Abstract
Introduction
at present, there are inconsistent research findings regarding the precise relationship between IL-6 gene polymorphisms (GPMs) and rheumatoid arthritis (RA). This work employed meta-analysis (MA) methodology to systematically evaluate the correlation between IL-6 GPMs and susceptibility to RA.
Material and methods
this study comprehensively searched multiple databases from inception to March 31, 2024. The search utilized keywords including “IL-6,” “Interleukin-6,” “Cytokines,” “Autoimmune diseases,” “Arthritis,” “rheumatoid arthritis,” “Inflammation,” “genetic polymorphism,” and “genetic variation.” Included studies focused on patients diagnosed with rheumatoid arthritis (RA), with healthy individuals or those without RA-related diseases as controls. The study designs encompassed cohort studies and case-control studies. Genetic frequency distributions of IL-6 gene rs1800795 (G-174C), rs1800796 (G-572C), and rs1800797 (G-597A) polymorphic sites were statistically analyzed. Quality of included studies was assessed. The preliminary assessment of literature heterogeneity was conducted. Quantitative assessment of heterogeneity results was performed using the I2 statistic in RevMan5.3. Publication bias (PB) assessment was conducted.
Result
the study included a total of 21 articles, comprising 9772 participants, with 4679 cases diagnosed with RA and 5093 individuals in the control (CT) group. The IL-6 gene allelic model (G vs C) exhibited a notable association with RA [OR = 0.66, 95%CI: 0.51–0.87, Z = 3.02, P = 0.003]. In Southeast Asian populations, IL-6 gene rs1800795 (G-174C) genotype (CC) demonstrated a considerable correlation with RA [OR = 15.23, 95%CI: 3.53–65.67, P = 0.00003]. IL-6 gene rs1800795 (G-174C) genotype (CG) was associated with RA [OR = 1.54, 95%CI: 1.10–2.17, Z = 2.48, P = 0.01], with only the Asian population showing an observable correlation [OR = 6.55, 95%CI: 1.28–33.45, P = 0.02]. Additionally, IL-6 rs1800795 (G-174C) genotype (GG) was associated with RA [OR = 0.66, 95%CI: 0.49–0.91, Z = 2.55, P = 0.01], with notable associations observed in the Asian [OR = 0.17, 95%CI: 0.03–0.83, P = 0.03] and mixed populations [OR = 1.29, 95%CI: 1.011.65, P = 0.04]. No correlation was found between rs1800796 (G-572C) and rs1800797 (G-597A) GPMs and RA (P > 0.05).
Conclusion
IL-6 gene rs1800795 (G-174C) allelic model and genotypes (CC, CG, GG) were all associated with susceptibility to RA, with the G allele model being susceptible in Southeast Asian populations, and genotypes (CG and GG) being susceptible in Asian populations. However, there was neglectable correlation between rs1800796 (G-572C) and rs1800797 (G-597A) GPMs and susceptibility to RA.
Keywords: IL-6, Genetic polymorphism, Rheumatoid arthritis, Correlation, Meta-analysis
1. Introduction
Rheumatoid arthritis (RA) is characterized by chronic arthritis, joint inflammation, and systemic inflammation, often leading to joint destruction and functional impairment. Its pathological features include progressive and persistent synovitis, resulting in cartilage and bone destruction, ultimately leading to joint deformity and functional impairment, greatly affecting patients’ quality of life [1,2]. The disease typically manifests as symmetric, persistent joint swelling and pain, often accompanied by morning stiffness. RA has a wide age range of onset, but the peak onset age is between 30 and 50 years, with a markedly higher incidence rate in females than in males [3]. RA pathogenesis is not fully understood, with current research generally suggesting that genetics, environment, and infection are closely related factors [4,5]. One of the recent focuses of research is the relationship between single nucleotide polymorphisms (SNPs) and susceptibility to RA [6], with numerous studies implying a notable association between SNPs and the onset of RA [7,8].
The role of cytokines in RA pathogenesis has been receiving increasing attention. Cytokines are a class of small molecular glycoproteins distributed in the extracellular space, participating in immune responses and playing crucial roles in the pathophysiological processes of arthritis-related diseases such as RA [9]. In the onset and progression of RA, cytokines are involved in joint bone and cartilage destruction, promoting the occurrence of osteoporosis. Among them, cytokines like IL-6 are considered to be one of the more important contributing factors in the pathogenesis of RA [10]. IL-6 is capable of regulating immune responses, inflammatory reactions, and tissue repair processes [11]. Past studies indicated that IL-6 acts in the onset of RA by promoting the production of inflammatory mediators, stimulating joint destruction, and influencing immune regulation [12]. The genetic polymorphism (PM) of IL-6 has been suggested to be associated with susceptibility to RA and its clinical manifestations [13]. However, there are inconsistent research findings regarding the precise relationship between IL-6 gene polymorphisms (GPMs) and RA. Some studies have reported a correlation between IL-6 GPMs and an increased risk of RA [14,15], while others have failed to observe this association [16,17]. To better understand the relationship between IL-6 GPMs and RA, this work conducted a meta-analysis (MA) to comprehensively assess the results of existing research and provide more reliable evidence. Through a systematic review and quantitative synthesis of existing literature, the aim was to demonstrate the correlation between IL-6 GPMs and the risk of RA, thus providing references for further research and clinical practice.
2. Materials and methodologies
2.1. Search
A comprehensive literature search was implemented using a keyword search methodology to obtain all relevant studies on correlation between the inflammatory factor IL-6 SNPs and RA. Keywords included “IL-6,” “Interleukin-6,” “Cytokines,” “Autoimmune diseases,” “Arthritis,” “rheumatoid arthritis,” “Inflammation,” “genetic polymorphism,” and “genetic variation”. The search utilized a combination of subject terms and their related expansion terms. Logical operators “or” and “and” were utilized to combine the keywords for joint searches, such as ((((IL-6) OR (Interleukin-6)) OR (Cytokines)) AND (Autoimmune diseases)) OR (Arthritis)) OR (rheumatoid arthritis)) AND (genetic polymorphism). A comprehensive search was conducted across CNKI, Wanfang, VIP, CBM, Google Scholar, Medline, Embase, PubMed, Cochrane Library, Nature, Web of Science, Springer, and Science Direct. The search period extended from inception to March 31, 2024, and tracking searches based on the references of retrieved articles were performed. During the search process, no language, ethnicity, or geographical restrictions were set to ensure the retrieval of the most comprehensive research literature possible.
2.2. Criteria
Inclusion criteria: i.) studies related to RA and IL-6 GPMs; ii.) all RA patients must meet the classification criteria and criteria for active RA established by the American College of Rheumatology (ACR), with no restriction on disease duration; iii.) the control (CT) group consisted of healthy individuals or those without RA-related diseases, while RA group comprised RA patients of any age, gender, or ethnicity; iv.) the exposure factor was IL-6 GPMs; v.) cohort studies and case-control studies; vi.) studies included IL-6 gene distribution proportions; vii.) observation indicators: distribution of SNP genetic models.
Exclusion criteria: i.) studies on other types of arthritis or immune diseases besides RA; ii.) studies where IL-6 GPM association with RA was not addressed in the original literature, or IL-6 GPM genotype frequencies and/or allele frequencies were not provided; iii.) studies lacking necessary data availability; iv.) non-randomized or non-case-control study designs; v.) reviews, individual case reports; vi.) literature focusing on animal subjects; vii.) duplicate publications.
2.3. Screening and data extraction
Two reviewers conducted literature screening independently. After keyword screening, EndNote X9 was employed for literature management, and duplicate literature was removed. The two reviewers individually read the article titles and abstracts, preliminarily excluding literature that was clearly unrelated to IL-6 GPMs and RA association. If there were disagreements between the two reviewers regarding the inclusion of certain literature, a third-party expert could be consulted for judgment, and the submitter could be contacted to supplement missing data. The literature full text was read, and inclusion in the study was further determined based on the full-text content.
Two reviewers independently conducted data extraction and cross-checked the extracted data to ensure accuracy. The extracted data included: year, author, country, ethnicity, sample size, age, gender, genotyping methods, genotypes, genotype frequencies in each group, and allele frequencies. The extracted data were organized in Excel.
2.4. Risk assessment of literature bias
The Newcastle-Ottawa Scale (NOS), a commonly utilized tool for assessing bias in observational studies, was employed. This tool aims to evaluate the quality of observational studies, including cohort and case-control studies. Two researchers independently assessed the risk of bias. The NOS consists of three aspects: selection of study subjects, comparability between the study group and CT group, and exposure or outcome assessment, each comprising multiple items. Evaluation criteria typically cover representativeness of study subjects, fairness of comparison between the study group and CT group, and accuracy of exposure or outcome assessment, among others. Based on the degree of compliance with each item, corresponding star ratings were assigned, and the scores were summed to obtain a total score. Each evaluator utilized the NOS to score each included study, with scores ranging 0–9 stars, where higher scores imply higher study quality, and studies with 6 or more stars are considered high quality. Evaluators utilized the NOS score to assess the quality of each study and determine whether to include it in the MA or systematic review. After independent scoring by the two evaluators, the results were cross-checked. In cases where there were discrepancies in scoring, evaluators resolved them through discussion or consulted a third-party expert to reach a consensus evaluation result.
2.5. Statistical methodologies
The included literature data were subjected to MA using RevMan5.3. Heterogeneity analysis: Initially, χ2 test was employed to conduct a preliminary examination of literature heterogeneity, with α = 0.05 and P < 0.05. Subsequently, I2 statistic in RevMan5.3 was utilized to quantitatively assess heterogeneity results. When I2<50 %, a fixed-effects model (FEM) was utilized for MA. When I2>50 %, a random-effects model (REM) was utilized for MA. Forest plots (FOPs) were generated, and Z values and P values were extracted from the results for determining the MA results, with all effect sizes expressed using 95 % confidence intervals (CI). P < 0.05 indicated statistically significant intergroup differences. Funnel plots (FUPs) were created using RevMan5.3, followed by Egger’s linear regression test for quantitative assessment of publication bias (PB) using STATA 10.0. After that, if the 95%CI included 0 and the corresponding P value was greater than 0.10, it indicated no PB.
3. Results
3.1. Search results of literature
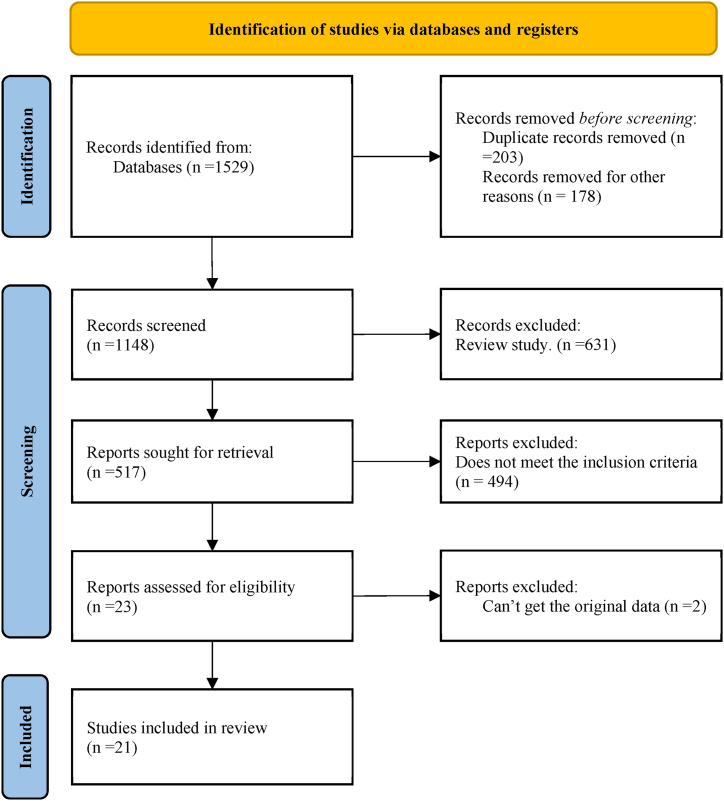
During the literature search, each keyword was entered separately into various online databases for searching, resulting in a total of 1529 articles retrieved. Initially, 203 duplicate articles were removed using EndNote X9, and an additional 178 reports, reviews, and commentaries were excluded. Further exclusions were made based on reading the titles and abstracts, resulting in the exclusion of 631 review articles. Additionally, 494 articles that did not address correlation between IL-6 GPMs and RA or failed to meet the inclusion criteria were excluded. This process left 23 articles remaining, but 2 of them could not be accessed to obtain original data through various channels. Finally, 21 articles were included for analysis. The literature search and screening process of this study are illustrated in Fig. 1.
