Abstract
Background
Numerous species of Ardisia are widely used for their medicinal and ornamental values in China. However, accurately identifying Ardisia species at the molecular level remains a challenge due to the morphological similarities among different species, the complexity of interspecific variation, and the limited availability of genetic markers. In this study, we reported 20 chloroplast genomes of Ardisia species from China and combined them with 8 previously published chloroplast genomes to conduct a comprehensive analysis for phylogenetic relationships and adaptive evolution.
Results
For the 28 Ardisia species analyzed in this study, the size of the chloroplast genomes ranged from 155,088 bp to 156,999 bp, and all exhibited a typical tetrad structure with conserved gene content and number. Each genome contained 85–88 protein-coding genes, 36–37 tRNA genes, and 8 rRNA genes. Comparative analysis showed that the genomic structures and gene order were relatively conserved with slight variations in the inverted repeat regions (IRs). Simple sequence repeats (SSRs) were predominantly single nucleotide repeats, while repeat sequences were mainly composed of palindromic and forward repeats. Twelve highly variable regions were identified as potential DNA barcodes for species identification and phylogenetic analysis of Ardisia. The phylogenetic tree supported the division of the subgenus Bladhia s.l. into two subgenera: Bladhia s.str. and Odontophylla (Yang) Huang. Further investigation revealed that two protein-coding genes (rbcL and rpoC2) were under positive selection and might be associated with the adaptation of Ardisia species to shaded environments.
Conclusion
Our study analyzed the chloroplast genomes of 20 Ardisia species from China to explore their phylogenetic relationships and adaptive evolution. By combining these results with data from eight previously published chloroplast genomes, the essential characteristics of Ardisia chloroplast genomes were clarified. The research establishes a theoretical basis for the classification, identification, and comprehension of the adaptive evolution of Ardisia species.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05892-x.
Keywords: Ardisia, Chloroplast genome, Phylogeny, Adaptive evolution
Background
Ardisia, the largest genus of Myrsinoideae (Primulaceae), is primarily found in tropical and subtropical regions and consists of over 700 accepted species [1]. The classification of Ardisia species in China follows the taxonomic system described in the Flora of China, including six sections: Sect. Pimelandra, Sect. Acrardisia, Sect. Tinus, Sect. Akosmos, Sect. Crispardisia, and Sect. Bladhia [2]. These species are primarily found in the southern regions of the Yangtze River, with approximately 65 recorded species and 12 varieties [3]. Ardisia species often used as ornamental plants in China are typically small trees or shrubs with brightly colored red fruits. In addition to their ornamental uses, several Ardisia plants, including A. japonica, A. crenata, A. gigantifolia, and A. crispa, have been used as traditional folk medicine herbs in southern China since ancient times, with various medicinal purposes, such as alleviating cough and phlegm, promoting blood circulation, reducing fatigue, and reducing swelling [4, 5]. Aidicha (the whole dried plant of A. japonica) and Zhushagen (the dried root of A. crenata) are included in the Pharmacopoeia of the People’s Republic of China (2020) (https://db.ouryao.com/yd2020/). Currently, Ardisia plants are used to produce several products, including compounded Aidicha tablets, Aidicha capsules, and Zhushagen dispensing granules, which are widely utilized in clinical applications in China [6]. Modern pharmacological studies have identified various chemical compounds in Ardisia plants, such as bergenin, ardisicrenoside, benzoquinone, triterpenes, and flavonoids [7, 8].
The morphological and clinical similarities between different Ardisia species often result in confusion and errors in taxonomic records, making accurate species identification challenging [9]. Ensuring the safety and quality of raw materials of Ardisia plants holds paramount importance in preserving the authenticity and efficacy of herbal products within the pharmaceutical supply chain. Four DNA barcodes (ITS, psbA-trnH, rbcL, and matK) were evaluated for Chinese Ardisia species, using a sample of 121 individuals from 33 species, and the results showed that the ITS fragment had a higher identification rate compared to the other three barcodes [10]. However, the accuracy of species identification using the ITS fragment was below 85%, indicating the need for further improvement in DNA barcoding techniques for Ardisia species. Additionally, the phylogenetic relationships between Asian Ardisia species and its relatives in the Myrsinoideae (Primulaceae) were analyzed using ITS, psbA-trnH and rpl32-trnL sequences [1]. The relationships among the subgenera remained poorly understood due to the low support rate of the inferred phylogenetic tree, highlighting the requirement for more powerful molecular markers.
In recent years, advances in sequencing assembly and annotation technology have significantly reduced the cost of chloroplast genome sequencing. Consequently, more and more chloroplast genome data has been successfully applied to plant phylogeny and evolutionary studies, including the reconstruction of phylogenetic relationships of plants [11]. The complete chloroplast genome along with its derived barcode has emerged as an ideal tool for species identification of economic plants. However, the number of published chloroplast genome sequences of Ardisia species is limited currently [12–14]. Comparative analysis of chloroplast genomes using a larger number of Ardisia species would greatly contribute to understand chloroplast genome evolution and reconstruct the phylogenetic relationships of Ardisia. The highly variable region of the chloroplast genome could provide potential genetic markers for species identification and phylogenetic analysis of Ardisia.
Adaptive evolution is considered to enhance the fitness of species to constantly changing environmental conditions [15]. Protein-coding genes of chloroplast genome often undergo adaptive evolution, for example, genes such as rbcL, ycf1, and accD, have been positively selected and are significantly correlated with environmental adaptations, including temperature, light, humidity, and atmospheric conditions [16]. In analysising six chloroplast genomes of Chrysosplenium (Saxifragaceae), 19 genes under positive selection were screened out, and most of them were involved in photosynthesis, which may be the adaptive response to shady and moist habitat [17]. Ardisia plants are also typically found in shaded habitats, such as the understory or near valleys and streams. These plants serve as an ideal model group for studying plant adaptations to low light conditions. Comparative chloroplast genome analysis of Ardisia species might provide insight into the effects of low light for angiosperms and enhance our understanding of the evolution of Ardisia species.
In this study, we sequenced the complete chloroplast genomes of 20 Ardisia species and conducted a comprehensive analysis along with 8 additional chloroplast genome data from GenBank. The aims of this study were: (a) to perform a comparative analysis of the structural characteristics of chloroplast genomes; (b) to identify highly variable regions for species identification and phylogenetic studies; (c) to conduct a phylogenetic analysis of Ardisia species in China; and (d) to examine the adaptive evolution of protein-coding genes. The complete genomes of the Ardisia species will serve as a theoretical foundation for the classification, identification, and understanding of the adaptive evolution of Ardisia species.
Results
General features of chloroplast genomes
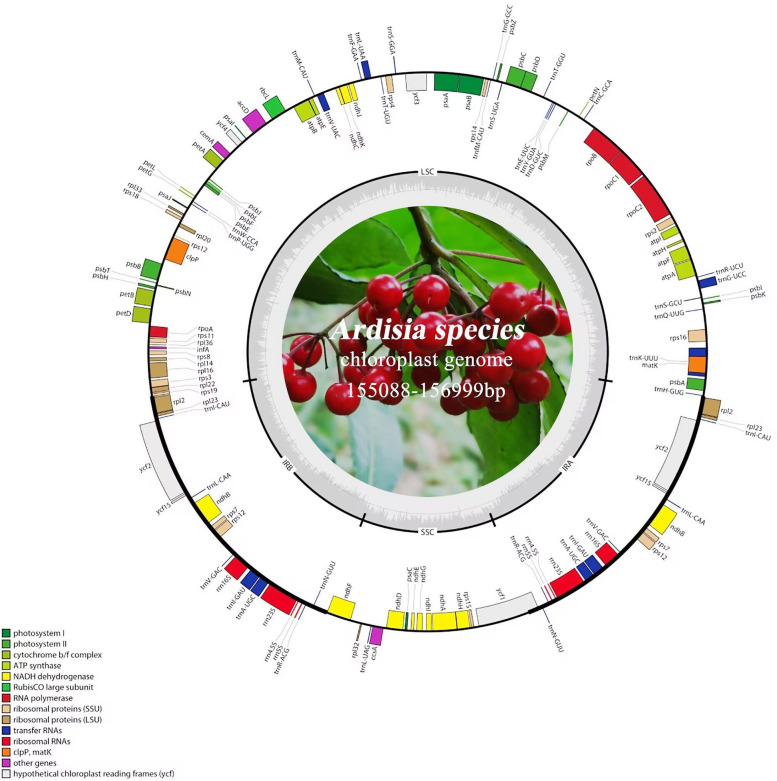
In each Ardisia species, the chloroplast genome consisted of a typical tetrad structure, which included one small single-copy (SSC), one large single-copy (LSC), and two inverted-repeat (IR) regions (Fig. 1). Among the 28 Ardisia species, A. bullata had the lowest GC content (36.00%), while A. gigantifolia had the highest GC content (37.30%). The lengths of the chloroplast genomes of the 28 Ardisia species ranged from 155,088 bp to 156,999 bp, including the SSC region (18,093 − 18,479 bp), LSC region (84,709 − 86,989 bp), and two IR regions (25,411 − 26,236 bp each) (Table 1). Each chloroplast genome of Ardisia species contained 85–88 protein-coding genes, 36–37 tRNA genes, and 8 rRNA genes (Table 1). Among these genes, 12 protein-coding genes and 6 tRNA genes contained introns, three of these genes (clpP, rps12, and ycf3) contained two introns, while the others contained only one intron (Table 2).
