Abstract
Background
Clear cell renal cell carcinoma (ccRCC) is a common histological subtype of malignant renal neoplasm. Protein lysine lactylation (Kla) plays a crucial role in tumor metabolic reprogramming. However, little is known regarding the distribution and potential biological functions of Kla in ccRCC. This study aimed to systematically investigate the role of Kla in ccRCC.
Methods
A total of 12 ccRCC samples were collected from 6 patients. Western blotting was performed to determine the trend of Kla-modified proteins in ccRCC. Liquid chromatography–tandem mass spectrometry was used to quantitatively analyze Kla in ccRCC. Gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and protein–protein interaction (PPI) network analyses were conducted to clarify the biological functions and interactional relationships of differentially lactylated proteins (DLPs).
Results
In total, 239 DLPs, including 441 lactylated sites, were identified by comparing ccRCC tissues with adjacent normal tissues. Kla-related enzymes have a higher affinity for alanine than for other amino acid residues in ccRCC. Subcellular localization analysis revealed that most DLPs were localized in the cytoplasm and mitochondria. GO enrichment analysis showed that most of the DLPs were enriched in metabolism-associated biological processes, including the purine ribonucleotide, monocarboxylic acid, ribonucleoside triphosphate, purine nucleoside triphosphate, and ATP metabolic processes. KEGG analysis indicated that most DLPs were also enriched in metabolism-related pathways, including glycolysis, amino acid (valine, leucine, and isoleucine) degradation, pyruvate metabolism, fatty acid degradation, and the citrate cycle. The top 20 hub proteins were screened from the PPI network based on their degree ranks.
Conclusions
This study revealed the role of Kla in ccRCC, which will extend our understanding of the potential molecular mechanisms underlying ccRCC formation and progression. These key Kla-modified proteins may be promising therapeutic targets for the treatment of ccRCC. However, further molecular experiments are required to validate these findings.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40001-024-02200-z.
Keywords: Clear cell renal cell carcinoma, Lysine lactylation, Gene ontology, Kyoto encyclopedia of genes and genomes, Bioinformatics
Introduction
Renal cancer is a common malignant tumor of the urinary system, characterized by rapid progression and high mortality. Recent cancer statistical data from the United States showed that the estimated number of new cases and deaths from renal cancer were 79,000 and 13,920, respectively, in 2022 [1]. Similarly, renal cancer is also one of the leading cancers, with high morbidity and mortality in China [2]; the estimated new cases and deaths were 77,410 and 31,172, respectively, in 2022 [2]. Clear cell renal cell carcinoma (ccRCC) is the most common histological subtype of renal cancer, with a high degree of malignancy [3]. Although therapeutic strategies for ccRCC have considerably progressed, the prognosis of patients with ccRCC remains unsatisfactory. Hence, the development of new treatment strategies to improve the prognosis of patients with ccRCC is urgently needed, and a deeper understanding of the pathogenesis and development of ccRCC will be helpful in achieving this goal.
Protein lysine lactylation (Kla) is a novel and important post-translational modification (PTM) that plays a vital role in tumor pathogenesis and development [4, 5]. Kla-modified proteins are widely involved in cancer metabolic reprogramming, immunoregulation, and cell differentiation via the regulation of protein activity, thereby changing molecular expression patterns [6–9]. For example, Kla-modified proteins affect tumorigenesis, progression, and metastasis by the Warburg effect [6, 10]. In addition, Kla-modified proteins can affect tumor development by regulating differentiation and infiltration of immune cells [4, 6]. Given the importance of Kla-modified proteins in cancer progression, they are also considered novel and promising targets for cancer treatment [4, 11–13]. Currently, the relationship between ccRCC progression and Kla modification of proteins remains unclear. Thus, comprehensive identification of lactylated proteins will provide insights into the potential mechanism of ccRCC development and help explore new therapeutic strategies for ccRCC.
Liquid chromatography–mass spectrometry technique (LC–MS) is a powerful tool for identifying the pattern of Kla in tissues [14, 15]. In this study, LC–MS was used to systematically and comprehensively identify the patterns of Kla modification in ccRCC. We found that there is an abundance of lactylated proteins in ccRCC, most of which are closely related to metabolic pathways. The results of this study provide a new perspective for understanding the relationship between protein Kla and the development of ccRCC. To the best of our knowledge, this is the first study to comprehensively explore the characteristics of Kla in ccRCC tissues.
Material and methods
ccRCC samples
All 12 resected ccRCC samples, including six cancer and six adjacent normal tissue samples, were collected from six patients diagnosed with ccRCC at the Haikou Affiliated Hospital of Central South University Xiangya School of Medicine. The histological subtypes of the tissues were definitively diagnosed by two pathologists. This study was approved by the Ethics Committee of the Haikou Affiliated Hospital of Central South University Xiangya School of Medicine.
Protein extraction and digestion
First, ccRCC samples were retrieved from the − 80 ℃ refrigerator and fully ground to powder with liquid nitrogen. Second, ccRCC samples were added to lysis buffer containing 1% Triton X-100, 1% protease inhibitor cocktail, 3 µM trichostatin A [TSA], and 50 mM nicotinamide [NAM]). The ccRCC samples were then sonicated three times on ice using a high-intensity ultrasonic processor. Next, the ccRCC samples were centrifuged at 2,000 × g for 10 min at 4 ℃ to remove the cell debris. Finally, the clear supernatant was extracted, and the protein concentration was determined using a bicinchoninic acid kit. The extracted proteins were zymolyzed using pancreatin, as described previously [10].
Western blot analysis
Total proteins were extracted from ccRCC and adjacent normal tissues and used to detect protein lactylation modifications. The experimental methods have been reported in a previous study [16]. Briefly, proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose filter membrane. After blocking with 5% milk, immunoblotting was performed using a pan-anti-Kla multiclonal antibody (WM101; Micrometer Biotech Company, Hangzhou, China).
