Abstract
Cardiomyocyte proliferation in adult Xenopus tropicalis during heart regeneration has remained largely contentious due to the absence of genetic evidence. Here, we generated a transgenic reporter line Tg(mlc2:H2C) expressing mCherry specifically in cardiomyocyte nuclei driven by the promoter of myosin light chain 2 (mlc2). Using the reporter line, we found that traditional whole-cell staining is not a rigorous way to identify cardiomyocytes in adult Xenopus tropicalis when using a cryosection with common thickness (5 μm) which leading to a high error, but this deviation could be reduced by increasing section thickness. In addition, the reporter line confirmed that apex resection injury greatly increased the proliferation of mlc2+ cardiomyocytes at 3-30 days post-resection (dpr), thereby regenerating the lost cardiac muscle by 30 dpr in adult Xenopus tropicalis. Our findings from the reporter line have rigorously defined cardiomyocyte proliferation in adult heart upon injury, thereby contributing heart regeneration in adult Xenopus tropicalis.
Subject terms: Cardiac regeneration, Regenerative medicine, Mitosis, Experimental models of disease
Introduction
In the past several decades, it was widely believed that mammalian heart is a terminally differentiated organ, which is incapable of regenerating after injury during adult period. Indeed, cardiovascular disease continues to be the leading cause of morbidity and mortality worldwide, due to the inability to regenerate the injured adult heart after myocardial infarction1. Although recent studies revealed that limited myocyte turnover occurs in the adult mammalian heart, it is insufficient to restore contractile function after cardiac injury2,3. It has been demonstrated that the complete cardiac regeneration occurs in neonatal mouse heart during the first week of postnatal life, but this capacity markedly diminishes by postnatal day 7 (p7)4, suggesting the gradually losing of regenerative capacity during mammalian heart development and maturation. In contrast, lower vertebrates such as zebrafish5 and urodele amphibian6 retain a remarkable capacity to regenerate the injured heart in adulthood, implying a requirement for comparative studies across species to better understand the mechanism of adult heart regeneration. In other words, exploring the differences in regenerative potential between different species is particularly valuable to find the regulators of heart regeneration7. Therefore, other alternative and suitable vertebrate model organisms are needed and useful for elucidating the molecular mechanism of adult heart regeneration.
Recently, we reported that adult western clawed frog (Xenopus tropicalis), one of the anuran amphibian frogs having closer evolutionary distance with mammalians, can replace the resected myocardium after removal of their ventricular apexes8,9. These events involve cardiomyocyte proliferation as mostly demonstrated by whole-cell staining of cardiomyocytes together with proliferating marker staining8,9. However, the cardiomyocyte proliferation determined by traditional staining method in adult X. tropicalis remains controversial. In the injured hearts of regenerative models, there is robust proliferation of non-cardiomyocytes including endothelial cells, hematopoietic cells, and fibroblasts10. However, proliferating cardiomyocytes are always in the minority. Thus, it is difficult to distinguish an endocardial cell nucleus proliferating alongside the muscle when using traditional whole-cell staining of cardiomyocytes. On the other hand, the commercial and specific antibodies against Xenopus are very limited, which greatly hindering the studies of protein localization and function in Xenopus11. However, antibodies produced against mouse or human proteins often do not strictly recognize Xenopus counterparts, because there can be considerable sequence diversity between orthologous proteins in Xenopus and mammals12. Therefore, cardiomyocyte proliferation during heart regeneration in adult X. tropicalis remains largely contentious due to the absence of genetic evidence. Whether cardiomyocyte proliferation is really induced and its contribution to adult heart regeneration in X. tropicalis should be readdressed rigorously by transgenic reporter line.
Specific promoter that targets Xenopus adult cardiomyocytes is critical to the transgenic reporter line. Previous studies have demonstrated that myosin light chain 2 (mlc2) is cardiac-specific gene that expresses in entire myocardium including ventricular and atrial cardiomyocytes from embryo to adult stage in lower vertebrates including zebrafish13 and Xenopus14,15. To rigorously define cardiomyocyte proliferation during heart regeneration in adult X. tropicalis, we generated a transgenic reporter line Tg(mlc2:H2C) that is capable of monitoring cardiomyocyte nuclei. This reporter line is unbiased for studying cardiomyocyte proliferation, as it specifically labeling cardiomyocyte nuclei which can completely overlap with cell proliferating markers such as Ki67, proliferating cell nuclear antigen (PCNA), phospho-Histone H3 (pH3), as well as nuclear incorporation of 5-ethynyl-2’-deoxyuridine (EdU)10. We used this reporter line to rigorously define cardiomyocyte proliferation during heart regeneration in adult X. tropicalis. Our data provided, for the first time, a direct genetic evidence supporting that adult X. tropicalis can really regenerate the injured myocardium through cardiomyocyte proliferation.
Results
Generation and characterization of Tg(mlc2:H2C) reporter line
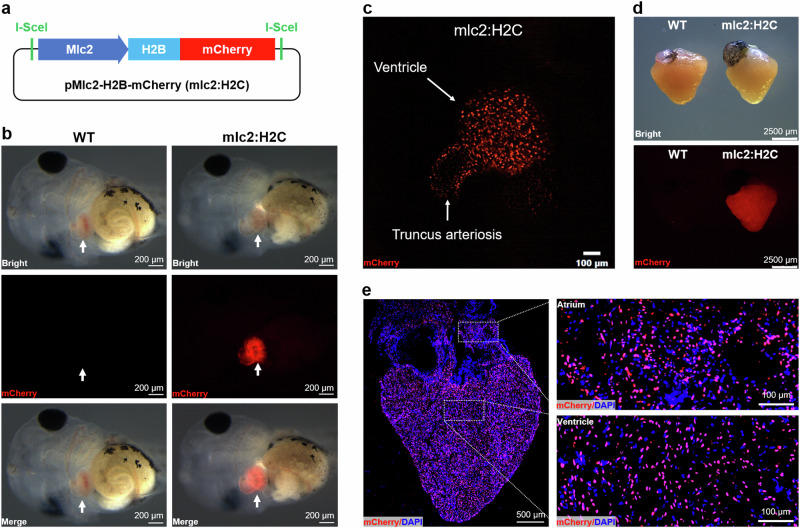
To label cardiomyocyte nucleus in X. tropicalis, we generated a transgenic plasmid (pMlc2-H2B-mCherry) that expresses the H2B-mCherry (H2C) fusion red fluorescent protein16 specifically in cardiomyocyte nuclei, which was driven by a 3 kb fragment of Xenopus mlc2 promoter14 (Fig. 1a). The F0 funders successfully passed their transgenes to the F1 generation, which were determined by cardiac-specific expression of mCherry in live tadpoles (Fig. 1b, Supplementary Video 1). Confocal microscope image (Fig. 1c) and video (Supplementary Video 2) revealed that the reporter gene mCherry specifically expressed in cardiomyocyte nuclei in F1 transgenic X. tropicalis tadpole. Transgene is also specifically expressed in heart of adult F1 X. tropicalis line (Fig. 1d). The cardiomyocyte nucleus-specific expression of mCherry reporter gene in ventricle and atrium was further determined in the adult F1 generation by confocal microscope (Fig. 1e). These data suggest that we successfully generated the heritable transgenic reporter line Tg(mlc2:H2C) which can be used to label all cardiomyocytes in the whole heart.
