Abstract
Background
Most Colorectal Cancer (CRC) patients exhibit limited responsiveness to anti-programmed cell death protein 1 (PD-1) therapy, with the underlying mechanisms remaining elusive. Circular RNAs (circRNAs) play a significant role in tumorigenesis and development, with potential applications in tumor screening and predicting treatment efficacy. However, there are few studies exploring the role of circRNAs in CRC immune evasion.
Methods
circRNA microarrays were used to identify circPHLPP2. RT-qPCR was used to examine the associations between the expression level of circPHLPP2 and the clinical characteristics of CRC patients. MTS assay, clone formation experiment, subcutaneous tumor implantation and multicolor flow cytometry were used to confirm the biological function of circPHLPP2. RAN-seq, RT-qPCR, and WB experiments were performed to investigate the downstream signaling pathways involved in circPHLPP2. RNA pull-down, RNA immunoprecipitation (RIP) and immunofluorescence staining were performed to identify the proteins associated with circPHLPP2.
Results
circPHLPP2 is up-regulated in CRC patients who exhibit resistance to anti-PD-1 based therapy. circPHLPP2 significantly promotes the proliferation and tumor growth of CRC cells. Knockdown of circPhlpp2 enhances the efficacy of anti-PD-1 in vivo. Mechanistically, the specific interaction between circPHLPP2 and ILF3 facilitates the nuclear accumulation of ILF3, which subsequently enhances the transcription of IL36γ. This process reduces NK cell infiltration and impairs NK cells’ granzyme B and IFN-γ production, thereby promoting tumor progression.
Conclusions
Overall, our findings reveal a novel mechanism by which circRNA regulates CRC immune evasion. circPHLPP2 may serve as a prognostic biomarker and potential therapeutic target for CRC patients.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12943-024-02192-8.
Keywords: CircPHLPP2, Colorectal cancer, NK cell, Immune evasion
Background
Colorectal cancer (CRC) is the third most common malignant tumor and the second-leading cause of cancer-associated mortality worldwide [1]. The absence of effective treatments greatly affects the quality of life and survival rates of patients with advanced CRC [2]. Fortunately, the emergence of immune checkpoint inhibitors (ICIs), including programmed cell death protein 1(PD-1) inhibitors, has brought new hope for the treatment of advanced CRC. As reported in the clinical studies, pembrolizumab monotherapy, a humanized monoclonal anti-PD1 antibody, significantly improved survival in advanced CRC patients with microsatellite instability-high/ deficient mismatch repair (MSI-H/ dMMR) [3]. However, CRC patients with microsatellite stable/ proficient mismatch repair (MSS/ pMMR) could not benefit from ICI monotherapy [4], and this group represents the majority, comprising 95% of individuals diagnosed with CRC [5]. Encouragingly, the REGONIVO clinical trial showed promising results with the combination of anti-PD-1 monoclonal antibody and tyrosine kinase inhibitors (TKIs) in treating metastatic CRC with MSS/pMMR [6]. Currently, there is still a lack of biomarkers to predict the efficacy of anti-PD-1 antibodies in advanced CRC patients with MSS/pMMR. Thus, it is urgent to further explore the molecular mechanism of immunotherapy resistance, aiming at finding new predictive indicators and therapeutic targets. These efforts could be beneficial to more rational clinical management and improve the survival of patients with advanced CRC.
Circular RNAs (circRNAs) are covalently closed, single-stranded RNA molecules that are ubiquitous and tissue-specific in humans [7]. Many pivotal circRNAs have been confirmed to be significantly dysregulated in tissues, cells, blood and exosomes of CRC, and are widely involved in the occurrence, metastasis or chemotherapy resistance of CRC [8, 9]. Notably, recent studies showed that circRNAs are involved in the regulation of tumor microenvironment and immune escape of tumor cells [10, 11]. For instance, CircCCAR1 deriving from exosomes promotes CD8 + T-cell dysfunction by stabilizing the PD-1 protein, thus leading to anti-PD1 resistance in hepatocellular carcinoma [12]. CircDLG1 promotes progression and resistance to anti-PD-1-based therapy of gastric cancer by increasing the expression of CXCL12 [13]. Nevertheless, the relationship between circRNAs and immunotherapy resistance of CRC has not yet been thoroughly explored. Besides, since the stabilization and the capacity to be detected in body fluids, circRNAs have the potential to be used as biomarkers for predicting the efficacy of anti-PD-1 therapy. In general, identification of circRNAs that function as key molecules driving immune evasion and immunotherapy resistance in CRC deserves investigation.
Acting as innate immune cells, natural killer (NK) cells can recognize tumor cells without sensitization [14]. NK cells could not only directly kill tumor cells through perforin or antibody-dependent cell-mediated cytotoxicity (ADCC), but also recruit and activate T cells by secreting cytokines and chemokines, thus participating in adaptive immunity [15, 16]. However, tumors could avoid being recognized and attacked by NK cells through several strategies [17], such as upregulating inhibitory receptors on the surface of NK cells, including PD-1. The blockade of PD-1 and PD-L1 elicited a strong NK cell response, indicating that NK cells were indispensable for the full therapeutic effect of immunotherapy [18]. Additionally, studies showed that numerous NK cells in the circulation or tumor tissues are negatively associated with tumor metastasis and poor prognosis in many cancers, including CRC [17, 19]. Although NK cells are recognized as crucial elements of multipronged immunotherapeutic strategies for cancer treatment [20], the underlying mechanisms contributing to anti-PD-1 resistance in CRC, particularly concerning NK cells, remain elusive.
Here, we identified that the highly expressed circPHLPP2 in CRC tissues is associated with anti-PD-1 resistance and poor survival in CRC. Mechanistically, circPHLPP2 up-regulates the transcription of IL36γ by binding ILF3, resulting in decreased infiltration of NK cells in tumor tissues and diminished production of granzyme B and interferon-γ (IFN-γ) of NK cells. This process ultimately facilitates the progression of CRC. Importantly, we demonstrated the enhanced efficacy of anti-PD-1 treatment in combination with the interference of circPhlpp2, highlighting circPHLPP2 as a promising biomarker and potential target for anti-PD-1 based CRC therapy.
Methods
Human tissues
For RNA microarray and real-time PCR analysis, fresh-frozen tissues and paraffin-embedded tissues were obtained from metastatic MSS/pMMR CRC patients who underwent colonoscopy with biopsies or palliative operation and received PD-1 inhibitor treatment at Sun Yat-sen University Cancer Center between August 2019 and October 2022. Response rates were determined based on the RECIST 1.1 guideline. Overall survival (OS) was assessed from the start of anti-PD-1 antibody treatment to the date of death from any cause or last contact. For immunohistochemical (IHC) staining, 381 cases of formalin-fixed paraffin-embedded CRC tissues along with the available clinicopathological information were obtained from Sun Yat-sen University Cancer Center between January 2007 and December 2012 with informed consent and agreement. OS was assessed from the date of surgery to the date of death from any cause or last contact.
CircRNA microarray assays
In order to identify circRNAs that may play a role in the resistance to anti-PD-1 therapy in MSS/pMMR CRC patients, a cohort of 8 patients who exhibited a positive response to anti-PD-1 based treatment manifesting partial response (PR), and 10 patients who did not respond to anti-PD-1 based therapy manifesting progressive disease (PD) were chosen for circRNA microarray analysis, encompassing primary tissues as well as adjacent normal tissues.
All sample preparation and microarray hybridization were performed according to the protocols of Arraystar (Rockville, MD, USA). Briefly, circRNAs were enriched by removing linear RNAs with Rnase R. The enriched circRNAs were then amplified and transcribed into fluorescent cRNA with a random priming method (Arraystar Super RNA Labeling Kit). The labeled cRNAs were hybridized onto Human circRNA Array V2 (8 × 15 k, Arraystar). Finally, the arrays were scanned by the Agilent Scanner G2505C and analyzed with Agilent Feature Extraction software (version 11.0.1.1). Quantile normalization and subsequent data analysis were conducted using R software.
Plasmid and cell transfection
Hsa_circ_0003681 (human circPHLPP2) overexpression vector (using pLC5-ciR vector) was constructed by GeneSeed (Guangzhou, China), EIF4A3 overexpression vector (using pLVXvector) was constructed by Tsingke Biotech (Beijing, China). Small interfering RNAs (siRNAs) for EIF4A3, HNRNPC, U2AF2 and their negative controls were synthesized by Genecefe (Jiangsu, China), si-ILF3 and si-NC were synthesized by Public Protein/Plasmid Library (Nanjing, China). Short hairpin RNAs (shRNAs) for hsa_circ_0003681, mmu_circ_0014549 (mouse circPhlpp2), mouse IL36γ and their negative controls were synthesized by GeneChem (Shanghai, China). The mmu_circ_0014549 lentivirus and control lentivirus were constructed by GenePharma (Shanghai, China). The sequences are listed in Table S1. Cell transfections and lentiviral transductions were conducted according to the manufacturer’s instructions and the reference [21].
Immunohistochemistry (IHC) assays
IHC assays were performed using standard methods as previously reported [22]. Paraffin-embedded tissue sections underwent deparaffinization and rehydration, followed by inhibition of endogenous peroxidase activity using 3% H2O2 for 10 min. Antigen retrieval was achieved by subjecting the sections to a pressure cooker with either sodium citrate buffer or EDTA at a sub-boiling temperature for 10 min. Subsequently, samples were blocked with 10% FBS for 1 h at room temperature to minimize nonspecific binding, incubated with primary antibody overnight at 4 °C, and then exposed to a biotinylated secondary antibody for 1 h at room temperature. The following antibodies were used: anti-IL36γ (1:200, Abclonal), anti-NKp46 (3 µg/mL, R&D). Nuclear counterstain was done using hematoxylin. The stained sections were assessed and graded individually according to their intensity levels: 0 (absent), 1 (low), 2 (moderate), and 3 (high). The cumulative score was calculated by multiplying the intensity grades of staining with the proportion of cells exhibiting positive staining.
