Abstract
Endometrial cancer (EC) with Mismatch Repair deficiency (MMRd) is characterized by the accumulation of insertions/deletions at microsatellite sites. These mutations lead to the synthesis of frameshift peptides (FSPs) that represent tumor-specific neoantigens (nAg) proved to be shared across patients/tumors with MMRd. In this study, we explored the feasibility of a nAg-based cancer vaccination design in EC with MMRd. We adopted a whole exome sequencing approach and ad hoc bioinformatics pipelines to characterize FSPs in 35 patients with EC. A mean of 146 mutated mononucleotide repeats (MNRs) was identified with enrichment in the patients’ group with MLH1 impairment. A high coverage emerged from the comparative analysis of the EC FSPs with the content of the previously validated NOUS-209 vaccine. We obtained pieces of evidence of FSPs translation as expressed proteins from Ribo-seq, supporting the potential as the target of vaccination. The development of a nAgs-based vaccine strategy in MMRd EC may be further explored.
Subject terms: Molecular medicine, Endometrial cancer
Introduction
Around 30% of patients with Endometrial Cancer (EC) are characterized by Mismatch Repair deficiency (MMRd)1. Mismatch Repair (MMR) is a highly conserved system with a key role in genomic stability maintenance, identifying and correcting DNA mismatches2. The MMRd status leads to the accumulation of insertion and deletion (indels) particularly at microsatellites DNA sites, repetitive regions of one to ten mono-, di-, tri-, or tetra-nucleotides units, leading to a high mutational burden. Among microsatellites, Mononucleotide repeats (MNRs) are the most abundant class. Indels accounted in open-reading-frame (ORF) are predicted to cause translational frameshifts and the generation of frameshift peptides (FSPs) encompass long amino acid stretches, potentially containing multiple immunologically relevant peptides (i.e. neoepitopes)3,4. In MMRd EC, the level of tumor-specific neoantigens (nAgs) derived from non-synonymous mutations exceeds 10-fold when compared to microsatellite stable tumors5,6. From a therapeutic perspective, in the advanced/recurrent setting, these patients have been recently shown to benefit the most from immune-checkpoint inhibitors (ICIs)7. Beyond ICIs, the landscape of immunotherapy is rapidly evolving exploring also therapeutic cancer vaccination. In this context, FSPs represent ideal molecular targets for cancer vaccine design. From the evaluation of MMRd-induced FSPs emerged the occurrence of “recurrent” mutational events, with genetic alterations involving coding MS that are shared across patients8,9. This typical feature of MSI-h tumors offers the opportunity to characterize shared expressed nAgs and develop an “off-the-shelf” nAgs-based cancer vaccine10,11. While FSPs have been tested as a cancer vaccine strategy in patients with MSI-h colorectal cancer in ongoing clinical trials (NCT01885702, NCT01461148, NCT04041310), few literature data are available regarding FSPs nAgs-based vaccine in MSI-h-related gynecologic cancers.
A cohort of patients with MMRd EC was evaluated by a Whole Exome Sequencing (WES) approach coupled with nAgs prediction bioinformatics pipelines to: (i) characterize the presence of MMRd-related FSPs; (ii) explore the potential of an “off-the-shelf” cancer vaccination design in EC.
Methods
Patients and histopathological Mismatch Repair evaluation
This was an observational, single-center study of Caucasian patients retrospectively enrolled at the Fondazione Policlinico Universitario “Agostino Gemelli” IRCCS (Rome, Italy) from the 1st of November 2018 to the 31st of April 2021. All women with a histological diagnosis of advanced EC were identified from the institutional registry of histopathology. The inclusion criteria were defined as follows: (i) histopathologically confirmed diagnosis of primary EC (FIGO stages III-IV); (ii) confirmed MMR deficiency pattern by standard diagnostic immunohistochemistry (IHC); (iii) formalin-fixed and paraffin-embedded (FFPE) tissue sample available. Exclusion criteria of enrollment were the diagnosis of any other malignancy in the previous 5 years or the diagnosis of a synchronous malignancy. MMR-IHC analysis was performed on FFPE tissue slides of primary tumors at the diagnosis by a dedicated gyneco-pathologist. The following clones were used: MSH2 (clone G219-1129; Ventana), MSH6 (clone SP93; Ventana), MLH1 (clone M1; Ventana), and PMS2 (clone A16-4; Ventana). The protein expression of MMR was considered as: “positive” if positive nuclei with mild to strong intensities were counted; “negative” if internal controls (stromal cells and lymphocytic infiltrates) resulted as positive and tumor cells were completely negative. EC tumors were classified as “MMR proficient” if informative results were obtained for all the investigated MMR proteins and as “MMRd” in the absence of expression of at least one of the four proteins.
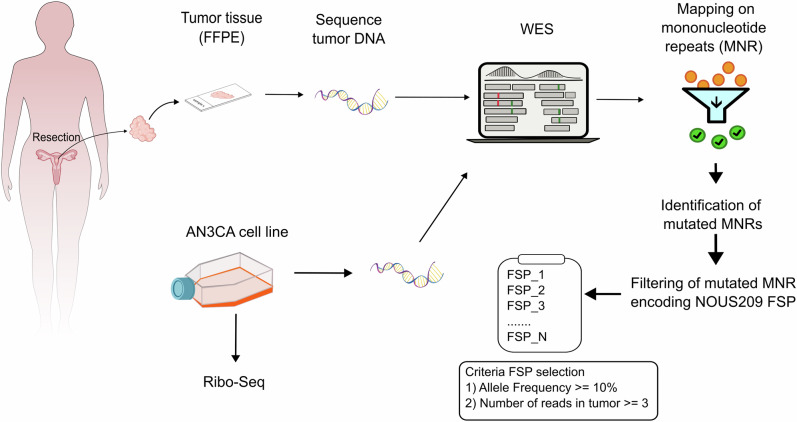
The study was approved by the institutional review board of Fondazione Policlinico Universitario “A. Gemelli” IRCS (ID 4558, Prot. N. 0041968/21). Relevant clinical data were collected and managed with REDCap electronic data capture tools hosted at Fondazione Policlinico Universitario “Agostino Gemelli” IRCCS (https://redcap‐irccs.policlinicogemelli.it). The experimental workflow of the study was reported in Fig. 1. Additional molecular data (MLH1-promoter methylation, PCR-based MSI status) were also collected.
Fig. 1. Workflow of the experimental design of the study.