Fig. 1.
Gene map of the A. argenticaulis chloroplast genome. Genes located outside the circle are transcribed counterclockwise, while genes inside the circle are transcribed clockwise. In the inner circle, the dark grey area represents GC content of the cp. genome, and the light grey area represents the AT content. Different color blocks represent genes that belong to different functional groups
Table 1.
Summary of the chloroplast genomesof 28 Ardisia species
| Species | Genome Length (bp) | LSC Length (bp) | SSC Length (bp) | IR Length (bp) | GC (%) | Total Genes | CDS | t RNA |
rRNA |
|---|---|---|---|---|---|---|---|---|---|
| A. argenticaulis | 156,940 | 86,068 | 18,400 | 26,236 | 37.03% | 131 | 87 | 36 | 8 |
| A. balansana | 155,307 | 85,673 | 18,342 | 25,646 | 37.21% | 132 | 88 | 36 | 8 |
| A. bullata | 155,088 | 84,709 | 18,367 | 26,006 | 36.00% | 132 | 88 | 36 | 8 |
| A. carnosicaulis | 156,548 | 86,102 | 18,346 | 26,050 | 37.05% | 131 | 87 | 36 | 8 |
| A. crenata | 156,540 | 86,093 | 18,347 | 26,050 | 37.10% | 132 | 87 | 36 | 8 |
| A. crispa | 156,709 | 86,300 | 18,411 | 25,999 | 37.06% | 131 | 87 | 36 | 8 |
| A. faberi | 156,104 | 86,989 | 18,293 | 25,411 | 36.96% | 130 | 85 | 37 | 8 |
| A. fordii | 156,999 | 86,131 | 18,404 | 26,232 | 37.02% | 132 | 88 | 36 | 8 |
| A. gigantifolia | 156,216 | 85,725 | 18,397 | 26,047 | 37.30% | 131 | 87 | 36 | 8 |
| A. japonica | 155,787 | 86,715 | 18,222 | 25,457 | 37.05% | 130 | 85 | 37 | 8 |
| A. lindleyana | 156,741 | 86,359 | 18,380 | 26,001 | 37.06% | 131 | 87 | 36 | 8 |
| A. maclurei | 155,751 | 86,725 | 18,104 | 25,467 | 37.02% | 129 | 85 | 36 | 8 |
| A. mamillata | 156,757 | 86,325 | 18,434 | 25,999 | 37.10% | 131 | 87 | 36 | 8 |
| A. merrillii | 156,746 | 86,351 | 18,387 | 26,004 | 37.08% | 131 | 87 | 36 | 8 |
| A. nutantiflora | 156,542 | 86,225 | 18,379 | 25,969 | 37.22% | 132 | 88 | 36 | 8 |
| A. obtusa | 156,626 | 86,168 | 18,346 | 26,056 | 37.07% | 131 | 87 | 36 | 8 |
| A. omissa | 156,761 | 86,335 | 18,428 | 25,999 | 37.05% | 132 | 87 | 37 | 8 |
| A. pedalis | 156,722 | 86,301 | 18,405 | 26,008 | 37.05% | 132 | 87 | 37 | 8 |
| A. perreticulata | 156,953 | 86,084 | 18,413 | 26,228 | 37.03% | 131 | 87 | 36 | 8 |
| A. polysticta | 156,506 | 86,078 | 18,328 | 26,050 | 37.07% | 131 | 87 | 36 | 8 |
| A. primulifolia | 156,728 | 86,318 | 18,402 | 26,004 | 37.05% | 131 | 87 | 36 | 8 |
| A. pseudocrispa | 156,744 | 86,354 | 18,308 | 25,999 | 37.04% | 131 | 87 | 36 | 8 |
| A. pusilla | 155,749 | 86,677 | 18,222 | 25,425 | 37.05% | 129 | 85 | 36 | 8 |
| A. quinquegona | 156,766 | 85,900 | 18,408 | 26,229 | 37.05% | 131 | 87 | 36 | 8 |
| A. replicata | 156,278 | 86,012 | 18,358 | 26,196 | 37.20% | 132 | 88 | 36 | 8 |
| A. sieboldii | 156,923 | 85,982 | 18,479 | 25,954 | 37.04% | 132 | 88 | 36 | 8 |
| A. solanacea | 156,518 | 86,033 | 18,093 | 26,231 | 37.14% | 132 | 88 | 36 | 8 |
| A. villosa | 156,720 | 86,353 | 18,339 | 26,014 | 37.07% | 131 | 87 | 36 | 8 |
LSC large single-copy, SSC small single-copy, IR inverted repeat
Table 2.
Genes in the chloroplast genome of 28 Ardisia species
| Category | Gene group | Gene name |
|---|---|---|
| Protein synthesis and DNA-replication | Ribosomal RNA genes | rrn4.5, rrn5, rrn16, rrn23 |
| Transfer RNA genes | trnA-UGC*, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC*, trnH-GUG(1, 2, 3, 4), trnI-CAU, trnI-GAU*, trnK-UUU*, trnL-CAA, trnL-UAA*, trnL-UAG, trnM-CAU, trnfM-CAU, trnN-GUU, trnP-UGG, trnQ-UUG, trnR-UCU, trnR-ACG, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC, trnV-UAC*, trnW-CCA, trnY-GUA | |
| Ribosomal protein genes (larger subunit) | rpl2*, rpl14, rpl16*, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36 | |
| Ribosomal protein genes (smaller subunit) | rps2, rps3, rps4, rps7, rps8, rps11, rps12**, rps14, rps15, rps16*, rps18, rps19 | |
| RNA polymerase | rpoA, rpoB, rpoC1*, rpoC2 | |
| Photosynthesis | Photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT | |
| Cytochrome b/f complex | petA, petB*, petD*, petG, petL, petN | |
| ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI | |
| Rubisco large subunit | rbcL | |
| NADH dehydrogenase | ndhA*, ndhB*, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Miscellaneous group | ATP-dependent protease | clpP** |
| Maturase | matK | |
| Acetyl-CoA carboxylase | accD | |
| Cytochrome c biogenesis | ccsA | |
| Inner membrane protein | cemA | |
| Translation initiation factor | infA | |
| Pseudogene unknown function | Hypothetical chloroplast reading frames (ycf) | ycf1, ycf2, ycf3**, ycf4, ycf15[2, 5, 6, 7] |
*—Gene containing a single intron; **—Gene containing two introns; ()—Gene exists in some species; []—Gene do not exist in some species. Species—1, A. fordii; 2, A. japonica; 3, A. omissa; 4, A. pedalis; 5, A. faberi; 6, A. maclurei; 7, A. pusilla
Boundary regions analysis
There are four boundaries between two inverted repeats (IR), large single copy (LSC), and small single copy (SSC) regions in the chloroplast genome, known as the IRb-LSC boundary (JLB line), IRb-SSC boundary (JSB line), IRa-SSC boundary (JSA line), and IRa-LSC boundary (JLA line). The chloroplast genome structures were conserved in 28 Ardisia species (Fig. 2). At the IRb-LSC boundary, the JLB line was located in the rps19 gene region of 28 Ardisia species. At the IRb-SSC boundary, the JSB line was located in the ndhF gene region of 23 Ardisia species, while the JSB line in 5 other species was 27–75 bp away from the ndhF gene. At the IRa-SSC boundary, the JSA line was located in the ycf1 gene region of 27 Ardisia species. In A. pseudocrispa, the JSA line was 170 bp away from the ycf1 gene, and the ycf1 gene was only 4,430 bp and located in the SSC region. At the IRa-LSC boundary, the JLA line was located between the rpl2 and psbA genes in 28 Ardisia species. Notably, it was observed that one of the two copies of rps19 gene was pseudogene in A. fordii, A. sieboldii, A. solanacea, A. replicata, A. nutantiflora, A. balansana, and A. bullata, which also presented at the LSC-IRb boundary. In A. faberi, A. japonica, and A. pedalis, trnH gene was located in the LSC region, 2–10 bp away from the JLA line.