Flow cytometry
Lysates were prepared with serum-free RPIM 1640, collagenase IV (0.2 mg/ml, Sigma-Aldrich) and DNase I (0.05 mg/ml, Roche), and then added to the tissue samples. Gentle ACS Octo Dissociator (Mitenyi G8H, Germany) was used to prepare single cell suspensions from tissues. Immune cells were isolated by Percoll gradient, and then stained with zombie (1:500, Biolegend) and surface markers (1:200). For intracellular protein staining, cells were stimulated with Cell Stimulation Cocktail (eBioscience) for 4 h prior and stained by intracellular markers (1:100). The antibodies utilized were listed in Table S3. Flow cytometry was performed on the CytoFlex (BD Biosciences, America) and analyzed using FlowJo V10 (FlowJo LLC).
RNA immunoprecipitation (RIP) assays
Magna RNA-binding protein immunoprecipitation kits (Millipore) were applied for RIP assays. Magnetic beads and antibodies were incubated at room temperature for 30 min in advance for conjunction. Anti-ILF3 (5 µg, Proteintech) and anti-EIF4A3 antibody (5 µg, Proteintech) were utilized in this study. IgG was used as a negative control. The cells were centrifuged and then lysed in an RIP buffer supplemented with a protease inhibitor cocktail and RNase inhibitor. The cell lysates were incubated with magnetic beads and rotated at 4 °C overnight. RNA was extracted from immobilized magnetic bead-bound complexes and subjected to RT-qPCR. RNA levels were normalized to the input.
GST pull-down assays
The RNA-binding proteins (RBPs) associated with circPHLPP2 were determined by using a circRNA pull-down assay with MS2-capturing protein [23]. The co-transfection of MS2-circPHLPP2 plasmid or MS2 plasmid and MS2-MCP-GST plasmid were conducted using lipo3000. The plasmid utilized in this experimental system was synthesized by GeneChem (Shanghai, China). Forty-eight hours after transfection, cells were lysed in RIPA buffer. Then the circPHLPP2-MS2-MCP-GST complex was purified by incubating crude lysates with glutathione Sepharose 4B (ThermoFisher). Subsequently, the captured products could be used for RT-qPCR, western blotting or mass spectrometry analyses as needed.
Fluorescence in situ hybridization (FISH) assay and immunofluorescence staining
FISH assays were performed using FISH probe reaction buffer (SA-Biotin system, GenePharma, Shanghai, China). Cy3-labeled circPHLPP2 probes were designed by GenePharma (Shanghai, China). HCT116 cells were fixed, permeabilized, and prehybridized. Subsequently, cells were then incubated with Cy3- labeled circPHLPP2 probes in hybridization mix overnight at 37 °C. After six washes using Buffer C, the cells were incubated with the blocking buffer (PBS containing 1%BSA and 0.05% TritonX-100) for 30 min at room temperature. Subsequently, the cells were incubated with primary antibody (1:100 dilution) for 1 h at room temperature, followed by incubation with Alexa Fluor 488 conjugated secondary antibodies (1:500, Invitrogen) for 45 min at 37 °C and DAPI (1:1000, GenePharma) for 15 min at room temperature. Confocal images were obtained with LSM880 (ZEISS, Germany).
In vivo tumorigenesis assays
NSG, BALB/c Nude, C57BL/6J and BALB/c mice were purchased from Vital River (Beijing, China). To generate subcutaneous tumor models, CRC cells (1.5 × 106 MC38; 5 × 105 CT26; 5 × 106 HCT116 or RKO) were injected subcutaneously into the right flanks of 6-week-old mice. Tumor volumes (V = width2 ×length/2) were measured every 3 days after 1 week of transplantation. Subcutaneous tumor tissues and blood were collected for flow cytometry analysis when the tumor volumes reached approximately 200 mm3. For drug treatment, when the tumor volumes reached 100 mm3, each mouse received 50 µg anti-PD-1 (BioXcell) or an equal volume of PBS every 3 days via i.p. injection. For NK cell depletion, 100µL anti-asialo-GM1 (Cell Biolabs) was injected i.p. into BALB/c mice once a week. 75 µg anti-NK1.1 (BioXcell) was injected into C57BL/6J mice through the tail vein one time every 3 days. All depletion treatments were initiated one day before tumor inoculation. Mice were euthanized when ethically defined endpoints had been reached. Subcutaneous tumor tissues were weighed and photographed, followed by being embedded in paraffin and sliced.
Statistical analysis
GraphPad Prism 9.0 software and SPSS 25.0 software were used for statistical analysis. Student’s t-tests and chi-square tests were used for the comparison between two groups and multiple groups, respectively. Survival curves were plotted using the Kaplan-Meier method and the significance was assessed using the log-rank test. In this article, * represents P < 0.05; ** represents P < 0.01; *** represents P < 0.001; **** represents P < 0.0001. All of them represent significant differences.
Results
High-level circPHLPP2 is associated with poor outcomes in anti-PD-1 resistant CRC patients
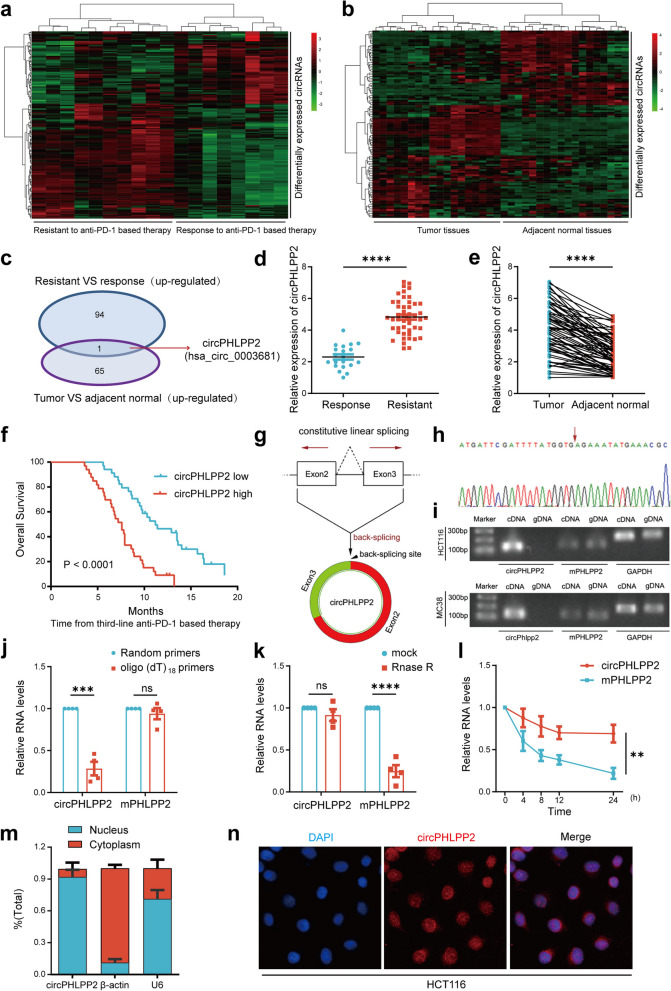
To identify circular RNAs correlated with the effectiveness of anti-PD-1 therapy in CRC patients with MSS/pMMR status, we performed a circRNA microarray analysis on 18 paired tissues (primary tissues and adjacent normal tissues) from MSS/pMMR CRC patients who were treated with anti-PD-1 based therapy. Among these patients, 10 participants exhibited PD by RECIST 1.1 [24] and were defined as patients resistant to anti-PD-1 therapy, while 8 participants showed PR and were defined as patients responsive to anti-PD-1 therapy. The heatmap showed that 94 circRNAs were significantly up-regulated in anti-PD-1 resistant tissues (Fig. 1a, Additional file 4), and 65 circRNAs were significantly up-regulated in tumor tissues as compared to normal tissues (Fig. 1b, Additional file 5). From the overlap of these two groups, we obtained a circRNA, namely hsa_circ_0003681, which is derived from PHLPP2, thus we designated it as circPHLPP2, whose biological function has not yet been reported (Fig. 1c). We next analyzed the relationship between circPHLPP2 and the clinical features and prognosis in 67 CRC patients who received anti-PD-1 based treatment by RT-qPCR. The results showed that circPHLPP2 was significantly up-regulated in anti-PD-1 resistant patients (Fig. 1d) as well as in CRC tissues compared to normal tissues (Fig. 1e), and the difference was statistically significant, indicating that the sequencing results were accurate and reliable. The patients were divided into two groups according to the median expression of circPHLPP2. Kaplan-Meier analysis showed that CRC patients with higher expression of circPHLPP2 had shorter overall survival and a worse prognosis (Fig. 1f). In conclusion, these results demonstrated that circPHLPP2 might play an important role in CRC progression and immune evasion.