Fig. 1. Generation and characterization of Tg(mlc2:H2C) transgenic X. tropicalis line.
a Schematic representation of the transgenic plasmid pMlc2-H2B-mCherry which harboring the expression cassette of H2B-mCherry (H2C) fusion protein under control of Xenopus mlc2 promoter. b Whole-mount bright-field (upper), epifluorescence (middle), and merged (lower) images showing mCherry expression in Wild-type (WT, left) and F1 Tg(mlc2:H2C) (right) tadpoles. Arrows indicate heart location. c Representative image of the living heart in F1 Tg(mlc2:H2C) tadpole. Red color means mCherry expression specifically in cardiomyocyte nuclei. d Whole-mount bright-field (upper) and epifluorescence (lower) images showing mCherry expression in the adult hearts from WT (left) and F1 Tg(mlc2:H2C) (right) frogs. e Immunostained images for mCherry (red) expression in the adult heart from F1 Tg(mlc2:H2C) frog. DAPI was used as a nuclear stain (blue). Left panel, whole cardiac section in low-magnification. Right panels, high-magnification images of the atrial (upper) and ventricular (lower) regions white boxed in the whole cardiac section. This figure was created by Photoshop Image 12 software using our own data in the work.
To evaluate the impact of reporter gene on heart development and function in X. tropicalis, we compared the expression of cardiac development-related genes (tbx5, tbx20, tnni3, tnnt2, hand1, hand2, bmp4, fbrsl1) between wild-type and transgenic tadpoles. Our data showed that there were no significant differences in the expressions of these cardiac development-related genes in transgenic line compared with that in wild-type tadpoles (Supplementary Figure 1a, b). Moreover, heart growth and cardiac contraction function in adult frogs were comparable between wild-type and transgenic line animals (Supplementary Figure 1c, d). In addition, the transgenic line was survived and heritable. Taken together, these findings suggest that the continued expression of H2B-mCherry specifically in cardiomyocyte nuclei has no significant impacts on heart development and function in X. tropicalis.
Comparation of the accuracy of cardiomyocyte identification by traditional staining and genetic labeling in adult X. tropicalis
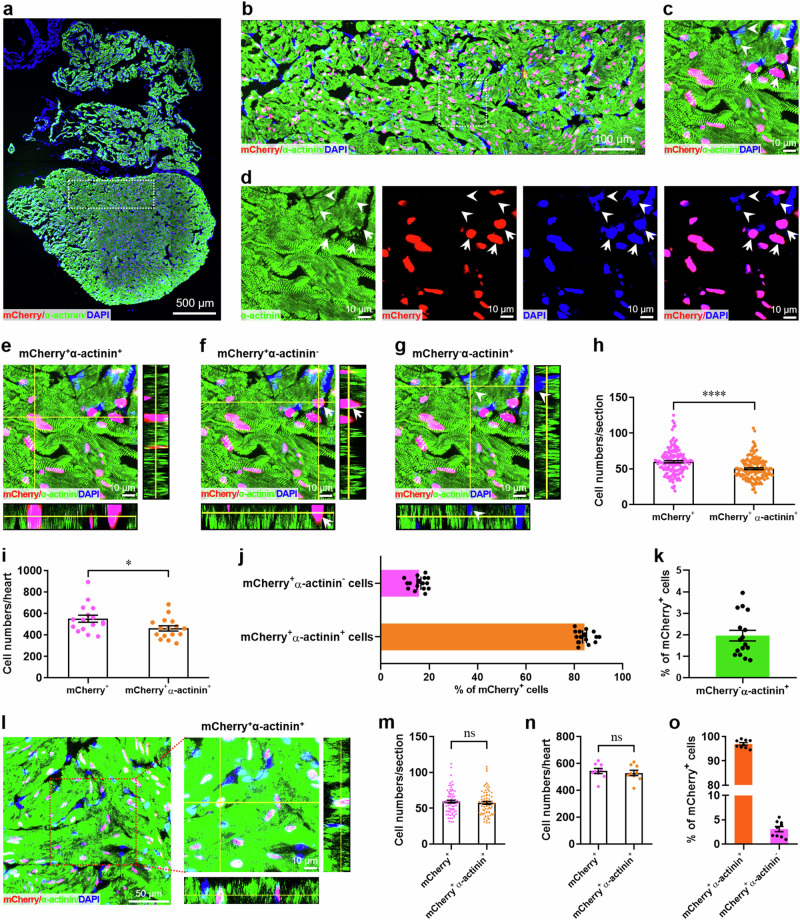
The popular method to determine cell proliferation is nuclear staining of cell cycle markers including Ki67, PCNA, pH3, as well as nuclear incorporation of EdU10. Although the ideal method for the evaluation of cardiomyocyte proliferation during heart regeneration is to label cardiomyocyte nuclei using antibody against Mef2c or PCM-117, some previous studies combined cell proliferating marker staining with whole-cell staining of cardiomyocytes using antibodies against cardiac contractile proteins such as cardiac troponin T (cTnT)4,18,19, α-actinin20–22, and myosin heavy chain (MHC)5,23,24. However, the whole-cell staining strategy is not rigorous because the proliferating non-cardiomyocyte nucleus alongside or overlapped with cardiac muscle cells may be included as proliferating cardiomyocytes. To evaluate the accuracy of traditional whole-cell staining of cardiomyocytes in adult X. tropicalis, the adult heart from F1 Tg(mlc2:H2C) line was firstly subjected to whole-cell staining of cardiomyocytes using the cryosection with common thickness (5 μm). Double staining of α-actinin (green) and mCherry (red) revealed the similar distribution patterns in the whole cardiac section (Fig. 2a). As expected, the high-magnification image of confocal microscopy showed that α-actinin expressed in whole cells, but mCherry expressed in nuclei (Fig. 2b). Although most mCherry-positive nuclei were completely and closely surrounded by α-actinin-positive cytoplasm (mCherry+α-actinin+), the mCherry+α-actinin- cell population (arrow) was detected, in which α-actinin-positive cytoplasm only partially surrounded the mCherry-positive nuclei. Moreover, some α-actinin-positive cells never expressed mCherry in nuclei (mCherry-α-actinin+, arrowhead) (Fig. 2c, d). The different spatial location of mCherry-positive nuclei and α-actinin-positive cytoplasm in these three cell populations were further confirmed by Z-stack confocal images (Fig. 2e–g). To quantify the ratio of these three cell populations, over 200 sections from sixteen adult hearts of Tg(mlc2:H2C) reporter line were examined. We found that the numbers of mCherry+α-actinin+ cardiomyocytes significantly lower than that of mCherry+ cardiomyocytes (Fig. 2h, i). Quantitatively, 84.23 ± 0.84% and 15.77 ± 0.84% cardiomyocytes were mCherry+α-actinin+ and mCherry+α-actinin- in Tg(mlc2:H2C) hearts, respectively (Fig. 2j). Moreover, 1.96 ± 0.25% cardiomyocytes were detected to be mCherry-α-actinin+ in in Tg(mlc2:H2C) hearts (Fig. 2k).
Fig. 2. Accuracy evaluation of whole-cell staining of cardiomyocytes in adult X. tropicalis.
a Immunostaining for mCherry (red) and α-actinin (green) expression in a whole cardiac section from the adult heart of F1 Tg(mlc2:H2C) frog using cryosections with common thickness (5 μm). DAPI was used as a nuclear stain (blue). b Magnified immunostaining image of the non-apical region white boxed in the whole cardiac section. c Magnified immunostaining image of the white boxed region in figure B showing that most mCherry-positive nuclei are completely and closely surrounded by α-actinin-positive cytoplasm (mCherry+α-actinin+). Arrow denotes the mCherry+α-actinin- cell in which α-actinin-positive cytoplasm only partially surrounds the mCherry-positive nucleus. Arrowhead denotes the α-actinin-positive cell which never expressed mCherry in nucleus (mCherry-α-actinin+ cell). d Single- and double-channel fluorescence images of figure C for α-actinin (green), mCherry (red), DAPI (blue), and mCherry/DAPI (pink) expression. e–g Representative Z-stack confocal images of mCherry+α-actinin+ (e), mCherry+α-actinin- (f), and mCherry-α-actinin+ (g) cells in figure c. h, i Quantification of mCherry+ and mCherry+α-actinin+ cell numbers in adult hearts of F1 Tg(mlc2:H2C) frogs. Data are presented as mean ± SEM (n = ~200 sections from 16 hearts for (h), n = 16 hearts for i). *p < 0.05, ****p < 0.0001 (Student’s t test). j Quantification of mCherry+α-actinin+ and mCherry+α-actinin- cells percentages in adult hearts of F1 Tg(mlc2:H2C) frogs (mean ± SEM, n = 16 hearts). k Quantification of mCherry-α-actinin+ cell percentage in adult hearts of F1 Tg(mlc2:H2C) frogs (mean ± SEM, n = 16 hearts). l–o Representative images (l) and quantification of mCherry+, mCherry+α-actinin+, and mCherry+α-actinin- cells (m–o) using thicker cryosections (10 μm). Data are presented as mean ± SEM (n = ~100 sections from 9 hearts for m, n = 9 hearts for n, o). Ns denotes no significant difference (Student’s t test).