Fig. 2.
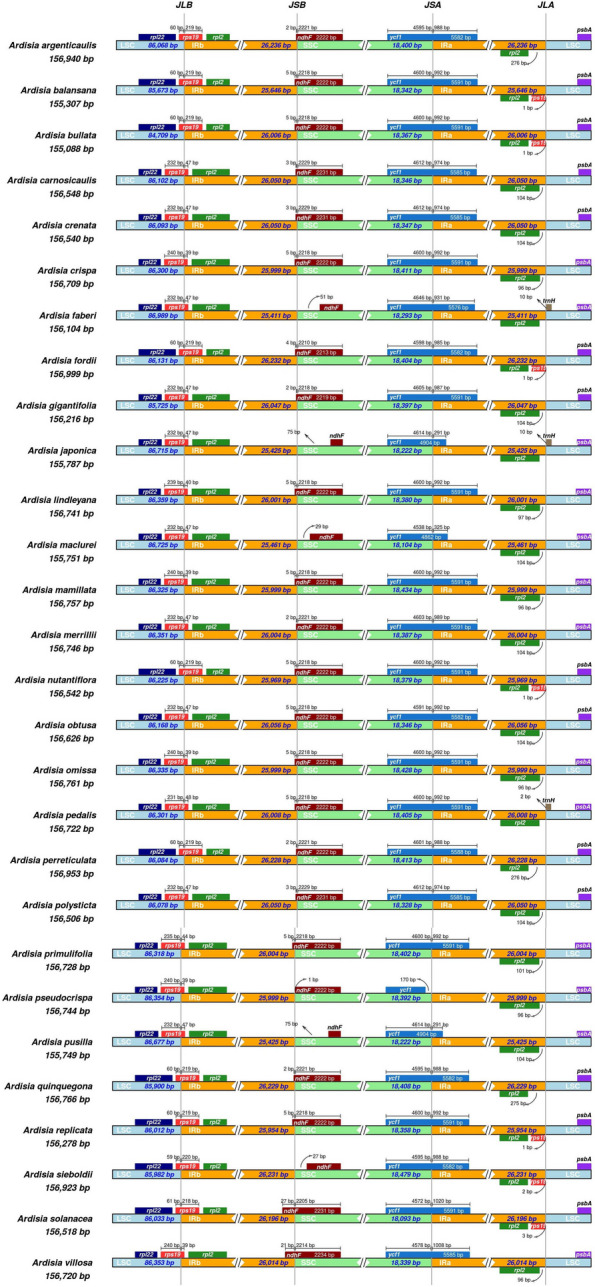
Comparison of the borders of the LSC, SSC, and IR regions among chloroplast genomes of 28 Ardisia species. JLB, JSB, JSA, and JLA denote the junction sites of LSC/IRb, IRb/SSC, SSC/IRa, and IRa/LSC respectively
Repeat sequence analysis
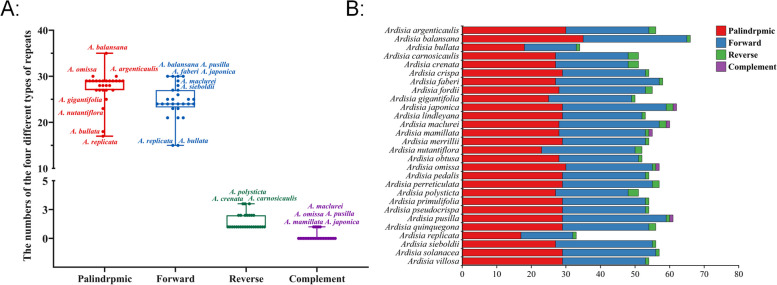
In this study, we employed Reputer software to analyze the repeat sequences in the chloroplast genomes of 28 Ardisia species. Each species displayed 33–66 repeat sequences, with palindromic repeats being the most prevalent (17–35 per species, accounting for 51.29% of total repeats), followed by forward repeats (15–30 per species, making up 45.65% of total repeats), reverse repeats (1–3 per species, constituting 2.72% of total repeats), and complementary repeats (0.33% of total repeats) being the least common (Fig. 3A, Table S2). Complementary repeat sequences were only found in five species (A. japonica, A. maclurei, A. mamillata, A. omissa, and A. pusilla), each with one such sequence. A. balansana had the highest number of palindromic repeats (35 sequences), followed by A. argenticaulis and A. omissa (30 sequences each), and A. replicata with the lowest count of 17 sequences. In terms of forward repeats, A. balansana, A. japonica, A. pusilla, and A. faberi showed the highest numbers (30 sequences each), while A. maclurei, A. mamillata, A. omissa, and A. pusilla displayed the fewest. A. bullata and A. replicata had the lowest number of forward repeats (15 sequences each), whereas A. carnosicaulis, A. crenata, and A. polysticta had the highest count of reverse repetitive sequences, each with three sequences (Fig. 3B, Table S2).
Fig. 3.
Analyses of repeat sequences. A Box plot showing the distribution of four different types of repeats among 28 Ardisia species. B Histogram showing the number of repeats in the chloroplast genomes of 28 Ardisia species
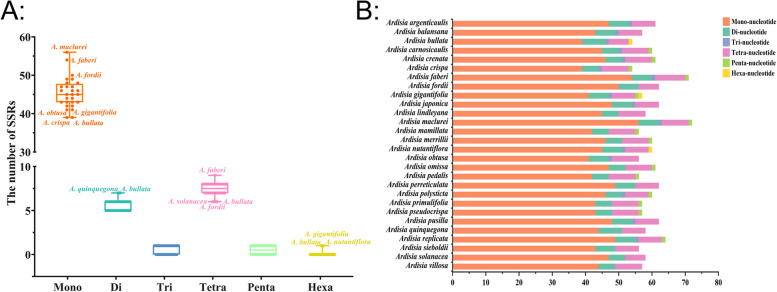
Microsatellites, also known as simple sequence repeats (SSRs), are continuous 1–6 bp nucleotide repeat units found in chloroplast genomes. A statistical analysis of 28 Ardisia species revealed a total of 1,669 SSRs, with each species containing between 54 and 72 SSRs (Table S3). Mononucleotide repeats accounted for 76.21% of all SSRs, with repeat numbers ranging from 39 to 56 among species. In addition, there were 157 dinucleotide repeats, 15 trinucleotide repeats, 208 tetranucleotide repeats, 14 pentanucleotide repeats, and 3 hexanucleotide repeats, representing 9.41%, 0.90%, 12.46%, 0.84%, and 0.18% of all SSRs, respectively. The repeat numbers for each type ranged from 5 to 7, 0 to 1, 6 to 9, 0 to 1, and 0 to 1, respectively (Table S3). Among the species, A. maclurei exhibited the highest number of mononucleotide repeats (56), while A. bullata and A. quinquegona had the most dinucleotide repeats (7 each), and A. faberi showed the highest count of tetranucleotide repeats (9). Trinucleotide and pentanucleotide repeats were only observed in 15 and 14 Ardisia species, respectively, with a single repeat identified in each species. Furthermore, only one hexanucleotide repeat was detected in the chloroplast genomes of A. bullata, A. gigantifolia, and A. nutantiflora (Fig. 4, Table S4).
Fig. 4.