Affinity enrichment
Peptides were dissolved in an immunoprecipitation (IP) buffer solution (100 mM NaCl, 1 mM EDTA, 50 mM Tris–HCl, and 0.5% NP-40, pH 8.0) and transferred to a pre-washed lactylation resin (PTM-1404, PTM Bio) for incubation at 4 °C overnight with gentle shaking. After incubation, the resin was washed four times with the IP buffer solution and twice with deionized water. Kla-modified peptides bound to the resin were eluted three times with 0.1% trifluoroacetic acid, and the eluted fractions were collected and vacuum-dried. The resulting peptides were desalted using C18 ZipTips according to the manufacturer’s instructions and then vacuum-dried again before LC–MS analysis.
LC–MS analysis
Peptides were dissolved in mobile phase A, which was a water-based solution containing 0.1% formic acid and 2% acetonitrile. Mobile phase B consisted of a mixture of acetonitrile and water containing 0.1% formic acid. A gradient elution was performed using the following conditions: 7% to 24% B from 0 to 42 min, 24% to 32% B from 42.0 to 54.0 min, 32% to 80% B from 54.0 to 57.0 min, and 80% B from 57.0 to 60.0 min, with a flow rate of 450 nl/min. The separated peptides were ionized in a capillary ion source and analyzed using a TimsTOF Pro mass spectrometer (Bruker). The ion source voltage was set to 2.0 kV, and both the parent ion and its fragments were detected and analyzed using high-resolution TOF. The second-level mass spectral scan range was set at 100–1700 m/z. The data acquisition mode used Parallel Accumulation Serial Fragmentation (PASEF), and ten PASEF mode acquisitions were performed after each primary mass spectrum acquisition to obtain second-level spectra for parent ions with charges ranging from 0 to 5. The dynamic exclusion time for the mass spectrometry scan was set to 30 s to avoid repeated scanning of the parent ions.
Database search
The tandem mass spectrometry results were examined using the MaxQuant search engine (version 1.6.15.0). The spectra were cross-referenced with the human SwissProt database, which contained 20422 entries, and a decoy database in reverse. Trypsin/P was used as the cleavage enzyme and up to two missed cleavages were permitted. In the first and primary searches, the precursor ion mass tolerance was set to 20 and 5 ppm, respectively, whereas the fragment ion mass tolerance was set to 0.02 Da. Protein N-terminal acetylation and methionine oxidation were defined as variable modifications, cysteine carbamidomethylation was defined as a fixed modification. The false discovery rate (FDR) was adjusted to < 1%.
Bioinformatics analysis
Identification and biological function analysis of differentially lactylated proteins (DLPs)
DLPs and differentially lactylated peptides were identified using the R software (version 4.1.2) based on a cutoff value (log fold change [FC] > 1.5 and P < 0.05). Subcellular localization annotations of DLPs were performed using the WolF Psort software [17]. Biological functions of DLPs were analyzed using online tools (Gene ontology (GO) (http://www.geneontology.org) [18] and Kyoto Encyclopedia of Genes and Genomes (KEGG) (https://www.genome.jp/kegg/) [19]), and P < 0.05 was considered statistically significant.
Protein–protein interaction (PPI) network analysis of the DLPs
PPI network analysis was conducted to investigate the interactional relationships among the DLPs using the STRING database (version 11.5) (https://string-db.org) [20]. Significant interactions were screened according to a confidence score of > 0.7 (high confidence). The results were downloaded and analyzed using the Cytoscape software (version 3.9.1) [21]. Hub DLPs were screened using the cytoHubba plug-in [22] of the Cytoscape software based on the degree rank of the nodes.
Results
Pattern of protein lactylation modification in ccRCC
