Abstract
Streptococcus pneumoniae and influenza A virus (IAV) are significant agents of pneumonia cases and severe respiratory infections globally. Secondary bacterial infections, particularly by Streptococcus pneumoniae, are common in IAV-infected individuals, leading to critical outcomes. Despite reducing mortality, pneumococcal vaccines have high production costs and are serotype specific. The emergence of new circulating serotypes has led to the search for new prevention strategies that provide a broad spectrum of protection. In this context, vaccination using antigens present in all serotypes, such as Pneumococcal Surface Protein A (PspA), can offer broad coverage regardless of serotype. Employing the reverse genetics technique, our research group developed a recombinant influenza A H1N1 virus that expresses PspA (Flu-PspA), through the replacement of neuraminidase by PspA. This virus was evaluated as a bivalent vaccine against infections caused by influenza A and S. pneumoniae in mice. Initially, we evaluated the Flu-PspA virus’s ability to infect cells and express PspA in vitro, its capacity to multiply in embryonated chicken eggs, and its safety when inoculated in mice. Subsequently, the protective effect against influenza A and Streptococcus pneumoniae lethal challenge infections in mice was assessed using different immunization protocols. Analysis of the production of antibodies against PspA4 protein and influenza, and the binding capacity of anti-PspA4 antibodies/complement deposition to different strains of S. pneumoniae were also evaluated. Our results demonstrate that the Flu-PspA virus vaccine efficiently induces PspA protein expression in vitro, and that it was able to multiply in embryonated chicken eggs even without exogenous neuraminidase. The Flu-PspA-based bivalent vaccine was demonstrated to be safe, stimulated high titers of anti-PspA and anti-influenza antibodies, and protected mice against homosubtypic and heterosubtypic influenza A and S. pneumoniae challenge. Moreover, an efficient binding of antibodies and complement deposition on the surface of pneumococcal strains ascribes the broad-spectrum vaccine response in vivo. In summary, this innovative approach holds promise for developing a dual-protective vaccine against two major respiratory pathogens.
Subject terms: Vaccines, Influenza virus
Introduction
Streptococcus pneumoniae is a Gram-positive bacterium, also known as pneumococcus, which has more than 100 different serotypes and is one of the leading causes of pneumonia and meningitis worldwide1–6. The influenza A virus (IAV) is an enveloped virus belonging to the Orthomyxoviridae family. It is responsible for causing seasonal epidemics every year, as well as severe pandemics resulting in high morbidity and mortality rates worldwide, especially in the elderly and children under 5 years old7–12. Moreover, it is essential to highlight that secondary bacterial infections, mainly caused by S. pneumoniae, are among the main complications in patients infected with the influenza virus, resulting in severe events such as intensive care unit hospitalization or death13–17.
Influenza vaccines consist of annually updated strains of influenza A viruses (subtypes H1N1 and H3N2) and influenza B viruses, either live attenuated or inactivated18. Flu vaccination acts mainly by inducing neutralizing antibodies against viral membrane surface proteins19–21. Currently, the most common flu vaccine platforms are based on virus propagation in embryonated chicken eggs, which allows large-scale production at low cost22,23. On the other hand, two types of vaccines are currently used against pneumococcal diseases. The first is the polysaccharide vaccine (PPSV), which is made up of different capsular polysaccharides. This has good coverage but does not protect children under 5 years of age (one of the highest risk groups). Secondly, we have conjugate vaccines (PCV), which have the most prevalent polysaccharides conjugated to proteins. These are capable of generating protection in children and the elderly, but provide limited coverage24,25. Despite reducing mortality rates, the current pneumococcal vaccines are serotype-specific, covering up to 23 serotypes, rendering them unable to protect against other serotypes and new bacterial strains26–29. Furthermore, these vaccines involve complex industrial manufacturing processes and high production costs30,31.
Thus, there is a clear need to develop new vaccines that can protect against a broad range of serotypes and can be produced at a lower cost31–33. In this context, vaccination using pneumococcal proteins present in all pneumococcal strains has the potential to offer broad coverage, independent of serotypes32,34,35. Among the different pneumococcal proteins, Pneumococcal surface protein A (PspA) is a potential candidate as a vaccine antigen36. PspA is a surface protein of pneumococcus that is present in all clinical isolates37,38. It is expressed in significant and constant quantities during colonization, carriage, and invasive disease, and its surface exposure (protruding beyond the capsule) allows efficient antibody binding39,40. Based on its variability in different isolates, PspA has been classified into three families based on sequence. These are subdivided into six clades: family 1 (clades 1 and 2), family 2 (clades 3, 4 and 5) and family 3 (clade 6), with 90% of all clinically-isolated pneumococcal strains belonging to families 1 and 238,41–46. Despite these variations, PspA antibodies are highly cross-reactive, especially when they are in the same family47–49.
In invasive infections, PspA impairs the deposition of C3 molecule fragments on the pneumococcal surface, thereby affecting complement-mediated opsonophagocytosis50–53. During the colonization phase, PspA acts by blocking the bactericidal action of apolactoferrin53,54. Additionally, it has been shown that PspA prevents the binding and death of S. pneumoniae mediated by neutrophil extracellular traps (NETs)55. Many studies have demonstrated that immunization with a recombinant PspA protein protects against invasive infection and nasal colonization in mice56–64. Furthermore, it has also been demonstrated that immunization with PspA attenuates early secondary pneumococcal pulmonary infections65. Moreover, in a phase 1 clinical trial, PspA proved to be safe and immunogenic for human immunization and was capable of inducing antibodies that passively protect against invasive pneumococcal infection in mice48,66.
Based on these findings, with the aim of creating a bivalent vaccine against infections caused by S. pneumoniae and influenza A, our research group developed a defective recombinant influenza virus (Flu-PspA) using reverse genetics, carrying the gene to express PspA. In this construct, part of the viral neuraminidase sequence was deleted and replaced by the PspA4Pro protein (clade 4), resulting in a recombinant influenza virus in which PspA4Pro is fused with the neuraminidase stalk. We demonstrated that the PspA protein of clade 4 was capable of inducing antibodies with greater reactivity in ELISA, western blot, and binding assays, as well as providing larger cross-protection against strains of S. pneumoniae with the PspA of other clades47,67. Thus, in this study, we evaluated the immune responses and the potential for protection of a heterologous prime-boost vaccination protocol using the Flu-PspA vaccine virus, boosted with recombinant PspA4 protein, as a bivalent vaccine strategy against infections caused by influenza A virus and S. pneumoniae in mice.
Materials and methods
Cells and viruses
MDCK (Madin-Darby canine kidney cells) and HEK 293T (Human Embryonic Kidney 293T) cells were grown in complete DMEM medium (Dulbecco’s Modified Eagle’s Medium) with high glucose concentration (4500 mg/L) (DMEM High Glucose, SIGMA, pH 7.2) (DMEM HG), containing 3.7 g/L of sodium bicarbonate (SIGMA), 50 mg/L of HEPES (SIGMA) and supplemented with 1% antibiotic solution (5 mg/mL of streptomycin and 5,000 U/mL of penicillin—Gibco®) and 10% fetal bovine serum (FBS-Gibco®). The cell cultures were kept in a humid incubator at 37 °C and in an atmosphere of 5% CO2. MDCK cells were used for viral amplification, preparation of recombinant influenza virus stocks, and evaluation of viral load by plaque assay. HEK 293T cells were used in coculture with MDCK cells for transfection using the reverse genetics technique to construct recombinant viruses.
Wild type A/Puerto Rico/8/1934 (H1N1) influenza virus was generated by eight plasmids driving reverse genetics, as previously described68. Wild type A/Scotland/74 (H3N2) influenza virus was generously provided by Dr. François Trottein from Université de Lille69.
Bacterial strains and growth conditions
Streptococcus pneumoniae EF3030 (serotype 19F, PspA clade 1), D39 (serotype 2, PspA clade 2), M10 (serotype 11A, PspA clade 3), 3JYP2670 (serotype 3, PspA clade 4) and ATCC6303 (serotype 3, PspA clade 5) strains were used in this work. These bacterial strains were inoculated on blood agar plates (Brain Heart Infusion agar—Kasvi, supplemented with 5% defibrinated sheep blood) and kept at 37 °C overnight in a dry incubator. Subsequently, they were grown in Todd-Hewitt broth (Kasvi) supplemented with 0.5% yeast extract (THY) at 37 °C without agitation until an optical density (O.D.) of 0.3–0.4 at a wavelength of 600 nm was attained. All stocks were stored in a THY medium containing 20% glycerol and stored at –80 °C.
Animals
Female C57BL/6 mice, specific pathogen-free (SPF), seven to eight weeks old, were acquired from the animal facility of the Federal University of Minas Gerais and kept in the animal facility of the René Rachou Institute (IRR/FIOCRUZ) according to institutional guidelines. The Fundação Oswaldo Cruz (FIOCRUZ) Animal Ethics Committee previously approved the research protocol, with the license number LW-9/17. The experiments were performed with at least 5 animals per group.
Plasmids
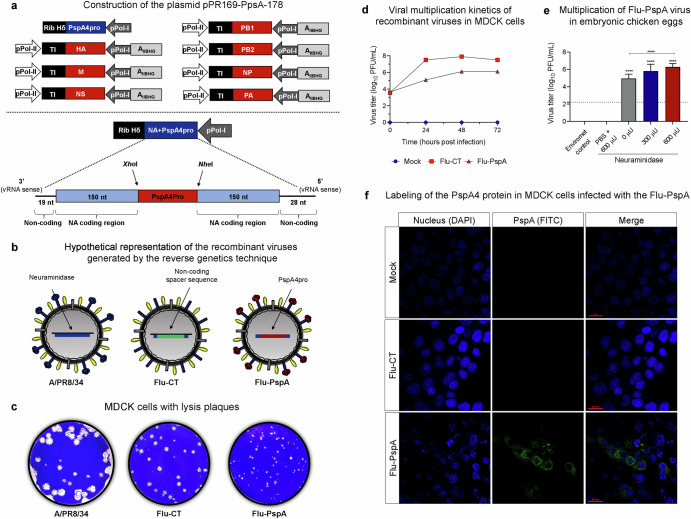
The pPRNA plasmid encodes the neuraminidase (NA) segment of influenza A/WSN/33 virus (H1N1), in which the NA segment was cloned in negative orientation in the pPR7 transfer plasmid, between the truncated sequence of the human polymerase I promoter (pPol-I) and the hepatitis delta virus ribozyme sequence (Rib Hδ)70,71. This plasmid was modified, giving rise to the plasmid pPRNA169x178, which encodes a truncated segment of NA that comprises the first 169 nucleotides of region 3’ of the neuraminidase segment (19 nucleotides of the non-coding region + 150 nucleotides of the coding region) and the last 178 nucleotides of the 5’ regions of the NA segment72. The sequence comprising the mature N-terminal α-helical surface-exposed region and the proline-rich region of PspA4 of isolate 255/00 (serotype 14) of S. pneumoniae, herein named PspA4Pro (GenBank EF649969.1), was cloned into pPRNA169x178, resulting in plasmid pPR169-PpsA-178 (Fig. 1a)47. It is important to point out that in this construction the PspA4Pro protein is produced in the form of fusion protein with the neuraminidase stalk, remaining harbored on the surface of the viral particle (Fig. 1b). As a control, we generated a plasmid (pPR-CT) encoding a spacer sequence, which was cloned into pPR169-X-178 as described above, resulting in the plasmid named pPR169-CT-17872.
Fig. 1. Construction and characterization of recombinant Influenza virus Flu-PspA.
a Schematic representation of the plasmids that encode the segments of the influenza virus used in reverse genetics and the sequence of the plasmid pPR169-PspA-178, which encodes part of the neuraminidase segment together with the PspA4Pro protein. b Hypothetical representation of the different viruses used in the study: wild-type influenza virus A/PR8/34 (PR8), Flu-CT (a recombinant influenza virus that carries a non-coding spacer sequence) and Flu-PspA (a recombinant influenza virus carrying the PspA4Pro protein from S. pneumoniae). c Phenotype of lysis plaques observed in MDCK cells after titration under agarose of A/PR8/34 and the recombinant viruses Flu-CT and Flu-PspA. d In vitro viral multiplication kinetics of recombinant influenza viruses in MDCK cells up to 72 h after infection. e Viral titers obtained with the multiplication of the Flu-PspA recombinant virus in embryonated chicken eggs for 48 h in different concentrations (300 µU or 600 µU) of exogenous neuraminidase. The bars represent the means ± standard deviation of the data, and ANOVA determined differences between groups. **** indicates a statistically significant difference at p value <0.0001, represented above the bars or connecting lines. f Labeling of PspA4Pro protein (in green/FITC) in uninfected MDCK cells (Mock) or infected with the Flu-CT or Flu-PspA recombinant viruses evaluated by confocal microscopy at 20 h after infection.
