Abstract
Background
Low back pain (LBP) is predominantly caused by degeneration of the intervertebral disc (IVD) and central nucleus pulposus (NP) region. Conservative treatments fail to restore disc function, motivating the exploration of nucleic acid therapies, such as the use of microRNAs (miRNAs). miRNAs have the potential to modulate expression of discogenic factors, while silencing the catabolic cascade associated with degeneration. To deliver these miRNAs, nonviral cell penetrating peptides (CPPs) are gaining favor given their low immunogenicity and strong targeting ability. Single miRNA therapies have been investigated for IVD repair, however dual miRNA delivery strategies have not been commonly examined and may augment regeneration.
Materials and methods
Transfection of four pro‐discogenic miRNAs (miRNA mimics:140‐5p; 149‐5p and inhibitors: 141‐3p; 221‐3p) and dual delivery of six miRNA pairings was performed using two CPPs, RALA and GET peptide (FLR), in primary rat NP monolayer culture, and in an ex vivo organ culture model of rat caudal discs. Protein expression of discogenic (aggrecan, collagen type II, and SOX9) and catabolic markers (ADAMTS5 and MMP13) were assessed.
Results
Monolayer investigations signified enhanced discogenic marker expression following dual miRNA delivery, signifying a synergistic effect when compared to single miRNA transfection. Utilization of an appropriate model was emphasized in our ex vivo organ culture experiment, revealing the establishment of a regenerative microenvironment characterized by reduced catabolic enzyme activity and enhanced matrix deposition, particularly following concurrent delivery of FLR‐miRNA‐149‐5p mimic and miRNA‐221‐3p inhibitor. Bioinformatics analysis of miRNA‐149‐5p mimic and miRNA‐221‐3p inhibitor identified distinct targets, pathways, and interactions, suggesting a mode of action for this amplified response.
Conclusion
Our findings suggest the potential of FLR‐miRNA‐149‐5p + miRNA‐221‐3p inhibitor to create an anti‐catabolic niche within the disc to foster regeneration in moderate cases of disc degeneration, which could be utilized in further studies with the overarching aim of developing treatments for LBP.
Keywords: anti‐catabolism, cell penetrating peptides, cell therapies, microRNA, regeneration
There is a current unmet need in the development of regenerative therapies for degeneration of the intervertebral disc. Here microRNAs were delivered with enhanced cell penetrating peptides, both individually and in pairs, with the aim of correcting the homeostatic anabolic:catabolic imbalance. We find that in an ex vivo organ model of the mildly degenerate disc, an anti‐catabolic niche can be generated that can foster discogenic potential.
1. INTRODUCTION
The incidence of low back pain (LBP) is on the rise, affecting an estimated 600 million people worldwide creating a growing socioeconomic burden, particularly in aging populations. 1 Though the causes are pleiotropic in nature, there is consensus that degeneration of the nucleus pulposus (NP) region of the intervertebral disc (IVD) through degradation of the glycosaminoglycan (GAG)‐rich extracellular matrix (ECM) and its key components is a primary cause. This is exemplified by a disruption to the catabolic:anabolic homeostasis, with a loss of matrix‐related transcription factors (TFs) such as Sry‐Box TF 9 (SOX9), and proteins, for example, aggrecan and collagen type II. 2 , 3 Concurrently there is an escalation in levels of matrix degrading enzymes, namely members of the matrix metalloproteinases (MMP) and a disintegrin and metalloprotease with thrombospondin motifs (ADAMTS) families. 4 , 5 , 6 , 7 , 8 Treatments for LBP are primarily conservative in nature, involving physiotherapy and pain management, progressing to spinal surgeries when degeneration increases in severity. 9 Surgeries such as discectomy and spinal fusion offer short term relief however this effect is not sustained, 10 indicating a need for more effective therapeutic options.
With this in mind, the introduction of regenerative nucleic acids has gained ground over the last decade, with microRNAs (miRNAs) offering the potential to treat a range of disorders. 11 , 12 , 13 , 14 miRNAs function in the post‐transcriptional regulation of target genes, of which for each miRNA hundreds of targets are applicable, 15 , 16 and this activity results in the stimulation or dampening of protein expression. 17 , 18 , 19 From a regenerative standpoint, we have selected four pro‐discogenic miRNAs that have demonstrated therapeutic potential, both in terms of predicted targets and those experimentally validated in the literature, namely: miRNA‐140‐5p mimic, miRNA‐141‐3p inhibitor, miRNA‐149‐5p mimic, and miRNA‐221‐3p inhibitor.
miRNA‐140‐5p has been shown to have a positive impact on driving chondrogenesis in models of osteoarthritis (OA), with similar key markers as those observed in discogenesis; SOX9, aggrecan, and collagen type II, 20 , 21 and in catabolic enzyme expression, specifically ADAMTS5 and MMP13, 22 , 23 indicating its potential usage in a regenerative capacity in the disc. The presence of miRNA‐141 has been implicated as a driver of catabolic degeneration in human NP cells, stimulating ADAMTS5 and MMP13 expression. 24 , 25 Xu et al. observed that by inhibiting miRNA‐141 there was a return toward healthy levels of collagen II and aggrecan deposition, with decreases in gene expression of ADAMTS and MMP family members. 26 The transduction of rat NP cells cultured in cytokine‐stimulated media with miRNA‐149‐5p restored gene and protein expression of aggrecan and collagen type II toward non‐stimulated, healthy levels. 27 In chondrocytes, a study by Wang et al. found evidence indicating an anti‐apoptotic role and silencing of MMP13, 28 which could aid in the stimulation of NP cells toward a regenerative behavior. Finally, the silencing of miRNA‐221 has been linked to enhancement of a discogenic‐like phenotype for human NP cells in vitro. 29 , 30 Dubbed chondroprotective, the inhibition of this miRNA may function in driving the deposition of ECM‐related markers. Taken together, there is experimental evidence to suggest the applicability of these miRNAs for IVD regeneration.
To exploit this regenerative potential of miRNAs, a method of delivery must be chosen to taxi these miRNAs into the cells. Though efficacious, the viral delivery of nucleic acids has decreased in popularity due to safety concerns and immunogenicity. 31 , 32 , 33 To this end, nonviral cell penetrating peptides (CPPs) that retain internalization capabilities while proving less stimulatory of the immune system are being explored. In this study, two CPPs were identified for investigation, cationic 30mer amphipathic peptide (RALA) and GAG‐binding enhanced transduction (GET) peptide FGF2B‐LK12‐8R (FLR), both of which function by complexing nucleic acids through electrostatic interactions. RALA, an amphipathic cationic peptide, has arginine residues incorporated throughout the structure to enhance the cationic charge and improve membrane transduction and internalization. 34 , 35 , 36 A modified GET peptide, FLR contains a fibroblast growth factor 2 heparin‐binding domain that facilitates homing to the cell membrane conjugated to an amphipathic and cationic peptide sequence. 37 , 38
The overall objective of this work was to explore the regenerative utility of six combined miRNA pairings in vitro and in an ex vivo organ culture model, while simultaneously assessing the suitability of CPPs for miRNA delivery to the NP region.