In the figure is reported the workflow utilized to perform the look-up analysis in the 35 EC MSI patients and in the AN3CA cell line. EC endometrial cancer, MSI-h microsatellite instability-high, MNR mononuclear repeats, FSP frameshift peptides, WES Whole Exome Sequencing.
MLH1-promoter methylation testing
The SALSA MS-MLPA kit ME011 mismatch repair genes (MMR) (MRC-Holland, Amsterdam, The Netherlands) was used to study aberrant CpG island methylation in the promoter of MMR genes, including MLH1 according to the manufacturer’s instruction. The method is based on specific methylation-specific probes designed to target DNA sequences that contain a restriction site for the methylation-sensitive restriction enzyme HhaI: MLH1 1 (−659 bp distance to ATG start); MLH1 2 (−383 bp distance to ATG start); MLH1 3 (246 bp distance to ATG start); MLH1 4 (−13 bp distance to ATG start); MLH1 5 (+208 bp distance to ATG start). Briefly, 200 ng of extracted DNA was used in each reaction. DNA was first denatured and subsequently cooled down to 25 °C, followed by the addition of MS-MLPA probes and a 16-h hybridization step. MS-MLPA assay is then split into two reactions: one tube is processed as a standard MLPA reaction, providing information on the copy number status of the target DNA; the other tube is incubated with the methylation-sensitive HhaI endonuclease. In the case of an unmethylated DNA target, the methylation-specific probe will be ligated and simultaneously digested by HhaI, consequently, it will not generate a peak signal because it cannot be amplified. In contrast, if the target sequence is methylated, the methyl group will prevent HhaI digestion, resulting in an amplification and a normal peak signal. After the ligation and ligation-digestion of the hybridized probes, a PCR step was performed according to the protocol. Then, PCR fragments were separated by capillary electrophoresis on an ABI 3500 Genetic Analyzer (Life Technologies, Gaithersburg, USA). Methylation status was calculated using the COFFALYSER.NET analysis software (MRC-Holland, Amsterdam, The Netherlands). At least three reference samples should be included in each MS-MLPA experiment.
MSI testing by PCR
The Microsatellite Instability status was evaluated using the AmoyDx® Microsatellite Instability (MSI) Detection Kit (AmoyDx, Singapore) according to the manufacturer’s instructions. The AmoyDx® MSI Detection Kit is a melting curve analysis (MCA)-based real-time PCR assay for the qualitative detection of microsatellite instability status in eight mononucleotide markers: EIF4E3, IFT140, PPP1CC, UBAC2, PRR5-ARHGAP8, ACVR2A, TAOK3, RBM14-RBM4. Briefly, 110 ng of DNA sample was used in each PCR reaction. The assay contains specific primers for each marker and specific FAM/CY5-labeled fluorescent probes targeting mutant sequences. PCR reactions and subsequent melting curves analyses of PCR amplicon were performed on the SLAN-96S Real-time PCR system (Zeesan Biotech, Xiamen City, Fujian Province, China). When there is a microsatellite unstable template in the sample, a melting peak will be generated in a specific melting temperature value range. For interpretation purposes, microsatellite instability at ≥2 loci was defined as MSI-high, instability at a single locus or no instability at any of the loci tested was defined as MSI-low/MSS. The limit of detection is set at 20% tumor cells. Each PCR run contained one positive control, one negative control, and one No Template Control, as required by the manufacturer.
Whole exome sequencing and mutational analysis at mononucleotide repeats
Eosin-stained histology tissue slides were examined by a dedicated pathologist to assess eligible FFPE samples with a minimum of 20% of tumor cell content. DNA was extracted from a 10 μm thick unstained FFPE slide using the MagCore® Genomic DNA FFPE One-Step kit (RBC Bioscience, New Taipei City, Taiwan) on the automated platform MagCore® HF16Plus (Diatech Lab Line, Jesi, Italy) according to manufacturer’s procedures. DNA quality was evaluated by using the Illumina Infinium FFPE QC kit (Illumina®, San Diego, USA) on the CFX Connect Real-Time PCR Detection System instrument (Bio-Rad®, Hercules, USA). The quantitation of the extracted DNA was assessed using the Qubit HS dsDNA fluorimetric assays (Thermo Fisher®, Waltham, MA, USA). The WES was carried out using the Illumina DNA Prep with Enrichment (S) Tagmentation kit on the Next Generation Sequencing (NGS) NovaSeq6000® platform (Illumina®, San Diego, USA) in paired-end 2 × 101 modes. The total sequenced read median is 301 million [Min:156; Max:687].
FastQ files were generated using the bcl2fastq2 Conversion Software (Illumina®, San Diego, USA). Preliminary quality control of the raw WES data was performed by filtering out reads of low quality with Trimmomatic-0.33 (LEADING: 5; TRAILING: 5; SLIDINGWINDOW: 4:20; MINLEN: 50)12. The remaining DNA read pairs were then aligned against the human reference genome version GRCh37/hg19 using BWA-mem13. Read pairs for which only one read was mapped and paired reads aligning to more than one genomic locus with the same mapping score were filtered out. Exomeseq alignments were then further processed by optimizing the local alignment around small indels, marking duplicated reads, and recalibrating the final base quality score in the realigned regions (program, parameters) with Picard tools (http://broadinstitute.github.io/picard/). Genomic coordinates of MNRs with lengths 6–23 bp were retrieved with MISA software14. The total list of MNRs was mapped on RefSeq transcriptome to retrieve the final subset of 30678 MNRs located within the ORF of protein-coding genes. A Perl script was used to perform a look-up analysis in Exomeseq data to detect mutations in the subset of 30678 MNR. The script is a wrapper that determines for each MNR locus the number of reads with/without mutations by launching iteratively the pileup software included in samtools 1.19.2. The output of pileup was then parsed to retrieve a measure of the coverage at each position and the mutation allele frequency (number of mutated reads/total number of reads). MNRs were considered mutated if have a mutation allele frequency ≥ 10% with a number of reads with a frameshift indels of 1 bp or 2 bp ≥38. The analysis was further restricted to 204 MNRs loci comparing the EC WES dataset to the FSPs content of the NOUS-209 vaccine8. The Kolmogorov–Smirnov statistic test (KS test) was applied to evaluate the FSPs distribution in the cohort of EC.
Frameshift mutations at the 30678 MNR in The Cancer Genome Atlas (TCGA) MSI endometrial cancers were determined by reanalyzing the Protected Mutation Annotation Format files available from TCGA (release date 40.0—March 29, 2024).
