Abstract
The abundance of life on Earth is almost entirely due to biological photosynthesis, which depends on light energy. The source of light in natural habitats has heretofore been thought to be the sun, thus restricting photosynthesis to solar photic environments on the surface of the Earth. If photosynthesis could take place in geothermally illuminated environments, it would increase the diversity of photosynthetic habitats both on Earth and on other worlds that have been proposed to possibly harbor life. Green sulfur bacteria are anaerobes that require light for growth by the oxidation of sulfur compounds to reduce CO2 to organic carbon, and are capable of photosynthetic growth at extremely low light intensities. We describe the isolation and cultivation of a previously unknown green sulfur bacterial species from a deep-sea hydrothermal vent, where the only source of light is geothermal radiation that includes wavelengths absorbed by photosynthetic pigments of this organism.
Keywords: photosynthesis, anoxygenic, green sulfur bacterium, evolution, habitat
Light energy from the sun drives photosynthesis to provide the primary source of nearly all of the organic carbon that supports life on Earth (1). An exception to this are deep-sea hydrothermal vents, such as black smokers located far below the photic zone in the oceans, where unusual microbial and invertebrate populations exist on organic material from CO2 reduction by chemotrophic bacteria that oxidize inorganic compounds (2). Hydrothermal vents may resemble the environment in which life evolved (3), and the discovery of geothermal light at otherwise dark deep-sea vents led to the suggestion that such light may have provided a selective advantage for the evolution of photosynthesis from a chemotrophic microbial ancestor that used light-sensing molecules for phototaxis toward nutrients associated with geothermal light (4, 5). However, it was not clear whether the photon flux emanating from hydrothermal vents could support the existence of an obligately photosynthetic organism.
A bacterium that appears to use light as an auxiliary source of energy to supplement an otherwise chemotrophic metabolism was isolated from the general vicinity of a deep-sea hydrothermal vent (6, 7), but we wished to determine whether an obligately photosynthetic microbe might exist in close proximity to a vent orifice. The discovery of such an organism in this environment would indicate that volcanic or geothermal light is harvested to drive photosynthetic reactions in the absence of light from the sun. The possibility of geothermal light-driven photosynthesis on Earth relates to speculations about the existence of extraterrestrial life on planets and moons far from the sun in the solar system (8) and, conceivably, in other galaxies.
Bulk DNA may be isolated from natural environments, and gene sequences may be used to infer the presence of microbial species and potential metabolic activities (9). Because these methods do not necessarily reveal the existence of viable cells or genuine physiological properties of an organism, we instead used a cultivation approach to search for living cells of obligately photosynthetic microbes at a deep-sea hydrothermal vent. Here, we report the capture and initial description of an anaerobic green sulfur phototrophic bacterium from a deep-sea black smoker.
Materials and Methods
Enrichment, Isolation, and Cultivation. Water samples (1 ml) were added to completely filled 16.5-ml screw-cap (anaerobic) tubes containing “Medium 1 for Cultivation of Green and Purple Sulfur Bacteria” as described for cultures from marine habitats (10), supplemented with 0.02% yeast extract and 0.1% thiosulfate. Tubes were incubated at ≈25°C on a bench top, and continuously illuminated with weak fluorescent light (from ceiling fixtures) supplemented with a 60-W incandescent lamp to yield a combined intensity of ≈10 μmole photons·m-2·s-1 (measured with a Li-Cor quantum sensor equipped with LI-190SB probe). In subsequent cultivations, GSB1 was grown anaerobically in saline SL10 medium, a minimal medium containing H2S as the sole electron donor and CO2 as the sole source of carbon (11), at 21°C and illuminated with 100 μmol photons·m-2·s-1 provided by incandescent lamps. Saline SL10 medium was solidified with agar (1.5%) to obtain colonies in tubes for purification of GSB1. A positive effect on growth was observed when the saline SL10 medium was supplemented with 5 mM acetate, 5 mM propionate, 0.05% peptone, or elemental sulfur (S0; a few milligrams per 10 ml). No growth stimulation was observed with any of 100 other substances added to the saline SL10 medium (see Supporting Text, which is published as supporting information on the PNAS web site).
