Abstract
Reverse micelles (RMs) have emerged as useful tools for the study of membrane associated proteins. With a nanoscale water core surrounded by surfactant and solubilized in a non-polar solvent, RMs stand apart as a unique membrane model. While RMs have been utilized as tools to investigate the physical properties of membranes and their associated water, RMs also effectively house membrane associated proteins for a variety of studies. High-resolution protein NMR revealed a need for development of improved RM formulations, which greatly enhanced the use of RMs for aqueous proteins. Protein-optimized RM formulations enabled encapsulation of challenging membrane associated protein types, including lipidated proteins, transmembrane proteins, and peripheral membrane proteins. Improvements in biological accuracy of RMs using phospholipid-based surfactants has advanced their utility as a membrane mimetic even further, better matching the chemistry of the most common cellular membrane lipids. Natural lipid extracts may also be used to construct RMs and house proteins, resulting in a membrane model that better represents the complexity of biological membranes. Recent applications in high-resolution investigations of protein-membrane interactions and inhibitor design of membrane associated proteins have demonstrated the usefulness of these systems in addressing this difficult category of protein. Further developments of RMs as membrane models will enhance the breadth of investigations facilitated by these systems and will enhance their use in biophysical, structural, and drug discovery pursuits of membrane associated proteins. In this review, we present the development of RMs as membrane models and their application to structural and biophysical study of membrane proteins.
Introduction
Among many functions, membrane proteins act as liaisons and gatekeepers of membranes, the primary barriers of cells [1]. Their central roles make them an important focus for a variety of studies, from basic biology to disease mechanisms and drug discovery. An assortment of membrane protein varieties exists, including transmembrane (TM) proteins that span the bilayer and are primarily of either α-helical bundle or β-barrel types [2], monotopic integral membrane proteins that are affixed to a single leaflet [3], proteins that are anchored to membranes through lipidations [4], as well as water-soluble proteins that interact with membranes [5]. In vitro investigations of membrane proteins require membrane models that simulate cellular membrane environments while retaining favorable properties for experimentation. Liposomes, nanodiscs, copolymer derived native nanodiscs, bicelles, micelles, amphipols, and other models have emerged as useful tools in the study of membrane proteins [6–8]. Among these, reverse micelles (RMs) stand out as an effective and unique tool in the arsenal of membrane models. RMs consist of nanoscale pools of water, which may contain a protein, solubilized within a hydrophobic solvent, often an alkane. Surfactants form the interface, with their hydrophilic headgroups oriented towards the RM core and the hydrophobic tails interacting with the solvent (Figure 1A)[9]. Due to a resemblance to membrane surfaces, membrane associated proteins may be housed within RMs.
Figure 1.
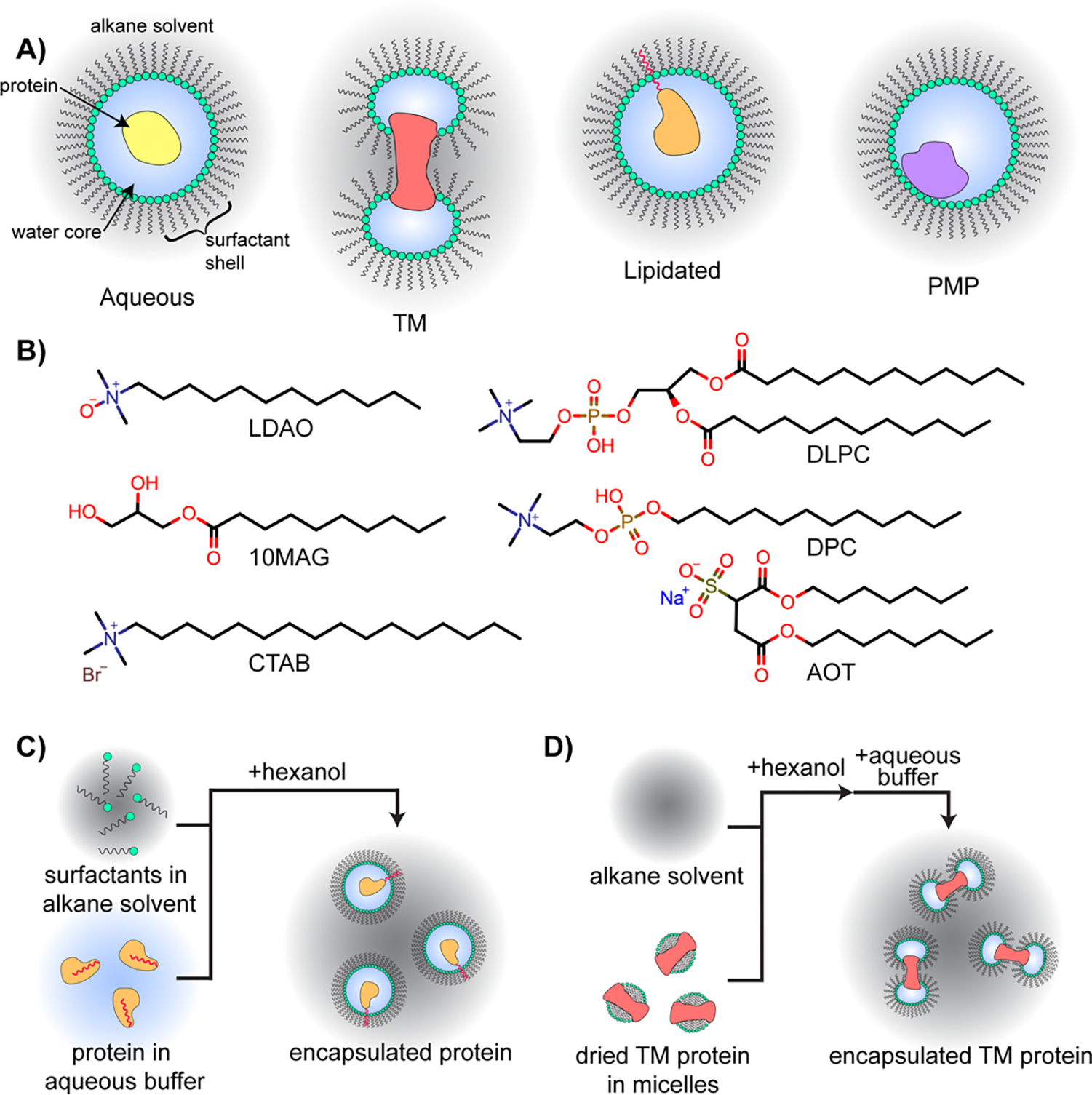
Depictions of multiple protein types encapsulated within RMs, commonly used RM surfactants, and RM construction workflows. A) RMs may house aqueous proteins within a water core surrounded by a surfactant shell, all solubilized in an alkane solvent. Transmembrane (TM) proteins may be housed within two RMs, each protecting the hydrophilic regions of the protein with the hydrophobic portions interfacing with surfactant tails or alkane solvent. Lipidated proteins may be encapsulated with their membrane anchors embedded into the surfactant shell. Peripheral membrane proteins (PMPs) may reside in the aqueous core of the protein with the membrane interface interacting with the surfactant shell. B) Optimized surfactants that are commonly used for aqueous protein encapsulation include the 10MAG/LDAO binary mixture and CTAB. The DLPC/DPC binary mixture effectively houses PMPs in their membrane embedded state. AOT was historically commonly used, but is now accepted as destabilizing to most proteins. C) General workflow for encapsulation of a water-solubilized proteins, which may be applicable to aqueous proteins, lipid-anchored proteins (depicted here), or PMPs. Small volume of aqueous buffer containing protein is added to the solvent containing lipids and mixed, with hexanol then added if necessary. A detailed protocol may be found in [13]. D) General workflow for TM protein encapsulation. A TM protein housed in a micelle is lyophilized from aqueous conditions, the alkane solvent is added, followed by hexanol, then the aqueous buffer. Details may be found in [17].
RMs comprise several tunable parameters, which provides adaptability for experimental applications. For example, size can be modified by altering water content or ionic strength of the aqueous core [10–12]. Chemical and physical properties of the inner RM surface may be tuned according to the identity of the surfactant (Figure 1B) [13,14]. Properties of the solvent, such as viscosity, may be adjusted according to the solvent identity [15,16]. While several RM formulations reliably encapsulate most proteins with minimal adjustments, optimization may be necessary [13]. A typical process for constructing RMs is by the addition of a relatively small volume of an aqueous buffer to surfactants that are suspended in an alkane solvent. The water content in RMs, typically called water loading (W0, molar ratio of water to surfactant), is often between 10 and 30 for protein applications. A cosurfactant, often hexanol, may be necessary to fully form the RMs, depending on the formulation. A typical strategy for encapsulating water-soluble proteins is by including them in aqueous buffer, which is introduced to surfactants in an alkane solvent and results in encapsulation upon RM formation (Figure 1C) [13]. For membrane proteins that are not water soluble, alternate strategies may be necessary such as drying water-solubilized TM proteins housed in micelles, followed by addition of solvent, cosolvent, then finally the aqueous buffer (Figure 1D) [17]. The variety of parameters and delivery methods available for RMs may necessitate an initial investment in optimization for some proteins. On the other hand, these parameters represent a versatility that enables RMs to be used as tunable membrane models in a wide range of studies.
