Abstract
Background
The high burden of malaria in Africa is largely due to the presence of competent and adapted Anopheles vector species. With invasive Anopheles stephensi implicated in malaria outbreaks in Africa, understanding the genomic basis of vector-parasite compatibility is essential for assessing the risk of future outbreaks due to this mosquito. Vector compatibility with P. falciparum arises from ancient coevolution and involves genes such as Pfs47 in P. falciparum and P47Rec in Anopheles. Questions remain about whether sub-continental vector variation is a selective pressure on current Plasmodium populations.
Methods
We analyzed the genetic diversity in parasite–vector interaction genes in P. falciparum and An. gambiae from 9 and 15 countries in Africa, respectively. Specifically, we looked for evidence of malaria vector-mediated selection within three P. falciparum genes (Pfs47, Pfs16, Pfs37) and conducted association analyses with occurrence probabilities of prominent malaria vectors.
Results
Higher protein haplotype diversities of Pfs47 and Pfs16 were associated with the probability of occurrence of An. arabiensis and An. funestus together. Only Pfs16 carried a signature of positive selection consistently (average Tajima’s D = −2.96), which was associated with the probability of occurrence of An. funestus. These findings support vector-mediated selection on the basis of vector species diversity that may be occurring within Africa. We also employed phylogenetic analyses of An. gambiae interaction genes (P47Rec, APN1, HPX15) to identify significant subspecies diversity as a prerequisite to vector-population-mediated selection. Anopheles gambiae HPX15 revealed significant within-species differentiation (multiple branches bootstrap > 70) compared with absence of variation in P47Rec, suggesting that further investigation into subspecies-mediated selection on the basis of HPX15 is needed. Finally, we observed five amino acid changes at P47Rec in invasive An. stephensi compared with dominant African Anopheles species, calling for further investigation of the impact these distinct P47Rec variants might have on local African P. falciparum Pfs47 diversity.
Conclusions
Overall, these findings suggest that vector variation within Africa could influence P. falciparum diversity and lay a genomic framework for future investigation of invasive An. stephensi’s impact on African malaria.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13071-024-06604-y.
Keywords: Vector–parasite interactions, Vector-mediated selection, Genetics, Coevolution
Background
In 2022, more than 249 million cases of malaria were reported, with the majority located in Africa [1]. Efforts to control this disease, which impacts half of the world’s population, have reached a critical point in the last few years following the 10% increase in cases observed in 2020 [2]. Most of these cases are due to the unicellular eukaryotic parasite Plasmodium falciparum adapted to spread through Anopheles gambiae s.l. mosquitoes present throughout most of the African continent [3]. Most recently, Anopheles stephensi, common to South Asia and the Middle East, has invaded the Horn of Africa and several other African countries, further exasperating malaria control [4–6]. Vectorial competence, breeding habitats, and behaviors vary even among the members of the native An. gambiae s.l. complex [7]. With the invasion of An. stephensi now contributing to increased complexity of the already diverse vector species composition in Africa, it is important to determine the new transmission dynamics in An. stephensi-invaded areas.
Interactions between Plasmodium and the mosquito midgut serve as the critical gateway for malaria transmission. The parasite invasion of the mosquito midgut requires an interaction between both parasite and mosquito proteins [8]. Previous studies have shown that the level of compatibility of interacting proteins between malaria vector species and parasite species varies depending on the haplotypes of the genes coding for these proteins, especially during the midgut invasion [9]. Many of the Anopheles and Plasmodium genes responsible for these interactions are currently being studied for their potential use in development of transmission blocking vaccines [10, 11].
In addition to within-species genetic diversity, the haplotype diversity of genes involved in interactions with malaria parasites across different vector species may influence the genetic diversity of key genes that mediate vector–parasite interactions in Plasmodium falciparum. Different mosquito species, such as An. gambiae and An. funestus, have varying ecological niches, behaviors, and interactions with the malaria parasite, which can lead to differential selective pressures on the parasite’s genes [12]. For instance, variations in the vector’s immune response, feeding habits, and geographical distribution can drive genetic diversity in the parasite as it adapts to survive and thrive in different vector environments [13]. Consequently, regions with diverse vector species compositions are likely to exhibit higher nucleotide and haplotype diversity in P. falciparum genes associated with vector interactions, reflecting the parasite’s adaptation to a range of vector-related selective pressures.
A well-studied example for parasite–vector interaction genes is the Pfs47-P47Rec complex in P. falciparum and An. gambiae [11, 14, 15]. The mosquito midgut protein P47Rec and parasite protein Pfs47 work as a receptor–ligand pair during the Plasmodium invasion by playing a role in the immune evasion of parasites to make the parasite “undetectable” to the mosquito immune system. Silencing P47Rec expression has reduced the infection of P. falciparum in An. gambiae mosquitoes [16].
With this “lock-and-key” type mechanism, the ability of P. falciparum strains to invade the Anopheles midgut cells is dependent on the correct matching of Pfs47 surface protein haplotype (“the key”) with the Anopheles midgut receptor P47Rec (“the lock”) [16]. Previous functional studies demonstrated that replacing Pfs47 haplotype in African P. falciparum with a different haplotype from another continent is sufficient to change the compatibility between the vector and parasite [9]. Later studies have shown that Pfs47 is important for the adaptation of P. falciparum to different malaria vectors in different continents [17]. This vector-mediated selective pressure at the continental level in Pfs47 resulted in significant population structure between different continents, particularly in domain 2 of the protein [14, 18, 19]. Subcontinental selective pressure on the Pfs47 has been observed in previous studies in Nigeria, Brazil, and Malaysia [19]. Still, significant knowledge gaps remain about the level of vector-mediated selective pressure on Pfs47 at a subcontinental level in Africa. This is important to evaluate given the multiple Anopheles vector species that exist sympatrically across Africa, some quite divergent from one another (e.g., the An. gambiae complex versus Anopheles funestus) [20].
In addition to the Pfs47-P47Rec system, there are several other protein coding genes being studied as transmission blocking vaccine (TBV) targets on the basis of their role in parasite–vector compatibility. Therefore, like Pfs47 and P47Rec, these parasite and vector genes may also involve vector-mediated selection [10, 15]. In our study we selected two more genes from P. falciparum and another two genes from An. gambiae, which are important for vector–parasite interactions. In Plasmodium parasites, Pfs16 (PF3D7_0406200) and Pfs37 (PF3D7_1204400) have been recognized to be important for vector–parasite interactions because of their significant upregulated expression in the sexual stages and interactions with mosquito midgut proteins [15]. Knocking out Pfs16 or Pfs37 has shown a reduction in the number of oocysts generated in the mosquito midgut during the parasite invasion [15, 21]. Initially Pfs16 was suspected to be required for optimal production of sexual stage parasites [22].
In addition to Pfs16 and Pfs37, other genes such as Pfs25 (PF3D7_1031000) and Pfs28 (PF3D7_1030900) have also been studied in the context of parasite–vector interactions. Pfs25 encodes a protein expressed on the surface of zygotes and ookinetes, playing a critical role in the development of oocysts in the mosquito midgut. Disruption of Pfs25 results in significantly reduced transmission efficiency of the parasite [23]. Similarly, Pfs28, which is co-expressed with Pfs25, has been implicated in ookinete development and midgut invasion [24]. While Pfs25 and Pfs28 are known to be essential in the later stages of parasite development in the mosquito, we chose Pfs16 and Pfs37 for their earlier involvement in the interaction, particularly during gametocyte and ookinete stages, which are critical for initial midgut invasion. By focusing on Pfs16 and Pfs37, our study aims to investigate earlier-stage interactions between P. falciparum and An. gambiae, complementing existing knowledge of transmission-blocking candidates such as Pfs25 and Pfs28. This approach provides a more comprehensive understanding of the genes involved across different stages of the parasite’s lifecycle within the mosquito.
