Abstract
Reelin mRNA and protein levels are reduced by ≈50% in various cortical structures of postmortem brain from patients diagnosed with schizophrenia or bipolar illness with psychosis. In addition, the mRNA encoding the methylating enzyme, DNA methyltransferase 1, is up-regulated in the same neurons that coexpress reelin and glutamic acid decarboxylase 67. We have analyzed the extent and pattern of methylation within the CpG island of the reelin promoter in genomic DNA isolated from cortices of schizophrenia patients and nonpsychiatric subjects. Ten (The Stanley Foundation Neuropathology Consortium) and five (Harvard Brain Collection) schizophrenia patients and an equal number of nonpsychiatric subjects were selected from each brain collection. Genomic DNA was isolated, amplified (from base pair -527 to base pair +322) after bisulphite treatment, and sequenced. The results show that within the promoter region there were interesting regional variations. There was increased methylation at positions -134 and 139, which is particularly important for regulation, because this portion of the promoter is functionally competent based on transient transfection assays. This promoter region binds a protein present in neuronal precursor nuclear extracts that express very low levels of reelin mRNA; i.e., an oligonucleotide corresponding to this region and that contains methylated cytosines binds more tightly to extracts from nonexpressing cells than the nonmethylated counterpart. Collectively, the data show that this promoter region has positive and negative properties and that the function of this complex cis element relates to its methylation status.
Keywords: DNA methyltransferase, epigenetics, gene regulation, methylation, psychiatric disorder
Schizophrenia is a devastating disorder with a population-wide morbidity approaching 1%. The genetics of the disease are perhaps one of the most studied facets of schizophrenia, but the results of multiple linkage analyses have not provided a clarification of underlying etiological factors that define the symptomatology of the disease (1, 2). However, insight has come from recent reports that examined biological markers that appear to be aberrantly regulated in postmortem brains of patients diagnosed with schizophrenia (3–5). It was noted that among >100 different markers examined, reelin and glutamic acid decarboxylase (GAD)67 are the most abnormal in the context of schizophrenia and bipolar illness (6). These two mRNAs and their cognate proteins are selectively coexpressed in GABAergic neurons of the mammalian cortex. The observation that reelin and GAD67 are down-regulated has been one of the more consistently replicated findings observed in postmortem cortex from schizophrenia patients (SZP). We previously reported reelin mRNA and protein levels are quantitatively reduced by ≈50% in various cortical structures of postmortem brain from SZP and bipolar patients (3, 4). The results obtained with respect to reelin were independently confirmed immunohistochemically in the hippocampus (7) and more recently in the cortex by using in situ hybridization (8, 9). The decrease in GAD levels were originally noted some years earlier (10). The reelin and GAD67 mRNAs colocalize and are largely decreased in the same GABAergic neurons that show a coincident increased expression of the DNA methylating enzyme DNA methyltransferase 1 (Dnmt1) (11).
The above findings are consistent with our working hypothesis that schizophrenia arises from a primary defect in GABAergic transmission. It seems clear that defective GABAergic signaling could result in increased glutamatergic transmission that might account for the observed psychosis common to schizophrenia and bipolar disorder (12). The concept of schizophrenia arising from a defect of GABAergic neurons and its impact on additional transmitter receptor systems has been previously discussed (5, 13–16). To investigate the mechanism responsible for the down-regulation of reelin expression, we first cloned and characterized the human reelin promoter (17). It was subsequently shown that reelin is secreted constitutively from cerebellar granule neurons (18) and that it functions in regulating dendritic spine resident mRNA translation in purified synaptoneurosome preparations (19).
More recently, we developed an epigenetic murine model in which reelin mRNA and protein levels are down-regulated in response to a chronic methionine treatment (20). The model is predicated on observations made in several labs that demonstrated that methionine administration to SZP exacerbated symptomatology in some 70% of patients receiving high doses of the amino acid (21). By using a methionine treatment paradigm, we have been able to down-regulate reelin and GAD67 in vivo (22) and in cultured neurons maintained in vitro (23). By using antisense oligonucleotides directed against Dnmt1, we have also been able to provide a link between the methionine-mediated down-regulation and the expression of Dnmt1 (23).
Our studies of the human reelin promoter lead us to suggest that a relationship exists between schizophrenia and the methylation status of the reelin promoter CpG island (17). This relationship is particularly relevant in light of the up-regulation of Dnmt1 mRNA and protein in cortical GABAergic neurons (11). More precisely, we hypothesize that the overexpression of Dnmt1 leads to the hypermethylation of the reelin promoter (and others as well) and that this process is likely responsible for the reduced expression of the corresponding mRNA(s). A preliminary report supporting this hypothesis showed that the reelin promoter was hypermethylated in SZP (9). To study the role of promoter methylation in down-regulating reelin expression, we have analyzed, thus far, the extent and pattern of methylation within the CpG island of the reelin promoter in genomic DNA isolated from two different brain regions from two collections of SZP and nonpsychiatric subjects (NPS). The data show a significant difference (P < 0.001) in the frequency of the number of methylated bases at specific sites in the promoter. Two positions in particular, -134 and -139, overlap with a functionally defined cis element in the promoter (17). Collectively, our findings allow us to propose a multistate transcriptional cascade that provides regulation through positive and negative factors acting at or very near the same site, depending on the methylation status of this sequence.