2. MATERIALS AND METHODS
2.1. Primary NP cell isolation and culture
Rat tissue was obtained from discarded tissue of animals that underwent procedures in the Comparative Medicine Unit (CMU) of Trinity College Dublin following approval by the animal research ethics committee (AREC) of Trinity College Dublin and the Health Products Regulatory Authority (HPRA) in Ireland (Approval – AE19136/P149). NP cells were harvested under sterile conditions from adult, skeletally mature, female Wistar rats (10–20 weeks) immediately following euthanasia, with three tails pooled to represent a donor. Due to the ease of access and high prevalence of use in IVD degeneration studies, 39 caudal discs were employed for all cell culture studies. NP tissue was enzymatically digested simultaneously with pronase (70 U/mL, Millipore, Merck Life Science Ltd., UK) and collagenase type II (400 U/mL, Molecular Probes, Bio‐Sciences, Ireland) for 6 h at 37°C at 10 rpm. Once cells were digested to a single cell level, they were plated in flasks for expansion at a density of 5 × 103 cells/cm3. Cells were expanded in expansion media (XPAN) comprised of low glucose Dulbecco's Modified Eagle Medium (LG DMEM, Sigma‐Aldrich, Merck Life Science Ltd., UK), 2% penicillin/streptomycin (Pen/Strep), and 10% fetal bovine serum (FBS, all Invitrogen, Bio‐Sciences Ltd., Ireland). XPAN culture media was measured within a range of 300–325 mOsm, which correlates to that of the degenerative IVD. 40 For ease of culture, all initial screenings were carried out under normoxic (20% O2) conditions in a humidified atmosphere at 37°C. NP cells were used between passage 1 and 3, as no morphological changes were visually observed.
2.2. 2D miRNA‐vector complex formation
miRNA complexes were generated through electrostatic interactions of FLR (Dixon Laboratory, University of Nottingham, UK) at predefined nitrogen:phosphate (N:P) charge ratios of 5, 37 , 41 and RALA (GenScript, Netherlands) at an N:P ratio of 10, 42 , 43 for miRNA mimics‐140‐5p and 149‐5p, and 141‐3p and 221‐3p inhibitors (all miRNAs were purchased from Horizon Discovery, UK). For assessment of dual miRNA delivery, the following miRNA pairings were formed: miRNA‐140‐5p mimic + 141‐3p inhibitor, 140‐5p + 149‐5p mimics, 140‐5p mimic + 221‐3p inhibitor, 141‐3p inhibitor + 149‐5p mimic, 141‐3p + 221‐3p inhibitors, and 149‐5p mimic + 221‐3p inhibitor. For all monolayer experiments an miRNA concentration of 10 ng/μL was used, which was previously optimized in our lab.
2.3. Cell transfections in monolayer
For immunofluorescence of protein markers cells were seeded at a density of 1 × 104 in 18‐well μ‐well slides (Ibidi GMBH, Germany) and allowed to attach for 24 h prior to transfection. For biochemical analysis of aggrecan and collagen production, NP cells were seeded at 1 × 105 in 6‐well plates, again allowing 24 h for attachment. In all cases, complexes were formed for 30 min at room temperature (RT) as described above in serum free OptiMEM (Invitrogen, Bio‐Sciences Ltd., Ireland). Complexes were added to cells following a phosphate buffered saline (PBS) wash and transfection allowed to proceed for 5–6 h at 37°C. At the end of incubation, complexes were removed, and cells cultured for 3 days for immunofluorescence analysis, and 14 days for biochemical analysis with media changes performed biweekly where appropriate. For all transfection experiments a non‐transfected (NT) control receiving OptiMEM alone was utilized. Preliminary transfection efficiency had previously been shown in our lab by flow cytometry as being above 90% for RALA and FLR at a dose of 10 ng/μL, with viability 24 h post‐transfection above 80% as assessed by Calcein staining. To ensure transfection was not impacting on ECM protein production, a negative mimic control (Horizon Discovery, UK) was employed as per manufacturer's instructions. For monolayer cultures, between three and five independent biological replicates were performed, as indicated in the figure legends. Table 1 outlines the specific cell culture experimental set‐up for each experiment.
TABLE 1.
Experimental outline of nucleus pulposus cell monolayer experiments.
| Assay | Single/dual miRNA | Time point (days) | Plate size | Seeding density | Cytokine insult | Output |
|---|---|---|---|---|---|---|
| Immunofluorescence | Single | 3 | 18‐well μ‐well slide (Ibidi) | 1 × 104 | No | Protein: aggrecan, collagen type II |
| Immunofluorescence | Dual | 3 | 18‐well μ‐well slide (Ibidi) | 1 × 104 | Yes | Protein: aggrecan, collagen type II |
| Immunofluorescence | Dual | 3 | 18‐well μ‐well slide (Ibidi) | 1 × 104 | Yes | Protein: ADAMTS5, MMP13 |
| Biochemical quantification | Dual | 14 | 6‐well | 1 × 105 | Yes | DNA, sGAGs, collagen |
Abbreviations: miRNA, microRNA; sGAGs, sulfated glycosaminoglycans.
2.4. Pro‐inflammatory cytokine challenge
To better represent physiological conditions a cytokine challenge was employed for assessing dual miRNA transfections; 24 hours post‐seeding, cells were starved of serum for 24 h and subsequently insulted with TNF‐α (50 ng/mL) and IL‐1β (10 ng/mL) for a further 24 h as previously described. 44 , 45 After 24 h of incubation, cytokine media was removed, cells were washed with PBS, and miRNA‐vector complexes were incubated as previously described.
2.5. Immunohistochemistry of monolayer cultures
Three days post‐transfection, cells were washed with PBS and fixed in 4% paraformaldehyde (PFA) for 12 min, cell membranes were permeabilized in 0.1% Triton X‐100 for 20 min, and nonspecific binding was blocked with 1% bovine serum albumin in PBS for 30 min (BSA, all Sigma‐Aldrich, Merck Life Science Ltd., Ireland). Primary antibody solutions: rabbit‐anti‐collagen II (PA599159), rabbit‐anti‐aggrecan (MA532695), rabbit‐anti‐ADAMTS5 (PA532142), and mouse‐anti‐MMP13 (MA514247, all Invitrogen, Bio‐Sciences Ltd., Ireland), were incubated overnight at 4°C. A concentration‐matched IgG control was used for all staining. Species‐matched AlexaFluor secondary antibodies: goat anti‐rabbit 488 (ab150077, Abcam, UK) and goat anti‐mouse 488 (A11001, Invitrogen, Bio‐Sciences Ltd., Ireland) were incubated for 1.5 h, with nuclear counterstaining with Hoechst Bisbenzimide 33258 (Sigma‐Aldrich, Merck Life Sciences Ltd., UK). Images for each independent biological replicate were captured in triplicate on a Leica SP8 scanning confocal microscope (Lecia Microsystems, Germany), with analysis of protein expression quantified using a custom CellProfiler pipeline (CellProfiler 4.2.1), which measures the signal intensity and normalizes per cell number. Fluorescence intensity was normalized to NT control to determine the degree of fold change between groups, with three to five independent biological replicates performed.
2.6. Quantitative biochemical analysis
Cells were digested for 18 h at 60°C in 350 μL 100 mM sodium phosphate/5 mM Na2EDTA buffer, with 3.88 U/mL papain enzyme and 5 mM l‐cysteine (both Sigma‐Aldrich, Merck Life Science Ltd., Ireland). Media samples were pooled and stored at −80°C prior to lyophilization following a standard freeze‐drying protocol (500 mTorr, −10°C, 16–18 h, Harvest Right™, USA) and incubated in activated papain solution as above. DNA content was assessed using Hoechst bisbenzimide 33258 dye assay (DNA QF Kit, Sigma‐Aldrich, Merck Life Science Ltd., Ireland) with a calf thymus DNA standard. Determination of sulfated glycosaminoglycans (sGAGs) was carried out using the dimethylmethylene blue binding assay (DMMB) (Blyscan, Biocolor Ltd., Northern Ireland) with a chondroitin sulfate (CS) standard. Total collagen content was assessed through hydroxyproline measurement, with samples hydrolyzed in concentrated hydrochloric acid (38% HCl) at 110°C for 18 h. A chloramine‐T assay was performed and collagen content was estimated using a hydroxyproline:collagen ratio of 1:7.69. 46 All biochemical measurements were normalized to the NT control, with five independent biological replicates performed.
