Abstract
The limited but recurrent outbreaks of the zoonotic Nipah virus (NiV) infection in humans, its high fatality rate, and the potential virus transmission from human to human make NiV a concerning threat with pandemic potential. There are no licensed vaccines to prevent infection and disease. A recombinant Hendra virus soluble G glycoprotein vaccine (HeV-sG-V) candidate was recently tested in a Phase I clinical trial. Because NiV outbreaks are sporadic, and with a few cases, licensing will likely require an alternate regulatory licensing pathway. Therefore, determining a reliable vaccine correlate of protection (CoP) will be critical. We assessed the immune responses elicited by HeV-sG-V in African Green monkeys and its relationship with protection from a NiV challenge. Data revealed values of specific binding and neutralizing antibody titers that predicted survival and allowed us to establish a mechanistic CoP for NiV Bangladesh and Malaysia strains.
Subject terms: Protein vaccines, Viral infection
Introduction
Nipah disease is caused by the bat-borne zoonotic Nipah virus (NiV) and can affect humans and animals. In humans, NiV infection is usually associated with a severe respiratory and encephalitic syndrome referred to as henipavirus disease that can often lead to death1. NiV is a member of the paramyxovirus family and, together with Hendra virus (HeV), is the prototype member of the genus henipavirus and to date remain the only henipaviruses that can cause henipavirus disease2. NiV was first isolated in 1999 after an outbreak in pigs and humans in Malaysia (1998 and 1999)3 and Singapore (1999)4. Since then, sporadic outbreaks have been reported in neighboring countries, including the Philippines, Bangladesh, and India5–9, with the latest outbreaks reported in 2024 in Bangladesh and India10,11.
NiV is shed in bat urine and saliva, and transmission to humans can occur from infected bats (the virus’s natural reservoir) via consumption of contaminated fruit or date sap12,13 or from infected domestic animals such as pigs and horses3–5. In addition, human-to-human transmission has been reported during some outbreaks in the Philippines, Bangladesh, and India5,6,12,14,15.
NiV infection in humans can be associated with mild to severe respiratory symptoms and encephalitis that often leads to death. The clinical diagnosis of NiV infection is challenging as at the onset of the disease, most patients present general malaise symptoms such as fever, headache, myalgia, and sometimes gastrointestinal disorders before the respiratory or neurological manifestations become apparent, leading to a fatality rate of above 50%16–18.
There are no licensed vaccines to prevent NiV infection in humans19; however, there are several vaccine candidates currently in preclinical and clinical development using different platform technologies, including viral vectors, protein subunits, virus-like particles, mRNA, and plasmid DNA targeting the glycoprotein (G) or the fusion glycoprotein (F) of Nipah or Hendra viruses20. Only three NiV vaccine candidates have reached the stage of clinical trials in the United States21,22: a vectored vaccine using a recombinant Vesicular Stomatitis Virus (rVSV)-based Ebola vaccine vector expressing the full length NiV G glycoprotein23 (NCT05178901); an mRNA vaccine encoding and expressing NiV F and G glycoproteins24 (NCT05398796) and a recombinant subunit NiV vaccine candidate using a soluble Hendra virus G glycoprotein (HeV-sG) (NCT04199169), which was first described and tested 20 years ago25,26 and further developed by Auro Vaccines LLC. The recombinant HeV-sG glycoprotein subunit NiV vaccine (HeV-sG-V) has been extensively evaluated in several animal species, including cats, ferrets, horses, and African Green Monkeys (AGM), where it elicits a potent and virus-neutralizing humoral immune response that is protective against lethal NiV and HeV challenge19,27,28. Anti-NiV and anti-HeV antibody responses correlate after vaccination with the HeV-sG antigen, and immunized animals are protected against both NiV and HeV lethal challenges28. The first-in-human Phase I clinical trial recently concluded, and no safety problems were identified. Furthermore, the results demonstrated that HeV-sG-V elicited a potent immune response characterized by a high titer of specific binding as well as neutralizing antibodies against two NiV strains, NiV Bangladesh (NiVB) and NiV Malaysia (NiVM)29.
The advancement of vaccines against emerging pathogens such as NiV can be hindered due in part to the small size and unpredictability of the intermittent outbreaks of the disease that makes the design of randomized controlled clinical studies such as a Phase III efficacy trial exceedingly difficult or even unfeasible30,31. Therefore, the establishment of a correlate of protection (CoP) in a relevant NiV animal model has the potential to inform and facilitate vaccine development and to define a non-classical licensure pathway against infection and disease caused by NiV32. Here we aimed to determine an immune CoP for NiV infection in non-human primates (NHP) using the HeV-sG-V in a dose escalation study approach.
In this study, we assessed HeV-sG-V-induced anti-NiV antibody responses in NHP and determined if these elicited responses protected vaccinated animals against a NiVB challenge. We used African Green Monkeys (Chlorocebus sabaeus), the most relevant animal species in recapitulating the outcome of NiV infection in humans33,34. The similarity between the disease observed in the AGM and the one observed in humans is critical in providing relevance to any potential immune CoP established in an animal model35.
Results
Survival and clinical signs
There was a dose-dependent vaccine effect as demonstrated by animals immunized with the higher HeV-sG-V doses showing a higher percentage of survival (Fig. 1A). However, there was an exception with one of the animals that received the highest dose (100 mcg) and still succumbed to the NiV infection. Table 1 lists the individual clinical observations for controls and HeV-sG-V-vaccinated AGMs showing clinical signs of infection upon NiVB challenge. Clinical signs in animals that succumbed to infection occurred between the 6th and 8th day post-challenge and included depression, loss of appetite, lethargy, increased abdominal breathing, and labored breathing. One of the AGMs in the 30 mcg group (male, NiVCoP30-2) presented a mild increased respiratory rate on day 8 post-challenge and a transient reduction in food intake between days 8 and 10 post-challenge but completely recovered by day 12 post-challenge. All surviving animals were healthy and free from any visual clinical signs until they were euthanized at the end of the study (Study Day 56).
Fig. 1. Survival curves, body weights, and body temperatures after NiVB challenge.
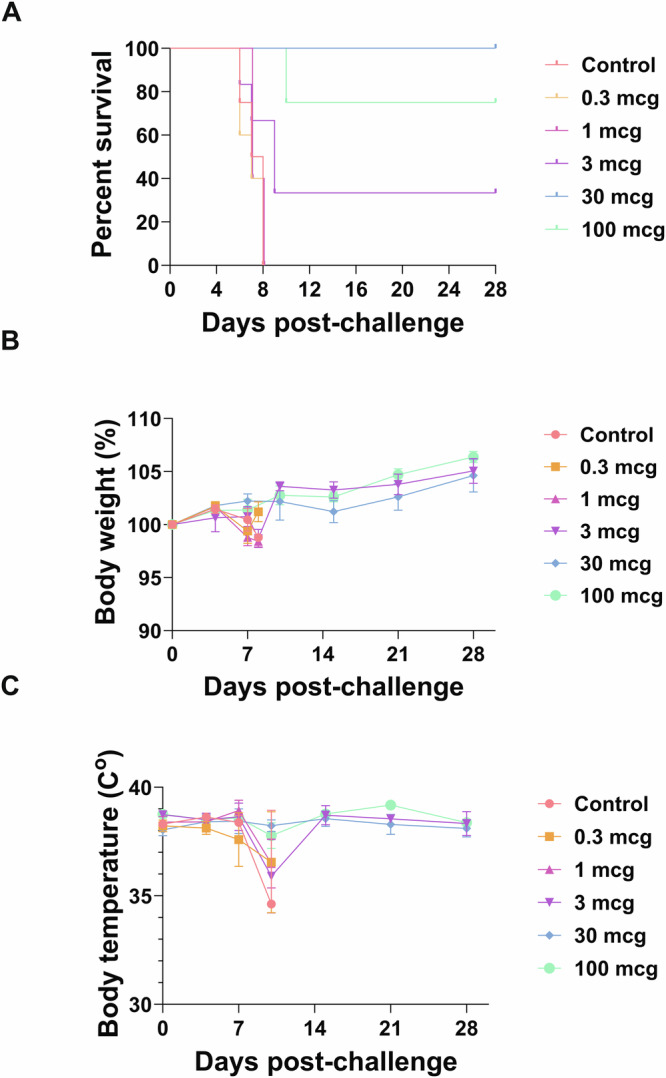
African Green monkeys were vaccinated with different doses of Hendra virus soluble glycoprotein vaccine (HeV-sG-V) or vehicle alone (alum) and challenged with a Nipah virus Bangladesh strain (NiVB) 28 days after vaccination (Study Day 28). The animals were monitored for clinical signs of infection for 28 days (Study Day 56). A Kaplan-Meier survival curves. B Body weight changes. C Body temperature changes after the NiVB challenge. The results combine the data obtained during the two iterations of the study. (*) Indicates a significant difference (**p < 0.01) compared to the control group (alum only). Error bars represent SE.