Fig. 1.
Basic process of literature search.
3.2. Basic information included in the literature
Twenty-one articles [[18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38]] were selected. Basic information from the included literature was compiled and summarized in Table 1. In total, 9772 samples were included, with 4679 cases in RA group and 5093 cases in CT group.
Table 1.
Basic information of included literature.
| Number of cases |
Age |
Sex (male/female) |
GPM detection methods | Locus |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| First author | Year | Country | Ethnicity | Total number of cases | RA group | CT group | RA group | CT group | RA group | CT group | ||
| Emonts M [18] | 2011 | Netherlands | Mixed | 839 | 376 | 463 | 59.3±13.7 | 59.3±13.7 | 99/277 | 233/230 | SBE | rs1800795(G-174C) |
| Arman A [19] | 2012 | Türkiye | Caucasus | 425 | 178 | 247 | 53.12±12.7 | 54.3±11.8 | 29/149 | 87/160 | PCR-RFLP | rs1800795(G-174C) |
| Chen J [20] | 2021 | China | Asia | 1002 | 508 | 494 | 54.34±12.01 | 54.03±8.83 | 134/374 | 124/370 | SBE | rs1800796(G-572C) |
| Dar SA [21] | 2017 | India | Southeast Asia | 114 | 34 | 80 | 51.39±2.27 | 36±2.9 | 15/19 | 32/48 | PCR-SSP | rs1800795(G-174C) |
| Li X [22] | 2014 | China | Asia | 1550 | 752 | 798 | 52.3±16.3 | 52.1±17.1 | 398/354 | 431/367 | PCR-RFLP | rs1800795(G-174C) |
| Li F [23] | 2014 | China | Asia | 587 | 256 | 331 | 50.26±12.86 | 48.08±13.92 | 60/196 | 92/239 | PCR-HRM | rs1800795(G-174C, rs1800796(G-572C), rs1800797(G-597A) |
| Shafia S [24] | 2014 | India | Asia | 350 | 150 | 200 | Unreported | Unreported | 19/131 | 27/173 | PCR-RFLP | rs1800795(G-174C) |
| You CG [25] | 2013 | China | Asia | 825 | 452 | 373 | 47.08±15.36 | 47.35±14.37 | 104/348 | 102/271 | PCR-HRM | rs1800797(G-597A),rs1800796(G-572C), rs1800795(G-174C) |
| Trajkov D [26] | 2009 | Macedonia | Europe | 386 | 85 | 301 | Unreported | Unreported | Unreported | Unreported | PCR-SSP | rs1800795(G-174C) |
| Palomino-Morales R [27] | 2009 | Spain | Europe | 537 | 311 | 226 | Unreported | Unreported | 83/228 | Unreported | TaqMan | rs1800795(G-174C) |
| Panoulas VF [28] | 2009 | England | Europe | 805 | 383 | 422 | Unreported | Unreported | Unreported | Unreported | Real-time PCR | rs1800795(G-174C) |
| Huang XZ [29] | 2007 | China | Asia | 288 | 120 | 168 | 48.48±14.98 | 45.18±8.11 | 20/100 | 67/101 | PCR-SSP | rs1800795(G-174C), rs1800796(G-572C) |
| Pawlik A [30] | 2005 | Poland | Europe | 203 | 98 | 105 | 48.5 | Unreported | 37/61 | Unreported | PCR-RFLP | rs1800795(G-174C) |
| Pascual M [31] | 2000 | Spain | Europe | 320 | 163 | 157 | 50.7±14 | Unreported | Unreported | Unreported | PCR-RFLP | rs1800795(G-174C) |
| Dahlqvist SR [32] | 2002 | Sweden | Europe | 440 | 257 | 183 | 62±12.9 | Unreported | 70/187 | Unreported | PCR-SSP | rs1800795(G-174C) |
| Amr K [33] | 2016 | Egypt | Southeast Asia | 198 | 99 | 99 | Unreported | Unreported | 9/90 | 41/58 | PCR-SSP | rs1800795(G-174C), rs1800796(G-572C) |
| Ad’hiah AH [34] | 2018 | Iraq | Southeast Asia | 96 | 51 | 45 | 44.9±10.7 | 41.3±8.7 | 22/29 | 15/30 | PCR-RFLP | rs1800796(G-572C), |
| Yucel B [35] | 2020 | Türkiye | Caucasus | 145 | 49 | 96 | 52±12 | Unreported | Unreported | Unreported | PCR-SSP | rs1800795(G-174C), |
| Nisar H [36] | 2021 | Pakistan | Southeast Asia | 315 | 150 | 165 | 43.53±11.11 | Unreported | 31/119 | Unreported | TaqMan | rs1800795(G-174C) |
| Zavaleta‐Muñiz SA [37] | 2016 | Mexico | Mixed | 239 | 137 | 102 | 50±9 | 48±10 | 2/135 | 6/96 | PCR-RFLP | rs1800795(G-174C) |
| Melo TS [38] | 1992 | Brazil | Mixed | 108 | 70 | 38 | 50.9±11.4 | 52.3±12.2 | Unreported | Unreported | TaqMan | rs1800795(G-174C) |
RA: rheumatoid arthritis; PCR: polymerase chain reaction; SBE: single base extension; RFLP: restriction fragment length polymorphism; SSP: sequence specific primers.
3.3. Quality evaluation of included literature
All 23 articles included in this study were observational studies, and their quality was assessed. The evaluation results showed that the NOS scores of all articles were 6 or above, meeting the quality requirements (Table 2).
Table 2.
Basic information of patients in the literature.
| First author | Year | Selection of research population | Intergroup comparability | Measurement of exposure factors | NOS score (points) |
|---|---|---|---|---|---|
| Emonts M | 2011 | ★★★ | ★★ | ★★★ | 8 |
| Arman A | 2012 | ★★★ | ★★ | ★★★ | 8 |
| Chen J | 2021 | ★★★ | ★★ | ★★★ | 8 |
| Dar SA | 2017 | ★★★ | ★★ | ★★★ | 8 |
| Li X | 2014 | ★★★ | ★★ | ★★★ | 8 |
| Li F | 2014 | ★★★ | ★★ | ★★★ | 8 |
| Shafia S | 2014 | ★★ | ★★ | ★★★ | 7 |
| Zavaleta-Muñiz SA | 2013 | ★★★ | ★★ | ★★★ | 8 |
| You CG | 2013 | ★★★ | ★★ | ★★ | 7 |
| Trajkov D | 2009 | ★★ | ★★ | ★★★ | 7 |
| Palomino-Morales R | 2009 | ★★ | ★★ | ★★ | 6 |
| Panoulas VF | 2009 | ★★ | ★★ | ★★ | 6 |
| Huang XZ | 2007 | ★★★ | ★★ | ★★★ | 8 |
| Pawlik A | 2005 | ★★★ | ★★ | ★★ | 7 |
| Pascual M | 2000 | ★★ | ★★ | ★★★ | 7 |
| Dahlqvist SR | 2002 | ★★ | ★★ | ★★★ | 7 |
| Amr K | 2016 | ★★ | ★★ | ★★★ | 7 |
| Ad’hiah AH | 2018 | ★★★ | ★★ | ★★★ | 8 |
| Yucel B | 2020 | ★★ | ★★ | ★★★ | 7 |
| Nisar H | 2021 | ★★ | ★★ | ★★★ | 7 |
| Zavaleta‐Muñiz SA | 2016 | ★★★ | ★★ | ★★★ | 8 |
| Melo TS | 1992 | ★★ | ★★ | ★★★ | 7 |
| Jahan T | 2024 | ★★ | ★ | ★★★ | 6 |
3.4. Correlation analysis between IL-6 gene rs1800795 (G-174C) and RA
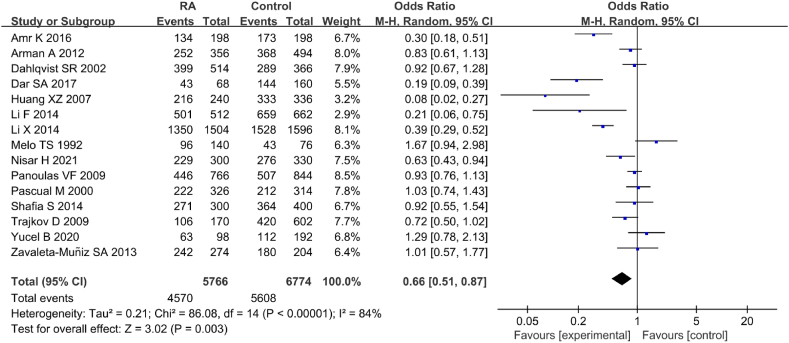
A total of 15 articles included in the literature examined association between IL-6 gene rs1800795(G-174C) allelic model (G vs C) and RA (Fig. 2). Marked heterogeneity was observed in the statistical results of the SNP allelic model (G vs C) of IL-6 gene rs1800795(G-174C) in both RA and CT groups (I2 = 84 %, P < 0.00001). Therefore, a REM was employed. Results indicated a considerable correlation between IL-6 gene rs1800795(G-174C) PM and RA [OR = 0.66, 95%CI: 0.51–0.87, Z = 3.02, P = 0.003]. To ensure the accuracy of this result, a sensitivity analysis (SA) was further conducted on the included literature.
Fig. 2.
FOP of allelic model (G vs C) and its correlation with RA.
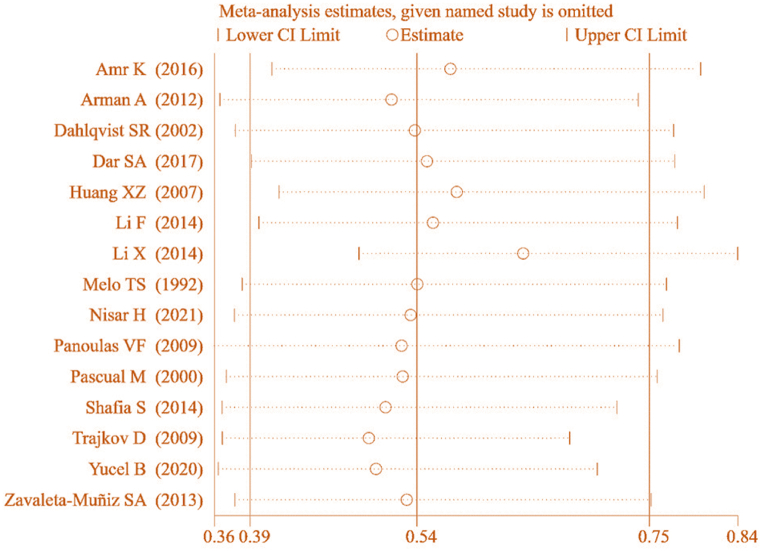
The results in Fig. 3 demonstrated that the MA of IL-6 gene rs1800795(G-174C) allelic model (G vs C) and its correlation with RA remained considerable [combined effect: OR = 0.541, 95%CI: 0.388–0.754, P < 0.05], implying a stable correlation between IL-6 gene rs1800795(G-174C) PM and RA. This suggests good stability of the obtained result.