The ambisense plasmids (Phw2000), which encode the other seven segments of the A/Puerto Rico/8/1934 virus (H1N1) (herein named PR8), were kindly provided by Dr. Ron Fouchier of the Erasmus Institute of Rotterdam (Netherlands)73.
Construction of recombinant virus
Recombinant Flu-PspA and Flu-CT viruses were generated by reverse genetics as previously described72. Briefly, 12-well cell culture plates containing subconfluent monolayer cocultures of HEK 293T (5 × 105 cells/well) and MDCK (3 × 105 cells/well), grown in high glucose DMEM medium for 24 h, were respectively co-transfected with the plasmid pPR-PspA or pPR-CT, and the other seven PR8 segments (250 ng of each plasmid) with FuGENE HD transfection® reagent (Promega). The cells were kept in complete DMEM supplemented with 2 µg/ml of Trypsin-TPCK, 0.3% of bovine serum (BSA), and 500 µU/mL of type III Vibrio cholerae neuraminidase (SIGMA). Three days after incubation, the supernatants were recovered, and the virus particle was purified twice by picking lysis plaques on MDCK cells under agarose overlay and titrated on MDCK cells as described before74. Viral work stocks were prepared by infecting MDCK cells at a multiplicity of infection (M.O.I) of 0.01 as described above and further titrated.
Biological and molecular characterization of the recombinant virus
Flu-PspA viral stocks were characterized by their genetic stability by RT-PCR and further by Sanger sequencing, as previously described70,75. The recombinant Flu-PspA, Flu-CT, and wild-type influenza A/PR8/34 (PR8) virus were evaluated for lysis plaque phenotype and infectious titers75. This viral multiplication kinetics of recombinant viruses was made in MDCK cells using MOI 1:750.
The production of the PspA4Pro fusion protein in MDCK cells infected with the Flu-PspA virus was evaluated by confocal microscopy, adapted from76. For this purpose, the recombinant Flu-PspA and Flu-CT viruses were amplified (M.O.I of 0.5) for 20 h in MDCK cells previously cultured on a Thermo Scientific™ Nunc™ Lab-Tek™ Chamber Slide. The cells were then incubated with polyclonal anti-PspA4Pro antibody (in-house produced antibody; diluted 1:100) and further labeled with FITC-conjugated anti-mouse IgG secondary antibody (Goat F(ab) Anti-Mouse IgG FITC—Abcam, diluted 1:1000). The nuclei were labeled with a solution containing 1 µg/mL of 4’,6’-diamino-2-phenyl-indole dichloride (DAPI—Biolegend code 422801). Finally, the cell monolayer was fixed, the wells were removed, the slide was mounted with ProLong™ Gold Antifade Mountant (Thermo Scientific™), and the cell monolayer was visualized under a Nikon confocal microscope (model C2 Plus) to assess the presence of fluorescence.
Multiplication capacity of Flu-PspA in embryonated chicken eggs
The ability of recombinant influenza virus Flu-PspA to multiply in embryonated chicken eggs was determined by inoculating these viruses into 9-day-old specific pathogen-free (SPF) embryonated eggs. The allantoic cavity was inoculated with 100 μL of a suspension containing 10³ PFU of the virus, or just PBS. The eggs were then incubated at 37 °C for 48 h. After this period, allantoic fluid was collected, and the viral titer was determined by plaque assay.
Expression and purification of recombinant PspA4 protein
The expression and purification of recombinant PspA4 protein have been described previously47. Briefly, Escherichia coli BL21 DE3 transformed with pAE-pspA4 was used to express the protein. Purification was performed by affinity chromatography (His-Tag), and the protein was treated with Triton X-114 for the removal of lipopolysaccharides, as previously described77, analyzed by SDS-PAGE, and then quantified by Bradford using Bradford Reagent for 0.1–1.4 mg/ml protein (Sigma-Aldrich).
Immunization of the animals
Mice were lightly anesthetized with 80–100 μL of a PBS solution containing 1.4 mg/mL of xylazine hydrochloride (Sytenc) and 10.3 mg/mL of ketamine hydrochloride (Sytenc). They were then immunized with heterologous prime-boost protocols via inoculation by the intranasal route (IN) with 105 plaque-forming unit (PFU) of recombinant influenza viruses (Flu-PspA or Flu-CT) diluted in 40 µL of PBS. Three weeks later, the animals were intranasally boosted with 5 µg of PspA4 recombinant protein in the presence or absence of alum adjuvant, diluted in PBS to 40 µL. Another group of animals was primed with Flu-CT and boosted with recombinant protein in the presence or absence of alum adjuvant. In the control groups, the animals were immunized twice by the intranasal route with PBS (PBS/PBS group) or Flu-CT virus (Flu-CT/Flu-CT group).
Safety of immunization using Flu-PspA and Flu-CT viruses
To compare the pathogenicity of the Flu-PspA and Flu-CT recombinant viruses with that of the wild-type influenza virus A/PR8/34 (PR8; H1N1), previously anesthetized animals were inoculated intranasally with 105 PFU of Flu-PspA or Flu-CT, or with 103 PFU of PR8 virus. The mock control group received PBS intranasally. Subsequently, the animals were monitored for 20 days to assess weight loss and survival. In another experimental set, the lungs of the inoculated animals were aseptically collected on days 1, 4 and 7 after infection for viral load quantification by plaque assay.
Survival and weight loss of the immunized animals after lethal challenges with S. pneumoniae (ATCC6303) or influenza virus A/PR8/34 (H1N1) or A/Scotland/74 (H3N2)
As described above, the immunized animals were anesthetized and challenged intranasally with 40 µL of a 5 × 104 CFU suspension of isolate ATCC6303 (7xLD50) per mouse, 21 days after the last immunization (day 42). Subsequently, the animals were monitored daily to assess weight loss and survival for 10 days. Deaths were recorded for up to 10 days post-inoculation. LD50 was calculated using the Reed-Muench method78.
For the lethal challenge with the influenza virus, mice were anesthetized and immunized intranasally (40 µL/animal) with 105 PFU/animal of Flu-PspA or Flu-CT virus, or with 40 µL of PBS, in a single dose (homosubtypic challenge) or two doses (heterosubtypic challenge). Three weeks after immunization, mice were challenged intranasally with 100xLD50 (1 × 105 PFU) of A/PR8/34 (H1N1) virus or approximately 5xLD50 (1 × 104 PFU) of A/Scotland/74 (H3N2) virus. Survival and weight loss of challenged animals were monitored daily for 10 days and 21 days, respectively.
Antibody titers against the PspA4 protein and influenza virus in the serum and bronchoalveolar lavage fluid (BALF)
To determine the antibody titers induced against the PspA4 protein and influenza virus, blood samples were collected (0.5 mL) 14 days after each dose and centrifuged at approximately 700 × g for 10 min at 18 °C to obtain the serum. For the harvesting of bronchoalveolar lavage fluid (BALF), the immunized mice were euthanized with an overdose of ketamine/xylazine solution. Subsequently, BALF was harvested by washing the lungs three times with 1 mL of PBS.
Specific antibodies were detected using the indirect immunoenzymatic method (ELISA), as adapted from Moreno et al.47. Briefly, 96-well polystyrene microplates (Nunc MaxiSorp™—Thermo Scientific™) were coated overnight at 4 °C with 0.1 μg/well of PspA4 protein or with 0.5 μg/well of purified PR8 virus in a carbonate/bicarbonate buffer (0.1 M pH 9.6). After washing and blocking with 1% bovine serum albumin (Fisher), serially diluted serum was added to the wells and incubated. Anti-mouse IgG peroxidase-conjugated antibody (Anti-Mouse IgG-Peroxidase antibody produced in rabbit—Sigma) or anti-IgG1 (BD Pharmingen™ HRP Rat Anti-Mouse IgG1—BD Biosciences) or anti-IgG2c (HRP Goat Anti-Mouse IgG2c, Human Adsorbed—Southern Biotech) were further incubated to the plates. After 3,3,5,5-tetra methyl benzidine substrate incubation, followed by 1N sulfuric acid stop solution, absorbance was determined at 450 nm in a plate reader (Thermo Scientific Multiskan™ GO). For the analyses, the antibody titer was defined as the reciprocal Log2 of the largest dilution in which the absorbance at 450 nm presented a value higher than or equal to 0.1.
Binding of IgG antibodies and deposition of complement on the pneumococcal surface
Five different isolates of S. pneumoniae (ATCC6303, 3JYP 2670, M10, D39 and EF3030) were thawed and grown as described above. Then, the bacteria were centrifuged, washed twice, and fixed overnight with a fixing solution. The O.D. was adjusted to approximately 0.4 at 600 nm. For antibody binding evaluation, 10 µL of bacteria was incubated with 5% serum (10 µL of serum diluted 1:10) from immunized mice in PBS for 30 min at 37 °C. After three washes with PBS, the mixture was incubated with 50 µL of FITC-conjugated anti-mouse IgG antibody (Goat F(ab) Anti-Mouse IgG FITC—Abcam, 1:1000) for 30 min on ice in the dark.
To evaluate complement deposition, the serum from the immunized animals was previously incubated at 56 °C for 30 min to inactivate the complement system proteins. Subsequently, 10 µL of bacteria was incubated with 10% serum (10 µL of serum diluted 1:5) from immunized mice in PBS for 30 min at 37 °C. The samples were washed three times with PBS and incubated with 20 µL of 10% normal mouse serum (source of complement) diluted 1:10 in 1% BSA solution (Fisher) at 37 °C for 30 min. Subsequently, the samples were washed three times with PBS and incubated with 50 µL of FITC-conjugated mouse anti-C3 antibody (MP Biochemicals, 1:1000) on ice in the dark for 30 min.
After three further washes, for both experiments, the samples were fixed with 100 µL of fixing solution (10 g/L of paraformaldehyde, 1% of sodium cacodylate, 6.65 g/L of sodium chloride, pH 7.2) and incubated at 4 °C overnight. The following day, samples were acquired on a BD FACScalibur™ flow cytometer using CellQuest™ software (10,000 events). The results were analyzed using FlowJo™ software and the percentage of positive cells was used for comparison between the groups. The analysis strategy used in FACS is represented in Supplementary Fig. 1.
Statistical analysis
The results obtained in the comparison between the groups were submitted to analysis of variance (ANOVA), followed by the Tukey test for multiple comparisons, and the data presented a normal distribution. On the other hand, when the data did not follow the Gaussian distribution, the Mann–Whitney test for independent samples was used. Survival curves were compared using the Log-rank (Mantel-Cox) and Gehan-Breslow-Wilcoxon tests for survival. The differences between the weight loss curves were calculated from the area under the curve (AUC). The analysis was performed using the GraphPad Prism 8 program with a statistical significance of ρ<0.05.
Results
Construction and characterization of recombinant Influenza virus Flu-PspA
The Flu-PspA virus is a defective recombinant influenza virus generated by reverse genetics, in which the gene that encodes the catalytic part of viral neuraminidase is deleted and replaced by the PspA4Pro gene (clade 4), which is fused with the neuraminidase stalk when expressed (Fig. 1b). The phenotype of lysis plaques in MDCK cells of Flu-PspA was compared to the recombinant viruses Flu-CT and the wild-type virus A/PR8/34. The Flu-PspA virus presents smaller lysis plaques than those seen by the PR8 and the recombinant Flu-CT virus (Fig. 1c).
The amplification of Flu-PspA in MDCK cells reaches ten times lower virus titers than Flu-CT (Fig. 1d). Despite the lower replication in MDCK cells, the Flu-PspA virus was able to multiply in embryonic chicken eggs, even without exogenous neuraminidase supplementation (Fig. 1e). In the test performed in the absence of neuraminidase, the average titer obtained in PFU/mL was ≅ 5 (log10), while in the amplifications performed in the presence of neuraminidase, the titers obtained were ≅ 5.8 (log10) and ≅ 6.3 (log10) using 300 μU and 600 μU of exogenous neuraminidase, respectively. In contrast, no viral titers were detected in the PBS + 600 μU group or the environmental control, which was expected.
To further characterize the recombinant virus, the expression efficiency of the PspA4Pro protein in MDCK cells infected with the Flu-PspA virus was evaluated by confocal microscopy after 20 h of infection. The PspA antigen was detected on the cell surface, where the virus buds, only in the MDCK cells infected with the Flu-PspA (Fig. 1f).
