Abstract
Background
Tramway Ridge, a geothermal Antarctic Specially Protected Area (elevation 3340 m) located near the summit of Mount Erebus, is home to a unique community composed of cosmopolitan surface-associated micro-organisms and abundant, poorly understood subsurface-associated microorganisms. Here, we use shotgun metagenomics to compare the functional capabilities of this community to those found elsewhere on Earth and to infer in situ diversity and metabolic capabilities of abundant subsurface taxa.
Results
We found that the functional potential in this community is most similar to that found in terrestrial hydrothermal environments (hot springs, sediments) and that the two dominant organisms in the subsurface carry high rates of in situ diversity which was taken as evidence of potential endemicity. They were found to be facultative anaerobic heterotrophs that likely share a pool of nitrogenous organic compounds while specializing in different carbon compounds.
Conclusions
Metagenomic insights have provided a detailed understanding of the microbe-based ecosystem found in geothermally heated fumaroles at Tramway Ridge. This approach enabled us to compare Tramway Ridge with other microbial systems, identify potentially endemic taxa and elucidate the key metabolic pathways that may enable specific organisms to dominate the ecosystem.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40793-024-00655-5.
Keywords: Endemic, Hydrothermal, Geothermal, Volcano, Fumarole, Metabolism, Sediments
Background
Mt. Erebus, Victoria Land, Antarctica, is the highest, most southern, isolated geothermal feature on the planet [1]. Since its original submarine eruption approximately 1.3 million years ago [2], the Antarctic Circumpolar current has kept Mt. Erebus relatively isolated from volcanoes found elsewhere on Earth [3]. This isolation has likely limited the colonization of Mt. Erebus by microorganisms to those transported by upper atmospheric winds [4]. Its biogeographical isolation is supported by previous research on the soil microbial communities at Tramway Ridge, a small geothermal feature on the summit plateau of Mount Erebus [4–9]. Tramway Ridge is currently recognized as an Antarctic Specially Protected Area [10] that contains unique, deep-branching, and potentially endemic lineages of Bacteria and Archaea within thermally stratified fumaroles (Fig. 1) [4–6, 11, 12].
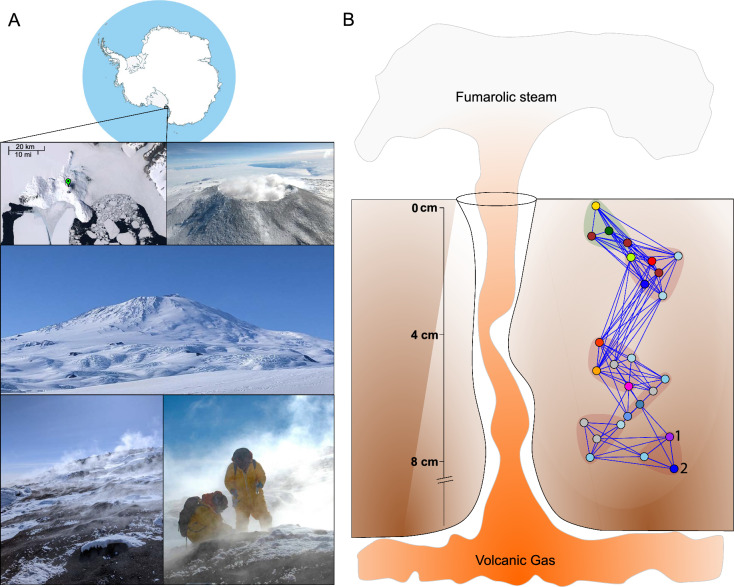
Fig. 1.
Overview of Tramway Ridge, Mt. Erebus. a Tramway Ridge is located near the summit of Mt. Erebus, a volcano on Ross Island, in the Ross Sea region of Antarctica (indicated by green marker on the satellite image). Photos from top to bottom: aerial image of the main crater of Mt. Erebus, which harbours an active lava lake; looking up towards the volcano from the shore of Ross Island; images of Tramway Ridge hot soils, including sampling in Tyvek suits (photos courtesy of Jon Tyler and Stephen Noell). Satellite imagery is from the Antarctic Digital Database Map Viewer https://www.add.scar.org/, Open Source. b Simple cutaway schematic of a fumarole. Water vapour and gasses heated underneath the surface are released at fumaroles, which host diverse and unique microbial lineages. The network on the right is the “intra-correlated groups” (ICGs) of Herbold et al., 2014 [4]. Nodes correspond to OTUs formed from 16S ribosomal RNA gene amplicons. Vertical placement of ICGs indicate an estimate of the location of maximal ICG abundance, based on data in Herbold et al., 2014 [4]. Blue lines indicate a positive correlation between OTUs, with the width of the line corresponding to the strength of the correlation. ICGs are background-shaded with the surface-associated ICG background-shaded in green. OTUs indicated with numbers 1 & 2 represent abundant, potentially endemic organisms discussed at length in this manuscript
The fumaroles of Tramway Ridge differ in many ways from other terrestrial hydrothermal features. They are characterized by hot (65 °C) CO2-rich steam venting through slightly alkaline (pH 8) hydrothermally altered mineral soils [13, 14]. Oxygen levels within fumarolic sediments are approximately 1 mg L−1 (roughly 25% saturated for the temperature) indicative of subsurface hypoxia [7, 15]. All Erebus hydrothermal features are driven by a phonolite magmatic source [9, 16]. Off-gasses tend to be composed primarily of steam, with low levels of sulfur and elevated concentrations of methane, hydrogen and CO [17, 18]. The high pH, moderate temperature, lack of standing water, and hypoxia distinguish them from other well-studied terrestrial hot springs and mud volcanoes and provide a unique geothermally-driven range of micro-environments for resident microbiota. Steam is vented through concentrated hotspots, generating steep temperature (− 20 to 65 °C) and pH (3.5–8) gradients over less than a meter that are major determinants of the composition of the thick cyanobacterial mats and associated microbial communities observed on the surface [5, 6]. Within fumaroles, the temperature is a relatively constant 65 °C, even at > 5 cm depth, but can decrease suddenly to less than 20 °C and stay low for 24 h or more at a time [6, 7]. Beyond its unique physical characteristics, the hot fumarolic soils of Tramway Ridge also have an extremely low total C:N ratio (ranging from 1:3 to 3:1) [4, 6], which suggests that the microbial community experiences continual carbon-limitation relative to nitrogen [19].
The isolation of Tramway Ridge, its unique geochemical environment, and microbiota make it an exciting site for evaluating potential endemism and for identifying novel metabolic pathways. Driven by the novelty of the taxa encountered there and the lack of information regarding their metabolic potential, we launched a detailed metagenomic study of high-altitude Antarctic fumaroles. First, we used functional profiles to contextualize the functional repertoire of this particular community with respect to other types of microbial communities. Second, we improved on our previous effort to identify endemic taxa [4], where we relied on matching partial 16S rRNA gene amplicon sequences with database entries by developing a novel metric that is based on synonymous polymorphisms within reconstructed environmental populations to define/circumscribe potentially endemic taxa based on in situ diversity. Finally, we sought to elucidate possible novel metabolic processes encoded by abundant taxa with high in situ diversity that are specifically localized to the fumarolic subsurface (depths > 2 cm).
Methods
Sample collection
Soil samples were collected within the Tramway Ridge Antarctic Specially Protected Area (ASPA 130) in February 2009 from two sites (site A–77° 31.103′ S, 167° 6.682′ S and site B–77° 31.306′ S, 167° 6.668′ E). Sites were chosen based on measuring a surface temperature of 65 °C with a stainless steel Checktemp1 temperature probe (Hanna Instruments, Rhode Island, USA) sterilized with 70% ethanol immediately prior to use. Temperature measurements were repeated for each layer sampled. Surface soil crusts were carefully set aside prior to collecting samples. Samples were collected by carefully removing 2 cm of soil in an approximately 5 cm × 5 cm square area using an autoclaved stainless steel spatula wiped with 70% ethanol just prior to sampling. Soil was placed into a fresh 50 mL Falcon tube and immediately frozen at − 20 °C. Sampling continued with the collection of a second layer (2–4 cm depth). Samples were assigned NCBI Biosample IDs SAMEA2163752 (Site A, 0–2 cm), SAMEA2163753 (Site A, 2–4 cm), SAMEA2163761 (Site B, 0–2 cm) and SAMEA2163755 (Site B 2–4 cm) under NCBI Bioproject PRJNA431961.
