Abstract
Background
The occurrence of post-traumatic stress disorder (PTSD) following a traumatic event is associated with biological differences that can represent the susceptibility to PTSD, the impact of trauma, or the sequelae of PTSD itself. These effects include differences in DNA methylation (DNAm), an important form of epigenetic gene regulation, at multiple CpG loci across the genome. Moreover, these effects can be shared or specific to both central and peripheral tissues. Here, we aim to identify blood DNAm differences associated with PTSD and characterize the underlying biological mechanisms by examining the extent to which they mirror associations across multiple brain regions.
Methods
As the Psychiatric Genomics Consortium (PGC) PTSD Epigenetics Workgroup, we conducted the largest cross-sectional meta-analysis of epigenome-wide association studies (EWASs) of PTSD to date, involving 5077 participants (2156 PTSD cases and 2921 trauma-exposed controls) from 23 civilian and military studies. PTSD diagnosis assessments were harmonized following the standardized guidelines established by the PGC-PTSD Workgroup. DNAm was assayed from blood using Illumina HumanMethylation450 or MethylationEPIC (850 K) BeadChips. Within each cohort, DNA methylation was regressed on PTSD, sex (if applicable), age, blood cell proportions, and ancestry. An inverse variance-weighted meta-analysis was performed. We conducted replication analyses in tissue from multiple brain regions, neuronal nuclei, and a cellular model of prolonged stress.
Results
We identified 11 CpG sites associated with PTSD in the overall meta-analysis (1.44e − 09 < p < 5.30e − 08), as well as 14 associated in analyses of specific strata (military vs civilian cohort, sex, and ancestry), including CpGs in AHRR and CDC42BPB. Many of these loci exhibit blood–brain correlation in methylation levels and cross-tissue associations with PTSD in multiple brain regions. Out of 9 CpGs annotated to a gene expressed in blood, methylation levels at 5 CpGs showed significant correlations with the expression levels of their respective annotated genes.
Conclusions
This study identifies 11 PTSD-associated CpGs and leverages data from postmortem brain samples, GWAS, and genome-wide expression data to interpret the biology underlying these associations and prioritize genes whose regulation differs in those with PTSD.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13073-024-01417-1.
Keywords: PTSD, Trauma, DNA methylation, Postmortem brain, GWAS, Gene expression
Background
Posttraumatic stress disorder (PTSD) is a serious psychiatric disorder characterized by intrusive memories of the traumatic event(s), avoidance of or numbing to situations that trigger those memories, and hyperarousal symptoms that can disturb mental and physical health [1]. These symptoms are associated with lower levels of self-care, lower compliance with medical treatment, and higher rates of substance use [2, 3]. Thus, it is not surprising that PTSD increases the risk for chronic medical conditions, such as cardiovascular disorders, independent of lifestyle factors (e.g., substance use and sleep quality) [4, 5]. Although most individuals experience at least one traumatic event, only a small fraction develop PTSD [6]. Genetic and environmental factors contribute to this differential susceptibility in PTSD development upon trauma exposure [7, 8].
Genome-wide association studies (GWAS) of PTSD demonstrated remarkable success at identifying relevant genes, many of which are involved in the stress response or immune function (see reviews[9, 10]). The recent Psychiatric Genomics Consortium PTSD Workgroup (PGC-PTSD) Freeze 3 GWAS identified 95 genomic loci associated with PTSD, implicating genes involved in stress, immune, fear, and threat-related processes [11]. Nonetheless, genetic differences cannot fully account for an individual’s susceptibility to PTSD. Trauma exposure has been shown to alter epigenetic patterns in both animal and human studies, prompting the need to conduct epigenetic studies of PTSD in addition to genetic studies [12, 13]. Epigenetic mechanisms are chemical modifications that can dictate the timing and magnitude of gene expression without altering the DNA sequence [14]. The most widely studied epigenetic mechanism is DNA methylation (DNAm), which is defined as the addition of a methyl group to cytosine bases, particularly at cytosine-guanine dinucleotides (CpG sites). DNAm patterns respond to changes in the environment, are potentially reversible, and can be targeted for disease therapies [15, 16]. Environmental influences on DNAm are apparent across the lifespan and may provide insight into the biological response to trauma [17].
Which specific DNAm sites differ across individuals and how they correlate with exposures and gene expression can vary across tissues [18]. DNAm in human brain tissue, which is most relevant to the study of psychiatric disorders, is not easily accessible in living patients and hence is not a viable PTSD biomarker for clinical use. However, correlation has been observed between peripheral tissues (e.g., blood) and brain DNAm levels at specific genomic loci, and thus, blood DNAm can potentially serve as a robust biomarker for implementing early intervention and developing improved preventative or therapeutic strategies for PTSD [19, 20]. Moreover, PTSD symptoms have been linked to the components of the peripheral immune system [21, 22] that can be readily assessed in blood DNAm. Multiple peripheral epigenome-wide association studies (EWASs) of PTSD identified CpGs in genes related to the immune system and neurotransmission [23–28]. While prior EWASs of PTSD have reported promising results, the small sample sizes and variability of analysis methods across studies make it difficult to combine and interpret the findings effectively. Recent meta-analyses led by the PGC-PTSD Epigenetics Workgroup minimized these issues by increasing sample size, increasing sample diversity, and using a consortium-supplied quality control and analysis pipelines [29–33]. These meta-analyses identified multiple new loci associated with PTSD, including NRG1, AHRR, MAD1L1, and TBXAS1, implicating immune dysregulation in those with PTSD [30–33].
Building on the prior work by Smith et al. [31], which conducted an EWAS meta-analysis in 1896 participants from 10 cohorts, this study includes 13 additional cohorts with a denser and more comprehensive DNAm array, bringing the sample up to 5077 participants from 23 civilian and military cohorts. Our current investigation replicated the findings of the initial PGC-PTSD epigenome-wide meta-analysis, reporting lower AHRR methylation in those with PTSD, and identified 9 new (11 total) loci associated with PTSD, as well as 14 CpGs associated in analyses of specific strata (military vs civilian cohort, sex, and ancestry). We also leveraged data from postmortem brain samples, a cellular model of prolonged stress, GWAS, and genome-wide gene expression studies to interpret the biology underlying these associations and prioritize genes whose regulation differs in those with PTSD.
Methods
Cohorts and post-traumatic stress disorder assessments
The study includes 2156 current PTSD cases and 2921 trauma-exposed controls from 9 civilian cohorts: BEAR, DNHS, DCHS, GTP, NIU, Shared Roots, AURORA, H3A_Rwanda, WTC; and 9 military cohorts: GMRFQUT, MRS, PRISMO, Army STARRS, PROGrESS, NCPTSD/TRACTS, INTRuST, and VA cohorts (VA-M-AA and VA-M-EA). For DNHS, GTP, MRS, PRISMO, and Army STARRS, two different datasets were available based on the DNAm array. Two different datasets for these five cohorts did not have any overlapping samples and were treated as independent studies. Sample characteristics for the 23 studies that participated in the meta-analysis are summarized in Table 1. Detailed descriptions of each cohort were presented in Additional file 1: eMethods.
Table 1.
Overview of the studies
| Cohort | Array | N | Cases N (%) |
Controls N (%) |
Female N (%) |
European N (%) |
African N (%) |
Age Mean (SD) |
|---|---|---|---|---|---|---|---|---|
| Civilian | ||||||||
| BEAR | EPIC | 162 | 36 (22%) | 126 (78%) | 119 (73%) | 112 (69%) | 3 (2%) | 15.16 (1.45) |
| DNHS-1 | EPIC | 423 | 26 (6%) | 397 (94%) | 255 (60%) | 23 (5%) | 384 (91%) | 54.54 (16.87) |
| DCHS | EPIC | 95 | 46 (48%) | 49 (52%) | 95 (100%) | 0 (0%) | 54 (57%) | 26.81 (5.2) |
| GTP-1 | EPIC | 479 | 158 (33%) | 321 (67%) | 340 (71%) | 12 (3%) | 448 (94%) | 42.22 (12.25) |
| NIU | EPIC | 140 | 18 (13%) | 122 (87%) | 140 (100%) | 110 (79%) | 19 (14%) | 26.01 (1.74) |
| Shared Roots | EPIC | 120 | 61 (51%) | 59 (49%) | 85 (71%) | 0 (0%) | 120 (100%) | 43.15 (10.77) |
| AURORA | EPIC | 206 | 57 (28%) | 149 (72%) | 154 (75%) | 67 (33%) | 131 (64%) | 39.24 (14.17) |
| H3A_Rwanda | EPIC | 73 | 32 (44%) | 41 (56%) | 73 (100%) | 0 (0%) | 73 (100%) | 45.54 (7.29) |
| DNHS-2 | 450 K | 100 | 40 (40%) | 60 (60%) | 60 (60%) | 13 (13%) | 87 (87%) | 53.6 (14.01) |
| GTP-2 | 450 K | 265 | 74 (28%) | 191 (72%) | 187 (71%) | 16 (6%) | 249 (94%) | 41.95 (12.37) |
| WTC | 450 K | 180 | 84 (47%) | 96 (53%) | 0 (0%) | 138 (77%) | 7 (4%) | 49.72 (8.25) |
| Civilian total | 2243 | 632 (28%) | 1611 (72%) | 1508 (67%) | 491 (22%) | 1575 (70%) | 41.89 (16.27) | |
| Military | ||||||||
| GMRFQUT | EPIC | 96 | 48 (50%) | 48 (50%) | 0 (0%) | 96 (100%) | 0 (0%) | 68.67 (4.36) |
| MRS-1 | EPIC | 127 | 64 (50%) | 63 (50%) | 0 (0%) | 88 (69%) | 5 (4%) | 23.07 (2.18) |
| PRISMO-1 | EPIC | 89 | 24 (27%) | 65 (73%) | 9 (10%) | 74 (83%) | 3 (3%) | 27.51 (8.63) |
| Army STARRS-1 | EPIC | 216 | 106 (49%) | 110 (51%) | 0 (0%) | 149 (69%) | 22 (10%) | 25.13 (4.82) |
| PROGrESS | EPIC | 140 | 112 (80%) | 28 (20%) | 14 (10%) | 89 (64%) | 40 (29%) | 34.77 (8.33) |
| NCPTSD/TRACTS | EPIC | 1028 | 638 (62%) | 390 (38%) | 231 (22%) | 706 (69%) | 123 (12%) | 44.06 (13.7) |
| MRS-2 | 450 K | 126 | 63 (50%) | 63 (50%) | 0 (0%) | 72 (57%) | 10 (8%) | 22.2 (3.04) |
| PRISMO-2 | 450 K | 62 | 32 (52%) | 30 (48%) | 0 (0%) | 62 (100%) | 0 (0%) | 27.1 (9.23) |
| Army STARRS-2 | 450 K | 102 | 51 (50%) | 51 (50%) | 0 (0%) | 102 (100%) | 0 (0%) | 23.79 (4.25) |
| INTRuST | 450 K | 303 | 116 (38%) | 187 (62%) | 102 (34%) | 206 (68%) | 58 (19%) | 34.09 (11.68) |
| VA-M-AA | 450 K | 369 | 183 (50%) | 186 (50%) | 184 (50%) | 0 (0%) | 369 (100%) | 38.36 (9.36) |
| VA-M-EA | 450 K | 176 | 87 (49%) | 89 (51%) | 38 (22%) | 176 (100%) | 0 (0%) | 34.87 (9.89) |
| Military total | 2834 | 1524 (54%) | 1310 (46%) | 578 (20%) | 1820 (64%) | 630 (22%) | 37.08 (14.33) | |
| Total | 5077 | 2156 (42%) | 2921 (58%) | 2086 (41%) | 2311 (46%) | 2205 (43%) | 39.2 (15.4) | |
Participating civilian cohorts: biomarkers, social, and affective predictors of suicidal thoughts and behaviors in adolescents (BEAR), Detroit Neighborhood Health Study (DNHS), Drakenstein Child Health Study (DCHS), Grady Trauma Project (GTP), Northern Illinois University Trauma Study (NIU), Shared Roots, Advancing Understanding of RecOvery afteR traumA (AURORA), Human Heredity and Health in Africa, Rwanda (H3A_Rwanda), World Trade Center 9/11 Responders (WTC). Participating military cohorts: Gallipoli Medical Research Foundation Queensland University of Technology (GMRFQUT), Marine Resiliency Study (MRS), Prospective Research in Stress-related Military Operations (PRISMO), Army Study to Assess Risk and Resilience in Servicemembers (Army STARRS), PROlonGed ExpoSure and Sertraline Trial (PROGrESS), Boston VA—National Center for PTSD/ Translational Research Center for TBI and Stress Disorders (NCPTSD/TRACTS), Injury and Traumatic Stress study (INTRuST), and Veterans Affairs’ Mental Illness Research, Education and Clinical Centers (VA-M-AA and VA-M-EA). Note: For DNHS, GTP, MRS, PRISMO, and Army STARRS cohorts, EPIC and 450 k datasets represent different sets of participants
Biomarkers, social, and affective predictors of suicidal thoughts and behaviors in adolescents (BEAR) [34] involved a sample of 194 adolescents who had been hospitalized for suicidal thoughts/behaviors. Of those, 163 provided a blood sample, and 162 samples that passed the DNAm quality control (QC) were included in the meta-analysis. PTSD diagnosis was assessed by the Clinician Administered PTSD Scale (CAPS) for Children and Adolescents for DSM-5 [35].