RMs have long been used as models to understand the physical properties of cellular membranes, such as the nature of hydration dynamics at membrane interfaces [18–23], partitioning of small molecules [24–27], and the effect of crowding or confinement near membrane surfaces on molecular conformation [28–30]. In addition, RMs have been used to encapsulate aqueous proteins which permits studies that are otherwise difficult or inaccessible. Low-viscosity alkane solvents may be leveraged to increase rotational diffusion in RMs, essentially breaking the size limit in protein NMR without the need for deuteration or transverse relaxation-optimized spectroscopy (TROSY) techniques [31]. The nanoscale water core provides a means to study the effect of confinement on proteins [32–35] and the impact of solvation on protein motions [36]. RMs have been used as platforms for investigating long-standing open questions in protein biophysics such as cold-denaturation [37–39] and hydration dynamics [40–42]. While recent development has been driven by a small number of groups, mostly in the protein NMR field, demonstration of RMs as platforms for studying membrane proteins have highlighted their potential. Here, we review the use and development of RMs for studying membrane proteins, including recent advances that promise to improve biological accuracy and facilitate new lines of investigation to this challenging class of protein.
Reverse micelles as platforms to house and study membrane proteins
Foundational research focused on understanding the formation and stability of RMs in various solvent systems [43,44]. RMs are unique membrane models due to the bulk solvent being hydrophobic, often an alkane, which may provide advantages. For example, the use of RMs to promote lipase catalysis takes advantage of this property. [45–49] RMs allowed access to the hydrophobic lipid substrate while stably housing the water-soluble protein within the RM water core, preventing direct exposure to alkanes which may be destabilizing. Solubilizing membrane associated proteins in an in vitro membrane model also requires access to hydrophobic and hydrophilic phases through an amphipathic interface. Early studies used RMs to house membrane associated myelin basic protein (MBP) or myelin proteolipid protein (PLP) in a membrane mimetic environment to study the structure of the protein-membrane complex [50–52]. MBP changes to a more ordered α-helical structure within RMs compared to aqueous conditions, reflecting its natural conformation in the membrane [51]. When housed in RMs, specific regions of PLP, such as the ε-amino groups of lysine residues, remained accessible to water, while others, like a key tryptophan residue, became shielded from the solvent, mirroring the expected interactions with biological membranes [50]. These observations highlight that RMs may house membrane proteins and preserve their structural integrity. While many models were initially proposed for how proteins were housed within RMs, the exact mode was not known [53]. It was not known if the hydrophobic regions of membrane proteins are directly solvated by the alkane solvent, if proteins may be adsorbed into the RM surface, or if a protein is solubilized by a single or multiple RMs. Small-angle X-ray scattering (SAXS) measurements helped resolve these questions in regard to transmembrane proteins and revealed the structure of the protein and RM complex [54]. The proteins are encapsulated within two RMs with the hydrophilic portions covered and protected by the aqueous core while the hydrophobic portions interact with either the surfactant tails or the solvent, forming a small bilayer-like environment (Figure 1A) [54,55].
RMs provide means to model interactions between proteins or peptides with membrane surfaces. The chemokine receptor CXCR1 N-terminal domain has a secondary structure similar to its membrane-bound form when encapsulated within bis(2-ethylhexyl) sulfosuccinate (AOT) RMs, as revealed through fluorescence and circular dichroism [56]. Upon increasing the water content, tryptophan residues became more exposed to water, revealing that structure and dynamics of this domain are intertwined with the level of hydration. Monomeric, misfolded “seed” structures of amyloid beta (Aβ) proteins are typically difficult to study due to aggregation effects [57]. However, encapsulation within AOT RMs stabilized these structures. Fourier-transform infrared spectroscopy (FTIR) revealed that extended β-strands formed and may mimic a conformation that is induced in crowded cellular conditions and the presence of membranes. In silico models agree with the structural stability and conformational changes conferred when protein is encapsulated within an RM [58]. Alanine-rich peptide AKA2 reveals a rigid helical conformation within an RM, unlike the protein in aqueous conditions which forms flexible random coils. Simulations confirmed that RMs are expected to help stabilize membrane associated helices that contain polar and apolar faces. The surface and interfacial properties of RMs well-mimic the amphipathic properties of membranes and aid study of membrane associated proteins and peptides.
High-resolution studies of membrane associated proteins in RMs
While these early studies established RMs as membrane models, detailed structural information regarding encapsulated proteins was still lacking. An early demonstration of RMs as membrane mimics for high-resolution NMR is encapsulation of the neurotransmitter pentapeptide Leu-enkephalin. NMR experiments produced high quality spectra of the peptide encapsulated within AOT RMs where 3D and nuclear Overhauser effect spectroscopy (NOESY) data was collected [59]. The structure of the peptide was similar to that obtained by crystallography. The peptide was adsorbed into the RM surface, mimicking the interaction observed with other membrane models [60]. Another study used NMR to observe gramicidin A (gA), a peptide that forms a dimeric ion-channel in membranes. The peptide was successfully encapsulated within AOT RMs and subsequently NMR diffusion and interresidue NOE experiments showed gA bridging two RMs as expected for the transmembrane dimer [61]. The use of NMR to observe these membrane-associated peptides revealed detail in the structural aspects of membrane interactions and showed promise for utilizing RMs for high-resolution observations of membrane associated proteins.
Using high-resolution NMR with RM encapsulation reveals details about the overall fold and functional competence of a protein. A striking observation from NMR is that the commonly used surfactant AOT unfolds most proteins [13,62]. While it is possible that small membrane-associated peptides may be tolerant to AOT, these observations demonstrated the necessity for alternate RM systems for encapsulating proteins (Figure 1B). Improved surfactant systems were developed and applied to membrane proteins, with careful consideration for retaining fold and function upon encapsulation. The 54kDa transmembrane potassium channel, KcsA, was encapsulated using several different surfactant systems while retaining its native homotetrameric structure [63]. Best spectral conditions were achieved with a mixture of cetyltrimethylammonium bromide (CTAB) and dihexadecyldimethylammonium bromide (DHAB). Retention of function was confirmed by 15N-HSQC, which showed shifting of key residues upon titration with K+ ions (Figure 2A).
Figure 2.
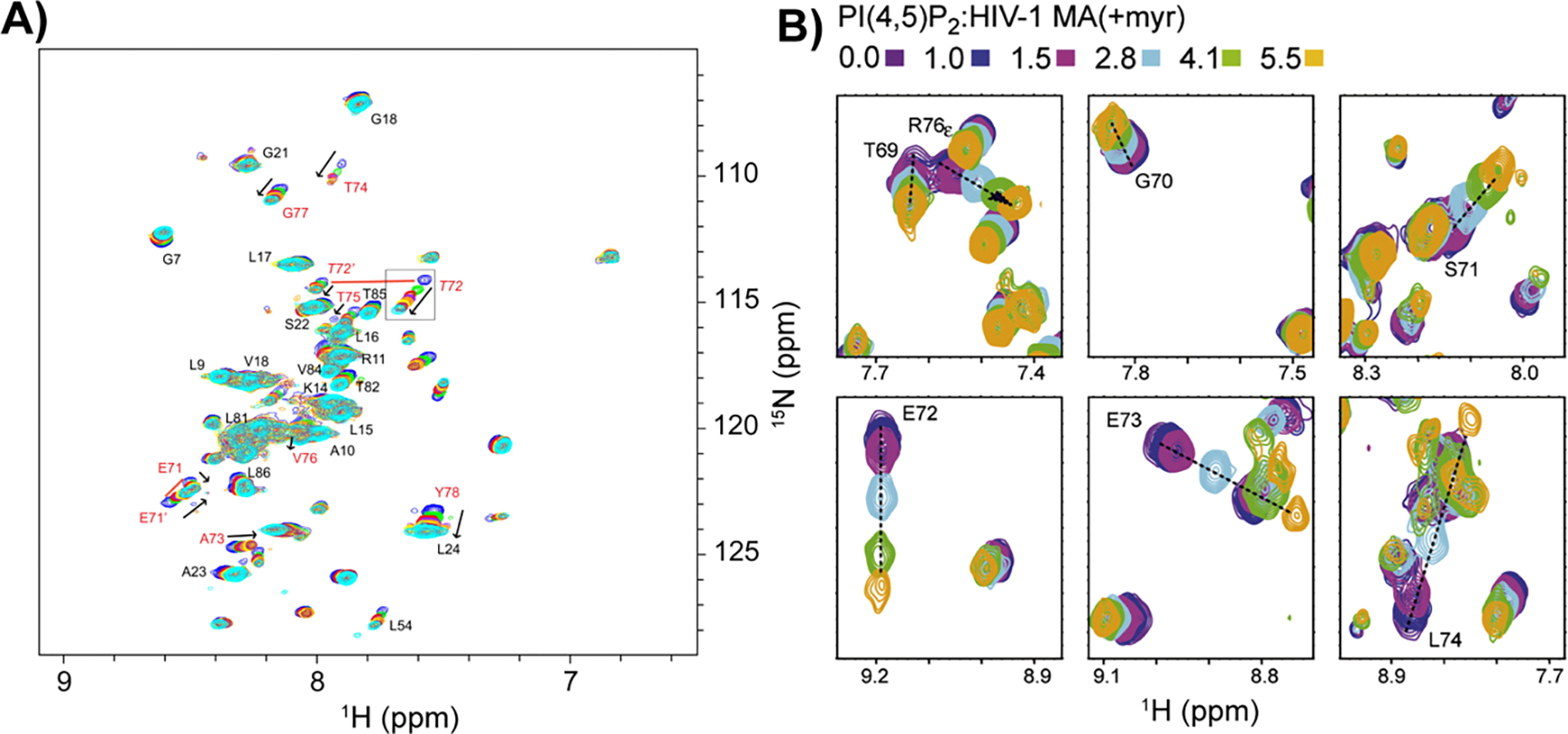
Reverse micelle encapsulation of functionally competent transmembrane and lipidated proteins. A) Overlay of 1H-15N HSQC spectra of a titration of encapsulated KcsA with potassium, from 0 mM to 28 mM K+. Arrows indicate increasing concentrations. Residues known to be associated with the KcsA selectivity filter are labeled in red, and other assigned residues are labeled in black. Resonances corresponding to the multiple conformers of residues T72 and E71 are connected by red lines. Reprinted from Kielec et al, Reverse micelles in integral membrane protein structural biology by solution NMR spectroscopy. Structure. 2009 Mar 11;17(3):345–51, Copyright 2009, with permission from Elsevier. B) Zoomed regions of the overlay of 1H-15N HSQC spectra of a titration of encapsulated myristoylated HIV matrix protein with phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2). Color of spectra corresponding to molar rations of lipid to protein are given. Reprinted from Valentine et al, Reverse Micelle Encapsulation of Membrane-Anchored Proteins for Solution NMR Studies. Structure. 2010 Jan 13;18(1):9–16, Copyright 2010, with permission from Elsevier.