The high number of mCherry+α-actinin- cells (15.77% of mCherry+ cells) might result from the misplacement of nucleus and cytoplasm of cardiomyocytes induced by cardiac sectioning due to the big size of myofibrils. To resolve the potential issue, we further performed this experiment using much thicker sections (10 μm). Indeed, there was no significant differences in the numbers of mCherry+α-actinin+ populations compared with that of mCherry+ cells, when used the thicker sections (Fig. 2l–n). Moreover, our new data using thicker sections showed an increased population of mCherry+α-actinin+ cells (96.92 ± 1.56% of total mCherry+ cells) and a decreased population of mCherry+α-actinin- cells (3.08 ± 1.55% of total mCherry+ cells) in the adult heart of Tg(mlc2:H2C) transgenic line (Fig. 2o), when compared with the thin sections (5 μm) (Fig. 2j). These findings reveal that the transgenic reporter line is really reliable to specifically identify cardiomyocytes. In addition, these data also imply that the accuracy of traditional whole-cell staining of cardiomyocytes is largely dependent on imaging depth.
Subsequently, the cryosection with common thickness (5 μm) of the adult heart from F1 Tg(mlc2:H2C) line was subjected to nuclear staining of cardiomyocytes using antibody against mammalian Mef2c. Nuclear double staining of Mef2c and mCherry showed the similar distribution patterns of Mef2C and mCherry in the whole cardiac section of X. tropicalis heart (Supplementary Figure 2a). The high-magnification image showed that all mCherry and most Mef2c were expressed in nuclei in cardiac section of F1 Tg(mlc2:H2C) line (Supplementary Figure 2b). Although most mCherry+ nuclei were co-labeled by Mef2c (mCherry+Mef2c+), many Mef2c signals were detected outside the cell nucleus (trigon) and in mCherry- nuclei (arrowhead), which indicating the non-specificity of Mef2c for cardiomyocyte nuclei in X. tropicalis (Supplementary Figure 2c, d). Z-stack confocal images further determined the individual spatial localization of mCherry+ and/or Mef2c+ nuclei in these different cell populations (Supplementary Figure 2e–g). Quantitatively, there was no significant differences between the numbers of mCherry+ and mCherry+Mef2c+ cells after examination of over 150 sections from eleven hearts of F1 Tg(mlc2:H2C) line (Supplementary Figure 2h, i). Moreover, the percentages of mCherry+Mef2c+ and mCherry+Mef2c- nuclei were 94.79 ± 0.62% and 5.21 ± 0.62% in Tg(mlc2:H2C) hearts, respectively (Supplementary Figure 2j). However, the percentage of mCherry+Mef2c+ cell population was significantly higher than that of mCherry+α-actinin+ cells (5 μm sections) in whole-cell staining of cardiomyocytes (Supplementary Figure 2k).
Transgenic reporter line confirms the newly forming myocardium in adult X. tropicalis hearts upon injury
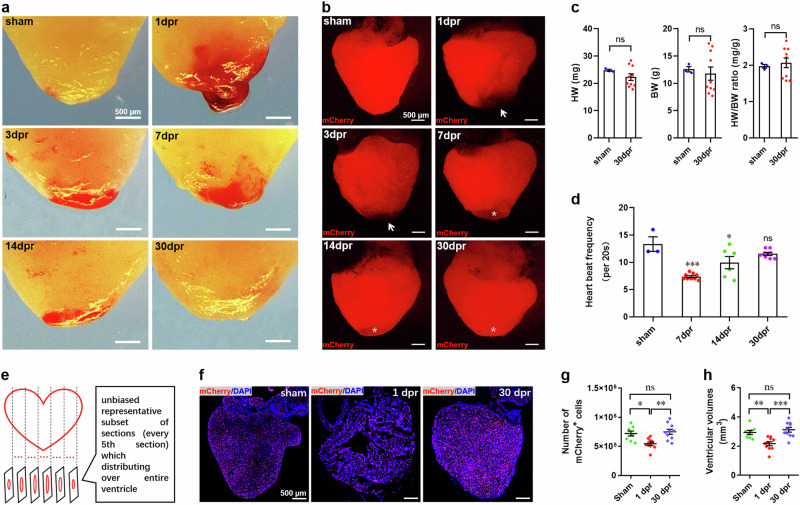
We next examined the restoration of ventricular myocardium in adult F1 Tg(mlc2:H2C) line upon apical resection injury. Adult transgenic frogs were subjected to apical resection injury as previously reported8,9. Morphological analysis of heart revealed the progressive restoration of the injured apex within 30 days post-resection (dpr). A large blood clot was firstly detected in the apex at 1 dpr. The gradual resorption of the apical blood clot and its replacement by normal myocardium were observed in the subsequent time points. By 30 dpr, the blood clot completely disappeared as determined by morphological analysis (Fig. 3a). Through the detection of mCherry fluorescence in Tg(mlc2:H2C) line, we found that mCherry was largely present in the adult ventricle. However, after resection of the ventricular apex, mlc2-driven mCherry was absent in the areas of removed apical tissues (arrow), which was gradually present along with apical blood clot resorption and myocardium restoration (asterisk) during a period from 0 to 30 dpr (Fig. 3b). These findings suggest that the adult Tg(mlc2:H2C) frog can restore the injured apex within 30 days, in the similar manner of the wild-type X. tropicalis as previously reported8,9. In consistent with these results, there was no significant difference in heart weight (HW), body weight (BW), and HW/BW ratio between the sham and the resected Tg(mlc2:H2C) frogs at 30 dpr (Fig. 3c). As expected, although the systolic properties of beating heart at 7 and 14 dpr were significantly lower than that in sham group, which was restored to a normal level at 30 dpr (Fig. 3d).
Fig. 3. Regeneration of ventricular myocardium of adult Tg(mlc2:H2C) transgenic X. tropicalis line.
a Representative bright-field images of adult hearts from F1 Tg(mlc2:H2C) frogs at the indicated time points after apical resection injury. b Representative epifluorescence images of adult hearts from F1 Tg(mlc2:H2C) frogs at the indicated time points after apical resection injury. Arrow marks the removed apex. Asterisk marks the restored apex. c Quantification of heart weight (HW), body weight (BW), and HW/BW ratio of frogs in sham-operated and 30 dpr groups. Data are presented as mean ± SEM (n = 3 ~ 10 frogs). Ns, no significant difference (Student’s t test). d Quantification of heart rate at 30 dpr indicating the restoration of normal systolic function of regenerated hearts. Data are presented as mean ± SEM (n = 3 ~ 10 frogs). *p < 0.05, ***p < 0.001 versus control, ns denotes no significant difference (one-way ANOVA test). e Schematic for quantification of ventricular cardiomyocyte numbers and ventricular volumes. f Representative images of adult hearts at the indicated time points after apical resection injury. g, h Quantification of ventricular cardiomyocyte numbers (g) and ventricular volumes (h) at the indicated time points. Data are presented as mean ± SEM (n = 8 ~ 10 hearts per group). *p < 0.05, **p < 0.01, ***p < 0.001 versus control, ns denotes no significant difference (one-way ANOVA test). This figure was created by Photoshop Image 12 software using our own data in the work.
