Abstract
Background
Sperm-associated antigen 5 (SPAG5) is a mitotic spindle protein crucial for coordinating the separation of sister chromatids into daughter cells. Increasing evidence suggests that SPAG5 is overexpressed in various malignancies, functioning as an oncogene. However, research specifically examining SPAG5 in esophageal cancer remains limited.
Methods
In this research, we leveraged bioinformatics techniques to evaluate the expression and prognostic significance of SPAG5 in a variety of cancer types. We conducted Gene Set Enrichment Analysis (GSEA) to elucidate the relationship between SPAG5 and cancer characteristics. Additionally, we investigated the correlation between SPAG5 expression and immune cell infiltration utilizing the TIMER2.0 platform. The TIDE platform was used to assess the impact of SPAG5 on the effectiveness of immunotherapy and to screen for potential therapeutic drugs. We employed qRT-PCR and immunohistochemistry staining to ascertain the expression of SPAG5 in esophageal cancer tissue. Through cellular functional experiments, we examined the influence of SPAG5 expression on the proliferation, apoptosis, invasion, and migration of esophageal cancer cells. The Pathscan Stress Signaling Antibody Array was utilized to probe the potential molecular mechanisms of SPAG5.
Results
SPAG5 exhibits high levels of expression in various cancers, encompassing esophageal cancer, and its presence indicates an unfavorable prognosis. SPAG5 is primarily enriched in pathways associated with cellular proliferation and demonstrates a correlation with immune gene expression as well as the infiltration of immune cells. Suppression of SPAG5 expression in esophageal cancer cells not only inhibits cell proliferation, but also attenuates cell invasion and migration while inducing cellular apoptosis. The depletion of SPAG5 results in a decline in the levels of critical signaling proteins.
Conclusion
SPAG5 plays a pivotal role in esophageal cancer cell proliferation, apoptosis, and metastasis within the tumor microenvironment, making it a promising therapeutic target.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40001-024-02182-y.
Keywords: SPAG5, Esophageal squamous cell carcinoma, Pan-cancer
Introduction
Esophageal cancer is ranked sixth in terms of global cancer incidence rate and is a highly aggressive and lethal tumor [1]. In China, esophageal squamous cell carcinoma (ESCC) represents 70% of global cases and is the fourth most prevalent cancer in the country [2]. Although surgery can provide certain therapeutic benefits for early-stage esophageal squamous cell carcinoma, many patients are typically diagnosed after missing the opportunity for surgical intervention [3]. Curative concurrent chemoradiotherapy is the recommended treatment for locally advanced esophageal squamous cell carcinoma that is inoperable [4]. Furthermore, immunotherapy, a promising treatment approach, is expected to enhance long-term survival rates in patients [5, 6]. Despite significant progress in esophageal cancer treatment, the overall prognosis remains unfavorable, with a 5-year survival rate below 25% [7]. Consequently, it is crucial to comprehend the potential molecular mechanisms involved in the occurrence and development of esophageal cancer, identify new prognostic molecular markers, and develop novel treatment strategies. These endeavors are vital for improving the survival duration of esophageal cancer patients and represent essential avenues for future research.
The sperm-associated antigen 5 (SPAG5), also known as astrin, maps to Ch17q11.2 and encodes a protein that binds to microtubules and localizes to the centrosome, playing an important role in promoting the fidelity of the mitotic process [8, 9]. In normal cells, SPAG5 forms a molecular switch by interacting with other proteins, regulating the kinetochore-microtubule dynamics to ensure the proper separation of sister chromatids into daughter cells [10]. Loss of SPAG5 leads to spindle checkpoint arrest, resulting in the formation of multipolar spindles and the disruption of sister chromatid cohesion [11]. SPAG5 has also been shown to be involved in the development of various tumors. Studies have reported the overexpression of SPAG5 in multiple cancers, including gastric cancer [12], breast cancer [13], gliomagenesis [14], lung adenocarcinomas [15] and bladder cancer [16], with a strong correlation to tumor growth, metastasis, and poor prognosis. Yuan found that downregulation of SPAG5 can significantly inhibit cell proliferation and growth by blocking the G2 phase/mitosis phase (G2/M) of the cell cycle and inducing cell apoptosis [17]. Similarly, in hepatocellular carcinoma, Yu-Feng Yang discovered that SPAG5 interacts with the centrosomal protein 55 (CEP55), significantly promoting tumor growth and metastasis through the phosphatidylinositol 3-kinase/protein kinase B (PI3K/AKT) signaling pathway [18]. Our previous research demonstrated that in ESCC, the expression of miR-363 is significantly downregulated, which subsequently regulates the proliferation and invasion of esophageal cancer cells by negatively modulating SPAG5 expression [19]. Although these findings imply that SPAG5 serves as a critical oncogene, its precise role in ESCC remains unclear and warrants further investigation.
This study systematically analyzed the potential role and mechanisms of SPAG5 in pan-cancer, focusing particularly on the association between SPAG5 expression and tumor prognosis, immune infiltration, and its biological functions and pathways. The results suggest that SPAG5 has significant potential as a biomarker for tumor prognosis and as a therapeutic target. Given the current lack of research on SPAG5 in esophageal cancer, we examined its effects on the proliferation, apoptosis, invasion, and metastasis of esophageal cancer cells. We also conducted enrichment analysis, immune cell infiltration analysis, and drug sensitivity assessments. The findings indicate that SPAG5 expression enhances the proliferation and invasion of esophageal cancer cells and effectively predicts their response to immunotherapy. Our study contributes to a better understanding of the mechanisms underlying SPAG5’s role in cancer, serving as a reference for future targets in tumor therapy and the prediction of immunotherapy efficacy.
Methods
Data processing and gene expression analysis
RNA sequencing (RNA-seq) data and clinical information of 33 tumor types were acquired from the Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/) and the Genotype-Tissue Expression (GTEx) database (https://commonfund.nih.gov/GTEx) through UCSC XENA (https://xena.ucsc.edu/) [20]. The data were downloaded and subjected to log2 transformation and standardization for the purpose of assessing the differential expression of SPAG5 between tumor samples and normal control groups. Statistical analysis was conducted using the R software, and the resulting outcomes were visualized through the utilization of the “ggplot2” package.
Prognostic analysis of SPAG5
The prognostic evaluation of SPAG5 expression was performed using Kaplan–Meier and univariate Cox regression analyses. The Cox regression analysis utilized continuous variables, while the Kaplan–Meier analysis classified SPAG5 expression levels into binary categories. Subsequently, log-rank p-values and hazard ratios (HRs) were visualized through a heatmap.
Identification of DEGs and enrichment analysis in pan-cancer
Tumor patients were categorized into high-expression and low-expression groups based on the median expression level of SPAG5, utilizing pan-cancer RNA-seq data obtained from TCGA database. “DESeq2” package was utilized to perform differential expression analysis and obtain the log2-fold change (log2FC) and adjusted p-values for each gene across different cancer types. Genes meeting the criteria of log2FC > log2 and p < 0.05 were designated as differentially expressed genes (DEGs). Subsequently, Gene Set Enrichment Analysis (GSEA) was conducted using the “ClusterProfiler” package in R to identify the key signaling pathways potentially involving SPAG5 in various cancer types [21]. Obtain the GSE53625 dataset from the Gene Expression Omnibus (GEO) database (“https://www.ncbi.nlm.nih.gov/geo/”) to perform gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis, in order to further investigate the impact of SPAG5 expression on cancer-related pathway activity in esophageal cancer. Data visualization was performed using the “ggplot2” package.
Analysis of immune-related characteristics of SPAG5
Download pan-cancer immune cell infiltration data from the Tumor Immune Estimation Resource 2.0 (TIMER2.0) database (http://timer.cistrome.org/) in TCGA for analyzing the correlation between SPAG5 expression and infiltration levels of various immune cell types in pan-cancer, including B cells, CD4+ T cells, CD8+ T cells, dendritic cells (DCs), macrophages, neutrophils, natural killer cells, monocytes, and mast cells [22]. Furthermore, the TIMER2.0 database was used to analyze the correlation between SPAG5 expression and immune checkpoint genes, along with other immune-related genes including major histocompatibility complex genes, chemokines, and their receptors. The findings were visually represented using heatmaps. The relative abundance of 22 immune cell populations in the cohort of esophageal cancer patients was estimated using the cell-type identification by estimating relative subsets of RNA transcripts (CIBERSORT) algorithm. Comparisons were made to evaluate the variations in immune cell infiltration between the high and low-expression groups of SPAG5. Furthermore, a spearman correlation analysis was employed to examine the relationship between SPAG5 expression and immune cell infiltration in esophageal cancer. The obtained results will be visualized using box plots and bubble plots, generated using the “ggplot2” package.