Detroit Neighborhood Health Study (DNHS) [23] involved 1547 participants whose PTSD symptoms were assessed by the PTSD checklist (PCL-C) [36] at the baseline wave. The meta-analysis included 523 participants with available DNAm data that passed the DNAm QC.
Drakenstein Child Health Study (DCHS) [37] is a population-based birth cohort that recruited 1000 pregnant people between 20–28 weeks gestation. DNAm data was available for 98 participants, and 95 samples that passed the DNAm QC were included in the meta-analysis. For the purpose of this study, PTSD was assessed using The Mini International Neuropsychiatric Interview (MINI) [38, 39].
Grady Trauma Project (GTP) is a large-scale ongoing study with > 10,000 participants. The meta-analysis included 744 participants with available DNAm data that passed the DNAm QC. Current PTSD was assessed using the Clinician-Administered PTSD Scale for DSM IV (CAPS-4) [40, 41] or the modified PTSD Symptomatic Scale (PSS) [42].
Northern Illinois University Trauma Study (NIU) involved 812 participants recruited to participate in a federally-funded study (NIH 5R21MH085436-02) to examine risk and protective factors following the NIU campus shooting on February 14th, 2008. Of those, 140 provided blood samples and were included in the meta-analysis. Current PTSD diagnosis at the first post-shooting assessment was assessed by self-report on the Distressing Events Questionnaire (DEQ) [43].
Shared Roots SHRS or “Understanding the SHARED ROOTS of Neuropsychiatric Disorders and Modifiable Risk Factors for Cardiovascular Disease” is a matched case–control study (N = 120) examining the factors that contribute to the comorbidity of metabolic syndrome and neuropsychiatric disorders [44]. The CAPS-5 [45] was administered by clinicians to assess PTSD over the prior month.
Advancing Understanding of RecOvery afteR traumA (AURORA) is a large cohort study of women and men presenting to the ED within 72 h after exposure to psychological trauma [46]. PTSD diagnosis at 6 months was defined using the PTSD Checklist for DSM-5 (PCL-5) [47–49]. The meta-analysis included a subset of the AURORA cohort with available phenotypic, DNA methylation, and RNA sequencing data at the 6-month follow-up after trauma exposure (N = 206).
Human Heredity and Health in Africa, Rwanda (H3A_Rwanda). Data from the H3A_Rwanda cohort were obtained from a subset of participants in a previously published pilot study (n = 50) [50, 51] and newly recruited participants via support from the H3Africa Consortium (n = 40) [52]. All study participants were women of Tutsi ethnicity who were pregnant during the genocide. PTSD was assessed by PCL-17 [53] for the pilot study and by PCL-5 [54] for the H3Africa-affiliated study. The meta-analysis included 73 participants with available phenotype and DNAm data.
World Trade Center Responders (WTC) [55, 56]. PTSD in relation to WTC exposures was assessed by the Structured Clinical Interview for DSM-IV Disorders (SCID) [57]. The sample (N = 180) providing blood samples for the epigenetics assays was restricted to men (the vast majority of the responders) and oversampled for posttraumatic stress disorder (PTSD) [25].
Gallipoli Medical Research Foundation Queensland University of Technology (GMRFQUT) is a large cohort of veterans who have been or are currently being treated for PTSD at the Keith Payne Unit within the Greenslopes Private Hospital in Queensland, Australia [58]. The CAPS-5 [40, 41] was used to assess current PTSD. The meta-analysis included 96 participants with available DNAm data.
Marine Resiliency Study (MRS). In the MRS [59, 60], PTSD was diagnosed by CAPS-5 [40, 41] up to 3 times, once before deployment and 3 and/or 6 months post-deployment. Samples of PTSD cases (N = 127) were selected from the 3- or 6-month post-deployment visits depending on which visit had the highest CAPS score. Combat-exposed controls with low to no PTSD-symptoms (N = 126) were selected from post-deployment visits, matching for age, ancestry, and time of post-deployment visit.
Prospective Research in Stress-related Military Operations (PRISMO) is a large prospective study of Dutch military soldiers [61, 62]. Current PTSD symptoms were assessed using the Self-Report Inventory for PTSD (SRIP) [63], and blood samples were collected approximately 1 month before and at both 1 and 6 months after deployment. PTSD cases (N = 56) were selected for this DNA methylation study from the 1- or 6-month post-deployment visits, depending on which visit had the highest SRIP score. All controls (N = 95) were combat exposed and had low PTSD symptoms [26].
The Army Study to Assess Risk and Resilience in Servicemembers (Army STARRS) is a multicomponent prospective study among US Army personnel [64]. PTSD diagnosis was assigned using multiple imputation methods that relied on PCL and CIDI-SC data [65]. Whole blood for methylation assays was collected approximately 6 weeks before deployment and 1-month post-deployment. PTSD cases (N = 157) were selected based on their PTSD diagnosis at 6 months post-deployment. Controls (N = 161) were participants without PTSD, matched on age, deployment stress, and childhood adversity.
PROlonGed ExpoSure and Sertraline Trial (PROGrESS) is a randomized-controlled trial (RCT; N = 223) designed to examine the comparative effectiveness of multiple treatment strategies across 24 weeks. PTSD is assessed by the CAPS-IV [40, 41]. This study included 112 PTSD cases and 28 trauma-exposed controls selected from the pre-treatment visit.
Boston VA National Center for PTSD/Translational Research Center for TBI and Stress Disorders (NCPTSD/TRACTS) included participants from the NCPTSD study [66], the PTSD and Accelerated Aging study [67], and the TRACTS study [68]. For all three studies, PTSD diagnosis was determined based on the CAPS for DSM-IV [40] or DSM-5 [69]. The NCPTSD and TRACTS cohort DNAm (N = 1028) data were jointly cleaned and analyzed together.
Injury and Traumatic Stress (INTRuST). This cohort included participants in studies of the INTRuST Consortium [70]. PTSD diagnosis was assessed by study-specific measures, including CAPS-IV, CAPS-5 [40, 41], and PCL-17 [53]. The meta-analysis included 303 participants with available phenotype and DNAm data.
Mid-Atlantic Mental Illness Research Education and Clinical Center PTSD Study (VA-M-AA & VA-M-EA) [71–73]. PTSD was diagnosed using the SCID [57]. The meta-analysis included 369 participants from VA-M-AA and 87 participants from VA-M-EA with available phenotype and DNAm data.
The sample is heterogeneous in terms of sex (41% female), ancestry (46% European, 43% African, and 11% of other ancestries), and cohort type (56% military cohort). Civilian cohorts skew towards being more female (67%) and African (70%), whereas military cohorts are predominantly male (80%) and European (64%). In addition, the prevalence of PTSD is higher in military cohorts (54%) than in civilian cohorts (28%).
All participants were exposed to a traumatic event and 42% met the criteria for current PTSD. The current PTSD diagnosis was assessed by each study following the standardized guidelines established by the PGC-PTSD Workgroup [7]. Briefly, current PTSD diagnosis was determined based on the specific criteria set by the principal investigator of each study. Participants without current PTSD but with a history of PTSD (i.e., remitted PTSD), were excluded. Detailed descriptions of cohorts and PTSD assessments are provided in the Additional file 1: eMethods. All participants in these studies gave informed consent. The institutional review boards of each respective institution approved these studies.
DNA methylation
Whole blood DNAm was measured using the Illumina MethylationEPIC BeadChip (EPIC array) in 14 studies, and the HumanMethylation450 BeadChip (450 K array) in 9 studies (Table 1). All studies used a standardized consortium-developed QC pipeline that differed somewhat depending on which chip was used. The 450 K array pipeline [29, 31] is described in Additional file 1: eMethods.
The EPIC pipeline (available at https://github.com/PGC-PTSD-EWAS/EPIC_QC) [74] was similar to the 450 K array pipeline. Samples with probe detection call rates lower than 90% and average intensity values that were either less than 50% of the overall sample mean or below 2000 arbitrary units (AU) were excluded. Probes with detection p-values > 0.01 were considered low quality and treated as missing. Probes that were missing in > 10% of the samples within the studies and were cross-hybridizing were removed [75]. Data was normalized using single-sample Noob (ssNoob) implemented in R package minfi [76]. ComBat was used to account for batch effects of chip and position while preserving PTSD, age, and sex effects (if applicable) [77]. Blood-cell composition (i.e., the proportion of CD8 + T, CD4 + T, natural killer (NK), B cells, monocytes, and neutrophils) was estimated using the robust partial correlation (RPC) method in Epidish [78] with a reference data specific to EPIC array [79]. For studies without genome-wide genotype data (DNHS, NIU, Shared Roots, AURORA, H3A_Rwanda, GMRFQUT, PROGrESS), we estimated ancestry principal components (PCs) from DNAm data, using the method developed by Barfield et al. [80], as previously described [33]. PCs 2 and 3, which were the components that correlate most with self-reported ancestry, were included as covariates [33, 80]. In cohorts with available genome-wide genotype data, PCs 1–3 from GWAS were used to adjust for ancestry. We used R package bacon to control inflation, only if doing so results in the lambda being closer to 1 [81]. To predict smoking status, a DNAm-based smoking score was calculated, as previously described [27] for cohorts with EPIC array data. A detailed description of DNAm-based ancestry PC and smoking score calculation is provided in Additional file 1: eMethods.
Epigenome-wide association analysis
The association between PTSD and DNAm was tested using multivariable linear regression models for cohorts with balanced plate designs. For studies in which plate layouts were not balanced (Shared Roots, H3A_Rwanda, GMRFQUT), we conducted mixed-effect regression models, including chip as a random effect term. The CpGassoc R package was used to fit the models [82]. The models were adjusted for age, sex (if applicable), blood cell composition (i.e., CD8T, CD4T, NK, B cell, and monocyte cell proportions), and ancestry PCs. While not included initially because of concerns about multicoliniarity and collider bias, a post hoc sensitivity analysis was performed including a covariate for smoking: DNAm-based smoking score in studies with EPIC data and current smoking status for studies with 450 K data. Furthermore, we conducted stratified analyses for both sexes, ancestry (European and African ancestry), and cohort type (civilian and military cohorts).
To combine results across studies, we performed inverse-variance weighted (IVW) meta-analysis in meta [83]. Meta-analysis tested 411,786 CpGs common to 450 K and EPIC arrays (23 studies), and 404,794 EPIC array specific CpGs (14 studies). Epigenome-wide significance threshold recommended for the MethylationEPIC BeadChip (p < 9.0e − 08) was used to determine statistical significance [84]. Gene Ontology (GO) enrichment analyses were conducted using the top 1000 CpGs in missMethyl [85]. A false discovery rate (FDR) threshold of 5% was used to identify significant GO terms.
Gene regulation
Correlations between PTSD-associated CpG sites’ DNAm levels and expression levels of the corresponding gene (as determined by the EPIC v1 array annotation) were tested in whole-blood RNA-sequencing (RNA-seq) data from participants in the BEAR (n = 127), AURORA (n = 173), NCPTSD Merit (n = 204), and MRS cohorts (n = 128 with multiple visits totaling 357 samples). The results were meta-analyzed using the IVW method using a Bonferroni correction for the CpGs examined. Detailed information about cohort-specific RNA-seq data generation is described in Additional file 1: eMethods.