Fig. 3.
SA of the allelic model and its correlation with RA.
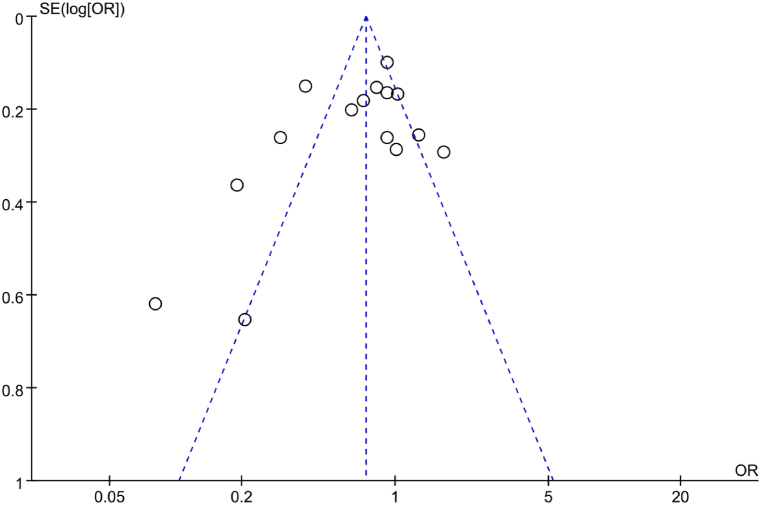
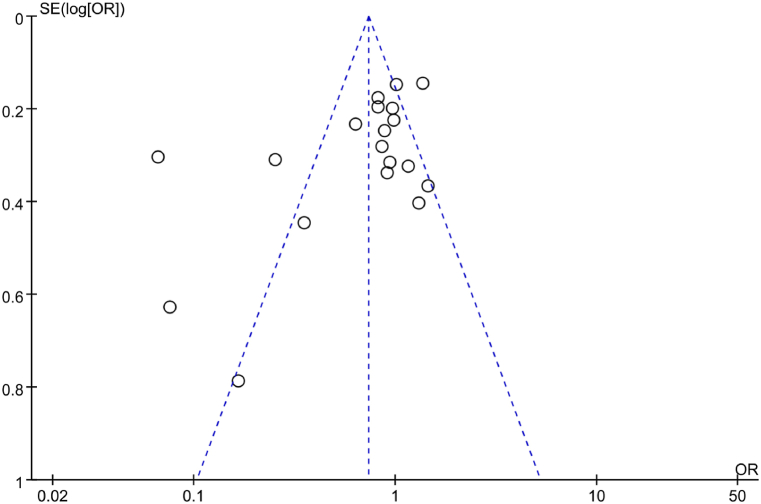
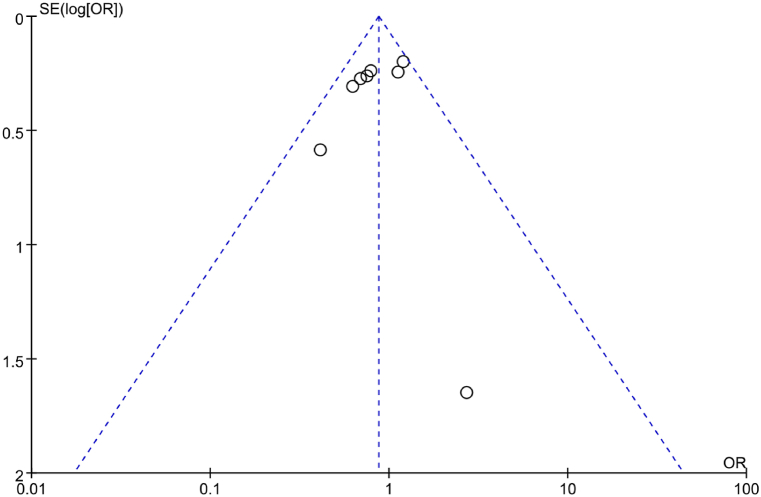
A funnel plot was utilized to analyze PB regarding correlation between IL-6 gene rs1800795(G-174C) allelic model (G vs C) and RA in the included literature. The results in Fig. 4 indicated a symmetric distribution of scatter points in the funnel plot, suggesting neglectable PB. Additionally, Egger’s test was conducted to further analyze PB. The results revealed that t = −0.60, P = 0.557, implying neglectable PB in the included studies.
Fig. 4.
FUP of association between the allelic model (G vs C) and RA.
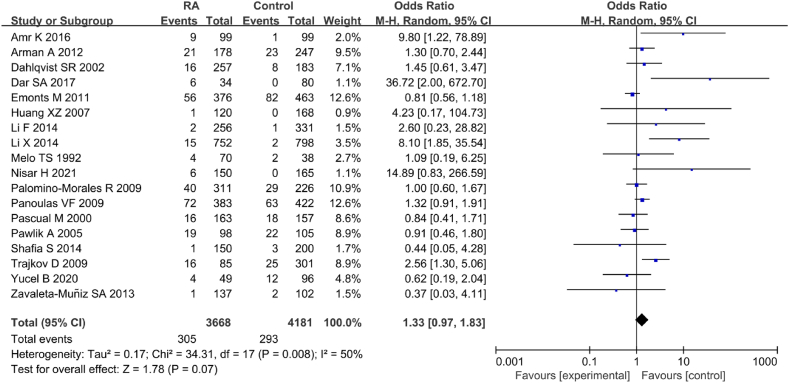
A total of 18 articles included in the literature examined correlation between IL-6 gene rs1800795(G-174C) genotype (CC) and RA (Fig. 5). Marked heterogeneity was observed in the statistical results of IL-6 rs1800795(G-174C) genotype (CC) between RA and CT groups (I2 = 50 %, P = 0.008). Hence, a REM was employed for analysis. The results indicated no correlation between IL-6 gene rs1800795(G-174C) PM and RA [OR = 1.33, 95%CI: 0.97–1.83, Z = 1.78, P = 0.07].
Fig. 5.
FOP of correlation between IL-6 gene rs1800795(G-174C) genotype (CC) and RA.
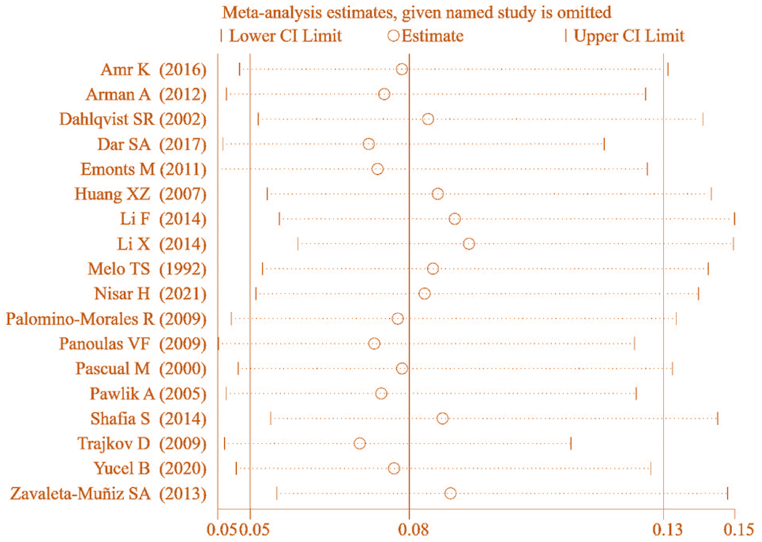
To ensure the accuracy of this result, a SA was further conducted on the included literature. The results (Fig. 6) demonstrated that the MA of correlation between IL-6 gene rs1800795(G-174C) genotype (CC) and RA was conducted by sequentially excluding individual studies. The combined effect [OR = 0.083, 95%CI: 0.516–0.132, P < 0.05] indicated a notable correlation between IL-6 gene rs1800795(G-174C) genotype (CC) and RA, suggesting that this result exhibits a certain degree of heterogeneity.
Fig. 6.
SA of association between IL-6 gene rs1800795(G-174C) genotype (CC) and RA.
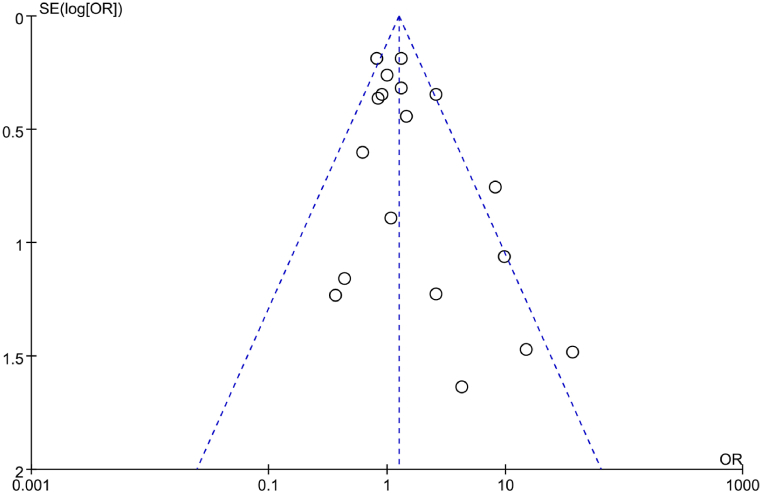
A funnel plot was utilized to analyze PB regarding correlation between IL-6 gene rs1800795(G-174C) genotype (CC) and RA in the included literature. The results, as depicted in Fig. 7, revealed a symmetric distribution of scatter points in the funnel plot, but some studies were located in the narrow part of the funnel plot, implying the potential presence of PB. Additionally, Egger’s test was conducted to further analyze PB. It indicated a considerable t = −2.46 with a corresponding P = 0.028, further suggesting the presence of PB in the included studies.
Fig. 7.
FUP of correlation between IL-6 gene rs1800795(G-174C) genotype (CC) and RA.
Based on different ethnicities, the groups were categorized into Asian, European, Caucasian, Southeast Asian, and mixed. MA revealed that among different ethnicities, only in the Southeast Asian population, IL-6 gene rs1800795(G-174C) genotype (CC) showed a marked correlation with RA [OR = 15.23, 95%CI: 3.53–65.67, P = 0.00003] (Table 3).
Table 3.
