Summary
The efficacy of hepatic arterial infusion chemotherapy (HAIC) with oxaliplatin (OXA) and 5-fluorouracil (5-Fu) for treating advanced perihilar cholangiocarcinoma (pCCA) has been demonstrated, yet the survival benefits of HAIC for pCCA patients vary. Here, we aimed to screen out HAIC resistance-related bile microRNAs (miRNAs) and explore the functions of specific bile miRNAs in pCCA based on high-throughput sequencing. Levels of bile miR-532-3p, miR-1250-5p, and miR-4772-5p were related to the survival of advanced pCCA patients after HAIC. However, only overexpression of miR-532-3p promoted OXA/5-Fu resistance, and downregulation of its expression improved sensitivity to OXA/5-Fu. Mechanistic investigations revealed secreted protein acidic and rich in cysteine (SPARC) as the direct target of miR-532-3p. Our study reveals that bile miR-532-3p, miR-1250-5p, and miR-4772-5p may serve as survival biomarkers in advanced pCCA patients after HAIC and that bile miR-532-3p promotes resistance to HAIC with OXA and 5-Fu via negatively regulating SPARC expression.
Subject areas: Molecular biology, Cell biology, Cancer
Graphical abstract
Highlights
-
•
Bile miR-532-3p, miR-1250-5p, and miR-4772-5p serve as survival biomarkers in pCCA
-
•
MiR-532-3p promotes HAIC resistance via negatively regulating SPARC expression
-
•
MiR-532-3p and SPARC may be utilized as therapeutic targets against HAIC resistance
Molecular biology; Cell biology; Cancer
Introduction
Cholangiocarcinoma (CCA) is a rare type of malignancy that originates from the gastrointestinal tract and accounts for 15% of primary liver malignancies.1 Over the last three decades, the incidence of CCA has increased.2 CCA is generally divided into intrahepatic, perihilar, and distal CCA.3 Of the three types, perihilar cholangiocarcinoma (pCCA) is a highly aggressive malignant neoplasm and accounts for 50%–70% of all CCA cases.4 At present, resection is the only possible curative treatment for pCCA. However, most pCCA patients have non-specific symptoms in the early stages of this disease and are not candidates for surgery when diagnosed, as diagnosis is often delayed and is commonly established only when jaundice occurs.5,6 For advanced CCAs, gemcitabine plus cisplatin (GemCis) remains the mainstay first-line systemic treatment, with a median overall survival (OS) of 11.7 months, as reported in the ABC-02 trial.7
Besides GemCis regimen, other adjuvant systemic treatments for CCA have been extensively explored.8,9,10 Recently, the combination of durvalumab or pembrolizumab with GemCis regimen has been demonstrated to prolong the survival of advanced CCA patients in the TOPAZ-1 and KEYNOTE-966 studies; however, the improvements in survival were also limited, with median OS (mOS) of 12.9 and 12.7 months, respectively.11,12 In a prospective phase II trial, we demonstrated the efficacy of hepatic arterial infusion chemotherapy (HAIC) comprised of oxaliplatin (OXA) and 5-fluorouracil (5-Fu) for unresectable pCCA, finding it to be superior to traditional first-line chemotherapy.13,14 However, the survival benefits of HAIC vary among different pCCA patients, and the underlying molecular mechanisms contributing to HAIC resistance have not been elucidated. Therefore, the identification of predictive biomarkers, the mechanism of HAIC resistance, and therapeutic targets against HAIC resistance will contribute to the improvement of HAIC efficiency and will help achieve better tumor control and longer survival for advanced pCCA patients.
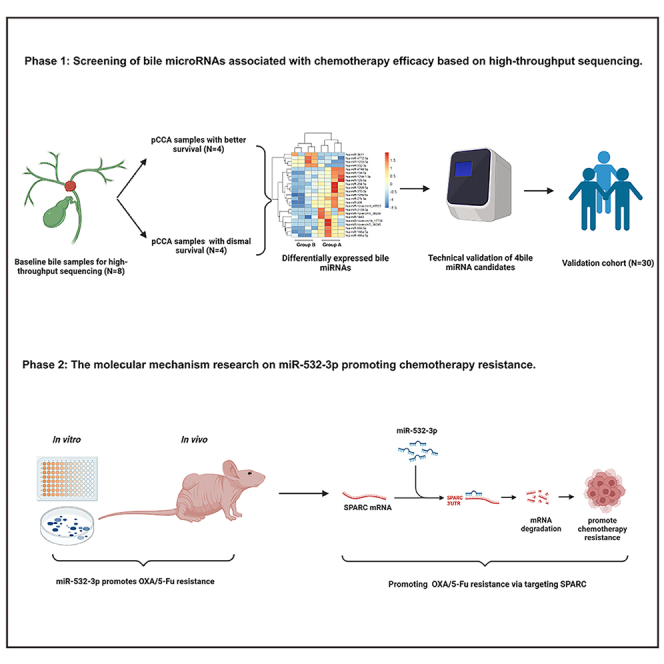
Previous studies have defined microRNAs (miRNAs) as small noncoding RNAs with a length ranging from 21 to 23 bp that can pair with 3′-untranslated regions (3′-UTRs) of genes to negatively influence gene expression and ultimately mediate target gene silencing.15,16,17 Multiple recent reports have identified miRNAs as tumor suppressors or oncogenic biomarkers.18,19 The roles of miRNAs in carcinogenesis have been well-defined, revealing that they act as oncogenes in CCA.20 Chemotherapy resistance reduces the efficacy of CCA treatment. The roles of miRNAs in chemotherapy resistance have also been explored in CCA.21,22,23,24,25 Moreover, it has been demonstrated that miRNAs can be isolated from biofluids, and thus they can serve as ideal clinical biomarkers.26,27 The aim of our research was to explore the roles of miRNAs in the treatment response to HAIC in advanced pCCA patients. We performed high-throughput sequencing, as well as quantitative polymerase chain reaction (qPCR) validation, on bile samples of HAIC-treated advanced pCCA patients and then screened out specific HAIC resistance-related miRNAs via cell viability assay. The biological functions of these miRNAs were then explored in vitro and in vivo via transfection of mimics/inhibitors or virus. mRNA sequencing, target prediction tools, dual-luciferase reporter assays, and rescue experiments were performed to recognize the target genes of specific miRNAs.
Results
Patients’ characteristics
The characteristics of patients for bile miRNA and mRNA sequencing at baseline are presented in Table 1. The mOS of group A patients (n = 4) was longer than that of group B patients (n = 4) (25 months [95% confidence interval (CI): 15.7–34.3 months] vs. 3 months [95% CI: 0.6–5.9 months], p = 0.007). The median progression-free survival (mPFS) of patients in group A was also longer than that of patients in group B (12 months [95% CI: 3.2–20.8 months] vs. 1.9 months [95% CI: 1.5–2.3 months], p = 0.007). There were no differences in the other characteristics between group A and B. Table 2 displays the baseline characteristics of the other 30 pCCA patients who were included in the validation cohort.
Table 1.
Summary of patient baseline characteristics
| characteristics | Patients in group A | Patients in group B | p value |
|---|---|---|---|
| Age (Year) | 68.50 ± 17.07 | 52.75 ± 14.08 | 0.205 |
| Sex (Male/Female) | 3 (75%)/1(25%) | 1(25%)/3(75%) | 0.157 |
| HBV infection (No/Yes) | 3 (75%)/1(25%) | 3 (75%)/1(25%) | 1.000 |
| Child-Pugh class (A/B) | 2 (50%)/2 (50%) | 0 (0%)/4 (100%) | 0.102 |
| CEA (ng/mL) | 3.47 ± 1.73 | 2.41 ± 1.20 | 0.356 |
| CA19-9 (U/mL) | 358.13 ± 290.97 | 731.25 ± 379.37 | 0.060 |
| Total bilirubin (μmol/L) | 73.18 ± 31.48 | 106.36 ± 18.68 | 0.131 |
| ECOG performance status (0/1) | 3 (75%)/1 (25%) | 2 (50%)/2 (50%) | 0.465 |
| Extent of disease (N0M0/N1M0/N2M0) | 4 (100%)/0 (0%)/0 (0%) | 2 (50%)/1 (25%)/1 (25%) | 0.264 |
| Treatment responsea (CR/PR/SD/PD) | 0 (0%)/3 (75%)/1 (25%)/0 (0%) | 0 (0%)/0 (0%)/0 (0%)/4 (100%) | 0.018 |
| mOS (months) | 25.00(15.66–34.34) | 3.00(0.06–5.94) | 0.007 |
| mPFS (months) | 12.00(3.18–20.82) | 1.90(1.51–2.29) | 0.007 |
HBV: hepatitis B virus; CEA: carcinoembryonic antigen; CA19-9: carbohydrate antigen 19-9; ECOG: Eastern Cooperative Oncology Group; CR: complete response; PR: partial response; SD: stable disease; PD: progressive disease; mOS: median overall survival; mPFS: median progression-free survival.
Treatment response was assessed according to the Response Evaluation Criteria in Solid Tumors guidelines (version 1.1).
Table 2.
Summary of baseline characteristics of patients in validation cohort
| Characteristic | Value | Characteristic | Value |
|---|---|---|---|
| Age (y) | 60.0 ± 13.0 | Extent of disease | |
| Sex | Locally advanced | 14 (46.7%) | |
| Male | 22 (73.3%) | N1 lymph node metastasis | 11 (36.7%) |
| Female | 8 (26.7%) | N2 or extrahepatic distant metastasis | 5 (16.7%) |
| HBV infection | Macroscopic growth patterns | ||
| No | 23 (76.7%) | PI | 15 (50.0%) |
| Yes | 7 (23.3%) | MF | 15 (50.0%) |
| Child-Pugh class | CEA (ng/mL) | ||
| A | 13 (43.3%) | <10 ng/mL | 27 (90.0%) |
| B | 7 (56.7%) | >10 ng/mL | 3 (10.0%) |
| ALBI grade | CA19-9 (U/mL) | ||
| 1 | 7 (23.3%) | <200 U/mL | 9 (30.0%) |
| 2 | 22 (73.3%) | >200 U/mL | 21 (70.0%) |
| 3 | 1 (3.3%) | ECOG performance status | |
| Total bilirubin (μmol/L) | 0 | 15 (50.0%) | |
| Median | 61.3 | 1 | 13 (43.3%) |
| Range | 14.9–100 | 2 | 2 (6.7%) |
| Albumin (g/L) | HAIC cycles | ||
| Median | 41.31 | Median | 4.87 |
| Range | 32.6–49.7 | Range | 2–6 |
| Survival (months) | |||
| mOS | 16.4 (10.77–22.2) | ||
| mPFS | 11.2 (8.55–13.9) |
PI: periductal infiltrating; MF: mass-forming; CEA: carcinoembryonic antigen; HBV: hepatitis B virus; CA19-9: carbohydrate antigen 19-9; ALBI: albumin-bilirubin; ECOG: Eastern Cooperative Oncology Group; HAIC: hepatic arterial infusion chemotherapy; mOS: median overall survival; mPFS: median progression-free survival.