Materials and Methods
Genomic DNA was isolated from two distinct brain collections. Ten occipital cortex samples were obtained from NPS and 10 were from SZP procured through The Stanley Foundation Neuropathology Consortium (National Institute of Mental Health, St. Elizabeth's Hospital, Washington, D.C.) (hereafter referred to as The Stanley Foundation) (24). Only limited amounts of tissue were available, which precluded an analysis of DNA from all patients available through this resource. To supplement these studies, we obtained prefrontal cortices (Brodman areas 9 or 10) from an additional five NPS and five SZP from the Harvard Brain Collection (Harvard Brain Tissue Resource Center, Belmont, MA). The demographics associated with each patient population are presented in Table 2, which is published as supporting information on the PNAS web site. We found no significant correlations between total methylation and postmortem interval, gender, age of death, age of onset (for SZP), drug equivalents (for SZP), or brain hemisphere.
Bisulfite Modification of Genomic DNA. Genomic DNA was isolated (25), and yield was determined by using optical density measurements (OD260/280). DNA (5 μg from each subject) was digested with 10 units of EcoRI at 37°C overnight and subsequently denatured by treatment with 0.2 N NaOH for 30 min at 37°C. After neutralization, DNA was extracted, and the ethanol was precipitated. Bisulfite modification of genomic DNA was carried out by using a CpGenome DNA modification kit (Intergen, Purchase, NY). Preliminary experiments were conducted to determine the optimal modification time and temperature, which was 5 h at 45°C. In general, we typically used one half of the original modified DNA for subsequent PCR amplification and cloning.
Nested PCR and Sequencing. After bisulphite modification, the strands (designated the A and B strands) are no longer complementary, and each strand requires a different set of primers for amplification. Primers designed to amplify the A strand never produced sufficient amounts of product even after performing nested PCR. The primers designed to amplify the B strands are as described in ref. 17. The reaction products of the B strand were amplified for 42 cycles by using an Advantage GC genomic PCR kit (BD Biosciences, Franklin Lakes, NJ). For the second amplification round, 1/50th of the product of the first round was used as starting template, and the second amplification was performed for 30 cycles. PCR products from each sample set were purified, and individual clones were sequenced in both directions at the University of Chicago Core Sequencing Facility.
Transient Transfections and Reporter Analysis. Transfections of the reelin DNA fragments to assess promoter function were performed in Ntera 2 (NT2) cells as described in ref. 17. The -514 promoter fragment was used as a template to generate the additional templates by RT-PCR. Our previous studies had shown that the -514 promoter was capable of full promoter activity in multiple cells lines. We used the dual luciferase reporter assay system (Promega/Fisher) to cotransfect the reelin promoter/luciferase vectors with the pRL-CMV vector to normalize for transfection efficiency. Transfections, performed in triplicate, were carried out independently a minimum of three times to obtain the data. Data are expressed as a ratio to the signal obtained from the simian virus 40 promoter/luciferase vector transfected in parallel (pGL3-Control Vector, Promega/Fisher).
Gel Shift Binding Assays. To determine the affinity of nuclear binding proteins to the methylated and nonmethylated sites, we synthesized oligonucleotides corresponding to this region (Integrated DNA Technologies, Coralville, IA). The oligos spanned the sequence of interest and extended from base pair -149 to base pair -127. The nonmethylated oligo was 5′-CGC TTT CCC AGG CCT GGC CGA GG-3′ (forward), and the complimentary sequence was 5′-CCT CGG CCA GGC CTG GGA AAG CG-3′ (reverse). The methylated counterparts were 5′-CGC TTT CC(5mC) AGG C(5mC)T GGC CGA GG-3′ (forward) and 5′-CCT CGG C(5mC)A GGC (5mC)TG GGA AAG CG-3′ (reverse). The oligos were mixed in equimolar portions and heated for 30 min at 90°C in a heating block. The heating block was turned off, and the block was allowed to come to room temperature overnight to facilitate annealing. Extracts were prepared from NT2- and retinoic acid (RA)-treated NT2 cells by using NE-PER nuclear extraction reagents (Pierce). The yield of nuclear proteins obtained from a starting packed cell pellet of 50 μl was typically 300 μg.
Double-stranded oligos were end-labeled with 32P and polynucleotide kinase to a specific activity of 7.5 × 107 cpm/μg. Approximately 1 × 105 cpm were incubated with nuclear extract, and gel shift assays were performed as described in ref. 26. After electrophoresis, gels were dried and exposed to phosphor screens for analysis on the Storm PhosphorImager (Molecular Dynamics). The density of each of the shifted bands was quantified. Dissociation rates were calculated from the intensities of the shifted bands after fitting the data to the one phase exponential decay: y = span·e(-kx) (26), where span represents the intensity of the shifted band in the absence of competition.
