This comparison was made using a 1‐tailed t test (test: “failed”/“unclear” > “yes”).
Bias Assessment
Our bias assessment strategy comprehensively addressed three critical dimensions: comparability of included studies, generalizability of results, and reproducibility of findings.
First, when retrieved studies were screened, we included only studies with a clearly defined AI method, with neuroimaging features, in a distinct cohort of clinically diagnosed PD patients, to address clinically relevant diagnostic, prognostic, or interventional aims. Whereas studies without neuroimaging features were excluded, those that complemented neuroimaging with other clinical features were not excluded. Second, we investigated the generalizability of study results by applying defined MQC, as described in the “PD‐ versus HC‐Specific Results” section, and by recording sample sizes. We expanded the investigation of generalizability beyond our five MQC by documenting metrics (balanced accuracy or sensitivity + specificity) or practices (eg, subsampling) appropriate for handling imbalanced data. 21 We assessed whether external test sets were used for validation and analyzed accuracy drops between test and external test sets using a paired t test. Finally, reproducibility with respect to study setup was scrutinized, including positron emission tomography (PET) tracer and scanner type, magnetic resonance imaging (MRI) scanner type, field strength, sequence, and electrode system and recorder type for electroencephalography (EEG) and magnetoencephalography (MEG). In the cases of functional imaging, information regarding activity (ie, resting or task) and eye status (ie, eyes open or closed) was required.
Results
Identification of Studies for Current Review
We included 244 of the initially retrieved 810 studies in the current systematic review (Fig. 1). Most commonly, we excluded articles that were not original (n = 108), did not focus on PD/include PD as a separate entity (n = 118), or did not use AI methodology (n = 147).
Description of the Categorization and Content of Studies
A total of 86% of studies reported at least one diagnostic aim, and 8% and 9% reported a prognostic and an interventional aim, respectively. In total, 284 papers were allocated to appropriate subcategories (several subcategories per study were possible). PD versus HC was the most prominent subcategory, with 54% (n = 131) of studies assigned to this aim, followed by PD versus APS (25% of studies, n = 60). Much fewer studies were categorized as diagnostic, prognostic, or interventional (<8% of studies per subcategory, n ≤ 18; Fig. 2). All studies in the individual subcategories, including outcomes, implementation details, and performance, along with the MQC ratings can be accessed in the Supplementary Materials, Tables R3–R14 (diagnosis: R3–R8, prognosis: R9–R12, intervention: R13–R14). The most frequently used model types were support vector machines (SVMs, 46% of studies, n = 131), convolutional neural networks (CNNs, 17%, n = 47), and ensemble learning (14%, n = 39). 88% (n = 250) of subcategory entries used unimodal, and 12% (n = 34) used multimodal neuroimaging data as input.
FIG. 2.
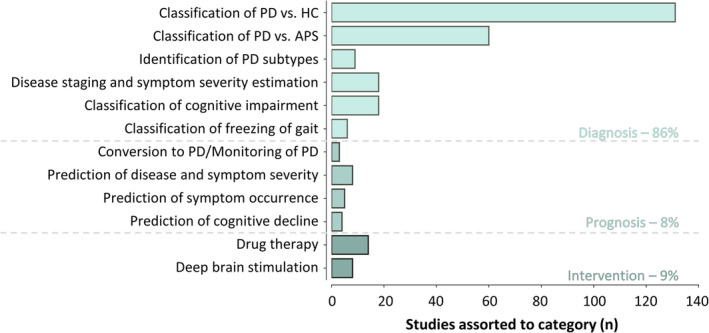
Quantity of studies categorized under diagnosis, prognosis, and intervention, including subcategories. Some studies had multiple aims and were thus assorted to several subcategories. APS, atypical parkinsonian syndromes; HC, healthy control; PD, parkinson's disease [Color figure can be viewed at wileyonlinelibrary.com]
Diagnosis
Studies categorized under “classification of PD versus HC” achieved accuracies of up to 100%, 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 which is further discussed in section "PD‐ versus HC‐Specific Results". The subcategory “classification of PD versus APS” included mainly binary approaches (67%), aiming to differentiate PD from specific or multiple combined other APS. In a comprehensive study, Huppertz et al demonstrated that PD can be distinguished not only from individual APS (multiple systems atrophy‐cerebellar type, multiple systems atrophy‐parkinsonian type, progressive supranuclear palsy‐richardson syndrome) dichotomously (balanced accuracy ≥85%) but also in a multiclass paradigm (all class accuracies ≥75%). 30 Another study utilized an ensemble of probabilistic binary classifiers to build a multiclass paradigm, which successfully distinguished between PD and APS forms (accuracy = 88%–94%). 31 Studies labeled as “identification of PD subtypes” applied either supervised methods to classify known clinical phenotypes of PD (eg, postural/gait vs. tremor dominant) 32 or unsupervised methods to identify new subtypes associated with the clinical characteristics of PD. 33 Moreover, multiple studies demonstrated the potential of using AI for “disease staging and symptom severity estimation,” with accuracies of up to 99% for classifying different stages (Hoehn & Yahr) 34 and correlation coefficients of up to 0.75 between true and estimated current symptom severity. 35 In the “classification of cognitive impairment” subcategory, studies showed that clinical stages of cognitive impairment can be classified with high accuracy (accuracy = 92%–100%), 34 , 36 , 37 , 38 , 39 and severity of global cognitive impairment (Mini‐Mental State Examination) can be estimated (r = 0.55). 40 Finally, studies demonstrated the potential of using AI on EEG data for real‐time “classification of freezing of gait” with accuracies of up to 86%. 41 , 42 In summary, neuroimaging‐based AI methods accurately captured different clinical aspects and may support differential diagnosis, disease subtyping, and staging in PD.
Prognosis
Two studies categorized under “conversion to PD/monitoring of PD” outlined that [123I]‐FP‐CIT SPECT scans may contain information to determine if subjects without evidence of dopaminergic deficit (SWEDD) will be recategorized to biomarker‐confirmed PD in the future. 43 , 44 Moreover, CNNs based on [18F]‐FDG‐PET may predict whether individuals with rapid eye movement behavioral disorder (RBD) will progress to PD with moderate accuracy (AUC = 72%). 45 Similarly, multiple studies labeled with “prediction of disease and symptom severity” predicted either motor symptom progression after 4 years (UPDRS‐III decline, MAE ≤4.7) 46 , 47 or overall disease progression after 3 years (Hoehn & Yahr stages, accuracy = 84%). 48 Additionally, five studies investigated “prediction of symptom occurrence.” Three studies predicted (accuracy = 73%–77%) the occurrence of FOG within 5 seconds based on EEG. 49 , 50 , 51 Prediction into the more distant future was achieved for the development of FOG 52 or RBD. 53 Finally, the development of mild cognitive impairment (accuracy = 87%) 54 or the conversion from mild cognitive impairment to dementia (accuracy = 74% 55 /AUC = 88% 56 ) was anticipated using imaging in the “prediction of cognitive decline” subcategory. To summarize, neuroimaging‐based AI anticipated short‐ and long‐term symptom and disease development, illustrating great potential for prognosis and intervention.