Table 1.
Individual Clinical Observations and Survival
| Group/Dose | Animal ID | Sex | Survival/Deatha | Clinical Signs | Clinical Pathology |
|---|---|---|---|---|---|
| Control | NiVCoPCx-1 | F | Day 8 | W,D,R,HP,Dy,RQ,NE, | NLK, Thr, ↑Glu,BUN,C,CRP,ALT |
| NiVCoPCx-2 | M | Day 6 | W,D,A,R,HP,Dy,RQ,NE | NLK, Thr, ↑Glu,BUN,C,CRP,ALT | |
| NiVCoPCx-3 | M | Day 8 | D,A,R,HP,Dy,RQ | NLK, Thr, ↑Glu,BUN,C,CRP,ALT | |
| NiVCoPCx-4 | F | Day 7 | D,A,W,R,Dy,RQ | NLK, Thr, ↑Glu,BUN,C,CRP,ALT | |
| 0.3 mcg | NiVCoP03-1 | F | Day 6 | A,W,HP,Dy,RQ | NLK, Thr, ↑Glu,BUN,C,CRP,ALT |
| NiVCoP03-2 | M | Day 8 | D,A,R,HP,Dy,RQ | NLK, Thr, ↑Glu,BUN,C,CRP,ALT | |
| NiVCoP03-3 | F | Day 6 | D,A,R,HP,Dy,RQ | NLK, Thr, ↑Glu,BUN,C,CRP,ALT | |
| NiVCoP03-4 | M | Day 7 | D,A,R,Dy,RQ | NLK, Thr, ↑Glu,BUN,C,CRP,ALT | |
| NiVCoP03-5 | F | Day 8 | D,A,R,Dy,RQ | NLK, Thr, ↑Glu,BUN,C,CRP,ALT | |
| 1 mcg | NiVCoP1-1 | M | Day 7 | D,A,R,HP,Dy,RQ | NLK, Thr, ↑Glu,BUN,C,CRP,ALT |
| NiVCoP1-2 | F | Day 7 | D,A,R,HP,Dy,RQ | NLK, Thr, ↑Glu,BUN,C,CRP,ALT | |
| NiVCoP1-3 | M | Day 8 | D,A,R,Dy,RQ | NLK, Thr, ↑Glu,BUN,C,CRP,ALT | |
| NiVCoP1-4 | F | Day 7 | D,A,W,R,Dy,RQ | NLK, Thr, ↑Glu,BUN,C,CRP,ALT | |
| NiVCoP1-5 | M | Day 8 | D,A,R,HP,Dy,RQ | NLK, Thr, ↑Glu,BUN,C,CRP,ALT | |
| 3 mcg | NiVCoP3-1 | M | Survived | NOCS | None |
| NiVCoP3-2 | M | Survived | NOCS - Except for TRA (D7,8,11) | None | |
| NiVCoP3-3 | M | Day 9 | D,A,R,HP,Dy,RQ | NLK, Thr, ↑Glu,BUN,C,CRP,ALT | |
| NiVCoP3-4 | F | Day 6 | D,W,R,Dy,RQ | NLK, Thr, ↑Glu,BUN,C,CRP,ALT | |
| NiVCoP3-5 | F | Day 7 | D,A,R,HP,Dy,RQ | NLK, Thr, ↑Glu,BUN,C,CRP,ALT | |
| NiVCoP3-6 | F | Day 9 | D,A,R,Dy,RQ,NE | NLK, Thr, ↑Glu,BUN,C,CRP,ALT | |
| 30 mcg | NiVCoP30-1 | M | Survived | NOCS | None |
| NiVCoP30-2 | M | Survived | A,Dy,RQ,TRA (D7 – D10) | NLK (D7), ↑ALT, CRP (D7,D10) | |
| NiVCoP30-3 | F | Survived | NOCS | None | |
| NiVCoP30-4 | F | Survived | NOCS | None | |
| 100 mcg | NiVCoP100-1 | M | Survived | NOCS | None |
| NiVCoP100-2 | M | Day 10 | D,A,W,R,Dy,RQ,NE | NLK, Thr, ↑Glu,BUN,C,CRP,ALT | |
| NiVCoP100-3 | F | Survived | NOCS | None | |
| NiVCoP100-4 | F | Survived | NOCS Except for TRA (D5,8,22) | None |
a Days after NiVB Challenge.
NOCS No Overt Clinical Signs, D Depression, A Anorexia, W Weakness, R Recumbency, HP Hunched Posture, Dy Dyspnea, RQ Respiration Quality, NE Nasal Exudate, TRA Transient reduced food intake, NLK Neutrophilic leukocytosis, Thr Thrombocytopenia, Glu Glucose, BUN Blood urea nitrogen, C Creatinine, CRP C-reactive protein, ALT Alanine transaminase.
Body weight and temperature
No significant changes in body weight (loss of ≥10% of initial body weight) were observed in this study (Fig. 1B). Although some animals presented a loss of appetite, none of the animals lost more than 5% of their starting body weight. No significant increases in body temperature indicating the presence of a fever (increase >1.5 ◦C) were recorded except for one animal from the 30 mcg group (NiVCoP30-2), which showed a transient fever 7 days post-challenge (Fig. 1C). As expected, most of the animals succumbing to infection presented hypothermia before reaching the humane endpoint and being euthanized.
Hematology
No significant differences were observed in any of the analyzed values between the baseline (before vaccination) and Study Day 28 (time of challenge). In all surviving animals, no significant differences were observed compared to the baseline throughout the study (Table 1). Only animal NiVCoP30-2 from the 30 mcg group showed transient neutropenia 7 days post-challenge evaluation (Study Day 35). All the animals that succumbed to infection presented leukocytosis related to an increase in neutrophils with a reduction in the number of platelets.
Blood chemistry
While no significant differences were observed in any of the analyzed blood biochemistry parameters when comparing the baseline (before vaccination) with Study Day 28 (time of challenge), the following changes were detected in animals immediately before succumbing to infection: Slightly elevated levels for glucose, blood urea nitrogen, creatinine, C-reactive protein, and alanine transaminase (Table 1). In all surviving animals, no significant differences in comparison to baseline were observed through the length of the study, except for animal NiVCoP30-2 which showed a slight but transient (7 and 10 days post-challenge) increase in alanine transaminase and C-reactive protein levels in blood (Table 1).
Viremia
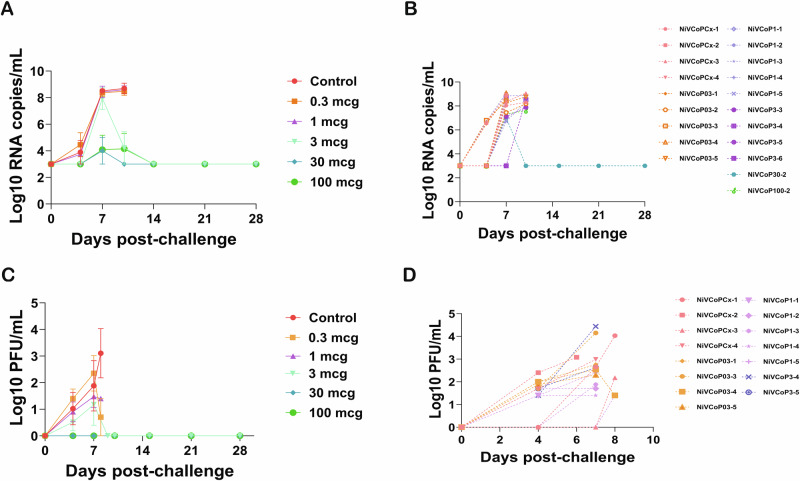
Viral RNA was detected by RT-qPCR in the blood of all the AGMs that succumbed to infection as early as 4 days post-challenge (Fig. 2A, B) and at all subsequent sampling dates until the animals died (= reaching the humane endpoint and being euthanized). Values ranged from 6.51 log10 copies/ml to 9.09 log10 copies/ml. None of the animals that survived the NiVB challenge had detectable viral RNA in their plasma except for NiVCoP30-2 which had detectable viral RNA (7.03 log10 copies/ml) on the day 7 post-challenge blood sample but was viral RNA-free in all the subsequent samples tested (Fig. 2B and Supplementary Table 2).