Fig. 1.
circRNA expression profiles in MSS/pMMR CRC and characterization of circPHLPP2. a Heatmap of circRNA expression in MSS/pMMR CRC tumor tissues resistant (n = 10) or responsive (n = 8) to anti‑PD‑1 therapy. b Heatmap of circRNA expression in MSS/pMMR CRC tumor tissues (n = 18) and paired adjacent normal tissues (n = 18), cut off is Log2 (Fold change) > 1 or < − 1, P value < 0.05. c A Venn diagram illustrating the overlapping circRNAs that are up-regulated in tissues resistant to anti-PD-1 therapy and in colorectal cancer tissues. d Relative expression of circPHLPP2 in tissues resistant (n = 20) and responsive to anti‑PD‑1 therapy (n = 47) detected by RT-qPCR. e Relative expression of circPHLPP2 in primary CRC tissues and paired adjacent normal tissues measured by RT-qPCR (n = 67). f Kaplan–Meier analysis of overall survival in CRC patients with different circPHLPP2 expression levels (n = 67, log-rank test). g Schematic illustration of circPHLPP2 formation. h The backsplice junction site of circPHLPP2 was identified by Sanger sequencing. i The existence of circPHLPP2 was validated in HCT116 and MC38 cell lines by RT-qPCR and gel electrophoresis. j The relative RNA levels of circPHLPP2 and mPHLPP2 in reverse transcription experiments using Random hexamers or oligo (dT)18 primers. k Resistance of circPHLPP2 and mPHLPP2 to digestion with RNase R was detected by RT-qPCR. l RNA abundance of circPHLPP2 and mPHLPP2 treated with Actinomycin D (2 µg/ml) was detected by RT-qPCR. m The intracellular localization of circPHLPP2 was detected by RNA cytoplasm/ nucleus fractionation assay. n The intracellular localization of circPHLPP2 was detected by FISH in HCT116 cells. Nuclei were stained with DAPI. Data in d, e, j and k were calculated by Student’s t test. Data in l were calculated by two‑way ANOVA test. **P < 0.01, ***P < 0.001, ****P < 0.0001
Characteristics of circPHLPP2 in CRC cells
According to the circBase database (http://www.circbase.org/), circPHLPP2 is generated from the second and third exons of PHLPP2 gene located on chr16 (71736500–71748704) with a length of 424 nt (Fig. 1g). The backsplice junction site of circPHLPP2 was amplified using divergent primers and confirmed by Sanger sequencing (Fig. 1h). Moreover, PCR analysis indicated that divergent primers could amplify circPHLPP2 from complementary DNA (cDNA) but not from genomic DNA (gDNA), while convergent primers could amplify the linear isoform of PHLPP2 from both cDNA and gDNA in HCT116 cells and MC38 cells (Fig. 1i). In addition, circPHLPP2 could be amplified by random primers, but not oligo(dT)18 primers (Fig. 1j), indicating that circPHLPP2 lacks a poly-A tail. RNase R digestion assays showed that circPHLPP2 was more resistant to RNase R digestion (a degrader of the linear RNA) than PHLPP2 mRNA (Fig. 1k). Actinomycin D (an inhibitor of transcription) treatment further showed that circPHLPP2 was more stable in comparison to PHLPP2 mRNA (Fig. 1l), which confirmed that circPHLPP2 harbors a stable closed-loop structure. Results of RNA cytosolic/nuclear fraction assay and FISH showed that circPHLPP2 was predominantly expressed and localized in the nucleus (Fig. 1m-n). Collectively, these results indicate that circPHLPP2 is a bona fide circRNA.
CircPHLPP2 promotes CRC progression in vivo
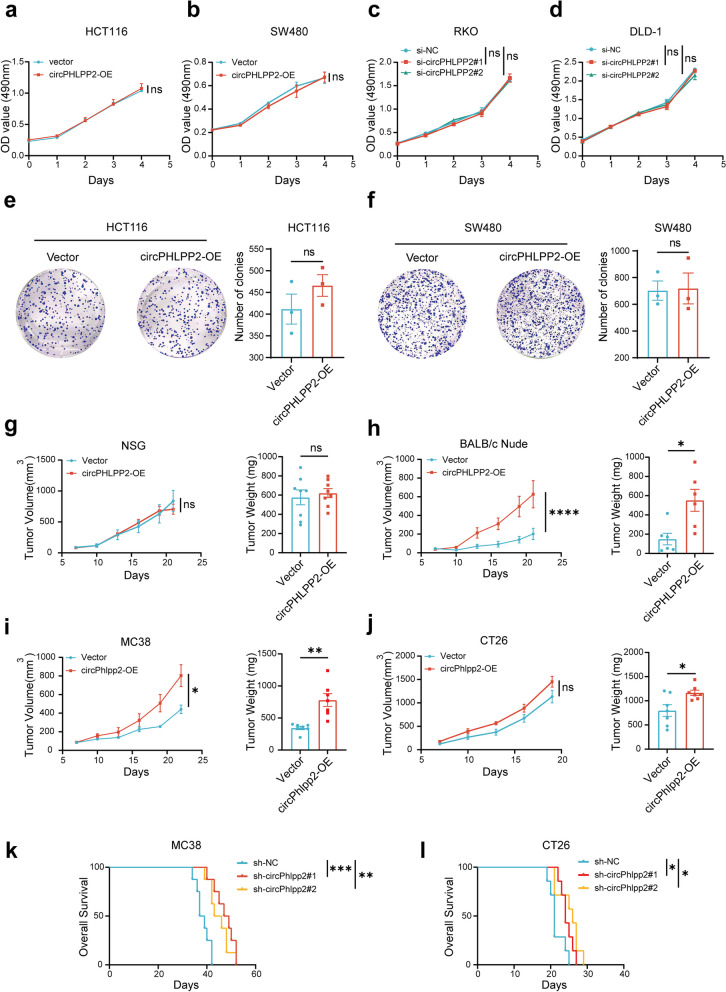
To study the role of circPHLPP2 in CRC progression, we constructed circPHLPP2 overexpression plasmid and siRNAs targeting the backsplice region of circPHLPP2 and confirmed circPHLPP2 was overexpressed or knocked down efficiently in CRC cells (Fig. S1a-b). In vitro MTS and colony formation assays indicated that overexpression or knockdown of circPHLPP2 did not influence the proliferation and colony formation ability of human CRC cells (Fig. 2a-f). To investigate the role of circPHLPP2 in vivo, human CRC cells stably overexpressing or knocking down circPHLPP2 (Fig. S1c-d) were inoculated into NSG mice subcutaneously. NSG mice are highly immunodeficient, lacking mature T, B, and NK cells and unable to produce immunoglobulins [25]. The results showed that circPHLPP2 did not appear to impair xenograft growth of CRC cells in NSG (Fig. 2g and Fig. S2a, c). Then we generated xenograft models using BALB/c Nude mice. Interestingly, up-regulation of circPHLPP2 significantly enhanced the proliferation of CRC cells (Fig. 2h and Fig. S2b), whereas down-regulation of circPHLPP2 resulted in the opposite effect (Fig. S2d). Nude mice exhibit a mutation in the Foxn1 gene located on chromosome 11, leading to congenital thymic dysplasia and impairment of T cell functionality while B cells and a robust NK cell response are retained [26]. Therefore, circPHLPP2 demonstrated a potential role in promoting tumorigenesis that could be associated with immune cells, specifically NK or B cells. Additionally, we subcutaneously inoculated MC38 (MSI-H CRC) with or without stable circPhlpp2 overexpression (Fig. S1e) into immunocompetent C57 mice. The phenotypic outcomes were in alignment with the results exhibited in nude mice (Fig. 2i, Fig. S2e), suggesting that circPHLPP2 notably promoted the proliferation of CRC cells in vivo. We next performed the experiment in CT26 (MSS CRC) [27], while circPhlpp2 overexpression did not exhibit a statistically significant impact on the tumor growth curve, it did demonstrate statistical significance in tumor weight (Fig. 2j and Fig. S2f). Then, we constructed MC38 or CT26 cells with circPhlpp2 stably knockdown (Fig. S1f) and found that circPhlpp2 knockdown drastically inhibited tumor proliferation (Fig. S2g-h) and significantly prolonged the survival of tumor-bearing mice (Fig. 2k-l). The above results indicated that circPHLPP2 promotes CRC cell proliferation by influencing immune microenvironment.
Fig. 2.
