Highlights
-
•
Conserved IAV antigens are often poorly immunogenic.
-
•
Displaying IAV antigens on VLPs profoundly improved their immunogenicity.
-
•
Diverse biological factories are available to effectively produce IAV VLPs.
-
•
Pain-free IAV immunizations can be accomplished by using VLP technology.
-
•
Nanostructure of VLPs imparts immunological advantage for intranasal vaccination.
Keywords: Virus-like particle, Influenza A virus, Vaccine, Conserved antigens, Pain-free, Production system, Broadly reactive, Clinical trial
Abstract
The threat of influenza A virus (IAV) remains an annual health concern, as almost 500,000 people die each year due to the seasonal flu. Current flu vaccines are highly dependent on embryonated chicken eggs for production, which is time consuming and costly. These vaccines only confer moderate protections in elderly people, and they lack cross-protectivity; thereby requiring annual reformulation to ensure effectiveness against contemporary circulating strains. To address current limitations, new strategies are being sought, with great emphasis given on exploiting IAV's conserved antigens for vaccine development, and by using different vaccine technologies to enhance immunogenicity and expedite vaccine production. Among these technologies, there are growing pre-clinical and clinical studies involving virus-like particles (VLPs), as they are capable to display multiple conserved IAV antigens and augment their immune responses. In this review, we outline recent findings involving broadly effective IAV antigens and strategies to display these antigens on VLPs. Current production systems for IAV VLP vaccines are comprehensively reviewed. Pain-free methods for administration of IAV VLP vaccines through intranasal and transdermal routes, as well as the mechanisms in stimulating immune responses are discussed in detail. The future perspectives of VLPs in IAV vaccine development are discussed, particularly concerning their potentials in overcoming current immunological limitations of IAV vaccines, and their inherent advantages in exploring intranasal vaccination studies. We also propose avenues to expedite VLP vaccine production, as we envision that there will be more clinical trials involving IAV VLP vaccines, leading to commercialization of these vaccines in the near future.
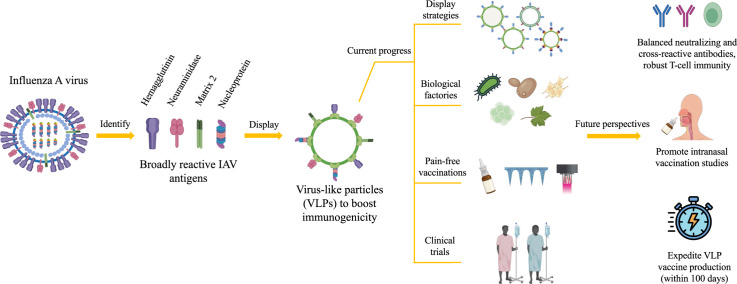
Graphical abstract
1. Introduction
In 1918, influenza A virus (IAV) attained global spotlight when it caused the devastating influenza pandemic, claiming the lives of over 50 million people (Honigsbaum, 2020). The virus caused more pandemics in 1957, 1968 and 2009, killing up to 1 million, 4 million and 200 000 people, respectively, in each series (Honigsbaum, 2020; Simonsen et al., 2013). Now, in 2024, the threat of an IAV pandemic has been greatly mitigated with continuous surveillance, introduction of public health measures (such as wearing masks), and annual flu vaccination. Nonetheless, each year, IAV still infects 5–15 % of the world population, and kills 500 000 people through the infamous seasonal influenza (Petrova and Russell, 2018). Therefore, the battle to minimize IAV menace remains ongoing and steadfast.
IAV of the family Orthomyxoviridae, is a single-stranded, negative-sense, enveloped RNA virus, whose genome has 8 segments that encode for P1, P2, P3, hemagglutinin (HA), nucleocapsid (NP), neuraminidase (NA), matrix (M) and non-structural protein 1 (NS1) (McGeoch et al., 1976). Among these proteins, HA and NA are of major interest as they determine the IAV subtypes, host tropism and immunity (Sam, 2015). So far, 19 different HA and 11 different NA proteins have been identified, theoretically leading to 209 possible subtypes of IAV, with 131 have been detected in nature till date (Fereidouni et al., 2023; Fukuyama et al., 2020; Tong et al., 2013). However, only H1N1 and H3N2 have been the most prevalent subtypes in humans while other subtypes are more dominant in aquatic birds (Kapoor and Dhama, 2014; Tyrrell et al., 2021). Even so, the constant incidence of zoonotic transmissions from avian or swine to humans is indeed an alarming event, as the lack of immunity towards these novel IAV subtypes may commence a new pandemic if a novel IAV develops gain-of-function mutations, enabling effective human-to-human transmission.
Currently, IAV in human population is effectively controlled through vaccination. These vaccines are mostly produced using eggs, either by inactivating (inactivated influenza virus, IIV) or attenuating the IAV (live-attenuated influenza virus, LAIV) (Houser and Subbarao, 2015; Krietsch Boerner, 2020). For those with egg allergies, recombinant HA (rHA) vaccines can be opted, as they are produced using insect cell lines (Houser and Subbarao, 2015). Nevertheless, current IAV vaccine technologies have several limitations. The production of IIV and LAIV are time consuming (6 to 8 months), and highly relies on the availability of embryonated eggs (Chen et al., 2020). Besides, IIV and rHA mainly contain HA proteins derived from contemporary circulating IAVs, and they only induce antibody production in strain-specific manners (Houser and Subbarao, 2015). As IAV HA mutates rapidly, these vaccines require annual reformulation to match with the contemporary IAV strains, with which the general populations are advised to be immunized yearly (Houser and Subbarao, 2015; Strohmeier et al., 2022). Moreover, IIV is less effective in stimulating T-cell immunity, which is crucial for protecting the elderly population (He et al., 2006; Hoft et al., 2011; Liu et al., 2023; Pillet et al., 2019). Unlike IIV and rHA, LAIV comprises all the native antigens of IAV, hence offering relatively greater breadth of protections (Subbarao, 2021). Yet, LAIV is recommended only for young and healthy people due to concerns on possible reversion of the attenuated IAVs to pathogenic strains (Houser and Subbarao, 2015; Soema et al., 2015). To overcome current limitations, researchers are in pursuit for new technologies to rapidly produce broad spectrum IAV vaccines, with great focus on harnessing the conserved antigens of IAVs. However, these conserved antigens are often poorly immunogenic, hence strategies to augment their immunogenicity are being investigated.
Display of foreign antigens on virus-like particles (VLPs) is one of the approaches to enhance the antigens’ immunogenicity. VLPs are nanoparticles constituted of self-assembling viral proteins, devoid of their viral genomes (Tan et al., 2023). The first VLP vaccine was based on the non-infectious particles purified from the sera of Australia antigen (later defined as hepatitis B virus surface antigen, HBsAg) positive patients in 1960s (Beyer et al., 1968). The production of a safer version of this VLP vaccine was actively pursued in the 1970s using recombinant DNA technology (Hofschneider and Murray, 2001). This approach is cheaper, faster and less laborious, which was then adopted to produce recombinant VLP vaccines to combat cervical cancer and hepatitis E caused by human papilloma virus (HPV) and hepatitis E virus (HEV), respectively (Mohsen et al., 2017b; Zhang et al., 2014). Over half a century, many strategies are now available to load antigens/cargoes onto VLPs, and diverse expression systems have been explored to produce them. Moreover, many pain-free methods have been developed to deliver VLPs into human bodies.
Over the years, VLPs have been harnessed to develop IAV vaccines, with special emphasis on generating broadly protective immune responses and circumventing vaccine-escape variants. This is achieved by determining conserved epitopes or antigens of IAVs, displaying them on VLPs, and producing them using diverse expression systems. In this review, we underscore recent findings on conserved IAV epitopes/antigens, methods of displaying them on VLPs, production systems to manufacture these VLPs, and pain-free methods of administering them. Moreover, we also highlight current IAV VLP vaccines in clinical trials, and the future perspectives of this platform in strengthening the strategies to develop mutation-resistant IAV vaccines.
2. Recent findings on broadly reactive IAV antigens
In vaccine designing processes, antigen selection plays a pivotal role in determining a vaccine's effectiveness as the process influences the type of immune responses stimulated, and the breadth of protection offered. For IAV, either through conventional or reverse vaccinology approaches, a few antigens have been identified from the virus that serve as immunogens in vaccine developments. Among these antigens, some of them are prone to antigenic drifts, while the rest are relatively conserved. The latest developments concerning the capacity of HA, NA, matrix 2 (M2) and NP in eliciting broad immune responses in animals are described below and summarized in Table 1, while the immune responses elicited by these antigens are depicted in Fig. 1.
Table 1.
Broadly reactive antigens of influenza A viruses (IAVs), their conserved sites, cross-reactivity with other IAVs and reported immune stimulations.
| Antigen | Region | Conserved sites | Cross-reactivity with other IAVs | Immune stimulations | References |
|---|---|---|---|---|---|
| Hemagglutinin (HA) | Head domain | Residues 87–120 of E site from A/Catalonia/63/2009 or A/South Carolina/1/18 H1N1 strain | H1N1, H3N2, H5N1 | B cell and CD4 T cell | López-Serrano et al. (2021); Vergara-Alert et al. (2012) |
| Receptor-binding sites (positions 134–140, 156–162, 187–191, 224–230) of A/Solomon Islands/03/2006 H1N1 strain | H1N1 (different strain) | B cell | Caradonna et al. (2022) | ||
| Receptor-binding sites (residues Y98, G134, S136, W153, E190, L194) of H1, H2 and H3 IAVs | H1N1, H2N2, H3N2, H9N2, H12N5 | B cell | Ekiert et al. (2012) | ||
| Receptor-binding sites (residues G134, S136, W153, L194) of H3 IAVs | H1N1, H2N2, H3N2, H3N8, H13N6 | B cell | Lee et al. (2014) | ||
| 90-loop and 220-loop of A/Solomon Islands/3/2006 H1N1 strain or A/Hong Kong/3/1968 H3N2 strain | H1N1, H2N2, H3N2, H4N6, H5N1, H6N1, H7N7, H8N4, H9N2, H10N7, H11N9, H12N5, H14N5, H15N9 | B cell | Bangaru et al. (2019) | ||
| 220-loop and residues 99–105 of H3 and H1 IAVs | H1N1, H2N2, H3N2, H4N6, H5N1, H6N1, H7N9, H8N4, H9N2, H10N8, H14N6 | B cell | McCarthy et al. (2021) | ||
| Stem domain | Residues 344–357 of HA from A/New Caledonia/20/1999 H1N1 strain (NCBI accession no: AAP34324) | H1N1, H2N8, H3N3, H4N6, H5N1, H6N1, H7N2, H8N4, H9N2, H10N4, H11N1, H12N5, H13N5 | B cell | Chun et al. (2008) | |
| HA from A/California/07/2009 H1N1 strain | H1N1 (different strain), H5N1 | B cell, CD4 T cell and CD8 T cell | Bliss et al. (2022) | ||
| Stem of A/New Caledonia/20/1999 H1N1 strain and A/Jiangxi-Donghu/346/2013 H10N8 strain | H1N1, H3N2, H5N1, H7N9, H10N8 | B cell | Yassine et al. (2015); Moin et al. (2022) | ||
| Neuraminidase (NA) | N1 head region (computationally optimized) | Different strains of H1N1 and H5N1 | B cell | Skarlupka et al. (2021) | |
| Matrix 2 (M2) | Ectodomain of M2 from A/Puerto Rico/8/1934 H1N1 strain | H6N6, H9N2, H10N8 | B cell, CD4 T cell | Yao et al. (2019) | |
| Ectodomain of M2 from human, swine and avian IAVs | H3N2, H5N1 | B cell, CD4 T cell and CD8 T cell | Lee et al. (2018) | ||
| Nucleoprotein (NP) | Residues 147–155 of NP from H3N2 | H1N1 | CD8 T cell | McGee and Huang (2022) | |
| NP from A/Wilson-Smith/1933 H1N1 strain | H1N1 (different strain), H3N2 | B cell, CD4 T cell and CD8 T cell | Del Campo et al. (2019) | ||
Fig. 1.
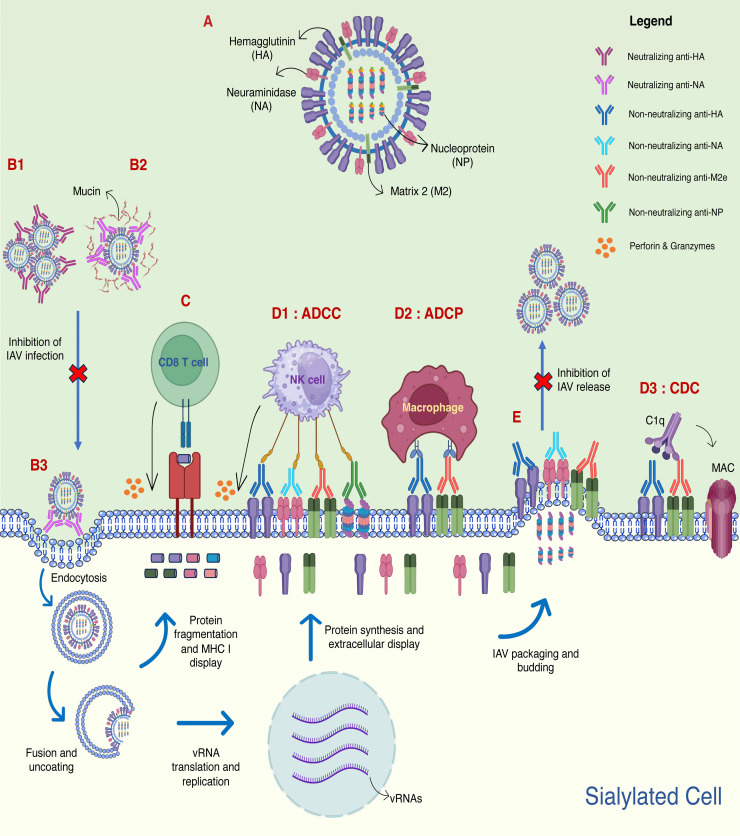
The immune responses induced by the hemagglutinin (HA), neuraminidase (NA), matrix 2 (M2) and nucleoprotein (NP) of influenza A virus (IAV). A schematic diagram depicting HA, NA, M2 and NP in IAV (A). The immune responses stimulated by the antigens include virus neutralization (B1, B2, B3), epitope-specific CD8 T cell cytotoxicity (C), Fc-mediated antibody effector functions (D1, D2, D3), and prevention of virus budding (E). Neutralizing anti-HA antibodies bind to HA, causing viral aggregations that prevent interaction with host's sialic acid (SA) receptor for viral infection (B1). Neutralizing anti-NA antibodies interact with NA's catalytic sites, inhibiting the virus release from sialylated mucins (B2) and sialylated “decoy” receptors on the cell surface (B3), thereby preventing efficient IAV infection. IAV epitopes originated from HA, NA, M2e and NP are displayed on major histocompatibility complex (MHC I), and its interaction with the epitope-specific CD8 T cells (C) causes cell lysis through cytolytic molecules (perforins and granzymes). Non-neutralizing anti-HA, anti-NA, anti-M2e and anti-NP interact with respective antigens displayed on the surface of the infected cell. The fragment crystallizable (Fc) region of the antibodies interacts with Fc receptors on natural killer cells (NK cells), resulting in antibody-dependent cellular cytotoxicity (ADCC) (D1) through secretion of perforins and granzymes. Fc region of non-neutralizing anti-HA and anti-M2e antibodies can also interact with macrophages to eliminate the infected cells through antibody-dependent cellular phagocytosis (ADCP) (D2), or engage with C1q protein that leads to complement-dependent cytotoxicity (CDC) (D3) through membrane attack complex (MAC). The binding of non-neutralizing anti-HA, anti-NA or anti-M2e antibodies on their respective antigens on the cell surface inhibits virus release (E) by preventing NA cleavage of sialic acid from HA (anti-NA), prevention of M2 mediated membrane scission (anti-M2e) and HA cross-linking (anti-HA).