Intervention
Studies in the subcategory “drug therapy” used neuroimaging‐based AI models to identify characteristic changes in brain activity (EEG/MEG) in response to dopaminergic treatment, 25 , 57 , 58 , 59 , 60 to classify or predict the occurrence of levodopa (l‐dopa)–induced dyskinesias, 61 , 62 , 63 or to classify or predict motor improvement from l‐dopa intake. 33 , 64 , 65 , 66 , 67 Similarly, UPDRS‐III improvement after “deep brain stimulation” was highly accurately classified 68 , 69 or even predicted from preoperative data 70 in several studies. 70 , 71 Notably, SVM based on preoperative T1‐weighted MRI and demographic data predicted motor, cognitive, and behavioral improvements after DBS. 72 Moreover, AI showed potential for the localization of stimulation sites from EEG, 73 functional MRI (fMRI), 74 or microelectrode recordings. 75
PD‐ versus HC‐Specific Results
Because PD versus HC was the most studied aim, we considered all 113 studies reporting accuracy in this subcategory to further explore which models and modalities best captured PD‐specific brain changes. Frequently used model types were SVMs (41% of studies), CNN (24%), ensemble learning (14%), and logistic regression (7%), and their mean sample sizes were 155, 466, 213, and 204 individuals, respectively. Weighted by sample size, ensemble learning methods (μweighted = 94.1%, SDweighted = 4.9%) outperformed other methods in the classification of PD, as assessed using weighted t tests with Bonferroni correction (Fig. 3A; Supplementary Materials Figure R1 and Table R1). The most frequently used modalities were T1‐weighted MRI (20% of studies), EEG (17%), [123I]‐FP‐CIT SPECT (13%), and resting‐state fMRI (10%), with average sample sizes of 354, 73, 662, and 102 individuals, respectively. Studies using [123I]‐FP‐CIT SPECT (μweighted = 97.4%, SD = 2.1%) were significantly more accurate compared to any other modality after Bonferroni correction (all P < 0.005; Fig. 3B; Supplementary Materials Figure R2 and Table S2).
FIG. 3.
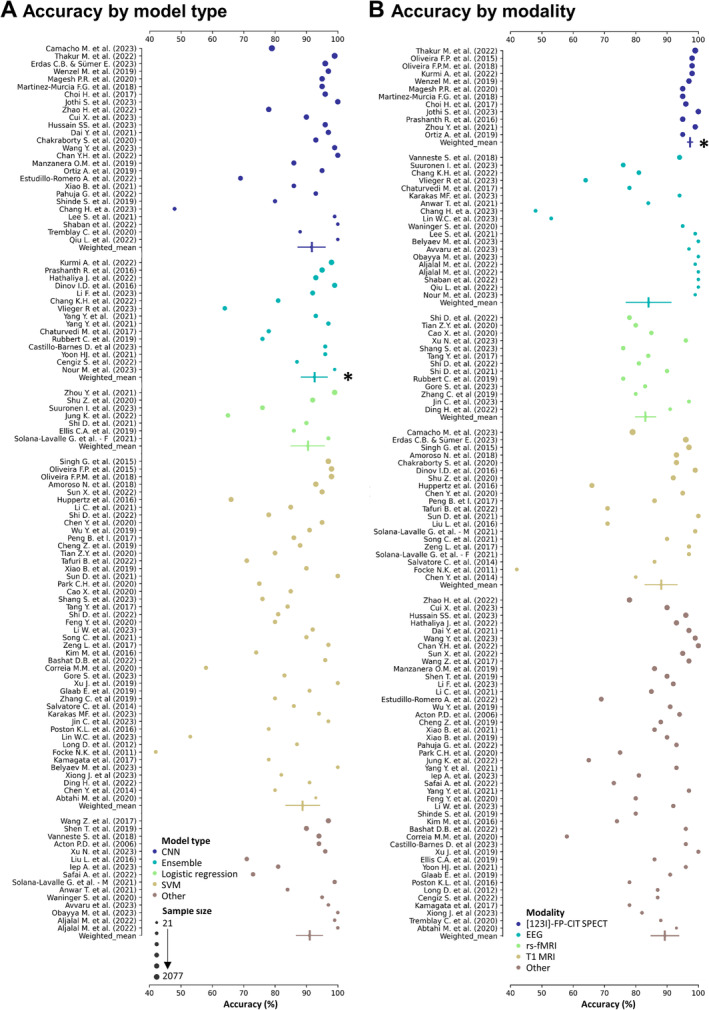
Accuracy of PD (Parkinson's disease) versus HC (healthy control) classification studies by (A) model type and (B) modality. (A) Accuracy of studies, sorted by model type (color) and sample size (size). Model types are sorted in alphabetical order. Studies of the same model type were also summarized to a weighted mean and weighted standard deviation, with sample size serving as weights (indicated by vertical lines). *Ensemble models performed significantly better (surviving Bonferroni correction, P < 0.005) than the remaining model types. (B) Accuracy of studies, sorted by modality type (color) and sample size (size, same as in A). Modalities are sorted in alphabetical order. Studies of the same modality were also summarized to a weighted mean and weighted standard deviation, with sample size serving as weights. *Significant differences (surviving Bonferroni correction, 10 comparisons ‐ P < 0.005) were detected between accuracies obtained using [123I]‐FP‐CIT SPECT and any other modality using weighted t tests. Notes for (A) and (B): sample sizes ranged from 21 to 2077 and were log transformed for plotting. Legend shows true (nontransformed) sample sizes. Only studies that reported accuracy were considered. If ranges were reported by studies, this figure shows the mean accuracy of these ranges. Only four most frequent modalities/models are shown explicitly, whereas the remaining ones were grouped as “others.” Studies of all model and modality types (including those summarized as “other”) are shown in Supplementary Materials Figures 1 and 2. CNN, convolutional neural network; SVM, support vector machine.
Differences in accuracy between other modalities were not significant. However, most (15 of 17) [123I]‐FP‐CIT studies used PPMI data, where (ab)normality is among the inclusion/exclusion criteria for both groups, leading to a strong circularity. The remaining two studies used clinically diagnosed PD patients without biomarker confirmation and still yielded accuracies of 96% 76 and 100%, 77 respectively. Supplementary Materials section R1 exhibits the performance of all model or modality types, that is, including those summarized as “other.”