PCR Amplification and Sequence Analyses. DNA was obtained from pure cultures of GSB1 using standard methods, and used in PCR amplification with degenerate oligonucleotide primers FMO1fd (5′-WCWAAHGACRYNACVACCGC-3′) and FMO4rd (5′-CGCTCCAGCGRTAYTCYTCRAGG-3′) for the FMO gene. The PCR conditions were 30 cycles of 98°C for 1 min, 52°C for 1 min, and 72°C for 2 min. The 16S rRNA gene segment was amplified by using primers 27f (5′-GAGTTTGATCCTGGCTCAG-3′) and 1525r (5′-AGAAAGGAGGTGATCCAGCC-3′) (12). The PCR conditions were 10 cycles of 94°C for 30 s, 59°C for 45 s, and 72°C for 60 s, followed by 20 cycles of 94°C for 30 s, 54°C for 45 s, and 72°C for 60 s. The PCR products were sequenced, the sequences were analyzed by blast of the NCBI databases, and alignments were created by using clustalw in bioedit for FMO or the online RDP-II alignment tool for 16S rRNA (13-15). MEGA2 was used to construct neighbor-joining trees based on the Jukes and Cantor model (16). The GenBank accession numbers of FMO/16S rRNA sequences used as representatives are as follows: Prosthecochloris aestuarii 2K, AJ290823/AJ290835; P. aestuarii DSM 271, AJ391151/Y07837; Chlorobium phaeovibriodes DSM 1678, AJ391163/AJ290833; Chlorobium vibrioforme DSM 260T, AJ391145/M62791; Chlorobium limicola f. thios. DSM 249, X83529/Y08102; Chlorobium phaeobacteroides 1549, AJ306184/AJ299413; Chlorobium tepidum ATCC 49652, L13700/M58468; Chlorobium vibrioforme f. thios. NCIB 8346, AJ391161/AJ290830; Chlorobium limicola UdG 6044, AJ306190/Y10645; Chlorobium limicola DSM 246, AJ391142/AJ290824; Chlorobium phaeobacteroides DSM 266, AJ391148/Y08104; Pelodictyon luteolum DSM 273, AJ391152/Y08107; Chlorobium phaeovibrioides DSM 269, AJ391150/Y08105; and Pelodictyon phaeoclathratiforme DSM 5477, AJ290822/Y08108. Additional accession numbers of 16S rRNA sequences are as follows: Chlorobium vibrioforme DSM 262, Y08103; Chlorobium limicola. f. thios. 9330, AJ290827; Chlorobium ferrooxidans DSM 13031, Y18253; Clathrochloris sulfurica, X53184; and Chloroherpeton thalassium, AF170103.
Pigment Analyses. Absorption and fluorescence emission (excitation at 460 nm) spectra of intact cells were obtained at room temperature with a Shimadzu UV-2501PC spectrophotometer and Photon Technology International fluorometer, respectively. Pigments were extracted from cells in acetone/methanol (7:2) overnight at -20°C and injected onto a 4.6 × 150 mm Waters Symmetry C8 (3.5 μm) column attached to an Agilent Technologies Model 1100 HPLC equipped with diode array absorption and scanning fluorometer detectors. Pigment spectra of individual HPLC peaks were taken from the data stored by the detector by using software provided by Agilent Technologies. Carotenoids were collected from HPLC peaks, dried under N2, and analyzed by MALDI-TOF mass spectrometry using terthiophene as the matrix (17).