2.7. Rat ex vivo organ culture
The top three caudal discs were isolated from rat tails, removing the skin and tendons to allow for easier access to the disc section. To avoid swelling of the NP, the CEP and vertebral elements were left intact. Following successive washes in povidone iodine (Duggan Veterinary Supplies Ltd., Ireland) and 70% industrial methylated spirit (IMS) the discs were incubated in LG DMEM, supplemented with 2% Pen/Strep, 0.2% Primocin (InvivoGen), and 0.25 μg/mL Amphotericin B (Sigma‐Aldrich, Merck Life Sciences Ltd., Ireland) for 24 h, before culture for 7 days in XPAN at physioxic (5% O2) to more accurately simulate the native IVD. At day 7, discs were injected with 2 μL of 0.025 U chondroitinase ABC (cABC, Sigma‐Aldrich, Merck Life Sciences Ltd., Ireland) using a 30G needle and degeneration allowed to progress for 7 days. Complexes of the miRNA combinations were formed for 30 min at a concentration of 25 ng/μL and 2 μL was delivered directly into the NP region. Fourteen days post‐transfection the discs were fixed overnight in 4% PFA and decalcified in decalcification solution‐lite (Sigma‐Aldrich, Merck Life Sciences, Ireland) until soft (~7 days). Each experiment was performed independently with three biological replicates. NP cell viability was assessed by live/dead staining with Calcein AM (Biotium) and ethidium homodimer (Sigma‐Aldrich, Merck Life Sciences Ltd., Ireland) for four independent biological replicates and imaged on a Leica SP8 scanning confocal microscope (Leica Microsystems, Germany).
2.8. Histology and immunocytochemistry
Following fixation, discs were dehydrated sequentially in increasing ethanol solutions and wax embedded. Disc sections were sliced in the sagittal orientation to a 10 μm thickness on a microtome (Leica Microsystems, Germany) with staining carried out for picrosirius red to demonstrate collagen deposition, and alcian blue to detect the presence of sGAGs. Imaging was performed on an Aperio Scanscope (Leica Microsystems, Germany). For immunocytochemistry, antigen retrieval was carried out using 35 U/mL pronase at 37°C for 30 min, followed by membrane permeabilization with 0.1% Triton X‐100 and blocking for 1 h with 5% BSA in PBS. Primary antibody solutions for rabbit‐anti‐SOX9, rabbit‐anti‐ADAMTS5 (PA532142, Invitrogen, Bio‐Sciences Ltd., Ireland), and mouse‐anti‐MMP13 (MA514247, Invitrogen, Bio‐Sciences Ltd., Ireland) were prepared in 1% BSA in PBS, with a concentration‐matched IgG control, and incubated overnight at 4°C. Species‐matched AlexaFluor secondary antibodies: goat anti‐rabbit 488 (ab150077, Abcam, UK) and goat anti‐mouse 488 (A11001, Invitrogen, Bio‐Sciences Ltd., Ireland) were incubated for 1.5 h at RT, with nuclear counterstaining using Hoechst bisbenzimide 33258 (Sigma‐Aldrich, Merck Life Sciences, Ireland). Samples were mounted with Fluoromount Aqueous Mounting Media (Sigma‐Aldrich, Merck Life Sciences, Ireland) and imaged on a Leica SP8 scanning confocal microscope. To assess protein expression, regions of interest (ROIs) of equal size were captured and fluorescence measured. The corrected total cell fluorescence was then determined as follows: integrated density – (area of selected cell mean × mean fluorescence of background readings). All read outs were normalized to the NT control to express fold changes of protein expression.
2.9. Bioinformatics analysis
To further investigate the roles of miRNA‐149‐5p mimic and miRNA‐221‐3p inhibitor in the degeneration of the IVD, strongly evidenced miRNA–gene interactions (those identified through reporter assays, Western blotting, or quantitative polymerase chain reaction (qPCR)) were determined using the online repositories miRNATarBase, 47 and MIENTURNET. 48 A miRNA‐gene target list was achieved with a cut‐off of p < 0.05. The lists for both miRNAs were visualized in Cytoscape, Version 3.10, 49 and enriched utilizing the Gene Ontology Biological Process Tool, removing redundant results. Relevant pathways were isolated, and protein–protein interaction (PPI) networks were visualized through the Search Tool for the Retrieval of Interacting Genes (STRING) database, 50 , 51 employing a 0.8 confidence score cut‐off. Nodes were displayed as being larger with higher levels of expression in the extracellular space, given the importance of ECM production in the IVD. The datasets obtained from miRTarBase and MIENTURNET are available Data S1.
2.10. Statistical analysis
Statistical analysis was carried out using GraphPad Prism (version 9.4.1) with a minimum of three biological replicates analyzed for each experimental group. For analysis between groups of three or more, a one‐way ANOVA with Tukey's post‐hoc test was performed. p‐values below 0.05 were considered statistically significant. Data is presented as mean ± standard error of mean (SEM).
3. RESULTS
3.1. Delivery of individual miRNAs demonstrated favorable influence on the modulation of ECM proteins in monolayer culture
As identified from literature, the four selected miRNAs, miRNA mimics 140‐5p and 149‐5p, and inhibitors of miRNAs 141‐3p and 221‐3p, were delivered individually to NP cells in monolayer by both RALA and FLR vectors, and their effect on regenerative ECM markers aggrecan and collagen II was assessed. Immunofluorescence quantification demonstrated transfection with miRNA‐140 mimic significantly enhanced aggrecan deposition following transfection with FLR (p = 0.015), and collagen II protein production was significantly increased by RALA delivery (p = 0.014), with a positive trend for FLR (p = 0.081) (Figure 1A,B). Some increases were also exhibited for miRNA‐149 mimic (Figure 1C,D) and miRNA‐221‐inhibitor (Figure 1E,F), although these were not found to be statistically significant. RALA‐miRNA‐221‐3p inhibitor stimulated significant aggrecan expression (p = 0.046), with collagen II showing increases (Figure 1G,H). Generally, improvements to matrix protein expression were approximately 1.5–2‐fold range in comparison to the NT control.
FIGURE 1.
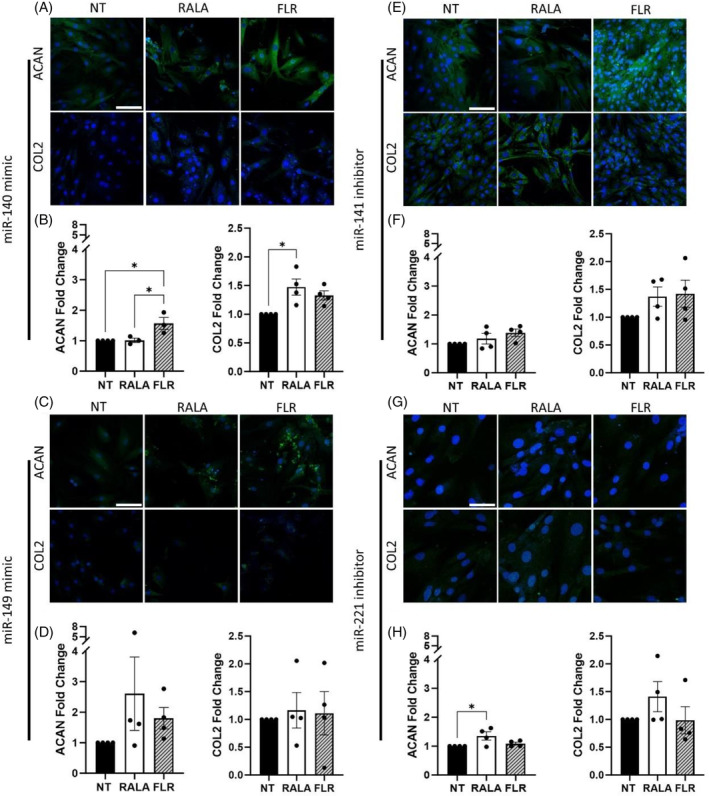
Transfection of rat nucleus pulposus cells with individual miRNA mimics and inhibitors with cell penetrating peptides demonstrated positive trends toward increased matrix production in monolayer culture. (A and B) miRNA 140 mimic transfection significantly increased aggrecan (ACAN) and collagen type II (COL2) protein expression. (C and D) 149 mimic, (E and F) 141 inhibitor, and (G and H) 221 inhibitor demonstrated instances of increased protein expression of ACAN and COL2 3 days post‐transfection. * (p < 0.05) indicates statistical significance, N = 3–4. Scale bars are 100 μm. miRNA, microRNA.