MQC Results
Only 20% of subcategory entries passed all MQC, with MQC 2 (no data leakage, 49% passed), MQC 5 (biological plausibility, 57%), and MQC 3 (minimal model complexity, 57%) passed least frequently (Fig. 4A). MQC 1 and 4 were passed by 95% and 92% of subcategory entries, respectively. Failure to pass implementation‐level MQC (MQC 1–3, 33% passed) was more frequently observed compared to failure to pass interpretation‐level MQC (MQC 4 and 5, 53% passed). Most subcategory entries passed between three and four MQC (Fig. 4B).
FIG. 4.
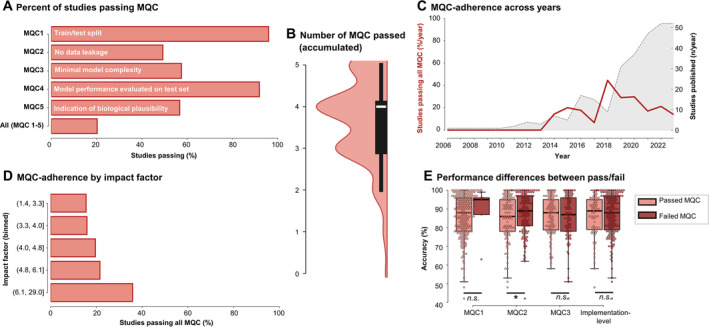
Minimal quality criteria (MQC) analysis. (A) Percentage of studies passing individual or all MQC. (B) Violin plot of the accumulated number of MQC passed by each individual study. (C) Number of total studies published (gray shaded area) and percentage of studies passing all MQC (red line) over the years. (D) Percentage of studies passing all MQC by impact factor. (E) Differences in accuracy between studies passing or failing individual or all implementation‐level MQC. [Color figure can be viewed at wileyonlinelibrary.com]
MQC 2 frequently failed due to test data influencing feature selection, for example, because feature selection was performed prior to splitting the data into training and test sets, prior to cross‐validation, or as recursive feature elimination in the absence of a test set. In studies where multiple samples were acquired per subject, such as EEG and fMRI, MQC 2 was also often failed when authors did not report whether data of the same subject was exclusively allocated to the training or test set. MQC 3 was failed in numerous cases due to incomplete descriptions of feature extraction/selection workflows. MQC 5 was failed in many cases where authors listed the selected features before model training but did not explain their importance to the final model performance (eg, through feature weights or permutation testing).
Investigating study quality over time, we observed that there was no increase in the percentage of studies passing all MQC per year, whereas there was a steep increase in the quantity of studies that were published (Fig. 4B). We also assessed whether MQC adherence covaried with the general level of quality control, as indicated by the impact factor of the journals. This analysis showed that, whereas there was a trend toward more studies passing MQC with higher‐impact factors, MQC adherence was generally low across bins (Fig. 4C). Finally, we investigated whether model accuracy was significantly different between study aims passing implementation‐level MQC. N = 225 study aims reported an accuracy of their results. We found that studies with data leakage yielded significantly higher accuracies compared to studies where no data leakage was identified (87.5 ± 10.9% vs. 84.6 ± 12.1%), indicating data leakage was a primary source of overfitting (t = −1.8, P = 0.035 [1 tailed], not significant after correction for multiple comparison [α = 0.017]). Other differences were not significant.
Bias Assessment beyond MQC
A total of 244 studies were deemed eligible for bias assessment. Importantly, we excluded 11 studies as they did not specify any research aim. Of the included studies, sample sizes ranged from 4 to 3188, with mean sample sizes for CNN, SVM, and ensemble learning approaches being 445, 128, and 246, respectively.
Generalizability was compromised due to few studies passing our MQC and because some classification aims (16%, n = 49) did not address data imbalance. Additionally, only 7% (n = 22) of aims validated their findings in external test sets. Unsurprisingly, mean performance in external test sets was significantly lower than that in test sets from the original dataset (mean difference = 5%, t = 3.1, P < 0.01).
Regarding reproducibility, several studies lacked essential details; 46% (n = 32) of PET/SPECT, 20% (n = 23) of structural MRI, and 6% (n = 2) of fMRI did not indicate the scanner type, of which 91%, 87%, and 100% used PPMI data, respectively. The tracer was not specified in 6% (n = 4 [75%, n = 3 PPMI]) of PET/SPECT studies. Field strength and sequences were not provided in 16% (n = 18) and 19% (n = 21) of structural, as well as 6% (n = 2) and 9% (n = 3) of fMRI studies. For fMRI studies, 9% (n = 3) lacked activity status, and 44% (n = 14) did not report eye status. Similarly, these details were missing in 5% (n = 2) and 21% (n = 8) of EEG and MEG studies, respectively. Additionally, 21% (n = 8) of EEG/MEG studies lacked information about the electrode system.
Discussion
Recent years have seen rapid development and increased accessibility of AI methods, leading to their widespread application in neuroimaging research. In this review, we systematically summarized and rated the quality of research papers using AI methods on neuroimaging data in PD, specifically focusing on diagnosis, prognosis, and therapeutic intervention outcomes. 54% of studies were concerned with the differentiation of PD and HC, whereas differential diagnosis and especially prognostic or intervention applications were rarely investigated. To the best of our knowledge, we are the first to assess the quality of such studies, revealing that the majority of studies—an alarming 80%—failed to pass all common and minimal quality standards of AI.
Methodology Misalignment and Clinical Impact of PD‐AI Research
With more than 50% of studies covering the “issue” of differentiating PD from HC, the potential of AI and its actual fields of application appears to be strongly misaligned. Multiple studies claimed that PD versus HC classification could support the early diagnosis of PD, that is, the early identification of PD‐related brain changes in asymptomatic individuals. However, PD patients included in such studies were often already in advanced stages and showed clinically overt symptoms. To evaluate the effectiveness of PD versus HC models in identifying asymptomatic individuals at risk for PD, clinical follow‐up for diagnosis verification is necessary. However, none of these studies included longitudinal assessments. The clinical utility of papers in this category is therefore severely limited. In our analyses, we showed that ensemble learning as a model and [123I]‐FP‐CIT SPECT as modality achieve almost perfect accuracy. Importantly, when stratification into PD or HC groups is already supported using [123I]‐FP‐CIT SPECT, as is the case in the PPMI study (https://www.ppmi-info.org/study-design/study-cohorts#overview/), their distinction based on the same modality is highly circular, and clinical relevance is not granted. Moreover, HCs in studies hardly represent a population likely to consult a neurologist for a potential diagnosis of PD. 78 Among individuals presenting to movement disorders clinics, major challenges rely on correct diagnosis of a particular parkinsonian syndrome, that is, differential diagnosis, 4 , 5 personalized assessments/subtyping and prognostic assessments, 79 , 80 or the prediction of therapeutic efficiency. 81 Unfortunately, such clinically relevant and more challenging aims were rare in the current literature. The potential of AI for multimodal integration and pattern recognition could be more strongly exploited for these tasks. PD is known to affect various levels of brain organization 82 , 83 ; thus, unimodal neuroimaging information, used by the majority of studies assessed here, may restrict information content on individual patients compared to multimodal assessment.