2.1. The whole hemagglutinin (HA) molecule
HA is the major immunogen of IAV as it is highly abundant in number (approximately 500 HA per virion) and relatively larger in size compared to the other viral transmembrane proteins (Bouvier and Palese, 2008; Mancini et al., 2011). When a person is infected with IAV, the body will produce neutralizing antibodies that bind to HA and impedes the virus infectivity (Bouvier and Palese, 2008; Houser and Subbarao, 2015). On this basis, conventional IAV vaccines such as AFLURIA® Quadrivalent and Flublok® Quadrivalent are formulated using the whole HA molecule. However, the whole glycoprotein does not only contain multiple antigenic sites, but it also contains non-antigenic sites that can disrupt an efficient immune stimulation. These non-antigenic peptides may compete with antigenic peptides for loading on MHC class I and II, which can eventually reduce presentations of the antigenic peptides (Adorini and Nagy, 1990). Besides, it is also evident that the whole HA molecule is less efficiently processed by dendritic cells (DCs) compared to peptides (Rosalia et al., 2013). With these reasons and constant antigenic drifts of HA glycoprotein, researchers are more attentive towards studying conserved epitopes within the HA glycoprotein for vaccine design, instead of using the whole HA molecule.
2.1.1. Globular head domain of HA
The head domain of HA contains the receptor binding site (RBS) for infection, and proximal to this site are five antigenic determinant sites (A, B, C, D, E) that are often targeted by anti-IAV neutralizing antibodies (Wang et al., 2022). Despite the high plasticity of this domain, some amino acid residues are well conserved such as those from positions 87 to 120 of the E site from the A/Catalonia/63/2009 H1N1 strain (López-Serrano et al., 2021). By vaccinating pigs with the respective peptide sequence, known as NG34, it was shown to confer protections against homologous and heterologous IAV challenges (López-Serrano et al., 2021). Besides, the RBS is also known to be relatively conserved (mainly the key residues W153, H183, L194, Y195) as mutations at this site may deter the viral infectivity (Wu and Wilson, 2020). Antibodies are usually elicited against this epitope; however, they only make up a smaller fraction of the antibody's repertoire, hence preventing broad protections (Caradonna et al., 2022). Recently, Caradonna et al. (2022) developed an IAV vaccine with enriched RBS epitopes to stimulate RBS-specific humoral responses. The RBS-specific antibodies were able to neutralize a strain-matched virus, and partially protected mice against a lethal H1N1 infection. Nevertheless, its protectiveness against other strains and subtypes was not determined. Despite that, anti-RBS antibodies, such as C05 and F045–092 have shown to neutralize different IAV subtypes in vitro (Ekiert et al., 2012; Lee et al., 2014; Ohshima et al., 2011). Besides, by studying broadly reactive antibodies, more conserved epitopes have been determined such as the HA-220 loop, HA residues 99–105 and HA-90 loop, which are located at the trimer interface of the HA head (Bangaru et al., 2019; McCarthy et al., 2021). These conserved epitopes can be used for vaccine development, instead of the whole HA molecule.
2.1.2. Stem domain of HA
The stem domain of HA is preserved from antigenic mutations as some of the residues act as the pH-threshold for membrane fusion (Wu and Wilson, 2020). However, the stem region is less efficient in eliciting antibody response following natural infections as it is poorly immunogenic (Wu and Wilson, 2020). With the whole HA-based commercial IAV vaccines, anti-stem cross protective antibodies can be generated, but their magnitude is highly dependent on the structural organization of HA and the age of recipients (Myers et al., 2023; Sánchez-de Prada et al., 2023). To harness sufficient anti-stem antibodies, Chun et al. (2008) developed a 14-residue fusion peptide vaccine that is known to be highly conserved across IAVs. The peptide vaccine generated high titer of monospecific antibody, known as Uni-1, and was capable of neutralizing IAVs in vitro, and protected mice against lethal challenge of diverse IAVs and IBVs through antibody-dependent cellular cytotoxicity (ADCC) (Muralidharan et al., 2022). Despite its promising potential as a universal vaccine, the hydrophobicity of the peptide presents a daunting challenge for further studies. Bliss et al. (2022) employed a different approach in eliciting anti-stem immune response, in which the researchers constructed an adenoviral vector encoding a secreted form of H1N1’s HA protein, inferring that this can increase the exposure of stem-derived epitopes. Immunization of mice with this protein provided 100 % protection from homologous, heterologous and heterosubtypic viral challenge. Likewise, to study the protectiveness of stem-directed immune response, Moin et al. (2022) co-immunized mice, ferrets and non-human primates with the HA stem trimers derived from group 1 and 2 IAVs. The stem trimers, which were displayed separately on ferritin nanoparticles, yielded IAV cross-group immunity owing to the broad neutralizing antibodies produced. Overall, these studies indicate that the conserved epitopes within the stem domain can be exploited to induce cross-protective immunity against diverse IAVs, attesting their potential to be further developed into a universal IAV vaccine.
2.2. The whole neuraminidase (NA) molecule
Each IAV virion has approximately 40 to 50 molecules of NA on its surface, making it the second most abundant glycoprotein after HA (Air, 2012; Rajendran et al., 2021). However, conventional IIV vaccine formulation gives less emphasis to this antigen as the vaccine's efficacy is usually measured with hemagglutination inhibition assay (HAI) (Monto et al., 2015). Therefore, elicitation of HA antibody in a host garner relatively more interest (Rosu et al., 2022). But, in the context of cross protections against IAVs, human and animal studies had shown that NA molecule has a lot to offer in the immunity landscape as it is subjected to lesser immune selection pressure compared to HA (Rajendran et al., 2021). Within NA, antigenic sites are located either in the highly conserved catalytic site or outside of this site with nearly 15 different broadly reactive mAbs were mapped within these regions (Creytens et al., 2021; Sun et al., 2022a). A vaccine based on the catalytic site may elicit mAbs that prevent NA's catalytic activity, thereby impeding effective IAV infections from the beginning, while the inclusion of epitopes outside the catalytic site tends to occlude viral egress from infected cells (Creytens et al., 2021). However, most of the currently developed NA vaccines are designed to mainly include the head domain of NA, which constitutes the catalytic site. By using a computational approach, Skarlupka et al. (2021) designed an NA vaccine (N1-I COBRA NA) that cross-reacted with human, avian and swine IAVs. The vaccine induced neutralizing antibodies against a broad panel of HXN1 IAVs, and protected mice against IAV challenges from different origins. With the promising result, the researchers suggested that the NA-based vaccine can be used in concert with HA antigen for the development of a universal IAV vaccine. However, Strohmeier et al. (2021) demonstrated that the NA and HA admixed vaccine may suppress the production of anti-NA antibodies, unless both immunogens are given separately at different inoculation sites. Despite hosting many epitopes that can generate broadly protective antibodies, there are limited number of studies on NA-based vaccines that have been reported in the last five years, which is likely due to the instability of the NA molecule, or it requires proper folding for maximal efficacy (McMahon et al., 2020; Strohmeier et al., 2021).
2.3. Matrix 2 (M2) protein
M2 is a tetrameric IAV membrane protein, with approximately 5–15 copies per virion (Rossman et al., 2010). The relative scarcity of the protein renders it poorly immunogenic. However, when cells are infected with IAVs, high levels of M2 protein will be expressed on the cellular surface (Rossman et al., 2010). Besides, its ectodomain (M2e) is long known to be conserved among human IAVs (including bats’ H17N10 and H18N11), especially its first nine amino acids (Kavishna et al., 2022). These underlying reasons prompted researchers to explore M2e's potential in developing IAV vaccines for broad protections. Yao et al. (2019) developed a H1N1 M2e-based DNA vaccine, and tested its efficacy in conferring cross-protection against other subtypes. The vaccine (p-tPA-p3M2e), designed with a signalling peptide (tPA) to increase the protein secretion, generated immune responses that completely protected BALB/c mice against homosubtypic challenge, and varying degree of protections (80 %-H9N2, 40 %-H6N6, and 20 %-H10N8) against heterosubtypic challenges. The study also demonstrated that the protective efficacy is dependent on the homology between the M2e sequence used in the vaccine and that of the challenged virus. Practically, the limited protection can be addressed by designing heterologous M2e into a single vaccine formulation, as demonstrated by Lee et al. (2018). The VLP-based vaccine displaying M2e sequences derived from human, swine and avian IAVs completely (100 %) protected mice from heterosubtypic challenges.
2.4. Nucleoprotein (NP)
NP does not possess high degree of variability; hence it is ideal for developments of universal IAV vaccines (McGee and Huang, 2022). However, unlike the aforementioned molecules (HA, NA and M2e), NP is located within the viral structure. Thus, it mainly stimulates cytotoxic T lymphocyte (CTL) responses, and contributes less to antibody-mediated reactions. Within the NP, region NP137–182 is evolutionarily conserved, and CTL-response epitope is located in NP147–155 (McGee and Huang, 2022). Empirically, mice immunized with NP147–155 (derived from H3N2) generated CTL responses, resulting in cross-protection against H1N1 challenge, but the protection was rather modest and dependent on the adjuvants used (McGee and Huang, 2022). Instead, NP-based cross-protection can be augmented by formulating it in a more complex form as demonstrated by Del Campo et al. (2019), in which the oligomeric NP vaccine (OVX836) completely protected mice against multiple IAV challenges, thereby paving its way into a phase II clinical trial (NCT05060887) (Leroux-Roels et al., 2023).
Currently, these antigens are present in the commercial vaccine formulations. However, the immunodominance of HA (due to its molecular size and quantity) results in dampened immune stimulation towards other antigens, and limits the breadth of cross-protection. To overcome this, vaccine should be tailored to display these antigens in proportionate quantity or engineer a multi-epitope vaccine (MEV) (based on HA, NA, M2e and NP broadly effective epitopes), in which both can be achieved through recombinant DNA technology. Nonetheless, the elicitation of neutralizing antibodies (contributed by HA and NA) should be of paramount importance, as allowing IAVs to infect and replicate in a host will generate more vaccine-escape variants.
3. Dynamics of displaying IAV antigens on VLPs
The existence of different vaccine technologies (e.g. mRNA, DNA, viral vector, subunit and VLP) facilitates IAV vaccine development, and addresses the shortcomings of contemporary vaccines (Lee et al., 2023; Matsuda et al., 2021; Ong et al., 2019; Saleh et al., 2020; Werninghaus et al., 2023). Among these technologies, the VLP platform often falls within the spotlight as it employs safe, biocompatible, and versatile antigen carriers. Importantly, VLPs are profoundly known to make an antigen more immunogenic through repetitive surface display, which is essential to generate strong B-lymphocyte stimulations.
3.1. VLPs displaying IAV antigens through genetic fusion
Genetic fusion involves the ligation of the coding region of an or a few antigens with the gene of a viral protein that can self-assemble into VLPs, which results in displaying the antigens on the outer surface of the particles. This method ensures high ligation efficiency between the antigens and the VLP monomer, maximizing antigen displayed. Through this approach, up to five copies of IAV's M2e were genetically fused to the C-terminal end of Macrobrachium rosenbergii nodavirus (MrNV) capsid protein, which self-assembled into chimeric VLPs (Yong et al., 2015). Displaying M2e on MrNV VLPs boosted its immunogenicity, and induced M2e specific immune response that fully protected BALB/c mice from IAVs H1N1 and H3N2 challenges (Ong et al., 2019; Yong et al., 2015). Besides, Ramirez et al. (2018) genetically fused the HA stalk (from H1N1) and three copies of M2e (each from H1N1, H5N1 and H11N9), and displayed them using the hepatitis B virus tandem core (HBc) platform. However, instead of genetically fusing these antigens in tandem with each VLP monomer, the monomers were instead designed to display either HA stalk or M2e antigens, otherwise the VLP cannot assemble properly. This alternative strategy ensured proper VLP formation, which allowed different IAV antigens to be displayed properly. The resulting chimeric VLPs, Tandiflu 1, fully protected BALB/c mice from heterologous H1N1 IAV challenge. Nevertheless, genetic fusion can be a laborious and time-consuming process, and the fusion partners may cause steric hindrance in VLP formation. To reduce the steric hindrance, it is recommended to use a small sized fusion partner (McGonigle et al., 2015). Besides, fusing foreign antigens to the viral protein monomers may reduce the yield of VLPs.
3.2. VLPs displaying IAV antigens through co-expression with viral protein monomers
Instead of genetically fusing IAV antigens to VLP monomers, this approach involves separate expression of a backbone protein and IAV antigens within a single expression cell. The protein monomers and IAV antigens will then self-assemble to form VLPs. There are a few viral proteins that can act as monomers, which include the Gag protein of retroviruses (e.g. bovine immunodeficiency virus, BIV; murine leukemia virus, MLV; and simian/human immunodeficiency virus, SHIV) and matrix 1 (M1) protein of IAVs (Guo et al., 2003; Haynes et al., 2009; Sun et al., 2022b; Tretyakova et al., 2013). Sun et al. (2022b) exploited the Gag protein of BIV to co-express hemagglutinins HA3 and HA9, and these independent proteins self-assembled into 180 nm-chimeric VLPs in Spodoptera frugiperda (Sf9) cells. Likewise, the M1 protein of IAV has been used as a protein monomer to display five copies of heterologous M2e and recombinant HA1, NA1, HA3 and NA2 proteins, in which these co-expressions led to the formation of VLPs (Lee et al., 2018; Liu et al., 2023). One major advantage of this method is that it can display IAV antigens in its native conformation (e.g. trimeric HA, tetrameric NA), which imparts less concern on steric hindrance for VLP formation. Nonetheless, these VLPs will be heterogenous in nature, as each particle may display different amount of IAV antigens.
