Abstract
Background
Sox domain containing genes are important metazoan transcriptional regulators implicated in a wide rage of developmental processes. The vertebrate B subgroup contains the Sox1, Sox2 and Sox3 genes that have early functions in neural development. Previous studies show that Drosophila Group B genes have been functionally conserved since they play essential roles in early neural specification and mutations in the Drosophila Dichaete and SoxN genes can be rescued with mammalian Sox genes. Despite their importance, the extent and organisation of the Group B family in Drosophila has not been fully characterised, an important step in using Drosophila to examine conserved aspects of Group B Sox gene function.
Results
We have used the directed cDNA sequencing along with the output from the publicly-available genome sequencing projects to examine the structure of Group B Sox domain genes in Drosophila melanogaster, Drosophila pseudoobscura, Anopheles gambiae and Apis mellifora. All of the insect genomes contain four genes encoding Group B proteins, two of which are intronless, as is the case with vertebrate group B genes. As has been previously reported and unusually for Group B genes, two of the insect group B genes, Sox21a and Sox21b, contain introns within their DNA-binding domains. We find that the highly unusual multi-exon structure of the Sox21b gene is common to the insects. In addition, we find that three of the group B Sox genes are organised in a linked cluster in the insect genomes. By in situ hybridisation we show that the pattern of expression of each of the four group B genes during embryogenesis is conserved between D. melanogaster and D. pseudoobscura.
Conclusion
The DNA-binding domain sequences and genomic organisation of the group B genes have been conserved over 300 My of evolution since the last common ancestor of the Hymenoptera and the Diptera. Our analysis suggests insects have two Group B1 genes, SoxN and Dichaete, and two Group B2 genes. The genomic organisation of Dichaete and another two Group B genes in a cluster, suggests they may be under concerted regulatory control. Our analysis suggests a simple model for the evolution of group B Sox genes in insects that differs from the proposed evolution of vertebrate Group B genes.
Background
The family of Sox-domain containing proteins encompass a group of metazoan transcriptional regulators first identified by their similarity with the mammalian testis-determining factor SRY. Membership of the Sox family is conferred by the presence of an HMG1-type DNA-binding domain sharing greater than 60% amino-acid sequence identity to that of SRY [1]. Mammalian genome sequencing projects indicate that in humans and mice there are twenty Sox genes [2], divided into eight subgroups (A-H) on the basis of sequence identity within and outwith the HMG-domain. Aside from mammals, Sox genes have been identified in all metazoans examined to date, including birds, fish amphibians, basal chordates, insects and nematodes [3].
The B subgroup is of particular interest since members of this group are most closely related to SRY and appear to be functionally conserved during evolution. Sequence analysis and functional studies suggest that, in vertebrates, the five members of the B subgroup can be subdivided into two further groups; B1; Sox1, Sox2 and Sox3; [4] and B2; Sox14 and Sox21; [5]. It has been suggested from studies in the chick that the three group B1 proteins act as gene activators whereas the B2 proteins act as gene repressors [6]. In terms of genomic organization, all five of the group B genes are devoid of introns. Sox3 is located on the mammalian X chromosome and is believed to be the ancestor of Sry [7,8]. In humans, the remaining four autosomal group B genes are arranged in two pairs, each comprising one B1 gene and one B2 gene: Sox2 and Sox14 map together on chromosome 3 [9,10] and Sox1 and Sox21 map together on chromosome 13 [5,11]. This organization is conserved, at least in part, in other vertebrates with Sox2-Sox14 mapping together in the chick and the monotreme, O. anatinus, and Sox1-Sox21 mapping together in the chick [12,13]. There is, however, no linkage of Group B Sox genes in the mouse genome [14,15]. A model suggesting the evolution of group B genes and Sry from a single ancestor has been proposed, which suggests that pairs of B1 and B2 genes arose by a tandem duplication and then a chromosomal duplication [13].
The fruitfly, Drosophila melanogaster, has proved to be a tractable system for studying conserved aspects of eukaryotic gene function and, with the production of other insect genome sequences, a useful baseline for evolutionary studies of gene organisation [16]. Whole-genome sequence is now available for three insects, Drosophila melanogaster, Drosophila pseudoobscura (which diverged from melanogaster some 46 million years ago) and Anopheles gambiae, which diverged from melanogaster approximately 250 million years ago [17,18]. Sequencing and assembly of a further ten Drosophila species is currently underway [19] promising an unparalleled data source for evolutionary studies. In addition to the diptera, the sequencing of the Hymenoptera, Apis mellifera (honey bee ~280 million years from Drosophila), is now well underway, allowing fragments of a fourth insect genome to be assessed. In functional terms, Drosophila is a useful model for studying SOX gene function due to its genetic tractability. For example, we have previously shown that, in the case of the Drosophila group B gene Dichaete, there is functional conservation between insect and mammalian genes [20]. In addition, we, and others, have demonstrated a degree of in vivo functional redundancy between Dichaete and SoxN [21,22] as had been proposed for the mammalian group B genes [23]. Of particular interest is the fact that the expression patterns and functional studies of group B genes suggest that they participate in the earliest events of CNS differentiation in all organisms that have been studied to date including Drosophila, Xenopus, chick, mouse, ascidians and hemichordates [24].
To further explore the relationship between group B Sox genes we examined the extent and organization of the family in insects. Our studies show that group B Sox gene organisation is similar in four different insects. We find conservation in the sequence and genome organization of the group B genes in D. melanogaster, D. pseudoobscura, A. gambiae and A. melifora. In contrast to mammals and in agreement with a previous report [25], we find that two group B2 genes contain introns and are organized as a single genomic cluster along with the intronless Dichaete gene. Our studies indicate a potentially different evolutionary path for members of the group B family in insects and vertebrates.
Results
To explore the structure of the group B Sox genes in insects we first accurately determined the extent and structure of the family in Drosophila melanogaster. The group B genes, Dichaete and SoxNeuro (SoxN) have already been well described in the literature [26-28]. Two other group B gene fragments have been identified [25], Sox21a and Sox21b, but their structure and genomic organisation have not been reported. Using a combination of database searching and DNA sequencing we characterised both of these genes in detail. We find no evidence for any other group B genes in Release 3.2 or Release 4 of the Drosophila genome sequence, indicating that there are a total of four in the D. melanogaster genome.
Sox21a
Blast searches of the Drosophila genome identified a group B HMG-domain interrupted by a 1655-bp intron in the 70D region of chromosome arm 3L. Using primers designed against each of these predicted exons we amplified a fragment of 1238 bp from the LD cDNA library produced by the BDGP [29]. For reasons that are as yet unclear, we have been unable to recover a clone from this or any of the several other cDNA libraries that we have screened. The fragment amplified from the library was sequenced in its entirety and found to contain a long open reading frame encoding a 389 amino acid Sox domain protein. The predicted polypeptide initiates with a methionine and probably contains the entire coding sequence for the gene. When aligned with the genome sequence we predict a gene with two exons spanning 2.8 kb. Blast searches with the predicted protein find over 90% identity with a range of group B Sox proteins in the HMG DNA-binding domain. The best scores are with the DNA-binding domains of the vertebrate Sox21 and Sox14 proteins; however, there is little significant similarity outside of the DNA-binding domain. The Sox21a gene has previously been reported as SoxB2-3 (CG7345) and it has been suggested that it may represent a pseudogene [25]. As we show below, RT-PCR and in situ hybridisation studies indicate that Sox21a is expressed in both D. melanogaster and D. pseudoobscura indicating that it is not a pseudogene.