Distribution maps of simple sequence repeats (SSRs) in the chloroplast genomes of 28 Ardisia species. A Box plot showing distribution of six SSR types among 28 Ardisia species. B Classification of SSRs in 28 Ardisia species by repeat type: mono-, mononucleotides; di-, dinucleotides; tri-, trinucleotides; tetra-, tetranucleotides; penta-, pentanucleotides; and hexa-, hexanucleotides
Comparative genomic analysis
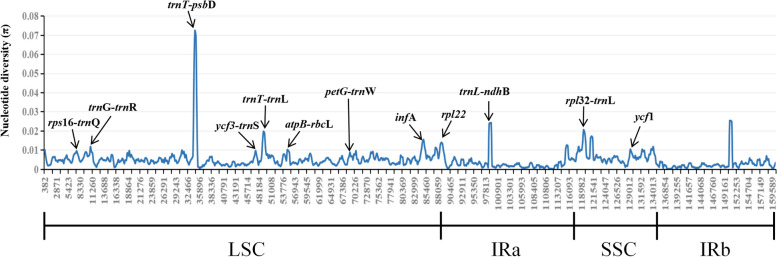
Using the Mauve Multiple Genome Comparison method, this study investigated rearrangements and colinearities in the chloroplast genomes of 28 Ardisia species (Fig. S2). The analysis identified two Locally Collinear Blocks (LCBs), indicating a high degree of similarity among the species. Nucleotide diversity (π) was also assessed using sliding window analysis with DnaSP to identify hotspot regions in the chloroplast genomes of Ardisia species (Fig. 5). The results revealed that intergenic regions displayed significantly higher levels of polymorphism compared to protein-coding regions. Specifically, the SSC region showed the highest nucleotide variation (average π = 0.00699), followed by the LSC region (average π = 0.00551) and the IR region (average π = 0.00271). In terms of protein-coding genes, infA, rpl22, and ycf1 exhibited higher π values (> 0.008) compared to other protein-coding genes. Nine intergenic regions (rps16-trnQ, trnG-trnR, trnT-psbD, ycf3-trnS, trnT-trnL, atpB-rbcL, petG-trnW, trnL-ndhB, and rpl32-trnL) exhibited high π values (π > 0.008).
Fig. 5.
Nucleotide diversity (π) of shared various regions in chloroplast genomes of 28 Ardisia species
Phylogenetic analysis
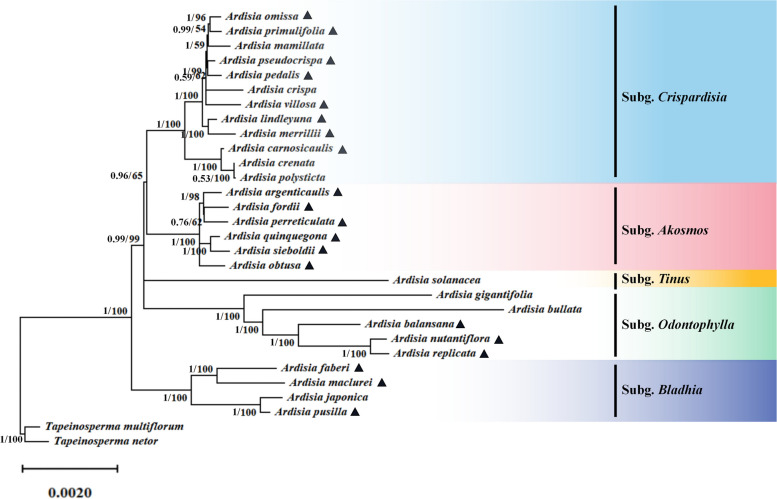
To assess the phylogenetic relationships among Ardisia species in China, 30 complete chloroplast genomes including the 28 Ardisia species and 2 outgroup species (Tapeinosperma multiflorum and T. netor), were used for phylogenetic analysis (Fig. 6, Table S1). Phylogenetic trees were constructed utilizing both maximum likelihood (ML) and Bayesian inference (BI) methods, considering five different data sets: complete chloroplast genome sequences, large single-copy (LSC) regions, inverted repeat (IR) regions, small single-copy (SSC) regions, and CDS datasets (Fig. 6, Fig. S3, Fig. S4, Fig. S5, Fig. S6). Analyses of the phylogenetic data from various datasets revealed subtle differences in topology, with phylogenetic trees based on complete chloroplast genomes showing the highest support. The results based on the complete chloroplast genome indicated that the 28 Ardisia species formed a monophyletic clade (BS = 100, PP = 1) which further divided into five smaller clades (Fig. 6). Among them, the subgenus Crispardisia was sister to the subgenus Akosmos, with both being sister to a group that included the species A. solanacea from the subgenus Tinus. Subgenus Odontophylla was sister to a monophyletic clade containing all remaining subgenera. Additionally, the subgenus Bladhia was identified as basal and sister to all other subgenera.
Fig. 6.
The phylogenetic tree is based on 30 complete chloroplast genome sequences using Bayesian inference (BI) and Maximum likelihood (ML) analyses. The number on the branches were Bayesian inference posterior probability/maximum likelihood bootstrap support values. The species marked with ▲ were newly collected in this study
Analysis of adaptive evolution
The nonsynonymous and synonymous substitution ratios (Ka/Ks) were calculated for the chloroplast genomes of 28 Ardisia species based on 79 common protein-coding genes, with Tapeinosperma netor used as a reference. Results showed that 22 genes had ratios that could not be calculated due to the absence of synonymous or nonsynonymous changes (Ka or Ks = 0), while the remaining 57 genes had average Ka/Ks ratios ranging from 0.009 (psbB) to 1.076 (rbcL). Notably, the average Ka/Ks ratio for the rbcL gene was above 1, and Ka/Ks ratios in 14 comparison groups were all greater than 1, indicating positive selection at specific chloroplast coding sites. Specifically, the Ka/Ks values for the rpoC1 gene in the comparison group between T. netor and A. crenata, A. japonica, and A. polysticta were 1.221, 1.221, and 1.003 respectively. For the rpoC2 gene in the comparison group between T. netor and A. argenticaulis, A. omissa, and A. pusilla, the Ka/Ks values were 1.202, 1.001, and 1.001 respectively. For the the rpl22 gene, the Ka/Ks values in the comparison groups of T. netor and A. balansan, A. gigantifolia, A. pedalis, and A. sieboldii were 1.422, 1.404, 1.233, and 1.123 respectively. Similarly, the Ka/Ks values of the rpl33 gene in T. netor and A. argenticaulis, A. bullata, A. crispa, A. lindleyana, A. maclureivs, A. omissa, and A. pusilla were all 1.147 (Table S5). It was observed that the Ka/Ks values of other genes were mostly less than or close to 1, suggesting that most protein-coding genes in the chloroplast genome of Ardisia species underwent purifying selection or experienced no selection pressure during the evolutionary process.
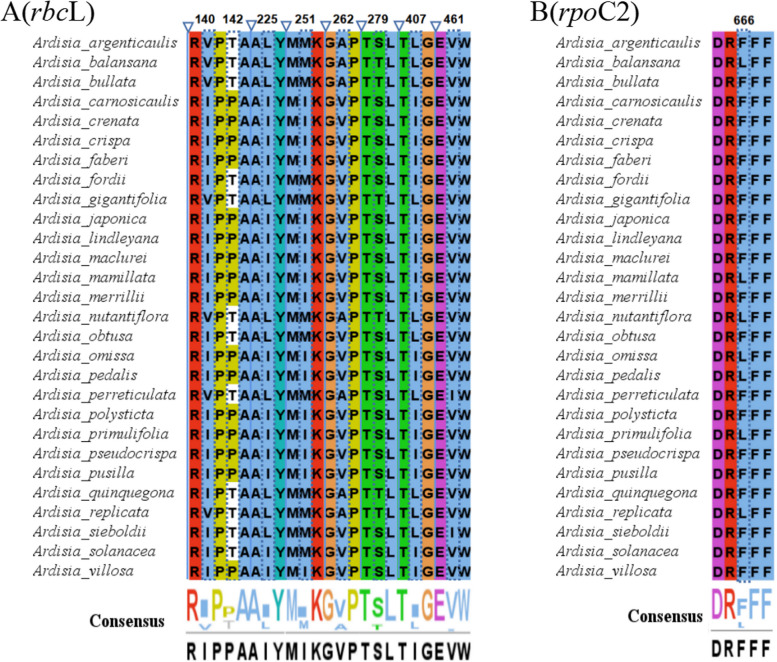
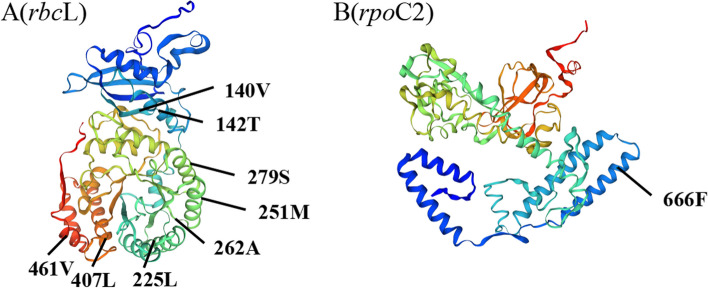
In addition, the protein-coding genes of the chloroplast genomes of 28 Ardisia species were analyzed using EasyCodeML v1.21 [18], and seven genes (cemA, ndhF, psbL, rbcL, rpoC2, ycf1, and ycf2) were identified under positive selection by a high posterior probability (> 95%) through the BEB test (Table S6). Therefore, only two genes (rbcL and rpoC2) were under positive selection using two selective pressure estimation strategies. Among the positively selected amino acid sites in the rbcL protein, two (140th and 262th) were located in the random coil, while the remaining six (142th, 225th, 251th, 279th, 407th and 461th) were located in the α-helix (Figs. 7A and 8A). In the RNA polymerase β subunit coding gene (rpoC2), one amino acid site (666th) was determined to be under positive selection (Fig. 7B), and spatial analysis showed that the site was situated in the α-helix (Fig. 8B).