3.3. VLPs displaying IAV antigens through protein conjugation
Protein conjugation is a method in which two or more independent polypeptides are ligated together through covalent bonds (Pihl et al., 2023). Through this method, VLPs can be produced in one expression system, while the IAV antigens in another system, and these two components can be ligated with the presence of a specific ligation site. For instance, the AP205 VLPs (or the AP-M2e VLPs) and the long alpha-helix (LAH; referred as HA tri-stalk protein) were produced separately in E. coli cells (Kirsteina et al., 2020). The HA tri-stalk was introduced with a sulfhydryl group, while the VLPs contain amine groups, allowing conjugation of these two molecules through amine-to-sulfhydryl crosslinking. The newly established AP/tri-stalk and AP-M2e/tri-stalk VLPs were able to protect mice from homologous and heterologous IAV challenges, of which the latter formulation protected against high-dose of H1N1 challenge (Kirsteina et al., 2020). Beside crosslinkers, IAV antigens can also be conjugated to VLPs through the formation of isopeptide bonds, which is a hallmark of the SpyTag-SpyCatcher platform. Sharma et al. (2020) displayed the globular head domain of HA protein on P22 VLPs, by genetically fusing the SpyTag (13 residues) on the VLPs, while the SpyCatcher (116 residue) was fused with the HA. The spontaneous isopeptide bond formed between the lysine of SpyCatcher and the aspartate of SpyTag allows the incorporation of up to 230 copies of HA head domain on the exterior of P22 VLPs. Using the same platform, the stem domain of HA and M2e antigen were displayed separately on norovirus VLPs, with the immune responses against the HA turned out to be more robust compared to M2e antigen (Heinimäki et al., 2022). Moreover, the conjugation efficiency of SpyCatcher-M2e and SpyTag-noro-VLPs was determined to be only 24 %, and it was postulated that the SpyCatcher is relatively immunogenic when short peptides (e.g. M2e ∼20 residue) are used as an immunogen (Heinimäki et al., 2022; Lampinen et al., 2023). The poor immunogenicity of M2e can be overcome by adding aluminium hydroxide adjuvant into the vaccine formulation, as demonstrated by Lampinen et al. (2023).
There are several strategies (Fig. 2) that can be employed to display IAV antigens on VLPs, offering alternatives for researchers. With this inherent flexibility, VLPs can be an ideal platform for IAV vaccine development, especially for poor immunogens like M2e. However, with surging demands for rapid vaccine production (within 100 days), current VLP displaying methods would likely fall behind, unless a rapid and highly efficient methods is available. In this context, a modular displaying platform (such as SpyTag-SpyCatcher) has a bright future, provided that the protein conjugation efficiency is improved beyond current capability.
Fig. 2.
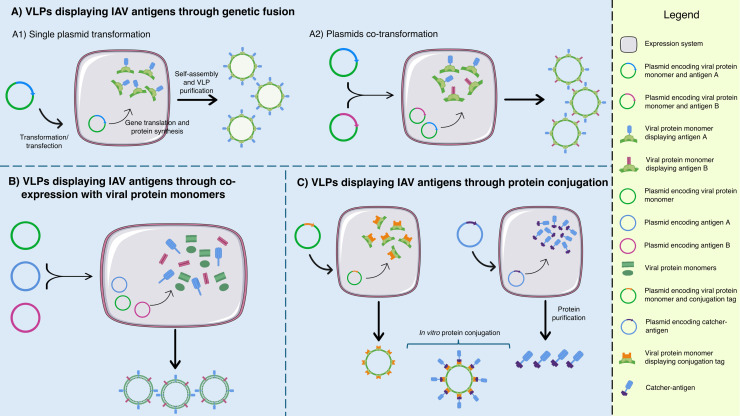
A schematic diagram summarizing the dynamics of displaying IAV antigens on VLPs. Through genetic fusion (A), single (A1) or different IAV antigens (A2) can be displayed on the viral protein monomers (which form the VLPs) by transforming the expression system with a single or different plasmids, respectively. Alternatively, IAV antigens can be displayed on VLPs by co-expression with viral protein monomers (B). This is succeeded by co-transforming the expression system with different plasmids encoding the viral protein monomer and those encoding IAV antigens. IAV antigens can also be displayed on VLPs through protein conjugation (C). Viral protein monomers displaying the conjugation tag are produced in one expression system, while the IAV antigens with complementary catcher are produced in a separate expression system. The VLPs and catcher-antigens are purified and conjugated in vitro.
4. Production systems for VLP-based IAV vaccines
VLP-based IAV vaccines can be produced using bacteria, yeasts, insect cells, mammalian cells and plants. The inherent properties of these biological factories are discussed below, and summarized in Table 2.
Table 2.
Comparison of VLP production in bacterial, yeast, insect cell, mammalian cell and plant expression systems based on specified parameters.
| Parameters | Bacteria | Yeasts | Insect cells | Mammalian cells | Plants |
|---|---|---|---|---|---|
| VLP yield | Up to 250 mg/L culture (Mohsen et al., 2022) | Up to 270 mg/L culture (Yang et al., 2020) | Up to 171 mg/L culture (Lin et al., 2015) | Up to 33.7 mg/L culture (Hirschberg et al., 2023) | Up to 200 μg/g fresh leaf tissue (Mardanova et al., 2022) |
| Growth period prior to VLP production | < 1 day (Sazegari et al., 2023; Yong et al., 2015) | ∼1 to 2 days (Sanchooli et al., 2020; Wetzel et al., 2019) | ∼2 to 3 days | ∼2 to 3 days | 28 days to 42 days (D'Aoust et al., 2008; Rutkowska et al., 2019) |
| VLP production period | 5 h – 18 h (Sazegari et al., 2023; Yong et al., 2015) |
66 h – 96 h (Sanchooli et al., 2020; Wetzel et al., 2019) | 72 h – 144 h (Tretyakova et al., 2013; Xu et al., 2019) | 48 h - 144 h (Buffin et al., 2019; Xu et al., 2020) | 96 h - 192 h (Mardanova et al., 2022; Rutkowska et al., 2019) |
| VLP complexity | Simple | Simple | Complex | Complex | Complex |
| Antigen displaying strategy | Genetic fusion, protein conjugation | Genetic fusion, protein conjugation | Genetic fusion, co-expression with viral protein monomers, protein conjugation | Genetic fusion, co-expression with viral protein monomers, protein conjugation | Genetic fusion, protein conjugation |
| Post-translational modification (PTM) pattern | Lacks PTM | High mannosylation (causes adverse immune reaction and reduces serum half-life) (Khan et al., 2017; Wildt and Gerngross, 2005) | Paucimannosylation and lacks terminal sialylation (reduces serum half-life) (Khan et al., 2017; Tomiya et al., 2004) | Highly resembles human cells (Khan et al., 2017) | Contains xylose and fucose residues (potential allergen), lacks terminal sialylation (Khan et al., 2017; Mercx et al., 2017) |
| Scalability | Easy | Easy | Difficult | Difficult | Easy |
| Production cost | Low | Low | High | High | Low |
4.1. Bacterial production system
Bacteria, mainly Escherichia coli (E. coli), have been widely used as a recombinant protein production system since 1970s, as they require simpler cultivation methods, proliferate rapidly, express high level of proteins, and have high scalability (Yang et al., 2021; Zhang et al., 2022). Beyond these features, the presence of diverse E. coli strains engineered to serve specific applications greatly benefits IAV VLP production through these cell factories. Using the TOP10 strain, which is known to be resilient to stress conditions (such as osmotic shock and acidic pH), Yong et al. (2015) produced MrNV VLPs displaying the M2e antigens. The purified chimeric VLPs protected mice against heterologous IAV challenges (Ong et. al 2019). Meanwhile, for BL21 (DE3) strain, it is one of the most salient strains of E. coli as it can grow relatively faster, with superior secretion capabilities (Rosano et al., 2019; Yoon et al., 2009). Additionally, this strain lacks proteases, which can extend the lifetime of recombinant proteins (Rosano et al., 2019). Using BL21(DE3), Kirsteina et al. (2020) produced AP205 VLP displaying M2e antigens (AP-M2e) and chemically coupled HA tri-stalk to form a separate VLP (AP-M2e/tri-stalk). The latter VLP was demonstrated to protect BALB/c mice completely from lethal homologous IAV challenge. A major immunological advantage of this expression system is that the VLPs produced tend to entrap prokaryotic RNAs (pRNAs). The pRNAs are known to act as ligands for toll-like receptor 7 (TLR7) and promote IgG2 class-switching, which plays an important role in protecting against IAV-derived mortality and morbidity (Gomes et al., 2019). Nonetheless, bacterial production system is only suitable to generate IAV VLPs with low complexity, as it lacks post-translational modifications (PTM) (Yang et al., 2021). Without PTM, the monomers of complex VLPs may not fold or assemble properly, resulting in unstable and insoluble particles.
4.2. Yeast production system
The first genetically engineered VLP-based hepatitis B vaccine (Recombivax HB) was produced in yeast and approved for use in West Germany and the USA in 1986 (Huzair and Sturdy, 2017). The advantages of the yeast production system include low production cost, short production time, easily scaled up, free from endotoxin issues, and products are post-translationally modified (Kim and Kim, 2017; Srivastava et al., 2023; Taguchi et al., 2015; Yang et al., 2021). Therefore, the yeast production system serves as an alternative to address solubility and misfolding issues commonly faced by the bacterial production system. On the industrial scale, Saccharomyces cerevisiae (S. cerevisiae) and Pichia pastoris (P. pastoris) are frequently used for production of clinically relevant proteins, as they are categorized as generally recognized as safe (GRAS) (Srivastava et al., 2023; Taguchi et al., 2015). Despite the abovementioned advantages, there are limited studies on the production of IAV VLP vaccines using this system in the past five years. Ionescu et al. (2006) produced human papilloma virus (HPV) VLPs in S. cerevisiae, and conjugated approximately 4000 copies of IAV M2e peptide containing an unnatural amino acid (6-aminohexanoic acid) onto the exterior of each VLP. This vaccine protected mice from IAV infections in dose-dependent manner (Ionescu et al., 2006). Meanwhile, Pietrzak et al. (2016) produced the extracellular domain of HA (from H5N1) in P. pastoris. The recombinant protein formed VLPs, and protected chickens from an H5N1 challenge (Pietrzak et al., 2016). Using P. pastoris, Kazaks et al. (2017) produced HBc VLPs displaying the conserved and hydrophobic domain of the HA stalk (known as LAH domain). The IAV VLP vaccine was produced in a 30-litre bioreactor, with a relatively low production yield, but compensated with an efficient purification process. This vaccine induced cross-reactive antibodies against both groups 1 and 2 HA proteins (Kazaks et al., 2017). Although the yeast production system offers PTM, the recombinant protein may succumb to hyper-mannosylated N-glycans, resulting in reduced in vivo half-life and tissue distribution (Li et al., 2022; Srivastava et al., 2023; Tariq et al., 2022). Fast clearance of IAV VLP particles may severely undermine the vaccine's immunogenicity.
4.3. Insect cell production system
Insect cells have gained popularity lately for VLP-based vaccine production, mainly contributed by their capability to synthesize complex proteins in their functional forms. In addition, this system can produce high protein yield in a short time (Tariq et al., 2022). The main component in this system is the baculovirus, which acts as an expression vector by infecting insect cells, which results in expression of recombinant proteins. By adopting this method, Shi et al. (2023) generated IAV VLPs displaying the stalk domain and four copies of M2e, with M1 protein as the backbone protein. The vaccine produced in Sf9 insect cells, conferred protection against homologous and heterologous viral challenges in mice (Shi et al., 2023). However, the progenies of baculovirus can be a common contaminant that is hard to separate from the VLPs (Matsuda et al., 2020; Tariq et al., 2022). Moreover, the infected cells will die upon baculovirus infection, hence requiring fresh insect cells for continuous vaccine production (Matsuda et al., 2020). To address this shortcoming, Matsuda et al. (2020) produced IAV VLPs in recombinant insect cells (Trichoplusia ni BTI-TN-5B1-4) without the baculovirus expression system. The cells were co-transfected with expression plasmids carrying the HA and M1 genes downstream of the Drosophila immunoglobulin heavy chain binding protein (BiP) signal sequence, which enhances recombinant protein production (Matsuda et al., 2020). Unlike the baculovirus-insect cell system, the recombinant insect cell line can be harnessed for continuous IAV VLP production. Although the insect cell production system is vastly used in pre-clinical VLP vaccine developments, it always associates with high production cost and low scalability (Gupta et al., 2023; Srivastava et al., 2023).
4.4. Mammalian cell production system
Mammalian cells have most of the machinery to produce highly complex VLPs, especially enveloped VLPs containing multiple structural proteins. Compared to the above production systems, the PTM of recombinant proteins in mammalian cells are deemed to be accurate, appropriate and human-like in their structures (Bryan et al., 2021; Gupta et al., 2023). Buffin et al. (2019) produced IAV VLPs by transfecting 293 T cells with the expression vectors encoding HA, NA and M1 proteins. The VLPs resembled authentic IAVs in terms of particle morphology, size and fine structure of the surface spikes (Buffin et al., 2019). Furthermore, the VLPs presented HA and NA at the proportion of 1:1, compared to that of 4:1 in native IAVs. Therefore, it is postulated that these IAV VLPs could generate more anti-NA antibodies compared to IIV vaccines, with which the former may offer better cross-protective effects. Although the researchers did not study such comparison, their immunogenicity study demonstrated that the HI and NI titers were relatively high even at lower doses (Buffin et al., 2019). Meanwhile, a study by Venereo-Sánchez et al. (2019) raised few possible issues with IAV VLP production in mammalian cells. The researchers produced the VLPs by transfecting HEK-293SF cells with plasmids encoding the HA, NA (IAVs) and Gag genes (HIV-1) (Venereo-Sánchez et al., 2019). Apart from the IAV VLPs, extracellular vehicles (EVs) of similar size were also co-produced (Venereo-Sánchez et al., 2019), possibly complicating the VLP purification process if a size-based purification method, such as size exclusion chromatography, is to be employed. Moreover, proteomic analysis revealed that the VLPs contained nucleolin, a host cell protein that interacts specifically with Gag protein (Venereo-Sánchez et al., 2019). As this host protein co-assembled with the VLPs; thus, a complete removal of the contaminant is a big challenge, in which the native structures and functions of the VLPs cannot be properly preserved. As a precaution, any production of VLPs involving the Gag protein should be designed carefully, as nucleolin can be an inevitable contaminant. Nevertheless, the mammalian cell production system is only practical to produce VLPs carrying big antigens (e.g. HA and NA molecules), as the system is expensive, prone to contaminations and very difficult to scale up (Gupta et al., 2023; Srivastava et al., 2023).
4.5. Plant production system
Molecular farming, the term used for production of recombinant proteins through plants, is relatively uncommon in IAV VLP vaccine production (Rybicki, 2020). There are few underlying reasons that discourage researchers from using the plant production system. Plants can take weeks, months or even years to reach the desired maturity (Srivastava et al., 2023). Besides, seasonal variations may affect this system (Srivastava et al., 2023), while the intrusion of plant insects can severely harm the sustainability of the plants. Unlike the aforementioned production systems, researchers have less control on the variables of the plant production system, thereby it is relatively more challenging. Nonetheless, a group of researchers produced HA5-VLPs by expressing HA from H5N1 in Nicotiana benthamiana (41–44 days old) through agroinfiltration (D'Aoust et al., 2008). The enveloped VLPs protected mice and ferret from homologous and heterologous IAV challenges in a dose-dependent manner (D'Aoust et al., 2008; Landry et al., 2010). The team then conducted a phase I clinical trial (NCT00984945) to demonstrate the vaccine's safety in healthy individuals aged 18–60 years old (Landry et al., 2010). Recently, the team developed quadrivalent IAV VLPs (QVLPs), and conducted phase I-II, phase II and phase III clinical trials to evaluate the vaccine's safety, immunogenicity and efficacy across a wide range of ages (Pillet et al., 2016; Pillet et al., 2019; Ward et al., 2020). Although the plants can produce up to 1500 doses of IAV vaccine per kg leaves, it took up to 48 days to harvest these VLPs (D'Aoust et al., 2008). The long production time will be a major drawback of this system, unless the plants can be grown faster through specific conditions.