Sox21b
Along with Sox21a, Blast searches indicated a second interrupted Sox domain in the same region of the genome. In this case, database searches found a potential cDNA clone from the BDGP (GH07353), which was obtained and sequenced in its entirety. The sequence of the clone revealed a long open reading frame, initiating with a methionine, encoding a predicted polypeptide of 571 amino acids. Alignment with the genome sequence indicates that the gene spans 19 kb of genomic DNA and is composed of 7 exons, the first of which is non-coding. The DNA-binding domain contains two introns; the first, 6388 bp in size, is in the middle of the DNA-binding domain and the second, of 59 bp, is in the same position and frame as the Sox21a intron described above. Blast searches with the predicted amino acid sequence find greater than 90% amino acid identity the DNA-binding domains of group B Sox proteins, the highest scores being with Dichaete. The sequence indicates that Sox21b corresponds to the SoxB2-2 gene fragment previously reported [25] and whole mount in situ hybridisation with probes derived from genomic DNA and the cDNA clone confirm the pattern of expression previously reported (Figure 2). Thus, both Sox21a and Sox21b are expressed group B Sox genes that have their DNA binding domains interrupted by introns.
Figure 2.
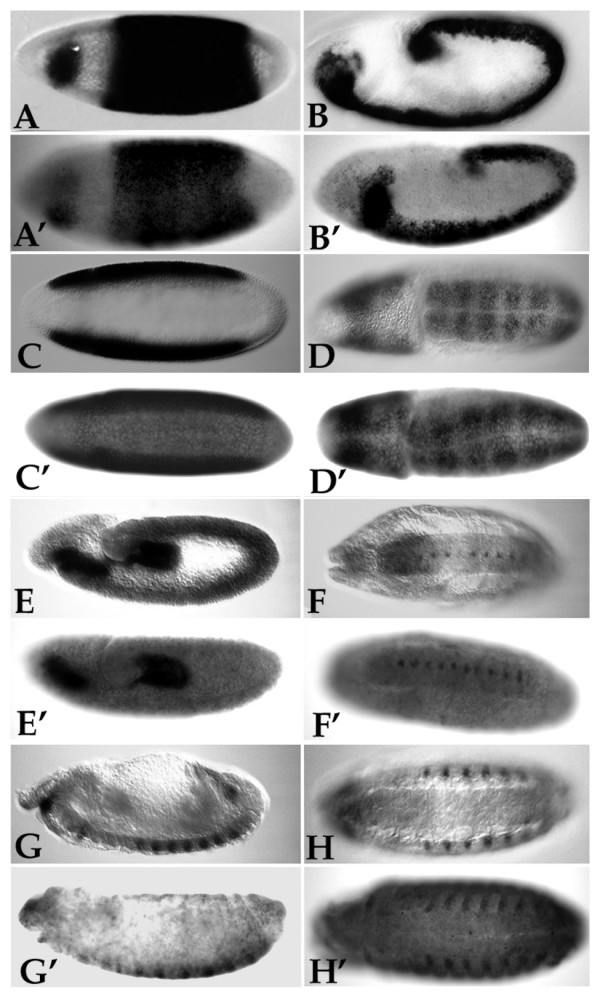
Embryonic expression of group B genes in D. melanogaster and D. pseudoobscura. A-H, D. melanogaster. A'-H', D. pseudoobscura; anterior is to the left in all cases. A-A') Lateral view of stage 5 embryos showing expression of Dichaete in a central domain and the cephalic neuroectoderm. B-B') Lateral views of stage 8 embryos showing extensive Dichaete expression in the developing CNS. C-C') Ventral view of stage 5 embryos showing SoxN expression is restricted from the presumptive mesoderm. D-D') Dorsal view of stage 8 embryos showing SoxN expression in the CNS. E-E') Lateral view of stage 9 embryos showing Sox21a expression in the anlage of the foregut and hindgut. F-F') Ventral views of stage 14 embryos showing Sox21a expression restricted to specific cells in the midline. G-G') Lateral views of stage 13 embryos showing Sox21b expression in abdominal epidermal stripes. H-H') Ventral view of stage 14 embryos showing Sox21b expression in abdominal epidermal stripes.
To verify the gene predictions and gain some insight into their possible biological functions we determined the developmental expression of each of the four group B genes by RT-PCR using RNA templates isolated from different stages of the Drosophila lifecycle. In the case of Sox21a and Sox21b, we used primers to the Sox-domain encoding exons spanning a predicted intron. All RT-PCR reactions included a reverse transcriptase minus reaction and the amplified products were verified by sequencing. The results of this analysis are presented in Table 1 and can be summarised as follows: the expression profiles of Dichaete and SoxN are very similar, they are expressed during embryonic, larval and pupal stages of development, the level of expression reducing during the later stages of pupal development. Both genes are expressed in adult male as well as female flies, with bodies showing stronger expression than heads. Sox21a is expressed throughout development and in adults it is stronger in heads than in bodies. In contrast to the other group B genes, Sox21b has a more complex expression pattern during development. It is strongly expressed in embryos but is below detectable levels for much of larval and pupal life. After eclosion it is weakly expressed in male heads but not bodies. Thus, in common with mammalian group B genes, all four D. melanogaster group B Sox are expressed during embryogenesis and at other stages throughout development.
Table 1.
representation of Sox expression in during Drosophila development assayed by RT-PCR.
| Dichaete | SoxNeuro | sox21a | sox21b | |
| Embryo | + | + | + | + |
| 1st instar | + | + | (+) | (+) |
| 2nd instar | + | + | (+) | - |
| Early 3rd instar | + | + | (+) | - |
| Late 3rd instar | + | + | + | - |
| Prepupa | + | + | + | - |
| 12 h pupa | + | + | + | - |
| 36 h pupa | (+) | (+) | + | - |
| Heads | (+) | (+) | + | (+) |
| Bodies | + | + | (+) | - |
| Male | + | + | + | (+) |
| Female | + | + | + | - |
+ = expression, - = no expression, (+) = weak expression.
Group B genes in other insects
Our findings show that Drosophila melanogaster has four group B Sox genes compared to the five found in vertebrates and, unlike vertebrates, two of the genes contain introns. To investigate whether this particular organization is unique to D. melanogaster we searched the available genome sequence of other insects to find potential Sox domain genes. Using the Dichaete DNA-binding domain as a query, we searched the Drosophila pseudoobscura, Anopheles gambiae and Apis mellifera genome and EST sequence databases using Blast-P and Blast-N (see materials and methods for EST and genome scaffold accessions). In all three cases we found evidence for four Group B genes and were able to build gene models, from the genome sequence alone or with the addition of EST data where available. The initial characterization of the insect group B genes, based on the HMG-domain sequence, suggests that there is a single orthologue of each Drosophila gene in the other three species.
SoxN
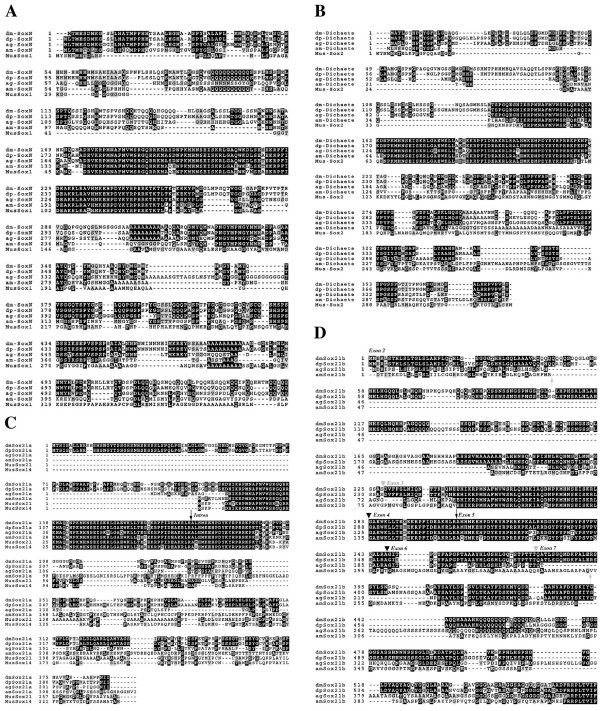
The alignment presented in Figure 1a. shows the similarity between the insect SoxN proteins and mouse Sox1. As previously reported, conservation between vertebrate and invertebrate Sox proteins is mostly restricted to the DNA-binding domains [26,27]. Between the insect proteins there are more extensive regions of homology outwith the DNA-binding domain. The Drosophila SoxN sequences show over 90% sequence identity over their entire length and, as expected from the phylogenies based on rDNA and protein coding sequences, the other insect sequences are more diverged [18,30]. A. gambiae is overall 64% identical to the melanogaster sequence with particularly well conserved regions in the N-terminal 50 amino acids and more patchy conservation C-terminal to the DNA-binding domain. A. mellifera is further diverged (52% identity with Drosophila). Conserved regions outside the DNA-binding domains among all four sequences are restricted to a stretch of amino acids C-terminal that may represent conserved functional motifs important in transcriptional regulation.