circPHLPP2 promotes proliferation of CRC cells in vivo. a-d The proliferation of CRC cells with circPHLPP2 overexpression or knockdown measured by MTS assay. e-f Colony formation assays for CRC cells with circPHLPP2 overexpression. g The volume and weight of tumor xenografts in NSG mice after subcutaneous inoculation with HCT116 cells stably overexpressing circPHLPP2 (n = 8). h The volume and weight of tumor xenografts in BALB/c Nude mice after subcutaneous inoculation with HCT116 cells stably overexpressing circPHLPP2 (n = 6). i The volume and weight of tumor allografts in C57 mice after subcutaneous inoculation with MC38 cells stably overexpressing circPhlpp2 (n = 7). j The volume and weight of tumor allografts in BALB/c mice after subcutaneous inoculation with CT26 cells stably overexpressing circPhlpp2 (n = 7). k-l Kaplan–Meier analysis of overall survival in MC38 tumor-bearing mice (n = 8) and CT26 tumor‐bearing mice (n = 7) (log-rank test). Data in a-d and g-j left were calculated by two‑way ANOVA test. Data in e-f and g-j right were calculated by Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
CircPHLPP2 promotes tumor progression by reducing NK cell infiltration and NK cells’ granzyme B and IFN-γ production
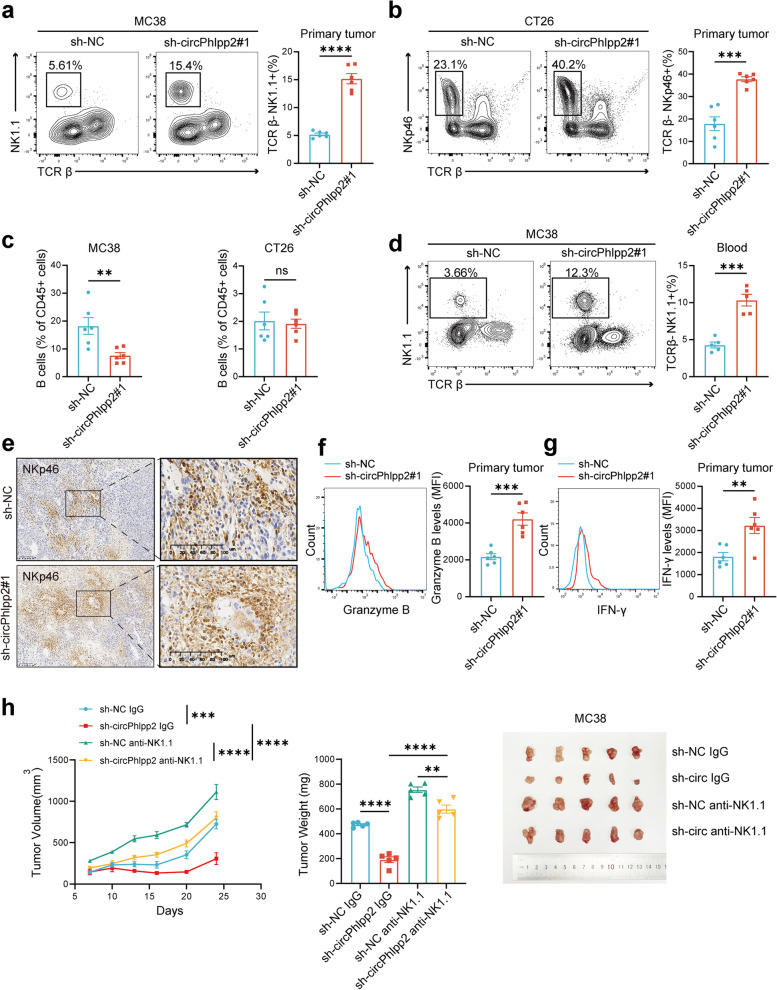
To further elucidate the impact of circPHLPP2 on tumor-infiltrating immune cells, tumors were gathered for multicolor flow analysis, the results revealed that knockdown of circPhlpp2 in both MC38 and CT26 cell lines led to a significant increase in the proportion of NK cells (Fig. 3a-b). The knockdown of circPhlpp2 in MC38 cells influenced B cell proportion either, whereas this effect was not evident in CT26 cells (Fig. 3c). Besides, no significant differences were observed in other immune cell populations (Fig. S4a-b), which indicated a potential role of circPhlpp2 in modulating NK cell infiltration. NK cells play a crucial role in the innate immune response, serving as the initial defense mechanism in the body’s anti-cancer defense. Further flow analysis showed that knockdown of circPhlpp2 resulted in an elevated percentage of NK cells not only in tumor tissues but also in peripheral blood (Fig. 3d). This suggests a systemic regulatory role of circPhlpp2, which means circPhlpp2 impacts the infiltration of NK cells in tumor immune microenvironment (TME) by reducing the proportion of NK cells in peripheral blood. IHC assay showed that the infiltration of NK cells in tissues increased after knocking down circPhlpp2, which also validated our findings (Fig. 3e). Besides, the knockdown of circPhlpp2 increased the expression levels of cytotoxic cytokines including granzyme B and IFN-γ in tumor-infiltrating NK cells (Fig. 3f-g), indicating that circPhlpp2 impairs the cytotoxic cytokine production capability of NK cells. To test whether circPhlpp2’s role in promoting CRC cell growth relied on NK cells, we depleted NK cells using pre-treatment with an anti-Asialo-GM1 antibody or anti-NK1.1 antibody before tumor cell inoculation (Fig. S4c). Depletion of NK cells significantly reversed the tumor suppressive phenotype induced by the knockdown of circPhlpp2. (Fig. 3h, Fig. S4d). The incomplete reversal effects by NK cell depletion observed in MC38 tumor models suggested that there might be other mechanisms beyond NK cells that contribute to tumor proliferation induced by circPhlpp2 (Fig. 3h). In summary, whatever in MSI-H or MSS tumors, circPhlpp2 plays a crucial role in immune evasion by modulating NK cell infiltration and cytotoxic cytokines expression, consequently facilitating tumor growth. Conversely, down-regulation of circPhlpp2 expression leads to increased infiltration of NK cells and elevated expression levels of granzyme B and IFN-γ, thereby augmenting anti-tumor immune response.
Fig. 3.
circPHLPP2 reduces NK cell proportion and granzyme B and IFN-γ expression. a-b Representative plots (left) and summary data (right) of NK cells in the primary tumors with or without circPhlpp2 knockdown from C57 mice (a) or BALB/c mice (b) as determined by flow cytometry (n = 6). c Frequency of B cells in tumors with or without circPhlpp2 knockdown from C57 mice (left) or BALB/c mice (right) as determined by flow cytometry (n = 6). d Representative plots (left) and summary data (right) of NK cells in blood of C57 mice bearing tumors with or without circPhlpp2 knockdown as determined by flow cytometry (n = 5). e Representative IHC images for NK cells (NKp46+) in tumors with or without circPhlpp2 knockdown from C57 mice, scale bar = 400 μm. f-g Expression levels of granzyme B (f) and IFN-γ (g) of NK cells in tumors with or without circPhlpp2 knockdown from C57 mice (n = 6). i The volume, weight and images of tumor allografts in NK cell-depleted C57 mice versus control C57 mice (n = 5) (left, two-way ANOVA test; middle, one-way ANOVA test). Data in a-d and f-g were calculated by Student’s t test. **P < 0.01, ***P < 0.001, ****P < 0.0001
IL36γ is a functional downstream mediator for circPHLPP2
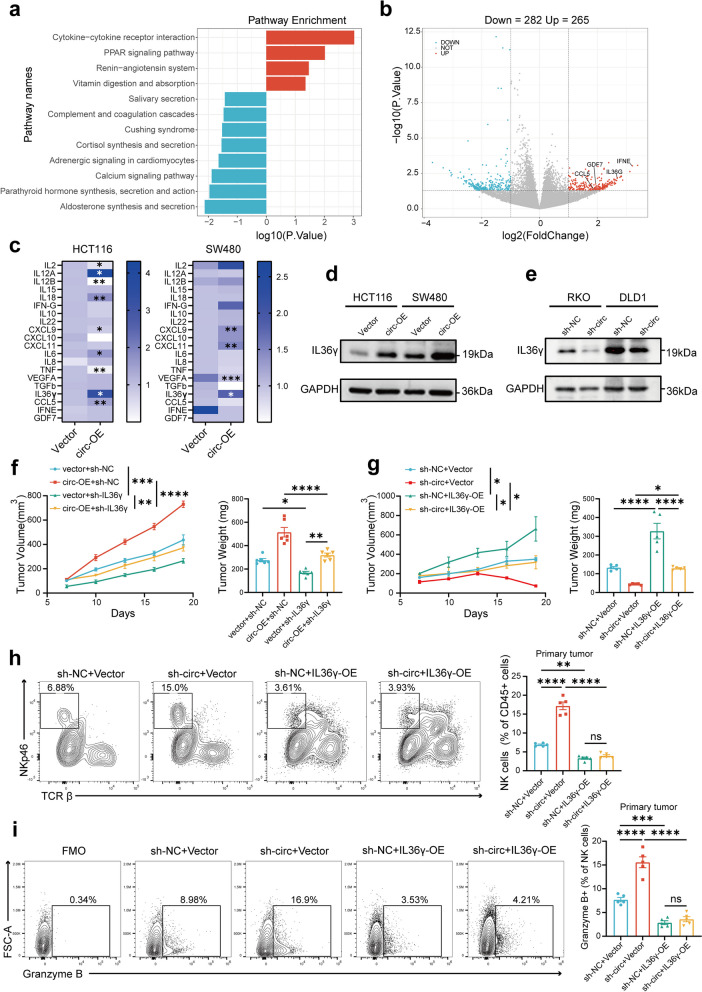
To investigate the downstream molecular mechanisms regulated by circPHLPP2, we performed RNA-seq-based mRNA expression profiling on HCT116 cells infected with circPHLPP2 overexpression or vector lentivirus (Additional file 6). KEGG pathways enrichment analysis indicated that circPHLPP2 was significantly involved in regulation of the cytokine-cytokine receptor interaction pathway (Fig. 4a), and differentially expressed genes (DEGs) including IL36γ, CCL5, IFNE and GDF7 were enriched in this pathway (Fig. 4b). Then we evaluated the correlation between these cytokines and immune infiltrates using the TCGA-COAD and TCGA-READ datasets. The results showed a significant negative correlation between IL36γ and NK cells (Fig. S5a), suggesting that IL36γ may serve as a downstream effector of circPHLPP2. Considering that circPHLPP2 may regulate TME via cytokines, we performed RT-qPCR for a variety of cytokines including IL36γ, CCL5, IFNE, and GDF7 in HCT116 and SW480 cells stably overexpressing circPHLPP2 and the control cells. The results indicated that circPHLPP2 overexpression changed the mRNA levels of multiple cytokines, but only IL36γ was up-regulated in both HCT116 and SW480 cells (Fig. 4c). Moreover, western blot analysis demonstrated that overexpression of circPHLPP2 increased the protein level of IL36γ while knockdown of circPHLPP2 decreased the protein level of IL36γ in CRC cells (Fig. 4d-e). Similar results were also found in mouse CRC cells (Fig. S5b). Thus, it can be inferred that circPHLPP2 plays a role in enhancing IL36γ transcription.
