Abstract
Dioecy in flowering plants has evolved independently many times, and thus the genetic mechanisms underlying sex determination are diverse. In hemp (Cannabis sativa), sex is controlled by a pair of sex chromosomes (XX for females and XY for males). In an attempt to understand the molecular mechanism responsible for sex expression in hemp plants, we carried out RNA sequencing of male and female plants at different developmental stages. Using a pipeline involving differential gene expression analysis and weighted gene co-expression network analysis, we identified genes important for male and female flower development. We also demonstrate that sex-biased expression is already established at very early vegetative stages, before the onset of reproductive development, and identify several genes encoding transcription factors of the REM, bZIP, and MADS families as candidate sex-determination genes in hemp. Our findings demonstrate that the gene regulatory networks governing male and female development in hemp diverge at a very early stage, leading to profound morphological differences between male and female hemp plants.
Keywords: bZIP, Cannabis sativa, differential gene expression, dioecy, hemp, sex determination, TM8, transcriptomics, REM, weighted gene co-expression network analysis
Differential gene expression analyses in young vegetative hemp plants identifies genes potentially crucial for sex determination in Cannabis sativa .
Introduction
Unlike animals, including humans, which are predominantly categorized as male or female, flowering plants are primarily bisexual (Sauquet et al., 2017). Within a single flower, both male and female reproductive organs coexist. Only approximately 12% of angiosperms produce unisexual flowers, with male and female flowers present either on the same plant (monoecious) or on different plants (dioecious) (Renner, 2014). Only 6% of flowering plants are dioecious and have distinct male and female individuals (Renner, 2014).
The molecular mechanisms governing dioecy remain elusive for the vast majority of species, but it appears likely that sex determination through a pair of sex chromosomes is common (Bachtrog et al., 2014; Renner, 2014; Masuda and Akagi, 2023). This can occur in the form of the common XY or ZW systems (Charlesworth, 2016). These systems show a high degree of evolutionary dynamics; for example, Populus euphratica has an XY system whereas the closely related Populus alba has a ZW system (Yang et al., 2021).
In a small number of species of flowering plants, a sex-linked region in which recombination is suppressed has been identified, and in an even smaller number, sex-determination genes within this region have been characterized (Charlesworth, 2016; Zhang et al., 2022; Masuda and Akagi, 2023). For a large number of plants, however, the sex-determination system remains largely uncharacterized.
Hemp is a primarily dioecious multipurpose crop that has gained renewed popularity as a crop for sustainable agriculture and as a medicinal plant (Schluttenhofer and Yuan, 2017; Schilling et al., 2021). Male and female hemp flowers differ morphologically, and male and female plants also possess different inflorescence structures (Spitzer-Rimon et al., 2019; Leme et al., 2020; Shi et al., 2024, Preprint). The female inflorescence of hemp plants is primarily associated with medicinal properties. The hemp inflorescence serves as a crucial source of cannabidiol (CBD), a non-psychoactive cannabinoid that offers various potential health benefits (Schluttenhofer and Yuan, 2017). CBD is exclusively extracted from female inflorescences and is utilized in a variety of products, including oils, tinctures, capsules, edibles, and topicals (Szalata et al., 2022). CBD-rich hemp plants are used for the production of medicinal products aimed at managing conditions such as pain, anxiety, epilepsy, and inflammation (Fasinu et al., 2016; Schilling et al., 2021).
Sex determination in hemp is complex and involves both genetic and environmental factors, although the primary determinant seems to be genetic (Schilling et al., 2021). Female plants have XX chromosomes, whereas male plants have XY chromosomes (Divashuk et al., 2014). The X and Y chromosomes are both 80–100 Mbps in size (Divashuk et al., 2014; Laverty et al., 2019; Grassa et al., 2021), and the non-recombining sex-linked region constitutes approximately 70% of the chromosomes, thus being relatively large compared with other dioecious species (Charlesworth, 2016; Prentout et al., 2020). The sex chromosomes are the oldest documented so far in plants, with an approximate age of 12–28 million years. It is likely that recombination between the X and Y chromosomes stopped millions of years ago, before Cannabis and its sister genus Humulus diverged from each other (Prentout et al., 2021). As a consequence, considerable sequence divergence has accumulated in the 70 Mbps non-recombining region, with hundreds of genes being sex linked (Prentout et al., 2020, 2021).
Many currently used methods to identify sex-determination genes rely on the identification of a sex-linked region on the X and Y chromosomes and the subsequent characterization of the most promising candidate sex-determination genes within this sex-linked region. This approach has proven successful in persimmon, kiwifruit, poplar, asparagus, date palms, and wild grapevine, in which sex-linked regions are relatively small (Akagi et al., 2018; Torres et al., 2018; Harkess et al., 2020; Massonnet et al., 2020; Müller et al., 2020). However, understanding the genetic basis of sex determination in hemp presents significant challenges primarily due to the large size of its sex-linked region, which contains thousands of genes (Laverty et al., 2019; Prentout et al., 2020; Grassa et al., 2021). In addition, the existing genome assemblies predominantly represent female plants, which presents challenges in identifying sex-determination genes that may be located on the Y chromosome (Feng et al., 2020; Grassa et al., 2021).
Gene expression analyses have identified sex-linked (i.e. putatively located on sex chromosomes) genes in hemp and have also found that a large number of hormone-related genes and developmental regulators are differentially expressed between mature male and female flowers (Prentout et al., 2020; Adal et al., 2021). Those approaches showed that substantial differences in gene expression exist between male and female hemp flowers, and that gene regulatory networks controlling male and female flower development might be quite divergent (Adal et al., 2021). As male and female hemp plants differ morphologically at the transition from vegetative to reproductive development (Shi et al., 2024, Preprint), it appears likely that molecular circuits governing male versus female development diverge very early during flower development or even before flower development starts (Adal et al., 2021; Shi et al., 2024, Preprint).
Here, we used transcriptomics data from early developmental stages (vegetative and early flowering stages) of hemp to undertake differential gene expression (DGE) analysis and weighted gene correlation network analysis (WGCNA) to identify candidate sex-determination genes. By combining both analyses, we detected a relatively small number of possible candidate genes among the hundreds of genes on the X and Y chromosomes. Among the candidates are genes encoding homologs of the transcription factors FLOWERING LOCUS D (FD) on the Y chromosome and REPRODUCTIVE MERISTEM 16 (REM16) on the X chromosome, homologs of both of which are implicated in reproductive development in other plants (Abe et al., 2005; Matias-Hernandez et al., 2010; Mantegazza et al., 2014; Mendes et al., 2016; Caselli et al., 2019; Manrique et al., 2023), thus constituting potential candidate sex-determination genes in hemp.
Materials and methods
Plant cultivation
Seeds of the photoperiod-insensitive hemp (Cannabis sativa L.) cultivar ‘FINOLA’ were sown in a mixture of one part perlite, one part vermiculite, and two parts compost (John Innes no. 2). After incubation in darkness for 2 d, the seedlings were moved to a greenhouse and cultivated under natural light conditions (Dublin, Ireland, from June to September 2021). The sex of all plants used for RNA sequencing (RNA-Seq) was confirmed by observing mature flowers.
RNA isolation and transcriptome sequencing
For the extraction of RNA, apical structures, including newly emerged leaves up to 10 mm in size, were collected from 20 individual hemp plants with 2, 4, and 9 leaf pairs (stages L2, L4, and L9), as previously described (Shi et al., 2024, Preprint). Tissues collected for RNA isolation from male and female hemp plants at different stages are shown in Fig. 1. RNA isolation was performed using the RNeasy® Plant Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions. Residual DNA was digested using RNase-free DNase I (Thermo Fisher Scientific, USA) following the manufacturer’s instructions. The amount of 1 μg RNA per sample for DNA digestion was calculated based on the RNA concentration examined from the NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, USA).
Fig. 1.
Apical tissue of male and female plants at different developmental stages was used for RNA-Seq. (A) Male hemp plants of the ‘FINOLA’ cultivar at a vegetative stage exhibit two true leaf pairs (stage L2M). (B) Male plants have transitioned into a reproductive stage and have a macroscopically visible apical inflorescence at four true leaf pairs (stage L4M) (B). (C, D) Female hemp plants of the ‘FINOLA’ cultivar with two (C) and four (D) true leaf pairs are in a vegetative stage (stages L2F and L4F). (E) Female plants start visibly flowering when they have approximately nine true leaf pairs (stage L9F). In each image, the white arrowhead points to the apex of the plant. Apical structures included the meristem and the most recent newly formed leaf pair not larger than 10 mm. Scale bars=10 mm. Staging was carried out according to Shi et al. (2024, Preprint).
Twenty RNA samples comprised four biological replicates (samples collected from four independent plants at the same growth stages) for five groups: two-leaf stage male (L2M) and female (L2F), L4M, L4F, and L9F. The RNA integrity numbers of all 20 RNA samples tested by the Qubit RNA IQ Assay Kit (Invitrogen, USA) were >8.5. RNA-Seq was performed using directional RNA-Seq (polyA library prep, PE150 sequencing, 8 GB data output per sample; Novogene, UK). Raw reads are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra), BioProject accession PRJNA1126191. Raw sequencing reads were processed using the Galaxy platform (Afgan et al., 2018). The quality of reads was inspected by using FastQC v. 0.11.8 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). RNA-Seq QC scores were ~36, indicating that the original RNA-Seq had high-quality sequences and did not require trimming.