Genetic effects
To evaluate the effect of nearby (< 1 MB) polymorphisms on DNAm levels of CpGs associated with PTSD, we used cis-methylation quantitative trait locus (cis-meQTL) data from GoDMC [86] and meQTL EPIC [87] databases. For both databases, their default multiple testing adjustment was utilized: an FDR threshold of 5% in meQTL EPIC and p < 1e − 08 in GoDMC. We checked the associations between the identified cis-SNPs and PTSD in the recent Freeze 3 GWAS from PGC-PTSD [11]. Finally, we evaluated genetic associations between the genes with PTSD-associated DNAm and PTSD, using the gene-based test results from the recent PGC-PTSD Freeze 3 GWAS [11].
Cross-tissue association analyses
Blood–brain correlations
The Blood Brain DNA Methylation Comparison Tool [19] was used to assess the correlations between methylation in blood and prefrontal cortex (PFC), entorhinal cortex (EC), superior temporal gyrus (STG), and cerebellum.
Postmortem brain DNAm
DNAm measured from post-mortem brains was obtained from two studies, each of which examined a unique but not necessarily distinct set of brain regions and cohorts: the National Center for PTSD Brain Bank cohort (NCPTSD-BB [88]) and the PsychENCODE Consortium for PTSD (PEC-PTSD) Brainomics cohort [89] (see Additional file 1: eMethods for details), both of which were sourced from the Lieber Institute for Brain Development.
Methylation at PTSD-associated CpGs identified in the EWAS was tested for association with PTSD in DNA extracted from postmortem dorsolateral prefrontal cortex (dlPFC, BA9/46), ventromedial prefrontal cortex (vmPFC, BA12/32), amygdala, and dentate gyrus (DG). DNAm in the post-mortem tissue was measured using the EPIC array. We examined the associations with PTSD in dlPFC and vmPFC of 42 PTSD cases and 30 controls from the NCPTSD-BB. The associations between DNAm and PTSD in amygdala and DG were tested in 77 PTSD cases and 77 controls from the PEC-PTSD. Hypergeometric tests were used to examine if the number of CpGs nominally associated with PTSD in both blood and brain tissues is more than expected by chance (see Additional file 1: eMethods for details).
Neuronal nuclei
We examined cross-tissue association from neuronal nuclei isolation from the orbitofrontal cortex (OFC) of 25 PTSD cases and 13 healthy controls collected at the VA’s NCPTSD-BB [90]. Fluorescence-Activated Nuclei Sorting (FANS) protocol was employed to isolate NeuN + cells and the nuclei underwent reduced representation oxidative bisulfite-sequencing (RRoxBS), as previously described [91] (see Additional file 1: eMethods for details). We examined whether there was differential methylation within 500 bp of CpGs from the epigenome-wide association analyses. Four CpG sites match between the EPIC array and RRoxBS within 500 bp (cg05575921, cg21161138, cg23576855, and cg26599989).
Cellular model of prolonged stress
Methylation at PTSD-associated CpGs identified in the EWAS was tested for association with prolonged stress in DNA extracted from fibroblasts obtained from the Coriell Institute Cell Repository. EPIC array was used to measure DNAm from a cellular model of prolonged stress in which fibroblasts were subjected to physiological stress hormone (cortisol) levels for a prolonged period (51 days) as previously described [92, 93]. Student’s t-test examined DNAm differences at PTSD-associated CpGs between cortisol (cellular model of prolonged stress) and vehicle (control) groups. Each treatment group included six biological replicates.
Results
Epigenome-wide association meta-analysis
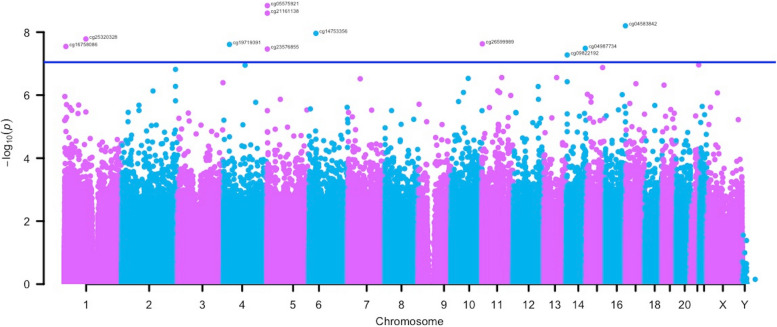
We identified 11 PTSD-associated CpGs that passed the epigenome-wide significance threshold (5.44 <|z|< 6.05, 5.3e − 08 < p < 1.4e − 09), Table 2, Fig. 1, Additional file 1: Fig. S1). Two of the CpGs near AHRR (cg05575921 and cg21161138) were associated with PTSD in the previous meta-analysis from the PGC-PTSD Epigenetics Workgroup [31], while the other 9 were novel. All CpG sites, except one site (cg21161138) near AHRR, remained nominally significant (3.17 <|z|< 5.19, 1.52e − 03 < p < 2.10e − 07) with the same direction of association in the sensitivity analysis adjusted for smoking (Additional file 1: Table S1 and Fig. S1).
Table 2.
CpG sites associated with current PTSD in the primary meta-analysis
| CpG | Position | Gene | β | SE | z | p-value |
|---|---|---|---|---|---|---|
| cg16758086* | chr1: 6,173,356 | CHD5 | 0.04 | 0.01 | 5.55 | 2.85e − 08 |
| cg25320328 | chr1:92,953,037 | GFI1 | − 0.03 | 0.01 | − 5.65 | 1.64e − 08 |
| cg19719391 | chr4:26,789,915 | Intergenic | 0.03 | 0.01 | 5.58 | 2.45e − 08 |
| cg23576855 | chr5:373,299 | AHRR | − 0.10 | 0.03 | − 5.52 | 3.44e − 08 |
| cg05575921 | chr5:373,378 | AHRR | − 0.04 | 0.01 | − 6.05 | 1.44e − 09 |
| cg21161138 | chr5:399,360 | AHRR | − 0.10 | 0.04 | − 5.96 | 2.50Ee − 09 |
| cg14753356 | chr6:30,720,108 | Intergenic | − 0.04 | 0.01 | − 5.72 | 1.09e − 08 |
| cg26599989 | chr11:1,297,087 | TOLLIP | 0.03 | 0.01 | 5.58 | 2.35e − 08 |
| cg04987734 | chr14:103,415,873 | CDC42BPB | 0.03 | 0.01 | 5.53 | 3.26e − 08 |
| cg09822192 | chr14:24,801,191 | ADCY4 | 0.04 | 0.01 | 5.44 | 5.30e − 08 |
| cg04583842 | chr16:88,103,117 | BANP | 0.04 | 0.01 | 5.81 | 6.29e − 09 |
Position is based on hg19. CpGs specific to EPIC-array were indicated with an asterisk (*). β, regression beta; SE, standard error
Fig. 1.
Manhattan plot of the epigenome-wide association meta-analyses. The x-axis depicts chromosomes and the location of each CpG site across the genome. The y-axis depicts the − log10 of the unadjusted p-value for the association with current PTSD. Each dot represents a CpG site. The solid blue line indicates the epigenome‐wide statistical significance at p < 9.0e − 8
Of the 11 PTSD-associated CpGs, 9 CpGs were annotated to a gene expressed in blood. Meta-analysis across 4 cohorts with RNAseq data identified 5 CpGs whose methylation levels were correlated with their annotated gene expression (p < 0.05), but only 3 CpGs remained significant after multiple test corrections for 9 CpGs (pBonferroni = 0.05/9 = 5.5e − 03). Specifically, methylation of cg05575921, cg21161138, and cg23576855 associated with AHRR expression (p < 5.5e − 03, − 16.69 ≤ z ≤ − 4.86; Additional file 1: Table S2). These findings highlight the potential regulatory impact of these CpGs on gene expression in the context of PTSD. Evaluation of the top 1000 CpGs did not result in any significant Gene Ontology enrichments.
Cross-tissue associations
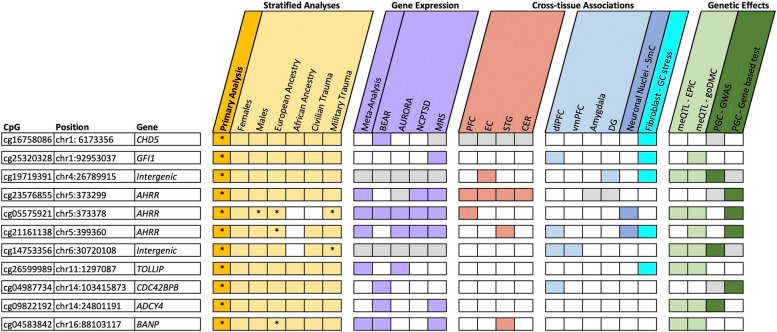
We next sought to evaluate whether these blood-based associations may reflect PTSD-associated differences in the brain. Of the 11 PTSD-associated CpGs, 5 appeared to demonstrate some degree of correlation in at least 1 brain region (Fig. 2, Additional file 1: Table S3) based on data from the Blood Brain DNA Methylation Comparison Tool database [19]. The strongest correlation was observed between the blood and PFC (r = 0.91) for cg23576855 (AHRR). Such correlation could result from either a parallel response to an environmental stimulus, such as stress, or underlying genetic variation. To investigate whether these correlations might be driven by a ubiquitous stress response, we leveraged data from a naturalistic model of stress [92]. Four CpGs exhibited significant methylation changes in fibroblasts when subjected to cortisol in a cellular model of prolonged stress after a Bonferroni correction for 11 CpGs examined (0.057 < ΔDNAm < 0.225, p < 4.5e − 03, Fig. 2, Additional file 1: Table S4). For example, methylation of cg16758086 in CHD5 increased more than 10% in response to cortisol (ΔDNAm = 0.101, p = 7.11e − 04).
Fig. 2.
Summary of all analyses and findings. The figure combines CpGs from the main analysis (gold) and stratified analyses for sex, ancestry, and trauma type (light gold); and summarizes the results of blood and brain correlations (rose); gene expression (purple); cross-tissue associations for multiple brain regions (light blue), neuronal nuclei (blue), and a fibroblast model of prolonged stress (aqua); and genetic effects, including methylation quantitative trait loci (meQTL) analyses (light green) and genetic associations from the recent PGC-PTSD GWAS (dark green). Positive findings (p < 0.05) are indicated with the specific color of the respective category. Asterisk (*) indicates epigenome-wide significance (p < 9e − 8). Gray represents the CpGs or genes that were not present in the respective datasets. PFC, prefrontal cortex; EC, entorhinal cortex; STG, superior temporal gyrus; CER, cerebellum; dlPFC, dorsolateral prefrontal cortex; vmPFC, ventromedial prefrontal cortex; DG, dentate gyrus; 5mC, 5-methylcytosine; GC, glucocorticoid
When examining the possibility of genetic-epigenetic effects, we noted that 8 CpGs were associated with at least one SNP within 1 MB according to GoDMC [86] and meQTL EPIC [94] databases (Additional file 1: Table S5). In total, we identified 26 lead meQTLs that were tested for association in the latest PGC-PTSD GWAS [11], of which 8 were nominally associated with PTSD (p < 0.05; Additional file 1: Table S5). For instance, lower methylation at the intergenic cg14753356 site is associated with PTSD (z = − 5.72, p = 1.09e − 08, Table 2) and higher methylation at cg14753356 is associated with rs28986310 T allele (beta = 0.31, p < 5e − 324), which increases the risk of PTSD (z = 6.73, p = 1.68e − 11, Additional file 1: Table S5). These data collectively suggest that some CpGs are under environmental influence, some are under genetic influence, and some are influenced by both genes and the environment. It is interesting to note that methylation of some CpGs, such as cg04987734 in CDC42BPB, did not appear to change in response to stress or to associate with underlying genetic variation. However, CDC42BPB did associate with PTSD in the recent PGC-PTSD gene-based analysis (z = 3.20, p = 6.94e − 04, Additional file 1: Table S6), suggesting that there are other mechanisms or perhaps tissue-specific regulation, underlying its association with PTSD.
We also investigated the cell type-specific expression patterns of the 7 genes identified in our study across various blood and brain cell types, using the Blood and Brain Atlas [95, 96]. The genes for which we observed a correlation between CpG methylation and expression in blood (AHRR, TOLLIP, BANP), and where methylation patterns were correlated between blood and brain (AHRR, BANP), were expressed across various immune cell types and brain regions. CHD5, GFI1, ADCY4, and CDC42BPB are expressed, albeit at low levels, across different immune cell types, and at high levels across all brain regions. In our study, the methylation of CpGs in these genes did not correlate with their expression in blood. Additionally, the methylation patterns of CpGs in these genes differed between blood and brain, potentially because these genes are not expressed in many blood cells.
