Abstract
Opium poppy is a crop of great commercial value as a source of several opium alkaloids for the pharmaceutical industries including morphine, codeine, thebaine, noscapine, and papaverine. Most enzymes involved in benzylisoquinoline alkaloid (BIA) biosynthesis in opium poppy have been functionally characterized, and opium poppy currently serves as a model system to study BIA metabolism in plants. BIA biosynthesis in opium poppy involves two biosynthetic gene clusters associated respectively with the morphine and noscapine branches. Recent reports have shown that genes in the same cluster are co-expressed, suggesting they might also be co-regulated. However, the transcriptional regulation of opium poppy BIA biosynthesis is not well studied. Opium poppy BIA biosynthesis involves three cell types associated with the phloem system: companion cells, sieve elements, and laticifers. The transcripts and enzymes associated with BIA biosynthesis are distributed across cell types, requiring the translocation of key enzymes and pathway intermediates between cell types. Together, these suggest that the regulation of BIA biosynthesis in opium poppy is multilayered and complex, involving biochemical, genomic, and physiological mechanisms. In this review, we highlight recent advances in genome sequencing and single cell and spatial transcriptomics with a focus on how these efforts can improve our understanding of the genomic and cell-specific regulation of BIA biosynthesis. Such knowledge is vital for opium poppy genetic improvement and metabolic engineering efforts targeting the modulation of alkaloid yield and composition.
Keywords: Alkaloid biosynthesis, benzylisoquinoline alkaloids, opium poppy, pangenome, single-cell and spatial muti-omics, transcriptional regulation
This review highlights how the biosynthesis of alkaloids in opium poppy is regulated—information that can be used for improving the yield and composition of commercially important compounds.
Introduction
Opium poppy (Papaver somniferum L.) is a multipurpose plant principally grown for the morphinan derivatives it produces, which are an indispensable part of modern medicine (Zohary et al., 2012; Guo et al., 2018; Singh et al., 2019; Xu et al., 2022). Poppy seed is a source of commercially important edible oils and is an essential ingredient in many cuisines, while the plant is also grown for ornamental purposes due to its attractive flowers (Tétényi, 1997; Bernáth and Németh, 2010; Zohary et al., 2012; Lančaričová et al., 2016; Butnariu et al., 2022). Recently, new applications of poppy have been proposed as a potential source for biodiesel production and nanomaterials (Gozmen Şanli et al., 2019; Tabatabaei majd et al., 2020; Kadhim et al., 2023). Opium poppy has also caused considerable challenges for human health and society due to the addictive properties of morphinan-derived medications and their illicit consumption as opium or heroin (Guo et al., 2018; Singh et al., 2019). Opium poppy is one of the earliest domesticated crops, but its geographic origin is unclear and remains a controversial topic, with origins in Mesopotamia or the western Mediterranean being the primary two theories (Kapoor, 1995; Salavert, 2017; Labanca et al., 2018; Jesus et al., 2021; Nencini, 2022). Today, opium poppy is legally cultivated only in designated regions to ensure the medicinal and non-medicinal material supply while preventing its addictive properties from causing public health and political issues (UNODC, 1961; Bernáth and Németh, 2010).
Opium poppy is diploid (2n=22) and primarily self-pollinated (Brandt et al., 1887; Kapoor, 1995; Tétényi, 1997; Evans, 2009). The evolutionary relationships between opium poppy and other species in the Papaver genus were established using morphological and cytological data (Pei et al., 2021). With an annual growth habit and the presence of anthocyanin in flowers, P. somniferum was established as one of the recently evolved species within the Papaver genus and grouped in the Papaver section with three other species: P. setigerum, P. glaucum, and P. gracile (Srivastava, 1989; Tétényi, 1997; Liscombe et al., 2005; Labanca et al., 2018; Lane et al., 2018; Y. Li et al., 2020). Cytological analysis indicated that P. setigerum (Troy poppy, n=22) is the only other species in the genus with a chromosome number that is a multiple of 11 (Tahara, 1915; Yasui, 1921, 1937; Sugiura, 1940; Kaul et al., 1979; Srivastava, 1989). The cross compatibility of P. somniferum and P. setigerum and homologous chromosome pairing during meiosis in interspecific hybrids of the two species also supported their close genetic relationships (Yasui, 1937; Hrishi, 1960; Malik et al., 1979; Espinasse and Dosba, 1982; Ojala and Rousi, 1986; Kadereit, 1987; Pyysalo et al., 1988). Furthermore, only P. somniferum and P. setigerum can produce morphine. Papaver setigerum was initially considered as the putative progenitor of the cultivated P. somniferum (Zohary et al., 2012; Salavert et al., 2018, 2020). However, recent advances in poppy genomic sequencing and data analyses indicated that P. somniferum and P. setigerum have a common ancestor and diverged approximately 5.0 million years ago (Yang et al., 2021; Zhang et al., 2023).
Benzylisoquinoline alkaloids (BIAs) are a diverse group of tyrosine-derived specialized metabolites with approximately 2500 known structures, found predominately in plants of the order Ranunculales (Hagel and Facchini, 2013; Beaudoin and Facchini, 2014; Singh et al., 2019; Y. Li et al., 2020; Yucebilgili Kurtoglu and Unver, 2021). Morphine was the first BIA isolated from opium poppy at the beginning of the 1800s, by Sertürner (Norn et al., 2005; Gach et al., 2011; Stefano et al., 2017). It was initially claimed to be a sleep-inducing agent and named after the Greek god of dreams, Morpheus. Morphine is currently regarded as a powerful painkiller that is widely used both orally and subcutaneously. The addictive properties of opium were also acknowledged in morphine, especially in chronic usage cases. Despite these side effects, morphine is an irreplaceable drug for relieving pain treatment (Gach et al., 2011; Catania et al., 2022; INCB, 2022; Singh et al., 2023). Besides morphine, opium poppy also produces other highly valuable pharmaceutical BIAs such as codeine, thebaine, papaverine, and noscapine. Codeine is a narcotic or opioid analgesic, in a similar manner to morphine, and is also commonly used to treat pain, coughing and diarrhoea (Dastmalchi et al., 2019b; Singh et al., 2019). Thebaine is used for the semi-synthesis of many non-addictive painkillers (Chen et al., 2018). Papaverine and noscapine are non-narcotic drugs. Papaverine is used for antispasmodic treatment, and noscapine is a potential anticancer drug (Winzer et al., 2012; Li and Smolke, 2016; Tamiru-Oli et al., 2018).
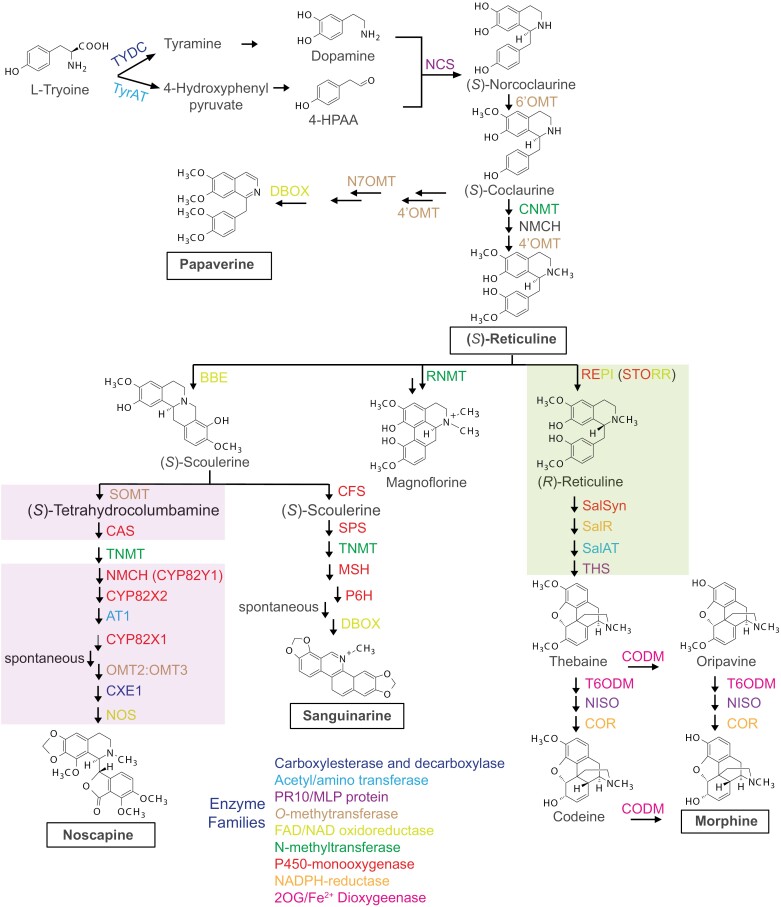
Opium poppy is used as a model system to study the BIA biosynthesis pathway because it is the only cultivated plant that produces morphine and a wealth of other bioactive BIAs (Tamiru-Oli et al., 2018; Pei et al., 2021; Catania et al., 2022). The BIA biosynthesis pathway has been successfully characterized through continuous efforts since the 1960s (Evans, 2009; Hagel and Facchini, 2013; Beaudoin and Facchini, 2014; Singh et al., 2019; Q. Li et al., 2020; Ozber and Facchini, 2022). We now know that BIAs share a common biosynthetic origin despite their marked structural diversity. More than 35 enzymes participating in the opium poppy BIA biosynthesis pathway have been functionally characterized (Singh et al., 2019; Agarwal et al., 2020) (Fig. 1). Biosynthesis begins with the conversion of two tyrosine derivatives, dopamine and 4-hydroxyphenyl acetaldehyde (4-HPAA), to the first committed intermediate (S)-norcoclaurine by norcoclaurine synthase (NCS) (Hagel and Facchini, 2013; Beaudoin and Facchini, 2014; Dastmalchi et al., 2018; Singh et al., 2019; Yucebilgili Kurtoglu and Unver, 2021; Ozber and Facchini, 2022) (Fig. 1). Next, (S)-norcoclaurine sequentially undergoes hydroxylation, O- and N-methylations to form the central branch-point intermediate, (S)-reticuline, which is shared among all species belonging to Papaveraceae family (Y. Li et al., 2020; Catania et al., 2022). (S)-Reticuline then goes through several structural rearrangements depending on Papaver species or chemotypes of opium poppy to yield different final products such as morphine, sanguinarine, noscapine, or magnoflorine (Fig. 1).
Fig. 1.
The benzylisoquinoline alkaloid (BIA) biosynthesis pathway in opium poppy. All enzymes shown have been functionally characterized, and enzyme families are shown with different colours. The noscapine (light purple box) and morphine gene clusters (light green box) are shown. 2OG, 2-oxoglutarate; 4-HPAA, 4-hydroxyphenylacetaldehyde; 4ʹOMT, 3ʹ-hydroxyl-N-methylcoclaurine 4ʹ-O-methyltransferase; 6ʹOMT, norcoclaurine 6-O-methyltransferase; AT1, 1,13-dihydroxy-N-methylcanadine 13-O-acetyltransferase; BBE, berberine bridge enzyme; CAS, canadine synthase; CFS, cheilanthifoline synthase; CNMT, coclaurine N-methyltransferase; CODM, codeine O-demethylase; COR, codeinone reductase; CXE1, 3-O-acetylpapaveroxine carboxylesterase; CYP82X1, 1-hydroxy-13-O-acetyl-N-methylcanadine 8-hydroxylase; CYP82X2, 1-hydroxy-N-methylcanadine 13-O-hydroxylase; DBOX, dihydrosanguinarine oxidase; MSH, N-methylstylopine 14-hydroxylase; N7OMT, norreticuline 7-O-methyltransferase; NCS, norcoclaurine synthase; NISO, neopinone isomerase; NMCH, N-methylcoclaurine 3ʹ-hydroxylase; NMCH (CYP82Y1), N-methylcanadine 1-hydroxylase; NOS, noscapine synthase; OMT2:OMT3, 4ʹ-O-desmethyl-3-O-acetylpapaveroxine 4ʹ-O-methyltransferase; P6H, protopine 6-hydroxylase; PR10/MLP: pathogenesis-related-10 family and major latex protein; REPI, reticuline epimerase; RNMT, reticuline N-methyltransferase; SalAT, salutaridinol 7-O-acetyltransferase; SalR, salutaridine reductase; SalSyn, salutaridine synthase; SanR, sanguinarine reductase; SOMT, scoulerine 9-O-methyltransferase; SPS, stylopine synthase; T6ODM, thebaine 6-O-demethylase; THS, thebaine synthase; TNMT, tetrahydroprotoberberine N-methyltransferase; TYDC, tyrosine decarboxylase; TyrAT, tyrosine aminotransferase (Beaudoin and Facchini, 2014; Singh et al., 2019; Ozber and Facchini, 2022).