To explore whether the restored apex results from the hypertrophy of cardiomyocytes located in the resection plane, we firstly assessed the size of cardiomyocytes in apex and revealed that there were no significant differences between sham and resected groups at 7 and 30 dpr (Supplementary Figure 3a, b). Moreover, no differences in the expression of hypertrophic genes in ventricles at 7 and 30 dpr compared with sham-operated group (Supplementary Figure 3c). These data imply that the restored apex might not result from cardiomyocyte hypertrophy. To further elucidate whether apex restoration is contributed by the newly formed cardiomyocytes, we subsequently quantified total cardiomyocyte numbers in the entire ventricles and ventricular volumes covered by mCherry-positive cells, using an unbiased stereological reconstruction method as described previously25 with some modification (Fig. 3e). We found that ventricular cardiomyocyte numbers were significantly lower at 1 dpr compared with sham-operated group, whereas resection injury-induced loss of cardiomyocytes were restored to the levels of pre-resection by 30 dpr (Fig. 3f, g). In line with cardiomyocyte numbers, apical resection-induced loss of ventricular volumes covered by mCherry-positive cells at 1 dpr were significantly restored by 30 dpr (Fig. 3h). Importantly, there were no significant differences in total cardiomyocyte numbers and ventricular volumes between 30 dpr and sham groups (Fig. 3g, h). Taken together, these findings from the reporter line genetically revealed the newly forming myocardium in adult X. tropicalis upon apical resection injury.
Transgenic reporter line verifies cardiomyocyte proliferation during heart regeneration in adult X. tropicalis
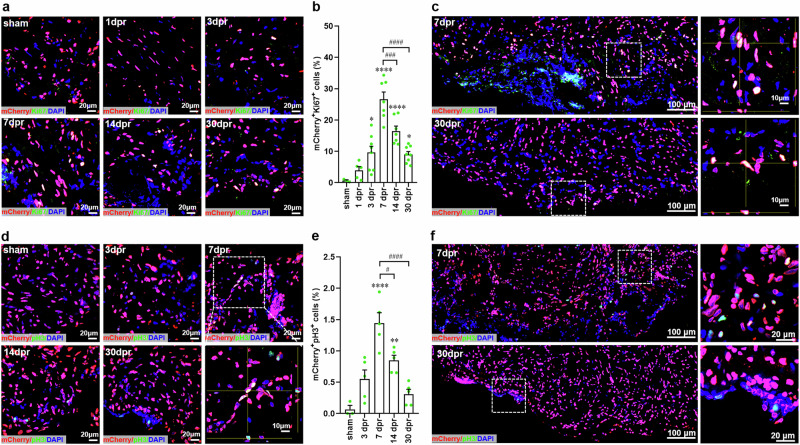
Using the whole-cell staining strategy, it has been reported that cardiomyocyte proliferation is vital to heart regeneration in adult X. tropicalis8,9. However, this conclusion might be contentious due to the defects of traditional whole-cell staining method and the non-specificity of antibodies. Thus, this potential issue should be clarified by more reliable genetic evidence. To further define the proliferation of cardiomyocytes during heart regeneration in adult X. tropicalis, we rigorously evaluated the proliferative capacity of cardiomyocytes using the Tg(mlc2:H2C) transgenic line. Ki67 is widely used as a general proliferation marker duo to its specific expression in the cell cycle stages of S, G2, and M in actively cycling cells10. We first examined cardiomyocyte proliferation in the apex of adult Tg(mlc2:H2C) transgenic frogs using colocalization of Ki67 with mCherry. In sham-operated apex, almost no mCherry+Ki67+ nuclei were detected. However, increased numbers of mCherry+Ki67+ nuclei were observed near the resection plane at 3-30 dpr when compared with sham group. The maximal level of mCherry+Ki67+ nuclei (26.58 ± 2.36%) was detected at 7 dpr, which was significantly decreased in the later period of regeneration at 14 and 30 dpr (Fig. 4a, b). By capturing the large-scale and Z-stack images of mCherry+Ki67+ nuclei, we observed the representative proliferating responses of cardiomyocytes in the apex at 7 and 30 dpr (Fig. 4c).
Fig. 4. Cardiomyocyte proliferation in adult Tg(mlc2:H2C) transgenic X. tropicalis line upon apical resection injury.
a Immunostaining for mCherry (red) and Ki67 (green) expression in the apex of adult hearts from F1 Tg(mlc2:H2C) frogs at the indicated time points after resection. DAPI was used as a nuclear stain (blue). b Quantification of mCherry+Ki67+ cells in ventricular apex during heart regeneration within 30 days. Data are presented as mean ± SEM (n = 3 hearts for sham, 5 ~ 8 hearts for injured groups). *p < 0.05, ****p < 0.0001 versus sham. ###p < 0.001, ####p < 0.0001 (one-way ANOVA test). c Representative images of mCherry+Ki67+ cells in the whole apical regions at 7 and 30 dpr. Magnified Z-stack confocal images of mCherry+Ki67+ cells are shown in right panel. d Immunostaining for mCherry (red) and pH3 (green, antibody from CST, #9701) expression in the apex of adult hearts from F1 Tg(mlc2:H2C) frogs at the indicated time points after resection. DAPI was used as a nuclear stain (blue). e Quantification of mCherry+pH3+ cells in ventricular apex during heart regeneration within 30 days. Data are presented as mean ± SEM (n = 3 hearts for sham, 5 hearts for injured groups). **p < 0.01, ****p < 0.0001 versus sham. #p < 0.05, ####p < 0.0001 (one-way ANOVA test). f Representative images of mCherry+pH3+ cells in the whole apical regions at 7 and 30 dpr. Magnified confocal images of mCherry+pH3+ cells are shown in right panel.
It has been demonstrated that cardiomyocytes can go through cell cycle phases but stop and not complete it, which may lead to false-positive results when using general proliferation markers expressed in early phases of cell cycle26. Therefore, the later cell cycle marker phospho-Histone H3 (pH3) was further used in this study. In agreement with Ki67 staining, immunofluorescent staining used the antibody against pH3 (CST, #3377) showed that apical resection injury greatly promoted cardiomyocyte proliferation in apex as demonstrated by the increased percentages of mCherry+pH3+ nuclei at 3-30 dpr (Supplementary Figure 4a, b). Consistently, the maximal level of pH3+mCherry+ nuclei (20.29 ± 2.15%) was detected at 7 dpr, which was remarkably decreased in the later period of regeneration at 30 dpr (Supplementary Figure 4a, b). The representative proliferating responses of cardiomyocytes in the apex at 7 and 30 dpr were confirmed by large-scale and Z-stack imaging of mCherry+pH3+ nuclei (Supplementary Figure 4c, d). Given that pH3 is a later marker of cell cycle, the number of pH3-positive cardiomyocytes should be much lower than that of Ki67-positive cardiomyocytes. The inappropriate high numbers of pH3-positive cardiomyocytes (20.29 ± 2.15%) observed above may result from the non-specific staining of pH3 antibody. Indeed, the reactivity of the above used antibody against pH3 (CST, #3377) includes zebrafish and mammalians rather than Xenopus. We think that this should be the main reason for the high numbers of pH3-positive cardiomyocytes in above observation. To resolve the potential problem, we further performed this experiment using a new antibody against pH3 (CST, #9701), which was predicted to react with Xenopus due to that the antigen sequence used to produce this antibody shares 100% sequence homology with Xenopus. As expected, staining used the new antibody against pH3 (CST, #9701) showed a much lower percentage of pH3-positive cardiomyocytes during adult heart regeneration in the transgenic line Tg(mlc2:H2C) (Fig. 4d–f) when compared with the original antibody (CST, #3377) (Supplementary Figure 4). Importantly, the variation trends of pH3-positive cardiomyocytes during heart regeneration were very similar between these two antibodies against pH3 (Fig. 4d–f and Supplementary Figure 4). These findings suggest that selection of mammalian antibodies for Xenopus studies should be very carefully because of the limitation of Xenopus-specific antibodies. Taken Ki67 and pH3 staining results together, our results from transgenic reporter line rigorously confirmed that apical resection injury indeed induces cardiomyocyte proliferation in adult X. tropicalis.