Associations in postmortem brain tissues
To evaluate whether the PTSD-associated CpGs from the blood-based analysis were also associated with PTSD in the brain regions of dlPFC, vmPFC, amygdala, and DG, we first examined overall patterns of association (p < 0.05) between blood-based PTSD-associated CpGs and those associated with PTSD in each of the respective brain regions using hypergeometric tests in probes with > 10% variability across tissues. The number of nominally significant CpGs associated with PTSD in both blood and the amygdala (p = 0.009) is more than expected by chance (Additional file 1: Table S7).
Next, we evaluated the specific PTSD-associated CpGs from blood-based analyses. Six CpGs were nominally associated with PTSD in at least one brain region (p < 0.05, Fig. 2, Additional file 1: Table S8). Notably, many of the CpG sites that changed in response to that naturalistic stress model associated with PTSD in at least one postmortem brain region (i.e., cg21161138 in AHRR, cg25320328 in GFI, and intergenic cg19719391). However, only cg04987734 (CDC42BPB) in the dlPFC and intergenic cg19719391 in the DG remained associated after applying a Bonferroni correction for the 11 CpGs examined (p < 4.5e − 3, Fig. 2, Additional file 1: Table S8). Specifically, PTSD cases had higher cg04987734 (CDC42BPB) methylation both in the blood (p = 3.26e − 8) and the dlPFC (p = 3.9e − 3), and higher and intergenic cg19719391 methylation both in the blood (p = 2.45e − 8) and the DG (p = 3.04e − 3) compared to trauma-exposed controls, suggesting a robust epigenetic alteration associated with PTSD that is detectable across different tissue types.
Finally, to more specifically examine the location of PTSD-associated CpGs in the brain, we leveraged data from FACS-sorted neuronal nuclei from the OFC of PTSD cases and controls. Of the 5 CpGs that associated with PTSD in any brain region from the bulk tissue, only CpGs in AHRR (cg05575921, cg21161138) appeared to differ in those with PTSD versus controls (Additional file 1: Table S9), suggesting that the other brain-based associations may be driven by other cell types, such as glia.
Stratified analyses for sex, ancestry, and cohort type
To identify DNAm differences specific to a sex, genetic ancestry, or cohort type, stratified analyses were performed (Table 3, Additional file 1: Fig. S2). We examined the direction of effects of 4937 CpG sites that nominally associated (p < 0.05) with PTSD in both the European and African ancestry strata (Additional file 1: Fig. S3A) and 4265 CpG sites that nominally associated (p < 0.05) with PTSD in both the male and female strata (Additional file 1: Fig. S3B). While there were significant correlations in effect sizes across analyses stratified by ancestry (r = 0.48, p < 2.2e − 16) and sex (r = 0.32, p < 2.2e − 16), and directions of associations of the 11 PTSD-associated CpGs from the primary analysis were largely consistent across strata, some unique associations emerged (Additional file 1: Table S10). We identified 1 epigenome-wide significant CpG site (cg25691167 in FERD3L) associated with PTSD in the female-stratified analysis (z = 5.48, p = 4.24e − 8; Additional file 1: Fig. S4). The 1 significant CpG site (cg05575921 in AHRR) in the male-stratified analysis (z = − 6.12, p = 9.3e − 10; Additional file 1: Fig. S5) was also identified in the primary analysis (Fig. 2). Five CpGs were associated with PTSD in European ancestry-stratified analysis (5.43 <|z|< 6.85, 5.7e − 08 < p < 7.2e − 12; Additional file 1: Fig. S6), of which 2 were unique and 3 were identified in the primary analysis (Fig. 2). One CpG (cg02003183 in CDC42BPB) was associated with PTSD in the African ancestry-stratified analysis (z = 5.48, p = 4.26e − 8; Additional file 1: Fig. S7). When examining cohort type, 1 CpG (cg27541344 in BCL11B) was associated with PTSD in the analysis of civilian cohorts (z = 5.39, p = 7.21e − 8; Additional file 1: Fig. S8). Finally, 5 CpGs were associated with PTSD in the military cohorts (5.36 <|z|< 6.22, 8.5e − 08 < p < 4.9e − 10; Additional file 1: Fig. S9), of which 3 were unique and 2 were identified in the primary analysis (Fig. 2).
Table 3.
CpG sites associated with current PTSD in the stratified analyses
| CpG | Position | Gene | β | SE | z | p-value |
|---|---|---|---|---|---|---|
| Stratified analysis for females | ||||||
| cg25691167 | chr7:19184961 | FERD3L | 0.09 | 0.03 | 5.48 | 4.24E-08 |
| Stratified analysis for males | ||||||
| cg05575921 | chr5:373378 | AHRR | -0.14 | 0.04 | -6.12 | 9.30E-10 |
| Stratified analysis for European ancestry | ||||||
| cg05575921 | chr5:373378 | AHRR | 0.05 | 0.02 | -6.85 | 7.24E-12 |
| cg21161138 | chr5:399360 | AHRR | -0.16 | 0.05 | -5.62 | 1.95E-08 |
| cg11256214 | chr12:110211642 | MGC14436 | 0.06 | 0.02 | 5.76 | 8.17E-09 |
| cg15977432 | chr19:56709655 | Intergenic | -0.05 | 0.02 | -5.46 | 4.68E-08 |
| cg04583842 | chr16:88103117 | BANP | 0.05 | 0.02 | 5.43 | 5.66E-08 |
| Stratified analysis for African ancestry | ||||||
| cg02003183 | chr14:103415882 | CDC42BPB | 0.11 | 0.04 | 5.48 | 4.26E-08 |
| Stratified analysis for civilian trauma | ||||||
| cg27541344* | chr14:99650422 | BCL11B | 0.02 | 0.04 | 5.39 | 7.21E-08 |
| Stratified analysis for military trauma | ||||||
| cg03329539 | chr2:233283329 | Intergenic | -0.03 | 0.01 | -5.36 | 8.53E-08 |
| cg21566642 | chr2:233284661 | Intergenic | -0.14 | 0.04 | -5.41 | 6.36E-08 |
| cg14753356 | chr6:30720108 | Intergenic | -0.04 | 0.02 | -5.47 | 4.53E-08 |
| cg05575921 | chr5:373378 | AHRR | -0.07 | 0.02 | -6.22 | 4.92E-10 |
| cg00774777* | chr11:76478902 | RP11-21L23.4 | 0.03 | 0.01 | 5.55 | 2.79E-08 |
Position is based on hg19. The sites that were also epigenome-wide significant (p < 9e-08) in the primary meta-analysis were shown in bold. CpGs specific to EPIC-array were indicated with an asterisk (*). β: Regression beta. SE: Standard error.
We identified 3 GO term enrichments in the female-stratified analysis (FDR < 0.05), including nervous system development (Additional file 1: Table S11). We did not identify any significant enrichments in other strata. These findings suggest that PTSD-associated DNAm patterns related to nervous system development may exhibit sex-specific patterns. The absence of significant GO term enrichments in the main analysis or other stratified analyses highlights the potential importance of considering sex-specific factors in epigenomic studies.
Discussion
In the present epigenome-wide meta-analysis of blood DNAm levels, we identified 11 CpG sites associated with PTSD, of which 2 had been identified in a prior meta-analysis of PTSD and 9 were novel. Because this study was conducted in blood, and PTSD is a brain-based disorder, we were interested in the degree to which we would observe the association of these blood-based CpGs in different brain regions implicated in PTSD along with other large-scale discoveries of PTSD. We noted overall enrichment of PTSD-associated CpGs in the amygdala and dentate gyrus. In addition, many loci showed blood–brain methylation correlations and cross-tissue associations with PTSD, with significant correlations between CpG site methylation levels and their respective gene expression levels.
We observed 3 CpGs (cg05575921, cg21161138, and cg23576855) in AHRR (aryl-hydrocarbon receptor repressor), 2 of which were identified in an earlier PGC-PTSD EWAS [31] and an independent study of US veterans [88]. DNA methylation at the AHRR CpGs is known to be influenced by smoking [97], and our effect sizes were attenuated when controlling for a DNAm-based smoking score. However, our previous study demonstrated that these associations were driven by non-smokers and were likely to be independent of smoking [31]. This is consistent with our finding that methylation of AHRR CpG sites changes in an in vitro model of naturalistic stress [92] and with the observation that AHRR associates with gene-based tests of PTSD from the PGC-PTSD GWAS [11]. The aryl hydrocarbon receptor (AhR) plays a role in immunomodulation, including the regulation of T lymphocytes, B cell maturation, and the activity of macrophages, dendritic cells, and neutrophils [98], supporting the link between the immune system and PTSD. Interestingly, AHRR methylation patterns in the blood of those with PTSD were associated with tryptophan metabolites, including the lower kynurenine and kynurenic acid levels [31]. Notably, cg21161138 DNAm was also associated with PTSD in postmortem dlPFC and neuronal nuclei from the OFC. Collectively, these data support that AHRR plays a role in PTSD that is independent of smoking status and warrants further mechanistic studies.
Our CDC42BPB (CDC42 binding protein kinase beta) findings are of particular interest. CDC42BPB is involved in the regulation of cytoskeletal rearrangement, cell migration, and neurodevelopment [99]. In the current study, increased CDC42BPB methylation at cg04987734 was associated with PTSD in both blood and the dlPFC. Notably, higher methylation at cg04987734 has been associated with depressive symptoms [100] and increased C-reactive protein (CRP) levels [101, 102], which is perhaps not surprising given the genetic correlation between PTSD and MDD [11] and the bi-directional genetic association between PTSD and CRP levels [103]. Multiple studies reported increased CRP levels and other inflammatory markers in those with PTSD, suggesting inflammation as an important component of PTSD [104–106]. Future studies are warranted to investigate the role of CDC42BPB in psychiatric disorders and the degree to which CDC42BPB methylation varies with respect to PTSD onset and treatment response.
We also identified PTSD-associated CpGs in genes GFI1, CHD5, TOLLIP, ADCY4, and BANP. While the precise mechanisms linking these genes to PTSD are not entirely understood, evidence suggests that they are responsive to stress and have been implicated in stress-related disorders, immune response, and other psychiatric disorders [107–111]. Collectively, these genes may contribute to PTSD through alterations in gene expression, synaptic and neural plasticity, and neuroimmune interactions. Future research should focus on elucidating the specific pathways and mechanisms by which these genes influence stress response and PTSD.
The stratified analyses identified DNAm-PTSD associations specific to sex, ancestry, and cohort type to identify DNAm differences that may be specific to strata, such as hormonal factors that underlie sex differences or occupational exposures related to military service. The PTSD-associated site cg25691167 (FERD3L) in females (p = 4.24e − 08) was not associated with PTSD in males (p = 0.57), suggesting that DNAm changes in cg25691167 might be sex-specific. The FERD3L (Fer3-like bHLH transcription factor) gene is a transcription factor involved in various developmental processes, particularly in neurogenesis [112], which is consistent with our pathway enrichment findings in the female-stratified analysis that identified nervous system development. Similarly, cg27541344 (BCL11B) was associated with PTSD in the civilian (p = 7.21e − 08), but not the military cohorts (p = 0.38), whereas 3 out of 5 PTSD-associated CpGs in the military cohorts were not significant in the civilian cohorts (p > 0.05). Though these associations are promising and warrant further study, it is important to note that the sex-stratified analyses may be confounded by the fact that many male-dominant cohorts are military cohorts, where specific factors such as circadian rhythm, age, and diet during military practices may drive the observed sex-specific findings.
