Abstract
Different theoretical frameworks have been invoked to guide the study of virus evolution. Three of the more prominent ones are (i) the evolution of virulence, (ii) life history theory, and (iii) the generalism–specialism dichotomy. All involve purported tradeoffs between traits that define the evolvability and constraint of virus-associated phenotypes. However, as popular as these frameworks are, there is a surprising paucity of direct laboratory tests of the frameworks that support their utility as broadly applicable theoretical pillars that can guide our understanding of disease evolution. In this study, we conduct a meta-analysis of direct experimental evidence for these three frameworks across several widely studied virus–host systems: plant viruses, fungal viruses, animal viruses, and bacteriophages. We extracted 60 datasets from 28 studies and found a range of relationships between traits in different analysis categories (e.g., frameworks, virus–host systems). Our work demonstrates that direct evidence for relationships between traits is highly idiosyncratic and specific to the host–virus system and theoretical framework. Consequently, scientists researching viral pathogens from different taxonomic groups might reconsider their allegiance to these canons as the basis for expectation, explanation, or prediction. Future efforts could benefit from consistent definitions, and from developing frameworks that are compatible with the evidence and apply to particular biological and ecological contexts.
Keywords: virus evolution, tradeoffs, virulence, life history theory, niche breadth
Introduction
The biological diversity of viruses—in morphology, natural history, and molecular mechanisms—can limit the development of theories that apply across diverse contexts. Nonetheless, the development of theoretical frameworks for virus evolution has helped set expectations, generate hypotheses, and to make more informed predictions. Three commonly used frameworks include (i) the evolution of virulence, (ii) life history theory, and (iii) the generalism–specialism dichotomy.
The evolution of virulence is perhaps the most popular framework in the field of pathogen evolution. The premise is that the use of within-host resources devoted to virus replication imposes a fitness cost (virulence) that necessarily increases host mortality, thereby reducing the virus’ ability to between hosts (Anderson and May 1982, Ewald 1991, Frank 1996). This framework has transformed how professionals across several fields—from the evolutionary sciences to clinical medicine—study the constraints surrounding the harmful viruses cause to their hosts. Several studies have shown evidence of virulence-transmission tradeoffs (Sacrist´an and Garc´ıa-Arenal 2008, Froissart et al. 2010, Goldhill and Turner 2014), while others have suggested this tradeoff is not always evident (Bull 1994, Levin and Bull 1994, Ebert and Bull 2003, Alizon et al. 2009, Doumayrou et al. 2013, Bull et al. 2014). The evolution of virulence remains a widely adopted framework that guides our expectations for how pathogens evolve towards more or less pathogenicity/virulence in host populations.
Life history theory is another framework that has been used to study virus evolution. It focuses on constraints on virus evolution driven by tradeoffs between traits associated with survival and reproduction, analogous to the expected tradeoffs in organisms imposed by energetic limitations (Stearns 1976, 1989), and has been applied to viruses. For example, “reproduction” or “fecundity” relates to the number of virus particles produced from infection and replication. Several studies have demonstrated a tradeoff between such traits and those associated with virus survival [virus particle stability outside the host, sometimes termed as free-living survival (Paepe et al. 2006, Heineman et al. 2012, Brandon Ogbunugafor et al. 2013)], while other studies have not found evidence for this tradeoff (Goldhill and Turner 2014, Handel et al. 2014). Even though life history theory was not developed specifically for the study of virus pathogens, its strength lies in its widespread application, which helps to bridge gaps between subfields of evolutionary biology.
The generalism–specialism dichotomy has a long history in the evolutionary ecology literature, applying to organisms across the biosphere (Futuyma and Moreno 1988, Poisot et al. 2011, Leggett et al. 2013). Specialists utilize relatively fewer resources, whereas generalists can access greater resources but are assumed to use them less efficiently (i.e. a “jack-of-all-trades is master of none”). In the context of pathogens, the theory is applied to host niche breadth, the range of hosts that a pathogen can successfully infect and propagate in (Garnick 1992, Fodor 2011). This frame- work has been useful because of its resonance with the ecological challenges associated with viral disease emergence, often defined by a viruses’ evolved ability to infect novel hosts (as in zoonosis or host shifts) (Elena et al. 2014, Longdon et al. 2014, Roche et al. 2014). Studies in viruses have demonstrated tradeoffs between generalism and specialism in some circumstances, where fitness decreases when a virus switches or expands its host range (Duffy et al. 2006, Elena et al. 2009, Moreno-P´erez et al. 2016, Bera et al. 2018). Like life history theory, this framework comes with the benefit of a long history applied to other organisms, which means that terms and metrics have not been invoked solely for the study of infectious diseases.
Though these frameworks are well-developed, there is dubious support for their continued relevance to virus evolution. Studies of tradeoff models are also limited by imprecise definitions for key terms, which affect how traits are measured and analyzed. For example, in plants, virulence is often defined as “aggressiveness” (Sacrist´an and Garc´ıa-Arenal 2008, Montarry et al. 2012), while in animals, virulence is defined as the harm done to the hosts (Bull 1994, Read 1994). Meanwhile, in theoretical studies, virulence is often defined as the mortality of host organisms (Anderson and May 1982, Day 2002b). While these differences may seem subtle, they can have powerful consequences for how we understand virulence evolution (Surasinghe et al. 2024). Unfortunately, the imprecision of these terms is often taken for granted in our collective adoption of the evolution of virulence (and other frameworks) in application to medicine and health policy.
In this study, we examine direct evidence from laboratory studies for the three tradeoff frameworks. Specifically, we examine the relationships between traits associated with the different frameworks: transmission versus virulence (evolution of virulence), survival versus reproduction (life history theory), and single-host fitness versus multiple host fitness (generalism-specialism dichotomy) frameworks. We focused only on experimental data and excluded data from theoretical, clinical studies, and observational studies because conditions in laboratory settings can be controlled and traits measured with relative ease. With regards to virus–host type, we focused on viruses that infect plants, animals, fungi, and bacteria, as these are systems where most experiments of this sort have been conducted. We demonstrate that direct support for any particular relationships between traits in these frameworks are scant, often specific to virus–host type and based on subjective definitions of traits (and methods used to measure them). Summarizing, we suggest that tradeoff frameworks applied to viruses be re-evaluated and used with caution in efforts to interpret or predict the direction of pathogenicity in virus evolution.
Methods
Note on the definition of “direct evidence”
Studies of virus evolution are widespread, and include studies from clinical and field settings (Holmes et al. 2005, Fraser et al. 2007, Fargette et al. 2008, Wasik et al. 2023), as well as mathematical and computational explorations (Gandon et al. 2001, Alizon and van Baalen 2005, Kucharski et al. 2015, Miller-Dickson et al. 2019, Gomez et al. 2020). While studies of various kinds have provided valuable contributions to our understanding, those that directly test the relationships between traits that correspond to various virus evolution frameworks are relatively scant. Our meta-analysis focused on direct evidence—studies where traits were measured, evolved, or manipulated in laboratory settings under controlled conditions. We argue that these studies provide an important window into the basis for these theoretical frameworks, and we sought to test whether such laboratory data supported the assumptions of the three theoretical frameworks under study. We note that observational studies have been very valuable in the study of virus evolution tradeoffs (especially in the evolution of virulence) (Fenner and Marshall 1957, Witter 1997, Fraser et al. 2007, McKay et al. 2020). Observational studies are, however, confounded by the myriad variables that could impact how traits associated with virulence and transmissions evolve and are measured in the natural world.
Systematic review of data for meta-analysis
Between 1 January 2023 and 31 March 2023, we searched for scholarly articles from Web of Science, Pub-Med, and Scopus databases. We focused only on these three databases because of their comprehensive coverage of international scholarship and reduced search biases. Searches using the Web of Science were limited to ecology, evolutionary biology, virology, microbiology, infectious diseases, immunology, tropical medicine, and parasitology. Similarly, searches using Scopus were limited to medicine, agricultural and biological sciences, biochemistry, genetics, molecular biology, immunology and microbiology, and pharmacology. The search terms used were “virulence transmission trade-offs viruses,” “virulence transmission tradeoffs viruses,” virulence transmission trade-offs bacteriophages,” “virulence transmission tradeoffs bacteriophages,” “virulence transmission trade-offs phages,” “virulence transmission trade-offs phages,” “survival reproduction trade-offs viruses,” “survival reproduction trade-offs viruses,” survival reproduction tradeoffs bacteriophages,” “survival reproduction trade-offs bacteriophages,” “survival reproduction trade-offs phages,” “survival reproduction trade-offs phages,” “host range/expansion/shift and fitness in viruses,” “host range/expansion/shift and fitness in bacteriophages,” and “host range/expansion/shift and fitness in phages.” The studies from our search and those identified from other sources were combined in a spreadsheet. Other sources included several reviews or studies cited in articles from our primary search.
Each study and its metadata were downloaded from the database, and all duplicates were removed, as indicated in the PRISMA chart (Supplementary Fig. S1). Some studies were excluded for various reasons, e.g. studies that did not focus on viruses, studies that were computational/mathematical, and those that were conference/opinion/perspectives or poster abstracts. Notably, many of the studies from our searches were computational/mathematical, where theoretical ideas in infectious disease have been studied for many decades. After this, the final studies were further screened, and if they did not directly test the frameworks or present data relevant to the frameworks, they were removed.
Our main inclusion criteria for meta-analysis were as follows: (i) the study served as primary literature and was published in a peer-reviewed academic journal, (ii) the study tested or measured the relationships within one or more of the three frameworks described for viruses (evolution of virulence, life history theory, generalism–specialism dichotomy), and (iii) correlational data were reported or could be calculated from the data reported. The remaining studies were screened under our inclusion criteria, and only those that met the criteria were used for data extraction. The inclusion criteria were carefully chosen to ensure that the studies in our analysis were relevant to our meta-analysis goal. Extracted data included the following: virus–host type, virus genome type, traits measured, statistics reported, sample size, and study reference (Supplementary Fig. S1).