Figure 1.
Full alignments of the insect Group B protein sequences. A) SoxN. B) Dichaete. C) Sox21a, the position of the conserved intron is indicated with an arrow. D) Sox21b, the location of the exons in the D. melanogaster sequence in indicated in italics above the alignment. Black arrowheads above the alignment indicate positions of introns conserved in all four species. The grey arrowheads indicate intron positions conserved in the diptera. The black arrow above the alignment indicates the drosophila specific intron and the grey arrows below the alignment indicates the apis-specific introns.
Dichaete
The situation with Dichaete is similar to that observed with SoxN, and the figures for amino acid identity are virtually identical (Figure 1b). Outside of the DNA-binding domain the Dichaete sequences show even less similarity comparing the Drosophila species and the other two insects; conservation between all four being restricted to limited regions C-terminal to the DNA-binding domain. Interestingly, we have shown that the C-terminal region of D. melanogaster Dichaete contains sequences required for activity in a context-specific manner [20] and C-terminal regions of the mouse and chicken Sox2 protein are believed to be involved in aspects of correct Sox2 function [31].
Sox21a
This gene is the least conserved between the four species and outside of the DNA-binding domain they show little similarity with vertebrate group B2 proteins (Figure 1c). There is extensive homology between the two Drosophila species, however, the Anopheles and Apis sequences are very diverged outside of the DNA binding domain. As with D. melanogaster, there are no EST sequences available that support the structure of Sox21a in the other insects.
Sox21b
The predicted Drosophila Sox21b proteins are again very similar, over 88% identical over their length. The other insect sequences are less well conserved, although the Anopheles sequence has a block of conservation C-terminal to the DNA-binding domain, including a Glutamic acid-rich domain (Figure 1d). The predicted Apis sequence is less well conserved, we note, however, that all four proteins are identical at the extreme C-terminus. With both the Anopheles and Apis proteins we cannot confidently predict the N-terminal exons and are unable to find any regions with amino acid similarity to the first 2 coding exons of the Drosophila sequences in the Anopheles or Apis genomic sequence between the end of Dichaete and the Sox21b Sox-domain encoding exons. Our current models are, however, supported by the available EST sequences for both species although the EST sequences are not full-length. Therefore, the definitive structure of these two insect Sox21b genes will require further investigation. Nevertheless, it is clear from the available sequence that orthologues of Sox21b are present in other insects.
To confirm the identification of four group B genes in both D. melanogaster and D. pseudoobscura, we performed whole-mount in situ hybridization to embryos of both species using exon-specific probes generated by PCR from genomic DNA. In all four cases we find very similar patterns of expression during embryogenesis. In the case of Dichaete, we find blastoderm expression including a broad central domain and a region of expression in the cephalic neuroectoderm (Figure 2A and 2A'). After gastrulation there is extensive expression in the developing CNS (Figure 2B and 2B') including the midline (not shown). With SoxN we find conserved blastoderm expression, including an identical restriction from the ventral region of the embryo, followed by extensive expression throughout the developing CNS (Figure 2C to 2D'). With Sox21a, we identified conserved expression in the anlage of the foregut and hindgut at stage 12 (Figure 2E and 2E') with later expression in specific cells of the midline after stage 14 (Figure 2F and 2F'). Sox21b shows conserved expression in abdominal epidermal stripes from stage 13 (Figure 2G to 2H'). These observations indicate that all four group B genes have conserved expression patterns during embryogenesis.
Genomic organisation of group B genes in Drosophila: the Dichaete complex
In some vertebrates the two classes of group B genes, B1 and B2, are linked on the same chromosome. In contrast, with Drosophila a single gene, SoxN, maps to the second chromosome and the remaining three all map to chromosome 3. We examined the organisation of the group B genes in the other insect genomes and found that the situation was very similar to that observed in Drosophila. In melanogaster, SoxN is intronless and sits alone in the middle of an 80 Kb island with no flanking genes for 35 Kb proximal and 45 Kb distal, an unusual organisation for a Drosophila gene. We have previously shown that Dichaete is controlled by extensive 3' regulatory sequences, suggesting that perhaps the paucity of genes flanking SoxN may also indicate the presence of extensive regulatory sequences. In support of this, we find several clusters of predicted transcription factor binding sites from 35 kb upstream to 20 kb downstream of SoxN when we use a stringent search criteria with CisAnalyst analysis software [32,33] (data not shown). Similar searches with Dichaete find previously identified regulatory sequences, suggesting that SoxN may indeed be subject to complex regulation. Comparative analysis of the melanogaster and pseudoobscura genomes with the Vista genome alignment viewer [34,35] indicates that the genomic organization is very similar in the two species. The Ensemble annotation of the Anophelese genome indicates that the region around SoxN is also sparsely populated, with only 2 short stretches of EST homology in the 150 kb flanking SoxN. Therefore, it is possible that SoxN is subject to complex regulatory control in Anopheles. There is currently insufficient contiguous genomic sequence from Apis to assess the organization of the SoxN region.
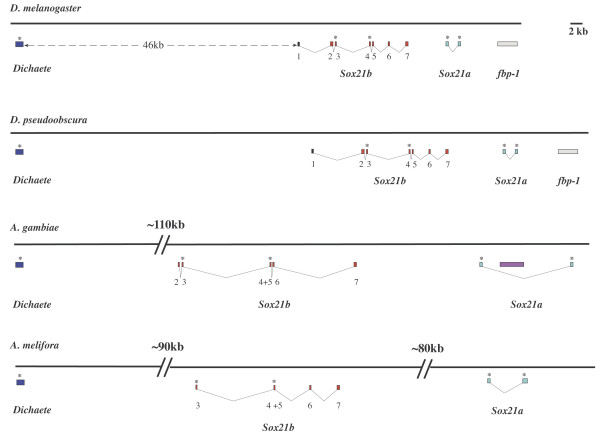
In the 70D region of Drosophila melanogaster chromosome arm 3L the remaining three group B genes are clustered within an 77 kb region (Fig 3). As we have previously reported, Dichaete is an intronless gene controlled by at least 30 Kb of regulatory sequence 3' to the transcription unit [36]. 16 kb further distal to these regulatory sequences we find the start of Sox21b and a further 28 kb distal to this the start of Sox21a. The region ends with the Fat Body Protein 1 (Fbp1) gene 6 kb downstream of Sox21a. The genomic organization of Sox21b is highly unusual for a group B Sox gene. It is split into seven exons, the first of which is non-coding and exons 3, 4 and 5 contain the DNA-binding domain. All of the predicted splice junctions have consensus GT-AG sequences. Sox21a, comprises 2 exons, each containing a portion of the DNA-binding domain. As we note above, the position of the intron, which has consensus splice junction sequences, is in the same position as the second DNA-binding domain intron of Sox21b (intron 4).
Figure 3.
The genomic organisation of the insect Dichaete regions. Exons are represented by shaded boxes and introns by the linking lines. A scale bar of 2 kb is indicated. The melanogaster and pseudoobscura sequences are to scale, the larger distance between Dichaete and Sox21b in Anopheles and Apis is indicated by a break in the line, the remainder of the diagram is to scale.