Strengths and limitations
To our knowledge, this is the largest EWAS of PTSD to date. Our sample is diverse in terms of sex, ancestry, and cohort type. We leveraged data from postmortem brain samples, a cellular model of prolonged stress, GWAS, and genome-wide expression data to support our findings. However, the study is not without limitations. First, methylation arrays only assess a subset of CpG sites in the genome; therefore, we may not capture all PTSD-associated CpG sites. Second, this is a cross-sectional study of participants with prior exposure to a traumatic event; thus, we were not able to assess whether the differences in DNAm between individuals with and without PTSD are a cause or consequence of PTSD or both. Third, our primary meta-analysis was performed using measures of blood DNAm. While this strategy provided valuable insights for future research on biomarkers of PTSD, it might not accurately represent the DNAm patterns within other tissues that are likely the most relevant to PTSD. However, the majority of the PTSD-associated CpG sites’ methylation levels were correlated between blood and at least one brain region. In addition, most PTSD-associated CpGs in blood were also associated with PTSD in one or more brain regions. Fourth, we do not have cell-type-specific DNAm and gene expression information necessary for further examination of cell-specific gene regulation across blood and brain cell types. Hence, we used bulk tissue and adjusted for cellular heterogeneity, which might have obscured some signals, given the alterations in cell composition in those with PTSD [113, 114]. Additionally, the brain regions examined varied between the online databases used to examine the blood–brain correlation of methylation values and the gene expression differences associated with PTSD, which can complicate interpretation. Finally, most cohorts that participated in the meta-analysis did not have detailed physical or psychiatric information on participants, including detailed information on chronicity, trauma type and timing, PTSD symptom course, and treatment, making it challenging to evaluate and adjust for potential confounders, including substance use, comorbidities, or medication use. The lack of detailed information on the types and timing of traumatic experiences across cohorts prevents us from making definitive conclusions about the impact of different trauma types on DNAm. Future studies are warranted to examine DNAm changes longitudinally, tracking participants both before and after trauma exposure to capture the dynamic epigenetic modifications that occur in response to traumatic events, providing a clearer understanding of how trauma influences DNAm over time.
Conclusions
Taken together, this study replicates our previous findings and identifies novel PTSD-associated CpGs. Supporting data from multiple sources suggest that epigenetic mechanisms, particularly methylation in AHRR and CDC42BPB, may contribute to the complex relationship between the immune system and PTSD.
Supplementary Information
Additional file 1: Table S1. PTSD-associated CpGs from the primary analysis in the sensitivity analysis adjusted for smoking score. Table S2. DNA methylation and cis-gene expression (RNAseq) correlations. Table S3. Correlation values of methylation between blood and brain regions from Blood Brain DNA Methylation Comparison Tool. Table S4. Results of PTSD-associated CpGs from blood in fibroblasts. Table S5. Significant SNPs from cis-meQTL analysis and their association with PTSD. Table S6. Genetic associations of the identified PTSD-associated genes from gene-based tests. Table S7. Results of hypergeometric tests in postmortem brain tissues. Table S8. Results of PTSD-associated CpGs from blood in the brain. Table S9. Results of PTSD-associated CpGs from blood in neuronal nuclei. Table S10. PTSD-associated CpGs from the primary analysis in the stratified analysis for sex, ancestry, and trauma type. Table S11. Gene Ontology Enrichment results in females. Fig S1. Forest Plots of PTSD-associated CpGs in each cohort. Fig S2. Manhattan plots of the stratified analyses. Fig S3. Correlation of effect sizes based on stratified analyses for ancestry and sex. Fig S4. Forest Plots of CpGs associated with PTSD in the stratified analysis for females. Fig S5. Forest Plots of CpGs associated with PTSD in the stratified analysis for males. Fig S6. Forest Plots of CpGs associated with PTSD in the stratified analysis for European ancestry. Fig S7. Forest Plots of CpGs associated with PTSD in the stratified analysis for African ancestry. Fig S8. Forest Plots of CpGs associated with PTSD in the stratified analysis for civilian trauma. Fig S9. Forest Plots of CpGs associated with PTSD in the stratified analysis for military trauma.
Additional file 2. Summary statistics of the main analysis.
Additional file 3. Summary statistics of the smoking-sensitivity analysis.
Additional file 4. Summary statistics of the stratified analysis for males.
Additional file 5. Summary statistics of the stratified analysis for females.
Additional file 6. Summary statistics of the stratified analysis for European ancestry.
Additional file 7. Summary statistics of the stratified analysis for African ancestry.
Additional file 8. Summary statistics of the stratified analysis for civilian cohorts.
Additional file 9. Summary statistics of the stratified analysis for military cohorts.
Acknowledgements
PGC-PTSD Epigenetics Workgroup: Reid S. Alisch, Ananda B. Amstadter, Don Armstrong, Archana Basu, Jean C. Beckham, Nicole L. Bjorklund, Barbara H. Chaiyachati, Judith B. M. Ensink, Segun Fatumo, Leland L. Fleming, Sandro Galea, Joel Gelernter, Ryan J. Herringa, Sonia Jain, Diana L. Juvinao-Quintero, Seyma Katrinli, Elizabeth Ketema, José J Martínez-Magaña, Burook Misganaw, Shiela Tiemi Nagamatsu, Danny M. Nispeling, John Pfeiffer, Christian Schmahl, Gen Shinozaki, Clara Snijders, Jennifer A. Sumner, Patricia C. Swart, Audrey Tyrka, Robert J. Ursano, Mirjam van Zuiden, Eric Vermetten, Jaqueline S. Womersley, Nagy A. Youssef, Yuanchao Zheng, Yiwen Zhu, Lea Zillich
PsychENCODE PTSD Brainomics Project: Dhivya Arasappan, Sabina Berretta, Rahul A. Bharadwaj, Frances A. Champagne, Leonardo Collado-Torres, Christos Chatzinakos, Nikolaos P. Daskalakis, Chris P. DiPietro, Duc M. Duong, Amy Deep-Soboslay, Nick Eagles, Louise Huuki, Thomas Hyde, Artemis Iatrou, Aarti Jajoo, Joel E. Kleinman, Charles B. Nemeroff, Geo Pertea, Deanna Ross, Nicholas T. Seyfried, Joo Heon Shin, Kerry J. Ressler, Clara Snijders, Ran Tao, Daniel R. Weinberger, Stefan Wuchty, Dennis Wylie
Traumatic Stress Brain Research Group: Victor E. Alvarez, David Benedek, Alicia Che, Dianne A. Cruz, David A. Davis, Matthew J. Girgenti, Ellen Hoffman, Paul E. Holtzheimer, Bertrand R. Huber, Alfred Kaye, John H. Krystal, Adam T. Labadorf, Terence M. Keane, Ann McKee, Brian Marx, Crystal Noller, Meghan Pierce, William K. Scott, Paula Schnurr, Krista DiSano, Thor Stein,
Douglas E. Williamson, Keith A. Young
Abbreviations
- CpG
Cytosine-guanine dinucleotides
- CRP
C-reactive protein
- DG
Dentate gyrus
- dlPFC
Dorsolateral prefrontal cortex
- DNAm
DNA methylation
- EC
Entorhinal cortex
- EWAS
Epigenome-wide association study
- FANS
Fluorescence-Activated Nuclei Sorting
- FDR
False discovery rate
- GWAS
Genome-wide association study
- IVW
Inverse-variance weighted
- meQTL
Methylation quantitative trait locus
- NK
Natural killer
- OFC
Orbitofrontal cortex
- PC
Principal component
- PFC
Prefrontal cortex
- PGC-PTSD
Psychiatric Genomics Consortium PTSD Workgroup
- PTSD
Posttraumatic stress disorder
- QC
Quality control
- RPC
Robust partial correlation
- RRoxBS
Reduced representation oxidative bisulfite-sequencing
- STG
Superior temporal gyrus
- vmPFC
Ventromedial prefrontal cortex
Authors’ contributions
PGC-PTSD writing group: S.K., M.W.L., C.M.N., A.K.S., and M.U. Study PI or co-PI: A.E.A., A.E.A.-K., D.G.B., J.C.B., E.B., C.F., S.G., E.G., G.G., M.A.H., S.J., R.C.K., N.A.K., J.E.K., K.C.K., P.-F.K., M.W.L., B.L., C.E.M., S.A.M., W.M., M.W.M., L.M., C.B.N., C.M.N., N.R.N., H.K.O., S.A.M.R., K.J.R., V.R., S.S., A.K.S., D.J.S., M.B.S., M.U., R.J.U., E.V., D.E.W., E.J.W., and R.M.Y. Obtained funding for studies: J.C.B., M.P.B., S.F., C.F., E.G., M.A.H., R.C.K., K.C.K., M.W.L., J.J.L., S.A.M., M.W.M., C.M.N., N.R.N., H.K.O., S.A.M.R., K.J.R., B.P.F.R., A.K.S., M.U., R.J.U., and E.V. Clinical: D.G.B., J.C.B., M.F.D., S.F., C.F., E.G., J.P.H., N.A.K., N.K., C.M., J.M., S.A.M.R., K.J.R., E.R., A.U., M.H.V., E.V., E.J.W., and L.L.V.D.H. Contributed data: A.E.A.-K., D.G.B., J.C.B., F.A.C., N.P.D., M.F.D., C.F., M.A.H., J.P.H., S.M.J.H., B.R.H., S.J., S.K., N.A.K., A.P.K., K.C.K., P.-F.K., I.L., A.L, B.L., J.J.L., D.M., W.M., M.W.M., J.M.-O., C.M., L.M., C.M.N., N.R.N., S.A.M.R., V.R., S.S., A.K.S., M.B.S., S.T., M.U., A.U., M.H.V., D.E.W., E.J.W., A.S.Z., and L.L.V.D.H. Statistical analysis: A.E.A.-K., D.A., M.P.B., L.B., C.-Y.C., S.D., N.P.D., S.F., M.E.G., A.J., S.K., A.P.K., I.L., M.W.L., A.X.M., D.M., M.S.M., C.M.N., D.L.N.-R., X-J.Q., A.R., B.P.F.R., A.K.S., C.H.V., A.H.W., E.B.W., A.S.Z., X.Z., and Y.Z. Bioinformatics: M.P.B., L.B., C.-Y.C., N.P.D., M.E.G., M.A.H., A.J., S.K., S.D.L., M.W.L., A.X.M., A.R., B.P.F.R., A.H.W., X.Z., and Y.Z. Genomics: M.P.B., M.A.H., S.D.L., A.X.M., B.P.F.R., C.H.V., A.S.Z., and X.Z. PI of the EWAS group: M.W.L., C.M.N., A.K.S., and M.U.
Funding
This work was supported by the National Institute of Mental Health (NIMH; R01MH108826 and R01MH106595). This work was also supported by I01BX003477, a Department of Veterans Affairs BLR&D grant to MWL; 1R03AG051877, 1R21AG061367-01, RF1AG068121, and 1I01CX001276-01A2 to EJW; R21MH102834, 5I01CX000431, 5R01MH079806 to MWM; R01MD011728 to MU; R01MH105379 to NRN; R01MH117291, R01MH117292, R01MH117293 to FAC; R01MH108826 to NPD; U01MH115485 to LM, SF, SJ, JM, AU, and DEW; R01MH117291, R01MH117292, R01MH117293 to JEK; CDC/NIOSH U01OH012466 to PFK; R01MH117291, R01MH117292, R01MH117293 to CBN; R01MH093500 to CMN; 1R15MH099521-01, 5R21MH085436-02, 1R15HD049907-01A1 to HKO; R01MH108826 to AKS; I01BX002577, IK2CX000525, and lK6BX003777 to NAK; U01MH110925 to SAM; R21DA050160 and 1DP1DA058737 to JMO; Department of Veterans Affairs B9254‐C to WM, B3001-C to CF; Department of Defense W81XWH-11–1-0073 and the National Center for Advancing Translational Sciences of the NIH UL1TR000433PEC-PTSD to SAMR; BX006186; BX005872 to VBR; VIDI award (91718336) from the Netherlands Scientific Organization to BPFR; Dutch Research Council (NWO) VIDI grant (09150171910042) to CHV. Brainomics work was supported by R01MH117291, R01MH117292, and R01MH117293. DJS and NK were supported by the South African Medical Research Council and the Bill and Melinda Gates Foundation (OPP 1017641). SS is supported by the South African Medical Research Council. SK is supported by the Building Interdisciplinary Research Careers in Women’s Health of the National Institutes of Health under Award Number K12HD085850.
Data availability
The main summary statistics data that support the findings of this study will be available within Supplementary Data upon publication. Individual-level data from the cohorts or cohort-level summary statistics will be made available to researchers following an approved analysis proposal through the PGC-PTSD Epigenetics Workgroup with the agreement of the cohort PIs. The raw data for the GTP cohort is available in the Gene Expression Omnibus database with the accession code GSE132203 [115]. Owing to limitations on data sharing as specified in the consent process and/or restrictions as part of the use agreement under which the data was accessed, data from the military/VA cohorts, VA MIRECC, MRS, Army STARRS, PRISMO, PROGrESS, and NCPTSD/TRACTS, cannot be publicly posted as part of this project. However, such data can be provided in de-identified form through a data use agreement following applicable guidelines on data sharing and privacy protection. For additional information on access to these data, including PI contact information for the cohorts accessed under a DUA, please contact the corresponding author.