Improved RM formulations have also enabled study of lipidated, membrane-anchored proteins via NMR (Figure 1A). The myristoyl anchor of recoverin embeds into the CTAB/hexanol RM shell upon encapsulation in its Ca2+-saturated, myristoyl-extruded state [64]. Importantly, only the myristoyl sequestered state is water-soluble; myristoyl extrusion typically results in aggregation. Similarly, HIV matrix protein was housed within CTAB/hexanol RMs with the myristoylation embedded in the surfactant shell [64]. Functional competence was demonstrated by binding the activating lipid phosphatidylinositol 4,5-bisphosphate (Figure 2B). Another example is the pH-dependent myristoyl switch in hisactophilin, which was studied by 15N-HSQC NMR [65]. Encapsulating hisactophilin in a CTAB/hexanol RM revealed that the protein is in the myristoyl-extruded state versus the solution state where the myristol is sequestered. The protective environment of the amphipathic RM shell allowed NMR experimentation for the myristoyl-extruded forms of these proteins, which is not possible in aqueous conditions due to aggregation induced by the exposed lipidations.
Peripheral membrane proteins (PMPs) are a class of membrane associated proteins that are water-soluble, but interact with membranes, often reversibly [5,66]. Due to the relative ease of experiments on water-solubilized proteins versus proteins associated with membranes, structural and in vitro experiments performed on PMPs tend to focus on the water-solubilized state. This leaves the functional, membrane bound form of most PMPs less well characterized. RMs have been applied to the study of membrane associated PMPs (Figure 1A). Cytochrome c is a PMP within the mitochondria and movement to the cytosol facilitates apoptosis [67]. Cytochrome c is highly compatible with the decylmonoacyl glycerol and lauryldimethylamino-N-oxide (10MAG/LDAO) surfactant system [68,69]. The NMR structure showed no conformational change upon encapsulation, demonstrating that the protein within RMs is identical to the water-solubilized state [67]. Titration of cardiolipin into the RM induced an interaction between the lipid and cytochrome c, a trigger for apoptosis. The mapped interactions localized to three sites; the previously characterized A- and L-site [70], and a new N-site observed through chemical shift perturbations (CSPs). This study demonstrates the use of RMs to study interactions between PMPs and membranes.
Improving the biological accuracy of RMs as models of cellular membranes.
While RMs were demonstrated to be powerful membrane mimetics for protein studies, often formulations used surfactants that did not fully replicate the chemistry of cellular membranes. To improve their biological accuracy, RMs have been formulated to more closely match lipids found in membranes. By using surfactants such as 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), the high phosphatidylcholine content found in many cellular membranes may be replicated. These formulations have been characterized with molecular dynamics (MD) simulations, SAXS, DLS, and terahertz spectroscopy revealing size, shape, and dynamical properties [71–74]. MD simulations of DOPC RMs were calibrated against experimental data and confirmed a small size that is tunable according to the water content and revealed a roughly spherical shape, qualities that are critical to certain studies (Figure 3)[73]. Size calibration allowed precise exploration of the nature of water dynamics in the aqueous core, which revealed slower hydration dynamics for smaller RMs. This finding that was subsequently reflected by time-domain terahertz spectroscopy combined with MD analysis of water in DOPC RMs that showed the water within hydration shell that contacts the inner RM surface has strong hydrogen bonding with the phosphocholine headgroups[75].
Figure 3.
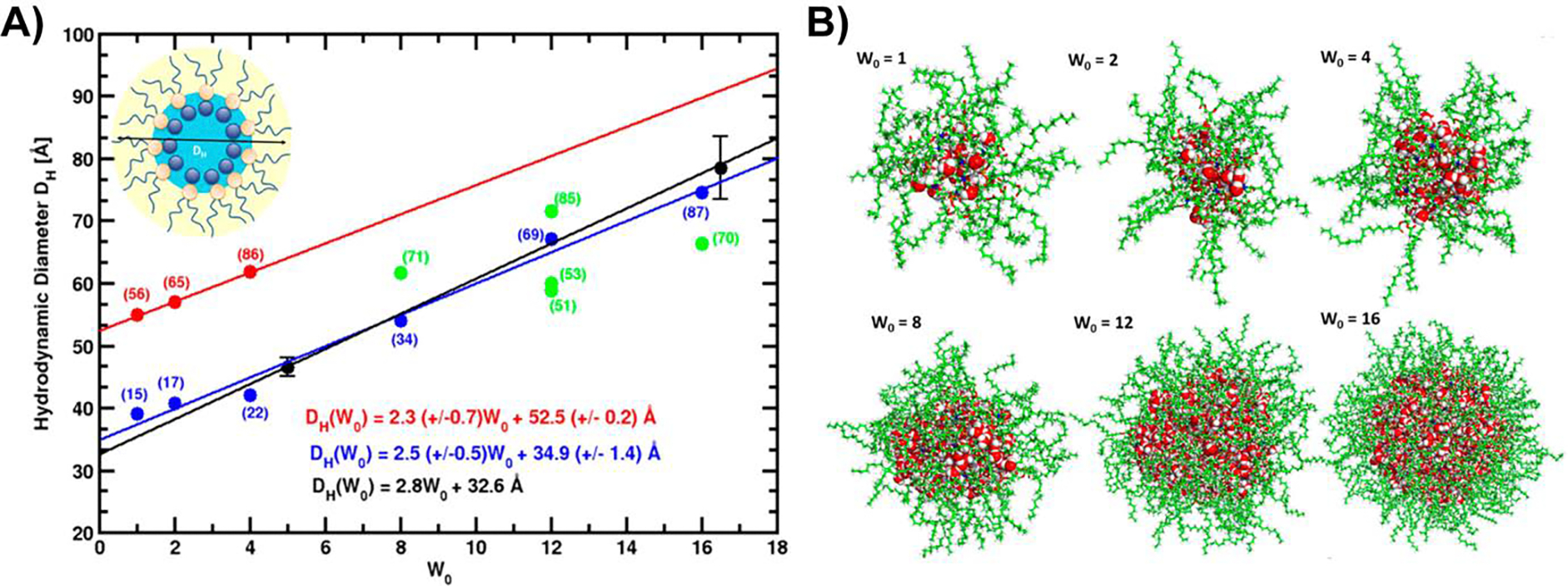
Experimentally calibrated molecular dynamics simulations of DOPC micelles. A) Hydrodynamic radius as a function of W0 from DLS measurements (black) [74], from initial MD trials using a fixed DOPC headgroup surface area to determine aggregation number (red), from intermediate trial-and-error attempts to calibrate aggregation number (green), and optimized MD simulations (blue) that reflect experimental measurements. Linear regressions are shown as lines and reported in the figure. Number of DOPC per RM is shown in parentheses. B) Snapshots of typical configurations from optimized MD simulations reflect somewhat ellipsoidal RMs at lower W0 values and more spherical shapes at larger W0 values. DOPC is in green, water oxygens are in red, water hydrogens are in white. Adapted with permission from Abel et al, On the structural and dynamical properties of DOPC reverse micelles. Langmuir. 2016 Oct 18;32(41):10610–20. Copyright 2016 American Chemical Society.
