Abstract
Background
Cancer stem cells (CSCs) have unique metabolic characteristics and are hypothesized to contribute significantly to the recurrence and drug resistance of glioblastoma multiforme (GBM). However, the reliance on mitochondrial metabolism and the underlying mechanism of glioblastoma stem cells (GSCs) remains to be elucidated.
Methods
To quantify differential mitochondrial protein expression between GSCs and differentiated cells, a mass spectrum screen was applied by the Stable Isotope Labeling with Amino Acids in Cell Culture (SILAC) technique. Functional experiments including CCK8, neurosphere formation, flow cytometry, transwell, and wound healing assays were conducted to evaluate GBM cell malignant phenotype. The potential molecular mechanism of FDFT1 was screened by RNA-seq analyses. The candidate target genes were validated through RT-qPCR and western blot analyses.
Results
As a top candidate, FDFT1 protein expression in GSCs was elevated relative to their differentiated counterparts. Functionally, the knockdown of FDFT1 suppressed the GBM cell proliferation and migration, while simultaneously enhancing sensitivity to temozolomide. Treatment with both the FDFT1 inhibitor (YM-53601) and simvastatin (an HMG-CoA reductase inhibitor) induced apoptosis in GSCs. Mechanistically, FDFT1 was transcriptionally regulated by SREBP2 but not SREBP1. Furthermore, FDFT1 activates the AKT pathway to regulate tumor metabolism and maintain the stemness of tumor cells.
Conclusions
GSCs exhibit a dependency on FDFT1-mediated mevalonate metabolism. Inhibition of FDFT1 could represent a potent strategy to eliminate GSCs.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13287-024-04102-7.
Keywords: Glioblastoma stem cell, FDFT1, YM-53601, cholesterol
Background
Glioblastoma multiforme (GBM), classified as a World Health Organization (WHO) grade IV astrocytoma, is one of the most prevalent tumors of the central nervous system. GBM is characterized by rapid cell proliferation, high malignancy, pronounced invasiveness, and a high mortality rate [1]. Although immunotherapy centered on immune checkpoint inhibitors has revolutionized the landscape of cancer treatment in recent years, its impact on neurological malignancies has been less pronounced [2, 3]. The primary approach to treating glioblastoma still relies on a conventional combination of surgical resection, radiotherapy, and chemotherapy. Previous research has established that glioblastoma stem cells (GSCs) possess resistance to standard chemotherapeutic agents such as carmustine (bis-chloroethylnitrosourea, BCNU) and temozolomide (TMZ), along with radiation therapy [4, 5]. Given that GSCs may drive the recurrence of GBM [6, 7], there is an urgent need to further uncover the mechanisms of GSCs in recurrence and metastasis, thereby providing a convincing strategy for the development of targeted therapies against these cells to combat this highly aggressive form of cancer.
Abnormal metabolic state is considered as one of the biological characteristics of cancer stem cells. As the critical hub of cellular metabolism, mitochondrial metabolism is the essential condition that determines the fate of stem cells [8]. Our previous studies revealed that glioblastoma stem cells exhibit an altered mitochondrial metabolism compared to differentiated tumor cells [9–11]. These stem cells rely on aerobic glycolysis, or the Warburg effect, to generate energy in the absence of oxygen, a phenomenon commonly observed in cancer cells. Conversely, differentiated tumor cells use oxidative phosphorylation to produce ATP. This metabolic shift in glioblastoma stem cells enables them to adapt to hypoxic environments and acquire a survival advantage. Importantly, the aberrant mitochondrial metabolism of GSCs is implicated in their resistance to standard therapeutic interventions, thereby complicating treatment strategies. Nevertheless, the specific molecules that influence mitochondria in regulating the fate and function of GSCs require further elucidation. Recently, we employed stable isotope labeling with amino acids in cell culture (SILAC) technique and identified FDFT1 to be elevated in GSCs.
FDFT1 is an enzyme that encodes squalene synthetase (SQS) and is implicated in the mevalonate (MVA) metabolic pathway. Recently, FDFT1 appears as an inhibitory factor or a cancer-promoting factor in different tumors [12–14]. For instance, FDFT1 has been shown to inhibit the malignant progression of colorectal cancer and ovarian cancer [12, 13], while promoting the carcinogenesis of tongue squamous cell carcinoma [14]. However, the biological function and mechanisms of FDFT1 in GSCs remain unclear. Here, we identified that FDFT1 plays a significant role in maintaining stemness and its inhibition led to the suppression of stem cell markers and an increased susceptibility to conventional chemotherapy. FDFT1 was transcriptionally regulated by SREBP2, which in turn activates the AKT pathway to regulate tumor metabolism. These findings underscore the promise of FDFT1 as a potential therapeutic target for the regulation of both metabolic activities and stemness characteristics in GSCs.
Methods
Cell culture
The glioblastoma cancer stem cell lines, GSC11 and GSC23, initially isolated from human glioblastoma tissues and utilized in several prior studies [10, 15, 16], were maintained in Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F-12; Gibco), enriched with B-27 supplement (Gibco), 2 mM L-glutamine (Mediatech), and supplemented with 20 ng/mL of both recombinant human epidermal growth factor (EGF; R&D Systems) and basic fibroblast growth factor (bFGF; R&D Systems). The established glioblastoma cell lines U87 and U251, procured from the American Type Culture Collection (ATCC; Rockville, MD, USA), were routinely maintained in DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS; Serana). All cell lines were incubated at 37 °C in a humidified atmosphere containing 5% CO2. Detailed stem cell culture method was previously reported by Fathi et al. [17].
Gene knockdown and overexpression
The transfection of siRNA, shRNA and overexpression plasmids were performed as described previously [18]. The Small RNA interference (siRNAs) against FDFT1, SREBP1, SREBP2 or scrambled siRNA were synthesized from IBSBIO (Shanghai, China), siRNA oligos were listed in Table S1. Knockdown of FDFT1 expression in U87 or U251 cells was performed using jetPRIME in vitro DNA& siRNA transfection Reagent. Briefly, 3 × 105 cells per well were seeded into six-well plates and allowed to settle overnight. Subsequently, the medium in each well was replaced with 2 ml of fresh medium containing 200 µl of a transfection mixture comprising jetPRIME Buffer, siRNA or scrambled RNA, and jetPRIME Reagent. Following a 48-hour incubation period, the transiently transfected cells were harvested, and RNA was extracted for subsequent analysis via quantitative reverse transcription polymerase chain reaction (qRT-PCR). For overexpression studies, the pCDNA3.1 lentiviral vector system was employed to enhance FDFT1 expression in U87 cells. The coding sequence of FDFT1 (NM_004462.5) was inserted into the pCDNA3.1 vector, which also included a green fluorescent protein (GFP) marker for tracking transfection efficiency. The efficiency of FDFT1 overexpression was assessed using both qRT-PCR and Western blot analysis.
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
PCR assay was conducted as described before [19]. U87 cells were lysed using Trizol reagent (Invitrogen) to obtain total RNA. From this RNA, 1 µg was reverse-transcribed into complementary DNA (cDNA) using the HiScript III RT SuperMix (Vazyme). The cDNA, diluted to 1/50 of the initial reverse transcription product, was then quantified in triplicate using a SYBR green master mix on a Roche LightCycler 480 real-time PCR system. β-actin served as an endogenous control to normalize the expression levels. The relative mRNA expression of the target gene was calculated using the 2−ΔΔCt method, which accounts for variations in the initial amount of RNA and efficiency of the reverse transcription reaction. Primers for each gene were listed in Table S2.