At the other end of the vector–parasite interaction equation, Anopheles midgut proteins AnAPN1 and HPX15 have been recognized for their importance in vector–parasite interactions and for their significant impact on the survival of the parasite. HPX15 is an immune-related protein with pattern-recognition molecules, and previous studies indicate that it promotes malaria transmission [10]. Specifically, HPX15 plays a role in the preservation of the functionality of stored sperm and long-term fertility in An. gambiae [25]. In An. stephensi mosquitoes, RNA interference-mediated silencing of midgut AsHPX15 gene has drastically reduced the number of developing P. berghei oocysts [26]. Alanyl aminopeptidase N (AnAPN1) is a protein that can elicit transmission-blocking antibodies, which is believed to be highly conserved among Anopheles vectors [27], though not thoroughly investigated across Africa. The effectiveness of antibodies targeting AnAPN1 against P. falciparum and P. vivax across distantly related Anopheles species is well studied [28].
While there is strong support of ancient vector-mediated selection on Plasmodium by continentally structured Anopheles species, questions remain about the potential for ongoing vector-mediated selection within a continental region. The goal of this study was to evaluate the potential for vector-mediated selection on parasite populations within African countries by examining the patterns of diversity in vector–parasite interacting genes. To better understand the subcontinental dynamics of vector-mediated selection, we aim to investigate the genetic diversity and selection signals in P. falciparum interaction genes, along with the vector species composition and subspecies variation in An. gambiae interaction genes. This study will specifically address the implications of An. stephensi invasion by providing insights into the nature of these interactions on a finer scale.
Methods
All the command line-based programs were run on the operating system Rocky Linux 8.8 (Green Obsidian), Architecture: × 86–64. The rest of the steps were carried out on a Windows (version 11 Education, 64-bit operating system) PC.
Data selection
We needed gene sequences from P. falciparum and An. gambiae from the malaria-endemic regions for this study. Therefore, Ag1000 and PF6K datasets shared by MalariaGen data-sharing network were used [29]. In this study, abiding to the Ag1000 terms of use, we did not use more than 10% of the genome data and we did not report any genome-wide statistics. When downloading genomes of vectors and parasites, sample sets were selected as separate populations, where at most 25 samples were collected from the same country in the same year. This number of samples (n = 25) was selected to hold the balance between representation of mosquitoes from a particular region and computational power required to perform the analysis. Another reason to select the same number (or close to 25) of samples for every population was to avoid the increment of number of haplotypes due to the large number of samples. The P. falciparum samples with high probability of multiple infections (Fws ≤ 0.95) were removed from the dataset using the Fws values calculated by the authors of the Pf6K dataset [29]. Abiding to above criteria, 418 P. falciparum genomes and 625 An. gambiae genomes were downloaded for 9 and 15 African countries, respectively. The European Nucleotide Archive (ENA) accession identifiers with country and year data are saved in Supplementary Table 1 and 2 CSV files.
Quality control
The downloaded genome sequences were subjected to quality control using FastQC v0.12.1 [30] to assess the overall quality of the raw reads. Specifically, we evaluated key metrics including basic statistics (e.g., total sequence length, GC content), per-base sequence quality (to check for base-specific errors), per-base N content (to identify unknown bases), and sequence length distribution (to detect potential adapter contamination or other sequencing artifacts). Any anomalies observed, such as significant drops in sequence quality at specific positions or elevated N content, were flagged for further investigation.
To improve the quality of the dataset, we used Trimmomatic v0.39 [31] for trimming the low-quality regions. This included trimming sequences with Phred scores below 20, removing adapter sequences, and filtering out reads shorter than 36 bases post-trimming. The impact of these filtering steps was reassessed by generating a second round of FastQC reports to ensure that all quality anomalies were adequately addressed before proceeding with alignment.
Sequence alignment to the reference genome.
The quality-filtered sequences were aligned to the An. gambiae and P. falciparum reference genomes, which were downloaded from the VectorBase (https://vectorbase.org/vectorbase/app) and PlasmoDB (https://plasmodb.org/plasmo/app) databases, respectively. Bowtie2 (version 2.5.1) [32] was used to perform the alignment on a Linux platform.
For the alignment, Bowtie2 was set to default parameters, which allow for end-to-end alignment. A maximum of two mismatches per read were permitted to balance sensitivity and accuracy. The resulting SAM files were converted to BAM format using SAMtools v1.10, followed by sorting and indexing to facilitate downstream variant calling and read depth analysis. Post-alignment quality checks, including alignment rates and mapping quality, were conducted to ensure that the majority of reads were correctly aligned to the reference genomes. Unmapped reads were excluded from further analysis.
Variant calling and gene sequence handling
The variants were called from the aligned .bam files using the mpileup option in BCFtools (version 1.17) program [33]. Variants were normalized and filtered to get the highest quality variant call respective to the reference genomes. Only the biallelic variants were filtered out with Phred-scaled quality scores greater than 30, read depth greater than 10, and frequencies higher than 1%. The sequences of the interested g enomic regions were extracted from the VCF files using SAMtools program (version 1.18) and consensus option in BCFtools program. Extracted sequences were saved in FASTA format for the downstream analysis. VCF files of the mosquito genomes were phased using Segmented HAPlotype Estimation & Imputation Tool (shapeit2—version 2.r904) to address the ploidy level. The gene sequences were aligned using Clustal and Muscle programs [34, 35].
Diversity statistics calculation
For all the genes studied here, for each population in both vectors and parasites, we calculated the Tajima’s D values, Fst values, nucleotide diversity, and haplotype diversity using the Pegas package (version 1.3) in R statistics (version 4.3.2) [36]. Additionally, genome-wide Fst and Tajima’s D values were calculated for comparisons. Values were recorded in tables for further analyses and visualized using the ggplot2 package in R statistics. Tajima’s D values were calculated according to the method described by Tajima in 1989 [37]. Tajima’s D values were recorded with the corresponding beta P-values for each population to facilitate selecting statistically significant signals of selection. Nucleotide diversity and haplotype diversities for each population pair were calculated as described in Nei in 1987 and Nei and Tajima in 1981, respectively [38, 39]. Pairwise Fst for each population pair was calculated using gene.dist() function in hierfstat (version 0.5.11) package in R statistics using the method described by Weir and Cockerham in 1984 [40]. Genome-wide Tajima’s D value and Fst values were calculated using VCFtools (version 0.1.16).
Investigation of relationships between vector occurrence probabilities and parasite gene haplotype diversities
In this step we investigated the relationships between amino acid haplotype diversities of the genes important for interacting with the vectors in parasites and the probability of occurrence of prominent malaria vectors in Africa. We downloaded predicted vector occurrence probabilities (VOP) for both 2010 and 2017 from the Malaria Atlas Project, corresponding to the locations where parasite samples were collected [41, 42]. Correlation analyses were performed between the amino acid haplotype diversities of parasite genes and VOP using cor.test() function on R statistics platform. For the vector species that had a statistically significant relationship with the haplotype diversity of parasite genes, regression models were fitted using lm() function to examine the interaction between vector species occurrence probabilities on haplotype diversities of parasite genes. Furthermore, to reduce the uncertainty and noise inherent in VOP data that were used in linear regressions, the probabilities were converted to a binomial variable of presence or absence of the vector species. We employed three distinct cutoff values for occurrence probabilities (0.5, 0.75, and 0.95) to assess vector presence, as a definitive rationale for selecting a single threshold was not available. Regression models were fitted to predict the amino acid haplotype diversities of the parasite genes against binomial vector occurrence of significantly correlated vector species as the predictor variables. Results of all the regression models were tabulated in an Excel sheet (Supplementary data sheet 2—Combined sheet). In addition to the regression analyses, we categorized and visualized the haplotype diversities and Tajima’s D values of parasite populations on the basis of the presence or absence of different combinations of vector species significantly associated with parasite gene haplotype diversity (Supplementary Figs. 4 and 5). This approach was specifically designed to identify patterns in haplotype diversities across various vector combinations that were not captured by the linear regression models.