When opting for a production system, researchers need to consider the developmental and manufacturing phases. In the developmental phase, the complexity of VLPs will determine the suitable production system. Complex VLPs can be produced suitably in insect cells, mammalian cells or plants. Meanwhile, to produce simple VLPs (which would not require complex protein folding or display of large antigens), bacteria and yeasts are more desirable as these production systems offer high yield, rapid productions and ease of scaling up; which are suitable for manufacturing purposes. Besides, the PTM patterns of these production systems are another aspect to consider as they might affect vaccines’ effectiveness in end-users. For this, apart from mammalian cells, parasite-based system (Leishmania tarentolae) also possesses similar PTM patterns to human cells, and are growingly utilized to produce VLPs (de Oliveira et al., 2019; Khan et al., 2017; Panasiuk et al., 2021; Zimna et al., 2023). To the best of our knowledge, there is still no IAV VLP vaccine produced using the Leishmania expression system (LEXSY).
5. Pain-free administration methods for IAV VLP vaccines
The conventional method for vaccination has been through injection, either intramuscular or subcutaneous (Tizard, 2021), as it is easy to execute and ensures sufficient stimulation of immune responses. However, this method is painful for most individuals, thereby causing vaccine hesitancy due to trypanophobia. With technological advancements, novel vaccine delivery methods (Fig. 3) have been scrutinized, wherein VLP-based IAV vaccines have been embracing these innovations through pre-clinical studies.
Fig. 3.
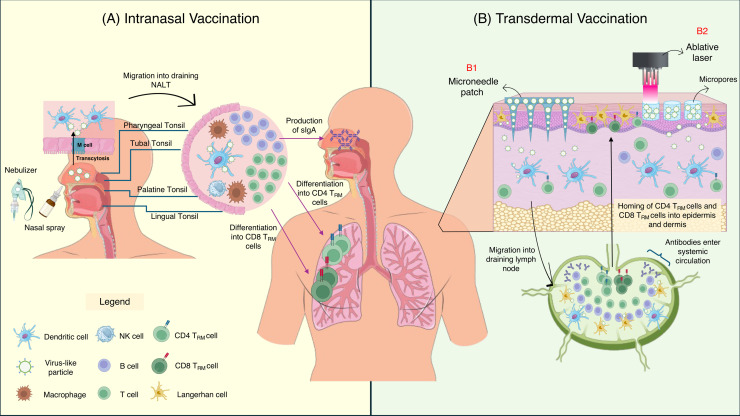
Pain-free IAV VLP vaccine administrations through intranasal and transdermal routes. In intranasal vaccination (A), IAV VLPs are inoculated into the nostrils using either a nasal spray or nebulizer. Upon inoculation, the VLPs are transported across the follicle-associated epithelium (FAE) via transcytosis by microfold (M) cells, and engulfed by nearby dendritic cells (DCs). The activated DCs then migrate into the draining nasal-associated lymphoid tissues (NALT), comprising of pharyngeal tonsil, tubal tonsil, palatine tonsil and lingual tonsil. The DCs then stimulated epitope-specific T cells and B cells, leading to lymphocyte proliferation and differentiation. A subset of CD4 and CD8 T cells differentiates into tissue-resident memory T cells (TRM), which reside at the nasal tissues and lungs. Upon next encounter with the same antigen, these TRM will rapidly proliferate and eliminate the IAV-infected cells. The activation of B cells results in production of secretory IgA (sIgA), which is secreted into the mucous lining to bind with IAV and prevent viral infection right at the viral entry site. Meanwhile, in transdermal vaccination (B), IAV VLP vaccines can be inoculated either through a microneedle patch (MP) (B1) or an ablative laser (B2). For MP, IAV VLPs are loaded onto the MP and applied onto the skin, which releases them slowly into the dermis and epidermis. Meanwhile, an ablative laser is used to create micropores on the epidermis, which acts as depots for sustained IAV VLPs release into the skin. Upon VLPs entry into the epidermis and dermis, the particles are engulfed by Langerhan cells (LCs) and dermal dendritic cell (dDC), which are abundantly present in these regions. The activated LCs and dDCs migrate towards the draining lymph node (LN), and results in activation, proliferation and differentiation of epitope-specific T cells and B cells. Subsets of CD4 and CD8 T cells differentiate into TRM, which migrate and reside in the skin, while other subsets such as T-effector memory cells (TEM) migrate to the peripheral organs for immunosurveillance. Meanwhile, activated B cells differentiate into plasma cells (to produce antibodies) and memory B cells (migrate to the peripheral organs for immunosurveillance).
5.1. Intranasal
Intranasal vaccinations are usually done using either nasal spray or nebulizer (Xi et al., 2021). This route of vaccination is highly suitable to combat respiratory viruses, as the mucosal immune response stimulated can prevent the viral infection and transmission from the viral entry site itself. Intranasal immunization of mice with chimeric cytokine (CC) and HA-incorporated VLP (CCHA-VLP) provided broad protective effects against IAVs of human and avian origins (Imagawa et al., 2023). Although the immunogenicity was relatively low (compared to intraperitoneal injection), the presence of adjuvant (sesame oil) was shown to improve the vaccine's efficacy (Imagawa et al., 2023). The subpar immune response has been the major stumbling block of intranasal vaccination, but now it is evident that adjuvants may help to overcome this issue. Meanwhile, Lee et al. (2018) vaccinated mice with M2e5x VLPs through the intranasal route, and the mucosal immune response cross-protected the mice against heterosubtypic IAV challenge. Despite protecting the mice, the anti-M2e antibody was unlikely to prevent IAV infections in the respiratory tract and viral transmission to other organisms as this antibody has no neutralizing potential (Sedova et al., 2019). Therefore, IAV antigens that can stimulate neutralizing antibodies, such as HA, are of high preference for intranasal vaccination. Nevertheless, intranasal inoculation of an IAV VLP vaccine displaying M2e (or other antigens such as NA and NP) may enhance the breadth of protection in the respiratory tract.
5.2. Microneedle patch
Microneedle patch (MP), which looks like band-aid patches, is made of micron sized needles (50–900 μm) to deliver vaccines into the epidermis and dermis, devoid of any pain (Menon et al., 2021). As these regions contain many Langerhans cells and dendritic cells (Menon et al., 2021), the antigen uptake and subsequent immune stimulations can be effectively boosted. Quan et al. (2010) substantiated this concept by showing that the mice vaccinated with IAV VLPs through MP elicited superior immunity compared to intramuscular shots. In more recent study, Gomes et. al (2023) developed dissolvable MP to deliver M2e5x VLPs. As the patch was applied on the mice, the microneedles dissolved into the mice's skin, releasing the M2e5x VLPs. Although this formulation is immunogenic, but to boost it further, the M2e5x VLPs were encapsulated with poly(lactic-co-glycolic) acid (PLGA) nanoparticles to support a sustained release into the skin (Gomes et al., 2023). Beyond the immunogenicity standpoint, MP can be a remarkable platform in the global vaccination arena, mainly driven by its feasibility and lower vaccination costs (Avcil and Çelik, 2021). However, the stability of IAV VLPs in MP needs to be studied thoroughly before it can be adopted to humans.
5.3. Ablative laser
Ablative laser is a novel method to deliver vaccines through the skin (transdermal) by creating micropores of desired size and depth, using precise laser epidermal system (P.L.E.A.S.E), in a painless manner (Joshi et al., 2021; Scheiblhofer et al., 2013). This micropores can then be inoculated with a vaccine to stimulate the skin-associated lymphoid tissue (SALT) (Scheiblhofer et al., 2013). Through this approach, strong immune responses can be stimulated, as the micropores are believed to act as vaccine depots for a sustained antigen supply (Chen et al., 2012; Scheiblhofer et al., 2013). Thereby, Gomes et al. (2022) tested this method by pipetting a vaccine suspension [M2e5x VLP-loaded bovine serum albumin (BSA) microparticles with adjuvants] into the laser-made micropores (2 mm-depth) on the mice's skin. The VLP-microparticle vaccine yielded comparable immune response to an IIV vaccine, which was inoculated with the same way (Gomes et al., 2022).
It must be notable that not all vaccine platforms can support different methods or routes of vaccination. For instance, LAIV (Fluenz® Tetra) is only indicated for intranasal vaccination and contraindicated for any injections as the attenuated viruses within the vaccine are designed to replicate at the cooler temperature of the upper respiratory tract (AstraZeneca 2022; Subbarao, 2021). Besides, IIV (Fluzone® Quadrivalent) is not advisable to be given via any other routes apart from intramuscular (Sanofi Pasteur, 2022). On this note, VLPs are more versatile and can be administered via the pain-free administration methods. We forecast that more in-depth studies will take place for painless IAV vaccinations, particularly those based on VLPs.
6. Broadly effective VLP-based IAV vaccines in the clinical trials
The current paradigm in IAV vaccine development is to develop a broadly protective vaccine against different IAV subtypes and strains. Besides, the vaccines can be used by a wide range of age groups, and induce humoral and cell-mediated immune responses, which are often lacking in current vaccines. With these expectations, VLPs are being engineered to display conserved IAV epitopes, and some of which have entered clinical trials.
6.1. HA-based VLP vaccine
Despite having the highest mutation rate, HA still contains many conserved epitopes, hence rendering its relevance for IAV vaccine development. Recently, a quadrivalent HA VLP (QVLP) vaccine was produced from the plant Nicotiana benthamiana (Pillet et al., 2016). In a phase I-II clinical trial (NCT01991587), the vaccine induced cross-reactive antibodies and T-cell responses against heterologous HA antigens. Unlike commercial IAV vaccines that usually constitute 15 μg HA/strain for licensure, the QVLP vaccine fulfilled licensure criteria (including HI titer > 40) at much lower doses (3 μg HA/strain), thereby demonstrating the potency of the VLP in boosting immune responses (Pillet et al., 2016). Even so, to induce a consistent immune response in a wider age group (18–49 and ≥50 years old), 30 μg HA/strain was found to be the ideal dosage from two phase II trial studies (NCT02233816 and NCT02236052) (Pillet et al., 2019). With these results, Ward et al. (2020) conducted two phase III clinical trials (NCT03301051 and NCT03739112) using the QVLP vaccine. Relative to commercial IAVs (quadrivalent influenza vaccine, QIV), the QVLP vaccine demonstrated comparable protective efficacy in 18–64 years old cohort. But for older population (≥ 65 years old), the QVLP induced significantly greater CD4 T cell responses compared to QIV, with which people of this age group highly rely on for protection (Ward et al., 2020). The QVLP vaccine can be produced within 6–8 weeks upon identification of a new IAV strain, hence offering a viable alternative for influenza vaccine that is safe and cross-protective, including for elderly populations.
6.2. HA, NA and M1-based VLP vaccine
The VLP vaccine formed by co-expression of M1 (H5N1), the whole HA and NA (H7N9) glycoproteins in Sf9 insect cells protected mice from homologous viral challenge and produced cross-reactive antibodies against heterologous virus (H7N3) (Smith et al., 2013). With this pre-clinical result, a phase I clinical trial was conducted (NCT01897701) involving 284 adults (≥18 years old) (Fries et al., 2013). By inoculating 15 μg and 45 μg of the VLP vaccine, 5.7 % and 15.6 % recipients, respectively, developed hemagglutination-inhibition (HAI) reciprocal antibody titers ≥40. However, with the adjuvant (ISCOMATRIX), 60 % of the recipients developed HAI antibody titers ≥40. The best formulation was 5 μg VLP with 60 units adjuvant, with which 80.6 % recipients showed HAI antibody titers ≥40. Interestingly, the VLP vaccine also induced significant NA-inhibiting antibodies in 71.9 % (non-adjuvanted vaccine), 92.0 % (with 30 units of adjuvant) and 97.2 % (with 60 units of adjuvant) of the recipients, which is expected to enhance the breadth of antibody-mediated protections (Fries et al., 2013). The vaccine can be ready for human inoculation within 3 months from initial determination of novel HA and NA protein sequences, fulfilling the 100-day goal of international epidemic preparedness coalition (Coalition for Epidemic Preparedness Innovations, CEPI).
Using the same IAV antigen combination (HA, NA, M1) and the Sf9 expression system, CPL Biologicals produced a trivalent influenza vaccine and commercialized it under the trademark name Cadiflu-S® (Carascal et al., 2022; Charlton Hume et al., 2019; Sparrow et al., 2021). The vaccine contains the HA from H1N1, H3N2 and influenza B virus (Cadila, 2016). Not much information is available for the clinical trial of this vaccine, but it is learnt that the vaccine conferred ≥70 % seroprotection rate from its phase III clinical study (Cadila, 2016).
6.3. M2e-based VLP vaccine
M2e is known to be a highly conserved antigen of IAV, but it is poorly immunogenic in its native form. By displaying the antigen on the VLP of HBc protein, and in the presence of entrapped nucleic acids, the immunogenicity was greatly augmented (Ibañez et al., 2013). The vaccine (VLP-1965) was shown to induce a strong Th-1 biased immune response in BALB/c mice, resulting in 100 % survival rate, and reduced morbidity upon challenge with IAV (Ibañez et al., 2013). Moreover, the research team also demonstrated that the presence of antibodies toward the carrier (HBc) neither affected the stimulation of anti-M2e antibodies nor the protective efficacy of the vaccine (De Filette et al., 2008). A phase I clinical trial of this vaccine (renamed as ACAM-FLU-A; NCT00819013) that involved 87 participants aged 18–40 years old, revealed that it was safe and immunogenic (NIH, 2012). Seroconversion was seen in all vaccinated groups, with the highest seen in the group vaccinated with the presence of an investigational adjuvant (QS-21) (NIH, 2012).
In a completely independent study, four copies of M2e (originated from human IAV and highly pathogenic avian IAV H5N1) were displayed on the HBc VLPs (Tsybalova et al., 2015). The HBc/4M2e vaccine conferred 75–100 % protection in mice challenged with lethal doses of IAV. Importantly, this study demonstrated that the HBc VLP displaying human or avian M2e induced better cross-reactive immune responses against heterologous IAVs compared to the VLP displaying both the human and avian M2e (Tsybalova et al., 2015). The vaccine (called Uniflu) was then extended to phase I clinical trial (NCT03789539), involving 54 participants aged 18–60 years old (Mezhenskaya et al., 2019). The outcomes of this human trial are highly anticipated.
The progression of VLP-based IAV vaccines into clinical trials indicates their potential as alternatives for IIV vaccines, which currently accounts for 89.6 % of global influenza vaccine production and consumption (Sparrow et al., 2021). Despite only a few VLP-based IAV vaccines have advanced into clinical stages in the past decade, the pre-clinical studies involving VLPs have increased significantly. Hence, we foresee more VLP-based IAV vaccines will enter clinical trials in the near future, and eventually lead to more commercializations.