Results
In the present study, we performed an analysis of the human reelin promoter by using genomic DNA isolated from NPS and SZP to test whether there were differences in the total number of methylated bases or in the pattern of methylation in these two conditions. As indicated above, this study was predicated on the previous analysis of the reelin promoter CpG island (17), the observation that reelin mRNA is reduced in various cortical regions of SZP patients (3) and the finding that reelin mRNA expression increased when the promoter region showed less methylation (17). After bisulphite modification of genomic DNA from each individual, the promoter region was amplified and products were subcloned and subsequently sequenced. Data from at least six clones each from The Stanley Foundation samples were obtained, and eight subclones were used to evaluate the Harvard Brain Collection data. The bisulphite modification/sequencing approach allows one to examine changes at individual bases along the region of interest. This method is particularly relevant when nonstandard methylation sites exist (e.g., CpNpG) such as what we encountered at positions -139 (CpApG) and -134 (CpTpG). Methylation of non-CpG sequences has been reported to be involved in the regulation of other genes, such as human synaptotagmin XI, which is also expressed in neurons (27).
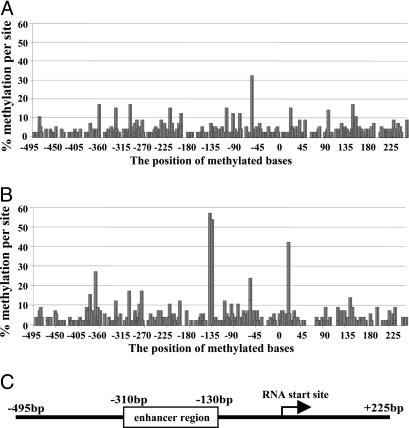
The methylation data obtained from the individuals from The Stanley Foundation are summarized in Fig. 1 A and B. A schematic representation of the reelin promoter is shown in Fig. 1C and is in scale with the positions of the methylated bases. The data are displayed as the percent of total methylated sites along the reelin promoter, numbering relative to the RNA start site (17). The data were obtained from genomic DNA isolated from occipital cortices from individual subjects from each group (Fig. 1 A, NPS; Fig. 1B, SZP). For the demographics associated with each sample subject, see Table 2. The maximum number of methylated cytosine residues for either group at any one position is 60, which would represent 100% methylation (six clones per subject with 10 subjects per group). This represents the situation in which all cytosine residues from all clones from each individual are methylated. In some cases (see Fig. 1B), the actual values approached half the maximum (positions -139 and -134). In general, the amounts of methylated residues in the NPS (Fig. 1 A) are lower, and this low-level sporadic methylation likely represents a background level of methylation that appears to accumulate at some positions more than others. This same level of background noise is also present in the SZP (Fig. 1B), although there are a few positions in which the increased methylation level reaches statistical significance (two-tailed t tests). In particular, both positions, -139 and -134, are significant (P < 0.001) with respect to these same sites in the NPS group (Fig. 1, compare A with B). A few other sites appear to be relevant as well, including positions -370, -58, and +21. Interestingly, position -370 is the site previously identified as being more heavily methylated in SZP as compared with NPS and is close to the cAMP response element consensus site (9). A comparison between the levels in the two conditions for each position (The Stanley Foundation) in terms of the raw numbers is shown in Table 1.
Fig. 1.
Methylation profile of the reelin promoter in NPS and SZP from The Stanley Foundation brain collection. (A and B) Methylation percentages determined by sequencing bisulphite modified genomic DNA from occipital cortices of NPS (A) and SZP (B) are plotted against the position in the reelin promoter (for numbering, see ref. 17). Six clones from each of 10 patients from each group (NPS vs. SZP) were included in the analysis, yielding a theoretical maximum number of 60 methylated bases at any one position. (C) A linear representation of the reelin promoter (17) aligned with the methylation positions shown in A and B.
Table 1. Reelin promoter methylation summary.
| Methylated bases | NPS | SZP | P value |
|---|---|---|---|
| The Stanley Foundation | |||
| Upstream of RNA start site | 32.7 ± 4.9 | 40.8 ± 4.2 | 0.227 |
| Reelin promoter (–527–+322) | 50.8 ± 6.9 | 55.6 ± 5.7 | 0.598 |
| –370 | 0.2 ± 0.2 | 1.6 ± 0.3 | 0.002* |
| –139 | 0.4 ± 0.2 | 3.4 ± 0.5 | <0.001* |
| –134 | 0.3 ± 0.2 | 3.2 ± 0.5 | <0.001* |
| –58 | 2.1 ± 0.2 | 1.4 ± 0.3 | 0.052 |
| +21 | 0.9 ± 0.4 | 2.5 ± 0.4 | 0.012* |
| Harvard Brain Collection | |||
| Upstream of RNA start site | 51.0 ± 2.6 | 66.8 ± 1.5 | <0.001* |
| Reelin promoter (–527 to +322) | 71.4 ± 2.6 | 91.4 ± 3.2 | 0.001* |
| –330 | 2.8 ± 1.2 | 4.0 ± 0.8 | 0.436 |
| –302 | 2.6 ± 1.2 | 4.0 ± 0.8 | 0.358 |
| –222 | 2.6 ± 1.2 | 4.0 ± 0.8 | 0.358 |
| –200 | 2.6 ± 1.2 | 4.0 ± 0.8 | 0.358 |
| –139 | 0.8 ± 0.4 | 2.6 ± 0.5 | 0.021* |
| –134 | 0.8 ± 0.2 | 2.6 ± 0.5 | 0.011* |
| –109 | 2.6 ± 1.2 | 4.2 ± 0.9 | 0.302 |
| –96 | 2.6 ± 1.2 | 4.0 ± 0.8 | 0.358 |
| –83 | 2.6 ± 1.2 | 4.0 ± 0.8 | 0.358 |
| –58 | 2.2 ± 0.7 | 2.0 ± 0.5 | 0.822 |
| +21 | 0.6 ± 0.4 | 2.2 ± 0.4 | 0.019* |
| +145 | 2.4 ± 1.2 | 3.4 ± 1.0 | 0.538 |
Values are means ± SEM. The maximum at any single position is 6 for The Stanley Foundation specimens (n = 10 NSP and 10 SZP) and 8 for the Harvard Brain Collection specimens (n = 5 NSP and 5 SZP). P values were determined from a paired t test of the data by using the means and SEM between the two groups. *, P < 0.05.
