Abstract
Myeloproliferative neoplasms (MPN) are chronic hematological disorders marked by the abnormal proliferation of bone marrow cells. The most commonly encountered forms are polycythemia vera (PV), primary myelofibrosis (PMF), and essential thrombocythemia (ET). These disorders are generally associated with increases in blood components, which can lead to conditions like splenomegaly, thrombosis, bleeding tendencies, and a heightened risk of progressing to acute leukemia. Previous research has indicated a possible link between immune cells and MPN, yet this association is still poorly understood. This study seeks to elucidate the causal relationship between immune cell characteristics and the development of MPN. In this study, we employed Mendelian randomization (MR) to investigate potential causal links between 731 immune cell traits and the risk of developing MPN, leveraging data from genome-wide association studies (GWAS). To ensure the robustness of our findings, we conducted extensive sensitivity analyses to assess heterogeneity and detect any pleiotropic effects. Moreover, we implemented a false discovery rate (FDR) correction to mitigate the risk of false positives that may result from the multiple hypothesis testing, thereby adjusting for any statistical biases due to multiple comparisons. The immune phenotype IgD on IgD+ CD24- B cells demonstrated a statistically significant protective effect against MPN (PFDR = 0.047). Upon adjusting the significance threshold to PFDR < 0.20, 16 immune cell phenotypes were significantly associated with MPN. Among these, 11 were found to exert a protective effect against MPN, 5 phenotypes were associated with an elevated risk of MPN. This research highlights a significant association between various immune cell phenotypes and the risk of developing MPN, thereby advancing our understanding of the intricate interplay between immune cell traits and the progression of MPN.
Keywords: causal inference, immunity, MR analysis, Myeloproliferative neoplasms, sensitivity
1. Introduction
Myeloproliferative neoplasms (MPN) constitute a category of clonal hematologic cancers that arise from hematopoietic stem.[1,2] These malignancies are characterized by the abnormal proliferation of 1 or more blood cell lineages within the bone marrow. The primary types of these neoplasms include polycythemia vera (PV), primary myelofibrosis (PMF), essential thrombocythemia (ET).[3] Patients with MPN typically exhibit abnormal peripheral blood cell counts, such as erythrocytosis, leucocytosis, or thrombocytosis, and may present with clinical manifestations like splenomegaly, thrombosis, or bleeding. Moreover, a subset of patients may progress to acute myeloid leukaemia, significantly increasing the lethality associated with MPN.[4]
The epidemiology of MPN varies by region. In Western countries, the annual incidence of MPN is approximately 1 to 3 per 100,000 people. Specifically, the incidence rates PV range from 0.4 to 2.8 per 100,000, PMF from 0.5 to 1.5 per 100,000, and ET from 1.5 to 2.5 per 100,000.[5] The incidence in East Asian regions is generally lower than in Western countries.[6] With the aging population and advancements in diagnostic technology, the incidence of MPN is gradually increasing.[7] Although MPN is relatively rare, its long disease course and treatment challenges significantly impact patients’ quality of life, underscoring the importance of research into their pathogenesis and potential treatment options.
The precise causes and mechanisms underlying MPN remain unclear. Current research suggests that immune function abnormalities are present in patients with MPN, impacting various components of both innate and adaptive immunity.[1,2] The success of immunosuppressive therapies, such as interferon-α, in treating MPN highlights the crucial role of the immune system in these disorders. Research has identified several immune cells, including T cells, B cells, and natural killer (NK) cells, as key players in the pathogenesis of MPNs, largely due to their involvement in the dysregulated activation of the JAK-STAT signaling pathway.[4] Nevertheless, the complexity of the immune system, coupled with the heterogeneity of MPN, complicates our understanding of the precise role that immune cells play in the development of these diseases.
Mendelian randomization (MR) is an analytical approach rooted in Mendel’s law of independent assortment, primarily used in epidemiology to infer causal relationships. Genome-wide association studies (GWAS) pinpoint genetic variants associated with heightened disease risk by examining millions of single nucleotide polymorphisms (SNPs) throughout the genome. GWAS is instrumental in uncovering associations between genetic variants and specific diseases or biological traits within populations. Recently, advancements in GWAS have enhanced the value of MR in uncovering causal relationships between immune traits and diseases.[8–10] Multiple studies have highlighted the significance of MR analysis in identifying causal links in hematologic diseases.[11–13] In this research, we employed 2-sample MR to explore the causal connections between immune cell characteristics and MPN, offering novel insights into the pathogenesis of MPN.