Safety and effectiveness of the immunization with Flu-PspA virus in inducing antibodies against PspA4 protein
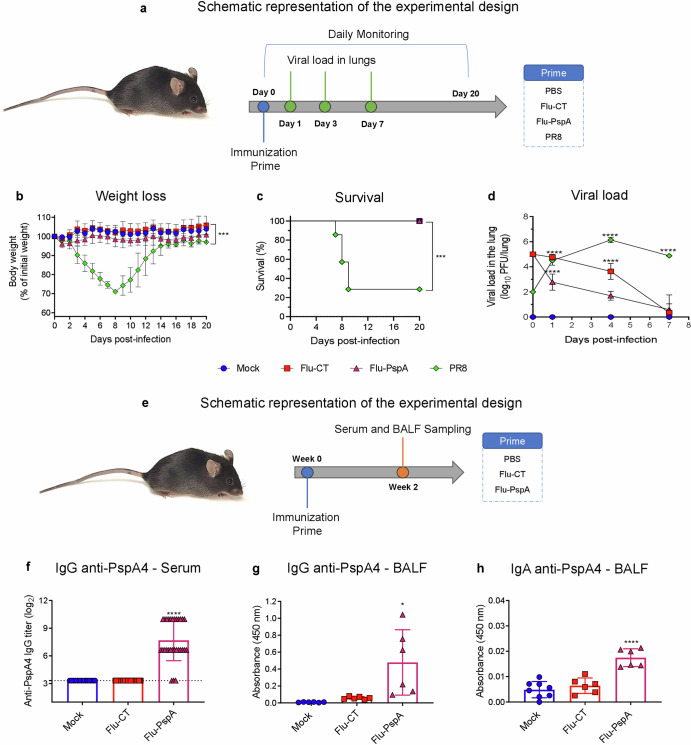
The safety of the recombinant Flu-PspA virus was evaluated in a murine model by comparing the virulence of the recombinant virus and the wild-type PR8 virus inoculated intranasally (Fig. 2a). Flu-PspA and Flu-CT were demonstrated to be safe, as the mice inoculated with these viruses did not present weight loss or mortality (Fig. 2b, c). In contrast, mice inoculated with wild-type virus PR8 presented intense weight loss (Fig. 2b) and 70% mortality until the 9th post-infection day with progressive weight recovery in mice that survived (Fig. 2c). When the viral loads were assessed in the lungs of animals inoculated with the viruses, we observed that mice infected with the PR8 virus presented a growing viral load, with about 5.7 (log10 PFU/lung) titer at day 7 post-infection. On the other hand, Flu-PspA or Flu-CT viral loads presented descending titers, reaching undetectable titers by the seventh day after infection. (Fig. 2d).
Fig. 2. Safety and effectiveness of the immunization with Flu-PspA virus in inducing antibodies against PspA4 protein.
a Schematic representation of the experimental design to evaluate the safety of the Flu-PspA virus. b Weight loss. c Survival. d Viral load in the lung observed after intranasal inoculation with the recombinant viruses (Flu-CT, Flu-PspA) or the wild-type (PR8) virus. e Schematic representation of the experimental design to evaluate the effectiveness of the Flu-PspA virus. f Reactivity of IgG antibodies specific for the PspA4 protein present in the serum. g IgG in the BALF and (h) IgA in the BALF of the animals after intranasal inoculation with PBS (Mock) or with Flu-CT or Flu-PspA recombinant viruses. Serum and BALF samples were collected 14 days after inoculation and serum antibody titers were represented as the log2 of the reciprocal of the highest dilution at which the O.D. at 450nm presenting a value ≥0.1. For BALF analyses, quantification was represented by the O.D. value at 450 nm using the pure sample. The survival curves were compared using the Log-rank (Mantel-Cox) and Gehan-Breslow-Wilcoxon tests. The differences between the weight loss curves of each group were calculated from the area under the curve (AUC). The bars represent the means ± standard deviation of the data from one independent experiment with at least 5 animals per group. Differences between groups were determined by ANOVA (p < 0.05). *, **, ***, and **** indicate statistically significant differences at p values <0.05, <0.01, <0.001 and <0.0001, respectively, and are represented above the bars or connecting lines.
To assess the effectiveness of the immunization with the Flu-PspA virus to induce antibodies, the animals were immunized with a single dose and specific antibodies against PspA4 protein were measured in the serum and BALF of the mice (Fig. 2e). Animals primed with Flu-PspA presented significant anti-PspA4 IgG titers in the serum (titer ≅ 7.7 (log2)), in contrast to animals immunized with PBS or Flu-CT virus (Fig. 2f). In BALF, specific IgG antibodies were detected, but only with significantly higher levels when compared to the Flu-CT group (Fig. 2g). Anti-PspA4 IgA antibodies were also detected in BALF of animals vaccinated with Flu-PspA (Fig. 2h).
Heterologous prime-boost protocol with Flu-PspA virus and PspA4 recombinant protein induced humoral immune response
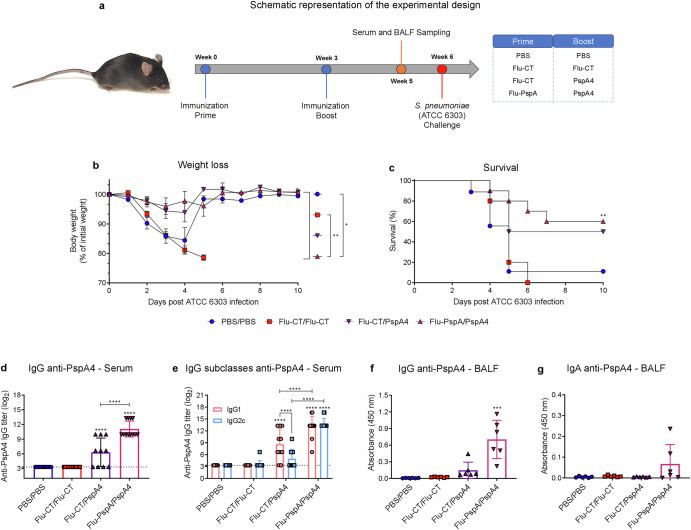
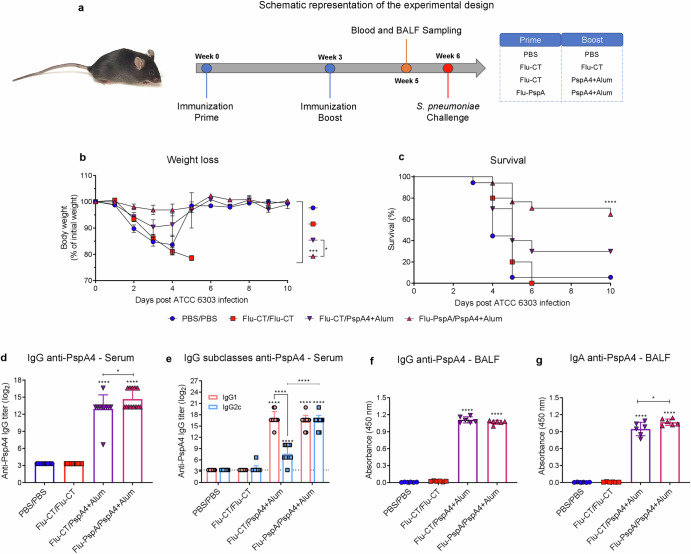
The heterologous prime-boost protocol with Flu-PspA virus and PspA4 recombinant protein was performed with a booster with 5 µg of PspA4 recombinant protein (Fig. 3a). The results showed that after the lethal challenge with pneumococci (S. pneumoniae - ATCC 6303), the animals in the PBS/PBS and Flu-CT/Flu-CT groups had high weight loss (Fig. 3b) and 11% and 0% survived, respectively (Fig. 3c). On the other hand, the group immunized with Flu-PspA/PspA4 had an important protection, with a 60% survival rate. In contrast, the animals in the Flu-CT/PspA4 control group had a survival rate of 50% (Fig. 3c). These two groups showed low weight loss (Fig. 3b).
Fig. 3. Heterologous prime-boost protocol with Flu-PspA virus and PspA4 recombinant protein.
a Schematic design to evaluate the efficacy of the heterologous prime-boost protocol with Flu-PspA virus and PspA4 protein, (b) weight loss. c Survival of mice after lethal challenge with pneumococcus (ATCC 6303 strain). d Reactivity of IgG anti-PspA4 antibodies and (e) IgG subclasses for the PspA4 protein in the serum. f Reactivity of IgG and (g) IgA antibodies specific for the PspA4 in BALF. PBS/PBS: animals that received two doses of PBS; Flu-CT/Flu-CT: animals that received two doses of 105PFU of control recombinant virus; Flu-CT/PspA4: animals that received 105PFU of control recombinant virus, in the first dose and boosted with 5 µg of PspA4 protein; Flu-PspA/PspA4: animals that received 105 PFU of Flu-PspA recombinant virus and boosted with 5 µg of PspA4 protein, all immunization intranasally. Serum and BALF were collected 14 days after the boost, and serum antibody titers were represented as log2 of the reciprocal of the highest dilution at which the O.D. at 450nm presenting a value ≥0.1. For BALF analyses, quantification was represented by the O.D. value at 450nm using the pure sample. The bars represent the means ± standard deviation of data from two independent experiments (a–e) or one experiment (3f and 3g), with at least 5 animals per group. Differences were determined by ANOVA (p < 0.05). *, **, ***, and **** indicate statistically significant differences at p values <0.05, <0.01, <0.001, and <0.0001, respectively. The differences between the weight loss curves were calculated from the area under the curve.
After the booster, high levels of total IgG anti-PspA4 antibodies were observed in the serum of animals in the Flu-PspA/PspA4 group (titer ≅ 11.1 (log2)), being significantly higher than those in the Flu-CT/PspA4 group (titer ≅ 6.3 (log2)) (Fig. 3d). For the IgG1 and IgG2c subclasses, after the second dose, the Flu-PspA/PspA4 group presented a mixed induction of IgG1 and IgG2c, with significantly equal titers between the subclasses. In contrast, the Flu-CT/PspA group only showed significant induction of IgG1, and both IgG1 and IgG2c titers were lower than those observed in the Flu-PspA/PspA4 immunized group (Fig. 3e). As expected, the PBS/PBS and Flu-CT/Flu-CT groups did not induce specific anti-PspA4 IgG antibodies in serum. In BALF, significant levels of specific IgG were observed after immunization in the Flu-PspA/PspA4 group (Fig. 3f) and the rise in IgA antibodies was not statistically significant (Fig. 3g).
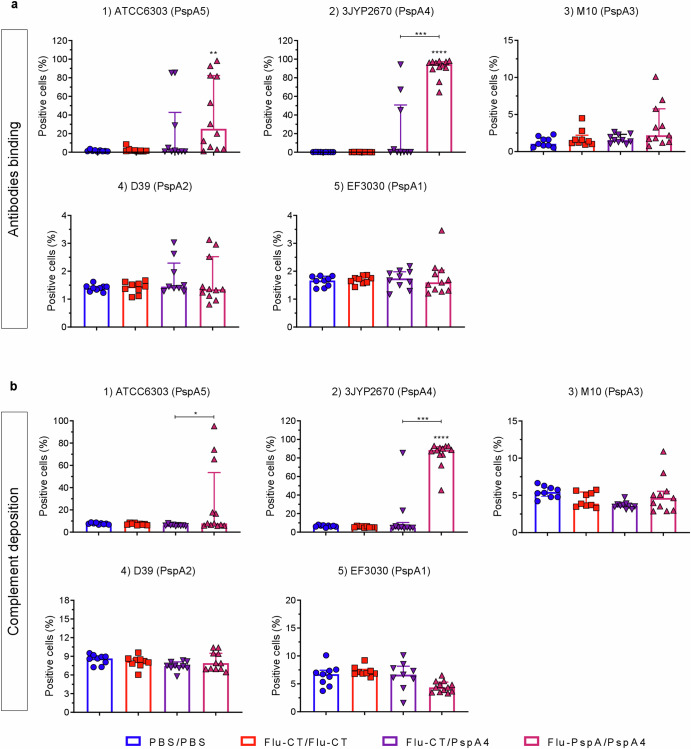
Binding of IgG antibodies induced through immunization with a heterologous prime-boost protocol onto the surface of pneumococcal strains expressing five different PspA clades (1 to 5), and its role in mediating complement (C3) deposition
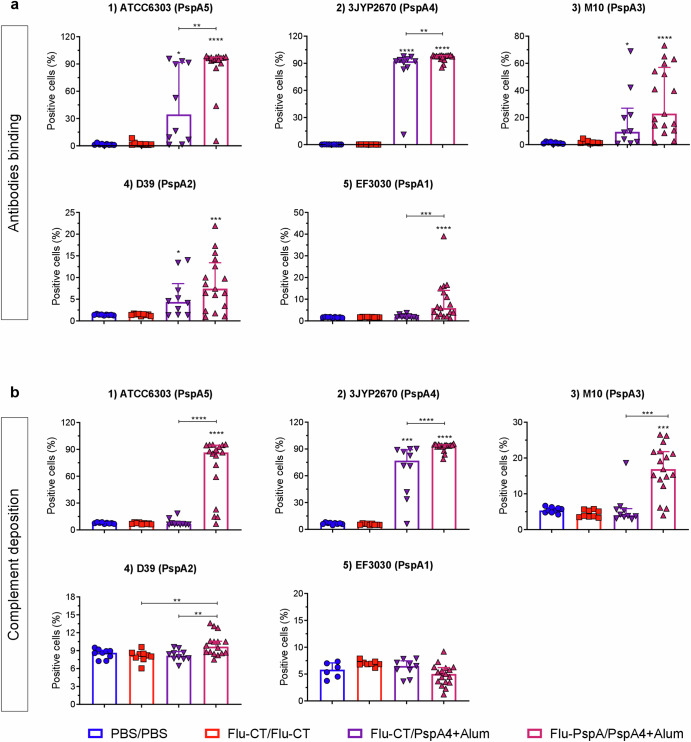
The capacity of antibodies to bind to the surface of pneumococcal strains expressing five different PspA clades (1 at 5), representing approximately 90% of all pneumococcal clinical isolates, was evaluated to investigate the effectiveness of antibodies induced by immunization. The ability of these antibodies to mediate complement (C3) deposition on the pneumococcal surface was also assessed. As seen in Fig. 4a, the serum from mice immunized with Flu-PspA/PspA4 exhibited a significantly higher antibody binding capacity to bacteria of the 3JYP2670 (PspA4) strain, which belongs to clade 4, the clade of the PspA protein used for immunization. This significant binding capacity was not detected in the serum from animals in the PBS/PBS, Flu-CT/Flu-CT, or Flu-CT/PspA4 groups. For this strain, Flu-PspA/PspA4 serum antibodies also demonstrated a significantly higher capacity to mediate complement deposition on the pathogen’s surface compared to the other groups (Fig. 4b).