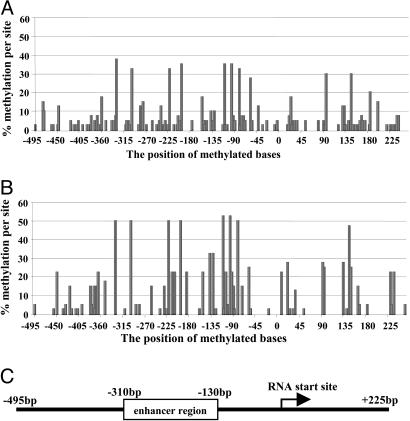
To confirm these data on a different brain set, we obtained prefrontal cortical samples from five NPS and five SZP from the Harvard Brain Collection. The demographics for these brain samples are also included in Table 2. Isolated genomic DNA was modified as described above, and eight clones from each subject were sequenced and analyzed (Fig. 2 A and B). A quick glance at the data does not show the striking difference that is readily apparent in Fig. 1. Instead, the Harvard Brain Collection samples showed a higher extent of background methylation and multiple sites appeared to be hypermethylated (Fig. 2 A and B). The reason for the difference in background methylation is not clear, although there is no clear relationship to, for example, postmortem interval or other factors associated with the brain collection. Although it remains a possibility that the difference could be related to the brain area, i.e., occipital cortex vs. prefrontal cortex, this remains to be established. Nevertheless, a number of sites in the promoter appeared higher than background in the Harvard Brain Collection samples. With respect to the data presented in Fig. 2, the maximum theoretical number of methylated sites was 40 (8 × 5 = 100%), as compared with 60 (6 × 10 = 100%) for The Stanley Foundation brain collection (Fig. 1). Although there were considerably more sites methylated in the Harvard Brain Collection samples, many of these sites were also elevated in both groups (i.e., NPS vs. SZP), and, hence, were not statistically significant. Those positions that were more heavily methylated in The Stanley Foundation SZP set were lower in the Harvard Brain Collection, but the trend toward increased methylation at sites -139 and -134 was still evident (see Fig. 3). A comparison between the levels in the two conditions for each position (Harvard Brain Collection) in terms of the raw numbers is shown in Table 1.
Fig. 2.
Methylation profile of the reelin promoter in NPS and SZP from the Harvard Brain Collection. (A and B) Methylation percentages determined by sequencing bisulphite modified genomic DNA from prefrontal cortices of NPS (A) and SZP (B) are plotted against the position in the reelin promoter. Eight clones from each of five patients from each group (NPS vs. SZP) were included in the analysis, yielding a theoretical maximum number of 40 (100%) methylated bases at any one position. (C) A linear representation of the reelin promoter (17) aligned with the methylation positions shown in A and B.
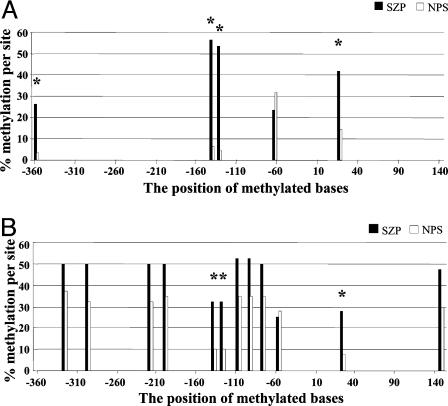
Fig. 3.
Methylation profile summary of The Stanley Foundation (A) and Harvard Brain Collection (B). Positions in which the total number of methylated bases exceeded 12 (or 25%) at any given position are included, with SZP (black bars) alongside NPS (white bars).
We identified those sites in the graphs in Figs. 1 and 2 at each place where the total number of methylated positions was 25% or more and redrew the data separately with the NPS and SZP data represented side-by-side. Fig. 3A shows a summary from The Stanley Foundation set and Fig. 3B shows the isolated data obtained from the Harvard Brain Collection. Each position in Fig. 3 in which there was a statistically significant difference is identified by an asterisk. For example, it seems evident that positions -139 and -134 are consistently increased in the SZP group. Position +21 is increased in genomic DNA from the occipital cortex of SZPs (The Stanley Foundation) and in frontal cortex of SZPs (Harvard Brain Collection), and the difference is significant (P < 0.012 and P < 0.019, respectively).
For either brain collection, we found no significant association between total methylation or methylation at specific sites and postmortem interval, age, age of onset, drug equivalents, method of death, gender, or brain hemisphere.
