To the editor:
Hearing loss is the most common sensory disorder. TSPEAR gene encodes thrombospondin‐type laminin G domain and epilepsy‐associated repeats containing protein. 1 While, patients with variants in the TSPEAR gene may present with different clinical phenotypes, including autosomal recessive nonsyndromic deafness (DFNB98, MIM614861); ectodermal dysplasia 14 of the hair/tooth type with or without hypohidrosis (ECTD14, MIM618180); or selective tooth agenesis‐10 (STHAG10, MIM 620173). 2 , 3 , 4 , 5 Here, we report a patient diagnosed with congenital sensorineural hearing loss, and a total of three unreported variants were detected.
The proband (IV‐1) is a 7‐year‐old girl. She didn't pass the neonatal hearing screening and underwent an auditory brainstem response (ABR) test 6 months after birth. Her hearing thresholds at 6 months were 65 dBnHL in both ears with normal acoustic impedance results, and then she was diagnosed with congenital sensorineural hearing loss. An ABR test conducted at the age of 6 years showed that hearing thresholds of both ears were 80 dBnHL. Distortion product otoacoustic emission was not elicited on both sides. The proband received bilateral hearing aids when she was 6 months old. According to her parents, she was able to respond normally to the minor sounds after wearing hearing aids. The proband also demonstrated dental caries (Figure 1A), ankyloglossia (Figure 1B), and heart‐shaped tongue (Figure 1C). After three corrective surgeries, her tongue was still difficult to stick out and her speech was still compromised. Her parents showed normal hearing, and after a strict physical examination, no abnormalities were found. She has a younger brother (IV‐2) who has undergone an amniocentesis without abnormality and passed neonatal hearing screening. There were two other members with hearing loss in her family (Figure 1D), but unfortunately, they have not been genetically analyzed.
FIGURE 1.
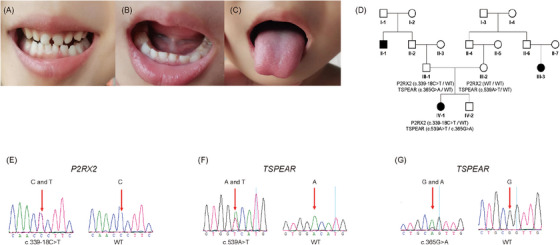
Clinical and genetic manifestations of the proband. (A) Dental caries. (B) Ankyloglossia. (C) Heart‐shaped tongue. (D) Pedigree of the proband. Black indicates family members with hearing loss. (E) Genetic sequencing result of c.339‐18C>T of P2RX2 gene. (F) Genetic sequencing result of c.539A>T of TSPEAR gene. (G) Genetic sequencing result of c.365G>A of TSPEAR gene. The red arrows indicate the mutated bases. WT means wild type.
The variant detection and analysis method has been described in detail in our previous studies. 6 , 7 , 8 We sampled peripheral blood from the proband and her parents for high‐throughput sequencing. Genomic DNA was isolated from the blood samples, and then fragmentation of the genomic DNA was performed to generate a paired‐end library. The amplified DNA library was sequenced on the BGISEQ‐500 platform. Sequencing data were compared with the human genome reference (GRCh37/hg19) to identify mutant genes and loci.
The proband carries heterozygous variant NM_012226.5 (P2RX2): c.339‐18C>T inherited from her father (Figure 1E). P2RX2 encodes the P2X2 receptor, which assembles as a trimer to form a channel gated by extracellular ATP. The variant was reported to have a minor allele frequency of 6.862e−7, while the allele frequency in the East Asian population is 0.00002230 in the gnomAD database. According to the American College of Medical Genetics and Genomics (ACMG)/Association for Molecular Pathology (AMP) guidelines, the evidence supports PM2 and BP4, and this variant was classified to have uncertain significance. Analysis using the RDDC database, MaxEntScan, and SpliceAI suggests that it may not affect splice.