2. Methods
2.1. Study design
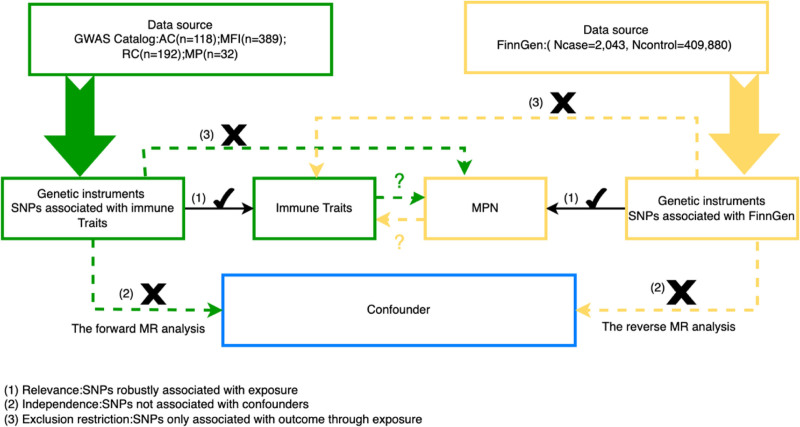
This study aims to explore the causal relationships between 731 immune cell traits and MPN. To ensure the validity of our findings, each MR analysis is conducted in accordance with 3 fundamental assumptions[14]: The Relevance Assumption, which requires that the instrumental variable (IV) is strongly associated with the exposure; The Independence Assumption, which necessitates that the IVs are independent of any confounders that may influence both the exposure and the outcome; and The Exclusion Restriction Assumption, which dictates that the IVs must impact the outcome exclusively through the exposure. SNPs are employed as IVs in this study. The methodological framework of the 2-sample bidirectional MR design applied to immune cell phenotypes and MPNs is depicted in Figure 1. Since this research involves the reanalysis of previously collected and publicly available data, additional ethical approval is not required.
Figure 1.
Principles of MR study design. AC = absolute cell, GWAS = genome-wide association study, MFI = median fluorescence intensities, MP = morphological parameters, MPN = Myeloproliferative neoplasms, MR = Mendelian randomization, RC = relative cell, SNP = Single nucleotide polymorphism.
To ensure the rigor and integrity of this observational MR study, the STROBE-MR (i.e., Strengthening the Reporting of Observational Studies in Epidemiology using Mendelian randomization) checklist was thoroughly followed and completed (Table S1, Supplemental Digital Content, http://links.lww.com/MD/O203).[15]
2.2. Sources of immunity-spanning GWAS data
The immune cell phenotype data utilized in this study were sourced from the publicly accessible GWAS Catalog database, available at https://www.ebi.ac.uk/gwas/.[9] The data encompass accession numbers from GCST0001391 to GCST0002121. The GWAS Catalog is a comprehensive public repository that aggregates and provides data from GWAS identifying associations between SNPs and various diseases or physiological traits. The dataset analyzed includes information from 3757 adult European Sardinians. After adjusting for gender and age, 22 million single SNP loci were retained for association analysis. The data encompass 731 immune phenotypes, including absolute cell (AC) counts (n = 118), median fluorescence intensities (MFI) reflecting surface antigen levels (n = 389), morphological parameters (MP) (n = 32), and relative cell (RC) counts (n = 192) (Table S2, Supplemental Digital Content, http://links.lww.com/MD/O203). To control for batch effects and time-dependent artifacts, MFIs were normalized for overall and daily changes by adjusting each value with the ratio of cohort mean to daily mean. MP were measured using forward scatter and side scatter to assess cell size and internal complexity. Notably, the MFI, AC, and RC traits represent a wide range of immune cell types, including B cells, cytotoxic T lymphocytes (CTLs), T cells at various maturation stages, monocytes, myeloid cells, and TBNK (T cells, B cells, NK cells). In contrast, the MP traits specifically focus on conventional dendritic cells (DCs) and TBNK panels.
2.3. Data sources for GWAS on MPN
The GWAS data for MPN were obtained from the FinnGen database (GWAS ID: finngen_R10_MYELOPROF_NONCML), which is accessible at https://r10.finngen.fi/. This dataset comprises genetic information from 411,923 individuals of European ancestry, including 2043 MPN patients and 409,880 healthy controls. The diagnoses of MPN in this study adhere to the WHO 2008 classification system for tumors of hematopoietic and lymphoid tissues. This includes PV, ET, and PMF, explicitly excluding chronic myeloid leukaemia. The detailed cohort design is available at the following website: https://r10.risteys.finregistry.fi/endpoints/MYELOPROF_NONCML. The genetic data within the FinnGen database primarily originate from participants’ blood samples, which are analyzed through whole-genome sequencing or genotyping, encompassing 16 million significant SNPs.[16] To maintain data integrity and ensure the accuracy of the IVs, stringent quality control protocols were applied to SNPs.