Fig. 2. Viral RNA and virus particles in plasma after NiVB challenge.
Plasma samples were collected from African Green monkeys vaccinated with Hendra virus soluble glycoprotein vaccine (HeV-sG-V) and challenged 28 days (Study Day 28) after vaccination with a Nipah virus Bangladesh strain (NiVB). Samples were collected on Study Days 28, 32, 35, 38, 43, 49, and 56 (corresponding to 0, 4, 7, 10, 15, 21, and 28 days post-challenge) or when animals reached the humane endpoint. A Viral RNA average levels per group. B Individual animal values at each sampling date. Viral RNA was detected in animals that succumbed to infection (reached the humane endpoint) except for animal NiVCoP30-2 from the 30 mcg group that only had transient viremia but survived the virus challenge. C Replicating virus particles in plasma. The virus was detected as early as 4 days post-challenge in some infected animals. D Individual animal virus titer in plasma. Only animals that succumbed to infection had detectable virus titer (plaque forming units or PFU) as determined by plaque assay. Of notice, not all the animals that succumbed to infection had detectable infectious virus particles in plasma. Error bars represent SE.
Plasma samples were tested for the presence of replicating NiVB by plaque assay (viremia). All AGMs in the control group showed detectable infectious NiVB with two of them having titers as early as 4 days post-challenge (Fig. 2C, D), and titers ranged from 1.7 log10 to 4.03 log10 PFU/ml. Of the vaccinated animals that succumbed to infection, 4 out of 5 in the 0.3 mcg, 5 out of 5 in the 1 mcg, and 2 out of 4 in the 3 mcg groups had detectable NiVB titers ranging from 1.4 to 4.4 log10 PFU/ml (Fig. 2D). Most of these animals had virus titers 4 days post-challenge and throughout the rest of the sampling time points. Four animals succumbing to infection had no detectable virus titers in plasma: NiVCoP03-2, NiVCoP3-3, NiVCoP3-6, and NiVCoP100-2. None of the animals that survived the challenge had detectable NiVB titers in plasma samples at any time points tested.
Total anti-NiV IgG antibody titers
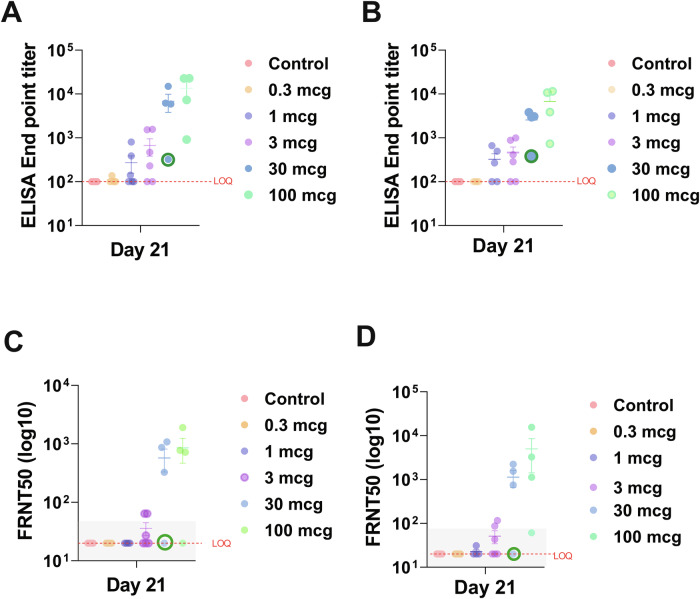
Total IgG titers were evaluated in samples collected on Study Days 0 (vaccination day), 14, and 21 (one week before challenge), while the animals were housed at ABSL-2. The lower limit of quantification for each ELISA was 100. Anti-NiVB and anti-NiVM antibody titers were detected in most vaccinated animals with a trend toward dose dependency when the group results were averaged (Fig. 3A, B; Supplementary Fig. 1A). No quantifiable anti-NiV-sG antibody titers against any of the two NiV strains tested (NiVB and NiVM) were detected in any of the control animals or in some of the animals that received lower doses of HeV-sG-V. Figure 3 shows the individual ELISA titers per group for NiVB (Fig. 3A) and NiVM (Fig. 3B) on Study Day 21. All the surviving animals had an antibody endpoint titer above 1100 against NiVB-sG (Fig. 4A) and above 800 against NiVM-G. Animal NiVCoP30-2 (30 mcg group) survived the NiV challenge with transient mild clinical signs of infection and had an ELISA endpoint titer of 322 against NiVB-sG and 381 against NiVM-sG on Study Day 21.
Fig. 3. Anti-NiV antibody response at Study Day 21 after HeV-sG-V vaccination.
Serum samples were collected from African Green monkeys vaccinated with different doses of Hendra virus soluble glycoprotein vaccine (HeV-sG-V) or vehicle alone (alum). Samples were collected on Study Days 0 (vaccination day), 14, and 21. Values of total IgG binding antibody titers against Nipah virus (NiV) glycoprotein Bangladesh strain (A) and Malaysia strain (B) from samples collected on Study Day 21 are shown. A trend for dose-dependance response is observed among the groups. All the animals from the control group had no detectable antibody titers (limit of quantification of LOQ). Anti-NiV neutralizing antibodies were measured on Study Day 21 against the Bangladesh strain (C) and the Malaysia strain (D). The animals with an FRNT-50 within the gray square succumbed to the virus challenge. Error bars represent SE. (Green circle): Animal NiVCoP30-2 from the 30 mcg group survived the NiVB challenge with transient, mild clinical signs of infection.
Fig. 4. Anti-NiV antibody response after HeV-sG-V vaccination presented per animal independent of the vaccine dose received.
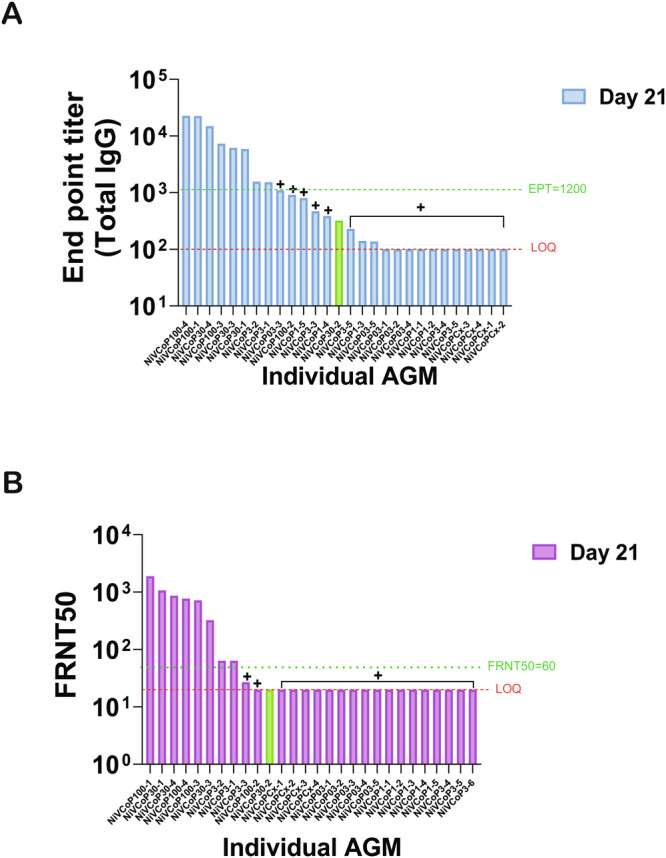
To better understand the role of the anti-NiV binding (A) and neutralizing (B) antibody titers in survival, the results from the immunogenicity studies are presented here with results from individual animals presented from highest to the lowest for samples obtained at Study Day 21 (21 days after vaccination). A Results indicate that animals with anti-Nipah virus Bangladesh strain (NiVB) binding antibody titers below 103 all succumbed to the NiVB challenge independently of the vaccine dose received. B All the animals with anti-NiVB neutralizing antibody titers response at Study Day 21 below an FRNT50 of 60, succumbed to infection. For both types of antibody responses (A, B), animal NiVCoP30-2 that received 30 mcg of vaccine (HeV-sG-V) had an antibody response below these thresholds and survived NiVB challenge with transient, mild clinical signs of infection (green bar).