Fig. 7.
Comparison of partial sites under positive selection of different genes
Fig. 8.
Spatial location of the positively selected sites in proteins of A. argenticaulis
Discussion
Sequence variation of chloroplast genomes
Within Angiosperms, it is commonly observed that the chloroplast genome adheres to a quadripartite structural arrangement, with typical sequence lengths falling within the range of 120–160 kb [19–21]. In this study, the length of complete chloroplast genomes of 28 Ardisia species ranged from 155,088 bp (A. bullata) to 156,999 bp (A. fordii) with an average length of 156,456 bp (Fig. 1; Table 1). The chloroplast genome across these species collectively comprised 129–132 genes, including 8 rRNA genes, 36–37 tRNA genes, and 85–88 protein-coding genes. The similar types and quantities of coding genes observed among these species suggest a degree of genetic preservation and stability within the chloroplast genomes of closely related plant species [22].
The inverted repeat (IR) region often undergoes contraction or expansion, which is a significant factor responsible for the variation in chloroplast genome length among angiosperm groups during evolution [23, 24]. By comparing analysis of the IR/SC boundary regions of the chloroplast genomes of the 28 Ardisia species, we observed dynamic changes in the IR regions, with some species (A. sieboldii, A. faberi, A. japonica, A. maclurei, and A. pusilla) showing noticeable expansion or contraction (Fig. 2). This phenomenon could be attributed to these species evolving at a faster rate or differentiating earlier, leading to a transformation of their genome structure during evolution, as observed in most other terrestrial plants [25].
Repeat sequences in chloroplast genome
Repeat sequences are commonly found in the chloroplast genome of plants, and their presence can result in duplication, deletion and rearrangement of segments, ultimately influencing species evolution and intraspecific genetic variation [26, 27]. In this study, we examined the chloroplast genomes of 28 Ardisia species and observed significant differences in the number and types of repeat sequences present in these genomes (Fig. 3, Table S2). Forward and palindromic repeats were the most prevalent, followed by reverse repeats, while complementary repeats were the least common. Not all species contained all four types of repeat sequences, and only six species had complementary repeats. Previous studies suggested that the number of repeat sequences could affect the stability of chloroplast genomes [28]. The variability in the number, type, and length of repeat sequences among species provides a potential basis for the development of new molecular genetic markers [29].
Simple Sequence Repeats (SSRs) in the chloroplast genome are highly variable in their internal specificity, making them valuable genetic markers in population genetic and evolutionary studies [30]. Our analysis revealed that the chloroplast genomes of 28 Ardisia species contained 54 to 72 SSR loci, most of which were single nucleotide repeat sequences composed of A/T repeat sequences (Fig. 4, Table S3, Table S4). The result is consistent with the sequence composition of SSRs in other angiosperm chloroplast genomes, further supporting the fact that SSRs primarily consist of short poly-A and poly-T repeats [31]. The variation in the distribution and number of SSRs among different Ardisia species may be attributed to mutations and deletions of gene sequences during plant evolution [32]. SSR markers with rich polymorphism could be utilized for variety purity detection, genetic diversity analysis, species identification and gene mapping of Ardisia plants in the future.
Identification of highly variable region
Due to the high conservation of protein-coding regions in the chloroplast genome, they are often not effective for species identification. Genes such as ycf1 and ycf2, which have high mutation rates and lengths over 5000 bp, present challenges for PCR amplification in practical applications. Therefore, identifying genic spacer regions with high variation rates and moderate lengths as potential DNA barcodes can assist in the identification of specific plant taxa [33–35]. In this study, nucleotide diversity (π) was calculated using the complete chloroplast genome sequences and identified 12 highly variable regions, including three protein-coding genes (infA, rpl22, and ycf1) and nine intergenic regions (rps16-trnQ, trnG-trnR, trnT-psbD, ycf3-trnS, trnT-trnL, atpB-rbcL, petG-trnW, trnL-ndhB, and rpl32-trnL) (Fig. 5). However, this method has limitations as it only considers differences after aligning all sequences, and these highly variable regions may not be suitable for identifying all species. Therefore, conducting pairwise comparisons of all species is necessary to ensure that the highly variable regions have a sequence length range that allows for conventional PCR product generation and contain enough variant sites to select the most appropriate sequences as DNA barcodes for accurate species identification.
Phylogenetic relationships
Ardisia includes a diverse group of species and varieties, making accurate identification challenging. The most comprehensive revision of Ardisia conducted by Mez identified 14 subgenera based on their habitats, leaf morphology, inflorescence position, and flower morphology in 1902 [36]. Since then, most taxonomic work on Ardisia has been limited to regional revisions [37–41]. Walker divided Ardisia into five sections based on various characters such as sepals, inflorescences, glandular dots, and leaf margins in his revision of the family Myrsinaceae of East Asia [41]. Chen et al. substantially followed Walker’s classification system, but included A. aberrans in section Pimelandra, and consequently, Ardisia species in China were ultimately divided into six Sect. [2]. Based on the phylogenetic relationship reconstructed using nuclear ITS and two chloroplast intergenic spaces (psbA-trnH and rpl32-trnL), the latest revised species classification indicates that Asian Ardisia was not a monophyletic group, and the relationships between most subgenera remained unresolved due to limited molecular markers [1].
Chloroplast genomes have proven successful in resolving phylogenetic relationships in various plant groups [42–45]. In this study, phylogenetic studies were conducted on the Ardisia plants in China based on chloroplast genome data of 28 Ardisia species and two outgroups (Tapeinosperma multiflorum and T. netor). Phylogenetic analysis based on different datasets, including LSC region, SSC region, IR region, coding sequences, and complete chloroplast genome, revealed that the 28 Ardisia species formed a monophyletic group, further divided into smaller clades: subgenus Crispardisia, subgenus Akosmos, subgenus Tinus, subgenus Odontophylla, and subgenus Bladhia (Fig. 6, Fig. S3, Fig. S4, Fig. S5, Fig. S6). The result supported the recent classification revision that the traditional subgenus Bladhia s.l. should be split into two clades: subgenus Bladhia s.str. and subgenus Odontophylla (Yang) Huang which could be distinguished based on distribution range and morphology [1, 46]. It is worth noting that the phylogenetic trees generated from different datasets reveal a polytomy at the root of the clade containing all subgenera except subgenus Bladhia. As a result, the relationships between certain clades remain unclear, including: (a) the clade consisting of the subgenera Crispardisia and Akosmos; (b) the subgenus Tinus; and (c) the subgenus Odontophylla.
In this study, the subgenus Crispardisia clade comprised 12 species (Fig. 7, Fig. S3). According to Mez’s taxonomic system, A. mamillata and A. primulifolia were initially placed in the subgenus Bladhia [36]. However, Pitard argued that the presence of marginal glandular dots on the leaves were the most distinctive feature of the subgenus Crispardisia, leading to the reclassification of these two species [47]. The molecular phylogenetic analysis in this study supported Pitard’s taxonomic treatment and suggested that assigning the two species to the subgenus Crispardisia was more appropriate. As a controversial species, A. argenticaulis was placed in the subgenus Bladhia by Walker [37]. However, Wang et al. argued that A. argenticaulis should be classified into the subgenus Akosmos [48]. The phylogenetic tree in this study supported that A. argenticaulis belongs to the subgenus Akosmos (Fig. 6, Fig. S3, Fig. S4, Fig. S5, Fig. S6). Overall, the phylogenetic analysis conducted in this study provided molecular evidence to support previous findings, and enhanced our understanding of the relationships between subgenus and species within Ardisia in China.
Adaptive selection on Ardisia chloroplast genes
Ardisia species are extremely shade-tolerant and primarily found in the understory of forests or in moist areas near valleys and streams. The influence of low light as a natural selective pressure may have resulted in adaptive changes in the chloroplast genes responsible for environmental adaptation [49–51]. In Chrysosplenium (Saxifragaceae), two genes related to photosynthesis (matK and ycf2) showed positive selective pressure, which might be associated with adaptation to low light conditions [17]. Similarly, we identified two genes that were under positively selection among the 28 Ardisia species, including rbcL and rpoC2 (Figs. 7 and 8, Table S5, Table S6).