DNA extraction, library preparation and sequencing
DNA was extracted from all samples using a modified CTAB (cetyltrimethylammonium bromide) bead-beating protocol [20] and quantified using the Quant-IT dsDNA BR Assay Kit (Invitrogen, Carlsbad, CA, USA). Portions of extracted metagenomic DNA for samples SAMEA2163752 (Site A, 0–2 cm), SAMEA2163761 (Site B, 0–2 cm) and SAMEA2163755 (Site B 2–4 cm) were frozen and sent to the sequencing facility at the University of California-Los Angeles (USA). Additionally, DNA from sample SAMEA2163752 (Site A, 0–2 cm) was sent to the University of Waikato (Hamilton, New Zealand). At each location, samples were processed and sequenced using standard protocols for the 454-Ti platform (Roche 454 Life Sciences, Branford, CT, USA). Initial analysis of the 454-generated data indicated that we would benefit from greater read depth. We then generated additional metagenomic data using Illumina. Frozen DNA from samples SAMEA2163752 (Site A, 0–2 cm) and SAMEA2163753 (Site A, 2–4 cm) were sent to the sequencing facility at University of California, Davis, where two paired-end libraries were prepared for each DNA sample (technical replicates) and sequenced using the Illumina Hi-Seq 1000 platform (Illumina, San Diego, CA, USA).
Metagenome assembly and binning
Our goal in binning was to produce a high number of metagenome assembled genomes (MAGs), which represent an average population genome estimator for each species. We used multiple (co-)assembly and binning methods in parallel as outlined below to leverage strengths of unique assembly and binning combinations which can affect the quantity and recovery of MAGs in a taxonomy-dependent manner [21]. Co-assemblies only combined merged datasets from the same sequencing platform as 454 and Illumina platforms vary in error profiles and are not typically assembled together [22].
Newbler v.3.0 (Roche) was used for assembling 454-generated data. Sff files from each 454 dataset were assembled independently and were also used in various pooled 454 assemblies in different combination (Table S1). Sff files were assembled using Newbler v.3.0 with a minimum overlap of 100 nucleotides and overlap identity of 98% (-mi 98 -ml 100 -minlen 45 -a 500 -l 2000).
Paired-end Illumina reads were pre-processed by removing any paired-end set for which identifying tags had at least one mismatch or for which the paired-end tags were not identical. Identifying tags were removed, adapters were removed, and reads were quality trimmed with BBDuk v.38.97 (ktrim = r k = 21 mink = 11 hdist = 2 minlen = 19 qtrim = r trimq = 15) [23]. Illumina data were assembled using three assemblers, two assemblers for metagenomic datasets, Megahit 1.2.9 (–k-min 27 –k-max 127 –k-step 10 –min-contig-len 500 –prune-level 3 –no-mercy –min-count 3 –no-local) [24] and MetaSPAdes v.3.15.3 (–meta –only-assembler) [25]. We also assembled metagenomes using SPAdes v.3.15.3 (–careful –only-assembler; –cov-cutoff auto –careful -k 25,55,65,75 –only-assembler)[26] due to its ability to produce high quality bins for rarer taxa [21]. Fastq files for each Illumina dataset were assembled independently and were also pooled in various combinations for additional assemblies (Table S1). In total, 36 unique assemblies were constructed (Table S1) and binned into MAGs separately.
BBMap v.38.97 [23] was used to map individual read sets against large (> 2 kb) contigs and scaffolds from each assembly. Maxbin v.2.2.7 [27] and Metabat 2 v.2. 15 [28] were both used to bin assemblies into an assembly-specific set of MAGs. CheckM v.1.2.3 was used to assess completeness and contamination of MAGs. MAGs from different assemblies were then de-replicated using dRep v.3.0.0 [29] with a completeness cutoff of 40%, contamination cutoff of 10%,a minimum genome size of 200 kb and otherwise default parameters. Each MAG produced was therefore the result of a unique combination of metagenomic dataset(s) used, assembly settings, and binner settings. For each final de-replicated MAG, these details may be found in Table S2. De-replicated MAGs (Table 1 and Table S2) were classified using GTDB-Tk v2.4.0 [30] with genome database release 220 [31], annotated (including tRNA and rRNA annotation) with the NCBI Prokaryotic Genome Annotation Pipeline [32] and screened for contamination with FCS-GX v0.5.3 [33]. Dereplicated MAGs were also clustered into species-level clusters in a final round of dereplication with dRep v.3.0.0 using gANI as the secondary clustering algorithm and 96.5% as the clustering threshold. MAGs discussed in the text were additionally annotated with eggNOG mapper v.2 [34], Cytochrome c oxidases were checked with the HCO classifier [35], hydrogenases were checked with HydDB [36], and CAZymes were checked with dbCAN3 [37].
Table 1.
Species-level representatives of metagenome-assembled genomes (MAGs) from Tramway Ridge fumarolic soils
| Name | Species group | # of strains in species group | Phylum | MIMAG reporting standard | GC |
|---|---|---|---|---|---|
| Blastocatellia bacterium ERB_27 | 27 | 2 | Acidobacteriota | High | 50.9 |
| Pyrinomonas sp. ERB_32 | 32 | 1 | Acidobacteriota | High | 60 |
| Acidimicrobiia bacterium ERB_23 | 23 | 3 | Actinobacteriota | Medium | 55.1 |
| Acidimicrobiia bacterium ERB_8 | 8 | 1 | Actinobacteriota | Medium | 69.4 |
| Thermoleophilia bacterium ERB_19 | 19 | 3 | Actinobacteriota | High | 69.5 |
| Armatimonadota bacterium ERB_24 | 24 | 1 | Armatimonadota | Medium | 60.8 |
| Armatimonadota bacterium ERB_33 | 33 | 1 | Armatimonadota | Medium | 60.5 |
| Armatimonadota bacterium ERB_34 | 34 | 2 | Armatimonadota | High | 58 |
| Armatimonadota bacterium ERB_6 | 6 | 3 | Armatimonadota | High | 61.1 |
| Candidatus Fervidibacter antarcticus ERB_15 | 15 | 1 | Armatimonadota | High | 56.3 |
| Chitinophagaceae sp. ERB_2 | 2 | 1 | Bacteroidota | Medium | 40.5 |
| Chitinophagaceae sp. ERB_3 | 3 | 2 | Bacteroidota | Medium | 39.3 |
| Ignavibacteria bacterium ERB_28 | 28 | 1 | Bacteroidota | High | 55.2 |
| Chloroflexota bacterium ERB_10 | 10 | 1 | Chloroflexota | Medium | 64.7 |
| Chloroflexota bacterium ERB_11 | 11 | 1 | Chloroflexota | Medium | 68.5 |
| Chloroflexota bacterium ERB_20 | 20 | 3 | Chloroflexota | High | 63.6 |
| Candidatus Nitrocaldera therma ERB_22 | 22 | 2 | Chloroflexota | High | 65.1 |
| Chloroflexota bacterium ERB_25 | 25 | 1 | Chloroflexota | Low-quality draft | 64.8 |
| Chloroflexota bacterium ERB_7 | 7 | 1 | Chloroflexota | Medium | 70.2 |
| Chloroflexota bacterium ERB_9 | 9 | 2 | Chloroflexota | Medium | 62.8 |
| Thermoflexus sp. ERB_21 | 21 | 3 | Chloroflexota | high | 69.6 |
| CSP 1-3 bacterium ERB_18 | 18 | 2 | CSP 1-3 | High | 70.1 |
| Leptolyngbya sp. ERB_1 | 1 | 2 | Cyanobacteriota | Medium | 47.2 |
| Mastigocladus sp. ERB_26 | 26 | 2 | Cyanobacteriota | High | 41.2 |
| Allomeiothermus sp. ERB_29 | 29 | 2 | Deinococcota | Medium | 66.3 |
| Meiothermus sp. ERB_30 | 30 | 2 | Deinococcota | Medium | 61.5 |
| Caldithermus sp. ERB_31 | 31 | 1 | Deinococcota | Medium | 68.5 |
| Thermus sp. ERB_17 | 17 | 5 | Deinococcota | High | 65.2 |
| Candidatus Dadabacteria bacterium ERB_12 | 12 | 1 | Desulfobacterota | Medium | 44.2 |
| Nitrospiraceae bacterium ERB_14 | 14 | 2 | Nitrospirota | Medium | 56.2 |
| Candidatus Lakebacteria bacterium ERB_C1 | C1 | 3 | Patescibacteria | High | 27.3 |