Anopheles stephensi mosquito collection and P47Rec sequence extraction
Anopheles stephensi DNA generated from previous studies [6, 43] were used for the analyses described below. These source specimens were part of a September–November 2018 collection from northeastern and eastern Ethiopian cities Semera and Kebridehar as a part of our previously published studies as previously described [6, 43]. Briefly, mosquitoes (n = 7) were collected using Centers for Disease Control and Prevention light traps and pyrethrum spray collection in houses, and larvae and pupae were sampled using the WHO dipping approach. The mosquito specimens were collected and handled following ethical guidelines as previously described by Balkew et al. in 2020 [6], and a materials and data-sharing agreement was established between Baylor University and Jigjiga University. DNA was extracted from the dissected heads and thoraxes of the mosquitos using the Qiagen DNeasy kit. Once the DNA was extracted, the P47Rec ortholog in An. stephensi was amplified using two primer pairs. The first pair (forward—5′-TGGCAAATGACTAACGTGGA-3′, reverse—5′-GTGTTGCCAGTTCGCTGTAA-3′) amplified the second and third exons, while the second pair (forward—5-GTGAGCAGCTGTACGTTGGA-3′, reverse—5-AAAACGGAAGGCATGTCATAA-3′) amplified the fourth exon. Sequences were aligned using the MUSCLE program and a maximum likelihood tree was generated using the RAxML version 2.0 program [44].
Results
Population structure and polymorphism in Plasmodium falciparum genes
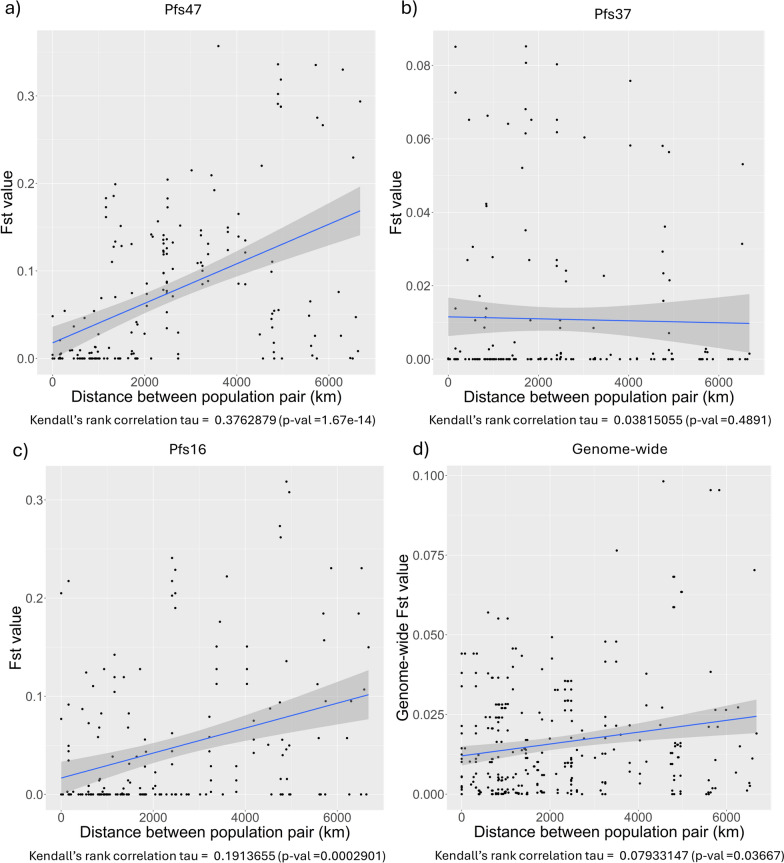
To investigate the population structure and its association with geographic distribution of P. falciparum genes, we measured the pairwise Fst between each population pair and tested for correlation with geographic distance between populations (see Fig. 1). For Pfs47, pairwise Fst values ranged between 0 and 0.3569732. The highest values were observed in the Malawi population against other populations. Malawi was also relatively (not statistically) isolated from other Central African countries including the Democratic Republic of Congo and Cameroon, which were relatively (not statistically) isolated from West African populations (see Supplementary Fig. 1). In Pfs47, a statistically significant but weak correlation was observed between pairwise Fst values and the geographic distances among the P. falciparum populations (see Fig. 1).
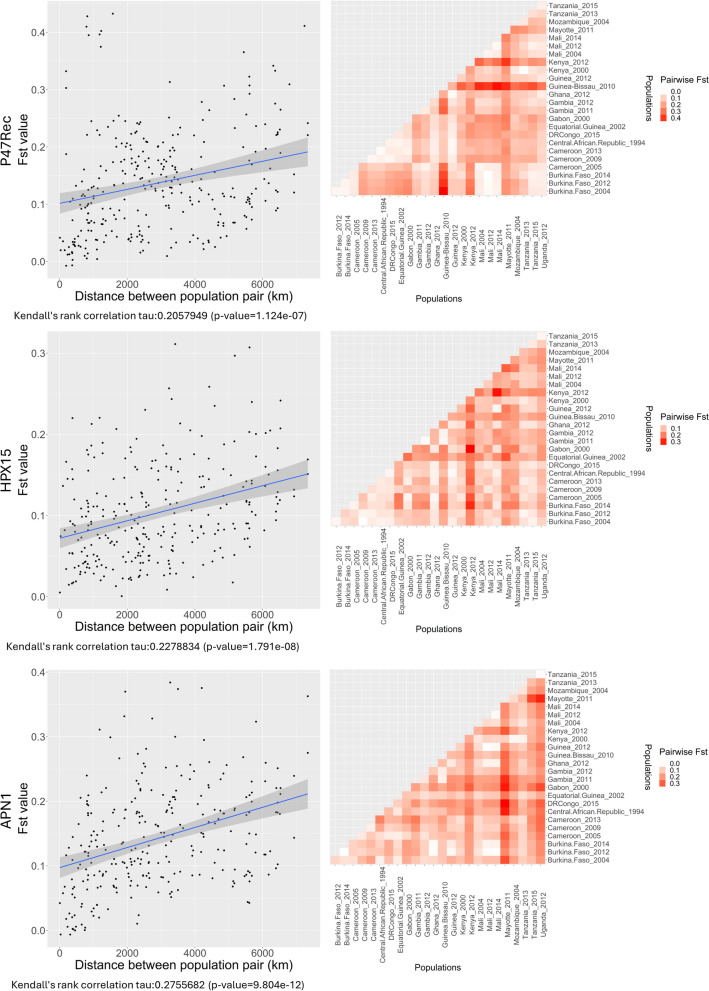
Fig. 1.
Analysis of genetic differentiation and geographic dispersion among populations of P. falciparum across Africa. Each panel depicts the correlation between pairwise fixation index (Fst) values and geographic distances, measured in kilometers, between population pairs for different genomic regions. The Fst value measures the genetic differentiation between populations, with higher values indicating greater divergence. The blue line represents a regression line fitted to the data points, illustrating the relationship between geographic separation and genetic diversity. a Displays Fst values for the Pfs47 gene, indicating a moderate positive correlation. b Shows Fst values for the Pfs37 gene, with a weak and nonsignificant correlation. c Illustrates the correlation for the Pfs16 gene, suggesting a mild positive correlation. d Presents the genome-wide correlation, indicating a slight but statistically significant correlation
In parallel to Pfs47, Pfs16 also showed a statistically significant but weak correlation between pairwise Fst values and the geographic distances among the P. falciparum populations (Kendall’s rank correlation tau = 0.1913655, P = 0.0002901). Again, Malawi had the highest pairwise Fst values, but the isolation patterns were different between Pfs47 and Pfs16 (see Supplementary Fig. 1). Both Pfs37 and genome-wide pairwise Fst values did not show statistically significant correlations with the distance between populations (Fig. 1).