Reads were mapped to the CBDRx reference transcriptome (https://www.ncbi.nlm.nih.gov/assembly/GCF_900626175.2/) (Grassa et al., 2021) using Salmon (Patro et al., 2017). Tximport was used to import transcript expression levels and calculate gene expression in transcripts per million (TPM) (Soneson et al., 2016).
Identification of putative Y chromosomal transcripts
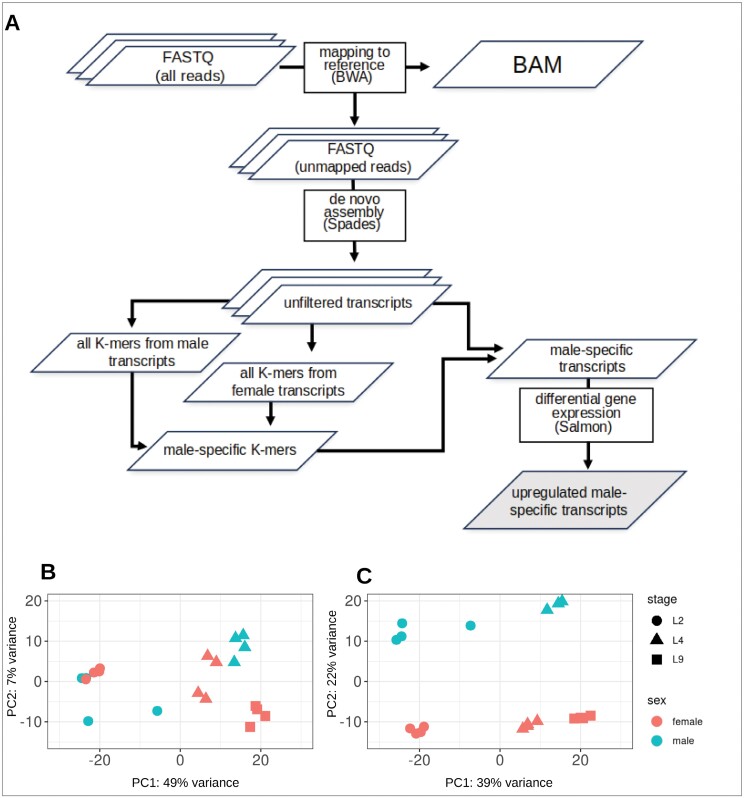
The Y chromosome sequence is missing from the CBDRx reference genome because the individual from which the genome was generated was a female plant with XX chromosomes (Grassa et al., 2021) and no reliable Y chromosome assembly at full chromosome level of a male C. sativa individual was available at the time of the study. This meant that RNA-Seq reads from male plants in our analyses that did not map to the female reference genome were composed of bacterial contamination and noise resulting from sequencing artefacts, as well as reads from the Y chromosome transcripts. Hence, unmapped male RNA-Seq reads can be used to generate Y-specific transcripts. After assembling those reads, identifying male-specific transcripts directly is challenging due to the variability introduced by single nucleotide polymorphisms, sequencing errors, and assembly artefacts, which make it difficult to directly identify transcripts that are conserved across all male samples. By focusing on conserved short sequences, in this case 16-mers, it is possible to more reliably detect male-specific transcripts, even with such variability, consequently improving the accuracy and efficiency of our analysis. Therefore, a K-mer approach was used to identify short sequences of the assembled transcripts conserved among male-derived samples, and subsequently to distinguish between Y chromosome-specific reads and contamination. Similar approaches have been used successfully in previous studies to identify sex-specific loci (Akagi et al., 2018; Torres et al., 2018). First, unmapped reads from male and female samples were assembled into longer transcripts using rnaSPAdes (Bushmanova et al., 2019). The transcripts were then used to generate K-mers (16-mers) using Jellyfish (Marçais and Kingsford, 2011). Subsequently, K-mers shared between all L4M male samples were selected. The same process was repeated to generate K-mers from samples derived from L4F female plants, and K-mers present in females were removed from the dataset containing male K-mers. As a result, K-mers were identified that were unique to males and not present in any female samples. These K-mers were then used to select the corresponding transcripts containing the identified male K-mers putatively derived from the Y chromosome. Those transcripts were further filtered to retain only the ones significantly overexpressed in males, with parameters of log2 fold change (log2FC) ≥1 and false discovery rate (FDR) P-value ≤0.05. Transcripts that we considered to be putatively encoded on the Y chromosome (hereafter termed ChrY transcripts) identified using the K-mer approach but displaying either low expression in both male and female samples or very high sequence similarity and at the same time no differential expression were not considered candidates. A limitation of this approach is that it produces only male transcripts, and our dataset may contain, for example, isoforms that originated from an autosomal chromosome but differ in sequence between male and female plants (e.g. through alternative splicing).
Differential gene expression analysis
Pre-processed RNA-Seq paired reads were mapped to the CBDRx transcriptome to which the candidate ChrY transcripts were added using Salmon (Patro et al., 2017). The transcript expression levels were imported and summarized at the gene level using the R package tximport (Soneson et al., 2016). The counts were then used to perform differential expression using DESeq2 (Love et al., 2014). Differentially expressed genes (DEGs) were identified based on the following criteria: an absolute log2FC ≥1, and a FDR P-value ≤0.05. Additionally, the DEGs were filtered by expression level, with an average TPM value >1. An online platform for data analysis and visualization (http://www.bioinformatics.com.cn) was used to generate heatmaps from TPM values, perform bi-directional hierarchical clustering, and create volcano plots.
Weighted gene co-expression network analysis
After quantification of the reads using Salmon and tximport, a variance-stabilizing transformation was applied to the expression matrix using DESeq2 (get VarianceStabilizedData). Subsequently, a signed network was constructed (power of 24). The power was chosen to ensure a scale-free topology model fit with a signed R2 variable >0.9 and mean connectivity <100. Following the clustering process, from the modules that were identified, the ones that exhibited expression correlated to sample sex were selected. The analysis was processed using the R package WGCNA (Zhang and Horvath, 2005; Langfelder and Horvath, 2008).
Functional annotation of identified candidate genes in regulating sex determination and flower development
The Mercator4 v6.0 platform (Schwacke et al., 2019) was employed to annotate the sex-biased genes from DGE analysis and the sex-related genes from weighted gene co-expression network analysis (WGCNA), and to identify candidate genes via protein sequences using default settings. The generated mapping files were then used as an input for metabolic pathways analysis in MapMan 3.7.0 using default settings (Usadel et al., 2009).
Results
RNA-Seq generated from male and female samples at different developmental stages
To determine differences in gene expression profiles between male and female hemp plants, we sampled material from different developmental stages of plants of both sexes to identify candidate genes involved in flower development and sex determination. In hemp, sex-specific flower development follows a distinctive pattern, in which male flowers do not show any signs of carpel initiation, whereas female flowers develop no stamens (Leme et al., 2020; Shi et al., 2024, Preprint). Flower development and timing, as well as overall plant and inflorescence structure, are morphologically markedly different in male and female hemp plants from the onset of reproductive development (Shi et al., 2024, Preprint), suggesting that genes governing sex determination may be active before reproductive development begins. Accordingly, our sampling strategy focused on the apical structure in the vegetative stages and just after the transition to reproductive development (Fig. 1).
Male ‘FINOLA’ hemp plants at the second true leaf stage (L2M) show no signs of flowering and thus are expected to be in a vegetative stage (Fig. 1A) (Shi et al., 2024, Preprint). Most male hemp plants start flowering shortly after L2M, at the fourth true leaf stage (L4M; Fig. 1B) (Shi et al., 2024, Preprint). To be able to compare gene expression profiles at similar ages and developmental stages between male and female hemp plants, we collected samples of female apical structures at the second, fourth and ninth true leaf stages (L2F, L4F, and L9F) (Fig. 1C–E). L2F and L4F are vegetative stages but are of similar age to their male counterparts L2M and L4M, whereas L9F corresponds to an early flowering stage and is thus of a similar developmental stage to L4M. For male and female hemp plants at different growth stages, we generated 20 samples (2 male and 3 female stages with 4 biological replicates each) with over 590 million high-quality paired-end reads in total (Supplementary Table S1).
Putative Y chromosome transcripts can be identified from unmapped RNA-Seq reads
Our RNA-Seq data mapped well to the C. sativa CBDRx reference genome (83.4–87.1%; Supplementary Table S1). However, the reference genome was generated from a female C. sativa plant and therefore lacked the sequence of the Y chromosome (Grassa et al., 2021), and no assembled Y chromosome was available at the time of our analysis. Consequently, important genes related to sex determination in hemp might have been missing from our analysis. To address this issue, we established a pipeline that identifies male-exclusive transcripts using a K-mer approach based on previously established methods (Fig. 2A) (Torres et al., 2018). RNA-Seq reads that failed to map to the reference genome were used to assemble approximately 30 000 transcripts per sample, corresponding to ~20 million 16-mers per sample. We identified approximately 100 000 16-mers belonging to 2500 transcripts that were present in L4M male samples and absent in L4F female samples. These transcripts were further filtered based on their expression level, to select only those with a male-biased expression. This process resulted in 379 transcripts that we consider to be putatively encoded on the Y chromosome (ChrY transcripts) (Supplementary Table S2). By contrast, using the same pipeline but with female-specific K-mers resulted in the identification of only four transcripts (Supplementary Table S2). This indicates that the pipeline works well in identifying ChrY-specific transcripts, as most of the female (ChrX-specific) transcripts were expected to be already mapped to the C. sativa reference genome and contamination with microorganisms, as well as sequencing noise, was presumed to be similar between male and female samples.