Categorization of data
We discretized the data into categories based on theoretical frameworks, combined virus–host type, and coupled categories. Datasets were placed in their respective frameworks based primarily on traits measured in the studies used in this meta-analysis. For virus–host type, separate categories were made for animal viruses and bacteriophages. For coupled categories, different analyses were done, as summarized in the results. We analyzed data by taking a gross look at the relationships between traits for all datasets (Supplementary Fig. S2) and then used one of the following methods: (i) a theoretical framework across virus-host types; (ii) a virus-host categorization across theoretical frameworks, and (iii) additional analyses of different coupled categories (Supplementary Fig. S3).
Statistical analyses
All meta-analyses were done following the methods described by previous studies (Schwarzer et al. 2015, Acevedo et al. 2019, Rafaluk-Mohr 2019, Harrer 2021). Correlations were extracted from studies and also calculated in instances where they were not directly reported. Since we were only interested in the relationship effect sizes, we pooled correlation coefficients, and all the meta-analyses were done using random effects models in the metacor function of the meta package (Balduzzi et al. 2019) of R version 4.1.1 (Posit 2023). Random effects models were useful here because, in addition to accounting for the sampling error of the pooled effect sizes, they also account for variation introduced by datasets drawn from different studies (Hedges and Vevea 1998). We used the Sidik–Jonkman (SJ) estimator (Sidik and Jonkman 2005) in the meta package to estimate between-study heterogeneity (R2) resulting from differences in sample size experimental design and virus–host systems However, regardless of the estimator used in a meta-analysis, there can be bias. To control for this, we used the Knapp–Hartung adjustments (Knapp and Hartung 2003) to calculate confidence intervals of pooled effect sizes because the number of datasets was small and the heterogeneity was high. Visualization of publication biases was conducted using contour-enhanced funnel plots shown in the Supplementary Material (Supplementary Fig. S4). In addition, after each meta-analysis was performed, the find.outliers function was used to identify outliers. After outliers were identified, additional meta-analyses were performed without outliers.
Results
Our meta-analysis examined direct experimental evidence for traits associated with the three theoretical frameworks (evolution of virulence, life history theory, and generalism–specialism dichotomy) of virus evolution. We extracted 60 datasets from 28 studies (Table 1, Supplementary Fig. S1) and compiled direct evidence for tradeoffs between traits measured in studies of virus evolution. Next, we explored a different possibility with regards to tradeoffs: perhaps relationship patterns between traits are less dependent on the model but are specific to virus–host system. To investigate this possibility, we examined tradeoffs between measured traits within virus–host types.
Table 1.
Datasets used in meta-analysis
| Dataset | Virus type | Host | Genome | Traits | Framework |
|---|---|---|---|---|---|
| 1 | Tomato spotted wilt virus | Plant | ssRNA | Transmission vs. virus titer (Rotenberg et al. 2009) | EoV |
| 2 | Tomato spotted wilt virus | Plant | ssRNA | Transmission vs. virus titer (Rotenberg et al. 2009) | EoV |
| 3 | Tomato spotted wilt virus | Plant | ssRNA | Transmission vs. virus titer (Rotenberg et al. 2009) | EoV |
| 4 | Tomato spotted wilt virus | Plant | ssRNA | Transmission vs. virus titer (Rotenberg et al. 2009) | EoV |
| 5 | Tomato spotted wilt virus | Plant | ssRNA | Transmission vs. virus titer (Rotenberg et al. 2009) | EoV |
| 6 | Cucumber mosaic virus | Plant | ssRNA | Accumulation vs. infection (Sacrist´an and Garc´ıa-Arenal 2008) effects | EoV |
| 7 | Cucumber mosaic virus | Plant | ssRNA | Growth effects on original host (Sacrist´an and Garc´ıa-Arenal 2008) | G/S |
| 8 | Cucumber mosaic virus | Plant | ssRNA | Original hosts vs. diversification (Sacrist´an and Garc´ıa-Arenal 2008) | G/S |
| 9 | Rice yellow mottle virus | Plant | ssRNA | Virus titer vs. weight loss (Poulicard et al. 2010) | EoV |
| 10 | Rice yellow mottle virus | Plant | ssRNA | Virus titer vs. weight loss (Poulicard et al. 2010) | EoV |
| 11 | Rice yellow mottle virus | Plant | ssRNA | Virus titer vs. weight loss (Poulicard et al. 2010) | EoV |
| 12 | Potato virus Y | Plant | ssRNA | Virus aggressiveness vs. virus loss (Montarry et al. 2012) | EoV |
| 13 | Potato virus Y | Plant | ssRNA | Virus aggressiveness vs. virus loss (Montarry et al. 2012) | EoV |
| 14 | Cauliflower mosaic virus | Plant | dsDNA | Leaf reduction vs. transmission (Doumayrou et al. 2013) | EoV |
| 15 | Cauliflower mosaic virus | Plant | dsDNA | Viral accumulation vs. leaf reduction (Doumayrou et al. 2013) | EoV |
| 16 | Cauliflower mosaic virus | Plant | dsDNA | Viral reduction vs. transmission (Doumayrou et al. 2013) | EoV |
| 17 | Cauliflower mosaic virus | Plant | dsDNA | Accumulation vs. virulence (Doumayrou et al. 2013) | EoV |
| 18 | Cauliflower mosaic virus | Plant | dsDNA | Accumulation vs. virulence (Doumayrou et al. 2013) | EoV |
| 19 | Cauliflower mosaic virus | Plant | dsDNA | Accumulation vs. transmission (Doumayrou et al. 2013) | EoV |
| 20 | Cauliflower mosaic virus | Plant | dsDNA | Accumulation vs. transmission (Doumayrou et al. 2013) | EoV |
| 21 | Cauliflower mosaic virus | Plant | dsDNA | Viral load vs. transmission (Doumayrou et al. 2013) | EoV |
| 22 | Cauliflower mosaic virus | Plant | dsDNA | Viral load vs. transmission (Doumayrou et al. 2013) | EoV |
| 23 | SpexNPV | Animal | dsDNA | Speed to kill vs. virus yield (Redman et al. 2016) | EoV |
| 24 | SpexNPV | Animal | dsDNA | Speed to kill vs. virus yield (Redman et al. 2016) | EoV |
| 25 | Cryphonectria hypovirus | Fungi | dsRNA | Colony size vs. sporulation (Brusini et al. 2017) | EoV |
| 26 | Cryphonectria hypovirus | Fungi | dsRNA | Colony size vs. spore size (Brusini et al. 2017) | EoV |
| 27 | Cryphonectria hypovirus | Fungi | dsRNA | Spore size vs. sporulation (Brusini et al. 2017) | EoV |
| 28 | West Nile virus | Animal | ssRNA | Attachment rate vs. alternating host (Deardorff et al. 2011) | EoV |
| 29 | West Nile virus | Animal | ssRNA | Survival vs. viral load (Ciota et al. 2013) | EoV |
| 30 | Vesicular stomatitis virus | Animal | ssRNA | Fitness vs. transmission time (Elena 2001) | G/S |
| 31 | Vesicular stomatitis virus | Animal | ssRNA | Fitness vs. transmission time (Elena 2001) | G/S |
| 32 | Vesicular stomatitis virus | Animal | ssRNA | Fitness vs. transmission time (Elena 2001) | G/S |
| 33 | Vesicular stomatitis virus | Animal | ssRNA | Fitness vs. transmission time (Elena 2001) | G/S |
| 34 | Vesicular stomatitis virus | Animal | ssRNA | Fecundity vs. survival (Brandon Ogbunugafor et al. 2013) | LHT |
| 35 | Vesicular stomatitis virus | Animal | ssRNA | Fecundity vs. survival (Brandon Ogbunugafor et al. 2013) | LHT |
| 36 | Vesicular stomatitis virus | Animal | ssRNA | Fitness vs. alternating hosts (Turner and Elena 2000) | LHT |
| 37 | Vesicular stomatitis virus | Animal | ssRNA | Survival vs. reproduction (Wasik et al. 2015) | LHT |
| 38 | Vesicular stomatitis virus | Animal | ssRNA | Generalists vs specialists (Alto and Turner 2010) | G/S |
| 39 | Vesicular stomatitis virus | Animal | ssRNA | Generalists vs specialists (Alto and Turner 2010) | G/S |
| 40 | Vesicular stomatitis virus | Animal | ssRNA | Generalists vs specialists (Alto and Turner 2010) | G/S |
| 41 | Vesicular stomatitis virus | Animal | ssRNA | Generalists vs specialists (Alto and Turner 2010) | G/S |
| 42 | Vesicular stomatitis virus | Animal | ssRNA | Robustness vs. thermostability (Presloid et al. 2016) | LHT |
| 43 | Vesicular stomatitis virus | Animal | ssRNA | Robustness vs. thermostability (Presloid et al. 2016) | LHT |
| 44 | Coliphages | Bacteria | NA | Multiplication rate vs. decay rate (Paepe et al. 2006) | LHT |
| 45 | ΦX174 | Bacteria | ssDNA | Growth rates vs. attachment rates (Crill et al. 2000) | EoV |
| 46 | Qβ | Bacteria | ssRNA | Adsorption rate vs. infectivity (Garc´ıa-Villada and Drake 2013) | EoV |
| 47 | Φ6 | Bacteria | dsRNA | Fitness vs. attachment rate (Ford et al. 2014) | EoV |
| 48 | Φ6 | Bacteria | dsRNA | Fitness vs. host range (Ferris et al. 2007) | G/S |
| 49 | Φ6 | Bacteria | dsRNA | Fitness vs. host range (Ferris et al. 2007) | G/S |
| 50 | Φ6 | Bacteria | dsRNA | Fitness vs. host range (Ferris et al. 2007) | G/S |
| 51 | Qβ | Bacteria | ssRNA | Growth vs. fitness (Domingo-Calap et al. 2010) | LHT |
| 52 | T7 | Bacteria | dsDNA | Adsorption rate vs. infectivity (Heineman et al. 2012) | EoV |
| 53 | P5 | Bacteria | dsDNA | Mortality vs. reproduction rate (Dessau et al. 2012) | LHT |
| 54 | ID11 | Bacteria | ssDNA | Fitness vs. binding affinity (Lee et al. 2011) | EoV |
| 55 | ID11 | Bacteria | ssDNA | Fitness vs. decay rate (Lee et al. 2011) | LHT |
| 56 | Qβ | Bacteria | ssDNA | Fitness vs. thermal adaptation (L´azaro et al. 2018) | G/S |
| 57 | Qβ | Bacteria | ssDNA | Fitness vs. thermal adaptation (L´azaro et al. 2018) | G/S |
| 58 | Qβ | Bacteria | ssDNA | Fitness vs. thermal adaptation (L´azaro et al. 2018) | G/S |
| 59 | Φ6 | Bacteria | dsRNA | Generalists vs. specialists (Bono et al. 2015) | G/S |
| 60 | Φ6 | Bacteria | dsRNA | Generalists vs. specialists (Bono et al. 2015) | G/S |
Datasets were not grouped even if they infect the same host or w from the same study. They were analyzed individually and restricted to one framework: evolution of virulence life history theory (LHT), and generalism–specialism dichotomy (G/S) (see “Methods” section). We note that the “Host does not provide any specific detail on species or subtaxa.” The hosts used in each study may or may be commonly associated with that virus in nature. We urge interested readers to consult individual those particulars.