Cardiomyocyte proliferation contributes to heart regeneration in adult Tg(mlc2:H2C) transgenic frogs
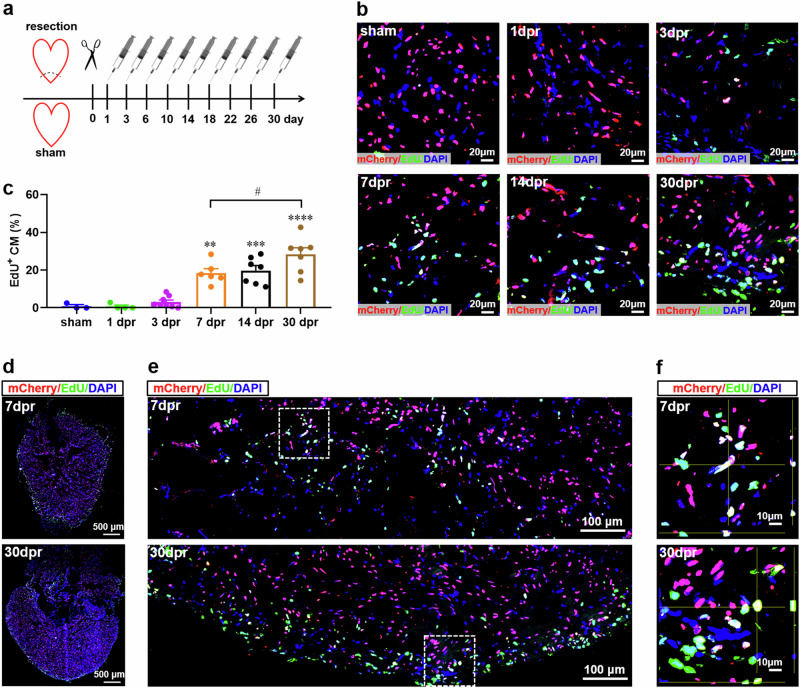
Finally, we assessed the cell cycle entry of cardiomyocytes in adult Tg(mlc2:H2C) transgenic frogs by measuring the nuclear incorporation of EdU, an efficient marker of DNA synthesis. To determine the proliferating cardiomyocytes during the whole period of heart regeneration within 30 dpr, EdU was injected once every three days for 30 days to label all proliferating cardiomyocytes during the whole period of cardiac regeneration (Fig. 5a). After injection, almost no mCherry+EdU+ nuclei were detected in the apex of sham-operated hearts. However, representative mCherry+EdU+ nuclei were observed from 3 to 30 dpr, and the percentages of mCherry+EdU+ nuclei were significantly increased at 7-30 dpr when compared with sham group. As expected, the percentage of mCherry+EdU+ nuclei at 30 dpr (28.30 ± 3.52%) was higher than that at 7 dpr (18.40 ± 2.36%) due to the accumulation of proliferating cardiomyocytes during the period from 7 to 30 dpr (Fig. 5b, c). Whole cardiac sections showed that the distributions of EdU-psotive mCherry signals were enriched in the apexes at 7 and 30 dpr, respectively (Fig. 5d). These expression patterns were further confirmed by the high-magnification images of apexes, which showed that proliferating cells including EdU+ cardiomyocytes were located near the resection plane (Fig. 5e). The EdU+ cardiomyocytes in apex were further verified by Z-stack confocal images at 7 and 30 dpr, respectively (Fig. 5f). These findings indicate that the newly forming myocardium in apex might result, at least in part, from the proliferating cardiomyocytes in adult X. tropicalis upon resection injury.
Fig. 5. Cardiomyocyte proliferation contributes to the newly formed myocardium in apex during heart regeneration in adult Tg(mlc2:H2C) transgenic X. tropicalis line.
a Schematic of EdU multiple injection experiment designed to label the proliferating cardiomyocytes in adult Tg(mlc2:H2C) transgenic X. tropicalis line during heart regeneration within 30 days. b Immunostaining for mCherry (red) and EdU (green) expression in the apex of adult hearts from F1 Tg(mlc2:H2C) frogs at the indicated time points after resection. DAPI was used as a nuclear stain (blue). c Quantification of mCherry+EdU+ cells in ventricular apex during heart regeneration within 30 days. Data are presented as mean ± SEM (n = 5 ~ 8 hearts per group). **p < 0.01, ***p < 0.001, ****p < 0.0001 versus sham (one-way ANOVA test). #p < 0.05 (Student’s t test). d Immunostaining for mCherry (red) and EdU (green) expression in a whole cardiac section from the adult heart of F1 Tg(mlc2:H2C) frog at 7 and 30 dpr. DAPI was used as a nuclear stain (blue). e Representative images of mCherry+EdU+ cells in the apical region at 7 and 30 dpr.. f Magnified Z-stack confocal images of mCherry+EdU+ cells white boxed in figure e. This figure was created by Photoshop Image 12 software using our own data in the work.
Discussion
In this study, we used a newly developed transgenic reporter line to define the cardiomyocyte proliferation and its contribution to heart regeneration in adult X. tropicalis. Here, we provided the direct genetic evidence that cardiomyocyte proliferation is indeed induced in adult X. tropicalis heart upon apical resection injury, thereby contributing to the restoration of the resected myocardium (Figs. 3–5). Comparation between traditional staining and transgenic reporter line revealed that whole-cell staining is not very rigorous way to identify cardiomyocytes in adult X. tropicalis, which leads to an error up to 17% when using the cryosection with common thickness (5 μm) (Fig. 2), raising caution on the determination of cardiomyocyte and its proliferation using traditional whole-cell staining approach in nonmammalian hearts.
Although adult mammals lost the capacity of heart regeneration, diverse studies on model organisms across various species have revealed that the outcome of heart injury is not always as detrimental as in adult mammals. Previous studies have shown that the adult heart in several fish and amphibian species has robust cardiac regeneration potential5,6, raising a requirement for comparative studies across species to better understand the regulators of adult heart regeneration. Therefore, more and more efforts have been invested in elucidating the mechanisms that determine heart regeneration and in screening approaches to improve the regenerative outcome in the injured adult mammalian heart. We previously reported, for the first time, that the resected ventricular apex in adult X. tropicalis can be regenerated in a scar-free manner8. Moreover, it seems that the heart regeneration in adult X. tropicalis might contribute to the restoration of resected ventricular apex8,9. These findings provide an alternative amphibian model organism for elucidating the mechanisms that determine adult heart regeneration. However, cardiomyocyte proliferation during adult heart regeneration in X. tropicalis was predominantly demonstrated by whole-cell staining of cardiomyocytes together with proliferating markers staining8,9. Previous studies have been demonstrated that more than half cardiac cells are non-cardiomyocytes including endothelial cells, hematopoietic cells, and fibroblast27,28. It is very difficult to distinguish an endocardial cell nucleus proliferating alongside the muscle using traditional whole-cell staining method, due to the robust proliferation of non-cardiomyocytes and the minor proliferation of cardiomyocytes during heart regeneration10. In addition, the lack of commercial and specific antibodies greatly hinders the studies of protein localization and function in Xenopus11. The most popular antibodies produced against mammalian proteins often do not specifically recognize Xenopus proteins due to the lower conservation of protein sequences between mammals and Xenopus12. Therefore, cardiomyocyte proliferation during heart regeneration in adult X. tropicalis needs to be further determined using more reliable genetic evidence.