AN3CA and Ribo-seq next-generation sequencing data analysis
Exome sequencing of the AN3CA cell line was performed at Genomix4Life s.r.l. (Via Salvador Allende, 43L, 84081 Salerno, Italy). Genomic DNA was fragmented and used for Illumina library construction (Illumina®, San Diego, USA). Exonic regions were captured using the Agilent human SureSelect All Exon kit (Agilent Technologies®, Inc., Santa Clara, CA). Paired-end sequencing, resulting in 100 bp from each end of fragments, was performed with the Hiseq2000 Genome Analyzer (Illumina®, San Diego, USA) at a target coverage of 120×. Ribo-seq was performed at OHMX.bio (Proeftuinstraat 86, B-9000 Ghent, Belgium). The look-up of NOUS-209 FSPs in Ribo-seq was performed with the same thresholds of minimum number of reads used for the 35 EC patients analysis. Normalized TPM was estimated as previously published15. Likelihood of translation for the mutated transcripts was performed at OHMX.bio by using the TIS transformer algorithm that is based on a deep learning model trained on 431,011,438 (96,655 true transcription initiation sites (0.022%)) RNA nucleotide positions annotated in the Ensemble v107 database16.
IFN-γ ELISpot assay
The immunogenicity of tested FSPs was measured by ex vivo ELISpot-forming cell assay after antigen-specific stimulation. Cryopreserved patients PBMC (NOUS-209 Ph1 trial NCT04041310) were thawed and rested overnight at 37 °C in R10 [RPMI 1640 (Gibco) supplemented with 10% heat-inactivated highly defined fetal bovine serum (FBS-HyClone), 2 mmol/L L-glutamine, 10 mmol/L HEPES buffer (N-2-hydroxyethylpiperazine-N-2-ethane sulfonic acid), 100 U/ml penicillin, and 100 µg/mL streptomycin (Gibco)]. Rested PBMC were stimulated with a set of covering peptides designed to cover the FSPs of interest, and tested for T cell response assessment as previously described17. More specifically, for each FSP, 15mer peptides overlapping by 11 amino acids covering the FSP sequence were used (3 μg/ml final concentration of each peptide). Spontaneous cytokine production (i.e., background) was assessed by incubating PBMCs with the medium only plus the peptide diluent DMSO (i.e., negative control) (Sigma–Aldrich Chemie GmbH, Germany), whereas the CEFX, a pool of known peptide epitopes for a range of human leukocyte antigen (HLA) subtypes and different infectious agents, was used as positive control. Results are expressed as SFC/106 PBMCs in stimulating cultures.
Results
Landscape of indels encoding potential neoantigens in MSI EC
A cohort of 35 MMRd, advanced, newly diagnosed EC patients was included in the study. Patients were young, with a median age of 58 years. Most patients (80%) had an endometrioid histotype and low-grade tumors. All patients received chemotherapy, in some cases coupled with radiotherapy. Only 2 patients were treated with surgery alone because of poor overall conditions. A total of 11 patients out of 35 (31.4%) relapsed and were treated with second-line chemotherapy. The IHC MMRd status resulted as: MLH1/PMS2 loss (n = 20); PMS2 loss (n = 7); MLH1 loss (n = 4); MSH6 loss (n = 2); MSH2/MSH6 loss (n = 1), and MSH2 loss (n = 1) (Table 1). The distribution of MMR phenotypes in our dataset aligned with the existing available data18. Additional molecular characterization results regarding PCR-based MSI status, MLH1-promoter methylation, and germline tests were reported in Supplementary Table 1.
Table 1.
Patients’ clinical characteristics at baseline
| Age at diagnosis (years, mean) | 58.17 |
|---|---|
| Histotype | |
| Endometrioid | 28 (80%) |
| Serous | 1 (2.8%) |
| Mixed | 4 (11.4%) |
| Dedifferentiated | 2 (5.7%) |
| Grade | |
| 1–2 | 24 (68.5%) |
| 3 | 10 (28.5%) |
| N.A. | 1 (2.8%) |
| LVSI | |
| Absent | 11 (31.4%) |
| Focal | 4 (11.4%) |
| Substantial | 20 (57.1%) |
| MMRd status | |
| MLH1/PMS2 loss | 20 (57.1%) |
| PMS2 loss | 7 (20%) |
| MLH1 loss | 4 (11.4%) |
| MSH6 loss | 2 (5.7%) |
| MSH2/MSH6 loss | 1 (2.8%) |
| MSH2 loss | 1 (2.8%) |
| 2018 FIGO stage | |
| IIIC1 | 15 (42.8%) |
| IIIC2 | 9 (25.7%) |
| IVA | 4 (11.4%) |
| IVB | 7 (20%) |
| First line treatment | |
| Surgery | 2 (5.7%) |
| Surgery + CHT | 12 (34.2%) |
| Surgery + CHT + EBRT | 13 (37.1%) |
| Surgery + CHT + EBRT + BRT | 8 (22.8%) |
| Status at follow-up | |
| NED | 26 |
| AWD | 2 |
| DOD | 6 |
| Lost | 1 |
LVSI lymph-vascular space invasion, CHT chemotherapy, EBRT external beam radiation therapy, BRT brachytherapy, NED no evidence of disease, AWD alive with disease, DOD died of disease.