The structure and components of the BIA biosynthetic pathway are now well characterized. Studies have indicated that the BIA metabolism in opium poppy is a spatially separated process involving three distinct cell types: companion cells, sieve elements, and laticifers of the phloem system (Onoyovwe et al., 2013; Beaudoin and Facchini, 2014; Singh et al., 2019; Ozber and Facchini, 2022). This results in a multilayered and complex regulation of opium poppy BIA biosynthesis, involving biochemical, genomic, and physiological mechanisms. Our understanding of these regulatory mechanisms is relatively limited, but increased knowledge could inform opium poppy breeding and biotechnology (Singh et al., 2019; Ozber and Facchini, 2022; Watkins and Facchini, 2022). Biochemical mechanisms of regulation have been well reviewed elsewhere (Singh et al., 2019). In this review, we focus on the genomic and cell-specific mechanisms of BIA biosynthesis regulation. We highlight gaps in knowledge and discuss how recent technical advances in whole genome sequencing and single cell and spatial transcriptomics can provide powerful tools to advance the field.
Structure of the benzylisoquinoline alkaloid biosynthetic pathway in opium poppy
The BIA biosynthetic pathway is divided into three main steps: (i) the formation of (S)-norcoclaurine, the common precursor to all other BIAs produced in plants, from two l-tyrosine derivatives, (ii) the conversion of (S)-norcoclaurine to the branch point intermediate (S)-reticuline, shared among all species belonging to the Papaveraceae family, and (iii) conversion of (S)-reticuline to the bioactive BIAs such as magnoflorine, noscapine, sanguinarine, codeine, and morphine (Fig. 1) (Tamiru-Oli et al., 2018; Singh et al., 2019). Although the BIA biosynthesis pathway produces numerous structurally diverse compounds, it only involves genes encoding a limited number of protein families (Dang et al., 2012; Hagel and Facchini, 2013; Beaudoin and Facchini, 2014; Dastmalchi et al., 2018) (Fig. 1). The cytochromes P450 (CYPs) are the major family with more than 11 members. Of these, the (S)- to (R)-reticuline (STORR) or reticuline epimerase (REPI) is a P450-oxidoreductase, which evolved from gene duplication, rearrangement, and fusion and is responsible for the gateway reaction directing metabolites towards the morphinan branch (Winzer et al., 2015; Guo et al., 2018; Y. Li et al., 2020; Yang et al., 2021; Catania et al., 2022). The other protein families include S-adenosylmethionine-dependent O- and N-methyltransferases, FAD/NAD oxidoreductases, amino/acetyltransferases, pathogenesis-related-10 family and major latex proteins (PR10/MLPs), 2-oxoglutarate/Fe(II)-dependent dioxygenases (ODDs), NADPH-reductase, decarboxylase, and carboxylesterases (Dang et al., 2012; Hagel and Facchini, 2013; Beaudoin and Facchini, 2014; Dastmalchi et al., 2018, 2019b).
Thebaine synthase (THS) and neopinone isomerase (NISO), two other members of the PR10/MLP protein family with NCS, catalyse two reaction steps in the morphine branch that were previously assumed to be spontaneous (Lee and Facchini, 2010; Chen et al., 2018; Dastmalchi et al., 2019b). Although most enzymes involved in the BIA biosynthesis pathway have been characterized, enzymes responsible for a small number of reactions steps are still unknown (Beaudoin and Facchini, 2014; Singh et al., 2019). The conversion of l-tyrosine to dopamine involves a tyrosine decarboxylase (TYDC), responsible for the decarboxylation of l-tyrosine to tyramine, and a yet to be identified hydroxylase that is required for the hydroxylation of tyramine to dopamine. Similarly, a second enzyme, thought to be a decarboxylase, required for converting l-tyrosine to 4-HPAA is also uncharacterized in opium poppy (Fig. 1). Additionally, many reactions that are currently assumed to be spontaneous could be catalysed by novel enzymes that are yet to be identified, as was the case with the reactions catalysed by NCS or THS (Samanani and Facchini, 2001; Chen et al., 2018). Advances in shotgun proteomics could be helpful to identify novel proteins and elucidate their function. Knowing all components of the BIA biosynthesis pathway is vital for studies aiming to unravel the transcriptional regulation BIA biosynthesis genes.
Transcriptional regulation of benzylisoquinoline alkaloid gene expression
Transcription factors (TFs) are the primary regulators of gene expression, and understanding their role is essential to characterize the transcriptional regulation of BIA biosynthesis in opium poppy (Kawano et al., 2012; Winzer et al., 2012; Mishra et al., 2013; Kakeshpour et al., 2015; Agarwal et al., 2016; Jia et al., 2023; Tan et al., 2023). However, only a handful of TFs with putative regulatory roles have been reported in opium poppy, and our understanding of the transcriptional regulation of BIA biosynthesis is still limited. To this end, we set out to identify additional candidate TFs that may potentially regulate the expression of BIA genes using publicly available RNA sequencing data.
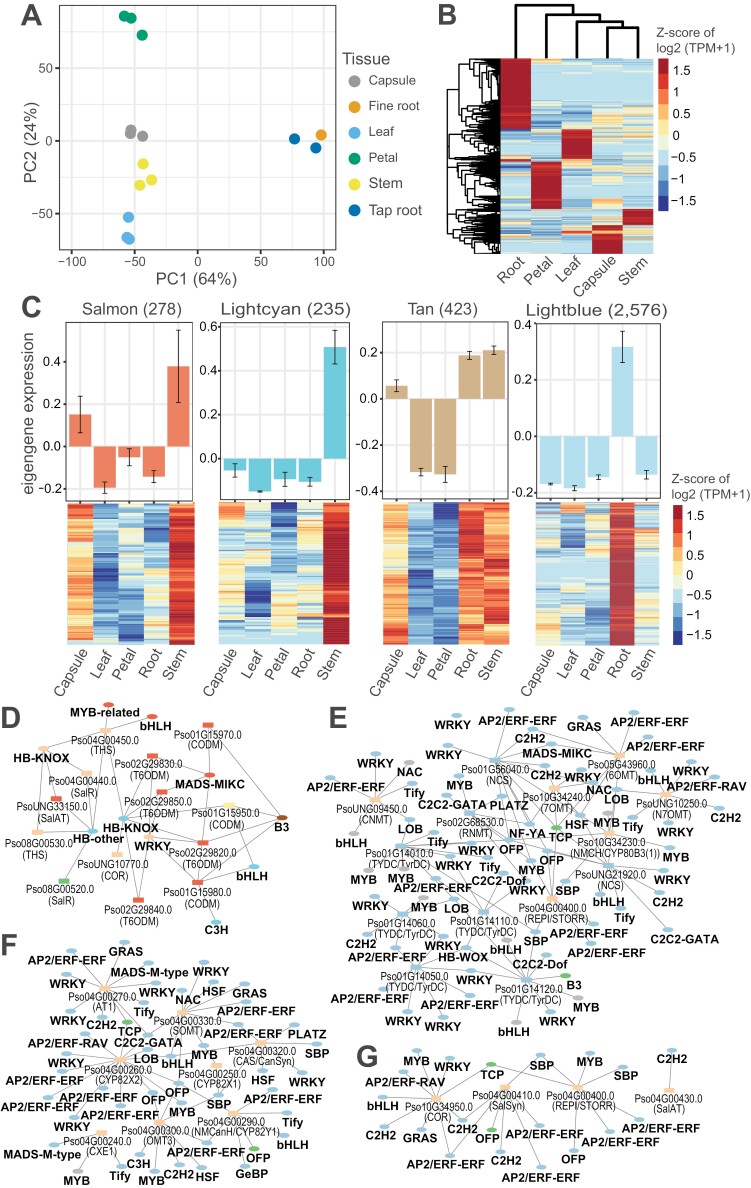
In the first instance, we used a recent RNA-seq dataset comprising gene expression data across six tissues of opium poppy: stem, capsule, root, leaf, petal, tap root, and fine root (Supplementary Table S1) (Jia et al., 2023). A principal component analysis of the top 500 most variable genes separated samples according to tissue type, similar to what was reported in the study (Fig. 2A). The tap and fine root samples clustered together, suggesting that these tissues have very similar gene expression profiles; these samples were therefore combined in the subsequent analyses. We then calculated an index of tissue specificity (tau; τ) for all expressed genes from the Jia et al. (2023) dataset (Supplementary Table S2); genes with a τ value greater than 0.8 were considered to be tissue-specific (Fig. 2B) (Kryuchkova-Mostacci and Robinson-Rechavi, 2017). Interestingly, the majority (71 out of 73) of the known and predicted BIA biosynthesis genes had the highest expression in either stem or root, supporting previous reports (Supplemantary Tables S2, S3) (Facchini and De Luca, 1994; Bird et al., 2003; Facchini and Park, 2003; Jia et al., 2023). More than half of these genes (43 out of 73) were associated with the three main branches of the BIA biosynthesis pathway, namely (S)-reticuline, noscapine, and morphine.
Fig. 2.
Identification of putative candidate regulators of BIA biosynthesis in opium poppy. (A) A principal component analysis plot of six RNA-seq libraries generated based on the 500 most variable genes and showing clustering according to tissues. Numbers in brackets correspond to the proportion of variance explained by the respective principal component. (B) Heatmap of tissue-specific gene expression, with red and blue indicating high and significantly low expression, respectively. (C) Stem and root gene co-expression modules identified using weighted correlation network analysis (WGCNA) (upper panel) and heatmap depicting the expression patterns of the genes included in each module (lower panel). Numbers in parentheses represent the number of genes and transcription factors in each module. (D) A stem gene regulatory sub-network highlighting morphine pathway genes and their predicted regulators (transcription factors). (E–G) Root gene regulatory sub-networks of S-reticuline, noscapine and morphine pathway genes and their predicted regulators, respectively. Ellipses and rectangles represent the predicted regulators and their target genes, respectively. Nodes are coloured according to their assigned WGCNA modules. For ease of visualization, subnetworks were filtered based on the following edge weight cut-offs: 0.5 (D) and 0.75 (E–G).
Genes and TFs operate in a concerted manner, with many interacting partners ultimately influencing gene expression outcomes. This has given rise to the concept of gene co-expression network analysis, which entails the identification of genes and TFs that cluster together with similar expression profiles at a point in time or upon exposure to perturbations (van Dam et al., 2017; Hurgobin and Lewsey, 2022). The co-expression of BIA genes has recently been reported (Q. Li et al., 2020). The authors also identified several uncharacterized MLP/PR10 proteins among the genes co-expressed with BIA genes, suggesting that these proteins may have important functions in the pathway. Another study reported the differential expression of BIA uptake permeases (BUPs), which act as alkaloid transporters, across the same tissues mentioned above (Dastmalchi et al., 2019a). Out of the nine homologues identified, eight homologues (BUP1, BUP2, BUP4–BUP9) exhibited tissue-specific expression based on RT-qPCR results. The genes encoding these transporters also clustered within the genome closely to known BIA biosynthetic genes. This observation implies that the transporters and biosynthetic genes may also be co-expressed, because genes that are located close to one another within the physical space of the genome may share topological associating domains, which can drive similar expression patterns (Hurst et al., 2004; Pombo and Dillon, 2015; Kustatscher et al., 2017). However, none of these studies identified TFs among the co-expressed genes.