Western blot
In line with previous practices [19], a modified RIPA lysis buffer with high potency, fortified with 1% protein phosphatase inhibitor, 1% protease inhibitor, and 1% phenylmethylsulfonyl fluoride (PMSF) from Fdbio, was employed for protein extraction. Cells were initially washed twice with ice-cold phosphate-buffered saline (PBS) and then lysed in 100–200 µl of the RIPA buffer for 30 min. Following lysis, cell debris was pelleted by centrifugation at 12,000 rpm for 15 min at 4 °C. The supernatants, enriched with soluble proteins, were carefully collected. Protein concentrations were subsequently quantified using the BCA Protein Assay Kit (Beyotime Biotechnology). Equivalent amounts of protein from each sample were loaded and resolved via electrophoresis on a denaturing 10% SDS-polyacrylamide gel. The resolved proteins were then transferred onto a polyvinylidene fluoride (PVDF) membrane. The membrane was immunodecorated with antibodies specific for FDFT1 (abcam, #ab236666), CD133 (Cell Signaling Technology, CST, #64326S), GFAP (CST, #80788S), SREBP1 (Abclonal, #A15586), SREBP2 (Abclonal, #A13049), PI3K (Abclonal, # A22730), Phospho-PI3K (Abclonal, #AP0427), AKT (Bioworld Technology, # AP0059), Phospho-Akt-T308 (Bioworld Technology, # AP0056), Phospho-Akt-S473 (Bioworld Technology, # BS4007) and β-actin (abcam, # ab8227), with β-actin serving as the loading control.
Cell viability assay
Cell proliferation was assessed using the CCK-8 assay as reported previously [20]. Initially, FDFT1 was suppressed by siRNA or pharmacologically inhibited with YM-53,601. Control cells were then plated in triplicate in 96-well plates at a density of 1000 cells per well and incubated for 24 h. Subsequently, 10% CCK-8 reagent (APE x BIO) diluted in complete medium was added to each well. The plates were then incubated at 37 °C for 3 h in the dark, after which the optical density (OD) at 450 nm was measured using a microplate reader. This measurement was taken daily for a period of 5 days to generate growth curves. The cell viability data were normalized to the OD value recorded on the first day to account for any initial variability.
Neurosphere formation assay
Consistent with our previous approach [10], for the neurosphere formation assay, glioblastoma stem cells GSC11 and GSC23 were plated at a density of 3 × 105 cells per well in 6-well plates. Following a 48-hour treatment with either simvastatin (STA) or YM-53,601, or their respective controls, the neurosphere formation was examined using a light microscope (Leica DMi 1). For the cell differentiation assay, GSC11 and GSC23 were plated similarly at a density of 3 × 105 cells per well in 6-well plates. The cells were induced to differentiate with or without the addition of 10% fetal bovine serum (FBS) for a period of 3 days. Subsequently, the morphological changes indicative of differentiation were photographed using an inverted microscope (Olympus IX71).
Flow cytometry analysis of cell apoptosis
Annexin V/propidium iodide (PI) staining was employed to evaluate the apoptotic effects induced by TMZ, STA and YM-53,601 in U87, GSC11 and GSC23 cells. Indicated cells were plated in 6-well plates and exposed to the respective drugs for 48 h. Post-treatment, cells were harvested into flow cytometry tubes, and suspended as single-cell preparations. The cells were then washed twice with phosphate-buffered saline (PBS). Subsequently, 5 µl of Annexin V–FITC and PI reagents were added to the cell suspension, which was incubated for 20 min at room temperature in the dark. The proportion of apoptotic cells was determined using flow cytometry.
Cell migration assay
Cell migration capacity was assessed using the transwell assay and wound healing assay as documented previously [21]. For the transwell migration assay, a total of 3 × 104 U87 cells or 2 × 104 U251 cells in 200 µl FBS free culture medium were seeded in triplicate onto the upper surface of 8.0-µm filter migration chambers. The lower compartment was filled with 800 ul DMEM medium with 10% FBS to serve as a chemoattractant. The cells were incubated at 37 °C in a 5% CO2 atmosphere for 24 h. Non-migrating cells on the upper surface were gently removed with cotton swabs. The cells that had migrated to the lower surface were fixed with methanol and stained with crystal violet. Images of three random fields were captured using an upright microscope (Nikon Ni-U) to quantify the migratory cells. For the wound healing assay, cells were plated into 6-well plates (8 × 105 cells/mL) using serum-free medium. After 24 h, a scratch was made through the cell monolayer with a pipette tip to create a ‘wound.’ The rate of cell migration into the wounded area was monitored over time using an inverted microscope (Olympus IX71), providing a visual representation of cell motility and healing capacity.
Mitochondrial isolation
Mitochondria were isolated from cells as described previously [22]. Here, mitochondria from GSC11 and GSC23 cells were extracted using the Q-proteome Mitochondria Isolation kit from Qiagen. The procedure began with washing the cells and suspending them in lysis buffer, followed by centrifugation at 1000 × g for 10 min to pellet the cells. The pellet was then resuspended in Disruption Buffer and homogenized using a syringe, passing it through 10 times to break up the cells. After homogenization, the mixture was centrifuged again at 1000 × g for 10 min to remove unbroken cells, nuclei, and debris. The supernatant, which contains mitochondria and microsomes, was then centrifuged at 6000 × g for 10 min to pellet the mitochondria. The supernatant was carefully removed, and the crude mitochondrial pellet was resuspended in Mitochondria Purification Buffer. Further purification was achieved through density gradient centrifugation to separate mitochondria from other organelles. The purified mitochondrial fraction was then resuspended in a lysis buffer containing 8 M urea, 4% CHAPS, 65 mM DTT, and 40 mM Tris to solubilize the mitochondrial proteins. The suspension was sonicated at 100 watts for 30 s to disrupt the mitochondrial membranes and release the proteins. Following sonication, the mixture was centrifuged at 25,000 × g for 30 min to pellet the insoluble material. The supernatant, which contains the solubilized mitochondrial proteins, was collected and used for subsequent proteomic analysis.
Cell culture with stable isotope labeling
The culture medium supplemented with 10% FBS and devoid of isotopic labeling, formulated with L-arginine (R0) at a concentration of 84 mg/ml or L-lysine (K0) at 146 mg/ml, was designated as the “light” (L) medium. Conversely, the “heavy” (H) medium was devoid of FBS but was enriched with isotopically labeled amino acids, specifically L-[13C6,15N4] arginine (R10) and L-[13C6,15N2] lysine (K8). The GSC11 and GSC23 stem cells were cultured in the heavy medium, whereas the differentiated cells were grown in the light medium for a minimum of five passages. This cultivation period was essential to ensure the comprehensive incorporation of the isotopic amino acids into the cellular proteins. Cells were then harvested and extracted protein for LC-MS analysis as previously [23].