Besides, the proband also carries compound heterozygous variants NM_144991.3 (TSPEAR): c.539A>T (p.Asp180Val) (Figure 1F) from her mother and NM_144991.3 (TSPEAR): c.365G>A, (p.Arg122Gln) (Figure 1G) from her father. TSPEAR c.539A>T (rs781821217) is a missense variant that occurs in Exon3, and there are no reports of pathogenicity at this locus. The variant was reported to have a filtering allele frequency of 0.00003357, while the allele frequency in the East Asian population is 0.0001342 in the gnomAD database. According to the ACMG/AMP guidelines (PM2_Supporting), this variant was classified to be uncertain significance as the ClinVar database showed. However, other prediction tools give different results. Polyphen‐2 suggests it may be benign, and the RDDC database proves this. While the PROVEAN score shows that it is deleterious. TSPEAR c.365G>A (rs141753295) is also a missense variant that occurs in Exon3 which is still unreported likewise. This variant was reported to have a filtering allele frequency of 0.001084 in the gnomAD database. In the ClinVar database, this variant has conflicting classifications of pathogenicity. According to the ACMG/AMP guidelines, the evidence supports BS1_Supporting, and therefore this variant was classified to be uncertain significance. PROVEAN score shows this variant is neutral. Polyphen‐2 suggests it may be a probably damaging variant.
TSPEAR belongs to a superfamily of proteins characterized by the presence of EAR repeats in tandem, which may form β‐propeller in proteins that may act as ligand‐binding structural domains. 1 , 9 Downregulation of TSPEAR in keratinocytes may affect Notch signaling. 10 TSPEAR is expressed in the base of the stereocilia of inner hair cells and outer hair cells, ganglion, stria vascularis, and vestibular in mice, 2 which suggests that TSPEAR may play an important role in cochlear cell differentiation and hearing maintenance. In our patient, we did not find other suspicious causes of hearing loss, such as pregnancy infections, use of ototoxic drugs, and perinatal hypoxic‐ischemic encephalopathy, so we believe that the hearing loss in our patient is mainly due to compound heterozygous variants in the TSPEAR gene. Our research hopes to expand the genetic spectrum of hearing loss and provide further evidence for the diagnosis and treatment of hearing loss. Recently, the success of OTOF gene therapy has given us hope for the treatment of hereditary hearing loss. 11 , 12 In addition, doctors and researchers should also actively look for other treatments for hearing loss, which may be promising to promote hair cell regeneration or find suitable Chinese herbal medicine. 13 – 15
CONSENT FOR PUBLICATION
Written informed consent was obtained from the parents of the patient for the publication of any potentially identifiable images or data included in this article.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
This study was funded by the Innovative Research Groups of Hubei Province (No. 2023AFA038), the National Key Research and Development Program of China (No. 2021YFF0702303, 2023YFE0203200), and the National Natural Science Foundation of China (No. 82071058). In addition, the authors would like to thank Xi Chen for her guidance on some of the methods mentioned in this manuscript.