Current State of Quality in PD‐AI Research
Beyond a descriptive representation, we performed quality assessments of all included studies in two areas, that is, methodological soundness at the implementation level and diligence of reporting at the interpretation level. At the implementation level, we demonstrated that studies frequently committed data leakage or failed to provide sufficient evidence of avoidance thereof (MQC 2). Moreover, numerous studies applied overly complex models or lacked a clear description of the number of features and participants used in their workflows (MQC 3). Although advanced techniques such as partial least squares regression have been proposed to handle high dimensionality in small training sets more efficiently than conventional AI algorithms, this remains a topic of debate. 84 Importantly, the application of more advanced methods may only partially resolve the issue. Therefore, minimal model complexity remains an important aspect when model validity is assessed. Studies passing implementation‐level MQC exhibited significantly lower accuracies than studies with (potential) data leakage. This finding underscores the assumption that data leakage induces overfitting; ie, it artificially inflates performance. 17 , 18 A major source of data leakage was the allocation of multiple samples per subject to both the training and test set. Indeed, a previous study showed that this practice can cause an overestimation of accuracy by about 50% in classifying PD versus HC. 18 Including overfitted AI models in clinical practice, thus, could decrease rather than increase diagnostic accuracy, as clinicians may not be aware of the erroneous behavior and adapt their diagnosis based on the model output 85 —a process known as “automation bias.” Beyond within‐study data leakage, the frequent usage of big databases such as PPMI also increases the risk for “cross‐literature data leakage”; that is, that feature selection for studies using some database is done based on prior literature employing partly the same data. However, this latter issue pertains to research in general and may even be less common in AI studies, where feature selection tends to be data rather than literature driven.
At the interpretation level, studies often did not report feature importance, thereby preventing the assessment of biological plausibility. Especially in clinical settings, it is important to understand and interpret the decision‐making process of models to confidently integrate AI‐generated information. 85 Moreover, feature importance is critical to form a conceptual bridge between AI and neuroscience, allowing neuroimaging‐based models to deepen mechanistic insights. 20 , 86
Recommendations for Future Research
Based on the issues described earlier, we could identify a tremendous gap between the clinical needs of PD management and the use of scientific resources in the realm of neuroimaging‐based AI research. This gap pertains to both study aims and quality standards. Therefore, we recommend fostering interdisciplinary collaboration between clinicians and engineers, wherein clinicians should be consulted to identify relevant research questions, and engineers should ensure correct and high‐quality solutions to these needs. Admittedly, a comprehensive assessment of quality standards in AI‐based studies requires considerable specific expertise even when provided with a catalog like the MQC we put forward here. It may still be surprising that even high‐impact journals did not predominantly publish high‐quality AI studies. Therefore, journal editorial boards should also promote awareness on the importance of AI quality standards during the review process. For example, reviewers could be prompted to indicate if they recommend an additional AI expert review.
We demonstrated that a fifth of studies did not account for data imbalance and only a minority of studies used external test sets. Whereas our MQC aimed to investigate the bare minimum of AI quality standards, AI applications aimed to translate into clinical tools are critical to account for both these issues to prove the generalizability of the results. One should account for data imbalance, for example, by subsampling, ie, reducing the number of individuals from one class to that of the smallest class, or by reporting adequate metrics (eg, balanced accuracy, F1 score, or area under the precision–recall curve). External test sets, that is, test sets acquired at a different site with potentially different scanning protocols or diagnostic criteria, are of central importance to demonstrate the generalizability. In cases where external test sets are unavailable, permutation testing provides an alternative to test whether a model is overfitted. 87 , 88 Some train–test division procedures are more prone to overestimations of accuracy than others. It was proposed to prefer nested cross‐validation or train–test split without cross‐validation, especially in small datasets (n < 1000). 16 Moreover, instead of a simple train–test split, multiple splits (training/validation/test) are preferable. 89 Generalizability is also constrained by different preprocessing/feature extraction techniques, which are often extensive and not well described. End‐to‐end AI solutions, wherein raw neuroimages are fed to AI models, would circumvent this source of bias. Finally, we recommend that analysis pipelines should be graphically visualized in AI publications, starting with feature selection, including train–test splitting or cross‐validation, and ending with the final outcome. This practice has immensely facilitated understanding different pipelines and assessing MQC adherence in our investigations.
Limitations
Some limitations of the current systematic review should be acknowledged. Even though we used a broad search term, we might have overlooked some new, highly specific neuroimaging‐based AI applications in this rapidly evolving field. Recent developments enabled non‐imaging PD biomarkers, such as from bodily fluids. Although neuroimaging offers the advantage of spatial information of abnormality, it may be worthwhile for future studies to extend the input modality (imaging or fluid biomarkers) depending on availability in clinical practice and the specific research objective. Although we provide a catalog of quality indicators, we recognize that the evaluation of MQC requires expertise, which may not be ubiquitously available. Given the extensive evaluation procedure, there is also a time lag between the conducted analyses and publication. Nonetheless, we have shown that quality remains low even in the most recent literature. Finally, our criteria are a first step and cover basic methodological requirements for AI studies rather than the whole picture. To ensure high‐quality research in the dynamic field of neuroimaging‐based AI research, there may soon be a need to update and complement such criteria.
Conclusion
In conclusion, we showed that there is a wide array of possible applications of neuroimaging‐based AI for PD, yet current resources are often used on tasks with questionable clinical relevance. It thus seems critical to enhance communication between engineers and clinicians. Moreover, in this first‐of‐its‐kind meta‐analysis, we demonstrated that the quality of neuroimaging‐based AI studies in PD is alarmingly low, especially at the implementation level. We therefore recommend to strengthen quality control of AI studies before they are published, optimally both during study design and at the review stage. By fostering high‐quality, interdisciplinary collaborations in AI research, more reliable, interpretable, and clinically relevant neuroimaging tools can ultimately advance clinical management of PD.