Evolution of virulence
In the evolution of virulence framework, we found a nonsignificant positive correlation between traits associated with transmission and virulence (Fig. 1, Table 2) [N = 32; R = 0.1541; 95% confidence interval (CI): −0.2178 − 0.4869; t = 0.84; P - value = .4068] consistent with an absence of a tradeoff. The test for heterogeneity was high (Q = 384.61; df = 27; P- value < .0001), and the I2 test was 93.0% with a 95% CI of 90.9–94.6%. Six outliers were identified (datasets 1, 4, 28, 29, 45, 52). Without the outliers, we found a positive correlation (N = 26; R = 0.2209; 95% CI: −0.0363 to 0.4506; t = 1.77; P- value = .0884). While the test for heterogeneity was lower (Q = 89.46; df = 21; P-value < .0001), and the I2 test was 76.5% with a 95% CI of 64.7–84.4%, these results were highly significant. Distributions of effect sizes are shown in (Fig. 1). In addition, because this framework had multiple studies with a sample size of N = 3, we conducted an additional analysis (Supplementary Fig. S5) to demonstrate that removing these studies of N = 3 did not affect the overall correlation found in the primary analysis.
Figure 1.
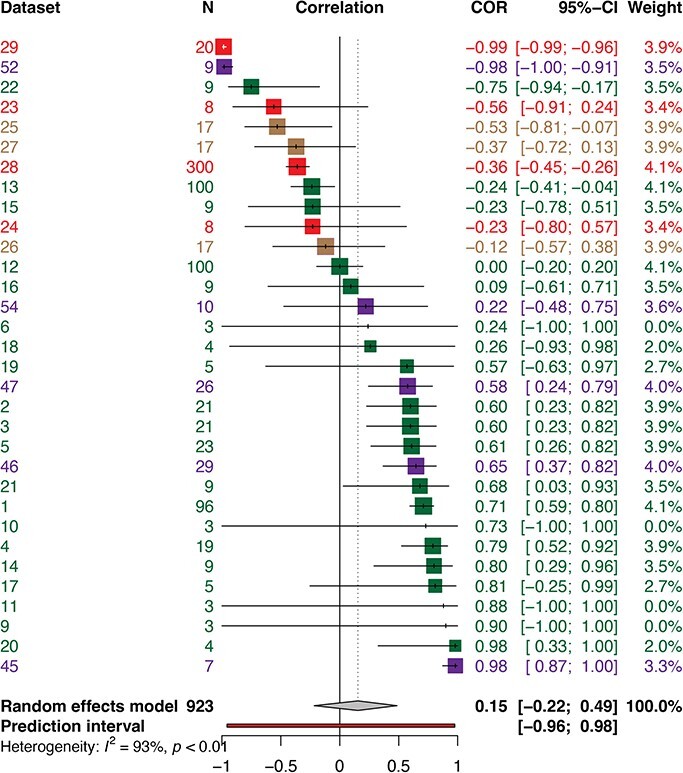
Forest plot showing the distribution of effect sizes for evolution of virulence datasets. The gray diamond represent the pooled correlation for evolution of virulence (R = 0.15). The colored squares represent the weight of the study on the pooled correlation, and the vertical black dash inside the colored squares shows the extracted correlation of the dataset. Datasets 6, 9, 10, and 11 have zero weight on overall correlation; therefore, they are not represented by any colored squares in this plot. Horizontal lines represent 95% confidence intervals for the dataset. Negative and positive correlations indicate values for individual datasets. The 95% confidence intervals and the weight of each dataset on the pooled correlation are indicated. The evolution of virulence framework is dominated mainly by plant viruses, represented by datasets 1, 2, 3, 4, 5, 6, 9, 10, 11, 12, 13, 14, 15, 16,17, 18, 19, 20, 21, 22 (green squares). Viruses that infect animals are represented in datasets 23, 24, 28, 29 (red squares), fungi datasets 25, 26, 27 (brown squares), and bacteriophages datasets 45, 46, 47, 52, 54 (purple squares).
Table 2.
Summary statistics from all analyses
| Analysis | Relationship | Correlation | Correlation P-value | Q | P-value | K |
|---|---|---|---|---|---|---|
| All three frameworks | Negative | −0.1818 | .1745 | 638.07 | .0001 | 60 |
| Evolution of virulence | Positive | 0.1541 | .4068 | 384.61 | .0001 | 32 |
| Life history theory | Negative | −0.6358 | .0863 | 173.32 | .0001 | 10 |
| Generalism–specialism | Negative | −0.5497 | .0001 | 15.9 | .1959 | 18 |
| Plant viruses | Positive | 0.4442 | .0027 | 110 | .0027 | 22 |
| Plant/Fungal viruses | Positive | 0.5444 | .0264 | 129.8 | <.0001 | 25 |
| Animal viruses | Negative | −0.6429 | .0065 | 145.07 | .0001 | 18 |
| Bacteriophages | Negative | −0.3535 | .0001 | 239.22 | .0001 | 17 |
| Evolution of virulence & life history theory | Negative | −0.0487 | .7841 | 597.54 | .0001 | 42 |
| Evolution of virulence & plant viruses | Positive | 0.4442 | .0043 | 110.32 | .0001 | 20 |
| Evolution of virulence & animal viruses | Negative | −0.7396 | .1692 | 70.422 | .0001 | 4 |
| Life history theory & animal viruses | Negative | −0.6267 | .2996 | 51.84 | .0001 | 6 |
| Generalism–specialism & animal viruses | Negative | −0.7145 | .0002 | 1.33 | .8569 | 8 |
| Evolution of virulence & bacteriophages | Positive | 0.3163 | .6816 | 63.75 | .0001 | 5 |
| Life history theory & bacteriophages | Negative | −0.6717 | .2107 | 68.54 | .0001 | 4 |
| Generalism–specialism & Bacteriophages | Negative | −0.5131 | .0069 | 13.25 | .0663 | 8 |
Effect sizes (correlations) with their P-values, tests for between-study heterogeneity (Q) with P-values, and the number of datasets combined in the analysis (K) are summarized.
Life history theory
Relatively, fewer studies were found for the life history theory framework (Fig. 2a, Table 2). We found a nonsignificant negative correlation between survival and reproduction (N = 10; R = −0.6358; 95% CI: −0.9266 to 0.1305; t = −1.93; P- value = .0863). The test for heterogeneity was high (Q = 173.32, df = 7; P- value = .0001), and the I2 test was 96.0% with a 95% CI of 93.9−97.3%. Running the analysis without the identified outliers (Datasets 36, 43, 53), we found a significant negative correlation implying tradeoffs between reproduction and survival (N = 7; R = −0.3217; 95% CI: −0.5504 to −0.0481; t = −2.86; P-value = .0288). The test for heterogeneity was reduced (Q = 10.36; df = 4; P- value = .0348), and the I2 test was 61.4% with a 95% CI 0.0–85.5%.
Figure 2.
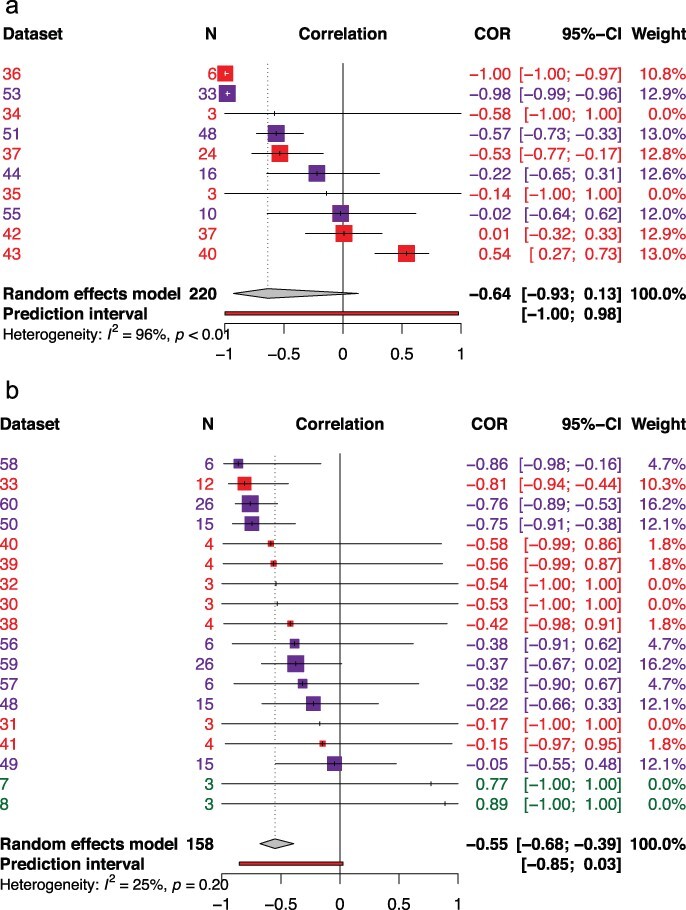
Forest plots displaying the distribution of effect sizes (correlations) for the life history theory (a), and the generalism–specialism dichotomy (b). The gray diamonds represent the pooled correlations, evolution of virulence (R = 0.15), life history theory (R = −0.64), and the generalism–specialism dichotomy (R = −0.55). The life history framework is mainly represented by negative correlations, which implies a tradeoff between survival and reproduction with animal viruses represented by datasets 34, 35, 36, 37, 42, and 43 and bacteriophages represented by datasets 44, 51, 53, and 55. Animal viruses in the generalism–specialism dichotomy are represented by datasets 30, 31, 32, 33, 38, 39, 40, and 41 while bacteriophages are represented by datasets 48, 49, 50, 56, 57, 58, 59, and 60. Datasets 7 and 8 represent plant viruses. Across panels, bacteriophage data sets are color-coded in purple, animal viruses in red, and plant viruses in green.