Correlation between IL-6 gene rs1800795(G-174C) genotype (CC) and RA across different ethnicities.
| Ethnicity | Number of studies included | Sample size | RA group(n/total) | CG (n/total) | Heterogeneity (I2) | OR [95%CI] | P |
|---|---|---|---|---|---|---|---|
| Asian | 4 | 2775 | 19/1278 | 6/1497 | 34 % | 2.85 [0.73, 11.04] | 0.13 |
| European | 6 | 2691 | 1791297 | 165/1394 | 31 % | 1.24 [0.92, 1.66] | 0.16 |
| Caucasian | 2 | 570 | 25/227 | 35/343 | 14 % | 1.08 [0.57,2.03] | 0.82 |
| Southeast Asian | 3 | 627 | 21/283 | 1/344 | 0 | 15.23 [3.53,65.67] | 0.0003 |
| Mixed | 3 | 1186 | 61/583 | 86/603 | 0 | 0.81 [0.57, 1.16] | 0.25 |
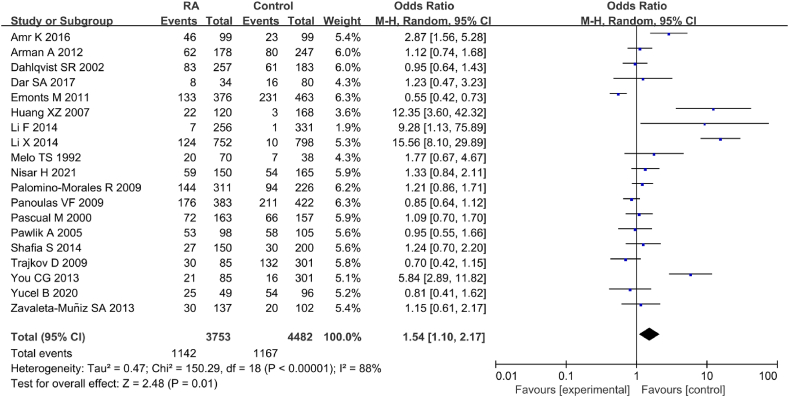
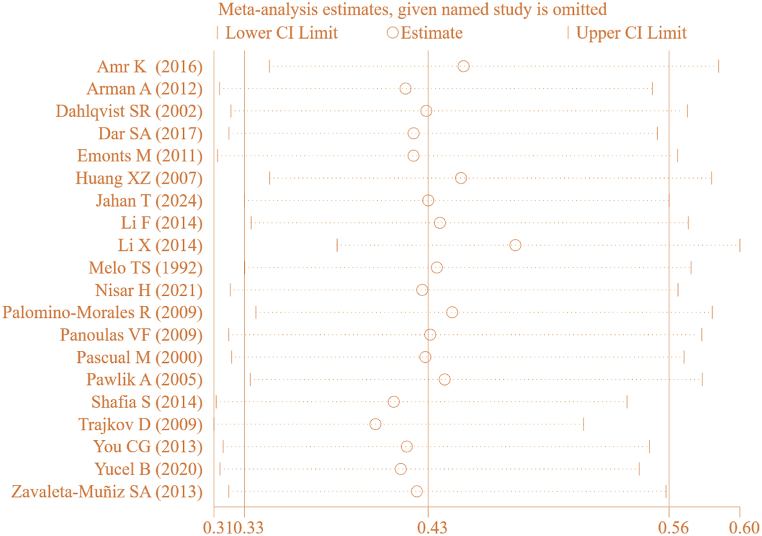
Incorporating a total of 19 articles, statistical analysis was conducted on correlation between IL-6 gene rs1800795(G-174C) genotype (CG) and RA (Fig. 8). The statistical results of IL-6 rs1800795(G-174C) genotype (CG) between RA and CT groups exhibited marked heterogeneity (I2 = 88 %, P < 0.00001). Therefore, a REM was employed, revealing an association between IL-6 gene rs1800795(G-174C) PM and RA [OR = 1.54, 95%CI: 1.10–2.17, Z = 2.48, P = 0.01].
Fig. 8.
FOP of correlation between IL-6 gene rs1800795(G-174C) genotype (CG) and RA.
To ensure the accuracy of this result, a SA was conducted by systematically excluding individual studies on correlation between IL-6 gene rs1800795(G-174C) genotype (CG) and RA (Fig. 9). The pooled effect size [OR = 0.371, 95%CI: 0.284–0.483, P < 0.05] remained substantial, implying a stable correlation between IL-6 gene rs1800795(G-174C) genotype (CG) and RA.
Fig. 9.
SA of association between IL-6 gene rs1800795(G-174C) genotype (CG) and RA.
A funnel plot was employed to analyze the PB of correlation between IL-6 gene rs1800795(G-174C) genotype (CG) and RA (Fig. 10). The scatter plot distribution of the funnel plot appeared symmetric, but some studies were located outside the funnel, suggesting the potential presence of PB. Furthermore, Egger’s test was conducted to further analyze the PB, yielding a result of t = −2.69, P = 0.015, further implying the existence of PB in the included studies.
Fig. 10.
FUP of correlation between IL-6 gene rs1800795(G-174C) genotype (CG) and RA.
Based on different ethnicities, including Asian, European, Caucasian, Southeast Asian, and mixed populations, MA revealed a considerable correlation between IL-6 gene rs1800795(G-174C) genotype (CG) and RA only in the Asian population [OR = 6.55, 95%CI: 1.28–33.45, P = 0.02] (Table 4).
Table 4.
Correlation between IL-6 gene rs1800795(G-174C) genotype (CG) and RA among different ethnicities.
| Ethnicity | Number of studies included | Sample size | RA group(n/total) | CG (n/total) | Heterogeneity (I2) | OR [95%CI] | P |
|---|---|---|---|---|---|---|---|
| Asian | 4 | 2775 | 180/1278 | 44/1497 | 92 % | 6.55 [1.28, 33.45] | 0.02 |
| European | 7 | 3077 | 579/1382 | 638/1695 | 79 % | 1.15 [0.81, 1.65] | 0.43 |
| Caucasian | 2 | 570 | 87/227 | 134/343 | 0 | 1.03 [0.72,1.46] | 0.88 |
| Southeast Asian | 3 | 627 | 113/283 | 93/344 | 54 % | 1.72 [0.99,2.97] | 0.05 |
| Mixed | 3 | 1186 | 183/583 | 258/603 | 77 % | 0.94 [0.46, 1.90] | 0.85 |
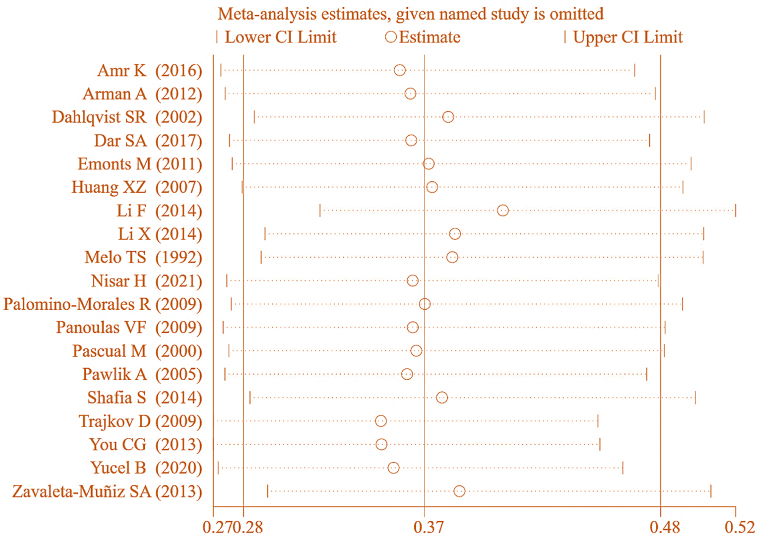
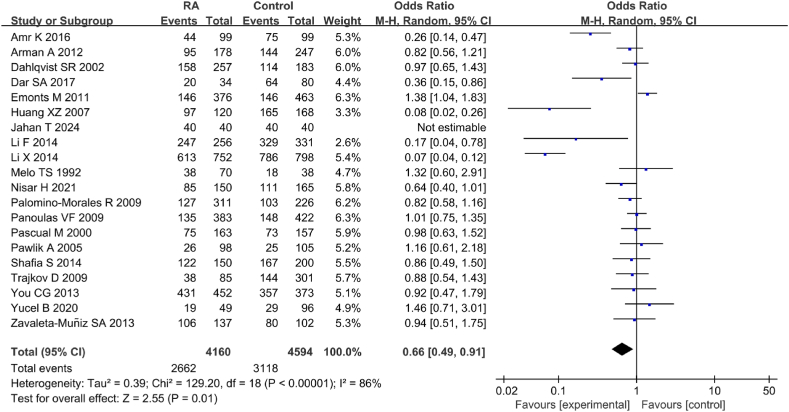
Twenty studies were included in correlation analysis between IL-6 gene rs1800795(G-174C) genotype (GG) and RA (Fig. 11). Considerable heterogeneity was observed in the statistical results between RA and CT groups (I2 = 86 %, P < 0.00001). Hence, a REM was applied for the analysis. The results revealed a notable correlation between IL-6 gene rs1800795(G-174C) PM and RA [OR = 0.66, 95%CI: 0.49–0.91, Z = 2.55, P = 0.01].
Fig. 11.
FOP of correlation between IL-6 gene rs1800795(G-174C) genotype (GG) and RA.
To ensure the accuracy of this result, a SA was conducted on the included studies. The results, as depicted in Fig. 12, indicated that upon sequentially excluding studies examining association between IL-6 gene rs1800795(G-174C) genotype (GG) and RA, the pooled effect size was [OR = 0.427, 95%CI:0.326–0.558, P < 0.05]. This suggests a stable correlation between IL-6 gene rs1800795(G-174C) genotype (GG) and RA.
Fig. 12.
SA of association between IL-6 gene rs1800795(G-174C) genotype (GG) and RA.
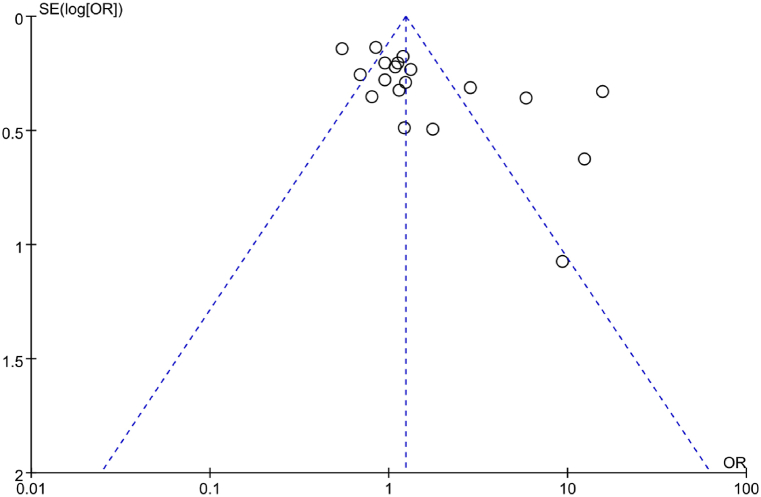
A funnel plot was employed to assess PB regarding correlation between IL-6 gene rs1800795(G-174C) genotype (GG) and RA. In Fig. 13, the scatter distribution of the funnel plot appeared symmetrical, implying neglectable PB. Additionally, an Egger’s test was conducted to further analyze PB. It revealed a t = 1.05 with a corresponding P = 0.310, further implying the absence of marked PB in the included studies.
Fig. 13.
FUP of correlation between IL-6 gene rs1800795(G-174C) genotype (GG) and RA.
Various ethnic groups were categorized into Asian, European, Caucasian, Southeast Asian, and mixed populations. MA revealed a notable correlation between IL-6 gene rs1800795(G-174C) genotype (GG) and RA in Asian populations [OR = 0.17, 95%CI: 0.03–0.83, P = 0.03]. Similarly, a marked correlation was observed in mixed populations [OR = 1.29, 95%CI: 1.01–1.65, P = 0.04] (Table 5).
Table 5.