Fig. 4.
IL36γ is functional downstream mediator for circPHLPP2. a KEGG enrichment analysis of differentially expressed mRNAs. DEGs were identified using the limma package in R software and the threshold was |log2 (foldchange) | > 1, p value < 0.05. b Volcano plot for upregulated and downregulated genes, the genes involved in cytokine-cytokine receptor interaction pathway were indicated. c Heatmap depicting relative expression of multiple cytokines in the HCT116 and SW480 cells infected with circPHLPP2 overexpression or vector lentivirus measured by RT-qPCR, data were normalized to vector group. d-e Western blots showing IL36γ protein expression levels upon circPHLPP2 overexpression (d) and knockdown (e) in human CRC cells. f The volume (left) and weight (right) of allografts in C57 mice after subcutaneous inoculation with MC38 cells with circPhlpp2 overexpression or IL36γ knockdown (n = 5). g The volume (left) and weight (right) of allografts in C57 mice after subcutaneous inoculation with MC38 cells with circPhlpp2 knockdown or IL36γ overexpression (n = 5). h Representative plots (left) and summary data (right) of NK cells in the primary tumors with circPhlpp2 knockdown or IL36γ overexpression from C57 mice as determined by flow cytometry (n = 5). i Expression levels of granzyme B of NK cells in tumors with circPhlpp2 knockdown or IL36γ overexpression as determined by flow cytometry (n = 5). Data in f-g left were calculated by two‑way ANOVA test. Data in f-i right were calculated by one‑way ANOVA test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
It has been reported that IL36γ is highly expressed in CRC tissues and is associated with worse patient prognosis. Furthermore, IL36γ has been shown to induce the up-regulation of various inflammatory mediators that play a critical role in modulating the immune microenvironment [28]. Our results showed that higher expression of IL36γ was associated with shorter overall survival of CRC patients (Fig. S5c-d). Besides, overexpression of IL36γ promoted MC38 proliferation (Fig. S5e) in vivo and reduced the proportion of NK cells in tumor microenvironment (Fig. S5f). In brief, IL36γ and circPHLPP2 exhibit similar phenotypes in promoting CRC progression. Next, we explored whether IL36γ functions as a downstream effector of circPHLPP2, as predicted, we found that IL36γ knockdown significantly rescued cell proliferation phenotype mediated by circPhlpp2 overexpression (Fig. 4f and Fig. S6c), the suppressed cell proliferation ability, increased NK proportion and granzyme B and IFN-γ expression levels caused by circPHLPP2 knockdown could be restored by IL36γ ectopic expression in MC38 cells as well (Fig. 4g-i and Fig. S6d-f). Collectively, these data indicated that circPHLPP2 exerts its effect by regulating IL36γ transcription.
CircPHLPP2 facilitates IL36γ transcription by binding to ILF3
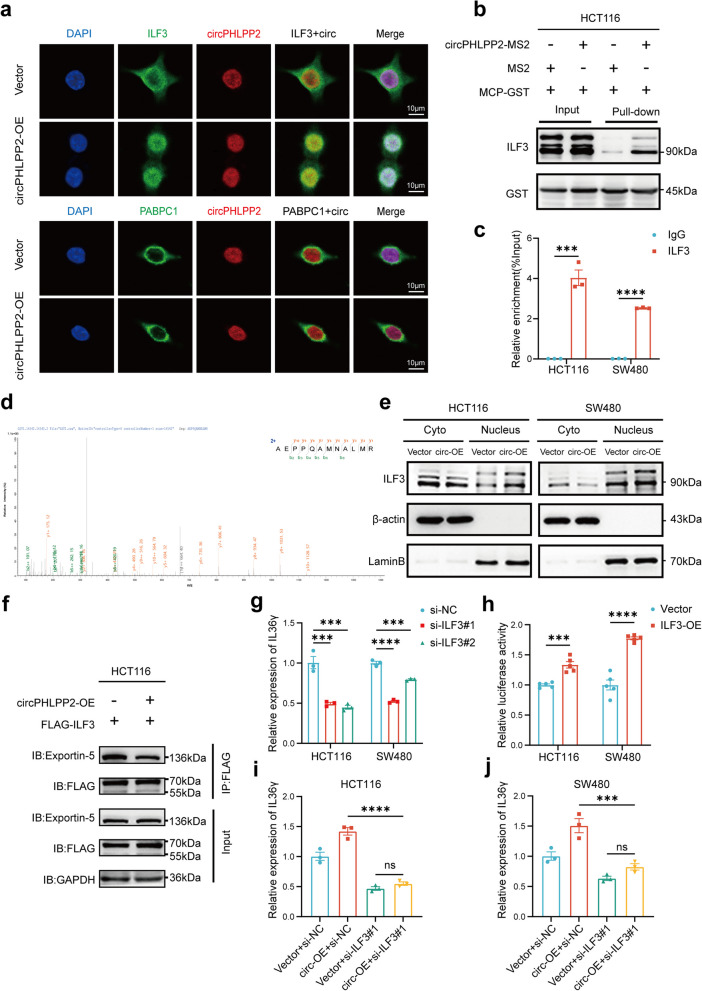
Then we attempted to explore how circPHLPP2 modulates IL36γ transcription. The Circinteractome database (https://circinteractome.nia.nih.gov/) indicates that circPHLPP2 contains multiple miRNA binding sites, yet does not exhibit binding to AGO2, a protein binding and guiding miRNAs to their target mRNA seed regions [29]. Therefore, we hypothesized that circPHLPP2 may function via binding proteins. We utilized MS2 tag-based RNA pull-down assays for the analysis of proteins bound to circPHLPP2. The circRNA pull-down products were enriched with the capture protein MCP-GST (Fig. S7a), and circPHLPP2 was highly abundant in the capture (Fig. S7b), both experiments indicated the successful specificity of the circRNA pull-down assay. According to mass spectrometry analysis in Additional file 7, the top-ranked protein, HNRNPC, has been reported to bind to a wide range of circRNAs and mainly facilitates the modulation of circRNA splicing to contribute to their formation, therefore, HNRNPC may not bind circPHLPP2 specifically. We performed FISH experiments on the second-ranked protein ILF3 and the third-ranked protein PABPC1 to verify whether they bind to circPHLPP2. The results revealed that ILF3 exhibited co-localization with circPHLPP2, and overexpression of circPHLPP2 increased the nuclear ILF3 (Fig. 5a). Co-expression of the circPHLPP2-MS2 and MCP-GST plasmids led to obvious enrichment of ILF3 compared with expression of the negative control (Fig. 5b), and a RIP assay verified that circPHLPP2 was specifically precipitated with anti-ILF3 antibody in CRC cells (Fig. 5c). These experiments indicated that circPHLPP2 interacts with ILF3. The spectrum of ILF3 by mass spectrometry analysis is shown in Fig. 5d. Nuclear-cytoplasmic separation assay further validated ILF3 expression was up-regulated in the nucleus after overexpression of circPHLPP2 (Fig. 5e).
Fig. 5.
CircPHLPP2 facilitates IL36γ transcription by binding to ILF3. a The co-localization assay of circPHLPP2 with ILF3 or PABPC1 in HCT116 cells was identified by RNA-FISH and immunofluorescence, scale bar = 10 μm. b Binding of circPHLPP2 with ILF3 was identified by RNA pull-down analysis using the MS2-tagging system. c RIP assay with an anti-ILF3 antibody or control IgG was performed in HCT116 and SW480 cells, followed by RT-qPCR to assess the enrichment of circPHLPP2. d Mass spectrometry analysis identified ILF3. e Western blotting analysis of cytoplasmic and nuclear ILF3 protein in HCT116 and SW480 cells with or without circPHLPP2 overexpression. f Coimmunoprecipitation (Co-IP) and western blot analysis of the interaction between ILF3 and exportin-5 in circPHLPP2 overexpression and control HCT116 cells. g Relative expression of IL36γ was measured by RT-qPCR in HCT116 and SW480 cells transfected with si-ILF3#1, si-ILF3#2 or si-NC. h Relative luciferase activity (FL/RL) was measured in HCT116 and SW480 cells after co-transfection of the IL36γ luciferase construct with ILF3 overexpression or control vector. i-j Relative expression of IL36γ was measured by RT-qPCR in HCT116 (i) and SW480 (j) cells with circPHLPP2 overexpression or ILF3 knockdown. Data in c and h were calculated by Student’s t test. Data in g and i-j were calculated by one‑way ANOVA test. ***P < 0.001, ****P < 0.0001
ILF3, also known as NF90/NF110, encodes a double-stranded RNA (dsRNA)-binding protein that complexes with other proteins, mRNAs, small noncoding RNAs, and dsRNAs to regulate gene expression and stabilize mRNAs [30], and intracellular localization of ILF3 appears to be associated with its functions [31]. As a shuttling protein between the nucleus and the cytoplasm, nuclear export of ILF3 is dependent on exportin-5, and exportin-5 cannot export RNA-bound ILF3 [32]. Furthermore, one study indicated that the formation of ILF3/circRNA complex may influence the subcellular localization of ILF3 and host immune response [33]. Thus, we proposed that circPHLPP2 competitively interacts with ILF3, consequently influencing ILF3 to interact with exportin-5 and causing the accumulation of ILF3 in the nucleus. The CO-IP assay proved that overexpression of circPHLPP2 inhibited exportin-5 binding to ILF3, thereby validating our hypothesis (Fig. 5f). ILF3 comprises a nuclear localization domain, two double-stranded RNA binding motifs (dsRBDs), an RGG RNA binding motif, and a C-terminal domain that is enriched with arginine and glycine residues [34]. One study has proved that exportin-5 mediates the export of ILF3 by binding to dsRBDs of ILF3, and dsRNA binding to this region inhibits exportin-5 binding [32]. We proposed that circPHLPP2 interacts with the dsRBDs region of ILF3, consequently disrupting the interaction between ILF3 and exportin-5. The GST pull-down assays showed that circPHLPP2 could no longer interact with ILF3 after deleting the ILF3 dsRBDs fragment (ILF3 △d), thereby confirming our hypothesis (Fig. S7c). Given the co-localization of circPHLPP2 and ILF3 in the nucleus, we hypothesized that ILF3 might regulate IL36γ transcription and designed two independent siRNAs targeting ILF3 to verify our hypothesis. Compared with the scramble control, siILF3#1 and siILF3#2 could effectively knock down ILF3 at the transcription level (Fig. S7d) and subsequently reduced IL36γ mRNA level in CRC cells (Fig. 5g). The analysis using IL36γ promoter-containing luciferase reporter plasmid further showed that overexpression of ILF3 increased luciferase activity compared to the control group (Fig. 5h). These results suggested that ILF3 regulates IL36γ transcription in CRC cells. Furthermore, ILF3 knockdown strikingly reversed the elevated IL36γ mRNA level caused by up-regulated circPHLPP2 (Fig. 5i-j), indicating that the transcriptional regulation of IL36γ by circPHLPP2 is dependent on the presence of ILF3. The above results revealed that circPHLPP2 regulates IL36γ transcription by combining with ILF3.