Fig. 2.
Identification of male-specific transcripts through K-mer analysis. (A) Schematic of the K-mers-based approach for detecting male-specific transcripts in hemp. A pipeline identifies transcripts exclusive to male samples. (B, C) Principal component analysis plots of RNA-Seq data based on normalized read count data from RNA-Seq samples excluding (B) and including (C) the Y chromosome-specific transcripts. The PCA plot was generated using the R function plot PCA from the package DESeq2 (Love et al., 2014).
Subsequently, we performed a principal component analysis (PCA) with the normalized reads of all 20 RNA-Seq samples. When analysed using reads mapped to the reference transcriptome only, samples were separated clearly by developmental stage, but to a lesser extent according to sex (Fig. 2B). The inclusion of the ChrY transcripts led to a clear separation of male and female samples in the PCA (Fig. 2C).
To generate supporting evidence for their Y chromosome origin, the putative ChrY transcripts were BLASTed against the ‘FINOLA’ genome (Laverty et al., 2019) (ASM341772v2), which was generated from a male plant and includes Y chromosome contigs. For comparison, ChrY transcripts were also BLASTed against the female reference genome ‘CBDRx’ (GCF_900626175.2) (Grassa et al., 2021) (Supplementary Fig. S1). The average identity for the best match of each transcript against the ‘FINOLA’ genome was 98%, whereas the best match against CBDRx averaged 92% identity. Because male transcripts originating from the Y chromosome are expected to diverge in sequence from transcripts originating from the X chromosome, this suggests that our pipeline indeed identified ChrY transcripts.
However, an alternative explanation for the higher identity of the ChrY transcripts with the ‘FINOLA’ genome as opposed to the CBDRx genome is that our RNA-Seq reads were derived from ‘FINOLA’ plants and thus the better match would be explained not by the presence of the Y chromosome but by the genetic between-cultivar variation between ‘FINOLA’ and CBDRx. To test this possibility, 379 transcripts were randomly sampled from the CBDRx transcriptome and BLASTed against the ‘FINOLA’ genome. This resulted in a high identity, averaging 98%. This finding further supports the assumption that the high identity of ChrY transcripts with the ‘FINOLA’ genome as opposed to the CBDRx genome is due to the Y chromosomal origin of these transcripts. The high identity of randomly selected ‘FINOLA’ transcripts with the CBDRx genome further supports the use of CBDRx as the reference genome to map and quantify our RNA-Seq reads.
Genes differentially expressed between male and female plants are detected in vegetative and flowering stages in hemp
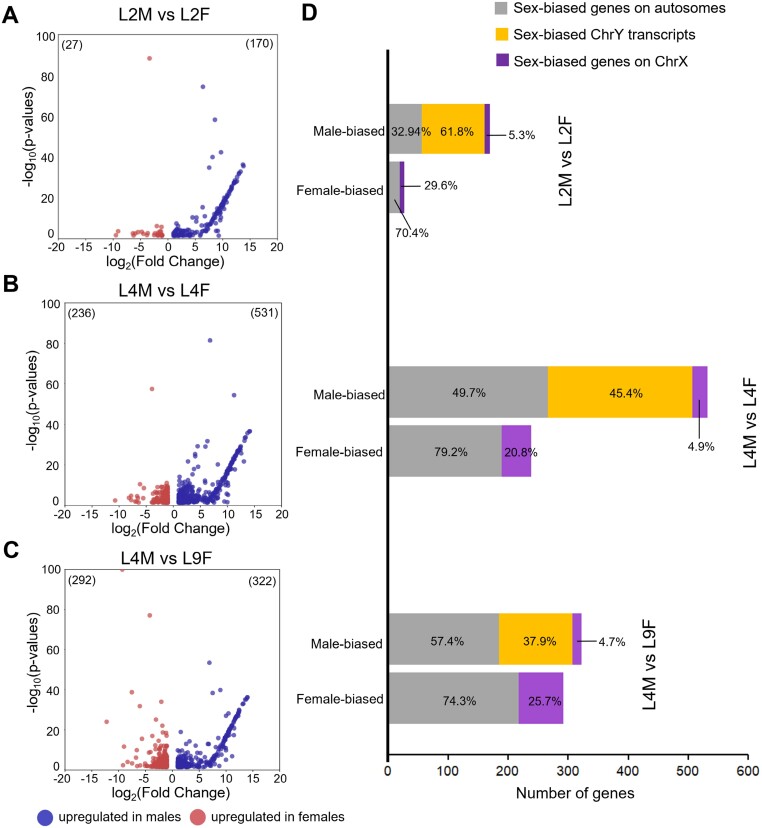
To identify transcripts specifically associated with male or female development, we conducted a DGE analysis between male and female hemp samples from different growth stages. The analysis primarily focused on pairwise comparisons between different developmental stages. With the cut-off value of |log2FC| ≥1 and FDR ≤0.05, the fewest DEGs (197 genes) were identified in the early vegetative stage comparison between male and female plants (L2M versus L2F) (Fig. 3A). Comparisons involving flowering male versus vegetative female plants (L4M versus L4F) exhibited the highest number of DEGs (767 genes) (Fig. 3B), while an intermediate number of DEGs was observed in the comparison of male versus female plants at flowering stage (L4M versus L9F) (614 genes) (Fig. 3C). For each comparison, there were more male-biased genes than female-biased genes. This difference was especially pronounced when comparing early developmental stages (L2M versus L2F, 86% male-biased), whereas the difference was markedly less at reproductive stages (L4M versus L9F, 52% male-biased) (Fig. 3D).
Fig. 3.
Genes are differentially expressed between male and female hemp plants at different stages of plant development. (A–C) Differential gene expression (DGE) analysis was conducted for male and female samples at vegetative stages (L2M versus L2F) (A), flowering male samples compared with vegetative female samples at stage L4 (L4M versus L4F) (B), and both sexes at flowering stages (L4M versus L9F) (C). Log10(P-values) represent the false discovery rate (FDR). Transcripts with |log2FC|≥1 and FDR of ≤0.05 [–log10(P-values) ≥1.30] were considered to be differentially expressed between male and female samples. The DEGs were also filtered by an average transcripts per million value >1. (D) The absolute number of DEGs is indicated for the three comparisons, including the proportion of sex-biased genes on autosomes (grey) and chromosome X (purple) and ChrY transcripts (yellow).
Presumably, genes involved in initiating sex determination should be located on the sex chromosomes. Of the DEGs and transcripts in vegetative stages (L2M versus L2F), 122 were derived from the sex chromosomes (ChrX and putative ChrY), constituting 62% of the total DEGs (Supplementary Table S3). More precisely, 61.8% of male-biased DEGs were putative ChrY transcripts, and 29.6% of female-biased genes were located on ChrX (Fig. 3D). In contrast, in the comparison of flowering samples (L4M versus L9F), a relatively smaller proportion of DEGs (34.8%) was located on the sex chromosomes (Supplementary Table S3), with 37.9% male-biased transcripts putatively on ChrY and 25.7% female-biased genes on ChrX (Fig. 3D).
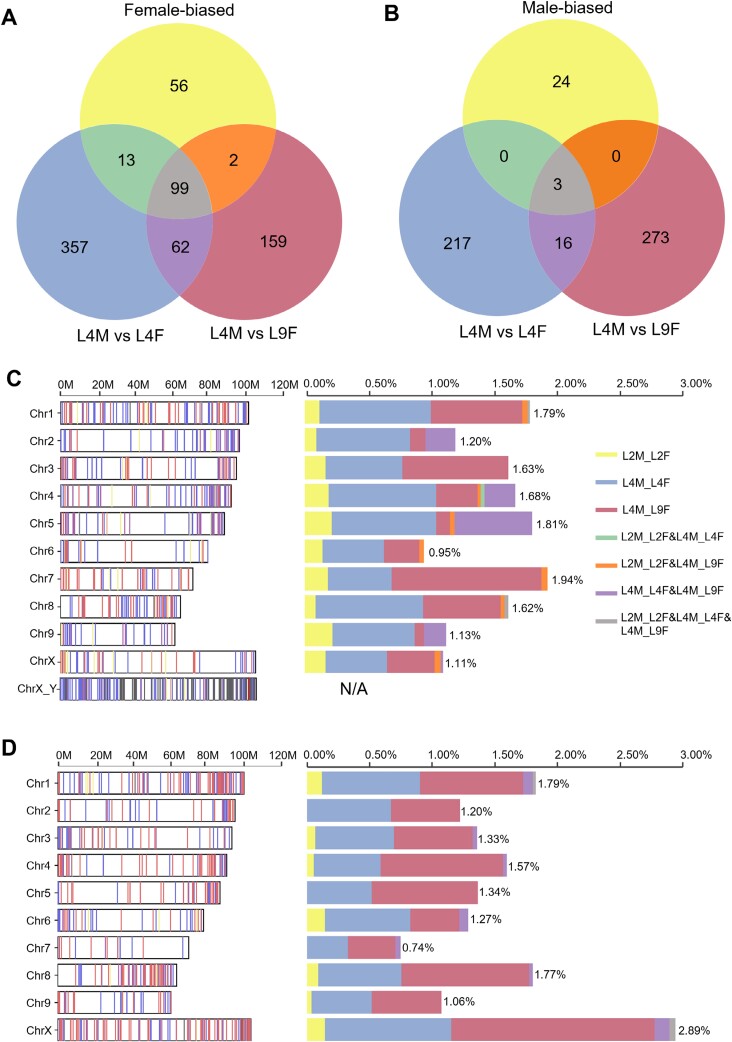
In addition, DGE appeared to be dynamic during plant development. Among the total of 1281 unique sex-biased genes (comprising 748 male-biased genes and 533 female-biased genes) identified from three comparisons (L2M versus L2F, L4M versus L4F, and L4M versus L9F), there were only 99 male-biased genes and 3 female-biased genes common to all three comparisons (Fig. 4A, B). Overall, sex-biased DEGs were distributed mostly equally on autosomes, with approximately 1–2% of genes of each chromosome being either male- or female-biased (Fig. 4C, D). However, the percentage of female-biased genes located on the X chromosome was considerably higher at almost 3% (Fig. 4D).