2.4. Selection of instrumental variables
This method relies on GWAS summary statistics, which have undergone rigorous quality control, and uses IVs to mitigate the impact of missing data and reduce bias from confounding variables. Consistent with recent research, the significance threshold for selecting IVs for each immune trait was set at 1 × 10−5 [10]. To minimize the influence of linkage disequilibrium (LD) among SNPs on the analysis outcomes, an LD r² threshold of <0.1 was applied, ensuring no other associated SNP within a 500 kb window exceeded this threshold.[17,18] The LDlink database (https://ldlink.nci.nih.gov/) was used to further verify the absence of confounding variables linked to the selected SNP loci. To prevent weak IV bias, only IVs with an F-statistic of ≥10 were included in the analysis. The F-statistic was calculated using the formula: F = R² × (n − k − 1) ÷ [k × (1 − R²)], where R² represents the proportion of variance explained by the IVs in relation to the exposure factor, n is the sample size, and k is the number of IVs considered.[19] Relevant data were extracted from the FinnGen database, with a focus on retaining only those SNPs that met the necessary assumptions for the analysis. Subsequently, the datasets for exposure and outcome were merged. During this process, palindromic sequences were excluded to avoid ambiguity in strand alignment, resulting in the final set of SNPs that were used as IVs for the exposure. The MR analysis employed in this study effectively addresses potential missing data (Table S2, Supplemental Digital Content, http://links.lww.com/MD/O203).
2.5. Statistical analysis
All statistical analyses in this study were performed using R software (version 4.3.3). To investigate the causal relationships between 731 immune phenotypes and MPN, we employed the “MR, TwoSampleMR, ggplot2” packages. The mentioned packages can be freely accessed on the official website of the R software. The methods employed in this study included Weighted Median (WM) analysis, mode-based estimation, and inverse variance weighted (IVW) analysis, with the IVW method serving as the primary analytical approach. WM and Mendelian randomization–Egger (MR-Egger) analyses were utilized as supplementary methods.[20] The false discovery rate (FDR) method was applied to adjust for multiple comparisons. For data showing significant causal links, sensitivity analyses were performed. These included Cochran’s Q test to assess heterogeneity among SNPs associated with immune cells that met the predefined assumptions.[21] MR-Egger analysis was used to detect potential horizontal pleiotropy.[22] Additionally, a leave-one-out analysis was conducted to determine whether any single SNP disproportionately influenced the results. The findings were considered robust if the overall error bars consistently remained on 1 side of zero, indicating no significant shift.[23]