Author Roles
1. Research project: A. Conception, B. Organization, C. Execution; 2. Statistical analysis: A. Design, B. Execution, C. Review and critique; 3. Manuscript preparation: A. Writing of the first draft, B. Review and critique
V.D. 1A, 1B, 1C, 2A, 2B, 2C, 3A, 3B
E.D. 1A, 1B, 1C, 2A, 2B, 2C, 3A, 3B
H.E. 1C
A.S. 1C, 2A, 3B
D.V. 1C, 2A, 3B
K.S. 1C, 2A, 3B
T.v.E. 1A, 1C, 2A, 2C, 3B
Full financial disclosures of all authors for the preceding 12 months
T.E. receives honoraria for consulting or advisory roles from Lundbeck Foundation, Lundbeck Pharma, Orion Pharma, and GT Gain Therapeutics SA. A.P.S. is supported by the Canadian Institutes of Health Research (CIHR) (PJT‐173540) and the Krembil‐Rossy Chair program. K.S. receives funding from the National Institutes of Health (R01NS088160, R01NS124228, R01DC011805, R01DC012545, R01DC019353, P50DC01990, and R01DE030464) and the Department of Defense, serves on the scientific advisory board of the Tourette Association of America and the Voice Foundation, and received consulting fees from Jazz Pharmaceuticals Inc. and AbbVie Inc.
Supporting information
Data S1. PRISMA 2020 Checklist.
Data S2. Supplementary Materials.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (German Research Foundation, project IDs 233886668 and 431549029). We thank the German Research Foundation for funding. Open Access funding enabled and organized by Projekt DEAL.
Verena Dzialas and Elena Doering have contributed equally to this study.
Relevant conflicts of interest/financial disclosures: T.E. (principal investigator) receives honoraria for consulting or advisory roles from Lundbeck Foundation, Lundbeck Pharma, Orion Pharma, and GT Gain Therapeutics SA. A.P.S. is supported by the Canadian Institutes of Health Research (CIHR) (PJT‐173540) and the Krembil‐Rossy Chair program. D.E.V. receives funding from the National Institutes of Health (U01 NS119562, R01 NS058487, R01 NS075012) and is the cofounder of Neuropacs. K.S. receives funding from the National Institutes of Health (R01NS088160, R01NS124228, R01DC011805, R01DC012545, R01DC019353, P50DC01990, R01DE030464) and the Department of Defense, serves on the scientific advisory board of the Tourette Association of America and the Voice Foundation, and received consulting fees from Jazz Pharmaceuticals Inc. and AbbVie Inc.
Funding agency: This work was supported by the Deutsche Forschungsgemeinschaft (German Research Foundation; project ID 233886668, GRK 1960, and project ID 431549029, SFB 1451).
Data Availability Statement
The data that supports the findings of this study are available in the supplementary material of this article.
References
- 1. Painous CM, Marti MJ. Cognitive impairment in Parkinson's disease: what we know so far. Res Rev Parkinson 2020;10:7–17. 10.2147/JPRLS.S263041 [DOI] [Google Scholar]
- 2. Bernheimer H, Birkmayer W, Hornykiewicz O, Jellinger K, Seitelberger F. Brain dopamine and the syndromes of Parkinson and Huntington. Clinical, morphological and neurochemical correlations. J Neurol Sci 1973;20(4):415–455. 10.1016/0022-510x(73)90175-5 [DOI] [PubMed] [Google Scholar]
- 3. Postuma RB, Berg D, Stern M, et al. MDS clinical diagnostic criteria for Parkinson's disease. Mov Disord 2015;30(12):1591–1601. 10.1002/mds.26424 [DOI] [PubMed] [Google Scholar]
- 4. Hughes AJ, Daniel SE, Lees AJ. Improved accuracy of clinical diagnosis of Lewy body Parkinson's disease. Neurology 2001;57(8):1497–1499. 10.1212/wnl.57.8.1497 [DOI] [PubMed] [Google Scholar]
- 5. Jankovic J, Rajput AH, McDermott MP, Perl DP, Parkinson Study Group . The evolution of diagnosis in early Parkinson disease. Arch Neurol 2000;57(3):369–372. 10.1001/archneur.57.3.369 [DOI] [PubMed] [Google Scholar]
- 6. Riederer PW, Wuketich ST. Time course of Nigrostriatal degeneration in Parkinson's disease: a detailed study of influential factors in human brain amine analysis. J Neural Transm 1976;38(3–4):277–301. 10.1007/BF01249445 [DOI] [PubMed] [Google Scholar]
- 7. Nigro S, Arabia G, Antonini A, et al. Magnetic resonance parkinsonism index: diagnostic accuracy of a fully automated algorithm in comparison with the manual measurement in a large Italian multicentre study in patients with progressive supranuclear palsy. Eur Radiol 2017;27(6):2665–2675. 10.1007/s00330-016-4622-x [DOI] [PubMed] [Google Scholar]
- 8. Bajaj S, Krismer F, Palma JA, et al. Diffusion‐weighted MRI distinguishes Parkinson disease from the parkinsonian variant of multiple system atrophy: a systematic review and meta‐analysis. PLoS One 2017;12(12):e0189897. 10.1371/journal.pone.0189897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tang CC, Poston KL, Eckert T, et al. Differential diagnosis of parkinsonism: a metabolic imaging study using pattern analysis. Lancet Neurol 2010;9(2):149–158. 10.1016/s1474-4422(10)70002-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dictionary OE. Artificial Intelligence. Oxford English Dictionary. Oxford: Oxford University Press; 2023. [Google Scholar]
- 11. Chen CW, Wang J, Pan D, et al. Applications of multi‐omics analysis in human diseases. MedComm 2023;4(4):e315. 10.1002/mco2.315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lamade A, Beekmann D, Eickhoff S, et al. Quality indicators artificial intelligence. Nervenarzt 2024;95(3):242–246. 10.1007/s00115-023-01573-6 [DOI] [PubMed] [Google Scholar]
- 13. Bonkhoff AK, Grefkes C. Precision medicine in stroke: towards personalized outcome predictions using artificial intelligence. Brain 2022;145(2):457–475. 10.1093/brain/awab439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mongan J, Moy L, Kahn CE Jr. Checklist for artificial intelligence in medical imaging (CLAIM): a guide for authors and reviewers. Radiology 2020;2(2):e200029. 10.1148/ryai.2020200029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Montesinos López OA, Montesinos López A, Crossa J. Overfitting, Model Tuning, and Evaluation of Prediction Performance. Multivariate Statistical Machine Learning Methods for Genomic Prediction. Cham: Springer International Publishing; 2022:109–139. [PubMed] [Google Scholar]