Identification of the differentially expressed bile miRNAs and mRNAs between groups A and B
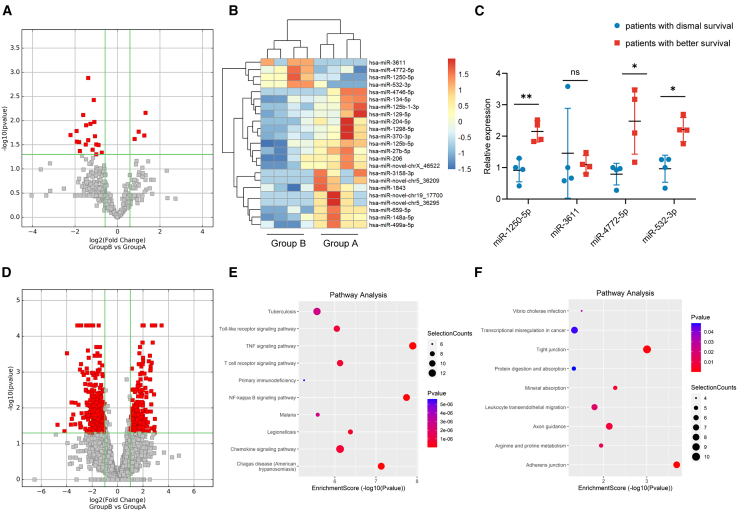
The differentially expressed miRNAs were screened between groups A and B. As shown in Figure 1A, compared with group A, there were four upregulated miRNAs and another 19 downregulated miRNAs in group B. Figure 1B shows the heatmap constructed according to differentially expressed miRNAs. qPCR was then performed to validate the miRNA sequencing results, which demonstrated increased expression of miR-1250-5p (2.15 ± 0.34-fold, p = 0.003), miR-4772-5p (2.48 ± 1.05-fold, p = 0.043), and miR-532-3p (2.21 ± 0.38-fold, p = 0.005) in group B (Figure 1C).
Figure 1.
High-throughput sequencing results
(A–C) Sequencing and qPCR validation of bile miRNAs.
(D) Sequencing of bile mRNAs.
(E and F) KEGG pathway analysis of the upregulated mRNAs in the bile of patients with dismal prognosis (E) and better prognosis (F). Data are represented as mean ± SD. n = 4, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns: no significance, miRNAs microRNAs, qPCR quantitative polymerase chain reaction, KEGG Kyoto Encyclopedia of Genes and Genomes.
For the mRNA sequencing, in group B, there were 221 upregulated and 407 downregulated mRNAs compared with group A (Figure 1D). Figures 1E and 1F show the results of pathway analysis.
miR-532-3p, miR-1250-5p, and miR-4772-5p serve as survival biomarkers for advanced pCCA patients after HAIC
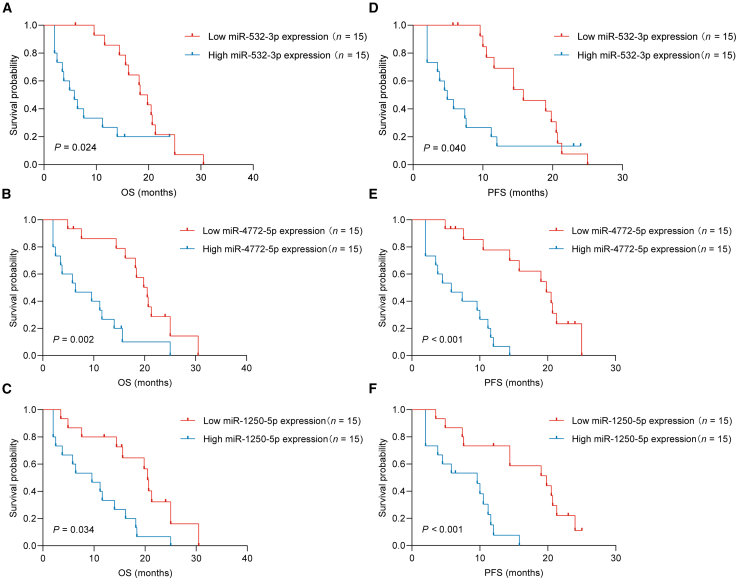
The miR-532-3p, miR-1250-5p, and miR-4772-5p level in bile were next validated in 30 other pCCA patients who received first-line HAIC treatment. Among these 30 pCCA patients, the mOS was 16.4 months (95% CI: 10.77–22.2), and mPFS was 11.2 months (95% CI: 8.55–13.9).
Patients in the entire cohort were then divided into the low miR-532-3p (or miR-1250-5p/miR-4772-5p) group (n = 15) and the high miR-532-3p (or miR-1250-5p/miR-4772-5p) group (n = 15) for further analysis. The mOS times of patients with low miR-532-3p, miR-1250-5p, and miR-4772-5p expression were 18.4 ± 1.48 months, 20.5 ± 0.76 months, and 20.5 ± 1.94 months, respectively. The mOS times of patients with high miR-532-3p, miR-1250-5p, and miR-4772-5p expression were 5.8 ± 1.68 months (p = 0.024), 11.2 ± 3.09 months (p = 0.034), and 6.4 ± 3.74 months (p = 0.002), respectively.
The mPFS times of patients with low miR-532-3p, miR-1250-5p, and miR-4772-5p expression were 15.8 ± 3.33 months, 19.8 ± 4.88 months, and 19.8 ± 2.79 months, respectively. The mPFS times of patients with high miR-532-3p, miR-1250-5p, and miR-4772-5p expression were 4.9 ± 1.29 months (p = 0.040), 9.6 ± 3.29 months (p = 0.034), and 5.8 ± 2.32 months (p = 0.002), respectively (Figures 2A–2F).
Figure 2.
miR-532-3p, miR-1250-5p, and miR-4772-5p serve as survival biomarkers for advanced pCCA patients after HAIC
(A–C) The relationship of miR-532-3p (A)/miR-1250-5p (B)/miR-4772-5p (C) expression with OS.
(D–F) The relationship of miR-532-3p (D)/miR-1250-5p (E)/miR-4772-5p (F) expression with PFS. pCCA perihilar cholangiocarcinoma, HAIC hepatic arterial infusion chemotherapy, OS overall survival, PFS progression-free survival.
In the validation cohort, the CEA level (HR, 1.138; 95% CI, 1.176–1.956; p = 0.041), CA19-9 level (HR, 1.777; 95% CI, 1.393–2.195; p = 0.009), miR-532-3p level (HR, 1.592, 95% CI: 1.286–2.129, p = 0.018), miR-1250-5p level (HR, 1.681; 95% CI, 1.282–2.098; p = 0.025), and miR-4772-5p level (HR, 1.585; 95% CI, 1.131–2.238; p = 0.032) were identified as risk factors for poor OS via univariate analysis. In the multivariate analysis, CA19-9 level (HR, 1.521; 95% CI, 1.192–2.490; p = 0.017), miR-532-3p level (HR, 1.540; 95% CI, 1.109–2.209; p = 0.024), miR-1250-5p level (HR, 1.472; 95% CI, 1.033–2.297; p = 0.028), and miR-4772-5p level (HR, 1.387; 95% CI, 1.080–2.454; p = 0.035) were identified as independent predictors of poor OS (Table 3).
Table 3.
Univariate and multivariate analyses of factors related to overall survival
| Characteristic | Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | |
| Age | 1.024(0.988–1.290) | 0.294 | ||
| Sex: Male/Female | 1.161(0.892–1.419) | 0.521 | ||
| HBV infection: No/Yes | 1.124(0722–1.395) | 0.858 | ||
| Child-Pugh class: A/B | 0.969(0.854–1.264) | 0.459 | ||
| ALBI: 1/2/3 | 1.211(0.902–1.813) | 0.263 | ||
| Total bilirubin | 1.115(0.977–1.121) | 0.294 | ||
| Albumin | 0.875(0.891–1.311) | 0.347 | ||
| ECOG performance status: 0/1/2 | 1.226(0.895–1.244) | 0.819 | ||
| Extent of disease: Locally advanced/N1/N2 or extrahepatic distant metastasis |
1.262(0.792–1.839) | 0.458 | ||
| Macroscopic growth patterns: PI/MF | 1.347(0.918–1.658) | 0.129 | ||
| CEA level: <10 ng/mL/>10 ng/mL | 1.318(1.176–1.956) | 0.041 | 1.228(0.803–1.839) | 0.245 |
| CA19-9 level: <200 U/mL/>200 U/mL | 1.777(1.393–2.195) | 0.009 | 1.521(1.192–2.490) | 0.017 |
| miR-532-3p level: Low/High | 1.592(1.286–2.129) | 0.018 | 1.540(1.109–2.209) | 0.024 |
| miR-1250-5p level: Low/High | 1.681(1.282–2.098) | 0.025 | 1.472(1.033–2.297) | 0.028 |
| miR-4772-5p level: Low/High | 1.585(1.131–2.338) | 0.032 | 1.387(1.080–2.454) | 0.035 |
HR: hazard ratio; CI: confidence interval; HBV: hepatitis B virus; ALBI: albumin-bilirubin; ECOG: Eastern Cooperative Oncology Group; PI: periductal infiltrating; MF: mass-forming; CEA: carcinoembryonic antigen; HBV: hepatitis B virus; CA19-9: carbohydrate antigen 19-9.
miR-532-3p promotes OXA/5-Fu resistance
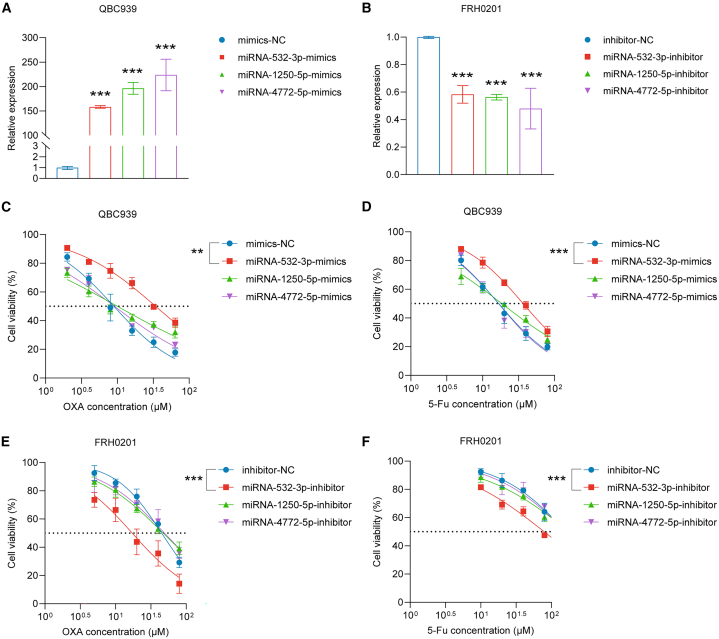
We next studied whether overexpression or decreased expression of miR-1250-5p, miR-4772-5p, or miR-532-3p can influence OXA/5-Fu resistance in human pCCA cell lines. The QBC939 cell line was transfected with miR-532-3p/miR-1250-5p/miR-4772-5p-mimics or mimics-NC, and the FRH0201 cell line was transfected with miR-532-3p/miR-1250-5p/miR-4772-5p-inhibitor or inhibitor-NC. The transfection efficiencies were determined by qPCR (Figures 3A and 3B).