Fig. 4.
Sex-biased genes across developmental stages and their chromosomal distribution. (A, B) Venn diagrams demonstrating the overlap of female-biased (A) and male-biased (B) genes across L2M versus L2F, L4M versus L4F, and L4M versus L9F comparisons. (C, D) Distribution of male-biased (C) and female-biased (D) genes on different chromosomes. For ChrX_Y the position of the closest BLAST hit of each putative Y transcript on chromosome X is plotted. The percentage bars indicate the percentage of sex-biased genes relative to the total number of genes on this chromosome.
Next, we explored how DEGs from pairwise comparisons were expressed across all developmental stages using heatmaps and bidirectionally clustering gene expression levels and developmental stages (Supplementary Fig. S2). Analysis of genes differentially expressed in vegetative stages (L2M versus L2F) showed that many genes that were up-regulated in male vegetative stages were also up-regulated in flowering male samples (L4M) (Supplementary Fig. S2A). Further, the clustering showed a clear separation of male samples (L2M and L4M) from female samples (L2F, L4F, and L9F) (Supplementary Fig. S2A). In flowering stages (L4M versus L9F), about half of the genes up-regulated in flowering male samples were also expressed in vegetative male samples, but these genes were expressed minimally or not at all in female samples (Supplementary Fig. S2B). Conversely, genes up-regulated in flowering female samples were mostly absent or only weakly expressed in other stages (Supplementary Fig. S2B). Regarding genes differentially expressed between flowering male and vegetative female plants at a similar plant age (L4F versus L4M), the pattern is more complex, which may be the result of the greater number of DEGs as well as the distinctions in both sex and development phases. Similar to the previous two comparisons, a subset of genes exhibited high expression exclusively in male plants (L2M and L4M) (Supplementary Fig. S2C).
Weighted gene co-expression network analysis reveals modules that show expression correlated with phenotypic sex expression in hemp
DGE analyses indicated that a large number of genes are differentially expressed in male versus female plants. To understand which genes shared a similar expression profile across the different developmental stages analysed, that is, which genes are up- and down-regulated together, we employed WGCNA. A total of 15 modules containing genes with similar expression profiles were identified. Among these, three modules appeared to be correlated with male (module I) and female (module II and III) development (Fig. 5; Supplementary Fig. S3; Supplementary Table S4).
Fig. 5.
Modules of co-expressed genes identified by weighted gene co-expression network analysis (WGCNA). Module I consists of genes up-regulated in male samples; module II consists of genes up-regulated late in female development (L4F and L9F); and module III consists of genes up-regulated in all female samples. Modules IV and V do not have a clear association with developmental stages. All modules identified through WGCNA can be found in Supplementary Fig. S3. Developmental stages are depicted at the bottom of the figure. Shades of blue represent a negative correlation, whereas shades of red represent a positive correlation, according to the key on the right.
Module I consisted of 656 genes and transcripts highly expressed in the vegetative and reproductive stages of male plants, but not in female plants (Fig. 5). Module II contained genes highly expressed in later developmental stages in female plants (L4F and L9F), while module III contained genes highly expressed in all stages of female development (Fig. 5). Modules II and III contained fewer genes than module I, with 168 and 154 genes, respectively. It is noteworthy that 75% of genes (116 out of 154) in module III (female-specific expression in all developmental stages) are located on the X chromosome of the reference genome. In contrast, only 46% of genes (71 out of 168) in module II (female-specific expression in later developmental stages) were located on the X chromosome. In other words, module III, which contains genes expressed in vegetative female samples (L2F), also contains more genes located on the X chromosome. Module XIII (which was not further analysed) showed a positive correlation with male samples, too, although not as strong as module I (Supplementary Fig. S3). The activity of genes in modules clearly correlated with sample sex as summarized by plotting eigengene values for modules I, II, and III, respectively (Supplementary Fig. S4). The eigengene values were obtained by taking the first principal component of the expression matrix of genes contained in those modules.
Further, WGCNA allows the identification of putative hub genes in each module (Supplementary Table S5); those are the genes with the highest connectivity in the WGCNA network. For module I, FE.chrY.t1, which is a putative homolog of Essential for poteXvirus Accumulation 1 (EXA1, AT5G42950) from Arabidopsis thaliana as it is the only significant BLAST match, was identified as a hub gene (Supplementary Table S5). We termed this gene CsEXA1. EXA1 was shown to interact with eukaryotic translation initiation factor 4E (eIF4E) in A. thaliana (Nishikawa et al., 2023) and is a possible repressor of translation (Wu et al., 2017). For module II, LOC115704295 was identified as a hub gene, encoding a homolog of the MADS-domain transcription factor TM8 (Steel et al., 2023) shown to be involved in flower development and carpel function in Solanum lycopersicum (Daminato et al., 2014). We termed this gene CsTM8. LOC115699937 is a candidate hub gene for module III. It encodes a B3 domain-containing transcription factor of the REPRODUCTIVE MERISTEM (REM) family (Mantegazza et al., 2014). We termed this gene CsREM16 owing to its closest BLAST match in A. thaliana being REM16 (AT4G33280). A phylogeny reconstruction using SHOOT.bio supports the phylogenetic affiliation (Supplementary Fig. S5). Interestingly, CsREM16 and CsTM8 are also two of the three genes that are differentially up-regulated across all DEG analyses (Fig. 3C).
Identification of core candidate genes for sex determination and sex-specific flower development in hemp
DGE analysis and WGCNA both identified potential sex-related genes for hemp. DGE analysis identifies genes significantly up- or down-regulated between male and female samples but without analysing their correlation across samples, whereas WGCNA identifies modules of co-expressed genes, helping to understand the broader network context by clustering genes with similar expression profiles across samples. We reasoned that integrating data from the two analyses would allow us to identify more reliable candidate genes involved in hemp sex determination and flower development, and annotate the function of these identified genes using MapMan (Supplementary Fig. S6; Supplementary Tables S6, S7) (Usadel et al., 2009). To further study the molecular responses associated with sex determination, we identified the top 10 bins containing the highest number of DEGs for the comparisons of L2M versus L2F, L4M versus L4F, and L4M versus L9F (Supplementary Fig. S7). Interestingly, ‘RNA biosynthesis’ consistently emerged as the largest bin of DEGs (Supplementary Fig. S7). Additionally, ‘RNA biosynthesis’, ‘RNA processing’, ‘plant organogenesis’, ‘solute transport’, and ‘protein homeostasis’ were present in all three comparisons, indicating that half of the top 10 bins are shared across different developmental stages.