Generalism–specialism
Studies in this framework showed a significant negative correlation of traits between host shift/expansion and fitness (N = 18; R = − 0.5497; 95% CI: −0.6769 to −0.3905; t = −6.34; P- value = .0001) (Fig. 2b, Table 2). The test for heterogeneity was low (Q = 15.90; df = 12; P-value = .1959), and the I2 test was 24.5% with a 95% CI of 0.0−60.8%. No outliers were detected in this framework.
Plant viruses
For plant viruses, we found a significant positive correlation between virulence and transmission (N = 22; R = 0.4442; 95% CI: 0.1838−0.6464; t = 3.41; P-value = .0027). A test of the between-study heterogeneity variance was estimated (Q = 110; df = 15; P-value = .0027); I2 = 86.4 with a 95% CI of 79.5–91.0%. Running the test without outliers (Datasets 13 and 22), we found a significant positive correlation (N = 20; R = 0.5658; 95% CI: 0.3910−0.7013; t = 5.88; P-value <.0001) with a reduced test of between-study heterogeneity variance (Q = 56.46; df = 13; P-value < .0001 and I2 = 77.0% and 95% CI 61.6–86.2%) (Fig. 3a, Table 2). In addition, we performed an analysis of viruses that infect plants with viruses that infect fungi (Table 2, Supplementary Fig. S6). We found a significant positive relationship (N = 25; R = 0.3150; 95% CI: 0.0417–0.5444; t = 2.37; P- value = .0264). A test for heterogeneity was also estimated (Q = 129.75; df = 18; P- value < .0001; I2 = 86.1 with a 95% CI 79.7−90.5%. Without outliers (Datasets 1, 13, 22, and 25), we found a positive correlation (N = 21; R = 0.4411; 95% CI: 0.2042−0.6292; t = 3.77; P-value < .0014; Q = 49.33; df = 14; P- < .0001; I2 = 71.6%; 95% CI: 52.2−83.1%).
Figure 3.
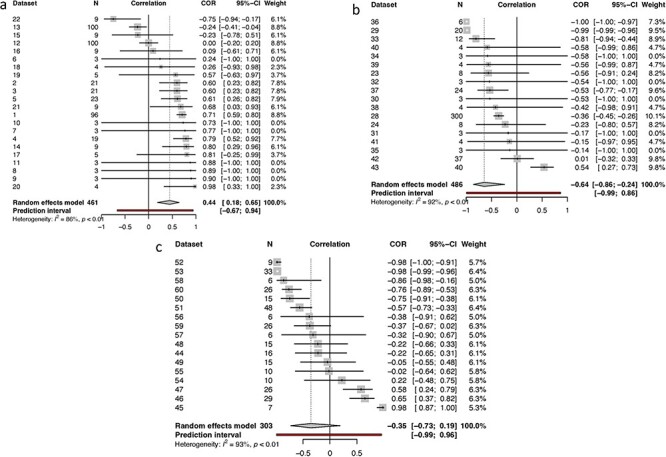
Forest plots showing the distribution of effect sizes for virus–host categories. Forest plot showing the distributions of effect sizes for viruses infecting plants (a), animals (b), and bacteriophages (c). The diamond indicates the pooled correlations. Viruses that infect plants indicate an overall positive correlation, which implies a positive relationship between virulence and transmission (R = 0.44). Most plant viruses tested the evolution of virulence framework, except for datasets 7 and 8 which tested the generalism–specialism dichotomy. No datasets explored life history theory in this category. Viruses that infect animals represented negative correlations with a pooled correlation of R = −0.6429. This category had a reasonable distribution of datasets across all three frameworks. The evolution of virulence is represented by datasets 23, 24, 28, and 29. The life history theory category is represented by datasets 34, 35, 36, 37, 42, 43, while the generalism–specialism dichotomy is represented by datasets 30, 31, 32, 33, 38, 39, 40, and 41. The pooled correlation for bacteriophages is R = −0.35 with most datasets identified for the generalism–specialism dichotomy (datasets 48, 49, 50, 56, 57, 58, 59, and 60). The evolution of virulence is represented by datasets 45, 46, 47, 52 and 54, and life history theory represented by datasets 44, 51, 53, and 55.
Animal viruses
For viruses infecting animals, we found a significant overall negative correlation (N = 18; R = −0.6429; 95% CI: −0.8571 to −0.2392; t = −3.10; P-value = .0065 (Fig. 3b; Table 2). A test of between-study heterogeneity variance was estimated (Q = 145.07; df = 12; P-value = .0001; I2 = 91.7%; 95% CI: 87.7−94.5%). Without outliers (Datasets 29, 36, and 43), we found a negative correlation between traits (N = 15; R = −0.3990; 95% CI: −0.5417 to −0.2340; t = −4.92; P-value < .0002). A test of between-study heterogeneity variance was estimated (Q = 13.22; df = 12; P-value = 0.3532; I2 = 9.2%; 95% CI: 0.0−47.6%).
Bacteriophages
For bacteriophages, we found a significant negative correlation (N = 17; R = − 0.3535; 95% CI: −0.7294 to 0.1863; t = −1.40; P-value = .0001. A test for between-study heterogeneity variance was estimated at (Q = 239.22; df = 16; P- = .0001), and I2 at 93.3%; 95% CI: 90.7−95.2%. Without outliers (Datasets 45, 46, 47, 52, and 53), we found a significant negative correlation between traits (N = 12; R = −0.4213; 95% CI: −0.6153 to 0.1792; t = − 3.69; P-value = .0036 and I2 at 47.6%. The test for heterogeneity was reduced (Q = 20.99; df = 11; P-value = .335; I2 = 47.6%; 95% CI: 0.0−73.1%). (Fig. 3c, Table 2.)
Analysis of coupled categories
The above analyses focus on the relationships between traits organized by tradeoff model or virus–host system. However, there is a possibility of interactions between these categories. That is, a trade-off framework (the evolution of virulence, for example) may have a statistical signature specific to certain virus–host systems Furthermore, we recognize that certain frameworks might be combined due to similarities in how traits are measured [e.g. some studies mention the evolution of virulence and life history theory in single studies (Brandon Ogbunugafor et al. 2013)]. It should be noted that Table 2 summarizes the results from analyses of frameworks and host types in detail and describes the results of datasets of the coupled categories. The analysis column specifies the type of analysis performed, and the nature of correlation is indicated along with corresponding P-values. Because of the different categories analyzed, we included the level of heterogeneity Q and the corresponding p − values with the number of datasets in the respective category. Below, we highlight some of the findings, also summarized in Table 2.
• The analysis of evolution of virulence that was combined with life history theory returned a nonsignificant negative correlation between traits (N = 42; R = − 0.0487; 95% CI: −0.3845 to 0.2985; t = −0.28; P-value = .7841; Q = 597.54; df = 35; P-value < .0001; I2 = 94.1%; 95% CI: 92.8−95.3%) (Supplementary Fig. S3).
• An analysis of traits associated with the evolution of virulence in only plant viruses produced a significant positive correlation between traits (N = 20; R = 0.4442; 95% CI: 0.1674 − 0.6561; t = 3.24; P-value = .0043; Q = 110.32; df = 15; P-value < .0001; I2 = 86.4%; 95% CI: 79.5− 91.0%).
• Studies of the evolution of virulence in animal viruses showed a nonsignificant negative correlation between traits (N = 7; R = −0.6036; 95% CI: −0.8970 to 0.0590; t = −2.26; P-value = .0649; Q = 72.52; df = 6; P-value < .0001; I2 = 91.7%; 95% CI: 85.5−95.3%).
• Studies using life history theory in animal viruses showed a nonsignificant negative correlation between traits (N = 6; R = − 0.6267; 95% CI: −0.9827 to 0.7161; t = − 1.16; P-value = .2996; Q = 51.84; df = 3; P-value < .0001; I2 = 94.2%; 95% CI: 88.3−97.1%).
• Those studies examining the generalism–specialism dichotomy analyzed in animal viruses showed a significant negative correlation between traits (N = 8; R = − 0.7145; 95% CI: −0.8312 to −0.5375; t = − 7.17; P-value = .0002; Q = 1.33; df = 4; P-value < .8569; I2 = 0%; 95% CI: 0.0−79.2%).
• Studies of the evolution of virulence analyzed in bacteriophages showed a nonsignificant positive correlation between traits (N = 5; R = 0.3163; 95% CI: −0.9393 to 0.9832; t = 0.44; P-value = .6816; Q = 63.75; df = 4; P-value < .0001; I2 = 93.7%; 95% CI: 88.2−96.7%).