Figure 3.
miR-532-3p influences the IC50 values of OXA and 5-Fu in vitro
(A) miR-1250-5p, miR-4772-5p, and miR-532-3p levels were upregulated in QBC939 cells following transient transfection.
(B) MiR-1250-5p, miR-4772-5p, and miR-532-3p levels were downregulated in FRH0201 cells following transient transfection.
(C and D) The IC50 values of OXA (C) and 5-Fu (D) in QBC939 cells.
(E andF) The IC50 values of OXA (E) and 5-Fu (F) in FRH0201 cells.
Data are represented as mean ± SD. n = 3, ∗∗p < 0.01, ∗∗∗p < 0.001, IC50 half inhibitory concentration, OXA oxaliplatin, 5-Fu 5-fluorouracil, NC negative control.
A CCK-8 assay was utilized to calculate the inhibitory concentration (IC50) values of OXA and 5-Fu. MiR-532-3p overexpression reduced the sensitivity to OXA/5-Fu (p < 0.05), and inhibition of miR-532-3p expression in the FRH0201 cell line significantly improved sensitivity (p < 0.05). However, transfection with mimics or inhibitors of miR-1250-5p/miR-4772-5p had no influence on the IC50 values of OXA or 5-Fu (Figures 3C–3F).
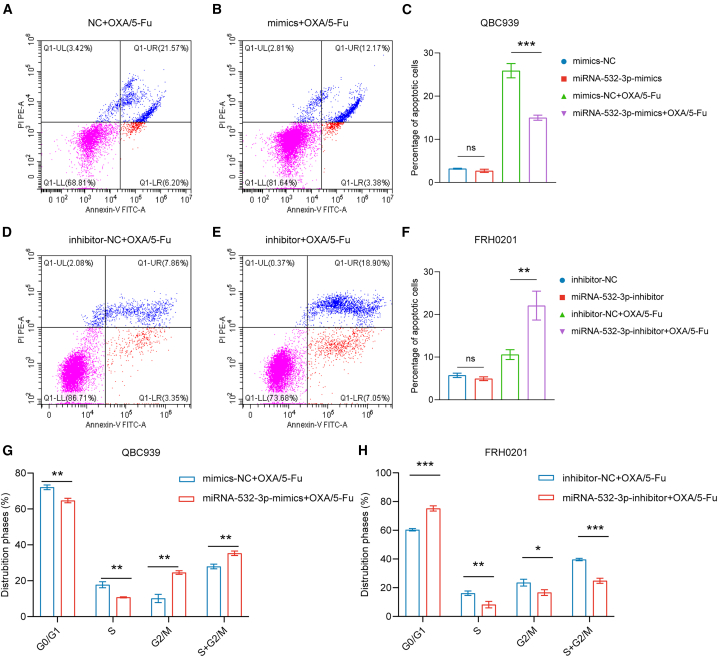
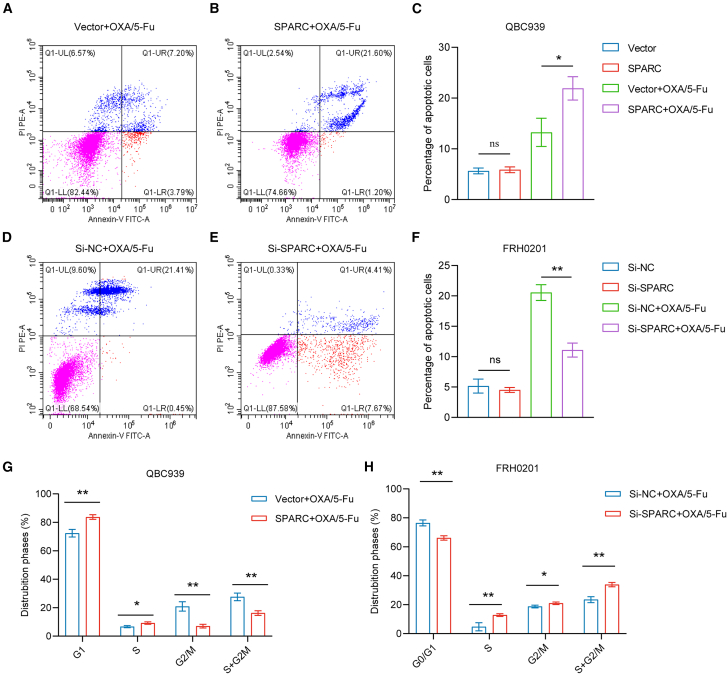
For the detection of cell apoptosis, following incubation of OXA and 5-Fu in the QBC939 cell line for 48 h, the proportion of total apoptotic cells was decreased in the miR-532-3p-mimic group (p < 0.05) (Figures 4A–4C). Similarly, following the application of OXA and 5-Fu in the FRH0201 cell line for 48 h, the proportion of total apoptotic cells were increased in the miR-532-3p-inhibitor group (p < 0.05) (Figures 4D–4F).
Figure 4.
miR-532-3p influences cell apoptosis and cell cycle distribution
(A–C) The percentage of total apoptotic QBC939 cells after incubation with OXA (5 μmol/L) and 5-Fu (10 μmol/L) for 48 h.
(D–F) The percentage of total apoptotic FRH0201 cells after incubation with OXA (20 μmol/L) and 5-Fu (80 μmol/L) for 48 h.
(G) Cell cycle distribution of QBC939 cells after incubation with OXA (5 μmol/L) and 5-Fu (10 μmol/L) for 48 h.
(H) Cell cycle distribution of FRH0201 cells after incubation with OXA (20 μmol/L) and 5-Fu (80 μmol/L) for 48 h.
Data are represented as mean ± SD. n = 3, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns: no significance, OXA oxaliplatin, 5-Fu 5-fluorouracil, NC negative control.
Cell cycle analysis revealed a significantly higher proportion (p < 0.05) of G2/M cells in the miR-532-3p group and a decrease in the proportion of cells in the G2/M phase (p < 0.05) in the miR-532-3p-inhibitor group after incubation with OXA and 5-Fu for 48 h, (Figures 4G and 4H).
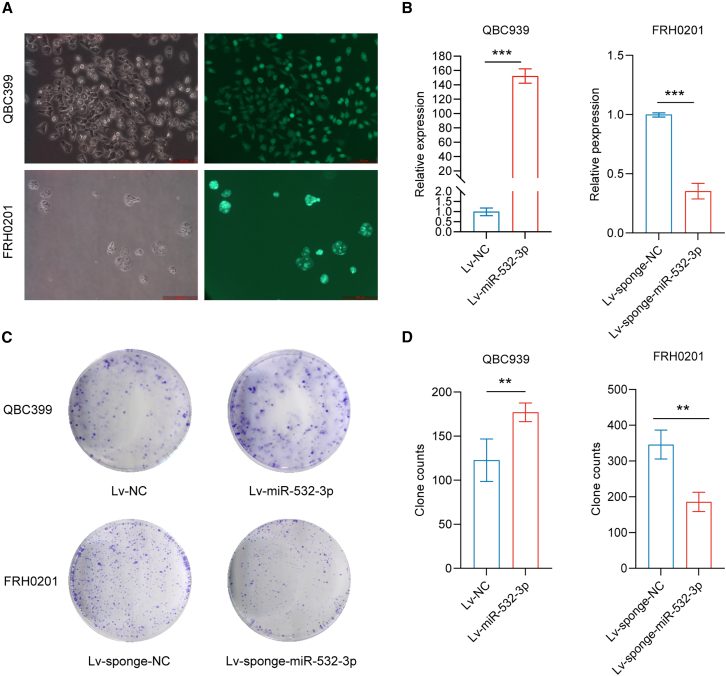
Lentivirus infection was utilized to influence miR-532-3p expression (Figures 5A and 5B), followed by a colony-forming assay and experiments in vivo. After incubation with OXA and 5-Fu for 14 days, the colony-forming assay revealed a significant increase in colony numbers in the miR-532-3p group in QBC939 cells and a decrease in the miR-532-3p-sponge group in FRH0201 cells (Figures 5C and 5D).
Figure 5.
miR-532-3p promotes colony formation
(A and B) The successful lentivirus infection of QBC939 (stable miR-532-3p overexpression) and FRH cells (stable miR-532-3p underexpression).
(C and D) Colony-forming assays on QBC cells (after incubation with OXA at a concentration of 5 μmol/L and 5-Fu at a concentration of 10 μmol/L for 14 days) and FRH0201 cells (after incubation with OXA at a concentration of 20 μmol/L and 5-Fu at a concentration of 80 μmol/L for 14 days).
Data are represented as mean ± SD. n = 3, ∗∗p < 0.01, ∗∗∗p < 0.001,OXA oxaliplatin, 5-Fu 5-fluorouracil, NC negative control.
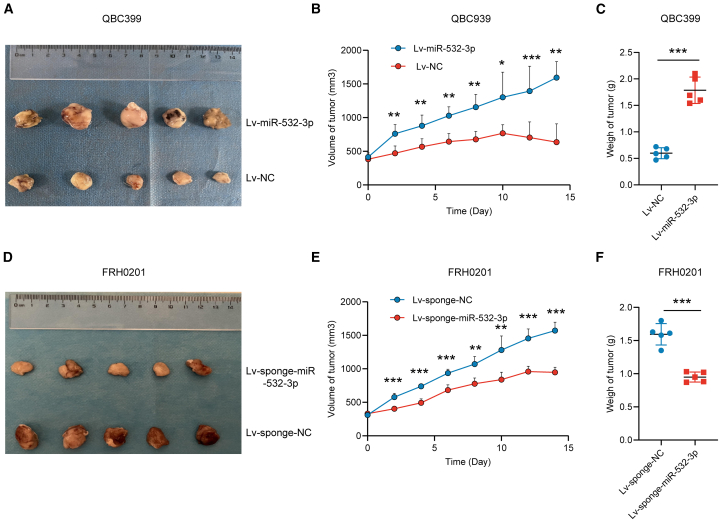
We subcutaneously implanted QBC939 cells stably overexpressing miR-532-3p and FRH0210 cells with stably decreased expression of miR-532-3p into the right flank of nude mice. Two weeks after OXA/5-Fu injection, all mice were sacrificed for further analysis. Mice bearing miR-532-3p QBC939 cells showed faster tumor growth. In contrast, mice bearing miR-532-3p-sponge FRH0201 cells showed slower tumor growth (Figure 6). These findings demonstrate miR-532-3p plays an important role in promoting OXA/5-Fu resistance.