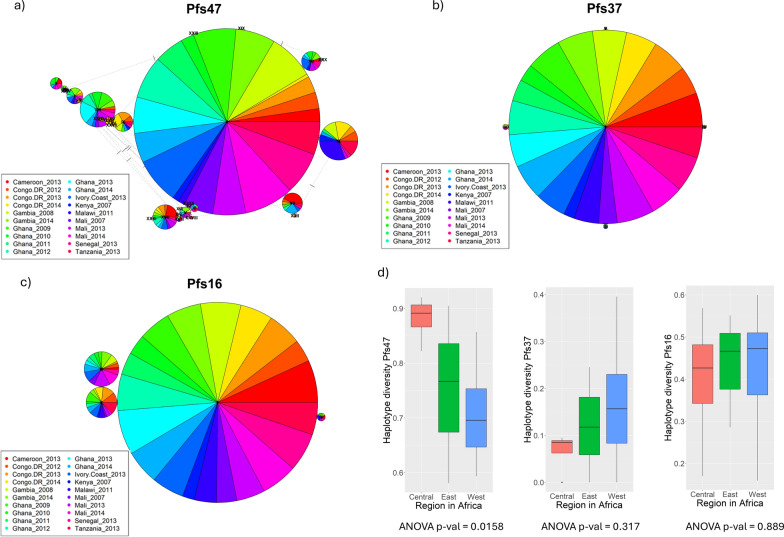
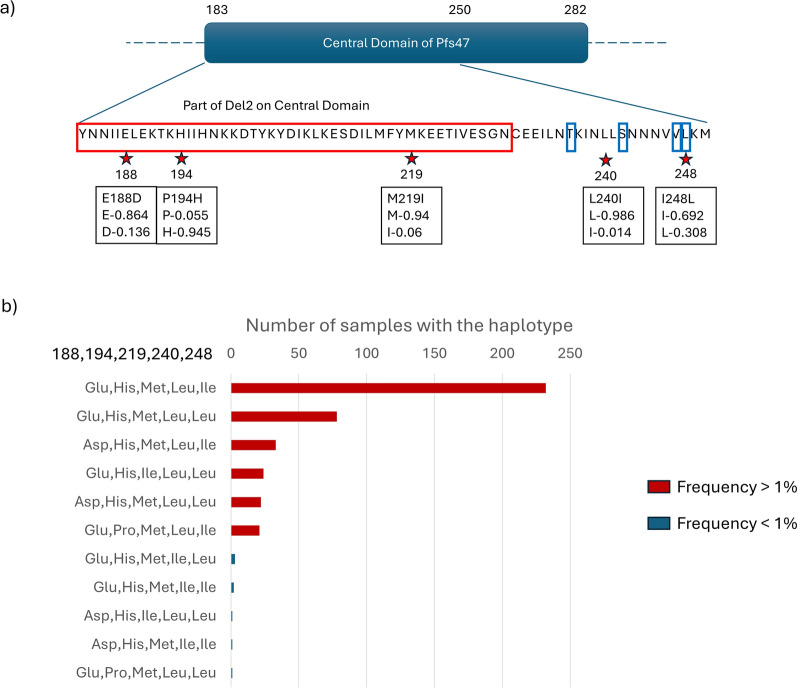
Among P. falciparum in the African countries we studied, the number of haplotypes for Pfs47, Pfs37, and Pfs16 were 32, 5, and 4, respectively (Fig. 2). We observed three polymorphic sites within the Del2 region in the central domain of Pfs47 (D2), which has been selected as a candidate antigen and which generates antibodies that block transmission (see Fig. 3) [11]. Secondly, we observed two amino acid polymorphisms between the two cysteines in Pfs47-D2, the region known to be important for mosquito infectivity (see Fig. 3). To understand the geographical distribution of haplotypes in parasite populations we generated haplotype networks for the three parasite genes. In all three haplotype networks, especially the Pfs47 central domain (D2) haplotype network, there was roughly equal representation from all the populations, indicating the presence of each haplotype in many parts of the continent (see Fig. 2 and Supplementary Figs. 2, 5). In Pfs47, haplotype II had the highest number of samples and represented most of the populations in the dataset. To find out which parts of the African continent harbor the highest number of parasite gene haplotypes, we divided the continent into three regions (East, Central, and West) and measured the haplotype diversity. Haplotype diversity level of Pfs47 was significantly higher in Central African countries [analysis of variance, ANOVA F(2, 17) = 5.344 P = 0.0158, Tukey’s HSD test F-values of West-Central and East-Central were 0.0119208 and 0.1825349 respectively, see Fig. 2d]. Compared with Pfs47, both Pfs37 and Pfs16 had haplotype diversity evenly distributed across the continent (see Fig. 2d box plot haplotype diversity means for Pfs37 and Pfs16). In Pfs47 there were nine single nucleotide polymorphisms (SNPs) and five of them had frequencies higher than 10% in the sample set we analyzed. Out of the nine SNPs, eight were non-synonymous mutations. There was a single SNP in Pfs16 coding sequence and an indel expanding from the 423rd base pair to the 428th base pair creating amino acid changes I85L, D140-, and K141-. All these variations had a frequency higher than 10%.
Fig. 2.
Haplotype diversity and distribution of P. falciparum genes Pfs47, Pfs37, and Pfs16 across Africa (based on nucleotide sequences). These haplotype networks graphically represent how each haplotype has been represented by different populations of parasites. The size of the pie represents the number of individuals with the haplotype and the size of each pie segment reflects the relative frequency of parasites originating from different populations for each haplotype. a Pfs47 had 32 haplotypes that are distributed among many populations. b Pfs37 had a single prominent haplotype, suggesting very low genetic variability. c Pfs16 also had a fewer number of haplotypes, suggesting less genetic variability compared with Pfs47 but higher than Pfs37. d Regional haplotype diversity analysis—box plots represent the distribution of haplotype diversity levels for Pfs47, Pfs37, and Pfs16 across three major regions: Central, East, and West Africa. Each box plot shows the median, quartiles, and potential outliers, providing a statistical summary of regional genetic diversity. ANOVA results below each plot indicate the statistical significance of differences in diversity across regions, with the P-values providing insights into regional variations in genetic diversity. (These calculations were based on nucleotide sequences and the calculations based on amino acid sequences are different from this.)
Fig. 3.
Analysis of non-synonymous mutations in the central domain of Pfs47 and their frequencies. a The locations of the mutations on domain 2 of Pfs47 protein. The red stars denote the polymorphic sites within the region (183rd aa to 250th aa) and red box denotes Del2 region, which is selected as antigen for TBV. The blue boxes show the previously identified mutations important for vector parasite interaction. Notation within the black boxes shows the mutation in the first line and frequency of each allele in second and third lines b This bar plot displays the distribution of haplotypes found within the 418 protein sequences of Pfs47’s central domain (D2), sampled from various populations across Africa. Haplotypes are categorized by the composition of their amino acid sequences at the polymorphic sites identified in panel (a). Bars are colored to distinguish haplotypes with a frequency greater than 1% (red) from those less frequent (blue), offering a visual summary of haplotype prevalence and diversity within the dataset
Signals of selection in parasite genes
One of the goals of this study was to see whether the parasite interacting genes are evolving under positive selection in any of the African populations investigated here. Therefore, we calculated the Tajima’s D values for each parasite gene for each population and compared it with the average Tajima’s D values of the entire genome (−0.783259075) for the samples analyzed in this study. Tajima’s D values were calculated for each population and tabulated (see Supplementary data sheet 1). Among parasite populations, the average Tajima’s D over the entire genome varied between −1.320805 and −0.2383106. For each population, Pfs47 and Pfs37 did not show any statistically significant signals of non-neutral evolution (averaged Tajima’s D values −0.420956776 and −1.167216138 respectively, beta P > 0.05). However, Pfs16 had significantly higher negative Tajima’s D values in all the individual populations (average Tajima’s D value = −2.960073673, beta P < 0.05), indicating that it is evolving non-neutrally in many parts of the continent. Since the Pfs16 Tajima’s D values were significantly negative compared with the genome-wide Tajima’s D value, this could be an indication of positive selection at this locus.
Population structure in An. gambiae genes
To investigate the potential influence of vector–parasite interactions on the population structure of Plasmodium parasites, we analyzed the population structure of genes (nucleotide sequences) coding for proteins known to be important for the survival of the parasite in An. gambiae s.s. in several African countries. We measured the pairwise Fst between each selected An. gambiae population pair and tested for correlation with the geographic distance between populations (see Fig. 4). The Fst values varied between 0 and 0.4327906, 0.3112532, and 0.3841258 for P47Rec, HPX15, and APN1, respectively. For the P47Rec gene, Guinea-Bissau and Kenya (2012) populations showed the highest level of isolation from other regions, while Central African populations had a trend of being differentiated from the other populations. In HPX15, the Kenya (2012) population was again the most isolated population compared with the rest of the populations, followed by Mayotte and Mozambique populations. However, in APN1 Mayotte population had the highest level of isolation followed by the Southeast African populations in Uganda, Tanzania, and Mozambique. Kenya (2012) was relatively (not statistically) different from the rest of the populations. All three genes studied here showed statistically significant but weak correlations between the pairwise Fst values and geographic distance among populations (see Fig. 4).