• Life history theory in bacteriophages showed a nonsignificant negative correlation between traits (N = 4; R = −0.6717; 95% CI: −0.9851 to 0.6741; t = −1.59; P-value = .2107; Q = 68.54; df = 3; P-value < .0001; I2 = 95.6%; 95% CI: 91.6−97.7%).
• The generalism–specialism dichotomy analyzed in bacteriophages produced a significant negative correlation of traits (N = 8; R = −0.5131; 95% CI: −0.7268 to −0.2089; t = −3.78; P-value = .0069; Q = 13.25; df = 7; P-value = .0663; I2 = 47.2%; 95% CI: 0.0−76.5%).
Discussion
In this study, we examined direct (experimental, laboratory-based) evidence for tradeoffs between traits across three frameworks: evolution of virulence, life history theory, and the generalism–specialism dichotomy. We find that evidence for relationships between traits differ according to the framework examined and host–virus type. In general, the direct evidence for tradeoffs between traits in any of the frameworks is mixed, with some evidence for correlation and anticorrelation depending on the tradeoff model and virus–host system. More broadly, our findings suggest that while such theory may be sound in conception, direct experimental evidence does not point to a singular framework offering consistent insight across a suite of virus–host types or scientific disciplines. That is, the support for tradeoff frameworks is highly idiosyncratic.
The evolution of virulence
This is likely the most widely adopted framework in the evolution of infectious disease is the evolution of virulence. Thus, it may qualify as a main focus of our study, as virulence studies constitute the majority of direct tests of tradeoff models focusing on some aspect of virus evolution. Because the evolution of virulence is applied so broadly, the direct evidence for it demonstrated great variation with respect to how traits (associated with virulence and transmission) are measured. For example, in Cauliflower mosaic virus (CMV), dry weight reduction in host stems and leaves is used as a proxy for virulence (Doumayrou et al. 2013), while in Tomato spotted wilt virus (TSWV), virulence is measured as the estimated number of virus titers in Frankliniella occidentalis (western flower thrips); similarly, transmission is also measured by the thrips ability to transmit viruses (Rotenberg et al. 2009). Until now, the evolution of virulence paradigm has often treated virulence as a trait driven by the biology of the pathogen and may undervalue the role of host biology (e.g. via the immune system), medical interventions, coinfection, and many other modulators (Casadevall et al. 1999). Virulence could be considered a complex trait composed of factors from the pathogen, host, other modifiers, and interactions between them. Therefore, theory governing how virulence evolves could be modernized to include these parameters. Relatedly, scientists could better explain how traits are measured and be more consistent with measures across systems.
Our findings reflect this complexity. As summarized in Table 2, the nature of relationships between traits differs, and our analyses show mixed results depending on the category of analysis performed. For example, in plant viruses, relationships between virulence and transmission appear positive; in animal viruses, negative; in bacteriophages, positive. That is, there is no consistent pattern for how virulence and transmission relate. Moreover, it may be specific to the system and setting. In addition, generalism–specialism has a significant (negative) relationship, the only framework with a strongly significant global pattern. When we look at other host–virus systems, more subtle patterns emerge, indicative of the differences in biology: plant viruses show a positive relationship, while bacteriophages and animal viruses show negative relationships. This might reflect differences in how frameworks are used and how data collection methods are applied across different host–virus systems. Furthermore, we show the presence of publication bias and significant between-study heterogeneity. These features all highlight the differences in how frameworks are considered and tested.
Life history theory
Traits associated with life history theory are relatively easy to define and measure, as “survival” and “reproduction” can be described and quantified as less ambiguous than traits involved in tradeoffs in other frameworks. However, there were few experimental tests of life history theory tradeoffs of viruses in our search and analysis. Most tests we analyzed in this study had negative correlations between traits, with wide variance across studies. Notably, one of the strengths of life history theory is how it includes the evolution of the ability to survive outside a host (free-living survival and fomite transmission). Indirect or fomite transmission can be relevant in natural histories where viruses must spend much of their life cycle in the extra-host environment. In this sense, there is a conceptual overlap between life history theory and the evolution of virulence, as “survival” in a life history framing relates to “transmission” in the evolution of virulence (Brandon Ogbunugafor et al. 2013).
The generalism–specialism dichotomy
Of the three frameworks we examined, the generalism–specialism dichotomy contains the data with the least heterogeneity between datasets. However, this may be due to one virus–host system being overrepresented in the experimental data: vesicular stomatitis virus (VSV). VSV is widely used in evolution studies due to its wide host range and relative tractability under experimental conditions (Hanson 1952, Turner and Elena 2000). Considering the potential disease implications (especially for humans and animals), one can expect studies of animal viruses to be overrepresented. Nonetheless, the relative consistency in measurements from study to study are not necessarily reflective of the relative robustness of the theory across virus–host systems The results of the more granular analysis of the generalism–specialism dichotomy (organized by virus–host type) demonstrate that, overall, there is a negative relationship between traits, indicative of a tradeoff (Fig. 1, Table 2). However, the low number of studies and the lack of virus–host diversity across studies caution against drawing large conclusions about the applicability of this framework.
Analyses by virus–host type
There are several noteworthy patterns that appeared in the analysis of particular virus–host types. Overall, datasets from studies of plant viruses were the most numerous, most of which examined tradeoffs within the evolution of virulence. This is appropriate, given the vastness of plant life in the biosphere (Bar-On et al. 2018). These studies demonstrated evidence for a positive relationship between transmission and virulence consistent with other studies (Lipsitch and Richard Moxon 1997, Sacrist´an and Garc´ıa-Arenal 2008). Patterns in animal viruses were profoundly influenced by data from VSV, which revealed a negative relationship between traits across tradeoff frameworks. Datasets of bacteriophages contained the most outliers and showed a significant negative relationship after they were removed. The presence of multiple outliers in these datasets here could be derived from studies where phages were exposed to widely different stressors. The stressors include elevated heat (Dessau et al. 2012), urea (Heineman et al. 2012), and differing times spent outside the host (Garc´ıa-Villada and Drake 2013), among other variables. Such diverse stressors can also obscure signals of correlational traits and differences in how traits are measured. In addition, another key contributing factor to the overall correlation may be the molecular basis of the viral genome (e.g. RNA vs. DNA, single-stranded vs. double-standard), which impacts life history traits of bacteriophages, particularly reproduction and survival (Kindt et al. 2001, Smith et al. 2001). Such differences can further confound our application of tradeoff frameworks in virus evolution.
Limitations
The limitations of our study reflect challenges in the greater literature of virus evolution that have offered unclear definitions and inconsistent measurements of traits. In particular, studies of this sort can suffer from being underpowered. We have attempted to address this issue by providing a transparent data pipeline and a detailed discussion of the inclusion criteria. Through this, we were able to extract datasets that facilitated a meaningful analysis that we believe contributes to the greater literature on virus evolution.
This specific issue aside, a more general question remains: what makes a study worth including in meta-analyses? We have attempted to explain the technical aspects of this study in the “Methods” section, but more broadly, the usefulness of a study relates to the metrics used in measuring tradeoffs and the clarity of the data reported in the study. Though translating theoretical variables into measurable laboratory parameters can be challenging, scientists can aim for consistency, clarity, and transparency in measuring and reporting experimental data. Relatedly, it is crucial that diverse sample of viruses are included in laboratory-based studies, ideally not solely viruses of clinical or agricultural importance to humans.
Our study aimed to test experimental evidence for the evolution of virulence, life history theory, and the generalism–specialism dichotomy frameworks by analyzing data across virus–host systems. We were focused only on experimental data where traits associated with the frameworks were directly tested. This is a stringent but necessary constraint. This criterion eliminates hundreds of studies of virus evolution (mostly with respect to the evolution of virulence) in theoretical, field and clinical research, many rigorously examining traits associated with, for example, clinical outcomes (virulence) and compound metrics related to transmissibility like the basic reproductive ratio (also known as the R0) (Chowell et al. 2004, Yang et al. 2009, Althaus 2014, Grubaugh et al. 2019, Althouse et al. 2020). These studies are important, and many offer insight into the practical manifestations of virus evolution (and clarify the shape of epidemics). Thus, our study does not imply that findings from these natural settings are irrelevant or incorrect. Within the definitions and structures outlined in those studies, patterns of virulence evolution very well may operate according to existing canon. Instead, our study aimed to test whether these virus evolution frameworks sensu stricto (many animated in field and clinical studies) are based on rigorous laboratory-derived findings. Future work may take an analogous approach to ours but based on clinical or field-based studies. Also, our study focuses strictly on viruses. There are, of course, analogues to the questions that we have asked in other pathogen types, where some studies have examined how virulence or cost of infection can vary based on tradeoffs with other factors (Budischak et al. 2018. Turner et al. 2021). While our study is a meta-analysis that aims to cover a breadth of studies, we recognize the vastness of the literature on tradeoff models in pathogen evolution. Though this study is not a review, we have attempted to engage a large breadth of literature on the application of tradeoff models in virus evolution. Nonetheless, there are many review, opinion, and perspective articles that have argued multiple perspectives on virulence and other frameworks, some challenging the paradigms not unlike ours (Day 2002a, 2003, Day and Proulx 2004, Alizon et al. 2009, Cressler et al. 2016, Acevedo et al. 2019). We encourage those interested to engage with this corpus.
Other study limits apply to specific virus-host systems, to different kinds of biases that may skew the literature, how they may impact the literature, and, by extension, the data analyzed in the meta-analysis. For example, many plant viruses in this study affect hosts of economic interest to humans and are not representative of the vast diversity of plant viruses in nature. More generally, studies of this sort are influenced by publication biases of various kinds (see Supplementary Fig. S4). Our meta-analysis did not aim to formally test hypotheses about the nature or direction of biases but rather analyze data from the literature based on a set of clear criteria (see “Methods” section). However, we did use funnel plots to assess the presence of publication bias in our meta-analysis. But our results highlight that published data isn’t necessarily reflective of the natural world, but rather, how the process of science may select certain sorts of objects (virus-host systems in our case) for data collection. This is a common challenge in meta-analyses, which undoubtedly influences data and the inferences drawn from them.