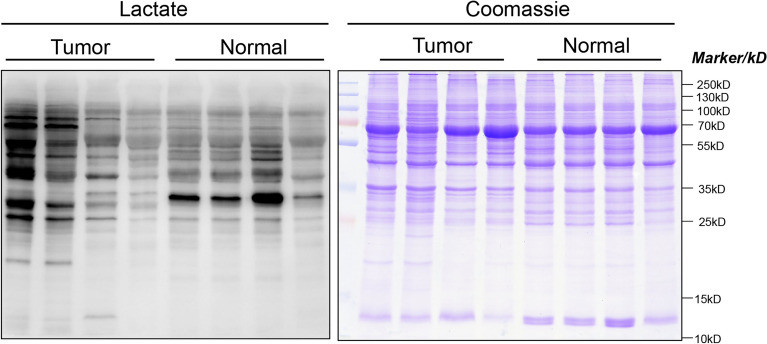
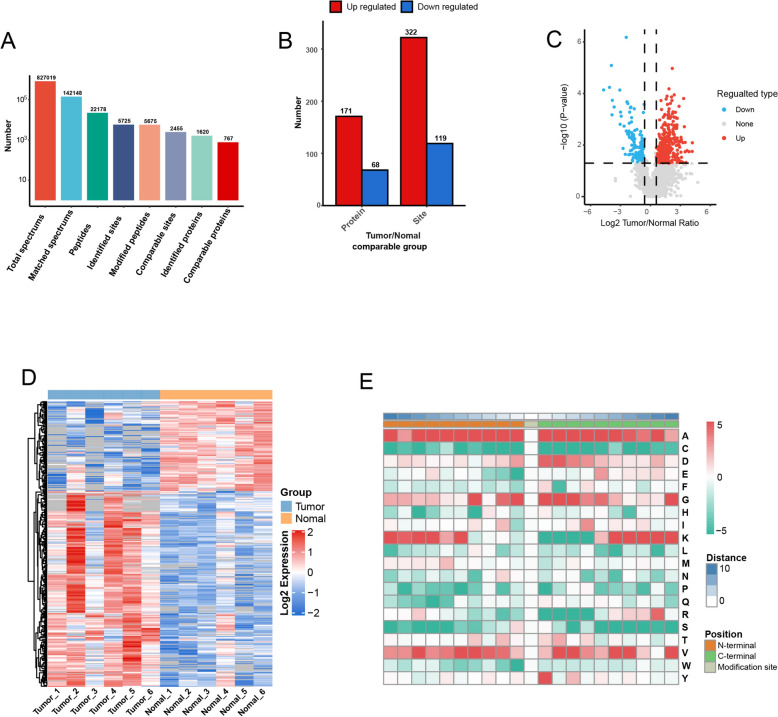
The pan anti-Kla antibody was used to identify the trend of Kla-modified proteins in four cancer tissues and four adjacent normal tissues. The results indicated that Kla-modified proteins were more abundant in ccRCC than in adjacent normal tissues (Figure 1). Quantitative proteomic analysis showed that the total number of identified spectra, matched spectra, peptides, identified sites, modified peptides, comparable sites, identified proteins, and comparable proteins were 827019, 142148, 22178, 5725, 5675, 2455, 1620, and 767, respectively, in ccRCC tissues (Figure 2A). Further analysis showed that all 171 Kla-modified proteins involving 322 lactylation sites were significantly upregulated in ccRCC tissues compared to adjacent normal tissues (log FC > 1.5 and P < 0.05) (Figure 2B–D). However, all 68 Kla-modified proteins involving 119 lactylation sites are significantly downregulated in ccRCC compared to adjacent normal tissues (log FC > − 1.5 and P < 0.05) (Figure 2B–D). Significantly upregulated or downregulated lactylated proteins were defined as DLPs. To understand the characteristics of the Kla-modified sites in ccRCC, motif analysis was used to identify ten N-terminal and C-terminal amino acids near the Kla-modified sites. The results showed that lysine (K) was most susceptible to modification by lactylation when the N-terminal (+ 10) or C-terminal (− 10) is alanine (Figure 2E).
Figure 1.
Lysine lactylation-modified proteins in cancer and adjacent normal tissues detected via Western blotting using pan anti-lactyllysine antibody. Coomassie blue staining was employed as a loading control to ensure equal protein loading across all lanes. The full blot images are shown in Figure S1
Figure 2.
Analysis of Kla-modified proteins in ccRCC. A Statistical summary of Kla-modified proteins. B Statistical summary of DLPs. C Volcano plots of DLPs. D Heat map visualizing the intensity of lactylation for DLPs. Red and green colors represent high and low levels of lactylation intensity, respectively; E heat map visualizing the characteristics of the Kla-modified sites. Red signifies high frequency, while green represents low frequency. ccRCC: clear cell renal cell carcinoma; DLPs: differentially lactylated proteins; Kla: lysine lactylation
The subcellular localization analysis of DLPs in ccRCC
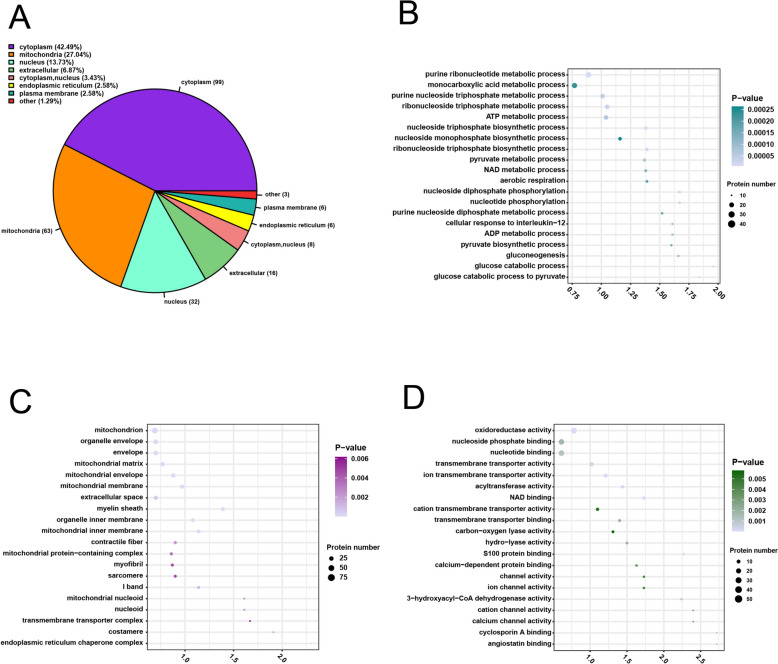
To identify the localization of DLPs in ccRCC, subcellular localization analysis was conducted. The results showed that the DLPs mainly localized in the cytoplasm (42.49%), followed by the mitochondria (27.4%), nucleus (13.73%), and extracellular matrix (6.87%) (Figure 3A).
Figure 3.
Subcellular localization and GO functional enrichment analyses of DLPs in ccRCC. A Subcellular localization analysis. B The top 20 biological process terms; C the top 20 cell component terms; D the top 20 molecular function terms. The vertical axis is GO functional description information, and the horizontal axis is the value of fold enrichment of DLPs in this functional type compared with the proportion of identified protein after Log2 conversion. The color of the dots indicates enrichment significance P value, dark color indicates strong enrichment significance, and the size of the dots indicates the number of DLPs in the GO functional class. GO: gene ontology; ccRCC: clear cell renal cell carcinoma; DLPs: differentially lactylated proteins.
Gene ontology (GO) functional enrichment analysis of DLPs
To understand the biological functions of the DLPs, GO functional enrichment analysis was performed based on three categories: biological process (BP), molecular function (MF), and cell components (CC). The top 20 results of the GO enrichment analysis of DLPs are shown in (Figure 3B–D and Table S1-S3).