The rbcL gene for the Rubisco large subunit was identified under positive selection (Figs. 7 and 8). Being the selection target of multiple environmental factors related to light, temperature, and carbon dioxide concentration, the rbcL gene is often under positive selection [52]. The positive selection of rbcL gene related with photosynthesis, suggested their role in the adaptation of Ardisia species to low-light habitats. Plastid-encoded RNA polymerase (PEP) is composed of four subunits (ɑ, β, β’, β’’) which are encoded by the rpoA, rpoB, rpoC1, and rpoC2 genes respectively [53–55]. Previous studies indicate that PEP is a crucial enzyme responsible for the transcription of photosynthesis genes in chloroplasts [56]. We identified an amino acid site under positive selection in the rpoC2 gene of Ardisia species in this study (Figs. 7 and 8), which suggested that PEP might play a significant role in expression of photosynthesis genes of adapting to the environment. In summary, two genes (rbcL and rpoC2) were under positive selection, which might have contributed to the adaptation of Ardisia species to various environmental conditions, particularly those characterized by low light levels.
Conclusions
The chloroplast genomes of 28 Ardisia species exhibited a typical tetrad structure, ranging in length from 155,088 bp to 156,999 bp. These genomes consisted of a large single-copy region (LSC) spanning 84,709 bp to 86,989 bp, a small single-copy region (SSC) ranging from 18,093 bp to 18,479 bp, and a pair of inverted repeat regions (IR) spanning 25,411 bp to 26,236 bp. The GC content ranged from 36.0 to 37.3%. The gene composition remained relatively conserved, comprising 129–132 coding genes including 85–88 protein-coding genes, 36–37 tRNA genes, and 8 rRNA genes, with 18 genes containing introns. Palindromic repeats were the most common among the repeat sequences, and the chloroplast genome sequences contained numerous SSR sites, with A/T being the predominant component. Comparative analysis of the chloroplast genome identified 12 highly variable regions, including 3 protein-coding genes (infA, rpl22, and ycf1) and 9 intergenic regions (rps16-trnQ, trnG-trnR, trnT-psbD, ycf3-trnS, trnT-trnL, atpB-rbcL, petG-trnW, trnL-ndhB, and rpl32-trnL), which could serve as potential DNA barcode markers for identifying Ardisia species. The phylogenetic tree supported the division of the subgenus Bladhia s.l. into subgenus Bladhia s.str. and subgenus Odontophylla (Yang) Huang. Notably, two genes (rbcL and rpoC2) exhibited positive selection, potentially linked to the adaptation of Ardisia species to low-light environments. This comprehensive study offers valuable insights into the chloroplast genome of Ardisia species, facilitating species identification and the utilization of germplasm resources.
Materials and methods
Plant materials and DNA extraction and sequencing
Twenty Ardisia species were collected in Guangxi, China and cultivated at the Guangxi Institute of Botany (Fig. S1, Table S1). Genomic DNA extraction was carried out from fresh green leaves using a modified CTAB method [57]. The extracted DNA was subjected to a series of meticulous procedures, including mechanical fragmentation, purification, terminal repair, and other necessary treatments prior to sequencing. Fragments of 350 bp were selected through agarose gel electrophoresis and subsequently amplified through PCR to establish a sequence library. High-throughput sequencing of the library was performed using the Illumina HiSeq 2000 sequencer (Illumina Biotechnology Company, San Diego, CA, USA) to generate paired-end (PE) reads. The genome was reassembled from the filtered data using NOVOPlasty (v.2.7.2) [58]. To ensure assembly accuracy, Bowtie2 (v2.0.1) was used to map all high-quality clean reads to the assembled genome sequence [59]. Finally, complete chloroplast genome sequences of 20 Ardisia species (accession number: OK054492-OK514747) were obtained, and a combined analysis of these genomes, along with the other 8 published chloroplast genomes acquired from GenBank was conducted (Table S1).
Genome annotation and sequence characterization
The annotation results were compared with the reference genome annotation information using Geneious v8.0.2 software to confirm the annotation. The positions of the stop and start codons of some protein-coding genes were manually adjusted. Geneious v8.0.2 software was also used for chloroplast genome boundary annotation [60]. A circular chloroplast genome map was created using the web program Organellar Genome DRAW [61].
Boundary regions and genome comparative analysis
The annotation of chloroplast genomes for 28 Ardisia species, including the boundaries of LSC, SSC, two IRs regions and genes near each boundary was mapped onto a simplified chloroplast genome structure map using IRscope (https://irscope.shinyapps.io/irapp/). Mauve Alignment in Geneious Prime was used for the chloroplast genome collinear analysis among the 28 Ardisia species [62]. Nucleotide polymorphisms (π) were calculated among the 28 Ardisia species using DnaSP v6.0 software, with specific parameter settings comprising a window length of 600 and a step length of 200 [63].
Repeat sequences analysis
Repeat sequences, encompassing forward, reverse, complement, and palindromic repeats, were quantified employing the online tool REPuter, accessible at https://bibiserv.cebitec.uni-bielefeld.de/reputer/manual.html, utilizing the following specific parameters: minimum repeat sequence length > 30 bp, repeat sequence similarity > 90%, and Hamming distance = 3 [64]. Simple sequence repeats were analyzed using the online tool MISA-web for identification and statistical analysis [65]. The parameters were set as follows: mononucleotide ≥ 10 repeat units, dinucleotide ≥ 5 repeat units, trinucleotide ≥ 4 repeat units, and tetra-, penta-, and hexanucleotides with at least 3 repeat units.
Phylogenetic Analysis
To infer phylogenetic relationships among Ardisia species, we independently analyzed the complete chloroplast genome and specific DNA fragments. The outgroup species (Tapeinosperma multiflorum and T. netor) were selected based on the research of Yan et al. [66]. The chloroplast genome sequences and specific DNA fragments were aligned using MAFFT (version 7.222) [67]. Maximum likelihood (ML) tree analysis of the aligned sequences was performed using RAxML 7.2.8 software with the best model of TVM + F + I + I + R4, determined through 1,000 guided repeat tests [68]. The Bayesian Inference (BI) tree was constructed using MrBayes v.3.2.6 software [69]. The Markov Chain Monte Carlo (MCMC) algorithm was run for 1,000,000 generations under the TPM1uf + I + G model, sampling every 1,000 generations. We ensured that the average standard deviation of the split frequencies remained below 0.01. Additionally, 25% of the samples from the burn-in phase were discarded before computing the consensus tree and determining the Bayesian posterior probabilities (PP).
Analysis of adaptive evolution
Twenty-eight Ardisia species and Tapeinosperma netor (used as a reference) were selected for the analysis of selection pressure on chloroplast genome protein-coding genes. The sequences of 79 shared chloroplast protein-coding genes were compared individually, and stop codons were removed. The ratio of paired nonsynonymous substitution rate (Ka) to synonymous substitution rate (Ks) was then calculated for all species using KaKs Calculator v2.0 [70]. To predict selection for each gene, we considered the ratios Ka/Ks. A ratio of Ka/Ks < 1 indicates purifying selection, a ratio of Ka/Ks = 1 indicates neutral selection, and a ratio of Ka/Ks > 1 indicates positive selection [71]. If Ks was 0, the Ka/Ks values were expressed as NA.
Furthermore, the site model (set to seqtype = 1, model = 0, NSsites = 0, 1, 2, 3, 7, 8) was used to perform the LRT (likelihood ratio test). The log-likelihoods of each model and the neutral model were compared using LRT to test for statistical significance. The alternative hypothesis model M8 was accepted if the p-value was less than 0.05 in the LRT results; otherwise, the null hypothesis model M7 was accepted. Genes with positive selection sites under the M8 model, where p < 0.05 and the Bayesian Empirical Bayes (BEB) posterior probability exceeded 0.95, were considered potential positively selected genes [72]. Upon detection of positive selection by LRT, the Bayesian Empirical Bayes (BEB) method was applied to estimate the posterior probability of each codon from the positive selection site category in models M2a and M8. The Bayesian Empirical Bayes method represents an improvement over the previous Naïve Empirical Bayes method, accounting for sampling errors of maximum likelihood estimates within the model [73–76]. Amino acid sequences were visualized using Jalview software to highlight positively selected sites [77]. To gain further insight into the structural characteristics of these genes, we utilized the online protein structure prediction tool SWISS-MODEL, with A. argenticaulis as an example [78].
Supplementary Information
Acknowledgements
Not applicable.