Fig. 4.
Genetic differentiation and the correlation between geographic distance and genetic differentiation for An. gambiae gene loci. Scatter plots: each scatter plot displays the correlation between pairwise Fst values and geographic distances among populations of An. gambiae s.s.. Fst values, measuring genetic differentiation, are plotted against the geographic distance in kilometers between each pair of sampled populations. The line represents a regression fit, indicating trends in genetic isolation by distance. Heat maps: These heat maps depict the pairwise Fst values between different populations of An. gambiae for the corresponding genes. Each cell in the matrix represents the Fst value between a pair of populations, with color intensity varying according to the scale of differentiation ranging from low (light) to high (dark). Populations are ordered and labeled on both axes, facilitating cross-reference between related populations
The number of SNPs that cause changes in amino acids were tabulated for each exon of the three genes (see Table 1). No non-synonymous mutations were detected in the P47Rec with a frequency higher than 1%. However, 32 and 72 nonsynonymous mutations with frequency higher than 1% were detected in the HPX15 and APN1, respectively. Given the amino acid variation in these two genes, we were interested in the phylogenetic relationship among the mosquitoes. Phylogenetic analysis revealed support for the presence of distinct clades within An. gambiae for HPX15 (bootstrap values > 70) but not in APN1 (Supplementary Figs. 6 and 7).
Table 1.
Number of non-synonymous mutations (NSMs) per exon in An. gambiae genes
| Gene | Exon | # of NSMs | # of NSMs with freq. > 0.01 |
|---|---|---|---|
| P47Rec | 1 | 0 | 0 |
| 2 | 2 | 0 | |
| 3 | 2 | 0 | |
| 4 | 5 | 0 | |
| Total | 9 | 0 | |
| HPX15 | 1 | 27 | 5 |
| 2 | 44 | 17 | |
| 3 | 38 | 10 | |
| Total | 109 | 32 | |
| APN1 | 1 | 23 | 7 |
| 2 | 18 | 10 | |
| 3 | 4 | 0 | |
| 4 | 25 | 3 | |
| 5 | 60 | 52 | |
| Total | 130 | 72 |
Relationships between vector occurrence probabilities and parasite gene haplotype diversity
We conducted individual association analysis of the P. falciparum diversity statistics and occurrence probabilities of eight commonly occurring malaria vector species in Africa (An. arabiensis, An. coluzzi, An. funestus, An. gambiae, An. melas, An. merus, An.0moucheti, and An. nili). Only Pfs47 and Pfs16 amino acid haplotype diversities had significant associations (Kendall’s rank correlation P < 0.05) with predicted occurrence probabilities of An. arabiensis, An. funestus, and An. moucheti (see Table 2). Regression models were fitted to explain the amino acid haplotype diversities of Pfs47 and Pfs16 using the VOPs of the previously mentioned three vector species as continuous and binomial variables separately (Supplementary data sheet 2—Separate and Combined sheets). Models fitted with VOPs as continuous variables had significant overall P-values (< 0.05) and adjusted R-squared values greater than 0.48 but estimates for the VOPs were close to zero. In the regression models fitted with VOPs as binomial variables, irrespective of the cutoff value (to determine the presence or absence of a vector species from occurrence probabilities), Pfs16 amino acid haplotype diversity was associated with An. arabiensis with a significant (< 0.05) P-value and an estimate close to zero while Pfs47 was associated with An. arabiensis only under a cutoff value of 0.5. Apart from the linear regression analyses between parasite gene haplotype diversities and vector occurrence probabilities, we visualized the haplotype diversities observed under occurrence of different combinations of three vector species (An. arabiensis, funestus, and moucheti—occurrence was determined by multiple threshold values as previously mentioned) that were significantly associated with higher haplotype diversities (Supplementary Fig. 4). We observed in most cases, irrespective of the threshold level used to define the probable presence or absence of the vector species on the basis of their predicted occurrence probability, that the combination of An. arabiensis and An. funestus was associated with higher levels of amino acid haplotype diversity in Pfs47 and Pfs16 (Supplementary data sheet 2—ANOVA sheet).
Table 2.
Observed significant Kendall’s rank correlation tau estimates and P-values for the amino acid haplotype diversities of parasite genes and predicted vector occurrence probabilities based on 2010 prediction model
| An. arabiensis | An. funestus | An. moucheti | |
|---|---|---|---|
| Pfs47 | |||
| Tau estimate | −0.4217757 | −0.3807641 | 0.434292 |
| P-value | 0.01082 | 0.02068 | 0.02132 |
| Pfs16 | |||
| Tau estimate | 0.6099269 | 0.5013643 | −0.4863484 |
| P-value | 0.0002694 | 0.002899 | 0.01087 |
| Pfs37 | |||
| NA | Nonsignificant | Nonsignificant | Nonsignificant |
We investigated the relationship between Tajima’s D values of Pfs16 and vector occurrences because it was the only gene that had signals of significant positive selection. We observed significant estimates for An. funestus VOP in both regression models fitted to explain Pfs16 Tajima’s D using vector occurrence probabilities as continuous variables and binomial variables (see Supplementary data sheet 2). It is important to mention that Pfs16 Tajima’s D values did not have a significant association with the region of Africa (Central, East, or West) as observed in the results of the one-way ANOVA table (Pfs16 Tajima’s D values against region—see supplementary data sheet 2). However, An. funestus vector occurrence (as a binomial variable) had a significant relationship with region (analysis of variance P = 0.0493). In addition to the regression analyses mentioned above, we created a violin plot of the Pfs47 haplotype diversity, Pfs16 haplotype diversity, and Pfs16 Tajima’s D values for different vector species combinations. In those plots, we observed that higher (i.e., less negative) Tajima’s D values and haplotype diversities at Pfs16 were associated with the combinations that included An. funestus (Supplementary Fig. 5, Supplementary data sheet—ANOVA sheet).
Comparative analysis of P47Rec in An. gambiae and An. stephensi
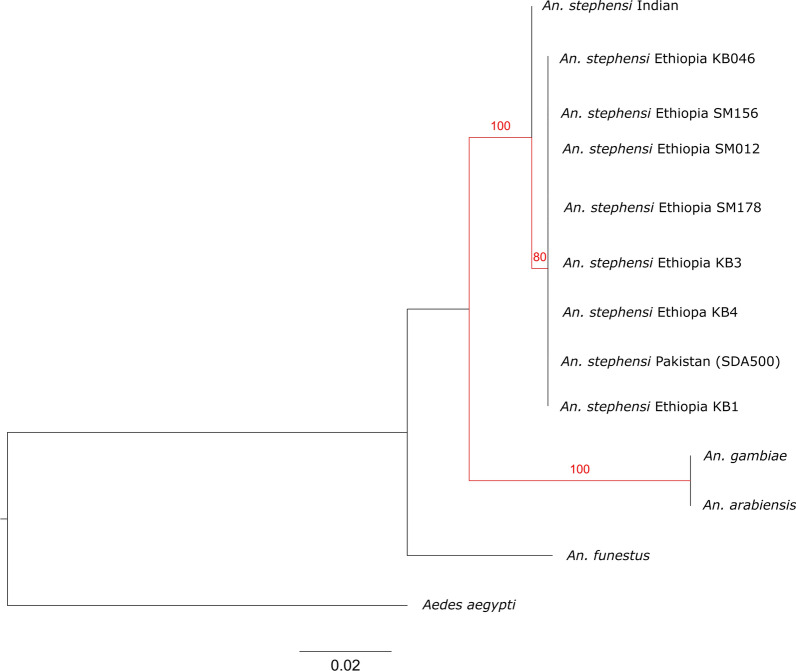
In An. gambiae s.s., the P47Rec amino acid sequence was very well conserved within the sequences we observed in Ethiopia. With the invasion of An. stephensi into the Horn of Africa (HOA) we wanted to investigate the differences in amino acid sequence of P47Rec ortholog in An. stephensi that could have an impact on compatibility between invasive vector and existing Plasmodium populations in the HOA. There are 28 amino acid changes among all the prominent malaria vectors in Africa (An. funestus, An. melas, An. quadriannulatus, An. arabiensis, An. merus, and An. gambiae) in P47Rec orthologs reported by Molina-cruz et al. in 2020 [16]. There were 18 amino acid changes (listed in Table 3) observed in P47Rec amino acid sequences between the An. stephensi collected in Ethiopia and the An. gambiae reference sequence compared with 25 observed between An. gambiae and An. funestus. Out of the 18 observed between An. gambiae and An. stephensi, 13 amino acid differences overlapped with the changes observed among all prominent African vectors mentioned above. There were five amino acid changes that are unique to An. stephensi (Table 3). No differentiation was observed between An. stephensi from Kebridehar and Semera. Further, we performed phylogenetic analysis of the coding sequence of the P47Rec gene (Fig. 5) including An. gambiae sl, An. funestus, An. arabiensis, and An. stephensi (sequences from Indian strain, Pakistani strain, and Ethiopia). The results indicated that the haplotype of the P47Rec ortholog in An. stephensi from Ethiopia is closer to that of An. stephensi from Pakistan (SDA500) as compared with the Indian one (separation was supported by bootstrap value = 80).