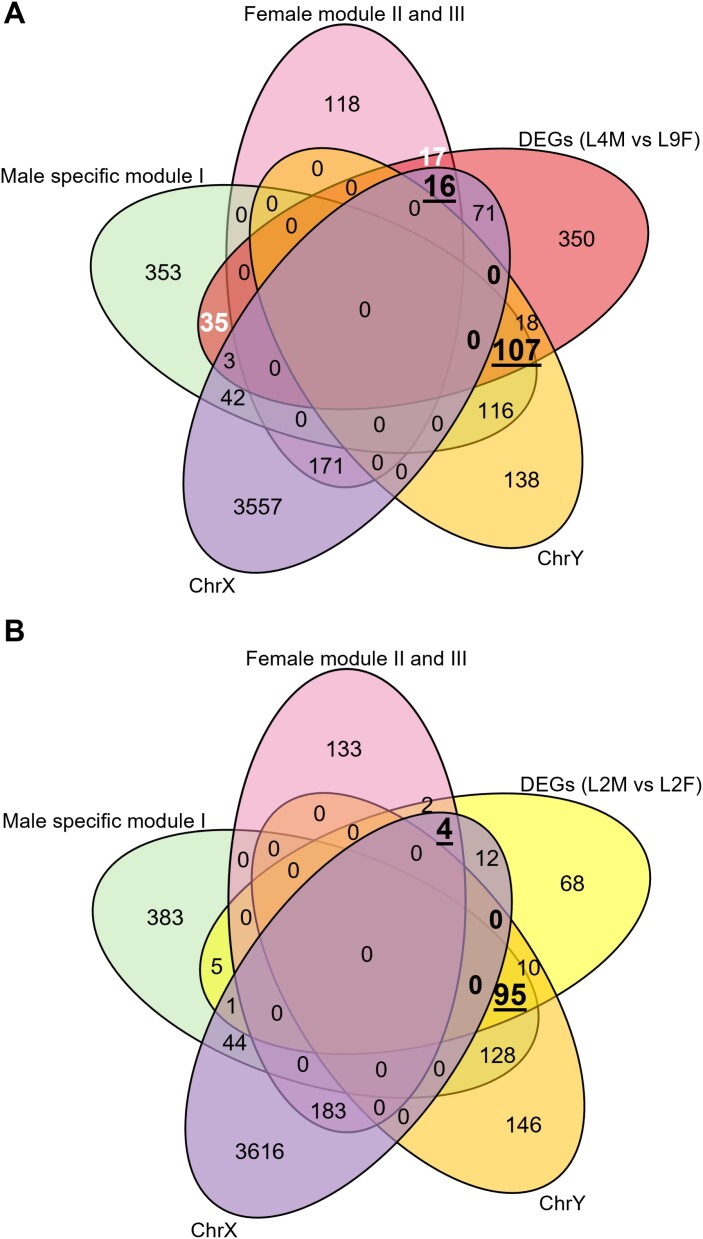
We hypothesized that genes involved specifically in male or female flower development would be differentially expressed in RNA-Seq data from flowering male and female samples (L4M versus L9F). At the same time, developmental regulators would be expected to be part of a male- or female-specific module determined through WGCNA. In total, there were 142 genes up-regulated in male flowers that also belonged to the male-specific module I, and 33 genes up-regulated in female flowers that were also part of the female-specific modules II or III (Fig. 6; Supplementary Fig. S8; Supplementary Table S8).
Fig. 6.
Identification of putative candidate genes for flower development and sex determination in hemp. The Venn diagrams illustrate the number of genes overlapping between the male- or female-specific modules identified through WGCNA, sex chromosome linkage, and DEGs identified in L4M versus L9F (A) and L2M versus L2F (B). Numbers in white text indicate putative flower development genes for male or female hemp plants that are not located on sex chromosomes. Numbers in black underlined text indicate putative sex-determination genes for male (overlap of putative ChrY, male-specific module from WGCNA, and male-biased genes from DEGs) or female (overlap of putative ChrX, female-specific module from WGCNA, and female-biased genes from DEGs) hemp plants. In the L4M versus L9F comparison (A), the combined counts of 17 and 16 represent all female flower development genes, whereas the combined counts of 35 and 107 represent all male flower development genes (see Supplementary Fig. S8).
We subsequently investigated the function of these genes using MapMan, and focused on genes involved in RNA biosynthesis, plant reproduction and plant organogenesis (MapMan bins 15, 28, and 29) (Supplementary Figs S9, S10). Approximately 11% (19 of 175) of genes identified belonged to one or more of those bins (Supplementary Table S8). Among those genes were several that are potential key regulators of flower development in hemp (Supplementary Fig. S11). One of the 142 male-related genes, CsPI (LOC115700653) is homologous to PISTILLATA (PI) from A. thaliana and belongs to the B-clade of the MADS-box gene family (Wan et al., 2021; Steel et al., 2023). PI is known to play a crucial role in stamen development, and thus CsPI is expected to be expressed exclusively in males. Likewise, CsAP3 (LOC115714657) is the homolog of APETALA3 (AP3) from A. thaliana and also part of the B-clade MADS-box gene family (Wan et al., 2021). AP3 and PI determine stamen identity in A. thaliana (Theißen et al., 2016). Meanwhile, CsTM8 (LOC115704295) was among the 33 female-related genes. As previously mentioned, CsTM8 serves as a hub gene in module II, which corresponds to later developmental stages in female plants (L4F and L9F). These results indicate that our approach can identify genes associated with flower development in either male or female flowers.
We next reasoned that candidate sex-determination genes might be differentially expressed during flower development (LM4 versus LF9), and be part of a female- or male-specific WGCNA module but also be located on the sex chromosomes (Fig. 6A). Given the profound morphological differences between male and female hemp flowers, it is also conceivable that key regulatory genes involved in sex determination act early in development, at or before the onset of flowering. We therefore also included genes differentially expressed in RNA-Seq data from male and female vegetative samples (L2M versus L2F), overlapping with the male- or female-specific modules in the WGCNA analysis as well as located on the sex chromosomes, as candidate sex-determination genes (Fig. 6B). Together, these considerations resulted in a total of 129 candidate sex-determination genes and transcripts. Of those, 18 are located on the X chromosome and 111 are putative Y chromosomal transcripts (Supplementary Table S9).
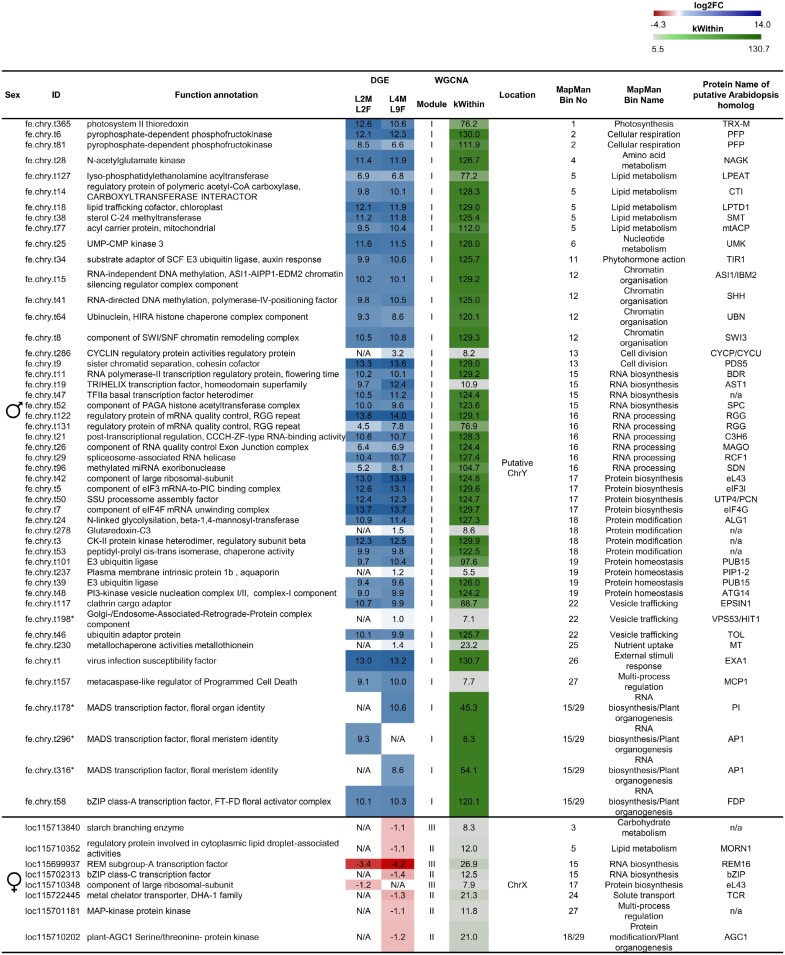
For 56 of these genes, a putative function was assigned by MapMan (Fig. 7; Supplementary Fig. S12; Supplementary Table S10). The remaining genes were not annotated in the Mercator4 dataset. Among those genes are several that have homologies to well-known regulators of reproductive development. One putative transcriptional regulator is encoded by FE.chrY.t58. The best BLAST match of FE.chrY.t58 on the X chromosome is LOC115723488 (named CsbZIP47 in Lu et al., 2022), which exhibits homology with FD (AT4G35900) (Lu et al., 2022), a bZIP transcription factor known to promote flowering in A. thaliana (Abe et al., 2005). Phylogeny reconstructions also indicate that FE.chrY.t58 encodes a group A bZIP transcription factor like FD from A. thaliana (Supplementary Fig. S5) (Dröge-Laser et al., 2018); we therefore termed FE.chrY.t58 CsFD.
Fig. 7.
Differential expression, putative function, and A. thaliana homologs of potential sex-determination genes in C. sativa. Genes that were significantly differentially expressed, highly correlated in weighted gene co-expression network analysis (WGCNA), and located on sex chromosomes were considered putative candidates. Expression values from differential gene expression (DGE) analysis are represented as log2 fold change (log2FC), with colour scales of red indicating down-regulation and blue indicating up-regulation. N/A indicates genes that are not differentially expressed at a certain stage. WGCNA results are indicated as kWithin values, where light grey indicates lower values and dark green indicates higher values, signifying a stronger correlation within the module. Putative functional annotation and A. thaliana homologs were identified by using Mercator4 and MapMan. Genes clustered into ‘35 - No Mercator4 annotation’ are unclassified proteins that cannot be assigned or annotated and are not presented here. Asterisks mark putative ChrY transcripts that, when BLASTed, have a query coverage and percentage identity of ≥90% with an autosomal gene. In those cases, the possibility cannot be excluded that the transcripts are false positives that are actually encoded on autosomes.