In addition, the “ESTIMATE” algorithm in the “IOBR” package is used to estimate the content of stromal cells and immune cells in tumor tissues [23]. Through the computation of immune scores and stromal scores, the level of tumor purity is predicted for each tumor sample. Spearman correlation analysis is employed to assess the correlation between SPAG5 and ESTIMATE scores, as well as tumor purity. Furthermore, the obtained results are visualized using the “ggplot2” package in R.
Immunotherapy prediction of SPAG5 in ESCC
We retrieved an independent esophageal cancer cohort from the GEO database and calculated the tumor immune dysfunction and exclusion (TIDE) score for this cohort using an online tool (http://tide.dfci.harvard.edu/login/) [24, 25]. A higher TIDE score is associated with a reduced response to immune checkpoint inhibition therapy. By comparing the differences in TIDE scores between patients with high and low SPAG5 expression, we assessed the ability of SPAG5 to predict the effectiveness of immune checkpoint inhibition therapy.
Estimation of drug response
The “oncoPredict” package was used to predict the therapeutic response of patients with esophageal cancer to a total of 538 distinct anticancer drugs [26]. The half-maximal inhibitory concentration (IC50) was used to assess the sensitivity of these drugs. Furthermore, the correlation between drug response and SPAG5 expression was determined using Pearson correlation, focusing on the top 20 drugs.
Cell culture
Human esophageal squamous carcinoma cell lines (TE-1, EC9706, and Eca-109) were cultured in Dulbecco’s modified Eagle’s medium (Corning Cellgro, Manassas, Virginia) supplemented with 10% fetal bovine serum (FBS) (Gibco BRL, Grand Island, New York).All cells were incubated at 37 °C in a humidified incubator with 5% CO2.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA was extracted using Trizol reagent (Pufei Biotechnology, Shanghai, China), followed by messenger RNA (mRNA) purification. Then, reverse transcription was performed utilizing the Moloney murine leukemia virus (MMLV) (Promega, Madison, Wisconsin). qRT-PCR analysis was conducted on the LightCycler 480 II RT-PCR system using SYBR Premix Ex Taq (Takara, Kyoto, Japan) to quantitatively determine the abundance of SPAG5 mRNA, with GAPDH serving as the internal control. The reaction conditions were as follows: 95 °C for 15 s, 95 °C for 5 s, 60 °C for 30 s, for a total of 45 cycles. The primers for SPAG5 were 5ʹ-TTGAGGCCCGTTTAGATACCA-3ʹ (forward) and 5ʹ-GCTTTCCTTGGAGCAATGTAGTT-3ʹ (reverse), and for GAPDH: 5ʹ-TGACTTCAACAGCGACACCCA-3ʹ (forward) and 5ʹ-CACCCTGTTGCTGTAGCCAAA-3ʹ (reverse). The 2−ΔΔCt method was used to calculate relative gene expression levels.
Lentivirus packaging and infection
We designed short hairpin RNA (shRNA) sequences that specifically target the SPAG5 mRNA (NM_006461) sequence, represented as: 5ʹ-CCGGCCATGCAACTGGATTATACAACTCGAGTTGTATAATCCAGTTGCATGGTTTTTG-3ʹ. A scrambled shRNA was used as a negative control. A single-stranded DNA oligonucleotide was utilized to construct the shRNA sequence, which was later cloned into the GV115 plasmid vector (GeneChem Corporation, Shanghai, China). The recombinant plasmid was validated through DNA sequencing. Then, following this, the pHelper1.0 and pHelper2.0 vectors, along with the recombinant plasmid, were co-transfected into 293T cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and incubated for 48 h. The infectious lentivirus vector (LV) was collected, centrifuged, and filtered. In our study, we designated the recombinant LV expressing SPAG5-shRNA as shSPAG5, and the LV expressing scrambled shRNA as shCtrl. To determine the infection titer, we performed a serial dilution titer assay, yielding viral titers of 5 × 108 TU/ml for shSPAG5 and 6 × 108 TU/ml for shCtrl in the medium. TE-1 cells were seeded at a density of 2 × 105 cells/well in a six-well plate and then infected with shSPAG5 or shCtrl until reaching 80–90% confluence. The infection efficiency was evaluated 72 h post-infection using a fluorescence microscope.
Immunohistochemical analysis
The expression of SPAG5 was assessed immunohistochemically in 14 pairs of ESCA tissues and adjacent tissues. The tissue samples were initially fixed in neutral buffered formalin and then embedded in paraffin. Sequential tissue sections, measuring 4 μm in thickness, were then mounted on positively charged glass slides and underwent deparaffinization and rehydration. A rabbit monoclonal anti-SPAG5 antibody (Sigma-Aldrich), diluted at 1:500, was employed in the staining process. As a negative control, an unrelated rabbit serum was utilized. Two independent observers, unaware of the clinical and follow-up data, objectively evaluated the staining results and resolved any discrepancies through consensus. Five random fields were examined using an optical microscope, assigning scores based on the percentage of positive cells: 0 denoted 0%, 1 represented 1–25%, 2 stood for 26–50%, 3 indicated 51–75%, and 4 signified > 76%. Staining intensity was graded as 0 for no staining, 1 for straw yellow, 2 for brown, and 3 for dark brown. The final staining index was calculated as the multiplication of the staining intensity score and the positive cell proportion score.
Western blot assay
After thawing on ice for 15 min, TE-1 cells were centrifuged at 4 °C for 15 min. The protein concentration of the samples was determined using the BCA protein assay kit (Beyotime, Shanghai, China) and adjusted to 2 μg/μl. The protein lysates were subjected to separation using a 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), followed by their transfer onto polyvinylidene difluoride (PVDF) membranes (Millipore, Burlington, MA). The membranes were then blocked with 5% non-fat milk for 1 h, followed by an overnight incubation at 4 °C with a 1:2000 dilution of mouse anti-Flag antibody (Sigma-Aldrich, St. Louis, MO) and mouse anti-GAPDH antibody (SantaCruz, Santa Cruz, CA). Subsequently, the membranes were washed with TBST, and after 2 h of incubation, they were further processed with a 1:2000 dilution of goat anti-mouse IgG antibody (SantaCruz). Finally, the protein bands were detected using pierce electrochemiluminescence (ECL) substrate (Thermo Fisher Scientific, Rockford, IL).
Cell growth assay
TE-1 cells infected with shSPAG5 and shCtrl were seeded at a density of 2,000 cells per well in a 96-well plate and incubated for 1–5 days. Daily live cell counts were performed with the Celigo imaging cytometer (Nexcelom Bioscience, Lawrence, MA) over 5 consecutive days. Finally, the obtained cell count data generated a comprehensive cell growth curve.
MTT assay
Two thousand infected cells were seeded into a 96-well plate and cultured for 5 days. Each well received daily treatment with a 5 mg/ml concentration of L3-(4,5-dimethylthiazol-2-yl)−2,5-diphenyltetrazolium bromide (MTT) (Genview, Craigieburn, Victoria, Australia). Subsequently, the plate was incubated for 4 h under optimal conditions. Afterward, 100 μl of dimethyl sulfoxide (DMSO) was added to dissolve the formazan crystals generated by viable cells. After shaking for 5 min, the absorbance at 490 nm was measured using an ELISA reader (Tecan Infinite, Tecan GmbH, Groedig, Austria).
Annexin V assay
TE-1 cells infected with lentivirus were cultured in a 6-well plate until reaching a confluence of over 70%. Subsequently, the cells were collected and washed with ice-cold phosphate buffered saline (PBS) at 4 °C, followed by centrifugation to remove the supernatant. The resulting cell pellets were resuspended in 200μL of binding buffer. Next, 10 μl of V-APC membrane-associated protein (eBioscience, San Diego, California, USA) was added to the suspension. The cells were incubated at room temperature in the dark and subsequently analyzed using a machine.
Caspase-3/7 activity
TE-1 cells infected with lentivirus were inoculated into a 96-well plate and cultured for 5 days in a CO2 incubator at a temperature of 37 °C. Subsequently, each well was treated with 100 μl of Caspase-Glo reagent and incubated at room temperature for a period of 2 h. Finally, luminescence readings were obtained using an enzyme-linked immunosorbent assay (ELISA) reader (Tecan Infinite, Tecan GmbH, Groedig, Austria).
Wound-healing assay
TE-1 cells infected with lentivirus were seeded into a 96-well plate in three rounds, each containing 5 × 104 cells. Upon reaching 90% confluence, the growth medium was replaced with a solution containing 0.5% FBS. A scratch was created at the center of the bottom of each well using a designated scratch tool. Subsequently, the cell layer underwent gentle washing with PBS. Fluorescence microscopy images were captured at time points 0 and 8 h post-scratching and later used to calculate the ability of cell migration.