3.2. Transfection of therapeutic miRNA pairings enhanced the deposition of matrix proteins in 2D monolayer cultures
While some upward trends were identified from individual miRNA delivery, we next explored if a combinatorial approach may amplify the regenerative response while also dampening the catabolic cascade associated with degeneration. Here, all groups, including NT controls were stimulated with pro‐inflammatory cytokines to induce catabolic enzyme production in order to evaluate how miRNA pairings could influence the promotion of matrix proteins and the suppression of degradative catabolic factors. With regards to aggrecan production, miRNA‐140 mimic + miRNA‐141 inhibitor markedly improved protein expression (RALA‐group p = 0.002, FLR‐group p = 0.045), with RALA‐miRNA inhibitors 141 + 221 demonstrating similar promise (p = 0.046, Figure 2A,B). FLR‐miRNA mimics 140 + 149 approached significance (p = 0.051) for aggrecan deposition. Among the remaining three combined groups, increases approaching a 4‐fold increase were observed, in comparison to approaching 1.5–2‐fold with single miRNA transfections, signifying the enhanced potential of dual miRNA delivery. Immunofluorescence analysis of collagen displayed less altered protein expression, though increases approached a mean 5‐fold increase for a number of groups, such as miRNA mimics 140 + 149, miRNA‐141 inhibitor + miRNA‐149 mimic, miRNA inhibitors 141 + 221, and miRNA‐149 mimic and 221 inhibitors (Figure 2C,D), again this was enhanced compared to the delivery of individual miRNAs (Figure 1A–H).
FIGURE 2.
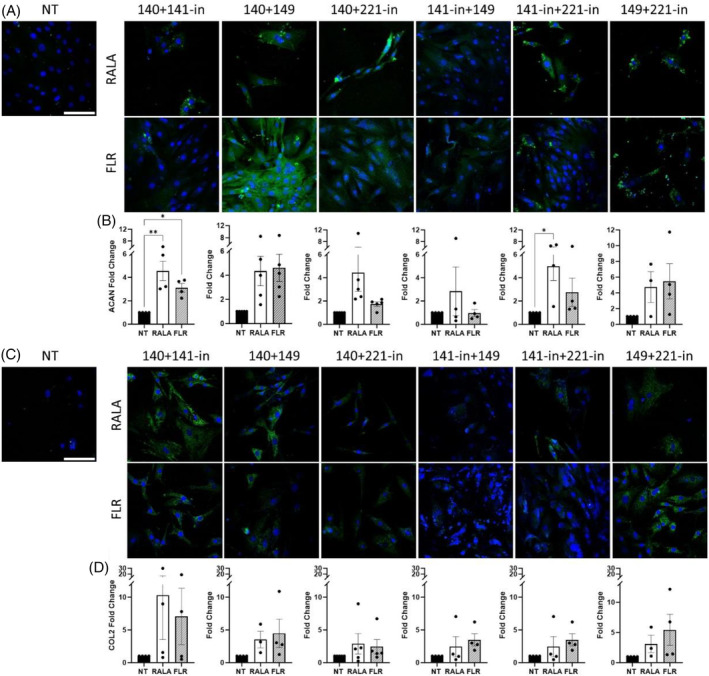
Delivery of miRNAs pairings with cell penetrating peptide‐mediated delivery increased extracellular matrix protein deposition in rat nucleus pulposus monolayer culture. (A and B) Immunofluorescence analysis 3 days post‐transfection exhibited increases in aggrecan (ACAN) expression for miRNA‐140‐mimic + miRNA‐141‐inhibitor, and miRNA‐141‐inhibitor + miRNA‐221‐inhibitor. Deposition of aggrecan demonstrated some positive trends toward increased deposition after 14 days in monolayer culture. (C and D) Collagen type II (COL2) protein expression demonstrated similar nonsignificant increases in production at 3 days post‐transfection. * (p < 0.05) and ** (p < 0.01) indicate statistical significance, N = 4–5. Scale bars are 100 μm. miRNA, microRNA; NT, non‐transfected.
Upon ascertaining that delivery of a negative control scramble miRNA did not significantly alter DNA, sGAG, or collagen content (Figure S1), biochemical analysis was performed on each miRNA pairing. No significant differences were found between the NT control and transfected groups (Figure S2A). Collagen deposition in 2D culture demonstrated limited changes in comparison to NT groups (Figure S2B), potentially representing the unsuitability of monolayer culture systems for identification of ECM factors and their modulation. Values of the samples prior to normalization are displayed in Figure S3.
3.3. Impact of combined vector‐miRNA delivery on catabolic enzyme expression in monolayer culture
As mentioned, cytokine stimulation was included for all groups to drive catabolic enzymes such as ADAMTS5 and MMP13 and screen the prospective downregulation of tandem miRNA delivery. RALA‐mediated delivery substantially increased ADAMTS5 above that of the NT and/or FLR groups, in the case of miRNA groupings 140 + 149 mimics, miRNA‐140 mimic + miRNA‐221 inhibitor, miRNA inhibitors 141 + 221, and miRNA‐149 mimic + 221 inhibitor (Figure 3A,B). Similar increases were identified for MMP13 in particular for miRNA‐140 mimic + miRNA‐141 inhibitor, and miRNA mimics 140 + 149 (p = 0.069), and to a greater extent in the RALA groups (Figure 3C,D). Some nonsignificant subtle decreases for FLR groups miRNA‐141 inhibitor + 149 mimic, and miRNA inhibitors 141 and miRNA‐221 were also observed.
FIGURE 3.
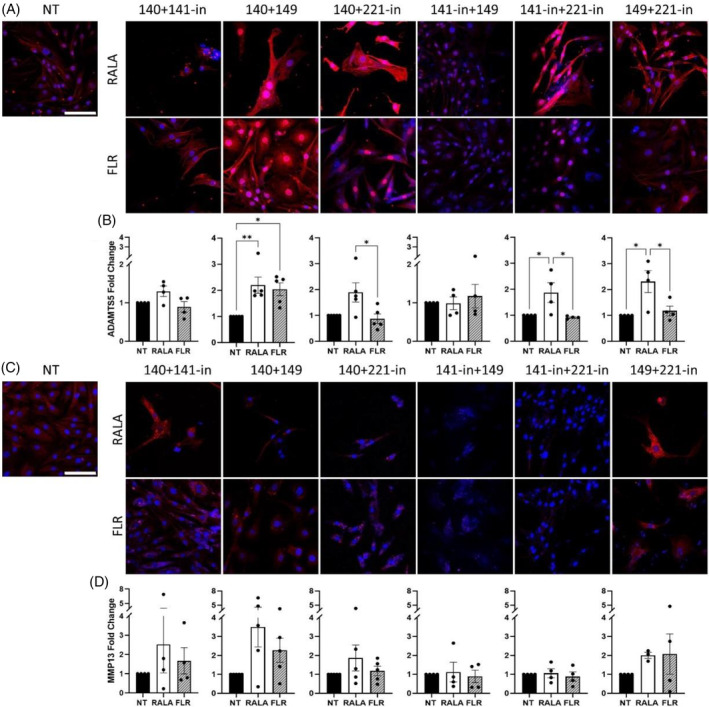
Effect of vector‐miRNA pairings on the production of catabolic enzymes in rat nucleus pulposus monolayer culture. (A and B) ADAMTS5 expression 3 days post‐transfection was significantly increased for miRNA‐140 + miRNA‐149 mimics, miRNA‐140 mimic + miRNA‐221‐inhibitor, miRNA‐141‐inhibitor + miRNA‐221‐inhibitor, and miRNA‐149 mimic + miRNA‐221‐inhibitor in comparison to non‐transfected (NT) and FLR‐transfected cells. (C and D) MMP13 protein expression showed some instances of increases though none to a significant degree. *p < 0.05 and **p < 0.01 indicate statistical significance, N = 4–5. Scale bars are 100 μm. miRNA, microRNA.