- 16. Vabalas A, Gowen E, Poliakoff E, Casson AJ. Machine learning algorithm validation with a limited sample size. PLoS One 2019;14(11):e0224365. 10.1371/journal.pone.0224365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kernbach JM, Staartjes VE. Foundations of machine learning‐based clinical prediction modeling: part II‐generalization and overfitting. Acta Neurochir Suppl 2022;134:15–21. 10.1007/978-3-030-85292-4_3 [DOI] [PubMed] [Google Scholar]
- 18. Yagis E, Atnafu SW, Seco G, de Herrera A, et al. Effect of data leakage in brain MRI classification using 2D convolutional neural networks. Sci Rep 2021;11(1):22544. 10.1038/s41598-021-01681-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hua J, Xiong Z, Lowey J, Suh E, Dougherty ER. Optimal number of features as a function of sample size for various classification rules. Bioinformatics 2005;21(8):1509–1515. 10.1093/bioinformatics/bti171 [DOI] [PubMed] [Google Scholar]
- 20. Kohoutova L, Heo J, Cha S, et al. Toward a unified framework for interpreting machine‐learning models in neuroimaging. Nat Protoc 2020;15(4):1399–1435. 10.1038/s41596-019-0289-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luque A, Carrasco A, Martín A, de las Heras A. The impact of class imbalance in classification performance metrics based on the binary confusion matrix. Pattern Recogn 2019;91:216–231. 10.1016/j.patcog.2019.02.023 [DOI] [Google Scholar]
- 22. Sun D, Wu X, Xia Y, et al. Differentiating Parkinson's disease motor subtypes: a radiomics analysis based on deep gray nuclear lesion and white matter. Neurosci Lett 2021;760:136083. 10.1016/j.neulet.2021.136083 [DOI] [PubMed] [Google Scholar]
- 23. Martins R, Oliveira F, Moreira F, et al. Automatic classification of idiopathic Parkinson's disease and atypical parkinsonian syndromes combining [(11)C]raclopride PET uptake and MRI grey matter morphometry. J Neural Eng 2021;18(4). 10.1088/1741-2552/abf772 [DOI] [PubMed] [Google Scholar]
- 24. Shiiba T, Arimura Y, Nagano M, Takahashi T, Takaki A. Improvement of classification performance of Parkinson's disease using shape features for machine learning on dopamine transporter single photon emission computed tomography. PLoS One 2020;15(1):e0228289. 10.1371/journal.pone.0228289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aljalal M, Aldosari SA, Molinas M, AlSharabi K, Alturki FA. Detection of Parkinson's disease from EEG signals using discrete wavelet transform, different entropy measures, and machine learning techniques. Sci Rep 2022;12(1):22547. 10.1038/s41598-022-26644-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chan YH, Wang C, Soh WK, Rajapakse JC. Combining neuroimaging and omics datasets for disease classification using graph neural networks. Front Neurosci 2022;16:866666. 10.3389/fnins.2022.866666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sarica A, Quattrone A, Quattrone A. Explainable machine learning with pairwise interactions for the classification of Parkinson's disease and SWEDD from clinical and imaging features. Brain Imaging Behav 2022;16(5):2188–2198. 10.1007/s11682-022-00688-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Belyaev M, Murugappan M, Velichko A, Korzun D. Entropy‐based machine learning model for fast diagnosis and monitoring of Parkinson's disease. Sensors 2023;23(20). 10.3390/s23208609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jothi S, Anita S, Sivakumar S. Modified exigent features block in JAN net for Analysing SPECT scan images to diagnose early‐stage Parkinson's disease. Curr Med Imaging 2023. 10.2174/1573405620666230605092654 [DOI] [PubMed] [Google Scholar]
- 30. Huppertz HJ, Möller L, Südmeyer M, et al. Differentiation of neurodegenerative parkinsonian syndromes by volumetric magnetic resonance imaging analysis and support vector machine classification. Mov Disord 2016;31(10):1506–1517. 10.1002/mds.26715 [DOI] [PubMed] [Google Scholar]
- 31. Morisi R, Manners DN, Gnecco G, et al. Multi‐class parkinsonian disorders classification with quantitative MR markers and graph‐based features using support vector machines. Parkinsonism Relat Disord 2018;47:64–70. 10.1016/j.parkreldis.2017.11.343 [DOI] [PubMed] [Google Scholar]
- 32. Pang H, Yu Z, Yu H, et al. Use of machine learning method on automatic classification of motor subtype of Parkinson's disease based on multilevel indices of rs‐fMRI. Parkinsonism Relat Disord 2021;90:65–72. 10.1016/j.parkreldis.2021.08.003 [DOI] [PubMed] [Google Scholar]
- 33. Zhou C, Wang L, Cheng W, et al. Two distinct trajectories of clinical and neurodegeneration events in Parkinson's disease. npj Parkinson's Dis 2023;9(1):111. 10.1038/s41531-023-00556-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kaplan E, Altunisik E, Ekmekyapar Firat Y, et al. Novel nested patch‐based feature extraction model for automated Parkinson's disease symptom classification using MRI images. Comput Methods Programs Biomed 2022;224:107030. 10.1016/j.cmpb.2022.107030 [DOI] [PubMed] [Google Scholar]
- 35. Chen Y, Zhu G, Liu D, et al. Brain morphological changes in hypokinetic dysarthria of Parkinson's disease and use of machine learning to predict severity. CNS Neurosci Ther 2020;26(7):711–719. 10.1111/cns.13304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen B, Xu M, Yu H, et al. Detection of mild cognitive impairment in Parkinson's disease using gradient boosting decision tree models based on multilevel DTI indices. J Transl Med 2023;21(1):310. 10.1186/s12967-023-04158-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Parajuli M, Amara AW, Shaban M. Deep‐learning detection of mild cognitive impairment from sleep electroencephalography for patients with Parkinson's disease. PLoS One 2023;18(8):e0286506. 10.1371/journal.pone.0286506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu C, Jiang Z, Liu S, et al. Frequency‐dependent microstate characteristics for mild cognitive impairment in Parkinson's disease. IEEE Trans Neural Syst Rehabil Eng 2023;31:4115–4124. 10.1109/tnsre.2023.3324343 [DOI] [PubMed] [Google Scholar]
- 39. Morales DA, Vives‐Gilabert Y, Gómez‐Ansón B, et al. Predicting dementia development in Parkinson's disease using Bayesian network classifiers. Psychiatry Res 2013;213(2):92–98. 10.1016/j.pscychresns.2012.06.001 [DOI] [PubMed] [Google Scholar]
- 40. Choi H, Kim YK, Yoon EJ, Lee JY, Lee DS. Cognitive signature of brain FDG PET based on deep learning: domain transfer from Alzheimer's disease to Parkinson's disease. Eur J Nucl Med Mol Imaging 2020;47(2):403–412. 10.1007/s00259-019-04538-7 [DOI] [PubMed] [Google Scholar]