Electron Microscopy. Negatively stained (2% aqueous uranyl acetate) cells were examined in a Hitachi H-7600 TEM. For thin sections, cells were fixed in 2.5% glutaraldehyde buffered with 0.1 M cacodylate buffer, pH 7.4, postfixed in cacodylate buffered 1% osmium tetroxide, dehydrated through an ethanol series, then infiltrated in a Spurr-Epon resin mix and polymerized at 60°C overnight. The processing was performed by using a Pelco Laboratory Microwave as described (www.emlab.ubc.ca/protocol.htm). Thin sections were cut with a diamond knife on a Leica Ultracut T microtome and put onto 300-mesh uncoated copper grids, stained in 2% uranyl acetate for 12 min and lead citrate for 6 min, and examined by using a Hitachi H-7600 TEM. For scanning electron microscopy, cells were fixed as above and collected on a 0.4-μm nucleopore filter. The cells were then postfixed in cacodylate buffered 1% osmium tetroxide, dehydrated through an ethanol series, critical point-dried by using liquid carbon dioxide, and examined in a Hitachi S-4700 scanning electron microscope.
Survival of GSB1 During Exposure to Air. Before exposure to air, all manipulations were conducted under an atmosphere of 95% N2/5% H2. Cells were harvested by centrifugation and resuspended in an anoxic, H2S-free, artificial seawater medium (18) buffered with 10 mM Hepes, pH 6.7, to ≈108 cells per ml. For air exposure times of up to 24 h, 80 ml of cell suspension was transferred to 125-ml glass serum bottles that were sealed with butyl rubber stoppers in an anaerobic chamber. The serum bottles were incubated without agitation at room temperature in the dark, and the experiment was started by continuously sparging the suspension with filter-sterilized synthetic air (Messer-Griessheim, A bis Zet, Nürnberg, Germany). Aliquots of 200 μl were taken aseptically with a syringe at time intervals and immediately transferred to saline SL10 medium supplemented with 1 mM sodium acetate. These samples were serially diluted 1:10 in screw-cap tubes containing the same medium, for enumeration by the most probable number (MPN) method (19, 20). After dilution, the inoculated MPN tubes were kept in the dark at 15°C overnight to allow for complete reduction of O2 carried over during inoculation, and then incubated at 21°C with illumination as described above. For long-term (up to 2 weeks) exposure to air, 80-ml cell suspensions in the artificial seawater medium were incubated in 250-ml baffled Erlenmeyer flasks closed with cotton plugs. After withdrawing the initial sample, the flasks were incubated at 250 rpm on a rotary shaker in darkness at 15°C under ambient air. Samples were taken and serially diluted for MPN enumeration as described above. Tubes were incubated for 1 month, the number of positive tubes for each dilution was scored, and MPN values were calculated by using the tables of de Man (19, 20).
Results and Discussion
On this cruise, we visited the East Pacific Rise, which is an area of high volcanic activity with a variety of vents that support characteristic ecosystems (2, 21). A water sample was obtained directly from the effluent plume within 50 cm above the orifice of the TY black smoker (2,391 m in depth; 9° 49.63′ N, 104° 17.37′ W), using a 1-liter capacity Niskin sampler on the ALVIN submersible. After return to the ship, 1-ml portions of the sample were used to inoculate culture media designed to enrich for different types of bacteria. A phototrophic sulfur bacterial enrichment medium incubated anaerobically with illumination gave rise to green-pigmented turbidity, which appeared to be due to a small, nonmotile bacterium that is called GSB1. Twenty-two other samples collected on this cruise, at various depths ranging from the surface to the ocean floor, failed to yield microbial growth in this enrichment medium, although growth of other organisms was obtained in other media.