RNA sequencing and analysis methodology
Total RNA was extracted from U87 and U251 cells post-transfection using TRIzol reagent. Subsequent cDNA library synthesis and paired-end sequencing services were provided by Gene Denovo (Guangzhou, China). RNA integrity was assessed using the Agilent Bioanalyzer 2100 system. Paired-end sequencing reads were generated on the Illumina Novaseq 6000 platform and aligned to the human reference genome (hg38) with hisat2. Differential expression analysis was conducted using the R package DESeq2 (version 1.34.0), with statistical significance set at p < 0.05 to identify differentially expressed genes.
Statistical analysis
All experiments were performed in triplicate to ensure reproducibility and reliability of the results. GraphPad prism software was used to analyze the data. For pairwise comparisons, Student’s t-test was employed to ascertain statistical differences. In cases where multiple groups were compared, one-way analysis of variance (ANOVA) was utilized. A p-value of less than 0.05 was considered the threshold for statistical significance.
Results
SILAC identifies FDFT1 overexpress in glioblastoma neurosphere cells
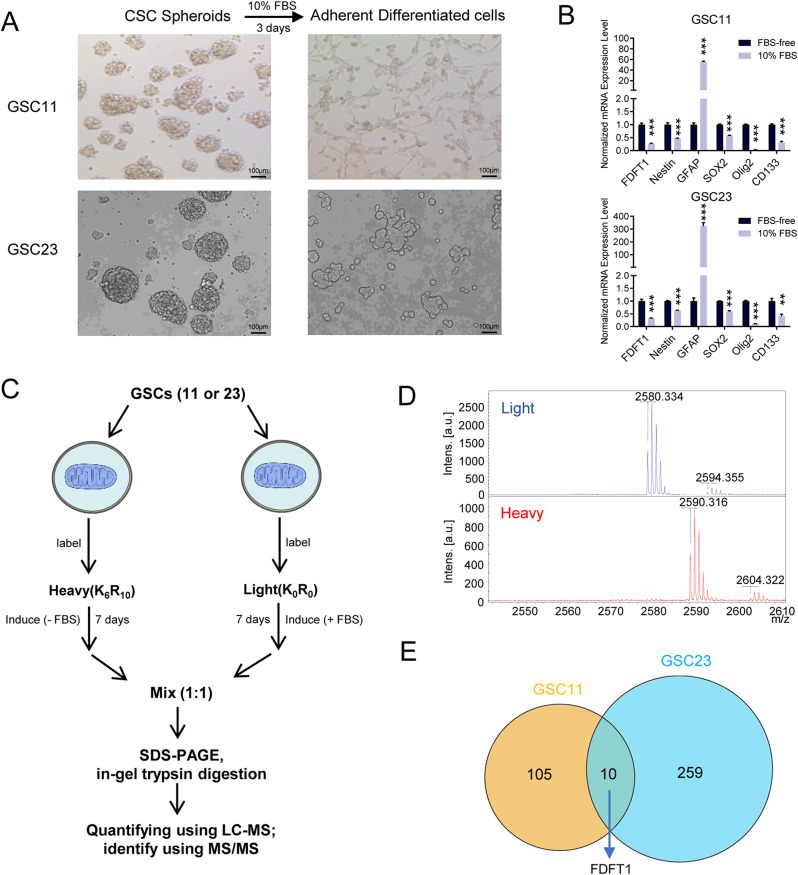
To study the underlying mechanism of differentiated metabolism relyment, two well-characterized, patient-derived glioblastoma stem cell-enriched spheroid lines, GSC11 and GSC23, were induced differentiation using FBS as described previously [10, 15]. After serum induction, the cells transitioned from a round neurospheric morphology to a more elongated, spindle-shaped adherent growth (Fig. 1A). Concurrently, there was a decrease in the expression of stemness markers such as CD133 and Nestin, and an increase in GFAP, indicative of neuronal differentiation (Fig. 1B). Since mitochondria is a crucial hub for cellular metabolism, we hypothesized the existence of possible proteomic shifts in cellular metabolism corresponding to the transition from undifferentiated to differentiated states. To investigate that, we employed SILAC, a robust quantitative proteomic technique, to compare mitochondrial proteins across both the undifferentiated neurospheric and differentiated adherent cell states. As depicted in Fig. 1C, glioblastoma neurosphere cells were cultured in a medium containing isotopically labeled (heavy) amino acids, while glioblastoma differentiated cells were grown in regular (light) medium. The different m/z ratios suggested successful isotopic labeling after several passages (Fig. 1D). Among the differential expressed proteins, we identified FDFT1 as being the most significantly overexpressed in glioblastoma neurosphere cells. In comparison, the gene was found to be overexpressed by about 22-fold in G11 and 3-fold in G23 stem cells relative to their corresponding differentiated cells, indicating a potential role in the maintenance of the stem cell phenotype (Table S3, Fig. 1E).
Fig. 1.
Mitochondrial Protein Expression Analysis in Glioblastoma Stem Cells. (A) Cellular morphological changes before and after differentiation of GSCs. (B) mRNA expression levels of stemness markers before and after serum induction in GSCs. (C) Schematic diagram of the workflow for mitochondrial proteome analysis using stable isotope labeling in cultured cells. (D) Validation of effective isotope labeling by mass spectrometry analysis. (E) Comparative analysis of differentially expressed proteins in GSCs before and after differentiation
FDFT1 promotes the proliferation and migration of glioma cells
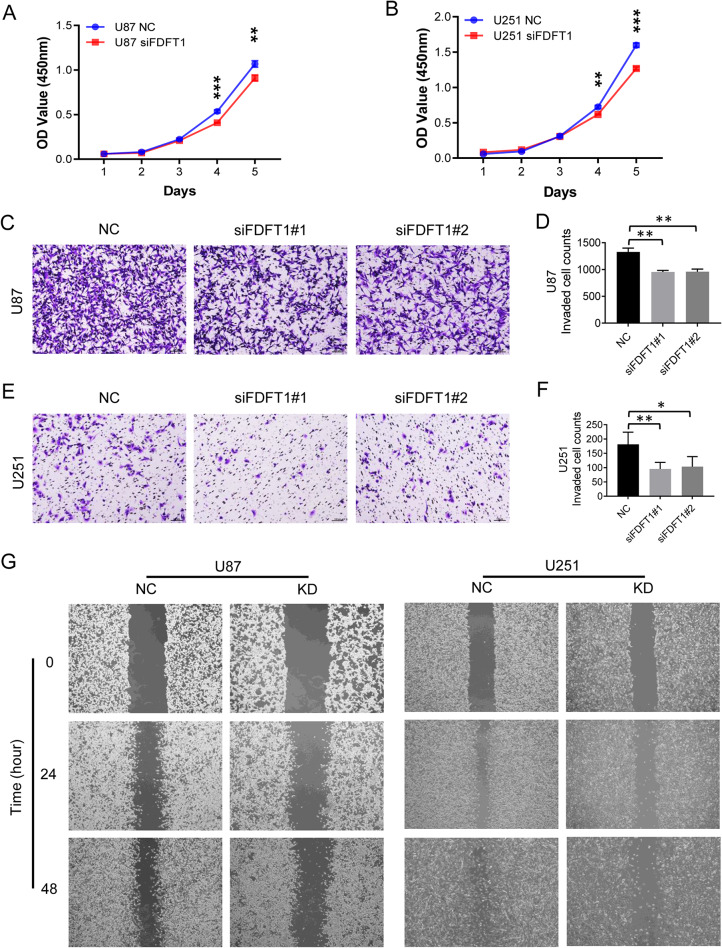
We next investigated the effects of FDFT1 perturbation on the proliferation and migration capabilities of glioma cells. Gene knockdown is confirmed to be effective by Western blot (Fig. S1). Our results indicated that downregulation of FDFT1 led to a significant reduction in the proliferation rate of U87 and U251 cells (Fig. 2A, B). The transwell assay was performed to evaluate the migration capacity. A marked decrease in the number of migrated cells was observed in FDFT1 knockdown U87 cells compared to the control group (Fig. 2C, D). Similar findings were observed in U251 cells (Fig. 2E, F). Additionally, the wound healing assay revealed a diminished migratory capacity in both glioma cell types when FDFT1 was downregulated (Fig. 2G).