| Gemmatales bacterium ERB_16 | 16 | 2 | Planctomycetota | High | 60.1 |
| Rhodanobacteraceae bacterium ERB_4 | 4 | 1 | Pseudomonadota | Medium | 62.8 |
| Candidatus Australarchaeum erebusense ERB_5 | 5 | 1 | Thermoproteota | High | 63.6 |
| Nitrososphaera sp. ERB_13 | 13 | 1 | Thermoproteota | Medium | 56.6 |
Lineage was assigned using GTDB-Tk v.2.4.0 [30] with release 220 of the Genome Taxonomy Database [31]. Completeness and contamination estimates were calculated in CheckM v.1.2.3 [55]. “Mimag reporting standard” follows the recommendations outlined previously [60]. For a comprehensive list of genome quality attributes for all binned strains, including completeness, contamination and tRNA/rRNA counts used for determining the MIMAG reporting standard, see Table S2
Endemicity index calculation
Quality-trimmed Illumina reads were mapped to each MAG using BBmap v.38.97 and further filtered using a hard 97% identity cutoff where identity = number of matches/length of alignment, with the additional requirement that at least 50 nucleotides mapped. Raw diversity was compiled using Samtools mpileup using option: -d 1000000 [38] for each MAG and each dataset independently. SNPs were determined using Varscan2 pileup2snp (options: –min-var-freq 0.01 –p-value 0.05) [39] and further filtered using the Benjamini–Hochberg multiple testing correction FDR = 0.01. SnpEff [40] was used to classify SNPs as synonymous with gff files produced with Prodigal [41]. The density of synonymous SNPs (DSynSNP) for a MAG was calculated as the number of Synonymous SNPs (NSynSNP) divided by MAG length in Mb (LMAG): DSynSNP = NSynSNP / LMAG. Because sensitivity increases as read depth increases, DSynSNP was corrected for read coverage of the MAG (CMAG) which was calculated as the number of reads mapped (NM) divided by MAG length (LMAG): CMAG = NM / LMAG. Endemicity Index (EI) was then the density of synonymous SNPs (DSynSNP) divided by read coverage (CMAG): EI = DSynSNP/CMAG. EI was calculated for each MAG/dataset combination only if NSynSNP ≥ 5 and reported EI values are the average of calculated values over four Illumina read sets representing two technical replicates each of two physical samples (Biosample SAMEA2163752 = SRR6519253 and SRR6519256. Biosample SAMEA2163753 = SRA accessions SRR6519254 and SRR6519255.).
Functional profile comparisons
Pfam [42] profiles were used to compare functional similarity between the metagenomic assemblies from Tramway Ridge and publicly available metagenomic assemblies in the Integrated Microbial Genomes (IMG) database [43]. A list of all assembled, published, and “unrestricted” environmental metagenomic datasets available through IMG was downloaded on 31 March, 2023. IMG-generated pfam profiles (counts of pfams present in metagenomic assembly) were downloaded for each available metagenome, resulting in 7652 total pfam profiles. The number of metagenomic datasets was reduced by removing datasets with total assembly length less 5 × 107 or greater than 1 × 109 bases. This subset was further reduced by removing metagenomic datasets with fewer than 3500 or greater than 7500 unique pfams. These filtering criteria resulted in a dataset of 4513 publicly sourced metagenomic datasets for comparative analysis. Profiles were analyzed in R 4.2.3. Jaccard dissimilarity was calculated using the vegdist() function from vegan 2.6–4 [44] based on presence/absence of pfams. Principle coordinate analysis (PCoA) was carried out using the pcoa() function from Version 5.7–1 of ape [45] and plotted in three dimensions using the plot3d() function from rgl 1.2.1 [46]. t-distributed stochastic neighbor embedding (tSNE) was calculated using the Rtsne() function from Rtsne 0.16 [47] and plotted with ggplot2 3.4.3 [48]. Rtsne settings were as follows: Rtsne(X, dims = 2,initial_dims = 5, perplexity = 300, theta = 0.5, check_duplicates = FALSE, pca = TRUE, partial_pca = FALSE, max_iter = 5,000,000, verbose = getOption("verbose",FALSE), is_distance = TRUE, Y_init = NULL, pca_center = TRUE, pca_scale = FALSE, normalize = TRUE, stop_lying_iter = 500,000, mom_switch_iter = 500,000, momentum = 0.5, final_momentum = 0.8, eta = 100, exaggeration_factor = 12).
Phylogenomic analysis
A collection of reference genomes for comparative phylogenomic analysis was assembled from representative species defined in release 214 (April, 2023) of the Genome Taxonomy Database [31]. GTDB classifications of MAGs from the current study were used to select from GTDB species representatives for an informative tree. For instance, in the case where a MAG was classified into an order but not a family, one representative taxon from each family was included, the phylogeny was calculated, and if the MAG was associated with a particular family, the process was repeated using genus representatives from that family. These genomes were supplemented with additional genomes from thermophilic environments [49, 50], thermophilic nitrifying enrichment cultures [51, 52], additional Nitrospirota [53] and additional Nitrososphaeria [54]. All genomes were downloaded and processed using CheckM v 1.2.3 [55] to generate concatenated alignments of 34 universal marker genes (43 marker HMMs). To be included in phylogenomic reconstruction, reference genomes were required to be at least 60% complete with less than 5% contamination and to have at least 4500 ungapped characters in the concatenated alignment (6988 total positions). All genomes used for phylogenetic analysis are listed in Table S3. Phylogenetic reconstruction with IQ-TREE 2 [56] included model selection with ModelFinder [57] and calculation of bootstraps with UFboots [58].
Results
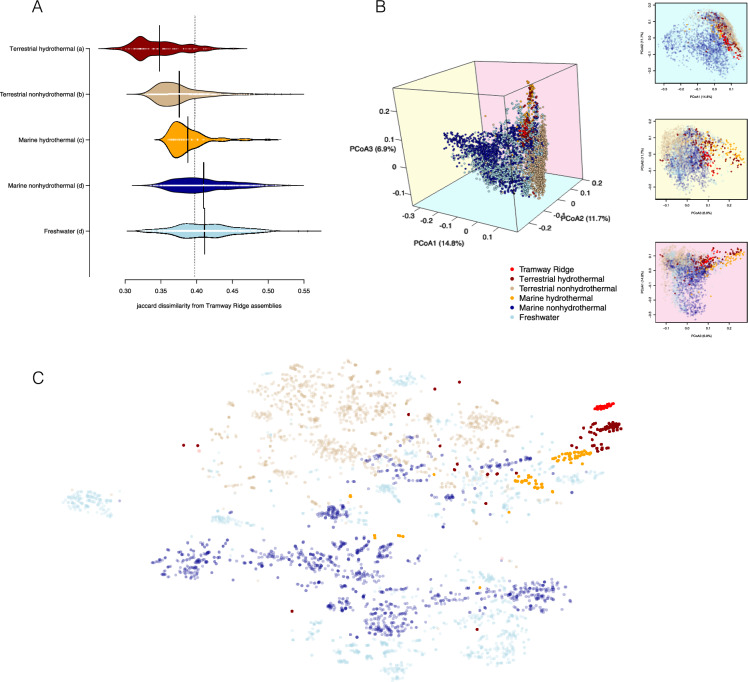
We found that the functional profiles from Tramway Ridge metagenomes resembled those from other thermally-influenced environments, in particular those from terrestrial hydrothermal systems. To learn this, we compared functional profiles of assembled Tramway Ridge metagenomes to 4513 publicly available and assembled environmental metagenomes broadly categorized as terrestrial hydrothermal / non-hydrothermal, freshwater, and marine hydrothermal / non-hydrothermal (Table S4). Functional profiles for metagenomes were constructed based on the presence-absence of Pfam protein family domains [59], and dissimilarities were calculated using the Jaccard index. A post-hoc Tukey’s HSD test (Fig. 2a) comparing Jaccard dissimilarity grouped according to the broad categories listed above showed that metagenomes from Tramway Ridge were most similar to terrestrial hydrothermal environments (Tukey category a) and least similar to metagenomes from non-hydrothermal marine and freshwater environments (Tukey category d). We continued with principal coordinate analysis (PCoA) and t-distributed Stochastic Neighbor Embedding (tSNE) visualizations to explore relationships that may not have been clear from grouped pairwise comparisons (Fig. 2bc). In both, Tramway Ridge microbial communities clustered loosely with microbial communities sourced from both terrestrial and marine hydrothermal environments to the exclusion of non-hydrothermal environments. t-SNE visualization (Fig. 2c) recovered several distinct but associated clusters from hydrothermal systems, one of which was composed exclusively of profiles from Tramway Ridge.