Table 3.
Amino acid differences between An. gambiae and An. stephensi coding sequences of P47Rec
| Exon | Amino acid change (from gambiae to stephensi) |
|---|---|
| Exon 2 | G23A ~ |
| I34V ~ | |
| Q53H* | |
| T67S* | |
| Exon 3 | S81G ~ |
| N90G ~ | |
| I105V ~ | |
| I119V* | |
| G131N ~ | |
| Exon 4 | V165I ~ |
| N169H ~ | |
| G173S ~ | |
| T181A ~ | |
| S195T ~ | |
| T217V ~ | |
| Q220N ~ | |
| S242T* | |
| A252T* |
The amino acid changes marked with * are changes unique to An. gambiae and An. stephensi collected in Ethiopia, and amino acid changes marked with ~ are observed among the other prominent malaria vectors (An. funestus, An. melas quadriannulatus, An. arabiensis, An. merus) and An. gambiae
Fig. 5.
Maximum likelihood tree for the P47Rec orthologs of prominent malaria vectors in Africa with P47Rec ortholog of An. stephensi collected in Ethiopia (500 bootstraps). The significant branches with bootstrap values higher than 70 are highlighted in red. It is important to note that all the sequences from Ethiopia are closely related to SDA500 strain and significantly divergent from Indian reference sequence
Discussion
Limited evidence of vector-mediated selective pressure on P. falciparum populations through P47 system within Africa
The P47 system was the best starting point to investigate vector-mediated selective pressure on parasite variation on a subcontinental scale because of the well-established molecular interaction of the P. falciparum Pfs47 and An. gambiae P47Rec proteins and their role in parasite invasion of the mosquito midgut. In this study, we observed 32 Pfs47 haplotypes overall but no correlation between haplotype distribution and geography. In addition, no evidence of selection was detected according to the Tajima’s D values observed in the parasite populations studied here. Furthermore, the P47Rec amino acid sequence is highly conserved among An. gambiae populations, indicating the absence of vector subpopulation variation necessary to drive selection by An. gambiae alone. Even though we see Pfs47 haplotype diversity within each population, the pattern of diversity in Pfs47 suggests only neutral processes at play. This does not necessarily contradict previous studies that show evidence of differing P. falciparum haplotype compatibility across continentally structured Anopheles species [9]. The study by Molina-Cruz et al. identified a broad diversity of Pfs47 haplotypes across East, Central, and West Africa, revealing substantial geographic variation in haplotype frequencies [17]. However, in our study, we intentionally limited the sample size, as one primary objective was to assess the relationship between P. falciparum gene haplotype diversity and VOP. Including disparate sample sizes from various populations may have inflated haplotype diversity estimates, particularly in regions with higher sampling densities. Therefore, we maintained a balanced sampling strategy to minimize such biases. While there are divergent Anopheles species that exist within Africa, the portions of the P47Rec that drive compatibility may be conserved in African Anopheles, leading to less restrictions on transmission specificity and the maintenance of diversity in Pfs47. Further functional analysis of P47Rec is needed to evaluate the precise genic regions at play in compatibility.
Interestingly, we also detected a new mutation (M219I) not yet reported in the central domain of the Pfs47 Del region, which has been chosen to use as target antigen for the development of a malaria TBV [11]. Both methionine and isoleucine are uncharged, hydrophobic amino acids with non-polar side chains. However, the substitution of methionine with isoleucine may still affect protein function and structure due to subtle differences in their physicochemical properties, such as side chain size and flexibility. Methionine contains sulfur, while isoleucine is a branched-chain amino acid, which could influence protein folding, stability, or interactions with other molecules. Further investigation is needed to fully understand the potential impact of this substitution. Thus, these findings may have implications for the efficacy of vaccines, given that the native vectors can transmit this strain with these variants.
Diversity in An. gambiae genes HPX15 and APN1
In contrast to P47Rec, both HPX15 and APN1 were highly diverse at the amino acid level. Phylogenetic analysis supported multiple distinct groups for HPX15 (bootstrap values > 70) but not for APN1 (bootstrap values < 70). The observed subspecies differentiation in HPX15 indicates the potential for this gene to serve as a driver of selection on its matching P. falciparum gene in the parasite. Further investigation of this locus coupled with the identification of its corresponding P. falciparum surface proteins will elucidate whether vector-population-mediated selective pressure is occurring. We anticipate detecting signatures of balancing selection in the HPX15 ligand(s) protein within P. falciparum populations across Africa due to the subpopulation variation observed at this locus on An. gambiae.
Potential signals in P. falciparum Pfs16
While the study of the P47 system revealed limited evidence of ongoing vector-population-mediated selection on the Pfs47 within Africa, differing patterns of diversity were observed at Pfs16. Fewer amino acid haplotypes were observed at this location and no geographic structure. Furthermore, a signal of positive selection or population expansion was observed in each study population at Pfs16. After comparing the Pfs16 Tajima’s D values with genome-wide values, we could rule out the possibility of observing negative Tajima’s D values due to population expansion. This indicates that selection has occurred more recently at this locus within Central Africa. We also observed two important mutations, a single nonsynonymous mutation and a nine nucleotide (three amino acid) indel. While the receptor for Pfs16 in vectors has not been identified yet, these findings support further investigation of this gene and potential receptors in the vector.
Association analysis supports vector-mediated selection
Nucleotide and haplotype diversity in P. falciparum genes crucial for vector–parasite interactions could be significantly influenced by the composition of vector species in malaria-endemic regions. However, we were unable to obtain vector composition data for all the locations where the P. falciparum sequences and samples used in this study originated. To test the above hypothesis, we downloaded the predicted vector occurrence probability values of the prominent malaria vectors for the locations of parasite samples from the Malaria Atlas Project (MAP) and investigated the relationships between haplotype diversities and Tajima’s D values of the genes important for the P. falciparum interaction with vectors in Africa [41, 42]. We observed a statistically significant correlation between amino acid haplotype diversities of parasite genes Pfs47 and Pfs16, and predicted occurrence probabilities of the vector species An. arabiensis, An. funestus, and An. moucheti individually. In addition to the individual associations, the above observation was confirmed by the association between higher haplotype diversities at Pfs16 and combined occurrence of An. arabiensis and An. funestus compared with the occurrence of a single vector. These signals could be an indication of vector-mediated selection on parasite populations. However, we cannot rule out the impact of geography-correlated variables (e.g., transmission intensity, ancient genetic variation clines) on diversity due to the significant F-values observed in one-way ANOVA between amino acid haplotype diversity and the region of the African continent (Central, East, and West).