Declarations
Ethics approval and consent to participate
All studies were approved by the institutional review boards (IRBs) of each respective institution and carried out in accordance with The Code of Ethics of the Helsinki Declaration. The BEAR study was approved by the Lifespan Hospitals IRB. DNHS was approved by the IRB at the University of Michigan and the University of North Carolina at Chapel Hill. DCHS was approved by the Faculty of Health Sciences, Human Research Ethics Committee, University of Cape Town, by Stellenbosch University, and by the Western Cape Provincial Health Research committee. GTP was approved by the IRBs of Emory University School of Medicine and the Research Oversight Committee of Grady Memorial Hospital. NIU study was approved by the Northern Illinois University IRB. Shared Roots study was approved by the IRB of Stellenbosch University (HREC: N13/08/115). AURORA is approved by the Biomedical IRB at UNC Chapel Hill through the office of Human Research Ethics, the central IRB for all study sites. H3_Rwanda was approved by the IRB of the College of Medicine and Health Sciences at the University of Rwanda (No.370/CMHS/IRB/2020). WTC study was approved by the Committees on Research Involving Human Subjects at Stony Brook University. GMRFQUT was approved by the Human Research Ethics Committee of the Queensland University of Technology and Greenslopes Private Hospital. MRS was approved by the IRBs of the University of California San Diego, VA San Diego Research Service, and Naval Health Research Center. PRISMO was approved by the IRB of the University Medical Center Utrecht. Army STARRS was approved by the Human Subjects Committees of the Uniformed Services University of the Health Sciences for the Henry M. Jackson Foundation (the primary grantee), the Institute for Social Research at the University of Michigan (the organization collecting the data), and all other collaborating organizations. PROGrESS was approved by the IRBs at VA Ann Arbor Healthcare System, the University of Michigan, Ralph H. Johnson VA Medical Center, VA San Diego Healthcare System, Massachusetts General Hospital, and the Department of Defense Human Research Protection Office. NCPTSD/TRACTS was approved by the IRBs of Human Studies Research at the VA Boston Healthcare System. INTRuST was approved by the Human Research Protection Program at the University of California San Diego. VA-M-AA and VA-M-EA were approved by the IRBs at the Salisbury VA, Hampton VA, Durham VA, and Duke University Medical Centers. All individuals provided written informed consent to participate in these studies.
Consent for publication
Not applicable.
Competing interests
CYC is an employee of Biogen Inc. NPD has served on scientific advisory boards for BioVie Pharma, Circular Genomics, and Sentio Solutions for unrelated work. NRN serves as an unpaid member of the Ilumivu advisory board. SAMR receives support from the Wounded Warrior Project (WWP), Department of Veterans Affairs (VA), National Institute of Health (NIH), McCormick Foundation, Tonix Pharmaceuticals, Woodruff Foundation, and Department of Defense (DOD). Dr. Rauch receives royalties from Oxford University Press and American. KJR serves as a consultant for Acer, Bionomics, and Jazz Pharma; SABs for Sage, Boehringer Ingelheim, and Senseye. DJS has received consultancy honoraria from Discovery Vitality, Johnson & Johnson, Kanna, L’Oreal, Lundbeck, Orion, Sanofi, Servier, Takeda and Vistagen. MBS has in the past 3 years received consulting income from Acadia Pharmaceuticals, Aptinyx, atai Life Sciences, BigHealth, Biogen, Bionomics, BioXcel Therapeutics, Boehringer Ingelheim, Clexio, Delix Therapeutics, Eisai, EmpowerPharm, Engrail Therapeutics, Janssen, Jazz Pharmaceuticals, NeuroTrauma Sciences, PureTech Health, Sage Therapeutics, Sumitomo Pharma, and Roche/Genentech. MBS has stock options in Oxeia Biopharmaceuticals and EpiVario. MBS has been paid for his editorial work on Depression and Anxiety (Editor-in-Chief), Biological Psychiatry (Deputy Editor), and UpToDate (Co-Editor-in-Chief for Psychiatry). MBS has also received research support from NIH, the Department of Veterans Affairs, and the Department of Defense. MBS is on the scientific advisory board for the Brain and Behavior Research Foundation and the Anxiety and Depression Association of America.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mark W. Logue, Email: loguem@bu.edu
PGC-PTSD Epigenetics Workgroup:
Seyma Katrinli, Jean C. Beckham, Segun Fatumo, Sandro Galea, Robert J. Ursano, Eric Vermetten, Reid S. Alisch, Ananda B Amstadter, Don Armstrong, Archana Basu, Nicole L Bjorklund, Barbara H Chaiyachati, Judith B M Ensink, Leland L Fleming, Joel Gelernter, Ryan J Herringa, Sonia Jain, Diana L Juvinao-Quintero, Elizabeth Ketema, José J Martínez-Magaña, Burook Misganaw, Shiela Tiemi Nagamatsu, Danny M Nispeling, John Pfeiffer, Christian Schmahl, Gen Shinozaki, Clara Snijders, Jennifer A Sumner, Patricia C Swart, Audrey Tyrka, Mirjam van Zuiden, Jaqueline S Womersley, Nagy A Youssef, Yuanchao Zheng, Yiwen Zhu, and Lea Zillich
PsychENCODE PTSD Brainomics Project:
Nikolaos P. Daskalakis, Frances A. Champagne, Aarti Jajoo, Joel E. Kleinman, Charles B. Nemeroff, Dhivya Arasappan, Sabina Berretta, Rahul A. Bharadwaj, Leonardo Collado-Torres, Christos Chatzinakos, Chris P. DiPietro, Duc M. Duong, Amy Deep-Soboslay, Nick Eagles, Louise Huuki, Thomas Hyde, Artemis Iatrou, Geo Pertea, Deanna Ross, Nicholas T. Seyfried, and Joo Heon Shin
Traumatic Stress Brain Research Group:
Bertrand Russel Huber, Victor E. Alvarez, David Benedek, Alicia Che, Dianne A. Cruz, David A. Davis, Matthew J. Girgenti, Ellen Hoffman, Paul E. Holtzheimer, Alfred Kaye, John H. Krystal, Adam T. Labadorf, Terence M. Keane, Ann McKee, Brian Marx, Crystal Noller, Meghan Pierce, William K. Scott, Paula Schnurr, Krista DiSano, Thor Stein, Douglas E. Williamson, and Keith A. Young
References
- 1.American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 2013.
- 2.Zen AL, Whooley MA, Zhao S, Cohen BE. Post-traumatic stress disorder is associated with poor health behaviors: findings from the heart and soul study. Health Psychol. 2012;31(2):194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacobsen LK, Southwick SM, Kosten TR. Substance use disorders in patients with posttraumatic stress disorder: a review of the literature. Am J Psychiatry. 2001;158(8):1184–90. [DOI] [PubMed] [Google Scholar]
- 4.O’Donovan A, Cohen BE, Seal KH, Bertenthal D, Margaretten M, Nishimi K, et al. Elevated risk for autoimmune disorders in iraq and afghanistan veterans with posttraumatic stress disorder. Biol Psychiatry. 2015;77(4):365–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryder AL, Azcarate PM, Cohen BE. PTSD and physical health. Curr Psychiatry Rep. 2018;20(12):116. [DOI] [PubMed] [Google Scholar]
- 6.Benjet C, Bromet E, Karam EG, Kessler RC, McLaughlin KA, Ruscio AM, et al. The epidemiology of traumatic event exposure worldwide: results from the World Mental Health Survey Consortium. Psychol Med. 2016;46(2):327–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Logue MW, Amstadter AB, Baker DG, Duncan L, Koenen KC, Liberzon I, et al. The psychiatric genomics consortium posttraumatic stress disorder workgroup: posttraumatic stress disorder enters the age of large-scale genomic collaboration. Neuropsychopharmacology. 2015;40(10):2287–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nievergelt CM, Ashley-Koch AE, Dalvie S, Hauser MA, Morey RA, Smith AK, et al. Genomic approaches to posttraumatic stress disorder: the psychiatric genomic consortium initiative. Biol Psychiatry. 2018;83(10):831–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polimanti R, Wendt FR. Posttraumatic stress disorder: from gene discovery to disease biology. Psychol Med. 2021;51(13):2178–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan LE, Cooper BN, Shen H. Robust findings from 25 years of PTSD Genetics Research. Curr Psychiatry Rep. 2018;20(12):115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nievergelt CM, Maihofer AX, Atkinson EG, Chen CY, Choi KW, Coleman JRI, et al. Genome-wide association analyses identify 95 risk loci and provide insights into the neurobiology of post-traumatic stress disorder. Nat Genet. 2024;56(5):792–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daskalakis NP, Rijal CM, King C, Huckins LM, Ressler KJ. Recent genetics and epigenetics approaches to PTSD. Curr Psychiatry Rep. 2018;20(5):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zannas AS, Provencal N, Binder EB. Epigenetics of posttraumatic stress disorder: current evidence, challenges, and future directions. Biol Psychiatry. 2015;78(5):327–35. [DOI] [PubMed] [Google Scholar]
- 14.Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330(6004):612–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker-Andresen D, Ratnu VS, Bredy TW. Dynamic DNA methylation: a prime candidate for genomic metaplasticity and behavioral adaptation. Trends Neurosci. 2013;36(1):3–13. [DOI] [PubMed] [Google Scholar]
- 16.Rasmi Y, Shokati A, Hassan A, Aziz SG, Bastani S, Jalali L, et al. The role of DNA methylation in progression of neurological disorders and neurodegenerative diseases as well as the prospect of using DNA methylation inhibitors as therapeutic agents for such disorders. IBRO Neurosci Rep. 2023;14:28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murgatroyd C, Wu Y, Bockmuhl Y, Spengler D. Genes learn from stress: how infantile trauma programs us for depression. Epigenetics. 2010;5(3):194–9. [DOI] [PubMed] [Google Scholar]
- 18.Lokk K, Modhukur V, Rajashekar B, Martens K, Magi R, Kolde R, et al. DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome Biol. 2014;15(4): r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hannon E, Lunnon K, Schalkwyk L, Mill J. Interindividual methylomic variation across blood, cortex, and cerebellum: implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics. 2015;10(11):1024–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daskalakis NP, Cohen H, Nievergelt CM, Baker DG, Buxbaum JD, Russo SJ, et al. New translational perspectives for blood-based biomarkers of PTSD: From glucocorticoid to immune mediators of stress susceptibility. Exp Neurol. 2016;284(Pt B):133–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrison FG, Miller MW, Logue MW, Assef M, Wolf EJ. DNA methylation correlates of PTSD: recent findings and technical challenges. Prog Neuropsychopharmacol Biol Psychiatry. 2019;90:223–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katrinli S, Oliveira NCS, Felger JC, Michopoulos V, Smith AK. The role of the immune system in posttraumatic stress disorder. Transl Psychiatry. 2022;12(1):313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, de Los SR, et al. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proc Natl Acad Sci U S A. 2010;107(20):9470–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith AK, Conneely KN, Kilaru V, Mercer KB, Weiss TE, Bradley B, et al. Differential immune system DNA methylation and cytokine regulation in post-traumatic stress disorder. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(6):700–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuan PF, Waszczuk MA, Kotov R, Marsit CJ, Guffanti G, Gonzalez A, et al. An epigenome-wide DNA methylation study of PTSD and depression in World Trade Center responders. Transl Psychiatry. 2017;7(6): e1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rutten BPF, Vermetten E, Vinkers CH, Ursini G, Daskalakis NP, Pishva E, et al. Longitudinal analyses of the DNA methylome in deployed military servicemen identify susceptibility loci for post-traumatic stress disorder. Mol Psychiatry. 2018;23(5):1145–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Logue MW, Miller MW, Wolf EJ, Huber BR, Morrison FG, Zhou Z, et al. An epigenome-wide association study of posttraumatic stress disorder in US veterans implicates several new DNA methylation loci. Clin Epigenetics. 2020;12(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammamieh R, Chakraborty N, Gautam A, Muhie S, Yang R, Donohue D, et al. Whole-genome DNA methylation status associated with clinical PTSD measures of OIF/OEF veterans. Transl Psychiatry. 2017;7(7): e1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ratanatharathorn A, Boks MP, Maihofer AX, Aiello AE, Amstadter AB, Ashley-Koch AE, et al. Epigenome-wide association of PTSD from heterogeneous cohorts with a common multi-site analysis pipeline. Am J Med Genet B Neuropsychiatr Genet. 2017;174(6):619–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uddin M, Ratanatharathorn A, Armstrong D, Kuan PF, Aiello AE, Bromet EJ, et al. Epigenetic meta-analysis across three civilian cohorts identifies NRG1 and HGS as blood-based biomarkers for post-traumatic stress disorder. Epigenomics. 2018;10(12):1585–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith AK, Ratanatharathorn A, Maihofer AX, Naviaux RK, Aiello AE, Amstadter AB, et al. Epigenome-wide meta-analysis of PTSD across 10 military and civilian cohorts identifies methylation changes in AHRR. Nat Commun. 2020;11(1):5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snijders C, Maihofer AX, Ratanatharathorn A, Baker DG, Boks MP, Geuze E, et al. Longitudinal epigenome-wide association studies of three male military cohorts reveal multiple CpG sites associated with post-traumatic stress disorder. Clin Epigenetics. 2020;12(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katrinli S, Maihofer AX, Wani AH, Pfeiffer JR, Ketema E, Ratanatharathorn A, et al. Epigenome-wide meta-analysis of PTSD symptom severity in three military cohorts implicates DNA methylation changes in genes involved in immune system and oxidative stress. Mol Psychiatry. 2022;27(3):1720–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nugent NR, Armey M, Boker S, Brick L, Knopik V, McGeary JE, et al. Adolescents hospitalised for suicidality: biomarkers, social and affective predictors: a cohort study. BMJ Open. 2022;12(10): e056063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pynoos R, Weathers F, Steinberg A, Marx B, Layne C, Kaloupek D, et al. Clinician-administered PTSD scale for DSM-5 - child/adolescent version. 2015.