A phosphoethanolamine-headgroup containing surfactant showed promise in encapsulating proteins for NMR spectroscopy, providing a RM surface that more closely resembles biological membranes [76]. 1H-NOESY NMR experiments concluded that cytochrome c was natively folded after several months in this system, demonstrating robust sample stability. A membrane mimicking RM (mmRM) system was recently developed for use in protein NMR, a mixture of 1,2-dilauroyl-sn-glycero-3-phosphocholine with n-dodecylphosphocholine (DLPC:DPC), allows PMPs to be encapsulated in their membrane embedded state with sample stability spanning months (Figure 4) [77]. A strongly membrane associating PMP, glutathione peroxidase 4 (GPx4), was encapsulated within the NMR optimized 10MAG/LDAO system in its water-solubilized form, with no detectible interaction with the RM shell (Figure 4A–B). The mmRM formulation facilitated embedment of GPx4 onto the interior surface, reflecting the known interaction with membranes (Figure 4C–F), which was also observed with phosphatidylethanolamine binding protein 1 (PEBP1). These mmRMs have been validated by dynamic light scattering (DLS) and SAXS as small and approximately spherical, important characteristics for studies such as protein NMR. Lipid-like surfactants provide an interior RM surface that more closely matches the chemistry of biological membranes, though the use of more diverse lipids could enhance the biological relevance of these homogenous RM systems.
Figure 4.
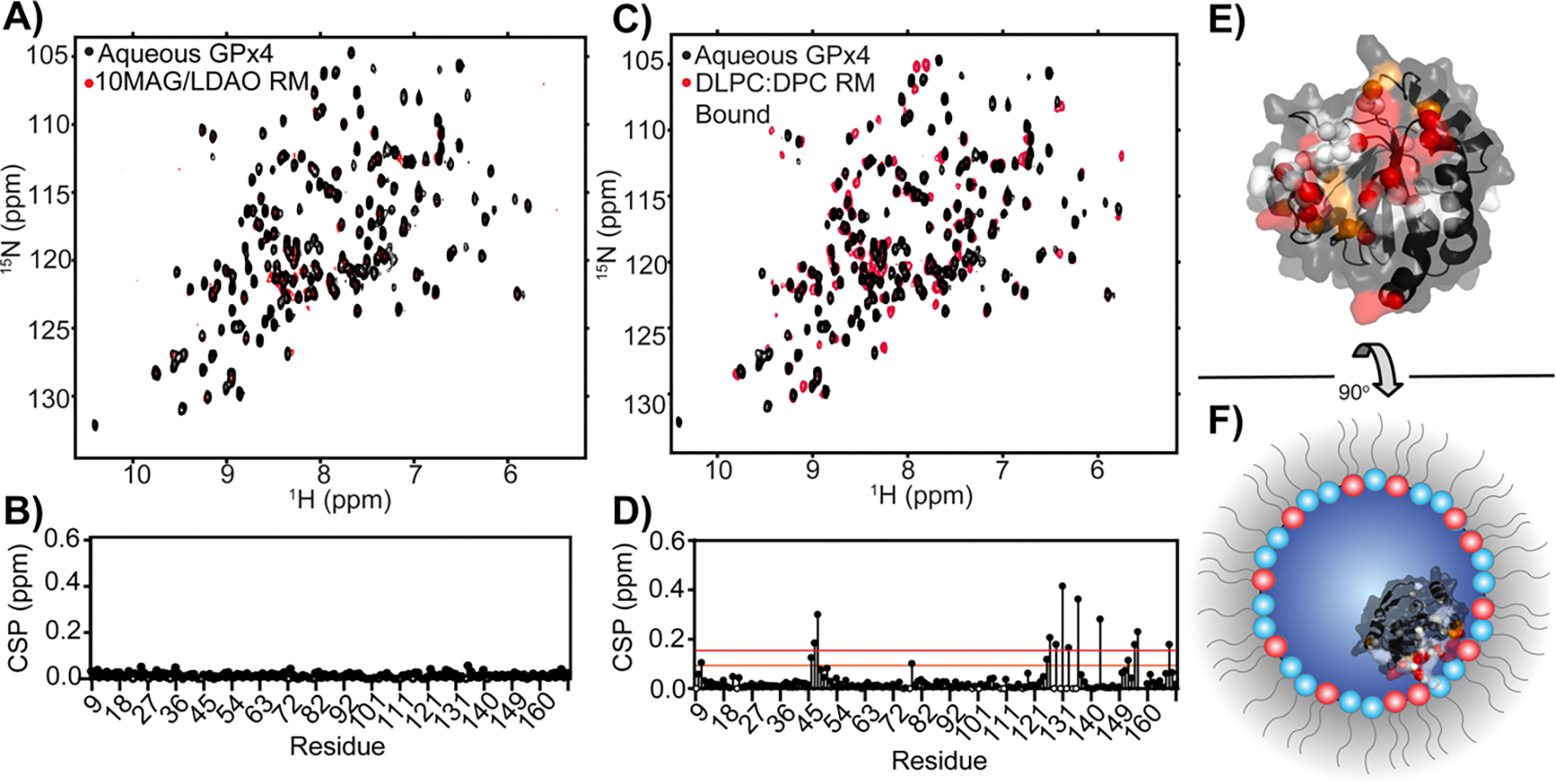
Demonstration of PMP encapsulation in a RM formulation optimized for aqueous proteins versus a RM formulation optimized to mimic membranes. A) Overlay of 1H-15N HSQCs of aqueous GPx4 (black) and GPx4 encapsulated in the 10MAG/LDAO mixture (red) and B) associated CSPs calculated per residue showing no interaction with the surfactant shell. C) Overlay of 1H-15N HSQCs of aqueous GPx4 (black) and GPx4 encapsulated in the DLPC/DPC mixture (red) and D) associated CSPs calculated for each residue. E) Mapping the residues that have CSPs greater than 1σ (orange) and 2σ (red) above a trimmed mean gives a structural map that may be used to F) estimate the interface of GPx4 with the mmRM which corresponds with the known membrane interface. Unobservable residues are depicted in white and residues with CSPs lower than 1σ are depicted in black. Adapted with permission from Labrecque et al, Membrane-mimicking reverse micelles for high-resolution interfacial study of proteins and membranes. Langmuir. 2022 Mar 17;38(12): 3676–86. Copyright 2022 American Chemical Society.
Enhancing the complexity of RMs to better mimic natural membranes.
The use of lecithin from various plant and animal sources achieves a more biologically accurate model by better representing the diversity of membrane lipids. Lecithin is an extract of phospholipids from a natural source, often soybeans, and contains a complex mixture with various phospholipid headgroups and fatty acid tails [78]. RMs may be constructed using lecithin and, despite the complex make-up of the lipid extract, size and shape parameters of lecithin RMs are tunable [79]. For example, in addition to water content driving lecithin RM size [80,81], the addition of oleic acid promotes the water core size to expand [82]. Salts are also known to change the structure of lecithin RMs from spherical to cylindrical and finally to reverse vesicles as shown by the use of DLS, transmission electron microscopy (TEM), UV-Vis spectroscopy, and SAXS [83]. Lecithin RMs support physical measurements of membrane properties, such as probing the nature of water dynamics against membrane surfaces using femtosecond fluorescence and absorption [80]. These experiments revealed that the lecithin RM has three water types: strongly bound, bound, and free, reflecting expected hydration behavior near membranes. Lecithin also shows similar characteristics to DOPC RMs with hydration dynamics being slower compared to AOT RMs [73,80]. This is attributed to the lipids having a stronger interaction with the water molecules than AOT. NMR relaxation measurements of water revealed the presence of a faster and a slower relaxing population, corresponding to water that interacts strongly to the lecithin headgroups and water that is more interior in the RMs [84]. Lecithin RMs have also been used to understand location preferences of encapsulated molecules, modeling their interactions with cellular membranes. Melatonin was encapsulated within lecithin and AOT and in both instances melatonin associated with the surfactant headgroups [85].
Demonstrating a breadth of lipid combinations that may form stable RMs, formulations using native polar lipids extracted from soy lecithin, porcine brain, and bovine heart all form RMs capable of encapsulating proteins for NMR spectroscopy [86,87]. Each formulation contained differing proportions of at least 7 different phospholipid types while also including fatty acids, cholesterol, and other components, yet native RMs (nRMs) made from each lipid extraction were of similar size and had similar water dependent tunability of size. Protein NMR revealed that interactions of PMPs with the interior surface of the nRMs are similar to the known membrane interactions[87]. Along with phospholipid-based RMs and mmRMs, nRMs are more biologically accurate membrane models which may be used for investigations of protein-membrane interactions.
Use of RMs to screen peripheral membrane proteins for fragment-based drug discovery.
A recent advance in the utility of RMs in the study of membrane associated proteins is applications in early-stage drug discovery. Membrane proteins are a particularly challenging class of protein for target-based drug development. Discovery efforts typically use screening methods that were developed for aqueous protein targets and are therefore limited in application to membrane protein targets. Fragment-based drug discovery (FBDD) is a powerful approach which is based on discovering small, weakly binding molecules (fragments) that may be combined to yield higher affinity inhibitors [88]. Biophysical methods are necessary for fragment screening due to their inherently weak binding, with protein NMR being a particularly useful method due to the structural information it provides [89,90]. The fragment-based approach is attractive for targets such as membrane associated proteins, where new classes of inhibitors may be desirable and the biases common to high-throughput screening libraries may need to be avoided. Existing NMR compatible membrane models such as bicelles and nanodiscs are large and often require extensive deuteration, making them cost prohibitive for screening large numbers of samples. Micelles, while useful, often destabilize proteins and the high concentrations of fragments required for screening can disrupt the micelle structure [91]. Reverse micelles offer a promising solution to these challenges.