Correlation between IL-6 gene rs1800795(G-174C) genotype (GG) and RA across different ethnicities.
| Ethnicity | Number of studies included | Sample size | RA group(n/total) | CG (n/total) | Heterogeneity (I2) | OR [95%CI] | P |
|---|---|---|---|---|---|---|---|
| Asian | 4 | 2775 | 1079/1278 | 1447/1497 | 93 % | 0.17 [0.03, 0.83] | 0.03 |
| European | 7 | 3516 | 990/1749 | 964/1767 | 0 | 0.95 [0.81, 1.11] | 0.50 |
| Caucasian | 2 | 570 | 114/227 | 173/343 | 48 % | 0.93 [0.66,1.31] | 0.69 |
| Southeast Asian | 3 | 627 | 149/283 | 150/344 | 98 % | 1.19 [0.07,20.23] | 0.90 |
| Mixed | 3 | 1186 | 290/583 | 244/603 | 0 | 1.29 [1.01, 1.65] | 0.04 |
3.5. Correlation analysis between IL-6 gene rs1800796 (G-572C) and RA
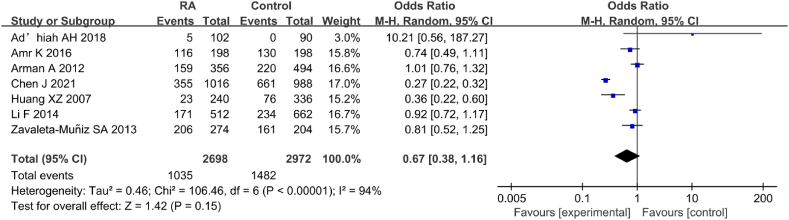
Seven studies were included in the analysis of IL-6 gene rs1800796(G-572C) allele model (G vs C) and its correlation with RA (Fig. 14). Notable heterogeneity was observed in the statistical results of the SNP allele model (G vs C) of IL-6 gene rs1800796(G-572C) between the RA and CT groups (I2 = 94 %, P < 0.00001). Therefore, a REM was employed. Neglectable correlation existed between IL-6 gene rs1800796(G-572C) PM and RA [OR = 0.67, 95%CI: 0.52–1.25, Z = 1.42, P = 0.15].
Fig. 14.
FOP of association between the allele model (G vs C) and RA.
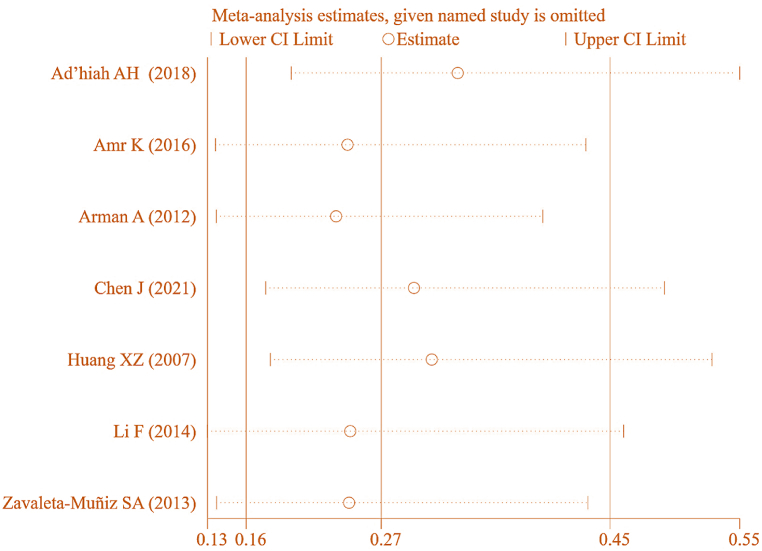
To ensure the accuracy of this result, a SA was conducted on the included studies. The results in Fig. 15 demonstrated that the MA of correlation between IL-6 gene rs1800796(G-572C) allele model (G vs C) and RA, with one study removed at a time, yielded a combined effect [OR = 0.265, 95%CI: 0.156–0.450, P < 0.05]. This suggests a correlation between IL-6 gene rs1800796(G-572C) PM and RA. However, it was noted that this result exhibited some heterogeneity, warranting further analysis.
Fig. 15.
SA of association between IL-6 gene rs1800796(G-572C) allele model (G vs C) and RA.
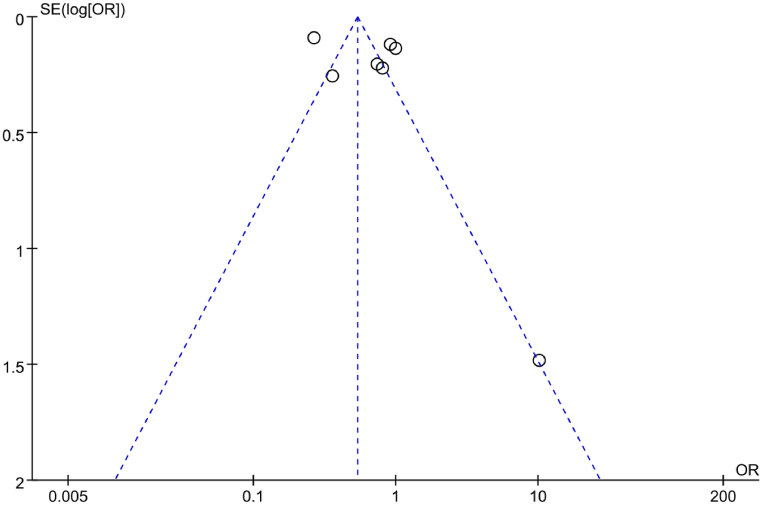
A funnel plot was utilized to analyze the PB regarding correlation between IL-6 gene rs1800796(G-572C) allele model (G vs C) and RA. The results, depicted in Fig. 16, indicated a symmetrical distribution of data points on the funnel plot, suggesting the absence of PB. Additionally, an Egger’s test was conducted for further analysis of PB. The results revealed a t = −1.02 and a corresponding P = 0.365, further confirming the absence of PB in the included studies.
Fig. 16.
FUP of association between the allele model (G vs C) and RA.
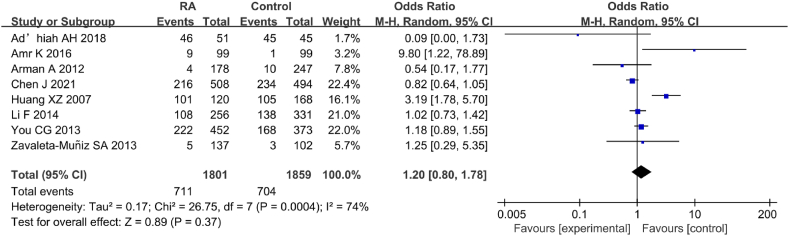
Eight studies were included in the analysis of correlation between IL-6 gene rs1800796(G-572C) genotype (CC) and RA (Fig. 17). The statistical results revealed considerable heterogeneity in the statistical outcomes between RA and CT groups (I2 = 74 %, P = 0.008). Consequently, a REM was employed for the analysis. The findings indicated neglectable correlation between IL-6 gene rs1800796(G-572C) PM and RA [OR = 1.20, 95%CI: 0.80–1.78, Z = 0.89, P = 0.37].
Fig. 17.
FOP of correlation between IL-6 gene rs1800796(G-572C) genotype (CC) and RA.
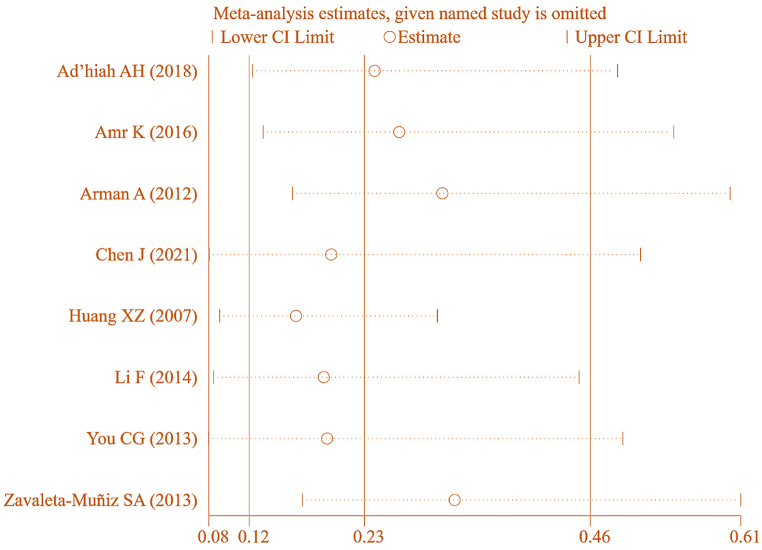
To ensure the accuracy of these findings, a SA was conducted by sequentially excluding studies on correlation between IL-6 gene rs1800796(G-572C) genotype (CC) and RA (Fig. 18). The pooled effect size [OR = 0.233, 95%CI: 0.118–0.458, P < 0.05] suggested a considerable correlation between IL-6 gene rs1800796(G-572C) genotype (CC) and RA, implying some degree of heterogeneity in these results.
Fig. 18.
SA of association between IL-6 gene rs1800796(G-572C) genotype (CC) and RA.
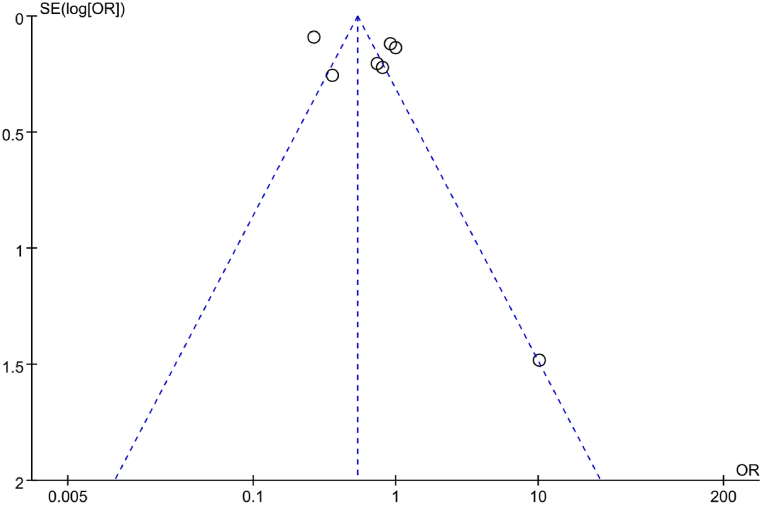
The PB regarding correlation between IL-6 gene rs1800796(G-572C) genotype (CC) and RA was assessed using a funnel plot (Fig. 19). Although the scatter distribution in the funnel plot appeared symmetrical, some studies were located outside the funnel plot, implying potential PB. Furthermore, Egger’s test was employed to further analyze the PB. The results revealed a considerable Egger’s regression intercept (t = −5.13, P = 0.004), implying the presence of PB in the included studies.
Fig. 19.
FUP of correlation between IL-6 gene rs1800796(G-572C) genotype (CC) and RA.
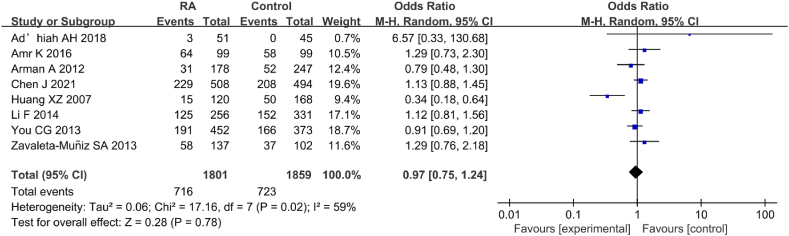
Eight studies were included in analysis of correlation between IL-6 gene rs1800796(G-572C) genotype (CG) and RA (Fig. 20). Great heterogeneity was observed in the statistical results of IL-6 rs1800796(G-572C) genotype (CG) between the RA and CT groups (I2 = 59 %, P = 0.02). Hence, a REM was applied for the analysis. The results revealed neglectable correlation between IL-6 gene rs1800796(G-572C) PM and RA (OR = 0.97, 95%CI: 0.75–1.24, Z = 0.288, P = 0.78).
Fig. 20.
FOP of correlation between IL-6 gene rs1800796(G-572C) genotype (CG) and RA.