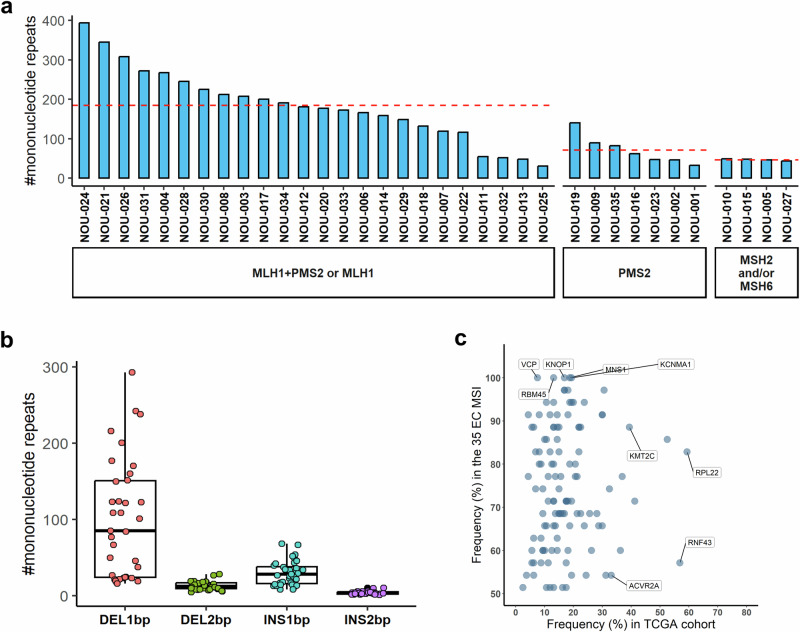
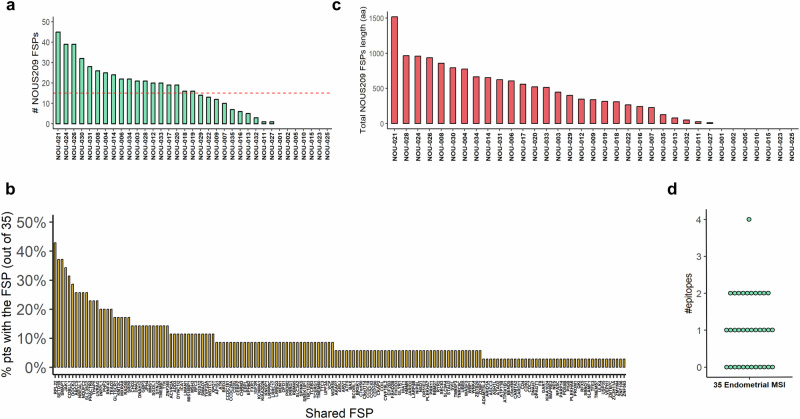
To investigate the presence of MNRs mutations encoding FSPs in our cohort of MMRd EC, we applied a multistep mapping procedure to align the raw Exomeseq data to the human genome (Gchr37). No blood or matched healthy tissue was available for this cohort therefore we apply on tumor NGS data a “look-up” analysis previously validated to determine the mutation landscape of 30,678 MNR sites located within the ORF of human protein-coding genes8. Overall, a mean of 146 mutated MNRs was identified (min: 31; max: 394) corresponding to the 0.47% of the loci monitored. This percentage is not statistically different from what it is observed in EC MSI-h patients annotated in TCGA (Supplementary Fig. 1). FSMs shared in at least 50% of patients of our cohort occur in a group of 117 genes that include ACVR2A, RNF43, RPL22, KMT2C, SETD1B, PDS5B already reported as targets frequently mutated in MSI tumors9,10,19. Interestingly we observed an overall higher sharedness of FSMs in our cohort compared to the TCGA dataset especially for 5 genes (MNS1, KCNMA1, RBM45, VCP) that are mutated in all the patients of our cohort but have a reduced frequency of mutation (7%–18%) in the TCGA EC MSI cohort (Fig. 2c). We observed an enrichment of the number of mutated MNRs loci in the patients that show an impairment of MLH1 (with or without PMS2), with a mean of 184 mutations (min: 31; max: 394) compared to the patients’ groups that have impairment of MSH2 and/or MSH6 (mean: 46; min: 44; max: 49) or only PMS2 (mean: 71; min: 33; max: 140) (Fig. 2a). As previously observed, most mutations are del1 bp (mean in patients: 85; min: 16; max: 293) followed by insertion of 1 bp and di-nucleotide indels (Fig. 2b). To investigate the potential of an “off-the-shelf” vaccination strategy in EC, we determined the presence of mutations encoding the FSPs targeted by the NOUS-209 genetic vaccine. Overall, 80% of patients (28 out of 35) share with the NOUS-209 vaccine at least 1 FSP induced by the mutated MNRs. Specifically, an average of 16 FSPs for patients (max: 45) shared with the NOUS-209 were detected in the analyzed tumor samples (Fig. 3a). In total, 163 different FSPs out of the 209 targeted by the vaccine were present in at least one patient. On average, each FSP was present in 9% of patients (min: 3%; max: 43%) (Fig. 3b; Supplementary Table 2).
Fig. 2. Landscape of mutated MNR identified in the 35 MSI-h EC cohort.
a Blue bars indicate the total number of mutated MNRs identified in the cohort of 35 EC patients, according to the IHC MMR results, with mean for each group. b Boxplots represent the distribution and the abundance of the out-of-frame indels detected at mutated MNRs. The EC patients are shown as NOU-1 to NOU-35. c Scatterplot reporting the distribution of genes with respect to their frequency of mutation in the analyzed cohort of 35 patients and in the TCGA EC MSI cohort. Only the genes targeted by FSMs shared ≥50% of the cohort of 35 MSI EC are reported MNR mononuclear repeats, EC endometrial cancer, MSI microsatellite instability, IHC immunohistochemistry.
Fig. 3. Enrichment of vaccine-targeted FSPs in the MSI-H EC cohort.
The figure shows the profile of the NOUS-209 targeted FSPs identified in the cohort of patients: a total number of mutations encoding NOUS-209 FSPs found in the 35 patients; b sharedness of the 163 mutations found in at least one patient; c cumulative length in amino acids of the identified FSPs in each patient; d number of potentially immunogenic epitope identified for the patient. The EC patients are shown as NOU-1 to NOU-35. EC endometrial cancer; MSI-h microsatellite instability-high, FSP frameshift peptides.
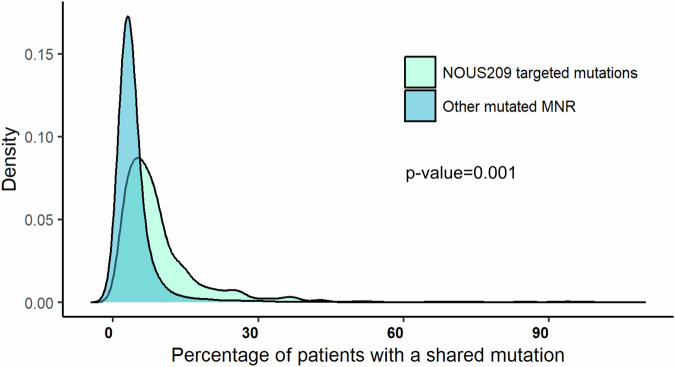
We estimate that the NOUS-209 FSPs present in each patient encode for an average of 402 non-self amino acids (min: 0; max: 1517aa) (Fig. 3c). From an immunologic point of view, the non-self neopeptides induced by indels have an antigenic potential similar to viral antigens. By reviewing published data of healthy volunteers vaccinated with viral vectors targeting polyantigens from HIV and HCV proteins, we determined that a non-self sequence of 400 aa targeted by a viral vector-based vaccine, was able to elicit an immune response against an average of 3 CD8 epitopes (min 1, max 12; Supplementary Fig. 2). We used this estimate to derive a rough “400 aa rule” that approximate the number of potential immunogenic epitopes induced by the FSPs by dividing the total length of non-self amino acid sequence by 400. On the basis of this rule, we estimated that the NOUS-209 vaccination has the potential to induce 1–4 immunogenic epitopes (Fig. 3d)8,20,21. Additionally, we compared the distribution of the frequency of detection of the 209 FSPs of the NOUS-209 vaccine against the frequency of detection of all the other FSPs detected in the EC patients. We estimate that the NOUS-209 FSPs selection enriches mutations more shared among the EC patients, with a median of 6% compared to 3% for the mutations detected in the other MNRs (KS test p-value = 0.001) (Fig. 4).