Previous studies on encapsulated aqueous proteins allowed for detection of very weak binding fragments and demonstrated stability of RMs in the presence of large concentrations of fragments or other small-molecules [92,93]. A recent development applied mmRMs to successfully screen a PMP for fragment binders [94]. GPx4 is a highly sought after PMP drug target whose inhibition has been implicated in treating a number of cancer types [95]. Current inhibitors are highly reactive, non-specific electrophiles that are not suitable as drug leads; non-covalent inhibition of GPx4 may circumvent this problem. GPx4 is in its active form when embedded in membranes, where it reduces lipid hydroperoxides [96]. Therefore, non-covalent inhibition would therefore target the membrane-engaged form of GPx4 to block processing of the lipid substrate. A fragment screen was performed on GPx4 in mmRMs to understand whether fragment binding could be detected [94]. The screen was successful and yielded a series of hits that bound to GPx4 within the membrane interface (Figure 5). Importantly, the fragments did not bind to water-solubilized GPx4, even at high concentrations, highlighting that embedment onto the mmRM surface was essential for formation of fragment binding sites. A fragment hit with an affinity of ~220 μM was improved by a secondary screen to an affinity of ~15 μM, demonstrating that hits can be developed towards inhibitors and eventually drug leads. The mmRM fragment screening approach serves as an effective entry point for inhibitor and drug development for membrane associated proteins.
Figure 5.
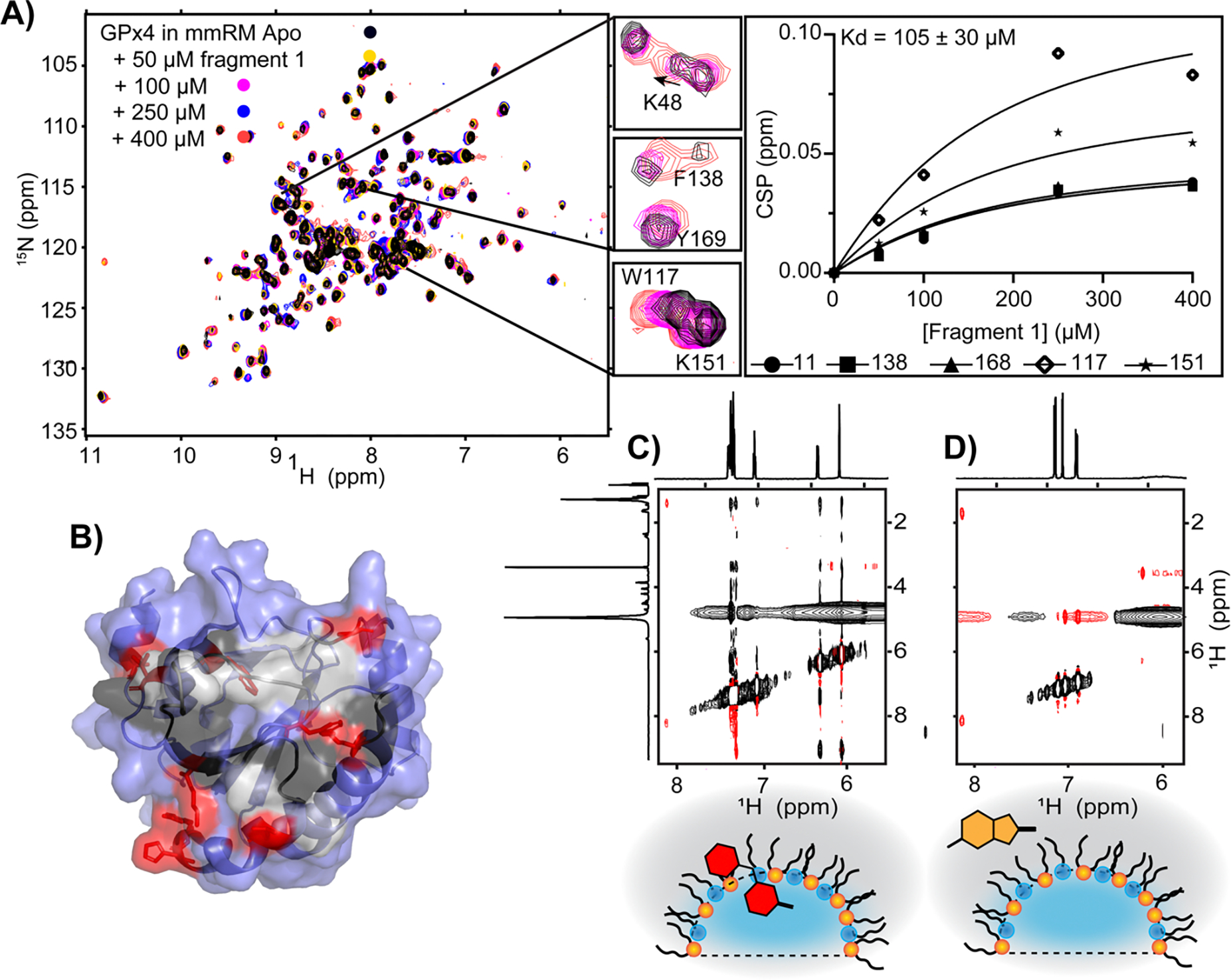
Use of mmRMs to screen for small-molecule binders for a PMP to initiate inhibitor design. A) A hit, discovered in a screen of a 1911-member library, was titrated against GPx4 encapsulated in mmRMs to characterize affinity and B) reveals binding within the membrane interface. Residues having significant CSPs upon fragment binding are depicted in red, other membrane interface residues in black, unassigned interface residues in gray, and non-interfacial residues are in blue. C) Example of a fragment hit that shows partitioning within the surfactant shell and D) a fragment hit that partitions into the alkane phase in the absence of protein, both demonstrated by 1H-1H NOESY spectra. Both are hits and reveal that multiple modes of partitioning within membranes may acceptable starting points for inhibitor design. All panels are adapted from [94].
Conclusions
These studies collectively highlight advancements in using RMs to house membrane proteins, to investigate protein-membrane interactions, and to initiate drug development efforts. They demonstrate the utility of RMs in providing a biologically relevant environment for detailed structural and biophysical analyses. While RMs have many benefits and unique applications, they are not without their drawbacks. As compared to bicelles or nanodiscs, the degree of curvature in RMs is high, which limits their accuracy as biological membrane models for some applications [97]. Initial optimization of conditions and additional production steps make use of RMs more time-consuming than micelles or bicelles. Finally, from our experience, a small fraction (roughly 10%) of proteins that are otherwise NMR compatible are unsuitable for use with RMs, possibly due to destabilization from exposure to surfactants or alkane. Despite some disadvantages, RMs are a potent addition to the set of tools available for membrane protein studies.
Recently, development of RMs as membrane mimics has been driven by a small number of groups and has mostly been limited to applications in protein NMR. We are optimistic that the latest improvements in biological accuracy of RMs will prove their usefulness for a larger and more diverse community of researchers. Distinctive aqueous protein studies that have been allowed by RMs, such as measurements of protein hydration dynamics and observations of confinement effects, may now be applied to entirely new classes of membrane associated proteins with the recent development of more accurate membrane mimetic RMs. The RM-based fragment screening approach promises to open a range of difficult membrane associated targets to inhibitor development. This body of work underscores the continued evolution and application of RM technology in membrane protein research, offering new insights and methodologies that enhance our understanding of these complex biological systems. Further developments in biological accuracy of RMs as membrane models and applications to new proteins and drug targets promise to advance membrane protein studies.
Perspectives.
Study of membrane associated proteins is dependent on the available membrane models, all of which have strengths and weaknesses. Reverse micelles are a promising and unique addition to the toolset for investigating of this challenging class of proteins.
Reverse micelles provide an adaptable membrane model useful for studying transmembrane, lipid-anchored, and peripheral membrane proteins. Recent work has revealed versatility in the lipids that may be used to construct RMs and has highlighted their use as screening platforms in fragment-based drug discovery for peripheral membrane proteins.
Promising future directions include leveraging new formulations to further improve biological accuracy of reverse micelles as membrane models and extending use of RMs to new areas in biophysical, structural, and inhibitor development studies for membrane associated proteins.
Acknowledgments
We thank members of the Fuglestad lab for helpful discussions.