Lastly, we should highlight what might be a glaring omission: we did not include tradeoffs in the evolution of resistance to antivirals, antibodies, and their associated evolutionary processes because resistance is often associated with reduced replication. There is a large and growing literature in these arenas, especially on HIV (Pennings 2013), influenza (Holmes et al. 2021), and SARS-CoV-2 (Segala et al. 2021). But while tradeoffs associated with resistance are of immense biomedical significance, these apply to specific circumstances of viruses evolving against small molecules or other antagonists, and do not qualify as a grander explanatory theory of virus evolution in the same way as the three foci of this study. In fact, one can argue that the evolution of HIV resistance to antiviral drugs (for example) is a special case of virus evolution that could be modeled using one of the three large frameworks in our study (evolution of virulence, life history theory, the generalism–specialism dichotomy).
Ideas and speculation
While our findings introduce skepticism that may undermine our confidence in existing frameworks, we emphasize that this could start a productive effort to generate new perspectives, theories, and tests. Future work could offer ways for the three frameworks examined in this study to be compatible. One hypothetical reconciliation between the different frameworks and our findings involves the notion that different frameworks apply to different scales of the interaction between virus and host. Previous studies have suggested how the evolution of virulence may manifest differently across scales of the disease emergence process (Geoghegan and Holmes 2018, Visher et al. 2021) and other temporal aspects of infection (Day 2003). We can extend this concept to include settings other than the disease emergence process. Figure 4 offers one such hypothetical scenario: that life history theory may apply to viruses in the intra-host setting, that the evolution of virulence dictates constraints at the between-host scale, and generalism–specialism operating at the between-species scales. We acknowledge that this overview is pure conjecture: we offer no data-driven reason to suggest any specific relationship between frameworks and scales exists. We offer it to suggest the perspectives that might explain our results and the general complexity of theory in virus evolution. Future efforts can directly test these theories or establish new multiscale theories.
Figure 4.
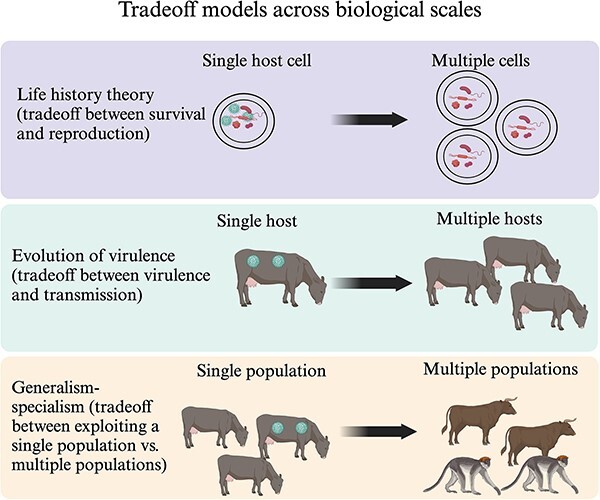
A hypothetical illustration of the evolutionary frameworks at different biological scales. Here, we present a scenario where the existence of three frameworks analyzed in this study–life history theory, evolution of virulence, and generalism–specialism–could be reconciled through application to different scales of virus–host interactions. Life history theory could be applied to the problem of reproduction within a cell versus surviving transmission between cells. The evolution of virulence might apply to virulence traits associated with growth within a host at the organismal scale, and transmission to other (generally conspecific) hosts. Lastly, for the generalism–specialism dichotomy, the purported tradeoff exists between growth fitness within a single species or population and the ability to propagate across different species or populations. This entire model represents a speculative view on tradeoff frame- works whereby they could all operate and have relevance for our efforts to understand the constraints underlying virus evolution.
Conclusion
The quest for unified theoretical frameworks to understand and predict virus evolution has a long history, a robust present, and a promising future. Current technological and computational advances offer new ways to measure virus traits at a large scale, with accompanying tools allowing scientists to uncover the genetic bases for such changes (Grubaugh et al. 2019, Black et al. 2020, Vogels et al. 2021). However, large data approaches alone cannot serve as a replacement for theoretical ambiguity. Scientists who study topics related to the evolution of infectious diseases can challenge themselves with basic questions about assumptions built into the theoretical frameworks used to study and discuss virus evolution.
Our findings indicate that the picture offered from direct tests of virus evolution frameworks is cloudy. This ambiguous picture implores us to revisit and possibly reconfigure theories that guide the study of virus evolution. Such a new picture could consider how variation in host–parasite systems, definitions of terms, measurements, analysis methods, and modeling approaches can profoundly complicate our expectations for virus evolution. Theory that accommodates this pluralism might (paradoxically) clarify our picture of virus evolution, with implications for evolutionary biology, disease ecology, evolutionary medicine, and epidemiology.
Supplementary Material
Acknowledgements
The authors would like to thank two anonymous peer reviewers for comments on a prior draft of the manuscript. We also thank V. Ezenwa, N. Grubaugh, P. Pennings, S. Almagro-Moreno, S. Scarpino, J. Lewis, L. Zaman, and members of the Pennings and Ogbunu labs for useful comments on topics related to this manuscript. Figure 4 was created with BioRender.com. Lastly, the authors would like to thank the organizers and participants in the workshop “Modeling and Theory in Population Biology” meeting at the Banff International Research Station (May 2024), where ideas related to this manuscript were discussed.
Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Contributor Information
Ketty Kabengele, Department of Ecology and Evolutionary Biology, Yale University, 165 Prospect Street, New Haven, CT 06511, United States.
Wendy C Turner, U.S. Geological Survey, Wisconsin Cooperative Wildlife Research Unit, Department of Forest and Wildlife Ecology, University of Wisconsin-Madison, 1630 Linden Drive, Madison, WI 53706, United States.
Paul E Turner, Department of Ecology and Evolutionary Biology, Yale University, 165 Prospect Street, New Haven, CT 06511, United States; Microbiology Program, Yale School of Medicine, 333 Cedar Street, New Haven, CT 06510, United States.
C. Brandon Ogbunugafor, Department of Ecology and Evolutionary Biology, Yale University, 165 Prospect Street, New Haven, CT 06511, United States; Public Health Modeling Unit, Yale School of Public Health 60 College Street , New Haven CT 06510, United States; Santa Fe Institute, 1399 Hyde Park Road, Santa Fe, NM 87501, United States.
Author contributions
Project conception: K.K., C.B.O, Collected data: K.K, Analyzed data: K.K., C.B.O, Integrated and interpreted results: K.K., W.C.T., P.E.T., C.B.O, Supervision: C.B.O. Writing, first drafts: K.K., C.B.O, and Revisions, editing, and commenting: K.K., W.C.T., P.E.T., C.B.O.
Supplementary data
Supplementary data is available at VEVOLU online.
Conflict of interest:
None declared.
Funding
The authors acknowledge support from the National Institutes of Health (grants R01AI168166 to C.B.O.) K.K. acknowledges support from the Training Program in Genetics, Yale School of Medicine. C.B.O. acknowledges support from the MLK Visiting Scholars and Professors Program at the Massachusetts Institute of Technology.
Data availability
Data and code can be found on GitHub: https://github.com/OgPlexus/MetaKetty1
Supplementary information
In the Supplementary Information, we offer additional figures and analyses that support findings from the main text. These include the PRISMA flow diagram (Supplementary Fig. S1), an analysis of all datasets from the meta-analysis (Supplementary Fig. S2), an analysis of the evolution of virulence and life history theory combined (Supplementary Fig. S3), an analysis of publication bias (Supplementary Fig. S4), an analysis of the evolution of virulence framework without N =3 datasets (Supplementary Fig. S5), an analysis of plant viruses combined with mycoviruses (viruses that infect fungi) (Supplementary Fig. S6), statistics from all analyses (Table 1), forest plots with very high contrasts of analyses of frameworks, and virus–host types (Supplementary Figs S7–9).