7. Future perspectives of VLPs in IAV vaccine development
The future perspective of VLPs in IAV vaccine development greatly relies on the strategy to alleviate influenza A disease. Currently, commercial flu vaccines mainly aim to generate neutralizing antibodies against seasonal IAVs, during which this approach causes immune pressure, and results in emergence of vaccine escape variants. To overcome this issue, IAV vaccines in pre-clinical studies are tailored to confer a broad protection against heterologous IAVs. At times, these vaccines give less attention on the importance of neutralizing antibodies. We opine that IAV vaccines should be formulated to generate neutralizing antibodies (to prevent the viral infection), and also with other conserved antigens, as to safeguard against newly emerging variants and eliminate the viral infected cells via antibody-dependent cellular cytotoxicity (ADCC), antibody-dependent cellular phagocytosis (ADCP) or complement-dependent cytotoxicity (CDC). Preventing IAV infection will disable the virus capacity to generate new variants from its own error-prone RNA polymerase, while immune responses derived from the conserved antigens will act as a safety net against novel variants by eliminating IAV infected cells, thereby inhibiting virus release and preventing subsequent infections. We strongly believe that VLPs hold potentials in materializing this strategy.
Seasonal flu vaccines, such as IIV and LAIV, contain all the IAV antigens in the formulation, yet HA is more immunodominant relative to other antigens (NA, M2e, NP) due to its abundance and larger size. As a result, immune responses are biased towards HA (Skarlupka et al., 2021). VLPs can be tailored to display multiple antigens; therefore, they can be designed to display different combinations of IAV molecules (HA, NA, M2e, NP) at equal proportions, which will induce a more balanced immune response, manifesting both neutralizing potential and broader cross-reactivity. To display these antigens proportionally, a simple, rapid, robust and modular approach is a good option. There are several inventions in the Catcher/Tag toolbox that can fulfil these criteria including the DogCatcher-DogTag, Jo-In and SpyCatcher003-SpyTag003 (Bonnet et al., 2017; Fan and Aranko, 2024; Keeble et al., 2019; Keeble et al., 2022). If adjuvants are needed as part of the vaccine formulation, the internal cavity of the VLPs can be packaged with adjuvants such as CpG, while VLPs with lipid bilayers can be incorporated with cytokine adjuvants (GPI-GM-CSF and GPI-IL-12) through protein transfer (Mohsen et al., 2017a; Park et al., 2023). The seamless incorporation of adjuvants into IAV VLPs using these latest inventions will maximise their capability as a vaccine platform, effectively boosting vaccine immunogenicity through a more economical approach.
Immunogenicity wise, VLPs are commonly known to induce balanced humoral and cell-mediated immune (CMI) responses (Hodgins et al., 2017). The latter response is essential for people who experience immunosenescence, as they are incapable of producing high level of protective antibodies against IAVs (Cadar et al., 2023; Liu et al., 2023; Pillet et al., 2019). Moreover, the effective induction of T-cell immunity is imperative to protect against heterologous IAVs, and offers long-term immunity against the viruses compared to the antibody-derived protection (Liu et al., 2023). Unlike the seasonal flu vaccine (QIV/split virus vaccine), VLP-based IAV vaccines have shown superior CMI induction in pre-clinical and clinical studies, without undermining humoral immune responses (Hodgins et al., 2017; Ward et al., 2020). Besides, the immunological superiority of VLPs spans beyond parenteral administration, with similar potency observed through intranasal delivery. This has been consistently proven in mice and ferret (Perrone et al., 2009), wherein Calzas and Chevalier (2019) and Nian et al. (2022) opined that VLPs are parts of the adjuvant/delivery system for the intranasal vaccine delivery. As there are growing interest on intranasal vaccination for IAV, we foresee that VLPs will be among the leading vaccine platform, mainly due to their nanoparticulate conformation which is essential for enhancing nasal associated lymphoid tissue (NALT) stimulation and inducing long-lasting protective immune response right at the IAV entry site.
With respect to vaccine production, the production time and cost are the two key players. Previously, the mumps vaccine was the fastest to get its approval from the Food and Drug Administration (FDA), which is four years from development to approval (Ball, 2021). But in 2020, the Covid-19 vaccine (Comirnaty®) received its approval in 326 days (CEPI, 2022). To prepare for future IAV outbreaks, current vaccine production paradigm aims to make IAV vaccines available for public within 100 days, upon determination of the pathogen's genetic sequence (CEPI, 2022; Saville et al., 2022). Currently, average VLP vaccine platform takes longer than recombinant protein platform (at least 84 days) to generate the first batch of experimental vaccines (CEPI, 2022; Tariq et al., 2022). This pace needs to be accelerated to 30 days, comprising of vaccine development and parallel large-scale manufacturing (CEPI, 2022). To achieve this goal, we believe that current VLP prototypes need to be extensively studied to determine their capacity to display antigens with different physicochemical properties (e.g. size, shape, hydrophobicity, surface charges) without affecting the VLPs’ stability. Upon emergence of new IAV variants, VLP prototypes with high flexibility can be harnessed to display the viral antigens through conjugation (using Catcher/Tag method) and immediately tested to determine the best VLP candidate. This may dramatically reduce vaccine developmental period and facilitate towards faster first batch production. Alternatively, as mRNA vaccines can be developed in relatively faster speed, VLPs displaying conserved IAV antigens (e.g. M2e, NP) can be used to encapsidate mRNAs encoding mutation prone IAV antigens or large antigens (such as HA or NA) in their internal cavity. This strategy may resolve the stability issues affecting current mRNA platform and expedite the pace of VLP vaccine production, as any mutations in IAV can be quickly addressed by changing the mRNA sequence, instead of conducting the lengthy cloning process. Meanwhile, at commercial scale, VLP-based vaccines can offer the cheapest production cost compared to inactivated virus vaccines, protein subunit vaccines and mRNA-based technology (Cheng and Peterson, 2023). Through bioprocess simulation, murine polyomavirus (MuPyV) VLP vaccine can be produced using E. coli at the cost of less than 1 cent per dose (with 50 μg protein per dose) using 1500-litre fed-batch fermentation (Chuan et al., 2014). For comparison, egg-based IIV vaccines, which dominate flu vaccine administration globally, cost $15.77 (from Sanofi) to $16.94 (from GlaxoSmithKline) per dose (Krietsch Boerner, 2020). With rapid production and at relatively low production cost, VLPs can serve as an ideal vaccine platform to protect the global community more instantly, and propel IAV vaccine equity more effectively.
8. Conclusions
The battle to alleviate the burden of influenza A has been ongoing for decades. Even so, each year, IAVs still infect and kill numerous people across the world. We believe that a shift in paradigm is needed, by focusing on vaccines that can generate both neutralizing and cross-reactive antibodies (derived from conserved IAV antigens) in a proportionate magnitude. Imbalance in these antibodies may compromise either the neutralizing potential or their cross-protection against new variants. Besides, the IAV vaccines should also elicit strong T-cell immunity, as this arm of immune response offers durable and better cross-protections against IAV variants, even for vulnerable groups. Moreover, the vaccines should be rapidly produced and offer an affordable price to attract the public to get vaccinated. For this to happen, VLPs hold a great prospective. With an increased number of ongoing pre-clinical studies, IAV VLPs will constantly evolve and overcome the shortcomings of current vaccines. We believe that these progresses will eventually culminate in influenza A as a trivial concern of global healthcare system, in coming years.
Funding
This work was supported by the Ministry of Higher Education of Malaysia under the Fundamental Research Grant Scheme (FRGS/1/2024/STG01/UPM/01/3).
CRediT authorship contribution statement
Jaffar Ali Muhamad Norizwan: Conceptualization, Investigation, Writing – original draft, Writing – review & editing. Wen Siang Tan: Conceptualization, Resources, Supervision, Writing – review & editing, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
JAMN was supported by the Graduate Research Fellowship (GRF GS60266) from Universiti Putra Malaysia (UPM).
Data availability
No experimental data were generated for this review article.
References
- Adorini L., Nagy Z.A. Peptide competition for antigen presentation. Trends Immunol. 1990;11(1):21–24. doi: 10.1016/0167-5699(90)90006-U. [DOI] [PubMed] [Google Scholar]
- Air G.M. Influenza neuraminidase. Influenza Other Respir. Viruses. 2012;6(4):245–256. doi: 10.1111/J.1750-2659.2011.00304.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AstraZeneca . 2022. FluenzⓇ Tetra product insert.https://www.ema.europa.eu/en/documents/product-information/fluenz-tetra-epar-product-information_en.pdf (accessed 7 May 2024) [Google Scholar]
- Avcil M., Çelik A. Microneedles in drug delivery: progress and challenges. Micromachines. 2021;12(11):1321. doi: 10.3390/MI12111321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball P. The lightning-fast quest for COVID vaccines - and what it means for other diseases. Nature. 2021;589:16–18. doi: 10.1038/D41586-020-03626-1. [DOI] [PubMed] [Google Scholar]
- Bangaru S., Lang S., Schotsaert M., Ward A.B., Wilson I.A., Crowe J.E., Vanderven H.A., Zhu X., Kose N., Bombardi R., Finn J.A., Kent S.J., Gilchuk P., Gilchuk I., Turner H.L., García-Sastre A., Li S. A site of vulnerability on the influenza virus hemagglutinin head domain trimer interface. Cell. 2019;177(5):1136–1152. doi: 10.1016/j.cell.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer M.E., Blumberg B.S., Werner B. Particles associated with Australia antigen in the sera of patients with leukaemia, Down's syndrome and hepatitis. Nature. 1968;218:1057–1059. doi: 10.1038/2181057a0. [DOI] [PubMed] [Google Scholar]
- Bliss C.M., Freyn A.W., Caniels T.G., Leyva-Grado V.H., Nachbagauer R., Sun W., Tan G.S., Gillespie V.L., McMahon M., Krammer F., Hill A.V.S., Palese P., Coughlan L. A single-shot adenoviral vaccine provides hemagglutinin stalk-mediated protection against heterosubtypic influenza challenge in mice. Mol. Ther. 2022;30(5):2024–2047. doi: 10.1016/J.YMTHE.2022.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet J., Cartannaz J., Tourcier G., Contreras-Martel C., Kleman J.P., Morlot C., Vernet T., Di Guilmi A.M. Autocatalytic association of proteins by covalent bond formation: a bio molecular welding toolbox derived from a bacterial adhesin. Sci. Rep. 2017;7:43564. doi: 10.1038/srep43564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier N.M., Palese P. The biology of influenza viruses. Vaccine. 2008;26(Suppl 4):D49–D53. doi: 10.1016/j.vaccine.2008.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan L., Clynes M., Meleady P. The emerging role of cellular post-translational modifications in modulating growth and productivity of recombinant Chinese hamster ovary cells. Biotechnol. Adv. 2021;49 doi: 10.1016/j.biotechadv.2021.107757. [DOI] [PubMed] [Google Scholar]
- Buffin S., Peubez I., Barrière F., Nicolaï M.C., Tapia T., Dhir V., Forma E., Sève N., Legastelois I. Influenza A and B virus-like particles produced in mammalian cells are highly immunogenic and induce functional antibodies. Vaccine. 2019;37(46):6857–6867. doi: 10.1016/j.vaccine.2019.09.057. [DOI] [PubMed] [Google Scholar]
- Cadar A.N., Martin D.E., Bartley J.M. Targeting the hallmarks of aging to improve influenza vaccine responses in older adults. Immun. Ageing. 2023;20:23. doi: 10.1186/S12979-023-00348-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadila, 2016. CadiFlu-S summary of product characteristics. https://cdsco.gov.in/opencms/resources/UploadCDSCOWeb/2018/UploadSmPC/4vacain.pdf (accessed 7 May 2024).
- Calzas C., Chevalier C. Innovative mucosal vaccine formulations against influenza A virus infections. Front. Immunol. 2019;10:1605. doi: 10.3389/fimmu.2019.01605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caradonna T.M., Ronsard L., Yousif A.S., Windsor I.W., Hecht R., Bracamonte-Moreno T., Roffler A.A., Maron M.J., Maurer D.P., Feldman J., Marchiori E., Barnes R.M., Rohrer D., Lonberg N., Oguin T.H., Sempowski G.D., Kepler T.B., Kuraoka M., Lingwood D., Schmidt A.G. An epitope-enriched immunogen expands responses to a conserved viral site. Cell Rep. 2022;41(6) doi: 10.1016/j.celrep.2022.111628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carascal M.B., Pavon R.D.N., Rivera W.L. Recent progress in recombinant influenza vaccine development toward heterosubtypic immune response. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.878943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CEPI . 2022. Delivering pandemic vaccines in 100 days, what will it takes?https://static.cepi.net/downloads/2024-02/CEPI-100-Days-Report-Digital-Version_29-11-22.pdf (accessed 7 May 2024) [DOI] [PubMed] [Google Scholar]
- Charlton Hume H.K., Vidigal J., Carrondo M.J.T., Middelberg A.P.J., Roldão A., Lua L.H.L. Synthetic biology for bioengineering virus-like particle vaccines. Biotechnol. Bioeng. 2019;116(4):919–935. doi: 10.1002/bit.26890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.R., Liu Y.M., Tseng Y.C., Ma C. Better influenza vaccines: an industry perspective. J. Biomed. Sci. 2020;27:33. doi: 10.1186/s12929-020-0626-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Shah D., Kositratna G., Manstein D., Anderson R.R., Wu M.X. Facilitation of transcutaneous drug delivery and vaccine immunization by a safe laser technology. J. Control Release. 2012;159(1):43–51. doi: 10.1016/j.jconrel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, J., Peterson, C., 2023. Modeling the economics of vaccine manufacturing. https://www.sigmaaldrich.com/deepweb/assets/sigmaaldrich/marketing/global/documents/366/324/vaccines-cost-modeling-wp12251en-mk.pdf (accessed 7 May 2024).