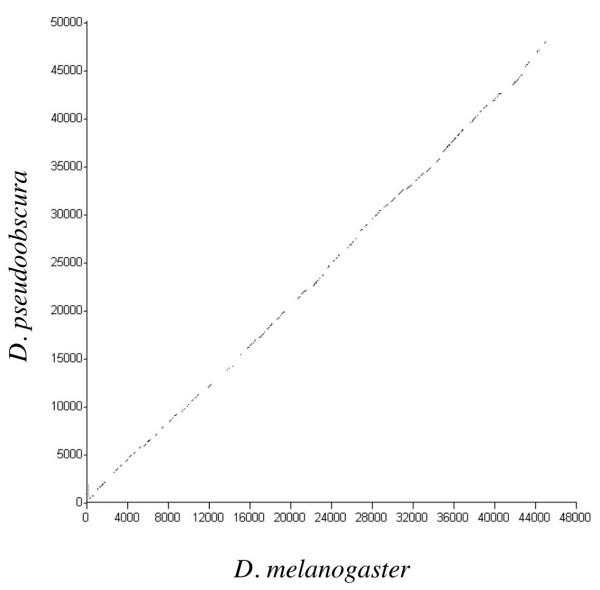
In the case of D. pseudoobscura, the homology extends from upstream of the Dichaete coding region to at least the Fat body protein 1 gene downstream of Sox21a. The organization of the three Sox genes is virtually identical comparing the two species and we could construct gene models including all of the Sox21a and Sox21b exons. There is absolute conservation of the intron position between both Drosophila species, furthermore, the sizes of the introns is also similar, although nucleotide similarity is lower than in coding sequences ranging from 40 – 75%. As with melanogaster, we find no evidence for additional genes in the intergenic region between Dichaete and Sox21b. We used the OWEN sequence alignment programme to plot the conservation between the Dichaete – Sox21b intergenic region in both species (Figure 4). Throughout the entire region we see that there is a high degree of sequence conservation, since we know that at least 30 kb of this region contains essential Dichaete regulatory sequences in melanogaster, we predict that regulation in the region will be similar in both species. A suggestion supported by the in situ hybridization data presented above (Figure 2).
Figure 4.
OWEN alignment of the region between Dichaete and Sox21b in D. melanogaster and D. pseudoobscura showing extensive sequence similarity throughout the 45 kb region.
The organization of the Dichaete region in the Anopheles genome is very similar to that in the Drosophila species with three genes found in a 190 kb region of chromosome arm 3L. Dichaete is intronless and Sox21b is located approximately 110 kb downstream of this. There are no other predicted genes in the region. The Sox21b has a similar structure to those of the Drosophilids, however, it is not identical. We have been unable to find a 5' non-coding exon and, as we note above, the second intron found in the DNA-binding domains of the Drosophila Sox21b genes is absent in Anopheles with exons 4 and 5 fused. The other introns are, however, conserved in position (figure 1d). With the Anopheles Sox21a gene, the single intron position is conserved with the Drosophila species, however, the intron is considerably larger and contains an insertion of a Q-class retrotransposon in the sequenced strain [37]. We find no evidence for an Fbp1 orthologue in the vicinity, the nearest similar sequence being some 5 Mb away on the same chromosome arm.
The available sequence in the region is more fragmentary in the case of Apis. Here we find an intronless Dichaete gene and can define two sets of exons corresponding to the split DNA binding domains of Sox21a and Sox21b. Overall, the organization is similar to the other insects; like Anopheles, the intergenic region between Dichaete and Sox21b is large (~90 kb), however, unlike the other insects the distance between Sox21b and Sox21a is also large (~80 kb). In the case of Sox21b we have used EST sequence to support the gene model we have derived. The EST confirms the first four exons and we predict the terminal exon on the basis of homology with the other species, particularly the terminal 30 amino acids. As with Anopheles, the Apis Sox21b gene has a single DNA-binding domain intron in the same position of the first Drosophila DNA-binding domain intron. The intron immediately downstream of the DNA-binding domain is also conserved in all four insects, however, the remaining two intron positions differ between Apis and the other insects. Although the Apis assembly is preliminary in this region, with several gaps still present in the sequence, the fact that the gene models are very similar to the other insects and that Dichaete and Sox21b predictions are supported by EST data suggests that the gene models we propose are likely to be accurate for the majority of the coding sequence.
We compared the Dichaete to Sox21b intergenic regions of Anopheles and Apis to the melanogaster sequence with the OWEN alignment tool and failed to detect any significant stretches of similarity, even at relatively low stringency. This suggests that if there is conservation in gene regulatory sequences between these diverged insects it may be difficult to detect or have undergone extensive rearrangement.
Evolutionary perspective on insect group B genes
To attempt a classification of the insect group B Sox genes, we performed a multiple sequence alignment with the DNA-binding domains of the predicted proteins along with representative group B-like sequences from other organisms (Figure 5). The aligned ClustalX output suggests that the insect Sox domains may be subdivided into 3 classes. The first clearly groups the SoxN proteins from each of the insects with the mammalian Sox1, 2 and 3 proteins. Along with these we find representative sequences from nematodes (C. elegans, S. ratti, and W. bancrofti), hemichordate Acorn worms (S. kowalevski and P. flava) and the sea squirt (H. roretzi). Thus, together these are likely to represent a single class, orthologous to vertebrate group B1 proteins. The second class, the Sox21a proteins, have sequences similar to the mammalian group B2 proteins, Sox14 and 21 and may represent an insect group B2 protein. The third class, containing Dichaete and Sox21b, are clearly differentiated from all other group B proteins by the presence of a Leucine/Isoleucine residue at position 18, an Isoleucine residue at position 23 and a divergent set of C-terminal amino acids. These two insect proteins may represent an insect-specific group B family. This suggests that a single group B1 protein, represented by SoxN-Sox3 like sequences, was present in a common ancestor before the divergence of vertebrates and invertebrates. Similarly, the close association of the insect Sox21a proteins with nematode and vertebrate Sox14 proteins suggests that these were also present in a common ancestor. The alignments clearly highlight the distinction between the Dichaete-Sox21b pair and other group B proteins, emphasizing a distinct evolutionary history for these proteins in the insects.
Figure 5.
Group B Sox-domain alignment. Clustal X alignment of DNA-binding domain sequences from the insect proteins and representative group B proteins from other species. The insect sequences are highlighted in grey. Accession numbers of protein sequences are as follows: SOX15 Human, O60248; SOX15 Mouse, P43267; Dichaete melanogaster, Q24533; Dichaete pseudoobscura, TR; Dichaete Anophelese, TR; Sox21b melanogaster, Q9VUD3; Sox21b pseudoobscura, TR; Sox21b Anophelese, TR; Sox21b Apis, TR; Dichaete Apis, TR; SOX1 Human, O00570; SOX1 Mouse, P53783; SOX1 Chicken, O57401; SOX3 Human, P41225; SOX3 Mouse, P53784; SOX3 Chicken, P48433; SOX3 Xenopus, P55863; SOX3 Medaka, Q9PT76; SOX2 Chicken, P48430; SOX2 Xenopus, O42569; SOX2 Human, P48431; SOX2 Mouse, P48432; SOX2 Sheep, P54231; SOX1 S. kowalevski, Q7YTD4; SOXB1 P. flava, (Taguchi et. al. 2002); SOXB1 Sea Urchin, Q9Y0D7; SoxN Apis, TR; SoxN melanogaster, Q9U1H5; SoxN pseudoobscura, TR; SoxN Anopheles, TR; SoxB s. ratti1, BI323817; SoxB s. ratti2, BI323817; SoxB W. bancrofti, CD455919; SOX2 C. elegans, Q21305; SOXB1 H. roretzi, Q86SB8; SOX19 Zebra Fish, P47792; SOX21 Zebra Fish, Q9YH21; SOX14 Chicken, Q9W7R6; SOX14 Human, O95416; SOX14 Mouse, Q04892; SOX14 Platypus, Q8MIP4; SOX21 Human, Q9Y651; SOX21 Mouse, Q811W0; SOX21 Chicken, Q9W7R5; SOXB2 P. flava, (Taguchi et. al. 2002); Sox21a melanogaster, Q9VUD1; Sox21a pseudoobscura, TR; Sox21a Anopheles, TR; Sox21a Apis, TR; SOXB2 Sea Urchin, Q9Y0D8; SoxB C. virginica, CD648628; SOX3 C. elegans, Q20201; SoxB T. spiralis, BG302262; SoxB M. hapla, BU095063; SRY Human, Q05066; SRY Sea Lion, AAR10360; SRY Mouse, Q05738. TR = This report.