Fig. 2.
FDFT1 Regulation on Glioma Cell Proliferation and Invasion Capacity. (A) CCK-8 assay evaluating the impact of FDFT1 on the proliferation of U87 cells. (B) CCK-8 assay assessing the effect of FDFT1 on the proliferation of U251 cells. (C) Transwell assay examining the influence of FDFT1 on the invasive capacity of U87 cells (magnification X10). (D) Quantitative analysis of invaded U87 cells. (E) Transwell assay investigating the effect of FDFT1 on the invasive capacity of U251 cells (magnification X10). (F) Quantitative analysis of invaded U251 cells. (G) Wound-healing assay determining the influence of FDFT1 on the invasive abilities of both U87 and U251 cells. Abbreviations: NC, negative control using scrambled siRNA; KD, knockdown; OE, overexpression; TMZ, temozolomide; *, p < 0.05; **, p < 0.01; ***, p < 0.001
FDFT1 overexpression causes drug resistance of glioma cells
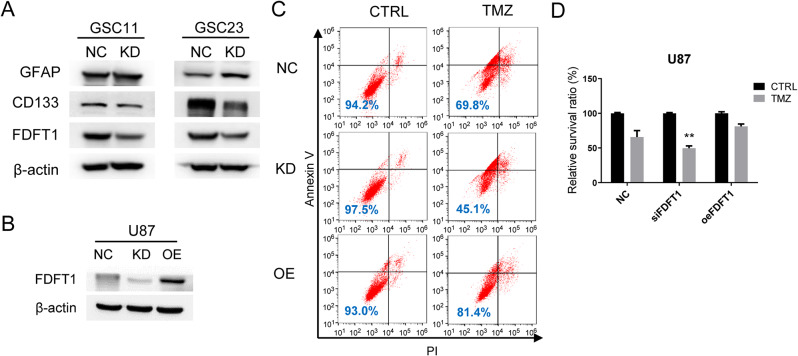
To determine whether FDFT1 functions as a stemness regulator in glioblastoma. We used siRNA to knock down FDFT1 in GSC11 and GSC23. Western blot analysis revealed that FDFT1 knockdown resulted in downregulation of the stemness marker CD133 and upregulation of the differentiation marker GFAP, findings that corroborate the effects of FBS induction (Fig. 3A). Given the association of CSCs with drug resistance, we further investigated whether altered FDFT1 expression influences the susceptibility of glioma cells to chemotherapy. We generated U87 cell lines with stable knockdown and overexpression of FDFT1, and Western blot confirmed the efficient modulation of FDFT1 levels (Fig. 3B). The chemosensitivity assays demonstrated that FDFT1 knockdown sensitized U87 cells to temozolomide (TMZ) treatment, whereas FDFT1 overexpression conferred resistance to the drug (Fig. 3C, D).
Fig. 3.
FDFT1 Enhances Expression of stemness marker and Drug Resistance in glioblastoma cells. (A) Protein expression levels of CD133 following the knockdown of FDFT1. (B) Protein expression levels in response to FDFT1 knockdown or overexpression. (C) A representative flow cytometry apoptosis assay result showing sensitivity of U87 cells to TMZ after FDFT1 knockdown or overexpression. (D) Statistical analysis of apoptosis induced by TMZ. Abbreviations: NC, negative control using scrambled siRNA; KD, knockdown; OE, overexpression; TMZ, temozolomide; **, p < 0.01
YM-53,601 inhibits proliferation and induces apoptosis in GSCs
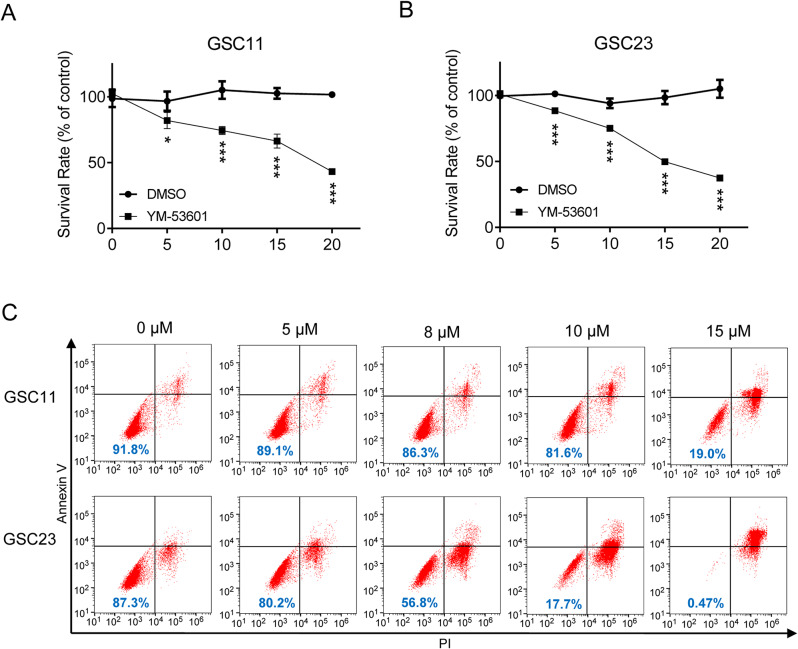
Based on the above observation that FDFT1 downregulation significantly sensitized glioma cells to chemotherapeutic drug, we reasoned that it can also be done by FDFT1 inhibitors. For this purpose, YM-53,601, a novel squalene synthase inhibitor, which leads to reduced cholesterol biosynthesis was used in this study. To test the effects of YM-53,601, GSC11 and GSC23 cells were treated with YM-53,601 alone or together with TMZ. As shown in Fig. 4A and B, YM-53,601 significantly inhibited the proliferation of GSC11 and GSC23 cells in a dose-dependent manner. Also, the pro-apoptotic effect of YM-53,601 was confirmed by Annexin V/PI staining using flow cytometry (Fig. 4C). To evaluate the potential of YM-53,601 to augment the efficacy of conventional chemotherapy, a combination treatment with YM-53,601 and TMZ was tested. We found the combination therapy induced a higher rate of apoptosis compared to the individual agents alone (Fig. S2).
Fig. 4.