References
- Acevedo MA, Dillemuth FP, Flick AJ. et al. Virulence-driven trade-offs in disease transmission: a meta-analysis. Evolution 2019;73:636–47. doi: 10.1111/evo.13692 [DOI] [PubMed] [Google Scholar]
- Alizon S, Hurford A, Mideo N. et al. Virulence evolution and the trade-off hypothesis: history, current state of affairs and the future. J Evol Biol 2009;22:245–59. doi: 10.1111/j.1420-9101.2008.01658.x [DOI] [PubMed] [Google Scholar]
- Alizon S, van Baalen M. Emergence of a convex trade-off between transmission and virulence. Am Nat 2005;165:E155–67. doi: 10.1086/430053 [DOI] [PubMed] [Google Scholar]
- Althaus CL. Estimating the reproduction number of Ebola virus (EBOV) during the 2014 outbreak in West Africa. PLoS Curr 2014;6:2157–3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althouse BM, Wenger EA, Miller JC. et al. Superspreading events in the transmission dynamics of SARS-CoV-2: opportunities for interventions and control. PLoS Biol 2020;18:e3000897. doi: 10.1371/journal.pbio.3000897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alto BW, Turner PE. Consequences of host adaptation for performance of vesicular stomatitis virus in novel thermal environments. Evol Ecol 2010;24:299–315. doi: 10.1007/s10682-009-9307-3 [DOI] [Google Scholar]
- Anderson RM, May RM. Coevolution of hosts and parasites. Parasitology 1982;85:411–26. doi: 10.1017/S0031182000055360 [DOI] [PubMed] [Google Scholar]
- Balduzzi S, Rücker G and Schwarzer G. How to perform a meta-analysis with R: a practical tutorial Evid Based Mental Health 2019;22:153–160 doi: 10.1136/ebmental-2019-300117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-On YM, Phillips R, Milo R. The biomass distribution on Earth. Proc Natl Acad Sci 2018;115:6506–11. doi: 10.1073/pnas.1711842115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bera S, Fraile A, Garc´ıa-Arenal F. Analysis of fitness trade-offs in the host range expansion of an rna virus, tobacco mild green mosaic virus. J Virol 2018;92:e01268–18. doi: 10.1128/JVI.01268-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black A, MacCannell DR, Sibley TR. et al. Ten recommendations for supporting open pathogen genomic analysis in public health. Nat Med 2020;26:832–41. doi: 10.1038/s41591-020-0935-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono LM, Gensel CL, Pfennig DW. et al. Evolutionary rescue and the coexistence of generalist and specialist competitors: an experimental test. Proc R Soc B 2015;282:20151932. doi: 10.1098/rspb.2015.1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon Ogbunugafor C, Alto BW, Overton TM. et al. Evolution of increased survival in RNA viruses specialized on cancer-derived cells. Am Nat 2013;181:585–95. doi: 10.1086/670052 [DOI] [PubMed] [Google Scholar]
- Brusini J, Wayne ML, Franc A. et al. The impact of parasitism on resource allocation in a fungal host: the case of Cryphonectria parasitica and its mycovirus, Cryphonectria Hypovirus 1. Ecol Evol 2017;7:5967–76. doi: 10.1002/ece3.3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budischak SA, O’Neal D, Jolles AE. et al. Differential host responses to parasitism shape divergent fitness costs of infection. Funct Ecol 2018;32:324–33. doi: 10.1111/1365-2435.12951 [DOI] [Google Scholar]
- Bull JJ. Virulence. Evolution 1994;48:1423–37. doi: 10.1111/j.1558-5646.1994.tb02185.x [DOI] [PubMed] [Google Scholar]
- Bull JJ, Lauring AS, Condit RC. Theory and empiricism in virulence evolution. PLoS Pathog 2014;10:e1004387. doi: 10.1371/journal.ppat.1004387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, Pirofski L-A, Fischetti VA. Host-pathogen interactions: redefining the basic concepts of virulence and pathogenicity. Infect Immun 1999;67:3703–13. doi: 10.1128/IAI.67.8.3703-3713.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowell G, Hengartner NW, Castillo-Chavez C. et al. The basic reproductive number of Ebola and the effects of public health measures: the cases of Congo and Uganda. J Theor Biol 2004;229:119–26. doi: 10.1016/j.jtbi.2004.03.006 [DOI] [PubMed] [Google Scholar]
- Ciota AT, Ehrbar DJ, Matacchiero AC. et al. The evolution of virulence of West Nile virus in a mosquito vector: implications for arbovirus adaptation and evolution. BMC Evol Biol 2013;13:71. doi: 10.1186/1471-2148-13-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cressler CE, McLEOD DV, Rozins C. et al. The adaptive evolution of virulence: a review of theoretical predictions and empirical tests. Parasitology 2016;143:915–30. doi: 10.1017/S003118201500092X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crill WD, Wichman HA, Bull JJ. Evolutionary reversals during viral adaptation to alternating hosts. Genetics 2000;154:27–37. doi: 10.1093/genetics/154.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day T. On the evolution of virulence and the relationship between various measures of mortality. Proc R Soc B 2002a;269:1317–23. doi: 10.1098/rspb.2002.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day T. Virulence evolution via host exploitation and toxin production in spore-producing pathogens. Ecol Lett 2002b;5:471–76. doi: 10.1046/j.1461-0248.2002.00342.x [DOI] [Google Scholar]
- Day T. Virulence evolution and the timing of disease life-history events. Trends Ecol Evol 2003;18:113–18. doi: 10.1016/S0169-5347(02)00049-6 [DOI] [Google Scholar]
- Day T, Proulx SR. A general theory for the evolutionary dynamics of virulence. Am Nat 2004;163:E40–63. doi: 10.1086/382548 [DOI] [PubMed] [Google Scholar]
- Deardorff ER, Fitzpatrick KA, Jerzak GVS. et al. West Nile virus experimental evolution in vivo and the trade-off hypothesis. PLoS Pathog 2011;7:e1002335. doi: 10.1371/journal.ppat.1002335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessau M, Goldhill D, McBride RL. et al. Selective pressure causes an RNA virus to trade reproductive fitness for increased structural and thermal stability of a viral enzyme. PLoS Genet 2012;8:e1003102. doi: 10.1371/journal.pgen.1003102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo-Calap P, Pereira-g´omez M, Sanju´an R. Selection for thermostability can lead to the emergence of mutational robustness in an RNA virus. J Evol Biol 2010;23:2453–60. doi: 10.1111/j.1420-9101.2010.02107.x [DOI] [PubMed] [Google Scholar]
- Doumayrou J, Avellan A, Froissart R. et al. An experimental test of the transmission-virulence trade-off hypothesis in a plant virus. Evolution 2013;67:477–86. doi: 10.1111/j.1558-5646.2012.01780.x [DOI] [PubMed] [Google Scholar]
- Duffy S, Turner PE, Burch CL. Pleiotropic costs of niche expansion in the RNA bacteriophage phi 6. Genetics 2006;172:751–57. doi: 10.1534/genetics.105.051136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert D, Bull JJ. Challenging the trade-off model for the evolution of virulence: is virulence management feasible? Trends Microbiol 2003;11:15–20. doi: 10.1016/S0966-842X(02)00003-3 [DOI] [PubMed] [Google Scholar]
- Elena SF. Evolutionary history conditions the timing of transmission in vesicular stomatitis virus. Infect Genet Evol 2001;1:151–59. doi: 10.1016/S1567-1348(01)00022-3 [DOI] [PubMed] [Google Scholar]
- Elena SF, Agudelo-Romero P, Lali´c J. The evolution of viruses in multi-host fit- ness landscapes. Open Virol J 2009;3:1–6. doi: 10.2174/1874357900903010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena SF, Fraile A, Garc´ıa-Arenal F. Chapter three—evolution and emergence of plant viruses. Adv Virus Res 2014;88:161–91. [DOI] [PubMed] [Google Scholar]
- Ewald PW. Transmission modes and the evolution of virulence. Hum Nat 1991;2:1–30. doi: 10.1007/BF02692179 [DOI] [PubMed] [Google Scholar]
- Fargette D, Pinel-Galzi A, Sérémé D. et al. Diversification of rice yellow mottle virus and related viruses spans the history of agriculture from the neolithic to the present. PLoS Pathog 2008;4:e1000125. doi: 10.1371/journal.ppat.1000125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenner F, Marshall ID. A comparison of the virulence for European rabbits (Oryctolagus cuniculus) of strains of myxoma virus recovered in the field in Australia, Europe and America. Epidemiol Infect 1957;55:149–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris MT, Joyce P, Burch CL. High frequency of mutations that expand the host range of an RNA Virus. Genetics 2007;176:1013–22. doi: 10.1534/genetics.106.064634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor E. Ecological niche of plant pathogens. Ann For Res 2011;54:3–21. [Google Scholar]
- Ford BE, Sun B, Carpino J. et al. Frequency and fitness consequences of bacteriophage 6 host range mutations. PLoS One 2014;9:e113078. doi: 10.1371/journal.pone.0113078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SA. Models of parasite virulence. Q Rev Biol 1996;71:37–78. doi: 10.1086/419267 [DOI] [PubMed] [Google Scholar]
- Fraser C, Hollingsworth TD, Chapman R. et al. Variation in HIV-1 set-point viral load: epidemiological analysis and an evolutionary hypothesis. Proc Natl Acad Sci 2007;104:17441–46. doi: 10.1073/pnas.0708559104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froissart R, Doumayrou J, Vuillaume F. et al. The virulence–transmission trade-off in vector-borne plant viruses: a review of (non-) existing studies. Philos Trans R Soc B 2010;365:1907–18. doi: 10.1098/rstb.2010.0068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futuyma DJ, Moreno G. The evolution of ecological specialization. Annu Rev Ecol Syst 1988;19:207–33. doi: 10.1146/annurev.es.19.110188.001231 [DOI] [Google Scholar]
- Gandon S, Mackinnon MJ, Nee S. et al. Imperfect vaccines and the evolution of pathogen virulence. Nature 2001;414:751–56. doi: 10.1038/414751a [DOI] [PubMed] [Google Scholar]
- Garc´ıa-Villada L, Drake JW. Experimental selection reveals a trade-off between fecundity and lifespan in the coliphage Qß. Open Biol 2013;3:130043. doi: 10.1098/rsob.130043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnick E. Niche breadth in parasites: an evolutionarily stable strategy model, with special reference to the protozoan parasite Leishmania. Theor Popul Biol 1992;42:62–103. doi: 10.1016/0040-5809(92)90005-E [DOI] [PubMed] [Google Scholar]
- Geoghegan JL, Holmes EC. The phylogenomics of evolving virus virulence. Nat Rev Genet 2018;19:756–69. doi: 10.1038/s41576-018-0055-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldhill DH, Turner PE. The evolution of life history trade-offs in viruses. Curr Opin Virol 2014;8:79–84. doi: 10.1016/j.coviro.2014.07.005 [DOI] [PubMed] [Google Scholar]