KEGG pathway analysis of DLPs
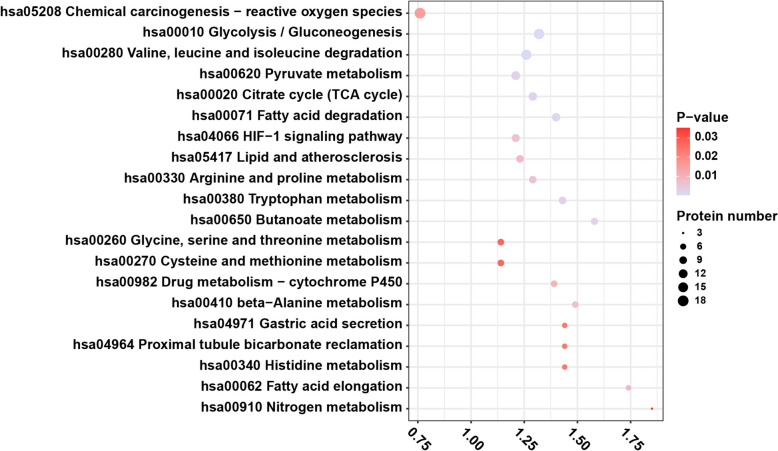
To identify the DLPs involved in potential pathways, KEGG pathway analysis was performed. The results showed that DLPs were mainly enriched in metabolism-related pathways, including glycolysis/gluconeogenesis, amino acid (valine, leucine, and isoleucine), pyruvate metabolism, fatty acid degradation, and the citrate cycle. The top 20 results of the KEGG enrichment analysis of DLPs are shown in Figure 4 and Table S4.
Figure 4.
Top 20 terms from KEGG enrichment analysis of DLPs in ccRCC. The vertical axis is KEGG functional description information, and the horizontal axis is the value of fold enrichment of DLPs in this functional type compared with the proportion of identified protein after Log2 conversion. The color of the dots indicates enrichment significance P value, dark color indicates strong enrichment significance, and the size of the dots indicates the number of DLPs in the KEGG functional class. KEGG: Kyoto Encyclopedia of Genes and Genomes; ccRCC: clear cell renal cell carcinoma; DLPs: differentially lactylated proteins
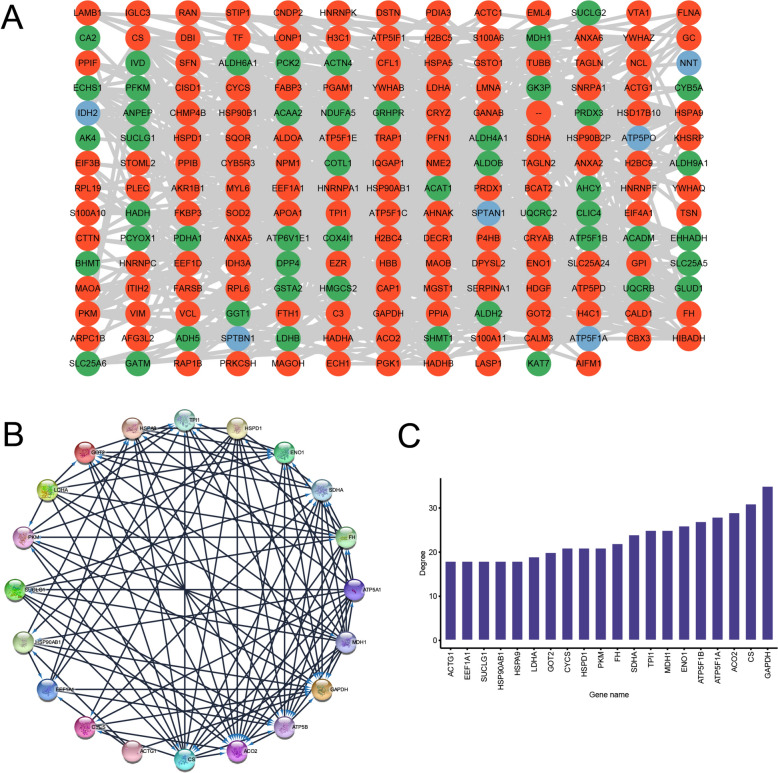
PPI networks of the DLPs
To understand the potential relationships among DLPs, PPI network analysis was conducted using the STRING database. According to a confidence score > 0.7 (high confidence) [23, 24], a total of 193 DLPs were screened and used to construct a final PPI network. The results from the STRING database analysis were downloaded and analyzed using the Cytoscape software (Figure 5A). The top 20 hub DLPs were identified using the CytoHubba plug-in according to their degree values (Figure 5B, C). The lactylated proteins identified were GAPDH, CS, ACO2, ATP5F1A, ATP5F1B, ENO1, TPI1, MDH1, SDHA, FH, CYCS, HSPD1, PKM, GOT2, LDHA, ACTG1, EEF1A1, SUCLG1, HSP90AB1, and HSPA9. GO and KEGG enrichment analyses were performed to determine the potential biological functions of the top 20 hub lactylated proteins. The top 20 hub lactylated proteins were mainly enriched in catalytic activity (MF), mitochondrial matrix (CC), Generation of precursor metabolites and energy (BP), metabolic pathways (KEGG) (Table 1).
Figure 5.
PPI network analysis of DLPs in ccRCC. A PPI network of DLPs. The red nodes denote upregulated DLPs, green nodes denote downregulated DLPs, and blue nodes denote DLP sites in both cancer and normal tissues. B PPI of top 20 hub proteins. C The degree rank of top 20 hub proteins. PPI: protein–protein interaction; ccRCC: clear cell renal cell carcinoma; DLPs: differentially lactylated proteins
Table 1.