- 36.Blanchard EB, Jones-Alexander J, Buckley TC, Forneris CA. Psychometric properties of the PTSD checklist (PCL). Behav Res Ther. 1996;34(8):669–73. [DOI] [PubMed] [Google Scholar]
- 37.Stein DJ, Koen N, Donald KA, Adnams CM, Koopowitz S, Lund C, et al. Investigating the psychosocial determinants of child health in Africa: the Drakenstein child health study. J Neurosci Methods. 2015;252:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lecrubier Y, Sheehan DV, Weiller E, Amorim P, Bonora I, Sheehan KH, et al. The Mini International Neuropsychiatric Interview (MINI). A short diagnostic structured interview: reliability and validity according to the CIDI. European psychiatry. 1997;12(5):224–31. [Google Scholar]
- 39.Sheehan DV, Lecrubier Y, Sheehan KH, Amorim P, Janavs J, Weiller E, et al. The Mini-International Neuropsychiatric Interview (M.I.N.I): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. The Journal of Clinical Psychiatry. 1998;59(Suppl 20):22–33. [PubMed] [Google Scholar]
- 40.Blake DD, Weathers FW, Nagy LM, Kaloupek DG, Gusman FD, Charney DS, et al. The development of a clinician-administered PTSD scale. J Trauma Stress. 1995;8(1):75–90. [DOI] [PubMed] [Google Scholar]
- 41.Weathers FW, Keane TM, Davidson JR. Clinician-administered PTSD scale: a review of the first ten years of research. Depress Anxiety. 2001;13(3):132–56. [DOI] [PubMed] [Google Scholar]
- 42.Falsetti SA, Resnick HS, Resick PA, Kilpatrick DG. The modified PTSD symptom scale: a brief self-report measure of posttraumatic stress disorder. The Behavior Therapist. 1993;16:161–2. [Google Scholar]
- 43.Kubany ES, Leisen MB, Kaplan AS, Kelly MP. Validation of a brief measure of posttraumatic stress disorder: the Distressing Event Questionnaire (DEQ). Psychol Assess. 2000;12(2):197–209. [DOI] [PubMed] [Google Scholar]
- 44.van den Heuvel LL, Stalder T, du Plessis S, Suliman S, Kirschbaum C, Seedat S. Hair cortisol levels in posttraumatic stress disorder and metabolic syndrome. Stress. 2020;23(5):577–89. [DOI] [PubMed] [Google Scholar]
- 45.Weathers FW, Bovin MJ, Lee DJ, Sloan DM, Schnurr PP, Kaloupek DG, et al. Clinician-administered PTSD scale for DSM-5. Psychol Assess. 2015; 30(3):383–95 [DOI] [PMC free article] [PubMed]
- 46.McLean SA, Ressler K, Koenen KC, Neylan T, Germine L, Jovanovic T, et al. The AURORA study: a longitudinal, multimodal library of brain biology and function after traumatic stress exposure. Mol Psychiatry. 2020;25(2):283–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blevins CA, Weathers FW, Davis MT, Witte TK, Domino JL. The posttraumatic stress disorder checklist for DSM-5 (PCL-5): development and initial psychometric evaluation. J Trauma Stress. 2015;28(6):489–98. [DOI] [PubMed] [Google Scholar]
- 48.Kessler RC, Ressler KJ, House SL, Beaudoin FL, An X, Stevens JS, et al. Socio-demographic and trauma-related predictors of PTSD within 8 weeks of a motor vehicle collision in the AURORA study. Mol Psychiatry. 2021;26(7):3108–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bovin MJ, Marx BP, Weathers FW, Gallagher MW, Rodriguez P, Schnurr PP, et al. Psychometric properties of the PTSD checklist for diagnostic and statistical manual of mental disorders-fifth edition (PCL-5) in veterans. Psychol Assess. 2016;28(11):1379–91. [DOI] [PubMed] [Google Scholar]
- 50.Perroud N, Rutembesa E, Paoloni-Giacobino A, Mutabaruka J, Mutesa L, Stenz L, et al. The Tutsi genocide and transgenerational transmission of maternal stress: epigenetics and biology of the HPA axis. World J Biol Psychiatry. 2014;15(4):334–45. [DOI] [PubMed] [Google Scholar]
- 51.Musanabaganwa C, Wani AH, Donglasan J, Fatumo S, Jansen S, Mutabaruka J, et al. Leukocyte methylomic imprints of exposure to the genocide against the Tutsi in Rwanda: a pilot epigenome-wide analysis. Epigenomics. 2022;14(1):11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mulder N, Abimiku A, Adebamowo SN, de Vries J, Matimba A, Olowoyo P, et al. H3Africa: current perspectives. Pharmgenomics Pers Med. 2018;11:59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weathers F, Ford J, Stamm B. Measurement of stress, trauma, and adaptation. In: Psychometric review of PTSD checklist (PCL-C, PCL-S PCL-M, PCL-PR). 1996. p. 250–1.
- 54.Weathers F, Litz B, Keane T, Palmieri P, Marx B, Schnurr P. The PTSD checklist for DSM-5 (PCL-5)–LEC-5 and extended criterion a [Measurement instrument]. 2013. Avaliable from https://www.ptsd.va.gov/.
- 55.Herbert R, Moline J, Skloot G, Metzger K, Baron S, Luft B, et al. The World Trade Center disaster and the health of workers: five-year assessment of a unique medical screening program. Environ Health Perspect. 2006;114(12):1853–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bromet EJ, Hobbs MJ, Clouston SA, Gonzalez A, Kotov R, Luft BJ. DSM-IV post-traumatic stress disorder among world trade center responders 11–13 years after the disaster of 11 September 2001 (9/11). Psychol Med. 2016;46(4):771–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.First MB, Spitzer RL, Gibbon M, Williams JBW. Structured clinical interview for DSM-IV-TR axis I disorders, research version, patient edition. (SCID-I/P) New York: Biometrics Research, New York State Psychiatric Institute; 2002.
- 58.Mehta D, Bruenig D, Carrillo-Roa T, Lawford B, Harvey W, Morris CP, et al. Genomewide DNA methylation analysis in combat veterans reveals a novel locus for PTSD. Acta Psychiatr Scand. 2017;136(5):493–505. [DOI] [PubMed] [Google Scholar]
- 59.Nievergelt CM, Maihofer AX, Mustapic M, Yurgil KA, Schork NJ, Miller MW, et al. Genomic predictors of combat stress vulnerability and resilience in U.S. Marines: a genome-wide association study across multiple ancestries implicates PRTFDC1 as a potential PTSD gene. Psychoneuroendocrinology. 2015;51:459–71. [DOI] [PubMed] [Google Scholar]
- 60.Baker DG, Nash WP, Litz BT, Geyer MA, Risbrough VB, Nievergelt CM, et al. Predictors of risk and resilience for posttraumatic stress disorder among ground combat marines: methods of the marine resiliency study. Prev Chronic Dis. 2012;9:E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reijnen A, Rademaker AR, Vermetten E, Geuze E. Prevalence of mental health symptoms in Dutch military personnel returning from deployment to Afghanistan: a 2-year longitudinal analysis. Eur Psychiatry. 2015;30(2):341–6. [DOI] [PubMed] [Google Scholar]
- 62.Eekhout I, Reijnen A, Vermetten E, Geuze E. Post-traumatic stress symptoms 5 years after military deployment to Afghanistan: an observational cohort study. Lancet Psychiatry. 2016;3(1):58–64. [DOI] [PubMed] [Google Scholar]
- 63.Hovens JE, Bramsen I, van der Ploeg HM. Self-rating inventory for posttraumatic stress disorder: review of the psychometric properties of a new brief Dutch screening instrument. Percept Mot Skills. 2002;94(3 Pt 1):996–1008. [DOI] [PubMed] [Google Scholar]
- 64.Ursano RJ, Colpe LJ, Heeringa SG, Kessler RC, Schoenbaum M, Stein MB, et al. The Army study to assess risk and resilience in servicemembers (Army STARRS). Psychiatry. 2014;77(2):107–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kessler RC, Santiago PN, Colpe LJ, Dempsey CL, First MB, Heeringa SG, et al. Clinical reappraisal of the Composite International Diagnostic Interview Screening Scales (CIDI-SC) in the army study to assess risk and resilience in servicemembers (Army STARRS). Int J Methods Psychiatr Res. 2013;22(4):303–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Logue MW, Baldwin C, Guffanti G, Melista E, Wolf EJ, Reardon AF, et al. A genome-wide association study of post-traumatic stress disorder identifies the retinoid-related orphan receptor alpha (RORA) gene as a significant risk locus. Mol Psychiatry. 2013;18(8):937–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wolf EJ, Miller MW, Hawn SE, Zhao X, Wallander SE, McCormick B, et al. Longitudinal study of traumatic-stress related cellular and cognitive aging. Brain Behav Immun. 2024;115:494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McGlinchey RE, Milberg WP, Fonda JR, Fortier CB. A methodology for assessing deployment trauma and its consequences in OEF/OIF/OND veterans: The TRACTS longitudinal prospective cohort study. Int J Methods Psychiatr Res. 2017;26(3):e1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weathers FW, Bovin MJ, Lee DJ, Sloan DM, Schnurr PP, Kaloupek DG, et al. The clinician-administered PTSD scale for DSM-5 (CAPS-5): development and initial psychometric evaluation in military veterans. Psychol Assess. 2018;30(3):383–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McAllister TW, Zafonte R, Jain S, Flashman LA, George MS, Grant GA, et al. Randomized placebo-controlled trial of methylphenidate or galantamine for persistent emotional and cognitive symptoms associated with PTSD and/or traumatic brain injury. Neuropsychopharmacology. 2016;41(5):1191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ashley-Koch AE, Garrett ME, Gibson J, Liu Y, Dennis MF, Kimbrel NA, et al. Genome-wide association study of posttraumatic stress disorder in a cohort of Iraq-Afghanistan era veterans. J Affect Disord. 2015;184:225–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rasmusson AM, Marx CE, Jain S, Farfel GM, Tsai J, Sun X, et al. A randomized controlled trial of ganaxolone in posttraumatic stress disorder. Psychopharmacology. 2017;234(15):2245–57. [DOI] [PubMed] [Google Scholar]
- 73.Lepage C, de Pierrefeu A, Koerte IK, Coleman MJ, Pasternak O, Grant G, et al. White matter abnormalities in mild traumatic brain injury with and without post-traumatic stress disorder: a subject-specific diffusion tensor imaging study. Brain Imaging Behav. 2018;12(3):870–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Katrinli S, PGC-PTSD-EWAS. EPICv2 pipeline (v.1.0). 2024. Available from: https://github.com/PGC-PTSD-EWAS/EPIC_QC.