Consequently, we set out to identify TFs that may be involved in the transcriptional regulation of BIA biosynthesis via weighted gene co-expression network analysis (WGCNA) (Langfelder and Horvath, 2008). Our analysis identified 13 modules of co-expressed genes and TFs (Supplementary Fig. S1; Supplementary Table S4). We identified 57 of the known BIA biosynthesis genes in these modules; the majority (55 genes) of these genes resided in four modules (salmon, lightcyan, tan, and lightblue) which were predominantly stem- or root-specific. (Fig. 2C; Supplementary Table S4). A significant number of TFs (263 root-specific and 26 stem-specific) were also identified in the 13 modules, with the majority (238 root-specific and 24 stem-specific) being in the salmon, lightcyan, tan, and lightblue modules (Supplementary Table S4).
Co-expression of a group of genes suggests, but does not prove, that they may be regulated by a common TF or set of TFs. This is termed co-regulation. Potential regulatory interactions may be examined further by the construction of gene regulatory networks (GRNs) (Karlebach and Shamir, 2008). This approach identifies putative interactions between TFs and their downstream target genes (interaction edge), which allows the identification of key candidate regulators of traits of interest (Van den Broeck et al., 2020). In this respect, we used the co-expressed TFs and target genes (including known BIA biosynthesis genes) identified by WGCNA to construct a stem-specific GRN and a root-specific GRN. We did so using the SCION method and additional RNA-seq data generated by Guo and colleagues (Supplementary Protocol S1; Supplementary Tables S5, S6; Guo et al., 2018; Clark et al., 2021). Sixteen BIA biosynthesis genes that were missing from the co-expression modules were also included among the targets as they could potentially be regulated by the TFs identified in this study (Supplementary Table S3). The stem GRN consisted of 26 TFs and 437 target genes, with a total of 1906 TF-target interactions (edges) (Supplementary Table S5). Similarly, the root GRN contained 263 TFs and 2190 target genes, connected by 374 894 edges (Supplementary Table S6). Next, we extracted a stem-specific morphine subnetwork and root-specific (S)-reticuline, noscapine, and morphine subnetworks from the two GRNs (Supplementary Table S5). The stem-specific morphine subnetwork consisted of 17 TFs, 14 BIA target genes and a total of 72 TF-target interactions, and predominantly contained TFs from the bHLH, C2H2, HB-KNOX, and MADS-MIKC families (Fig. 2D). The root-specific (S)-reticuline subnetwork contained 263 TFs, 14 BIA target genes, and 2364 TF-target interactions. Similarly, the root-specific noscapine subnetwork contained 258 TFs and eight BIA targets connected by 1312 edges. The root-specific morphine subnetwork contained 263 TFs, four BIA targets and 637 edges (Supplementary Table S6). The TFs (regulators) predicted in all the three root-specific subnetworks were predominantly from the AP2/ERF-ERF, WRKY, MYB, bHLH, NAC, and C2H2 TF families (Fig. 2E, F).
Our combined analyses of tissue specificity, WGCNA, and GRN identified many potential regulators of BIA biosynthesis in opium poppy. While useful, it is important to remember that such type of in silico predictions often harbour false positives. Therefore, wet-lab validation is key to ensuring the credibility of these predictions. To this end, DNA affinity purification and sequencing (DAP-seq) is a cheap and rapid assay to generate genome-wide in vitro TF–DNA interaction maps for candidate TFs (Bartlett et al., 2017). Virus-induced gene silencing has been instrumental for functional validation of BIA biosynthesis genes in opium poppy (Hileman et al., 2005). The same approach can be used to target candidate TFs and, in combination with RNA-seq, provide vital data on the expression patterns of predicted downstream target BIA genes.
Overall, our findings support what has been reported in the literature. Some of the TF families mentioned above were previously predicted to have a role in the regulation of BIA biosynthesis based on the analysis of TF binding sites and expression profiles of BIA biosynthesis genes. Members of the 10-gene cluster for noscapine biosynthesis all have similar WRKY and MYB binding motifs in their promoter regions and are also shown to be co-expressed, implicating these two TF families in the co-regulation of the gene cluster (Winzer et al., 2012; Kakeshpour et al., 2015). The W-box cis-element, a known WRKY binding site, has also been identified in the promotor regions of TYDC, norcoclaurine 6-O-methyltransferase (6OMT), coclaurine N-methyltransferase (CNMT), reticuline 7-O-methyltransferase (7OMT), salutaridinol 7-O-acetyltransferase (SalAT), and codeinone reductase (COR) genes (Mishra et al., 2013; Agarwal et al., 2016). Recently, HB6, a member of the HB–HD–ZIP TF family that likely regulates 19 genes involved in (S)-reticuline, morphinan, noscapine, sanguinarine, and laudanine biosynthesis, has been identified using assay for transposase-accessible chromatin using sequencing (ATAC-seq) and transcriptome analyses (Kryuchkova-Mostacci and Robinson-Rechavi, 2017; Van den Broeck et al., 2020; Jia et al., 2023).
Another important type of gene expression regulator is microRNAs (miRNAs) (Jones-Rhoades et al., 2006). These endogenous single-stranded non-coding small RNAs that are 18–30 bp in length are involved in post-transcriptional regulation in eukaryotes either by targeted-mRNA cleavage or by affecting translation (Filipowicz et al., 2008). In plants, they have been shown to play a crucial role in regulating the biosynthesis of secondary metabolites such as flavonoids, terpenoids, alkaloids, and phenolic acid compounds (Hossain et al., 2022; Jeena et al., 2022; Sun et al., 2022; Zhang et al., 2022). Pso-miRNA13, miRNA408, and pso-miRNA2161 are three miRNAs identified to be involved in BIA biosynthesis regulation in opium poppy (Boke et al., 2015; Singh et al., 2019; Jeena et al., 2022). While pso-miRNA13 and pso-miRNA2161 have been found only in opium poppy, miRNA408 is present in other species, for example, in red sage (Salvia miltiorrhza), playing a regulatory role in salvianolic acid biosynthesis. Pso-miRNA13 is supposed to cleave transcripts of 7OMT, which is responsible for the conversion of the central precursor (S)-reticuline to (S)-laudanine, which is then converted to papaverine (Fig. 1). Therefore, 7OMT silencing could indirectly enhance the biosynthesis of other BIAs, such as morphine, noscapine, or sanguinarine. Pso-miRNA2161 is predicted to target transcripts of 3ʹ-hydroxyl-N-methylcoclaurine 4ʹ-O-methyltransferase and is likely involved in regulating BIA accumulation in stem and capsule tissues. On the other hand, miRNA408 possibly silences the berberine bridge enzyme (BBE), which is responsible for the conversion of (S)-reticuline to (S)-scoulerine. Moreover, transcripts of other BIA biosynthetic genes, TYDC, COR, and SalAT, might be silenced by these miRNAs through in silico analyses. Future works are required to elucidate the regulation mechanism of miRNAs in BIA biosynthesis.
Compartmentalization of benzylisoquinoline alkaloid biosynthesis between different tissue and cell types
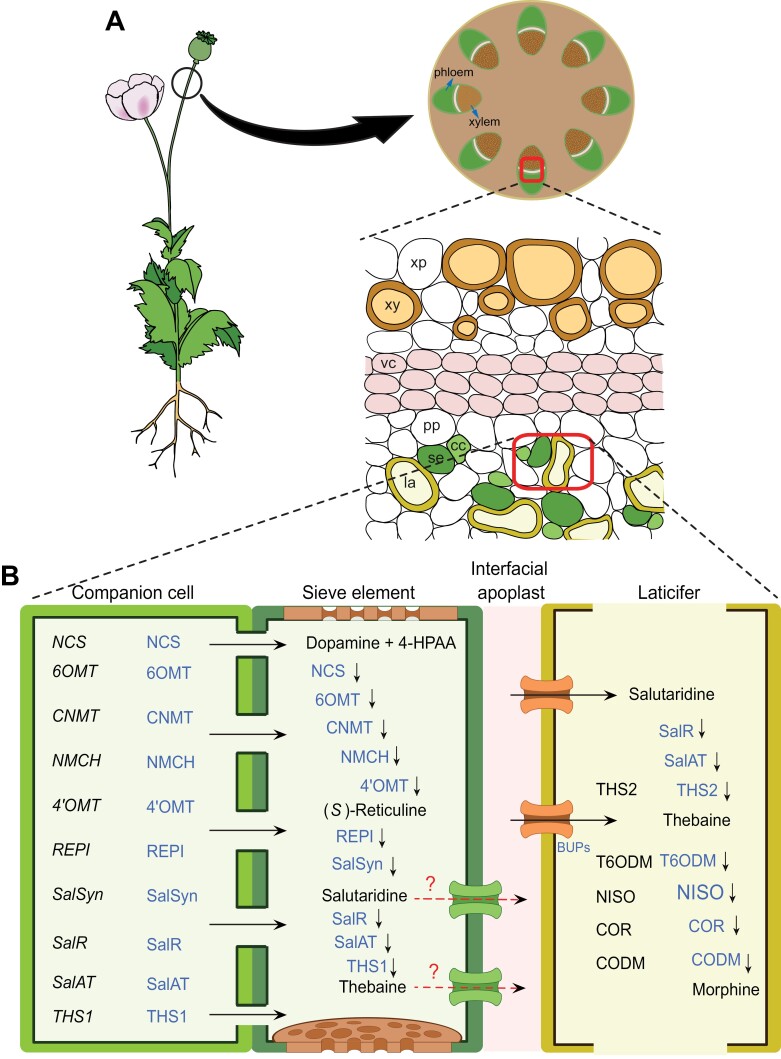
In opium poppy, BIA biosynthetic gene transcription, translation, and enzyme activity involve three distinct cell types: companion cells, sieve elements, and laticifers of the phloem system (Fig. 3) (Bird et al., 2003; Weid et al., 2004; Samanani et al., 2006; Liscombe and Facchini, 2008; Lee et al., 2013; Onoyovwe et al., 2013; Singh et al., 2019; Ozber and Facchini, 2022). Sieve elements that are enucleate and incapable of RNA and protein synthesis are kept alive by the connection with neighbouring companion cells via numerous plasmodesmata, resulting in the sieve element–companion cell complex (Fukuda et al., 2005; Faulkner, 2018; Kim and Frommer, 2023). This complex is a mandatory component of the phloem in all vascular plants. In contrast, laticifers are highly specialized cells and present only in the phloem system of several phylogenetically unrelated groups (Castelblanque et al., 2016; Ramos et al., 2019; Johnson et al., 2021). They form a tube-like network throughout the plant body and produce and store latex. Most BIAs of opium poppy are accumulated in latex of laticifers (Weid et al., 2004; Onoyovwe et al., 2013; Beaudoin and Facchini, 2014; Singh et al., 2019; Ozber and Facchini, 2022).
Fig. 3.