Transwell migration and invasion assay
The TE-1 cells, infected with lentivirus, were seeded at a density of 1 × 105 per well in culture dishes with or without matrix gel (Corning Costar, Cambridge, MA, USA). After incubating for 48 h, non-migratory and non-invasive cells were eliminated using a cotton swab. Following this, migrating and invasive cells were fixed, stained with crystal violet solution (Shanghai Yuan Ye Biotechnology Co., Ltd., China), and quantified using a microscope.
Pathscan intracellular signaling array
A cell lysis buffer was formulated to attain a concentration between 0.2 and 1 mg/ml upon cell collection and lysis. The PathScan Antibody Array Kit (Danvers, MA) was then employed at room temperature to quantify the levels of crucial signal transduction proteins in TE-1 cells exhibiting reduced SPAG5 expression.
Statistical analysis
Each experiment was replicated three times. The t-test was utilized to determine the significance of differences between the shSPAG5 and shCtrl groups in cellular assays. The prognosis of SPAG5 was evaluated using univariate Cox regression analysis. Survival analysis was conducted using the Kaplan–Meier method and the log-rank test. The statistical significance was determined at a level of p < 0.05. The statistical analyses were performed using SPSS software (version 22.0) and GraphPad Prism software (version 8.0).
Results
Differential expression levels of SPAG5 in pan-cancer
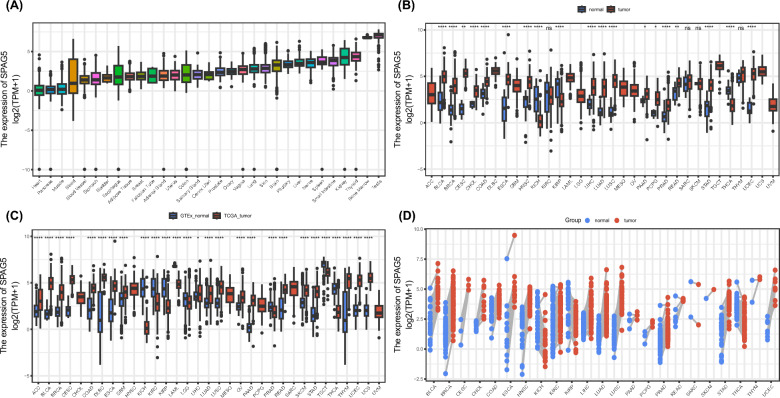
As shown in Fig. 1A, the data from the GTEx database indicate that SPAG5 is expressed in the majority of normal tissues, with the highest expression levels observed in the testes and bone marrow. Further comparison of SPAG5 expression in tumor tissues and normal tissues was conducted using the TCGA database. The findings exhibit a significant upregulation of SPAG5 expression in tumor tissues compared to their normal counterparts in bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cholangiocarcinoma (CHOL), cervical and endocervical cancers (CESC), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), head and neck squamous carcinoma (HNSC), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), pancreatic adenocarcinoma (PAAD), pheochromocytoma and paraganglioma (PCPG), prostate adenocarcinoma (PRAD), uterine corpus endometrial carcinoma (UCEC), and stomach adenocarcinoma (STAD) (Fig. 1B). Notably, higher SPAG5 levels were observed in normal tissues in kidney chromophobe (KICH), kidney renal papillary cell carcinoma (KIRP), and thyroid carcinoma (THCA). Furthermore, the analysis of paired samples indicated an increased expression of SPAG5 in cancerous tissues when compared to neighboring normal tissues (Fig. 1D). We conducted an integration of the TCGA and GTEx databases, revealing a significant upregulation of SPAG5 in various cancer types, such as adrenocortical carcinoma (ACC), BRCA, BLCA, COAD, CESC, lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), ESCA, glioblastoma multiforme (GBM), LUAD, LIHC, lung squamous cell carcinoma (LUSC), ovarian serous cystadenocarcinoma (OV), PAAD, PRAD, STAD, melanoma (SKCM), thymoma (THYM), uterine carcinosarcoma (UCS) and UCEC (Fig. 1C). In conclusion, SPAG5 is highly expressed in most tumors.
Fig. 1.
Differential expression levels of SPAG5 in pan-cancer. A Comparison of SPAG5 expression in different normal tissues based on GTEx database. B Comparison of SPAG5 expression between tumor and normal samples in pan-cancer based on TCGA database. C Comparison of SPAG5 expression between tumor and normal samples based on TCGA and GTEx database. D Comparison of SPAG5 expression between tumor and paired normal samples based on TCGA database (ns, p ≥ 0.05; *p < 0.05, **p < 0.01, ***p < 0.001)
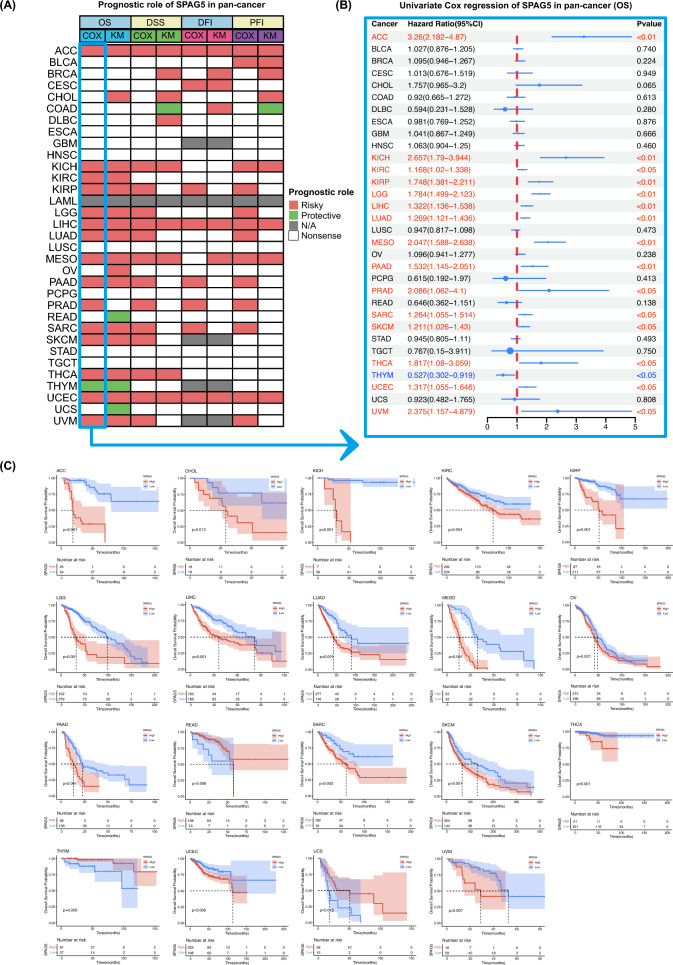
SPAG5 expression is correlated with prognosis
The pan-cancer prognosis analysis heatmap reveals a strong correlation between SPAG5 expression and cancer outcomes across various tumor types (Fig. 2A). Univariate Cox regression analysis demonstrates that elevated SPAG5 levels pose a risk for patients with ACC, KICH, KIRP, kidney renal clear cell carcinoma (KIRC), LIHC, lower grade glioma (LGG), LUAD, mesothelioma (MESO), PAAD, PRAD, SKCM, sarcoma (SARC), THCA, UCEC and uveal melanoma (UVM) (Fig. 2B). Surprisingly, SPAG5 acts as a protective factor in THYM patients. Additionally, Kaplan–Meier survival analysis confirms that increased SPAG5 expression is significantly associated with shortened overall survival (OS) in several cancers, including ACC, CHOL, KICH, KIRP, KIRC, LIHC, LUAD, LGG, MESO, OV, PAAD, SKCM, SARC, THCA, UVM, and UCEC (Fig. 2C). In contrast, it indicates improved prognosis in READ, THYM, and UCS. For progression-free interval (PFI), high SPAG5 expression is associated with a poor prognosis in ACC, BLCA, KIRP, KICH, LIHC, LGG, LUAD, PAAD, PRAD, MESO, SARC, UCEC, and UVM (Supplemental Fig. 1). The analysis results for disease-specific survival (DSS) and disease-free interval (DFI) also demonstrate that elevated SPAG5 expression is a significant risk factor for various cancers. In summary, high SPAG5 expression is associated with unfavorable outcomes in various cancers.
Fig. 2.