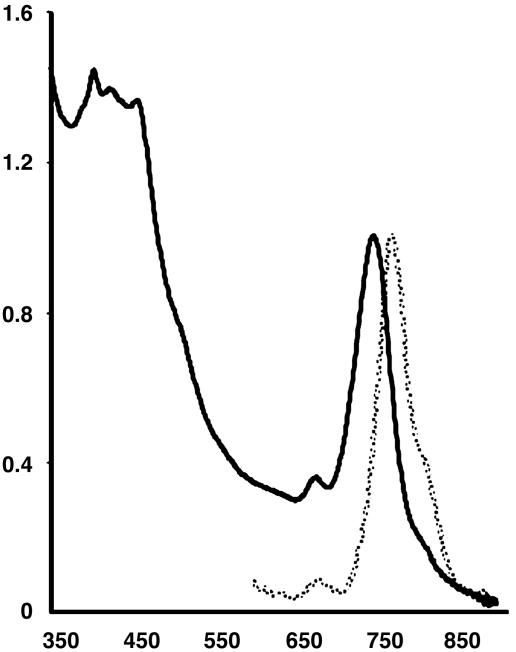
After isolation of GSB1 in pure culture, the in vivo absorption spectrum (Fig. 1) was found to be similar to that of green sulfur bacteria such as those in the genera Chlorobium and Prosthecochloris (22), with a major peak at ≈750 nm indicating the presence of light-harvesting bacteriochlorophyll (BChl) c, and absorption in the 450 nm region due to a BChl Soret band and light-harvesting carotenoid pigments (23). The in vivo fluorescence emission spectrum (Fig. 1) contained a major peak at ≈775 nm, indicative of BChl c (23). The GSB1 pigments were extracted into an organic solvent, resolved in HPLC, and determined to be very similar to the pigments of a Chlorobium tepidum control (24). Thus, the quantitatively major chlorophylls of GSB1 are BChls c on the basis of absorption/fluorescence spectra and HPLC elution times, and mass spectrometry indicated that the major carotenoid is chlorobactene (data not shown).
Fig. 1.
Absorption (solid line) and fluorescence emission (broken line) spectra of GSB1 intact cells. Vertical axis gives absorbance/fluorescence (arbitrary units) and horizontal axis gives wavelengths in nanometers.
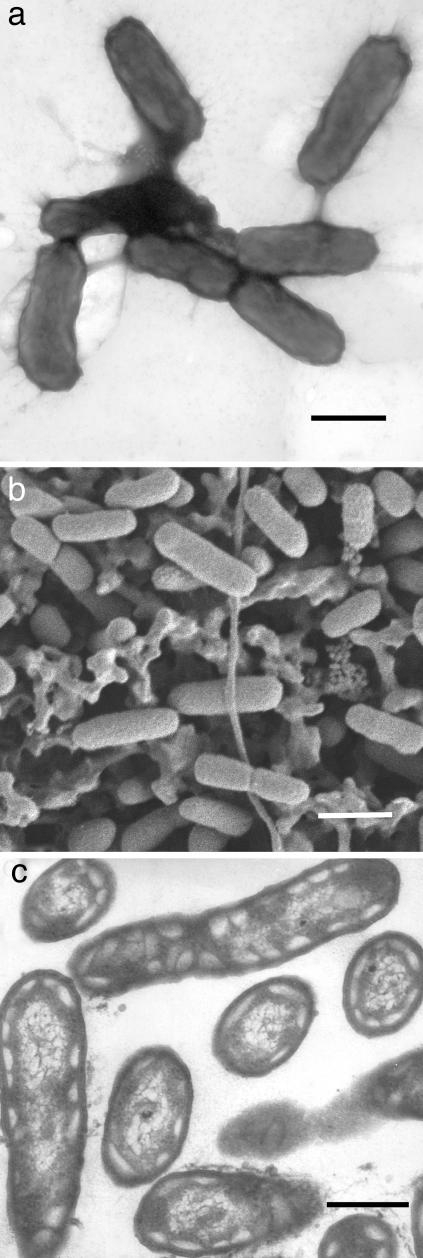
Electron microscopy (Fig. 2) showed that GSB1 is rod-shaped (≈0.3 × 1 μm), and revealed the presence of chlorosomes (light-harvesting structures found in green sulfur bacteria) (22) appressed against the inner membrane of these Gram-negative cells. The constrictions of some cells indicate that cell division occurs by binary transverse fission, and the absence of flagella is consistent with the lack of motility in liquid media.
Fig. 2.
Morphology and ultrastructure of GSB1 cells. (a) Negatively stained cells viewed by transmission electron microscopy. (Bar, 500 nm.) (b) Cells deposited on a filter and viewed by scanning electron microscopy. (Bar, 800 nm.) (c) Thin section through cells viewed by transmission electron microscopy with electron-transparent structures characteristic of chlorosomes. (Bar, 300 nm.)