Schematic illustration of the cellular localization of morphine biosynthesis in opium poppy. (A) Morphine biosynthesis involves three cell types: companion cells (cc), sieve elements (se), and laticifers (la) of the phloem system. (B) The enzymes synthesized in companion cells are transported to sieve elements (black horizontal arrows) through plasmodesmata. The intermediate compounds of salutaridine and thebaine are transferred from sieve elements to latifer cells (red horizontal arrows) through apoplast by a family of benzylisoquinoline uptake permeases (BUPs). The enzymes required for the final stages of alkaloid biosynthesis are mainly localized in laticifers. Transcripts and proteins primarily detected in each cell type are shown in black italic and in blue, respectively. vc, vascular cambium; pp, phloem parenchyma; xy, xylem vessels. 4ʹOMT, 3ʹ-hydroxyl-N-methylcoclaurine 4ʹ-O-methyltransferase; 6OMT, norcoclaurine 6-O-methyltransferase; CNMT, coclaurine N-methyltransferase; CODM, codeine O-demethylase; COR, codeinone reductase; NCS, norcoclaurine synthase; NISO, neopinone isomerase; NMCH, N-methylcoclaurine 3ʹ-hydroxylase; REPI, reticuline epimerase; SalAT, salutaridinol 7-O-acetyltransferase; SalR, salutaridine reductase; SalSyn, salutaridine synthase; T6ODM, thebaine 6-O-demethylase; THS, thebaine synthase (Liscombe and Facchini, 2008; Lee et al., 2013; Beaudoin and Facchini, 2014; Ozber and Facchini, 2022).
Previous research based on in situ hybridization and immunofluorescence suggested that most BIA-related genes are transcribed and translated into enzymes in companion cells, then transported to sieve elements and laticifers (Bird et al., 2003; Weid et al., 2004; Samanani et al., 2006; Liscombe and Facchini, 2008; Lee et al., 2013). Recently, with advances in transcriptomic and proteomic sequencing, transcripts of some genes and their mature enzymes were also found in laticifers, indicating translation likely occurs in those cells (Onoyovwe et al., 2013; Chen et al., 2018; Dastmalchi et al., 2019b; Ozber et al., 2022). The enzymes involved in the early stages of the morphinan pathway (converting dopamine and 4-HPAA to salutaridine) are mainly present in sieve elements, while those catalysing the final steps are primarily found in laticifer cells (Fig. 3) (Beaudoin and Facchini, 2014; Ozber and Facchini, 2022). For example, transcripts of four genes responsible for producing morphine from thebaine [thebaine 6-O-demethylase (T6ODM), NISO, COR, and codeine O-demethylase (CODM)] and their corresponding enzymes are detected in laticifers (Onoyovwe et al., 2013). Similar models are observed for noscapine and papaverine biosynthesis in opium poppy (Ozber et al., 2022). Two genes [3-O-acetylpapaveroxine carboxylesterase (CXE) and noscapine synthase (NOS)] involved in the two final steps of noscapine biosynthesis are expressed in laticifers, whereas the remaining genes in the noscapine cluster are exclusively transcribed and translated in companion cells.
BIA biosynthesis related components, composed of enzymes, precursors, and final products, are mainly transported between companion cells, sieve elements, and laticifers through symplast and apoplast pathways (Fig. 3) (Bird et al., 2003; Weid et al., 2004; Samanani et al., 2006; Onoyovwe et al., 2013; Beaudoin and Facchini, 2014; Ozber and Facchini, 2022). The numerous plasmodesmata between companion cells and sieve elements establish the symplast transport, facilitating the movement of BIA biosynthetic enzymes from companion cells to sieve elements. However, not all proteins from companion cells are transferred to sieve elements, and the distribution of BIA biosynthetic enzymes has been shown differently between parts of poppy plants (Bird et al., 2003; Weid et al., 2004; Fukuda et al., 2005; Beaudoin and Facchini, 2014; Faulkner, 2018; Kim and Frommer, 2023). The tissue-specific mechanism of BIA biosynthetic enzyme distribution between companion cells and sieve elements remains to be studied in detail.
Symplastic transport of pathway intermediates between sieve elements and laticifers is also suggested based on the presence of plasmodesmata connecting them (Facchini and De Luca, 2008). Recently, an uptake transporter family, known as benzylisoquinoline uptake permeases (BUPs), has been identified to transfer BIAs from sieve elements to laticifers in opium poppy (Fig. 3) (Dastmalchi et al., 2019a). This indicated the presence of apoplast transport for BIAs from sieve elements to laticifers. Interestingly, although BUPs were found in BIA gene clustering, including noscapine, none had an importing capacity for noscapine or papaverine (Dastmalchi et al., 2019a; Ozber and Facchini, 2022). This suggests the existence of additional transporters for carrying BIAs to laticifers. Proteins of the ATP-binding cassette (ABC) family or multi-antimicrobial extrusion (MATE) family are responsible for the translocation of berberine in Coptis japonica, and are therefore potential transporter candidates for future research in opium poppy (Shitan et al., 2003; Takanashi et al., 2017).
Future perspectives: new approaches that could deepen the understanding of benzylisoquinoline alkaloid biosynthesis regulation
Single-cell multi-omics and spatial transcriptomics
The majority of studies conducted to date to identify metabolic pathway-associated genes and enzymes in plants have relied on the use of whole tissues/organs (Giacomello, 2021; Tenorio Berrío et al., 2021; Zhang et al., 2022; Depuydt et al., 2023). In poppy, for example, global BIA gene expression has been surveyed at the organ-level, predominantly in the capsules, but also in leaves, roots, and stem (Winzer et al., 2012; Guo et al., 2018; Zhao et al., 2019; Q. Li et al., 2020; Yang et al., 2021; Xu et al., 2022; Jia et al., 2023). However, the localization of BIAs is not limited to distinct organs but also to specific cells within these organs. The cell-type-specific biosynthesis and accumulation of BIAs in poppy has been documented to a certain extent, but knowledge gaps remain in terms of the underlying mechanisms that enable expression of BIA pathway genes to be controlled in a cell-type-specific manner (Samanani et al., 2006; Onoyovwe et al., 2013; Chen et al., 2018; Dastmalchi et al., 2019b; Ozber and Facchini, 2022; Ozber et al., 2022). The application of single-cell ‘omics’ technologies provides an opportunity to bridge these gaps. One such application is single-cell RNA-seq (scRNA-seq) whereby the transcriptional activity of single cells can be mapped and quantified (Shaw et al., 2021). The coordination of BIA biosynthesis involves three distinct cell types (companion cells, laticifers, and sieve elements) but it remains unknown whether all cells within each of these cell types behave uniformly or if, within cell-types, there are functional subpopulations of cells. Many plant species have been reported to have more than one type of companion cell, which differ in their structural features as well as their degree of plasmodesmatal connectivity with neighbouring cells (Kim and Frommer, 2023). It is possible that this could also be the case for opium poppy. ScRNA-seq could be employed to investigate this and potentially identify marker genes for each subpopulation within the companion cells.
Additionally, given that BIA biosynthesis also occurs in organs other than the capsules, it would be interesting to investigate whether the expression profile of the pathway genes differs between organs and their cells, shedding more light on the biology of the pathway. The differential accumulation of BIAs across organs is well-known, with sanguinarine located exclusively in the roots, papaverine and noscapine found only in the shoot latex, and morphine and codeine found in roots, but to a greater extent in shoot latex (Facchini and De Luca, 1994). The association between tissue-specific gene expression and accumulation of BIAs was recently reported (Q. Li et al., 2020). The authors identified the genes involved in the sanguinarine pathway and the later stages of the morphine and noscapine portion of the BIA biosynthesis pathway to be root-specific and latex-specific, respectively. An extension of this finding would be to identify which cell subpopulations within these distinct organs exhibit this pattern of gene expression.
The organ-level gene network analyses (co-expression and regulation) that have been performed in opium poppy have improved our understanding of the basis for BIA gene transcriptional regulation (Q. Li et al., 2020; Xu et al., 2022; Jia et al., 2023). Extending these organ-level analyses to the single-cell resolution would allow cell-specific modules and their constituent genes and corresponding TFs to be identified. Additionally, the analysis of cell-specific GRNs could potentially identify TFs that drive the expression of key enzymes and transporters in a targeted manner and encourage the commercial production of opioids using synthetic biology. A step further would be to compare cell-specific gene expression and regulation between different poppy lineages to gain a better understanding of the differences in BIA variation among cultivars; this could assist with the selection of elite varieties in breeding programmes.
The study of the poppy epigenome is likely to complement transcriptional studies in this species and offer novel insights into the epigenetic regulation of BIA biosynthesis. This can be achieved via ATAC-seq, which enables the identification of accessible chromatin regions (ACRs) in the genome (Buenrostro et al., 2015). These regions tend to be associated with promoters of genes that are actively expressed and harbour cis-regulatory elements (CREs) that are associated with these genes. CREs are the DNA recognition sequences (motifs) that TFs bind to, allowing TFs to find the genes that they regulate. The application of ATAC-seq in plants is in its infancy, with only one such study reported in opium poppy (Lu et al., 2016; Farmer et al., 2021; Jia et al., 2023). Recently, Jia et al. (2023) mapped the open chromatin landscape of poppy across six tissues (leaf, stem, capsule, petal, tap root, and fine root), and integration with tissue-specific RNA-seq data revealed the presence of ACRs in the BIA genes exclusively in the capsule, stem, and root (Jia et al., 2023). Analysis of the CREs within the ACRs highlighted common DNA motifs, which suggested that these tissues share several TFs that regulate the tissue-specific expression of BIA biosynthesis genes. The combined application of scRNA-seq and scATAC-seq could further expand on these findings by potentially revealing whether there are subpopulations of cells within each tissue type that are preferentially associated with the BIA gene-specific ACRs and how this differs among poppy cultivars with varying BIA contents.
The cell-specific accumulation of BIAs in poppy has been investigated using traditional approaches such as immunofluorescence and RNA in situ hybridization, which have been applied to whole tissues (Bird et al., 2003; Samanani et al., 2006; Lee and Facchini, 2010; Beaudoin and Facchini, 2014). In this respect, single-cell metabolomics (SCM), which is the high-throughput analysis of metabolites at the single-cell level, could prove beneficial for confirming the findings from previous studies as well as uncovering novel information (Guo et al., 2021; Hu et al., 2023). For instance, SCM could be used to investigate the differential accumulation of BIAs in cell subpopulations. It may also be possible to capture the presence of minor intermediate metabolites or novel ones that might be otherwise undetectable in the more heterogeneous mixtures of cells from whole organ samples; this will not only provide a more complete picture of the metabolic diversity of the pathway but also present opportunities for synthesizing new drugs. Q. Li et al. (2020) recently reported that co-variation patterns between gene expression and alkaloid levels were similar across tissues and time points. However, given that these measurements were performed using bulk tissues and were hence averaged, it is possible that subtle differences at the single-cell level were missed. Therefore, the integration of SCM with scRNA-seq would allow for more accurate correlations between cell-type-specific expression of biosynthetic enzymes and the presence of the corresponding metabolites across tissues and developmental stages.