NiV-neutralizing antibody titers
NiV neutralization titers were tested in samples collected on Study Day 0 (vaccination day), 14, and 21 (one week before the challenge) using psVSV-NiV-B and psVSV-NiV-M reporter viruses, and results are expressed as FRNT50 with the lower limit of quantification for each test of 20. Similar to the total IgG results by ELISA, sera from all animals in the control group did not show quantifiable (< 20) FRNT50 antibody titers against any of the two reporter viruses tested (Fig. 3C, D). All the surviving animals had an FRNT50 titer above 60 against psVSV-NiV-B (Fig. 4B) and above 80 against psVSV-NiV-M. Serum from animal NiVCoP30-2 that survived the NiVB challenge did not show a quantifiable FRNT50 antibody titer at any time point tested against any of the neutralization reporter viruses (Fig. 3C, D).
Determination of a correlate of protection
The first step in the analyses was to assess how well the assay results from the Study Day 21 bleeds (7 days before the NiVB challenge) fulfill the requirements of the Prentice Criteria. This assessment involved four analyses for each one of the measured Study Day 21 immune responses (NiVB and NiVM ELISA; NiVB and NiVM FRNT50). Since the first analysis seeks to correlate dose with the outcome, it does not involve the Study Day 21 immune measures and, therefore, is the same analysis for all four candidate surrogates. The results of the significant logistic regression of mortality on dose are summarized in Table 2. The results demonstrate a significant effect of the Day 21 samples in relation to survival, fulfilling the requirements of the Prentice Criteria.
Table 2.
Significant Logistic regression of mortality on dose candidate. CoP: [(Est/SE)/(P-Value)]
| Criteria | NiVB ELISA Titer on Study Day 21 | NiVM ELISA Titer on Study Day 21 | NiVB FRNT50 | NiVM FRNT50 |
|---|---|---|---|---|
| Treatment (Dose) affecting endpoint | (2.16)/(0.032) | (2.16)/(0.032) | (2.16)/(0.032) | (2.16)/(0.032) |
| Treatment (Dose) affecting CoP | (5.12)/( < 0.0001) | (5.15)/( < 0.0001) | (6.21)/( < 0.0001) | (7.48)/(0.0001) |
| CoP affecting endpoint | (2.17)/(0.027) | (2.00)/(0.046) | (2.23)/(0.026) | (2.37)/(0.018) |
| Treatment offers no more information than CoPa | 0.028/0.85 | 0.10/0.37 | 0.06/0.14 | 0.06/0.48 |
a.- Summary P-Values for Correlate of Protection (CoP) and Dose (CoP/Dose).
Study Day 21 immune measures satisfy the first three of the Prentice Criteria to various degrees. The Study Day 21 ELISA NiVM titer is the weakest, but valid predictor of survival, while the other three immune measures are strong survival predictors. Only the Study Day 21 NiVB ELISA titer results satisfy the requirement of eliminating the need for information from the treatment (the fourth Prentice Criteria). The results from the analyses determined the value of the surrogate needed to attain various levels of protection and the obtained values are presented in Table 3.
Table 3.
Correlate of Protection Values
| Probability of survival | ELISA Day 21 (Lower-upper 95% limit) |
VN Day 21 (FRNT50) (Lower-upper 95% limit) |
||
|---|---|---|---|---|
| NiVB | NiVM | NiVB | NiVM | |
| 50 |
844 (285–4381) |
837 (450–1558) |
30.4 (15.6–65631) |
65 (25–8825) |
| 80 |
1937 (852–121269) |
1460 (792-THC) |
49.8 (23.7–3.50e + 7) |
143 (53–2.7e + 6) |
| 90 |
3149 (1251–1.1e + 6) |
2021 (1013-THC) |
67 (28.8–1.4e + 9) |
229 (74–8.62e + 7) |
Discussion
The regular occurrence of NiV infection outbreaks in humans, the high mortality rate, and the possibility for NiV to be transmitted from person to person make NiV a concerning threat with pandemic potential1,36. Currently, there are no licensed vaccines to prevent infection in humans exposed to the virus; however, there are several promising vaccine candidates under preclinical and clinical development19,21.
The recombinant HeV-sG glycoprotein subunit NiV vaccine (HeV-sG-V) that was used in the AGM immune CoP studies described here was recently tested in a first-in-human Phase I clinical trial and no adverse effects were observed. In addition, the vaccine demonstrated an excellent immunogenicity profile22,29. Because NiV outbreaks are sporadic and the number of cases is low, a traditional licensing pathway involving large Phase III/ efficacy clinical trials is not feasible. Therefore, non-classical regulatory vaccine licensing strategies must be explored. The United States Food and Drug Administration as well as other international regulatory agencies offer different alternative pathways for the approval of drugs that treat serious conditions and address an unmet medical need: (a) an approval under the animal rule or (b) an accelerated approval relying on the use of surrogate endpoint(s) (https://www.fda.gov/). The vaccine dose-down study described here aimed to determine an immune CoP against a lethal NiVB challenge in AGMs. This non-human primate model is considered the gold standard for NiV-related pre- and non-clinical studies as it closely recapitulates what is clinically observed in NiV-infected humans33,34. An immune CoP is defined as an immunological marker that reliably relates to protection and can be used as a predictor of clinical/non-clinical outcomes35,37,38. Both humoral and cellular immune responses are believed to play an important role in protection against NiV infection39,40; however, the vast majority of the available evidence suggests that the presence of neutralizing antibodies is associated with protection; this applies to several NiV animal models and is also supported by limited studies looking at human infections during past outbreaks18,28,41–43. Furthermore, passive transfer studies in AGMs using the neutralizing human monoclonal antibody m102.4 to the NiV and HeV G glycoproteins demonstrated the ability to protect the animals from a lethal NiV or HeV infection44–46, and more recently, the application of neutralizing humanized monoclonal antibodies to the NiV and HeV F glycoproteins has also proved to be potently protective47,48. Hence, for this study, we measured the humoral immune response (total IgG and neutralizing antibodies against NiVB and NiVM strains) in AGM upon a single IM vaccination with the HeV-sG-V. The goal was to correlate the NiV specific antibody responses with protection against death in this lethal NiVB challenge model. The selected dose-down study design demonstrated dose-dependency on the protection from lethality as animals in the highest dose groups survived the NiVB challenge. Previous studies using HeV-sG-V have shown similar protection in different animal models using either single or prime-boost vaccine regimens27,49,50. In addition, a recent vaccine study using the AGM model demonstrated the longevity of the immune response as HeV-sG-V protected animals challenged with NiV one year after a single HeV-sG-V dose28.