Among the X chromosomal candidate sex-determination genes are two genes encoding putative transcription factors: CsREM16 (LOC11569937), which was also identified as a hub gene in the female-specific WGCNA module, and LOC115702313 (named CsbZIP13 in Lu et al., 2022), which has homologies to AtbZIP9, a leucine zipper transcription factor gene from A. thaliana (Lu et al., 2022).
Discussion
In this study, a comprehensive analysis of male and female hemp transcriptomes was carried out across both vegetative and flowering stages. Through a combination of DGE analysis, WGCNA, and knowledge of the chromosomal locations of genes, a list of 129 candidate sex-determining genes in C. sativa was established (Supplementary Table S9). Previous studies have shown that similar approaches can successfully identify sex-determination genes (Akagi et al., 2018; Torres et al., 2018; Massonnet et al., 2020). Clearly, some potential candidates might be missed because the difference in their expression between male and female plants is too small, or sequence divergence between X and Y alleles rather than differences in expression might be responsible for sex determination. However, the observation that numerous plausible transcriptional regulators were identified in our analysis supports the notion that candidate sex-determination genes can be identified using the pipeline established here.
X chromosomal candidate sex-determination genes
The sex-linked region on the X chromosome of C. sativa is approximately 70 Mbp in size and encodes approximately 2069 genes (Prentout et al., 2020; Grassa et al., 2021). Our analysis identified 18 X chromosomal candidate sex-determination genes (Prentout et al., 2020) (Supplementary Table S9). Among these 18 genes, we consider CsREM16 to be a strong candidate for female function development, as it was identified as strongly female-biased at both the vegetative and flowering stages (Fig. 7). Additionally, CsREM16 is a hub gene showing a high correlation with other genes in female module III and is located in the non-recombining region of the X chromosome.
In A. thaliana, the REM family of transcription factors has been implicated in reproductive development (Mantegazza et al., 2014). Some inconsistencies exist in the literature with respect to naming REM genes. CsREM16 identified here appears to be orthologous to REM16 (At4g33280) from A. thaliana described by Franco-Zorrilla et al. (2002). However, At4g33280 was termed REM6 by Mantegazza et al. (2014), and REM16 was used as the gene name for At3g53310 by Mantegazza et al. (2014) and Yu et al. (2020).
REM genes function in male and female gametophyte development (Matias-Hernandez et al., 2010; Mendes et al., 2016; Caselli et al., 2019) as well as in flowering time (Levy et al., 2002; Richter et al., 2019; Yu et al., 2020). It does not therefore appear implausible that a REM gene has been co-opted to control sex determination in C. sativa.
Another gene that constitutes a candidate sex-determination gene is CsbZIP13 (LOC115702313). CsbZIP9 belongs to the group C/S1 bZIP transcription factors, which have been implicated in the stress response and energy homeostasis (Dröge-Laser and Weiste, 2018; Dröge-Laser et al., 2018). A role of C/S1 bZIP transcription factors in plant development has also been proposed (Dröge-Laser and Weiste, 2018). It has recently been shown that C/S1 bZIP transcription factors are involved in controlling plant architecture in A. thaliana and spikelet number in wheat (Glenn et al., 2023; Kreisz et al., 2024). It will be interesting to see whether CsbZIP9 has a role in reproductive development and sex determination in hemp.
Putative Y chromosomal candidate sex-determination genes
The vast majority of candidate sex-determination genes identified in this study are putative Y chromosomal transcripts (111 Y chromosomal versus 18 X chromosomal) (Supplementary Table S9). This discrepancy between the number of X and Y chromosomal candidate genes may be due to the absence of the Y chromosome in female plants and hence the higher likelihood of detecting sex-biased expression for Y chromosomal genes (Fig. 3). During vegetative stages (L2F versus L2M), 86.3% of DEGs exhibited male bias, while 13.7% showed female bias. In flowering stages, this ratio shifted to 52.4% male-biased genes and 47.6% female-biased genes (Fig. 3). This aligns with previous studies on sex determination in hemp, which found a higher abundance of male-biased genes (Prentout et al., 2020; Adal et al., 2021). This pattern of male-biased genes being more numerous than female-biased genes is commonly suggested in dioecious plants (Cossard et al., 2019; Prentout et al., 2020).
One noteworthy putatively Y chromosome-encoded sex-determination gene is CsFD (FE.chrY.t58). CsFD shows sequence similarity to AtFD, a transcription factor that is known to facilitate flowering in A. thaliana (Abe et al., 2005). It is conceivable that CsFD is responsible for the well-documented earlier flowering of male C. sativa plants (Steel et al., 2023). This supports the idea that CsFD could play an evident role in sex determination in hemp and lead to the development of male flowers.
Another gene of interest from the candidate Y chromosome transcripts is CsEXA1 (FE.chrY.t1), previously mentioned as a hub gene in module I and a homolog of A. thaliana EXA1. In A. thaliana, EXA1 was shown to interact with the initiator of translation eIF4E (Nishikawa et al., 2023), and it is supposed to repress the translation of genes in a targeted manner (Wu et al., 2017). One possible hypothesis is that in C. sativa CsEXA1 inhibits the translation of genes necessary for female function development. Intriguingly, a putative interaction partner of CsEXA1, a homolog of the eukaryotic translation initiation factor 4G (LOC115710319), is encoded on the non-recombining region of chromosome X and is overexpressed in females.
It should be noted that the presence of male-exclusive K-mers in transcripts is not definitive evidence of the Y chromosomal origin of those transcripts. Alternative splicing with isoforms exclusive to males will cause the presence of such K-mers, or zero expression of autosomal transcripts in females will result in transcripts present only in males and thus containing male-exclusive K-mers. Another possibility is the occurrence of single nucleotide polymorphisms by chance only in males, although this risk is lessened the bigger the sample size is. Indeed, some transcripts identified by the pipeline might be false positives (Fig. 7). However, the aforementioned transcripts CsEXA1 and CsFD, selected as being possibly related to sex expression, all have a diverged counterpart on chromosome X and are thus likely to actually be on the Y chromosome.
A MADS-domain transcription factor could play a role in sex expression in female C. sativa
As a potential downstream gene involved in female flower development, we have identified the MADS-box gene CsTM8 (LOC115704295), which is homologous to TM8 from tomato. Although the function of TM8 is currently not well characterized (Heijmans et al., 2012), in S. lycopersicum it is also expressed in flowers, and its overexpression results in a reduction of pollen viability whereas the expression of a dominant negative version of TM8 results in the development of seedless fruits (Daminato et al., 2014). In Nicotiana benthamiana, virus-induced gene silencing of NbTM8 can lead to an increase in floral organ numbers (Coenen et al., 2018). Interestingly, TM8-like genes have been lost in a number of angiosperm lineages and, unlike other MADS-box gene subfamilies, duplicates were generally not retained after whole-genome duplication events (Gramzow and Theißen, 2015; Coenen et al., 2018). Overall, the picture emerges that TM8-like genes are involved in reproductive development, but it was also hypothesized that the function of TM8-like genes has diverged throughout angiosperm evolution (Coenen et al., 2018). Together, this makes CsTM8 an interesting potential candidate as a sex-determination gene in hemp. Moreover, although according to the CBDRx reference genome CsTM8 is positioned on chromosome 1, a BLAST search on the more recent Pink Pepper assembly (ASM2916894v1, https://www.ncbi.nlm.nih.gov/datasets/genome/GCA_029168945.1/) shows that it is located on the non-recombining region of the X chromosome in a region 3 million nucleotides from the sex-determination candidate CsREM16, which makes the involvement of CsTM8 in sex determination or sex expression even more plausible.
Sex-determination pathways may already be activated during vegetative stages in hemp
Our results show that sex-biased genes are associated with different developmental stages in hemp (Fig. 3). During the vegetative phase, core sex-determination genes on the sex chromosomes are expected to be activated. As the flowering phase begins, more genes involved in processes downstream of sex determination will be expressed. In terms of quantity, the DGE analysis revealed 614 DEGs when comparing flowering male and female plants, whereas there were 197 DEGs when comparing male and female plants in the vegetative stage (Fig. 3). This observation is in line with the expectation of increased activity of sex-biased genes during the reproductive phase and the involvement of multiple structures in specific floral organ development and metabolic processes (Cossard et al., 2019; Prentout et al., 2020).
It is noteworthy that the ratio of sex-biased genes encoded on sex chromosomes decreases while the total number of sex-biased genes increases from vegetative to flowering stages. In vegetative stages, 61.9% of DEGs are located on sex chromosomes. However, in flowering stages, only 34.5% of DEGs are located on sex chromosomes (Supplementary Table S3). These results suggest that core sex-determination genes on the sex chromosomes are likely activated early in the vegetative phase. This is similar to what has been observed in Mercurialis annua, a wind-pollinated dioecious annual herb, where males and females exhibited differences in gene expression even at the first leaf stage, with the expression of sex-biased genes peaking just before and after flowering (Cossard et al., 2019). This indicates that sex bias in gene expression in M. annua as well as in hemp begins early in vegetative development, long before the flowering stage. Of course, numerous genes showing sex-biased expression may not be involved in sex determination but may show biased expression just because they are located on the sex chromosomes. However, we consider the transcriptional regulators identified here to be plausible candidates for sex-determination genes.