3.4. Vector‐miRNA pairings fostered a regenerative niche in rat ex vivo organ culture
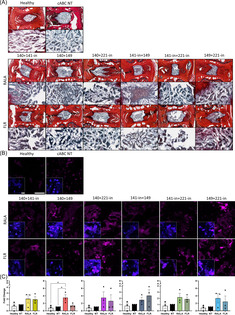
Though useful in initial screenings, in vitro studies do not robustly represent a physiological environment. To this end, a mildly degenerated model was established in ex vivo rat caudal organ cultures to evaluate the effects of tandem vector‐miRNA treatments. NP viability following 21 days in culture was found to be 48.32% for the healthy control, 47.51% for the NT control, and 49.16% for the vector‐miRNA‐pair treatment, with no significant differences between groups (Figure S4). Histological appraisal of sGAG presence revealed marked increases in a number of the groups, such as RALA‐miRNA‐140 mimic + miRNA‐221 inhibitor and miRNA‐141 + miRNA‐221 inhibitor. Transfection using FLR led to increased sGAG deposition for miRNA‐140 mimic + miRNA‐141 inhibitor, and miRNA‐140 + 149 mimics. Overall, the strongest staining was shown for miRNA‐149 mimic + miRNA‐221 inhibitor, with the FLR group supporting the largest increase in sGAG staining (Figure 4A). A significant increase in the key TF SOX9 was observed following RALA‐miRNA‐140 + 149 mimics delivery in comparison to both the healthy and NT control, with evidence of increasing expression in the other groups for both vector‐mediated transfections (Figure 4B,C).
FIGURE 4.
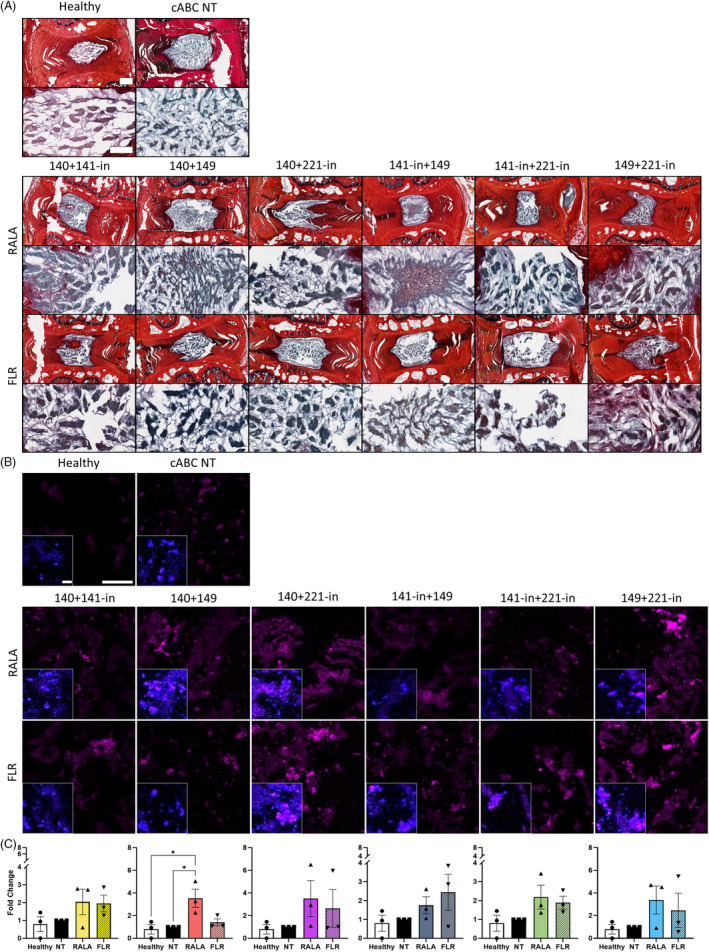
Histological staining of miRNA pairings delivered with cell penetrating peptides (RALA and FLR) in ex vivo rat caudal organ cultures 14 days post‐transfection demonstrated instances of increases to glycosaminoglycan (GAG) and transcription factor SOX9. (A) Collagen was detected with picrosirius red staining (red), and (GAG) presence with alcian blue (blue/purple). There is evidence of increased GAG deposition in comparison to the controls in groups 140 + 141‐in, 140 + 149, and 141‐in + 149, with more intense GAG staining observed for FLR 149 + 221‐in. (B) Antibody labeling of SOX9 (magenta) revealed elevated expression levels across the majority of groups with a significant upregulation after transfection with RALA‐miRNA‐140 + 149. (C) Semi‐quantitative analysis of SOX9 expression. * (p < 0.05) indicates statistical significance; N = 3 independent biological replicates. Healthy controls did not receive cABC treatment, with non‐transfected (NT) controls degenerated with cABC and injected with a sham PBS injection. Scale bar for histology images of whole discs is 500 μm, and images of GAG staining are 150 μm. Scale bar for fluorescence images is 100 μm. Scale bar for insets with nuclear (blue) staining are 20 μm. miRNA, microRNA; PBS, phosphate buffered saline.
In addition to increases in sGAGs, an encouraging suppression of key ECM‐degrading proteins was evidenced with immunofluorescence imaging in the FLR‐miRNA‐149 mimic + miRNA‐221 inhibitor group. A significant decrease in ADAMTS5 (p = 0.036) protein expression was identified (Figure 5A,B).
FIGURE 5.
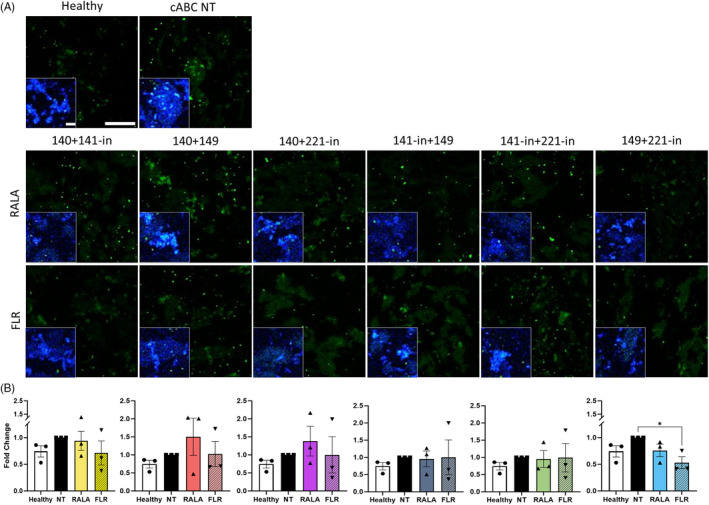
Combined delivery of FLR‐miRNA‐149 mimic and miRNA‐221‐inhibitor markedly decreased protein expression of the catabolic enzyme ADAMTS5 in rat caudal organ cultures 14 days post‐transfection. (A) Immunolabeling for ADAMTS5 (green) demonstrated significantly reduced expression following transfection of miRNA‐149 mimic and miRNA‐221‐inhibitor using FLR, lower than that of the non‐transfected (NT) control. (B) * (p < 0.05) indicates statistical significance; N = 3 independent biological replicates. Scale bar is 100 μm. Scale bar for insets with nuclear (blue) staining are 20 μm. miRNA, microRNA.