In the present study, one animal receiving the highest dose of 100 mcg HeV-sG (NiVCoP100-2) succumbed to infection 10 days after NiVB challenge. Furthermore, this animal presented clinical signs similar to the unvaccinated animals in the control group, although clinical signs and time to death were delayed. A retrospective analysis of the vaccination process did not reveal any deviation from the protocol. Compared to other animals of the same dose group, NiVCoP100-2 had only low anti-NiV antibody titers as measured on Study Day 21, one week before the NiVB challenge. The reason(s) why this animal had a low antibody response upon HeV-sG-V vaccination is not clear. Although unexpected due to the high dose given (100 mcg), studies with other vaccines often reveal a low percentage of vaccinees with no overt underlying conditions that do not develop an immune response to the vaccine51. This outcome for animal NiVCoP100-2 was the first indicator that, independently of the vaccine dose, the magnitude of the humoral immune response (antibody titers) may serve as a predictor of protection. On the other hand, animal NiVCoP30-2, which received the mid-range dose of 30 mcg HeV-sG-V, also showed a weak total IgG antibody titer and no detectable neutralizing antibody titer on Study Day 21 and yet survived the otherwise lethal NiVB challenge. While all other surviving animals did not show any clinical NiV disease signs and viral RNA was undetectable in any of their blood samples, surviving NiVCoP30-2 showed some respiratory distress between 7 and 10 days after the NiVB challenge. In agreement with the clinical signs this animal also presented measurable viral RNA in blood 7 days post-challenge; subsequent samples later in the study were devoid of NiV genomic RNA. The reason(s) why this animal survived the NiVB challenge with just a transient mild clinical disease when having developed a suboptimal humoral immune response characterized by low total IgG antibody titers and no specific NiV-neutralizing antibodies is unclear. Antibody-mediated control and clearance of infection can be achieved by other mechanisms besides neutralization52. Though not well established for NiV infection, recent preclinical NiV studies demonstrated that both antibody-dependent cellular phagocytosis (ADCP) and antibody-dependent complement deposition (ADCD) activities play a role in vaccine-mediated protection from NiV in small animal models53,54. We and others have shown in studies using pigs and AGMs that in addition to antibody-mediated protection, there is evidence for the role of the cellular immune response in protection from lethal NiVB challenge55–57. In these studies, the evidence of a cellular immune response was detected in animals after the virus challenge28,55. Furthermore, there is limited data from human NiV infection survivors, which further supports the idea that the cellular immune response may play a role in a survival outcome. In a longitudinal study of the immune status of two patients, the authors found that while the total numbers of CD8+ cells were not elevated in these two survivors, their CD8+ cells were activated39. Therefore, one or more of these non-nAb-related mechanisms of immune protection may explain the survival of the low-responder vaccinated animal NiVCoP30-2.
All surviving animals had a total IgG ELISA endpoint antibody titer in serum above 1000 against the NiV-sG-B coating antigen and above 800 against the NiV-sG-M coating antigen. Similarly, all the surviving animals had an FRNT50 titer above a certain threshold, i.e., 60 against an rVSVΔG-based NiVB reporter virus and 80 against an rVSVΔG-based NiVM reporter virus. As described above, the exception was animal NiVCoP30-2 (30 mcg HeV-sG-V dosing group) which survived the NiVB challenge with antibody levels below these thresholds. All immune monitoring results in conjunction with their respective clinical outcomes were finally analyzed to estimate the relationships between dose, immune response, and mortality to determine a potential immune CoP for NiV infection in this AGM animal model. A strong statistical correlation is necessary to establish a predictive immune indicator58. The various parameters defined by the Prentice Criteria59,60 were estimated to assess the strength of the relationships. The results from the analyses first determined the value of the surrogate needed to attain various levels of protection and then allowed us to conclude that all the immune assay results analyzed in this study - ELISA and neutralization titers can provide a predictor of survival. The preponderance of published data suggests that neutralizing antibodies against the NiV G and F glycoproteins are the key immune response that could be an appropriate measure for a potential CoP56,61,62; thereby, they are considered a mechanistic CoP (mCoP), which is preferable over a non-mechanistic CoP (nCoP)52,63. Here, we determined what can be considered a mCoP for NiVB and NiV based on measured total IgG binding and neutralizing antibody titers. In a recently published study using an rVSV-vectored vaccine design against NiV31, the authors tried to achieve a similar goal of establishing a CoP in AGMs by using similar immune assays but different reagents, e.g., live NiV for their neutralization assay of choice. Their initial dose-down study design failed to identify a threshold level for neutralizing titers that confers protection against death as all vaccinated AGMs with a detectable neutralizing titer survived the NiV challenge. Immune monitoring of a second study design, an onset of-protection study, confirmed this observation and concluded that any positive anti-NiV neutralization antibody titer can be used as putative CoP. Due to the limitations in the BSL4 space, the number of animals per group had to be kept small. Therefore, we split the study into two iterations to increase the group size, allowing us to have at least an n = 4 per group, which, based on our experience and that of others, is an acceptable number of animals to draw significant conclusions based on the study design. Our HeV-sG-V dose down/NiVB AGM challenge study design allowed us to establish CoPs for either total IgG ELISA titers or neutralizing antibody titers, in addition to the analyses of immune responses related to survival using well-accepted biostatistical criteria assigning CoP cut-offs values. These CoP cut-offs have been characterized for the high-throughput-capable ELISA and BSL-2-compatible neutralization assays described here, which rely on either using in-house produced NiVB-sG and NiVM-sG antigens or single-cycle rVSVΔG-based NiVB and NiVM reporter viruses, respectively. Furthermore, these assays have been further validated and used to analyze human serum samples from the Phase I clinical trial recently concluded29. Finally, we determined predictive values of CoP that correlate with various levels of protection from lethality. These results have been recently used to link the measured seroresponse rates in the Phase I clinical trial participants receiving the HeV-sG-V. Because of the limitations to the study size due to the BSL4 conditions, we used a high dose of the NiVB (a challenge dose that causes a uniform death between days six and eight post-challenge). This infectious dose is likely much higher than what the susceptible population encounters under natural virus exposure and transmission, suggesting that the protection observed here will be more efficacious in a natural exposure setting28. In conjunction with planned, qualitative passive transfer studies using human IgGs from responders in the HeV-sG-V’s Phase I clinical trial, the CoP data in AGMs presented here will be crucial to define a surrogate endpoint based on either binding or neutralizing antibody titers for NiV vaccine development at later clinical stages following a non-classical regulatory approval pathway.
Methods
Animals
Twenty-eight young adult African Green Monkeys (AGM) weighing 3–7.5 kg were divided into two iterations to accommodate this considerable number of animals. Males and females were randomly assigned to the different treatment groups, always trying to have equal numbers of males and females per treatment group. The first iteration (study 1) had a total of 12 animals (7 males and 5 females), while the second one (study 2) had a total of 16 animals (7 males and 9 females) (Table 4). Animals were anesthetized with Ketamine (5–20 mg/kg i.m.) or Telazol (2–9 mg/kg, i.m.) for all procedures. Animals that met the humane study endpoint based on clinical disease or that survived to the predetermined day 28 post-challenge endpoint were euthanized by intravenous or intracardiac injection of pentobarbital (≥80 mg/kg); death was ensured by bilateral thoracotomy. All research studies involving the use of animals were reviewed and approved by The University of Texas Medical Branch Institutional Animal Care and Use Committee. The studies were done following the Guide for the Care and Use of Laboratory Animals, 8th edition64.
Table 4.
Group Designations
| Study Iteration | Treatment Group | Animal ID | Sex |
|---|---|---|---|
| 1 | Control | NiVCoPCx-1 | F |
| NiVCoPCx-2 | M | ||
| 2 | NiVCoPCx-3 | M | |
| NiVCoPCx-4 | F | ||
| 2 | 0.3 mcg | NiVCoP03-1 | F |
| NiVCoP03-2 | M | ||
| NiVCoP03-3 | F | ||
| NiVCoP03-4 | M | ||
| NiVCoP03-5 | F | ||
| 2 | 1 mcg | NiVCoP1-1 | M |
| NiVCoP1-2 | F | ||
| NiVCoP1-3 | M | ||
| NiVCoP1-4 | F | ||
| NiVCoP1-5 | M | ||
| 1 | 3 mcg | NiVCoP3-1 | M |
| NiVCoP3-2 | M | ||
| 2 | NiVCoP3-3 | M | |
| NiVCoP3-4 | F | ||
| NiVCoP3-5 | F | ||
| NiVCoP3-6 | F | ||
| 1 | 30 mcg | NiVCoP30-1 | M |
| NiVCoP30-2 | M | ||
| NiVCoP30-3 | F | ||
| NiVCoP30-4 | F | ||
| 1 | 100 mcg | NiVCoP100-1 | M |
| NiVCoP100-2 | M | ||
| NiVCoP100-3 | F | ||
| NiVCoP100-4 | F |
Vaccine
HeV-sG-V also known as HenipaVaxTM is a recombinant subunit vaccine formulated as 0.1 mg/ml HeV-sG adjuvanted with 1 mg/ml of aluminum hydroxide (alum) in suspension. The vaccine and the alum alone (control group) were administered as two separate 0.5 mL intramuscular (IM) injections into the lumbar and/or thigh muscles. The vaccine for lower dose applications in this NiVB challenge was prepared by diluting HeV-sG-V with sterile phosphate-buffered saline.