Figure 6.
miR-532-3p promotes OXA/5-Fu resistance in vivo
(A–C) Tumor volumes and weights of mice bearing QBC939 cells (stably overexpressing miR-532-3p or control) 2 weeks following the first drug injection.
(D–F) Tumor volumes and weights of mice bearing FRH0201 cells (stably underexpressing miR-532-3p or control) after 2 weeks following the first drug injection. Data are represented as mean ± SD. n = 5, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, OXA oxaliplatin, 5-Fu 5-fluorouracil, NC negative control.
miR-532-3p promotes OXA and 5-fu resistance by targeting SPARC
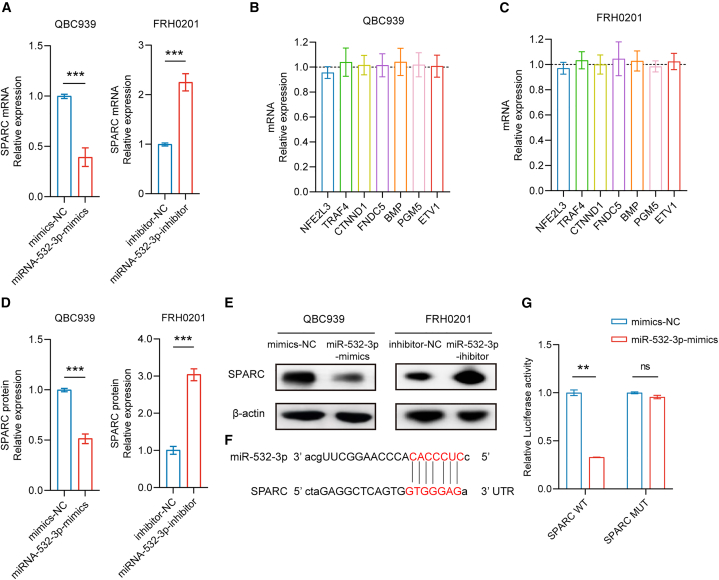
miRNAs exert their biological functions through mediating target gene silencing. Therefore, it is necessary to explore the molecular mechanisms miRNA-532-3p employs via identifying any directly targeted genes. The miRNA target prediction software miRanda and Targetscan (https://www.targetscan.org/vert_80/) identified 1037 probable target genes for miRNA-532-3p. Meanwhile, according to the sequencing results, there were 407 downregulated mRNAs in the bile samples of group B. Collectively, 25 genes were identified that intersected between the results from the prediction software and the sequencing results. After incorporating previous study findings on these 25 genes, a total of eight cancer-related genes, namely, nuclear factor (erythroid 2-like) factor 3 (NFE2L3), tumor necrosis factor receptor-associated factor 4 (TRAF4), catenin delta-1 (CTNND1), secreted protein acidic and rich in cysteine (SPARC), fibronectin type III domain-containing 5 (FNDC5), bone morphogenetic protein (BMP), phosphoglucomutase 5 (PGM5), and E-26 variant transcription factor 1 (ETV1), were preliminarily recognized as miRNA-532-3p target genes with higher probability.
The results of qPCR showed that miR-532-3p overexpression decreased SPARC mRNA levels, while decreased miR-532-3p expression increased SPARC mRNA levels (Figure 7A). However, overexpression or decreased expression of miR-532-3p had no influence on the other seven genes (Figures 7B and 7C). Downregulated SPARC protein levels were observed in the QBC939 cell line with increased miR-532-3p levels (p < 0.05), and upregulated SPARC protein levels were observed in FRH0201 cells with decreased miR-532-3p levels (p < 0.05) (Figures 7D and 7E). The miR-532-3p sequence and 3′ UTR of SPARC are shown in Figure 7F. According to luciferase reporter assays, miR-532-3p inhibited the luciferase activity of the pmriGLO-SPARC 3′UTR-wt group (p < 0.05), but there was no effect on the pmriGLO-SPARC 3′UTR-mut group in the 293T cell line (Figure 7G). Thus, it was confirmed that SPARC was the target gene of miR532-3p.
Figure 7.
miR-532-3p pairs with 3′-UTR of SPARC
(A–C) Overexpression or decreased expression of miR-532-3p affected SPARC mRNA levels but did not affect the mRNA expression of other genes.
(D and E) SPARC protein levels were affected by miR-532-3p.
(F) Binding sites between the SPARC 3′UTR and miR-532-3p.
(G) Results of luciferase reporter assays. Data are represented as mean ± SD. n = 3, ∗∗p < 0.01, ∗∗∗p < 0.001, ns: no significance, 3′-UTRs 3′-untranslated regions, SPARC secreted protein acidic and rich in cysteine, NC negative control.
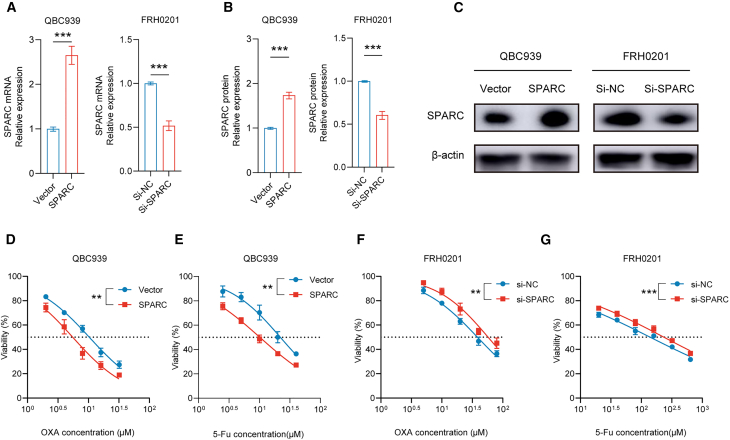
Next, the biological functions of SPARC were explored. SPARC expression increased (p < 0.05) after transfection with plasmids encoding SPARC and decreased after transfection with an siRNA against SPARC (Figures 8A–8C). Overexpression of SPARC significantly improved sensitivity to OXA and 5-Fu in the QBC939 cell line, and inhibition of SPARC via siRNA transfection in FRH0201 cells significantly reduced the sensitivity (Figures 8D–8G). With the combined use of OXA and 5-Fu for 48 h, the proportion of total apoptotic cells was increased in the SPARC group (p < 0.05) (Figures 9A–9C). Similarly, the proportion of total apoptotic cells was decreased in the si-SPARC group (p < 0.05) with the application of OXA and 5-Fu for 48 h (Figures 9D–9F). Cell cycle analysis revealed a decrease in the proportion of cells in the G2/M phase (p < 0.05) in the SPARC group and an increased G2/M phase proportion (p < 0.05) in si-SPARC group after incubation with OXA and 5-Fu for 48 h (Figures 9G and 9H).
Figure 8.
SPARC influences the IC50 values of OXA/5-Fu in vitro
(A–C) Successful transfection of SPARC plasmids on QBC939 cells and siRNA against SPARC on FRH cells.
(D and E) IC50 values of OXA (D) and 5-Fu (E) on QBC939 cells.
(F and G) IC50 values of OXA (F) and 5-Fu (G) on FRH0201 cells. Data are represented as mean ± SD. n = 3, ∗∗p < 0.01, ∗∗∗p < 0.001, SPARC secreted protein acidic and rich in cysteine, IC50 half inhibitory concentration, OXA oxaliplatin, 5-Fu 5-fluorouracil, siRNA small interfering RNA, NC negative control.
Figure 9.
miR-532-3p influences cell apoptosis and cell cycle distribution
(A–C) Percentage of total apoptotic QBC939 cells (following the transfection of SPARC plasmids or control) after incubation with OXA (5 μmol/L) and 5-Fu (10 μmol/L) for 48 h.
(D–F) Percentage of total apoptotic FRH0201 cells (following the transfection of siRNA against SPARC or control, 2 μg) after incubation with OXA (20 μmol/L) and 5-Fu (80 μmol/L) for 48 h.
(G) Cell cycle distribution of QBC939 cells (following the transfection of SPARC plasmids or control) after incubation with OXA (5 μmol/L) and 5-Fu (10 μmol/L) for 48 h.
(H) Cell cycle distribution of FRH0201 (following the transfection of siRNA of SPARC or control) cells after incubation with OXA (20 μmol/L) and 5-Fu (80 μmol/L) for 48 h. Data are represented as mean ± SD. n = 3, ∗p < 0.05, ∗∗p < 0.01, ns: no significance. SPARC secreted protein acidic and rich in cysteine, OXA oxaliplatin, 5-Fu 5-fluorouracil, siRNA small interfering RNA.
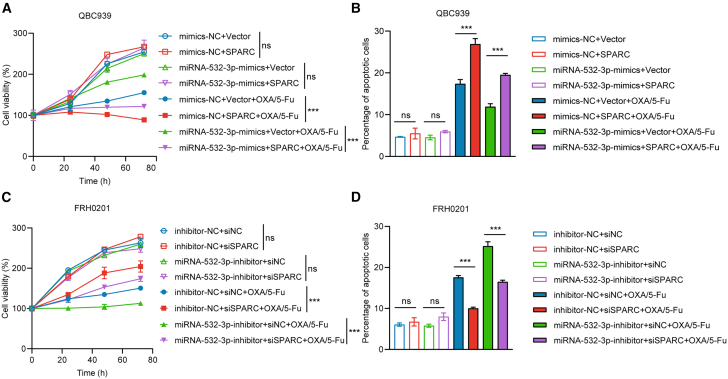
We also performed rescue experiments. In the QBC939 cell line, after incubation with OXA and 5-Fu, cell proliferation (p < 0.05) and cell apoptosis (p < 0.05) were partially reversed with SPARC co-transfection (Figures 10A and 10B). Similarly, in FRH0201 cells, after incubation with OXA and 5-Fu, cell growth (p < 0.05) and apoptosis (p < 0.05) were also partially reversed with si-SPARC co-transfection compared with transfection with miR-532-3p inhibitor alone (Figures 10C and 10D). In brief, the results revealed miR-532-3p promotes OXA/5-Fu resistance via targeting SPARC. The representative flow cytometry analysis of apoptosis in rescue experiments in shown in Figure S1.
Figure 10.