- 75.McCartney DL, Walker RM, Morris SW, McIntosh AM, Porteous DJ, Evans KL. Identification of polymorphic and off-target probe binding sites on the Illumina Infinium MethylationEPIC BeadChip. Genom Data. 2016;9:22–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fortin JP, Triche TJ Jr, Hansen KD. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017;33(4):558–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Teschendorff AE, Breeze CE, Zheng SC, Beck S. A comparison of reference-based algorithms for correcting cell-type heterogeneity in epigenome-wide association studies. BMC Bioinformatics. 2017;18(1):105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Salas LA, Koestler DC, Butler RA, Hansen HM, Wiencke JK, Kelsey KT, et al. An optimized library for reference-based deconvolution of whole-blood biospecimens assayed using the Illumina HumanMethylationEPIC BeadArray. Genome Biol. 2018;19(1):64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barfield RT, Almli LM, Kilaru V, Smith AK, Mercer KB, Duncan R, et al. Accounting for population stratification in DNA methylation studies. Genet Epidemiol. 2014;38(3):231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van Iterson M, van Zwet EW, Consortium B, Heijmans BT. Controlling bias and inflation in epigenome- and transcriptome-wide association studies using the empirical null distribution. Genome Biol. 2017;18(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barfield RT, Kilaru V, Smith AK, Conneely KN. CpGassoc: an R function for analysis of DNA methylation microarray data. Bioinformatics. 2012;28(9):1280–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Balduzzi S, Rucker G, Schwarzer G. How to perform a meta-analysis with R: a practical tutorial. Evid Based Ment Health. 2019;22(4):153–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mansell G, Gorrie-Stone TJ, Bao Y, Kumari M, Schalkwyk LS, Mill J, et al. Guidance for DNA methylation studies: statistical insights from the Illumina EPIC array. BMC Genomics. 2019;20(1):366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Phipson B, Maksimovic J, Oshlack A. missMethyl: an R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinformatics. 2016;32(2):286–8. [DOI] [PubMed] [Google Scholar]
- 86.Min JL, Hemani G, Hannon E, Dekkers KF, Castillo-Fernandez J, Luijk R, et al. Genomic and phenotypic insights from an atlas of genetic effects on DNA methylation. Nat Genet. 2021;53(9):1311–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Villicana S, Castillo-Fernandez J, Hannon E, Christiansen C, Tsai PC, Maddock J, et al. Genetic impacts on DNA methylation help elucidate regulatory genomic processes. Genome Biol. 2023;24(1):176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Logue MW, Miller MW, Wolf EJ, Huber BR, Morrison FG, Zhou Z, et al. An epigenome-wide association study of posttraumatic stress disorder in US veterans implicates several new DNA methylation loci. Clin Epigenetics. 2020; IN PRESS. [DOI] [PMC free article] [PubMed]
- 89.Daskalakis NP, Iatrou A, Chatzinakos C, Jajoo A, Snijders C, DiPietro CP, et al. Systems biology dissection of PTSD and MDD across brain regions, cell-types and blood. Science. In Revision. [DOI] [PMC free article] [PubMed]
- 90.Friedman MJ, Huber BR, Brady CB, Ursano RJ, Benedek DM, Kowall NW, et al. VA’s national PTSD brain bank: a national resource for research. Curr Psychiatry Rep. 2017;19(10):73. [DOI] [PubMed] [Google Scholar]
- 91.Rompala G, Nagamatsu ST, Martinez-Magana JJ, Nunez-Rios DL, Wang J, Girgenti MJ, et al. Profiling neuronal methylome and hydroxymethylome of opioid use disorder in the human orbitofrontal cortex. Nat Commun. 2023;14(1):4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zannas AS. Naturalistic stress hormone levels drive cumulative epigenomic changes along the cellular lifespan. Int J Mol Sci. 2021;22(16):8778. [DOI] [PMC free article] [PubMed]
- 93.Leung CS, Kosyk O, Welter EM, Dietrich N, Archer TK, Zannas AS. Chronic stress-driven glucocorticoid receptor activation programs key cell phenotypes and functional epigenomic patterns in human fibroblasts. iScience. 2022;25(9):104960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Villicaña S, Castillo-Fernandez J, Hannon E, Christiansen C, Tsai PC, Maddock J, et al. Genetic impacts on DNA methylation help elucidate regulatory genomic processes. bioRxiv. 2023:2023.03.31.535045. [DOI] [PMC free article] [PubMed]
- 95.Uhlen M, Karlsson MJ, Zhong W, Tebani A, Pou C, Mikes J, et al. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science. 2019;366(6472):eaax9198. [DOI] [PubMed] [Google Scholar]
- 96.Sjostedt E, Zhong W, Fagerberg L, Karlsson M, Mitsios N, Adori C, et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science. 2020;367(6482):eaay5947. [DOI] [PubMed] [Google Scholar]
- 97.Wilson R, Wahl S, Pfeiffer L, Ward-Caviness CK, Kunze S, Kretschmer A, et al. The dynamics of smoking-related disturbed methylation: a two time-point study of methylation change in smokers, non-smokers and former smokers. BMC Genomics. 2017;18(1):805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hao N, Whitelaw ML. The emerging roles of AhR in physiology and immunity. Biochem Pharmacol. 2013;86(5):561–70. [DOI] [PubMed] [Google Scholar]
- 99.Chen XQ, Tan I, Leung T, Lim L. The myotonic dystrophy kinase-related Cdc42-binding kinase is involved in the regulation of neurite outgrowth in PC12 cells. J Biol Chem. 1999;274(28):19901–5. [DOI] [PubMed] [Google Scholar]
- 100.Story Jovanova O, Nedeljkovic I, Spieler D, Walker RM, Liu C, Luciano M, et al. DNA methylation signatures of depressive symptoms in middle-aged and elderly persons: meta-analysis of multiethnic epigenome-wide studies. JAMA Psychiat. 2018;75(9):949–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ligthart S, Marzi C, Aslibekyan S, Mendelson MM, Conneely KN, Tanaka T, et al. DNA methylation signatures of chronic low-grade inflammation are associated with complex diseases. Genome Biol. 2016;17(1):255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wielscher M, Mandaviya PR, Kuehnel B, Joehanes R, Mustafa R, Robinson O, et al. DNA methylation signature of chronic low-grade inflammation and its role in cardio-respiratory diseases. Nat Commun. 2022;13(1):2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Muniz Carvalho C, Wendt FR, Maihofer AX, Stein DJ, Stein MB, Sumner JA, et al. Dissecting the genetic association of C-reactive protein with PTSD, traumatic events, and social support. Neuropsychopharmacology. 2021;46(6):1071–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Eraly SA, Nievergelt CM, Maihofer AX, Barkauskas DA, Biswas N, Agorastos A, et al. Assessment of plasma C-reactive protein as a biomarker of posttraumatic stress disorder risk. JAMA Psychiat. 2014;71(4):423–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang JJ, Jiang W. Immune biomarkers alterations in post-traumatic stress disorder: a systematic review and meta-analysis. J Affect Disord. 2020;268:39–46. [DOI] [PubMed] [Google Scholar]
- 106.Passos IC, Vasconcelos-Moreno MP, Costa LG, Kunz M, Brietzke E, Quevedo J, et al. Inflammatory markers in post-traumatic stress disorder: a systematic review, meta-analysis, and meta-regression. Lancet Psychiatry. 2015;2(11):1002–12. [DOI] [PubMed] [Google Scholar]
- 107.Carvalho Silva R, Martini P, Hohoff C, Mattevi S, Bortolomasi M, Menesello V, et al. DNA methylation changes in association with trauma-focused psychotherapy efficacy in treatment-resistant depression patients: a prospective longitudinal study. Eur J Psychotraumatol. 2024;15(1): 2314913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wille A, Amort T, Singewald N, Sartori SB, Lusser A. Dysregulation of select ATP-dependent chromatin remodeling factors in high trait anxiety. Behav Brain Res. 2016;311:141–6. [DOI] [PubMed] [Google Scholar]
- 109.Lohoff FW, Roy A, Jung J, Longley M, Rosoff DB, Luo A, et al. Epigenome-wide association study and multi-tissue replication of individuals with alcohol use disorder: evidence for abnormal glucocorticoid signaling pathway gene regulation. Mol Psychiatry. 2021;26(6):2224–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Baker LP, Nielsen MD, Impey S, Hacker BM, Poser SW, Chan MY, et al. Regulation and immunohistochemical localization of betagamma-stimulated adenylyl cyclases in mouse hippocampus. J Neurosci. 1999;19(1):180–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li S, Papale LA, Zhang Q, Madrid A, Chen L, Chopra P, et al. Genome-wide alterations in hippocampal 5-hydroxymethylcytosine links plasticity genes to acute stress. Neurobiol Dis. 2016;86:99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Verzi MP, Anderson JP, Dodou E, Kelly KK, Greene SB, North BJ, et al. N-twist, an evolutionarily conserved bHLH protein expressed in the developing CNS, functions as a transcriptional inhibitor. Dev Biol. 2002;249(1):174–90. [DOI] [PubMed] [Google Scholar]
- 113.Katrinli S, Smith AK. Immune system regulation and role of the human leukocyte antigen in posttraumatic stress disorder. Neurobiol Stress. 2021;15: 100366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim GS, Smith AK, Xue F, Michopoulos V, Lori A, Armstrong DL, et al. Methylomic profiles reveal sex-specific differences in leukocyte composition associated with post-traumatic stress disorder. Brain Behav Immun. 2019;81:280–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kilaru V, Knight AK, Katrinli S, Cobb D, Lori A, Gillespie CF, et al. Critical evaluation of copy number variant calling methods using DNA methylation. Genet Epidemiol. 2020;44(2):148–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. PTSD-associated CpGs from the primary analysis in the sensitivity analysis adjusted for smoking score. Table S2. DNA methylation and cis-gene expression (RNAseq) correlations. Table S3. Correlation values of methylation between blood and brain regions from Blood Brain DNA Methylation Comparison Tool. Table S4. Results of PTSD-associated CpGs from blood in fibroblasts. Table S5. Significant SNPs from cis-meQTL analysis and their association with PTSD. Table S6. Genetic associations of the identified PTSD-associated genes from gene-based tests. Table S7. Results of hypergeometric tests in postmortem brain tissues. Table S8. Results of PTSD-associated CpGs from blood in the brain. Table S9. Results of PTSD-associated CpGs from blood in neuronal nuclei. Table S10. PTSD-associated CpGs from the primary analysis in the stratified analysis for sex, ancestry, and trauma type. Table S11. Gene Ontology Enrichment results in females. Fig S1. Forest Plots of PTSD-associated CpGs in each cohort. Fig S2. Manhattan plots of the stratified analyses. Fig S3. Correlation of effect sizes based on stratified analyses for ancestry and sex. Fig S4. Forest Plots of CpGs associated with PTSD in the stratified analysis for females. Fig S5. Forest Plots of CpGs associated with PTSD in the stratified analysis for males. Fig S6. Forest Plots of CpGs associated with PTSD in the stratified analysis for European ancestry. Fig S7. Forest Plots of CpGs associated with PTSD in the stratified analysis for African ancestry. Fig S8. Forest Plots of CpGs associated with PTSD in the stratified analysis for civilian trauma. Fig S9. Forest Plots of CpGs associated with PTSD in the stratified analysis for military trauma.
Additional file 2. Summary statistics of the main analysis.
Additional file 3. Summary statistics of the smoking-sensitivity analysis.
Additional file 4. Summary statistics of the stratified analysis for males.
Additional file 5. Summary statistics of the stratified analysis for females.
Additional file 6. Summary statistics of the stratified analysis for European ancestry.
Additional file 7. Summary statistics of the stratified analysis for African ancestry.
Additional file 8. Summary statistics of the stratified analysis for civilian cohorts.
Additional file 9. Summary statistics of the stratified analysis for military cohorts.
Data Availability Statement
The main summary statistics data that support the findings of this study will be available within Supplementary Data upon publication. Individual-level data from the cohorts or cohort-level summary statistics will be made available to researchers following an approved analysis proposal through the PGC-PTSD Epigenetics Workgroup with the agreement of the cohort PIs. The raw data for the GTP cohort is available in the Gene Expression Omnibus database with the accession code GSE132203 [115]. Owing to limitations on data sharing as specified in the consent process and/or restrictions as part of the use agreement under which the data was accessed, data from the military/VA cohorts, VA MIRECC, MRS, Army STARRS, PRISMO, PROGrESS, and NCPTSD/TRACTS, cannot be publicly posted as part of this project. However, such data can be provided in de-identified form through a data use agreement following applicable guidelines on data sharing and privacy protection. For additional information on access to these data, including PI contact information for the cohorts accessed under a DUA, please contact the corresponding author.