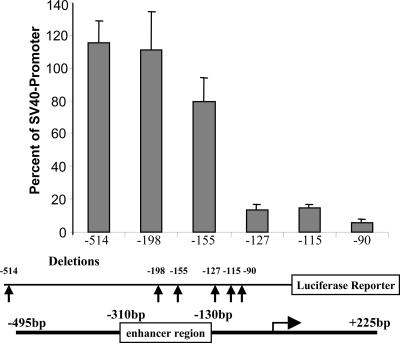
Promoter Analysis. Previously, we analyzed various portions of the reelin promoter to identify the existence and location of cis-acting elements involved in regulating expression by using a transient transfection approach (17). We revisited the region of the promoter containing the -139/-134 methylation hot spots after these sites proved to be relevant in the context of promoter hypermethylation. To determine whether loss of these sites decreased expression in a transient assay system, we tested constructs with various amounts of the promoter for their ability to direct expression in NT2 neuronal precursor cells. We had previously used this cell line to examine reelin promoter function (17). In the experiment presented here (Fig. 4), we used the -514 promoter construct for comparative purposes because this amount of the promoter had full function in NT2 cells. As sequences were deleted from the 5′ end, the promoter sequentially lost function, with a major drop-off occurring once sequences above -127 were deleted. That is, the -155 vector maintained 69% of full activity. However, when sequences between -155 and -127 were deleted, only 11% residual activity remained. Subsequent deletions (-115 and -90) did not further reduce activity (12% and 5%). These results indicate that when the region of the promoter containing the -139/-134 sites is deleted, significant function is lost with respect to the remaining sequences and their abilities to direct reelin promoter expression. In the absence of site-directed mutagenesis, it is difficult to define the precise effects of these specific bases; however, the results clearly show that this region of the promoter contains a positive cis-acting element.
Fig. 4.
Transfection of the reelin promoter -514 and deletion/luciferase reporter constructs (-198, -155, -127, -115, and -90) into NT2 cells. (Top) The amount of promoter activity is expressed as a function of 5′ flanking sequence present relative to the simian virus 40 promoter transfected in parallel. Data represent the mean obtained from three measurements made from a minimum of three separate experiments after correcting for transfection efficiency. (Middle) The line below the bar graph indicates the amount of 5′ flanking sequence present in the construct (arrows). (Bottom) A linear representation of the reelin promoter deletions aligned with the promoter.
Gel Mobility Shift Assays. To examine the ability of this portion of the promoter to bind nuclear proteins, we prepared two sets of oligonucleotides. The first contained sequences that were not methylated (WT) and the second set contained methylated cytosine residues (methyl) at the relevant positions (cytosine at -141 and cytosine at -137 bp: the -139/-134 sequences refer to the guanine positions identified from the B strand sequences). We prepared extracts from NT2 cells and used these extracts to probe for the presence of nuclear proteins that bind to the sequences of interest. Nuclear proteins in these extracts bound to the nonmethylated and methylated oligonucleotides corresponding to these sites. These cells are neuronal precursors that express only low levels of reelin mRNA. When we differentiate these cultures with RA, reelin transcription is induced and accompanied by a change in the chromatin structure in the vicinity of the promoter (17).
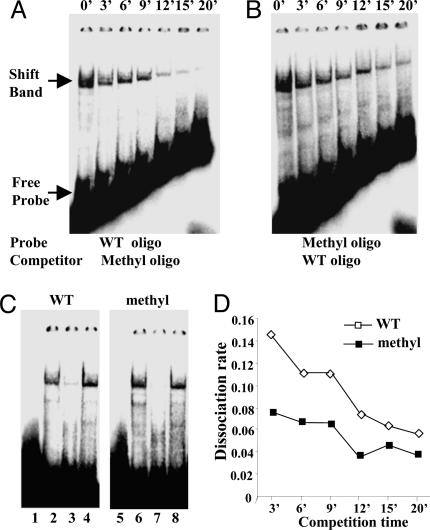
Cross-competition assays were performed with the corresponding oligonucleotide to determine relative dissociation rates. That is, binding to labeled WT oligo was competed by using a 300-fold molar excess of methyl oligo (Fig. 5A), and methyl oligo binding was competed at the same molar excess with WT oligo (Fig. 5B). At various times (0–20 min) after a 30-min incubation, unlabeled competitor was added, and aliquots were analyzed by gel mobility shift assay (Fig. 5 A and B). Interestingly, the dissociation rate constants calculated from the decay curves (Fig. 5D) showed that the binding affinity of the proteins present in NT2 cell nuclear extracts was higher for the methylated sites (kD = 0.054) than for the nonmethylated sites (kD = 0.094). Fig. 5C shows the WT oligo (Fig 5C Left) binding with NT2 cell extract and the methyl oligo (Fig. 5C Right) binding alongside homologous (specific) and nonspecific oligo competitions. Cross-competition experiments (data not shown) were also performed with extracts prepared from RA-differentiated NT2 cell extracts, and these extracts showed little difference in binding affinity for either oligonucleotide (kD values of 0.078 and 0.085, respectively). The above data demonstrate that the binding of proteins present in nuclear extracts that do not express reelin is tighter to the methylated site than the nonmethylated site.
Fig. 5.