SPARC partially reverses the function of miR-532-3p in vitro
(A) Cell proliferation was reversed with co-transfection of miR-532-3p mimics (50 nM) and SPARC (2 μg) on QBC939 cells after incubation with OXA (5 μmol/L) and 5-Fu (10 μmol/L).
(B) Similarly, cell apoptosis was reversed on QBC939 cells after incubation with OXA (5 μmol/L) and 5-Fu (10 μmol/L) for 48 h.
(C) Cell proliferation was reversed with co-transfection of miR-532-3p inhibitor (100 nM) and siRNA (100 nM) on FRH0201 cells after incubation with OXA (20 μmol/L) and 5-Fu (80 μmol/L).
(D) Cell apoptosis was reversed on FRH0201 cells after incubation with OXA (20 μmol/L) and 5-Fu (80 μmol/L) for 48 h. Data are represented as mean ± SD. n = 3, ∗∗∗p < 0.001, ns: no significance, SPARC secreted protein acidic and rich in cysteine, OXA oxaliplatin, 5-Fu 5-fluorouracil, siRNA small interfering RNA, NC negative control.
Discussion
We have previously demonstrated the encouraging results of HAIC with OXA and 5-Fu for advanced pCCA in a prospective phase II trial, with mPFS and mOS times of 12.2 and 20.5 months, respectively.14 However, the survival benefits of HAIC for pCCA patients vary markedly, and it is usually difficult to perform core needle biopsies to obtain pCCA tissues in clinical practice because of the particular perihilar anatomical site of the lesion and its periductal infiltration growth pattern along the bile duct wall. Thus, there is a clinical need for the identification of reliable circulating biomarkers that can assist in predicting treatment response and survival in HAIC-treated pCCA patients. Previous studies have demonstrated that miRNAs participate in CCA pathogenesis, which can be used as prognostic biomarkers and therapeutic targets for CCA patients.28 Noticeably, throughout the last decade, cell-free non-coding RNAs (cfRNAs), particularly miRNAs, have been envisioned as biofluid biomarkers and therapeutic targets because of their abundance and stability.29 Next-generation sequencing (NGS) is critical for identifying the liquid biopsy biomarkers of tumors, as most molecular signals derived from tumors in bodily fluids have low intensity. NGS improves the generation of high-resolution data for low-expressed transcripts and improves the ability to detect rare tumor-derived transcript products in circulation. Our study combined high-throughput sequencing of both miRNAs and mRNAs to investigate chemotherapy resistance in pCCA patients. Specifically, the identified bile miRNAs (such as miR-532-3p, miR-1250-5p, and miR-4772-5p) and the enriched pathways (e.g., NF-κB, Toll-like receptor, and TNF signaling) are closely associated with chemotherapy resistance, as previously documented in both clinical and experimental research.30,31,32
Circulating miRNAs have unique advantages in acting as liquid biomarkers. Compared with other biomarkers (such as mRNAs, proteins, and metabolites), miRNAs can be easily detected and are less resistant to degradation and modification.27 Several previous studies have explored the use of circulating miRNA in CCA patients.33,34,35 According to a meta-analysis that evaluated the diagnostic value of circulating miRNAs for CCA patients, bile showed higher diagnostic efficiency compared with serum, tissue, and urine.36 In addition, other studies have demonstrated that bile miRNAs can serve as reliable biomarkers for CCA patients,35,37,38 which is consistent with the results of our research. Bile-derived miRNAs and blood-derived miRNAs share similarities in their biogenesis, as both are packaged into extracellular vesicles such as exosomes and microvesicles, which can be released into various bodily fluids. However, the primary difference lies in their physiological and pathological roles, given that bile-derived miRNAs are often associated with liver function, cholestasis, and biliary tract diseases, while blood-derived miRNAs may reflect systemic conditions, including inflammatory responses and metabolic disorders.39 Moreover, there are studies showing that miR-532-3p and miR-1250-5p may serve as liquid biomarkers for patients with malignant cancer. Blood miR-532-3p level inversely correlates with rituximab-induced lymphodepletion and with the CD20 expression on CD19+ cells, which could predict the efficiency of rituximab-mediated lymphodepletion in patients with chronic lymphocytic leukemia.40 Apoptosis-associated tyrosine kinase (AATK)/miR-1250 methylation is absent in healthy peripheral blood, but is detectable in non-Hodgkin’s lymphoma (NHL) cell lines.41 In brief, as a noninvasive liquid biopsy marker, bile miRNA has an important application prospect for cancer diagnosis and prognosis prediction.
In the current study, in vitro and in vivo assays revealed that only miR-532-3p was related to chemotherapy resistance. Overexpression of miR-532-3p promoted OXA/5-Fu resistance, whereas downregulation of its expression improved sensitivity to OXA/5-Fu. Further, the other two circulating miRNAs (miR-1250-5p and miR-4772-5p) might be prognostic factors in pCCA patients. The functions of miR-532-3p have been explored previously. Aberrant high miR-532-3p expression has been found in several types of cancers40,42,43 For hepatobiliary malignancies, miR-532-3p can promote tumor progression via targeting receptor protein tyrosine phosphatase T (PTPRT).44 Conversely, miR-532-3p has also been recognized as a tumor suppressor in several types of human cancer.45,46,47 Regarding chemotherapy resistance, which reduces the efficacy of CCA therapies, the roles of miRNAs in CCA chemotherapy resistance have been explored.22,23,24,25 Previous research has indicated that miR-532-3p is involved in regulating chemosensitivity in other cancers.40,48 However, there are few studies evaluating the exact effect of bile miR-532-3p expression on chemotherapy response in pCCA. miR-532-3p may play a role in promoting or suppressing tumor development in different cancer species, which may be based on its different downstream target genes. In short, our research revealed that miR-532-3p in bile promoted HAIC resistance of advanced pCCA, and bile miR-532-3p/miR-1250-5p/miR-4772-5p might serve as circulating prognostic biomarkers for HAIC-treated advanced pCCA. However, when conducting experiments in conventional culture media, the local environment plays a crucial role in miRNA function and secretion. Bile contains specific lipids and proteins that may influence miRNA stability and packaging, and the absence of bile may alter miRNA behavior and target interactions. Therefore, results derived from such environments must be interpreted with caution because they may not fully represent the in vivo bile conditions.
Previous studies have revealed that miRNAs bind to the 3′-UTRs of genes to exert their biological influence.15 Therefore, we further explored the potential target genes of miR-532-3p via mRNA sequencing, miRNA target prediction software, dual-luciferase reporter assays, and rescue experiments. Specifically, SPARC was characterized as a target gene of miR-532-3p, which partially reversed miR-532-3p′s biological function.
Secreted protein SPARC, also known as osteonectin or BM-40, is a matricellular protein that plays an important role in manipulating cell adhesion, extracellular matrix production, growth factor activity, and cell cycle.49,50 The aberrant expression of SPARC at injury, regeneration, inflammation, and cancer sites, reveals its potential application as a prospective target and therapeutic indicator in the treatment of previous disease.49 The biological functions of SPARC have previously been studied in biliary tract cancers.51,52,53 For resectable iCCA, patients with SPARC-positive tumors have favorable OS rates.51 Loss of SPARC expression is related to reactive oxygen species accumulation in bladder cancer,52 and is also associated with the low differentiation state of biliary tract cancers.53 Moreover, SPARC functions as a chemosensitizer in several cancers.54,55,56,57,58 Specifically, SPARC negatively regulates glucose metabolism in hepatocellular carcinoma by downregulating the expression of key enzymes in glucose metabolism pathway. Overexpression of SPARC renders hepatocellular carcinoma cells tolerance to glucose deprivation and re-sensitizes 5-Fu resistant hepatocellular carcinoma cells through downregulation of glycolysis,55 which is in line with the results of our study. In conclusion, the present study demonstrates that bile miR-532-3p promotes HAIC (with OXA and 5-Fu) resistance of advanced pCCA via negatively regulating SPARC expression.
In summary, our study demonstrated that bile miR-532-3p, miR-1250-5p, and miR-4772-5p could serve as survival biomarkers for advanced pCCA patients after HAIC. Among them, only miR-532-3p was related to chemotherapy resistance and promoted HAIC resistance via targeting SPARC in advanced pCCA. Therefore, miR-532-3p/SPARC might serve as new therapeutic targets against HAIC resistance.
Limitations of the study
Our research has several limitations. First, due to the difficulty involved in obtaining pCCA tissues, the expression levels of miR-532-3p, miR-1250-5p, and miR-4772-5p in tissue were not explored to cross-validate their bile expression. Second, because this was a single-center study based on several bile samples, data on circulating miR-532-3p, miR-1250-5p, and miR-4772-5p levels in bile as prognostic biomarkers in HAIC-treated pCCA require further validation in a large pCCA patient cohort. Finally, the clinical application of miR-532-3p/SPARC as new therapeutic targets against HAIC resistance and potential downstream molecular signaling pathways of miR-532-3p/SPARC should be further explored. To ensure the relevance of our findings, we aim to conduct future studies in bile-containing media or in models that more accurately mimic the biliary environment, which will better translate these observations into applicable research.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Xiaodong Wang (xiaodongw75@yahoo.com).
Materials availability
This study did not generate new unique reagents.
Data and code availability
• miRNA sequencing data have been deposited at Mendeley data. mRNA sequencing data have been deposited in the Gene Expression Omnibus (GEO). miRNA and mRNA sequencing data are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
• This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
We thank LetPub (www.letpub.com) for linguistic assistance. We appreciated all the patients who contributed bile samples for this study. This study was funded by the National Natural Science Foundation of China (no. 82172039), the Beijing Hospitals Authority Clinical Medicine Development of Special Funding Support (no. ZYLX202117), and the Beijing Natural Science Foundation (no. 7212198).