YM-53,601 Inhibits the Viability of Glioblastoma Stem Cells. (A) MTT assay demonstrating the inhibitory effect of YM-53,601 on the proliferation of GSC11 cells. (B) MTT assay showing the suppressive impact of YM-53,601 on the growth of GSC23 cells. (C) Assessment of apoptosis induction in GSC11 and GSC23 cells treated with YM-53,601. *, p < 0.05; **, p < 0.01; ***, p < 0.001
GSCs are more dependent on mevalonate metabolism than differentiate cells
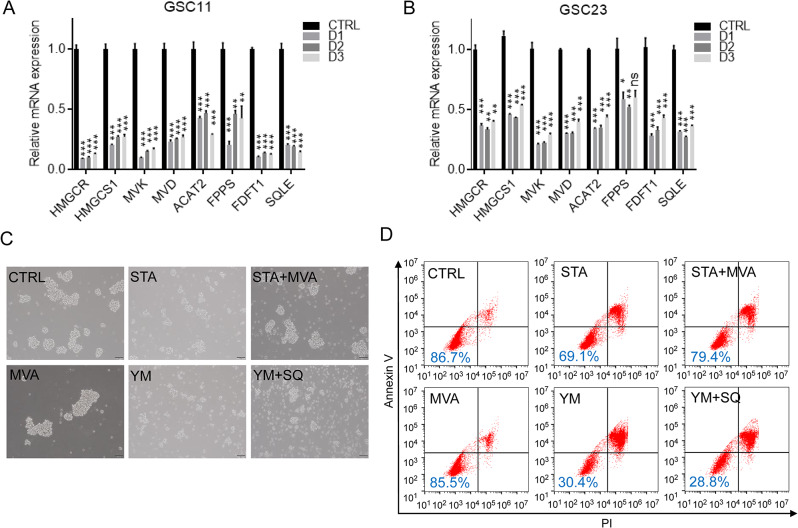
Since FDFT1 is a pivotal enzyme in mevalonate metabolism pathway, its overexpression in GSC11 and GSC23 cells suggested that GSCs might be addicted to mevalonate metabolism. Comparative analysis with differentiated cells revealed that mRNA expression levels of key mevalonate metabolism genes, including HMGCR, HMGCS1, MVK, and SQLE, were upregulated in CSC-enriched spheroids (Fig. 5A, B). This suggested a potential addiction to cholesterol biosynthesis in GSCs. To probe this cholesterol dependency, we utilized simvastatin (STA), a clinically available drug that inhibits HMGCR and lowers blood cholesterol concentration. As shown in Fig. 5C and Fig. S3A, the addition of STA to the cell culture medium significantly inhibited neurosphere formation of GSCs, an effect that was partially mitigated by the replenishment of mevalonate. However, the inhibition of neurosphere formation induced by YM-53,601 treatment did not show any significant response to mevalonate supplementation. In flow cytometry assays, STA induced apoptosis in GSCs, and this effect was partially attenuated by mevalonate. In contrast, apoptosis induced by YM-53,601 was found to be intractable (Fig. 5D and Fig. S3B). Notably, squalene, a downstream metabolite of FDFT1, was ineffective in counteracting apoptosis induced by YM-53,601 (Fig. 5D).
Fig. 5.
Dependency of Glioblastoma Stem Cells on Mevalonate Metabolism. (A) Alterations in mevalonate metabolic gene expression in GSC11 cells before and after differentiation. (B) Changes in mevalonate metabolic gene expression in GSC23 cells before and after differentiation. (C) Impact of statin drugs and YM-53,601 on the spheroid formation ability of GSC11 cells. (D) Effects of statin drugs and YM-53,601 on the viability of GSC11 cells. Abbreviations: CTRL, control; STA, simvastatin; MVA, mevalonic acid; YM, YM-53,601; SQ, squalene; ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001
FDFT1 is regulated by SREBP2 in glioma cells
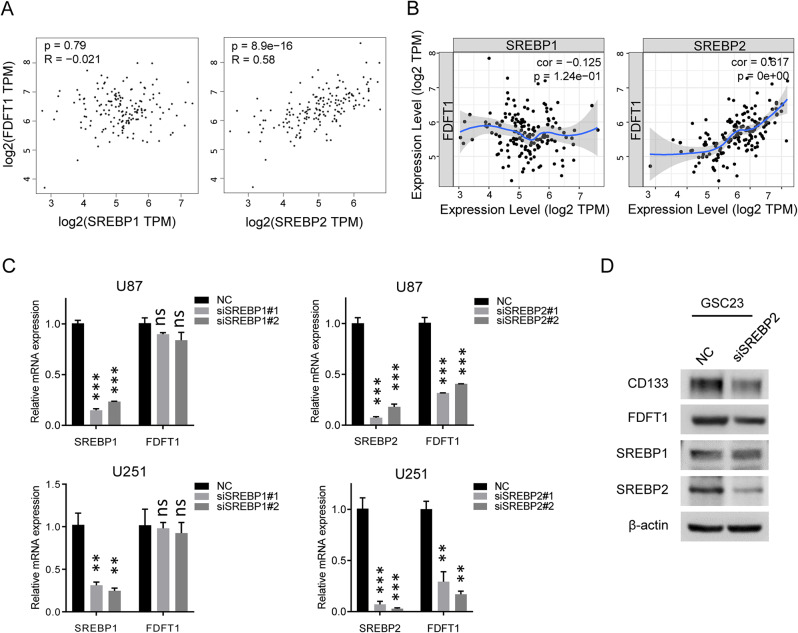
Sterol regulatory element-binding proteins (SREBPs) such as SREBP1 and SREBP2, belong to a family of transcription factors that mainly regulate the expression of genes involved in sterol and fatty acid biosynthesis. Emerging evidence indicate that SREBP1 and SREBP2 are associated with the regulation of cancer stemness [24–26]. Given this, we investigated whether FDFT1 was regulated by SREBPs. Correlation analysis utilizing both the TIMER and GEPIA databases demonstrated a positive correlation between FDFT1 expression and that of SREBP2, with no significant association observed with SREBP1 (Fig. 6A, B). Specifically, siRNA-mediated suppression of SREBP1 did not induce downregulation of FDFT1 in U87 and U251 cells. In contrast, knockdown of SREBP2 led to a significant reduction in FDFT1 expression (Fig. 6C). Furthermore, western blot analysis indicated that downregulation of SREBP2, but not SREBP1, substantially decreased the expression of FDFT1 as well as the stem cell marker CD133 in GSC23 cells (Fig. 6D).
Fig. 6.