Association between SPAG5 expression and prognostic. A Summary of the correlation between expression of SPAG5 with overall survival (OS), disease-specific survival (DSS), disease-free interval (DFI) and progression-free interval (PFI) based on the univariate Cox regression and Kaplan–Meier models. B The single-factor Cox regression analysis of SPAG5 in pan-cancer. C The Kaplan–Meier overall survival curves of SPAG5 in pan-cancer
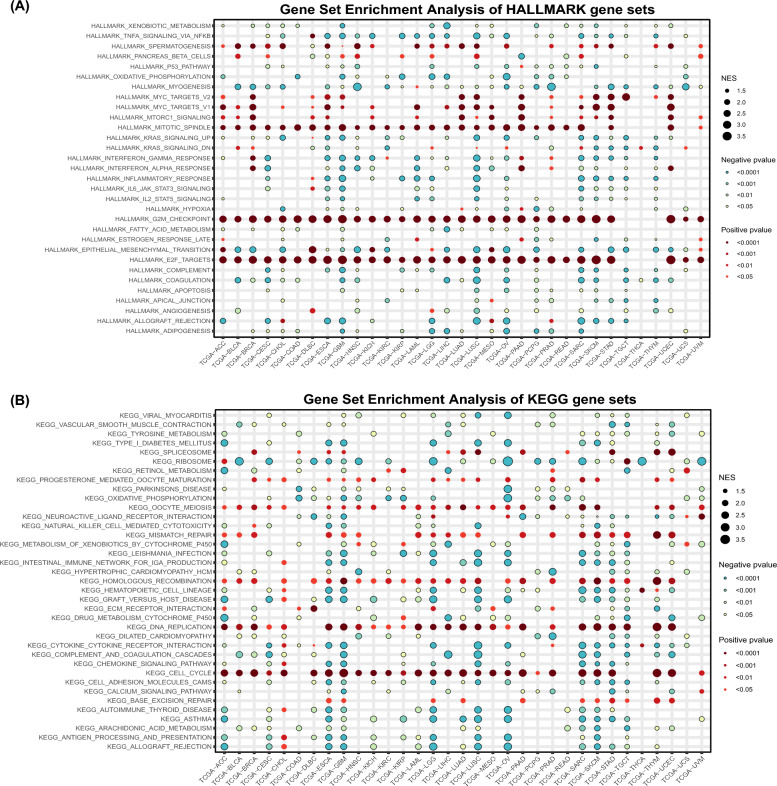
Gene set enrichment analysis of SPAG5 in pan-cancer
The relationship between SPAG5 expression and cancer-related signaling pathways was investigated using GSEA enrichment analysis to gain deeper insights into the molecular mechanisms underlying SPAG5's involvement in cancer development. The findings revealed a strong association between SPAG5 expression and cell proliferation pathways. Figure 3A demonstrates a significant positive correlation between SPAG5 expression and multiple key pathways in the majority of cancers, including DNA replication, cell cycle regulation, oocyte maturation, homologous recombination, G2/M checkpoint control, and the mechanistic target of rapamycin complex 1 (mTORC1) signaling pathway. It is worth noting that SPAG5 expression was enriched in pathways related to spermatogenesis and mitotic spindle, consistent with the origin and biological function of SPAG5. Furthermore, the GSEA analysis based on the KEGG signaling pathway validated the above results. The expression level of SPAG5 is significantly enriched in multiple pathways related to cellular proliferation, including DNA replication, the cell cycle, oocyte meiosis, mismatch repair, and homologous recombination (Fig. 3B). In summary, SPAG5 has the potential to promote the onset and progression of various tumors through its influence on cell proliferation.
Fig. 3.
Functional annotation of SPAG5 in pan-cancer. A The HALLMARK gene set enrichment analysis of SPAG5 in pan-cancer. B The KEGG gene set enrichment analysis of SPAG5 in pan-cancer
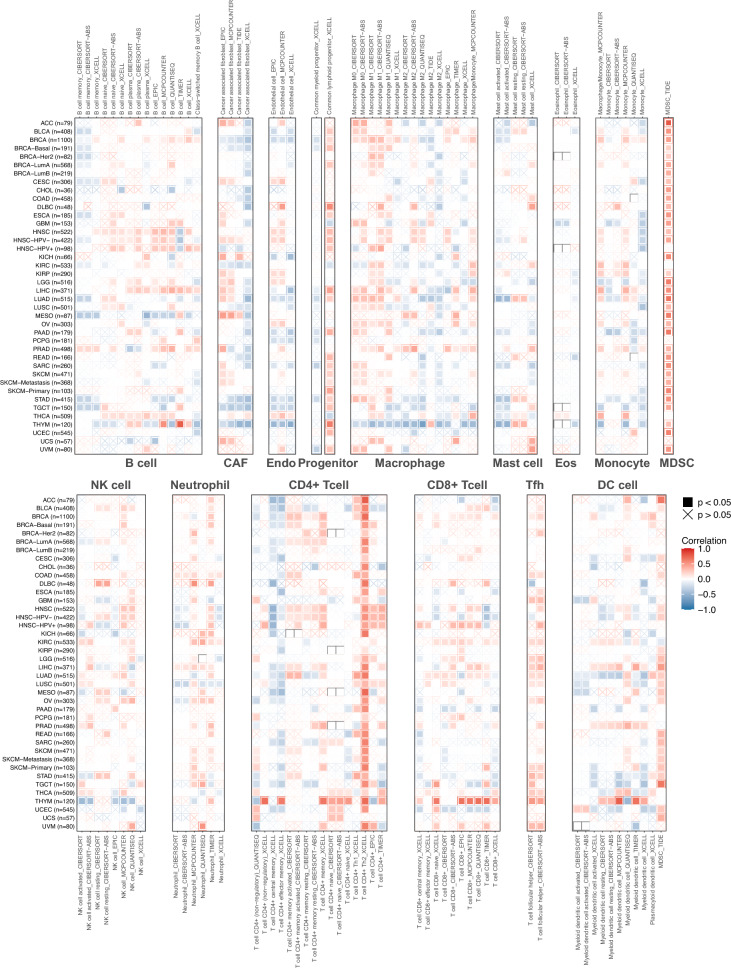
Correlation between SPAG5 expression and immune characteristics in pan-cancer
To investigate the relationship between SPAG5 expression and tumor immunity, we conducted a correlation analysis using the TIMER2.0 database to analyze the expression of SPAG5 and the infiltration level of immune cells in various cancer samples. The results revealed a significant correlation between SPAG5 expression and the infiltration of diverse immune cells across different tumor samples (Fig. 4). Interestingly, we observed a negative correlation between SPAG5 expression and the infiltration levels of CD4+ T cells, CD8+ T cells, neutrophils, B cells, and dendritic cells in most tumors, while a positive correlation was found with the infiltration levels of myeloid-derived suppressor cells (MDSCs). Notably, abnormal infiltration of distinct macrophage types was observed in various tumors. For instance, SPAG5 expression exhibited a positive correlation with the infiltration level of M1 macrophages in BRCA, HNSC, KIRC, LGG, LIHC, LUAD, LUSC, OV, PRAD, STAD, and THCA, and also with the infiltration level of M2 macrophages in BRCA, HNSC, and PRAD. We further employed the “ESTIMATE” algorithm to estimate the proportion of stromal cells and immune cells in pan-cancer samples. Additionally, we used this algorithm to predict the tumor purity of each individual tumor sample. The analysis of the results revealed that the expression of SPAG5 exhibited a negative correlation with the immune and stromal scores in the majority of tumor types (Supplemental Fig. 2). However, there was a significant positive correlation between the expression of SPAG5 and tumor purity.
Fig. 4.
Correlation between SPAG5 expression and the immune cell infiltration levels in pan-cancer
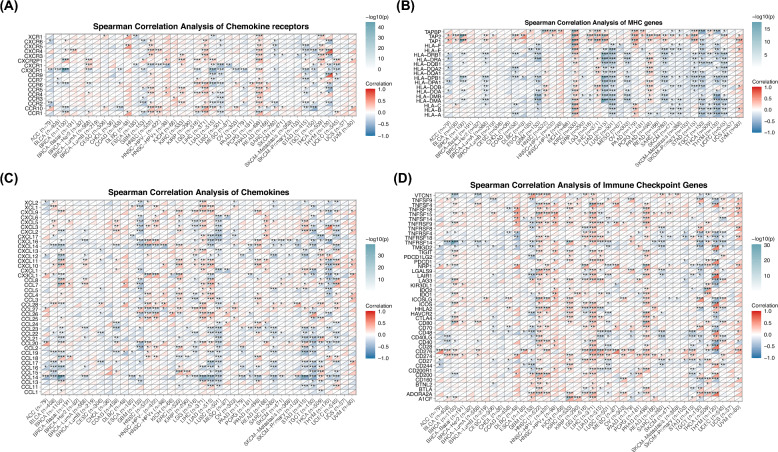
Moreover, we investigated the association between SPAG5 expression and immune-related gene expression, including immune checkpoint genes, major histocompatibility complex (MHC) genes, chemokines, and their receptors. Our research findings indicate that the expression of SPAG5 is positively correlated with immune checkpoint genes such as programmed cell death protein 1 (PDCD1), indoleamine 2,3 dioxygenase 1 (IDO1), programmed cell death 1 ligand 2 (PDCD1LG2), T cell immunoreceptor with immunoglobulin and ITIM domain(TIGIT) and CD276 in cancers including GBM, HNSC, KIRC, LGG, LIHC and PCPG (Fig. 5D). Furthermore, in most cancer types, the expression of SPAG5 exhibited a strong correlation with the expression of various chemokines, their receptors, and MHC genes (Fig. 5A–C). In conclusion, SPAG5 potentially exerts its role in shaping the tumor immune microenvironment through interaction with immune cells and its influence on the expression of immune-related molecules.