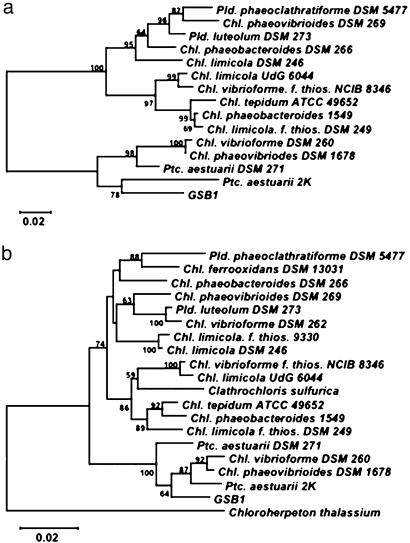
Green sulfur bacteria uniquely contain a light-harvesting protein called the Fenna-Matthews-Olson (FMO) protein (23). Oligonucleotide primers were designed by using conserved sequences of FMO genes, and GSB1 DNA was used to PCR-amplify a 970-bp DNA segment. The DNA sequence of this PCR product encoded a 323-aa sequence that was from 71% to 91% identical in alignments with FMO sequences from 14 species of green sulfur bacteria. A tree of FMO sequences (Fig. 3a) indicates that the GSB1 FMO protein is most closely related to the FMO proteins of Chlorobium and Prosthecochloris marine species. PCR was also used to amplify a ≈1.5-kb segment of the GSB1 16S rRNA gene, and a tree of 19 bacterial 16S rDNA sequences (Fig. 3b) again places GSB1 in a cluster that includes Chlorobium and Prosthecochloris marine species. We conclude that the GSB1 isolate is a previously unknown marine species of the green sulfur bacteria that is related to organisms classified as in the Chlorobium and Prosthecochloris genera (22). The capture of GSB1 from a deep-sea sample is unexpected, because viable green sulfur bacteria were thought to be found only in environments where light from the sun is available (22, 25).
Fig. 3.
Phylogenetic analyses of GSB1. (a) Tree of FMO protein amino acid sequences. (b) Tree of 16S rDNA sequences. Support values at nodes are given as percentages, and scale bars represent the expected number of changes per residue position.
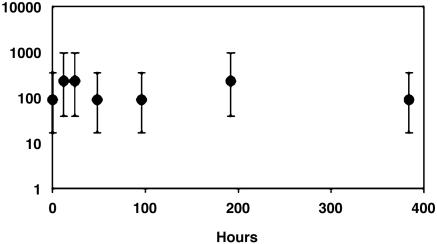
The growth of GSB1 requires anaerobiosis, light, H2S or elemental S, and CO2. Of 104 substances tested, only acetate, propionate, peptone, and elemental S stimulated photosynthetic growth in the minimal medium SL10 (see Materials and Methods and Supporting Text). Exposure of cultures to air in the presence of light and H2S reduced viability, but we found that GSB1 is resistant to exposure to air in the absence of light and H2S (Fig. 4). There was no significant loss in GSB1 viability after 2 weeks, as also found with an anoxic control. The resistance of GSB1 to the toxic effects of O2 in air is consistent with survival in the fluctuating environment of deep-sea hydrothermal vents (26), which could depend on the ability to survive translocation from decaying to nascent vents in the dark, oxygenated ocean depths (see below).
Fig. 4.
Survival of GSB1 during exposure to air in darkness and the absence of H2S. The vertical axis (log10 scale) gives percentages of viable cells based on most probable number (MPN) enumerations relative to microscopic counts (19, 20), and the horizontal axis gives the time of incubation. Points give average values, and vertical bars indicate 95% confidence limits.