Correlation Between SREBPs and FDFT1. (A) GEPIA database analysis illustrating the expression correlation between SREBP1/2 and FDFT1. (B) TIMER database analysis demonstrating the expression correlation between SREBP1/2 and FDFT1. (C) qPCR results showing the mRNA expression levels of FDFT1 following the knockdown of SREBP1/2. (D) Western blot (WB) results indicating the protein expression levels of FDFT1 upon SREBP2 knockdown in GSC23 cells. Abbreviations: ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001
FDFT1 regulates stemness through Akt pathway
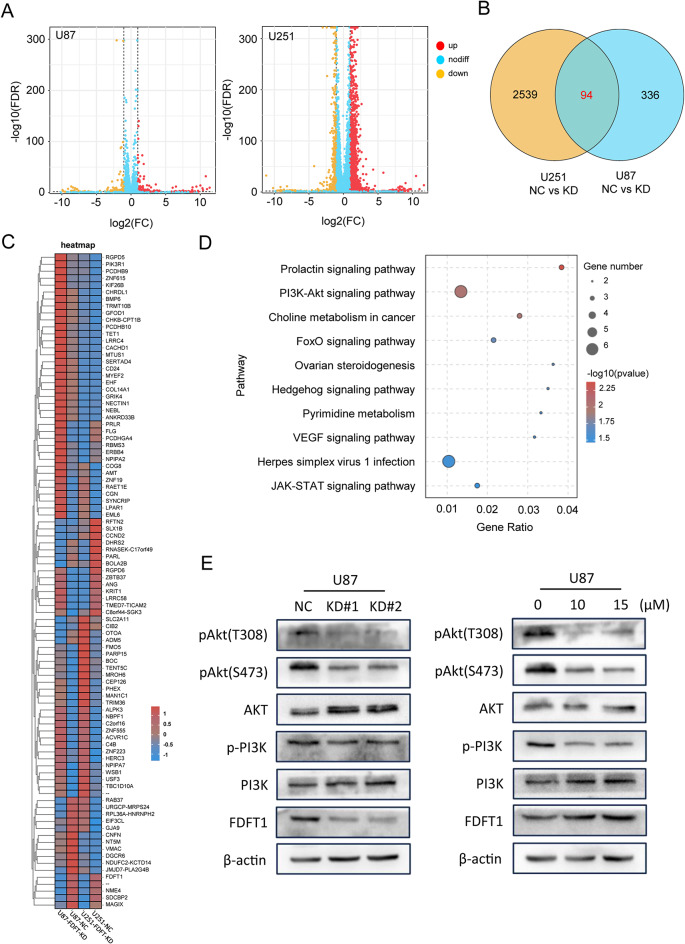
We further analyzed the potential biological functions of the differentially-expressed genes following FDFT1 downregulation. The number of upregulated and down-regulated genes was shown in Fig. 7A. U87 and U251 cells were intersected to obtain differentially-expressed genes, and 94 genes were listed in the heatmap (Fig. 7B, C). Subsequently, we performed GO and KEGG enrichment analysis on this gene set. The significant pathways associated with FDFT1 were predominantly enriched in the ‘Prolactin signaling pathway’, ‘PI3K/AKT signaling pathway’, and ‘Choline metabolism in cancer’ (Fig. 7D). Previous studies have indicated that Akt activation is a key regulator in cancer stemness [27–29]. Herein, our western blot results demonstrated that the downregulation of FDFT1 led to a reduction in Akt phosphorylation, a finding that was corroborated by the treatment with YM-53,601 (Fig. 7E). These results propose that Akt signaling may be implicated in FDFT1-mediated metabolic pathways.
Fig. 7.
Analysis of Downstream Effector Molecules of FDFT1. (A) Volcano plot depicting molecular changes before and after FDFT1 knockdown in U87 and U251 cells. (B) Venn diagram representing the intersection of genes affected in both U87 and U251 upon FDFT1 knockdown. (C) Expression matrix highlighting the expression patterns of the intersecting genes. (D) Overview of signaling pathways altered following FDFT1 knockdown. (E) Impact of FDFT1 knockdown and YM-53,601 treatment on the Akt signaling pathway
Discussion
Despite the pivotal role of mitochondria in cellular respiration is well acknowledged, the metabolic nuances between cancer stem cells (CSCs) and their differentiated cancer cell counterparts are not yet fully elucidated. Previous studies from our laboratory and other groups have delineated that CSCs possess a unique metabolic signature, notably marked by an upregulation of glycolysis and a concomitant decrease in oxygen consumption [10, 15, 30, 31]. Elevated cholesterol levels have been implicated as a significant contributor to the process of tumorigenesis [32–34]. Accumulation of cholesterol has been demonstrated to impair mitochondrial function, which in turn compels cells to rely more heavily on glycolysis as an alternative pathway for energy generation [35–37]. In the current study, FDFT1 gene expression was significantly elevated in glioma stem cells (GSCs) compared to differentiated cells. FDFT1 encodes squalene synthase, a key enzyme in cholesterol biosynthesis. This suggests that FDFT1-mediated cholesterol metabolism could potentially influence the behavior of stem cells, warranting further investigation into its role in the pathophysiology of cancer.
Indeed, a surge of recent studies have underscored the pivotal role of cholesterol metabolism in CSCs, revealing its intricate ties to tumorigenesis, development, and metastasis, as well as immune checkpoint blockade [38, 39]. Cancer cells were considered to reduce cholesterol efflux and increase uptake through transporter such as ABCA1, ABCG1 [39]. The mevalonate pathway is fundamental to cholesterol synthesis, and intriguingly in the current study, genes associated with this metabolic pathway are found to be upregulated in GSCs, suggesting an enhanced addiction to mevalonate metabolism. Prior research has predominantly concentrated on the inhibition of HMG-CoA reductase (HMGCR) by statins to reduce cholesterol levels, thereby potentially decreasing the risk of various cancers, mitigating the invasiveness and metastatic potential of CSCs, and even inducing their demise [40–43]. FDFT1 is another key enzyme for cholesterol synthesis. Although the FDFT1 gene has been implicated in the progression of several malignancies and is correlated with adverse outcomes [44–46], its specific relationship with the stemness of cancer cells has been less explored. In this study, we observed that overexpression of FDFT1 confers resistance to temozolomide, a phenomenon predominantly observed in GSCs. This resistance may be attributed to FDFT1’s role in stimulating cholesterol synthesis, which could subsequently shield cells from undergoing apoptosis [37, 47]. Additionally, the accumulation of squalene, a product of FDFT1’s enzymatic activity, has been shown to protect cells against ferroptosis by maintaining the appropriate balance of polyunsaturated fatty acids within the cell membrane [48]. These findings underscore the multifaceted role of FDFT1 in the survival and resistance mechanisms of GSCs.
The intricate relationship between FDFT1 and the maintenance of stemness in glioma cells is further illuminated by the discovery that SREBP2 is the key transcriptional regulator of FDFT1. Interestingly, recent studies shown that SREBP2 is the master regulator of cholesterol biosynthesis to maintain self-renewal capacity of glioblastoma cells [49]. Moreover, it caused drug resistance of CSCs by promoting cholesterol biosynthesis [47]. By modulating the activity of SREBP2, it may be possible to attenuate the expression of FDFT1, potentially reducing the tumorigenic and invasive capabilities of GSCs. Which is similar to the effects of FBS, the knockdown of FDFT1 leads to a decrease in the expression of stem cell markers and an upregulation of differentiation markers such as GFAP. This shift indicates a transition from a stem-like state to a more differentiated state. Additionally, the knockdown of FDFT1 results in a weakened migratory capacity of glioma cells, further emphasizing the gene’s importance in the invasive properties of these cells.
The exploration of the molecular underpinnings of FDFT1’s role in glioma stem cells has led to intriguing parallels with the mechanisms observed in prostate cancer, where the interplay between cholesterol metabolism and oncogenic signaling pathways is well-documented. Phosphatase and tensin homolog (PTEN) is a well-known tumor suppressor, and its loss in prostate cancer leads to the activation of the PI3K-Akt signaling pathway [50]. This activation is associated with an increase in cholesterol ester (CE) accumulation and promotes cancer development [38]. Our transcriptional analysis suggests that FDFT1 may regulate stemness through the PI3K/AKT signaling pathway. This is supported by Western blot data, which confirms the association between FDFT1 and the activation of the PI3K-Akt pathway. Interestingly, a recent study showed that enhanced levels of SQLE, the downstream enzyme of FDFT1, can activate Akt signalling, thereby increasing CE levels and facilitating cancer development [51]. Moreover, Akt can also control the expression and activity of SREBP2 to stimulate the production of cholesterol [52]. These findings suggest Akt plays a crucial role in the cholesterol synthesis.