To ensure the accuracy of this result, a SA was conducted on the included literature (Fig. 21). Each study examining correlation between IL-6 gene rs1800796(G-572C) genotype (CG) and RA was sequentially excluded from the MA. The combined effect revealed a notable correlation between IL-6 gene rs1800796(G-572C) genotype (CG) and RA (OR = 0.335, 95%CI: 0.233–0.482, P < 0.05), implying a certain level of heterogeneity in this result.
Fig. 21.
SA of association between IL-6 gene rs1800796(G-572C) genotype (CG) and RA.
The PB of the included literature on correlation between IL-6 gene rs1800796(G-572C) genotype (CG) and RA was assessed using a funnel plot (Fig. 22). The symmetrical distribution of data points in the funnel plot suggests the absence of considerable PB. Furthermore, an Egger’s test was employed for further analysis of PB. The results revealed a t = −1.46 and a P = 0.203, implying neglectable PB in the included studies.
Fig. 22.
FUP of correlation between IL-6 gene rs1800796(G-572C) genotype (CG) and RA.
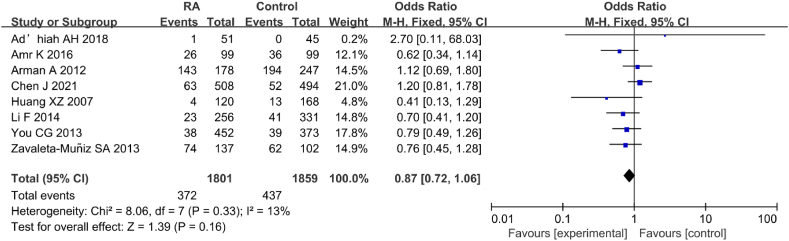
Eight studies analyzed correlation between IL-6 gene rs1800796(G-572C) genotype (GG) and RA (Fig. 23). The statistical analysis revealed neglectable heterogeneity in the statistical results between the RA and CT groups (I2 = 13 %, P = 0.33). Hence, a FEM was employed for the analysis. The findings indicated neglectable correlation between IL-6 gene rs1800796(G-572C) PM and RA [OR = 0.87, 95%CI: 0.72–1.06, Z = 1.39, P = 0.16].
Fig. 23.
FOP of correlation between IL-6 gene rs1800796(G-572C) genotype (GG) and RA.
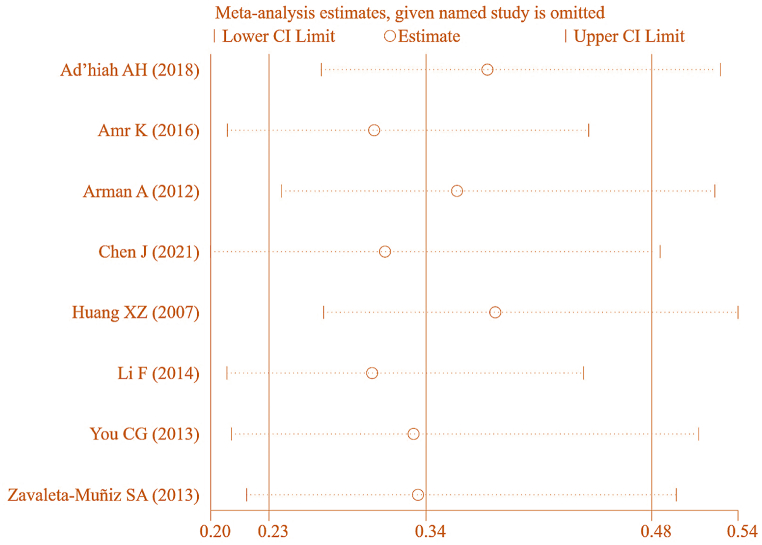
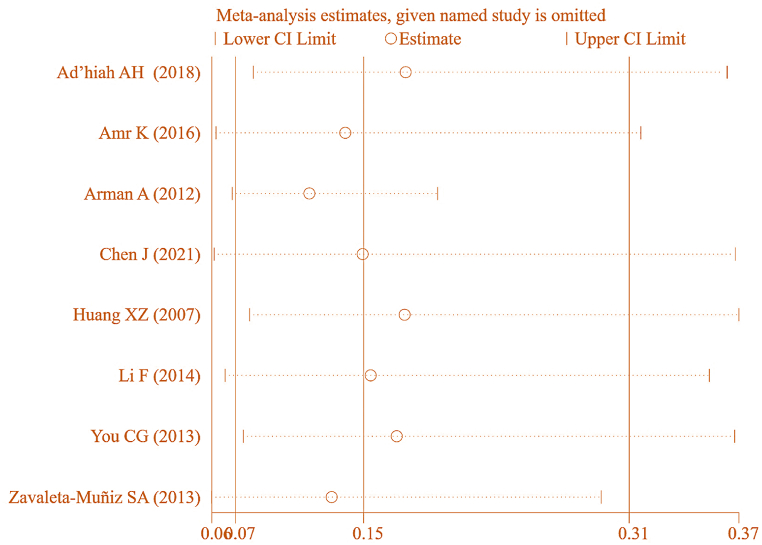
To ensure the accuracy of this result, further SA was conducted on the included literature, as depicted in Fig. 24. Each study examining correlation between IL-6 gene rs1800796(G-572C) genotype (GG) and RA was sequentially removed for MA. The combined effect yielded [OR = 0.148, 95%CI: 0.072–0.307, P < 0.05], implying a correlation between IL-6 gene rs1800796(G-572C) genotype (GG) and RA. This suggests a certain degree of heterogeneity in the results.
Fig. 24.
SA of association between IL-6 gene rs1800796(G-572C) genotype (GG) and RA.
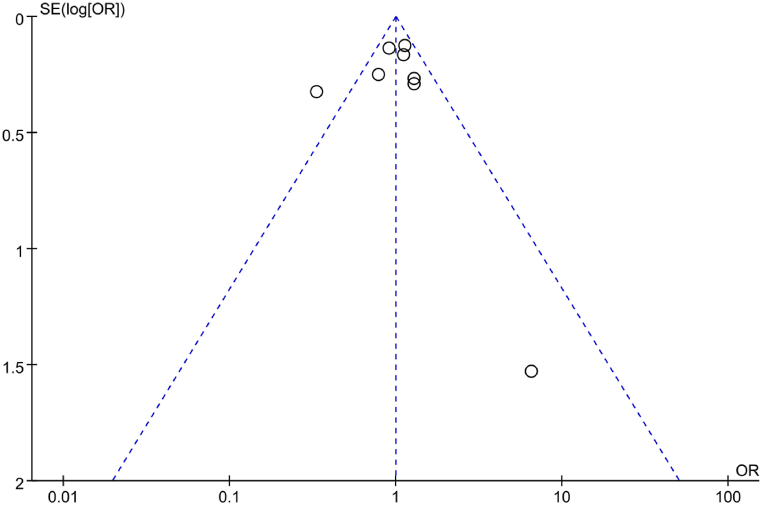
The PB regarding correlation between IL-6 gene rs1800796(G-572C) genotype (GG) and RA was analyzed using a funnel plot (Fig. 25). The symmetrical scatter distribution on the funnel plot indicates the absence of considerable PB. Furthermore, Egger’s test was employed for further analysis of PB. The results revealed a t = −0.49 with a corresponding P = 0.645, suggesting neglectable PB in the included studies.
Fig. 25.
FUP analysis of correlation between IL-6 gene rs1800796(G-572C) genotype (GG) and RA.
Different ethnic groups were categorized into four groups: Asian, Caucasian, Southeast Asian, and mixed. Only one study was available for the Caucasian and mixed groups and hence were not included in the analysis. MA revealed neglectable correlation between IL-6 gene rs1800796(G-572C) genotype (CC) and RA in both Asian and Southeast Asian populations (P > 0.05) (Table 6).
Table 6.
Association between IL-6 gene rs1800796(G-572C) genotype (CC) and RA across different ethnicities.
| Ethnicity | Number of studies included | Sample size | RA group (n/total) | CG (n/total) | Heterogeneity (I2) | OR [95%CI] | P |
|---|---|---|---|---|---|---|---|
| CC | |||||||
| Asian | 4 | 2702 | 647/1336 | 645/1366 | 84 % | 1.24 [0.83, 1.87] | 0.30 |
| Southeast Asian | 2 | 294 | 55/150 | 46/144 | 85 % | 1.07 [0.01,105.82] | 0.98 |
| CG | |||||||
| Asian | 4 | 2702 | 560/1336 | 576/1366 | 77 % | 0.87 [0.62, 1.24] | 0.45 |
| Southeast Asian | 2 | 294 | 67/150 | 58/144 | 10 % | 1.47 [0.61,3.53] | 0.38 |
3.6. Correlation analysis between IL-6 gene rs1800797 (G-597A) and RA
Three studies analyzed correlation between IL-6 gene rs1800797(G-597A) genotype and RA (Table 7). The genotype GG of IL-6 rs1800797(G-597A) exhibited heterogeneity in both the RA and CT groups (I2 = 74 %, P = 0.02), thus a REM was employed for the analysis. The results indicated neglectable association between IL-6 gene rs1800795(G-174C) PM and RA (OR = 0.57, 95%CI: 0.24–1.346, Z = 1.29, P = 0.20). Conversely, the genotype AA of IL-6 rs1800797(G-597A) showed no heterogeneity in the RA and CT groups (I2 = 0 %, P = 0.46), therefore a FEM was utilized for the analysis. The findings revealed neglectable correlation between IL-6 gene rs1800795(G-174C) PM and RA (OR = 1.28, 95%CI: 0.73–12.23, Z = 0.87, P = 0.38).
Table 7.
Correlation analysis between rs1800797 (G-597A) genotype and RA.
| Genotype | Research included | RA group (n/total) | CG (n/total) | Weight/Heterogeneity | OR [95%CI] | P |
|---|---|---|---|---|---|---|
| GG | ||||||
| Arman A | 97/178 | 133/247 | 47.0 % | 1.03 [0.70, 1.51] | ||
| Li F | 249/256 | 330/331 | 12.7 % | 0.11 [0.01,0.88] | ||
| You CG | 418/452 | 359/373 | 40.4 % | 0.48 [0.25, 0.91] | ||
| Merger effect | 74 % | 0.57 [0.24,1.34] | 0.20 | |||
| AA | ||||||
| Arman A | 22/178 | 28/247 | 93.6 % | 1.10 [0.61, 2.00] | ||
| Li F | 2/256 | 1/331 | 3.9 % | 2.60 [0.23,28.82] | ||
| You CG | 3/452 | 0/373 | 2.5 % | 0.48 [0.30,112.96] | ||
| Merger effect | 0 | 1.28 [0.73,2.23] | 0.38 | |||
4. Discussion
IL-6 regulates the onset and progression of arthritis through two distinct signaling pathways. The first pathway involves IL-6 binding to its membrane receptor, IL-6Rα, forming a complex. This complex subsequently influences tyrosine protein kinases 1 and 2, leading to the release of pro-inflammatory cytokines such as TNF-α and IL-17, which ultimately trigger arthritis. The second pathway involves IL-6 binding to the gp130 membrane protein, activating associated JAK family proteins. These JAK proteins then phosphorylate specific residues on gp130, activating downstream STAT proteins, particularly STAT3. Activated STAT3 translocates to the nucleus and regulates the expression of genes associated with inflammation and immune responses, including pro-inflammatory cytokines (e.g., IL-1, TNF-α) and factors related to immune cell proliferation and differentiation. Researchers noted that IL-6 and its receptor significantly stimulate RANKL secretion in synovial tissue, promoting osteoclast maturation and contributing to joint damage.