Dissociation rate analysis by electromobility shift competition assays. Labeled oligonucleotides were incubated on ice for 30 min with nuclear extracts prepared from NT2 cells. After a 30-min incubation, a 300-fold excess of unlabeled oligonucleotide was added, and aliquots were analyzed by gel mobility shift assay after the indicated times (0–20 min). (A) NT2 cell nuclear extract incubated with labeled WT (nonmethylated oligonucleotide competed by unlabeled methylated oligonucleotide. (B) NT2 cell nuclear extract incubated with labeled methylated oligonucleotide and competed by unlabeled WT oligonucleotide. (C) Specificity of WT and methyl shifts. (Left) WT oligo. Lanes: 1, control shift (no extract); 2, NT2 cell extract; 3, homologous competition (200-fold); 4, Sp1 oligo (nonspecific) competition (200-fold). (Right) Methyl oligo. Lanes: 5, control shift (no extract); 6, NT2 cell extract; 7, homologous competition (200-fold); 8, Sp1 (nonspecific) oligo competition. (D) The dissociation rate was calculated by scanning the shifted bands with a densitometer and fitting the data to an exponential regression (y = span·e-kx), where x is time, y is specific binding, and span is the binding at time 0 and kD is the dissociation rate constant (26). ▪, Methyl probe with nonlabeled WT oligo competition; ▪, WT probe with nonlabeled methyl oligo competition. The affinity for the methylated oligo (kD = 0.054) was ≈1.7-fold higher than that obtained for the nonmethylated oligo (kD = 0.094).
Discussion
We have previously examined the expression of the human reelin promoter in neuronal precursor (NT2) cells and their RA-induced counterparts (hNT neurons) (17). We have also shown that the regulation of reelin expression in this system depends on the methylation status of the promoter (17). This model is ideal for studying gene regulation as reelin mRNA is expressed at low levels in NT2 cells and is increased nearly 100-fold upon exposure to RA. We examined the methylation status of the reelin promoter CpG island and found that it was more heavily methylated when the gene is silent and that methylation decreases as transcription increases. Moreover, the change in methylation status is accompanied by changes in local chromatin structure in the vicinity of the promoter. RA, through an incompletely defined mechanism, likely indirectly stimulates reelin promoter demethylation, which initiates a cascade that ultimately results in chromatin opening and recruitment of trans-acting factors to the transcription complex.
More recently, we have been able to show that drugs, such as valproic acid (VPA), activate reelin expression through a mechanism that is independent of the action of RA. VPA acts as a histone deacetylase inhibitor resulting in increased histone acetylation and a more relaxed chromatin configuration (28). We have previously shown that VPA treatment of NT2 cells increases reelin expression in a dose- and time-dependent manner (17). Moreover, the treatment increases the extent of predominantly histone H3 lysine acetylation and results in a subsequent decrease in the methylation status of the reelin promoter CpG island (29). We have also established that treatment of primary murine cortical neurons in vitro (23) or administration of methionine to mice in vivo (22) induces a down-regulation of reelin mRNA and protein. The methionine treatment increases the extent of methylation of the mouse reelin promoter both in vitro and in vivo (22, 23). The increased methylation in vitro is prevented by coincubating the cultures with Dnmt1 antisense oligonucleotides (23). Interestingly, VPA not only prevents this decrease in mRNA from occurring but also corrects the schizophrenia-like behaviors associated with this treatment. That is, coadministration of methionine and VPA normalizes the methionine-induced reelin mRNA down-regulation and corrects the disruption of prepulse inhibition of startle and the methionine-induced social interaction deficits (30).
A recent paper (9) has reported that the reelin promoter is hypermethylated in the prefrontal cortex of postmortem SZP as compared with NPS. This group used a combination of bisulphite modification and methylation-specific PCR to show that particular regions of the promoter were more heavily methylated in genomic DNA isolated from SZP. Although the authors argue that their study was preliminary, the data are convincing and do not differ in substance from what we have presented here. The main difference was the region examined in the two studies. The first group (9) focused on a region further upstream (from base pair -440 to base pair -360; numbering is according to ref. 17) than our study, which encompassed the entire promoter region and part of the first exon. We see some hypermethylation in the same region that is proximal to the cAMP-response element and the upstream Sp1 site they refer to, particularly with The Stanley Foundation brain collection (Fig. 1, approximately at base pair -370). Interestingly, the other group had analyzed the Harvard Brain Collection for their study, and we do not see significant methylation in this region with the Harvard Brain Collection as presented here (Fig. 2). This difference may simply reflect that different brains were used in the two studies or that the regions dissected for each study were different, and it also points out the need for additional replication to provide further validation. Our previous deletion studies (17) did not provide evidence of a functional correlate in the region methylation uncovered by this group (9). It should be noted that these investigators did not look at the entire promoter region and so did not detect methylation in the regions where we found the highest degree of difference between the SZP and NPS groups. However, this group independently found significant differences in the amount of methylation in genomic DNA from SZP and NPS and, hence, confirmed the hypothesis we put forward with respect to the reelin down-regulation in SZP (17, 20).