Sex determination in hemp—a multi-gene affair?
In most of the dioecious angiosperms studied in detail so far, one or two sex-determination genes have been identified (Akagi et al., 2014, 2018; Torres et al., 2018; Harkess et al., 2020; Massonnet et al., 2020; Müller et al., 2020). However, the sex chromosomes of hemp are the oldest identified so far in flowering plants (Prentout et al., 2021), which means that significant time has passed to allow the accumulation of genes and alleles that contribute to sexual dimorphism. Flowering time, inflorescence architecture, and floral morphology differ between male and female hemp plants (Leme et al., 2020; Dowling et al., 2024; Shi et al., 2024, Preprint), and it remains to be seen whether a single locus instigates those differences or whether several distinct genes located on the sex chromosomes contribute together to the sexual dimorphism observed today. The genes identified in this study may well contribute to sexual dimorphism, but whether they constitute bona fide sex-determination genes remains to be seen. Future functional analyses involving mutant and overexpression lines will hopefully provide additional evidence in this direction.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. Sequence comparison of ChrY transcripts to ‘CBDRx’ and ‘FINOLA’ genomes.
Fig. S2. Hierarchical clustering of DEGs in hemp.
Fig. S3. Modules of co-expressed genes identified by WGCNA.
Fig. S4. Eigengene values for the sex-related modules.
Fig. S5. SHOOT.bio homology association.
Fig. S6. Summary of MapMan metabolic pathways from the DEGs.
Fig. S7. Top 10 MapMan metabolic pathways from the DEGs in hemp.
Fig. S8. Identification of putative candidate genes for flower development in hemp.
Fig. S9. DEGs potentially involved in sex determination and floral development.
Fig. S10. Sex-related genes identified by WGCNA with potential function in RNA biosynthesis, plant organogenesis, and plant reproduction.
Fig. S11. Functional annotation of putative candidate genes for flower development.
Fig. S12. Identification of putative candidate genes for early sex determination.
Table S1. RNA-Seq paired-read counts and alignment statistics for all samples used for transcriptome analysis of each flower sex type and developmental stage.
Table S2. Putative male-specific transcripts generated through de novo assembly and the K-mer approach.
Table S3. Summary of the counts of DEGs.
Table S4. List of genes and modules identified through WGCNA.
Table S5. Hub genes of sex-correlated modules in WGCNA.
Table S6. Summary of MapMan metabolic pathways from five modules identified from the WGCNA.
Table S7. MapMan result of key DEGs potentially involving sex determination and floral development (belongs to Bin 15, 28, 29) in L2F versus L2F, L4M versus L4F, and L4MF versus L9F.
Table S8. List of candidate sex-specific flower development genes in hemp.
Table S9. List of candidate sex-related genes in hemp.
Table S10. Supplementary information for Fig. 7. Candidate sex determination genes in hemp.
Acknowledgements
We thank Nina Trubanová for informative discussions on the phylogeny of MADS-box genes in C. sativa.
Contributor Information
Jiaqi Shi, School of Biology and Environmental Science and Earth Institute, University College Dublin, Dublin, Ireland.
Matteo Toscani, School of Biology and Environmental Science and Earth Institute, University College Dublin, Dublin, Ireland.
Caroline A Dowling, School of Biology and Environmental Science and Earth Institute, University College Dublin, Dublin, Ireland.
Susanne Schilling, School of Biology and Environmental Science and Earth Institute, University College Dublin, Dublin, Ireland.
Rainer Melzer, School of Biology and Environmental Science and Earth Institute, University College Dublin, Dublin, Ireland.
Ricarda Jost, La Trobe University, Australia.
Author contributions
JS: conceptualization, formal analysis, funding acquisition, investigation, methodology, visualization, writing—original draft; MT: conceptualization, formal analysis, methodology, software, visualization; writing—original draft; CAD: methodology; SS: conceptualization, funding acquisition, project administration, resources, supervision, writing—review & editing; RM: conceptualization, funding acquisition, project administration, resources, supervision, writing—review & editing.
Conflict of interest
No conflict of interest is declared.
Funding
The research conducted in this publication was funded by the Irish Research Council under grant number IRCLA/2022/3294. JS is supported by a China Scholarship Council Postgraduate Scholarship (grant no.: CSC no. 201908300031). CAD is supported by an Irish Research Council–Environmental Protection Agency Government of Ireland Postgraduate Scholarship (grant no. GOIPG/2019/1987).
Data availability
All raw sequencing files generated in this study are available in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA1126191.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T.. 2005. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. [DOI] [PubMed] [Google Scholar]
- Adal AM, Doshi K, Holbrook L, Mahmoud SS.. 2021. Comparative RNA-Seq analysis reveals genes associated with masculinization in female Cannabis sativa. Planta 253, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afgan E, Baker D, Batut B, et al. 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Research 46, W537–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T, Henry IM, Ohtani H, Morimoto T, Beppu K, Kataoka I, Tao R.. 2018. A Y-encoded suppressor of feminization arose via lineage-specific duplication of a cytokinin response regulator in kiwifruit. The Plant Cell 30, 780–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T, Henry IM, Tao R, Comai L.. 2014. Plant genetics. A Y-chromosome-encoded small RNA acts as a sex determinant in persimmons. Science 346, 646–650. [DOI] [PubMed] [Google Scholar]
- Bachtrog D, Mank JE, Peichel CL, et al. 2014. Sex determination: why so many ways of doing it? PLoS Biology 12, e1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushmanova E, Antipov D, Lapidus A, Prjibelski AD.. 2019. rnaSPAdes: a de novo transcriptome assembler and its application to RNA-Seq data. GigaScience 8, giz100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caselli F, Beretta VM, Mantegazza O, et al. 2019. REM34 and REM35 control female and male gametophyte development in Arabidopsis thaliana. Frontiers in Plant Science 10, 1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. 2016. Plant sex chromosomes. Annual Review of Plant Biology 67, 397–420. [DOI] [PubMed] [Google Scholar]
- Coenen H, Viaene T, Vandenbussche M, Geuten K.. 2018. TM8 represses developmental timing in Nicotiana benthamiana and has functionally diversified in angiosperms. BMC Plant Biology 18, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossard GG, Toups MA, Pannell JR.. 2019. Sexual dimorphism and rapid turnover in gene expression in pre-reproductive seedlings of a dioecious herb. Annals of Botany 123, 1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daminato M, Masiero S, Resentini F, Lovisetto A, Casadoro G.. 2014. Characterization of TM8, a MADS-box gene expressed in tomato flowers. BMC Plant Biology 14, 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divashuk MG, Alexandrov OS, Razumova OV, Kirov IV, Karlov GI.. 2014. Molecular cytogenetic characterization of the dioecious Cannabis sativa with an XY chromosome sex determination system. PLoS One 9, e85118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling CA, Shi J, Toth JA, Quade MA, Smart LB, McCabe PF, Schilling S, Melzer R.. 2024. A FLOWERING LOCUS T ortholog is associated with photoperiod-insensitive flowering in hemp (Cannabis sativa L.). The Plant Journal 119, 383–403. [DOI] [PubMed] [Google Scholar]
- Dröge-Laser W, Snoek BL, Snel B, Weiste C.. 2018. The Arabidopsis bZIP transcription factor family—an update. Current Opinion in Plant Biology 45, 36–49. [DOI] [PubMed] [Google Scholar]
- Dröge-Laser W, Weiste C.. 2018. The C/S1 bZIP network: a regulatory hub orchestrating plant energy homeostasis. Trends in Plant Science 23, 422–433. [DOI] [PubMed] [Google Scholar]
- Fasinu PS, Phillips S, ElSohly MA, Walker LA.. 2016. Current status and prospects for cannabidiol preparations as new therapeutic agents. Pharmacotherapy 36, 781–796. [DOI] [PubMed] [Google Scholar]
- Feng G, Sanderson BJ, Keefover-Ring K, Liu J, Ma T, Yin T, Smart LB, DiFazio SP, Olson MS.. 2020. Pathways to sex determination in plants: how many roads lead to Rome? Current Opinion in Plant Biology 54, 61–68. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Cubas P, Jarillo JA, Fernández-Calvín B, Salinas J, Martínez-Zapater JM.. 2002. AtREM1, a member of a new family of B3 domain-containing genes, is preferentially expressed in reproductive meristems. Plant Physiology 128, 418–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn P, Woods DP, Zhang J, Gabay G, Odle N, Dubcovsky J.. 2023. Wheat bZIPC1 interacts with FT2 and contributes to the regulation of spikelet number per spike. Theoretical and Applied Genetics 136, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramzow L, Theißen G.. 2015. Phylogenomics reveals surprising sets of essential and dispensable clades of MIKCc-group MADS-box genes in flowering plants. Journal of Experimental Zoology. Part B. Molecular and Developmental Evolution 324, 353–362. [DOI] [PubMed] [Google Scholar]