Studies showed that serum IL-6 levels are elevated in RA patients, and its levels can reflect changes in the disease status of RA [39]. High levels of IL-6 can stimulate B lymphocytes to produce autoantibodies such as rheumatoid factors, thus enhancing the inflammatory response [40,41]. In RA, the synthesis and serum IL-6 are influenced by multiple factors, including PMs in the promoter or coding region of IL-6 gene [42]. IL-6 gene is on chromosome 7p21, and its genetic PMs may affect IL-6 expression levels, thereby influencing the proliferation and secretion of immune cells, leading to worsening of RA symptoms and related tissue damage [43]. Some studies indicated that specific PMs in IL-6 gene are related to susceptibility to RA in certain populations [44,45], and in patients with active RA, serum and synovial fluid levels of IL-6 are greatly elevated, positively correlating with disease activity and severity [46]. Therefore, PMs in IL-6 gene may influence the pathogenesis and clinical manifestations of RA. However, there is currently no consensus on the exact relationship between IL-6 GPMs and susceptibility to RA. Although some studies supported an association between IL-6 GPMs and RA, this correlation has not consistently been observed across different populations and studies, possibly due to population differences, variations in study design, and other unknown factors.
In this MA, a total of 21 studies were identified, comprising 9772 participants, with 4679 RA patients and 5093 controls. The focus was on the analysis of IL-6 GPMs rs1800795 (G-174C), rs1800796 (G-572C), and rs1800797 (G-597A) gene loci. The results showed a notable correlation between IL-6 gene rs1800795 (G-174C) PM (G vs C) and RA [OR = 0.66, 95%CI: 0.51–0.87, Z = 3.02, P = 0.003]. SA revealed a combined effect [OR = 0.541, 95%CI: 0.388–0.754, P < 0.05], implying the potential importance of IL-6 gene rs1800795 (G-174C) PM in the pathogenesis of RA. There was marked genetic PM at IL-6 gene rs1800795 (G-174C) locus in both RA patients and controls, and this PM was negatively correlated with the risk of RA occurrence. This suggests that individuals carrying specific alleles may be less susceptible to RA than others in certain circumstances. Additionally, it was found that only in the Southeast Asian population, IL-6 gene rs1800795 (G-174C) genotype (CC) was notably associated with RA [OR = 15.23, 95%CI: 3.53–65.67, P = 0.00003]. This finding indicates that individuals carrying IL-6 gene rs1800795 (G-174C) genotype (CC) are more likely to develop RA in the Southeast Asian population. This may imply a closer correlation between this genotype and the pathogenesis of RA in the Southeast Asian population compared to other ethnicities. The results indicated an association between IL-6 gene rs1800795 (G-174C) genotype (CG) and RA [OR = 1.54, 95%CI: 1.10–2.17, Z = 2.48, P = 0.01]. Interestingly, this correlation was found to be considerable only in the Asian population, where IL-6 gene rs1800795 (G-174C) genotype (CG) showed a strong correlation with RA [OR = 6.55, 95%CI: 1.28–33.45, P = 0.02]. This suggests a considerable difference in correlation of IL-6 gene rs1800795 (G-174C) genotype (CG) with RA among different ethnic groups. Particularly in the Asian population, individuals carrying IL-6 gene rs1800795 (G-174C) genotype (CG) are more susceptible to RA compared to other ethnicities. Zhang et al. (2022) [47] found in a Han Chinese population that the genotype distribution of IL-6 gene rs1800795 locus was GG > GC > CC, with the heterozygous GC and homozygous CC genotypes, as well as the frequency of the C allele, greatly higher in RA group than in CT group (P < 0.05). This finding, similar to the results of our study, supports association of rs1800795 with RA occurrence in Asian populations. However, researchers suggested that there is no correlation between IL-6 gene rs1800795 (G-174C) genotype and the occurrence of RA, further substantiating correlation of the rs1800795 (G-174C) genotype (CG) with RA in Asian populations [[48], [49], [50]]. Additionally, it was found that IL-6 gene rs1800795 (G-174C) genotype (GG) was associated with RA, with a notable correlation observed in the Asian population and also in the mixed population. The lack of correlation between IL-6 gene rs1800795 (G-174C) genotype (GG) and RA in other ethnicities (including European, Caucasian, and Southeast Asian populations) suggests that the genotype at IL-6 gene rs1800795 (G-174C) locus may have ethnicity-specific effects, leading to varying associations with RA among different ethnic groups.
The MA results indicated neglectable correlation between IL-6 gene rs1800796 (G-572C) and rs1800797 (G-597A) loci and the occurrence of RA. However, SA of multiple results suggests an association, which may be attributed to differences in study design, sample size, genotyping methods, characteristics of study subjects, and statistical methods. Therefore, further research is needed to explore correlation between IL-6 gene rs1800796 (G-572C) and rs1800797 (G-597A) loci and the occurrence of RA.
In summary, there is a notable correlation between IL-6 GPM rs1800795 (G-174C) and the occurrence of RA, while rs1800796 (G-572C) and rs1800797 (G-597A) show neglectable correlation with RA. Due to limitations in the included study data, this study did not identify associations between IL-6 GPMs and the clinical phenotype or serological markers of RA. Moreover, the correlation between IL-6 levels and factors such as comorbidities, disease duration, disease severity, body weight, and lifestyle has not been analyzed. These confounding variables may impact the development of RA. Future research should focus on expanding the sample size and further investigating the relationship between IL-6 GPMs and RA clinical phenotypes, serological markers, and relevant confounding factors. This will provide valuable insights for understanding the pathogenesis and treatment strategies of RA.
5. Conclusion
This study systematically evaluated correlation between IL-6 GPMs and susceptibility to RA using MA. Twenty-one articles were included, assessing the gene frequency distribution of IL-6 GPMs at rs1800795 (G-174C), rs1800796 (G-572C), and rs1800797 (G-597A) loci. The results revealed a notable correlation between the allelic model (G vs C) of IL-6 gene rs1800795 (G-174C) locus and RA. Among different ethnicities, notable correlations were observed between IL-6 gene rs1800795 (G-174C) genotype (GG) and RA in Asian and mixed populations, while the genotype (CC) in Southeast Asian populations was also related to RA. Neglectable correlations were found between rs1800796 (G-572C) and rs1800797 (G-597A) GPMs and RA. These findings are crucial for understanding the role of IL-6 gene in pathogenesis of RA, particularly with potential clinical implications in Asian populations.
CRediT authorship contribution statement
Xiaojuan Hao: Writing – review & editing, Writing – original draft, Visualization, Validation, Data curation, Conceptualization. Huani Zhao: Writing – review & editing, Writing – original draft, Methodology, Investigation, Data curation, Conceptualization. Linhui Zhu: Writing – original draft, Visualization, Validation, Data curation. Zhiteng Li: Writing – review & editing, Writing – original draft, Validation, Formal analysis, Data curation, Conceptualization. Jing Yang: Writing – review & editing, Writing – original draft, Methodology, Formal analysis, Data curation, Conceptualization. Qian Bai: Writing – review & editing, Writing – original draft, Visualization, Formal analysis, Data curation, Conceptualization.
Data availability statement
The data relevant to this study are available in accordance with applicable requirements.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Kerschbaumer A., Sepriano A., Bergstra S.A., et al. Efficacy of synthetic and biological DMARDs: a systematic literature review informing the 2022 update of the EULAR recommendations for the management of rheumatoid arthritis. Ann. Rheum. Dis. 2023;82(1):95–106. doi: 10.1136/ard-2022-223365. [DOI] [PubMed] [Google Scholar]
- 2.Smolen J.S., Aletaha D., McInnes I.B. Rheumatoid arthritis [published correction appears in Lancet. 2016 Oct 22;388(10055):1984] Lancet. 2016;388(10055):2023–2038. doi: 10.1016/S0140-6736(16)30173-8. [DOI] [PubMed] [Google Scholar]
- 3.Gibofsky A. Epidemiology, pathophysiology, and diagnosis of rheumatoid arthritis: a Synopsis. Am. J. Manag. Care. 2014;20(7 Suppl):S128–S135. [PubMed] [Google Scholar]
- 4.Rheumatoid arthritis. Nat. Rev. Dis. Prim. 2018;4 doi: 10.1038/nrdp.2018.2. Published 2018 Feb 8. [DOI] [Google Scholar]
- 5.Jang S., Kwon E.J., Lee J.J. Rheumatoid arthritis: pathogenic roles of diverse immune cells. Int. J. Mol. Sci. 2022;23(2):905. doi: 10.3390/ijms23020905. Published 2022 Jan 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin Y.J., Anzaghe M., Schülke S. Update on the pathomechanism, diagnosis, and treatment options for rheumatoid arthritis. Cells. 2020;9(4):880. doi: 10.3390/cells9040880. Published 2020 Apr 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tornero Molina J., Balsa Criado A., Blanco García F., et al. Expert Recommendations on the Interleukin 6 Blockade in Patients with Rheumatoid Arthritis. Recomendaciones de experto sobre el bloqueo de la interleucina 6 en pacientes con artritis reumatoide. Reumatol. Clínica. 2020;16(4):272–281. doi: 10.1016/j.reuma.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Schoels M.M., van der Heijde D., Breedveld F.C., et al. Blocking the effects of interleukin-6 in rheumatoid arthritis and other inflammatory rheumatic diseases: systematic literature review and meta-analysis informing a consensus statement. Ann. Rheum. Dis. 2013;72(4):583–589. doi: 10.1136/annrheumdis-2012-202470. [published correction appears in Ann Rheum Dis. 2013 Jun;72(6):1110. Murikama, Miho M [corrected to Murakami, Miho]] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandolfi F., Franza L., Carusi V., Altamura S., Andriollo G., Nucera E. Interleukin-6 in rheumatoid arthritis. Int. J. Mol. Sci. 2020;21(15):5238. doi: 10.3390/ijms21155238. Published 2020 Jul 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeuchi T., Yoshida H., Tanaka S. Role of interleukin-6 in bone destruction and bone repair in rheumatoid arthritis. Autoimmun. Rev. 2021;20(9) doi: 10.1016/j.autrev.2021.102884. [DOI] [PubMed] [Google Scholar]
- 11.Li B., Xiao Y., Xing D., Ma X.L., Liu J. Circulating interleukin-6 and rheumatoid arthritis: a Mendelian randomization meta-analysis. 2016 Jul 18;95(28) doi: 10.1097/MD.0000000000003855. [published correction appears in Medicine (Baltimore) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishimoto N. Interleukin-6 in rheumatoid arthritis. Curr. Opin. Rheumatol. 2006;18(3):277–281. doi: 10.1097/01.bor.0000218949.19860.d1. [DOI] [PubMed] [Google Scholar]