One possible mechanism by which this modification could lead to gene silencing is if methylation prevents the interaction of positive trans-acting factors from binding to their cognate recognition sites, which is a formal possibility. Our transient transfection data support this concept, because deletion of this region of the promoter results in a loss of function. Another equally plausible mechanism is that methylation may recruit the binding of negative transcriptional repressors, such as MeCP2, MBD2, and/or Dnmt1, to the methylated base. This stable repressor complex would shut down mRNA transcription by inducing chromatin condensation in this region, which may also involve histone deacetylation and other histone tail modifications. Determining whether both or either mechanism is operative will require additional studies. However, our gel shift data suggest that methylation recruits binding of repressor proteins that have a higher affinity for the methylated sequence than the nonmethylated promoter.
Of particular interest in light of our data are that two of the three sites that are more heavily methylated in SZP in both brain sets are nonstandard or atypical CpNpG sites rather than traditional CpG sites. That is, the methylation we observed at positions -139 (CpApG) and -134 (CpTpG) are considered to be atypical with respect to the CpG sites normally methylated. There is evidence to suggest that Dnmt3A may methylate non-CpG sites in embryonic stem cells (31); however, we have not been able to detect the expression of Dnmt3a in tissue isolated from mouse or human brain (W. Rudzicka and A. Guidotti, personal communication). This atypical methylation has relevance to other genes expressed in neurons, such as synaptotagmin XI, which has also been implicated both genetically and pharmacologically to the onset of schizophrenia (27). Non-CpG methylation of a region proximal to the synaptotagmin XI promoter reduces binding activity of an Sp family member and correlates with reduced expression of the gene. Although Dnmt1 has been shown to be more highly expressed in GABAergic neurons of the schizophrenia brain (11), it is unclear whether Dnmt1 is able to methylate these sites, so this question awaits further studies.
Collectively, our findings are consistent with promoter methylation being important in the context of the reduced expression of reelin in schizophrenia. Moreover, we would like to suggest that methylation might promote the binding of repressor factors to functionally defined cis-acting elements, hence compromising promoter function. Collectively, these studies should provide the foundations for a new pharmacology for the treatment of schizophrenia by identifying targets, such as Dnmt1 and histone deacetylases, for pharmacological manipulation. Research into Rett syndrome, which has a clear neurological phenotype, may provide insight into schizophrenia because the protein deficient in Rett syndrome, namely MeCP2 (32), is likely relevant to understanding the repression of genes shut down through promoter hypermethylation. It should not be overlooked that if schizophrenia is caused by a progressive increase in methylation, then it may be possible to prevent disease onset by the prophylactic intervention of at-risk adolescents at an early enough stage of the disease. Although there are now currently two reports in the literature suggesting that promoter methylation is associated with the etiology of schizophrenia, these studies need additional replication to provide additional confidence in the findings.
Supplementary Material
Acknowledgments
We thank Dr. Gabrielle D'Arcangelo (The Cain Foundation Laboratories, Baylor College of Medicine, Houston), Dr. Moshe Szyf (Department of Pharmacology and Therapeutics, McGill University, Montreal), and Dr. Boris P. Sokolov (Molecular Neurobiology Branch, National Institutes of Drug Abuse, National Institutes of Health, Baltimore) for constructive criticism and suggestions in the preparation of the manuscript. This work was supported by National Institutes of Mental Health Grants MH62682 (to D.R.G.), MH062188 (to A.G.), and MH062090 (to E.C.).
Author contributions: D.R.G., A.G., and E.C. designed research; D.R.G., X.J., Y.C., R.P.S., and C.P.M. performed research; D.R.G. contributed new reagents/analytic tools; D.R.G., X.J., and Y.C. analyzed data; and D.R.G. wrote the paper.
Abbreviations: Dnmt, DNA methyl transferase; GAD, glutamic acid decarboxylase; NPS, nonpsychiatric subject(s); NT2, Ntera 2; RA, retinoic acid; SZP, schizophrenia patient(s).
References
- 1.Petronis, A. (2001) Trends Genet. 17, 142-146. [DOI] [PubMed] [Google Scholar]
- 2.Petronis, A, Gottesman, I. I., Kan, P., Kennedy, J. L., Basile, V. S., Paterson, A. D. & Popendikyte, V. (2003) Schizophr. Bull. 29, 169-178. [DOI] [PubMed] [Google Scholar]