The vaccine used for the study was derived from the same drug product lot used in the now-completed first-in-human Phase I clinical trial.
Challenge virus
The NiVB challenge stock used in both studies was derived from NiV Bangladesh strain #200401066 obtained in 2004 from a fatal human case during the outbreak in Rajbari, Bangladesh.
Enzyme-linked immunoabsorbent assay (ELISA)
Recombinant soluble glycoproteins from NiVB (NiV-sG-B) and NiVM (NiV-sG-M) were produced in-house using Chinese hamster ovary cells constitutively expressing either NiV-sG-B or NiV-sG-M. Immulon HB 96-well high-binding plates (Thermo Fisher Scientific, VWR Cat. No. 6240-972) were coated with NiV-sG-B (for NiV-sG-B IgG ELISA) or NiV-sG-M (for NiV-sG-M IgG ELISA) overnight at 4°C. The next day, the antigen-coated plates were washed and blocked for 2 h at room temperature with 5% non-fat dry milk in Dulbecco’s phosphate buffered saline with 0.1% Tween 20. After another washing step, serum serial dilutions (3-fold serial dilutions, starting at 1:100 dilution) were added and incubated for 1 h. Then the plates were washed and incubated with a peroxidase-conjugated goat anti-monkey IgG (Fitzgerald, Acton, MA, Cat. No. 43R-IG020HRP) solution for 1 hr at room temperature. After a final washing step, the binding of anti-NiV-sG total IgG antibodies to either NiV-sG-B or NiV-sG-M was revealed by a colorimetric reaction using 3,3,5,5 Tetramethylbenzidine (TMB) liquid substrate (Thermo Fisher Scientific, Wheltman, MA, Cat. No. 002023). The reaction was stopped with sulfuric acid, and the absorbance for each well was measured at 450 nm using a microplate reader (SpectraMax PC340 model, Molecular Devices LLC, San Jose, CA). Data was evaluated using the SoftMax Pro v 5.4 software (Molecular Devices) and tabulated in Excel calculation worksheets (Microsoft Corp., Seattle, WA). Each sample was tested in duplicates and analyzed using a 4-parameter logistic curve fit. Final data was reported as end-point titer, defined as the reciprocal of the highest serum sample dilution after background subtraction (blank wells) resulting in an OD ≥ the assay cut-off. Assays were performed by personnel blinded to the study groups. Both ELISAs were verified by characterizing and documenting the reliability and suitability of the method in terms of precision, accuracy, linearity, and specificity65,66.
Neutralization assay
Single-cycle rVSV-ΔG-based neutralization reporter viruses pseudotyped with the F and G glycoproteins from NiVB or NiVM (psVSV-NiV-B and psVSV-NiV-M, respectively) and expressing eGFP were produced in-house. The neutralization assay used here determined the ability of test serum samples to bind to psVSV-NiV-B or psVSV-NiV-M and to prevent a corresponding infection of Vero cells in vitro. Briefly, Vero cells (ATCC, Manassas, VA, Cat No. CCL-81) were grown on black clear bottom 96-wells culture plates (Corning, Corning, NY, Cat. No. 3904) to produce a confluent cell monolayer. Serial dilutions of test serum samples were combined with solutions containing a known quantity of psVSV-NiV-B or psVSV-NiV-M and incubated for 2 h at 37 °C to allow for serum antibody/virus binding and neutralization. The serum antibody/virus mixture was transferred to the Vero cell monolayer and incubated for 20 h at 32 °C to allow non-neutralized psVSV to express eGFP in infected Vero cells. After incubation, the cell monolayers were fixed with 4% formaldehyde for 20 min at room temperature in the dark, and the formaldehyde solution was discarded. Finally, the plates were washed and scanned before the fluorescent foci were counted using the ImmunoSpot S6 Universal Analyzer (CTL, Cleveland, OH) within 2 h. NiV neutralization data are reported as 50% fluorescent foci reduction neutralization (FRNT50) titers, which is the reciprocal of the highest dilution needed to inhibit the number of GFP signals by 50% compared with the mean value relative to the virus control wells in the absence of serum. The lower limit of quantification for each test was an FRNT50 of 20. If the FRNT50 value was less than 20, it was assigned a value of 10 for statistical analysis. Assays were performed by personnel blinded to the study groups. Both neutralization assays using the two different reporter viruses were verified by characterizing and documenting the reliability and suitability of the method in terms of precision, accuracy, linearity, and specificity65,66.
Vaccination and challenge
Male and female AGMs were randomly divided into four groups for the vaccine phase of each study (Table 4). On Study Day 0, animals in Study 1 received a single dose of either 100 micrograms (mcg), 30 mcg, or 3 mcg of HeV-sG-V, while the control group received alum alone. Study 2 animals were divided into four groups and received on Study Day 0 a single dose of either 3 mcg, 1 mcg, 0.3 mcg of HeV-sG-V, respectively, or alum alone. Vaccination of the animals was performed under anesthesia. Vaccine preparations were thoroughly mixed to achieve a homogeneous suspension just before the desired dose volume was withdrawn. AGMs were immunized while housed in an Animal Biosafety Level 2 (ABSL-2) facility and transferred to an ABSL-4 for the challenge phase of the studies. On Study Day 28, each animal was inoculated with 5 × 105 plaque-forming units (PFU) of NiVB, with the inoculum split equally and delivered via intratracheal and intranasal routes67,68. The animals were observed for clinical signs of disease for 28 days (Study Day 56) when all surviving animals were euthanized for necropsy and tissue harvest. Body weights and temperatures measured rectally under anesthesia were recorded for each monkey during the challenge phase on days 0 (Study Day 28), 4, 7, 10, 15, 21, and 28 (Study Day 56) post-challenge or at the time of euthanasia for animals that succumbed to infection.
Hematology and serum biochemistry
Blood specimens were collected from anesthetized animals via the femoral artery (alternating sides after each bleeding) into serum separator tubes and tubes containing ethylenediaminetetraacetic acid (EDTA; 1 mL). Blood was collected on Study Days 0, 7, 14, 21, 28, 32, 35, 38, 43, 49, and 56 (=28 days post-challenge) or at the time of euthanasia for animals that succumbed to infection. Hematology analysis was performed in blood samples collected in tubes containing EDTA using a laser-based hematologic analyzer (Vetscan HM5, Zoetis, Parsippany, NJ). Serum biochemistry was analyzed using a Piccolo point-of-care blood analyzer and Biochemistry Panel Plus discs (Abaxis, Sunnyvale, CA, USA).
RNA isolation from NiVB-Infected AGMs
Whole blood samples were collected during the challenge phase on Study Days 28, 32, 35, 38, 43, 49, and 56 ( = 28 days post-challenge) or at the time of euthanasia. Immediately after blood collection, RNA was extracted from 100 µL of whole blood utilizing 600 µl of AVL viral lysis buffer and the QIAamp Viral RNA Mini Kit (Qiagen, Germantown, MD).
Quantification of viral load
NiV loads were quantified in collected blood samples by quantitative detection of NiV genomic RNA by RT-qPCR using primers/probes targeting the NiVB N gene and the intergenic region between NiVB N and P as previously described in ref. 64. Threshold cycle (CT) values representing the number of NiV genomes were analyzed using CFX Manager Software and data are reported as genome equivalents (GEq).