3. Results
3.1. Exploration of the causal effect of immunophenotypes on MPN
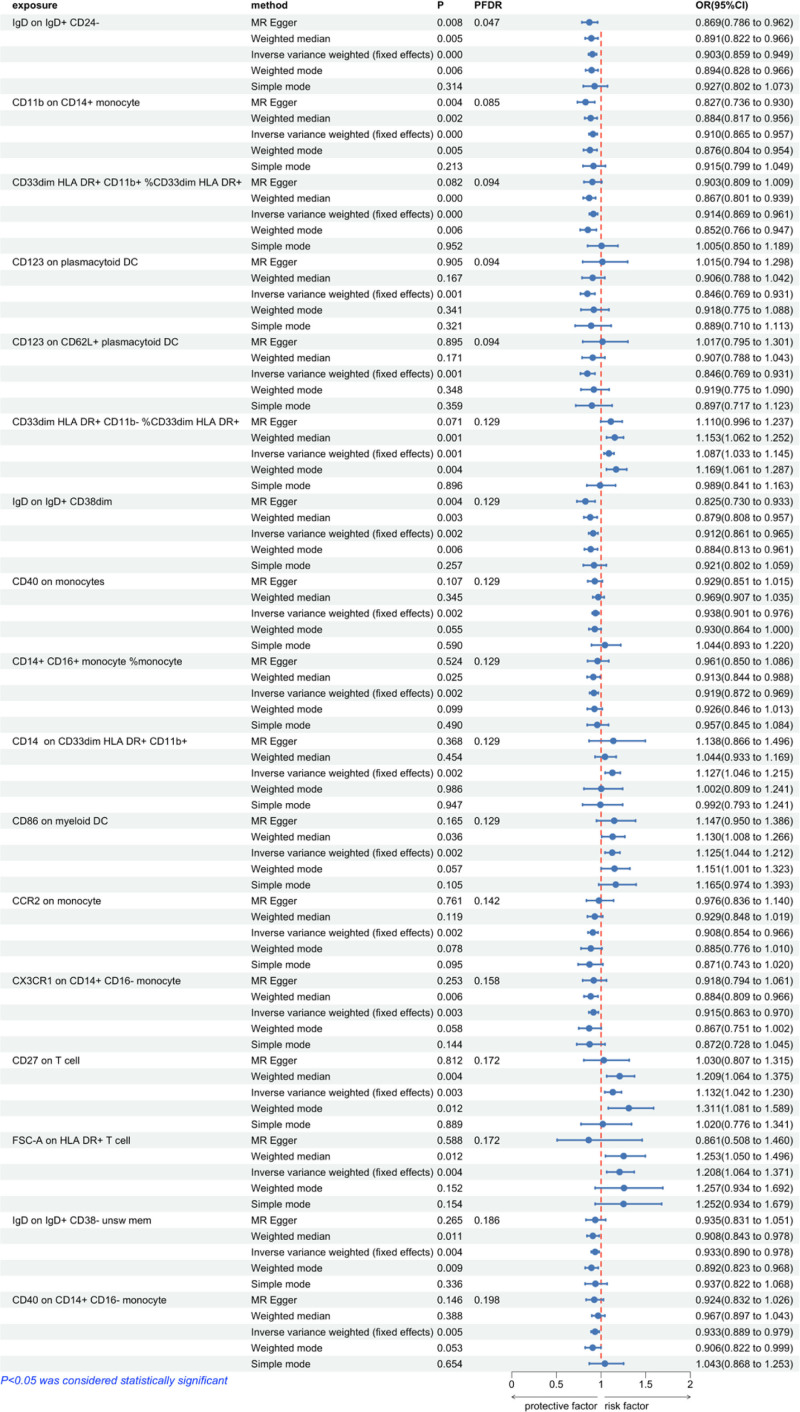
To investigate the causal relationship between MPN and immunophenotypes, we primarily employed the IVW method within a 2-sample MR analysis. After adjusting for multiple comparisons using the FDR method, the immunophenotype IgD on IgD+ CD24- B cells demonstrated a statistically significant protective effect on MPN (PFDR = 0.047). The odds ratio (OR) estimates for the risk of MPN associated with this immunophenotype was 0.903 (95% confidence interval [CI]: 0.859–0.949). When the significance threshold was adjusted to PFDR < 0.20, 16 immunophenotypes were identified as significantly associated with MPN. Among these, 11 immunophenotypes demonstrated a protective effect against MPN: CD11b on CD14+ monocytes, CD33dim HLA-DR+ CD11b+ %CD33dim HLA-DR+, CD123 on plasmacytoid DCs, CD123 on CD62L+ plasmacytoid DCs, IgD on IgD+ CD38dim B cells, CD40 on monocytes, CD14+ CD16+ monocytes % monocytes, CCR2 on monocytes, CX3CR1 on CD14+ CD16− monocytes, IgD on IgD+ CD38− unswitched memory B cells, and CD40 on CD14+ CD16− monocytes. Conversely, 5 immunophenotypes were associated with an increased risk of MPN: CD33dim HLA-DR+ CD11b− %CD33dim HLA-DR+, CD14 on CD33dim HLA-DR+ CD11b+ monocytes, CD86 on myeloid DCs, CD27 on T cells, and FSC-A on HLA-DR+ T cells. Using the IVW method, the OR estimates for the risk of MPN were as follows: 0.910 (95% CI: 0.865–0.957, PFDR = 0.085) for CD11b on CD14+ monocytes, 0.914 (95% CI: 0.869–0.961, PFDR = 0.094) for CD33dim HLA-DR+ CD11b+ %CD33dim HLA-DR+, 0.846 (95% CI: 0.769–0.931, PFDR = 0.094) for CD123 on plasmacytoid DCs, 0.846 (95% CI: 0.769–0.931, PFDR = 0.094) for CD123 on CD62L+ plasmacytoid DCs, 0.912 (95% CI: 0.861–0.965, PFDR = 0.129) for IgD on IgD+ CD38dim B cells, 0.938 (95% CI: 0.901–0.976, PFDR = 0.129) for CD40 on monocytes, 0.919 (95% CI: 0.872–0.969, PFDR = 0.129) for CD14+ CD16+ monocytes % monocytes, 0.908 (95% CI: 0.854–0.966, PFDR = 0.142) for CCR2 on monocytes, 0.915 (95% CI: 0.863–0.970, PFDR = 0.158) for CX3CR1 on CD14+ CD16− monocytes, 0.933 (95% CI: 0.890–0.978, PFDR = 0.186) for IgD on IgD+ CD38− unswitched memory B cells, and 0.933 (95% CI: 0.889–0.979, PFDR = 0.198) for CD40 on CD14+ CD16− monocytes. The OR estimates for an increased risk of MPN were as follows: 1.087 (95% CI: 1.033–1.145, PFDR = 0.129) for CD33dim HLA-DR+ CD11b− %CD33dim HLA-DR+, 1.127 (95% CI: 1.046–1.215, PFDR = 0.129) for CD14 on CD33dim HLA-DR+ CD11b+, 1.125 (95% CI: 1.044–1.212, PFDR = 0.129) for CD86 on myeloid DCs, 1.132 (95% CI: 1.042–1.230, PFDR = 0.172) for CD27 on T cells, and 1.208 (95% CI: 1.064–1.371, PFDR = 0.172) for FSC-A on HLA-DR+ T cells (Fig. 2).