Top 5 terms from GO and KEGG enrichment analysis for hub proteins in ccRCC
| Category | Term name | Gene count | Description | Genes | p-value |
|---|---|---|---|---|---|
| BP | GO:0044238 | 12 | Primary metabolic process | ACO2, GAPDH, TPI1, GOT2, ATP5B, SDHA, CYCS, PKM, CS, FH, ATP5A1, MDH1 | 7.72E-06 |
| BP | GO:0044237 | 12 | Cellular metabolic process | ACO2, GAPDH, TPI1, GOT2, ATP5B, SDHA, CYCS, PKM, CS, FH, ATP5A1, MDH1 | 1.03E-05 |
| BP | GO:0071704 | 12 | Organic substance metabolic process | ACO2, GAPDH, TPI1, GOT2, ATP5B, SDHA, CYCS, PKM, CS, FH, ATP5A1, MDH1 | 1.51E-05 |
| BP | GO:0006091 | 11 | Generation of precursor metabolites and energy | ACO2, GAPDH, TPI1, ATP5B, SDHA, CYCS, PKM, CS, FH, ATP5A1, MDH1 | 4.12E-18 |
| BP | GO:0044281 | 11 | Small molecule metabolic process | ACO2, GAPDH, TPI1, GOT2, ATP5B, SDHA, PKM, CS, FH, ATP5A1, MDH1 | 2.20E-11 |
| CC | GO:0005739 | 9 | Mitochondrion | ACO2, GOT2, ATP5B, SDHA, CYCS, PKM, CS, FH, ATP5A1 | 3.11E-08 |
| CC | GO:0070062 | 9 | Extracellular exosome | GAPDH, TPI1, GOT2, ATP5B, PKM, CS, FH, ATP5A1, MDH1 | 3.11E-07 |
| CC | GO:0005759 | 6 | Mitochondrial matrix | ACO2, GOT2, ATP5B, CS, FH, ATP5A1 | 1.82E-07 |
| CC | GO:0031967 | 6 | Organelle envelope | GAPDH, GOT2, ATP5B, SDHA, CYCS, ATP5A1 | 3.84E-05 |
| CC | GO:0005743 | 5 | Mitochondrial inner membrane | GOT2, ATP5B, SDHA, CYCS, ATP5A1 | 6.27E-06 |
| MF | GO:0003824 | 12 | Catalytic activity | ACO2, GAPDH, TPI1, GOT2, ATP5B, SDHA, CYCS, PKM, CS, FH, ATP5A1, MDH1 | 2.38E-07 |
| MF | GO:0016835 | 3 | Carbon-oxygen lyase activity | ACO2, TPI1, FH | 1.16E-05 |
| MF | GO:0043532 | 2 | Angiostatin binding | ATP5B, ATP5A1 | 5.16E-06 |
| MF | GO:0046933 | 2 | Proton-transporting ATP synthase activity, rotational mechanism | ATP5B, ATP5A1 | 2.68E-05 |
| MF | GO:0042288 | 2 | MHC class I protein binding | ATP5B, ATP5A1 | 7.90E-05 |
| KEGG | hsa01100 | 11 | Metabolic pathways | ACO2, GAPDH, TPI1, GOT2, ATP5B, SDHA, PKM, CS, FH, ATP5A1, MDH1 | 4.22E-12 |
| KEGG | hsa01200 | 9 | Carbon metabolism | ACO2, GAPDH, TPI1, GOT2, SDHA, PKM, CS, FH, MDH1 | 3.07E-18 |
| KEGG | hsa01230 | 6 | Biosynthesis of amino acids | ACO2, GAPDH, TPI1, GOT2, PKM, CS | 3.22E-12 |
| KEGG | hsa00020 | 5 | Citrate cycle (TCA cycle) | ACO2, SDHA, CS, FH, MDH1 | 9.12E-12 |
| KEGG | hsa05010 | 5 | Alzheimer disease | GAPDH, ATP5B, SDHA, CYCS, ATP5A1 | 1.46E-06 |
GO: gene ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; ccRCC: clear cell renal cell carcinoma; DLPs: differentially lactylated proteins
Discussion
Glycolysis is an important metabolic biological process that maintains cell viability [25]. Lactate is an important metabolite of the cellular glycolytic pathway, particularly under anaerobic conditions [25–29]. Additionally, high lactic acid accumulation promotes protein lactylation [4, 10, 30]. Furthermore, previous studies have shown that the tumor microenvironment is hypoxic, and cancer cell glycolysis is active, with a large amount of lactate production [29, 31–33]. Moreover, high concentrations of lactate contribute to promoting tumor growth, invasion, angiogenesis, immunosuppression, and metastasis [29, 34–36]. ccRCC is the most common histological subtype of renal cell carcinoma, characterized by high rates of recurrence and progression. Protein lactylation also plays a crucial role in regulating the progression of ccRCC cells [13, 37]. In the present study, we conducted quantitative proteomic analysis to identify the protein lactylation modification atlas of ccRCC using mass spectrometric detection. The results of this study revealed that there are extensive protein lactylation modifications in ccRCC. Biological function analysis of the DLPs indicated that most DLPs were enriched in metabolism-related biological functions and metabolism-related pathways. The findings of this study preliminarily illustrate that the influence of protein lactylation on ccRCC progression might be closely related to the metabolic regulation of cancer cells.
Protein lactylation is an important epigenetic modification widely observed in various cancers [4, 30, 38, 39]. For example, a recent study used mass spectrometry to detect the lactylation modifications of proteins in hepatocellular carcinoma and found a large number of lactylation-modified proteins that were mainly involved in several metabolism-related biological processes [16]. Likewise, a recent study also reported a similar phenomenon [10]. Additionally, protein lactylation modification has been shown to be closely related to ccRCC progression [13]. Yang et al. [13] reported that high levels of histone H3 lactylation modification were associated with poor prognosis in patients with ccRCC; mechanically, lactylated histone H3 mainly promoted ccRCC progression by enhancing the transcription of PDGFRβ and its signaling pathways. Although the findings of Yang et al. are valuable, it remains unknown whether large-scale lactylation modifications of proteins occur in ccRCC. Therefore, it is necessary to comprehensively identify the characteristics of lactylation modifications of proteins in ccRCC. Exploring the pattern of global protein lactylation modification in ccRCC will expand our understanding of the role of lactylation modifications of proteins and provide insights into the underlying mechanism of ccRCC progression. In this study, we preliminarily analyzed the abundance of Kla-modified proteins in ccRCC. Subsequently, 239 DLPs involving 441 sites were identified, of which 171 (322) were upregulated and 68 (119) were downregulated. Notably, motif analysis proved that alanine is the most common amino acid surrounding lysine (K)-lactylation modification sites in ccRCC. This finding also suggests that lysine (K)-lactylation modification-related enzymes have a higher affinity for alanine residues than for other residues in ccRCC. Additionally, the results of subcellular localization analysis suggested that most DLPs function in the cytoplasm, mitochondria, and nucleus. Taken together, these preliminary findings illustrate that lactylated proteins might play an important role in the development and progression of ccRCC.