Taken together, the analysis presented here shows that the genomic organization and sequence of group B Sox genes have been conserved during insect evolution. Particularly striking is the clustering of three genes in a small region of the genome. The structure of these genes and their relationship with vertebrate Group B genes suggest that SoxN and Sox21a are homologous to vertebrate group B1 and B2 genes respectively, whereas Dichaete and Sox21b may represent insect-specific group B genes.
Discussion
The sequence alignments of the HMG DNA-binding domains from insect and mammalian group B Sox proteins suggests that the insect proteins may be separated into three distinct groups. The first, containing SoxN, aligns with the vertebrate Sox1, 2 and 3 proteins and most likely represents an orthologue of the vertebrate group B1 class. This conclusion, based on sequence, is supported by the functional analysis of group B1 proteins in vertebrates and Drosophila. In both cases, group B1 genes are expressed from the earliest stages of CNS development and are implicated in regulating early neural specification [21,22,38,39]. In addition, we have evidence that mammalian Sox1 genes can rescue SoxN phenotypes in the Drosophila CNS, supporting the view that these proteins are functionally conserved (P. Overton and S.R. unpublished observations). The group B sequences isolated from the basal chordates, acorn worm and sea squirt, have also been shown to be expressed early in the specification of the CNS [40,41]. Thus, it appears that all metazoans studied to date have at least one group B gene with expression marking neural lineages early in development. Further studies of primitive invertebrates will determine whether group B Sox expression is a universal marker for CNS development.
In a previously published phylogenetic studies it was suggested that Dichaete be classified as a Group B2 protein [3]. However, while the analysis clearly differentiates between the group B proteins and other fly Sox proteins it could not unambiguously resolve the relationship between each of the group B proteins. In terms of function and expression, the Dichaete gene behaves very much like a group B1 gene, it is expressed early during CNS development and is required for neural differentiation [20,42]. We have previously shown that the mouse Sox2 gene efficiently rescues Dichaete phenotypes, further supporting a functionally similarity between Dichaete and vertebrate group B1 genes [20,42]. In contrast to the conclusion based on functional studies, the sequence analysis suggests that insect Dichaete DNA-binding domain sequences are markedly different from other group B1 proteins and are more similar to group B2 proteins. The conservation of the insect sequences indicates that a Dichaete-like sequence was present at least 300 My years ago, when Apis and the Diptera last shared a common ancestor [18]. We believe that the functional evidence is more convincing than the arguments based on sequence alignments and therefore suggest that Dichaete represents a group B1 function that has diverged from the canonical group B1 sequence, presumably due to selection for insect-specific functions. For example, Dichaete is required for early segmentation in the Drosophila embryo, a highly derived function, and it may be that sequence changes in the HMG-domain have been selected for such a function while still allowing a role in CNS-specification. As with Drosophila, both Anopheles and Apis are long germ insects that share some aspects of early development such as the early appearance of striped domains of even skipped expression [43,44]. Thus it is possible that insect Dichaete genes have a common role in early patterning events. It will be of considerable interest to examine the complement of group B Sox genes in Coleoptera, Homoptera or Orthoptera to see if the HMG domain sequence and gene organisation is the same as the insects so far sequenced. To investigate this we used the Dichaete DNA-binding domain to search the available sequence of the silk moth Bombyx mori. [45] and found a single Group B gene that was clearly an orthologue of the Dichaete genes discussed here, containing the diagnostic Leucine and Isoleucine residues described here.
As with vertebrate group B1 genes, SoxN and Dichaete are expressed in broadly overlapping domains and act partially redundantly in CNS specification [21,22]. The close similarity between the expression and function of SoxN and Dichaete in the CNS raises the possibility that they arose from a common ancestor by a duplication event and may thus share some common regulatory sequences. However, when we compared the sequences 5' or 3' to SoxN with the Dichaete 3' sequence we could not detect any sequence similarity indicating that any conservation in regulatory sequences is not visible at a large scale; this is not entirely surprising since we cannot detect any sequence similarity between the Dichaete regulatory sequences from Drosophila and Anopheles, while our analysis indicates the divergence of SoxN and Dichaete predates the Drosophila-Anopheles divergence.
Based on the sequence alignment of insect Sox21a DNA-binding domains with those of vertebrate Sox14 proteins, it is possible that Sox21a may be an orthologue of the group B2 class. It has been suggested that in chicken Sox14 and Sox21 act as antagonists of group B1 function in a subset of the developing CNS [6]. The function of Sox21a in Drosophila is not known at present, however, Sox21a is expressed late in the development of the embryonic CNS midline, a site of SoxN and Dichaete expression, indicating there is the potential for the type of antagonistic interaction proposed for vertebrates. The Sox21b DNA-binding domain sequence indicates that it is closely related to Dichaete. Both these proteins have a set of unique residues in their DNA-binding domains that are not found in any other group B proteins identified to date. The Sox21b gene is conserved between the insects and its close similarity to Dichaete suggests that both genes arose from a common origin in the ancestor of the arthropods after their divergence from the nematodes since there is no close sequence in C. elegans or its relatives. In terms of expression, Sox21b is expressed in the large hindgut along with Dichaete, supporting the possibility that it may also antagonise the activity of Dichaete. In this respect then Sox21b may represent a group B2 function. It is therefore possible that insects contain 2 group B1 class activities, involved in early CNS development, and two B2 class genes. Again we emphasise that the functional assignment of the insect genes may contrast with the data derived from sequence analysis, which predicts a single group B1 gene and three group B2 genes. We suggest that the separation of group B Sox domains into a B1 class and B2 class based solely on sequence does not reflect meaningful functional differences in insects. We have initiated a functional analysis of Sox21a and Sox21b in the hope that we can clarify this issue.
The genome organisation of the Dichaete cluster is unusual, not only are three genes clustered together in the genome but two of them, Sox21a and Sox21b, have introns within the HMG-domain. The single Sox21a intron is conserved in all four of the insect genes suggesting that it is ancestral to the insects. Sox21b is more complex, there are six introns in melanogaster and pseudoobscura, four of these are conserved in Anopheles and two are conserved in Apis. In the Drosophila species, there are two introns in the DNA-binding domain, the first of which is present in all four insects. The second intron, in an identical location to the Sox21a intron, is only found in the two Drosophila species. A simple model of a single intron loss is therefore unlikely to account for this since both Apis and Anopheles do not have the intron. It is possible that Apis and Anophelese lost the intron independently or, alternatively, that the common ancestor of the Drosophila species gained the intron, perhaps via a gene conversion event with Sox21a. Interestingly, the two group B genes from C. elegans also contain introns in the DNA-binding domain, in identical positions in both genes, but they are in different positions to the Sox21a and Sox21b introns. This suggests that the common ancestor of insects and nematodes did not contain DNA-binding domain introns and that these have been acquired independently in both lineages.