- Gomez LM, Meszaros VA, Turner WC. et al. The epidemiological signature of pathogen populations that vary in the relationship between free-living parasite survival and virulence. Viruses 2020;12:1055. doi: 10.3390/v12091055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh ND, Ladner JT, Lemey P. et al. Tracking virus outbreaks in the twenty-first century. Nat Microbiol 2019;4:10–19. doi: 10.1038/s41564-018-0296-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel A, Lebarbenchon C, Stallknecht D. et al. Trade-offs between and within scales: environmental persistence and within-host fitness of avian influenza viruses. Proc R Soc B 2014;281:20133051. doi: 10.1098/rspb.2013.3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson RP. The natural history of vesicular stomatitis. Bacteriol Rev 1952;16:179–204. doi: 10.1128/br.16.3.179-204.1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrer M, Cuijpers P, Furukawa T, and Ebert D. Doing Meta-Analysis with R: A Hands-On Guide (New YorK: Chapman and Hall/CRC; )doi: 10.1201/9781003107347. 2021. [DOI] [Google Scholar]
- Hedges LV, Vevea JL. Fixed- and random-effects models in meta-analysis. Psychol Methods 1998;3:486–504. doi: 10.1037/1082-989X.3.4.486 [DOI] [Google Scholar]
- Heineman RH, Brown SP, Brockhurst MA. Experimental evolution of a bacteriophage virus reveals the trajectory of adaptation across a Fecundity/Longevity Trade-Off. PLoS One 2012;7:e46322. doi: 10.1371/journal.pone.0046322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC, Ghedin E, Miller N. et al. Whole-genome analysis of human Influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol 2005;3:e300. doi: 10.1371/journal.pbio.0030300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC, Hurt AC, Dobbie Z. et al. Understanding the impact of resistance to Influenza Antivi- rals. Clin Microbiol Rev 2021;34:10–1128. doi: 10.1128/CMR.00224-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindt J, Tzlil S, Ben-Shaul A. et al. DNA packaging and ejection forces in bacteriophage. Proc Natl Acad Sci U S A 2001;98:13671–74. doi: 10.1073/pnas.241486298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp G, Hartung J. Improved tests for a random effects meta-regression with a single covariate. Stat Med 2003;22:2693–710. doi: 10.1002/sim.1482 [DOI] [PubMed] [Google Scholar]
- Kucharski AJ, Lessler J, Read JM. et al. Estimating the Life Course of Influenza A(H3N2) antibody responses from cross-sectional data. PLoS Biol 2015;13:e1002082. doi: 10.1371/journal.pbio.1002082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- L´azaro E, Arribas M, Cabanillas L. et al. Evolutionary adaptation of an RNA bacteriophage to the simultaneous increase in the within-host and extracellular temperatures. Sci Rep 2018;8:8080. doi: 10.1038/s41598-018-26443-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Miller CR, Nagel AC. et al. First-step mutations for adaptation at elevated temperature increase capsid stability in a virus. PLoS One 2011;6:e25640. doi: 10.1371/journal.pone.0025640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett HC, Buckling A, Long GH. et al. Generalism and the evolution of parasite virulence. Trends Ecol Evol 2013;28:592–96. doi: 10.1016/j.tree.2013.07.002 [DOI] [PubMed] [Google Scholar]
- Levin BR, Bull JJ. Short-sighted evolution and the virulence of pathogenic microorganisms. Trends Microbiol 1994;2:76–81. doi: 10.1016/0966-842X(94)90538-X [DOI] [PubMed] [Google Scholar]
- Lipsitch M, Richard Moxon E. Virulence and transmissibility of pathogens: what is the relation-ship? Trends Microbiol 1997;5:31–37. doi: 10.1016/S0966-842X(97)81772-6 [DOI] [PubMed] [Google Scholar]
- Longdon B, Brockhurst MA, Russell CA. et al. The evolution and genetics of virus host shifts. PLoS Pathog 2014;10:e1004395. doi: 10.1371/journal.ppat.1004395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay B, Ebell M, Dale AP. et al. Virulence-mediated infectiousness and activity trade-offs and their impact on transmission potential of influenza patients. Proc R Soc B 2020;287:20200496. doi: 10.1098/rspb.2020.0496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Dickson MDMeszaros VA, Junior FB. et al. Waterborne, abiotic and other indirectly transmitted (W.A.I.T.) infections are defined by the dynamics of free-living pathogens and environmental reservoirs. Tech. rep. Section: New Results Type: article. bioRxiv 2019: 525089.
- Montarry J, Cartier E, Jacquemond M. et al. Virus adaptation to quantitative plant resistance: erosion or breakdown? J Evol Biol 2012;25:2242–52. doi: 10.1111/j.1420-9101.2012.02600.x [DOI] [PubMed] [Google Scholar]
- Moreno-P´erez MG, García-Luque I, Fraile A. et al. Mutations that determine resistance breaking in a plant RNA virus have pleiotropic effects on its fitness that depend on the host environment and on the type, single or mixed, of infection. J Virol 2016;90:9128–37. doi: 10.1128/JVI.00737-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paepe MD, Taddei F, Kirkwood T. Viruses’ life history: towards a mechanistic basis of a trade- off between survival and reproduction among phages. PLoS Biol 2006;4:e193. doi: 10.1371/journal.pbio.0040193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennings PS. HIV drug resistance: problems and perspectives. Infect Dis Rep 2013;5:e5. doi: 10.4081/idr.2013.s1.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poisot T, Bever JD, Nemri A. et al. A conceptual framework for the evolution of ecological specialisation. Ecol Lett 2011;14:841–51. doi: 10.1111/j.1461-0248.2011.01645.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posit . A Better Way to Deploy R & Python. 2023. https://www.posit.co/ (17 October 2023, date last accessed).
- Poulicard N, Pinel‐galzi A, Hebrard E. et al. Why Rice yellow mottle virus, a rapidly evolving RNA plant virus, is not efficient at breaking rymv1-2 resistance. Mol Plant Pathol 2010;11:145–54. doi: 10.1111/j.1364-3703.2009.00582.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presloid JB, Mohammad TF, Lauring AS. et al. Antigenic diversification is correlated with increased thermostability in a mammalian virus. Virology 2016;496:203–14. doi: 10.1016/j.virol.2016.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafaluk-Mohr C. The relationship between parasite virulence and environmental persistence: a meta-analysis. Parasitology 2019;146:897–902. doi: 10.1017/S0031182019000015 [DOI] [PubMed] [Google Scholar]
- Read AF. The evolution of virulence. Trends Microbiol 1994;2:73–76. doi: 10.1016/0966-842X(94)90537-1 [DOI] [PubMed] [Google Scholar]
- Redman EM, Wilson K, Cory JS. Trade-offs and mixed infections in an obligate- killing insect pathogen. J Anim Ecol 2016;85: 1200–09. doi: 10.1111/1365-2656.12547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche B, Broutin H, Choisy M. et al. The niche reduction approach: an opportunity for optimal control of infectious diseases in low-income countries? BMC Public Health 2014;14:753. doi: 10.1186/1471-2458-14-753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotenberg D, Krishna Kumar NK, Ullman DE. et al. Variation in Tomato spotted wilt virus Titer in Frankliniella occidentalis and Its Association with Frequency of Transmission. Phytopathology® 2009;99: 404–410 doi: 10.1094/PHYTO-99-4-0404 [DOI] [PubMed] [Google Scholar]
- Sacrist´an S, Fraile A, Malpica JM. et al. An analysis of host adaptation and its relationship with virulence in cucumber mosaic virus. Phytopathology® 2005;95:827–33. doi: 10.1094/PHYTO-95-0827 [DOI] [PubMed] [Google Scholar]
- Sacrist´an S, Garc´ıa-Arenal F. The evolution of virulence and pathogenicity in plant pathogen populations. Mol Plant Pathol 2008;9:369–84. doi: 10.1111/j.1364-3703.2007.00460.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer G, Carpenter JR, Ru¨cker G. Meta-Analysis with R. Use R! Cham: Springer International Publishing, 2015. [Google Scholar]
- Segala FV, Bavaro DF, Di Gennaro F. et al. Impact of SARS-CoV-2 epidemic on antimicrobial resistance: a literature review. Viruses 2021;13:2110. doi: 10.3390/v13112110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidik K, Jonkman JN. Simple heterogeneity variance estimation for meta-analysis. J R Stat Soc 2005;54:367–84. [Google Scholar]
- Smith DE, Tans SJ, Smith SB. et al. The bacteriophage 29 portal motor can package DNA against a large internal force. Nature 2001;413:748–52. doi: 10.1038/35099581 [DOI] [PubMed] [Google Scholar]
- Stearns SC. Life-history tactics: a review of the ideas. Q Rev Biol 1976;51:3–47. doi: 10.1086/409052 [DOI] [PubMed] [Google Scholar]
- Stearns SC. Trade-offs in life-history evolution. Funct Ecol 1989;3:259. doi: 10.2307/2389364 [DOI] [Google Scholar]
- Surasinghe S, Kabengele K, Turner PE. et al. Evolutionary invasion analysis of modern epidemics highlights the context-dependence of virulence evolution. Bull Math Biol 2024;86:88. doi: 10.1007/s11538-024-01313-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner PE, Elena SF. Cost of host radiation in an RNA virus. Genetics 2000;156:1465–70. doi: 10.1093/genetics/156.4.1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner WC, Kamath PL, van Heerden H. et al. The roles of environmental variation and parasite survival in virulence-transmission relationships. R Soc Open Sci 2021;8:210088. doi: 10.1098/rsos.210088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visher E, Evensen C, Guth S. et al. The three Ts of virulence evolution during zoonotic emergence. Proc R Soc B 2021;288:20210900. doi: 10.1098/rspb.2021.0900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels CBF, Breban MI, Ott IM. et al. Multiplex qPCR discriminates variants of concern to enhance global surveil- lance of SARS-CoV-2. PLoS Biol 2021;19:e3001236. doi: 10.1371/journal.pbio.3001236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasik BR, Bhushan A, Ogbunugafor CB. et al. Delayed transmission selects for increased survival of vesicular stomatitis virus. Evolution 2015;69:117–25. doi: 10.1111/evo.12544 [DOI] [PubMed] [Google Scholar]
- Wasik BR, Rothschild E, Voorhees IEH. et al. Understanding the divergent evolution and epidemiology of H3N8 influenza viruses in dogs and horses. Virus Evol 2023;9:vead052. doi: 10.1093/ve/vead052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witter RL. Increased virulence of Marek’s disease virus field isolates. Avian Dis 1997;41:149–63. doi: 10.2307/1592455 [DOI] [PubMed] [Google Scholar]
- Yang Y, Sugimoto JD, Halloran ME. et al. The transmissibility and control of pandemic Influenza A (H1N1) Virus. Science 2009;326:729–33. doi: 10.1126/science.1177373 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and code can be found on GitHub: https://github.com/OgPlexus/MetaKetty1