The reduction in ADAMTS5 protein release was concomitant with a marked reduction in MMP13 (p = 0.001) (Figure 6A,B). In both cases, the fold change were equivalent to expression in the healthy, non‐degenerated discs. Interestingly, mirroring somewhat what was observed in monolayer, RALA‐miRNA‐140 mimic + miRNA‐221 inhibitor elevated MMP13 expression and was found to be significant (p = 0.022).
FIGURE 6.
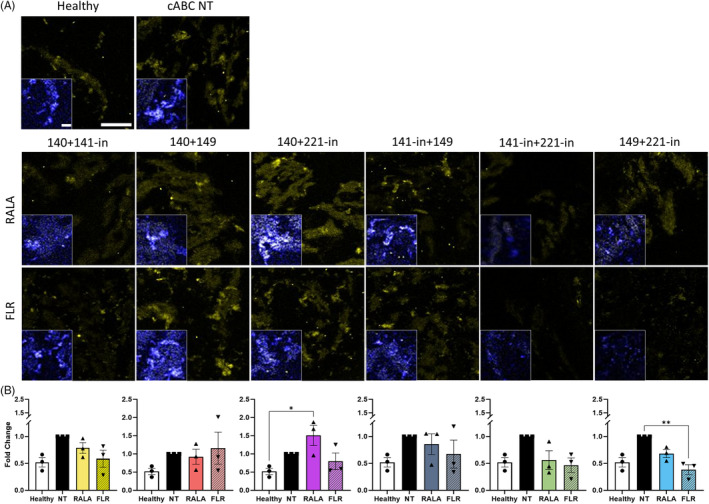
FLR‐miRNA‐149 mimic and miRNA‐221‐inhibitor combined delivery significantly reduced protein expression of the catabolic enzyme MMP13 in rat caudal ex vivo organ culture explants 14 days post‐transfection. (A) Immunofluorescence detected significantly reduced MMP13 (yellow) expression following transfection of FLR‐miRNA‐149 mimic and miRNA‐221‐inhibitor in comparison to the non‐transfected (NT) control. (B) RALA‐mediated miRNA‐140 mimic and miRNA‐221‐inhibitor transfection significantly increased MMP13 production. * (p < 0.05) and ** (p < 0.01) indicate statistical significance; N = 3 independent biological replicates. Scale bar is 100 μm. Scale bar for insets with nuclear (blue) staining are 20 μm. miRNA, microRNA.
3.5. Enrichment analysis identified distinct pathways and targets that interact with miRNA‐149‐5p mimic and miRNA‐221‐3p inhibitor
To further investigate the mechanism behind miRNA‐149‐5p mimic and miRNA‐221‐3p inhibitor in the downregulation of catabolic factors and stimulation of anabolic factors in the IVD, bioinformatic tools were utilized. A total of 54 strongly validated enriched targets were identified for the two miRNAs. miRNA‐149‐5p mimic was significantly implicated in a number of relevant pathways, including Response to Lipopolysaccharide (LPS), Execution Phase of Apoptosis, Response to Peptidoglycan, and Regulation of Signal Transduction (Figure 7A–D). A table of all gene name abbreviations is provided in Table S1.
FIGURE 7.
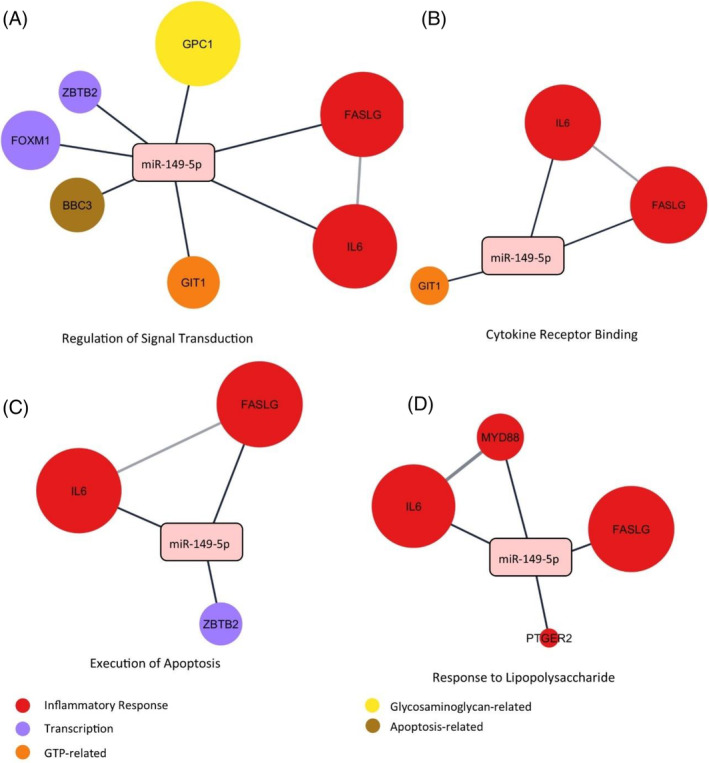
Gene ontology (GO) pathway enrichment analysis of miRNA‐149‐5p and experimentally validated protein targets. (A) Regulation of Signal Transduction. (B) Cytokine Receptor Binding. (C) Execution of Apoptosis. (D) Response to Lipopolysaccharide. Expression levels within the extracellular compartment are visually presented, where larger nodes correspond to a greater degree of expression within the extracellular space. Nodes are connected if an interaction is known to occur. Network constructed in Cytoscape Version 3.10. GTP, guanosine triphosphate; miRNA, microRNA.
Conversely, miRNA‐221‐3p was enriched for pathways that relate to IDD, including Response to Hypoxia, Signal Transduction, Response to Cytokine, and Regulation of Protein Serine/Threonine Kinase Activity (Figure 8A–D). Interestingly, except for BCL2 Binding Component 3 (BBC3), there was no overlap in the target protein interactions, with both miRNAs having distinct targets and pathways with which they interact. A full list of abbreviated gene terms is provided in Table S2.
FIGURE 8.
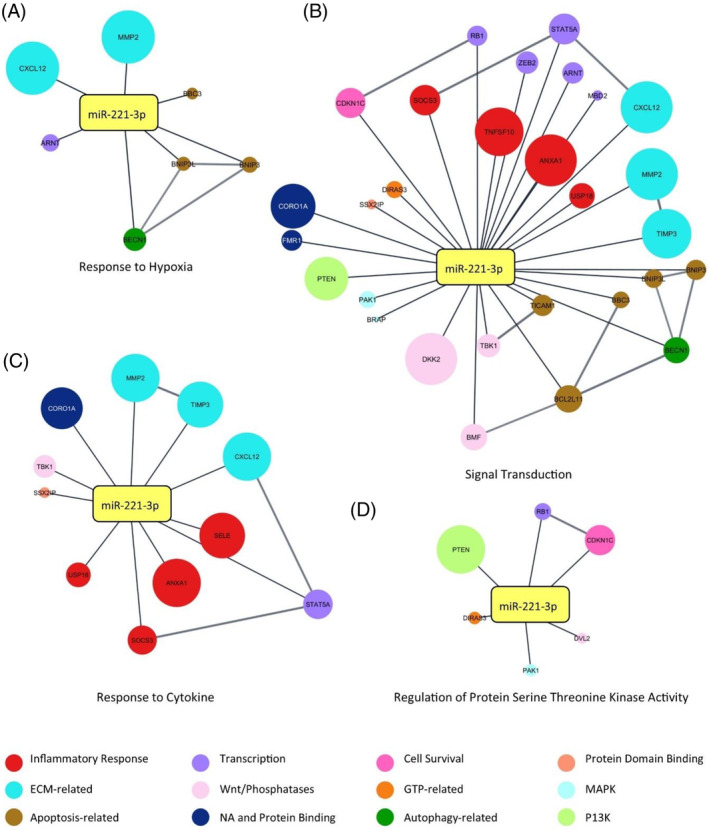
Gene ontology (GO) pathway enrichment analysis of miRNA‐221‐3p and experimentally validated protein targets. (A) Response to Hypoxia. (B) Signal Transduction. (C) Response to Cytokine. (D) Regulation of Protein Serine Threonine Kinase Activity. Expression levels within the extracellular compartment are visually presented, where larger nodes correspond to greater degree of expression within the extracellular space. Nodes are connected if an interaction is known to occur. Network constructed in Cytoscape Version 3.10. GTP, guanosine triphosphate; MAPK, mitogen‐activated protein kinases; NA, nucleic acid; P13K, phosphoinositide 3‐kinase.