Although the sample that contained GSB1 was obtained directly from a black smoker plume, it could be argued that this bacterium grows in the surrounding bulk water. We view this possibility as unlikely because the surrounding water is oxygenated and lacks a source of reduced S and light. The nearest locale (on the coast of Costa Rica) that could provide solar light and H2S to support the anaerobic photosynthetic growth of green sulfur bacteria is ≈2,250 km distant from the TY black smoker, and the chance that GSB1 was directly transported by currents over that distance to be captured in a vent plume sample at >2 km depth appears to be vanishingly small. Instead, we suggest that the nonmotile GSB1 was shed from a microbial mat (22, 25) or similar microenvironment within centimeters of the TY vent orifice, and was swept into the turbulent plume. In such a microenvironment, microbial respiration or spontaneous chemical reactions between reduced substances present in the vent effluent and O2 in the surrounding seawater could provide anaerobiosis, and a steep temperature gradient from the ≈2°C surrounding water and the ≈300°C interior of a vent could allow for survival close enough to geothermal light by harvesting of photons for photosynthesis. The source of reduced S could be either SO4-reducing bacteria in a mat, or the vent effluent. Multiple samples obtained at several locations and depths, and a second 1-ml portion of the 1-liter sample that contained GSB1 did not yield growth under the same enrichment conditions. Thus, GSB1-like bacteria were not found at distances from a vent, although it appears that GSB1 was a minor component of the microbial community at the TY black smoker at the time of this cruise.
The properties of GSB1 are consistent with its deep-sea survival being enhanced by geothermal light that has been reported at several hydrothermal vents (27). Although there are vent-to-vent differences, in general the light intensity detected was greatest at wavelengths in excess of 700 nm (i.e., thermal or blackbody radiation) (27). The photon flux at 750 ± 50 nm (corresponding to the long wavelength absorption peak of light-harvesting BChl c in GSB1) at the orifice of a 370°C black smoker was ≈108 photons·cm-2·s-1·sr-1 (28); the flux in the 400- to 500-nm range (short wavelength BChl and chlorobactene absorption peaks) was ≈104 photons·cm-2·s-1·sr-1 (27, 29) (6 × 1013 photons·cm-2·s-1 = 1 μmole photons·m-2·s-1; the term sr refers to a solid angle measured in steradians). Within 1-2 cm of 332°C flange pools on black smoker chimneys, the total photon flux (≈1011 photons·cm-2·s-1·sr-1) over the 600- to 1,000-nm range was estimated to be of the same order of magnitude as the solar photon availability for a green sulfur bacterium living at 80 m depth in the Black Sea (28, 30). The Black Sea bacterium is a brown-colored strain of the green sulfur bacteria, and contains BChl e and isorenieratene carotenoids that are thought to improve the harvesting of solar green light that penetrates to such depths (31, 32). There is relatively little light in the green (≈550 nm) region of spectra measured at deep-sea hydrothermal vents, and so the absence of isorenieratene carotenoids and the presence of BChl c in GSB1 are in accordance with the geothermal light wavelengths that have been measured at vents. The in situ cell division time of the Black Sea bacterium was calculated to be 2.8 years (30). Because of similarly low light intensities at deep-sea vents, GSB1 may be thought of as eking out an existence by infrequent harvesting of rare geothermal photons; this is in keeping with current ideas about the survival of bacteria in oligotrophic habitats, and in contrast to the relatively vigorous growth obtained under laboratory conditions.
Although GSB1 was captured from the TY black smoker effluent, it is unlikely to be the direct descendent of a line of photosynthetic organisms that have continuously occupied this deep-sea hydrothermal vent since the appearance of anoxygenic photosynthesis on Earth >3 × 109 years ago (33), before the evolution of oxygenic photosynthesis that led to the accumulation of O2 in the atmosphere ≈2 × 109 years ago (34, 35). This is because individual hydrothermal vents are ephemeral relative to geological time scales, as observations from repeated visits to black smokers indicated major structural changes over the course of days to decades (26). There is evidence that isorenieratene carotenoid-containing green sulfur bacteria thrived in the North Atlantic Ocean in the Cenomanian/Turonian (C/T) age ≈108 years ago during poorly understood “oceanic anoxic events” (36), which may have also occurred in the Pacific Ocean. The oceans have risen and subsided, and land masses have shifted over such time scales, and so the possibility exists of vent-to-shore as well as vent-to-vent exchange of green sulfur bacteria in the past.