The conservation of genome structure with the insect Dichaete cluster suggests that there may be functional constraints on the organisation. We suggest that this is likely to be a reflection of shared regulatory sequence since the region between Dichaete and Sox21b in melanogaster contains extensive regulatory sequences essential for correct Dichaete expression. We note that both Sox21a and Sox21b have expression domains that overlap with Dichaete, in the midline for Sox21a and the hindgut with Sox21b. These expression domains may therefore be controlled by common regulatory sequences and the need to maintain coordinated regulation of the three genes has maintained the integrity of the cluster in the insects. The conservation in expression between D. melanogaster and D. pseudoobscura is consistent with this view; it will be of interests to examine the expression of the all of the Sox genes in Anopheles to further explore this hypothesis.
Conclusion
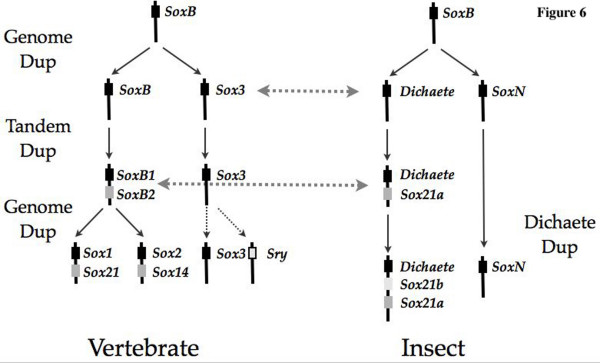
Taking our observation together, we propose a simple model for the evolution of group B SOX genes (Figure 6). We base our model on the proposal of Kirby et. al. [13] who suggest that a single group B gene underwent a duplication to generate two Sox3- like genes. We propose that these are represented by SoxN and Dichaete in the insects. A further tandem duplication of one of these genes generated linked group B1 and group B2 genes. We propose that this is represented by Dichaete duplicating to generate Sox21a. Sox21a would then acquire the sequence changes characteristic of the group B2 class of proteins. We suggest that these events predate the Protostome-Deuterostome divergence over 650 My ago [46] and provide the basal Sox gene complement of the Bilateria of three group B Sox genes. After the divergence of the lineages leading to vertebrates and invertebrates, Dichaete diverged from the canonical group B DNA-binding domain sequence and then underwent further duplication event, at least predating the divergence of the holometabolous insects, to generate Sox21b. An analysis of the group B family in other insects and basal chordates will be required to definitively describe the ancestral situation.
Figure 6.
A model for the evolution of Group B Sox genes in insects following the proposal of Kirby et al (2002) for vertebrates. In this view an ancestral group B gene is duplicated during an ancient genome duplication event to generate Dichaete and SoxN. A tandem duplication of Dichaete generates Sox21a; these events would be common to the ancestor of vertebrates and invertebrates. In insects, a further duplication of Dichaete gives rise to Sox21b.
Methods
Genome sequences
The following sources were used to obtain genome sequence: D. melanogaster (Release 3.2, [47,48]) from FlyBase [49] and the following scaffolds were used; AE003535 for the Dichaete region and AE003622 for the SoxN region. D. pseudoobscura (Freeze_1 assembly) was obtained from the Human Genome Sequencing Center, Baylor College of Medicine (HGSC-BCM [50]) and the following scaffolds used; Contig5946_Contig6670 for the Dichaete region and Contig1741_Contig5707 for the SoxN region. Anopheles gambiae genome sequence release 19.2a.1, compiled by the International Anopheles Genome Project [51], was obtained from the Ensembl server at the Wellcome Trust Sanger Institute [52]. In the Ensemble annotation the Sox genes have the following accessions: SoxN (ENSANGG00000019842), Dichaete (ENSANGG00000010137), Sox21a (ENSANGG00000010002) and Sox21b (ENSANGG00000009947). Anopheles EST sequences representing Dichaete (TC44994) and Sox21b (TC45155) were obtained from The Institute for Genome Research [53]. Apis mellifera Genome assembly Amel_1.1 was obtained from HGSC-BCM [54] and the following scaffolds used: for the Dichaete region, Group8.12 (Dichaete and Sox21b) was found to overlap by 4.5 kb with GroupUn.570 containing Sox21a and the sequences were combined into a single contig. SoxN was contained within Group17.6. In addition a search of the Honey Bee Brain EST project [55,56] uncovered two EST sequences corresponding to Dichaete (BB170009A10D01) and Sox21b (BB170011B20A11). These were used to verify the exon predictions from the genome sequence. Vertebrate group B sequences were obtained from Uniprot. Nematode sequences were recovered by Blast searches of the EST collections at Nematode.net (Genome Sequencing Center, Washington University, St Louis, [57]).
Informatics tools
Homology searching was performed using the Blast algorithm [58] at Sanger, HGSC-BCM and Berkeley Drosophila Genome Project [59] web sites. Genomic sequences were imported into Artemis v5 [60,61] and annotated manually using the Blast output as a guide. Multiple Sequence alignments were performed locally using ClustalXv1.8 [62] and graphically represented with BoxShade [63]. The alignment of intergenic regions was performed with OWEN [64].
Molecular biology
A cDNA clone for Sox21b (GH07353) was obtained from the Drosophila gene collection [29] and sequenced on both strands using an ABI prism kit in the Genetics Department sequencing core. PCR and RT-PCR amplifications were carried out using minor modifications to standard techniques [65] using the following primer combinations:
Melanogaster primers for RT-PCR
Dichaete F ACAATCCATTCCATCAACTACC
Dichaete R TTGGTGTTCCCTCCTTACTC
Sox21B F AGTCTCATGAACAGCGGAAG
Sox21B R GGAGTTGCTCAGATACGACG
SoxN F CAGCAGCAACAGCAACACTAC
SoxN R TTTCATCGCCTCGCCACAAC
Pseudoobscura primers for in situ probes:
Dp-Dichaete F CGAACTACGGATTCCACCT
Dp-Dichaete R CATTCCGTTGGCCTGCAT
Dp-SoxN F AGCTGAGTCACCATAACCAC
Dp-soxN R GTCATGTGATGGCTACCAA
Dp-Sox21A Exon1 F GAGCATCTCGACGCTACTAC
Dp-Sox21A Exon 1 R GGAATTGGAGTGGCTATGAT
Dp-Sox21A Exon 2 F CTAAGGACATGCAGTCACAG
Dp-Sox21A Exon 2 R GACTTCACGCAGCCGTAGGAT
Dp-Sox21B F CGTCTATCCACACACCTGTC
Dp-Sox21B R GACGATGTCTGCTGCTGTT
Whole-mount in situ hybridisation to Drosophila embryos was performed using minor modifications to a standard protocol [66].
All genetic nomenclature is according to FlyBase [49].
Authors' contributions
C.McK. performed the sequencing, mapping of the Drosophila Sox21a and Sox21b genes in and the in situ hybridisation experiments. G.W. carried out the RT-PCR analysis. S.R. designed the experiments, carried out the genomic analysis and wrote the paper.
Acknowledgments
Acknowledgements
This work was supported by an UK-Medical Research Council programme grant to S.R., M. Ashburner and D. Gubb. We are grateful to M. Ashburner for comments on the manuscript and to S. Marcellini and J. Roote for assistance with the D. pseudoobscura husbandry.
Contributor Information
Carol McKimmie, Email: cm10019@mole.bio.cam.ac.uk.
Gertrud Woerfel, Email: gw236@mole.bio.cam.ac.uk.
Steven Russell, Email: s.russell@gen.cam.ac.uk.