- Chuan Y.P., Wibowo N., Lua L.H.L., Middelberg A.P.J. The economics of virus-like particle and capsomere vaccines. Biochem. Eng. J. 2014;90:255–263. doi: 10.1016/j.bej.2014.06.005. [DOI] [Google Scholar]
- Chun S., Li C., Van Domselaar G., Wang J., Farnsworth A., Cui X., Rode H., Cyr T.D., He R., Li X. Universal antibodies and their applications to the quantitative determination of virtually all subtypes of the influenza A viral hemagglutinins. Vaccine. 2008;26(48):6068–6076. doi: 10.1016/j.vaccine.2008.09.015. [DOI] [PubMed] [Google Scholar]
- Creytens S., Pascha M.N., Ballegeer M., Saelens X., de Haan C.A.M. Influenza neuraminidase characteristics and potential as a vaccine target. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.786617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust M.A., Lavoie P.O., Couture M.M.J., Trépanier S., Guay J.M., Dargis M., Mongrand S., Landry N., Ward B.J., Vézina L.P. Influenza virus-like particles produced by transient expression in Nicotiana benthamiana induce a protective immune response against a lethal viral challenge in mice. Plant Biotechnol. J. 2008;6(9):930–940. doi: 10.1111/j.1467-7652.2008.00384.x. [DOI] [PubMed] [Google Scholar]
- De Filette M., Martens W., Smet A., Schotsaert M., Birkett A., Londõ No-Arcila P., Fiers W., Saelens X. Universal influenza A M2e-HBc vaccine protects against disease even in the presence of pre-existing anti-HBc antibodies. Vaccine. 2008;26(51):6503–6507. doi: 10.1016/j.vaccine.2008.09.038. [DOI] [PubMed] [Google Scholar]
- de Oliveira T.A., Silva W.da, da Rocha Torres N., Badaró de Moraes J.V, Senra R.L., de Oliveira Mendes T.A., Júnior A.S., Bressan G.C., Fietto J.L.R. Application of the LEXSY Leishmania tarentolae system as a recombinant protein expression platform: A review. Process Biochem. 2019;87:164–173. doi: 10.1016/j.procbio.2019.08.019. [DOI] [Google Scholar]
- Del Campo J., Pizzorno A., Djebali S., Bouley J., Haller M., Pérez-Vargas J., Lina B., Boivin G., Hamelin M.E., Nicolas F., Le Vert A., Leverrier Y., Rosa-Calatrava M., Marvel J., Hill F. OVX836 a recombinant nucleoprotein vaccine inducing cellular responses and protective efficacy against multiple influenza A subtypes. npj Vaccines. 2019;4:4. doi: 10.1038/s41541-019-0098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert D.C., Kashyap A.K., Steel J., Rubrum A., Bhabha G., Khayat R., Lee J.H., Dillon M.A., O'Neil R.E., Faynboym A.M., Horowitz M., Horowitz L., Ward A.B., Palese P., Webby R., Lerner R.A., Bhatt R.R., Wilson I.A. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. 2012;489:526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan R., Aranko A.S. Catcher/Tag toolbox: Biomolecular click-reactions for protein engineering beyond genetics. ChemBioChem. 2024;25(1) doi: 10.1002/cbic.202300600. [DOI] [PubMed] [Google Scholar]
- Fereidouni S., Starick E., Karamendin K., Genova C.D., Scott S.D., Khan Y., Harder T., Kydyrmanov A. Genetic characterization of a new candidate hemagglutinin subtype of influenza A viruses. Emerg. Microbes Infec. 2023;12(2) doi: 10.1080/22221751.2023.2225645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries L.F., Smith G.E., Glenn G.M. A recombinant viruslike particle influenza A (H7N9) vaccine. N. Engl. J. Med. 2013;369(26):2564–2566. doi: 10.1056/nejmc1313186. [DOI] [PubMed] [Google Scholar]
- Fukuyama H., Shinnakasu R., Kurosaki T. Influenza vaccination strategies targeting the hemagglutinin stem region. Immunol. Rev. 2020;296(1):132–141. doi: 10.1111/imr.12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes A.C., Roesti E.S., El-Turabi A., Bachmann M.F. Type of RNA packed in VLPs impacts IgG class switching—implications for an influenza vaccine design. Vaccines. 2019;7(2):47. doi: 10.3390/vaccines7020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes K.B., Menon I., Bagwe P., Bajaj L., Kang S.M., D'Souza M.J. Enhanced immunogenicity of an influenza ectodomain matrix-2 protein virus-like particle (M2e VLP) using polymeric microparticles for vaccine delivery. Viruses. 2022;14(9):1920. doi: 10.3390/v14091920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes K.B., Vijayanand S., Bagwe P., Menon I., Kale A., Patil S., Kang S.M., Uddin M.N., D'Souza M.J. Vaccine-induced immunity elicited by microneedle delivery of influenza ectodomain matrix protein 2 virus-like particle (M2e VLP)-loaded PLGA nanoparticles. Int. J. Mol. Sci. 2023;24(13):10612. doi: 10.3390/ijms241310612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Lu X., Kang S.M., Chen C., Compans R.W., Yao Q. Enhancement of mucosal immune responses by chimeric influenza HA/SHIV virus-like particles. Virology. 2003;313(2):502–513. doi: 10.1016/S0042-6822(03)00372-6. [DOI] [PubMed] [Google Scholar]
- Gupta R., Arora K., Roy S.S., Joseph A., Rastogi R., Arora N.M., Kundu P.K. Platforms, advances, and technical challenges in virus-like particles-based vaccines. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1123805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes J.R., Dokken L., Wiley J.A., Cawthon A.G., Bigger J., Harmsen A.G., Richardson C. Influenza-pseudotyped Gag virus-like particle vaccines provide broad protection against highly pathogenic avian influenza challenge. Vaccine. 2009;27(4):530–541. doi: 10.1016/j.vaccine.2008.11.011. [DOI] [PubMed] [Google Scholar]
- He X.-S., Holmes T.H., Zhang C., Mahmood K., Kemble G.W., Lewis D.B., Dekker C.L., Greenberg H.B., Arvin A.M. Cellular immune responses in children and adults receiving inactivated or live attenuated influenza vaccines. J. Virol. 2006;80(23):11756–11766. doi: 10.1128/jvi.01460-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinimäki S., Lampinen V., Tamminen K., Hankaniemi M.M., Malm M., Hytönen V.P., Blazevic V. Antigenicity and immunogenicity of HA2 and M2e influenza virus antigens conjugated to norovirus-like, VP1 capsid-based particles by the SpyTag/SpyCatcher technology. Virology. 2022;566:89–97. doi: 10.1016/j.virol.2021.12.001. [DOI] [PubMed] [Google Scholar]
- Hirschberg S., Ghazaani F., Ben Amor G., Pydde M., Nagel A., Germani S., Monica L., Schlör A., Bauer H., Hornung J., Voetz M., Dwai Y., Scheer B., Ringel F., Kamal-Eddin O., Harms C., Füner J., Adrian L., Pruß A., Schulze-Forster K., Hanack K., Kamhieh-Milz J. An efficient and scalable method for the production of immunogenic SARS-CoV-2 virus-like particles (VLP) from a mammalian suspension cell line. Vaccines. 2023;11(9):1469. doi: 10.3390/vaccines11091469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgins B., Yam K.K., Winter K., Pillet S., Landry N., Ward B.J. A single intramuscular dose of a plant-made virus-like particle vaccine elicits a balanced humoral and cellular response and protects young and aged mice from influenza H1N1 virus challenge despite a modest/absent humoral response. Clin. Vaccine Immunol. 2017;24(12) doi: 10.1128/cvi.00273-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofschneider P.H., Murray K. In: Recombinant Protein Drugs. Milestones in Drug Therapy. Buckel P., editor. Birkhäuser; Basel: 2001. Combining science and business: from recombinant DNA to vaccines against hepatitis B virus. [DOI] [Google Scholar]
- Hoft D.F., Babusis E., Worku S., Spencer C.T., Lottenbach K., Truscott S.M., Abate G., Sakala I.G., Edwards K.M., Buddy Creech C., Gerber M.A., Bernstein D.I., Newman F., Graham I., Anderson E.L., Belshe R.B. Live and inactivated influenza vaccines induce similar humoral responses, but only live vaccines induce diverse T-cell responses in young children. J. Infect. Dis. 2011;204(6):845–853. doi: 10.1093/infdis/jir436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigsbaum M. Revisiting the 1957 and 1968 influenza pandemics. Lancet. 2020;395(10240):1824–1826. doi: 10.1016/S0140-6736(20)31201-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser K., Subbarao K. Influenza vaccines: challenges and solutions. Cell Host Microbe. 2015;17(3):295–300. doi: 10.1016/j.chom.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huzair F., Sturdy S. Biotechnology and the transformation of vaccine innovation: the case of the hepatitis B vaccines 1968–2000. Stud. Hist. Phi. Part C. 2017;64:11–21. doi: 10.1016/j.shpsc.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibañez L.I., Roose K., De Filette M., Schotsaert M., De Sloovere J., Roels S., Pollard C., Schepens B., Grooten J., Fiers W., Saelens X. M2e-Displaying virus-like particles with associated RNA promote T helper 1 type adaptive immunity against influenza A. PLoS One. 2013;8(3):e59081. doi: 10.1371/journal.pone.0059081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imagawa T., Arasaki Y., Maegawa K., Sugita S., Nerome K. Advancing usability of an influenza hemagglutinin virus-like particle vaccine expressing a chimeric cytokine. Virol. J. 2023;20:102. doi: 10.1186/s12985-023-02076-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionescu R.M., Przysiecki C.T., Liang X., Garsky V.M., Fan J., Wang B., Troutman R., Rippeon Y., Flanagan E., Shiver J., Shi L. Pharmaceutical and immunological evaluation of human papillomavirus viruslike particle as an antigen carrier. J. Pharm. Sci. 2006;95(1):70–79. doi: 10.1002/jps.20493. [DOI] [PubMed] [Google Scholar]
- Joshi D., Gala R.P., Uddin M.N., D’souza M.J. Novel ablative laser mediated transdermal immunization for microparticulate measles vaccine. Int. J. Pharm. 2021;606:120882. doi: 10.1016/j.ijpharm.2021.120882. [DOI] [PubMed] [Google Scholar]
- Kapoor S., Dhama K. Springer Science and Business Media. 2014. Insight into influenza viruses of animals and humans; pp. 1–222. [DOI] [Google Scholar]
- Kavishna R., Kang T.Y., Vacca M., Chua B.Y.L., Park H.Y., Tan P.S., Chow V.T.K., Lahoud M.H., Alonso S. A single-shot vaccine approach for the universal influenza A vaccine candidate M2e. Proc. Natl. Acad. Sci. U. S. A. 2022;119(13) doi: 10.1073/pnas.2025607119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazaks A., Lu I.N., Farinelle S., Ramirez A., Crescente V., Blaha B., Ogonah O., Mukhopadhyay T., de Obanos M.P., Krimer A., Akopjana I., Bogans J., Ose V., Kirsteina A., Kazaka T., Stonehouse N.J., Rowlands D.J., Muller C.P., Tars K., Rosenberg W.M. Production and purification of chimeric HBc virus-like particles carrying influenza virus LAH domain as vaccine candidates. BMC Biotechnol. 2017;17:79. doi: 10.1186/S12896-017-0396-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeble A.H., Turkki P., Stokes S., Anuar I.N.A.K., Rahikainen R., Hytönen V.P., Howarth M. Approaching infinite affinity through engineering of peptide-protein interaction. Proc. Natl. Acad. Sci. U. S. A. 2019;116(52):26523–26533. doi: 10.1073/pnas.1909653116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeble A.H., Yadav V.K., Ferla M.P., Bauer C.C., Chuntharpursat-Bon E., Huang J., Bon R.S., Howarth M. DogCatcher allows loop-friendly protein-protein ligation. Cell Chem. Biol. 2022;29(2):339–350.e10. doi: 10.1016/j.chembiol.2021.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A.H., Bayat H., Rajabibazl M., Sabri S., Rahimpour A. Humanizing glycosylation pathways in eukaryotic expression systems. World J. Microbiol. Biotechnol. 2017;33:4. doi: 10.1007/s11274-016-2172-7. [DOI] [PubMed] [Google Scholar]
- Kim H.J., Kim H.J. Yeast as an expression system for producing virus-like particles: what factors do we need to consider? Lett. Appl. Microbiol. 2017;64(2):111–123. doi: 10.1111/LAM.12695. [DOI] [PubMed] [Google Scholar]
- Kirsteina A., Akopjana I., Bogans J., Lieknina I., Jansons J., Skrastina D., Kazaka T., Tars K., Isakova-Sivak I., Mezhenskaya D., Kotomina T., Matyushenko V., Rudenko L., Kazaks A. Construction and immunogenicity of a novel multivalent vaccine prototype based on conserved influenza virus antigens. Vaccines. 2020;8(2):197. doi: 10.3390/vaccines8020197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krietsch Boerner L. The flu shot and the egg. ACS Cent. Sci. 2020;6(2):89–92. doi: 10.1021/acscentsci.0c00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampinen V., Gröhn S., Soppela S., Blazevic V., Hytönen V.P., Hankaniemi M.M. SpyTag/SpyCatcher display of influenza M2e peptide on norovirus-like particle provides stronger immunization than direct genetic fusion. Front. Cell. Infect. Microbiol. 2023;13 doi: 10.3389/fcimb.2023.1216364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry N., Ward B.J., Trépanier S., Montomoli E., Le Dargis M., Lapini G., Vézina L.P. Preclinical and clinical development of plant-made virus-like particle vaccine against avian H5N1 influenza. PLoS One. 2010;5(12):e15559. doi: 10.1371/journal.pone.0015559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I.T., Nachbagauer R., Ensz D., Schwartz H., Carmona L., Schaefers K., Avanesov A., Stadlbauer D., Henry C., Chen R., Huang W., Schrempp D.R., Ananworanich J., Paris R. Safety and immunogenicity of a phase 1/2 randomized clinical trial of a quadrivalent, mRNA-based seasonal influenza vaccine (mRNA-1010) in healthy adults: interim analysis. Nat. Commun. 2023;14:3631. doi: 10.1038/s41467-023-39376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.S., Ohshima N., Stanfield R.L., Yu W., Iba Y., Okuno Y., Kurosawa Y., Wilson I.A. Receptor mimicry by antibody F045–092 facilitates universal binding to the H3 subtype of influenza virus. Nat. Commun. 2014;5:3614. doi: 10.1038/ncomms4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.T., Ko E.J., Lee Y., Kim K.H., Kim M.C., Lee Y.N., Kang S.M. Intranasal vaccination with M2e5x virus-like particles induces humoral and cellular immune responses conferring cross-protection against heterosubtypic influenza viruses. PLoS One. 2018;13(1) doi: 10.1371/journal.pone.0190868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux-Roels I., Willems P., Waerlop G., Janssens Y., Tourneur J., De Boever F., Alhatemi A., Jacobs B., Nicolas F., Leroux-Roels G., Le Vert A. Immunogenicity, safety, and preliminary efficacy evaluation of OVX836, a nucleoprotein-based universal influenza A vaccine candidate: a randomised, double-blind, placebo-controlled, phase 2a trial. Lancet Infect. Dis. 2023;23(12):1360–1369. doi: 10.1016/s1473-3099(23)00351-1. [DOI] [PubMed] [Google Scholar]