Fig. 5.
Correlation between SPAG5 and immune genes expression in pan-cancer. The Spearman correlation heatmap shows the correlation between SPAG5 expression and chemokine receptors (A), major histocompatibility complex genes (B), chemokines (C) and immune checkpoint genes (D) in pan-cancer. Red represents positive correlation and blue represent negative correlation (*p < 0.05, **p < 0.01, ***p < 0.001)
Value of SPAG5 in the screening, diagnosis and treatment of ESCC
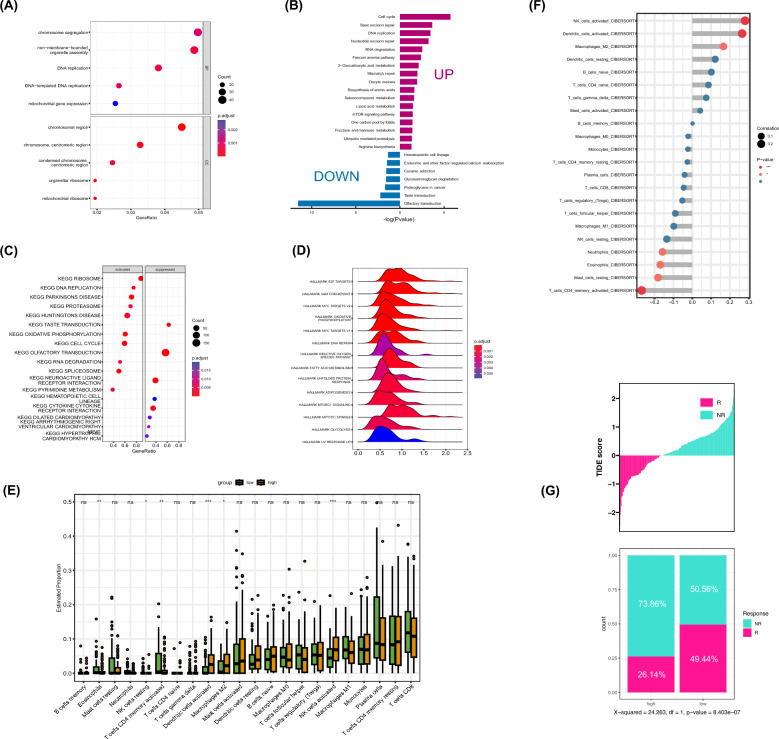
This study investigates the role of SPAG5 expression in the pathogenesis and progression of ESCC, with the aim of determining its potential value in ESCC screening, diagnosis, and treatment. We first evaluated SPAG5 expression levels in ESCC tissues through immunohistochemistry, revealing a higher expression in ESCC tissues compared to adjacent non-tumor tissues (Fig. 7A). Subsequently, we examined the potential roles of SPAG5 in ESCC. GO enrichment analysis showed significant enrichment of SPAG5 expression in biological processes such as chromosome segregation, DNA replication, and organelle assembly (Fig. 6A). This was corroborated by KEGG and GSEA enrichment analyses, which indicated a strong association between SPAG5 expression and cell proliferation-related pathways, including DNA replication, the cell cycle, and the mTOR signaling pathway (Fig. 6B–D). These findings suggest that SPAG5, being highly expressed in ESCC, may contribute to ESCC pathogenesis and progression by promoting cell proliferation.
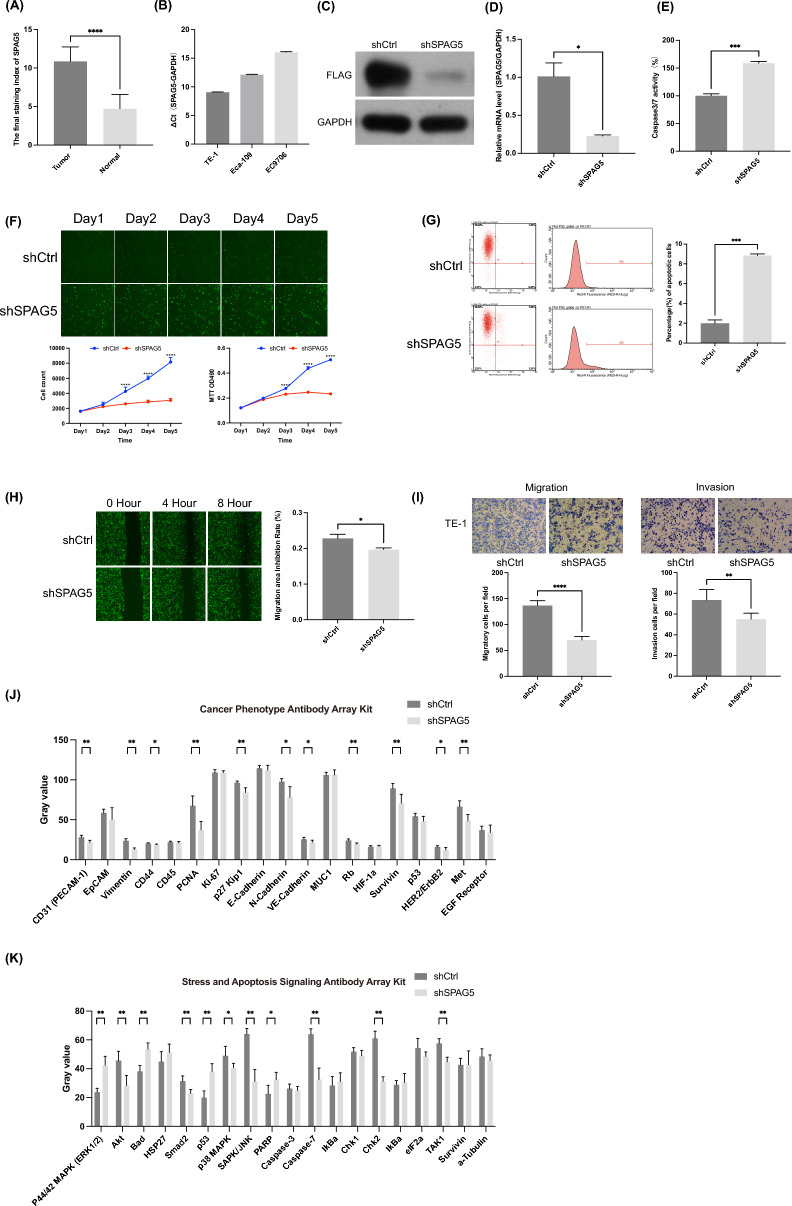
Fig. 7.
Exploring the impact of SPAG5 on ESCC cell function. A Immunohistochemical expression of SPAG5 in ESCC and corresponding adjacent normal tissues. B Expression of SPAG5 mRNA in TE-1, Eca-109, and EC9706. The expression of SPAG5 in TE-1 cells transfected with LV‐shSPAG5 was measured by Western blotting (C) and qRT-PCR (D). E The effect of SPAG5 knockout on apoptosis of TE-1 cells was studied by caspase-3/7 assay. F The effect of SPAG5 knockdown on the viability of TE-1 cells was investigated by the Celigo assay and MTT assay. G The percentage of apoptotic cells in TE-1 cells transfected with SPAG5-shRNA-LV was detected by FACS analysis. H The impact of SPAG5 knockdown on the migration ability of ESCC cells was investigated through wound healing experiments. I The effect of SPAG5 knockdown on the migration and invasive ability of TE-1 cells was investigated through Transwell assays. J The changes in cancer phenotype-related signaling pathways in TE-1 cells after SPAG5 knockdown. K The changes in stress and apoptosis-related signaling pathways in TE-1 cells after SPAG5 knockdown (ns, p ≥ 0.05; *p < 0.05, **p < 0.01, ***p < 0.001)
Fig. 6.