Fig. 2.
Exploration of pairwise Jaccard distance between Pfam-based functional profiles of public metagenomes. Profiles were constructed as vectors of Pfam presence-absence, and distance was calculated using the Jaccard Index (see methods for details) a Bean plot showing the distribution of dissimilarity between functional profiles from Tramway Ridge metagenomes and other broad environmental categories. The dashed vertical line indicates the median dissimilarity of all metagenomes. The categorical labels from a post hoc Tukey’s test are indicated for each environmental category. b 3-D principal coordinate analysis (PCoA) plot based on pairwise Jaccard distance between publicly available metagenomic assemblies and 2-D projections comparing each of the three first coordinate axes. Each point represents a single dataset. c t-distributed stochastic neighbor embedding (tSNE) projection of the data in b
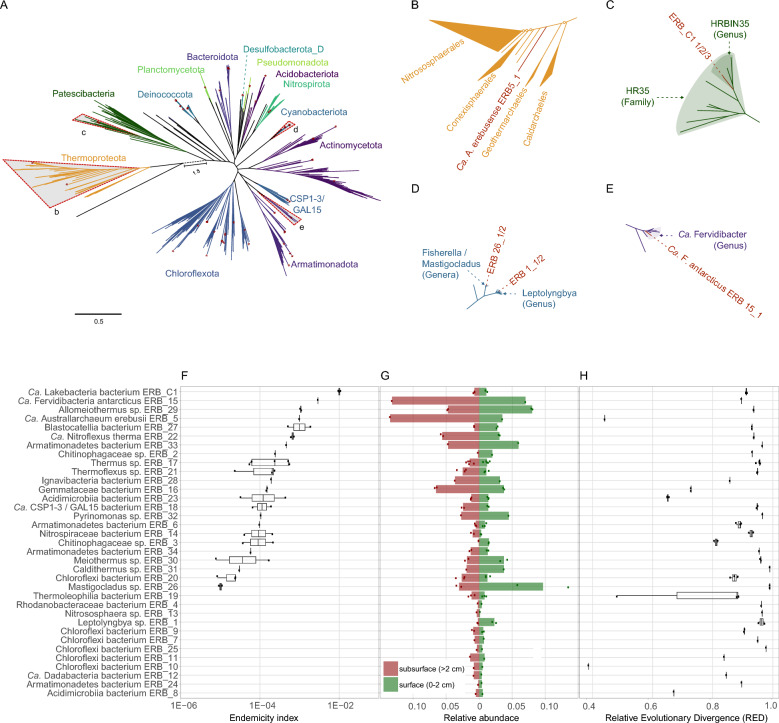
Metagenome-assembled genomes (MAGs) were constructed from DNA extracted from soils at two 65 °C fumaroles at Tramway Ridge (Table S5). We recovered 63 MAGs at the strain level (99% average nucleotide identity, ANI), which clustered into 35 species-level representatives (> 96.5% average nucleotide identity) (Fig. 3, Table 1, Table S2). A total of 16 species were represented by MAGs that met the MIMAG standard for high-quality drafts, and 18 species were represented by MAGs met the standard for medium-level drafts [60]; these include nearly complete (75–96%) genome bins for a novel order of Archaea within the Nitrososphaeria (Fig. 3b, 3h) and novel lineages of Bacteria including Armatimonadota, Chloroflexota, Actinobacteriota, and Candidate division CSP1-3. Additional MAGs of interest include those belonging to the Candidatus Patescibacteria phylum (Fig. 3c), Mastigocladus genus (Fig. 3d) and Candidatus Fervidibacter genus (Fig. 3e). MAGs were classified using GTDB-Tk v.2.4.0 [30] with release 220 of the Genome Taxonomy Database [31], and novel taxa of note were given proposed names (Table 1, Table S2).
Fig. 3.
a Phylogenomic reconstruction of all MAGs recovered from Tramway Ridge, coloured according to phylum-level classification by GTDB-Tk v.2.4.0 using GTDB release 220. Trees were reconstructed with IQtree2 using an alignment of 34 concatenated marker genes generated with CheckM v.1.2.3. ModelFinder identified the best-fit model as LG + F + I + I + R10. Only MAGs with > 60% completeness and < 5% contamination as determined with CheckM v.1.2.3 were used for phylogenetic reconstruction. Bootstrap values have been omitted and location of Tramway MAGs are denoted with a red asterisk. Four clades of interest have been marked with grey triangles. The branch separating Archaea and Bacteria has been truncated and the actual branch length is given as 1.8. A complete, annotated tree is provided as Figure S1. b–e Subtrees from a that are bound by grey triangles. The majority of bipartitions shown exceed 95% UFboot support. Those marked with an open circle are those with less than 95% UFBoot support. Relevant taxonomic clades defined in GTDB release 220 are shaded and the location of Tramway MAGs are indicated as necessary for clarity. f Endemicity index (EI) of species-level collections of MAGs. Each line presents data for one species-level collection of MAGs (> 96.5% gANI). Strain-level MAGs within each species are shown as individual dots. Vertically, species are ordered by EI, with a higher EI indicating a larger diversity at synonymous coding positions. g Read abundance is reported as the relative proportion of metagenomic reads in Illumina datasets from the subsurface (brown–red) and near-surface (green) samples that map to each MAG. The middle line represents 0 and deviations to the left represent abundance in the subsurface and deviations to the right represent abundance in the near-surface. h Relative Evolutionary Divergence (RED) values for each MAG as determined with GTDB-Tk. Low RED values correspond to deep branches within the reference tree
Tramway Ridge occupies a unique location on Earth that makes it ideal for the study of the effects of isolation on microbial communities due to its remote location and the geothermal enrichment that prevents intrusion of nearby psychrophilic and mesophilic microorganisms. Here, we quantified in situ diversity for each of the MAGs we generated. We interpreted the in situ diversity of a given taxon as being proportional to the degree of endemism of that taxon, reasoning that endemic species have had an extended opportunity to diversify on-site, as opposed to recent arrivals that would be subject to founder effects. We developed a simple metric, which we called the endemicity index (EI) (Fig. 3F) that measured the in situ diversity of a MAG by calculating the frequency of synonymous mutations in a MAG accounting for read depth. High values (e.g. 10–2) of EI indicate high diversity whereas low values (e.g. 10–6) indicate low diversity.
The lowest median EI value (1 × 10–5) for a single species was calculated for ERB_26, (Fig. 3f), which represented two individual strains of a cyanobacterium belonging to the Fischerella / Mastigocladus genus (Table S2) and dominated the near-surface (5–10% of near-surface reads). The highest median EI value (1 × 10–2) for a single species was calculated for ERB_C1 (Fig. 3f), which represented three individual strains of Candidatus Patescibacteria (Table S2). Ca. Lakebacteria bacterium ERB_C1 classified into the HRBIN35 genus in the HR35 family (Fig. 3c). Based on the percentage of reads recruited, it was slightly more abundant in the near-surface (0–2 cm depth) than in the subsurface (> 2 cm, Fig. 3g). Due to this distribution pattern and the fact that an accelerated evolutionary process is a hallmark feature of members of this lineage [31, 61, 62], we did not pursue further analysis of this genome, which would be best accomplished in a comprehensive comparison with other Ca. Patescibacteria.