Abbreviations
- Cp
Chloroplast
- LSC
Large single-copy region
- IR
Inverted repeat region
- SSC
Small single-copy region
- CDS
Protein-coding sequences
- tRNAs
Transport RNAs
- rRNAs
Ribosomal RNAs
- SSRs
Simple sequence repeats
- π
Nucleotide diversity
- Ka/Ks
The rate of non-synonymous substitutions to the rate of synonymous substitutions
- RSCU
Relative synonymous codon usage
- Pi
Nucleotide diversity
- ML
Maximum-likelihood
- BEB
Bayes empirical bayes
- NCBI
National Center for Biotechnology Information
Authors’ contributions
ZJ: conceived the study, performed data analysis and drafted the manuscript; NY: collected and identifed the species of sample, designed the experiments, analyzed the data; LJ, DY and WL: assisted in conceptualizing and designing experiments; MS: collected and identifed the species of sample, data curation; ZB: funding acquisition, reviewed the manuscript critically. All authors have read and agreed with the contents of the manuscript
Funding
This work was supported by National Natural Science Foundation of China (32060090), Natural Science Foundation of Guangxi Province (2021JJA130119) and Science and technology plan project of Guangzhou Construction Group ([2022]-KJ019), ([2021]-KJ014).
Data availability
All data generated or analysed during this study are included in this manuscript. All the annotated chloroplast sequences data reported here were deposited in GenBank (https://www.ncbi.nlm.nih.gov/) with accession numbers and voucher number shown in Table S1.
Declarations
Ethics approval and consent to participate
The collection of all samples completely complies with national and local legislation permission. Plant samples used in the study were not included in the list of national key protected plants and were not collected from the national park or nature reserve when we collected them. According to national and local legislation, no specifc permission was required for collecting these plants when we collected them. Voucher specimens were prepared and deposited at the Herbarium of Guangxi Institute of Botany, Chinese Academy of Sciences (IBK).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jin Zhang and Yangyang Ning contributed equally to this work.
Contributor Information
Shizhong Mao, Email: 943437887@qq.com.
Bo Zhao, Email: 122017017@glmc.edu.cn.
References
- 1.Yang CJ, Hu JM. Molecular phylogeny of Asian Ardisia (Myrsinoideae, Primulaceae) and their leaf-nodulated endosymbionts, Burkholderia s.l. (Burkholderiaceae). PLoS ONE. 2022;17(1):e0261188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walker EH. A revision of the eastern Asiatic Myrsinaceae. Philip J Sci. 1940;73(1/2):1–258. [Google Scholar]
- 3.Chen J, Pipoly JJ, Myrsinaceae. In: Wu Z, Raven PH, editors. Flora of China. Beijing: Science; 1996. pp. 1–38. [Google Scholar]
- 4.Mu LH, Bai L, Dong XZ, Yan FQ, Guo DH, Zheng XL, Liu P. Antitumor activity of triterpenoid saponin-rich Adisia gigantifolia extract on human breast adenocarcinoma cells in vitro and in vivo. Biol Pharm Bull. 2014;37(6):1035–41. [DOI] [PubMed] [Google Scholar]
- 5.de Mejía EG, Ramírez-Mares MV. Ardisia: health-promoting properties and toxicity of phytochemicals and extracts. Toxicol Mech Methods. 2011;21(9):667–74. [DOI] [PubMed] [Google Scholar]
- 6.Xin X, Yu D, Zhu L, Gu ZX, Yuan L, Huang S. Qualitative and quantitative method for compound Aidicha tablets. Cent South Pharma. 2015;13:410–3. [Google Scholar]
- 7.Jasamai M, Jalil J, Jantan I. Molecular docking study on platelet-activating factor antagonistic activity of bioactive compounds isolated from Guttiferae and Ardisia species. Nat Prod Res. 2015;29(11):1055–8. [DOI] [PubMed] [Google Scholar]
- 8.Liu B, Liu R, Liu Q, Ashby CR Jr, Zhang H, Chen ZS. The ethnomedicinal and functional uses, phytochemical and pharmacology of compounds from Ardisia species: an updated review. Med Res Rev. 2022;42(5):1888–929. [DOI] [PubMed] [Google Scholar]
- 9.Liu YM, Wang K, Liu Z, Luo K, Chen SL, Chen KL. Identification of medical plants of 24 Ardisia species from China using the matK genetic marker. Pharmacogn Mag. 2013;9:331–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiong C, Sun W, Wu L, Xu R, Zhang Y, Zhu W, Panjwani JHE, Liu Z, Zhao B. Evaluation of four commonly used DNA barcoding loci for Ardisia species Identification. Front Plant Sci. 2022;13:860778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang XF, Landis JB, Wang HX, Zhu ZX, Wang HF. Comparative analysis of chloroplast genome structure and molecular dating in Myrtales. BMC Plant Biol. 2021;21(1):219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Q, Zhao B, Zhang J, Lu ZC, Liang JS, Li JJ. High resolution melting of chloroplast mini-barcode in star anise (Illicium verum) authentication. Ind Crops Prod. 2023;197:116626. [Google Scholar]
- 13.Hou N, Li M, Shen J, Chen Z, Luo Y, Deng L. The complete chloroplast genome of Ardisia mamillata (Myrsinaceae). Mitochondrial DNA B Resour. 2019;4(2):3441–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi Y, Liu B. Complete chloroplast genome sequence of Ardisia Gigantifolia (Myrsinaceae), a vulnerable medicinal plant. Mitochondrial DNA B Resour. 2019;4(2):4037–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z, Dos RM. Statistical properties of the branch-site test of positive selection. Mol Biol Evol. 2011;28(3):1217–28. [DOI] [PubMed] [Google Scholar]
- 16.Dong WL, Wang RN, Zhang NY, Fan WB, Fang MF, Li ZH. Molecular evolution of Chloroplast genomes of Orchid species: insights into phylogenetic relationship and adaptive evolution. Int J Mol Sci. 2018;19(3):716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Z, Liao R, Yang T, Dong X, Lan D, Qin R, Liu H. Analysis of six chloroplast genomes provides insight into the evolution of Chrysosplenium (Saxifragaceae). BMC Genomics. 2020;21(1):621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao F, Chen C, Arab DA, Du Z, He Y, Ho SYW. EasyCodeML: a visual tool for analysis of selection using CodeML. Ecol Evol. 2019;9(7):3891–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Price M, Su DM, et al. Phylogeny and Comparative Analysis for the Plastid Genomes of Five Tulipa (Liliaceae). Biomed Res Int. 2021;2021:6648429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han C, Ding R, Zong X, Zhang L, Chen X, Qu B. Structural characterization of Platanthera Ussuriensis chloroplast genome and comparative analyses with other species of Orchidaceae. BMC Genomics. 2022;23(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang H, Tian J, Yang J, Dong X, Zhong Z, Mwachala G, Zhang C, Hu G, Wang Q. Comparative and phylogenetic analyses of six Kenya Polystachya (Orchidaceae) species based on the complete chloroplast genome sequences. BMC Plant Biol. 2022;22(1):177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan X, Wang W, Wagutu GK, Li W, Li X, Chen Y. Fifteen complete chloroplast genomes of Trapa species (Trapaceae): insight into genome structure, comparative analysis and phylogenetic relationships. BMC Plant Biol. 2022;22(1):230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maréchal A, Brisson N. Recombination and the maintenance of plant organelle genome stability. New Phytol. 2010;186(2):299–317. [DOI] [PubMed] [Google Scholar]
- 24.Jin G, Li W, Song F, Yang L, Wen Z, Feng Y. Comparative analysis of complete Artemisia subgenus Seriphidium (Asteraceae: Anthemideae) chloroplast genomes: insights into structural divergence and phylogenetic relationships. BMC Plant Biol. 2023;23(1):136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang F, Wang T, Shu X, Wang N, Zhuang W, Wang Z. Complete chloroplast genomes and comparative analyses of L. Chinensis, L. Anhuiensis, and L. Aurea (Amaryllidaceae). Int J Mol Sci. 2020;21(16):5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong PB, Wang RN, Afzal N, Liu ML, Yue M, Liu JN, Tan JL, Li ZH. Phylogenetic relationships and molecular evolution of woody forest tree family Aceraceae based on plastid phylogenomics and nuclear gene variations. Genomics. 2021;113(4):2365–76. [DOI] [PubMed] [Google Scholar]
- 27.Qin HH, Cai J, Liu CK, Zhou RX, Price M, Zhou SD, He XJ. The plastid genome of twenty-two species from Ferula, Talassia, and Soranthus: comparative analysis, phylogenetic implications, and adaptive evolution. BMC Plant Biol. 2023;23(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo YY, Yang JX, Bai MZ, Zhang GQ, Liu ZJ. The chloroplast genome evolution of Venus slipper (Paphiopedilum): IR expansion, SSC contraction, and highly rearranged SSC regions. BMC Plant Biol. 2021;21(1):248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Provan J, Corbett G, McNicol JW, Powell W. Chloroplast DNA variability in wild and cultivated rice (Oryza spp.) revealed by polymorphic chloroplast simple sequence repeats. Genome. 1997;40(1):104–10. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Y, Yin J, Guo H, Zhang Y, Xiao W, Sun C, Wu J, Qu X, Yu J, Wang X, Xiao J. The complete chloroplast genome provides insight into the evolution and polymorphism of Panax ginseng. Front Plant Sci. 2015;5:696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ping J, Feng P, Li J, Zhang R, Su Y, Wang T. Molecular evolution and SSRs analysis based on the chloroplast genome of Callitropsis Funebris. Ecol Evol. 2021;11(9):4786–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo H, Wang L, Xu W, Huo Z, Yang P, Zhang Q, Wang H, Li P, Lu X. The complete chloroplast genome sequence of Cyathula officinalis and comparative analysis with four related species. Gene. 2022;839:146728. [DOI] [PubMed] [Google Scholar]
- 33.Abdullah, Henriquez CL, Croat TB, Poczai P, Ahmed I. Mutational dynamics of Aroid Chloroplast genomes II. Front Genet. 2021;11:610838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu J, Fu J, Fang Y, Xiang J, Dong H. Complete chloroplast genomes of Rubus species (Rosaceae) and comparative analysis within the genus. BMC Genomics. 2022;23(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gong L, Ding X, Guan W, Zhang D, Zhang J, Bai J, Xu W, Huang J, Qiu X, Zheng X, Zhang D, Li S, Huang Z, Su H. Comparative chloroplast genome analyses of Amomum: insights into evolutionary history and species identification. BMC Plant Biol. 2022;22(1):520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mez C, Myrsinaceae In: A,. Das pflanzenreich, heft 9, IV. Fam. 236. Leipzig: Verlag von Wilhelm Engelmann; 1902. pp. 1–473. [Google Scholar]
- 37.Swartz MD. Nova Genera et Species Plantarum seu Prodromus, Vol. 3. Swederi: Holmiae, Upsaliae, & Aboae. 1788:1–48.