Value of SPAG5 in the screening, diagnosis and treatment of ESCC. A Enrichment of gene ontology (GO) terms for SPAG5 in ESCC. B Enrichment of Kyoto Encyclopedia of Genes and Genomes (KEGG) terms for SPAG5 in ESCC. C Gene set enrichment analysis of KEGG gene sets for SPAG5 in ESCC. D Gene set enrichment analysis of HALLMARK gene sets for SPAG5 in ESCC. E Differences of 22 immune cell subtypes in the high and low SPAG5 expression groups in ESCC. F Correlation between SPAG5 expression and the degree of immune cell infiltration in ESCC. G Proportional differences in treatment response rates to anti-PD-1 therapy between high and low SPAG5 groups in ESCC patients (ns, p ≥ 0.05; *p < 0.05, **p < 0.01, ***p < 0.001)
Given the emerging importance of immunotherapy in esophageal cancer treatment, we further investigated the relationship between SPAG5 expression and immune cell infiltration. CIBERSORT analysis revealed that high SPAG5 expression is associated with decreased infiltration of activated CD4+ T cells and increased infiltration of activated DCs, activated NK cells, and M2 macrophages (Fig. 6E). Correlation analysis confirmed these associations (Fig. 6F), suggesting that high SPAG5 expression may induce an immunosuppressive state in ESCC, potentially affecting immunotherapy outcomes. We also assessed the impact of SPAG5 expression on the response to immune checkpoint inhibitors (ICIs) using the TIDE database, finding that high SPAG5 expression is associated with a lower immunotherapy response rate (Fig. 6G). Additionally, our analysis of the relationship between SPAG5 expression and the efficacy of various chemotherapy drugs suggested that SPAG5 may enhance sensitivity to drugs such as docetaxel, paclitaxel, erlotinib, gemcitabine, and sorafenib (Supplementary Fig. 3). In conclusion, SPAG5 may serve as a promising biomarker for cancer treatment, playing a pivotal role in guiding clinical drug selection and predicting the efficacy of immunotherapy in esophageal cancer.
Exploring the impact of SPAG5 on ESCC cell function
SPAG5 mRNA is expressed in TE-1, EC9706, and Eca-109 cell lines, with the highest expression observed in TE-1 cells (Fig. 7B). Consequently, lentivirus-mediated RNA interference (RNAi) was employed to silence SPAG5 in TE-1 cells. Western blot analysis (Fig. 7C) and qRT-PCR (Fig. 7D) confirmed the effective reduction of SPAG5 expression through lentivirus-mediated RNAi. To further investigate the impact of SPAG5 expression on cell growth, lentivirus-infected TE-1 cells were cultured for 5 consecutive days, and proliferation was assessed daily using Celigo imaging. The results indicated a significant decrease in the growth curve of the shSPAG5 group compared to the shCtrl group (Fig. 7F). Moreover, MTT assays revealed a significant inhibition of cell viability upon SPAG5 silencing (Fig. 7F), confirming the inhibitory effect of SPAG5 knockdown on TE-1 cell proliferation. Caspase-3/7 activity analysis was performed to assess the influence of SPAG5 on cell apoptosis. As depicted in Fig. 7E, caspase-3/7 activity significantly increased in the LV-shSPAG5-infected group compared to the LV-shCtrl-infected group. FACS analysis also demonstrated a significant increase in the percentage of apoptotic cells in TE-1 cells transfected with SPAG5-shRNA-LV, indicating that SPAG5 knockdown can induce apoptosis in ESCC cells (Fig. 7G).
Scratch wound healing and Transwell migration assays were then utilized to evaluate the impact of SPAG5 on cell migration. The wound healing assay results showed that SPAG5 depletion significantly attenuated the migration capability of TE-1 cells (Fig. 7H). The Transwell migration assay also revealed a substantial decrease in invasion and migration ability in TE-1 cells infected with LV-shSPAG5 (Fig. 7I), suggesting that SPAG5 can promote invasion and migration of ESCC cells.
Lastly, the PathScan Antibody Array Kit was used to explore the potential mechanisms through which SPAG5 may contribute to esophageal cancer. The results demonstrated that SPAG5 knockdown significantly reduced the expression of various proteins involved in cancer phenotype-associated signaling pathways, including platelet endothelial cell adhesion molecule-1 (PECAM-1), vimentin, CD44, proliferating cell nuclear antigen (PCNA), p27 Kip1, neural cadherin (N-cadherin), vascular endothelial-cadherin (VE-cadherin), retinoblastoma protein (Rb), survivin, human epidermal growth factor receptor 2 (HER2) and mesenchymal–epithelial transition factor (Met) (Fig. 7J). Conversely, the expression of proteins involved in stress and apoptosis signaling pathways, such as stress-activated protein kinases/Jun amino-terminal kinases (SAPK/JNK), SMAD family member 2 (Smad2), checkpoint kinase 2 (Chk2), p38 mitogen-activated protein kinase (p38 MAPK) and phosphorylated transforming growth factor-β-activated kinase 1 (p-TAK1) showed a significant decrease, while the expression of Bcl-2-associated death promoter (Bad), extracellular signal-regulated kinase 1/2 (ERK1/2), tumor protein p53 (p53) and cleaved poly ADP-ribose polymerase (PARP) showed a significant increase (Fig. 7K). In conclusion, SPAG5 may exert its effects by modulating proteins involved in cell proliferation and apoptosis.
Discussion
In this study, we conducted a comprehensive analysis to examine the expression patterns and prognostic relevance of SPAG5 in various types of tumors. The findings revealed a significant upregulation of SPAG5 expression in the majority of tumors, demonstrating a strong association with unfavorable prognosis. Previous studies have reported that a rise in copy number variations contributes to the upregulation of SPAG5, which is linked to a decrease in overall survival rate among breast cancer patients [27]. Furthermore, heightened expression of SPAG5 was observed in tissues of cervical and gastric cancers, showing a close correlation with decreased patient survival [17, 28]. Immunohistochemical analysis further validated a substantial increase in SPAG5 expression in esophageal cancer tissues. Therefore, it can be inferred that SPAG5 plays a role as an oncogene in esophageal cancer and has the potential to serve as a promising target for cancer gene therapy.
Sperm-associated antigen 5 (SPAG5) is vital for the functionality and dynamic regulation of the mitotic spindle, mitotic progression, and chromosomal segregation accuracy [29]. Current studies have confirmed that SPAG5 is significantly overexpressed in HCC tissues, and its suppression inhibits HCC cell multiplication and induces apoptosis [30]. Similarly, knocking down SPAG5 in glioblastoma inhibits cell proliferation and colony formation while promoting apoptosis [14]. Nevertheless, investigations into SPAG5's mechanism within esophageal cancer remain scarce. Therefore, we conducted further investigations on the influence of SPAG5 on the proliferation and migration of esophageal cancer cells. Our findings indicate that suppressing the expression of SPAG5 impedes both the proliferation and invasion of ESCC cells while promoting apoptotic cell death. Through a series of experimental analyses using Celigo and MTT methods, we found that the growth curve of the shSPAG5 group decreased significantly, providing strong evidence that SPAG5 expression promoted cell proliferation. Similarly, our Transwell experiments established that knockdown of SPAG5 adversely affects the migration and invasion capacity of ESCC cells. Moreover, our investigations utilizing Caspase-3/7 activity detection and FACS analysis confirm that downregulation of SPAG5 induces apoptotic cell death specifically in ESCC cells. Furthermore, enrichment analysis reveals the connection between the dysregulation of SPAG5 expression and multiple biological pathways associated with cell proliferation, including cell cycle regulation, oocyte meiosis, DNA replication, G2/M checkpoint, and mTORC1 signaling. These findings collectively support the crucial contribution of SPAG5 dysregulation in the growth, invasion, and migration processes of esophageal cancer cells.
Then we used the PathScan Antibody Array Kit to explore the potential mechanisms of SPAG5 in promoting esophageal cancer progression. Notably, SPAG5 downregulation considerably decreased the expression of multiple proteins in ESCC cells, including PECAM-1, Vimentin, CD44, p-TAK1, PCNA, Smad2, p27 Kip1, N-Cadherin, VE-Cadherin, Rb, Survivin, HER2/ErbB2, Met, SAPK/JNK, p38 MAPK and Chk2. Among these, p27 Kip1 is a cell cycle regulatory protein that primarily suppresses the transition from G1 to S phase and also regulates the G2/M process and cell division completion through CDK-dependent or independent mechanisms, thereby influencing essential functions such as cell cycle regulation, cell migration, and cell proliferation [31, 32]. Survivin, a cellular apoptosis protein family constituent, protects cells from apoptosis and regulates mitosis [33]. Abnormal overexpression of survivin in cancer is associated with advanced diseases, increased tumor recurrence rates, shortened overall survival, and resistance to chemotherapy and radiotherapy [34]. Furthermore, HER2 overexpression has been detected in gastric and esophageal cancers, making it a prominent target for anticancer therapy [35]. These results suggest that SPAG5 may exert its function by regulating proteins associated with cell proliferation and apoptosis.