Author contributions
X.W., Y.K., and S.F. conceptualized the study. G.C. collected the bile samples. S.F., H.D., Y.D., L.X., and C.N. analyzed the data. S.F., Y.Z., and K.Z. performed the cell and animal experiments. S.F. and H.D. drafted the manuscript, which was critically revised by X.W. and Y.K.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-β-actin | Abcam | Cat#ab6276; RRID: AB_2223210 |
| Rabbit monoclonal anti-SPARC | Abcam | Cat#ab207743; RRID: AB_2924283 |
| Bacterial and virus strains | ||
| Lv-miR-532-3p | Genechem | N/A |
| Lv-sponge-miR-532-3p | Genechem | N/A |
| Biological samples | ||
| Bile samples | Peking university cancer hospital (China) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Oxaliplatin | MedChemexpress | Cat#HY-17371 |
| 5-fluorouracil | MedChemexpress | Cat#HY-90006 |
| Critical commercial assays | ||
| High-throughput sequencing of miRNAs and mRNAs | CloudSeq Biotech Inc | N/A |
| Lipofectamine RNAiMAX | Invitrogen | Cat#13778030 |
| Lipofectamine 3000 | Invitrogen | Cat#L3000001 |
| MicroRNA Reverse Transcription Kit | EZBioscience | Cat#EZB-miRT3 |
| PrimeScript™ RT Reagent Kit | Takara Bio USA | Cat#RR037A |
| bicinchoninic acid protein assay kit | Servicebio | Cat#G2026-200T |
| ECL Plus Western Blotting Detection Reagents | Solarbio | Cat#PE0010 |
| Counting Kit-8 (CCK-8) kit | Solarbio | Cat#CA1210 |
| Trizol reagent | Thermo Fisher | Cat#15596018CN |
| Deposited data | ||
| miRNA-seq count data | This paper | https://data.mendeley.com/datasets/6rx2k6xhjg/2 |
| mRNA-seq count data | This paper | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE280797 |
| Experimental models: Cell lines | ||
| QBC939 cells | Qingqi (Shanghai) Biotechnology development Co., LTD | Cat#BFN60810642 |
| FRH0201 cells | Otwo Biotech | Cat#HTX2093 |
| 293T cell line | Cell Bank of the Chinese Academy of Sciences | SCSP-502 |
| Oligonucleotides | ||
| See Table S1 for sequences of miRNA mimics, inhibitor, and siRNA | N/A | N/A |
| See Table S2 for primer sequences in qPCR | N/A | N/A |
| Recombinant DNA | ||
| pcDNA3.1-secreted protein acidic and rich in cysteine (SPARC) | Genechem | N/A |
| control vector plasmids | Genechem | N/A |
| Software and algorithms | ||
| SPSS 20.0 software | IBM | https://www.ibm.com/support/pages/spss-statistics-20-available-download |
| GraphPad Prism 9 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
Experimental model and study participant details
A retrospective review of the Hospital Information System identified a total of 31 advanced pCCA patients who received first-line HAIC treatment from June 2015 to June 2016 at Peking University Cancer Hospital, of which bile samples were obtained from 18 patients prior to HAIC treatment. According to the complete survival follow-up, all of the patients died, with the following survival distribution: OS < 6 months, n = 7; OS between 6 months and 12 months, n = 4; and OS > 12 months, n = 7. We ultimately selected the bile samples of four patients among the seven with better prognosis (OS > 12 months; group A) and four patients among the seven with dismal prognosis (OS < 6 months; group B) from the biobank according to the patients’ baseline characteristics and results of RNA quality control. This ensured that there were fewer statistically significant differences between the two groups (other than survival), and bile RNAs were reliable for miRNA and mRNA sequencing. Another 30 baseline bile samples from pCCA patients who had received first-line HAIC treatment from June 2019 to June 2020 at our center served as the validation cohort. The study was approved by institutional review board of Peking University Cancer Hospital (approval protocol number: 2021KT144). All methods were performed in accordance with the Declaration of Helsinki. All patients provided signed consent for participation. All human participants were Chinese. HAIC was performed as previously described.59 The HAIC regimen comprised OXA (40 mg/m2 for 2 h), 5-Fu (800 mg/m2 for 22 h), and intravenous folinic acid (200 mg/m2), and was administered on days 1–3 every 4 weeks. The bile samples (15 mL) were obtained by percutaneous transhepatic cholangial drainage (PTCD) within 7 days before the first cycle of the HAIC procedure. Centrifugation of the fresh bile at 2,000 rpm for 10 min yielded a supernatant, which was then stored in a freezer at −80°C. Patients signed informed consent forms for clinical specimen collection prior to PTCD.
Two human perihilar CCA cell lines (QBC939/FRH0201) and a 293T cell line were used in our study. The cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Cat#G4524-500ML, Servicebio, Wuhan, China) with 10% fetal bovine serum (Cat#G8003-500ML, Servicebio, Wuhan, China) and 1% penicillin/streptomycin (Cat#G4003-100ML, Servicebio, Wuhan, China). The cells were cultured in a humidified incubator at 37°C and supplemented with 5% CO2.
Female 4–6-week-old nude mice were purchased from HFK Bio-Technology (Beijing, China) and kept in specific pathogen-free conditions. QBC cells (5×106) stably overexpressing miR-532-3p, FRH0201 cells (5×106) stably underexpressing miR-532-3p, or negative controls were suspended in 0.2 mL DMEM and then subcutaneously inoculated into the right flank of nude mice (n = 5 mice/group). When the tumor’s long diameter was approximately 10 mm, OXA (8 mg/kg) and 5-Fu (10 mg/kg) were injected peritumorally every day for a week to simulate perfusion chemotherapy. Tumor sizes were measured every 2 days until the end of observation. Tumor volumes were calculated as: V = (L × W2)/2 (V, volume; L, length; W, width of tumor). All mice were sacrificed 2 weeks after the first drug injection. The animal study was approved by institutional review board of Peking University Cancer Hospital (approval protocol number: EAEC 2021-08).
Method details
High-throughput sequencing
The sequencing of miRNAs and mRNAs was performed by CloudSeq Biotech Inc. (Shanghai, China). Briefly, total RNA was extracted to prepare the miRNA sequencing library. An Illumina HiSeq 4000 sequencer was used for high-throughput sequencing. Following processing with cutadapt software (v1.9.2), the adaptor-trimmed reads (≥15 nt) were then aligned to the merged pre-miRNA databases. Tag counts per million aligned miRNAs were used to normalize the read counts. Differentially expressed miRNAs were filtered by |Fold change| ≥ 1.5 and p value <0.05. miRNA targets were identified by miRanda and Targetscan (https://www.targetscan.org/vert_80/).
For mRNA sequencing, after removing rRNA, a NEBNext Ultra II Directional RNA Library Prep Kit (New England Biolabs, Inc., MA, USA) was used to construct RNA libraries. High-throughput sequencing was performed using an Illumina HiSeq instrument. Differentially expressed mRNAs were filtered by |Fold change| ≥ 2.0 and p value <0.05. Finally, Gene Ontology (GO) and pathway enrichment analyses were also performed for differentially expressed mRNAs.
Transfection and virus infection
Cells were transfected with transient miRNA mimics, an inhibitor, or a negative control (NC) (Hanbio, Shanghai, China) via Lipofectamine RNAiMAX (Cat#13778030, Invitrogen, Carlsbad, CA, USA) in a six-well plate (1.5×105 cells/well). The pcDNA3.1-secreted protein acidic and rich in cysteine (SPARC) or control vector plasmids (Genechem, Shanghai, China) were transfected with Lipofectamine 3000 (Cat#L3000001, Invitrogen, Carlsbad, CA, USA). si-RNA (Hanbio, Shanghai, China) against SPARC or negative controls was transfected with Lipofectamine RNAiMAX (Cat#13778030, Invitrogen, Carlsbad, CA, USA).
Stable miR-532-3p overexpressing and underexpressing cells were created through lentiviral infection (Genechem, Shanghai, China). The transfection efficiency was analyzed by qPCR. The SPARC expression after transfection with plasmids or si-RNA was analyzed by qPCR and western blot. The miRNA mimic, inhibitor, and small interfering RNA (siRNA) sequences are shown in Table S1.
RNA expression analyses
RNA (including miRNAs) was extracted using Trizol reagent (Cat#15596018CN, Thermo Fisher, Waltham, MA, USA). A Nanodrop 8000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) was utilized to measure the RNA concentration. OD260/OD280 ratios ranging from 1.9 to 2.0 were defined as being of good quality for further analysis. miRNA and mRNA reverse transcription were performed using a microRNA Reverse Transcription Kit (Cat#EZB-miRT3, EZBioscience, California, USA) and a PrimeScript RT Reagent Kit (Cat#RR037A, Takara Bio USA, Inc), respectively. RNU6B and β-actin were used as internal controls. All data were subsequently analyzed by the 2−ΔΔCt method. The primers sequences are listed in Table S2.
Western blot
Radioimmunoprecipitation assay (RIPA) buffer was used to extract protein. Proteins were separated in 10% NuPAGE Bis-Tris SDS/polyacrylamide gel electrophoresis (PAGE) protein gels (Cat#P1200, Solarbio, Beijing, China) after quantification using a bicinchoninic acid protein assay kit (Cat#G2026-200T, Servicebio, Wuhan, China). Subsequently, the separated proteins were transferred to polyvinylidene difluoride membranes (Cat#YA1701, Solarbio, Beijing, China), which were then blocked with 5% nonfat milk powder for 2 h. Next, the membranes were blotted with antibodies against β-actin (Cat#ab6276, Abcam, Cambridge, UK) or SPARC (Cat#ab207743, Abcam, Cambridge, UK) at 4°C overnight, followed by incubation with the secondary antibody. ECL Plus Western Blotting Detection Reagents (Cat#PE0010, Solarbio, Beijing, China) were utilized to visualize proteins.
OXA and 5-fu treatment in vitro
OXA (Cat#HY-17371) and 5-Fu (Cat#HY-90006) were purchased from MedChemexpress (NJ, USA) and dissolved in dimethyl sulfoxide (DMSO; Cat#D8371; Merck, Germany). To calculate the half-maximal inhibitory concentration (IC50) values of OXA and 5-Fu, QBC939 cells were incubated in 96-well plates with OXA at concentrations of 2, 4, 6, 8, 16, 32, and 64 μmol/L or with 5-Fu at concentrations of 5, 10, 20, 40, and 80 μmol/L for 24 h. FRH0201 cells were incubated with OXA at concentrations of 5, 10, 20, 40, and 80 μmol/L or with 5-Fu at concentrations of 10, 20, 40, 80, 160, 320, and 640 μmol/L in 96-well plates for 24 h. For cell cycle and apoptosis analyses, QBC939 cells were incubated with OXA at 5 μmol/L and with 5-Fu at 10 μmol/L in 6-well plates for 48 h. FRH0201 cells were incubated with OXA at 20 μmol/L and with 5-Fu at 80 μmol/L in 6-well plates for 48 h. For the colony-forming assay, QBC939 cells were incubated with OXA at 5 μmol/L and with 5-Fu at 10 μmol/L in 6-well plates for 14 days. FRH0201 cells were incubated with OXA at 20 μmol/L and with 5-Fu at 80 μmol/L for 14 days.
Cell viability assay to evaluate OXA and 5-fu sensitivity
After transient transfection for 48 h, the cells were plated in 96-well plates to evaluate OXA and 5-Fu sensitivity (5000 cells/well). Next, the cells were incubated with OXA/5-Fu in a concentration gradient for 24 h. Then, absorbance readings at 450 nm were obtained via a Counting Kit-8 (CCK-8) kit (Cat#CA1210, Solarbio, Beijing, China). For rescue experiments, QBC939 and FRH0201 cell lines were incubated with OXA and 5-Fu (QBC939: OXA at 5 μmol and 5-Fu at 10 μmoll; FRH0201: OXA at 20 μmol and 5-Fu at 80 μmol) in 96-well plates (5000 cells/well) for 72 h. The absorbance reading at 450 nm was measured every 24 h. For the colony-forming assay, after cell transfection with miRNA-expressing virus for 48 h, 1000 cells per well were plated to 6-well plates and incubated with OXA and 5-Fu. Colonies were counted fourteen days later.