4. DISCUSSION
The introduction of nonvirally delivered nucleic acids is an attractive therapeutic avenue, whereby key proteins and pathways can be regulated in a manner that drives regeneration or suppresses catabolic factors. For IVD treatment, this work highlights the potential of dual miRNA delivery when combined with CPPs and demonstrated that FLR‐miRNA‐149‐5p mimic + miRNA‐221‐3p inhibitor stimulated the production of sGAGs and ECM proteins, while encouraging the downregulation of matrix degrading enzymes (ADAMTS5 and MMP3). Although there were promising results observed in monolayer culture, the adoption of an ex vivo organ culture model underscored the significance of using biologically relevant models. In this model, a notable reduction in catabolic enzyme activity was observed, further emphasizing its relevance and potential.
Within our monolayer investigations, we identified increases in aggrecan and type II collagen protein expression above 4‐fold in our miRNA pair groupings, suggesting that dual‐transfection is amplifying the regenerative response. In vitro protein expression increases following miR delivery above 1.5‐fold of both aggrecan, and collagen type II have been linked to improvements in disc height index (DHI), GAG production, and restoration of the catabolic:anabolic balance in rodent models. 25 , 52 The potential and efficacy of dual miRNA pairings has also been observed in other fields. In the context of diabetic wounds, Wang et al. identified that while transfection using miRNAs‐129 and ‐335 individually promoted healing, when delivered in tandem this effect was strengthened. 53 In a similar manner, it was identified that dual delivery of let‐7b and miRNA‐34a reduced the proliferation of non‐small cell lung cancer lines to a greater degree than single delivery alone, subsequently reducing tumor size. 54 Building on a 2009 study, 55 Mariner et al. differentiated human MSCs toward an osteogenic lineage with co‐delivery of miRNA‐148b mimic and miRNA‐489 inhibitor, to a greater extent than individual delivery. 56 Loading of collagen‐nanohydroxyapatite scaffolds with a combination of miRNA‐210 mimic and miRNA‐16 inhibitor resulted in significantly increased vascularization and bone density in a rat calvarial defect model, further indicating the potential of dual miRNA delivery. 57 These examples offer insight into the potential strengthening of positive stimuli with the correct combination of miRNAs and their cooperative impact.
In relation to the expression of catabolic proteins in monolayer, ADAMTS5 expression was, for the majority of the miRNA‐pair groups, enhanced in the RALA groups, with FLR‐transfected combinations at times being significantly reduced in comparison. A similar, though not as pronounced effect was seen for MMP13. The FLR CPP was designed to include a GAG‐binding peptide which fosters interaction with the lipid bilayer of the target cell and enhances cellular uptake of the given cargo. 58 Potentially, the homing effect of FLR to GAG‐rich domains, which is abundant in the NP, facilitated targeted delivery of the miRNA pairings, resulting in more efficient downregulation of ADAMTS5 and MMP13. The monolayer parameters of these screening experiments should be taken into consideration however, and the inherent lack of a protective ECM. 59 These conditions may have exacerbated the effect of the cytokine challenge, whereby any potential anabolic effect of the vector‐miRNA pairings may have been reduced. The variability identified may lie in the inherent inter‐donor differences, with rats being from different donors and preparations. Added to this, cells used for in vitro studies often lose their circadian synchronicity, which may impact upon their responses to treatments, 60 , 61 prompting further investigations in a more relevant ex vivo organ culture model. As a limitation to our 2D screenings, the implementation of normoxic culture conditions may have impacted upon matrix production and influenced the phenotype of NP cells; however, for our ex vivo organ culture experiments we adopted physioxic oxygen levels.
Similar to our 2D experiments, when examining ex vivo organ cultures, we observed elevated deposition of ADAMTS5 and MMP13 in the RALA groups compared to the FLR groups. For most FLR‐miRNA pairings, a decrease in enzyme production was observed, which was comparable to the levels seen in the non‐degenerated, healthy control group. However, the strongest decreases for both ADAMTS5 and MMP3 were identified in the FLR‐miRNA‐149‐5p mimic + miRNA‐221‐3p group. We also identified increases in the expression of the key TF SOX9 in all groups, with the strongest degree of sGAG deposition observed histologically in the FLR‐miRNA‐149‐5p mimic + miRNA‐221‐3p inhibitor group. Deemed an anti‐chondrogenic miRNA, the inhibition of miRNA‐221‐3p has been shown to promote chondrogenesis through stimulation of aggrecan and collagen type II production. 62 , 63 Recently, the application of acetone extracts from Violina pumpkin leaves to IVD cells demonstrated the suppression of miRNA‐221 expression and promoted the restoration of a healthy disc phenotype. This was notably characterized by a significant increase in the levels of aggrecan, collagen type II, and SOX9, in conjunction with enhanced resilience to damage induced by reactive oxygen species (ROS). 64 miRNA‐149‐5p was identified as a promoter of discogenic behavior under LPS‐stimulated culture conditions. 27 Its role in positively regulating the inflammatory response has been extensively documented in disease models of OA. 65 , 66 Elevated levels of pro‐inflammatory cytokines, specifically IL‐1β and TNF‐α, are observed in degenerated discs, exacerbating the catabolic cascade. 67 , 68 , 69 , 70 To further elucidate the potential of FLR‐miRNA‐149‐5p mimic + miRNA‐221‐3p inhibitor, future investigations will integrate anti‐inflammatory mechanisms and their role in IVD degeneration. While beyond the scope of this work, future work may also benefit from the inclusion of NP‐specific markers, such as members of the Cytokeratin family, Brachyury T, and CD24.
The enhanced modulation of regenerative outcomes with dual miRNA delivery may be explained by the targeting of distinct sets of pathways by both miRNA‐149‐5p mimic + miRNA‐221‐3p inhibitor. Through the analysis of enriched and experimentally validated miRNA–target interactions, various pathways and targets of both miRNA‐149‐5p mimic and miRNA‐221‐3p inhibitor were identified. miRNA‐149‐5p was primarily associated with a role in inflammatory processes, such as validated interactions with interleukin‐6 (IL‐6), Fas Ligand (FASLG), and myeloid differentiation primary response protein 88 (MyD88), all of which were highly expressed in the extracellular compartment. A meta‐analysis conducted by Deng et al. demonstrated a significant increase in IL‐6 protein expression in patients with IDD compared to controls. 71 Additionally, a separate study established a correlation between IL‐6 concentration and LBP. 72 Clinically, the incidence of reported pain and higher levels of IL‐6 has been identified, 73 suggesting the importance of this cytokine in the degenerative cascade involved in IDD and the associated clinical relevance. As a member of the TNF family, polymorphisms in FASLG have been associated with degeneration of the IVD, 74 with FASLG driving NP cell apoptosis through the binding of FAS and recruitment of pro‐inflammatory cytokines. 75 , 76 The toll‐like receptor MyD88 has previously been linked with miRNA‐149 in LPS‐stimulation, whereby its silencing facilitated a dampening of the inflammatory milieu and stimulated a return toward a healthy expression of aggrecan and collagen type II. 27 MyD88 expression has been identified as being driven by pro‐inflammatory cytokines in the disc 77 along with the immune response in cardiovascular and neurological disorders. 78 , 79 Considering the role of inflammation in IDD, the potential of miRNA‐149‐5p to mitigate these effects holds significant promise. Future research endeavors will explore this potential while striving to more accurately replicate the intricate interactions that drive the degenerative cascade and impede regenerative efforts.