- 3.Guidotti, A., Auta, J., Davis, J. M., DiGiorgi-Gerenini, V., Dwivedi, J., Grayson, D. R., Impagnatiello, F., Pandey, G. N., Pesold, C., Sharma, R. F., et al. (2000) Arch. Gen. Psychiatry 57, 1061-1069. [DOI] [PubMed] [Google Scholar]
- 4.Impagnatiello, F., Guidotti, A., Pesold, C., Dwivedi, Y., Caruncho, H., Pisu, M., Uzunov, D., Smalheiser, N., Davis, J., Pandey, G., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 15718-15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costa, E., Grayson, D. R., Mitchell, C. P., Tremolizzo, L., Veldic, M. & Guidotti, A. (2004) Crit. Rev. Neurobiol. 15, 121-142. [DOI] [PubMed] [Google Scholar]
- 6.Torrey, E. F., Barci, B. M., Webster, M. J., Bartko, J. J., Meador-Woodruff, J. H. & Knable, M. B. (2005) Biol. Psychiatry 57, 252-260. [DOI] [PubMed] [Google Scholar]
- 7.Fatemi, S. H., Earle, J. A. & McMenomy, T. (2000) Mol. Psych. 5, 654-663. [DOI] [PubMed] [Google Scholar]
- 8.Eastwood, S. L. & Harrison, P. J. (2003) Mol. Psychiatry 8, 821-831. [DOI] [PubMed] [Google Scholar]
- 9.Abdolmaleky, H. M., Cheng, H.-H., Russo, A., Smith, C. L., Faraone, S. V., Wilcox, M., Shafa, R., Glatt, S. J., Nguyen, G., Ponte, J. F., et al. (2005) Am. J. Med. Genet. B 134, 60-66. [DOI] [PubMed] [Google Scholar]
- 10.Akbarian, S., Kim, J. J., Potkin, G., Hagman, J. O., Tafazzoli, A., Bunney, W. E., Jr., & Jones, E. G. (1995) Arch. Gen. Psychiatry 552, 258-266. [DOI] [PubMed] [Google Scholar]
- 11.Veldic, M., Caruncho, H. J., Liu, W. S., Davis, J., Satta, R., Grayson, D. R., Guidotti, A. & Costa, E. (2004) Proc. Natl. Acad. Sci. USA 101, 348-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalkman, H. O. & Loetscher, E. (2003) J. Neural. Transm. 110, 803-812. [DOI] [PubMed] [Google Scholar]
- 13.Volk, D. W. & Lewis, D. A. (2002) Physiol. Behav. 77, 501-505. [DOI] [PubMed] [Google Scholar]
- 14.Heckers, S., Stone, D., Walsh, J., Shick, J., Koul, P. & Benes, F. M. (2002) Arch. Gen. Psych. 59, 521-529. [DOI] [PubMed] [Google Scholar]
- 15.Guidotti, A., Auta, J., Davis, J. M., Dong, E., Grayson, D. R., Veldic, M., Zhang, X. & Costa, E. (2005) Psychopharmacology, in press. [DOI] [PubMed]
- 16.Lewis, D. A., Hashimoto, T. & Volk, D. W. (2005) Nat. Rev. Neurosci. 6, 312-324. [DOI] [PubMed] [Google Scholar]
- 17.Chen. Y., Sharma, R., Costa, R. H., Costa, E. & Grayson, D. R. (2002) Nucl. Acids Res. 3, 2930-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacor, P. N., Grayson, D. R., Auta, J., Sugaya, I., Costa, E. & Guidotti, A. (2000) Proc. Natl. Acad. Sci. USA 97, 3556-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong, E., Caruncho, H., Liu, W. S., Smalheiser, N. R., Grayson, D. R., Costa, E. & Guidotti, A. (2003) Proc. Natl. Acad. Sci. USA 18, 235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costa, E., Grayson, D. R. & Guidotti, A. (2002) Mol. Int. 3, 220-229. [DOI] [PubMed] [Google Scholar]
- 21.Antun, F. T., Burnett, G. B., Cooper, A. J., Daly, R. J., Smythies, J. R. & Zeally, A. K. (1971) J. Psychiatry Res. 8, 63-71. [DOI] [PubMed] [Google Scholar]
- 22.Tremolizzo, L., Carboni, G., Ruzicka, W. B., Mitchell, C. P., Sugaya, I., Tueting, P., Sharma, R., Grayson, D. R., Costa, E. & Guidotti, A. (2002) Proc. Natl. Acad. Sci. USA 99, 10795-17100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noh, J. S., Sharma, R. P., Veldic, M., Salvacion, A., Jia, X., Chen, Y., Costa, E. & Grayson, D. R. (2005) Proc. Natl. Acad. Sci. USA 102, 1749-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torrey, E. F., Webster, M., Knable, M., Johnston, N. & Yolkin, R. H. (2000) Schizophr. Res. 44, 151-155. [DOI] [PubMed] [Google Scholar]
- 25.Zuccotti, M. & Monk, M. (1995) Nat. Genet. 9, 316-320. [DOI] [PubMed] [Google Scholar]
- 26.Chen, H., Banerjee, A. K. & Hannapel, D. J. (2004) Plant J. 38, 276-284. [DOI] [PubMed] [Google Scholar]
- 27.Inoue, S. & Oishi, M. (2005) Gene 348, 123-134. [DOI] [PubMed] [Google Scholar]
- 28.Phiel, C. J., Zhang, F., Huang, E. Y., Guenther, M. G., Lazar, M. A. & Klein, P. S. (2001) J. Biol. Chem. 276, 36734-36741. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell, C. P., Chen, Y., Kundakovic, M., Costa, E. & Grayson, D. R. (2005) J. Neurochem. 93, 483-492. [DOI] [PubMed] [Google Scholar]
- 30.Tremolizzo, L., Doueiri, M. S., Dong, E., Grayson, D. R., Davis, J., Pinna, G., Tueting, P., Rodriguez-Menendez, V., Costa, E. & Guidotti, A. (2005) Biol. Psych. 57, 500-509. [DOI] [PubMed] [Google Scholar]
- 31.Ramsahoye, B. H., Biniszkiewicz, D., Lyko, F., Clark, V., Bird, A. P. & Jaenisch, R. (2000) Proc. Natl. Acad. Sci. USA 97, 5237-5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amir, R. E., Van Den Veyver, I. B., Wan, M., Tran, C. Q., Francke, U. & Zoghbi, H. Y. (1999) Nat. Genet. 23, 185-188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.