Plasma was obtained from the blood collected in tubes containing EDTA. NiV titration in plasma samples (collected during the challenge phase of the experiment) was performed by plaque assay using Vero E6 cells (ATCC CRL-1587). Briefly, increasing 10-fold dilutions of the samples were adsorbed to Vero E6 cell monolayers in duplicate wells (200 μl/well) and overlaid with 0.8% agarose in 1X Minimum Essentials Medium (MEM) with 5% FBS and 1% penicillin/streptomycin. After 2 to 3 days of incubation at 37 °C/5% CO2, neutral red stain was added, and plaques were counted following an additional 24-hour incubation. The limit of quantification for this assay was defined as 25 PFU/mL67.
Statistical analysis to determine a correlate of protection
The analyses described here aimed to assess whether the four measured Study Day 21 antibody titers corresponding to the two NiV strains (NiVB and NiVM) and two assays (ELISA and Neutralization) might provide a candidate immune CoP against death due to the NiVB challenge. The statistical analysis plan and the assessment were predetermined and guided by the Prentice Criteria59,60,69, which defines four relationships for validation (Supplementary Table 1). Detailed analyses were done on each candidate surrogate (virus strain and serological assay). These methods used standard logistic regression for three of the four analyses and standard regression for the second criterion (treatment must affect the CoP).
The calculated results were then used to assess the value of the surrogate needed to attain various levels of protection from NiVB challenge, which required an inverse prediction of the fitted regression. The quantities of the regression, such as the standard error of the parameters, were then used to create a confidence interval for the predicted dose. This last step required the use of Fieller’s Theorem70. In all the analyses, one-half of the value was used as the observation when an assay produced a value below the lower limit. All calculations were performed in JMP software version 16.1 (JMP Statistical Discovery, Cary, NC).
All other indicated statistical analyses were performed with Prism 9 (GraphPad, San Diego, CA).
Supplementary information
Acknowledgements
The authors thank the UTMB Animal Resource Center for husbandry support of laboratory animals. This research was supported by the Coalition for Epidemic Preparedness Innovations (CEPI) under a grant to Auro Vaccines LLC. Additional support was provided by the Department of Health and Human Services, National Institutes of Health grants U19AI142764 to CCB and UC7AI094660 for BSL-4 operations support of the Galveston National Laboratory. The opinions and assertions expressed herein are those of the authors and do not reflect the official policy or position of Auro Vaccines LLC, University of Texas Medical Branch, Coalition for Epidemic Preparedness Innovations, the Uniformed Services University of the Health Sciences, the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. (Bethesda, Maryland) or any other agency of the U.S. Government.
Author contributions
V.H.L.G.- Study design, supervision, data analysis, writing the original draft of the paper, reviewing, and editing. D.P.- Performed immune assays, data analysis, reviewing, and editing the paper. K.N.A.- Performed the PCR assays, clinical pathology assays, and data analysis. G.G.L.- Performed immune assays and data analysis. V.B.- Performed the clinical pathology assays, the infectivity assays, and data analysis. D.J.D.- Performed the NHP vaccination and infection experiments. A.L.-Performed immune assays and data analysis. M.E.- Study design, supervision, data analysis. A.S.D. Data analysis, reviewing, and editing the paper. B.S.- Formal statistical analysis. C.C.B.- Data analysis, reviewing, and editing the paper. R.W.C.- Performed animal procedures and clinical observations, data analysis, reviewing, and editing the paper. S.H. Project administration, supervision, data analysis, reviewing, and editing the paper. T.W.G.- Study design, performed animal procedures and clinical observations, data analysis, reviewing, and editing the paper.
Data availability
All substantial data is available in the text and the Supplementary Figures and Tables. Certificates of analysis and origins, material data sheets, and detailed procedures are available upon request to the corresponding author. Materials generated will be available after the appropriate material transfer agreement.
Competing interests
V.H.L.G., D.P., G.G.L., and S.H. are Auro Vaccines LLC employees. C.C.B. is a US federal employee and co-inventor on US and foreign patents pertaining to soluble forms of the Hendra virus and Nipah virus G glycoproteins whose assignee is the United States as represented by the Henry M. Jackson Foundation for the Advancement of Military Medicine. Soluble forms of the Hendra virus and Nipah virus G glycoproteins are licensed to Zoetis Inc. and Auro Vaccines LLC. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41541-024-01036-2.
References
- 1.Moore, K. A. et al. Measures to prevent and treat Nipah virus disease: research priorities for 2024-29. Lancet Infect Dis. Published online July 2024. 10.1016/S1473-3099(24)00262-7 (2024). [DOI] [PubMed]
- 2.Eaton, B. T., Broder, C. C., Middleton, D. & Wang, L. F. Hendra and Nipah viruses: different and dangerous. Nat. Rev. Microbiol.4, 23–35 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chua, K. B. et al. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet354, 1257–1259 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Chew, M. H. et al. Risk factors for Nipah virus infection among abattoir workers in Singapore. J. Infect. Dis.181, 1760–1763 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Ching, P. K. et al. Outbreak of henipavirus infection, Philippines, 2014. Emerg. Infect. Dis.21, 328–331 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chadha, M. S. et al. Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg. Infect. Dis.12, 235–240 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uwishema, O. et al. A short communication of Nipah virus outbreak in India: An urgent rising concern. Ann. Med Surg. (Lond.)82, 104599 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.World Health Organization. Nipah virus outbreak(s) in Bangladesh, January-April 2004 = Flambee(s) d’infection à virus Nipah au Bangladesh, janvier-avril 2004. Wkly. Epidemiological Rec.79, 168–171 (2004).
- 9.Yadav, P. D. et al. Nipah Virus Outbreak in Kerala State, India Amidst of COVID-19 Pandemic. Front Public Health10, 818545 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization. Nipah virus infection- Bangladesh. https://www.who.int/emergencies/disease-outbreak-news/item/2024-DON508 (2024).
- 11.Thiagarajan, T. Nipah virus: Kerala reports second death in four months. Br.J.Med.38610.1136/bmj.q2058 (2024). [DOI] [PubMed]
- 12.Rahman, M. & Chakraborty, A. Nipah virus outbreaks in Bangladesh: a deadly infectious disease. WHO Southeast Asia J. Public Health1, 208–212 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Anderson, D. E. et al. Isolation and Full-Genome Characterization of Nipah Viruses from Bats, Bangladesh. Emerg. Infect. Dis.25, 166–170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas, B. P. et al. “Nipah Virus Infection in Kozhikode, Kerala, South India, in 2018: Epidemiology of an Outbreak of an Emerging Disease. Indian J. Community Med44, 383–387 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikolay, B. et al. A Framework to Monitor Changes in Transmission and Epidemiology of Emerging Pathogens: Lessons From Nipah Virus. J. Infect. Dis.221, S363–S369 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alam, A. M. Nipah virus, an emerging zoonotic disease causing fatal encephalitis. Clin. Med (Lond.)22, 348–352 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thakur, V. et al. Nipah Outbreak: Is it the beginning of another pandemic in the era of COVID-19 and Zika. Brain Behav. Immun.99, 25–26 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bruno, L. et al. Nipah Virus Disease: Epidemiological, Clinical, Diagnostic and Legislative Aspects of This Unpredictable Emerging Zoonosis. Animals (Basel)13, 159 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amaya, M. & Broder, C. C. Vaccines to Emerging Viruses: Nipah and Hendra. Annu Rev. Virol.7, 447–473 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomez Roman, R. et al. Medical countermeasures against henipaviruses: a review and public health perspective. Lancet Infect. Dis.22, e13–e27 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodrigue, V. et al. Current progress towards prevention of Nipah and Hendra disease in humans: A scoping review of vaccine and monoclonal antibody candidates being evaluated in clinical trials. Trop. Med Int Health29, 354–364 (2024). [DOI] [PubMed] [Google Scholar]
- 22.National Library of Medicine. Clinicaltrials.gov https://clinicaltrials.gov/ (2022).