Regardless, the capture of GSB1 at a deep-sea hydrothermal vent, but not from surrounding waters, indicates that geothermal light and associated reduced S compounds are sufficient to at least enhance the survival of green sulfur bacteria in the otherwise dark, oxygenated ocean depths. This discovery expands the range of possible environments that could harbor life forms which use light energy to drive endergonic biochemical reactions (1, 25), and frees the thinking of the scientific community from the constraint that any form of life that depends on light energy is necessarily limited to solarly illuminated habitats.
Supplementary Material
Acknowledgments
We thank the ALVIN Group and the captain and crew of R/V Atlantis for assistance in collecting samples, the University of British Columbia Imaging Facility for assistance in electron microscopy, and D. Brune and J. Bickmeier for assistance with mass spectrometry. This research was supported by the Natural Sciences and Engineering Research Council (Canada), the U.S. National Aeronautics and Space Administration Astrobiology and Exobiology programs, the U.S. Department of Energy Biosciences program, and the U.S. National Science Foundation RIDGE program.
Author contributions: J.T.B., J.O., R.E.B., C.L.V.D., and F.G.P. designed research; J.T.B., J.O., M.T.L., A.K.M., A.S.L., R.E.B., C.L.V.D., T.A.M., and F.G.P. performed research; J.T.B. and R.E.B. analyzed data; and J.T.B., R.E.B., and C.L.V.D. wrote the paper.
Abbreviation: BChl, bacteriochlorophyll.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AY627684 (FMO) and AY627756 (16S rRNA)].
References
- 1.Blankenship, R. E. (2002) Molecular Mechanisms of Photosynthesis (Blackwell Science, Oxford).
- 2.Van Dover, C. L. (2000) The Ecology of Deep-Sea Hydrothermal Vents (Princeton Univ. Press, Princeton).
- 3.Martin, W. & Russell, M. J. (2003) Philos. Trans. R. Soc. London B 358, 59-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Dover, C. L., Reynolds, G. T., Chave, A. D. & Tyson, J. A. (1996) Geophys. Res. Lett. 23, 2049-2052. [Google Scholar]
- 5.Nisbet, E. G., Cann, J. R. & VanDover, C. L. (1995) Nature (London) 373, 479-480. [Google Scholar]
- 6.Yurkov, V. V., Krieger, S., Stackebrandt, E. & Beatty, J. T. (1999) J. Bacteriol. 181, 4517-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beatty, J. T. (2002) Photosynth. Res. 73, 109-114. [DOI] [PubMed] [Google Scholar]
- 8.Chyba, C. F. & Hand, K. P. (2001) Science 292, 2026-2027. [DOI] [PubMed] [Google Scholar]
- 9.Venter, J. C., Remington, K., Heidelberg, J. F., Halpern, A. L., Rusch, D., Eisen, J. A., Wu, D., Paulsen, I., Nelson, K. E., Nelson, W., et al. (2004) Science 304, 66-74. [DOI] [PubMed] [Google Scholar]
- 10.Pfennig, N. & Trüper, H. G. (1981) in The Prokaryotes: A Handbook on Habitats, Isolation, and Identification of Bacteria, eds. Starr, M. P., Stolp, H., Trüper, H. G., Balows, A. & Schlegel, H. G. (Springer, New York), Vol. 1, pp. 279-289. [Google Scholar]
- 11.Widdel, F., Kohring, G. W. & Mayer, F. (1983) Arch. Microbiol. 134, 286-294. [Google Scholar]
- 12.Rainey, F. A., Ward-Rainey, N., Kroppenstedt, R. M. & Stackebrandt, E. (1996) Intl. J. Syst. Bact. 46, 1088-1092. [DOI] [PubMed] [Google Scholar]