Knockdown of circPhlpp2 enhanced the efficacy of anti-PD-1 treatment in vivo
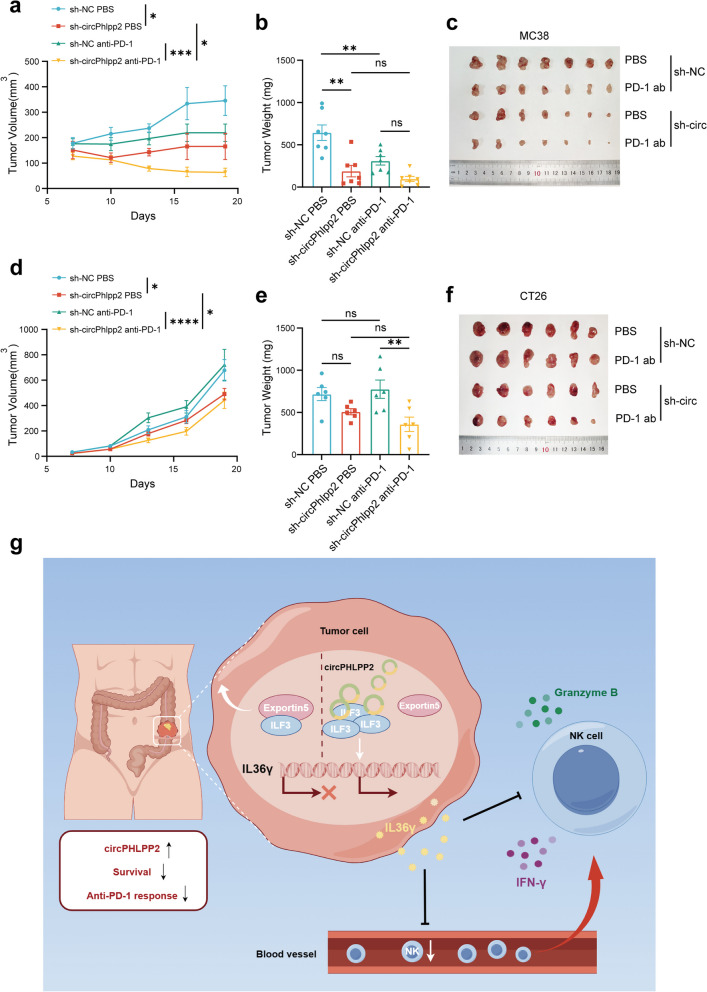
Our results demonstrated that circPhlpp2 could promote CRC cell proliferation in vivo and knockdown of circPHLPP2 could reverse its suppression of NK cells and enhance body’s anti-tumor immune response. Next, we sought to determine whether inhibiting circPHLPP2 could be used to potentiate the efficacy of anti-PD-1 therapy. We evaluated the effects of circPhlpp2 knockdown, anti-PD-1 therapy, and their combined application on tumor growth in immunocompetent murine models. In MC38 model, which represents the MSI-H form of CRC and is particularly responsive to immunotherapy [35], we found that anti-PD-1 treatment inhibited tumor progression, and circPhlpp2 knockdown could inhibit tumor progression even better than anti-PD-1 treatment (Fig. 6a-c). In CT26 model characterized by MSS and inherent resistance to immunotherapy [36], anti-PD-1 treatment was ineffective as expected, while circPhlpp2 knockdown exhibited a certain trend in inhibiting tumor proliferation (Fig. 6d-f). Additionally, anti-PD-1 treatment in mice bearing sh-circPhlpp2 tumors was more effective than sh-NC groups in both models, especially in tumor volume (Fig. 6a, d), suggesting that knockdown of circPhlpp2 significantly augments the efficacy of anti-PD-1 treatment.
Fig. 6.
Knockdown of circPHLPP2 enhances the efficacy of anti-PD-1 treatment in vivo. a-c The volume (a) and weight (b) of tumor allografts in C57 mice after subcutaneous inoculation with MC38 cells with or without circPhlpp2 knockdown. After the treatment with anti-PD-1 antibody, tumors were extracted and photographed (c) (n = 7). d-f The volume (d) and weight (e) of tumor allografts in BALB/c mice after subcutaneous inoculation with CT26 cells with or without circPhlpp2 knockdown. After the treatment with anti-PD-1 antibody, tumors were extracted and photographed (f) (n = 6). g Schematic diagram illustrating that circPHLPP2 facilitates CRC cell progression by enhancing the expression of IL36γ through its interaction with ILF3, ultimately reducing NK cell infiltration as well as granzyme B and IFN-γ expression levels of NK cells. Targeting circPHLPP2 may enhance the efficacy of anti-PD-1 therapy in CRC patients (By Figdraw). Data in a and d were calculated by two‑way ANOVA test. Data in b and e were calculated by one‑way ANOVA test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
Discussion
Cancer immunotherapy has recently shown promising antitumor effects in various types of tumors including CRC. However, resistance to immune therapy remains to be solved, particularly among CRC patients with MSS/pMMR status [37]. Increasing evidence has indicated that circRNAs are involved in tumor progression [38], and a subset of circular RNAs has been demonstrated to exhibit a correlation with the effectiveness of anti-PD-1 therapy [39]. The exploration of circRNAs that modulate the immune microenvironment has the potential to yield valuable insights into predictive markers for immunotherapy efficacy and identify therapeutic targets for cancer patients. In this study, we identified circPHLPP2 is associated with anti-PD-1 efficacy in MSS/pMMR-type advanced CRC patients and we first reported the functional mechanism of circPHLPP2 involved in immune evasion. Moreover, we proposed a rationale for enhancing the efficacy of anti-PD-1 therapy in CRC.
By detecting the expression level of circPHLPP2 in a cohort of CRC patients undergoing anti-PD-1 based therapy, we found that compared with paraneoplastic normal tissues, circPHLPP2 was significantly increased in CRC tissues, and circPHLPP2 was notably up-regulated in anti-PD1-resistant CRC patients. In addition, the overall survival rate of patients with higher expression of circPHLPP2 was worse, indicating that circPHLPP2 may function as a potential tumor progression factor. Further biological experiments demonstrated that circPHLPP2 did not impact CRC cell proliferation in vitro, but did enhance tumor growth in nude mice and immunocompetent mice, suggesting that circPHLPP2 may play a role in interactions between tumor cells and immune cells. Here, we exhibited that circPhlpp2 diminished the presence of NK cells in the peripheral blood and their infiltration into tumor tissues as well as the expression levels of granzyme B and IFN-γ of NK cells, thereby facilitating immune evasion. Considering that NK cells possess the capability to kill tumor cells through ADCC and other mechanisms, further investigation is warranted to explore the potential impact of circPhlpp2 on NK cell functions beyond cytotoxic cytokine production. Even if T cells are considered the key mediators of the impressive efficacy of checkpoint inhibitors, blockade of the CTLA-4 or PD-1 pathway might also enhance the anti-tumor responses of NK cells [40]. Previous studies have also reported that circUHRF1 drives resistance to anti-PD1 immunotherapy by inducing NK cell dysfunction in HCC [41]. Thus, circPHLPP2 plays a significant role in inducing resistance to anti-PD1 therapy in MSS/pMMR-type advanced CRC patients by modulating NK cells. Our study demonstrated that inhibition of circPhlpp2 enhanced the efficacy of anti-PD-1 therapy in CRC and circPHLPP2 may serve as a promising therapeutic target.