- Li X., Shen J., Chen X., Chen L., Wan S., Qiu X., Chen K., Chen C., Tan H. Humanization of yeasts for glycan-type end-products. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.930658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.Y., Yeh C.T., Li W.H., Yu C.P., Lin W.C., Yang J.Y., Wu H.L., Hu Y.C. Enhanced enterovirus 71 virus-like particle yield from a new baculovirus design. Biotechnol. Bioeng. 2015;112(10):2005–2015. doi: 10.1002/bit.25625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhao T., Wang L., Yang Z., Luo C., Li M., Luo H., Sun C., Yan H., Shu Y. A mosaic influenza virus-like particles vaccine provides broad humoral and cellular immune responses against influenza A viruses. npj Vaccines. 2023;8:132. doi: 10.1038/s41541-023-00728-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Serrano S., Cordoba L., Pérez-Maillo M., Pleguezuelos P., Remarque E.J., Ebensen T., Guzmán C.A., Christensen D., Segalés J., Darji A. Immune responses to pandemic H1N1 influenza virus infection in pigs vaccinated with a conserved hemagglutinin HA1 peptide adjuvanted with CAF®01 or CDA/αGalCerMPEG. Vaccines. 2021;9(7):751. doi: 10.3390/vaccines9070751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini N., Solforosi L., Clementi N., De Marco D., Clementi M., Burioni R. A potential role for monoclonal antibodies in prophylactic and therapeutic treatment of influenza. Antiviral Res. 2011;92(1):15–26. doi: 10.1016/j.antiviral.2011.07.013. [DOI] [PubMed] [Google Scholar]
- Mardanova E.S., Kotlyarov R.Y., Stuchinskaya M.D., Nikolaeva L.I., Zahmanova G., Ravin N.V. High-yield production of chimeric hepatitis E virus-like particles bearing the M2e influenza epitope and receptor binding domain of SARS-CoV-2 in plants using viral vectors. Int. J. Mol. Sci. 2022;23(24):15684. doi: 10.3390/ijms232415684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K., Migueles S.A., Huang J., Bolkhovitinov L., Stuccio S., Griesman T., Pullano A.A., Kang B.H., Ishida E., Zimmerman M., Kashyap N., Martins K.M., Stadlbauer D., Pederson J., Patamawenu A., Wright N., Shofner T., Evans S., Jason Liang C., Candia J., Biancotto A., Fantoni G., Poole A., Smith J., Alexander J., Gurwith M., Krammer F., Connors M. A replication-competent adenovirus-vectored influenza vaccine induces durable systemic and mucosal immunity. J. Clin. Invest. 2021;131(5) doi: 10.1172/jci140794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T., Tanijima T., Hirose A., Masumi-Koizumi K., Katsuda T., Yamaji H. Production of influenza virus-like particles using recombinant insect cells. Biochem. Eng. J. 2020;163 doi: 10.1016/j.bej.2020.107757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy K.R., Lee J., Watanabe A., Kuraoka M., Robinson-Mccarthy L.R., Georgiou G., Kelsoe G., Harrison S.C. A prevalent focused human antibody response to the influenza virus hemagglutinin head interface. mBio. 2021;12(3) doi: 10.1128/mbio.01144-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee M.C., Huang W. Evolutionary conservation and positive selection of influenza A nucleoprotein CTL epitopes for universal vaccination. J. Med. Virol. 2022;94(6):2578–2587. doi: 10.1002/jmv.27662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D., Fellner P., Newton C. Influenza virus genome consists of eight distinct RNA species. Proc. Natl. Acad. Sci. U. S. A. 1976;73(9):3045–3049. doi: 10.1073/pnas.73.9.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonigle R., Yap W.B., Ong S.T., Gatherer D., Bakker S.E., Tan W.S., Bhella D. An N-terminal extension to the hepatitis B virus core protein forms a poorly ordered trimeric spike in assembled virus-like particles. J. Struct. Biol. 2015;189(2):73–80. doi: 10.1016/j.jsb.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon M., Strohmeier S., Rajendran M., Capuano C., Ellebedy A.H., Wilson P.C., Krammer F. Correctly folded - but not necessarily functional - influenza virus neuraminidase is required to induce protective antibody responses in mice. Vaccine. 2020;38(45):7129–7137. doi: 10.1016/j.vaccine.2020.08.067. [DOI] [PubMed] [Google Scholar]
- Menon I., Bagwe P., Gomes K.B., Bajaj L., Gala R., Uddin M.N., D'souza M.J., Zughaier S.M. Microneedles: a new generation vaccine delivery system. Micromachines. 2021;12(4):435. doi: 10.3390/mi12040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercx S., Smargiasso N., Chaumont F., De Pauw E., Boutry M., Navarre C. Inactivation of the β(1,2)-xylosyltransferase and the α(1,3)-fucosyltransferase genes in Nicotiana tabacum BY-2 cells by a multiplex CRISPR/Cas9 strategy results in glycoproteins without plant-specific glycans. Front. Plant Sci. 2017;8:403. doi: 10.3389/fpls.2017.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezhenskaya D., Isakova-Sivak I., Rudenko L. M2e-based universal influenza vaccines: a historical overview and new approaches to development. J. Biomed. Sci. 2019;26:76. doi: 10.1186/s12929-019-0572-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohsen M.O., Balke I., Zinkhan S., Zeltina V., Liu X., Chang X., Krenger P.S., Plattner K., Gharailoo Z., Vogt A.C.S., Augusto G., Zwicker M., Roongta S., Rothen D.A., Josi R., Costa J.J.da, Sobczak J.M., Nonic A., Brand L.A., Nuss K., Martina B., Speiser D.E., Kündig T., Jennings G.T., Walton S.M., Vogel M., Zeltins A., Bachmann M.F. A scalable and highly immunogenic virus-like particle-based vaccine against SARS-CoV-2. Allergy. 2022;77(1):243–257. doi: 10.1111/all.15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohsen M.O., Gomes A.C., Cabral-Miranda G., Krueger C.C., Leoratti F.M., Stein J.V., Bachmann M.F. Delivering adjuvants and antigens in separate nanoparticles eliminates the need of physical linkage for effective vaccination. J. Control Release. 2017;251:92–100. doi: 10.1016/j.jconrel.2017.02.031. [DOI] [PubMed] [Google Scholar]
- Mohsen M.O., Zha L., Cabral-Miranda G., Bachmann M.F. Major findings and recent advances in virus–like particle (VLP)-based vaccines. Semin. Immunol. 2017;34:123–132. doi: 10.1016/j.smim.2017.08.014. [DOI] [PubMed] [Google Scholar]
- Moin S.M., Boyington J.C., Boyoglu-Barnum S., Gillespie R.A., Cerutti G., Cheung C.S.F., Cagigi A., Gallagher J.R., Brand J., Prabhakaran M., Tsybovsky Y., Stephens T., Fisher B.E., Creanga A., Ataca S., Rawi R., Corbett K.S., Crank M.C., Karlsson Hedestam G.B., Gorman J., McDermott A.B., Harris A.K., Zhou T., Kwong P.D., Shapiro L., Mascola J.R., Graham B.S., Kanekiyo M. Co-immunization with hemagglutinin stem immunogens elicits cross-group neutralizing antibodies and broad protection against influenza A viruses. Immunity. 2022;55(12):2405–2418.e7. doi: 10.1016/j.immuni.2022.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monto A.S., Petrie J.G., Cross R.T., Johnson E., Liu M., Zhong W., Levine M., Katz J.M., Ohmit S.E. Antibody to influenza virus neuraminidase: an independent correlate of protection. J. Infect. Dis. 2015;212(8):1191–1199. doi: 10.1093/infdis/jiv195. [DOI] [PubMed] [Google Scholar]
- Muralidharan A., Gravel C., Harris G., Hashem A.M., Zhang W., Safronetz D., Van Domselaar G., Krammer F., Sauve S., Rosu-Myles M., Wang L., Chen W., Li X. Universal antibody targeting the highly conserved fusion peptide provides cross-protection in mice. Hum. Vacc. Immunother. 2022;18(5):2083428. doi: 10.1080/21645515.2022.2083428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers M.L., Gallagher J.R., Kim A.J., Payne W.H., Maldonado-Puga S., Assimakopoulos H., Bock K.W., Torian U., Moore I.N., Harris A.K. Commercial influenza vaccines vary in HA-complex structure and in induction of cross-reactive HA antibodies. Nat. Commun. 2023;14:1763. doi: 10.1038/s41467-023-37162-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nian X., Zhang J., Huang S., Duan K., Li X., Yang X. Development of nasal vaccines and the associated challenges. Pharmaceutics. 2022;14(10):1983. doi: 10.3390/pharmaceutics14101983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH . 2012. Safety study of recombinant M2e influenza-A vaccine in healthy adults (FLU-A)https://clinicaltrials.gov/study/NCT00819013#study-overview (accessed 31 July 2024) [Google Scholar]
- Ohshima N., Iba Y., Kubota-Koketsu R., Asano Y., Okuno Y., Kurosawa Y. Naturally occurring antibodies in humans can neutralize a variety of influenza virus strains, including H3, H1, H2, and H5. J. Virol. 2011;85(21):11048–11057. doi: 10.1128/JVI.05397-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong H.K., Yong C.Y., Tan W.S., Yeap S.K., Omar A.R., Razak M.A., Ho K.L. An influenza A vaccine based on the extracellular domain of matrix 2 protein protects BALB/c mice against H1N1 and H3N2. Vaccines. 2019;7(3):91. doi: 10.3390/vaccines7030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panasiuk M., Zimmer K., Czarnota A., Grzyb K., Narajczyk M., Peszyńska-Sularz G., Żołędowska S., Nidzworski D., Hovhannisyan L., Gromadzka B. Immunization with Leishmania tarentolae-derived norovirus virus-like particles elicits high humoral response and stimulates the production of neutralizing antibodies. Microb. Cell Fact. 2021;20:186. doi: 10.1186/S12934-021-01677-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B.R., Bommireddy R., Chung D.H., Kim K.H., Subbiah J., Jung Y.J., Bhatnagar N., Pack C.D., Ramachandiran S., Reddy S.J.C., Selvaraj P., Kang S.M. Hemagglutinin virus-like particles incorporated with membrane-bound cytokine adjuvants provide protection against homologous and heterologous influenza virus challenge in aged mice. Immun. Ageing. 2023;20:20. doi: 10.1186/s12979-023-00344-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone L.A., Ahmad A., Veguilla V., Lu X., Smith G., Katz J.M., Pushko P., Tumpey T.M. Intranasal vaccination with 1918 influenza virus-like particles protects mice and ferrets from lethal 1918 and H5N1 influenza virus challenge. J. Virol. 2009;83(11):5726–5734. doi: 10.1128/jvi.00207-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova V.N., Russell C.A. The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 2018;16:47–60. doi: 10.1038/nrmicro.2017.118. [DOI] [PubMed] [Google Scholar]
- Pietrzak M., Macioła A., Zdanowski K., Protas-Klukowska A.M., Olszewska M., Śmietanka K., Minta Z., Szewczyk B., Kopera E. An avian influenza H5N1 virus vaccine candidate based on the extracellular domain produced in yeast system as subviral particles protects chickens from lethal challenge. Antiviral Res. 2016;133:242–249. doi: 10.1016/j.antiviral.2016.08.001. [DOI] [PubMed] [Google Scholar]
- Pihl R., Zheng Q., David Y. Nature-inspired protein ligation and its applications. Nat. Rev. Chem. 2023;7:234–255. doi: 10.1038/s41570-023-00468-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillet S., Aubin É., Trépanier S., Bussière D., Dargis M., Poulin J.F., Yassine-Diab B., Ward B.J., Landry N. A plant-derived quadrivalent virus like particle influenza vaccine induces cross-reactive antibody and T cell response in healthy adults. Clin. Immunol. 2016;168:72–87. doi: 10.1016/j.clim.2016.03.008. [DOI] [PubMed] [Google Scholar]
- Pillet S., Couillard J., Trépanier S., Poulin J.F., Yassine-Diab B., Guy B., Ward B.J., Landry N. Immunogenicity and safety of a quadrivalent plant-derived virus like particle influenza vaccine candidate—Two randomized Phase II clinical trials in 18 to 49 and ≥50 years old adults. PLoS One. 2019;14(6) doi: 10.1371/journal.pone.0216533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan F.-S., Kim Y.-C., Vunnava A., Yoo D.-G., Song J.-M., Prausnitz M.R., Compans R.W., Kang S.-M. Intradermal vaccination with influenza virus-like particles by using microneedles induces protection superior to that with intramuscular immunization. J. Virol. 2010;84(15):7760–7769. doi: 10.1128/jvi.01849-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran M., Krammer F., McMahon M. The human antibody response to the influenza virus neuraminidase following infection or vaccination. Vaccines. 2021;9(8):846. doi: 10.3390/vaccines9080846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez A., Morris S., Maucourant S., D'Ascanio I., Crescente V., Lu I.N., Farinelle S., Muller C.P., Whelan M., Rosenberg W. A virus-like particle vaccine candidate for influenza A virus based on multiple conserved antigens presented on hepatitis B tandem core particles. Vaccine. 2018;36(6):873–880. doi: 10.1016/j.vaccine.2017.12.053. [DOI] [PubMed] [Google Scholar]
- Rosalia R.A., Quakkelaar E.D., Redeker A., Khan S., Camps M., Drijfhout J.W., Silva A.L., Jiskoot W., van Hall T., van Veelen P.A., Janssen G., Franken K., Cruz L.J., Tromp A., Oostendorp J., van der Burg S.H., Ossendorp F., Melief C.J.M. Dendritic cells process synthetic long peptides better than whole protein, improving antigen presentation and T-cell activation. Eur. J. Immunol. 2013;43(10):2554–2565. doi: 10.1002/eji.201343324. [DOI] [PubMed] [Google Scholar]
- Rosano G.L., Morales E.S., Ceccarelli E.A. New tools for recombinant protein production in Escherichia coli: A 5-year update. Protein Sci. 2019;28(8):1412–1422. doi: 10.1002/pro.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman J.S., Jing X., Leser G.P., Balannik V., Pinto L.H., Lamb R.A. Influenza virus M2 ion channel protein is necessary for filamentous virion formation. J. Virol. 2010;84(10):5078–5088. doi: 10.1128/jvi.00119-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosu M.E., Kok A., Bestebroer T.M., de Meulder D., Verveer E.P., Pronk M.R., Dekker L.J.M., Luider T.M., Richard M., van den Brand J.M.A., Fouchier R.A.M., Herfst S. Contribution of neuraminidase to the efficacy of seasonal split influenza vaccines in the ferret model. J. Virol. 2022;96(6) doi: 10.1128/jvi.01959-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowska D.A., Mokoena N.B., Tsekoa T.L., Dibakwane V.S., O'Kennedy M.M. Plant-produced chimeric virus-like particles - A new generation vaccine against African horse sickness. BMC Vet. Res. 2019;15:432. doi: 10.1186/s12917-019-2184-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybicki E.P. Plant molecular farming of virus-like nanoparticles as vaccines and reagents. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020;12(2):e1587. doi: 10.1002/wnan.1587. [DOI] [PubMed] [Google Scholar]
- Saleh M., Nowroozi J., Farahmand B., Fotouhi F. An approach to the influenza chimeric subunit vaccine (3M2e-HA2-NP) provides efficient protection against lethal virus challenge. Biotechnol. Lett. 2020;42:1147–1159. doi: 10.1007/s10529-020-02822-3. [DOI] [PubMed] [Google Scholar]
- Sam J.I.C. The burden of human influenza in Malaysia. Med. J. Malaysia. 2015;70(3):127–130. PMID: 26248773. [PubMed] [Google Scholar]
- Sánchez-de Prada L., Sanz-Muñoz I., Sun W., Palese P., Ortiz de Lejarazu R., Eiros J.M., García-Sastre A., Aydillo T. Group 1 and group 2 hemagglutinin stalk antibody response according to age. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1194073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchooli A., Aghayipour K., Naghlani S.K., Samiee Z., Kiasari B.A., Makvandi M. Production of human papillomavirus type-16 L1 VLP in Pichia pastoris. Appl. Biochem. Microbiol. 2020;56:51–57. doi: 10.1134/S0003683820010147. [DOI] [Google Scholar]
- Sanofi Pasteur, 2022. Fluzone® Quadrivalent product insert. https://www.fda.gov/media/119856/download (accessed 7 May 2024).