- 38.Stone BC. New and noteworthy Malaysian Myrsinaceae. I Malays for. 1982;45(1):101–21. [Google Scholar]
- 39.Chase MW, Reveal JL. A phylogenetic classification of the land plants to accompany APG III. Bot J Linn Soc. 2009;161(2):122–7. [Google Scholar]
- 40.Chase MW, Christenhusz MJM, Fay MF. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 2016;181(1):1–20. [Google Scholar]
- 41.Walker EH. A revision of the eastern Asiatic Myrsinaceae. Philippine J Sci. 1940;73:1–258. [Google Scholar]
- 42.Tang C, Chen X, Deng Y, Geng L, Ma J, Wei X. Complete chloroplast genomes of Sorbus Sensu Stricto (Rosaceae): comparative analyses and phylogenetic relationships. BMC Plant Biol. 2022;22(1):495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Javaid N, Ramzan M, Khan IA, Alahmadi TA, Datta R, Fahad S, Danish S. The chloroplast genome of Farsetia Hamiltonii Royle, phylogenetic analysis, and comparative study with other members of Clade C of Brassicaceae. BMC Plant Biol. 2022;22(1):384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li E, Liu K, Deng R, Gao Y, Liu X, Dong W, Zhang Z. Insights into the phylogeny and chloroplast genome evolution of Eriocaulon (Eriocaulaceae). BMC Plant Biol. 2023;23(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang L, Deng S, Zhu Y, Da Q. Comparative chloroplast genomics of 34 species in subtribe Swertiinae (Gentianaceae) with implications for its phylogeny. BMC Plant Biol. 2023;23(1):164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang YP, Dwyer JD. Taxonomy of subgenus Bladhia of Ardisia (Myrsinaceae). Taiwania. 1989;34(2):192–298. [Google Scholar]
- 47.Pitard J, Myrsinaceae H, Lecomte. Fl Indo-Chine. 1930;3:765–877. [Google Scholar]
- 48.Wang J, Xia HN. New synonym of Chinese Ardisia (Myrsinaceae), with critical notes on the status of the subgenus Chinensia. Trop Subtropical J Bot. 2009;17(1):83–5. [Google Scholar]
- 49.Ivanova Z, Sablok G, Daskalova E, Zahmanova G, Apostolova E, Yahubyan G, Baev V. Chloroplast Genome Analysis of Resurrection Tertiary Relict Haberlea rhodopensis highlights genes important for desiccation stress response. Front Plant Sci. 2017;8:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Piot A, Hackel J, Christin PA, Besnard G. One-third of the plastid genes evolved under positive selection in PACMAD grasses. Planta. 2018;247(1):255–66. [DOI] [PubMed] [Google Scholar]
- 51.Li B, Liu T, Ali A, Xiao Y, Shan N, Sun J, Huang Y, Zhou Q, Zhu Q. Complete chloroplast genome sequences of three aroideae species (Araceae): lights into selective pressure, marker development and phylogenetic relationships. BMC Genomics. 2022;23(1):218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Architecture of the photosynthetic oxygen-evolving center. Science. 2004;303(5665):1831–8. [DOI] [PubMed] [Google Scholar]
- 53.Loll B, Kern J, Saenger W, Zouni A, Biesiadka J. Towards complete cofactor arrangement in the 3.0 a resolution structure of photosystem II. Nature. 2005;438(7070):1040–4. [DOI] [PubMed] [Google Scholar]
- 54.Guskov A, Kern J, Gabdulkhakov A, Broser M, Zouni A, Saenger W. Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride. Nat Struct Mol Biol. 2009;16(3):334–42. [DOI] [PubMed] [Google Scholar]
- 55.Lu Y. Identification and roles of Photosystem II Assembly, Stability, and repair factors in Arabidopsis. Front Plant Sci. 2016;7:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong L, Zhou W, Wang H, Ding S, Lu Q, Wen X, Peng L, Zhang L, Lu C. Chloroplast small heat shock protein HSP21 interacts with plastid nucleoid protein pTAC5 and is essential for chloroplast development in Arabidopsis under heat stress. Plant Cell. 2013;25(8):2925–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Doyle JJ. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–5. [Google Scholar]
- 58.Dierckxsens N, Mardulyn P, Smits G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017;45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramirez-Gonzalez RH, Bonnal R, Caccamo M, Maclean D. Bio-samtools: Ruby bindings for SAMtools, a library for accessing BAM files containing high-throughput sequence alignments. Source Code Biol Med. 2012;7(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Greiner S, Lehwark P, Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47(W1):W59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5(6):e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 2017;34(12):3299–302. [DOI] [PubMed] [Google Scholar]
- 64.Lei W, Ni D, Wang Y, Shao J, Wang X, Yang D, Wang J, Chen H, Liu C. Intraspecific and heteroplasmic variations, gene losses and inversions in the chloroplast genome of Astragalus Membranaceus. Sci Rep. 2016;6:21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beier S, Thiel T, Münch T, Scholz U, Mascher M. MISA-web: a web server for microsatellite prediction. Bioinformatics. 2017;33(16):2583–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yan X, Liu T, Yuan X, Xu Y, Yan H, Hao G. Chloroplast genomes and comparative analyses among Thirteen Taxa within Myrsinaceae s.str. Clade (Myrsinoideae, Primulaceae). Int J Mol Sci. 2019;20(18):4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–90. [DOI] [PubMed] [Google Scholar]
- 69.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteom Bioinf. 2010;8(1):77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21(1):139–76. [DOI] [PubMed] [Google Scholar]
- 72.Anisimova M, Bielawski JP, Yang Z. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol Biol Evol. 2001;18(8):1585–92. [DOI] [PubMed] [Google Scholar]
- 73.Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148(3):929–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang Z, Swanson WJ. Codon-substitution models to detect adaptive evolution that account for heterogeneous selective pressures among site classes. Mol Biol Evol. 2002;19(1):49–57. [DOI] [PubMed] [Google Scholar]
- 75.Yang Z, Wong WS, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22(4):1107–18. [DOI] [PubMed] [Google Scholar]
- 76.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–91. [DOI] [PubMed] [Google Scholar]
- 77.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this manuscript. All the annotated chloroplast sequences data reported here were deposited in GenBank (https://www.ncbi.nlm.nih.gov/) with accession numbers and voucher number shown in Table S1.