- 41. Ly QT, Ardi Handojoseno AM, Gilat M, et al. Detection of gait initiation failure in Parkinson's disease based on wavelet transform and support vector machine. Annu Int Conf IEEE Eng Med Biol Soc 2017;2017:3048–3051. 10.1109/embc.2017.8037500 [DOI] [PubMed] [Google Scholar]
- 42. Quynh Tran L, Ardi Handojoseno AM, Gilat M, et al. Detection of turning freeze in Parkinson's disease based on S‐transform decomposition of EEG signals. Annu Int Conf IEEE Eng Med Biol Soc 2017;2017:3044–3047. 10.1109/embc.2017.8037499 [DOI] [PubMed] [Google Scholar]
- 43. Choi H, Ha S, Im HJ, Paek SH, Lee DS. Refining diagnosis of Parkinson's disease with deep learning‐based interpretation of dopamine transporter imaging. NeuroImage: Clin 2017;16:586–594. 10.1016/j.nicl.2017.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Khachnaoui H, Khlifa N, Mabrouk R. Machine learning for early Parkinson's disease identification within SWEDD group using clinical and DaTSCAN SPECT imaging features. J Imaging 2022;8(4). 10.3390/jimaging8040097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tripathi S, Mattioli P, Liguori C, Chiaravalloti A, Arnaldi D, Giancardo L. Brain hemisphere dissimilarity, a self‐supervised learning approach for alpha‐synucleinopathies prediction with FDG PET. Proc IEEE Int Symp Biomed Imaging 2023;2023:1–5. 10.1109/isbi53787.2023.10230560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Leung KH, Rowe SP, Pomper MG, Du Y. A three‐stage, deep learning, ensemble approach for prognosis in patients with Parkinson's disease. EJNMMI Res 2021;11(1):52. 10.1186/s13550-021-00795-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Salmanpour MR, Shamsaei M, Saberi A, et al. Machine learning methods for optimal prediction of motor outcome in Parkinson's disease. Phys Med 2020;69:233–240. 10.1016/j.ejmp.2019.12.022 [DOI] [PubMed] [Google Scholar]
- 48. Shu ZY, Cui SJ, Wu X, et al. Predicting the progression of Parkinson's disease using conventional MRI and machine learning: An application of radiomic biomarkers in whole‐brain white matter. Magn Reson Med 2021;85(3):1611–1624. 10.1002/mrm.28522 [DOI] [PubMed] [Google Scholar]
- 49. Handojoseno AM, Shine JM, Nguyen TN, Tran Y, Lewis SJ, Nguyen HT. Using EEG spatial correlation, cross frequency energy, and wavelet coefficients for the prediction of freezing of gait in Parkinson's disease patients. Annu Int Conf IEEE Eng Med Biol Soc 2013;2013:4263–4266. 10.1109/embc.2013.6610487 [DOI] [PubMed] [Google Scholar]
- 50. Handojoseno AM, Shine JM, Nguyen TN, Tran Y, Lewis SJ, Nguyen HT. The detection of freezing of gait in Parkinson's disease patients using EEG signals based on wavelet decomposition. Annu Int Conf IEEE Eng Med Biol Soc 2012;2012:69–72. 10.1109/embc.2012.6345873 [DOI] [PubMed] [Google Scholar]
- 51. Handojoseno AMA, Naik GR, Gilat M, et al. Prediction of freezing of gait in patients with Parkinson's disease using EEG signals. Stud Health Technol Inform 2018;246:124–131. [PubMed] [Google Scholar]
- 52. Li Y, Huang X, Ruan X, et al. Baseline cerebral structural morphology predict freezing of gait in early drug‐naive Parkinson's disease. npj Parkinson's Dis 2022;8(1):176. 10.1038/s41531-022-00442-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cesari M, Christensen JAE, Muntean ML, et al. A data‐driven system to identify REM sleep behavior disorder and to predict its progression from the prodromal stage in Parkinson's disease. Sleep Med 2021;77:238–248. 10.1016/j.sleep.2020.04.010 [DOI] [PubMed] [Google Scholar]
- 54. Huang X, He Q, Ruan X, et al. Structural connectivity from DTI to predict mild cognitive impairment in de novo Parkinson's disease. NeuroImage: Clin 2024;41:103548. 10.1016/j.nicl.2023.103548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Booth S, Park KW, Lee CS, Ko JH. Predicting cognitive decline in Parkinson's disease using FDG‐PET‐based supervised learning. J Clin Invest 2022;132(20). 10.1172/jci157074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shin NY, Bang M, Yoo SW, et al. Cortical thickness from MRI to predict conversion from mild cognitive impairment to dementia in Parkinson disease: a machine learning‐based model. Radiology 2021;300(2):390–399. 10.1148/radiol.2021203383 [DOI] [PubMed] [Google Scholar]
- 57. Aljalal M, Aldosari SA, AlSharabi K, Abdurraqeeb AM, Alturki FA. Parkinson's disease detection from resting‐state EEG signals using common spatial pattern, entropy, and machine learning techniques. Diagnostics 2022;12(5). 10.3390/diagnostics12051033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shaban M, Amara AW. Resting‐state electroencephalography based deep‐learning for the detection of Parkinson's disease. PLoS One 2022;17(2):e0263159. 10.1371/journal.pone.0263159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shah SAA, Zhang L, Bais A. Dynamical system based compact deep hybrid network for classification of Parkinson disease related EEG signals. Neural Networks 2020;130:75–84. 10.1016/j.neunet.2020.06.018 [DOI] [PubMed] [Google Scholar]
- 60. Peña E, Mohammad TM, Almohammed F, et al. Individual magnetoencephalography response profiles to short‐duration L‐Dopa in Parkinson's disease. Front Hum Neurosci 2021;15:640591. 10.3389/fnhum.2021.640591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Herz DM, Haagensen BN, Christensen MS, et al. The acute brain response to levodopa heralds dyskinesias in Parkinson disease. Ann Neurol 2014;75(6):829–836. 10.1002/ana.24138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Herz DM, Haagensen BN, Nielsen SH, Madsen KH, Løkkegaard A, Siebner HR. Resting‐state connectivity predicts levodopa‐induced dyskinesias in Parkinson's disease. Mov Disord 2016;31(4):521–529. 10.1002/mds.26540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Luo Y, Chen H, Gui M. Radiomics and hybrid models based on machine learning to predict levodopa‐induced dyskinesia of Parkinson's disease in the first 6 years of levodopa treatment. Diagnostics 2023;13(15). 10.3390/diagnostics13152511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xie Y, Gao C, Wu B, Peng L, Wu J, Lang L. Morphologic brain network predicts levodopa responsiveness in Parkinson disease. Front Aging Neurosci 2022;14:990913. 10.3389/fnagi.2022.990913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yang B, Wang X, Mo J, et al. The amplitude of low‐frequency fluctuation predicts levodopa treatment response in patients with Parkinson's disease. Parkinsonism Relat Disord 2021;92:26–32. 10.1016/j.parkreldis.2021.10.003 [DOI] [PubMed] [Google Scholar]