References
- Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex determining region encodes a protein with homology to a conserved DNA binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- Schepers GE, Teasdale RD, Koopman P. Twenty pairs of Sox: extent, homology, and nomenclature of the mouse and human Sox transcription factor gene families. Mol. Cell. 2002;3:167–170. doi: 10.1016/S1534-5807(02)00223-X. [DOI] [PubMed] [Google Scholar]
- Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- Collignon J, Sockanathan S, Hacker A, Cohen-Tannoudji M, Norris D, Rastan S, Stevanovic M, Goodfellow PN, Lovell-Badge R. A comparison of the properties of SOX3 with SRY and two related genes SOX1 and SOX2. Development. 1996;122:509–520. doi: 10.1242/dev.122.2.509. [DOI] [PubMed] [Google Scholar]
- Malas S, Duthies S, Deloukas P, Episkopou V. The isolation and high-resolution mapping of human SOX14 and SOX21 ; two members of the SOX gene family related to SOX1, SOX2 and SOX3. Mamm Genome. 1999;10:934–937. doi: 10.1007/s003359901118. [DOI] [PubMed] [Google Scholar]
- Uchikawa M, Kamachi Y, Kondoh H. Two distinct group B SOX genes for transcriptional activators and repressors: their expression during embryonic organogenesis of the chicken. Mech Dev. 1999;84:103–120. doi: 10.1016/S0925-4773(99)00083-0. [DOI] [PubMed] [Google Scholar]
- Stevanovic M, Lovell-Badge R, Collignon J, Goodfellow PN. SOX3 is an X-linked gene related to SRY. Hum Mol Genet. 1993;2:2013–2018. doi: 10.1093/hmg/2.12.2013. [DOI] [PubMed] [Google Scholar]
- Foster JW, Graves JA. An SRY-related sequence on the marsupial X chromosome: implications for the evolution of the mammalian testis-determining gene. Proc Nat Acad Sci USA. 1994;91:1927–1931. doi: 10.1073/pnas.91.5.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsic N, Rajic T, Stanojcic S, Goodfellow PN, Stevanovic. M. Characterisation and mapping of the human SOX14 gene. Cytogenet Cell Genet. 1998;83:139–146. doi: 10.1159/000015149. [DOI] [PubMed] [Google Scholar]
- Stevanovic M, Zuffardi O, Collignon J, Lovell-Badge R, Goodfellow PN. The cDNA sequence and chromosomal location of the human SOX2 gene. Mamm Genome. 1994;5:640–642. doi: 10.1007/BF00411460. [DOI] [PubMed] [Google Scholar]
- Malas S, Duthie SM, Mohri F, Lovell-Badge R, Episkopou V. Cloning and mapping of human SOX1: a highly conserved gene expressed in the developing brain. Mamm Genome. 1997;8:866–868. doi: 10.1007/s003359900597. [DOI] [PubMed] [Google Scholar]
- Kuroiwa A, Uchikawa M, Kamachi Y, Kondoh H, Nishida-Umehara C, Masabanda J, Griffin DK, Matsuda Y. Chromosome assignment of eight SOX family genes in chicken. Cytogenet. Genome Res. 2002;98:189–193. doi: 10.1159/000069803. [DOI] [PubMed] [Google Scholar]
- Kirby PJ, Waters PD, Delbridge M, Svartman M, Stewart AN, Nagai K, Graves JAM. Cloning and mapping of platypus SOX2 and SOX14: Insights into SOX group B evolution. Cytogenetic and Genome Research. 2002;98:96–100. doi: 10.1159/000068539. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Informatics: http://www.informatics.jax.org/
- Blake JA, Richardson JE, Bult CJ, Kadin JA, Eppig JT. MGD: The mouse genome database. Nucleic Acids Res. 2003;31:193–195. doi: 10.1093/nar/gkg047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman CM, Pfeiffer BD, Rincon-Limas DE, Hoskins RA, Gnirke A, Mungal CJ, Wang AM, Kronmiller B, Pacleb J, Park S, Stapleton M, Wan K, George R, Jong PJ, Botas J, Rubin GM, Celniker SE. Assessing the impact of comparative genomic sequence data on the functional annotation of the Drosophila genome. Genome Biology. 2002;3:RESEARCH0086.. doi: 10.1186/gb-2002-3-12-research0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR. Progress and Prospects in Evolutionary Biology: The Drosophila Model. Oxford, Oxford University Press.; 1997. [Google Scholar]
- Gaunt MW, Miles MA. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol Biol Evol. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- Annotation of 12 Drosophila Genomes: http://rana.lbl.gov/drosophila/multipleflies.html
- Sanchez-Soriano N, Russell S. The Drosophila Sox-domain protein Dichaete is required for the development of the central nervous system midline. Development. 1998;125:3989–3996. doi: 10.1242/dev.125.20.3989. [DOI] [PubMed] [Google Scholar]
- Buescher M, Hing FS, Chia W. Formation of neuroblasts in the embryonic central nervous system of Drosophila melanogaster is controlled by SoxNeuro. Development. 2002;129:4193–4203. doi: 10.1242/dev.129.18.4193. [DOI] [PubMed] [Google Scholar]
- Overton PM, Meadows LA, Urban J, Russell S. Evidence for differential and redundant function of the Sox genes Dichaete and SoxN during CNS development in Drosophila. Development. 2002;129:4219–4228. doi: 10.1242/dev.129.18.4219. [DOI] [PubMed] [Google Scholar]
- Nishiguchi S, Wood H, Kondoh H, Lovell-Badge R, Episkopou V. Sox1 directly regulates the g-crystallin genes and is essential for lens development in mice. Genes & Development. 1998;12:776–781. doi: 10.1101/gad.12.6.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai Y. Roles of Sox factors in neural determination: conserved signaling in evolution? Int J Dev Biol. 2001;45:321–326. [PubMed] [Google Scholar]
- Cremazy F, Berta P, Girard F. Genome-wide analysis of Sox genes in Drosophila melanogaster. Mech Dev. 2001;109:371–375. doi: 10.1016/S0925-4773(01)00529-9. [DOI] [PubMed] [Google Scholar]
- Russell SRH, Sanchez-Soriano N, Wright CR, Ashburner M. The Dichaete gene of Drosophila melanogaster encodes a Sox-domain protein required for embryonic segmentation. Development. 1996;122:3669–3676. doi: 10.1242/dev.122.11.3669. [DOI] [PubMed] [Google Scholar]
- Cremazy F, Berta P, Girard F. SoxNeuro, a new Drosophila Sox gene expressed in the developing central nervous system. Mech Dev. 2000;93:215–219. doi: 10.1016/S0925-4773(00)00268-9. [DOI] [PubMed] [Google Scholar]
- Nambu PA, Nambu JR. The Drosophila fish-hook gene encodes an HMG domain protein essential for segmentation and CNS development. Development. 1996;122 doi: 10.1242/dev.122.11.3467. [DOI] [PubMed] [Google Scholar]
- Stapleton M, Liao G, Brokstein P, Hong L, Carninci P, Shiraki T, Hayashizaki Y, Champe M, Pacleb J, Wan K, Yu C, Carlson J, George R, Celniker SE, Rubin GM. The Drosophila gene collection: identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res. 2002;12:1294–1300. doi: 10.1101/gr.269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich M, Tautz D. Evolution and phylogeny of the Diptera: a molecular phylogenetic analysis using 28S rDNA sequences. Syst Biol. 1997;46:674–698. doi: 10.1093/sysbio/46.4.674. [DOI] [PubMed] [Google Scholar]
- Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes & Dev. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- Cis-analyst: http://rana.lbl.gov/cis-analyst/
- Berman BP, Nibu Y, Pfeiffer BD, Tomancak P, Celniker SE, Levine M, Rubin GM, Eisen MB. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc Natl Acad Sci U S A. 2002;99:757–762. doi: 10.1073/pnas.231608898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vista Genome Browser: http://pipeline.lbl.gov/pseudo