Two MAGs, ERB_15_1 and ERB_5_1, with relatively high EI values (3 × 10–3 and 1 × 10–3, respectively) dominated the subsurface community at depths greater than 2 cm (Fig. 3f, Fig. 3g). ERB_5_1 shared less than 40% average amino acid identity (AAI) with any other Nitrososphaeria (formerly Thaumarchaeota) and formed a singular deep branch of the Nitrososphaeria in phylogenetic trees (Fig. 3b) with relative evolutionary divergence (RED) values of 0.44 (Fig. 3h) indicating that it represents a novel order of Archaea. ERB_15_1 shared 69% AAI and 77% ANI with Candidatus Fervidibacter sacchari, establishing it as a novel species of this genus (Fig. 3c). Here we propose the names Candidatus Fervidibacter antarcticus (ERB_15_1) and Candidatus Australarchaeum erebusense (ERB_5_1). We picked these two MAGs for further analyses based on their high EI values, subsurface abundance, and taxonomic novelty.
Candidatus A. erebusense (ERB_5_1) branches near the base of the Nitrososphaeria (Fig. 3b). Like other deeply diverging lineages, it lacked marker genes for cobalamin biosynthesis (cob/cbi/bluB) and ammonia oxidation (archaeal amoABC, HAO) [63–65]. Consistent with previous observations of deeply diverging Nitrososphaeria, the genome of Ca. A. erebusense encoded genes for the beta oxidation of fatty acids and peptide degradation and several genes that may be used for amino acid-based oxidation (Table S6). Ca. A. erebusense encoded aminotransferases and glutamate dehydrogenase that could be used to produce NAD(P)H by converting glutamate to 2-oxoglutaric acid. A 2-oxoglutarate:ferredoxin oxidoreductase could then convert the 2-oxoglutaric acid to succinyl-CoA, which could then be converted to succinate with the production of ATP via ADP-forming succinate-CoA ligase (Table S6). Unlike other deep-branching Nitrososphaeria, Ca. A. erebusense also additionally encoded two aa3-type (low-affinity) cytochrome C-oxidases (Table S6). We identified a putative aerobic carbon monoxide dehydrogenase (CODH) cluster of genes (coxMLS) that may confer the ability to use CO as an additional energy source. However, the large subunit of the CODH in Ca. A. erebusense contained a hallmark motif, AYXGAGR, of type II CODH, which oxidizes CO only very slowly and possibly incidentally [66]. No predicted carbon fixation pathways were identified.
Candidatus Fervidibacter antarcticus (ERB_15_1) shared 69% AAI and 77% ANI with Candidatus Fervidibacter sacchari, establishing it as a novel species of this genus (Fig. 3c). Based on conservative annotations, the Ca. F. antarcticus genome encoded 144 CAZymes, including 69 glycosyl hydrolase (GH) domains spanning 32 GH families (Table S7, Table S8), and an impressive 15 variants of the unusual and poorly characterized GH109 family. Furthermore, when we group the Ca. F. antarcticus CAZymes by substrate utilization capacity, the GH109s belong to the largest cohort (35.25%) which appear to be involved in depolymerisation cascades of N-acetylhexosamide glycans such as chitins and chitosan (Table S8). Other major polysaccharide utilization cohorts include cellulose and hemicellulose (19.8%), pectin (14.3%) and ß-glucans (19.8%), with minor cohorts for ɑ-glucans, ɑ-mannans and ɑ-fucans. Ca. Fervidibacter genomes, including Ca. F. antarcticus additionally encoded numerous aminotransferases, glutamate dehydrogenase, indole-pyruvate:ferrodoxin oxidoreductase and ADP-forming succinate-CoA ligase (Table S7) which may explain the ability of Ca. Fervidibacter sacchari to grow solely on casamino acids [67]. The Ca. F. antarcticus genome also encoded Group 2a NiFe hydrogenases which are high-affinity hydrogenases that enable the survival of soil heterotrophs under carbon limitation [68] and enable hydrogenotrophic growth of autotrophic nitrite-oxidizing bacteria under nitrite limitations [69]. The Ca. F. antarcticus genome also encoded a cluster of putative CODH genes (coxMLS), which contained the hallmark motif, AYXGAGR, of type II CODH, indicating a low, incidental activity [66]. No carbon fixation pathways were found.
Discussion
Metagenome functional profiles
Mt. Erebus has evolved over time, starting with seafloor rifting and growing as a subaerial volcano into a modern-day stratovolcano [2]. The current conditions at Tramway Ridge differ remarkably from other extant non-Antarctic terrestrial and marine hydrothermal systems, being primarily driven by the unique phonolite magmatic source resulting in alkaline fumaroles with low sulphidic content. It is unknown how long geothermal features such as the fumaroles at Tramway Ridge have been present on Mt. Erebus; however, volcanism was once widespread across the Ross Island massif [1]. Extant geothermal features may represent a once widespread ecosystem of similar features. Given this complex history of Mt. Erebus, we questioned whether the Tramway Ridge community has retained a legacy signature of its origin on the seafloor or if the modern-day Tramway Ridge community better resembles other terrestrial hydrothermal sites.
To answer this question, we compared functional profiles of assembled Tramway Ridge metagenomes to a large set of publicly available metagenomes. Metagenomes from Tramway Ridge were distinct from all others (Fig. 2c) but showed the most similarity to terrestrial hydrothermal environments such as hot springs and associated sediments. This indicates that the legacy of a sea floor origin is less important than the extant conditions at Tramway ridge in defining the microbial community. This is also reflected in the prevalence of taxa at Tramway Ridge that are found in other terrestrial hydrothermal environments but not in seafloor hydrothermal systems, such as the genera Mastigocladus, Caldithermus, Meiothermus and phyla such as the Chloroflexota, Armatimonadota and Actinobacteriota.
The unique nature of Tramway Ridge metagenomic profiles likely reflects the site’s relatively low diversity [4] and the type of functional profile analysis used here. First, pfams were used, which are very coarse functional units. Second, gene abundance was not available for most metagenomes, so we were forced to use presence-absence as data. Third, we found that the use of pfam presence-absence was sensitive to metagenome assembly size as well as the total number of unique pfams detected. In combination, these features had the potential to obfuscate any meaningful relationships. Our analysis was optimized to be as permissive as possible, allowing as many metagenomes into the analysis while still comparing like against like. Using this strategy, we were able to compare Tramway Ridge metagenomic datasets to 4513 out of 7652 publicly available and unrestricted metagenomes.
Within continental Antarctica, only three known surface-expressed active geothermal areas exist (Mt. Rittmann, Mt. Melbourne and Mt Erebus), each of which is separated by vast ice fields [7]. It is thought that intercontinental transport of microbe-bearing particulates into Antarctica occurs much less frequently than intracontinental transport [70], suggesting that the introduction of exogenous microbes is relatively infrequent. Recent studies have used database searches to identify potentially endemic species [71], and a lack of sequence identity to database entries has been used in the past to suggest that novel sequences indicate endemism [4]. However, defining endemism based on whether sequence matches exist within a database can be problematic as this definition is sensitive to database composition. Conclusions drawn may not withstand the inevitable growth in database size and the diversity it holds. For this study, we took an alternative approach and attempted to define the degree of endemism of a given taxon as being proportional to the in situ diversity of that taxon. For these inferences, we assumed that rare colonization events have been limited to single clones due to the extreme isolation of Mt. Erebus. Therefore, populations arising from recently introduced taxa would be expected to exhibit relatively low levels of genetic polymorphism and endemic microbial populations would be expected to show high levels of genetic polymorphism.
We developed the endemicity index (EI) (Fig. 3F), focused on accumulations of synonymous mutations, which were assumed to be under reduced selection pressure. We used the EI to assess the diversity of a microbial population represented by a MAG. Similar calculations have been successfully applied to approximate effective population size and genomic fluidity [72]. High values (e.g. 10–2) of EI indicate high diversity which we interpreted as reflecting a neutral evolutionary process occurring under minimal contemporary selection pressures. Low values (e.g. 10–6) indicate low diversity, which we interpreted as possible evidence of a relatively recent arrival, a local population bottleneck, or a recent selective sweep.