Figure 2.
Forest plot showing the causal associations between immune cell phenotypes and Myeloproliferative neoplasms explored using different methods. CI = confidence interval, FDR = false discovery rate, MR = Mendelian randomization, OR = odds ratio.
The Cochran’s Q test yielded a P-value > .05, indicating no significant heterogeneity in the results. Additionally, the MR-Egger analysis also produced a P-value greater than .05, suggesting no significant evidence of pleiotropy (Table 1). The leave-one-out analysis revealed no outliers (Fig. S1, Supplemental Digital Content, http://links.lww.com/MD/O204). Moreover, both scatter plots and funnel plots were utilized to verify the stability and consistency of the results (Figs. S2 and S3, Supplemental Digital Content, http://links.lww.com/MD/O205 and http://links.lww.com/MD/O206).
Table 1.
Heterogeneity and pleiotropy analysis in forward Mendelian randomization.
| Panel | Immune traits | Test of heterogeneity | MR-Egger | ||
|---|---|---|---|---|---|
| Q | P-value | Intercept | P-value | ||
| B cell | IgD on IgD + CD24− | 40.815 | .978 | 0.010 | .395 |
| Myeloid cell | CD11b on CD14+ monocyte | 23.645 | .746 | 0.034 | .088 |
| Myeloid cell | CD33dim HLA-DR + CD11b+ %CD33dim HLA-DR+ | 25.431 | .604 | 0.004 | .817 |
| cDC | CD123 on plasmacytoid DC | 16.608 | .411 | −0.042 | .136 |
| cDC | CD123 on CD62L+ plasmacytoid DC | 16.641 | .409 | −0.042 | .133 |
| Myeloid cell | CD33dim HLA-DR + CD11b− %CD33dim HLA-DR+ | 24.547 | .545 | −0.007 | .677 |
| B cell | IgD on IgD + CD38dim | 35.345 | .680 | 0.030 | .080 |
| Monocyte | CD40 on monocytes | 85.145 | .177 | 0.003 | .815 |
| Monocyte | CD14+ CD16+ monocyte %monocyte | 67.708 | .231 | −0.012 | .434 |
| Myeloid cell | CD14 on CD33dim HLA-DR + CD11b+ | 23.409 | .136 | −0.003 | .942 |
| cDC | CD86 on myeloid DC | 34.871 | .289 | −0.004 | .822 |
| cDC | CCR2 on monocyte | 38.349 | .499 | −0.021 | .329 |
| Monocyte | CX3CR1 on CD14+ CD16− monocyte | 50.615 | .410 | −0.001 | .961 |
| B cell | CD27 on T cell | 23.709 | .255 | 0.025 | .426 |
| TBNK | FSC-A on HLA-DR + T cell | 17.341 | .299 | 0.054 | .215 |
| B cell | IgD on IgD + CD38− unsw mem | 56.829 | .236 | 0.000 | .980 |
| Monocyte | CD40 on CD14+ CD16−monocyte | 51.554 | .300 | 0.003 | .842 |
3.2. Examination of the causal effect of MPN on immunophenotypes
To evaluate the potential for a reverse causal relationship, we treated MPN as the exposure and the 731 immune cell phenotypes as the outcomes within a 2-sample MR framework using the IVW method. After applying the FDR correction, no significant reverse causal effects were detected at the PFDR < 0.05 threshold. Even when the significance threshold was relaxed to PFDR < 0.20, no significant correlations were observed. These findings suggest that there is no evidence for a reverse causal relationship between MPN and the 731 immune cell phenotypes analyzed (Table S3, Supplemental Digital Content, http://links.lww.com/MD/O203).
4. Discussion
This study marks the first systematic investigation into the causal relationships between various immune cell phenotypes and MPN using a 2-sample MR approach. Our findings demonstrate that, after adjusting for the FDR, the immune cell phenotype IgD on IgD+ CD24− B cells exhibits a statistically significant protective effect against MPN. When the significance threshold was relaxed to PFDR < 0.20, 16 immune phenotypes were significantly associated with MPN. Of these, 11 phenotypes exhibited a protective effect, while 5 were linked to an increased risk of developing MPN. These results underscore the potentially critical protective role of B cells, particularly the IgD+ B cell subset, in modulating the humoral immune response and influencing the pathogenesis of MPN. Additionally, immune cells such as CD11b on CD14+ monocytes and CD123 on plasmacytoid DCs also showed a potential protective effect against MPN. The study also uncovered significant associations between specific immune cell traits and an increased risk of developing MPN, particularly traits such as CD33dim HLA-DR+ CD11b− %CD33dim HLA-DR+ and CD86 expression on myeloid DCs. These findings suggest that certain markers on DCs may contribute to an elevated risk of MPN, potentially playing a role in the disease’s progression.