Fig. 4. Binding of IgG antibodies induced through immunization with a heterologous prime-boost protocol onto the surface of pneumococcal strains expressing five different PspA clades (1 to 5), and its role in mediating complement (C3) deposition.
a Antibody binding and (b) complement deposition on the surface of pneumococcus from strains ATCC6303 (1), 3JYP2670 (2), M10 (3), D39 (4), and EF3030 (5). PBS/PBS: animals that received two doses of PBS, intranasally; Flu-CT/Flu-CT: animals that received two doses of 105 PFU of control recombinant virus, intranasally; Flu-CT/PspA4: animals that received 105 PFU of control recombinant virus, intranasally, in the first dose and boosted with 5 µg of PspA4 protein, also intranasally; Flu-PspA/PspA4: animals that received 105 PFU of Flu-PspA recombinant virus, intranasally, in the first dose and boosted with 5 µg of PspA4 protein, also intranasally. Serum samples were collected 14 days after the second dose of immunization. Bars represent the medians ± interquartile range of data from two independent experiments with at least 5 animals per group. Differences in the percentage of positive cells between groups were determined by the Mann–Whitney test (p < 0.05). *, **, ***, and **** indicate statistically significant differences at p values <0.05, <0.01, <0.001, and <0.0001, respectively, and are represented above the bars or connecting lines.
Serum from the prime-boost Flu-PspA/PspA4 group also showed a significant IgG binding capacity (Fig. 4a) and mediated complement deposition (Fig. 4b) to the pneumococcal strain used in the challenge experiments, ATCC6303 (clade 5), but not to the strains EF3030 (clade 1), D39 (clade 2) or M10 (clade 3).
Optimization of the heterologous prime-boost protocol with Alum adjuvant, aiming to increase vaccine efficacy
Despite the promising results obtained with the vaccine protocol employing the Flu-PspA virus plus PspA4 (5 μg) recombinant protein previously used, potential cross-protection between strains with heterologous PspAs, assessed by antibody binding and complement deposition, was still partial. Therefore, in an effort to enhance the broad-spectrum response of the immunization protocol, the adjuvant Alum was added to the PspA4 protein, maintaining all other conditions of the previous protocol (Fig. 5a).
Fig. 5. Optimization of the heterologous prime-boost protocol with Alum adjuvant.
a Schematic design to evaluate the efficacy of the optimized heterologous prime-boost protocol, b weight loss, c survival mice after lethal challenge with pneumococcus (ATCC 6303), d reactivity of IgG, e IgG subclasses for the PspA4 protein in the serum, f reactivity IgG and (g) IgA antibodies specific for the PspA4 protein in BALF. PBS/PBS: animals that received two doses of PBS; Flu-CT/Flu-CT: animals that received two doses of 105 PFU of control recombinant virus; Flu-CT/PspA4+Alum: animals that received 105 PFU of control recombinant virus and boosted with 5 µg of PspA4 adjuvanted with alum; Flu-PspA/PspA4+Alum: animals that received 105PFU of Flu-PspA recombinant virus and boosted with 5 µg of PspA4 protein adjuvanted with alum, all immunization intranasally. Serum and BALF were collected 14 days after the boost, and serum antibody titers were represented as the log2 of the reciprocal of the highest dilution at which the O.D. at 450 nm presenting a value ≥0.1. For BALF analyses, quantification was represented by the O.D. value at 450 nm using the pure sample. The bars represent the means ± standard deviation of three independent experiments (b–e) or one experiment (f and g), with at least 5 animals per group. Differences between were determined by ANOVA (p < 0.05). *, **, ***, and **** indicate significant differences at p values <0.05, <0.01, <0.001, and <0.0001, respectively. The survival curves were compared using the Log-rank (Mantel-Cox) and Gehan-Breslow-Wilcoxon tests. The differences between the weight loss curves were calculated from the area under the curve.
After the pneumococcal lethal challenge, a higher weight loss was observed in the control groups PBS/PBS and Flu-CT/Flu-CT (Fig. 5b) until the fourth post-infection day, which presented a 5% and 0% survival rate, respectively (Fig. 5c). On the other hand, 65% of the animals immunized with Flu-PspA/PspA4+Alum survived after the pneumococcal lethal challenge. In comparison, 30% of the group immunized with Flu-CT/PspA4+Alum survived (Fig. 5c).
Corroborating the high protection rate observed in the animals of the Flu-PspA/PspA4+Alum vaccine group, a high titer of anti-PspA4 IgG antibodies was observed in the serum of the animals in this group (titer ≅ 15 (log2)) after the boost. This titer was significantly higher than that observed in the Flu-CT/PspA4+Alum control group, which had a mean titer of approximately 13 (log2). As expected, no specific anti-PspA4 IgG antibodies were detected in the serum of the animals in the PBS/PBS or Flu-CT/Flu-CT groups (Fig. 5d).
Furthermore, in the serum of the animals of the Flu-PspA/PspA4+Alum group, a significant induction of IgG1 and IgG2c subclasses was observed, with no statistical differences between them (Fig. 5c). On the other hand, animals immunized with Flu-CT/PspA4+Alum had higher IgG1 titers compared to IgG2c, and significantly lower IgG2c titers than those observed in the Flu-PspA/PspA4+Alum group. The PBS/PBS and Flu-CT/Flu-CT control groups did not induce anti-PspA4 IgG1 or IgG2c antibodies (Fig. 5e).
In BALF, high levels of specific IgG antibodies in the Flu-PspA/PspA4+Alum immunized group and the Flu-CT/PspA4+Alum control group were observed, with no statistical differences between the two groups (Fig. 5f). Furthermore, high levels of specific IgA antibodies were also observed in these two groups, with significantly higher levels in the Flu-PspA/PspA4+Alum vaccine group compared to the Flu-CT/PspA4+Alum control group. No specific IgG and IgA antibodies were detected in the PBS/PBS or Flu-CT/Flu-CT control groups (Fig. 5g).
Binding of the IgG antibodies onto the surface of pneumococcal strains expressing five different PspA clades (1 at 5) and mediation of complement (C3) deposition after immunization with optimized protocol
To investigate the effectiveness of antibodies induced by immunization employing the optimized protocol, we evaluated the capacity of the antibodies to bind to the surface of strains expressing five different PspA clades (1 at 5), and the ability of these antibodies to mediate complement (C3) deposition on the bacterial surface. In general, IgG antibodies from serum from the immunized animal group led to higher binding to the surface of different pneumococcal strains and a higher capacity for complement deposition (Fig. 6a).
Fig. 6. Binding of the IgG antibodies onto surface of pneumococcal strains expressing five different PspA clades (1 at 5) and mediation of complement (C3) deposition after immunization with optimized protocol.
a Antibody binding and (b) complement deposition on the surface of pneumococcus from strains ATCC6303 (1), 3JYP2670 (2), M10 (3), D39 (4), and EF3030 (5). PBS/PBS: animals that received two doses of PBS, intranasally; Flu-CT/Flu-CT: animals that received two doses of 105 PFU of control recombinant virus, intranasally; Flu-CT/PspA4+Alum: animals that received 105 PFU of control recombinant virus, intranasally, in the first dose and boosted with 5 µg of PspA4 protein adjuvanted with alum, also intranasally; Flu-PspA/PspA4+Alum: animals that received 105 PFU of Flu-PspA recombinant virus, intranasally, in the first dose and boosted with 5 µg of PspA4 protein adjuvanted with alum, also intranasally. Serum samples were collected 14 days after the second dose of immunization. Bars represent the medians ± interquartile range of data from two independent experiments with at least 5 animals per group. Differences in the percentage of positive cells between groups were determined by the Mann–Whitney test (p < 0.05). *, **, ***, and **** indicate statistically significant differences at p values <0.05, <0.01, <0.001, and <0.0001, respectively, and are represented above the bars or connecting lines.
The strain ATCC6303 (clade 5), which was used in lethal pneumococcal challenges, had high binding when tested with the serum of the mice immunized with Flu-PspA/PspA4+Alum. This group exhibited significantly greater antibody binding compared to the PBS/PBS and Flu-CT/Flu-CT control groups, as well as the Flu-CT/PspA4+Alum group (Fig. 6a). Furthermore, the antibodies in the serum of the vaccine group also displayed a significantly higher capacity for complement deposition compared to that observed in the serum of animals from the other groups (Fig. 6b), which did not exhibit efficient deposition. The control group Flu-CT/PspA4+Alum showed significantly higher antibody binding than the PBS/PBS and Flu-CT/Flu-CT groups (Fig. 6a), but these antibodies were unable to mediate complement deposition on the surface of bacteria from this strain (Fig. 6b).
For the 3JYP2670 strain (clade 4), antibody binding in the Flu-PspA/PspA4+Alum immunized group was significantly higher than the Flu-CT/PspA4+Alum, PBS/PBS, and Flu-CT/Flu-CT groups (Fig. 6a). Similarly, complement deposition on bacteria from this strain was also significantly higher when compared to the other groups (Fig. 6b). The Flu-CT/PspA4+Alum control group also exhibited higher antibody binding to this strain when compared to the PBS/PBS and Flu-CT/Flu-CT groups, and this strain was the only one that displayed effective complement deposition mediated by the antibodies induced by this group (p<0.001).
As for the M10 strain (clade 3), the Flu-PspA/PspA4+Alum group showed higher binding than the PBS/PBS and Flu-CT/Flu-CT groups, but with no significant difference compared to the control group Flu-CT/PspA4+Alum (p = 0.1035) (Fig. 6a). Regarding complement deposition, the Flu-PspA/PspA4+Alum group displayed higher deposition than all control groups (Fig. 6b). For the D39 strain (PspA clade 2), antibodies induced by the Flu-PspA/PspA4+Alum group showed significantly higher binding than those in the PBS/PBS and Flu-CT/Flu-CT groups, but with no significant differences compared to the control group Flu-CT/PspA4+Alum (p = 0.2044) (Fig. 6a). Additionally, the Flu-PspA/PspA4+Alum group exhibited greater complement deposition than the Flu-CT/Flu-CT and Flu-CT/PspA4+Alum (Fig. 6b). In contrast, although the control group Flu-CT/PspA4+Alum showed significantly greater antibody binding to pneumococcal from the M10 and D39 strains than the other control groups (Fig. 6a), these antibodies were not able to efficiently mediate complement deposition on the surface of the bacteria (Fig. 6b).
Regarding the EF3030 strain (clade 1), antibodies from the Flu-PspA/PspA4+Alum group exhibited significantly higher binding than the PBS/PBS and Flu-CT/Flu-CT control and the Flu-CT/PspA4+Alum group (Fig. 6a). However, for this strain, the antibodies were unable to mediate complement deposition effectively (Fig. 6b).