Shi X, Liu X, Zhao Z, Zong Y, Sun Y. Novel compound heterozygous variants in the TSPEAR gene causing autosomal recessive hearing loss in a Chinese family. Pediatr Investig. 2024;8:313–315. 10.1002/ped4.12454
REFERENCES
- 1. Scheel H, Tomiuk S, Hofmann K. A common protein interaction domain links two recently identified epilepsy genes. Hum Mol Genet. 2002;11:1757‐1762. DOI: 10.1093/hmg/11.15.1757 [DOI] [PubMed] [Google Scholar]
- 2. Delmaghani S, Aghaie A, Michalski N, Bonnet C, Weil D, Petit C. Defect in the gene encoding the EAR/EPTP domain‐containing protein TSPEAR causes DFNB98 profound deafness. Hum Mol Genet. 2012;21:3835‐3844. DOI: 10.1093/hmg/dds212 [DOI] [PubMed] [Google Scholar]
- 3. Jackson A, Lin SJ, Jones EA, Chandler KE, Orr D, Moss C, et al. Clinical, genetic, epidemiologic, evolutionary, and functional delineation of TSPEAR‐related autosomal recessive ectodermal dysplasia 14. HGG Adv. 2023;4:100186. DOI: 10.1016/j.xhgg.2023.100186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bowles B, Ferrer A, Nishimura CJ, Pinto E Vairo F, Rey T, Leheup B, et al. TSPEAR variants are primarily associated with ectodermal dysplasia and tooth agenesis but not hearing loss: a novel cohort study. Am J Med Genet A. 2021;185:2417‐2433. DOI: 10.1002/ajmg.a.62347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Song JS, Bae M, Kim JW. Novel TSPEAR mutations in non‐syndromic oligodontia. Oral Dis. 2020;26:847‐849. DOI: 10.1111/odi.13316 [DOI] [PubMed] [Google Scholar]
- 6. Chen S, Jin Y, Xie L, Xie W, Xu K, Qiu Y, et al. A novel spontaneous mutation of the SOX10 gene associated with waardenburg syndrome type II. Neural Plast. 2020;2020:9260807. DOI: 10.1155/2020/9260807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jin Y, Liu XZ, Xie L, Xie W, Chen S, Sun Y. Targeted next‐generation sequencing identified novel compound heterozygous variants in the PTPRQ gene causing autosomal recessive hearing loss in a Chinese family. Front Genet. 2022;13:884522. DOI: 10.3389/fgene.2022.884522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sun Y, Xiang J, Liu Y, Chen S, Yu J, Peng J, et al. Increased diagnostic yield by reanalysis of data from a hearing loss gene panel. BMC Med Genomics. 2019;12:76. DOI: 10.1186/s12920-019-0531-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Staub E, Pérez‐Tur J, Siebert R, Nobile C, Moschonas NK, Deloukas P, et al. The novel EPTP repeat defines a superfamily of proteins implicated in epileptic disorders. Trends Biochem Sci. 2002;27:441‐444. DOI: 10.1016/s0968-0004(02)02163-1 [DOI] [PubMed] [Google Scholar]
- 10. Peled A, Sarig O, Samuelov L, Bertolini M, Ziv L, Weissglas‐Volkov D, et al. Mutations in TSPEAR, encoding a regulator of Notch signaling, affect tooth and hair follicle morphogenesis. PLoS Genet. 2016;12:e1006369. DOI: 10.1371/journal.pgen.1006369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Qi J, Tan F, Zhang L, Lu L, Zhang S, Zhai Y, et al. AAV‐mediated gene therapy restores hearing in patients with DFNB9 deafness. Adv Sci. 2024;11:e2306788. DOI: 10.1002/advs.202306788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lv J, Wang H, Cheng X, Chen Y, Wang D, Zhang L, et al. AAV1‐hOTOF gene therapy for autosomal recessive deafness 9: a single‐arm trial. Lancet. 2024;403:2317‐2325. DOI: 10.1016/S0140-6736(23)02874-X [DOI] [PubMed] [Google Scholar]
- 13. Qi J, Huang W, Lu Y, Yang X, Zhou Y, Chen T, et al. Stem cell‐based hair cell regeneration and therapy in the inner ear. Neurosci Bull. 2024;40:113‐126. DOI: 10.1007/s12264-023-01130-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu Y, Zhang J, Liu Q, Miao Z, Chai R, Chen W. Development of Chinese herbal medicine for sensorineural hearing loss. Acta Pharm Sin B. 2024;14:455‐467. DOI: 10.1016/j.apsb.2023.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun Q, Zhang L, Chen T, Li N, Tan F, Gu X, et al. AAV‐mediated Gpm6b expression supports hair cell reprogramming. Cell Prolif. 2024;57:e13620. DOI: 10.1111/cpr.13620 [DOI] [PMC free article] [PubMed] [Google Scholar]