Metabolic reprogramming, an essential biological pathway for the survival of cancer cells, is a primary characteristic of the Warburg effect and is closely correlated with epigenetic modification [11, 38–40]. Previous studies proved that protein lactylation modification and metabolic rewriting interact together to regulate the fate of cancer cells [29, 38, 41]. In this study, we utilized the GO enrichment analysis to expound the biological functions of the DLPs and found that most of the DLPs principally enriched on binding-associated MF and metabolism-associated BP such as nucleotide binding (MF), nucleoside phosphate binding (MF), purine ribonucleotide metabolic process (BP), monocarboxylic acid metabolic process (BP), ribonucleoside triphosphate metabolic process (BP), purine nucleoside triphosphate metabolic process (BP), and ATP metabolic process (BP). These findings suggest that DLPs may promote ccRCC progression by regulating the metabolic processes of cancer cells. Subsequent KEGG analysis confirmed the aforementioned hypothesis and showed that most DLPs were mainly enriched in metabolism-related pathways, including glycolysis, amino acid (valine, leucine, and isoleucine) degradation, pyruvate metabolism, fatty acid degradation, and the citrate cycle. These metabolism-related pathways are closely associated with cancer progression [42–45]. In brief, the above results from ccRCC are similar to existing research findings on other cancer types in which high lactate concentrations in the tumor microenvironment accelerate protein lactylation modifications in cancer cells, thereby regulating metabolic reprogramming and promoting their own development and progression [29, 32, 34, 46, 47].
PPI are crucial proteins that perform biological functions and play a vital role in tumor progression [48–50]. Expounding the relationship of the interaction among Kla-modified proteins contributes to providing new insights into understanding the mechanism of progression and seeking therapeutic targets for ccRCC [48, 51]. The results of the PPI analysis in this study indicate that there is a complex interaction network among DLPs that promotes ccRCC progression by controlling metabolic remodeling. Subsequent results from the identification of hub proteins showed that almost all hub Kla-modified proteins were closely related to metabolic processes, such as GAPDH [52], CS [53], ACO2 [54], ATP5F1A [55], and ATP5F1B [56]. Based on these findings, Kla-modified hub proteins may represent promising therapeutic targets for ccRCC; however, further molecular and functional studies are necessary to confirm their therapeutic potential.
However, this study has some limitations. First, the components of the ccRCC microenvironment are complex, comprising cancer, immune, and stromal cell populations [57, 58], but the Kla-modified proteins were detected in whole ccRCC tissues in this study. Therefore, the Kla-modified landscape is yet to precisely map every ccRCC cell type. Additionally, the identified Kla-modified proteins are involved in key metabolic pathways, such as glycolysis, which are critical for cancer cell survival. While these findings suggest potential therapeutic relevance, further in vitro and in vivo experiments are required to confirm their roles as actionable therapeutic targets. Moreover, this study did not account for variations in overall protein expression, which may confound the effects attributed to lactylation modifications. Future work will incorporate quantitative proteomics to distinguish between lactylation-specific changes and those due to altered protein abundance. Finally, because the effects of lactylation modifications on the function of different proteins are different, further molecular experiments are necessary to examine the role of these proteins in the formation and progression of ccRCC.
In conclusion, this study revealed the landscape of protein lactylation modifications in ccRCC using a whole-proteome lactylation detection technique. The results showed that there were abundant protein lactylation modifications in ccRCC and normal tissues, and alanine residues were the most concomitant to lysine (K)-lactylation modifications in ccRCC. Notably, DLPs may promote ccRCC progression by regulating metabolic remodeling. These findings improve our understanding of the mechanism underlying ccRCC progression and provide a new direction for the identification of therapeutic targets for ccRCC. Further molecular and biological experiments are required to verify the findings of this study.
Supplementary Information
Acknowledgments
Not applicable.
Author contributions
Bangbei Wan and Cai Lv designed the study and analyzed the data; Bangbei Wan, Yuan Huang, Binghao Gong, and Yaohui Zeng revised the images; Bangbei Wan, Yuan Huang, Binghao Gong, and Yaohui Zeng performed the literature search and collected data for the manuscript; Bangbei Wan and Cai Lv revised the manuscript. All authors have read and approved the final manuscript.
Funding
This project was supported by the Hainan Province Clinical Medical Center (QWYH202175), the National Natural Science Foundation of China (82160544 and 82260304), the Specific Research Fund of The Innovation Platform for Academicians of Hainan Province (YSPTZX202311), the Natural Science Foundation of Hainan Province (820RC771), the Key R&D Projects of Hainan Province (ZDYF2022SHFZ074, ZDYF2022SHFZ280, ZDYF2017086, and ZDYF2019157), and the Research and Cultivation Fund of Hainan Medical University (HYPY2020015).
Availability of data and materials
Data supporting the findings of this study are available in Supplementary material.
Declarations
Ethics approval and consent to participate
All methods were carried out in accordance with relevant guidelines and regulations or Declaration of Helsinki. The research protocol and informed consent were approved by the Ethics Committee of the Haikou Affiliated Hospital of Central South University Xiangya School of Medicine. Informed Consent was obtained from the participants.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Bangbei Wan and Yuan Huang have contributed equally to this work and should be regarded as co-first authors.
Contributor Information
Bangbei Wan, Email: 939313612@qq.com.