Efficacy of Flu-PspA immunization in stimulating antibody production against Flu (PR8) and conferring protection against infection challenges posed by both homosubtypic (H1N1) and heterosubtypic (H3N2) influenza strains
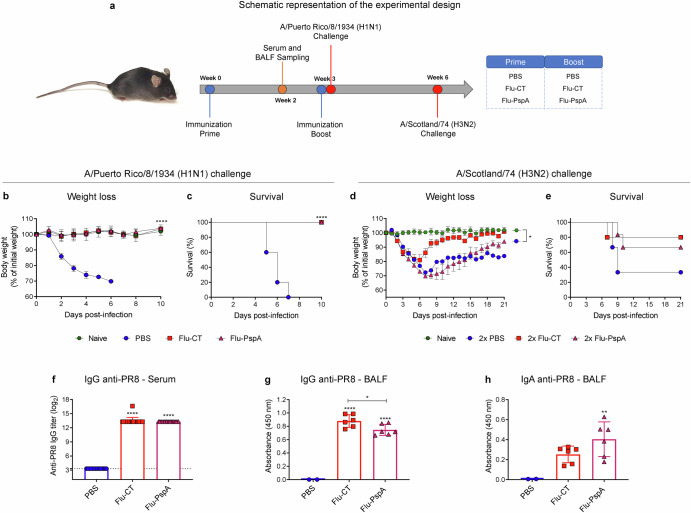
To assess protection against the challenge of influenza virus infection, the mice that were prime immunized with Flu-PspA virus were subjected to a homosubtypic challenge with the A/PR8/34 (H1N1) virus (Fig. 7a). In this protocol, we observed that animals immunized with a single dose of Flu-CT or Flu-PspA virus, as well as the animals in the uninfected naive group, did not lose weight or show any other clinical signs that are characteristic of the viral infection (Fig. 7b). They also had a survival rate of 100% (Fig. 7c). On the other hand, the animals inoculated with PBS showed significant weight loss, which resulted in the death of 100% of the animals within 7 days.
Fig. 7. Efficacy of Flu-PspA immunization in stimulating antibody production against Flu (PR8) and conferring protection against infection challenges posed by both homosubtypic (H1N1) and heterosubtypic (H3N2) influenza strains.
a Schematic representation of the experimental design. b Weight loss and (c) survival of the immunized mice after lethal challenge with PR8 influenza virus (A/Puerto Rico/8/1934-H1N1). d Weight loss and (e) survival of the immunized mice after sublethal challenge with Scotland H3N2 virus. f Reactivity of the IgG antibodies for the PR8 influenza virus in the serum. g Reactivity of the IgG and (h) IgA antibodies for the PR8 influenza virus in BALF. The animals were inoculated with PBS or 105 PFU of the Flu-CT or Flu-PspA recombinant viruses. Serum and BALF were collected 14 days after the inoculation and serum antibody titers were represented as the log2 of the reciprocal of the highest dilution at which the O.D. at 450 nm presenting a value ≥0.1. For BALF analyses, quantification was represented by the O.D. value at 450 nm using the pure sample. The bars represent the means ± standard deviation of data from one independent experiment with at least 5 animals per group. Differences between groups were determined by ANOVA (p < 0.05). *, **, ***, and **** indicate significant differences at p values <0.05, <0.01, <0.001, and <0.0001, respectively, and are represented above the bars or connecting lines. The survival curves were compared using the Log-rank (Mantel-Cox) and Gehan-Breslow-Wilcoxon tests. The differences between the weight loss curves of each group were calculated from the area under the curve.
In addition, we evaluated whether a homologous prime boost with the recombinant viruses could confer protection against a heterosubtypic challenge with the A/Scotland/74 (H3N2) virus. 21 days after the infection challenge, animals from the naive group had not lost weight and showed 100% survival, whereas the PBS/PBS group showed significant weight loss and presented only 33.3% survival (Fig. 7d, e). Interestingly, 80% and 66.7% of the experimental vaccinated groups, those in the 2x Flu-CT and 2x Flu-PspA, respectively, survived after the heterosubtypic challenge (Fig. 7e). Although there is a clear difference in the survival curves of the groups, they were not statistically different in any of the tests used (Log-rank (Mantel-Cox) and Gehan-Breslow-Wilcoxon tests).
To assess the production of specific antibodies against the PR8 influenza virus, the serum antibody titers (total IgG) induced after immunization with the Flu-PspA virus were measured by ELISA (Fig. 7f). After immunization, a significant induction of specific antibodies against the PR8 influenza virus (mean titer ≅ 13 (log2)) was observed in both the serum of animals that were inoculated with the recombinant Flu-CT virus and the animals immunized with Flu-PspA (Fig. 7f). In BALF, significant levels of specific IgG were detected in the animals immunized with the recombinant viruses, with higher levels in the Flu-CT group (Fig. 7g). Specific IgA indices were also observed in the BALF of the animals inoculated with Flu-PspA, but only with a statistical difference concerning the PBS group (Fig. 7h). The presence of specific anti-influenza antibodies was not observed in the serum of the animals of the PBS group (Fig. 7f, g, h).
Discussion
Although antibiotic treatments and a variety of vaccines are available, pneumococcal infections still result in high mortality rates, particularly among high-risk groups. Additionally, the increasing resistance to antibiotics such as penicillin and the limitations of current vaccines, which are serotype-specific, underscore the need for new prevention strategies. One such approach is the use of conserved pneumococcal proteins, such as PspA, which have been shown to provide broad coverage, independent of serotypes28,32,33,79,80. Previous studies have demonstrated the potential of PspA, particularly from clade 4, in offering cross-protection against different strains of S. pneumoniae, making it an important alternative antigen for vaccines as opposed to conventional polysaccharide-based vaccines31,32,34–36,63,81. To explore this strategy further, our research group developed a replication-defective recombinant influenza virus (Flu-PspA), in which the catalytic portion of the viral neuraminidase was replaced with the PspA protein (clade 4), aiming to create a bivalent vaccine effective against both S. pneumoniae and influenza A. The PspA protein from clade 4 was chosen because previous studies had demonstrated that this clade provides greater cross-protection against strains of S. pneumoniae with PspA from other clades, making it a strong candidate for serotype-independent immunity against pneumococcal infections47,67.
After its construction, the Flu-PspA virus proved to be capable of replicating in MDCK cells through the addition of exogenous neuraminidase, efficiently expressing the heterologous PspA protein in virus-infected cells. Moreover, it was able to replicate in embryonated chicken eggs, the primary substrate used for seasonal flu vaccine production worldwide82–84. In eggs, it was able to replicate even in the when exogenous neuraminidase was not added, achieving infectious titers similar to those routinely obtained with MDCK cell culture and slightly lower titers in the absence of neuraminidase. Since Flu-PspA is a defective recombinant virus that does not display neuraminidase activity, the addition of exogenous NA is necessary for it to multiply. This sialidase acts by cleaving sialic acid molecules present in the membrane glycoproteins of infected cells, allowing the release of newly produced viral particles85. Thus, since the Flu-PspA was able to multiply in embryonated eggs without the addition of exogenous neuraminidase, it is possible that another enzyme with sialidase activity was present in the embryonated eggs to perform this function. A possible candidate sialidase, present in chicken embryos, would be lysosomal sialidase (Neuraminidase-1 – Neu1), which acts in the elastogenesis of the embryo, responsible for the formation of elastic fibers in developing tissues, mainly in the lungs and arteries86,87. Therefore, the presence of Neu1 on the cell surface during this process may have been responsible for the cleavage of sialic acid molecules present in infected cells, resulting in the release of viral particles synthesized after viral multiplication. However, tests are needed to confirm this hypothesis.
As Flu-PspA can replicate in embryonated chicken eggs, it can be scaled up for industrial production, which can be done without the addition of exogenous neuraminidase, resulting in even lower production costs. Furthermore, the Flu-PspA virus demonstrated attenuation in mice, as inoculated animals showed no weight loss or mortality, and they cleared the virus after a few days, confirming that the virus lost the ability to cause productive infection and is safe for immunization. This is because the Flu-PspA recombinant virus had the PspA4Pro protein sequence inserted into the viral neuraminidase coding region, replacing its catalytic center. In this way, the resulting virus does not have neuraminidase activity and is not, therefore, capable of leaving the host cell after multiplication. As it is a replication-defective virus, the vaccine is safe for use in humans, as is the case with other attenuated influenza virus vaccines88–92. This fact demonstrates its potential to be used as a recombinant vaccine, consistent with results previously obtained by our group and other researchers72,93–95.
Since S. pneumoniae primarily invades the mucosal tissues of the respiratory tract, intranasal vaccination is a promising alternative for inducing local immunity against respiratory pathogens such as S. pneumoniae96–98. Therefore, to evaluate the ability of the Flu-PspA virus to induce protective immunity against S. pneumoniae, we assessed the virus’s efficacy in producing specific antibodies against the PspA protein in a murine model via intranasal immunization. Our results showed that immunization with just one dose of Flu-PspA was able to induce high titers of anti-PspA antibodies in both serum and BALF. However, a single dose of this virus was not able to confer protection against lethal pneumococcal challenge. Performing a homologous prime and then boosting with two doses of Flu-PspA did not result in higher titers of specific antibodies against the PspA4 protein, as there was not adequate stimulation by the booster doses (data not shown). Although inoculation with the Flu-PspA vaccine virus induced high levels of antibodies, a second immunization with the Flu-PspA virus does not stimulate the immune system against PspA, probably due to neutralization of the vector by the high levels of antibodies against hemagglutinin induced after the first inoculation. Therefore, homologous immunization with two doses of the Flu-PspA virus did not result in protection, so we decided to perform a boost with the PspA4 protein.
Given this, we evaluated a heterologous prime-boost protocol based on priming with Flu-PspA and boosting with 5 µg of recombinant PspA protein. In this protocol, we observed an induction of specific antibodies against PspA and an important protection against lethal pneumococcal challenge. Although the results using this heterologous prime-boost protocol were encouraging, demonstrating for the first time that our vaccine formulation would be able to confer protective immunity against pneumococcal infection in vaccinated animals, when we evaluated the antibodies and complement binding in different pneumococcal strains, we did not observe an effective response against strains expressing heterologous PspAs. In our next stage, aiming to achieve broad-spectrum response, we optimized the vaccination protocol by adding the adjuvant Alum in PspA4 protein in the vaccine boost. The use of Alum intranasally has been shown to induce an intense IgA response99–102. This change allowed us to increase the levels of specific IgG antibodies in serum and IgA in BALF, in addition to maintaining high protection rates. This resulted in 65% protection for the Flu-PspA/PspA4+Alum vaccine group and 30% for the control group Flu-CT/PspA4+Alum.
In general, animals that were primed with the Flu-PspA virus and received a booster dose with 5 μg of recombinant PspA protein (whether adjuvanted or not), showed higher protection rates compared with control groups boosted with recombinant PspA protein (Flu-CT/PspA4 and Flu-CT/PspA4+Alum). This indicates that priming with Flu-PspA intensified protection rates against lethal pneumococcal challenge. It is essential to note that in the two vaccination protocols tested, the survival curves of all groups were statistically different after analyses using the Log-rank (Mantel-Cox) and Gehan-Breslow-Wilcoxon tests. Therefore, it can be asserted that these two vaccine protocols not only increased the survival rates of immunized mice, but also enhanced their survival in the early stages of pneumococcal infection103,104, potentially allowing more time for intervention in treating patients with the disease.
It is important to emphasize that the induction of specific antibodies to PspA is a crucial protective mechanism, allowing increased complement activation and C3b deposition on bacteria, leading to pneumococcal death by IgG-mediated opsonophagocytosis, which is an essential mechanism for eliminating pneumococci105–108. Therefore, in these two protocols, we also investigated the profile of IgG anti-PspA subclasses and observed that prior inoculation with Flu-PspA promotes a change from the IgG1 isotype to IgG2, with a preferential induction of IgG2c anti-PspA4 and an increase in the production of this subclass. This fact led to a mixed and balanced induction of IgG1 (related to the preferential development of a Th2 response) and IgG2c (associated with the preferential development of a Th1/Th17 response)109,110. In contrast, this induction is very different from that observed with the inoculation of recombinant PspA (alone or adjuvanted), in which an immune response with a Th2 profile with high levels of IgG1 predominates59,111–115.
It is known that the response pattern of IgG subclasses can affect, through different mechanisms, bacterial clearance during pneumococcal infections. IgG2 antibodies, for instance, represent the isotype with the most remarkable capacity to mediate complement deposition on the pneumococcal surface in mice, as their Fc domain presents a solid connection to the C1q component, an activator of the classical complement pathway116–118. On the other hand, the IgG1 subclass has a limited affinity with this component and is therefore not as effective in activating complement deposition through the classical pathway116–118. Thus, if large amounts of IgG1 are induced, there may be competition between the subclasses, which may compromise the binding of IgG2 and, consequently, affect the deposition of complement on the bacterial surface119,120. However, high levels of IgG2 can result in uncontrolled IgG-induced inflammation119,121–123. Furthermore, since pneumococcus is an extracellular bacterium, complement-mediated phagocytosis enhanced by specific antibodies (classical pathway of complement activation) is essential for pneumococcal clearance from the host107,108,124,125.