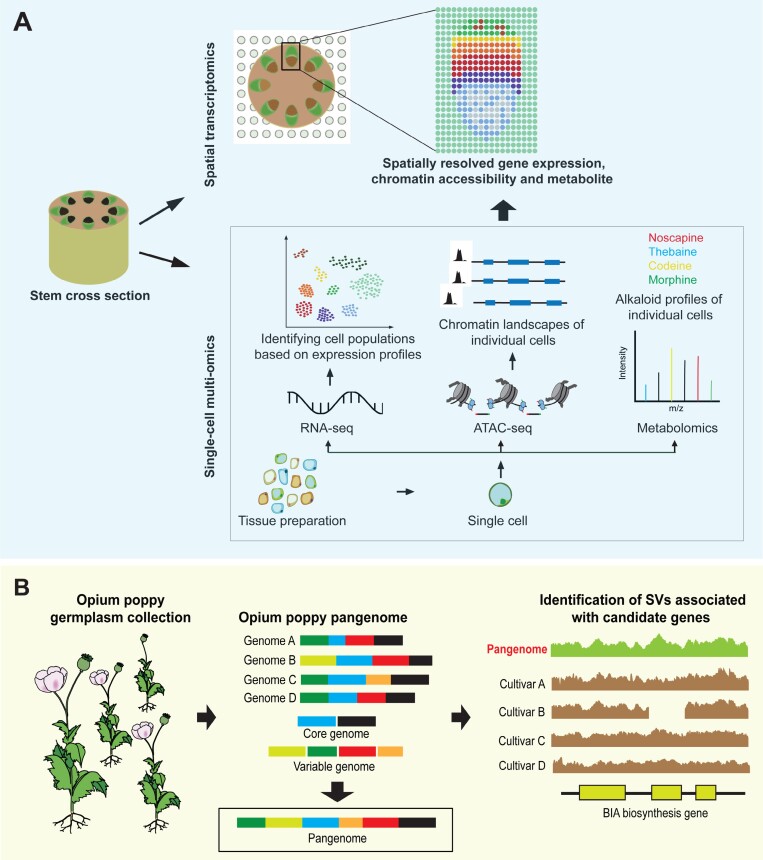
Application of the single-cell ‘omics’ technologies discussed above requires the relevant tissues to be dissociated into individual cells, resulting in the cells losing their spatial information. The knowledge of where cells are located and how close they are to one another is crucial for understanding intercellular communication (Longo et al., 2021). Spatial transcriptomics addresses this issue by localizing transcripts to precise regions in native tissues, and when combined with scRNA-seq and scATAC-seq would allow associations to be identified between the transcriptomic and epigenetic regulation of single cells and their spatial location, shedding light on the interactions between cell subpopulations (Fig. 4A) (Longo et al., 2021; Nobori et al., 2023; Peirats-Llobet et al., 2023). The integration of these modalities would be valuable in gaining an understanding of the spatiotemporal regulation of BIA metabolism; in particular it would be interesting to know how the cells interact with each other during development or under stress. It will also be interesting to see how this correlates with the accumulation of BIAs via the application of SCM, hence furthering our understanding of the differential accumulation of these metabolites in poppy (Hu et al., 2023).
Fig. 4.
Proposed schemes for applying the latest omics tools to study the regulation of alkaloid biosynthesis in opium poppy. (A) Integration of single-cell ‘omics’ technologies with spatial transcriptomics in opium poppy. A spatial barcoded map of the vascular tissue is obtained; this map represents the gene expression of localized mRNA transcripts across the tissue section. In parallel, the vascular tissue undergoes tissue dissociation for the application of single-cell modalities. This includes single-cell RNA sequencing (scRNA-seq) for obtaining the gene expression profiles of single cells, single-cell assay for transposase-accessible chromatin using sequencing (scATAC-seq) for surveying the open chromatin landscape of the cells, and single-cell metabolomics (SCM) for assessing the metabolite content of the cells. Finally, the individual single-cell ‘omics’ measurements are integrated with spatial information to annotate the cells. (B) An overview of opium poppy pangenome construction. Diverse germplasm collections from different geographic regions representing landraces, modern cultivars, and their wild relatives are used for whole genome re-sequencing. The data generated are compared to the current opium poppy reference genome sequence to identify genome-wide structural variations (SVs), based on which a non-redundant poppy pangenome is constructed. The pangenome is used for an in-depth analysis of SVs associated with the BIA biosynthetic genes; as an example, the presence of a deletion in the second exon of a candidate BIA biosynthesis gene is shown. Such information will inform genetic improvement of traits of commercial interest including total alkaloid yield and composition.
Pangenomics and natural variation
The presence of major genome structural variations (SVs) makes plant genomes highly complex and diverse, with considerable intra- and inter-species divergence between genomes (Buckler et al. 2006, Soltis and Soltis 2021). Consequently, single reference genomes do not fully capture the entire genetic diversity of the species, and hence the development of pangenomes as new references (Hurgobin and Edwards, 2017; Bayer et al., 2020). Pangenomes represent the full complement of the DNA sequence of a species, which now also captures major SVs such as presence/absence (PAV) and copy number (CNVs) variations, chromosomal rearrangements and translocations as a result of recent advances in long-read DNA sequencing technologies (Ho et al., 2020). This emerging approach has been instrumental in dissecting SVs associated with yield and quality related traits in multiple species (Zhou et al., 2022; Li et al., 2023; Wang et al., 2023). Pangenomes are often constructed from hundreds or even thousands of phylogenetically and geographically diverse accessions that include landraces, modern varieties, and their wild relatives (Gao et al., 2019). Including crop wild relatives in developing pangenomes would provide the genetic diversity required for breeding improved and new crop varieties.
Plant secondary metabolites are formed by stepwise enzymatic reactions that together form biosynthetic pathways. The genes associated with such pathways are sometimes found as biosynthetic gene clusters (BGCs) (Polturak and Osbourn, 2021). The existence of BGCs might be important to ensure co-inheritance and co-regulation, as well as to avoid the accumulation of intermediate compounds to toxic levels (Nützmann et al., 2018; Z. Liu et al., 2020). Such clusters have been reported in opium poppy including for the genes associated with BIA biosynthesis (Winzer et al., 2012; Guo et al., 2018; Conneely et al., 2022; Zhou and Liu, 2022) (Fig. 1). An increasing body of evidence shows that gene and genome duplications have provided the basis for the diversification of plant secondary metabolism in multiple species including in opium poppy (Itkin et al., 2013; Wang et al., 2020; Liu et al., 2021).
The single reference opium poppy genome was first assembled in 2018 (Guo et al., 2018). This genomic resource has been vital for getting deeper insight into the genomic arrangement of key BIA biosynthesis genes, revealing gene clusters for noscapine and morphine biosynthesis spanning a 584-kbp genomic region on chromosome 11 (Guo et al., 2018). The reference genome was further improved with Hi-C sequencing, generating chromosome-scale scaffolds that allowed the anchoring of 35 additional BIA genes (Q. Li et al., 2020). It was also shown that CNVs of key BIA genes significantly correlate with alkaloid profiles and that co-expression of BIA genes increases with BGCs (Q. Li et al., 2020). Comparison of three chromosomal scale genome assemblies representing P. somniferum, P. setigerum and P. rhoeas revealed that SVs involving gene duplication, translocation, and fusion have contributed to the generation and maintenance of BIA gene clusters in opium poppy (Yang et al., 2021). Independently, genome and transcriptome comparisons between a Chinese landrace and the reference opium poppy cultivar implicated genome expansion as one of the major drivers of high BIA biosynthesis (Pei et al., 2021). A recent study involving the re-sequencing of 10 opium poppy cultivars reported CNVs in 63 out of the 109 BIA genes and showed that the CNV of key BIA biosynthesis genes was responsible for the diversity in alkaloid yield and composition (Q. Li et al., 2020). The same authors reported that BIA genes in the same cluster are highly co-expressed, suggesting co-regulation.
Opium poppy breeding has involved extensive selection for plants with increased alkaloid yield and varying alkaloid compositions, generating a considerable number of varieties with diverse alkaloid profiles (Singh et al., 2014). As such, the genetic diversity in BIA biosynthetic genes has provided the molecular basis for much of the observed variation in alkaloid profiles (Millgate et al., 2004; Hagel and Facchini, 2010; Winzer et al., 2012, 2015; Pathak et al., 2013; Agarwal et al., 2016). A large deletion spanning the 10-gene noscapine cluster region has led to the development of non-noscapine producing cultivars with significantly elevated morphine and/or thebaine contents (Winzer et al., 2012). Similarly, the high papaverine-accumulating variety pap1 and thebaine oripavine poppy 1 variety top1 (also known as Norman) are mutants harbouring variations in regulatory and structural genes of the BIA biosynthesis pathway (Millgate et al., 2004; Pathak et al., 2013; Agarwal et al., 2016). Despite the strict international regulation that limited licit opium poppy production and germplasm movement, germplasm collections of considerable sizes representing P. somniferum and other related species are currently maintained by various international and national gene banks (Bajpai et al., 1999; Brezinova et al., 2009; Dittbrenner et al., 2012; Celik et al., 2016). Several studies have characterized a subset of these resources to show the existence of considerable morphological and chemical diversity (Dittbrenner et al., 2012; Celik et al., 2016; Verma et al., 2016; Hong et al., 2022).
The development of an opium poppy pangenome that integrates the existing germplasm would be vital for revealing the true extent of genetic variability in the genome as well as forming the basis for opium poppy breeding aiming to improve traits of commercial interest (Fig. 4B). With long-read sequencing becoming increasingly accessible, it will be possible to construct a graph-based, non-redundant pangenome that incorporates SVs from individual genomes, facilitating the visualization of accession-specific sequences in a single resource and making comparisons between accessions easier (Garrison et al., 2018; Bayer et al., 2020; Zanini et al., 2022). The availability of a poppy pangenome promises to shed light on fundamental aspects of genetic variation pertaining to the BIA biosynthesis genes. The majority of plant pangenomics studies have focused on the variations within the coding regions of genes (Golicz et al., 2016; Montenegro et al., 2017; Hurgobin et al., 2018; Y. Liu et al., 2020; Walkowiak et al., 2020). However, the presence of genetic variants in CREs that are associated with genes of interest, such as the BIA biosynthesis genes, should not be overlooked; these can affect gene expression while reducing the effect of pleiotropy. As such, variations in CREs could be exploited for expanding the allelic diversity of BIA biosynthesis genes in poppy. More specifically, it may be possible to apply genome editing technologies such as CRISPR/Cas9 to create different types and strengths of CRE mutations in genes of interest to investigate how these regulatory variations affect BIA biosynthesis genes.
Concluding remarks
The considerable advances in poppy research over the last decades have led to remarkable improvements in our knowledge of the BIA biosynthesis pathway and the subcellular localization of pathway components. Improved genome assemblies and genome comparisons across related Papaver species have also shed light on the genomic organization of BIA biosynthesis genes and the contribution of major genome SVs to the generation and maintenance of BIA gene clusters. Current reports showing the co-expression of BIA pathway gene clusters point to the possibility that these genes are also co-regulated. However, our understanding of how opium poppy BIA biosynthesis is regulated is still very limited.
Elucidation of the transcriptional and regulatory mechanisms of the BIA pathway, especially at the single-cell level, may aid with the identification of key regulators that can be targeted for improvement of traits of interest and for biotechnological applications. To this end, the contribution of single-cell ‘omics’ technologies discussed above will be significant. Gene expression analysis at the single-cell resolution has proved powerful in studying transcriptome dynamics associated with the regulation of important biological processes including secondary metabolism. The identification of ACRs and CREs that are associated with genes of interest will complement these transcriptional studies by providing a comprehensive view of the epigenetic regulation of BIA biosynthesis at the single-cell level. Last, but not least, the analysis of metabolites at cell-type resolution would shed more light on the differential accumulation of BIAs in key cell types and how this process differs between tissues and poppy lineages of varying BIA content. The integration of these individual modalities with spatial transcriptomics is likely to generate crucial data that can further our understanding of how the expression of BIA genes is coordinated in the different cells/tissues.
Considerable genetic diversity exists in the global poppy germplasm collection. This collection provides the genetic and chemical diversity that underpins poppy research and genetic improvement. The wild relatives of opium poppy are also potential sources useful genes that can be deployed to improve key traits in commercial poppy cultivars, but these resources are underexploited. An opium poppy pangenome that captures the diversity in landraces, commercial cultivars, and their wild relatives is crucial to unravel the genome structural variants associated with alkaloid yield and composition.
Supplementary data
The following supplementary data are available at JXB online.
Protocol S1. Identification of tissue-specific genes, weighted gene co-expression network analysis (WGCNA) and gene regulatory network analysis.
Fig. S1. Co-expression modules identified by WGCNA.
Table S1. Accessions of RNA-seq data used in this study.
Table S2. Tau values assigned to expressed opium poppy genes.
Table S3. List of BIA biosynthesis used in this study.
Table S4. List of genes assigned to co-expression modules by WGCNA.
Table S5. Stem-specific gene regulatory network.
Table S6. Root-specific gene regulatory network.