Cai Lv, Email: 198312170@csu.edu.cn.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33. [DOI] [PubMed] [Google Scholar]
- 2.Xia C, Dong X, Li H, Cao M, Sun D, He S, Yang F, Yan X, Zhang S, Li N, et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J (Engl). 2022;135(5):584–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373(9669):1119–32. [DOI] [PubMed] [Google Scholar]
- 4.Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y, Liu W, Kim S, Lee S, Perez-Neut M, et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574(7779):575–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang L, Wang X, Liu J, Liu X, Li S, Zheng F, Dong Q, Xu S, Xiong J, Fu B. Prognostic and tumor microenvironmental feature of clear cell renal cell carcinoma revealed by m6A and lactylation modification-related genes. Front Immunol. 2023;14:1225023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen AN, Luo Y, Yang YH, Fu JT, Geng XM, Shi JP, Yang J. Lactylation, a novel metabolic reprogramming code: current status and prospects. Front Immunol. 2021;12: 688910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Yang P, Yu T, Gao M, Liu D, Zhang J, Lu C, Chen X, Zhang X, Liu Y. Lactylation of PKM2 suppresses inflammatory metabolic adaptation in pro-inflammatory macrophages. Int J Biol Sci. 2022;18(16):6210–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong J, He J, Zhu J, Pan J, Liao W, Ye H, Wang H, Song Y, Du Y, Cui B, et al. Lactylation-driven METTL3-mediated RNA m(6)A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol Cell. 2022;82(9):1660–16771610. [DOI] [PubMed] [Google Scholar]
- 9.Gu J, Zhou J, Chen Q, Xu X, Gao J, Li X, Shao Q, Zhou B, Zhou H, Wei S, et al. Tumor metabolite lactate promotes tumorigenesis by modulating MOESIN lactylation and enhancing TGF-beta signaling in regulatory T cells. Cell Rep. 2022;39(12): 110986. [DOI] [PubMed] [Google Scholar]
- 10.Yang Z, Yan C, Ma J, Peng P, Ren X, Cai S, Shen X, Wu Y, Zhang S, Wang X, et al. Lactylome analysis suggests lactylation-dependent mechanisms of metabolic adaptation in hepatocellular carcinoma. Nat Metab. 2023;5(1):61–79. [DOI] [PubMed] [Google Scholar]
- 11.Chen L, Huang L, Gu Y, Cang W, Sun P, Xiang Y. Lactate-Lactylation Hands between Metabolic Reprogramming and Immunosuppression. Int J Mol Sci. 2022. 10.3390/ijms231911943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Izzo LT, Wellen KE. Histone lactylation links metabolism and gene regulation. Nature. 2019;574(7779):492–3. [DOI] [PubMed] [Google Scholar]
- 13.Yang J, Luo L, Zhao C, Li X, Wang Z, Zeng Z, Yang X, Zheng X, Jie H, Kang L, et al. A positive feedback loop between inactive VHL-triggered histone lactylation and PDGFRbeta signaling drives clear cell renal cell carcinoma progression. Int J Biol Sci. 2022;18(8):3470–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun Y, Chen Y, Peng T. A bioorthogonal chemical reporter for the detection and identification of protein lactylation. Chem Sci. 2022;13(20):6019–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong H, Zhang J, Zhang H, Han Y, Lu C, Chen C, Tan X, Wang S, Bai X, Zhai G, et al. YiaC and CobB regulate lysine lactylation in Escherichia coli. Nat Commun. 2022;13(1):6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong H, Chen X, Wang H, Gu X, Yuan Y, Zhang Z. Global profiling of protein lysine lactylation and potential target modified protein analysis in hepatocellular carcinoma. Proteomics. 2023. 10.1002/pmic.202200432. [DOI] [PubMed] [Google Scholar]
- 17.Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucl Acid Res. 2007;35(Web Server issue):W585-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25(1):25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucl Acid Res. 2000;28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: a database of predicted functional associations between proteins. Nucl Acid Res. 2003;31(1):258–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. Cytoscape stringapp: network analysis and visualization of proteomics data. J Proteome Res. 2019;18(2):623–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, Jouffre N, Huynen MA, Bork P. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucl Acid Res. 2005. 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, Gable AL, Fang T, Doncheva NT, Pyysalo S, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucl Acid Res. 2023;51(D1):D638–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brooks GA. Lactate as a fulcrum of metabolism. Redox Biol. 2020;35: 101454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brooks GA. The science and translation of lactate shuttle theory. Cell Metab. 2018;27(4):757–85. [DOI] [PubMed] [Google Scholar]
- 28.Rabinowitz JD, Enerback S. Lactate: the ugly duckling of energy metabolism. Nat Metab. 2020;2(7):566–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Yang Y, Zhang B, Lin X, Fu X, An Y, Zou Y, Wang JX, Wang Z, Yu T. Lactate metabolism in human health and disease. Signal Transduct Target Ther. 2022;7(1):305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan L, Feng F, Wu J, Fan S, Han J, Wang S, Yang L, Liu W, Wang C, Xu K. Demethylzeylasteral targets lactate by inhibiting histone lactylation to suppress the tumorigenicity of liver cancer stem cells. Pharmacol Res. 2022;181: 106270. [DOI] [PubMed] [Google Scholar]