A cyanobacterium belonging to either the Fischerella or Mastigocladus genus recovered the lowest median EI value (1 × 10–5) for a single species. This species dominated the near-surface. Although difficult to distinguish these two genera based on 16S rRNA gene sequence and GTDB classifies all members of the genera Fischerella and Mastigocladus as Fisherella, the distinction of Mastigocladus is recognized as a distinct genus by the List of Prokaryotic names with Standing in Nomenclature (LPSN). Therefore we classified this MAG as a member of the Mastigocladus genus to be consistent with the classification of specimens collected from Tramway Ridge in the past [73]. The low in situ diversity observed for this taxon was consistent with previous studies that showed that the global phylogeography of Mastigocladus reflects a geologically recent radiation from Yellowstone National Park, USA [74] and that the surface-associated microbial community at Tramway Ridge is likely dominated by aeolian-distributed cosmopolitan members of non-Antarctic temperate and terrestrial hydrothermal soil communities [4].
We identified Candidatus Australarchaeum erebusense (ERB_5_1) and Candidatus Fervidibacter antarcticus (ERB_15_1) as two MAGs with relatively high EI values and abundance in the subsurface at depths greater than 2 cm (Fig. 3f, Fig. 3g). In our earlier amplicon-based study, these two taxa were similarly identified as dominant, potentially endemic, and associated with the subsurface, but were referred to as “Thaumarchaeota-like archaeon” and OCtSpA1-106 respectively [4].
The most abundant subsurface-associated organism recovered was Candidatus A. erebusense (ERB_5_1), a member of the Nitrososphaeria (formerly Thaumarchaeota). This class of Archaea is a globally distributed group that is best known for the chemolithoautotrophic oxidation of ammonia [75] and for the apparent universal synthesis of cobalamin [76]. However, several deeply divergent lineages of Nitrososphaeria identified through the construction of MAGs [50, 63, 77] and a single cultivated species, Candidatus Conexivisphaera calidus NAS-02 [78] have been shown to lack these hallmark attributes. These deeply diverging lineages have been predicted to be predominantly anaerobic heterotrophs [63, 77] capable of beta oxidation of fatty acids and protein/peptide degradation [78, 79]. Candidatus A. erebusense (ERB_5_1) is one of the deepest-branching members of the Nitrososphaeria (Fig. 3b) and like other deeply diverging lineages, it encodes genes for the beta oxidation of fatty acids and peptide degradation while lacking marker genes for cobalamin biosynthesis and ammonia oxidation (Table S6). Candidatus A. erebusense may also employ amino acid-based oxidation, similar to a pathway used by Thermococcus kodakarensis [80] and a proposed alternative metabolism for Ca. Nitrosocaldus islandicus [64], a representative of thermophilic ammonia oxidizing archaea (AOA). In this proposed metabolism, glutamate could be utilized to generate both reducing power and ATP to power the cell.
However, Ca. A. erebusense is also predicted to respire oxygen, a unique prediction among its closest, presumably anaerobic relatives. It encodes two aa3-type (low-affinity) cytochrome C-oxidases which could presumably drive beta oxidation of fatty acids. However it is unclear whether aerobic respiration could be coupled with an amino acid degradation pathway, which is typically thought to be an anaerobic metabolism [64, 80]. It is unclear if these energy-generating pathways are mutually exclusive and operate under specific oxidative conditions. At least during summer, oxygen levels in the subsurface are around 30% saturation [4] and therefore, no organisms inhabiting the fumaroles are likely to be obligate anaerobes. However, it is also reasonable to assume that the wet, steamy subsurface experiences anoxia at least at small spatial scales. Therefore, we hypothesize that Ca. A. erebusense switches between metabolic pathways depending on the oxic environment, using beta-oxidation of fatty acids when oxygen levels are sufficiently high and peptide fermentation when oxygen levels are low. CO metabolism is likely to be used for maintenance during times of nutritional stress, as has previously been shown for Antarctic soil microbes [54].
Another abundant subsurface MAG examined in detail belongs to the thermophilic genus Candidatus Fervidibacter, which was first discovered in Octopus Spring, Yellowstone National Park (clone OctSpA1-106, [81]. The genus is named after Ca. Fervidibacter sacchari, which encodes a large repertoire of carbohydrate-active enzymes (CAZymes) [82] and which has recently been isolated and described [67]. Like other Ca. Fervidibacter, Ca. F. antarcticus (ERB_15_1) encodes a significant number and diversity of CAZymes including a large, diverse cohort of the unusual and poorly characterized GH109 family. Previous findings suggest that these novel enzymes are involved in extracellular polysaccharide metabolism unique to thermophilic systems [67, 83, 84] with the diversity of Ca. F. antarcticus GH109s suggesting a diverse and unique polysaccharide utilization profile typical of this genus. Interestingly, Ca. Fervidibacter genomes, including Ca. F. antarcticus appear to also encode mechanisms that enable growth on casamino acids [67], [67]. The encoded hydrogenase and CODH are likely maintenance mechanisms to enable survival under carbon limitation with growth primarily supported through aerobic heterotrophic growth on saccharides and anaerobic growth on amino acids.
Conclusion
In the current study, we have used metagenomics to assess several aspects of the microbial community inhabiting the fumarolic soils of Tramway Ridge, Mt. Erebus, Antarctica. We observed a shared functional repertoire between Tramway Ridge and other geothermal systems, specifically terrestrial hydrothermal systems such as hot springs. We then assessed metagenome-assembled genomes (MAGs) using a novel measure to identify two potentially endemic taxa that are more abundant in the Tramway Ridge subsurface (> 2 cm depth) than the near-surface (< 2 cm depth). We named these two taxa Candidatus Australarchaeum erebusense and Candidatus Fervidibacter antarcticus to reflect that they were first observed at Mt. Erebus (Ca. A. erebusense) or that this is the first time this genus has been describved from Antarctica (Ca. F. antarcticus). A close examination of the metabolic repertoire of these taxa revealed that they are likely both facultative anaerobic heterotrophs that specialize in using different carbon sources under aerobic conditions, but that use similar organic compounds during anaerobic growth. Like other deep-branching non-AOA Nitrososphaeria, Ca. A. erebusense possesses a putative pathway for the beta-oxidation of fatty acids. Like other Candidatus Fervidibacter, Ca. F. antarcticus is predicted to utilize sugars and scavenge hydrogen gas under aerobic conditions [82, 85]. Under oxygen-limited conditions, both may utilize similar peptides and amino acids for energy and carbon acquisition. We hypothesize that this pattern of metabolic utilization may reflect the extreme and carbon-limiting C:N ratios (1:3 to 3:1) encountered at Tramway Ridge. Dominant taxa may share nitrogen-rich compounds such as peptides and amino acids since the demand for such compounds may not be as high as for carbon-rich compounds. Instead, different taxa specialize in utilizing specific carbon compounds (sugars vs. fatty acids) through selective exclusion, allowing both to co-exist by carving out specific nutritional niches. Each is also equipped with form II aerobic carbon monoxide dehydrogenase genes that may provide maintenance energy during times of starvation. Together, these insights provide an unprecedented view into the dominant metabolic processes that may sustain life in this harsh, isolated environment.
Supplementary Information
Acknowledgements
Antarctic logistic support for Event K-023 was provided by Antarctica New Zealand.
Author contributions
Planning and field sampling by CWH, CJV, SCC and IRM. Data analysis by CJV and CWH. All authors contributed to the interpretation of data and writing of the manuscript.
Funding
Financial support was provided by grant UOW0802 from the New Zealand Marsden Fund to SCC and IRM and a CRE award from the National Geographic Society to S.C.C.