- Grassa CJ, Weiblen GD, Wenger JP, Dabney C, Poplawski SG, Timothy Motley S, Michael TP, Schwartz CJ.. 2021. A new Cannabis genome assembly associates elevated cannabidiol (CBD) with hemp introgressed into marijuana. New Phytologist 230, 1665–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkess A, Huang K, Van Der Hulst R, Tissen B, Caplan JL, Koppula A, Batish M, Meyers BC, Leebens-Mack J.. 2020. Sex determination by two Y-linked genes in garden asparagus. The Plant Cell 32, 1790–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans K, Morel P, Vandenbussche M.. 2012. MADS-box genes and floral development: the dark side. Journal of Experimental Botany 63, 5397–5404. [DOI] [PubMed] [Google Scholar]
- Kreisz P, Hellens AM, Fröschel C, et al. 2024. S1 basic leucine zipper transcription factors shape plant architecture by controlling C/N partitioning to apical and lateral organs. Proceedings of the National Academy of Sciences, USA 121, e2313343121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S.. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laverty KU, Stout JM, Sullivan MJ, et al. 2019. A physical and genetic map of Cannabis sativa identifies extensive rearrangements at the THC/CBD acid synthase loci. Genome Research 29, 146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leme FM, Schönenberger J, Staedler YM, Teixeira SP.. 2020. Comparative floral development reveals novel aspects of structure and diversity of flowers in Cannabaceae. Botanical Journal of the Linnean Society 193, 64–83. [Google Scholar]
- Levy YY, Mesnage S, Mylne JS, Gendall AR, Dean C.. 2002. Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 297, 243–246. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Meng XX, Zhang YM, et al. 2022. Genome-wide identification and expression profiles of bZIP genes in Cannabis sativa L. Cannabis and Cannabinoid Research 7, 882–895. [DOI] [PubMed] [Google Scholar]
- Manrique S, Caselli F, Matías-Hernández L, Franks RG, Colombo L, Gregis V.. 2023. Assessing the role of REM13, REM34 and REM46 during the transition to the reproductive phase in Arabidopsis thaliana. Plant Molecular Biology 112, 179–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantegazza O, Gregis V, Mendes MA, Morandini P, Alves-Ferreira M, Patreze CM, Nardeli SM, Kater MM, Colombo L.. 2014. Analysis of the Arabidopsis REM gene family predicts functions during flower development. Annals of Botany 114, 1507–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, Kingsford C.. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massonnet M, Cochetel N, Minio A, et al. 2020. The genetic basis of sex determination in grapes. Nature Communications 11, 2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Akagi T.. 2023. Evolution of sex in crops: recurrent scrap and rebuild. Breeding Science 73, 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias-Hernandez L, Battaglia R, Galbiati F, Rubes M, Eichenberger C, Grossniklaus U, Kater MM, Colombo L.. 2010. VERDANDI is a direct target of the MADS domain ovule identity complex and affects embryo sac differentiation in Arabidopsis. The Plant Cell 22, 1702–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes MA, Guerra RF, Castelnovo B, Silva-Velazquez Y, Morandini P, Manrique S, Baumann N, Groß-Hardt R, Dickinson H, Colombo L.. 2016. Live and let die: a REM complex promotes fertilization through synergid cell death in Arabidopsis. Development 143, 2780–2790. [DOI] [PubMed] [Google Scholar]
- Müller NA, Kersten B, Leite Montalvão AP, et al. 2020. A single gene underlies the dynamic evolution of poplar sex determination. Nature Plants 6, 630–637. [DOI] [PubMed] [Google Scholar]
- Nishikawa M, Katsu K, Koinuma H, et al. 2023. Interaction of EXA1 and eIF4E family members facilitates potexvirus infection in Arabidopsis thaliana. Journal of Virology 97, e0022123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C.. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nature Methods 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentout D, Razumova O, Rhoné B, Badouin H, Henri H, Feng C, Käfer J, Karlov G, Marais GAB.. 2020. An efficient RNA-seq-based segregation analysis identifies the sex chromosomes of Cannabis sativa. Genome Research 30, 164–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentout D, Stajner N, Cerenak A, Tricou T, Brochier-Armanet C, Jakse J, Käfer J, Marais GAB.. 2021. Plant genera Cannabis and Humulus share the same pair of well-differentiated sex chromosomes. New Phytologist 231, 1599–1611. [DOI] [PubMed] [Google Scholar]
- Renner SS. 2014. The relative and absolute frequencies of angiosperm sexual systems: dioecy, monoecy, gynodioecy, and an updated online database. American Journal of Botany 101, 1588–1596. [DOI] [PubMed] [Google Scholar]
- Richter R, Kinoshita A, Vincent C, Martinez-Gallegos R, Gao H, van Driel AD, Hyun Y, Mateos JL, Coupland G.. 2019. Floral regulators FLC and SOC1 directly regulate expression of the B3-type transcription factor TARGET OF FLC AND SVP 1 at the Arabidopsis shoot apex via antagonistic chromatin modifications. PLoS Genetics 15, e1008065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauquet H, von Balthazar M, Magallón S, et al. 2017. The ancestral flower of angiosperms and its early diversification. Nature Communications 8, 16047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling S, Dowling CA, Shi J, Ryan L, Hunt D, O’Reilly E, Perry AS, Kinnane O, McCabe PF, Melzer R.. 2021. The cream of the crop: biology, breeding, and applications of Cannabis sativa. Annual Plant Reviews Online 4. doi: https://doi.org/ 10.1002/9781119312994.apr0740 [DOI] [Google Scholar]
- Schluttenhofer C, Yuan L.. 2017. Challenges towards revitalizing hemp: a multifaceted crop. Trends in Plant Science 22, 917–929. [DOI] [PubMed] [Google Scholar]
- Schwacke R, Ponce-Soto GY, Krause K, Bolger AM, Arsova B, Hallab A, Gruden K, Stitt M, Bolger ME, Usadel B.. 2019. MapMan4: a refined protein classification and annotation framework applicable to multi-omics data analysis. Molecular Plant 12, 879–892. [DOI] [PubMed] [Google Scholar]
- Shi J, Schilling S, Melzer R.. 2024. Morphological and genetic analysis of inflorescence and flower development in hemp (Cannabis sativa L.). BioRxiv doi: 10.1101/2024.01.25.577276. [Preprint]. [DOI] [Google Scholar]
- Soneson C, Love MI, Robinson MD.. 2016. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research 4, 1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer-Rimon B, Duchin S, Bernstein N, Kamenetsky R.. 2019. Architecture and florogenesis in female Cannabis sativa plants. Frontiers in Plant Science 10, 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel L, Welling M, Ristevski N, Johnson K, Gendall A.. 2023. Comparative genomics of flowering behavior in Cannabis sativa. Frontiers in Plant Science 14, 1227898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalata M, Dreger M, Zielińska A, Banach J, Szalata M, Wielgus K.. 2022. Simple extraction of cannabinoids from female inflorescences of hemp (Cannabis sativa L.). Molecules 27, 5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theißen G, Melzer R, Rümpler F.. 2016. MADS-domain transcription factors and the floral quartet model of flower development: linking plant development and evolution. Development 143, 3259–3271. [DOI] [PubMed] [Google Scholar]
- Torres MF, Mathew LS, Ahmed I, Al-Azwani IK, Krueger R, Rivera-Nuñez D, Mohamoud YA, Clark AG, Suhre K, Malek JA.. 2018. Genus-wide sequencing supports a two-locus model for sex-determination in Phoenix. Nature Communications 9, 3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Poree F, Nagel A, Lohse M, Czedik-Eysenberg A, Stitt M.. 2009. A guide to using MapMan to visualize and compare omics data in plants: a case study in the crop species, Maize. Plant, Cell & Environment 32, 1211–1229. [DOI] [PubMed] [Google Scholar]
- Wan Z, Lu M, Wu S, Mi Y, Zhai J.. 2021. Identification and expression analysis of the MIKC-type MADS-box gene family in the original plant of traditional Chinese medicine hemp seed. Acta Pharmaceutica Sinica 56, 3173–3183. [Google Scholar]
- Wu F, Sedivy EJ, Price WB, Haider W, Hanzawa Y.. 2017. Evolutionary trajectories of duplicated FT homologues and their roles in soybean domestication. The Plant Journal 90, 941–953. [DOI] [PubMed] [Google Scholar]
- Yang W, Wang D, Li Y, et al. 2021. A general model to explain repeated turnovers of sex determination in the Salicaceae. Molecular Biology and Evolution 38, 968–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Qiao L, Chen J, Rong Y, Zhao Y, Cui X, Xu J, Hou X, Dong C-H.. 2020. Arabidopsis REM16 acts as a B3 domain transcription factor to promote flowering time via directly binding to the promoters of SOC1 and FT. The Plant Journal 103, 1386–1398. [DOI] [PubMed] [Google Scholar]
- Zhang B, Horvath S.. 2005. A general framework for weighted gene co-expression network analysis. Statistical Applications in Genetics and Molecular Biology 4, Article17. [DOI] [PubMed] [Google Scholar]
- Zhang X, Pan L, Guo W, Li Y, Wang W.. 2022. A convergent mechanism of sex determination in dioecious plants: distinct sex-determining genes display converged regulation on floral B-class genes. Frontiers in Plant Science 13, 953445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequencing files generated in this study are available in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA1126191.