- Saville M., Cramer J.P., Downham M., Hacker A., Lurie N., Van der Veken L., Whelan M., Hatchett R. Delivering pandemic vaccines in 100 days — what will it take? N. Engl. J. Med. 2022;387(2):e3. doi: 10.1056/nejmp2202669. [DOI] [PubMed] [Google Scholar]
- Sazegari S., Akbarzadeh Niaki M., Afsharifar A., Niazi A., Derakhshandeh A., Moradi Vahdat M., Hemmati F, Eskandari M.H. Chimeric hepatitis B core virus-like particles harboring SARS-CoV2 epitope elicit a humoral immune response in mice. Microb. Cell Fact. 2023;22:39. doi: 10.1186/s12934-023-02043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiblhofer S., Thalhamer J., Weiss R. Laser microporation of the skin: prospects for painless application of protective and therapeutic vaccines. Expert Opin. Drug Deliv. 2013;10(6):761–773. doi: 10.1517/17425247.2013.773970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedova E.S., Scherbinin D.N., Lysenko A.A., Alekseeva S.V., Artemova E.A., Shmarov M.M. Non-neutralizing antibodies directed at conservative influenza antigens. ActaNaturae. 2019;11(4):22–32. doi: 10.32607/20758251-2019-11-4-22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma J., Shepardson K., Johns L.L., Wellham J., Avera J., Schwarz B., Rynda-Apple A., Douglas T. A self-adjuvanted, modular, antigenic VLP for rapid response to influenza virus variability. ACS Appl. Mater. Interfaces. 2020;12(16):18211–18224. doi: 10.1021/acsami.9b21776. [DOI] [PubMed] [Google Scholar]
- Shi L., Long Y., Zhu Y., Dong J., Chen Y., Feng H., Sun X. VLPs containing stalk domain and ectodomain of matrix protein 2 of influenza induce protection in mice. Virol. J. 2023;20:38. doi: 10.1186/s12985-023-01994-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen L., Spreeuwenberg P., Lustig R., Taylor R.J., Fleming D.M., Kroneman M., Van Kerkhove M.D., Mounts A.W., Paget W.J., the GLaMOR Collaborating Teams Global mortality estimates for the 2009 influenza pandemic from the GLaMOR project: A modeling study. PLoS Med. 2013;10(11) doi: 10.1371/journal.pmed.1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarlupka A.L., Bebin-Blackwell A.-G., Sumner S.F., Ross T.M. Universal influenza virus neuraminidase vaccine elicits protective immune responses against human seasonal and pre-pandemic strains. J. Virol. 2021;17(95) doi: 10.1128/jvi.00759-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G.E., Flyer D.C., Raghunandan R., Liu Y., Wei Z., Wu Y., Kpamegan E., Courbron D., Fries L.F., Glenn G.M. Development of influenza H7N9 virus like particle (VLP) vaccine: Homologous A/Anhui/1/2013 (H7N9) protection and heterologous A/chicken/Jalisco/CPA1/2012 (H7N3) cross-protection in vaccinated mice challenged with H7N9 virus. Vaccine. 2013;31(40):4305–4313. doi: 10.1016/j.vaccine.2013.07.043. [DOI] [PubMed] [Google Scholar]
- Soema P.C., Kompier R., Amorij J.P., Kersten G.F.A. Current and next generation influenza vaccines: formulation and production strategies. Eur. J. Pharm. Biopharm. 2015;94:251–263. doi: 10.1016/j.ejpb.2015.05.023. [DOI] [PubMed] [Google Scholar]
- Sparrow E., Wood J.G., Chadwick C., Newall A.T., Torvaldsen S., Moen A., Torelli G. Global production capacity of seasonal and pandemic influenza vaccines in 2019. Vaccine. 2021;39(3):512–520. doi: 10.1016/j.vaccine.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava V., Nand K.N., Ahmad A., Kumar R. Yeast-based virus-like particles as an emerging platform for vaccine development and delivery. Vaccines. 2023;11(2):479. doi: 10.3390/vaccines11020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohmeier S., Amanat F., Campbell J.D., Traquina P., Coffman R.L., Krammer F. A CpG 1018 adjuvanted neuraminidase vaccine provides robust protection from influenza virus challenge in mice. npj Vaccines. 2022;7(1):81. doi: 10.1038/s41541-022-00486-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohmeier S., Amanat F., Zhu X., McMahon M., Deming M.E., Pasetti M.F., Neuzil K.M., Wilson I.A., Krammer F. A novel recombinant influenza virus neuraminidase vaccine candidate stabilized by a measles virus phosphoprotein tetramerization domain provides robust protection from virus challenge in the mouse model. mBio. 2021;12(6) doi: 10.1128/mbio.02241-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K. Live attenuated cold-adapted influenza vaccines. Cold Spring Harb. Perspect. Med. 2021;11 doi: 10.1101/cshperspect.a038653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Ling Z., Yang Z., Sun B. Broad neutralizing antibody-based strategies to tackle influenza. Curr. Opin. Virol. 2022;53 doi: 10.1016/j.coviro.2022.101207. [DOI] [PubMed] [Google Scholar]
- Sun Y.X., Li Z.R., Zhang P.J., Han J.H., Di H.Y., Qin J.Y., Cong Y.L. A single vaccination of chimeric bivalent virus-like particle vaccine confers protection against H9N2 and H3N2 avian influenza in commercial broilers and allows a strategy of differentiating infected from vaccinated animals. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.902515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi S., Ooi T., Mizuno K., Matsusaki H. Advances and needs for endotoxin-free production strains. Appl. Microbiol. Biotechnol. 2015;99:9349–9360. doi: 10.1007/s00253-015-6947-9. [DOI] [PubMed] [Google Scholar]
- Tan J.S., Jaffar Ali M.N.B., Gan B.K., Tan W.S. Next-generation viral nanoparticles for targeted delivery of therapeutics: Fundamentals, methods, biomedical applications, and challenges. Expert Opin. Drug Deliv. 2023;20(7):955–978. doi: 10.1080/17425247.2023.2228202. [DOI] [PubMed] [Google Scholar]
- Tariq H., Batool S., Asif S., Ali M., Abbasi B.H. Virus-like particles: Revolutionary platforms for developing vaccines against emerging infectious diseases. Front. Microbiol. 2022;12 doi: 10.3389/fmicb.2021.790121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tizard, I.R., 2021. The administration of vaccines. Vaccines for veterinarians. 87–104. 10.1016/b978-0-323-68299-2.00017-4. [DOI]
- Tomiya N., Narang S., Lee Y.C., Betenbaugh M.J. Comparing N-glycan processing in mammalian cell lines to native and engineered lepidopteran insect cell lines. Glycoconjugate J. 2004;21(6):343–360. doi: 10.1023/b:glyc.0000046275.28315.87. [DOI] [PubMed] [Google Scholar]
- Tong S., Zhu X., Li Y., Shi M., Zhang J., Bourgeois M., Yang H., Chen X., Recuenco S., Gomez J., Chen L.M., Johnson A., Tao Y., Dreyfus C., Yu W., McBride R., Carney P.J., Gilbert A.T., Chang J., Guo Z., Davis C.T., Paulson J.C., Stevens J., Rupprecht C.E., Holmes E.C., Wilson I.A., Donis R.O. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9(10) doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretyakova I., Pearce M.B., Florese R., Tumpey T.M., Pushko P. Intranasal vaccination with H5, H7 and H9 hemagglutinins co-localized in a virus-like particle protects ferrets from multiple avian influenza viruses. Virology. 2013;442(1):67–73. doi: 10.1016/j.virol.2013.03.027. [DOI] [PubMed] [Google Scholar]
- Tsybalova L.M., Stepanova L.A., Kuprianov V.V, Blokhina E.A., Potapchuk M.V, Korotkov A.V, Gorshkov A.N., Kasyanenko M.A., Ravin N.V, Kiselev O.I. Development of a candidate influenza vaccine based on virus-like particles displaying influenza M2e peptide into the immunodominant region of hepatitis B core antigen: Broad protective efficacy of particles carrying four copies of M2e. Vaccine. 2015;33(29):3398–3406. doi: 10.1016/j.vaccine.2015.04.073. [DOI] [PubMed] [Google Scholar]
- Tyrrell C.S., Allen J.L.Y., Gkrania-Klotsas E. Influenza: epidemiology and hospital management. Medicine. 2021;49(12):797–804. doi: 10.1016/j.mpmed.2021.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venereo-Sánchez A., Fulton K., Koczka K., Twine S., Chahal P., Ansorge S., Gilbert R., Henry O., Kamen A. Characterization of influenza H1N1 Gag virus-like particles and extracellular vesicles co-produced in HEK-293SF. Vaccine. 2019;37(47):7100–7107. doi: 10.1016/j.vaccine.2019.07.057. [DOI] [PubMed] [Google Scholar]
- Vergara-Alert J., Argilaguet J.M., Busquets N., Ballester M., Martín-Valls G.E., Rivas R., López-Soria S., Solanes D., Majó N., Segalés J., Veljkovic V., Rodríguez F., Darji A. Conserved synthetic peptides from the hemagglutinin of influenza viruses induce broad humoral and T-cell responses in a pig model. PLoS One. 2012;7(7):e40524. doi: 10.1371/journal.pone.0040524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tang C.Y., Wan X.F. Antigenic characterization of influenza and SARS-CoV-2 viruses. Anal. Bioanal. Chem. 2022;414(9):2841–2881. doi: 10.1007/s00216-021-03806-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward B.J., Makarkov A., Séguin A., Pillet S., Trépanier S., Dhaliwall J., Libman M.D., Vesikari T., Landry N. Efficacy, immunogenicity, and safety of a plant-derived, quadrivalent, virus-like particle influenza vaccine in adults (18–64 years) and older adults (≥65 years): two multicentre, randomised phase 3 trials. Lancet. 2020;396(10261):1491–1503. doi: 10.1016/s0140-6736(20)32014-6. [DOI] [PubMed] [Google Scholar]
- Werninghaus I.C., Hinke D.M., Fossum E., Bogen B., Braathen R. Neuraminidase delivered as an APC-targeted DNA vaccine induces protective antibodies against influenza. Mol. Ther. 2023;31(7):2188–2205. doi: 10.1016/j.ymthe.2023.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel D., Barbian A., Jenzelewski V., Schembecker G., Merz J., Piontek M. Bioprocess optimization for purification of chimeric VLP displaying BVDV E2 antigens produced in yeast Hansenula polymorpha. J. Biotechnol. 2019;306:203–212. doi: 10.1016/j.jbiotec.2019.10.008. [DOI] [PubMed] [Google Scholar]
- Wildt S., Gerngross T.U. The humanization of N-glycosylation pathways in yeast. Nat. Rev. Microbiol. 2005;3(2):119–128. doi: 10.1038/nrmicro1087. [DOI] [PubMed] [Google Scholar]
- Wu N.C., Wilson I.A. Influenza hemagglutinin structures and antibody recognition. Cold Spring Harb. Perspect. Med. 2020;10:a038778. doi: 10.1101/cshperspect.a038778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi J., Lei L.R., Zouzas W., April Si X. Nasally inhaled therapeutics and vaccination for COVID-19: Developments and challenges. MedComm. 2021;2(4):569–586. doi: 10.1002/mco2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Shi M., Li J., Song P., Li N. Construction of SARS-CoV-2 virus-like particles by mammalian expression system. Front. Bioeng. Biotechnol. 2020;8:862. doi: 10.3389/fbioe.2020.00862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Ma S., Huang Y., Chen F., Chen L., Ding D., Zheng Y., Li H., Xiao J., Feng J., Peng T. Virus-like particle vaccines for poliovirus types 1, 2, and 3 with enhanced thermostability expressed in insect cells. Vaccine. 2019;37(17):2340–2347. doi: 10.1016/j.vaccine.2019.03.031. [DOI] [PubMed] [Google Scholar]
- Yang J., Zhang L., Zhang C., Lu Y. Exploration on the expression and assembly of virus-like particles. Biotechnol. Notes. 2021;2:51–58. doi: 10.1016/j.biotno.2021.08.003. [DOI] [Google Scholar]
- Yang Z., Gao F., Wang X., Shi Likang, Zhou Z., Jiang Y., Ma X., Zhang C., Zhou C., Zeng X., Liu G., Fan J., Mao Q., Shi Li. Development and characterization of an enterovirus 71 (EV71) virus-like particles (VLPs) vaccine produced in Pichia pastoris. Hum. Vacc. Immunother. 2020;16(7):1602–1610. doi: 10.1080/21645515.2019.1649554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Wang H., Chen Jianjun, Shao Z., He B., Chen Jie, Lan J., Chen Q., Chen Z. Protection against homo and hetero-subtypic influenza A virus by optimized M2e DNA vaccine. Emerg. Microbes Infect. 2019;8(1):45–54. doi: 10.1080/22221751.2018.1558962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassine H.M., Boyington J.C., McTamney P.M., Wei C.J., Kanekiyo M., Kong W.P., Gallagher J.R., Wang L., Zhang Y., Joyce M.G., Lingwood D., Moin S.M., Andersen H., Okuno Y., Rao S.S., Harris A.K., Kwong P.D., Mascola J.R., Nabel G.J., Graham B.S. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 2015;21(9):1065–1070. doi: 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]
- Yong C.Y., Yeap S.K., Ho K.L., Omar A.R., Tan W.S. Potential recombinant vaccine against influenza A virus based on M2e displayed on nodaviral capsid nanoparticles. Int. J. Nanomed. 2015;10(1):2751–2763. doi: 10.2147/ijn.s77405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S.H., Jeong H., Kwon S.K., Kim J.F. Genomics, biological features, and biotechnological applications of Escherichia coli B: “Is B for better?!”. Syst. Biol. Biotechnol. E. coli. 2009:1–17. doi: 10.1007/978-1-4020-9394-4_1. [DOI] [Google Scholar]
- Zhang X., Wei M., Pan H., Lin Z., Wang K., Weng Z., Zhu Y., Xin L., Zhang J., Li S., Xia N., Zhao Q. Robust manufacturing and comprehensive characterization of recombinant hepatitis E virus-like particles in Hecolin®. Vaccine. 2014;32(32):4039–4050. doi: 10.1016/j.vaccine.2014.05.064. [DOI] [PubMed] [Google Scholar]
- Zhang Z.X., Nong F.T., Wang Y.Z., Yan C.X., Gu Y., Song P., Sun X.M. Strategies for efficient production of recombinant proteins in Escherichia coli: Alleviating the host burden and enhancing protein activity. Microb. Cell Fact. 2022;21(1):191. doi: 10.1186/s12934-022-01917-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimna M., Brzuska G., Salát J., Svoboda P., Baranska K., Szewczyk B., Růžek D., Krol E. Functional characterization and immunogenicity of a novel vaccine candidate against tick-borne encephalitis virus based on Leishmania-derived virus-like particles. Antiviral Res. 2023;209 doi: 10.1016/j.antiviral.2022.105511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No experimental data were generated for this review article.