Therefore, in these two vaccination protocols, we also investigated the ability of the antibodies generated by immunization to bind and deposit complement on the surface of different pneumococcal strains. Aiming to investigate the broad-spectrum response of the vaccine, we selected strains for these experiments that express PspAs from different clades (clades 1, 2, 3, 4 and 5), which comprise about 90% of all pneumococcal clinical isolates44,126. Thus, in the optimized vaccine protocol (Flu-PspA/PspA4+Alum), we observed that immunizations associating Flu-PspA with recombinant PspA4 induced a significant production of antibodies with a high ability to bind and deposit complement on different pneumococcal isolates specifically. There was a higher antibody binding to PspA proteins from clades 3, 4 and 5 and lower binding to strains with lower amino acid identity (especially from clade 1). Therefore, these results suggest a broad-spectrum and serotype-independent pneumococcal vaccine response. Although we did not evaluate protection against lethal challenge using these serotypes from different PspA clades, based on the strong antibody response/complement deposition, the vaccination protocol would result in protection against these different strains, especially against clade 4 strains, as they have PspA homologous to that used as the vaccine antigen.
Some studies have demonstrated that antibody binding to PspA increases the ability to eliminate bacteria through the bactericidal action of apolactoferrin during the colonization phase, thus preventing bacterial invasion53,127,128. Additionally, Martinez and collaborators (2019)55 showed that the presence of anti-PspA antibodies increases the induction of NETs by activated neutrophils and enhances the capture of pneumococci in these traps. Therefore, it is possible that antibodies induced by immunization, when specifically binding to PspA, blocked its anti-complement function and amplified complement system activation. This resulted in more effective opsonization and phagocytosis, and consequently prevented invasion and sepsis108,129. In line with these findings, several studies using the PspA protein as an antigen have demonstrated that it can induce high levels of specific antibodies, which can bind to the pneumococcal surface and promote effective complement deposition47,108,130–134.
It is noteworthy that in this work we also demonstrate that in addition to conferring protective immunity against pneumococcal infection, the tested immunization protocols were also capable of inducing high antibody titers and protecting against lethal challenge with the homologous A/PR8/34 (H1N1) influenza virus with just one dose of the Flu-PspA virus. The homosubtypic protection observed is mainly due to the presence of neutralizing antibodies that target the head region of the hemagglutinin (HA) protein on the Influenza virus. These antibodies neutralize the virus by blocking its ability to recognize and bind to receptors on the surface of target cells, thereby preventing viral entry and infection. As a result, the onset of clinical symptoms is also prevented135. Additionally, T cell immunity, particularly the role of CD8+ T lymphocytes (CTLs), may contribute to this protection. These CTLs are crucial in fighting infections by directly eliminating infected cells through perforin- and granzyme-mediated cell lysis, and by secreting cytokines that help activate other components of the immune system136,137.
Furthermore, we evaluated whether an additional dose of recombinant viruses would provide any protection against a lethal heterosubtypic challenge with influenza virus A/Scotland/74 (H3N2). However, despite having observed higher protection rates, we did not observe a significant difference between the curves. Thus, although immunization with recombinant viruses was not able to immediately prevent infection with an H3N2 subtype virus that resulted in high weight loss and characteristic clinical signs of the disease, it appears that it was clearly capable of increasing the survival rates of infected mice. Several studies have demonstrated that prior infection with antigenically distinct influenza viruses induces cross-protective immune responses against challenge with heterologous virus strains. This protection is often correlated with the presence of cross-reactive (but not neutralizing) antibodies and cellular immune responses against conserved regions of viral proteins such as the HA stalk, M2 protein and nucleoprotein (NP)138,139. Based on this, we believe that the induction of non-neutralizing cross-reactive antibodies and the action of CD8+ T lymphocytes were responsible for the increased protection after heterosubtypic lethal challenge observed in animals immunized with recombinant influenza viruses. In general, due to all the pneumococcal and influenza protection results that we obtained in experiments using the Flu-PspA vaccine, it leads us to believe that it will also be effective in protecting in cases of coinfection with influenza A virus and S. pneumoniae.
Two other studies that use recombinant influenza viruses carrying PspA as a vaccine approach have already been described in the literature. In the first strategy, the viral hemagglutinin was replaced by PspA140, while in the second, the replacement was carried out in the PB2 protein of the viral replication complex141. In these two approaches, recombinant viruses were amplified in cells that constitutively produced the respective deleted protein. Although both studies showed high protection rates against a lethal pneumococcal challenge, the S. pneumoniae used in the challenge has a PspA with high identity (at least 97.4%) with the PspA introduced in the influenza virus (both clade 2)142. Therefore, with this high similarity of the amino acid sequence of the PspA proteins, high protection rates after challenge were expected due to a greater number of epitopes in the region of high variability of the PspA α-helical domain48,126,143,144.
Furthermore, none of these studies evaluated whether the induced antibodies could bind to the surface of strains representative of different PspA clades. This means that it remains unclear whether these recombinant viruses can protect against isolates expressing PspAs from other clades. It is important to note that, for the replication of these recombinant viruses, they must be cultivated in constitutively transfected cells that produce the viral protein the recombinant virus is unable to generate. While these are elegant and interesting strategies for scientific research, they have limited viability for industrial-scale application. This is because the use of these cells as a platform for virus replication depends on ensuring that there will be no reversion to the wild-type phenotype. Furthermore, these recombinant viruses could not be replicated in embryonated eggs, which are the main substrate for influenza vaccine production worldwide23,145.
One limitation of this work is that the evaluated protocols still depend on the association of Flu-PspA with the recombinant PspA4 protein. This problem could be overcome by boosting with a recombinant virus expressing PspA4 with a different hemagglutinin (HA) subtype. Thus, this strategy could result in lower neutralization of the recombinant virus in the booster dose and, consequently, better antigen stimulation. This would eliminate the need to boost with a recombinant protein. Additionally, boosting with a heterologous subtype of influenza HA could further improve heterosubtypic vaccine response, increasing cross-reactivity between different Influenza subtypes20,138. Due to this, we are constructing a recombinant influenza virus with type 3 hemagglutinin (H3) instead of type 1 hemagglutinin (H1), so that we can use it in a heterologous prime boost protocol with H1-PspA in the first dose, and H3-PspA in the boost, aiming to minimize neutralization of the vector and intensify the response against PspA. In addition, this would add protection against type 3 hemagglutinin to the vaccine, as observed in vaccines against seasonal influenza. However, this immunization regimen would not result in “universal protection” against the seasonal influenza virus, as the immunogenicity of influenza vaccines is based on the recognition of the highly variable portions of hemagglutinin and neuraminidase, which are in constant evolution and must be updated annually. A partial homologous protection against the circulating Influenza A strains could be achieved by this mixed H1-PspA/H3-PspA protocol.
In summary, our results highlight the potential use of the recombinant Flu-PspA virus associated with recombinant PspA4 protein in heterologous prime-boost protocols (Prime: Flu-PspA; boost: recombinant protein) as a tool for the development of new vaccines against infections caused by S. pneumoniae and influenza A.
Supplementary information
Acknowledgements
This study was financially supported by Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Coordenadoria de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). R.T.G, R.C.R. and M.S.S.A. are grateful to CNPq for their research fellowship. Our thanks to the Program for Technological Development in Tools for Health-RPT-FIOCRUZ for the use of their flow cytometry, confocal microscopy and sequencing facilities.
Author contributions
K.F.C., A.V. M., E. N. M. and M.S.S.A. conceived and designed the research protocol. A.V.M. was responsible for acquiring the financing. K.F.C., L.R.A.S., B.S.A.S.S., K. R. A. C., S.G.S.M., A.P.F.G., N.R.S.M., F.S.K., A.V.M. and M.S.S.A. conducted the experiments and acquired data. K.F.C., P.A.A., M.A.S.C., M.P.X., C.C.G., R.C.R., R.T.G., E.A.C., E.N.M., A.V.M. and M.S.S.A. analyzed and interpreted the data. K.F.C., C.C.G, A.V.M. and M.S.S.A. wrote and reviewed the manuscript. All authors read, reviewed and approved the final version of the manuscript.
Data availability
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Competing interests
None of the authors have a financial or personal relationship with others or organizations that could inappropriately influence or bias the paper’s content. K.F.C., A.V.M., M.S. S.A., K.R.A., B.S. A. S.S., S.G.S., F.S.K., P.A.A., A.P.F.G., M.A. P.X., E.A.C., N. R. S.M., R.T.G. and E.N.M. are co-inventors of the potential vaccine evaluated in this study. The application number of the patent evaluation process is BR1020220151636.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41541-024-01033-5.
References
- 1.Naucler, P., Darenberg, J., Morfeldt, E., Örtqvist, Å. & Henriques Normark, B. Contribution of host, bacterial factors and antibiotic treatment to mortality in adult patients with bacteraemic pneumococcal pneumonia. Thorax68, 571 LP–571579 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Cartwright, K. Pneumococcal disease in western Europe: burden of disease, antibiotic resistance and management. Eur. J. Pediatr.161, 188–195 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Troeger, C. et al. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis.18, 1191–1210 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peyrani, P., Mandell, L., Torres, A. & Tillotson, G. S. The burden of community-acquired bacterial pneumonia in the era of antibiotic resistance. Expert Rev. Respir. Med13, 139–152 (2019). [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization. Pneumonia in children. https://www.who.int/news-room/fact-sheets/detail/pneumonia (2022).
- 6.Ganaie, F. et al. A New Pneumococcal Capsule Type, 10D, is the 100th Serotype and Has a Large cps Fragment from an Oral Streptococcus. mBio11, e00937–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasparini, R., Amicizia, D., Lai, P. L. & Panatto, D. Clinical and socioeconomic impact of seasonal and pandemic influenza in adults and the elderly. Hum. Vaccin Immunother.8, 21–28 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Bouvier, N. M. & Palese, P. The biology of influenza viruses. Vaccine26, D49–D53 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knipe, D. M. & Howley, P. Fields Virology. (Wolters Kluwer Health, 2013).