To investigate the impact of VOP on selection more directly, we examined the relationship between Tajima’s D values of Pfs16 (Pfs16 was the only gene with significant Tajima’s D values) and VOP of mosquito species that had a statistically significant correlation with parasite gene haplotype diversities individually. We found an association between An. funestus and Pfs16 Tajima’s D values, such that the presence of An. funestus was associated with a lower signal of positive selection. Anopheles funestus is genetically distinct from An. arabiensis and An. moucheti, both members of the An. gambiae complex. This genetic diversity may have driven P. falciparum to develop a broader array of Pfs16 haplotypes, enhancing its ability to be transmitted by both the An. gambiae complex and An. funestus. Consequently, the presence of multiple haplotypes could lead to a lower signal of positive selection in P. falciparum Pfs16. A limitation in this analysis is the availability of data on the lower end of An. funestus occurrence probability distribution. We have a single data point that is coming from Kilifi in Kenya (the only data point from Kenya). According to both the Malaria Atlas Project prediction of 2010 [41] and 2017 [42], Kenya has a low occurrence probability of An. funestus. It is also important to mention that the VOPs in the Malaria Atlas Project were predicted values based on geographical and environmental factors, which could add more ambiguity/noise to the estimate values of correlation analysis and regression analysis. Ultimately, these results indicate that there could be an influence from the vector species composition on the selection of the parasite genes important for the interaction with vectors.
Implications for An. stephensi invasion in Africa
Given that Pfs47 in African P. falciparum populations exhibited signals of neutral evolution in relation to the current sympatric vector populations (An. gambiae, An. funestus, etc.), we aimed to investigate how the evolution of Pfs47 might be influenced by the introduction of An. stephensi. As an initial step, we examined the P47Rec ortholog in An. stephensi. Since the P47Rec coding sequence in An. gambiae in Africa was fully conserved, we wanted to investigate the number of amino acid changes in P47Rec ortholog in invasive An. stephensi. We compared the amino acid sequence of the P47Rec ortholog in An. stephensi from Ethiopia with the An.gambiae sequence and found 18 amino acid differences. These findings combined with phylogenetic analysis indicating differentiation between An. stephensi and the African species (An. gambiae bootstrap value = 100 and An. funestus bootstrap value = 100) support the potential for new P. falciparum haplotype compatibilities in Africa with the arrival, spread, and establishment of the invasive An. stephensi. In addition, the phylogenetic analysis revealed a close relationship between P47Rec in the invasive An. stephensi and the SDA500 An. stephensi strain (bootstrap = 100). The SDA500 strain is known to be highly susceptible to both I248L haplotypes in Pfs47 in P. falciparum [18]. If the Pf47Rec was the gene that underwent artificial selection leading to higher susceptibility, it is possible the same patterns of susceptibility would be observed in An. stephensi with the similar P47Rec sequence. Therefore, the presence of similar P47Rec sequences (leading to high susceptibility) in the invasive An. stephensi may facilitate the gradual emergence of more Pfs47 haplotypes in Africa. However, other genes may also influence the susceptibility of the SDA500 strain. To accurately determine the characteristics of An. stephensi—Plasmodium compatibility in Ethiopia, experimental infections are necessary to validate these hypotheses.
Future directions
In this study we focused on several genes known to be important for vector–parasite interactions of malaria and their role in shaping the population structure of P. falciparum parasites through selective forces exerted from vector populations. There could be many other genomic factors that can influence the vector–parasite interactions and studies should include investigation of additional genes such as Pfs25, Pfs28, and circumsporozoite and TRAP-related protein (CTRP). This study did not include samples from all the malaria endemic countries in Africa. In addition, the samples were collected in multiple years that expanded over two decades. In a future study we expect to broaden our list of genes used in the analyses and to include samples from other malaria endemic countries in Africa.
Conclusions
This study provides preliminary insight into the potential for vector subspecies level and multiple vector species selective pressure impacting Plasmodium–Anopheles compatibility within Africa. Notably, these findings support the notion that compatibility is complex and in addition functional, and population genetic investigations are needed. Our objective was to explore genetic diversity and related metrics as an initial step. It is crucial to recognize that definitive conclusions about compatibility cannot be drawn without experimental infection data. Furthermore, this study provides the first analysis to explore how occurrence of multiple vectors and invasive An. stephensi could change parasite diversity in multiple African countries. Finally, the current structure of diversity revealed that these transmission-relevant loci have major implications for the design and efficacy of vaccines and antimalarial treatments in An. stephensi-invaded regions.
Supplementary Information
Supplementary Material 1: Figure 1: Pairwise Fst values for Pfs47, Pfs37 and Pfs16. Values indicated by color (white = low, red = high, grey = NaN).
Supplementary Material 2: Figure 2: The number of samples for each haplotype observed in P. falciparum populations for Pfs47, Pfs37 and Pfs16 genes. Bars are colored by the populations.
Supplementary Material 3: Figure 3: Haplotype network of Pfs47 central domain (D2).
Supplementary Material 4: Figure 4: Haplotype diversities of parasite genes (Pfs47 and Pfs16) against presence or absence (determined by VOP cutoff values 0.5, 0.75, and 0.95 separately) of combinations of only the vector species significantly associated with haplotype diversities. Some comparison categories were dropped due to lack of data points. Note: There are many other vector species that could be present in the same location and that were not considered in this analysis.
Supplementary Material 5: Figure 5: Tajima’s D values of Pfs16 against presence or absence (determined by VOP cutoff values 0.5, 0.75, and 0.95 separately) of combinations of vector species significantly associated with haplotype diversities. Some comparison categories were dropped due to lack of data points. Note: There are many other vector species that could be present in the same location and that were not considered in this analysis.
Supplementary Material P47Rec ortholog Coding Sequences.
Acknowledgement
We sincerely thank the Baylor University Department of Biology for providing the essential resources and institutional support that made this research possible. We also extend our gratitude to Dr. Solomon Yared and his team for their efforts in mosquito collection. Additionally, we appreciate the collaboration of the President’s Malaria Initiative (PMI) VectorLink project. Lastly, we acknowledge Elizabeth Waymire for her invaluable assistance with proofreading.
Author contributions
I.G. and T.E.C. conceptualized and designed the study. I.G. conducted the data downloading, bioinformatics analyses, and statistical analyses. J.D.S. contributed to PCR amplifying P47Rec ortholog sequence from An. stephensi mosquitoes collected in Ethiopia. T.E.C. provided oversight and guidance throughout the project and was responsible for securing funding. I.G. wrote the initial manuscript draft, and J.D.S. and T.E.C. provided critical revisions. All authors reviewed and approved the final manuscript.
Funding
This work was funded by the National Institutes of Health Research Enhancement Award (1R15AI151766) and the Department of Biology at Baylor University.
Availability of data and materials
No datasets were generated or analyzed during the current study.
Declarations
Ethics approval and consent to participate
Wild mosquitoes used for this study were collected from dwellings and animal houses, following homeowners’ verbal consent. The study protocol for the collection was reviewed by the Centers for Disease Control and Prevention, USA, and determined to be research not involving human subjects (2017-227). A material transfer agreement was established with Baylor University and the US PMI/Abt Associates for molecular analysis of mosquitoes.
Competing interests
The authors declare no competing interests.
Consent for publication
Not applicable.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Isuru Gunarathna and Tamar E. Carter have equally contributed to this work. Joseph D. Spear amplified the P47Rec ortholog sequences from An. stephensi mosquitos.
References
- 1.WHO. World malaria report 2023. Geneva: World Health Organization; 2023. [Google Scholar]
- 2.WHO. World malaria report 2022. Geneva: World Health Organization; 2022.