- 31.Doherty JR, Cleveland JL. Targeting lactate metabolism for cancer therapeutics. J Clin Invest. 2013;123(9):3685–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ippolito L, Comito G, Parri M, Iozzo M, Duatti A, Virgilio F, Lorito N, Bacci M, Pardella E, Sandrini G, et al. Lactate rewires lipid metabolism and sustains a metabolic-epigenetic axis in prostate cancer. Cancer Res. 2022;82(7):1267–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Bari L, Atlante A. Including the mitochondrial metabolism of L-lactate in cancer metabolic reprogramming. Cell Mol Life Sci. 2018;75(15):2763–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.San-Millan I, Brooks GA. Reexamining cancer metabolism: lactate production for carcinogenesis could be the purpose and explanation of the Warburg effect. Carcinogenesis. 2017;38(2):119–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirschhaeuser F, Sattler UG, Mueller-Klieser W. Lactate: a metabolic key player in cancer. Cancer Res. 2011;71(22):6921–5. [DOI] [PubMed] [Google Scholar]
- 36.Wang ZH, Peng WB, Zhang P, Yang XP, Zhou Q. Lactate in the tumour microenvironment: From immune modulation to therapy. EBioMedicine. 2021;73: 103627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Chen P, Zhou J, Li H, Pan Z. Prognostic impact of lactylation-associated gene modifications in clear cell renal cell carcinoma: Insights into molecular landscape and therapeutic opportunities. Environ Toxicol. 2024;39(3):1360–73. [DOI] [PubMed] [Google Scholar]
- 38.Sun L, Zhang H, Gao P. Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein Cell. 2022;13(12):877–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai X, Lv X, Thompson EW, Ostrikov KK. Histone lactylation: epigenetic mark of glycolytic switch. Trend Genet. 2022;38(2):124–7. [DOI] [PubMed] [Google Scholar]
- 40.Lv X, Lv Y, Dai X. Lactate, histone lactylation and cancer hallmarks. Expert Rev Mol Med. 2023;25: e7. [DOI] [PubMed] [Google Scholar]
- 41.Wang JH, Mao L, Wang J, Zhang X, Wu M, Wen Q, Yu SC. Beyond metabolic waste: lysine lactylation and its potential roles in cancer progression and cell fate determination. Cell Oncol (Dordr). 2023. 10.1007/s13402-023-00775-z. [DOI] [PubMed] [Google Scholar]
- 42.Ganapathy-Kanniappan S, Geschwind JF. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer. 2013;12:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Z, Zhang H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell Mol Life Sci. 2016;73(2):377–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gray LR, Tompkins SC, Taylor EB. Regulation of pyruvate metabolism and human disease. Cell Mol Life Sci. 2014;71(14):2577–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson NM, Mucka P, Kern JG, Feng H. The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell. 2018;9(2):216–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Certo M, Tsai CH, Pucino V, Ho PC, Mauro C. Lactate modulation of immune responses in inflammatory versus tumour microenvironments. Nat Rev Immunol. 2021;21(3):151–61. [DOI] [PubMed] [Google Scholar]
- 47.Ippolito L, Morandi A, Giannoni E, Chiarugi P. Lactate: a metabolic driver in the tumour landscape. Trend Biochem Sci. 2019;44(2):153–66. [DOI] [PubMed] [Google Scholar]
- 48.Cheng SS, Yang GJ, Wang W, Leung CH, Ma DL. The design and development of covalent protein-protein interaction inhibitors for cancer treatment. J Hematol Oncol. 2020;13(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hao Y, Zhao S, Wang Z. Targeting the protein-protein interaction between IRS1 and mutant p110alpha for cancer therapy. Toxicol Pathol. 2014;42(1):140–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rajagopal T, Talluri S, Akshaya RL, Dunna NR. HOTAIR LncRNA: a novel oncogenic propellant in human cancer. Clin Chim Acta. 2020;503:1–18. [DOI] [PubMed] [Google Scholar]
- 51.Song Y, Wang S, Zhao M, Yang X, Yu B. Strategies targeting protein tyrosine phosphatase SHP2 for cancer therapy. J Med Chem. 2022;65(4):3066–79. [DOI] [PubMed] [Google Scholar]
- 52.Tarze A, Deniaud A, Le Bras M, Maillier E, Molle D, Larochette N, Zamzami N, Jan G, Kroemer G, Brenner C. GAPDH, a novel regulator of the pro-apoptotic mitochondrial membrane permeabilization. Oncogene. 2007;26(18):2606–20. [DOI] [PubMed] [Google Scholar]
- 53.Berge M, Pezzatti J, Gonzalez-Ruiz V, Degeorges L, Mottet-Osman G, Rudaz S, Viollier PH. Bacterial cell cycle control by citrate synthase independent of enzymatic activity. Elife. 2020. 10.7554/eLife.52272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sawant Dessai A, Dominguez MP, Chen UI, Hasper J, Prechtl C, Yu C, Katsuta E, Dai T, Zhu B, Jung SY, et al. Transcriptional repression of SIRT3 potentiates mitochondrial aconitase activation to drive aggressive prostate cancer to the bone. Cancer Res. 2021;81(1):50–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chouhan S, Sawant M, Weimholt C, Luo J, Sprung RW, Terrado M, Mueller DM, Earp HS, Mahajan NP. TNK2/ACK1-mediated phosphorylation of ATP5F1A (ATP synthase F1 subunit alpha) selectively augments survival of prostate cancer while engendering mitochondrial vulnerability. Autophagy. 2023;19(3):1000–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bruggemann M, Gromes A, Poss M, Schmidt D, Klumper N, Tolkach Y, Dietrich D, Kristiansen G, Muller SC, Ellinger J. Systematic analysis of the expression of the mitochondrial ATP synthase (complex V) subunits in clear cell renal cell carcinoma. Transl Oncol. 2017;10(4):661–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krishna C, DiNatale RG, Kuo F, Srivastava RM, Vuong L, Chowell D, Gupta S, Vanderbilt C, Purohit TA, Liu M, et al. Single-cell sequencing links multiregional immune landscapes and tissue-resident T cells in ccRCC to tumor topology and therapy efficacy. Cancer Cell. 2021;39(5):662-677 e666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y, Narayanan SP, Mannan R, Raskind G, Wang X, Vats P, Su F, Hosseini N, Cao X, Kumar-Sinha C, et al. Single-cell analyses of renal cell cancers reveal insights into tumor microenvironment, cell of origin, and therapy response. Proc Natl Acad Sci U S A. 2021. 10.1073/pnas.2103240118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are available in Supplementary material.