Previous studies have shown that circRNAs may induce either immune activation or tolerance through the regulation of molecules such as cytokines and chemokines. For example, circCYP24A1 could affect tumor microenvironment by promoting the paracrine and distal secretion of CCL5 [42]. In this study, we demonstrated that circPHLPP2 was involved in the regulation of the cytokine-cytokine receptor interaction pathway, and IL36γ served as a downstream functional molecule of circPHLPP2. Indeed, IL36γ has been proven to be involved in the progression and metastasis of various cancers. For instance, IL-36γ promotes non-small cell lung cancer progression by enhancing GSH biogenesis and inhibiting oxidative stress-induced cell death [43]. It has also been proved that IL-36γ promotes colon inflammation and tumorigenesis by directly modulating the cell-matrix adhesion network to regulate Wnt signaling [28]. Additionally, it has been shown that high levels of IL-36α and IL-36β or high IL-36α with low IL-36γ are associated with longer survival in CRC patients, indicating their potential as biomarkers for prognosis. These researches suggested that IL36γ promotes CRC development, which is consistent with our findings. However, the effects of IL- 36γ in regulating immune responses are complex. A study demonstrated that IL-36γ promotes antitumor immune responses in melanoma by enhancing IFN-γ production by CD8 + T cells and NK cells [44], while another study showed that IL-36γ exerts a pathogenic role in colitis by inhibiting regulatory T cell (Treg) differentiation and promoting Th9 polarization [45]. Our study confirmed that akin to the phenotype induced by circPhlpp2, IL36γ facilitates CRC cell proliferation and diminishes NK cell infiltration and granzyme B and IFN-γ expression levels. The effect induced by knockdown of circPhlpp2 can be reversed by overexpression of IL36γ. Thus, we supposed that circPhlpp2 participates in the progression of CRC partly by secretion of IL36γ, and the impact of IL36γ on tumor progression seems to be dependent on the specificity of the tissues and diseases.
Previous studies indicated that circRNAs can act as miRNA sponges, interact with RBPs, and potentially translate peptides. The function of circRNAs likely depends on their specific subcellular localization [46], and circRNAs in the nucleus are involved in the regulation of transcription, alternative splicing and chromatin looping [38]. In our study, FISH revealed that circPHLPP2 was predominantly localized in the nucleus. Additionally, circPHLPP2 could diretly bind to ILF3, causing its nuclear accumulation. A study has demonstrated that the export of ILF3 necessitates its binding to exportin-5, a process that may be impeded by the interaction of RNA with ILF3 [32]. We showed that the combination of circPHLPP2 and ILF3 decreased ILF3’s interaction with exportin-5, consequently leading to an increased accumulation of ILF3 within the nucleus. In fact, ILF3 was tightly correlated with pathogenesis, progression, drug resistance, and prognosis in various cancers [30, 47, 48]. Through the ability to bind both DNA and RNA, ILF3 can regulate transcription, translation, mRNA stability and primary microRNA processing [49, 50]. Within the nucleus, ILF3 has the capacity to regulate the transcriptional expression of specific genes, such as TGF-β2 and uPA [47, 51]. Our data demonstrated that ILF3 can enhance IL36 transcription in the nucleus, and the interaction between circPHLPP2 and ILF3 facilitates the aggregation of ILF3 in the nucleus, leading to the up-regulation of IL36γ expression.
Our study has several limitations. First, since the sample size is small, our results need to be validated in a larger cohort of patients. Second, the specific mechanism by which IL36γ regulates the infiltration and function of NK cells has not been fully elucidated and requires further investigation. Third, the mouse model could not fully reflect the tumor immune microenvironment of the patients, and further study using humanized mouse models is needed to confirm our results.
Conclusion
In conclusion, circPHLPP2 promotes CRC progression via modulating TME. Specifically, circPHLPP2 facilitates CRC cell progression by enhancing the expression of IL36γ through its interaction with ILF3, ultimately reducing NK cell infiltration as well as granzyme B and IFN-γ expression levels of NK cells (Fig. 6g). Targeting circPHLPP2 may enhance the efficacy of anti-PD-1 therapy in CRC patients.
Supplementary Information
Acknowledgements
We thank all the patients enrolled in this study.
Abbreviations
- CRC
Colorectal Cancer
- PD-1
Programmed cell death protein 1
- circRNAs
Circular RNAs
- RIP
RNA immunoprecipitation
- ICIs
Immune checkpoint inhibitors
- MSI-H
Microsatellite instability-high
- dMMR
Deficient mismatch repair
- MSS
Microsatellite stable
- pMMR
Proficient mismatch repair
- TKIs
Tyrosine kinase inhibitors
- NK cells
Natural killer cells
- ADCC
Antibody-dependent cell-mediated cytotoxicity
- IFN-γ
Interferon-γ
- OS
Overall survival
- IHC
Immunohistochemistry
- PR
Partial response
- PD
Progressive disease
- RT-qPCR
Real-time quantitative polymerase chain reaction
- RBPs
RNA-binding proteins
- FISH
Fluorescence in situ hybridization
- cDNA
Complementary DNA
- gDNA
Genomic DNA
- TME
Tumor immune microenvironment
- DEGs
Differentially expressed genes
- dsRNA
Double-stranded RNA
- dsRBDs
double-stranded RNA binding motifs
Authors’ contributions
CDL designed the study. HY, CZR, HRZ and WDS collected the data. HY, CZR, HRZ and WDS analyzed and interpreted the data. HY, CZR and HRZ performed the statistical analysis. HY, JHQ, and CDL wrote the manuscript. HY and CDL contributed to discussion and data interpretation. HY and CDL revised the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by National Natural Science Foundation of China (No.82373376) and Natural Science Foundation of Guangdong Province (No. 2023A1515030257).
Data availability
All the data supporting these findings are available within the article and additional files or from the corresponding authors upon reasonable request.
Declarations
Ethics approval and consent to participate
The study was approved by the Medical Ethics Committee of Sun Yat-sen University Cancer Center (SYSUCC) with written informed consents from all the patients (SZR2022-008). The animal studies were ethically approved by the Institutional Ethics Committee for Clinical Research and Animal Trials of the SYSUCC (L102022022003L).
Consent for publication
Informed consent was obtained from all patients before the use of these clinical materials for research purposes.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yan Hu, Ren-Ze Huang and Ze-Rong Cai contributed equally to this work.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 2.Guo S, Sun Y. OTOP2, inversely modulated by miR-3148, inhibits CRC Cell Migration, Proliferation and epithelial-mesenchymal transition: evidence from Bioinformatics Data Mining and Experimental Verification. Cancer Manag Res. 2022;14:1371–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diaz LA Jr., Shiu KK, Kim TW, Jensen BV, Jensen LH, Punt C, Smith D, Garcia-Carbonero R, Benavides M, Gibbs P, et al. Pembrolizumab versus chemotherapy for microsatellite instability-high or mismatch repair-deficient metastatic colorectal cancer (KEYNOTE-177): final analysis of a randomised, open-label, phase 3 study. Lancet Oncol. 2022;23:659–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He X, Xu C. Immune checkpoint signaling and cancer immunotherapy. Cell Res. 2020;30:660–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manca P, Corti F, Intini R, Mazzoli G, Miceli R, Germani MM, Bergamo F, Ambrosini M, Cristarella E, Cerantola R, et al. Tumour mutational burden as a biomarker in patients with mismatch repair deficient/microsatellite instability-high metastatic colorectal cancer treated with immune checkpoint inhibitors. Eur J Cancer. 2023;187:15–24. [DOI] [PubMed] [Google Scholar]
- 6.Fukuoka S, Hara H, Takahashi N, Kojima T, Kawazoe A, Asayama M, Yoshii T, Kotani D, Tamura H, Mikamoto Y, et al. Regorafenib Plus Nivolumab in patients with Advanced gastric or colorectal Cancer: an Open-Label, Dose-Escalation, and dose-expansion phase ib trial (REGONIVO, EPOC1603). J Clin Oncol. 2020;38:2053–61. [DOI] [PubMed] [Google Scholar]
- 7.Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long F, Lin Z, Li L, Ma M, Lu Z, Jing L, Li X, Lin C. Comprehensive landscape and future perspectives of circular RNAs in colorectal cancer. Mol Cancer. 2021;20:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ju HQ, Zhao Q, Wang F, Lan P, Wang Z, Zuo ZX, Wu QN, Fan XJ, Mo HY, Chen L, et al. A circRNA signature predicts postoperative recurrence in stage II/III colon cancer. EMBO Mol Med. 2019;11:e10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang M, Peng X, Yang L, Yang S, Li X, Tang S, Li B, Jin H, Wu B, Liu J, Li H. Non-coding RNA derived from extracellular vesicles in cancer immune escape: Biological functions and potential clinical applications. Cancer Lett. 2021;501:234–46. [DOI] [PubMed] [Google Scholar]