- 66. Chen Y, Zhu G, Liu D, et al. The morphology of thalamic subnuclei in Parkinson's disease and the effects of machine learning on disease diagnosis and clinical evaluation. J Neurol Sci 2020;411:116721. 10.1016/j.jns.2020.116721 [DOI] [PubMed] [Google Scholar]
- 67. Ballarini T, Mueller K, Albrecht F, et al. Regional gray matter changes and age predict individual treatment response in Parkinson's disease. NeuroImage Clin 2019;21:101636. 10.1016/j.nicl.2018.101636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chen Y, Zhu G, Liu Y, et al. Predict initial subthalamic nucleus stimulation outcome in Parkinson's disease with brain morphology. CNS Neurosci Ther 2022;28(5):667–676. 10.1111/cns.13797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yang B, Wang X, Mo J, et al. The altered spontaneous neural activity in patients with Parkinson's disease and its predictive value for the motor improvement of deep brain stimulation. NeuroImage Clin 2023;38:103430. 10.1016/j.nicl.2023.103430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chang B, Xiong C, Ni C, et al. Prediction of STN‐DBS for Parkinson's disease by uric acid‐related brain function connectivity: a machine learning study based on resting state function MRI. Front Aging Neurosci 2023;15:1105107. 10.3389/fnagi.2023.1105107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Geraedts VJ, Koch M, Kuiper R, et al. Preoperative electroencephalography‐based machine learning predicts cognitive deterioration after subthalamic deep brain stimulation. Mov Disord 2021;36(10):2324–2334. 10.1002/mds.28661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Peralta M, Haegelen C, Jannin P, Baxter JSH. PassFlow: a multimodal workflow for predicting deep brain stimulation outcomes. Int J Comput Assist Radiol Surg 2021;16(8):1361–1370. 10.1007/s11548-021-02435-9 [DOI] [PubMed] [Google Scholar]
- 73. Sand D, Arkadir D, Abu Snineh M, et al. Deep brain stimulation can differentiate subregions of the human subthalamic nucleus area by EEG biomarkers. Front Syst Neurosci 2021;15:747681. 10.3389/fnsys.2021.747681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Boutet A, Madhavan R, Elias GJB, et al. Predicting optimal deep brain stimulation parameters for Parkinson's disease using functional MRI and machine learning. Nat Commun 2021;12(1):3043. 10.1038/s41467-021-23311-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lu CW, Malaga KA, Chou KL, Chestek CA, Patil PG. High density microelectrode recording predicts span of therapeutic tissue activation volumes in subthalamic deep brain stimulation for Parkinson disease. Brain Stimul 2020;13(2):412–419. 10.1016/j.brs.2019.11.013 [DOI] [PubMed] [Google Scholar]
- 76. Ben Bashat D, Thaler A, Lerman Shacham H, et al. Neuromelanin and T(2)*‐MRI for the assessment of genetically at‐risk, prodromal, and symptomatic Parkinson's disease. npj Parkinson's Dis 2022;8(1):139. 10.1038/s41531-022-00405-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Augimeri A, Cherubini A, Cascini GL, et al. CADA‐computer‐aided DaTSCAN analysis. EJNMMI Phys 2016;3(1):4. 10.1186/s40658-016-0140-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Parkinson Progression Marker I . The Parkinson progression marker initiative (PPMI). Prog Neurobiol 2011;95(4):629–635. 10.1016/j.pneurobio.2011.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tolosa E, Garrido A, Scholz SW, Poewe W. Challenges in the diagnosis of Parkinson's disease. Lancet Neurol 2021;20(5):385–397. 10.1016/s1474-4422(21)00030-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Marras C, Lang A. Parkinson's disease subtypes: lost in translation? J Neurol Neurosurg Psychiatry 2013;84(4):409–415. 10.1136/jnnp-2012-303455 [DOI] [PubMed] [Google Scholar]
- 81. Lang AE, Espay AJ. Disease modification in Parkinson's disease: current approaches, challenges, and future considerations. Mov Disord 2018;33(5):660–677. 10.1002/mds.27360 [DOI] [PubMed] [Google Scholar]
- 82. McGregor MM, Nelson AB. Circuit mechanisms of Parkinson's disease. Neuron 2019;101(6):1042–1056. 10.1016/j.neuron.2019.03.004 [DOI] [PubMed] [Google Scholar]
- 83. Zaman V, Shields DC, Shams R, et al. Cellular and molecular pathophysiology in the progression of Parkinson's disease. Metab Brain Dis 2021;36(5):815–827. 10.1007/s11011-021-00689-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Rönkkö M, McIntosh CN, Antonakis J, Edwards JR. Partial least squares path modeling: time for some serious second thoughts. J Oper Manag 2016;47‐48:9–27. 10.1016/j.jom.2016.05.002 [DOI] [Google Scholar]
- 85. Bond RR, Novotny T, Andrsova I, et al. Automation bias in medicine: the influence of automated diagnoses on interpreter accuracy and uncertainty when reading electrocardiograms. J Electrocardiol 2018;51(6S):S6–S11. 10.1016/j.jelectrocard.2018.08.007 [DOI] [PubMed] [Google Scholar]
- 86. Roscher R, Bohn B, Duarte MF, Garcke J. Explainable machine learning for scientific insights and discoveries. IEEE Access 2020;8:42200–42216. 10.1109/Access.2020.2976199 [DOI] [Google Scholar]
- 87. Desaire H. How (not) to generate a highly predictive biomarker panel using machine learning. J Proteome Res 2022;21(9):2071–2074. 10.1021/acs.jproteome.2c00117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Golland P, Fischl B. Permutation tests for classification: towards statistical significance in image‐based studies. Inf Process Med Imaging 2003;18:330–341. 10.1007/978-3-540-45087-0_28 [DOI] [PubMed] [Google Scholar]
- 89. An C, Park YW, Ahn SS, Han K, Kim H, Lee SK. Radiomics machine learning study with a small sample size: single random training‐test set split may lead to unreliable results. PLoS One 2021;16(8):e0256152. 10.1371/journal.pone.0256152 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. PRISMA 2020 Checklist.
Data S2. Supplementary Materials.
Data Availability Statement
The data that supports the findings of this study are available in the supplementary material of this article.