Research has demonstrated the pivotal role of infiltrating immune cells in the tumor microenvironment in initiating and advancing tumor progression, thereby exerting a notable impact on the prognosis of cancer patients [36]. Consequently, we conducted a comprehensive analysis to investigate the relationship between SPAG5 and immune cell infiltration. The outcomes revealed a significant association between aberrant infiltration of the majority of immune cells and the expression of SPAG5 across various malignancies. Notably, in esophageal cancer, there was a substantial positive correlation observed between the expression of SPAG5 and M2 macrophages. M2 macrophages are capable of secreting a range of anti-inflammatory and pro-tumor factors, thereby facilitating tumor proliferation, metastasis, angiogenesis, drug resistance, and contributing to the establishment of an immunosuppressive microenvironment [37]. Specifically, the secretion of matrix metallopeptidase 9 (MMP9) and vascular endothelial growth factor A (VEGFA) by M2 macrophages fosters neovascularization, while activation of the nuclear factor kB (NF-kB) signaling pathway through the cytokine interleukin-1beta (IL-1β) secretion enhances the epithelial–mesenchymal transition (EMT) process, promoting the migration and invasion of ESCC cells [38]. Additionally, we explored the association between SPAG5 and immune-related genes, revealing a significant positive correlation between SPAG5 expression and several immune checkpoint genes, including PDCD1, IDO1, PDCD1LG2, TIGIT and CD276. Programmed cell death protein 1 (PDCD1) is an immune checkpoint receptor expressed in activated T cells, involved in regulating the function of effector CD8+ T cells and promoting the differentiation of CD4 + T cells into regulatory T cells [39, 40]. Similarly, CD276 (B7-H3), a member of the B7 superfamily, plays a role in modulating T cell-mediated immune responses [41]. These findings highlight the possible association between SPAG5 and the regulation of the tumor microenvironment, potentially influencing anti-tumor immune-based therapies. Subsequent investigation further uncovered that patients with high SPAG5 expression in esophageal cancer exhibited diminished response rates to PD-L1 treatment compared to those with low expression levels. These findings underscore the involvement of SPAG5 in tumor immune regulation, promoting tumor development, and suggest its potential as a promising biomarker for immune checkpoint blockade therapy in ESCC.
Our research demonstrates that SPAG5 is significantly involved in the pathogenesis and development of esophageal cancer. However, the current study has several limitations. We have initially explored the potential mechanisms of SPAG5 in esophageal cancer; nevertheless, its specific molecular pathways and regulatory networks have not been completely clarified. Future research should include in vivo experiments with animal models of esophageal cancer to confirm the role of SPAG5 in disease progression. Despite using a large amount of pan-cancer RNA sequencing data, the sample size specific to esophageal squamous cell carcinoma is still limited. It is necessary to collect a wider range of samples from esophageal cancer patients in different geographical, genetic, and lifestyle contexts. This will help assess SPAG5’s expression patterns and prognostic implications in esophageal cancer more comprehensively.
Conclusion
In summary, SPAG5 is overexpressed in various cancers and is closely associated with poor patient prognosis. In ESCC, its overexpression is crucial for promoting cancer cell proliferation, metastasis, and resistance to apoptosis. Additionally, it influences the effectiveness of immunotherapy by altering the tumor immune microenvironment. These findings suggest that SPAG5 could be a highly promising therapeutic target.
Supplementary Information
Supplementary Material 1. Supplemental Figure 1. Univariate Cox regression and Kaplan–Meier methods were used to analyze the correlation between SPAG5 expression and disease-specific survival, disease-free intervaland progression-free interval. Supplemental Figure 2. The correlation between SPAG5 expression and ESTIMATE score, immune score, stromal score, and tumor purityin pan-cancer. Red represents positive correlation and blue represent negative correlation. Supplemental Figure 3. The IC50 differences between the high and low SPAG5 expression groups of the 20 selected chemotherapy drugs.
Acknowledgements
We appreciate the support of the TCGA, GTEx, UCSC XENA and other bioinformatics websites and database, as well as statistical analysis methods.
Abbreviations
- ACC
Adrenocortical carcinoma
- APC
Allophycocyanin
- Bad
Bcl-2-associated death promoter
- BLCA
Bladder urothelial carcinoma
- BRCA
Breast invasive carcinoma
- CEP55
Centrosomal protein 55
- CESC
Cervical and endocervical cancers
- Chk2
Checkpoint kinase 2
- CHOL
Cholangiocarcinoma
- COAD
Colon adenocarcinoma
- DCs
Dendritic cells
- DFI
Disease-free interval
- DLBC
Lymphoid neoplasm diffuse large B-cell lymphoma
- DMSO
Dimethyl sulfoxide
- DSS
Disease-specific survival
- ELISA
Enzyme-linked immunosorbent assay
- EMT
Epithelial–mesenchymal transition
- ERK1/2
Extracellular signal-regulated kinase 1/2
- ESCA
Esophageal carcinoma
- ESCC
Esophageal squamous cell carcinoma
- G2/M
G2 phase/mitosis phase
- GBM
Glioblastoma multiforme
- GEO
Gene Expression Omnibus
- GO
Gene Ontology
- GSEA
Gene Set Enrichment Analyses
- GTEx
The genotype-tissue expression
- HER2/ErbB2
Human epidermal growth factor receptor 2
- HNSC
Head and neck squamous carcinoma
- HR
Hazard ratio
- ICIs
Immune checkpoint inhibitors
- IDO1
Indoleamine 2,3 dioxygenase 1
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KICH
Kidney chromophobe
- KIRC
Kidney renal clear cell carcinoma
- KIRP
Kidney renal papillary cell carcinoma
- KM
Kaplan–Meier
- LAML
Acute myeloid leukemia
- LGG
Lower grade glioma
- LIHC
Liver hepatocellular carcinoma
- Log FC
Log2-fold change
- LUAD
Lung adenocarcinoma
- LUSC
Lung squamous cell carcinoma
- LV
Lentivirus vector
- MDSCs
Myeloid-derived suppressor cells
- MESO
Mesothelioma
- Met
Mesenchymal–epithelial transition factor
- MHC
Major histocompatibility complex
- mTORC1
Mechanistic target of rapamycin complex 1
- MTT
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- MMP9
Matrix metallopeptidase 9
- N-Cadherin
Neural cadherin
- NF-kB
Nuclear factor kB
- OS
Overall survival
- OV
Ovarian serous cystadenocarcinoma
- p38 MAPK
P38 mitogen-activated protein kinase
- p53
Tumor protein p53
- PAAD
Pancreatic adenocarcinoma
- PARP
Poly ADP-ribose polymerase
- PBS
Phosphate buffered saline
- PCNA
Proliferating cell nuclear antigen
- PCPG
Pheochromocytoma and paraganglioma
- PDCD1
Programmed cell death protein 1
- PDCD1LG2
Programmed cell death 1 ligand 2
- PECAM-1
Platelet endothelial cell adhesion molecule-1
- PFI
Progression-free interval
- PI3K/AKT
Phosphatidylinositol 3-kinase/protein kinase B
- PRAD
Prostate adenocarcinoma
- p-TAK1
Phosphorylated transforming growth factor-β-activated kinase 1
- qRT-PCR
Quantitative reverse transcription-polymerase chain reaction
- Rb
Retinoblastoma protein
- READ
Rectum adenocarcinoma
- SAPK/JNK
Stress-activated protein kinases/Jun amino-terminal kinases
- SARC
Sarcoma
- shRNA
Short hairpin RNA
- SKCM
Melanoma
- Smad2
SMAD family member 2
- SPAG5
Sperm-associated antigen 5
- STAD
Stomach adenocarcinoma
- TCGA
The Cancer Genome Atlas
- TGCT
Testicular germ cell tumors
- THCA
Thyroid carcinoma
- THYM
Thymoma
- TIDE
Tumor immune dysfunction and exclusion
- TIGIT
T cell immunoreceptor with immunoglobulin and ITIM domain
- TIMER2.0
Tumor immune estimation resource 2.0
- UCEC
Uterine corpus endometrial carcinoma
- UCS
Uterine carcinosarcoma
- UVM
Uveal melanoma
- VE-Cadherin
Vascular endothelial-cadherin
- VEGFA
Vascular endothelial growth factor A
- IL-1β
Cytokine interleukin-1beta
Author contributions
Xiaohui Z, Mingxin Z conceived and designed the experiments. Xiaohui Z, Lingmin Z, Yanan Z,Qian L, and Shiyu J performed the experiments and analyzed the data. Xiaohui Z, Lingmin Z, Manli C performed bioinformatics analysis. Xiaohui Z, Qian L, Mingxin Z wrote the paper. All authors read and approved the final manuscript.