Fig. 4. Frequency of NOUS-209 FSPs mutations compared to other mutations.
Kernel Density Estimation plot reporting the distribution of the frequency of observation of NOUS-209 detected mutations (light blue) compared to the distribution of all the other mutations at MNR loci (blue). The density distribution was estimated by using the density function included in R 4.2.1. Statistical significance was determined with the Kolomogorv–Smirnoff test. EC endometrial cancer, FSP frameshift peptides.
Evidence of translation of NOUS-209 FSPs detected in the 35 EC
To test the potential of NOUS-209 FSPs to be effectively translated as part of expressed proteins, we performed a look-up of the mutations encoding the FSPs identified in the 35 patients in Exomeseq and Ribo-seq data performed on the AN3CA microsatellite unstable EC human cell line.
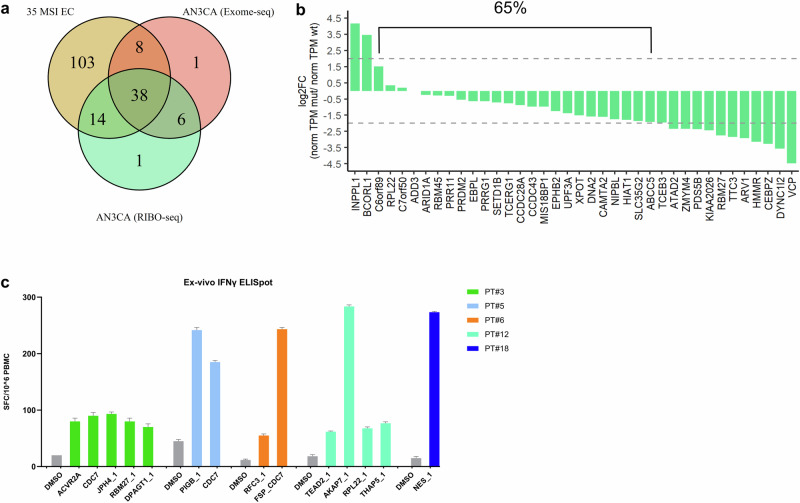
Overall, 46 NOUS-209 mutations out of the 163 detected in the 35 patients were also present in the Exomeseq of the cell line (Fig. 5a). Combining Exomeseq and Ribo-seq AN3CA data, 38 out of the 46 mutations (83%) were translated in mRNA and found bound to the ribosome (Fig. 5a). Additionally, 65% of mutations have an expression level comparable with the WT counterpart (log2 FC in the range ±2) (Fig. 5b)
Fig. 5. Detection of NOUS-209 FSMs in AN3CA cell line.
a Venn diagram showing the overlap among the FSPs detected in the 35 patients (brown) with Exomeseq (pink) and Ribo-seq (green) mutations detected in the AN3CA MSI EC cell line. b log2 fold change of the RNA expression of mutated vs the wild type counterparts for the 38 loci confirmed by Ribo-seq analysis. The expression is estimated by using normTPM measure (details in “Methods” section). c Immunogenicity of 14 FSPs in a cohort of MSI CRC vaccinated with NOUS-209. Ex vivo IFN-γ ELISpot on patients' PBMCs stimulated with pools of overlapping peptides covering the sequences of the tests FSPs. The number of SFCs per 106 PBMCs is reported. DMSO (peptide diluent represented the negative control).
We predicted the translatability of the 38 mutations found in Ribo-seq by using the TIS transformer pipeline to predict translation events that are more likely to occur16. According to this pipeline, 64 transcripts embedding 38 mutations were predicted with high translatability. Most of these transcripts were ranked with top prediction scores compared to the overall number of transcripts totally predicted, highlighting the higher coding potential of the 38 NOUS-209 FSPs in the EC cell line (Supplementary Fig. 3).
Immunogenicity of the FSPs detected in the 35 EC
Nous-209 vaccine is currently being investigated in a phase 1 trial in patients with metastatic gastric, colorectal, and gastro-esophageal junction MMRd tumors combined with αPD-1 pembrolizumab [17; NCT04041310]. Patients enrolled in this trial have received one Great Ape Adenoviruses (GAd) vaccination followed by three Modified Vaccinia Ankara (MVA) immunizations, in a “prime/repeated boost” protocol, combined with treatment with αPD-1 every 3 weeks. Immunogenicity was the secondary endpoint of the trial and was tested by ex vivo ELISpot on patients’ PBMCs with 16 pools of peptides corresponding to the 209 FSP targeted by NOUS-209.
Interestingly, among the immunogenic FSPs, we found 14 FSPs part of the 38 NOUS-209 FSPs identified in the cohort of the 35 EC. These data provide functional validation confirming the immunogenicity of the selected FSPs (Fig. 5c).
Discussion
In summary, the findings support the presence of nAgs in MMRd EC tumors which could be targeted by therapeutic “off-the-shelf” vaccination. In our MMRd EC cohort, we observed: (i) mutated MNRs loci with features consistent with previous data from TGCA8 and enrichment in MLH1 negative EC subset; (ii) MNRs encoding FSPs that are shared with those present in the cancer vaccination approach NOUS-209; (iii) experimental pieces of evidence of translatability and immunogenicity of the shared FSPs.