- 10.Paget, J. et al. Global mortality associated with seasonal influenza epidemics: New burden estimates and predictors from the GLaMOR Project. J. Glob. Health9, 20421 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iuliano, A. D. et al. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet391, 1285–1300 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taubenberger, J. K. & Morens, D. M. The 1918 Influenza Pandemic and Its Legacy. Cold Spring Harb. Perspect. Med.10, a038695 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hussell, T., Wissinger, E. & Goulding, J. Bacterial complications during pandemic influenza infection. Future Microbiol4, 269–272 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Metersky, M. L., Masterton, R. G., Lode, H., File, T. M. Jr & Babinchak, T. Epidemiology, microbiology, and treatment considerations for bacterial pneumonia complicating influenza. Int. J. Infect. Dis.16, e321–e331 (2012). [DOI] [PubMed] [Google Scholar]
- 15.MacIntyre, C. R. et al. The role of pneumonia and secondary bacterial infection in fatal and serious outcomes of pandemic influenza a(H1N1)pdm09. BMC Infect. Dis.18, 637 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morens, D. M., Taubenberger, J. K. & Fauci, A. S. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: Implications for pandemic influenza preparedness. J. Infect. Dis.198, 962–970 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sender, V., Hentrich, K. & Henriques-Normark, B. Virus-induced changes of the respiratory tract environment promote secondary infections with streptococcus pneumoniae. Front Cell Infect. Microbiol11, 199 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sparrow, E. et al. Global production capacity of seasonal and pandemic influenza vaccines in 2019. Vaccine39, 512–520 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reperant, L. A., Rimmelzwaan, G. F. & Osterhaus, A. D. M. E. Advances in influenza vaccination. F1000Prime Rep.6, 47 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soema, P. C., Kompier, R., Amorij, J.-P. & Kersten, G. F. A. Current and next generation influenza vaccines: Formulation and production strategies. Eur. J. Pharm. Biopharm.94, 251–263 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Houser, K. & Subbarao, K. Influenza vaccines: Challenges and solutions. Cell Host Microbe17, 295–300 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ping, J. et al. Development of high-yield influenza A virus vaccine viruses. Nat. Commun.6, 8148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krammer, F. et al. Influenza. Nat. Rev. Dis. Prim.4, 3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki, M. et al. Serotype-specific effectiveness of 23-valent pneumococcal polysaccharide vaccine against pneumococcal pneumonia in adults aged 65 years or older: a multicentre, prospective, test-negative design study. Lancet Infect. Dis.17, 313–321 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Kong, Y. et al. Immunogenicity and safety of a 23-valent pneumococcal polysaccharide vaccine in Chinese healthy population aged >2 years: A randomized, double-blinded, active control, phase III trial. Hum. Vaccin Immunother.11, 2425–2433 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu, T. et al. Incidence of invasive pneumococcal disease in children with commercial insurance or Medicaid coverage in the United States before and after the introduction of 7- and 13-valent pneumococcal conjugate vaccines during 1998–2018. BMC Public Health22, 1677 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andrews, N. J. et al. Serotype-specific effectiveness and correlates of protection for the 13-valent pneumococcal conjugate vaccine: a postlicensure indirect cohort study. Lancet Infect. Dis.14, 839–846 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Du, Q., Shi, W., Yu, D. & Yao, K. Epidemiology of non-vaccine serotypes of Streptococcus pneumoniae before and after universal administration of pneumococcal conjugate vaccines. Hum. Vaccin Immunother.17, 5628–5637 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunne, E. M. et al. Effect of pneumococcal vaccination on nasopharyngeal carriage of Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, and Staphylococcus aureus in Fijian children. J. Clin. Microbiol50, 1034–1038 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonten, M. J. M. et al. Polysaccharide conjugate vaccine against pneumococcal pneumonia in adults. N. Engl. J. Med.372, 1114–1125 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Moffitt, K. & Malley, R. Rationale and prospects for novel pneumococcal vaccines. Hum. Vaccin Immunother.12, 383–392 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moffitt, K. L. & Malley, R. Next generation pneumococcal vaccines. Curr. Opin. Immunol.23, 407–413 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darrieux, M., Goulart, C., Briles, D. & Leite, L. CdeC. Current status and perspectives on protein-based pneumococcal vaccines. Crit. Rev. Microbiol41, 190–200 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Malley, R. & Anderson, P. W. Serotype-independent pneumococcal experimental vaccines that induce cellular as well as humoral immunity. Proc. Natl Acad. Sci.109, 3623–3627 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pichichero, M. E., Khan, M. N. & Xu, Q. Next generation protein based Streptococcus pneumoniae vaccines. Hum. Vaccin Immunother.12, 194–205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alghamdi, S. et al. Pneumococcal surface protein A: A promising candidate for the next generation of pneumococcal vaccines. Cell Mol. Biol.67, 289–298 (2022). [DOI] [PubMed] [Google Scholar]
- 37.Crain, M. J. et al. Pneumococcal surface protein A (PspA) is serologically highly variable and is expressed by all clinically important capsular serotypes of Streptococcus pneumoniae. Infect. Immun.58, 3293–3299 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Croney, C. M., Coats, M. T., Nahm, M. H., Briles, D. E. & Crain, M. J. PspA family distribution, unlike capsular serotype, remains unaltered following introduction of the heptavalent pneumococcal conjugate vaccine. Clin. Vaccin. Immunol.19, 891–896 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orihuela, C. J. et al. Microarray analysis of pneumococcal gene expression during invasive disease. Infect. Immun.72, 5582–5596 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Talkington, D. F., Crimmins, D. L., Voellinger, D. C., Yother, J. & Briles, D. E. A 43-kilodalton pneumococcal surface protein, PspA: isolation, protective abilities, and structural analysis of the amino-terminal sequence. Infect. Immun.59, 1285–1289 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brandileone, M. C. C. et al. Typing of pneumococcal surface protein A (PspA) in Streptococcus pneumoniae isolated during epidemiological surveillance in Brazil: towards novel pneumococcal protein vaccines. Vaccine22, 3890–3896 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Bernard, B., Giovanni, G., R, F. R. & K, H. S. Pneumococcal pspA Sequence Types of Prevalent Multiresistant Pneumococcal Strains in the United States and of Internationally Disseminated Clones. J. Clin. Microbiol38, 3663–3669 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qian, J. et al. Diversity of pneumococcal surface protein A (PspA) and relation to sequence typing in Streptococcus pneumoniae causing invasive disease in Chinese children. Eur. J. Clin. Microbiol. Infect. Dis.31, 217–223 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Hollingshead, S. K., Becker, R. & Briles, D. E. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect. Immun.68, 5889–5900 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang, B. et al. Distribution and Variation of Serotypes and Pneumococcal Surface Protein A Clades of Streptococcus pneumoniae Strains Isolated From Adult Patients With Invasive Pneumococcal Disease in Japan. Front Cell Infect. Microbiol11, 617573 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang, H. et al. PspA diversity, serotype distribution and antimicrobial resistance of invasive pneumococcal isolates from paediatric patients in Shenzhen, China. Infect. Drug Resist14, 49–58 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moreno, A. T. et al. Immunization of mice with single PspA fragments induces antibodies capable of mediating complement deposition on different pneumococcal strains and cross-protection. Clin. Vaccin. Immunol.17, 439–446 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nabors, G. S. et al. Immunization of healthy adults with a single recombinant pneumococcal surface protein A (PspA) variant stimulates broadly cross-reactive antibodies to heterologous PspA molecules. Vaccine18, 1743–1754 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Lane, J. R., Tata, M., Briles, D. E. & Orihuela, C. J. A jack of all trades: The role of pneumococcal surface protein A in the pathogenesis of streptococcus pneumoniae. Front Cell Infect. Microbiol12, 826264 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren, B., Szalai, A. J., Thomas, O., Hollingshead, S. K. & Briles, D. E. Both family 1 and family 2 PspA proteins can inhibit complement deposition and confer virulence to a capsular serotype 3 strain of streptococcus pneumoniae. Infect. Immun.71, 75–85 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tu, A. H., Fulgham, R. L., McCrory, M. A., Briles, D. E. & Szalai, A. J. Pneumococcal surface protein A inhibits complement activation by Streptococcus pneumoniae. Infect. Immun.67, 4720–4724 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mukerji, R. et al. Pneumococcal surface protein A inhibits complement deposition on the pneumococcal surface by competing with the binding of C-reactive protein to cell-surface phosphocholine. J. Immunol.189, 5327–5335 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaper, M., Hollingshead, S. K., Benjamin, W. H. Jr & Briles, D. E. PspA protects Streptococcus pneumoniae from killing by apolactoferrin, and antibody to PspA enhances killing of pneumococci by apolactoferrin [corrected]. Infect. Immun.72, 5031–5040 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Håkansson, A. et al. Characterization of binding of human lactoferrin to pneumococcal surface protein A. Infect. Immun.69, 3372–3381 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martinez, P. J. et al. PspA facilitates evasion of pneumococci from bactericidal activity of neutrophil extracellular traps (NETs). Micro. Pathog.136, 103653 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Briles, D. E. et al. Immunizations with pneumococcal surface protein A and pneumolysin are protective against pneumonia in a murine model of pulmonary infection with streptococcus pneumoniae. J. Infect. Dis.188, 339–348 (2003). [DOI] [PubMed] [Google Scholar]
- 57.BRILES, D. E. et al. PspA and PspC: Their potential for use as pneumococcal vaccines. Microb. Drug Resistance3, 401–408 (1997). [DOI] [PubMed] [Google Scholar]
- 58.Wu, H.-Y., Nahm, M. H., Guo, Y., Russell, M. W. & Briles, D. E. Intranasal immunization of mice with PspA (pneumococcal surface protein A) can prevent intranasal carriage, pulmonary infection, and sepsis with streptococcus pneumoniae. J. Infect. Dis.175, 839–846 (1997). [DOI] [PubMed] [Google Scholar]
- 59.Ferreira, D. M. et al. Characterization of protective mucosal and systemic immune responses elicited by pneumococcal surface protein PspA and PspC nasal vaccines against a respiratory pneumococcal challenge in mice. Clin. Vaccin. Immunol.16, 636–645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Briles, D. E. et al. Intranasal immunization of mice with a mixture of the pneumococcal proteins PsaA and PspA is highly protective against nasopharyngeal carriage of streptococcus pneumoniae. Infect. Immun.68, 796–800 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamamoto, M. et al. Oral immunization with PspA elicits protective humoral immunity against Streptococcus pneumoniae infection. Infect. Immun.65, 640 LP–640644 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miyaji, E. N. et al. Evaluation of a vaccine formulation against streptococcus pneumoniae based on choline-binding proteins. Clin. Vaccin. Immunol.22, 213–220 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scott, N. R., Mann, B., Tuomanen, E. I. & Orihuela, C. J. Multi-valent protein hybrid pneumococcal vaccines: A strategy for the next generation of vaccines. Vaccines (Basel)9, 209 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu, J. et al. Comparison of immunogenicity and protection of two pneumococcal protein vaccines based on PsaA and PspA. Infect. Immun.86, e00916-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.King, Q. O., Lei, B. & Harmsen, A. G. Pneumococcal surface protein A contributes to secondary Streptococcus pneumoniae infection after influenza virus infection. J. Infect. Dis.200, 537–545 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Briles, D. E. et al. Immunization of humans with recombinant pneumococcal surface protein A (rPspA) elicits antibodies that passively protect mice from fatal infection with streptococcus pneumoniae bearing heterologous PspA. J. Infect. Dis.182, 1694–1701 (2000). [DOI] [PubMed] [Google Scholar]
- 67.Darrieux, M. et al. Recognition of pneumococcal isolates by antisera raised against PspA fragments from different clades. J. Med Microbiol57, 273–278 (2008). [DOI] [PubMed] [Google Scholar]
- 68.Wit, Ede et al. Efficient generation and growth of influenza virus A/PR/8/34 from eight cDNA fragments. Virus Res.103, 155–161 (2004). [DOI] [PubMed] [Google Scholar]
- 69.Barthelemy, A. et al. Influenza A virus-induced release of interleukin-10 inhibits the anti-microbial activities of invariant natural killer T cells during invasive pneumococcal superinfection. Mucosal Immunol.10, 460–469 (2017). [DOI] [PubMed] [Google Scholar]
- 70.Machado, A. V., Naffakh, N., Werf, Svander & Escriou, N. Expression of a foreign gene by stable recombinant influenza viruses harboring a dicistronic genomic segment with an internal promoter. Virology313, 235–249 (2003). [DOI] [PubMed] [Google Scholar]
- 71.Vieira Machado, A., Naffakh, N., Gerbaud, S., van der Werf, S. & Escriou, N. Recombinant influenza A viruses harboring optimized dicistronic NA segment with an extended native 5′ terminal sequence: Induction of heterospecific B and T cell responses in mice. Virology345, 73–87 (2006). [DOI] [PubMed] [Google Scholar]
- 72.Barbosa, R. P. A. et al. Protective immunity and safety of a genetically modified influenza virus vaccine. PLoS One9, e98685 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hoffmann, E., Neumann, G., Hobom, G., Webster, R. G. & Kawaoka, Y. “Ambisense” approach for the generation of influenza A virus: vRNA and mRNA synthesis from one template. Virology267, 310–317 (2000). [DOI] [PubMed] [Google Scholar]
- 74.Machado, A. V. et al. Prime and boost immunization with influenza and adenovirus encoding the Toxoplasma gondii surface antigen 2 (SAG2) induces strong protective immunity. Vaccine28, 3247–3256 (2010). [DOI] [PubMed] [Google Scholar]
- 75.Barbosa, R. P. A. et al. Vaccination Using Recombinants Influenza and Adenoviruses Encoding Amastigote Surface Protein-2 Are Highly Effective on Protection against Trypanosoma cruzi Infection. PLoS One8, e61795 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Castro Martin, I. F. et al. Influenza virus genome reaches the plasma membrane via a modified endoplasmic reticulum and Rab11-dependent vesicles. Nat. Commun.8, 1396 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aida, Y. & Pabst, M. J. Removal of endotoxin from protein solutions by phase separation using Triton X-114. J. Immunol. Methods132, 191–195 (1990). [DOI] [PubMed] [Google Scholar]
- 78.Reed, L. J. & Muench, H. A simple method of estimating fifty per cent endpoints12. Am. J. Epidemiol.27, 493–497 (1938). [Google Scholar]
- 79.Principi, N. & Esposito, S. Development of pneumococcal vaccines over the last 10 years. Expert Opin. Biol. Ther.18, 7–17 (2018). [DOI] [PubMed] [Google Scholar]
- 80.Mestrovic, T. et al. The burden of bacterial antimicrobial resistance in the WHO European region in 2019: a cross-country systematic analysis. Lancet Public Health7, e897–e913 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Duke, J. A. & Avci, F. Y. Emerging vaccine strategies against the incessant pneumococcal disease. NPJ Vaccines8, 122 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rajão, D. S. & Pérez, D. R. Universal vaccines and vaccine platforms to protect against influenza viruses in humans and agriculture. Front Microbiol9, 123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nogales, A. & Martínez-Sobrido, L. Reverse genetics approaches for the development of influenza vaccines. Int. J. Mol. Sci.18, 20 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wong, S.-S. & Webby, R. J. Traditional and new influenza vaccines. Clin. Microbiol Rev.26, 476–492 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wagner, R., Matrosovich, M. & Klenk, H.-D. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev. Med Virol.12, 159–166 (2002). [DOI] [PubMed] [Google Scholar]
- 86.Bennasroune, A. et al. Elastic fibers and elastin receptor complex: Neuraminidase-1 takes the center stage. Matrix Biol.84, 57–67 (2019). [DOI] [PubMed] [Google Scholar]
- 87.Hinek, A., Pshezhetsky, A. V., von Itzstein, M. & Starcher, B. Lysosomal sialidase (neuraminidase-1) is targeted to the cell surface in a multiprotein complex that facilitates elastic fiber assembly. J. Biol. Chem.281, 3698–3710 (2006). [DOI] [PubMed] [Google Scholar]
- 88.Bandell, A., Ambrose, C. S., Maniaci, J. & Wojtczak, H. Safety of live attenuated influenza vaccine (LAIV) in children and adults with asthma: a systematic literature review and narrative synthesis. Expert Rev. Vaccines20, 717–728 (2021). [DOI] [PubMed] [Google Scholar]
- 89.Tosh, P. K., Boyce, T. G. & Poland, G. A. Flu myths: Dispelling the myths associated with live attenuated influenza vaccine. Mayo Clin. Proc.83, 77–84 (2008). [DOI] [PubMed] [Google Scholar]
- 90.Carter, N. J. & Curran, M. P. Live attenuated influenza vaccine (FluMist®; FluenzTM). Drugs71, 1591–1622 (2011). [DOI] [PubMed] [Google Scholar]
- 91.Duffy, J. et al. Live attenuated influenza vaccine use and safety in children and adults with asthma. Ann. Allergy, Asthma Immunol.118, 439–444 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rose, M. A. et al. Effectiveness, tolerability and patient satisfaction of paediatric live-attenuated influenza immunization (LAIV) in routine-care in Germany: A case-control-study. Trials Vaccinol.2, 49–52 (2013). [Google Scholar]
- 93.Martina, B. E. E. et al. A recombinant influenza A Virus Expressing Domain III of West Nile Virus Induces Protective Immune Responses against Influenza and West Nile Virus. PLoS One6, e18995 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.de Goede, A. L. et al. Characterization of recombinant influenza A virus as a vector for HIV-1 p17Gag. Vaccine27, 5735–5739 (2009). [DOI] [PubMed] [Google Scholar]
- 95.Shinya, K., Fujii, Y., Ito, H., Ito, T. & Kawaoka, Y. Characterization of a neuraminidase-deficient influenza a virus as a potential gene delivery vector and a live vaccine. J. Virol.78, 3083–3088 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Camberlein, E. et al. Importance of bacterial replication and alveolar macrophage-independent clearance mechanisms during early lung infection with Streptococcus pneumoniae. Infect. Immun.83, 1181–1189 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smith, N. M. et al. Regionally compartmentalized resident memory T cells mediate naturally acquired protection against pneumococcal pneumonia. Mucosal Immunol.11, 220–235 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wilson, R. et al. Protection against Streptococcus pneumoniae lung infection after nasopharyngeal colonization requires both humoral and cellular immune responses. Mucosal Immunol.8, 627 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sasaki, E. et al. Nasal alum-adjuvanted vaccine promotes IL-33 release from alveolar epithelial cells that elicits IgA production via type 2 immune responses. PLoS Pathog.17, e1009890- (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xu, H. et al. Nasal aluminum (oxy)hydroxide enables adsorbed antigens to induce specific systemic and mucosal immune responses. Hum. Vaccin Immunother.13, 2688–2694 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Thakkar, S. G. et al. Intranasal immunization with aluminum salt-adjuvanted dry powder vaccine. J. Controlled Release292, 111–118 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen, L. et al. An intranasal vaccine targeting the receptor binding domain of SARS-CoV-2 elicits a protective immune response. Front Immunol.13, 1005321 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Machin, D., Cheung, Y. B. & Parmar, M. Survival Analysis: A Practical Approach. (Wiley, 2006).