- 3.Mbacham WF, Ayong L, Guewo-Fokeng M, Makoge V. Current situation of malaria in Africa. Methods Mol Biol. 2019;2013:29–44. [DOI] [PubMed] [Google Scholar]
- 4.Carter TE, Yared S, Gebresilassie A, Bonnell V, Damodaran L, Lopez K, et al. First detection of Anopheles stephensi Liston, 1901 (Diptera: culicidae) in Ethiopia using molecular and morphological approaches. Acta Trop. 2018;188:180–6. [DOI] [PubMed]
- 5.Faulde MK, Rueda LM, Khaireh BA. First record of the Asian malaria vector Anopheles stephensi and its possible role in the resurgence of malaria in Djibouti. Horn of Africa Acta Trop. 2014;139:39–43. [DOI] [PubMed] [Google Scholar]
- 6.Balkew M, Mumba P, Dengela D, Yohannes G, Getachew D, Yared S, et al. Geographical distribution of Anopheles stephensi in eastern Ethiopia. Parasit Vectors. 2020;13:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coetzee M, Craig M, le Sueur D. Distribution of African malaria mosquitoes belonging to the Anopheles gambiae complex. Parasitol Today. 2000;16:74–7. [DOI] [PubMed] [Google Scholar]
- 8.Vega-Rodriguez J, Ghosh AK, Kanzok SM, Dinglasan RR, Wang S, Bongio NJ, et al. Multiple pathways for Plasmodium ookinete invasion of the mosquito midgut. Proc Natl Acad Sci USA. 2014;111:E492-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molina-Cruz A, Canepa GE, Kamath N, Pavlovic NV, Mu J, Ramphul UN, et al. Plasmodium evasion of mosquito immunity and global malaria transmission: the lock-and-key theory. Proc Natl Acad Sci USA. 2015;112:15178–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui Y, Niu G, Li VL, Wang X, Li J. Analysis of blood-induced Anopheles gambiae midgut proteins and sexual stage Plasmodium falciparum interaction reveals mosquito genes important for malaria transmission. Sci Rep. 2020;10:14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molina-Cruz A, Barillas-Mury C. Pfs47 as a malaria transmission-blocking vaccine target. Am J Trop Med Hyg. 2022;107:27–31. [DOI] [PubMed] [Google Scholar]
- 12.Kahamba NF, Finda M, Ngowo HS, Msugupakulya BJ, Baldini F, Koekemoer LL, et al. Using ecological observations to improve malaria control in areas where Anopheles funestus is the dominant vector. Malar J. 2022;21:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oke CE, Ingham VA, Walling CA, Reece SE. Vector control: agents of selection on malaria parasites? Trends Parasitol. 2022;38:890–903. [DOI] [PubMed] [Google Scholar]
- 14.Molina-Cruz A, Canepa GE, Barillas-Mury C. Plasmodium P47: a key gene for malaria transmission by mosquito vectors. Curr Opin Microbiol. 2017;40:168–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niu G, Cui Y, Wang X, Keleta Y, Li J. Studies of the parasite-midgut interaction reveal Plasmodium proteins important for malaria transmission to mosquitoes. Front Cell Infect Microbiol. 2021;11:654216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molina-Cruz A, Canepa GE, Alves ESTL, Williams AE, Nagyal S, Yenkoidiok-Douti L, et al. Plasmodium falciparum evades immunity of anopheline mosquitoes by interacting with a Pfs47 midgut receptor. Proc Natl Acad Sci USA. 2020;117:2597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molina-Cruz A, Canepa GE, Dwivedi A, Liu W, Raytselis N, Antonio-Nkondjio C, et al. Role of Pfs47 in the dispersal of ancestral Plasmodium falciparum malaria through adaptation to different anopheline vectors. Proc Natl Acad Sci USA. 2023;120:e2213626120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Canepa GE, Molina-Cruz A, Barillas-Mury C. Molecular analysis of Pfs47-mediated Plasmodium evasion of mosquito immunity. PLoS ONE. 2016;11:e0168279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anthony TG, Polley SD, Vogler AP, Conway DJ. Evidence of non-neutral polymorphism in Plasmodium falciparum gamete surface protein genes Pfs47 and Pfs48/45. Mol Biochem Parasitol. 2007;156:117–23. [DOI] [PubMed] [Google Scholar]
- 20.Neafsey DE, Waterhouse RM, Abai MR, Aganezov SS, Alekseyev MA, Allen JE, et al. Mosquito genomics: highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science. 2015;347:1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keleta Y, Ramelow J, Cui L, Li J. Molecular interactions between parasite and mosquito during midgut invasion as targets to block malaria transmission. NPJ Vaccines. 2021;6:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kongkasuriyachai D, Fujioka H, Kumar N. Functional analysis of Plasmodium falciparum parasitophorous vacuole membrane protein (Pfs16) during gametocytogenesis and gametogenesis by targeted gene disruption. Mol Biochem Parasitol. 2004;133:275–85. [DOI] [PubMed] [Google Scholar]
- 23.Ochwedo KO, Onyango SA, Omondi CJ, Orondo PW, Ondeto BM, Lee MC, et al. Signatures of selection and drivers for novel mutation on transmission-blocking vaccine candidate Pfs25 gene in western Kenya. PLoS ONE. 2022;17:e0266394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shukla N, Tang WK, Tolia NH. Structural analysis of Plasmodium falciparum ookinete surface antigen Pfs28 relevant for malaria vaccine design. Sci Rep. 2022;12:19556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaw WR, Teodori E, Mitchell SN, Baldini F, Gabrieli P, Rogers DW, et al. Mating activates the heme peroxidase HPX15 in the sperm storage organ to ensure fertility in Anopheles gambiae. Proc Natl Acad Sci USA. 2014;111:5854–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kajla M, Kakani P, Choudhury TP, Kumar V, Gupta K, Dhawan R, et al. Anopheles stephensi heme peroxidase HPX15 suppresses midgut immunity to support Plasmodium development. Front Immunol. 2017;8:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathias DK, Plieskatt JL, Armistead JS, Bethony JM, Abdul-Majid KB, McMillan A, et al. Expression, immunogenicity, histopathology, and potency of a mosquito-based malaria transmission-blocking recombinant vaccine. Infect Immun. 2012;80:1606–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Armistead JS, Morlais I, Mathias DK, Jardim JG, Joy J, Fridman A, et al. Antibodies to a single, conserved epitope in Anopheles APN1 inhibit universal transmission of Plasmodium falciparum and Plasmodium vivax malaria. Infect Immun. 2014;82:818–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MalariaGen, Ahouidi A, Ali M, Almagro-Garcia J, Amambua-Ngwa A, Amaratunga C, et al. An open dataset of Plasmodium falciparum genome variation in 7000 worldwide samples. Wellcome Open Res. 2021;6:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrews S. FastQC: a quality control tool for high throughput sequence data. Retrieved on 11/08/2023, from https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 31.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, et al. Multiple sequence alignment with the Clustal series of programs. Nucl Acids Res. 2003;31:3497–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res. 2004;32:1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfeifer B, Wittelsburger U, Ramos-Onsins SE, Lercher MJ. PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol Biol Evol. 2014;31:1929–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- 39.Nei M, Tajima F. DNA polymorphism detectable by restriction endonucleases. Genetics. 1981;97:145–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–70. [DOI] [PubMed] [Google Scholar]
- 41.Sinka ME, Bangs MJ, Manguin S, Rubio-Palis Y, Chareonviriyaphap T, Coetzee M, et al. A global map of dominant malaria vectors. Parasit Vectors. 2012;5:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiebe A, Longbottom J, Gleave K, Shearer FM, Sinka ME, Massey NC, et al. Geographical distributions of African malaria vector sibling species and evidence for insecticide resistance. Malar J. 2017;16:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carter TE, Yared S, Getachew D, Spear J, Choi SH, Samake JN, et al. Genetic diversity of Anopheles stephensi in Ethiopia provides insight into patterns of spread. Parasit Vectors. 2021;14:602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edler D, Klein J, Antonelli A, Silvestro D. raxmlGUI 2.0: a graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol Evold. 2021;12:373–7. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Figure 1: Pairwise Fst values for Pfs47, Pfs37 and Pfs16. Values indicated by color (white = low, red = high, grey = NaN).
Supplementary Material 2: Figure 2: The number of samples for each haplotype observed in P. falciparum populations for Pfs47, Pfs37 and Pfs16 genes. Bars are colored by the populations.
Supplementary Material 3: Figure 3: Haplotype network of Pfs47 central domain (D2).
Supplementary Material 4: Figure 4: Haplotype diversities of parasite genes (Pfs47 and Pfs16) against presence or absence (determined by VOP cutoff values 0.5, 0.75, and 0.95 separately) of combinations of only the vector species significantly associated with haplotype diversities. Some comparison categories were dropped due to lack of data points. Note: There are many other vector species that could be present in the same location and that were not considered in this analysis.
Supplementary Material 5: Figure 5: Tajima’s D values of Pfs16 against presence or absence (determined by VOP cutoff values 0.5, 0.75, and 0.95 separately) of combinations of vector species significantly associated with haplotype diversities. Some comparison categories were dropped due to lack of data points. Note: There are many other vector species that could be present in the same location and that were not considered in this analysis.
Supplementary Material P47Rec ortholog Coding Sequences.
Data Availability Statement
No datasets were generated or analyzed during the current study.