In particular, the enrichment of mutated MNR coding for nAgs FSPs in MLH/MLH1 plus PMS2 EC subset we found, could be explained by the functional overlap between MMR proteins. The MMR system consists of the four major proteins MLH1, MSH2, MSH6, and PMS2. The MMR proteins work two by two, forming the MLH1/PMS2 MutLα complex and the MSH2/MSH6 MutSα complex. MutSα recognizes single base pair mismatch and binds the MutLα to perform the excision of the single mismatch and to resynthesize the DNA strand. The correction of other types of DNA errors occurs via the MutSα that preferably binds with other MutL homologs complex, such as MutLβ (MLH1-PMS1) and MutLγ (MLH1-MLH3)22,23. In the MMR system, essential roles could be attributed to MLH1 and MSH2, in contrast to PMS2 and MSH6. The latter may potentially be compensated by other homologous proteins24–26. In the case of MSH6 loss, MSH3 can interact with MSH2 to form MutSβ complex and partially replace the MutSα activity, resulting in the correction of some DNA mismatch errors27,28. At the same time, a lower cancer penetrance was observed in PMS2-mutated Lynch families probably explained by the compensatory role of MLH3, which can bind and partially function with MLH1 (MutLγ complex)29–32.
Secondly, to explore the effective applicability of an “off-the-shelf” vaccine design in EC, we further validated the subset of the FSPs targeted by NOUS-209 and identified in the cohort of patients. The presence of vaccine-encoded FSPs was confirmed to achieve high coverage in our cohort (163 EC FSPs out of the 209 targeted by the vaccine). Moreover, comparing the distribution of all the FSPs detected in our samples with the subgroup of FSPs that match with the NOUS-209 pool, we calculated a significant enrichment of highly shared MNR mutations in EC patients, supporting the strength of the bioinformatic platform adopted (Fig. 4)8. These findings also confirmed the hypothesis that MMRd status leads to the occurrence of FSPs that are “shared” across tumors, being detectable in EC as well as other MSI tumors, as previously described in TGCA cohorts and in clinical validation settings8. Additionally, the majority of patients in our cohort (80%) shared at least 1 FSP matched with the NOUS-209 pool of FSPs (Fig. 2a) supporting the consistency of an “across-tumors” and an “across-patients” vaccination approach based on FSPs nAgs induced by MMRd status.
Finally, we estimated that the identified EC FSPs coded for at least one immunogenic epitope in a consistent number of patients (46%), supporting the immunogenicity of the NOUS-209. Therefore, our comparative analysis highlighted a high potential coding of the FSPs identified in the study, supporting the strength of vaccine-based immunotherapy and the immunogenicity of the NOUS-209 in the poorly explored clinical context of EC. According to the Ribo-seq data obtained in the MSI endometrial cell line, we determined that a relevant fraction of vaccine-targeted FSPs are expressed at a translational level. Moreover, a subset of these FSPs was found immunogenic in patients, providing functional validation of the selected antigens supporting the therapeutic potential of vaccinations targeting these nAgs.
Our study has major limitations related to a possible selection bias of the enrolled patients as we used IHC as a surrogate method for MSI evaluation. From the integration of IHC MMRd data with other molecular tests, it emerged that the discordant IHC/MSI-PCR cases (characterized by loss of PMS2 or MSH6) fall, among others, into the groups with lower abundance of the out-of-frame indels detected at mutated MNRs and lower abundance of NOUS-209 targeted FSPs. These results could be related to the functional overlap of MMR proteins, as abovementioned. At the same time, integrated data contribute to the controversial discussion about the definitive methodological strategy and the complex relation between MMRd and MSI. In this context, the unusual MMR phenotype (i.e. isolated MLH1 and MSH2 loss) identified in the cohort, was confirmed to be related to MLH1-promoter hypermethylation or MSI-h. The MSI-PCR and MMR-IHC methods are generally considered equivalent in diagnostic performance, even if the common MSI-PCR strategies are optimized for a panel of loci selected for colorectal cancers33. According to the ESMO Clinical Practice Guideline and ASCO recommendations, IHC is sufficient for the diagnosis of MMRd EC and it is recommended as standard practice34,35. Additionally, several papers and meta-analyses showed that IHC is an accurate surrogate of MSI molecular testing in EC tumors36,37. Despite a substantial agreement, discrepancies were also described38–41. Overall, patients with MMRd EC should be referred for MLH1 gene promoter hypermethylation (in case of MLH1/PMS2 loss) and/or germline testing, regardless of the results of MSI analysis42. An additional limitation of the study lies in the lack of normal tissues for the identification of true positive mutations and the overall reduced accuracy of Exomeseq technology compared to other PCR-based analytic techniques for the identification of mutations at MNR loci43. From a technical point of view, it is unfeasible to monitor by PCR all the MNR loci located within the ORF of the protein-coding human genes. To overcome the lack of healthy tissues for comparison, we decided to adopt a previously published pipeline with thresholds validated for detecting a reduced number of false positive mutations8.
The rational development of an effective cancer vaccination mainly relies on the successful identification of potent nAgs, which may be theoretically recognized as “non-self” by the patient’s immune system, presented with HLA molecules on cell surfaces, trigger immune responses, and finally lead to nAgs-expressing cancer cells death32. In the last couple of years, several approaches have been adopted for genetic cancer vaccine design. Most of the studies focus on vaccines targeting nAgs identified from the patient’s tumor, generating an individual-specific vaccine. However, the adoption of a personalized single-tumor strategy is considered complex and expensive44–46. On the other hand, previous studies reported the widespread occurrence of nAgs that are common across MSI tumors leading to the development of “off-the-shelf”9,44.
Neoantigen-based vaccines represent an emergent immunotherapy approach evaluated in several clinical trials involving different solid tumors (NCT03502785, NCT03289962)45. The MicOryx was the first-in-human clinical trial to investigate an FSP vaccine in a cohort of participants with MMRd colorectal cancer (NCT01461148). The experimental vaccine was based on a combination of three recurrent frameshift-derived nAgs and it has proven to be safe, showing strong immune responses against FSPs in all patients vaccinated46,47. An ongoing Phase I/II clinical trial explores the utility of a dendritic cell-based vaccine loaded with nAgs in MMRd colorectal cancer patients (NCT01885702). Even though the analyzed patients are not endometrial patients, we expect that the immune response and the immunogenic potential of the FSP will be similar in endometrial patients because they share the hallmark of being MSI.