Previous research indicates that B cells are vital in regulating humoral immunity within chronic inflammatory environments. Moreover, they may influence cell proliferation and differentiation by secreting cytokines and other signaling molecules. This multifunctional role highlights their importance in both immune response modulation and the broader cellular processes within these environments.[24–27] Certain B cell subsets have been found to inhibit tumor cell expansion by mechanisms such as delivering inhibitory signals or engaging in competitive inhibition, thereby exerting a suppressive effect on disease progression.[28,29] Our MR analysis indicates that IgD+ B cell subsets may contribute to the progression of MPN, aligning with findings from previous observational studies. Monocytes and DCs, among other immune cell populations, well-recognized for their pivotal roles in regulating immune responses and maintaining the tumor microenvironment.[30–33] These cells can modulate inflammatory responses and help stabilize the tumor microenvironment by secreting various cytokines, such as IL-12,[34] IL-15,[35,36] and TNF-α.[37,38] These cytokines enhance the antitumor activity of other immune cells, potentially decelerating the progression of MPN.[29–32]
Myeloid DCs are essential for antigen presentation and immune activation, and their dysregulated activation can exacerbate bone marrow fibrosis and worsen the tumor microenvironment through pro-inflammatory signaling pathways, thereby contributing to the progression of MPN.[39–41] These findings align with our study’s outcomes, which suggest a dual role for the immune system in the pathogenesis of MPN. On 1 hand, immune cells like monocytes and DCs may offer protection against MPN by regulating immune responses and reducing inflammation. However, when these immune cells become dysfunctional or excessively activated, they can exacerbate pathological immune responses by releasing large amounts of pro-inflammatory cytokines, such as TNF-α and IFN-γ.[42,43] This, in turn, can contribute to the progression of MPN, underscoring the complex and ambivalent role of the immune system in the development of the disease. Overall, the interactions between immune cell subsets and MPN are complex and diverse. Monocytes not only regulate inflammatory responses but also influence the bone marrow microenvironment and promote the proliferation of pathological clones through complex intercellular interactions. DCs are essential for antigen presentation and immune response regulation, thereby impacting the progression of MPN. Additionally, they contribute to the overall immune status of patients by promoting inflammation and interacting with the tumor microenvironment, further influencing disease dynamics.
In conclusion, this study utilized 2-sample bidirectional MR analysis to investigate the causal relationship between immune cells and MPN, emphasizing the complex interactions within the immune system and MPN. While the precise mechanisms through which immune cells influence MPN pathogenesis remain partially understood, elucidating the roles of specific immune cell subsets in MPN could substantially improve our understanding of the disease’s pathological processes. Our findings serve as a crucial reference for future research into the dynamics between immune cells and MPN and could provide insights for developing novel therapeutic approaches.
This study employed 2-sample bidirectional MR analysis, leveraging data from large, published GWAS cohorts, which offered a substantial sample size and increased statistical power. Consequently, the findings are theoretically robust. However, there are notable limitations to consider. First, the use of public databases makes batch differences between the various datasets analyzed in this study unavoidable. Data from different populations may also introduce confounding factors. Despite multiple sensitivity analyses, the possibility of horizontal pleiotropy cannot be entirely excluded. Second, the diagnosis of MPN in this study follows the 2008 WHO classification of tumors of hematopoietic and lymphoid tissues, which includes PV, ET, and PMF, with chronic myeloid leukemia specifically excluded. However, no further subdivisions were made among PV, ET, and PMF, limiting our ability to compare different subtypes of MPN. Third, the study is confined to individuals of European ancestry, which may restrict the generalizability of the findings to other ethnic groups and limit the broader applicability of the conclusions. Furthermore, the use of a more lenient significance threshold to comprehensively evaluate the association between immune phenotypes and MPN may have increased the risk of false-positive results.
5. Conclusions
In summary, this study employs 2-sample MR analysis to investigate potential causal relationships between a range of immune phenotypes and MPN. These results deepen our comprehension of MPN pathogenesis and open up new possibilities for future therapeutic approaches. The findings suggest that B cells, monocytes, and DCs are pivotal in influencing MPN progression, potentially impacting both disease advancement and patient outcomes through intricate immune regulatory processes. Furthermore, this research highlights the significance of incorporating immune system variability into MPN studies. Subsequent research should aim to confirm these results and investigate further mechanisms to enhance MPN treatment strategies.
Acknowledgments
We are grateful to all the studies that have made the public GWAS summary data available.
Author contributions
Conceptualization: Yao Wang, Yang Fei.
Data curation: Yao Wang, Yang Fei.
Formal analysis: Yao Wang, Yang Fei.
Methodology: Yang Fei.
Software: Yao Wang, Yang Fei.
Writing – original draft: Yao Wang, Yang Fei.