It has been demonstrated that Xenopus myosin light chain 2 (mlc2) is cardiac-specific gene and expresses throughout the entire myocardium, from the onset of cardiac differentiation in embryo to the formation of mature and chambered heart in adult stage14,15. Using the mlc2 promoter, we newly developed a transgenic reporter line Tg(mlc2:H2C) which specifically labeling cardiomyocyte nuclei with mCherry (Fig. 1). Given that mlc2 promoter of Xenopus can faithfully drive reporter gene expression exclusively in cardiomyocytes14,15, it is reliable that the mCherry-labeling nuclei are the true cardiomyocytes in the heart of Tg(mlc2:H2C) transgenic line. To minimize the interference of whole-cell staining with evaluation of cardiomyocyte proliferation, traditional whole-cell staining strategy only quantitates the proliferating nuclei that were completely surrounded by whole-cell markers of cardiomyocytes. According to this principle, our genetic data from reporter line showed that about 15.77 ± 0.84% cardiomyocytes (mCherry+α-actinin-) might be omitted and that 1.96 ± 0.25% false positive cardiomyocytes (mCherry-α-actinin+) may be included when using the traditional whole-cell staining method and the cryosection with common thickness (5 μm) (Fig. 2a–k). In other words, the deviation of cardiomyocyte identification is more than 17% for whole-cell staining strategy. One possible reason for the higher population of mCherry+α-actinin- cells might be the non-specific staining of cardiac cells in X. tropicalis using antibodies against mammalian proteins. On the other hand, cardiac sectioning may result in the nucleocytoplasmic separation of cardiomyocytes due to the big size of myofibrils, thereby leading to the misplacement of nucleus and cytoplasm. Another possible reason is that α-actinin signals are not imaged or present in the tested section, but they would be found in close proximity to the nucleus higher “up” above the nucleus. This assumption was indeed verified by increasing the thickness of cryosection which could reduce the population of mCherry+α-actinin- cells (Fig. 2l–o).
To avoid the defect of whole-cell staining in detection of cardiomyocyte proliferation, increasing number of studies proposed to detect cardiomyocyte proliferation using the double staining for proliferating markers and cardiomyocyte nuclear markers including myocyte enhancer factor 2 C (Mef2c)29–32 and PCM-133–35. The nuclear co-staining of cardiomyocytes should be a more accurate method to identify proliferating cardiomyocytes when compared with the traditional whole-cell staining strategy. As expected, the transgenic reporter line showed that nuclear labeling is a more accurate approach during cardiomyocyte proliferation assessment in X. tropicalis when compared with whole-cell staining of cardiomyocytes (Fig. 2 and Supplementary Figure 2). Given the non-specificity of Mef2c antibody for cardiomyocyte nuclei in X. tropicalis (Supplementary Figure 2c–g), the nuclear staining of Mef2c is not a wonderful strategy for assessment of cardiomyocyte proliferation. Moreover, no signals were detected in X. tropicalis heart when using the antibody against mammalian PCM-1 (data not shown). In addition, we found that the antibody against pH3 (CST, #9701) rather than the one against pH3 (CST, #3377) could faithfully recognize proliferating cardiomyocytes in X. tropicalis (Fig. 4 and Supplementary Figure 4). These findings indicate that the non-specificity of mammalian antibodies for Xenopus endogenous proteins is a severe issue that should not be neglected. In the present study, our data from reporter line provided, for the first time, a direct genetic evidence showing that the resected apex can really be replaced by newly forming myocardium in adult X. tropicalis within 30 dpr (Fig. 3). Indeed, apex resection injury remarkably induced the proliferation of mlc2+ cardiomyocytes at 3-30 dpr, thereby regenerating the lost cardiac muscle at 30 dpr in adult X. tropicalis (Figs. 4 and 5).
In conclusion, we have rigorously defined the proliferation of cardiomyocytes during heart regeneration in adult X. tropicalis using a transgenic reporter line. Our data support an important role for cardiomyocyte proliferation in the adult heart’s regenerative potential in X. tropicalis. With knowledge of cardiomyocyte proliferation in adult X. tropicalis, the cellular and molecular regulatory interactions which regulate adult heart regeneration in X. tropicalis can now be more informatively pursued. Although mammalian cardiomyocytes are able to divide and renew in adulthood, they cannot regenerate the injured heart because of the very lower frequencies of cardiomyocyte proliferation2,36. Therefore, it is promising for mammalian heart regeneration to take advantage of mechanisms underlying adult cardiomyocyte proliferation in lower vertebrates. In addition, the newly established reporter line in this study provides a valuable genetic tool to evaluate cardiomyocyte proliferation in X. tropicalis. In particular, this line can be crossed with other genetic line to rigorously determine the effects of target gene on cardiomyocyte proliferation during heart regeneration in X. tropicalis. Given that X. tropicalis is a true diploid organism with many advantages including short generation time37, sequenced genome information38, and available genome editing approach39, our genetic evidences of adult cardiomyocyte proliferation further propose that X. tropicalis is an alternative and suitable amphibian model organism to clarify the molecular mechanism of adult heart regeneration.
Methods
Construct and Xenopus tropicalis transgenic line
To construct the transgenic plasmid pMlc2-H2B-mCherry (mlc2:H2C), the H2B-mCherry DNA fragment from p3-H2B-mCherry16 was cloned downstream of the 3-kilobase (kb) Xenopus mlc2 promoter14. The entire construct was flanked with I-SceI sites for transgenesis using the meganuclease method (Fig. 1a). To generate stable transgenic lines, the pMlc2-H2B-mCherry plasmid were injected into one-cell-stage embryos with I-SceI meganuclease as previously reported40. The full name of this transgenic line is Tg(mlc2:H2C). Positive founder (F0) tadpoles expressed red fluorescence specifically in heart were identified by Leica M205FA stereo fluorescence microscope and raised to sexual maturity. To confirm the successful transgenesis in F1 generation, male founder frogs were crossed with wild-type females to generate the stable transgenic line Tg(mlc2:H2C). The adult F1 Tg(mlc2:H2C) frogs at 10 ~ 12 months were used in this study. Both male and female frogs were used and were randomized into different treatment groups in all experiments. At the end of experiments, animals were anesthetized in ice for 2 minutes, followed by bleeding to induce euthanasia. All animal protocols and procedures were approved by the Institutional Animal Care and Use Committee of Jinan University. Data and methods are reported here in accordance with ARRIVE guidelines41.
Whole-mount fluorescence microscopy
The wild-type and F1 Tg(mlc2:H2C) adult frogs were anesthetized in ice for 2 min. The hearts quickly extracted and washed in phosphate-buffered saline (PBS). The isolated adult hearts or tadpoles were placed on an agar gel for whole-mount bright-field or fluorescence imaging using a Leica M205FA stereo fluorescence microscope. To determine the nuclear expression of transgene in cardiomyocytes, high-magnification fluorescent images and videos of the heart in the living F1 Tg(mlc2:H2C) tadpoles placed on an agar gel were further captured using the SpinSR10 spinning disk confocal super resolution microscope (Olympus).