In this context, the polyvalent viral vectored vaccine developed by Nouscom and named NOUS-209 has been demonstrated to be safe and highly immunogenic in MMRd/MSI-h patients affected by colorectal, gastric, or gastro-esophageal junction cancers in combination with Pembrolizumab (NCT04041310)8,48,49. The trial is now enrolling patients for a phase-2 study to assess the clinical efficacy50. NOUS-209 is a genetic vaccine designed to maximize the intra-patient and inter-patient antitumoral immune response in MSI tumors8. In MMRd EC, the benefit observed in patients receiving the anti-PD-1 ICIs provides indirect evidence that anti-tumor T cells, present in inflamed tumor tissue and likely recognizing the nAgs, could be reactivated. In this framework, our data represent a solid basis to warrant further evaluation of NOUS-209 in combination with immune-checkpoint inhibitors in EC patients. Finally, MSI-h status is reported in approximately 4% of all diagnosed cancers and is the molecular hallmark of Lynch Syndrome51. Lynch syndrome results from mutations in DNA mismatch repair genes (i.e. MLH1, MSH2, MSH6, or PMS2) inherited in an autosomal dominant manner. It occurs in 3% of patients with colorectal cancer and 6% of patients with EC, but it is also associated with other tumor types (renal pelvis, ovary, stomach, small bowel, and ureter)52. Individuals with Lynch Syndrome have a lifetime-increased risk of MSI cancer development mainly colorectal (40%–60% in women; up to 90% in men) and endometrial (40%-60%)53. The diagnosis of Lynch syndrome implies the adoption of screening and prevention strategies. Germline defects in DNA mismatch repair genes predispose carriers to frameshift mutations at coding mononucleotide repeats in the genome53. The occurrence of MSI-h cancers in the hereditary context of Lynch Syndrome provides the unique opportunity to evaluate such a vaccine approach as a cancer-preventive strategy in high-risk mutation carriers, identifying a valuable cohort of patients currently being studied (NCT05078866).
The typical features of MSI tumors, as here described, and the opportunity to develop an “off-the-shelf” nAgs-based cancer vaccine partially solve some of the most relevant barriers for cancer vaccination adoption, being based on shared expressed antigens that: (i) potentially cover cancer timing evolution; (ii) does not require a patient-specific pre-selection of potential nAgs subset. Several studies highlighted the relationship between the nAgs load (i.e. the number of unique nAgs) and the good clinical response to the ICIs for various cancer types6. It is reasonable to suppose that the combination of nAgs-based cancer vaccination with ICIs may represent a valuable approach aimed at improving response rates in MSI-h EC.
Supplementary information
Author contributions
E.D.P., C.N., E.M., and G.L. “Writing - Original Draft”; G.S., A.U., and F.F. “Project administration” and “Conceptualization”; E.D.P., C.N., and E.S. “Methodology”; E.M., G.L., A.M.D., and A. P. “Software” and “Formal Analysis”; E.D.P., M.D.B, R.T., A.P., and D.P. “Investigations”; T.P. and L.G. “Data Curation”; A.M, G.D., and G.Z. “supervision”.All authors discussed, edited, and contributed to the final manuscript, approving the submitted version.
Data availability
The datasets used and analyzed during the current study are available from the corresponding authors on reasonable request and with permission of the Data Protection Officer of the “Fondazione Policlinico Universitario A. Gemelli” IRCCS of Rome.
Competing interests
E.S. is the founder of Nouscom. E.M., L.G., E.S., and A.M.D, are employed by Nouscom. E.S., E.M., L.G., E.S., and A.M.D declare no financial or non-financial competing interests. All other authors declare no financial or non-financial competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Andrea Urbani, Francesco Fanfani.
Supplementary information
The online version contains supplementary material available at 10.1038/s41698-024-00779-4.
References
- 1.Ramchander, N. C. et al. Distinct immunological landscapes characterize inherited and sporadic mismatch repair deficient endometrial cancer. Front. Immunol.10, 3023 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mas-Ponte, D., McCullough, M. & Supek, F. Spectrum of DNA mismatch repair failures viewed through the lens of cancer genomics and implications for therapy. Clin. Sci.136, 383–404 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linnebacher, M. et al. Frameshift peptide-derived T-cell epitopes: a source of novel tumor-specific antigens. Int. J. Cancer93, 6–11 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Boland, C. R. et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res.58, 5248–5257 (1998). [PubMed] [Google Scholar]
- 5.Vogelstein, B. et al. Cancer genome landscapes. Science339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hodi, F. S. et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med.363, 711–723 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mirza, M. R. et al. Dostarlimab for primary advanced or recurrent endometrial cancer. N. Engl. J. Med.388, 2145–2158 (2023). [DOI] [PubMed] [Google Scholar]
- 8.Leoni, G. et al. A genetic vaccine encoding shared cancer neoantigens to treat tumors with microsatellite instability. Cancer Res.80, 3972–3982 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Kim, T. M. et al. The landscape of microsatellite instability in colorectal and endometrial cancer genomes. Cell155, 858–868 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roudko, V. et al. Shared immunogenic poly-epitope frameshift mutations in microsatellite unstable tumors. Cell183, 1634–1649.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin, M. J. et al. Cancer vaccines: the next immunotherapy frontier. Nat. Cancer3, 911–926 (2022). [DOI] [PubMed] [Google Scholar]
- 12.Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thiel, T. et al. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). TAG. Theor. Appl. Genet.106, 411–422 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Leoni, G. et al. VENUS, a novel selection approach to improve the accuracy of neoantigens’ prediction. Vaccines9, 880 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clauwaert, J., McVey, Z., Gupta, R. & Menschaert, G. TIS Transformer: remapping the human proteome using deep learning. NAR Genom. Bioinform.5, lqad021 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D’Alise, A. M., et al. Adenoviral-based vaccine promotes neoantigen-specific CD8+ T cell stemness and tumor rejection. Sci. Transl. Med.10.1126/scitranslmed.abo7604 (2022). [DOI] [PMC free article] [PubMed]
- 18.Salem, M. E. et al. Impact of MLH1, PMS2, MSH2, and MSH6 alterations on tumor mutation burden (TMB) and PD-L1 expression in 1,057 microsatellite instability-high (MSI-H) tumors. J. Clin. Oncol.36, 3572–3572 (2018). [Google Scholar]