Writing – review & editing: Yao Wang, Yang Fei.
Supplementary Material
Abbreviations:
- AC
- absolute cell
- CI
- confidence interval
- CTL
- cytotoxic T lymphocytes
- DC
- dendritic cell
- ET
- essential thrombocythemia
- FDR
- false discovery rate
- GWAS
- genome-wide association studies
- IV
- instrumental variable
- IVW
- inverse variance weighted
- LD
- linkage disequilibrium
- MFI
- median fluorescence intensities
- MP
- morphological parameters
- MPN
- Myeloproliferative neoplasms
- MR
- Mendelian randomization
- NK
- natural killer
- OR
- odds ratio
- PMF
- primary myelofibrosis
- PV
- polycythemia vera
- RC
- relative cell
- SNP
- single nucleotide polymorphism
- TBNK
- T cells, B cells, NK cells
- WM
- weighted median
The authors have given consent for publication.
This research is supported by Hwamei Research Foundation of Ningbo No.2 Hospital, Grant No.2024HMKYA10.
Ethics approval and consent to participate are not applicable to this study.
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplemental Digital Content is available for this article.
How to cite this article: Wang Y, Fei Y. Causal relationship between 731 immune cells and the risk of myeloproliferative neoplasms: A 2-sample bidirectional Mendelian randomization study. Medicine 2024;103:51(e40945).
References
- [1].Godfrey AL. Myeloproliferative neoplasms (MPNs). Blood Rev. 2020;42:100717. [DOI] [PubMed] [Google Scholar]
- [2].Spivak JL. Myeloproliferative neoplasms. N Engl J Med. 2017;376:2168–81. [DOI] [PubMed] [Google Scholar]
- [3].Grinfeld J, Nangalia J, Baxter EJ, et al. Classification and personalized prognosis in myeloproliferative neoplasms. N Engl J Med. 2018;379:1416–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Greenfield G, McMullin MF, Mills K. Molecular pathogenesis of the myeloproliferative neoplasms. J Hematol Oncol. 2021;14:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Titmarsh GJ, Duncombe AS, McMullin MF, et al. How common are myeloproliferative neoplasms? A systematic review and meta-analysis. Am J Hematol. 2014;89:581–7. [DOI] [PubMed] [Google Scholar]
- [6].Yap YY, Law KB, Sathar J, et al. The epidemiology and clinical characteristics of myeloproliferative neoplasms in Malaysia. Exp Hematol Oncol. 2018;7:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hultcrantz M, Ravn Landtblom A, Andréasson B, et al. Incidence of myeloproliferative neoplasms: trends by subgroup and age in a population-based study in Sweden. J Intern Med. 2020;287:448–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gu J, Yan G-M, Kong X-L, Zhang Y-Y, Huang L-H, Lu H-M. Assessing the causal relationship between immune traits and systemic lupus erythematosus by bi-directional Mendelian randomization analysis. Mol Genet Genomics. 2023;298:1493–503. [DOI] [PubMed] [Google Scholar]
- [9].Orrù V, Steri M, Sidore C, et al. Complex genetic signatures in immune cells underlie autoimmunity and inform therapy. Nat Genet. 2020;52:1036–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wang C, Zhu D, Zhang D, et al. Causal role of immune cells in schizophrenia: Mendelian randomization (MR) study. BMC Psychiatry. 2023;23:590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kjaergaard AD, Teumer A, Marouli E, et al. Thyroid function, pernicious anemia and erythropoiesis: a two-sample Mendelian randomization study. Hum Mol Genet. 2022;31:2548–59. [DOI] [PubMed] [Google Scholar]
- [12].Lu C, Chen Q, Tao H, et al. The causal effect of inflammatory bowel disease on diffuse large B-cell lymphoma: two-sample Mendelian randomization study. Front Immunol. 2023;14:1171446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang Q, Shi Q, Lu J, Wang Z, Hou J. Causal relationships between inflammatory factors and multiple myeloma: a bidirectional Mendelian randomization study. Int J Cancer. 2022;151:1750–9. [DOI] [PubMed] [Google Scholar]
- [14].Davey Smith G, Holmes MV, Davies NM, Ebrahim S. Mendel’s laws, Mendelian randomization and causal inference in observational data: substantive and nomenclatural issues. Eur J Epidemiol. 2020;35:99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Skrivankova VW, Richmond RC, Woolf BAR, et al. Strengthening the reporting of observational studies in epidemiology using Mendelian randomization: the STROBE-MR statement. JAMA. 2021;326:1614–21. [DOI] [PubMed] [Google Scholar]
- [16].Kurki MI, Karjalainen J, Palta P, et al. ; FinnGen. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613:508–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Auton A, Brooks LD, Durbin RM, et al. ; 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Fei Y, Yu H, Wu Y, Gong S. The causal relationship between immune cells and ankylosing spondylitis: a bidirectional Mendelian randomization study. Arthritis Res Ther. 2024;26:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fu S, Du Y, Pan T, Ma F, He H, Li Y. Causal role of immune cells in aplastic anemia: Mendelian randomization (MR) study. Sci Rep. 2024;14:18010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46:1734–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yan D, Liu G, Yin Y, et al. A Mendelian randomization study revealed a causal link between napping and deep vein thrombosis (DVT). Sleep Breath. 2024;28:665–71. [DOI] [PubMed] [Google Scholar]