- 11.Ma Z, Shuai Y, Gao X, Wen X, Ji J. Circular RNAs in the tumour microenvironment. Mol Cancer. 2020;19:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Z, Chen G, Zhao Y, Gao H, Li L, Yin Y, Jiang J, Wang L, Mang Y, Gao Y, et al. Exosome-derived circCCAR1 promotes CD8 + T-cell dysfunction and anti-PD1 resistance in hepatocellular carcinoma. Mol Cancer. 2023;22:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen DL, Sheng H, Zhang DS, Jin Y, Zhao BT, Chen N, Song K, Xu RH. The circular RNA circDLG1 promotes gastric cancer progression and anti-PD-1 resistance through the regulation of CXCL12 by sponging miR-141-3p. Mol Cancer. 2021;20:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimasaki N, Jain A, Campana D. NK cells for cancer immunotherapy. Nat Rev Drug Discov. 2020;19:200–18. [DOI] [PubMed] [Google Scholar]
- 15.Voskoboinik I, Smyth MJ, Trapani JA. Perforin-mediated target-cell death and immune homeostasis. Nat Rev Immunol. 2006;6:940–52. [DOI] [PubMed] [Google Scholar]
- 16.Fauriat C, Long EO, Ljunggren HG, Bryceson YT. Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood. 2010;115:2167–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.López-Soto A, Gonzalez S, Smyth MJ, Galluzzi L. Control of Metastasis by NK Cells. Cancer Cell. 2017;32:135–54. [DOI] [PubMed] [Google Scholar]
- 18.Hsu J, Hodgins JJ, Marathe M, Nicolai CJ, Bourgeois-Daigneault MC, Trevino TN, Azimi CS, Scheer AK, Randolph HE, Thompson TW, et al. Contribution of NK cells to immunotherapy mediated by PD-1/PD-L1 blockade. J Clin Invest. 2018;128:4654–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imai K, Matsuyama S, Miyake S, Suga K, Nakachi K. Natural cytotoxic activity of peripheral-blood lymphocytes and cancer incidence: an 11-year follow-up study of a general population. Lancet. 2000;356:1795–9. [DOI] [PubMed] [Google Scholar]
- 20.Melero I, Ochoa MC, Molina C, Sanchez-Gregorio S, Garasa S, Luri-Rey C, Hervas-Stubbs S, Casares N, Elizalde E, Gomis G, et al. Intratumoral co-injection of NK cells and NKG2A-neutralizing monoclonal antibodies. EMBO Mol Med. 2023;15:e17804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ju HQ, Ying H, Tian T, Ling J, Fu J, Lu Y, Wu M, Yang L, Achreja A, Chen G, et al. Mutant Kras- and p16-regulated NOX4 activation overcomes metabolic checkpoints in development of pancreatic ductal adenocarcinoma. Nat Commun. 2017;8:14437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin JF, Hu PS, Wang YY, Tan YT, Yu K, Liao K, Wu QN, Li T, Meng Q, Lin JZ, et al. Phosphorylated NFS1 weakens oxaliplatin-based chemosensitivity of colorectal cancer by preventing PANoptosis. Signal Transduct Target Ther. 2022;7:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng R, Zhang K, Tan S, Gao F, Zhang Y, Xu W, Wang H, Gu D, Zhu L, Li S, et al. Exosomal circLPAR1 functions in colorectal cancer diagnosis and tumorigenesis through suppressing BRD4 via METTL3-eIF3h interaction. Mol Cancer. 2022;21:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seymour L, Bogaerts J, Perrone A, Ford R, Schwartz LH, Mandrekar S, Lin NU, Litière S, Dancey J, Chen A, et al. iRECIST: guidelines for response criteria for use in trials testing immunotherapeutics. Lancet Oncol. 2017;18:e143–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gandara DR, Mack PC, Bult C, Li T, Lara PN Jr., Riess JW, Astrow SH, Gandour-Edwards R, Cooke DT, Yoneda KY, et al. Bridging tumor genomics to patient outcomes through an integrated patient-derived xenograft platform. Clin Lung Cancer. 2015;16:165–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawada M, Inoue H, Kajikawa M, Sugiura M, Sakamoto S, Urano S, Karasawa C, Usami I, Futakuchi M, Masuda T. A novel monoclonal antibody targeting coxsackie virus and adenovirus receptor inhibits tumor growth in vivo. Sci Rep. 2017;7:40400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hegde PS, Chen DS. Top 10 challenges in Cancer Immunotherapy. Immunity. 2020;52:17–35. [DOI] [PubMed] [Google Scholar]
- 28.Yang W, Dong HP, Wang P, Xu ZG, Xian J, Chen J, Wu H, Lou Y, Lin D, Zhong B. IL-36γ and IL-36Ra reciprocally regulate Colon inflammation and tumorigenesis by modulating the Cell-Matrix Adhesion Network and wnt signaling. Adv Sci (Weinh). 2022;9:e2103035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rowley JW, Chappaz S, Corduan A, Chong MM, Campbell R, Khoury A, Manne BK, Wurtzel JG, Michael JV, Goldfinger LE, et al. Dicer1-mediated miRNA processing shapes the mRNA profile and function of murine platelets. Blood. 2016;127:1743–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li K, Wu JL, Qin B, Fan Z, Tang Q, Lu W, Zhang H, Xing F, Meng M, Zou S, et al. ILF3 is a substrate of SPOP for regulating serine biosynthesis in colorectal cancer. Cell Res. 2020;30:163–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castella S, Bernard R, Corno M, Fradin A, Larcher JC. Ilf3 and NF90 functions in RNA biology. Wiley Interdiscip Rev RNA. 2015;6:243–56. [DOI] [PubMed] [Google Scholar]
- 32.Brownawell AM, Macara IG. Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J Cell Biol. 2002;156:53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, Wei J, Yao RW, Yang L, Chen LL. Coordinated circRNA Biogenesis and function with NF90/NF110 in viral infection. Mol Cell. 2017;67:214–e227217. [DOI] [PubMed] [Google Scholar]
- 34.Duchange N, Pidoux J, Camus E, Sauvaget D. Alternative splicing in the human interleukin enhancer binding factor 3 (ILF3) gene. Gene. 2000;261:345–53. [DOI] [PubMed] [Google Scholar]
- 35.Bao Y, Zhai J, Chen H, Wong CC, Liang C, Ding Y, Huang D, Gou H, Chen D, Pan Y, et al. Targeting m(6)a reader YTHDF1 augments antitumour immunity and boosts anti-PD-1 efficacy in colorectal cancer. Gut. 2023;72:1497–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang B, Yao K, Zhou E, Zhang L, Cheng C. Chr20q amplification defines a distinct molecular subtype of microsatellite stable colorectal Cancer. Cancer Res. 2021;81:1977–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ganesh K, Stadler ZK, Cercek A, Mendelsohn RB, Shia J, Segal NH, Diaz LA. Jr.: Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol. 2019;16:361–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21:475–90. [DOI] [PubMed] [Google Scholar]
- 39.Jiang W, Pan S, Chen X, Wang ZW, Zhu X. The role of lncRNAs and circRNAs in the PD-1/PD-L1 pathway in cancer immunotherapy. Mol Cancer. 2021;20:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guillerey C, Huntington ND, Smyth MJ. Targeting natural killer cells in cancer immunotherapy. Nat Immunol. 2016;17:1025–36. [DOI] [PubMed] [Google Scholar]
- 41.Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, Cai JB, Ke AW. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer. 2020;19:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu L, Sang Y, Nan X, Zheng Y, Liu F, Meng L, Sang M, Shan B. circCYP24A1 facilitates esophageal squamous cell carcinoma progression through binding PKM2 to regulate NF-κB-induced CCL5 secretion. Mol Cancer. 2022;21:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang P, Yang W, Guo H, Dong HP, Guo YY, Gan H, Wang Z, Cheng Y, Deng Y, Xie S, et al. IL-36γ and IL-36Ra reciprocally regulate NSCLC progression by modulating GSH Homeostasis and oxidative stress-Induced cell death. Adv Sci (Weinh). 2021;8:e2101501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X, Zhao X, Feng C, Weinstein A, Xia R, Wen W, Lv Q, Zuo S, Tang P, Yang X, et al. IL-36γ transforms the Tumor Microenvironment and promotes type 1 lymphocyte-mediated Antitumor Immune responses. Cancer Cell. 2015;28:296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harusato A, Abo H, Ngo VL, Yi SW, Mitsutake K, Osuka S, Kohlmeier JE, Li JD, Gewirtz AT, Nusrat A, Denning TL. IL-36γ signaling controls the induced regulatory T cell-Th9 cell balance via NFκB activation and STAT transcription factors. Mucosal Immunol. 2017;10:1455–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu CX, Chen LL. Circular RNAs: characterization, cellular roles, and applications. Cell. 2022;185:2016–34. [DOI] [PubMed] [Google Scholar]
- 47.Li P, Mi Q, Yan S, Xie Y, Cui Z, Zhang S, Wang Y, Gao H, Wang Y, Li J, et al. Characterization of circSCL38A1 as a novel oncogene in bladder cancer via targeting ILF3/TGF-β2 signaling axis. Cell Death Dis. 2023;14:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao L, Zhang Y, Luo Q, Guo C, Chen Z, Lai C. DHRS4-AS1 regulate gastric cancer apoptosis and cell proliferation by destabilizing DHX9 and inhibited the association between DHX9 and ILF3. Cancer Cell Int. 2023;23:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu J, Liu ZX, Li JJ, Zeng ZL, Wang JH, Luo XJ, Wong CW, Zheng JB, Pu HY, Mo HY, et al. The Macrophage-Associated LncRNA MALR facilitates ILF3 liquid-liquid phase separation to promote HIF1α signaling in Esophageal Cancer. Cancer Res. 2023;83:1476–89. [DOI] [PubMed] [Google Scholar]
- 50.Tan J, Sun M, Yin J, Zhou Q, Zhao R, Chen Q, Sun H, Jiang C, Li S, He Y. Hsa_circ_0005050 interacts with ILF3 and affects cell apoptosis and proliferation by disrupting the balance between p53 and p65. Chem Biol Interact. 2022;368:110208. [DOI] [PubMed] [Google Scholar]
- 51.Hu Q, Lu YY, Noh H, Hong S, Dong Z, Ding HF, Su SB, Huang S. Interleukin enhancer-binding factor 3 promotes breast tumor progression by regulating sustained urokinase-type plasminogen activator expression. Oncogene. 2013;32:3933–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data supporting these findings are available within the article and additional files or from the corresponding authors upon reasonable request.