Funding
Research reported in this publication was supported by the NIGMS of the National Institutes of Health under Award Number R35GM147221. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- 10MAG
decylmonoacyl glycerol
- AOT
bis(2-ethylhexyl) sulfosuccinate
- CSP
chemical shift perturbation
- CTAB
cetyltrimethylammonium bromide
- DHAB
dihexadecyldimethylammonium bromide
- DLPC
1,2-dilauroyl-sn-glycero-3-phosphocholine
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine
- DPC
n-dodecylphosphocholine
- FBDD
Fragment Based Drug Discovery
- gA
gramicidin A
- GPx4
glutathione peroxidase 4
- LDAO
lauryldimethylamino-N-oxide
- MD
molecular dynamics
- MBP
myelin basic protein
- mmRM
membrane mimicking Reverse Micelle
- NOESY
nuclear Overhauser effect spectroscopy
- nRM
native Reverse Micelle
- PLP
myelin proteolipid protein
- PMP
Peripheral Membrane Protein
- RM
Reverse Micelle
- SAXS
Small-angle X-ray scattering
- TEM
transmission electron microscopy
- TM
transmembrane
- TROSY
transverse relaxation-optimized spectroscopy
References
- 1.Engel A and Gaub HE (2008) Structure and Mechanics of Membrane Proteins. Annu Rev Biochem 77, 127–148 10.1146/annurev.biochem.77.062706.154450 [DOI] [PubMed] [Google Scholar]
- 2.Vinothkumar KR and Henderson R (2010) Structures of membrane proteins. Q Rev Biophys 43, 65–158 10.1017/S0033583510000041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen KN, Entova S, Ray LC and Imperiali B (2019) Monotopic Membrane Proteins Join the Fold. Trends Biochem Sci 44, 7–20 10.1016/j.tibs.2018.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nadolski MJ and Linder ME (2007) Protein lipidation. FEBS J 274, 5202–5210 10.1111/j.1742-4658.2007.06056.x [DOI] [PubMed] [Google Scholar]
- 5.Whited AM and Johs A (2015) The interactions of peripheral membrane proteins with biological membranes. Chem Phys Lipids 192, 51–59 10.1016/j.chemphyslip.2015.07.015 [DOI] [PubMed] [Google Scholar]
- 6.Majeed S, Ahmad AB, Sehar U and Georgieva ER (2021) Lipid Membrane Mimetics in Functional and Structural Studies of Integral Membrane Proteins. Membranes (Basel) 11, 685 10.3390/membranes11090685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dörr JM, Scheidelaar S, Koorengevel MC, Dominguez JJ, Schäfer M, van Walree CA, et al. (2016) The styrene–maleic acid copolymer: a versatile tool in membrane research. European Biophysics Journal 45, 3–21 10.1007/s00249-015-1093-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Popot J-L, Althoff T, Bagnard D, Banères J-L, Bazzacco P, Billon-Denis E, et al. (2011) Amphipols From A to Z. Annu Rev Biophys 40, 379–408 10.1146/annurev-biophys-042910-155219 [DOI] [PubMed] [Google Scholar]
- 9.Wong M, Thomas JK and Nowak T (1977) Structure and state of water in reversed micelles. 3. J Am Chem Soc 99, 4730–4736 10.1021/ja00456a034 [DOI] [Google Scholar]
- 10.Fuglestad B, Gupta K, Wand AJ and Sharp KA (2019) Water loading driven size, shape, and composition of cetyltrimethylammonium/hexanol/pentane reverse micelles. J Colloid Interface Sci 540, 207–217 10.1016/j.jcis.2019.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fathi H, Kelly JP, Vasquez VR and Graeve OA (2012) Ionic Concentration Effects on Reverse Micelle Size and Stability: Implications for the Synthesis of Nanoparticles. Langmuir 28, 9267–9274 10.1021/la300586f [DOI] [PubMed] [Google Scholar]
- 12.Eskici G and Axelsen PH (2016) The Size of AOT Reverse Micelles. J Phys Chem B 120, 11337–11347 10.1021/acs.jpcb.6b06420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuglestad B, Marques BS, Jorge C, Kerstetter NE, Valentine KG and Wand AJ (2019) Reverse Micelle Encapsulation of Proteins for NMR Spectroscopy. In: Methods in enzymology. Elsevier; 2019. p. 43–75. 10.1016/bs.mie.2018.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nucci NV, Valentine KG and Wand AJ (2014) High-resolution NMR spectroscopy of encapsulated proteins dissolved in low-viscosity fluids. Journal of Magnetic Resonance 241, 137–147 10.1016/j.jmr.2013.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nucci NV, Marques BS, Bédard S, Dogan J, Gledhill JM, Moorman VR, et al. (2011) Optimization of NMR spectroscopy of encapsulated proteins dissolved in low viscosity fluids. J Biomol NMR 50, 421 10.1007/s10858-011-9528-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peterson RW, Lefebvre BG and Wand AJ (2005) High-Resolution NMR Studies of Encapsulated Proteins in Liquid Ethane. J Am Chem Soc 127, 10176–10177 10.1021/ja0526517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kielec JM, Valentine KG, Babu CR and Wand AJ (2009) Reverse Micelles in Integral Membrane Protein Structural Biology by Solution NMR Spectroscopy. Structure 17, 345–351 10.1016/j.str.2009.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faeder J and Ladanyi BM (2000) Molecular Dynamics Simulations of the Interior of Aqueous Reverse Micelles. J Phys Chem B 104, 1033–1046 10.1021/jp993076u [DOI] [Google Scholar]
- 19.Bakulin AA, Cringus D, Pieniazek PA, Skinner JL, Jansen TLC and Pshenichnikov MS (2013) Dynamics of Water Confined in Reversed Micelles: Multidimensional Vibrational Spectroscopy Study. J Phys Chem B 117, 15545–15558 10.1021/jp405853j [DOI] [PubMed] [Google Scholar]
- 20.Harpham MR, Ladanyi BM, Levinger NE and Herwig KW (2004) Water motion in reverse micelles studied by quasielastic neutron scattering and molecular dynamics simulations. J Chem Phys 121, 7855–7868 10.1063/1.1792592 [DOI] [PubMed] [Google Scholar]
- 21.Costard R, Levinger NE, Nibbering ETJ and Elsaesser T (2012) Ultrafast Vibrational Dynamics of Water Confined in Phospholipid Reverse Micelles. J Phys Chem B 116, 5752–5759 10.1021/jp3039016 [DOI] [PubMed] [Google Scholar]
- 22.Vincent M, de Foresta B and Gallay J (2005) Nanosecond Dynamics of a Mimicked Membrane-Water Interface Observed by Time-Resolved Stokes Shift of LAURDAN. Biophys J 88, 4337–4350 10.1529/biophysj.104.057497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakulin AA and Pshenichnikov MS (2011) Reduced coupling of water molecules near the surface of reverse micelles. Physical Chemistry Chemical Physics 13, 19355 10.1039/c1cp22235j [DOI] [PubMed] [Google Scholar]
- 24.Crans DC, Rithner CD, Baruah B, Gourley BL and Levinger NE (2006) Molecular Probe Location in Reverse Micelles Determined by NMR Dipolar Interactions. J Am Chem Soc 128, 4437–4445 10.1021/ja0583721 [DOI] [PubMed] [Google Scholar]
- 25.Novaira M, Biasutti MA, Silber JJ and Correa NM (2007) New Insights on the Photophysical Behavior of PRODAN in Anionic and Cationic Reverse Micelles: From Which State or States Does It Emit? J Phys Chem B 111, 748–759 10.1021/jp065528q [DOI] [PubMed] [Google Scholar]
- 26.Novaira M, Moyano F, Biasutti MA, Silber JJ and Correa NM (2008) An Example of How to Use AOT Reverse Micelle Interfaces to Control a Photoinduced Intramolecular Charge-Transfer Process. Langmuir 24, 4637–4646 10.1021/la704004m [DOI] [PubMed] [Google Scholar]
- 27.Sengupta B, Guharay J and Sengupta PK (2000) Characterization of the fluorescence emission properties of prodan in different reverse micellar environments. Spectrochim Acta A Mol Biomol Spectrosc 56, 1433–1441 10.1016/S1386-1425(00)00245-6 [DOI] [PubMed] [Google Scholar]
- 28.Ho M-C and Chang C-W (2014) Cationic and anionic reverse micelles as the molecular crowding container for G-quadruplex structure. RSC Adv. 4, 20531–20534 10.1039/C4RA02141J [DOI] [Google Scholar]
- 29.Wiebenga-Sanford BP, Washington JB, Cosgrove B, Palomares EF, Vasquez DA, Rithner CD, et al. (2018) Sweet Confinement: Glucose and Carbohydrate Osmolytes in Reverse Micelles. J Phys Chem B 122, 9555–9566 10.1021/acs.jpcb.8b07406 [DOI] [PubMed] [Google Scholar]
- 30.Honegger P and Steinhauser O (2019) Towards capturing cellular complexity: combining encapsulation and macromolecular crowding in a reverse micelle. Physical Chemistry Chemical Physics 21, 8108–8120 10.1039/C9CP00053D [DOI] [PubMed] [Google Scholar]