Given the critical role of FDFT1 for GSCs, it is reasonable to test the antitumor effect of FDFT1 inhibitors. Previous studies mostly used statins to block HMG-CoA reductase to lower cholesterol. However, as the previous step of mevalonate metabolism, HMG-CoA reductase might have more side effect since it also lower the levels of ubiquinone and dolicol, both essential for cell growth and viability [53]. From this point of view, YM-53,601, has been proved more selective alternative to statins. Our study revealed that while simvastatin induced reversible apoptosis in GSCs via mevalonate supplementation, YM-53,601 induced irreversible cell death unaffected by metabolite supplementation, suggesting a distinct mechanism warranting further exploration. Additionally, YM-53,601 enhanced the efficacy of temozolomide, supporting the rationale for combining FDFT1 inhibitors with chemotherapy to potentially surmount drug resistance in GSCs, though further in vivo studies are imperative.
Conclusion
In summary, the metabolic state of GSCs differs substantially from the metabolic state of differentiated glioma cells, and it correlates with resistance to standard chemotherapeutic agents. FDFT1 overexpression maintain GSCs by AKT pathway. Targeting FDFT1-specific metabolic pathways in glioblastoma may spare GSCs.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to express our gratitude for the support from the National Natural Science Foundation of China (No. 81903043, 82203435), the Guangdong Basic and Applied Basic Research Foundation (2018A030310086, 2021A1515111138), the Guangzhou Science and Technology Plan Project Support (2023A04J2103), and the Medical Research Project of Foshan (20240394). The authors also thank Dr. Sha Fu and Dr. Peng Huang for helpful discussion and comments.
Abbreviations
- BCNU
Bis-chloroethylnitrosourea
- CSC
Cancer stem cell
- GBM
Glioblastoma multiforme
- GFAP
Glial fibrillary acidic protein
- GSC
Glioblastoma stem cells
- HMGCR
HMG-CoA reductase
- PBS
Phosphate-buffered saline
- PMSF
Phenylmethylsulfonyl fluoride
- PTEN
Phosphatase and tensin homolog
- SILAC
Stable isotope labeling with amino acids in cell culture
- siRNA
Small RNA interference
- SQS
Squalene synthase
- SREBP
Sterol regulatory element-binding protein
- STA
Simvastatin
- TMZ
Temozolomide
Author contributions
Study administration, validation, and design: Yongsheng Huang, Peng Wang, Jianwei Liao. Methodology, execution of the experiments, data collection and analysis, manuscript preparation: Hui Mo, Jiajia Shao, Zhun Li. Acquisition, interpretation of data: Peiting Zeng, Xinke Yin. Financial support, administrative support: Jiajia Shao, Yongsheng Huang, Jianwei Liao. Study supervision, final approval of the manuscript: Jianwei Liao. All authors read and approved the final manuscript. The authors declare that they have not used AI-generated work in this manuscript.
Funding
This study was supported by funding from the National Natural Science Foundation of China (No. 81903043, 82203435), the Guangdong Basic and Applied Basic Research Foundation (2018A030310086, 2021A1515111138), the Guangzhou Science and Technology Plan Project Support (2023A04J2103), and the Medical Research Project of Foshan (20240394).
Data availability
The datasets used during the current study are available in Mendeley Data, V1, doi: 10.17632/rv85k6jy6f.1.
Declarations
Ethics approval and consent to participate
The study meets the ethical guidelines and the legal requirements. The study was conducted in accordance with the Helsinki Declaration. This study was approved by the Institutional Review Board of Sun Yat-sen Memorial Hospital for the project “Study on the mechanism of cholesterol synthesis mediated by FDFT1 to maintain the stemness of glioma stem cells” (NO. SYSKY-2023-502-01), the date of approval is 06/09/2023.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hui Mo, Jiajia Shao and Zhun Li contributed equally to this work.
Contributor Information
Yongsheng Huang, Email: huangysh65@mail.sysu.edu.cn.
Peng Wang, Email: wangp49@mail.sysu.edu.cn.
Jianwei Liao, Email: liaojw8@mail.sysu.edu.cn.
References
- 1.Salvalaggio A, et al. Glioblastoma and brain connectivity: the need for a paradigm shift. Lancet Neurol. 2024;23(7):740–8. [DOI] [PubMed] [Google Scholar]
- 2.Reardon DA, et al. Effect of Nivolumab vs Bevacizumab in Patients With Recurrent Glioblastoma: The CheckMate 143 Phase 3 Randomized Clinical Trial. JAMA Oncol. 2020;6(7):1003–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Omuro A, et al. Radiotherapy combined with nivolumab or temozolomide for newly diagnosed glioblastoma with unmethylated MGMT promoter: An international randomized phase III trial. Neuro Oncol. 2023;25(1):123–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–60. [DOI] [PubMed] [Google Scholar]
- 5.Beier D, Schulz JB, Beier CP. Chemoresistance of glioblastoma cancer stem cells–much more complex than expected. Mol Cancer. 2011;10:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gangoso E, et al. Glioblastomas acquire myeloid-affiliated transcriptional programs via epigenetic immunoediting to elicit immune evasion. Cell. 2021;184(9):2454–e247026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muftuoglu Y, Pajonk F. Targeting Glioma Stem Cells. Neurosurg Clin N Am. 2021;32(2):283–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakrabarty RP, Chandel NS. Mitochondria as Signaling Organelles Control Mammalian Stem Cell Fate. Cell Stem Cell. 2021;28(3):394–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Y, et al. Metabolic alterations in highly tumorigenic glioblastoma cells: preference for hypoxia and high dependency on glycolysis. J Biol Chem. 2011;286(37):32843–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan S, et al. Effective elimination of cancer stem cells by a novel drug combination strategy. Stem Cells. 2013;31(1):23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu PP, et al. Metabolic regulation of cancer cell side population by glucose through activation of the Akt pathway. Cell Death Differ. 2014;21(1):124–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weng ML, et al. Fasting inhibits aerobic glycolysis and proliferation in colorectal cancer via the Fdft1-mediated AKT/mTOR/HIF1α pathway suppression. Nat Commun. 2020;11(1):1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu J, et al. 20(S)-Rg3 upregulates FDFT1 via reducing miR-4425 to inhibit ovarian cancer progression. Arch Biochem Biophys. 2020;693:108569. [DOI] [PubMed] [Google Scholar]
- 14.Chattopadhyay T, Mallick B. FDFT1 repression by piR-39980 prevents oncogenesis by regulating proliferation and apoptosis through hypoxia in tongue squamous cell carcinoma. Life Sci. 2023;329:121954. [DOI] [PubMed] [Google Scholar]
- 15.Yuan S, et al. Metabolic activation of mitochondria in glioma stem cells promotes cancer development through a reactive oxygen species-mediated mechanism. Stem Cell Res Ther. 2015;6:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He H, et al. Glycomic and transcriptomic response of GSC11 glioblastoma stem cells to STAT3 phosphorylation inhibition and serum-induced differentiation. J Proteome Res. 2010;9(5):2098–108. [DOI] [PubMed] [Google Scholar]
- 17.Fathi E, et al. Effect of Rat Bone Marrow Derived-Mesenchymal Stem Cells on Granulocyte Differentiation of Mononuclear Cells as Preclinical Agent in Cellbased Therapy. Curr Gene Ther. 2022;22(2):152–61. [DOI] [PubMed] [Google Scholar]