Cell cycle and cell apoptosis analyses
After transfection (48 h) and incubation with OXA and 5-Fu (48 h), the cells were collected from 6-well plates and then fixed in 70% cold ethanol for 12 h for cell cycle analysis. Then, the cells were stained with 50 μg/mL propidium iodide (PI) for 30 min and the cell cycle was analyzed using a FACS Calibur system (BD Biosciences, Erembodegem, Belgium). Cell apoptosis was measured via staining with Annexin V and PI for 15 min, followed by flow cytometry (BD Biosciences, Erembodegem, Belgium).
Luciferase reporter assays
Targetscan was used to predict binding sequences. The sequences of wild-type and mutant seed regions of SPARC were synthesized and cloned into pmriGLO (Miaolingbio, Wuhan, China). In 6-well plates, 293T cells were transfected with mimics NC + pmriGLO-SPARC 3′UTR-wt, miR-532-3p mimics + pmriGLO-SPARC 3′UTR-wt, mimics NC + pmriGLO-SPARC 3′UTR-mut, or miR-532-3p mimics + pmriGLO-SPARC 3′UTR-mut and pmriGLO. Luciferase activity assays were performed after transfection for 48 h.
Quantification and Statistical analysis
Statistical analyses were conducted using SPSS 20.0 software (IBM) and GraphPad Prism 9 (GraphPad). The significance level was set at p ≤ 0.05. The survival significance was determined by a log rank test. Continuous data were reported as mean ± standard deviation (SD) and were evaluated by two-tailed unpaired Student’s t tests. Categorical variables were presented as frequencies, and were analyzed by chi-square tests and Fisher’s exact tests.
Published: November 20, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.111437.
Contributor Information
Yan Kong, Email: k-yan08@163.com.
Xiaodong Wang, Email: xiaodongw75@yahoo.com.
Supplemental information
References
- 1.Greten T.F., Schwabe R., Bardeesy N., Ma L., Goyal L., Kelley R.K., Wang X.W. Immunology and immunotherapy of cholangiocarcinoma. Nat. Rev. Gastroenterol. Hepatol. 2023;20:349–365. doi: 10.1038/s41575-022-00741-4. [DOI] [PubMed] [Google Scholar]
- 2.Mauro E., Ferrer-Fàbrega J., Sauri T., Soler A., Cobo A., Burrel M., Iserte G., Forner A. New challenges in the management of cholangiocarcinoma: the role of liver transplantation, locoregional therapies, and systemic therapy. Cancers. 2023;15 doi: 10.3390/cancers15041244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rizvi S., Khan S.A., Hallemeier C.L., Kelley R.K., Gores G.J. Cholangiocarcinoma - evolving concepts and therapeutic strategies. Nat. Rev. Clin. Oncol. 2018;15:95–111. doi: 10.1038/nrclinonc.2017.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soares K.C., Jarnagin W.R. The landmark series: hilar cholangiocarcinoma. Ann. Surg Oncol. 2021;28:4158–4170. doi: 10.1245/s10434-021-09871-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan E., Berlin J. Biliary tract cancers: understudied and poorly understood. J. Clin. Oncol. 2015;33:1845–1848. doi: 10.1200/jco.2014.59.7591. [DOI] [PubMed] [Google Scholar]
- 6.Razumilava N., Gores G.J. Cholangiocarcinoma. Lancet. 2014;383:2168–2179. doi: 10.1016/S0140-6736(13)61903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valle J., Wasan H., Palmer D.H., Cunningham D., Anthoney A., Maraveyas A., Madhusudan S., Iveson T., Hughes S., Pereira S.P., et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N. Engl. J. Med. 2010;362:1273–1281. doi: 10.1056/NEJMoa0908721. [DOI] [PubMed] [Google Scholar]
- 8.Rizzo A., Brandi G. Neoadjuvant therapy for cholangiocarcinoma: A comprehensive literature review. Cancer Treat. Res. Commun. 2021;27 doi: 10.1016/j.ctarc.2021.100354. [DOI] [PubMed] [Google Scholar]
- 9.Rizzo A., Brandi G. Pitfalls, challenges, and updates in adjuvant systemic treatment for resected biliary tract cancer. Expet Rev. Gastroenterol. Hepatol. 2021;15:547–554. doi: 10.1080/17474124.2021.1890031. [DOI] [PubMed] [Google Scholar]
- 10.Ricci A.D., Rizzo A., Brandi G. Immunotherapy in biliary tract cancer: Worthy of a second look. Cancer Control. 2020;27 doi: 10.1177/1073274820948047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh D.Y., He A.R., Qin S., Chen L.T., Okusaka T., Vogel A., Kim J.W., Lee T., Lee M.A., Kitano M., et al. 78P Updated overall survival (OS) from the phase III TOPAZ-1 study of durvalumab (D) or placebo (PBO) plus gemcitabine and cisplatin (+ GC) in patients (pts) with advanced biliary tract cancer (BTC) Ann. Oncol. 2022;33:S1462–S1463. doi: 10.1016/j.annonc.2022.10.114. [DOI] [Google Scholar]
- 12.Kelley R.K., Ueno M., Yoo C., Finn R.S., Furuse J., Ren Z., Yau T., Klümpen H.-J., Chan S.L., Ozaka M., et al. Pembrolizumab in combination with gemcitabine and cisplatin compared with gemcitabine and cisplatin alone for patients with advanced biliary tract cancer (KEYNOTE-966): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2023;401:1853–1865. doi: 10.1016/S0140-6736(23)00727-4. [DOI] [PubMed] [Google Scholar]
- 13.Palmieri L.J., Lavolé J., Dermine S., Brezault C., Dhooge M., Barré A., Chaussade S., Coriat R. The choice for the optimal therapy in advanced biliary tract cancers: Chemotherapy, targeted therapies or immunotherapy. Pharmacol. Ther. 2020;210 doi: 10.1016/j.pharmthera.2020.107517. [DOI] [PubMed] [Google Scholar]
- 14.Wang X., Hu J., Cao G., Zhu X., Cui Y., Ji X., Li X., Yang R., Chen H., Xu H., et al. Phase II study of hepatic arterial infusion chemotherapy with oxaliplatin and 5-fluorouracil for advanced perihilar cholangiocarcinoma. Radiology. 2017;283:580–589. doi: 10.1148/radiol.2016160572. [DOI] [PubMed] [Google Scholar]
- 15.Bartel D.P. MicroRNAs. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 16.He L., Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 17.Saliminejad K., Khorram Khorshid H.R., Soleymani Fard S., Ghaffari S.H. An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019;234:5451–5465. doi: 10.1002/jcp.27486. [DOI] [PubMed] [Google Scholar]
- 18.Sheervalilou R., Ansarin K., Fekri Aval S., Shirvaliloo S., Pilehvar-soltanahmadi Y., Mohammadian M., Zarghami N. An update on sputum MicroRNAs in lung cancer diagnosis. Diagn. Cytopathol. 2016;44:442–449. doi: 10.1002/dc.23444. [DOI] [PubMed] [Google Scholar]
- 19.Mohammadian F., Pilehvar-Soltanahmadi Y., Alipour S., Dadashpour M., Zarghami N. Chrysin alters microRNAs expression levels in gastric cancer cells: possible molecular mechanism. Drug Res. 2017;67:509–514. doi: 10.1055/s-0042-119647. [DOI] [PubMed] [Google Scholar]
- 20.Salati M., Braconi C. Noncoding RNA in cholangiocarcinoma. Semin. Liver Dis. 2019;39:13–25. doi: 10.1055/s-0038-1676097. [DOI] [PubMed] [Google Scholar]
- 21.Lampis A., Carotenuto P., Vlachogiannis G., Cascione L., Hedayat S., Burke R., Clarke P., Bosma E., Simbolo M., Scarpa A., et al. MIR21 drives resistance to heat shock protein 90 inhibition in cholangiocarcinoma. Gastroenterology. 2018;154:1066–1079.e5. doi: 10.1053/j.gastro.2017.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asukai K., Kawamoto K., Eguchi H., Konno M., Asai A., Iwagami Y., Yamada D., Asaoka T., Noda T., Wada H., et al. Micro-RNA-130a-3p regulates gemcitabine resistance via PPARG in cholangiocarcinoma. Ann. Surg Oncol. 2017;24:2344–2352. doi: 10.1245/s10434-017-5871-x. [DOI] [PubMed] [Google Scholar]
- 23.Jiao D., Yan Y., Shui S., Wu G., Ren J., Wang Y., Han X. miR-106b regulates the 5-fluorouracil resistance by targeting Zbtb7a in cholangiocarcinoma. Oncotarget. 2017;8:52913–52922. doi: 10.18632/oncotarget.17577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y., Zhang W., Chen L., Chen W., Xu S., Tang L., Yang Y., Li Q., Jiang Q., Miao L. The ATO/miRNA-885-5p/MTPN axis induces reversal of drug-resistance in cholangiocarcinoma. Cell. Oncol. 2021;44:907–916. doi: 10.1007/s13402-021-00610-3. [DOI] [PubMed] [Google Scholar]
- 25.Silakit R., Kitirat Y., Thongchot S., Loilome W., Techasen A., Ungarreevittaya P., Khuntikeo N., Yongvanit P., Yang J.H., Kim N.H., et al. Potential role of HIF-1-responsive microRNA210/HIF3 axis on gemcitabine resistance in cholangiocarcinoma cells. PLoS One. 2018;13 doi: 10.1371/journal.pone.0199827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L., Peterson A., Noteboom J., O'Briant K.C., Allen A., et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macias R.I.R., Kornek M., Rodrigues P.M., Paiva N.A., Castro R.E., Urban S., Pereira S.P., Cadamuro M., Rupp C., Loosen S.H., et al. Diagnostic and prognostic biomarkers in cholangiocarcinoma. Liver Int. 2019;39:108–122. doi: 10.1111/liv.14090. [DOI] [PubMed] [Google Scholar]