- 19.Giannakis, M. et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat. Genet.46, 1264–1266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borthwick, N. et al. Vaccine-elicited human T cells recognizing conserved protein regions inhibit HIV-1. Mol. Ther. J. Am. Soc. Gene Ther.22, 464–475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swadling, L. et al. Highly-immunogenic virally-vectored T-cell vaccines cannot overcome subversion of the T-cell response by HCV during chronic infection. Vaccines4, 27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fishel, R. Mismatch repair. J. Biol. Chem.290, 26395–26403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pannafino, G. & Alani, E. Coordinated and independent roles for MLH subunits in DNA repair. Cells10, 948 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Acharya, S. et al. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc. Natl Acad. Sci. USA93, 13629–13634 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kondo, E., Horii, A., & Fukushige, S. The interacting domains of three MutL heterodimers in man: hMLH1 interacts with 36 homologous amino acid residues within hMLH3, hPMS1 and hPMS2. Nucleic Acids Res.29, 1695–1702 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yılmaz, A., Mirili, C., Bilici, M. & Tekin, S. B. Colorectal cancer in Lynch syndrome associated with PMS2 and MSH6 mutations. Int. J. Colorectal Dis.35, 351–353 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Montminy, E. M. et al. Shifts in the proportion of distant stage early-onset colorectal adenocarcinoma in the United States. Cancer Epidemiol. Biomarkers Prev.31, 334–341 (2022). [DOI] [PubMed] [Google Scholar]
- 28.Umar, A. et al. Functional overlap in mismatch repair by human MSH3 and MSH6. Genetics148, 1637–1646 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasela, M., Nyström, M. & Kansikas, M. PMS2 expression decrease causes severe problems in mismatch repair. Hum. Mutat.40, 904–907 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cannavo, E. et al. Expression of the MutL homologue hMLH3 in human cells and its role in DNA mismatch repair. Cancer Res.65, 10759–10766 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Korhonen, M. K., Raevaara, T. E., Lohi, H. & Nyström, M. Conditional nuclear localization of hMLH3 suggests a minor activity in mismatch repair and supports its role as a low-risk gene in HNPCC. Oncol. Rep.17, 351–354 (2007). [PubMed] [Google Scholar]
- 32.Senter, L. et al. The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology135, 419–428 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nádorvári, M. L. et al. Microsatellite instability and mismatch repair protein deficiency: equal predictive markers? Pathol. Oncol. Res.30, 1611719 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oaknin, A. et al. ESMO Guidelines Committee. Endometrial cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol.33, 860–877 (2022). [DOI] [PubMed] [Google Scholar]
- 35.Vikas, P. et al. Mismatch repair and microsatellite instability testing for immune checkpoint inhibitor therapy: ASCO Endorsement of College of American Pathologists Guideline. J. Clin. Oncol.41, 1943–1948 (2023). [DOI] [PubMed] [Google Scholar]
- 36.Raffone, A. et al. Diagnostic accuracy of immunohistochemistry for mismatch repair proteins as surrogate of microsatellite instability molecular testing in endometrial cancer. Pathol. Oncol. Res.26, 1417–1427 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Stelloo, E. et al. Practical guidance for mismatch repair-deficiency testing in endometrial cancer. Ann. Oncol.28, 96–102 (2017). [DOI] [PubMed] [Google Scholar]
- 38.McConechy, M. K. et al. Detection of DNA mismatch repair (MMR) deficiencies by immunohistochemistry can effectively diagnose the microsatellite instability (MSI) phenotype in endometrial carcinomas. Gynecol. Oncol.137, 306–310 (2015). [DOI] [PubMed] [Google Scholar]
- 39.McCarthy, A. J. et al. Heterogenous loss of mismatch repair (MMR) protein expression: a challenge for immunohistochemical interpretation and microsatellite instability (MSI) evaluation. J. Pathol. Clin. Res.5, 115–129 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Streel, S. et al. Diagnostic performance of immunohistochemistry compared to molecular techniques for microsatellite instability and p53 mutation detection in endometrial cancer. Int. J. Mol. Sci.24, 4866 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riedinger, C. J. et al. Characterization of mismatch-repair/microsatellite instability-discordant endometrial cancers. Cancer130, 385–399 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dedeurwaerdere, F. et al. Comparison of microsatellite instability detection by immunohistochemistry and molecular techniques in colorectal and endometrial cancer. Sci. Rep.11, 12880 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ballhausen, A. et al. The shared frameshift mutation landscape of microsatellite-unstable cancers suggests immunoediting during tumor evolution. Nat. Commun.11, 4740 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bolivar, A. M., Duzagac, F., Sinha, K. M. & Vilar, E. Advances in vaccine development for cancer prevention and treatment in Lynch Syndrome. Mol. Asp. Med.93, 101204 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reardon, D. A. et al. Intramuscular (IM) INO-5401 + INO-9012 with electroporation (EP) in combination with cemiplimab (REGN2810) in newly diagnosed glioblastoma. J. Clin. Oncol.40, 2004–2004 (2022).35108031 [Google Scholar]
- 46.Kloor, M. et al. A frameshift peptide neoantigen-based vaccine for mismatch repair-deficient cancers: a phase I/IIa clinical trial. Clin. Cancer Res.26, 4503–4510 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Reuschenbach, M. et al. A multiplex method for the detection of serum antibodies against in silico-predicted tumor antigens. Cancer Immunol. Immunother.63, 1251–1259 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.D’Alise, A. M. et al. Adenoviral-based vaccine promotes neoantigen-specific CD8+ T cell stemness and tumor rejection. Sci. Transl. Med.14, eabo7604 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fakih, M. et al. First clinical and immunogenicity results including all subjects enrolled in a phase I study of Nous-209, an off-the-shelf immunotherapy, with pembrolizumab, for the treatment of tumors with a deficiency in mismatch repair/microsatellite instability (dMMR/MSI). J. Clin. Oncol.40, 2515–2515 (2022).35724356 [Google Scholar]
- 50.Overman, M. J. et al. Results of phase I-II bridging study for Nous-209, a neoantigen cancer immunotherapy, in combination with pembrolizumab as first line treatment in patients with advanced dMMR/MSI-h colorectal cancer. J. Clin. Oncol.41, e14665–e14665 (2023). [Google Scholar]
- 51.Cortes-Ciriano, I., Lee, S., Park, W. Y., Kim, T. M. & Park, P. J. A molecular portrait of microsatellite instability across multiple cancers. Nat. Commun.8, 15180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papadopoulou, E. et al. Microsatellite instability is insufficiently used as a biomarker for Lynch syndrome testing in clinical practice. JCO Precis Oncol.8, e2300332 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meyer, L. A. et al. Endometrial cancer and Lynch syndrome: clinical and pathologic considerations. Cancer Control16, 14–22 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding authors on reasonable request and with permission of the Data Protection Officer of the “Fondazione Policlinico Universitario A. Gemelli” IRCCS of Rome.