- 18.Huang CS, et al. YTHDF2 promotes intrahepatic cholangiocarcinoma progression and desensitises cisplatin treatment by increasing CDKN1B mRNA degradation. Clin Transl Med. 2022;12(6):e848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farahzadi R, et al. Granulocyte differentiation of rat bone marrow resident C-kit(+) hematopoietic stem cells induced by mesenchymal stem cells could be considered as new option in cell-based therapy. Regen Ther. 2023;23:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai CY et al. Ionizing Radiation Induces Resistant Glioblastoma Stem-Like Cells by Promoting Autophagy via the Wnt/β-Catenin Pathway. Life (Basel), 2021;11(5). [DOI] [PMC free article] [PubMed]
- 21.Lin Z, et al. ANXA1 as a Prognostic and Immune Microenvironmental Marker for Gliomas Based on Transcriptomic Analysis and Experimental Validation. Front Cell Dev Biol. 2021;9:659080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schäfer JA, et al. Global mitochondrial protein import proteomics reveal distinct regulation by translation and translocation machinery. Mol Cell. 2022;82(2):435–e4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shevchenko A, et al. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1(6):2856–60. [DOI] [PubMed] [Google Scholar]
- 24.Wen YA, et al. Downregulation of SREBP inhibits tumor growth and initiation by altering cellular metabolism in colon cancer. Cell Death Dis. 2018;9(3):265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou C, et al. Resveratrol enhances the chemotherapeutic response and reverses the stemness induced by gemcitabine in pancreatic cancer cells via targeting SREBP1. Cell Prolif. 2019;52(1):e12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue L, et al. Targeting SREBP-2-Regulated Mevalonate Metabolism for Cancer Therapy. Front Oncol. 2020;10:1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, et al. AKT drives SOX2 overexpression and cancer cell stemness in esophageal cancer by protecting SOX2 from UBR5-mediated degradation. Oncogene. 2019;38(26):5250–64. [DOI] [PubMed] [Google Scholar]
- 28.Zhong C, et al. Remodeling cancer stemness by collagen/fibronectin via the AKT and CDC42 signaling pathway crosstalk in glioma. Theranostics. 2021;11(4):1991–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang YH et al. Attenuation of PI3K-Akt-mTOR Pathway to Reduce Cancer Stemness on Chemoresistant Lung Cancer Cells by Shikonin and Synergy with BEZ235 Inhibitor. Int J Mol Sci, 2024;25(1). [DOI] [PMC free article] [PubMed]
- 30.Yamashita N, et al. MUC1-C integrates aerobic glycolysis with suppression of oxidative phosphorylation in triple-negative breast cancer stem cells. iScience. 2023;26(11):108168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian M, et al. TIPE drives a cancer stem-like phenotype by promoting glycolysis via PKM2/HIF-1α axis in melanoma. eLife Sciences Publications, Ltd. 2024.
- 32.Garcia-Ruiz C, et al. MITOCHONDRIAL CHOLESTEROL AND CANCER. Semin Cancer Biol. 2021;73:76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King RJ, Singh PK, Mehla K. The cholesterol pathway: impact on immunity and cancer. Trends Immunol. 2022;43(1):78–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang B, Song BL, Xu C. Cholesterol metabolism in cancer: mechanisms and therapeutic opportunities. Nat Metab. 2020;2(2):132–41. [DOI] [PubMed] [Google Scholar]
- 35.Yu W, et al. Altered cholesterol metabolism in Niemann-Pick type C1 mouse brains affects mitochondrial function. J Biol Chem. 2005;280(12):11731–9. [DOI] [PubMed] [Google Scholar]
- 36.Solsona-Vilarrasa E, et al. Cholesterol enrichment in liver mitochondria impairs oxidative phosphorylation and disrupts the assembly of respiratory supercomplexes. Redox Biol. 2019;24:101214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goicoechea L, et al. Mitochondrial cholesterol: Metabolism and impact on redox biology and disease. Redox Biol. 2023;61:102643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu X, et al. Dysregulation of cholesterol metabolism in cancer progression. Oncogene. 2023;42(45):3289–302. [DOI] [PubMed] [Google Scholar]
- 39.Lu J, et al. Targeting cholesterol metabolism in Cancer: From molecular mechanisms to therapeutic implications. Biochem Pharmacol. 2023;218:115907. [DOI] [PubMed] [Google Scholar]
- 40.Sun J, et al. Statin use and risk of colorectal cancer in patients with inflammatory bowel disease. EClinicalMedicine. 2023;63:102182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singal AG, Kanwal F, Llovet JM. Global trends in hepatocellular carcinoma epidemiology: implications for screening, prevention and therapy. Nat Rev Clin Oncol. 2023;20(12):864–84. [DOI] [PubMed] [Google Scholar]
- 42.Maier CR, et al. USP28 controls SREBP2 and the mevalonate pathway to drive tumour growth in squamous cancer. Cell Death Differ. 2023;30(7):1710–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Centonze G, et al. p140Cap modulates the mevalonate pathway decreasing cell migration and enhancing drug sensitivity in breast cancer cells. Cell Death Dis. 2023;14(12):849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fukuma Y, et al. Role of squalene synthase in prostate cancer risk and the biological aggressiveness of human prostate cancer. Prostate Cancer Prostatic Dis. 2012;15(4):339–45. [DOI] [PubMed] [Google Scholar]
- 45.Yang YF, et al. Squalene synthase promotes the invasion of lung cancer cells via the osteopontin/ERK pathway. Oncogenesis. 2020;9(8):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biancur DE, et al. Functional Genomics Identifies Metabolic Vulnerabilities in Pancreatic Cancer. Cell Metab. 2021;33(1):199–e2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mok EHK, et al. Caspase-3-Induced Activation of SREBP2 Drives Drug Resistance via Promotion of Cholesterol Biosynthesis in Hepatocellular Carcinoma. Cancer Res. 2022;82(17):3102–15. [DOI] [PubMed] [Google Scholar]
- 48.Sun Q, et al. Cholesterol mediated ferroptosis suppression reveals essential roles of Coenzyme Q and squalene. Commun Biol. 2023;6(1):1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gu D, et al. Sterol regulatory element-binding protein 2 maintains glioblastoma stem cells by keeping the balance between cholesterol biosynthesis and uptake. Neuro Oncol. 2023;25(9):1578–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yue S, et al. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab. 2014;19(3):393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu R, et al. SQLE promotes pancreatic cancer growth by attenuating ER stress and activating lipid rafts-regulated Src/PI3K/Akt signaling pathway. Cell Death Dis. 2023;14(8):497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Porstmann T, et al. PKB/Akt induces transcription of enzymes involved in cholesterol and fatty acid biosynthesis via activation of SREBP. Oncogene. 2005;24(43):6465–81. [DOI] [PubMed] [Google Scholar]
- 53.Ugawa T, et al. YM-53601, a novel squalene synthase inhibitor, reduces plasma cholesterol and triglyceride levels in several animal species. Br J Pharmacol. 2000;131(1):63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used during the current study are available in Mendeley Data, V1, doi: 10.17632/rv85k6jy6f.1.