Contributor Information
Uyen Vu Thuy Hong, Australian Research Council Research Hub for Medicinal Agriculture, La Trobe University, AgriBio Building, Bundoora, VIC 3086, Australia; La Trobe Institute for Sustainable Agriculture and Food, Department of Plant, Animal and Soil Sciences, La Trobe University, AgriBio Building, Bundoora, VIC 3086, Australia.
Muluneh Tamiru-Oli, Australian Research Council Research Hub for Medicinal Agriculture, La Trobe University, AgriBio Building, Bundoora, VIC 3086, Australia; La Trobe Institute for Sustainable Agriculture and Food, Department of Plant, Animal and Soil Sciences, La Trobe University, AgriBio Building, Bundoora, VIC 3086, Australia.
Bhavna Hurgobin, Australian Research Council Research Hub for Medicinal Agriculture, La Trobe University, AgriBio Building, Bundoora, VIC 3086, Australia; La Trobe Institute for Sustainable Agriculture and Food, Department of Plant, Animal and Soil Sciences, La Trobe University, AgriBio Building, Bundoora, VIC 3086, Australia.
Mathew G Lewsey, Australian Research Council Research Hub for Medicinal Agriculture, La Trobe University, AgriBio Building, Bundoora, VIC 3086, Australia; La Trobe Institute for Sustainable Agriculture and Food, Department of Plant, Animal and Soil Sciences, La Trobe University, AgriBio Building, Bundoora, VIC 3086, Australia; Australian Research Council Centre of Excellence in Plants for Space, AgriBio Building, La Trobe University, Bundoora, VIC 3086, Australia.
Susanne Schilling, University College Dublin, Ireland.
Author contributions
UVTH, MTO, BH, and MGL: conceptualization of the review. BH and UVTH: performing bioinformatics analyses. All authors contributed to drafting the work, revised the final manuscript, and approved submission.
Conflict of interest
The authors declare no conflict of interest.
Funding
This work was supported by grants from the Australian Research Council to MGL (CE230100015, IH180100006). UVTH was supported by a La Trobe University Graduate Research Scholarship and a La Trobe University Full Fee Research Scholarship.
Data availability
The RNA sequencing data downloaded from public database and used in this manuscript are summarized in Supplementary Table S1, and the relevant analyses are detailed in Supplementary Protocol S1.
References
- Agarwal P, Pathak S, Kumar RS, Dhar YV, Shukla S, Asif MH, Trivedi PK.. 2020. Short-chain dehydrogenase/reductase, PsDeHase, from opium poppy: putative involvement in papaverine biosynthesis. Plant Cell, Tissue and Organ Culture 143, 431–440. [Google Scholar]
- Agarwal P, Pathak S, Lakhwani D, Gupta P, Asif MH, Trivedi PK.. 2016. Comparative analysis of transcription factor gene families from Papaver somniferum: identification of regulatory factors involved in benzylisoquinoline alkaloid biosynthesis. Protoplasma 253, 857–871. [DOI] [PubMed] [Google Scholar]
- Bajpai S, Prajapati S, Luthra R, Sharma S, Naqvi A, Kumar S.. 1999. Variation in the seed and oil yields and oil quality in the Indian germplasm of opium poppy Papaver somniferum. Genetic Resources and Crop Evolution 46, 435–439. [Google Scholar]
- Bartlett A, O’Malley RC, Huang S-C, Galli M, Nery JR, Gallavotti A, Ecker JR.. 2017. Mapping genome-wide transcription-factor binding sites using DAP-seq. Nature Protocols 12, 1659–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer PE, Golicz AA, Scheben A, Batley J, Edwards D.. 2020. Plant pan-genomes are the new reference. Nature Plants 6, 914–920. [DOI] [PubMed] [Google Scholar]
- Beaudoin GAW, Facchini PJ.. 2014. Benzylisoquinoline alkaloid biosynthesis in opium poppy. Planta 240, 19–32. [DOI] [PubMed] [Google Scholar]
- Bernáth J, Németh É.. 2010. Poppy. In: Vollmann J, Rajcan I, eds. Oil crops. New York: Springer New York, 449–468. [Google Scholar]
- Bird DA, Franceschi VR, Facchini PJ.. 2003. A tale of three cell types: alkaloid biosynthesis is localized to sieve elements in opium poppy. The Plant Cell 15, 2626–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boke H, Ozhuner E, Turktas M, Parmaksiz I, Ozcan S, Unver T.. 2015. Regulation of the alkaloid biosynthesis by miRNA in opium poppy. Plant Biotechnology Journal 13, 409–420. [DOI] [PubMed] [Google Scholar]
- Brandt W, Gürke M, Köhler FE, Pabst G, Schellenberg G, Vogtherr M.. 1887. Köhler’s Medizinal-Pflanzen in naturgetreuen Abbildungen mit kurz erläuterndem Texte: Atlas zur Pharmacopoea germanica, austriaca, belgica, danica, helvetica, hungarica, rossica, suecica, Neerlandica, British pharmacopoeia, zum Codex medicamentarius, sowie zur Pharmacopoeia of the United States of America. Gera-Untermhaus: Fr. Eugen Köhler. [Google Scholar]
- Brezinova B, Macak M, Eftimova J.. 2009. The morphological diversity of selected traits of world collection of poppy genotypes (genus Papaver). Journal of Central European Agriculture 10, 183–192. [Google Scholar]
- Buckler ES, Gaut BS, McMullen MD.. 2006. Molecular and functional diversity of maize. Current opinion in plant biology 9, 172–176. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, Greenleaf WJ.. 2015. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Current Protocols in Molecular Biology 109, 21.29.21–21.29.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butnariu M, Quispe C, Herrera-Bravo J, et al. 2022. Papaver plants: current insights on phytochemical and nutritional composition along with biotechnological applications. Oxidative Medicine and Cellular Longevity 2022, 2041769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelblanque L, Balaguer B, Martí C, Rodríguez JJ, Orozco M, Vera P.. 2016. Novel insights into the organization of laticifer cells: a cell comprising a unified whole system. Plant Physiology 172, 1032–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catania T, Li Y, Winzer T, et al. 2022. A functionally conserved STORR gene fusion in Papaver species that diverged 16.8 million years ago. Nature Communications 13, 3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celik I, Camci H, Kose A, Kosar FC, Doganlar S, Frary A.. 2016. Molecular genetic diversity and association mapping of morphine content and agronomic traits in Turkish opium poppy (Papaver somniferum) germplasm. Molecular Breeding 36, 46. [Google Scholar]
- Chen X, Hagel JM, Chang L, et al. 2018. A pathogenesis-related 10 protein catalyzes the final step in thebaine biosynthesis. Nature Chemical Biology 14, 738–743. [DOI] [PubMed] [Google Scholar]
- Clark NM, Nolan TM, Wang P, Song G, Montes C, Valentine CT, Guo H, Sozzani R, Yin Y, Walley JW.. 2021. Integrated omics networks reveal the temporal signaling events of brassinosteroid response in Arabidopsis. Nature Communications 12, 5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conneely LJ, Berkowitz O, Lewsey MG.. 2022. Emerging trends in genomic and epigenomic regulation of plant specialised metabolism. Phytochemistry 203, 113427. [DOI] [PubMed] [Google Scholar]
- Dang TTT, Onoyovwi A, Farrow SC, Facchini PJ.. 2012. Biochemical genomics for gene discovery in benzylisoquinoline alkaloid biosynthesis in opium poppy and related species. In: Hopwood DA, ed. Methods in enzymology, Vol. 515. Academic Press, 231–266. [DOI] [PubMed] [Google Scholar]
- Dastmalchi M, Chang L, Chen R, Yu L, Chen X, Hagel J, Facchini PJ.. 2019a. Purine permease-type benzylisoquinoline alkaloid transporters in opium poppy. Plant Physiology 181, 916–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastmalchi M, Chen X, Hagel JM, Chang L, Chen R, Ramasamy S, Yeaman S, Facchini PJ.. 2019b. Neopinone isomerase is involved in codeine and morphine biosynthesis in opium poppy. Nature Chemical Biology 15, 384–390. [DOI] [PubMed] [Google Scholar]
- Dastmalchi M, Park MR, Morris JS, Facchini P.. 2018. Family portraits: the enzymes behind benzylisoquinoline alkaloid diversity. Phytochemistry Reviews 17, 249–277. [Google Scholar]
- Depuydt T, De Rybel B, Vandepoele K.. 2023. Charting plant gene functions in the multi-omics and single-cell era. Trends in Plant Science 28, 283–296. [DOI] [PubMed] [Google Scholar]
- Dittbrenner A, Mock H-P, Börner A, Lohwasser U.. 2012. Variability of alkaloid content in Papaver somniferum L. Journal of Applied Botany and Food Quality 82, 103–107. [Google Scholar]
- Espinasse A, Dosba F.. 1982. Cytological analysis of hybrids between Papaver somniferum L. and Papaver bracteatum Lindl.; phylogenetic relationships between the two species. Agronomie 2, 281–286. [Google Scholar]
- Evans W. 2009. Trease and Evans’ pharmacognosy. Edinburgh, London, New York, Philadelphia, St Louis, Sydney, Toronto: Elsevier Health Sciences. [Google Scholar]
- Facchini PJ, De Luca V.. 1994. Differential and tissue-specific expression of a gene family for tyrosine/dopa decarboxylase in opium poppy. The Journal of Biological Chemistry 269, 26684–26690. [PubMed] [Google Scholar]
- Facchini PJ, De Luca V.. 2008. Opium poppy and Madagascar periwinkle: model non-model systems to investigate alkaloid biosynthesis in plants. The Plant Journal 54, 763–784. [DOI] [PubMed] [Google Scholar]
- Facchini PJ, Park S-U.. 2003. Developmental and inducible accumulation of gene transcripts involved in alkaloid biosynthesis in opium poppy. Phytochemistry 64, 177–186. [DOI] [PubMed] [Google Scholar]
- Farmer A, Thibivilliers S, Ryu KH, Schiefelbein J, Libault M.. 2021. Single-nucleus RNA and ATAC sequencing reveals the impact of chromatin accessibility on gene expression in Arabidopsis roots at the single-cell level. Molecular Plant 14, 372–383. [DOI] [PubMed] [Google Scholar]
- Faulkner C. 2018. Plasmodesmata and the symplast. Current Biology 28, R1374–R1378. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N.. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature Reviews Genetics 9, 102–114. [DOI] [PubMed] [Google Scholar]
- Fukuda A, Fujimaki S, Mori T, Suzui N, Ishiyama K, Hayakawa T, Yamaya T, Fujiwara T, Yoneyama T, Hayashi H.. 2005. Differential distribution of proteins expressed in companion cells in the sieve element-companion cell complex of rice plants. Plant and Cell Physiology 46, 1779–1786. [DOI] [PubMed] [Google Scholar]
- Gach K, Wyrębska A, Fichna J, Janecka A.. 2011. The role of morphine in regulation of cancer cell growth. Naunyn-Schmiedeberg’s Archives of Pharmacology 384, 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Gonda I, Sun H, et al. 2019. The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nature Genetics 51, 1044–1051. [DOI] [PubMed] [Google Scholar]
- Garrison E, Sirén J, Novak AM, et al. 2018. Variation graph toolkit improves read mapping by representing genetic variation in the reference. Nature Biotechnology 36, 875–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomello S. 2021. A new era for plant science: spatial single-cell transcriptomics. Current Opinion in Plant Biology 60, 102041. [DOI] [PubMed] [Google Scholar]
- Golicz AA, Bayer PE, Barker GC, et al. 2016. The pangenome of an agronomically important crop plant Brassica oleracea. Nature Communications 7, 13390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozmen Şanli B, Uludamar E, Özcanli M.. 2019. Evaluation of energetic-exergetic and sustainability parameters of biodiesel fuels produced from palm oil and opium poppy oil as alternative fuels in diesel engines. Fuel 258, 116116. [Google Scholar]
- Guo L, Winzer T, Yang X, et al. 2018. The opium poppy genome and morphinan production. Science 362, 343–347. [DOI] [PubMed] [Google Scholar]