- Couronne O, Poliakov A, Bray N, Ishkhanov T, Ryaboy D, Rubin E, Pachter L, Dubchak I. Strategies and Tools for Whole-Genome Alignments. Genome Res. 2003;13:73–80. doi: 10.1101/gr.762503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Soriano N, Russell S. Regulatory mutations of the Drosophila Sox gene Dichaete reveal new functions in embryonic brain and hindgut development. Dev Biol. 2000;129:1165–1174. doi: 10.1006/dbio.2000.9648. [DOI] [PubMed] [Google Scholar]
- Besansky NJ, Bedell JA, Mukabayire O. Q: a new retrotransposon from the mosquito Anopheles gambiae. Insect Mol Biol. 1994;3:49–56. doi: 10.1111/j.1365-2583.1994.tb00150.x. [DOI] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes & Development. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/S0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Taguchi S, Tagawa K, Humphreys T, Satoh N. Group B Sox genes that contribute to specification of the vertebrate brain are expressed in the apical organ and ciliary bands of hemichordate larvae. Zool Sci. 2002;19:57–66. doi: 10.2108/zsj.19.57. [DOI] [PubMed] [Google Scholar]
- Miya T, Nishida H. Expression pattern and transcriptional control of SoxB1 in embryos of the ascidian Halocynthia roretzi. Zool Sci. 2003;20:59–67. doi: 10.2108/zsj.20.59. [DOI] [PubMed] [Google Scholar]
- Zhao G, Skeath JB. The Sox-domain containing gene Dichaete/fish-hook acts in concert with vnd and ind to regulate cell fate in the Drosophila neuroectoderm. Development. 2002;129:1165–1174. doi: 10.1242/dev.129.5.1165. [DOI] [PubMed] [Google Scholar]
- Davis GK, Patel NH. Short, long and beyond: Molecular and embryological approaches to insect segmentation. Ann Rev Entomol. 2002;47:669–699. doi: 10.1146/annurev.ento.47.091201.145251. [DOI] [PubMed] [Google Scholar]
- Goltsev Y, Hsiong W, Lanzaro G, Levine M. Different combinations of gap repressors for common stripes in Anopheles and Drosophila embryos. Dev Biol. 2004;275 doi: 10.1016/j.ydbio.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Xia Q, Zhou Z, Lu C, Cheng D, Dai F, Li B, Zhao P, Zha X, Cheng T, Chai C, Pan G, Xu J, Liu C, Lin Y, Qian J, Hou Y, Wu Z, Li G, Pan M, Li C, Shen Y, Lan X, Yuan L, Li T, Xu H, Yang G, Wan Y, Zhu Y, M MY, Shen W, Wu D, Xiang Z, Yu J, Wang J, Li R, Shi J, Li H, Li G, Su J, Wang X, Li G, Zhang Z, Wu QW, Li J, Zhang Q, Wei N, Xu J, Sun H, Dong L, Liu D, Zhao S, Zhao X, Meng Q, Lan F, Huang X, Li Y, 0Fang L, Li C, D DL, Sun Y, Zhang Z, Yang Z, Huang Y, Xi Y, Qi Q, He D, Huang H, Zhang X, Wang Z, Li W, Cao Y, Yu Y, Yu H, Li J, Ye J, Chen H, Zhou Y, Liu B, Wang J, Ye J, Ji H, Li S, Ni P, Zhang J, Zhang Y, Zheng H, Mao B, Wang W, Ye C, Li S, Wang J, Wong GK, Yang H. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science. 2004;306:1937–1940. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- Ayala FJ, Rzhetsky A, Ayala FJ. Origin of the metazoan phyla: Molecular clocks confirm paleontolgical estimates. Proc Nat Acad Sci USA. 1998;95:606–611. doi: 10.1073/pnas.95.2.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S, Crosby MA, Mungall CJ, Matthews BB, Campbell KS, Hradecky P, Huang Y, Kaminker JS, Millburn GH, Prochnik SE, Smith CD, Tupy JL, Whitfied EJ, Bayraktaroglu L, Berman BP, Bettencourt BR, Celniker SE, AD ADG, Drysdale RA, Harris NL, Richter J, Russo S, Schroeder AJ, Shu SQ, Stapleton M, Yamada C, Ashburner M, Gelbart WM, Rubin GM, Lewis SE. Annotation of the Drosophila melanogaster euchromatic genome sequence: a systematic review. Genome Biol. 2002;3:RESEARCH0083. doi: 10.1186/gb-2002-3-12-research0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Wheeler DA, Kronmiller B, Carlson JW, Halpern A, Patel S, Adams M, Champe M, Dugan SP, Frise E, Hodgson A, George RA, Hoskins RA, Laverty T, Muzny DM, Nelson CR, Pacleb JM, Park S, Pfeiffer BD, Richards S, Sodergren EJ, Svirskas R, Tabor PE, Wan K, Stapleton M, Sutton GG, Venter C, Weinstock G, Scherer SE, Myers EW, Gibbs RA, Rubin CM. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 2002;3:RESEARCH0079.. doi: 10.1186/gb-2002-3-12-research0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale RA, Crosby MA, . TFBC. FlyBase: genes and gene models. Nucleic Acids Research. 2005;33::390–395. doi: 10.1093/nar/gki046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D.pseudoobscura sequencing project: http://www.hgsc.bcm.tmc.edu/projects/drosophila/
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R, Salzberg SL, Loftus B, Yandell M, Majoros WH, Rusch DB, Lai Z, Kraft CL, Abril JF, Anthouard V, Arensburger P, Atkinson PW, Baden H, Berardinis V, Baldwin D, Benes V, Biedler J, Blass C, Bolanos R, Boscus D, Barnstead M, Cai S, Center A, Chaturverdi K, Christophides GK, Chrystal MA, Clamp M, Cravchik A, Curwen V, Dana A, Delcher A, Dew I, Evans CA, Flanigan M, Grundschober-Freimoser A, Friedli L, Gu Z, Guan P, Guigo R, Hillenmeyer ME, Hladun SL, Hogan JR, Hong YS, Hoover J, Jaillon O, Ke Z, Kodira C, Kokoza E, Koutsos A, Letunic I, Levitsky A, Liang Y, Lin JJ, Lobo NF, Lopez JR, Malek JA, McIntosh TC, Meister S, Miller J, Mobarry C, Mongin E, Murphy SD, O'Brochta DA, Pfannkoch C, Qi R, Regier MA, Remington K, H HS, Sharakhova MV, Sitter CD, Shetty J, Smith TJ, Strong R, Sun J, Thomasova D, Ton LQ, Topalis P, Tu Z, Unger MF, Walenz B, Wang A, Wang J, Wang M, Wang X, Woodford KJ, Wortman JR, Wu M, Yao A, Zdobnov EM, Zhang H, Zhao Q, Zhao S, Zhu SC, Zhimulev I, Coluzzi M, Torre A, Roth CW, Louis C, Kalush F, Mural RJ, Myers EW, Adams MD, Smith HO, Broder S, Gardner MJ, Fraser CM, Birney E, Bork P, Brey PT, Venter JC, Weissenbach J, Kafatos FC, Collins FH, Hoffman SL. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Mosquito genome browser: http://www.ensembl.org/Anopheles_gambiae/
- Mosquito gene index: http://www.tigr.org/tigr-scripts/tgi/T_index.cgi?species=mosquito
- A. mellifera sequencing project: http://www.hgsc.bcm.tmc.edu/projects/honeybee/
- Honey Bee EST Project: http://titan.biotec.uiuc.edu/bee/honeybee_project.htm
- Whitfield CW, Band MR, Bonaldo MF, Kumar CG, Lui L, Pardinas JR, Robertson HM, Soares MB, Robinson GE. Annotated expressed sequence tags and cDNA microarrays for studies of brain and behaviour in the Honey Bee. Genome Res. 2002;4:555–566. doi: 10.1101/gr.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nematode.net: http://www.nematode.net/BLAST
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1006/jmbi.1990.9999. [DOI] [PubMed] [Google Scholar]
- Berkeley Drosophila Genome Project: http://www.fruitfly.org
- Artemis: http://www.sanger.ac.uk/Software/Artemis/
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualisation and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucl Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxshade: http://www.ch.embnet.org/software/BOX_form.html
- Ogurtsov AY, Roytberg MA, Shabalina SA, Kondrashov AS. OWEN: aligning long collinear regions of genomes. Bioinformatics. 2002;18:1703–1704. doi: 10.1093/bioinformatics/18.12.1703. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: a laboratory manual. New York, Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridisation method for the localisation of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]