Funding
This work was financially supported by the Natural Science Basic Research Program (2024JC-YBMS-664), Natural Science Basic Research Plan of Shaanxi Province (2022JM-502), Social Development Project of Shaanxi Province (2023-YBSF-072), the Xi’an Science and Technology Plan Project-Innovation Capability Support Program (23YXYJ0180), the Xi’an Science and Technology Plan Project-General medical research projects (2024JH-YLYB-0359), Supporting Fund Project of the First Affiliated Hospital of Xi’an Medical College (XYFYPT-2023-04, XYFYPT-2023-06, XYFYPT-2023-07), the Innovation Team of Xi’an Medical University (2021TD15), Yulin Science and Technology Association Youth Talent Support Program Project (20230555).
Availability of data and materials
The datasets obtained from web-based sources and subsequently analyzed in our study were: The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/), UCSC Xena (https://xena.ucsc.edu/) and Timer 2.0 database (http://timer.comp-genomics.org). Gene expression profile of GSE53625 dataset from Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE53625).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaohui Zhang and Lingmin Zhang contributed to the work equally.
Contributor Information
Qian Li, Email: liqian1103@126.com.
Mingxin Zhang, Email: zmx3115@xiyi.edu.cn.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. [DOI] [PubMed] [Google Scholar]
- 2.Pennathur A, Gibson MK, Jobe BA, et al. Oesophageal carcinoma. Lancet (London, England). 2013;381(9864):400–12. [DOI] [PubMed] [Google Scholar]
- 3.Pennathur A, Farkas A, Krasinskas AM, et al. Esophagectomy for T1 esophageal cancer: outcomes in 100 patients and implications for endoscopic therapy. Ann Thorac Surg. 2009;87(4):1048–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harada K, Rogers JE, Iwatsuki M, et al. Recent advances in treating oesophageal cancer. F1000Research. 2020;9:1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kudo T, Hamamoto Y, Kato K, et al. Nivolumab treatment for oesophageal squamous-cell carcinoma: an open-label, multicentre, phase 2 trial. Lancet Oncol. 2017;18(5):631–9. [DOI] [PubMed] [Google Scholar]
- 6.Janjigian YY, Bendell J, Calvo E, et al. CheckMate-032 study: efficacy and safety of nivolumab and nivolumab plus ipilimumab in patients with metastatic esophagogastric cancer. J Clin Oncol. 2018;36(28):2836–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349(23):2241–52. [DOI] [PubMed] [Google Scholar]
- 8.He J, Green AR, Li Y, et al. SPAG5: an emerging oncogene. Trends in Cancer. 2020;6(7):543–7. [DOI] [PubMed] [Google Scholar]
- 9.Gruber J, Harborth J, Schnabel J, et al. The mitotic-spindle-associated protein astrin is essential for progression through mitosis. J Cell Sci. 2002;115(Pt 21):4053–9. [DOI] [PubMed] [Google Scholar]
- 10.Ying Z, Yang J, Li W, et al. Astrin: a key player in mitosis and cancer. Front Cell Dev Biol. 2020;8:866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thein KH, Kleylein-Sohn J, Nigg EA, et al. Astrin is required for the maintenance of sister chromatid cohesion and centrosome integrity. J Cell Biol. 2007;178(3):345–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu G, Liu S, Cao G, et al. SPAG5 contributes to the progression of gastric cancer by upregulation of Survivin depend on activating the wnt/β-catenin pathway. Exp Cell Res. 2019;379(1):83–91. [DOI] [PubMed] [Google Scholar]
- 13.Li M, Li A, Zhou S, et al. SPAG5 upregulation contributes to enhanced c-MYC transcriptional activity via interaction with c-MYC binding protein in triple-negative breast cancer. J Hematol Oncol. 2019;12(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C, Su H, Cheng R, et al. SPAG5 is involved in human gliomagenesis through the regulation of cell proliferation and apoptosis. Front Oncol. 2021;11: 673780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao G, Xu X, Chen Z, et al. SPAG5 expression predicts poor prognosis and is associated with adverse immune infiltration in lung adenocarcinomas. Clin Med Insights Oncol. 2023;17:11795549231199916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu JY, Zeng QH, Cao PG, et al. SPAG5 promotes proliferation and suppresses apoptosis in bladder urothelial carcinoma by upregulating Wnt3 via activating the AKT/mTOR pathway and predicts poorer survival. Oncogene. 2018;37(29):3937–52. [DOI] [PubMed] [Google Scholar]
- 17.Yuan LJ, Li JD, Zhang L, et al. SPAG5 upregulation predicts poor prognosis in cervical cancer patients and alters sensitivity to taxol treatment via the mTOR signaling pathway. Cell Death Dis. 2014;5(5): e1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang YF, Zhang MF, Tian QH, et al. SPAG5 interacts with CEP55 and exerts oncogenic activities via PI3K/AKT pathway in hepatocellular carcinoma. Mol Cancer. 2018;17(1):117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Wang L, Lu N, et al. Micro RNA-363 inhibits esophageal squamous cell carcinoma progression by directly targeting sperm-associated antigen 5. J Int Med Res. 2020;48(6):300060520932795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldman MJ, Craft B, Hastie M, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38(6):675–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu T, Hu E, Xu S, et al. ClusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Cambridge (Mass)). 2021;2(3): 100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Can Res. 2017;77(21):e108–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng D, Ye Z, Shen R, et al. IOBR: multi-omics immuno-oncology biological research to decode tumor microenvironment and signatures. Front Immunol. 2021;12: 687975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu J, Li K, Zhang W, et al. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med. 2020;12(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang P, Gu S, Pan D, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24(10):1550–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maeser D, Gruener RF, Huang RS. oncoPredict: an R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief Bioinform. 2021;22(6):bbab260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abdel-Fatah TMA, Agarwal D, Liu DX, et al. SPAG5 as a prognostic biomarker and chemotherapy sensitivity predictor in breast cancer: a retrospective, integrated genomic, transcriptomic, and protein analysis. Lancet Oncol. 2016;17(7):1004–18. [DOI] [PubMed] [Google Scholar]
- 28.An J, Yang L, Pan Y, et al. SPAG5 activates PI3K/AKT pathway and promotes the tumor progression and chemo-resistance in gastric cancer. DNA Cell Biol. 2022;41(10):893–902. [DOI] [PubMed] [Google Scholar]
- 29.Manning AL, Bakhoum SF, Maffini S, et al. CLASP1, astrin and Kif2b form a molecular switch that regulates kinetochore-microtubule dynamics to promote mitotic progression and fidelity. EMBO J. 2010;29(20):3531–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H, Hu J, Wei R, et al. SPAG5 promotes hepatocellular carcinoma progression by downregulating SCARA5 through modifying β-catenin degradation. J Exp Clin Cancer Res CR. 2018;37(1):229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bencivenga D, Stampone E, Roberti D, et al. p27Kip1, an intrinsically unstructured protein with scaffold properties. Cells. 2021;10(9):2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bencivenga D, Caldarelli I, Stampone E, et al. p27 Kip1 and human cancers: a reappraisal of a still enigmatic protein. Cancer Lett. 2017;403:354–65. [DOI] [PubMed] [Google Scholar]
- 33.Siragusa G, Tomasello L, Giordano C, et al. Survivin (BIRC5): Implications in cancer therapy. Life Sci. 2024;350: 122788. [DOI] [PubMed] [Google Scholar]
- 34.Athanasoula KC, Gogas H, Polonifi K, et al. Survivin beyond physiology: orchestration of multistep carcinogenesis and therapeutic potentials. Cancer Lett. 2014;347(2):175–82. [DOI] [PubMed] [Google Scholar]
- 35.Roskoski R. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res. 2014;79:34–74. [DOI] [PubMed] [Google Scholar]
- 36.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mantovani A, Sozzani S, Locati M, et al. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23(11):549–55. [DOI] [PubMed] [Google Scholar]
- 38.Shapouri-Moghaddam A, Mohammadian S, Vazini H, et al. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol. 2018;233(9):6425–40. [DOI] [PubMed] [Google Scholar]
- 39.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei SC, Duffy CR, Allison JP. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 2018;8(9):1069–86. [DOI] [PubMed] [Google Scholar]
- 41.Zhou WT, Jin WL. B7–H3/CD276: an emerging cancer immunotherapy. Front Immunol. 2021;12: 701006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1. Supplemental Figure 1. Univariate Cox regression and Kaplan–Meier methods were used to analyze the correlation between SPAG5 expression and disease-specific survival, disease-free intervaland progression-free interval. Supplemental Figure 2. The correlation between SPAG5 expression and ESTIMATE score, immune score, stromal score, and tumor purityin pan-cancer. Red represents positive correlation and blue represent negative correlation. Supplemental Figure 3. The IC50 differences between the high and low SPAG5 expression groups of the 20 selected chemotherapy drugs.
Data Availability Statement
The datasets obtained from web-based sources and subsequently analyzed in our study were: The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/), UCSC Xena (https://xena.ucsc.edu/) and Timer 2.0 database (http://timer.comp-genomics.org). Gene expression profile of GSE53625 dataset from Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE53625).