Availability of data and material
All raw metagenomic sequence data and metagenome assembled genomes used in this manuscript are available under bioproject accession PRJNA431961 (https://www.ncbi.nlm.nih.gov/bioproject/431961). Data and scripts necessary to reproduce figures and calculate endemicity index are available at https://doi.org/10.5281/zenodo.14203511.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kyle PR, McIntosh WC, Schmidt-Thomé M, Mueller P, Tessensohn F, Noll MR, et al. A. McMurdo volcanic group western ross embayment. In: LeMasurier WE, Thomson JW, Baker PE, Kyle PR, Rowley PD, Smellie JL, et al., editors. Volcanoes of the antarctic plate and southern oceans. Washington, D. C.: American Geophysical Union; 1990. p. 18–145.
- 2.Esser RP, Kyle PR, McIntosh WC. 40Ar/39Ar dating of the eruptive history of Mount Erebus, Antarctica: volcano evolution. Bull Volcanol. 2004;66(8):671–86. [Google Scholar]
- 3.Barker PF, Thomas E. Origin, signature and palaeoclimatic influence of the Antarctic Circumpolar Current. Earth Sci Rev. 2004;66(1–2):143–62. [Google Scholar]
- 4.Herbold CW, Lee CK, McDonald IR, Cary SC. Evidence of global-scale aeolian dispersal and endemism in isolated geothermal microbial communities of Antarctica. Nat Commun. 2014;20(5):3875. [DOI] [PubMed] [Google Scholar]
- 5.Broady PA. Taxonomic and ecological investigations of algae on steam-warmed soil on Mt Erebus, Ross Island Antarctica. Phycologia. 1984;23(3):257–71. [Google Scholar]
- 6.Soo RM, Wood SA, Grzymski JJ, McDonald IR, Cary SC. Microbial biodiversity of thermophilic communities in hot mineral soils of Tramway Ridge, Mount Erebus Antarctica. Environ Microbiol. 2009;11(3):715–28. [DOI] [PubMed] [Google Scholar]
- 7.Herbold CW, McDonald IR, Cary SC. Microbial ecology of geothermal habitats in antarctica. In: Cowan DA, editor. Antarctic terrestrial microbiology. Berlin, Heidelberg: Springer Berlin Heidelberg; 2014. p. 181–215.
- 8.Herbold C, Cary SC, Connell L, Convey P, Poirot C. Geothermal environments in Antarctica. Antarctic environments portal. 2018
- 9.Noell SE, Baptista MS, Smith E, McDonald IR, Lee CK, Stott MB, et al. Unique geothermal chemistry shapes microbial communities on mt. erebus, antarctica. Front Microbiol. 2022. 10.3389/fmicb.2022.836943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parties of the Antarctic Treaty. Antarctic Treaty database [Internet]. ASPA 175 - high altitude geothermal sites of the Ross sea region. 2014 [cited 2018 Oct 25]. Available from: https://www.ats.aq/devAS/info_measures_listitem.aspx?lang=e&id=573
- 11.Ugolini FC, Starkey RL. Soils and Micro-organisms from Mount Erebus. Antarctica Nat. 1966;211(5047):440–1. [Google Scholar]
- 12.Vickers CJ, Herbold CW, Cary SC, Mcdonald IR. Insights into the metabolism of the high temperature microbial community of Tramway Ridge, Mount Erebus Antarctica. Antarctic Sci. 2016;28(4):241–9. [Google Scholar]
- 13.Wardell LJ, Kyle PR, Campbell AR. Carbon dioxide emissions from fumarolic ice towers, Mount Erebus volcano, Antarctica. Geological Soc, London, Spec Publications. 2003;213(1):231–46. [Google Scholar]
- 14.Ugolini FC. Soils of mount erebus, antarctica. NZ J Geol Geophys. 1967;10(2):431–42. [Google Scholar]
- 15.Vaquer-Sunyer R, Duarte CM. Thresholds of hypoxia for marine biodiversity. Proc Natl Acad Sci USA. 2008;105(40):15452–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sims KWW, Aster RC, Gaetani G, Blichert-Toft J, Phillips EH, Wallace PJ, et al. Mount Erebus. In: Smellie JL, Panter KS, Geyer A, editors. Volcanism in Antarctica: 200 Million Years of Subduction, Rifting and Continental Break-up. lyellcollection.org; 2021.
- 17.Sweeney D, Kyle PR, Oppenheimer C. Sulfur dioxide emissions and degassing behavior of Erebus volcano, Antarctica. J Volcanol Geoth Res. 2008;177(3):725–33. [Google Scholar]
- 18.Ilanko T, Fischer TP, Kyle P, Curtis A, Lee H, Sano Y. Modification of fumarolic gases by the ice-covered edifice of Erebus volcano, Antarctica. J Volcanol Geoth Res. 2019;381:119–39. [Google Scholar]
- 19.Mooshammer M, Wanek W, Hämmerle I, Fuchslueger L, Hofhansl F, Knoltsch A, et al. Adjustment of microbial nitrogen use efficiency to carbon:nitrogen imbalances regulates soil nitrogen cycling. Nat Commun. 2014;16(5):3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrett JE, Virginia RA, Wall DH, Cary SC, Adams BJ, Hacker AL, et al. Co-variation in soil biodiversity and biogeochemistry in northern and southern Victoria Land Antarctica. Antarctic Sci. 2006;18(4):535–48. [Google Scholar]
- 21.Gerner SM, Rattei T, Graf AB. Assessment of urban microbiome assemblies with the help of targeted in silico gold standards. Biol Direct. 2018;13(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laehnemann D, Borkhardt A, McHardy AC. Denoising DNA deep sequencing data-high-throughput sequencing errors and their correction. Brief Bioinformatics. 2016;17(1):154–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bushnell B. BBMap: A Fast, Accurate, splice-aware aligner. 2014;
- 24.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27(5):824–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31(10):1674–6. [DOI] [PubMed] [Google Scholar]
- 26.Nurk S, Bankevich A, Antipov D, Gurevich A, Korobeynikov A, Lapidus A, et al. Assembling Genomes and Mini-metagenomes from Highly Chimeric Reads. In: Deng M, Jiang R, Sun F, Zhang X, editors. Research in computational molecular biology. Berlin, Heidelberg: Springer Berlin Heidelberg; 2013. p. 158–70.