- 104.Pintilie, M. Competing Risks: A Practical Perspective. (Wiley, New York, NY, USA, 2006).
- 105.Joiner, K., Brown, E., Hammer, C., Warren, K. & Frank, M. Studies on the mechanism of bacterial resistance to complement-mediated killing. III. C5b-9 deposits stably on rough and type 7 S. pneumoniae without causing bacterial killing. J. Immunol.130, 845–849 (1983). [PubMed] [Google Scholar]
- 106.Vitharsson, G., Jonsdottir, I., Jonsson, S. & Valdimarsson, H. Opsonization and antibodies to capsular and cell wall polysaccharides of Streptococcus pneumoniae. J. Infect. Dis.170, 592–599 (1994). [DOI] [PubMed] [Google Scholar]
- 107.Paterson, G. K. & Orihuela, C. J. Pneumococci: immunology of the innate host response. Respirology15, 1057–1063 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ren, B., Szalai, A. J., Hollingshead, S. K. & Briles, D. E. Effects of PspA and Antibodies to PspA on activation and deposition of complement on the pneumococcal surface. Infect. Immun.72, 114–122 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Snapper, C. M. & Mond, J. J. Towards a comprehensive view of immunoglobulin class switching. Immunol. Today14, 15–17 (1993). [DOI] [PubMed] [Google Scholar]
- 110.Finkelman, F. D. et al. Lymphokine control of in vivo immunoglobulin isotype selection. Annu Rev. Immunol.8, 303–333 (1990). [DOI] [PubMed] [Google Scholar]
- 111.Vadesilho, C. F. M. et al. Characterization of the antibody response elicited by immunization with pneumococcal surface protein A (PspA) as recombinant protein or DNA vaccine and analysis of protection against an intranasal lethal challenge with Streptococcus pneumoniae. Micro. Pathog.53, 243–249 (2012). [DOI] [PubMed] [Google Scholar]
- 112.Hanniffy, S. B., Carter, A. T., Hitchin, E. & Wells, J. M. Mucosal delivery of a pneumococcal vaccine using lactococcus lactis affords protection against respiratory infection. J. Infect. Dis.195, 185–193 (2007). [DOI] [PubMed] [Google Scholar]
- 113.Rodrigues, T. C. et al. Mucosal immunization with PspA (Pneumococcal surface protein A)-adsorbed nanoparticles targeting the lungs for protection against pneumococcal infection. PLoS One13, e0191692 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Castro, J. T. et al. Evaluation of inactivated Bordetella pertussis as a delivery system for the immunization of mice with Pneumococcal Surface Antigen A. PLoS One15, e0228055 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bordon, J. et al. Understanding the roles of cytokines and neutrophil activity and neutrophil apoptosis in the protective versus deleterious inflammatory response in pneumonia. Int. J. Infect. Dis.17, e76–e83 (2013). [DOI] [PubMed] [Google Scholar]
- 116.Wood, B. L. & Levin, G. R. Interactions between mouse IgG2 antibodies are common and mediated by plasma C1q. Cytom. B Clin. Cytom.70B, 321–328 (2006). [DOI] [PubMed] [Google Scholar]
- 117.Leatherbarrow, R. J. & Dwek, R. A. Binding of complement subcomponent Clq to mouse IgGl, IgG2a AND IgG2b: A novel Clq binding assay. Mol. Immunol.21, 321–327 (1984). [DOI] [PubMed] [Google Scholar]
- 118.Aschermann, S., Lux, A., Baerenwaldt, A., Biburger, M. & Nimmerjahn, F. The other side of immunoglobulin G: suppressor of inflammation. Clin. Exp. Immunol.160, 161–167 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lilienthal, G.-M. et al. Potential of murine IgG1 and human IgG4 to inhibit the classical complement and Fcγ receptor activation pathways. Front Immunol.9, 958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Melis, J. P. M. et al. Complement in therapy and disease: Regulating the complement system with antibody-based therapeutics. Mol. Immunol.67, 117–130 (2015). [DOI] [PubMed] [Google Scholar]
- 121.Arulanandam, B. P., Lynch, J. M., Briles, D. E., Hollingshead, S. & Metzger, D. W. Intranasal vaccination with pneumococcal surface protein A and interleukin-12 augments antibody-mediated opsonization and protective immunity against Streptococcus pneumoniae infection. Infect. Immun.69, 6718–6724 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Neuberger, M. S. & Rajewsky, K. Activation of mouse complement by monoclonal mouse antibodies. Eur. J. Immunol.11, 1012–1016 (1981). [DOI] [PubMed] [Google Scholar]
- 123.Oishi, K., Koles, N. L., Guelde, G. & Pollack, M. Antibacterial and protective properties of monoclonal antibodies reactive with Escherichia coli O111:B4 lipopolysaccharide: relation to antibody isotype and complement-fixing activity. J. Infect. Dis.165, 34–45 (1992). [DOI] [PubMed] [Google Scholar]
- 124.Brown, E. J., Hosea, S. W. & Frank, M. M. The role of antibody and complement in the reticuloendothelial clearance of pneumococci from the bloodstream. Rev. Infect. Dis.5, S797–S805 (1983). [DOI] [PubMed] [Google Scholar]
- 125.Brown, J. S. et al. The classical pathway is the dominant complement pathway required for innate immunity to Streptococcus pneumoniae infection in mice. Proc. Natl Acad. Sci.99, 16969–16974 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.McDaniel, L. S., Sheffield, J. S., Delucchi, P. & Briles, D. E. PspA, a surface protein of Streptococcus pneumoniae, is capable of eliciting protection against pneumococci of more than one capsular type. Infect. Immun.59, 222–228 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McCool, T. L., Cate, T. R., Moy, G. & Weiser, J. N. the immune response to pneumococcal proteins during experimental human carriage. J. Exp. Med.195, 359–365 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bitsaktsis, C. et al. Mucosal immunization with an unadjuvanted vaccine that targets streptococcus pneumoniae PspA to human Fcγ receptor type I protects against pneumococcal infection through complement- and lactoferrin-mediated bactericidal activity. Infect. Immun.80, 1166–1180 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ren, B., Li, J., Genschmer, K., Hollingshead, S. K. & Briles, D. E. The absence of PspA or presence of antibody to PspA facilitates the complement-dependent phagocytosis of pneumococci in vitro. Clin. Vaccin. Immunol.19, 1574–1582 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Briles, D. E. et al. The potential to use PspA and other pneumococcal proteins to elicit protection against pneumococcal infection. Vaccine18, 1707–1711 (2000). [DOI] [PubMed] [Google Scholar]
- 131.Ferreira, D. M. et al. Protection against nasal colonization with Streptococcus pneumoniae by parenteral immunization with a DNA vaccine encoding PspA (Pneumococcal surface protein A). Micro. Pathog.48, 205–213 (2010). [DOI] [PubMed] [Google Scholar]
- 132.Oma, K. et al. Intranasal immunization with a mixture of PspA and a Toll-like receptor agonist induces specific antibodies and enhances bacterial clearance in the airways of mice. Vaccine27, 3181–3188 (2009). [DOI] [PubMed] [Google Scholar]
- 133.Campos, I. B. et al. Nasal immunization of mice with Lactobacillus casei expressing the Pneumococcal Surface Protein A: induction of antibodies, complement deposition and partial protection against Streptococcus pneumoniae challenge. Microbes Infect.10, 481–488 (2008). [DOI] [PubMed] [Google Scholar]
- 134.Xin, W., Li, Y., Mo, H., Roland, K. L. & Curtiss, R. PspA Family Fusion Proteins Delivered by Attenuated <em>Salmonella enterica</em> Serovar Typhimurium Extend and Enhance Protection against <em>Streptococcus pneumoniae</em>. Infect. Immun.77, 4518 LP–4514528 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Brandenburg, B. et al. Mechanisms of hemagglutinin targeted influenza virus neutralization. PLoS One8, e80034 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rimmelzwaan, G. F., Fouchier, R. A. M. & Osterhaus, A. D. M. E. Influenza virus-specific cytotoxic T lymphocytes: a correlate of protection and a basis for vaccine development. Curr. Opin. Biotechnol.18, 529–536 (2007). [DOI] [PubMed] [Google Scholar]
- 137.Sridhar, S. et al. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat. Med. 19, 1305–1312 (2013). [DOI] [PubMed] [Google Scholar]
- 138.Dong, W. et al. Cross-protective immune responses induced by sequential influenza virus infection and by sequential vaccination with inactivated influenza vaccines. Front Immunol.9, 2312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kirchenbaum, G. A., Carter, D. M. & Ross, T. M. Sequential infection in ferrets with antigenically distinct seasonal H1N1 influenza viruses boosts hemagglutinin stalk-specific antibodies. J. Virol.90, 1116–1128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Katsura, H. et al. A bivalent vaccine based on a replication-incompetent influenza virus protects against streptococcus pneumoniae and influenza virus infection. J. Virol.88, 13410–13417 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Uraki, R. et al. A bivalent vaccine based on a PB2-knockout influenza virus protects mice from secondary pneumococcal pneumonia. J. Infect. Dis.212, 1939–1948 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pimenta, F. C. et al. Genetic diversity of PspA types among nasopharyngeal isolates collected during an ongoing surveillance study of children in Brazil. J. Clin. Microbiol44, 2838–2843 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Darrieux, M. et al. Fusion proteins containing family 1 and family 2 pspa fragments elicit protection against streptococcus pneumoniae that correlates with antibody-mediated enhancement of complement deposition. Infect. Immun.75, 5930–5938 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Roche, H., Ren, B., McDaniel, L. S., Håkansson, A. & Briles, D. E. Relative roles of genetic background and variation in pspa in the ability of antibodies to PspA to protect against capsular type 3 and 4 strains of <em>Streptococcus pneumoniae</em>. Infect. Immun.71, 4498 LP–4494505 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Milián, E. & Kamen, A. A. Current and emerging cell culture manufacturing technologies for influenza vaccines. Biomed. Res. Int. 2015, 504831 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.