- 13.Cole, J. R., Chai, B., Marsh, T. L., Farris, R. J., Wang, Q., Kulam, S. A., Chandra, S., McGarrell, D. M., Schmidt, T. M., Garrity, G. M., et al. (2003) Nucleic Acids Res. 31, 442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall, T. A. (1999) Nucleic Acids Symp. Ser. 41, 95-98. [Google Scholar]
- 15.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403-410. [DOI] [PubMed] [Google Scholar]
- 16.Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244-1245. [DOI] [PubMed] [Google Scholar]
- 17.Brune, D. C. (1999) Rapid Commun. Mass Spectrom. 13, 384-389. [Google Scholar]
- 18.Coolen, M. J. L. & Overmann, J. (2000) Appl. Environ. Microbiol. 66, 2589-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Man, J. C. (1977) Eur. J. Appl. Microbiol. 4, 307-316. [Google Scholar]
- 20.De Man, J. C. (1983) Eur. J. Appl. Microbiol. Biotechnol. 17, 301-305. [Google Scholar]
- 21.Haymon, R. M., Fornari, D., Edwards, M., Carbotte, S., Wright, D. & Macdonald, K. C. (1991) Earth Planet Sci. Lett. 104, 513-534. [Google Scholar]
- 22.Overmann, J. (2004) in The Prokaryotes: An Evolving Electronic Resource for the Microbiological Community (Springer, New York), http://141.150.157.117:8080/prokPUB/chaprender/jsp/showchap.jsp?chapnum=323.
- 23.Blankenship, R. E. & Matsuura, K. (2003) in Light-Harvesting Antennas, eds. Green, B. R. & Parson, W. W. (Kluwer, Dordrecht, The Netherlands), pp. 195-217.
- 24.Frigaard, N.-U. & Bryant, D. A. (2004) Arch. Microbiol. 182, 265-276. [DOI] [PubMed] [Google Scholar]
- 25.Overmann, J. & Garcia-Pichel, F. (2004) in The Prokaryotes: An Evolving Electronic Resource for the Microbiological Community (Springer, New York), http://141.150.157.117:8080/prokPUB/chaprender/jsp/showchap.jsp?chapnum=239.
- 26.Fornari, D. J., Shank, T., Damm, K. L. V., Gregg, T. K. P., Lilley, M., Levai, G., Bray, A., Haymon, R. M., Perfit, M. R. & Lutz, R. (1998) Earth Planet Sci. Lett. 160, 419-431. [Google Scholar]
- 27.White, S. N., Chave, A. D., Reynolds, G. T. & VanDover, C. L. (2002) Geophys. Res. Lett. 29, 10/1029/2002GL014977. [Google Scholar]
- 28.White, S. N., Chave, A. D., Reynolds, G. T., Gaidos, E. J., Tyson, J. A. & VanDover, C. L. (2000) Geophys. Res. Lett. 29, 1151-1154. [Google Scholar]
- 29.White, S. N., Chave, A. D. & Reynolds, G. T. (2002) J. Geophys. Res. 107, doi: 10.1029/2000JB000015. [DOI]
- 30.Overmann, J., Cypionka, H. & Pfennig, N. (1992) Limnol. Oceanogr. 370, 150-155. [Google Scholar]
- 31.Pfennig, N. (1967) Annu. Rev. Microbiol. 21, 285-384. [DOI] [PubMed] [Google Scholar]
- 32.Overmann, J., Beatty, J. T., Hall, K. J. & Pfennig, N. (1991) Limnol. Oceanogr. 36, 846-859. [Google Scholar]
- 33.Des Marais, D. J. (2000) Science 289, 1703-1705. [PubMed] [Google Scholar]
- 34.Dismukes, G. C., Klimov, V. V., Baranov, S. V., Kozlov, Y. N., DasGupta, J. & Tyryshkin, A. (2001) Proc. Natl. Acad. Sci. USA 98, 2170-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kasting, J. F. & Siefert, J. L. (2002) Science 296, 1066-1068. [DOI] [PubMed] [Google Scholar]
- 36.Sinninghe Damsté, J. S. & Köster, J. (1998) Earth Planet Sci. Lett. 158, 165-173. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.