- 27.Wu Y-W, Simmons BA, Singer SW. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 2016;32(4):605–7. [DOI] [PubMed] [Google Scholar]
- 28.Kang DD, Froula J, Egan R, Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;27(3): e1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olm MR, Brown CT, Brooks B, Banfield JF. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 2017;11(12):2864–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2019;36(6):1925–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P-A, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36(10):996–1004. [DOI] [PubMed] [Google Scholar]
- 32.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44(14):6614–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Astashyn A, Tvedte ES, Sweeney D, Sapojnikov V, Bouk N, Joukov V, et al. Rapid and sensitive detection of genome contamination at scale with FCS-GX. Genome Biol. 2024;25(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol. 2017;34(8):2115–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sousa FL, Alves RJ, Pereira-Leal JB, Teixeira M, Pereira MM. A bioinformatics classifier and database for heme-copper oxygen reductases. PLoS ONE. 2011;6(4): e19117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Søndergaard D, Pedersen CNS, Greening C. HydDB: a web tool for hydrogenase classification and analysis. Sci Rep. 2016;27(6):34212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng J, Ge Q, Yan Y, Zhang X, Huang L, Yin Y. dbCAN3: automated carbohydrate-active enzyme and substrate annotation. Nucleic Acids Res. 2023;51(W1):W115–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;8(11):119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen I-MA, Chu K, Palaniappan K, Ratner A, Huang J, Huntemann M, et al. The IMG/M data management and analysis system vol 7: content updates and new features. Nucleic Acids Res. 2023;51(D1):D723-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oksanen J, Simpson GL, Blanchet FG, Kindt R, Legendre P, Minchin PR, et al. vegan: Community Ecology Package. 2022;
- 45.Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35(3):526–8. [DOI] [PubMed] [Google Scholar]
- 46.Murdoch D, Adler D. rgl: 3D Visualization using OpenGL. 2023;
- 47.Krijthe JH, Van der Maaten L. Rtsne: T-distributed stochastic neighbor embedding using Barnes-Hut implementation. R package version 013, URL https://github.com/jkrijthe/Rtsne. 2015;
- 48.Wickham H. ggplot2: elegant graphics for data analysis (Use R!). 2nd ed. Cham: Springer; 2016. [Google Scholar]
- 49.Lai D, Hedlund BP, Mau RL, Jiao J-Y, Li J, Hayer M, et al. Resource partitioning and amino acid assimilation in a terrestrial geothermal spring. ISME J. 2023;17(11):2112–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beam JP, Jay ZJ, Kozubal MA, Inskeep WP. Niche specialization of novel Thaumarchaeota to oxic and hypoxic acidic geothermal springs of Yellowstone National Park. ISME J. 2014;8(4):938–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kato S, Sakai S, Hirai M, Tasumi E, Nishizawa M, Suzuki K, et al. Long-term cultivation and metagenomics reveal ecophysiology of previously uncultivated thermophiles involved in biogeochemical nitrogen cycle. Microbes Environ. 2018;33(1):107–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spieck E, Spohn M, Wendt K, Bock E, Shively J, Frank J, et al. Extremophilic nitrite-oxidizing Chloroflexi from Yellowstone hot springs. ISME J. 2020;14(2):364–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mueller AJ, Daebeler A, Herbold CW, Kirkegaard RH, Daims H. Cultivation and genomic characterization of novel and ubiquitous marine nitrite-oxidizing bacteria from the Nitrospirales. ISME J. 2023;17(11):2123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ji M, Greening C, Vanwonterghem I, Carere CR, Bay SK, Steen JA, et al. Atmospheric trace gases support primary production in Antarctic desert surface soil. Nature. 2017;552(7685):400–3. [DOI] [PubMed] [Google Scholar]
- 55.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25(7):1043–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37(5):1530–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14(6):587–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Minh BQ, Nguyen MAT, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013;30(5):1188–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–30. [DOI] [PMC free article] [PubMed]
- 60.Bowers RM, Kyrpides NC, Stepanauskas R, Harmon-Smith M, Doud D, Reddy TBK, et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol. 2017;35(8):725–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, et al. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature. 2015;523(7559):208–11. [DOI] [PubMed] [Google Scholar]
- 62.Nelson WC, Stegen JC. The reduced genomes of Parcubacteria (OD1) contain signatures of a symbiotic lifestyle. Front Microbiol. 2015;21(6):713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ren M, Feng X, Huang Y, Wang H, Hu Z, Clingenpeel S, et al. Phylogenomics suggests oxygen availability as a driving force in Thaumarchaeota evolution. ISME J. 2019. 10.1038/s41396-019-0418-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daebeler A, Herbold CW, Vierheilig J, Sedlacek CJ, Pjevac P, Albertsen M, et al. Cultivation and genomic analysis of “candidatus nitrosocaldus islandicus”, an obligately thermophilic, ammonia-oxidizing thaumarchaeon from a hot spring biofilm in graendalur valley, iceland. Front Microbiol. 2018;14(9):193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kato S, Ohnishi M, Nagamori M, Yuki M, Takashina T, Ohkuma M, et al. Conexivisphaera calida gen. nov., sp. nov., a thermophilic sulfur- and iron-reducing archaeon, and proposal of Conexivisphaeraceae fam nov., Conexivisphaerales ord. nov., and Conexivisphaeria class. nov. in the phylum Thaumarchaeota. Int J Syst Evol Microbiol. 2021. 10.1099/ijsem.0.004595. [DOI] [PubMed] [Google Scholar]
- 66.King GM, Weber CF. Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat Rev Microbiol. 2007;5(2):107–18. [DOI] [PubMed] [Google Scholar]
- 67.Hedlund B, Nou N, Covington J, Lai D, Mayali X, Seymour C, et al. Genome-guided isolation of Fervidibacter sacchari, an aerobic, hyperthermophilic polysaccharide-degrading specialist. Res Sq. 2024.
- 68.Greening C, Berney M, Hards K, Cook GM, Conrad R. A soil actinobacterium scavenges atmospheric H2 using two membrane-associated, oxygen-dependent [NiFe] hydrogenases. Proc Natl Acad Sci USA. 2014;111(11):4257–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koch H, Galushko A, Albertsen M, Schintlmeister A, Gruber-Dorninger C, Lücker S, et al. Growth of nitrite-oxidizing bacteria by aerobic hydrogen oxidation. Science. 2014;345(6200):1052–4. [DOI] [PubMed] [Google Scholar]
- 70.Archer SDJ, Lee KC, Caruso T, Maki T, Lee CK, Cary SC, et al. Airborne microbial transport limitation to isolated Antarctic soil habitats. Nat Microbiol. 2019;4(6):925–32. [DOI] [PubMed] [Google Scholar]
- 71.Power JF, Carere CR, Welford HE, Hudson DT, Lee KC, Moreau JW, et al. A genus in the bacterial phylum Aquificota appears to be endemic to Aotearoa-New Zealand. Nat Commun. 2024;15(1):179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Andreani NA, Hesse E, Vos M. Prokaryote genome fluidity is dependent on effective population size. ISME J. 2017;11(7):1719–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Melick DR, Broady PA, Rowan KS. Morphological and physiological characteristics of a non-heterocystous strain of the cyanobacterium Mastigocladus laminosus Cohn from fumarolic soil on Mt Erebus Antarctica. Polar Biol. 1991. 10.1007/BF00234270. [Google Scholar]
- 74.Miller SR, Castenholz RW, Pedersen D. Phylogeography of the thermophilic cyanobacterium Mastigocladus laminosus. Appl Environ Microbiol. 2007;73(15):4751–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pester M, Schleper C, Wagner M. The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr Opin Microbiol. 2011;14(3):300–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Doxey AC, Kurtz DA, Lynch MDJ, Sauder LA, Neufeld JD. Aquatic metagenomes implicate Thaumarchaeota in global cobalamin production. ISME J. 2015;9(2):461–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hua Z-S, Qu Y-N, Zhu Q, Zhou E-M, Qi Y-L, Yin Y-R, et al. Genomic inference of the metabolism and evolution of the archaeal phylum Aigarchaeota. Nat Commun. 2018;9(1):2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kato S, Itoh T, Yuki M, Nagamori M, Ohnishi M, Uematsu K, et al. Isolation and characterization of a thermophilic sulfur- and iron-reducing thaumarchaeote from a terrestrial acidic hot spring. ISME J. 2019;13(10):2465–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lin X, Handley KM, Gilbert JA, Kostka JE. Metabolic potential of fatty acid oxidation and anaerobic respiration by abundant members of Thaumarchaeota and Thermoplasmata in deep anoxic peat. ISME J. 2015;9(12):2740–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yokooji Y, Sato T, Fujiwara S, Imanaka T, Atomi H. Genetic examination of initial amino acid oxidation and glutamate catabolism in the hyperthermophilic archaeon Thermococcus kodakarensis. J Bacteriol. 2013;195(9):1940–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Blank CE, Cady SL, Pace NR. Microbial composition of near-boiling silica-depositing thermal springs throughout Yellowstone National Park. Appl Environ Microbiol. 2002;68(10):5123–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499(7459):431–7. [DOI] [PubMed] [Google Scholar]
- 83.Liu QP, Sulzenbacher G, Yuan H, Bennett EP, Pietz G, Saunders K, et al. Bacterial glycosidases for the production of universal red blood cells. Nat Biotechnol. 2007;25(4):454–64. [DOI] [PubMed] [Google Scholar]
- 84.Teze D, Shuoker B, Chaberski EK, Kunstmann S, Fredslund F, Nielsen TS, et al. The catalytic acid-base in GH109 resides in a conserved GGHGG Loop and allows for comparable α-retaining and β-inverting activity in an N -Acetylgalactosaminidase from Akkermansia muciniphila. ACS Catal. 2020;9:3809–19. [Google Scholar]
- 85.Hedlund BP, Dodsworth JA, Murugapiran SK, Rinke C, Woyke T. Impact of single-cell genomics and metagenomics on the emerging view of extremophile “microbial dark matter.” Extremophiles. 2014;18(5):865–75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw metagenomic sequence data and metagenome assembled genomes used in this manuscript are available under bioproject accession PRJNA431961 (https://www.ncbi.nlm.nih.gov/bioproject/431961). Data and scripts necessary to reproduce figures and calculate endemicity index are available at https://doi.org/10.5281/zenodo.14203511.