Ventricular apex resection in adult X. tropicalis
Apical resection surgeries in adult F1 Tg(mlc2:H2C) frogs were performed as described previously9. In brief, the anaesthetized frogs were moved to plate with ventral side up, fixed by thumbtacks into limbs. A small incision penetrating the skin and pericardial sac was induced by iridectomy scissors. The ventricle was exposed by gentle pressure, ventricle apex was then removed by scissors. The incision was blotted by gossypium asepticum prior to quickly clotting of wounds. Body wall incisions were sutured, and frogs were waked up within several minutes in culture dish with litter water which does not submerge the body. After waking up, frogs were returned to water and stimulated to breathe by vigorously squirting water with a pipette. Sham operation was performed as above procedure without ventricle resection. The hearts were collected after surgery and briefly rinsed in phosphate-buffered saline (PBS) solution to remove residual blood from the ventricles. The ventricles without non-cardiac tissue and atria were blotted and weighed.
Immunofluorescence staining
Immunofluorescence staining was performed as described previously9, with some modification. Briefly, hearts were collected in PBS on ice and then fixed in 4% paraformaldehyde at 4 °C for 30 min. After three washes in PBS, the tissues were then embedded in optimum cutting temperature (OCT, Sakura) and stored at -80°C until sectioning. Cryosections (5 μm) were collected on positively slides and stored at -80°C until use. For immunostaining, tissue sections were blocked with PBST (0.1% Triton X-100 and 5% goat serum in PBS) for 1 h at room temperature, followed by incubation with the primary antibody overnight at 4 °C. The next day, tissue sections were washed three times with PBS, incubated with Alexa-Fluor-conjugated secondary antibodies (Jackson Immuno Research) for 30 min at room temperature. Sections were then rinsed three times in PBS, stained with DAPI (Sigma) at a concentration of 2 μg/mL to label nucleus and mounted with Antifade Mounting Medium (Jackson ImmunoResearch Laboratories). Antibodies to the following proteins were used: mCherry (Proteintech, 26765-1-AP, 1:400 dilution), α-Actinin (Sigma, SAB3300072, 1:200 dilution), Mef2c (Abcam, ab227085, 1:100 dilution), Ki67 (Abcam, ab16667, 1:250), and pH3 (CST, #3377 and #9701, 1:200 dilution). Images were captured by laser-scanning confocal microscope (LSM 700, Zeiss) in the ventricle areas nearby resection plane or apex, and analyzed by ZEN 2012 software (Zeiss). In addition, thicker cryosections (10 μm) were also used for α-actinin staining.
Quantification of labeled cardiomyocytes
Frog hearts were collected and embedded for serial sectioning. For each adult heart, about 10 ~ 15 serial sections of ventricles were stained for mCherry and cardiomyocyte marker proteins. To evaluate the accuracy of traditional whole-cell staining of cardiomyocytes, ~200 sections from 16 adult hearts were stained for mCherry and α-Actinin. To evaluate the accuracy of nuclear staining of cardiomyocytes, ~150 sections from 11 adult hearts were stained for mCherry and Mef2c. The percentage of individual cell population labeled with different fluorescence signals were calculated as the number of each population divided by the number of mCherry+ nuclei. DAPI was used as a nuclear stain. To quantify the proliferating cardiomyocytes during heart regeneration upon injury, ventricle areas nearby resection plane (no more than 200 μm) were imaged for resected hearts. For sham-operated and completely regenerated hearts, images were captured in the ventricle areas nearby apex.
Cardiomyocyte number and ventricular volume quantification
To determine cardiomyocyte number and ventricular volume in the whole ventricles, an unbiased stereological reconstruction method25 was used with some modification (Fig. 3e). In brief, the entire heart of transgenic reporter line Tg(mlc2:H2C) was subjected to serial section with 10 μm thickness. Unbiased representative subset of total sections (every 5th section) which distributing over entire ventricle was then used to determine ventricular cardiomyocyte numbers and ventricular areas covered by mCherry-positive cells. For cardiomyocyte number quantification, the total number of cardiomyocytes in the whole ventricle was calculated by multiplying the measured number of cardiomyocytes by 5 (1/5th of sections were analyzed). Total size of the ventricular area was measured by adding up the individual areas of all tested sections (every 5th section) representing 1/5th of the ventricle. Total ventricle volume was then calculated by multiplying the measured ventricular area by 5*10 μm.
EdU labeling
The EdU labeling of cardiomyocytes during heart regeneration was performed as previously described in ref. 9. In brief, frogs were injected intraperitoneally with 50 μl of a 2 mg/ml solution of EdU (RiboBio, Guangzhou, China) dissolved in sterile water. EdU was injected once every three days for 30 days to label all proliferating CMs during the whole period of cardiac regeneration. The last injection was performed 8 h prior to heart collection at 30 dpr. This method could monitor all proliferating cardiomyocytes during the periods from 0 dpr to the indicated time points. Sham-operated frogs underwent the same procedure without the apical resection. Hearts were embedded in optimal cutting temperature (OCT) (Sakura, USA) for frozen section (5 μm). Sections were rinsed three times in PBS and fixed in 4% paraformaldehyde for 30 min. After rinsing three times again, sections were then incubated with 2 mg/mL glycine solution for 10 min, permeabilized with 0.5% Triton X-100 in PBS for 10 minutes, and then rinsed with PBS once for 5 min. This was followed by incubation with Apollo®488 staining solution (1×) at room temperature for 30 min. Sections were then rinsed with methanol for 5 min, washed with PBS once for 5 min, blocked with 5% goat serum for 1 h, and followed by incubation with primary antibody against mCherry (Proteintech, 26765-1-AP, 1:400 dilution) overnight. The following day, incubation with Alexa-Fluor-conjugated secondary antibodies (Jackson Immuno Research) for 30 min at room temperature. Sections were washed three times in PBS, stained with DAPI for 10 min to label nuclei, and mounted in Antifade Mounting Medium. Images were captured by laser-scanning confocal microscope (LSM 700, Zeiss) in the ventricle areas nearby resection plane or apex as mentioned above, and analyzed by ZEN 2012 software (Zeiss).
Visualization of regenerated apex and systolic function quantification
Both sham-operated and resected frogs at 7, 14, and 30 dpr were anesthetized in ice for 2 minutes. Hearts were quickly extracted and cleaned to remove blood clot. Cleaning hearts were then cultured in the medium (DMEM plus 10% (vol/vol) FBS, 1% (vol/vol) MEM-NEAA, 100 U/ml penicillin, 100 μg/ml streptomycin and 50 μM 2-mercaptoethanol) as described previously42. To visualize apex regeneration at different time points, both bright-field and fluorescence images of the regenerated apex were captured using Leica M205FA stereo fluorescence microscope. To quantify cardiac systolic function during regeneration, heat beating frequency was counted within one minute for each heart. The beating frequency of heart was calculated by the beating number per twenty seconds.
Wheat germ agglutinin (WGA) staining
Wheat germ agglutinin (WGA) conjugated to Alexa Fluor 488 (Invitrogen, W11261, 5 μg/ml) was used to quantify cardiomyocyte size as previously reported9. In brief, following deparaffinized, rehydrated, slides were then incubated with WGA conjugated to Alexa Fluor 488 (Invitrogen, W11261, 5 μg/ml) for 10 minutes at room temperature. To quantify the cell size, 5 independent hearts per group (at least 300 cells) were captured near apex with laser-scanning confocal microscope (LSM 700, Zeiss). ZEN 2012 lite software (Zeiss) was used to quantify the size of each cell.
Quantitative Real-Time PCR (qPCR)
Total RNA was isolated using RNeasy Kit (Qiagen, Valencia, CA, USA) according to the protocol of the manufacturer, respectively. Reverse transcription to cDNA was performed with 30 ng of total RNA, random primers, and SuperScript III Reverse Transcriptase (Roche, USA). The qPCR was performed using a Light Cycler 480 SYBR Green I Master (Roche, USA) and the MiniOpticon qPCR System (Bio-Rad, CA, USA). After denaturation for 10 min at 95 °C, the reactions were subjected to 45 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s. GAPDH was used as the internal standard control to normalize gene expression using the 2-ΔΔCt method. The sequences of the qPCR primers were listed in supplementary Table 1.