- Guo S, Zhang C, Le A.. 2021. The limitless applications of single-cell metabolomics. Current Opinion in Biotechnology 71, 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagel JM, Facchini PJ.. 2010. Dioxygenases catalyze the O-demethylation steps of morphine biosynthesis in opium poppy. Nature Chemical Biology 6, 273–275. [DOI] [PubMed] [Google Scholar]
- Hagel JM, Facchini PJ.. 2013. Benzylisoquinoline alkaloid metabolism: a century of discovery and a brave new world. Plant and Cell Physiology 54, 647–672. [DOI] [PubMed] [Google Scholar]
- Hileman LC, Drea S, de Martino G, Litt A, Irish VF.. 2005. Virus-induced gene silencing is an effective tool for assaying gene function in the basal eudicot species Papaver somniferum (opium poppy). The Plant Journal 44, 334–341. [DOI] [PubMed] [Google Scholar]
- Ho SS, Urban AE, Mills RE.. 2020. Structural variation in the sequencing era. Nature Reviews Genetics 21, 171–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong UVT, Tamiru-Oli M, Hurgobin B, Okey CR, Abreu AR, Lewsey MG.. 2022. Insights into opium poppy (Papaver spp.) genetic diversity from genotyping-by-sequencing analysis. Scientific Reports 12, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain R, Quispe C, Saikat ASM, Jain D, Habib A, Janmeda P, Islam MT, Radha, Daştan SD, Kumar M, et al. 2022. Biosynthesis of secondary metabolites based on the regulation of MicroRNAs. Biomed Research International 2022, 9349897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrishi NJ. 1960. Cytogenetical studies on Papaver somniferum L. and Papaver setigerum DC. and their hybrid. Genetica 31, 1–130. [DOI] [PubMed] [Google Scholar]
- Hu T, Allam M, Cai S, Henderson W, Yueh B, Garipcan A, Ievlev AV, Afkarian M, Beyaz S, Coskun AF.. 2023. Single-cell spatial metabolomics with cell-type specific protein profiling for tissue systems biology. Nature Communications 14, 8260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurgobin B, Edwards D.. 2017. SNP discovery using a pangenome: has the single reference approach become obsolete? Biology 6, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurgobin B, Golicz AA, Bayer PE, et al. 2018. Homoeologous exchange is a major cause of gene presence/absence variation in the amphidiploid Brassica napus. Plant Biotechnology Journal 16, 1265–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurgobin B, Lewsey MG.. 2022. Applications of cell- and tissue-specific ‘omics to improve plant productivity. Emerging Topics in Life Sciences 6, 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Pál C, Lercher MJ.. 2004. The evolutionary dynamics of eukaryotic gene order. Nature Reviews Genetics 5, 299–310. [DOI] [PubMed] [Google Scholar]
- INCB. 2022. Narcotic drugs: estimated world requirements for 2023; statistics for 2021. Vienna: International Narcotics Control Board. https://www.incb.org/incb/en/narcotic-drugs/Technical_Reports/2022/narcotic-drugs-technical-report-2022.html
- Itkin M, Heinig U, Tzfadia O, et al. 2013. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341, 175–179. [DOI] [PubMed] [Google Scholar]
- Jeena GS, Singh N, Shikha, Shukla RK.. 2022. An insight into microRNA biogenesis and its regulatory role in plant secondary metabolism. Plant Cell Reports 41, 1651–1671. [DOI] [PubMed] [Google Scholar]
- Jesus A, Bonhomme V, Evin A, Ivorra S, Soteras R, Salavert A, Antolín F, Bouby L.. 2021. A morphometric approach to track opium poppy domestication. Scientific Reports 11, 9778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Xu Y, Wang B, Guo L, Guo M, Che X, Ye K.. 2023. The tissue-specific chromatin accessibility landscape of Papaver somniferum. Frontiers in Genetics 14, 1136736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AR, Moghe GD, Frank MH.. 2021. Growing a glue factory: open questions in laticifer development. Current Opinion in Plant Biology 64, 102096. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B.. 2006. MicroRNAs and their regulatory roles in plants. Annual Review of Plant Biology 57, 19–53. [DOI] [PubMed] [Google Scholar]
- Kadereit JW. 1987. Experimental evidence on the affinities of Papaver somniferum (Papaveraceae). Plant Systematics and Evolution 156, 189–195. [Google Scholar]
- Kadhim AA, Abbas NR, Kadhum HH, Albukhaty S, Jabir MS, Naji AM, Hamzah SS, Mohammed MKA, Al-Karagoly H.. 2023. Investigating the effects of biogenic zinc oxide nanoparticles produced using Papaver somniferum extract on oxidative stress, cytotoxicity, and the induction of apoptosis in the THP-1 cell line. Biological Trace Element Research 201, 4697–4709. [DOI] [PubMed] [Google Scholar]
- Kakeshpour T, Nayebi S, Rashidi Monfared S, Moieni A, Karimzadeh G.. 2015. Identification and expression analyses of MYB and WRKY transcription factor genes in Papaver somniferum L. Physiology and Molecular Biology of Plants 21, 465–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor L. 1995. Opium poppy: botany, chemistry, and pharmacology. Binghampton, NY: Food Products Press/Howarth Press. [Google Scholar]
- Karlebach G, Shamir R.. 2008. Modelling and analysis of gene regulatory networks. Nature Reviews Molecular Cell Biology 9, 770–780. [DOI] [PubMed] [Google Scholar]
- Kaul B, Tandon V, Choudhary D.. 1979. Cytogenetic studies in Papaver somniferum L. Proceedings of the Indian Academy of Sciences-Section B. Part 2, Plant Sciences 88, 321–325. [Google Scholar]
- Kawano N, Kiuchi F, Kawahara N, Yoshimatsu K.. 2012. Genetic and phenotypic analyses of a Papaver somniferum T-DNA insertional mutant with altered alkaloid composition. Pharmaceuticals 5, 133–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-Y, Frommer WB.. 2023. Companion cell and sieve elements. Encyclopedia of Life Sciences 3, doi: 10.1002/9780470015902.a0029552. [Google Scholar]
- Kryuchkova-Mostacci N, Robinson-Rechavi M.. 2017. A benchmark of gene expression tissue-specificity metrics. Briefings in Bioinformatics 18, 205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustatscher G, Grabowski P, Rappsilber J.. 2017. Pervasive coexpression of spatially proximal genes is buffered at the protein level. Molecular Systems Biology 13, 937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labanca F, Ovesnà J, Milella L.. 2018. Papaver somniferum L. taxonomy, uses and new insight in poppy alkaloid pathways. Phytochemistry Reviews 17, 853–871. [Google Scholar]
- Lančaričová A, Havrlentová M, Muchová D, Bednárová A.. 2016. Oil content and fatty acids composition of poppy seeds cultivated in two localities of Slovakia. Agriculture (Polnohospodárstvo) 62, 19–27. [Google Scholar]
- Lane AK, Augustin MM, Ayyampalayam S, Plant A, Gleissberg S, Di Stilio VS, Depamphilis CW, Wong GK-S, Kutchan TM, Leebens-Mack JH.. 2018. Phylogenomic analysis of Ranunculales resolves branching events across the order. Botanical Journal of the Linnean Society 187, 157–166. [Google Scholar]
- Langfelder P, Horvath S.. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E-J, Facchini P.. 2010. Norcoclaurine synthase is a member of the pathogenesis-related 10/Bet v1 protein family. The Plant Cell 22, 3489–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E-J, Hagel J, Facchini P.. 2013. Role of the phloem in the biochemistry and ecophysiology of benzylisoquinoline alkaloid metabolism. Frontiers in Plant Science 4, 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, He Q, Wang J, et al. 2023. Super-pangenome analyses highlight genomic diversity and structural variation across wild and cultivated tomato species. Nature Genetics 55, 852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Ramasamy S, Singh P, et al. 2020. Gene clustering and copy number variation in alkaloid metabolic pathways of opium poppy. Nature Communications 11, 1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Smolke CD.. 2016. Engineering biosynthesis of the anticancer alkaloid noscapine in yeast. Nature Communications 7, 12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Winzer T, He Z, Graham IA.. 2020. Over 100 million years of enzyme evolution underpinning the production of morphine in the Papaveraceae family of flowering plants. Plant Communications 1, 100029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscombe DK, Facchini PJ.. 2008. Evolutionary and cellular webs in benzylisoquinoline alkaloid biosynthesis. Current Opinion in Biotechnology 19, 173–180. [DOI] [PubMed] [Google Scholar]
- Liscombe DK, MacLeod BP, Loukanina N, Nandi OI, Facchini PJ.. 2005. Evidence for the monophyletic evolution of benzylisoquinoline alkaloid biosynthesis in angiosperms. Phytochemistry 66, 1374–1393. [DOI] [PubMed] [Google Scholar]
- Liu Y, Du H, Li P, et al. 2020. Pan-genome of wild and cultivated soybeans. Cell 182, 162–176.e13. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang B, Shu S, et al. 2021. Analysis of the Coptis chinensis genome reveals the diversification of protoberberine-type alkaloids. Nature Communications 12, 3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Cheema J, Vigouroux M, Hill L, Reed J, Paajanen P, Yant L, Osbourn A.. 2020. Formation and diversification of a paradigm biosynthetic gene cluster in plants. Nature Communications 11, 5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo SK, Guo MG, Ji AL, Khavari PA.. 2021. Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nature Reviews Genetics 22, 627–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Hofmeister BT, Vollmers C, DuBois RM, Schmitz RJ.. 2016. Combining ATAC-seq with nuclei sorting for discovery of cis-regulatory regions in plant genomes. Nucleic Acids Research 45, e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik C, Mary T, Grover I.. 1979. Cytogenetic studies in Papaver. V. Cytogenetic studies on P. somniferum × P. setigerum hybrids and amphiploids. Cytologia 44, 59–69. [Google Scholar]
- Millgate AG, Pogson BJ, Wilson IW, Kutchan TM, Zenk MH, Gerlach WL, Fist AJ, Larkin PJ.. 2004. Morphine-pathway block in top1 poppies. Nature 431, 413–414. [DOI] [PubMed] [Google Scholar]
- Mishra S, Triptahi V, Singh S, Phukan UJ, Gupta M, Shanker K, Shukla RK.. 2013. Wound induced tanscriptional regulation of benzylisoquinoline pathway and characterization of wound inducible PsWRKY transcription factor from Papaver somniferum. PLoS One 8, e52784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenegro JD, Golicz AA, Bayer PE, et al. 2017. The pangenome of hexaploid bread wheat. The Plant Journal 90, 1007–1013. [DOI] [PubMed] [Google Scholar]
- Nencini P. 2022. Facts and factoids in the early history of the opium poppy. The Social History of Alcohol and Drugs 36, 45–71. [Google Scholar]
- Nobori T, Oliva M, Lister R, Ecker JR.. 2023. Multiplexed single-cell 3D spatial gene expression analysis in plant tissue using PHYTOMap. Nature Plants 9, 1026–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norn S, Kruse PR, Kruse E.. 2005. History of opium poppy and morphine. Dansk Medicinhistorisk Arbog 33, 171–184. [PubMed] [Google Scholar]
- Nützmann H-W, Scazzocchio C, Osbourn A.. 2018. Metabolic gene clusters in eukaryotes. Annual Review of Genetics 52, 159–183. [DOI] [PubMed] [Google Scholar]
- Ojala A, Rousi A. 1986. Interspecific hybridization in Papaver: 1. F1 hybrids of P. somniferum with perennial species of sect. Oxytona. Annales Botanici Fennici 23, 289–303. [Google Scholar]