- [22].Lyu B, Ma J, Bai Y, Feng Z. Casual effects of gut microbiota on risk of infections: a two-sample Mendelian randomization study. Front Microbiol. 2023;14:1284723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Richardson TG, Leyden GM, Davey Smith G. Time-varying and tissue-dependent effects of adiposity on leptin levels: a Mendelian randomization study. ELife. 2023;12:e84646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].De Gruijter NM, Jebson B, Rosser EC. Cytokine production by human B cells: role in health and autoimmune disease. Clin Exp Immunol. 2022;210:253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Shin B, An G, Cockrell RC. Examining B-cell dynamics and responsiveness in different inflammatory milieus using an agent-based model. PLoS Comput Biol. 2024;20:e1011776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Syeda MZ, Hong T, Huang C, Huang W, Mu Q. B cell memory: from generation to reactivation: a multipronged defense wall against pathogens. Cell Death Discovery. 2024;10:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tsai DY, Hung KH, Chang CW, Lin KI. Regulatory mechanisms of B cell responses and the implication in B cell-related diseases. J Biomed Sci. 2019;26:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Aziz M, Holodick NE, Rothstein TL, Wang P. The role of B-1 cells in inflammation. Immunol Res. 2015;63:153–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Shang J, Zha H, Sun Y. Phenotypes, functions, and clinical relevance of regulatory B cells in cancer. Front Immunol. 2020;11:582657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Li D, Shao F, Yu Q, et al. The complex interplay of tumor-infiltrating cells in driving therapeutic resistance pathways. Cell Commun Signal. 2024;22:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].León B, Ardavín C. Monocyte-derived dendritic cells in innate and adaptive immunity. Immunol Cell Biol. 2008;86:320–4. [DOI] [PubMed] [Google Scholar]
- [32].Pourahmad R, Rezaei N. Role of monocyte-derived dendritic cells (MoDCs) in tumor immune response. In: Rezaei N. (ed). Handbook of Cancer and Immunology. Cham: Springer International Publishing; 2022. p. 1–18. [Google Scholar]
- [33].Verneau J, Sautés-Fridman C, Sun CM. Dendritic cells in the tumor microenvironment: prognostic and theranostic impact. Semin Immunol. 2020;48:101410. [DOI] [PubMed] [Google Scholar]
- [34].Mirlekar B, Pylayeva-Gupta Y. IL-12 family cytokines in cancer and immunotherapy. Cancers (Basel). 2021;13:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kurz E, Hirsch CA, Dalton T, et al. Exercise-induced engagement of the IL-15/IL-15Rα axis promotes anti-tumor immunity in pancreatic cancer. Cancer Cell. 2022;40:720–37.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ma S, Caligiuri MA, Yu J. Harnessing IL-15 signaling to potentiate NK cell-mediated cancer immunotherapy. Trends Immunol. 2022;43:833–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Montfort A, Colacios C, Levade T, Andrieu-Abadie N, Meyer N, Ségui B. The TNF paradox in cancer progression and immunotherapy. Front Immunol. 2019;10:1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zidi I, Mestiri S, Bartegi A, Amor NB. TNF-alpha and its inhibitors in cancer. Med Oncol. 2010;27:185–98. [DOI] [PubMed] [Google Scholar]
- [39].Bakhtiyari M, Liaghat M, Aziziyan F, et al. The role of bone marrow microenvironment (BMM) cells in acute myeloid leukemia (AML) progression: immune checkpoints, metabolic checkpoints, and signaling pathways. Cell Commun Signal. 2023;21:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chistiakov DA, Sobenin IA, Orekhov AN, Bobryshev YV. Myeloid dendritic cells: development, functions, and role in atherosclerotic inflammation. Immunobiology. 2015;220:833–44. [DOI] [PubMed] [Google Scholar]
- [41].Haas L, Obenauf AC. Allies or enemies-the multifaceted role of myeloid cells in the tumor microenvironment. Front Immunol. 2019;10:2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Masselli E, Pozzi G, Gobbi G, et al. Cytokine profiling in myeloproliferative neoplasms: overview on phenotype correlation, outcome prediction, and role of genetic variants. Cells. 2020;9:2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Van Loo G, Bertrand MJM. Death by TNF: a road to inflammation. Nat Rev Immunol. 2023;23:289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.