Statistical analysis
All statistics were calculated using GraphPad Prism 8 Software. All data are presented as the mean ± SEM. Among three or more groups, statistical analysis was performed using one-way ANOVA followed by Dunnett’s multiple comparison tests. Comparisons between two groups were analyzed using unpaired Student’s t-test. A p-value of less than 0.05 was considered statistically significant.
Supplementary information
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (82030039, 82230047, 82370247, 82070257, 81770240), the Fundamental Research Funds for the Central Universities (21623110), the Basic and Applied Basic Research Foundation of Guangdong Province (2023A1515012147), and the Research Grant of Key Laboratory of Regenerative Medicine, Ministry of Education, Jinan University (ZSYXM202402, ZSYXM202303), China.
Author contributions
X.L.L., J.H.L., and Y.C. performed most experiments including apical resection surgery, immunofluorescence staining, EdU labeling, and analyzed the data. H.Z., S.Y.H., and H.Y.W. contributed some experiments. X.F.Q. and X.L.L. designed and interpreted experiments. L.Z. and Z.B.Y. reviewed the manuscript and provided important suggestions. X.F.Q. wrote the manuscript with help from co-authors.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xiao-Lin Lin, Jin-Hua Lin, Yan Cao.
Contributor Information
Ze-Bing Ye, Email: bean9350@sina.com.
Li Zheng, Email: Zhengli8244@126.com.
Xu-Feng Qi, Email: qixufeng@jnu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41536-024-00384-w.
References
- 1.Tsao, C. W. et al. Heart Disease and Stroke Statistics-2023 Update: A Report From the American Heart Association. Circulation147, e93–e621 (2023). [DOI] [PubMed] [Google Scholar]
- 2.Bergmann, O. et al. Evidence for Cardiomyocyte Renewal in Humans. Science324, 98–102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Senyo, S. E. et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature493, 433–436 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porrello, E. R. et al. Transient Regenerative Potential of the Neonatal Mouse Heart. Science331, 1078–1080 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poss, K. D., Wilson, L. G. & Keating, M. T. Heart regeneration in zebrafish. Science298, 2188–2190 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Becker, R. O., Chapin, S. & Sherry, R. Regeneration of the ventricular myocardium in amphibians. Nature248, 145–147 (1974). [DOI] [PubMed] [Google Scholar]
- 7.Weinberger, M. & Riley, P. R. Animal models to study cardiac regeneration. Nat. Rev. Cardiol.21, 89–105 (2024). [DOI] [PubMed] [Google Scholar]
- 8.Liao, S. et al. Heart regeneration in adult Xenopus tropicalis after apical resection. Cell Biosci.7, 70 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu, H. Y. et al. Fosl1 is vital to heart regeneration upon apex resection in adult Xenopus tropicalis. NPJ Regen. Med. 6, 36 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kretzschmar, K. et al. Profiling proliferative cells and their progeny in damaged murine hearts. P Natl Acad. Sci. USA115, E12245–E12254 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willsey, H. R. Whole-Mount RNA In Situ Hybridization and Immunofluorescence of Xenopus Embryos and Tadpoles. Cold Spring Harb. Protoc.2021, 1–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horr, B. et al. Production and characterization of monoclonal antibodies to Xenopus proteins. Development150, dev201309 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rottbauer, W. et al. Cardiac myosin light chain-2: a novel essential component of thick-myofilament assembly and contractility of the heart. Circ. Res. 99, 323–331 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Latinkic, B. V. et al. Transcriptional regulation of the cardiac-specific MLC2 gene during Xenopus embryonic development. Development131, 669–679 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Breckenridge, R., Kotecha, S., Towers, N., Bennett, M. & Mohun, T. Pan-myocardial expression of Cre recombinase throughout mouse development. Genesis45, 135–144 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Love, N. R. et al. pTransgenesis: a cross-species, modular transgenesis resource. Development138, 5451–5458 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Auchampach, J. et al. Measuring cardiomyocyte cell-cycle activity and proliferation in the age of heart regeneration. Am. J. Physiol. Heart Circ. Physiol.322, H579–H596 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tao, G. et al. Pitx2 promotes heart repair by activating the antioxidant response after cardiac injury. Nature534, 119–123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahmoud, A. I. et al. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature497, 249–253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei, K. et al. Epicardial FSTL1 reconstitution regenerates the adult mammalian heart. Nature525, 479–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan, Y., Duan, X., Wang, B., Liu, X. & Zhan, Z. Murine neonatal cardiac B cells promote cardiomyocyte proliferation and heart regeneration. NPJ Regen. Med. 8, 7 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han, C. et al. Acute inflammation stimulates a regenerative response in the neonatal mouse heart. Cell Res. 25, 1137–1151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heallen, T. et al. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science332, 458–461 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boogerd, C. J. et al. Cardiomyocyte proliferation is suppressed by ARID1A-mediated YAP inhibition during cardiac maturation. Nat. Commun.14, 4716 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertozzi, A. et al. Is zebrafish heart regeneration “complete”? Lineage-restricted cardiomyocytes proliferate to pre-injury numbers but some fail to differentiate in fibrotic hearts. Dev. Biol.471, 106–118 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Lazar, E., Sadek, H. A. & Bergmann, O. Cardiomyocyte renewal in the human heart: insights from the fall-out. Eur. Heart J.38, 2333–2342 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Litvinukova, M. et al. Cells of the adult human heart. Nature588, 466–472 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinto, A. R. et al. Revisiting Cardiac Cellular Composition. Circ. Res. 118, 400–409 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao, C. et al. Endothelial Brg1 fine-tunes Notch signaling during zebrafish heart regeneration. NPJ Regen. Med. 8, 21 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao, C. et al. Chromatin-remodelling factor Brg1 regulates myocardial proliferation and regeneration in zebrafish. Nat. Commun.7, 13787 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu, B. et al. Origin and function of activated fibroblast states during zebrafish heart regeneration. Nat. Genet54, 1227–1237 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun, F. et al. Enhancer selection dictates gene expression responses in remote organs during tissue regeneration. Nat. Cell Biol.24, 685–696 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bywater, M. J. et al. Reactivation of Myc transcription in the mouse heart unlocks its proliferative capacity. Nat. Commun.11, 1827 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen, N. U. N. et al. A calcineurin-Hoxb13 axis regulates growth mode of mammalian cardiomyocytes. Nature582, 271–276 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen, Y. et al. Reversible reprogramming of cardiomyocytes to a fetal state drives heart regeneration in mice. Science373, 1537–1540 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Mollova, M. et al. Cardiomyocyte proliferation contributes to heart growth in young humans. Proc. Natl Acad. Sci. USA110, 1446–1451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grammer, T. C., Khokha, M. K., Lane, M. A., Lam, K. & Harland, R. M. Identification of mutants in inbred Xenopus tropicalis. Mech. Dev.122, 263–272 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Hellsten, U. et al. The genome of the Western clawed frog Xenopus tropicalis. Science328, 633–636 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mao, C. Z. et al. CRISPR/Cas9-mediated efficient and precise targeted integration of donor DNA harboring double cleavage sites in Xenopus tropicalis. Faseb J.32, 6495–6509 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Noble, A., Abu-Daya, A. & Guille, M. J. I-SceI-Mediated Transgenesis in Xenopus. Cold Spring Harb. Protoc.2022, Pdb prot107011 (2022). [DOI] [PubMed] [Google Scholar]
- 41.du Sert, N. P. et al. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. Plos Biol.18, e3000410 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao, J. & Poss, K. D. Explant culture of adult zebrafish hearts for epicardial regeneration studies. Nat. Protoc.11, 872–881 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.