- Onoyovwe A, Hagel JM, Chen X, Khan MF, Schriemer DC, Facchini PJ.. 2013. Morphine biosynthesis in opium poppy involves two cell types: sieve elements and laticifers. The Plant Cell 25, 4110–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozber N, Carr SC, Morris JS, Liang S, Watkins JL, Caldo KM, Hagel JM, Ng KKS, Facchini PJ.. 2022. Alkaloid binding to opium poppy major latex proteins triggers structural modification and functional aggregation. Nature Communications 13, 6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozber N, Facchini PJ.. 2022. Phloem-specific localization of benzylisoquinoline alkaloid metabolism in opium poppy. Journal of Plant Physiology 271, 153641. [DOI] [PubMed] [Google Scholar]
- Pathak S, Lakhwani D, Gupta P, Mishra BK, Shukla S, Asif MH, Trivedi PK.. 2013. Comparative transcriptome analysis using high papaverine mutant of Papaver somniferum reveals pathway and uncharacterized steps of papaverine biosynthesis. PLoS One 8, e65622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei L, Wang B, Ye J, et al. 2021. Genome and transcriptome of Papaver somniferum Chinese landrace CHM indicates that massive genome expansion contributes to high benzylisoquinoline alkaloid biosynthesis. Horticulture Research 8, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirats-Llobet M, Yi C, Liew Lim C, Berkowitz O, Narsai R, Lewsey Mathew G, Whelan J.. 2023. Spatially resolved transcriptomic analysis of the germinating barley grain. Nucleic Acids Research 51, 7798–7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polturak G, Osbourn A.. 2021. The emerging role of biosynthetic gene clusters in plant defense and plant interactions. PLoS Pathogens 17, e1009698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo A, Dillon N.. 2015. Three-dimensional genome architecture: players and mechanisms. Nature Reviews Molecular Cell Biology 16, 245–257. [DOI] [PubMed] [Google Scholar]
- Pyysalo H, Widen C-J, Salemink CA, Lewing E, Rousi A, Ojala A.. 1988. Interspecific hybridization in Papaver. 2. Alkaloid contents of P. somniferum and species of the section Oxytona and their interspecific hybrids. Annales Botanici Fennici 25, 1–10. [Google Scholar]
- Ramos MV, Demarco D, da Costa Souza IC, de Freitas CDT.. 2019. Laticifers, latex, and their role in plant defense. Trends in Plant Science 24, 553–567. [DOI] [PubMed] [Google Scholar]
- Salavert A. 2017. Agricultural dispersals in Mediterranean and temperate Europe. In: Shugart H, ed. The Oxford Research Encyclopedia of Agriculture and the Environment. Oxford: Oxford University Press. [Google Scholar]
- Salavert A, Martin L, Antolín F, Zazzo A.. 2018. The opium poppy in Europe: exploring its origin and dispersal during the Neolithic. Antiquity 92, e1. [Google Scholar]
- Salavert A, Zazzo A, Martin L, et al. 2020. Direct dating reveals the early history of opium poppy in western Europe. Scientific Reports 10, 20263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanani N, Alcantara J, Bourgault R, Zulak KG, Facchini PJ.. 2006. The role of phloem sieve elements and laticifers in the biosynthesis and accumulation of alkaloids in opium poppy. The Plant Journal 47, 547–563. [DOI] [PubMed] [Google Scholar]
- Samanani N, Facchini PJ.. 2001. Isolation and partial characterization of norcoclaurine synthase, the first committed step in benzylisoquinoline alkaloid biosynthesis, from opium poppy. Planta 213, 898–906. [DOI] [PubMed] [Google Scholar]
- Shaw R, Tian X, Xu J.. 2021. Single-cell transcriptome analysis in plants: advances and challenges. Molecular Plant 14, 115–126. [DOI] [PubMed] [Google Scholar]
- Shitan N, Bazin I, Dan K, Obata K, Kigawa K, Ueda K, Sato F, Forestier C, Yazaki K.. 2003. Involvement of CjMDR1, a plant multidrug-resistance-type ATP-binding cassette protein, in alkaloid transport in Coptis japonica. Proceedings of the National Academy of Sciences, USA 100, 751–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Menéndez-Perdomo IM, Facchini PJ.. 2019. Benzylisoquinoline alkaloid biosynthesis in opium poppy: an update. Phytochemistry Reviews 18, 1457–1482. [Google Scholar]
- Singh M, Chaturvedi C, Shasany A, Shukla A.. 2014. Impact of promising genotypes of Papaver somniferum L. developed for beneficial uses. Acta Horticulturae 1036, 29–41. [Google Scholar]
- Singh S, Husain D, Singh V, et al. 2023. Genetic variability for qualitative and quantitative characters and study of character association for their exploitation in genetic improvement of opium poppy (Papaver somniferum L.). Industrial Crops and Products 200, 116863. [Google Scholar]
- Soltis PS, Soltis DE.. 2021. Plant genomes: markers of evolutionary history and drivers of evolutionary change. Plants, People, Planet 3, 74–82. [Google Scholar]
- Srivastava S. 1989. Cytogenetic investigations in hyoscyamus L and papaver L. PhD thesis, Kanpur University. [Google Scholar]
- Stefano GB, Pilonis N, Ptacek R, Kream RM.. 2017. Reciprocal evolution of opiate science from medical and cultural perspectives. Medical Science Monitor 23, 2890–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura T. 1940. Chromosome studies on Papaveraceae with special reference to the phylogeny. Cytologia 10, 558–576. [Google Scholar]
- Sun M, Xu S, Mei Y, Li J, Gu Y, Zhang W, Wang J.. 2022. MicroRNAs in medicinal plants. International Journal of Molecular Sciences 23, 10477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabatabaei majd M, Ramezanzadeh M, Bahlakeh G, Ramezanzadeh B.. 2020. Probing molecular adsorption/interactions and anti-corrosion performance of poppy extract in acidic environments. Journal of Molecular Liquids 304, 112750. [Google Scholar]
- Tahara M. 1915. The chromosomes of Papaver. Shokubutsugaku Zasshi 29, 254–257. [Google Scholar]
- Takanashi K, Yamada Y, Sasaki T, Yamamoto Y, Sato F, Yazaki K.. 2017. A multidrug and toxic compound extrusion transporter mediates berberine accumulation into vacuoles in Coptis japonica. Phytochemistry 138, 76–82. [DOI] [PubMed] [Google Scholar]
- Tamiru-Oli M, Premaratna SD, Gendall AR, Lewsey MG.. 2018. Biochemistry, genetics, and genomics of opium poppy (Papaver somniferum) for crop improvement. Annual Plant Reviews Online 2, 1177–1219. [Google Scholar]
- Tan TG, Türktaş M, Güçlü G.. 2023. In silico comparative transcriptome analysis of Papaver somniferum cultivars. Acta Scientiarum Polonorum Hortorum Cultus 22, 69–78. [Google Scholar]
- Tenorio Berrío R, Verstaen K, Vandamme N, et al. 2021. Single-cell transcriptomics sheds light on the identity and metabolism of developing leaf cells. Plant Physiology 188, 898–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tétényi P. 1997. Opium poppy (Papaver somniferum): botany and horticulture. Horticultural Reviews 19, 373–408. [Google Scholar]
- UNODC. 1961. Single convention on narcotic drugs, 1961. Vienna: United Nations Office on Drugs and Crime. https://www.unodc.org/unodc/en/treaties/single-convention.html [Google Scholar]
- van Dam S, Võsa U, van der Graaf A, Franke L, de Magalhães JP.. 2017. Gene co-expression analysis for functional classification and gene–disease predictions. Briefings in Bioinformatics 19, 575–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Broeck L, Gordon M, Inzé D, Williams C, Sozzani R.. 2020. Gene regulatory network inference: connecting plant biology and mathematical modeling. Frontiers in Genetics 11, 510276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma N, Jena SN, Shukla S, Yadav K.. 2016. Genetic diversity, population structure and marker trait associations for alkaloid content and licit opium yield in India-wide collection of poppy (Papaver somniferum L.). Plant Gene 7, 26–41. [Google Scholar]
- Walkowiak S, Gao L, Monat C, et al. 2020. Multiple wheat genomes reveal global variation in modern breeding. Nature 588, 277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yang W, Zhang S, et al. 2023. A pangenome analysis pipeline provides insights into functional gene identification in rice. Genome Biology 24, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Luo Y, Huang J, et al. 2020. The genome evolution and domestication of tropical fruit mango. Genome Biology 21, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins JL, Facchini PJ.. 2022. Compartmentalization at the interface of primary and alkaloid metabolism. Current Opinion in Plant Biology 66, 102186. [DOI] [PubMed] [Google Scholar]
- Weid M, Ziegler J, Kutchan TM.. 2004. The roles of latex and the vascular bundle in morphine biosynthesis in the opium poppy, Papaver somniferum. Proceedings of the National Academy of Sciences, USA 101, 13957–13962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer T, Gazda V, He Z, et al. 2012. A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science 336, 1704–1708. [DOI] [PubMed] [Google Scholar]
- Winzer T, Kern M, King AJ, et al. 2015. Morphinan biosynthesis in opium poppy requires a P450-oxidoreductase fusion protein. Science 349, 309–312. [DOI] [PubMed] [Google Scholar]
- Xu T, Yang X, Jia Y, et al. 2022. A global survey of the transcriptome of the opium poppy (Papaver somniferum) based on single-molecule long-read isoform sequencing. The Plant Journal 110, 607–620. [DOI] [PubMed] [Google Scholar]
- Yang X, Gao S, Guo L, et al. 2021. Three chromosome-scale Papaver genomes reveal punctuated patchwork evolution of the morphinan and noscapine biosynthesis pathway. Nature Communications 12, 6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui K. 1921. On the behavior of chromosomes in the meiotic phase of some artificially raised Papaver hybrids. Shokubutsugaku Zasshi 35, en154–en167. [Google Scholar]
- Yasui K. 1937. Cytogenetic studies in artificially raised interspecific hybrids of Papaver. Cytologia 8, 331–342. [Google Scholar]
- Yucebilgili Kurtoglu K, Unver T.. 2021. Integrated omics analysis of benzylisoquinoline alkaloid (BIA) metabolism in opium poppy (Papaver somniferum L.). In: Tombuloglu H, Unver T, Tombuloglu G, Hakeem KR, eds. Oil crop genomics. Cham: Springer International Publishing, 291–315. [Google Scholar]
- Zanini SF, Bayer PE, Wells R, Snowdon RJ, Batley J, Varshney RK, Nguyen HT, Edwards D, Golicz AA.. 2022. Pangenomics in crop improvement—From coding structural variations to finding regulatory variants with pangenome graphs. The Plant Genome 15, e20177. [DOI] [PubMed] [Google Scholar]
- Zhang R-G, Lu C, Li G-Y, et al. 2023. Subgenome-aware analyses suggest a reticulate allopolyploidization origin in three Papaver genomes. Nature Communications 14, 2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Wu Y, Huang X, Wu W, Lyu L, Li W.. 2022. Research progress about microRNAs involved in plant secondary metabolism. International Journal of Biological Macromolecules 216, 820–829. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhang Z, Li M, Luo J, Chen F, Gong Y, Li Y, Wei Y, Su Y, Kong L.. 2019. Transcriptomic profiles of 33 opium poppy samples in different tissues, growth phases, and cultivars. Scientific Data 6, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Liu Z.. 2022. Unlocking plant metabolic diversity: a (pan)-genomic view. Plant Communications 3, 100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhang Z, Bao Z, et al. 2022. Graph pangenome captures missing heritability and empowers tomato breeding. Nature 606, 527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohary D, Weiss E, Hopf M.. 2012. Oil- and fibre-producing crops. Domestication of plants in the old world: the origin and spread of domesticated plants in Southwest Asia, Europe, and the Mediterranean Basin. Oxford: Oxford University Press, 100–113. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data downloaded from public database and used in this manuscript are summarized in Supplementary Table S1, and the relevant analyses are detailed in Supplementary Protocol S1.