- 31.Wand AJ, Ehrhardt MR and Flynn PF (1998) High-resolution NMR of encapsulated proteins dissolved in low-viscosity fluids. Proceedings of the National Academy of Sciences 95, 15299–15302 10.1073/pnas.95.26.15299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nucci NV, Fuglestad B, Athanasoula EA and Wand AJ (2014) Role of cavities and hydration in the pressure unfolding of T4 lysozyme. Proceedings of the National Academy of Sciences 111, 13846–13851 10.1073/pnas.1410655111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Senske M, Smith AE and Pielak GJ (2016) Protein Stability in Reverse Micelles. Angewandte Chemie International Edition 55, 3586–3589 10.1002/anie.201508981 [DOI] [PubMed] [Google Scholar]
- 34.Peterson RW, Anbalagan K, Tommos C and Wand AJ (2004) Forced Folding and Structural Analysis of Metastable Proteins. J Am Chem Soc 126, 9498–9499 10.1021/ja047900q [DOI] [PubMed] [Google Scholar]
- 35.Xu G, Cheng K, Wu Q, Liu M and Li C (2017) Confinement Alters the Structure and Function of Calmodulin. Angewandte Chemie International Edition 56, 530–534 10.1002/anie.201609639 [DOI] [PubMed] [Google Scholar]
- 36.Marques BS, Stetz MA, Jorge C, Valentine KG, Wand AJ and Nucci NV (2020) Protein conformational entropy is not slaved to water. Sci Rep 10, 17587 10.1038/s41598-020-74382-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pometun MS, Peterson RW, Babu CR and Wand AJ (2006) Cold Denaturation of Encapsulated Ubiquitin. J Am Chem Soc 128, 10652–10653 10.1021/ja0628654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Babu CR, Hilser VJ and Wand AJ (2004) Direct access to the cooperative substructure of proteins and the protein ensemble via cold denaturation. Nat Struct Mol Biol 11, 352–357 10.1038/nsmb739 [DOI] [PubMed] [Google Scholar]
- 39.Van Horn WD, Simorellis AK and Flynn PF (2005) Low-temperature studies of encapsulated proteins. J Am Chem Soc 127, 13553–13560 10.1021/ja052805i [DOI] [PubMed] [Google Scholar]
- 40.Nucci NV, Pometun MS and Wand AJ (2011) Mapping the Hydration Dynamics of Ubiquitin. J Am Chem Soc 133, 12326–12329 10.1021/ja202033k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nucci NV, Pometun MS and Wand AJ (2011) Site-resolved measurement of water-protein interactions by solution NMR. Nat Struct Mol Biol 18, 245–249 10.1038/nsmb.1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gallo PN, Iovine JC and Nucci NV (2018) Toward comprehensive measurement of protein hydration dynamics: Facilitation of NMR-based methods by reverse micelle encapsulation. Methods 148, 146–153 10.1016/j.ymeth.2018.07.008 [DOI] [PubMed] [Google Scholar]
- 43.Jean Y-C and Ache HJ (1978) Study of the micelle formation and the effect of additives on this process in reversed micellar systems by positron annihilation techniques. J Am Chem Soc 100, 6320–6327 10.1021/ja00488a004 [DOI] [Google Scholar]
- 44.Schulman JH, Stoeckenius W and Prince LM (1959) Mechanism of Formation and Structure of Micro Emulsions by Electron Microscopy. J Phys Chem 63, 1677–1680 10.1021/j150580a027 [DOI] [Google Scholar]
- 45.Choi Y and Chang P-S (2022) Kinetic modeling of lipase-catalysed hydrolysis of triacylglycerol in a reverse micelle system for the determination of integral stereoselectivity. Catal Sci Technol 12, 2819–2828 10.1039/D1CY02182F [DOI] [Google Scholar]
- 46.Hayes DG and Gulari E (1990) Esterification reactions of lipase in reverse micelles. Biotechnol Bioeng 35, 793–801 10.1002/bit.260350807 [DOI] [PubMed] [Google Scholar]
- 47.Rao AM, Murray MA, John VT and Abraham G (1991) Characteristics of Lipase Catalysis During Ester Synthesis in Reversed Micellar Systems. Biocatalysis 4, 253–264 10.3109/10242429109000689 [DOI] [Google Scholar]
- 48.Orlich B and Schomäcker R (2002) Enzyme Catalysis in Reverse Micelles. In: Dutta NN, Hammar F, Haralampidis K and Karanth NG, editors History and Trends in Bioprocessing and Biotransformation. Advances in Biochemical Engineering/Biotechnology. Berlin: Springer; 2002. p. 185–208. 10.1007/3-540-44604-4_6 [DOI] [PubMed] [Google Scholar]
- 49.Carvalho CM and Cabral JM (2000) Reverse micelles as reaction media for lipases. Biochimie 82, 1063–85 10.1016/s0300-9084(00)01187-1 [DOI] [PubMed] [Google Scholar]
- 50.Binks BP, Chatenay D, Nicot C, Urbach W and Waks M (1989) Structural parameters of the myelin transmembrane proteolipid in reverse micelles. Biophys J 55, 949–55 10.1016/S0006-3495(89)82893-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicot C, Vacher M, Vincent M, Gallay J and Waks M (1985) Membrane proteins in reverse micelles: myelin basic protein in a membrane-mimetic environment. Biochemistry 24, 7024–7032 10.1021/bi00345a041 [DOI] [PubMed] [Google Scholar]
- 52.Merdas A, Gindre M, Le Huérou J-Y, Nicot C, Ober R, Urbach W, et al. (1998) Bridging of Nonionic Reverse Micelles by a Myelin Transmembrane Protein. J Phys Chem B 102, 528–533 10.1021/jp9717498 [DOI] [Google Scholar]
- 53.Luisi PL (1985) Enzymes Hosted in Reverse Micelles in Hydrocarbon Solution. Angewandte Chemie International Edition in English 24, 439–450 10.1002/anie.198504393 [DOI] [Google Scholar]
- 54.Ramakrishnan VR, Darszon A and Montal M (1983) A small angle x-ray scattering study of a rhodopsin-lipid complex in hexane. Journal of Biological Chemistry 258, 4857–4860 10.1016/S0021-9258(18)32504-3 [DOI] [PubMed] [Google Scholar]
- 55.Wirz J and Rosenbusch JP (1984) The Formation of Reverse Mixed Micelles Consisting of Membrane Proteins and AOT in Isooctane. In: Luisi PL and Straub BE, editorsReverse Micelles. Boston, MA: Springer US; 1984. p. 231–238. 10.1007/978-1-4757-6424-6_22 [DOI] [Google Scholar]
- 56.Chaudhuri A, Basu P, Haldar S, Kombrabail M, Krishnamoorthy G, Rajarathnam K, et al. (2013) Organization and Dynamics of the N-Terminal Domain of Chemokine Receptor CXCR1 in Reverse Micelles: Effect of Graded Hydration. J Phys Chem B 117, 1225–1233 10.1021/jp3095352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yeung PS-W and Axelsen PH (2012) The Crowded Environment of a Reverse Micelle Induces the Formation of β-Strand Seed Structures for Nucleating Amyloid Fibril Formation. J Am Chem Soc 134, 6061–6063 10.1021/ja3004478 [DOI] [PubMed] [Google Scholar]
- 58.Martinez AV, DeSensi SC, Dominguez L, Rivera E and Straub JE (2011) Protein folding in a reverse micelle environment: The role of confinement and dehydration. J Chem Phys 134 10.1063/1.3545982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rudolph-Böhner S, Quarzago D, Czisch M, Ragnarsson U and Moroder L (1997) Conformational preferences of Leu-enkephalin in reverse micelles as membrane-mimicking environment. Biopolymers 41, 591–606 [DOI] [Google Scholar]
- 60.Liu S, Shibata A, Ueno S, Xu F, Baba Y, Jiang D, et al. (2006) Investigation of interaction of Leu-enkephalin with lipid membranes. Colloids Surf B Biointerfaces 48, 148–158 10.1016/j.colsurfb.2006.02.005 [DOI] [PubMed] [Google Scholar]
- 61.Van Horn WD, Ogilvie ME and Flynn PF (2008) Use of reverse micelles in membrane protein structural biology. J Biomol NMR 40, 203–211 10.1007/s10858-008-9227-5 [DOI] [PubMed] [Google Scholar]