- 28.Shi T., Morishita A., Kobara H., Masaki T. The role of microRNAs in cholangiocarcinoma. Int. J. Mol. Sci. 2021;22:7627. doi: 10.3390/ijms22147627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janssen F.W., Lak N.S.M., Janda C.Y., Kester L.A., Meister M.T., Merks J.H.M., van den Heuvel-Eibrink M.M., van Noesel M.M., Zsiros J., Tytgat G.A.M., Looijenga L.H.J. A comprehensive overview of liquid biopsy applications in pediatric solid tumors. npj Precis. Oncol. 2024;8:172. doi: 10.1038/s41698-024-00657-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grossmann L.D., Chen C.-H., Uzun Y., Thadi A., Wolpaw A.J., Louault K., Goldstein Y., Surrey L.F., Martinez D., Calafatti M., et al. Identification and characterization of chemotherapy resistant high-risk neuroblastoma persister cells. Cancer Discov. 2024 doi: 10.1158/2159-8290.Cd-24-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Lorenzo A., Bolli E., Ruiu R., Ferrauto G., Di Gregorio E., Avalle L., Savino A., Poggio P., Merighi I.F., Riccardo F., et al. Toll-like receptor 2 promotes breast cancer progression and resistance to chemotherapy. OncoImmunology. 2022;11 doi: 10.1080/2162402X.2022.2086752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang H., Zhou H., Zhang L., Tang T., Li N. Molecular mechanism of MLCK1 inducing 5-Fu resistance in colorectal cancer cells through activation of TNFR2/NF-κB pathway. Discov. Oncol. 2024;15:159. doi: 10.1007/s12672-024-01019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silakit R., Loilome W., Yongvanit P., Chusorn P., Techasen A., Boonmars T., Khuntikeo N., Chamadol N., Pairojkul C., Namwat N. Circulating miR-192 in liver fluke-associated cholangiocarcinoma patients: a prospective prognostic indicator. J. Hepatobiliary. Pancreat. Sci. 2014;21:864–872. doi: 10.1002/jhbp.145. [DOI] [PubMed] [Google Scholar]
- 34.Bernuzzi F., Marabita F., Lleo A., Carbone M., Mirolo M., Marzioni M., Alpini G., Alvaro D., Boberg K.M., Locati M., et al. Serum microRNAs as novel biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Clin. Exp. Immunol. 2016;185:61–71. doi: 10.1111/cei.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shigehara K., Yokomuro S., Ishibashi O., Mizuguchi Y., Arima Y., Kawahigashi Y., Kanda T., Akagi I., Tajiri T., Yoshida H., et al. Real-time PCR-based analysis of the human bile microRNAome identifies miR-9 as a potential diagnostic biomarker for biliary tract cancer. PLoS One. 2011;6 doi: 10.1371/journal.pone.0023584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang Z., Liu X., Zhang Q., Wang C., Zhao Y. Diagnostic value of microRNAs as biomarkers for cholangiocarcinoma. Dig. Liver Dis. 2016;48:1227–1232. doi: 10.1016/j.dld.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 37.Pan Y., Shao S., Sun H., Zhu H., Fang H. Bile-derived exosome noncoding RNAs as potential diagnostic and prognostic biomarkers for cholangiocarcinoma. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.985089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shu L., Li X., Liu Z., Li K., Shi A., Tang Y., Zhao L., Huang L., Zhang Z., Zhang D., et al. Bile exosomal miR-182/183-5p increases cholangiocarcinoma stemness and progression by targeting HPGD and increasing PGE2 generation. Hepatology. 2024;79:307–322. doi: 10.1097/hep.0000000000000437. [DOI] [PubMed] [Google Scholar]
- 39.Van der Auwera S., Ameling S., Wittfeld K., Bülow R., Nauck M., Völzke H., Völker U., Grabe H.J. Circulating miRNAs modulating systemic low-grade inflammation and affecting neurodegeneration. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2024;135 doi: 10.1016/j.pnpbp.2024.111130. [DOI] [PubMed] [Google Scholar]
- 40.Gagez A.-L., Duroux-Richard I., Leprêtre S., Orsini-Piocelle F., Letestu R., De Guibert S., Tuaillon E., Leblond V., Khalifa O., Gouilleux-Gruart V., et al. miR-125b and miR-532-3p predict the efficiency of rituximab-mediated lymphodepletion in chronic lymphocytic leukemia patients. A French innovative leukemia organization study. Haematologica. 2017;102:746–754. doi: 10.3324/haematol.2016.153189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang M.Y., Wang L.Q., Chim C.S. miR-1250-5p is a novel tumor suppressive intronic miRNA hypermethylated in non-Hodgkin’s lymphoma: novel targets with impact on ERK signaling and cell migration. Cell Commun. Signal. 2021;19:62. doi: 10.1186/s12964-021-00707-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao B.S., Liu S.G., Wang T.Y., Ji Y.H., Qi B., Tao Y.P., Li H.C., Wu X.N. Screening of microRNA in patients with esophageal cancer at same tumor node metastasis stage with different prognoses. Asian Pac. J. Cancer Prev. APJCP. 2013;14:139–143. doi: 10.7314/apjcp.2013.14.1.139. [DOI] [PubMed] [Google Scholar]
- 43.Zhou Y., Zheng X., Lu J., Chen W., Li X., Zhao L. Ginsenoside 20(S)-Rg3 inhibits the Warburg effect via modulating DNMT3A/MiR-532-3p/HK2 pathway in ovarian cancer cells. Cell. Physiol. Biochem. 2018;45:2548–2559. doi: 10.1159/000488273. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y., Yang Z., Wang L., Sun L., Liu Z., Li Q., Yao B., Chen T., Wang C., Yang W., et al. miR-532-3p promotes hepatocellular carcinoma progression by targeting PTPRT. Biomed. Pharmacother. 2019;109:991–999. doi: 10.1016/j.biopha.2018.10.145. [DOI] [PubMed] [Google Scholar]
- 45.Wang K., Gong D., Qiao X., Zheng J. MiR-532-3p inhibited the methylation of SOCS2 to suppress the progression of PC by targeting DNMT3A. Life Sci. Alliance. 2023;6 doi: 10.26508/lsa.202201703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tuo X., Zhou Y., Yang X., Ma S., Liu D., Zhang X., Hou H., Wang R., Li X., Zhao L. miR-532–3p suppresses proliferation and invasion of ovarian cancer cells via GPNMB/HIF-1α/HK2 axis. Pathol. Res. Pract. 2022;237 doi: 10.1016/j.prp.2022.154032. [DOI] [PubMed] [Google Scholar]
- 47.Liu Z.Y., Zhao C.G. miR-532-3p inhibits the progression of tongue squamous cell carcinoma by targeting podoplanin. Chin. Med. J. 2021;134:2999–3008. doi: 10.1097/CM9.0000000000001563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang H., Yan L., Zhong J., Hong L., Zhang N., Luo X. Circ_0025033 deficiency suppresses paclitaxel resistance and malignant development of paclitaxel-resistant ovarian cancer cells by modulating the miR-532-3p/FOXM1 network. Immunopharmacol. Immunotoxicol. 2022;44:275–286. doi: 10.1080/08923973.2022.2038194. [DOI] [PubMed] [Google Scholar]
- 49.Jiang S., Sun H.-F., Li S., Zhang N., Chen J.-S., Liu J.-X. SPARC: a potential target for functional nanomaterials and drugs. Front. Mol. Biosci. 2023;10 doi: 10.3389/fmolb.2023.1235428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bradshaw A.D. Diverse biological functions of the SPARC family of proteins. Int. J. Biochem. Cell Biol. 2012;44:480–488. doi: 10.1016/j.biocel.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng C.-T., Chu Y.-Y., Yeh C.-N., Huang S.-C., Chen M.H., Wang S.-Y., Tsai C.-Y., Chiang K.-C., Chen Y.-Y., Ma M.-C., et al. Peritumoral SPARC expression and patient outcome with resectable intrahepatic cholangiocarcinoma. OncoTargets Ther. 2015;8:1899–1907. doi: 10.2147/OTT.S78728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Said N., Frierson H.F., Sanchez-Carbayo M., Brekken R.A., Theodorescu D. Loss of SPARC in bladder cancer enhances carcinogenesis and progression. J. Clin. Invest. 2013;123:751–766. doi: 10.1172/JCI64782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aghamaliyev U., Gaitantzi H., Thomas M., Simon-Keller K., Gaiser T., Marx A., Yagublu V., Araos J., Cai C., Valous N.A., et al. Downregulation of SPARC is associated with epithelial-mesenchymal transition and low differentiation state of biliary tract cancer cells. Eur. Surg. Res. 2019;60:1–12. doi: 10.1159/000494734. [DOI] [PubMed] [Google Scholar]
- 54.Rahman M., Chan A.P.K., Tang M., Tai I.T. A peptide of SPARC interferes with the interaction between caspase8 and Bcl2 to resensitize chemoresistant tumors and enhance their regression in vivo. PLoS One. 2011;6 doi: 10.1371/journal.pone.0026390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hua H.-W., Jiang F., Huang Q., Liao Z.-J., Ding G. Re-sensitization of 5-FU resistance by SPARC through negative regulation of glucose metabolism in hepatocellular carcinoma. Tumor Biol. 2015;36:303–313. doi: 10.1007/s13277-014-2633-2. [DOI] [PubMed] [Google Scholar]
- 56.Fan X., Mao Z., Ma X., Cui L., Qu J., Lv L., Dang S., Wang X., Zhang J. Secreted protein acidic and rich in cysteine enhances the chemosensitivity of pancreatic cancer cells to gemcitabine. Tumor Biol. 2016;37:2267–2273. doi: 10.1007/s13277-015-4044-4. [DOI] [PubMed] [Google Scholar]
- 57.Ma J., Ma Y., Chen S., Guo S., Hu J., Yue T., Zhang J., Zhu J., Wang P., Chen G., Liu Y. SPARC enhances 5-FU chemosensitivity in gastric cancer by modulating epithelial-mesenchymal transition and apoptosis. Biochem. Biophys. Res. Commun. 2021;558:134–140. doi: 10.1016/j.bbrc.2021.04.009. [DOI] [PubMed] [Google Scholar]
- 58.Notaro A., Sabella S., Pellerito O., Vento R., Calvaruso G., Giuliano M. The secreted protein acidic and rich in cysteine is a critical mediator of cell death program induced by WIN/TRAIL combined treatment in osteosarcoma cells. Int. J. Oncol. 2016;48:1039–1044. doi: 10.3892/ijo.2015.3307. [DOI] [PubMed] [Google Scholar]
- 59.Hu J., Zhu X., Wang X., Cao G., Wang X., Yang R. Evaluation of percutaneous unilateral trans-femoral implantation of side-hole port-catheter system with coil only fixed-catheter-tip for hepatic arterial infusion chemotherapy. Cancer Imag. 2019;19:15. doi: 10.1186/s40644-019-0202-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
• miRNA sequencing data have been deposited at Mendeley data. mRNA sequencing data have been deposited in the Gene Expression Omnibus (GEO). miRNA and mRNA sequencing data are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
• This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.