- 23.Monath, T. P. et al. Recombinant vesicular stomatitis vaccine against Nipah virus has a favorable safety profile: Model for assessment of live vaccines with neurotropic potential. PLoS Pathog.18, e1010658 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.National Institutes of Health. NIH launches clinical trial of mRNA nipah virus vaccine.https://www.nih.gov/news-events/news-releases/nih-launches-clinical-trial-mrna-nipah-virus vaccine#:~:text=The%20National%20Institute%20of%20Allergy,prevent%20infection%20with%20Nipah%20virus (2022).
- 25.Bossart, K. N. et al. Receptor binding, fusion inhibition, and induction of cross-reactive neutralizing antibodies by a soluble G glycoprotein of Hendra virus. J. Virol.79, 6690–6702 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mungall, B. A. et al. Feline model of acute nipah virus infection and protection with a soluble glycoprotein-based subunit vaccine. J. Virol.80, 12293–12302 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bossart, K. N. et al. A Hendra virus G glycoprotein subunit vaccine protects African green monkeys from Nipah virus challenge. Sci. Transl. Med4, 146ra107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geisbert, T. W. et al. A single dose investigational subunit vaccine for human use against Nipah virus and Hendra virus. NPJ Vaccines6, 23 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frenck Jr., R. W. et al. Safety and immunogenicity of a Nipah virus vaccine (HeV-sG-V) in adults: A single centre, randomized, observed-blind, placebo-controlled, Phase 1 study. Lancet (2024).
- 30.Nikolay, B. et al. Assessing the feasibility of Nipah vaccine efficacy trials based on previous outbreaks in Bangladesh. Vaccine39, 5600–5606 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Monath, T. P. et al. Immunological correlates of protection afforded by PHV02 live, attenuated recombinant vesicular stomatitis virus vector vaccine against Nipah virus disease. Front Immunol.14, 1216225 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King, D. F. et al. Realising the potential of correlates of protection for vaccine development, licensure and use: short summary. NPJ Vaccines9, 82 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnston, S. C. et al. Detailed analysis of the African green monkey model of Nipah virus disease. PLoS One10, e0117817 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pigeaud, D. D. et al. Animal Models for Henipavirus Research. Viruses15, 1980 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finch, C. L. et al. Bridging Animal and Human Data in Pursuit of Vaccine Licensure. Vaccines (Basel)10, 1384 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.World Health Organization. Nipah research and development (R&D) roadmap. https://www.who.int/publications/m/item/nipah-research-and-development-(r-d)-roadmap (2019).
- 37.Longet, S. et al. Ebolavirus: Comparison of Survivor Immunology and Animal Models in the Search for a Correlate of Protection. Front Immunol.11, 599568 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plotkin, S. A. Recent updates on correlates of vaccine-induced protection. Front. Immunol.13, 1081107 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arunkumar, G. et al. Adaptive Immune Responses in Humans During Nipah Virus Acute and Convalescent Phases of Infection. Clin. Infect. Dis.69, 1752–1756 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Liew, Y. J. M. et al. The Immunobiology of Nipah Virus. Microorganisms10, 1162 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeBuysscher, B. L. et al. Single-dose live-attenuated Nipah virus vaccines confer complete protection by eliciting antibodies directed against surface glycoproteins. Vaccine32, 2637–2644 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shete, A. M. et al. Antibody response in symptomatic & asymptomatic Nipah virus cases from Kerala, India. Indian J. Med. Res.154, 533–535 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Doremalen, N. et al. ChAdOx1 NiV vaccination protects against lethal Nipah Bangladesh virus infection in African green monkeys. NPJ Vaccines7, 171 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bossart, K. N. et al. A neutralizing human monoclonal antibody protects against lethal disease in a new ferret model of acute nipah virus infection. PLoS Pathog.5, e1000642 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bossart, K. N. et al. A neutralizing human monoclonal antibody protects African green monkeys from Hendra virus challenge. Sci. Transl. Med.3, 105ra103 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geisbert, T. W. et al. Therapeutic treatment of Nipah virus infection in nonhuman primates with a neutralizing human monoclonal antibody. Sci. Transl. Med.6, 242ra82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mire, C. E. et al. A cross-reactive humanized monoclonal antibody targeting fusion glycoprotein function protects ferrets against lethal Nipah virus and hendra virus infection. J. Infect. Dis.221, S471–S479 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeitlin, L. et al. Therapeutic administration of a cross-reactive mAb targeting the fusion glycoprotein of nipah virus protects nonhuman primates. Sci. Transl. Med.16, eadl2055 (2024). [DOI] [PubMed] [Google Scholar]
- 49.Pallister, J. A. et al. Vaccination of ferrets with a recombinant G glycoprotein subunit vaccine provides protection against Nipah virus disease for over 12 months. Virol. J.10, 237 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Middleton, D. et al. Hendra virus vaccine, a one health approach to protecting horse, human, and environmental health. Emerg. Infect. Dis.20, 372–379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiedermann, U. et al. Primary vaccine failure to routine vaccines: Why and what to do? Hum. Vaccin Immunother.12, 239–243 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Britto, C. & Alter, G. The next frontier in vaccine design: blending immune correlates of protection into rational vaccine design. Curr. Opin. Immunol.78, 102234 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welch, S. R. et al. Single-dose mucosal replicon-particle vaccine protects against lethal Nipah virus infection up to 3 days after vaccination. Sci. Adv.9, eadh4057 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scholte, F. E. M. et al. Characterization of humoral responses to Nipah virus infection in the Syrian Hamster model of disease. J. Infect Dis.230, 438–443 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pickering, B. S. et al. Protection against henipaviruses in swine requires both, cell-mediated and humoral immune response. Vaccine34, 4777–4786 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lara, A. et al. Peripheral immune response in the African green monkey model following Nipah-Malaysia virus exposure by intermediate-size particle aerosol. PLoS Negl. Trop. Dis.13, e0007454 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cong, Y. et al. Loss in lung volume and changes in the immune response demonstrate disease progression in African green monkeys infected by small-particle aerosol and intratracheal exposure to Nipah virus. PLoS Negl. Trop. Dis.11, e0005532 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.World Health Organization. Correlates of vaccine-induced protection: methods and implications. https://www.who.int/publications/i/item/WHO-IVB-13.01 (2013).
- 59.Prentice, R. L. Surrogate endpoints in clinical trials: definition and operational criteria. Stat. Med.8, 431–440 (1989). [DOI] [PubMed] [Google Scholar]
- 60.Heller, G. Statistical controversies in clinical research: an initial evaluation of a surrogate end point using a single randomized clinical trial and the Prentice criteria. Ann. Oncol.26, 2012–2016 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woolsey, C. et al. Recombinant vesicular stomatitis virus-vectored vaccine induces long-lasting immunity against Nipah virus disease. J. Clin. Invest.133, 3164946 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, Z. et al. Potent monoclonal antibody-mediated neutralization of a divergent Hendra virus variant. Proc. Natl Acad. Sci. USA119, e2122769119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Plotkin, S. A. & Gilbert, P. B. Nomenclature for immune correlates of protection after vaccination. Clin. Infect. Dis.54, 1615–1617 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.National Research Council. Guide for the care and use of laboratory animals. 8th edition. The National Academy Press.(2011).
- 65.Andersson, U. et al. A Practical Guide to Immunoassay Method Validation. Front. Neurol.6, 1–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.The International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (2023). “Validation of Analytical procedures Q2(R2).
- 67.Mire, C. E. et al. Pathogenic Differences between Nipah Virus Bangladesh and Malaysia Strains in Primates: Implications for Antibody Therapy. Sci. Rep.6, 30916 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Foster, S. L. et al. A recombinant VSV-vectored vaccine rapidly protects nonhuman primates against lethal Nipah virus disease. Proc. Natl. Acad. Sci. USA119, e2200065119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Callegaro, A. & Tibaldi, F. Assessing Correlates of Protection in Vaccine Trials: Statistical Solutions in the Context of High Vaccine Efficacy. BMC Med. Res. Method19, 47 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fieller, E. C. Some problems in interval estimations. J. R. Soc., Ser. B16, 175–185 (1954). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All substantial data is available in the text and the Supplementary Figures and Tables. Certificates of analysis and origins, material data sheets, and detailed procedures are available upon request to the corresponding author. Materials generated will be available after the appropriate material transfer agreement.