- 62.Peterson RW, Pometun MS, Shi Z and Wand AJ (2005) Novel surfactant mixtures for NMR spectroscopy of encapsulated proteins dissolved in low-viscosity fluids. Protein Science 14, 2919–2921 10.1110/ps.051535405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kielec JM, Valentine KG, Babu CR and Wand AJ (2009) Reverse Micelles in Integral Membrane Protein Structural Biology by Solution NMR Spectroscopy. Structure 17, 345–351 10.1016/j.str.2009.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Valentine KG, Peterson RW, Saad JS, Summers MF, Xu X, Ames JB, et al. (2010) Reverse Micelle Encapsulation of Membrane-Anchored Proteins for Solution NMR Studies. Structure 18, 9–16 10.1016/j.str.2009.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.MacKenzie DWS, Schaefer A, Steckner J, Leo CA, Naser D, Artikis E, et al. (2022) A fine balance of hydrophobic-electrostatic communication pathways in a pH-switching protein. Proceedings of the National Academy of Sciences 119, e2119686119 10.1073/pnas.2119686119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moravcevic K, Oxley CL and Lemmon MA (2012) Conditional Peripheral Membrane Proteins: Facing up to Limited Specificity. Structure 20, 15–27 10.1016/j.str.2011.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.O’Brien ES, Nucci NV, Fuglestad B, Tommos C and Wand AJ (2015) Defining the Apoptotic Trigger: The Interaction of Cytochrome C and Cardiolipin. Journal of Biological Chemistry 290, 30879–30887 10.1074/jbc.M115.689406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dodevski I, Nucci NV, Valentine KG, Sidhu GK, O’Brien ES, Pardi A, et al. (2014) Optimized Reverse Micelle Surfactant System for High-Resolution NMR Spectroscopy of Encapsulated Proteins and Nucleic Acids Dissolved in Low Viscosity Fluids. J Am Chem Soc 136, 3465–3474 10.1021/ja410716w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stackhouse CI, Pierson KN, Labrecque CL, Mawson C, Berg J, Fuglestad B, et al. (2024) Characterization of 10MAG/LDAO reverse micelles: Understanding versatility for protein encapsulation. Biophys Chem 311, 107269 10.1016/j.bpc.2024.107269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kagan VE, Bayır HA, Belikova NA, Kapralov O, Tyurina YY, Tyurin VA, et al. (2009) Cytochrome c/cardiolipin relations in mitochondria: a kiss of death. Free Radic Biol Med 46, 1439–1453 10.1016/j.freeradbiomed.2009.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang J, Tang C, Wang Y, Chang C, Zhang J, Hu J, et al. (2019) The terahertz dynamics interfaces to ion–lipid interaction confined in phospholipid reverse micelles. Chemical Communications 55, 15141–15144 10.1039/C9CC07598D [DOI] [PubMed] [Google Scholar]
- 72.Lehtinen O-P, Nugroho RWN, Lehtimaa T, Vierros S, Hiekkataipale P, Ruokolainen J, et al. (2017) Effect of temperature, water content and free fatty acid on reverse micelle formation of phospholipids in vegetable oil. Colloids Surf B Biointerfaces 160, 355–363 10.1016/j.colsurfb.2017.09.050 [DOI] [PubMed] [Google Scholar]
- 73.Abel S, Galamba N, Karakas E, Marchi M, Thompson WH and Laage D (2016) On the Structural and Dynamical Properties of DOPC Reverse Micelles. Langmuir 32, 10610–10620 10.1021/acs.langmuir.6b02566 [DOI] [PubMed] [Google Scholar]
- 74.Levinger NE, Costard R, Nibbering ETJ and Elsaesser T (2011) Ultrafast Energy Migration Pathways in Self-Assembled Phospholipids Interacting with Confined Water. J Phys Chem A 115, 11952–11959 10.1021/jp206099a [DOI] [PubMed] [Google Scholar]
- 75.Folpini G, Siebert T, Woerner M, Abel S, Laage D and Elsaesser T (2017) Water Librations in the Hydration Shell of Phospholipids. J Phys Chem Lett 8, 4492–4497 10.1021/acs.jpclett.7b01942 [DOI] [PubMed] [Google Scholar]
- 76.Doussin S, Birlirakis N, Georgin D, Taran F and Berthault P (2006) Novel Zwitterionic Reverse Micelles for Encapsulation of Proteins in Low-Viscosity Media. Chemistry – A European Journal 12, 4170–4175 10.1002/chem.200501422 [DOI] [PubMed] [Google Scholar]
- 77.Labrecque CL, Nolan AL, Develin AM, Castillo AJ, Offenbacher AR and Fuglestad B (2022) Membrane-Mimicking Reverse Micelles for High-Resolution Interfacial Study of Proteins and Membranes. Langmuir 38, 3676–3686 10.1021/acs.langmuir.1c03085 [DOI] [PubMed] [Google Scholar]
- 78.Glonek T (1998) 31P nuclear magnetic resonance phospholipid analysis of anionic-enriched lecithins. J Am Oil Chem Soc 75, 569–573 10.1007/s11746-998-0067-y [DOI] [Google Scholar]
- 79.Walde P, Giuliani AM, Boicelli CA and Luisi PL (1990) Phospholipid-based reverse micelles. Chem Phys Lipids 53, 265–288 10.1016/0009-3084(90)90026-N [DOI] [PubMed] [Google Scholar]
- 80.Willard DM, Riter RE and Levinger NE (1998) Dynamics of Polar Solvation in Lecithin/Water/Cyclohexane Reverse Micelles. J Am Chem Soc 120, 4151–4160 10.1021/ja980086k [DOI] [Google Scholar]
- 81.Wachtel E, Federman S and Greenspoon N (1992) Interaction of Carbohydrates with Phosphatidylcholine Inverse Micelles. Isr J Chem 32, 113–119 10.1002/ijch.199200015 [DOI] [Google Scholar]
- 82.Penttilä PA, Vierros S, Utriainen K, Carl N, Rautkari L, Sammalkorpi M, et al. (2019) Phospholipid-Based Reverse Micelle Structures in Vegetable Oil Modified by Water Content, Free Fatty Acid, and Temperature. Langmuir 35, acs.langmuir.9b01135 10.1021/acs.langmuir.9b01135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oh E-J, Park D-G, Lim Y-S, Sik Jin K and Lee H-Y (2022) Structural transition of reverse cylindrical micelles to reverse vesicles by mixtures of lecithin and inorganic salts. J Colloid Interface Sci 615, 768–777 10.1016/j.jcis.2022.02.015 [DOI] [PubMed] [Google Scholar]
- 84.Boicelli CA, Conti F, Giomini M and Giuliani AM (1982) Interactions of small molecules with phospholipids in inverted micelles. Chem Phys Lett 89, 490–496 10.1016/0009-2614(82)83052-2 [DOI] [Google Scholar]
- 85.Ceraulo L, Ferrugia M, Tesoriere L, Segreto S, Livrea MA and Liveri VT (1999) Interactions of melatonin with membrane models: Portioning of melatonin in AOT and lecithin reversed micelles. J Pineal Res 26, 108–112 10.1111/j.1600-079X.1999.tb00570.x [DOI] [PubMed] [Google Scholar]
- 86.Walters SH and Fuglestad B (2024) An NMR Approach for Investigating Membrane Protein–Lipid Interactions Using Native Reverse Micelles. Bio Protoc 14 10.21769/BioProtoc.5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Walters SH, Castillo AJ, Develin AM, Labrecque CL, Qu Y and Fuglestad B (2023) Investigating protein-membrane interactions using native reverse micelles constructed from naturally sourced lipids. Protein Science 32 10.1002/pro.4786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Murray CW and Rees DC (2009) The rise of fragment-based drug discovery. Nat Chem 1, 187–192 10.1038/nchem.217 [DOI] [PubMed] [Google Scholar]
- 89.Davis BJ and Giannetti AM (2016) The Synthesis of Biophysical Methods In Support of Robust Fragment-Based Lead Discovery. In: Fragment-based Drug Discovery Lessons and Outlook. Wiley Online Library; 2016. p. 119–138. 10.1002/9783527683604.ch06 [DOI] [Google Scholar]
- 90.Shuker SB, Hajduk PJ, Meadows RP and Fesik SW (1996) Discovering High-Affinity Ligands for Proteins: SAR by NMR. Science (1979) 274, 1531–1534 10.1126/science.274.5292.1531 [DOI] [PubMed] [Google Scholar]
- 91.Li GC, Castro MA, Ukwaththage T and Sanders CR (2024) Optimizing NMR fragment-based drug screening for membrane protein targets. J Struct Biol X 9, 100100 10.1016/j.yjsbx.2024.100100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fuglestad B, Kerstetter NE and Wand AJ (2019) Site-Resolved and Quantitative Characterization of Very Weak Protein–Ligand Interactions. ACS Chem Biol 14, 1398–1402 10.1021/acschembio.9b00353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fuglestad B, Kerstetter NE, Bédard S and Wand AJ (2019) Extending the Detection Limit in Fragment Screening of Proteins Using Reverse Micelle Encapsulation. ACS Chem Biol 14, acschembio.9b00537 10.1021/acschembio.9b00537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Labrecque CL and Fuglestad B (2024) Ligandability at the Membrane Interface of GPx4 Revealed through a Reverse Micelle Fragment Screening Platform. JACS Au 4, 2676–2686 10.1021/jacsau.4c00427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Seibt TM, Proneth B and Conrad M (2019) Role of GPX4 in ferroptosis and its pharmacological implication. Free Radic Biol Med 133, 144–152 10.1016/j.freeradbiomed.2018.09.014 [DOI] [PubMed] [Google Scholar]
- 96.Ursini F and Maiorino M (2020) Lipid peroxidation and ferroptosis: The role of GSH and GPx4. Free Radic Biol Med 152, 175–185 10.1016/j.freeradbiomed.2020.02.027 [DOI] [PubMed] [Google Scholar]
- 97.Warschawski DE, Arnold AA, Beaugrand M, Gravel A, Chartrand É and Marcotte I (2011) Choosing membrane mimetics for NMR structural studies of transmembrane proteins. Biochimica et Biophysica Acta (BBA) - Biomembranes 1808, 1957–1974 10.1016/j.bbamem.2011.03.016 [DOI] [PubMed] [Google Scholar]