- 13.Choy E.H.S., Calabrese L.H. Neuroendocrine and neurophysiological effects of interleukin 6 in rheumatoid arthritis. Rheumatology. 2018;57(11):1885–1895. doi: 10.1093/rheumatology/kex391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X., Teichtahl A.J., Wicks I.P. Interleukin-6 in rheumatoid arthritis - from the laboratory to the bedside. Curr Pharm Des. 2015;21(17):2187–2197. doi: 10.2174/1381612821666150310143332. [DOI] [PubMed] [Google Scholar]
- 15.Liu X., Li L., Wang Q., et al. A novel humanized anti-interleukin-6 antibody HZ0408b with anti-rheumatoid arthritis therapeutic potential. Front. Immunol. 2022;12 doi: 10.3389/fimmu.2021.816646. Published 2022 Jan 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y.C., Chou Y.C., Chen H.C., Lu C.C., Chang D.M. Interleukin-6 and interleukin-17 are related to depression in patients with rheumatoid arthritis. Int J Rheum Dis. 2019;22(6):980–985. doi: 10.1111/1756-185X.13529. [DOI] [PubMed] [Google Scholar]
- 17.Yip R.M.L., Yim C.W. Role of interleukin 6 inhibitors in the management of rheumatoid arthritis. J. Clin. Rheumatol. 2021;27(8):e516–e524. doi: 10.1097/RHU.0000000000001293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emonts M., Hazes M.J., Houwing-Duistermaat J.J., et al. Polymorphisms in genes controlling inflammation and tissue repair in rheumatoid arthritis: a case control study. BMC Med. Genet. 2011;12:36. doi: 10.1186/1471-2350-12-36. Published 2011 Mar 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arman A., Coker A., Sarioz O., Inanc N., Direskeneli H. Lack of association between IL-6 gene polymorphisms and rheumatoid arthritis in Turkish population. Rheumatol. Int. 2012;32(7):2199–2201. doi: 10.1007/s00296-011-2057-x. [DOI] [PubMed] [Google Scholar]
- 20.Chen J., Zhang A., Yang Y., Si Y., Hao D. Assessment of interleukin 6 gene polymorphisms with rheumatoid arthritis. Gene. 2021;765 doi: 10.1016/j.gene.2020.145070. [DOI] [PubMed] [Google Scholar]
- 21.Dar S.A., Haque S., Mandal R.K., et al. Interleukin-6-174G > C (rs1800795) polymorphism distribution and its association with rheumatoid arthritis: a case-control study and meta-analysis. Autoimmunity. 2017;50(3):158–169. doi: 10.1080/08916934.2016.1261833. [DOI] [PubMed] [Google Scholar]
- 22.Li X., Chai W., Ni M., et al. The effects of gene polymorphisms in interleukin-4 and interleukin-6 on the susceptibility of rheumatoid arthritis in a Chinese population. BioMed Res. Int. 2014 doi: 10.1155/2014/265435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F., Xu J., Zheng J., et al. Association between interleukin-6 gene polymorphisms and rheumatoid arthritis in Chinese Han population: a case-control study and a meta-analysis. Sci. Rep. 2014;4:5714. doi: 10.1038/srep05714. Published 2014 Jul 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shafia S., Dilafroze Sofi FA., Rasool R., Javeed S., Shah Z.A. Rheumatoid arthritis and genetic variations in cytokine genes: a population-based study in Kashmir Valley. Immunol. Invest. 2014;43(4):349–359. doi: 10.3109/08820139.2013.879171. [DOI] [PubMed] [Google Scholar]
- 25.You C.G., Li X.J., Li Y.M., et al. Association analysis of single nucleotide polymorphisms of proinflammatory cytokine and their receptors genes with rheumatoid arthritis in northwest Chinese Han population. Cytokine. 2013;61(1):133–138. doi: 10.1016/j.cyto.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Trajkov D., Mishevska-Perchinkova S., Karadzova-Stojanoska A., Petlichkovski A., Strezova A., Spiroski M. Association of 22 cytokine gene polymorphisms with rheumatoid arthritis in population of ethnic Macedonians. Clin. Rheumatol. 2009;28(11):1291–1300. doi: 10.1007/s10067-009-1238-4. [DOI] [PubMed] [Google Scholar]
- 27.Palomino-Morales R., Gonzalez-Juanatey C., Vazquez-Rodriguez T.R., et al. Interleukin-6 gene -174 promoter polymorphism is associated with endothelial dysfunction but not with disease susceptibility in patients with rheumatoid arthritis. Clin. Exp. Rheumatol. 2009;27(6):964–970. [PubMed] [Google Scholar]
- 28.Panoulas V.F., Stavropoulos-Kalinoglou A., Metsios G.S., et al. Association of interleukin-6 (IL-6)-174G/C gene polymorphism with cardiovascular disease in patients with rheumatoid arthritis: the role of obesity and smoking. Atherosclerosis. 2009;204(1):178–183. doi: 10.1016/j.atherosclerosis.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 29.Huang X.Z., Zhuang J.H., Ren Y.G., Zhou L.J., Zhou Q. Association of interleukin-6 and interleukin-18 gene polymorphism with rheumatoid arthritis in Guangdong Han population. Nan Fang Yi Ke Da Xue Xue Bao. 2007;27(11):1661–1664. [PubMed] [Google Scholar]
- 30.Pawlik A., Wrzesniewska J., Florczak M., Gawronska-Szklarz B., Herczynska M. IL-6 promoter polymorphism in patients with rheumatoid arthritis. Scand. J. Rheumatol. 2005;34(2):109–113. doi: 10.1080/03009740510026373. [DOI] [PubMed] [Google Scholar]
- 31.Pascual M., Nieto A., Matarán L., Balsa A., Pascual-Salcedo D., Martín J. IL-6 promoter polymorphisms in rheumatoid arthritis. Genes Immun. 2000;1(5):338–340. doi: 10.1038/sj.gene.6363677. [DOI] [PubMed] [Google Scholar]
- 32.Dahlqvist S.R., Arlestig L., Sikström C., Linghult S. Tumor necrosis factor receptor type II (exon 6) and interleukin-6 (-174) gene polymorphisms are not associated with family history but tumor necrosis factor receptor type II is associated with hypertension in patients with rheumatoid arthritis from northern Sweden. Arthritis Rheum. 2002;46(11):3096–3098. doi: 10.1002/art.10592. [DOI] [PubMed] [Google Scholar]
- 33.Amr K., El-Awady R., Raslan H. Assessment of the -174G/C (rs1800795) and -572G/C (rs1800796) interleukin 6 gene polymorphisms in Egyptian patients with rheumatoid arthritis. Open Access Maced J Med Sci. 2016;4(4):574–577. doi: 10.3889/oamjms.2016.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ad’hiah A.H., Mahmood A.S., Al-kazaz A., Mayouf K. Gene expression and six single nucleotide polymorphisms of interleukin-6 in rheumatoid arthritis: a case-control study in Iraqi patients. Alexandria Journal of Medicine. 2018;54:639–645. [Google Scholar]
- 35.Yucel B., Sumer C., Gok I., Karkucak M., Alemdaroglu E., Ucar F. Associations between cytokine gene polymorphisms and rheumatoid arthritis in Turkish population. North Clin Istanb. 2020;7(6):563–571. doi: 10.14744/nci.2020.70845. Published 2020 Nov 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nisar H., Pasha U., Mirza M.U., et al. Impact of IL-17F 7488T/C functional polymorphism on progressive rheumatoid arthritis: novel insight from the molecular dynamic simulations. Immunol. Invest. 2021;50(4):416–426. doi: 10.1080/08820139.2020.1775642. [DOI] [PubMed] [Google Scholar]
- 37.Zavaleta-Muñiz S.A., Martín-Márquez B.T., Gonzalez-Lopez L., et al. The -174G/C and -572G/C interleukin 6 promoter gene polymorphisms in Mexican patients with rheumatoid arthritis: a case control study. Clin. Dev. Immunol. 2016;2013 doi: 10.1155/2013/959084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melo T.S., Silva M.L.E., Silva Júnior M.L.M., Duarte A.P., Gueiros L.A. Characterization of clinical, laboratory, IL-6 serum levels, and IL-6-174 G/C genetic polymorphisms in patients with rheumatoid arthritis and Sjögren's syndrome. Rev. Assoc. Med. Bras. 1992;67(11):1600–1604. doi: 10.1590/1806-9282.20210665. [DOI] [PubMed] [Google Scholar]
- 39.Popkova T.V., Novikova D.S., Nasonov E.L. Ter. Arkh. 2016;88(5):93–101. doi: 10.17116/terarkh201688593-101. [DOI] [PubMed] [Google Scholar]
- 40.Almeida-Santiago C., Quevedo-Abeledo J.C., Hernández-Hernández V., et al. Circulating interleukin-6 and cardiovascular disease risk in patients with rheumatoid arthritis with low disease activity due to active therapy. Clin. Exp. Rheumatol. 2023;41(7):1537–1543. doi: 10.55563/clinexprheumatol/mr4bka. [DOI] [PubMed] [Google Scholar]
- 41.Rönnelid J., Knight A., Lysholm J., et al. High levels of interleukin-6 in rheumatoid arthritis joint fluids can stimulate local production of C-reactive protein resulting in elevated circulating levels. Joint Bone Spine. 2021;88(3) doi: 10.1016/j.jbspin.2021.105159. [DOI] [PubMed] [Google Scholar]
- 42.Tornero Molina J., Balsa Criado A., Blanco García F., et al. Expert Recommendations on the Interleukin 6 Blockade in Patients with Rheumatoid Arthritis. Recomendaciones de experto sobre el bloqueo de la interleucina 6 en pacientes con artritis reumatoide. Reumatol. Clínica. 2020;16(4):272–281. doi: 10.1016/j.reuma.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 43.Ahmed S., Hussain S., Ammar A., Jahan S., Khaliq S., Kaul H. Interleukin 6 receptor (IL6-R) gene polymorphisms underlie susceptibility to rheumatoid arthritis. Clin. Lab. 2017;63(9):1365–1369. doi: 10.7754/Clin.Lab.2017.170216. [DOI] [PubMed] [Google Scholar]
- 44.Schoels M.M., van der Heijde D., Breedveld F.C., et al. Blocking the effects of interleukin-6 in rheumatoid arthritis and other inflammatory rheumatic diseases: systematic literature review and meta-analysis informing a consensus statement. Ann. Rheum. Dis. 2013;72(4):583–589. doi: 10.1136/annrheumdis-2012-202470. [published correction appears in Ann Rheum Dis. 2013 Jun;72(6):1110. Murikama, Miho M [corrected to Murakami, Miho]] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jarlborg M., Gabay C. Systemic effects of IL-6 blockade in rheumatoid arthritis beyond the joints. Cytokine. 2022;149 doi: 10.1016/j.cyto.2021.155742. [DOI] [PubMed] [Google Scholar]
- 46.Pacheco-Soto B.T., Porchia L.M., Lara-Vazquez W.C., Torres-Rasgado E., Perez-Fuentes R., Gonzalez-Mejia M.E. The association between interleukin-6 promoter polymorphisms and rheumatoid arthritis by ethnicity: a meta-analysis of 33 studies. Reumatol Clin (Engl Ed). 2021;17(8):447–455. doi: 10.1016/j.reumae.2020.03.003. [DOI] [PubMed] [Google Scholar]
- 47.Zhang M., Bai Y., Wang Y., et al. Cumulative evidence for associations between genetic variants in interleukin 6 receptor gene and human diseases and phenotypes. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.860703. Published 2022 Apr 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elkhawaga S.Y., Gomaa M.H., Elsayed M.M., Ebeed A.A. NFKB1 promoter -94 insertion/deletion ATTG polymorphism (rs28362491) is associated with severity and disease progression of rheumatoid arthritis through interleukin-6 levels modulation in Egyptian patients. Clin. Rheumatol. 2021;40(7):2927–2937. doi: 10.1007/s10067-021-05584-z. [DOI] [PubMed] [Google Scholar]
- 49.Lee Y.H., Song G.G. Associations between the interleukin-6 rs1800795 G/C and interleukin-6 receptor rs12083537 A/G polymorphisms and response to disease-modifying antirheumatic drugs in rheumatoid arthritis: a meta-analysis. Int. Immunopharm. 2022;112 doi: 10.1016/j.intimp.2022.109184. [DOI] [PubMed] [Google Scholar]
- 50.Lopez-Lasanta M., Julià A., Maymó J., et al. Variation at interleukin-6 receptor gene is associated to joint damage in rheumatoid arthritis. Arthritis Res. Ther. 2015;17(1):242. doi: 10.1186/s13075-015-0737-8. Published 2015 Sep. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data relevant to this study are available in accordance with applicable requirements.