As a chondrogenesis‐related miRNA, miRNA‐221‐3p was enriched for ECM‐related pathways, with highly expressed targets in MMP2, Tissue Inhibitor of Metalloproteinase 3 (TIMP3), and C‐X‐C Motif Chemokine Ligand 12 (CXCL12) within the extracellular space. As matrix degrading enzyme promoters and inhibitors, respectively, a dysregulation of members of the MMP and TIMP families exacerbates the catabolic imbalance at the crux of IDD. 7 , 80 MMP2 specifically acts through the degradation and remodeling of collagens, 81 and polymorphisms in the gene have been associated with a statistically significant increase in the risk of developing IDD. 82 TIMP3 is unique among the TIMP family as it can suppress all MMPs, and a range of ADAMTS, including −4 and −5 which are of particular relevance to IDD. 8 , 83 , 84 A recent study identified a suppression of the discogenic pain marker Substance P following stimulation with TIMP3 in a rodent model, possibly through a reduction in vascularization. 85 The upregulation of TIMP3 could therefore serve a therapeutic role when seeking regenerative therapies. Serum levels of the chemokine CXCL12, also referred to as stromal cell‐derived factor 1 (SDF‐1), have been found to be elevated with more severe levels of IVD degeneration and associated pain. 86 A connection has been identified between a dose‐dependent effect of CXCL12 upon MMP2 protein expression in cartilage endplate (CEP) cells; however, when delivered directly to the NP region in a rat tail model facilitated an increase in proteoglycan deposition, possibly via a stem cell homing mechanism. 87 Contrary to this amelioration of degeneration in the NP, CXCL12 has predominantly been linked to inflammation‐induced apoptosis and matrix destruction. 88 , 89 , 90 Given the potential relationships between TIMP3, MMP2, and CXCL12, their interactions with miRNA‐221‐3p may prove advantageous in restoring regenerative function to the damaged IVD.
Finally, this work highlights the importance of adopting an appropriate and relevant study model, particularly when working within the three‐dimensional constraints of the native ECM. While our monolayer investigations evidenced some trends, more meaningful results were demonstrated in the ex vivo organ culture model. In the context of the IVD, NP cells readily lose their native phenotype when cultured in monolayer. 91 Furthermore, rats belong to the subsection of species that retain an NP progenitor notochordal cell population throughout adulthood. However, previous work has identified the reduction of notochordal porcine cells upon passaging, whereby NP and notochordal cells were indistinguishable from one another. 92 In the rat ex vivo organ culture model, notochordal cells will be present and constitute up to 30% of the NP cell population. However, injury in the form of degeneration diminishes this number. 93 Notochordal cells have been linked to regenerative potential, 94 , 95 and their presence may have aided in some of the positive changes identified following miRNA delivery. Future research will explore miRNA pairs in animal models that experience a decrease in their notochordal cell population during development, thereby providing a more accurate representation of humans. Nonetheless, utilizing the entire organ imparts a more physiologically relevant niche upon which to assess therapeutics, with the native ECM providing paracrine signaling to better replicate the in vivo scenario. 96 Furthermore, ex vivo culture systems better reconstitute the metabolic microenvironment, offering more realistic systems to explore potential therapeutics. 97 The inclusion of a whole organ also offers the potential of employing loading pressure that more closely resembles the native mechanical stimuli experienced, 98 and warrants further investigation.
The reduction shown in pro‐catabolic enzymes possibly signifies the beginning of the creation of a microenvironmental niche with reduced catabolic activity that would facilitate regeneration. The salient biochemical characteristics of the degenerated NP are defined by a destabilization of homeostasis causing reductions in vital discogenic proteins such as aggrecan and type II collagen, stimulation of degradative enzymes, including members of the ADAMTS and MMP families, and an increased inflammatory response. 99 , 100 , 101 The importance of ameliorating this degradative cascade has been well documented. 80 , 102 , 103 , 104 , 105 There could potentially be an inflammatory response to the carrier; however, such a reaction has not previously been reported for RALA or FLR. 36 , 106 Additionally, inflammation at the site of the injection could occur, as has been documented in in vivo rodent models. 107 , 108 The marked reduction in ADAMTST5 and MMP13 identified in the ex vivo organ culture for FLR‐miRNA‐149‐5p mimic + 221‐3p inhibitor may signify a calming of this catabolic environment, creating a niche whereby regenerative effects can proceed, as observed with the increased sGAG deposition. This promising result demonstrates how the delivery of exogenous therapeutic vector‐miRNA pairings can influence and engineer a suitable microenvironment to facilitate the return toward a homeostatic balance.
5. CONCLUSION
Through our screening of six potentially regenerative miRNA pairings delivered by employing CPPs, we identified FLR‐miRNA‐149‐5p mimic + miRNA‐221‐3p inhibitor pairing as being the most suitable for promoting the deposition of sGAGs, and simultaneously dampening of pro‐catabolic factors. A bioinformatics approach to analyze the experimentally validated targets of these two miRNAs demonstrated distinct targets and pathways that may be beneficial for IVD regeneration, with dual delivery amplifying those expected when delivered in isolation. Overall, this work suggests a therapeutic vector‐miRNA pairing that may foster regeneration by the creation of an anti‐catabolic niche within the IVD.
AUTHOR CONTRIBUTIONS
Tara Ní Néill and Conor T. Buckley contributed substantially to the conception and design of the work. Tara Ní Néill performed the acquisition and interpretation of literature, data analysis, presentation and interpretation of results, drafting of the article, revising it critically, and final approval. Marcos N. Barcellona and Niamh Wilson contributed to acquisition of laboratory data. Fergal J. O'Brien, James E. Dixon, and Caroline M. Curtin provided contributions to study design, technical support, and interpretation of data. Conor T. Buckley, as the overall project funding holder, takes responsibility for the integrity of the work from inception to finalized article, and provided substantial contribution to data interpretation and presentation. Tara Ní Néill and Conor T. Buckley drafted the manuscript. All authors critically revised the manuscript and approved the final version.
CONFLICT OF INTEREST STATEMENT
Conor T. Buckley is an Editorial Board member of JOR Spine and co‐author of this article. They were excluded from editorial decision‐making related to the acceptance of this article for publication in the journal. The remaining authors declare no conflicts of interest.
Supporting information
Data S1. Supporting information.
Figure S1. Transfection of scramble negative control miR did not significantly alter the production of DNA, sulfated glycosaminoglycans (sGAGs), or collagen in monolayer. Delivery of a negative control miR showed no significant changes to biochemical quantification 14 days post‐transfection. N = 5; ns, nonsignificant.
Figure S2. Biochemical analysis of rat nucleus pulposus monolayer cultures following vector‐miR pairing delivery demonstrated some positive increases 14 days post‐transfection. (A) Sulfated glycosaminoglycans (sGAG) and (B) collagen (COL). Data is displayed as sGAG or COL normalized to DNA content and the non‐transfected (NT) control. N = 5.
Figure S3. Biochemical quantification of DNA, sulfated glycosaminoglycan (sGAG), and collagen (COL) levels 14 days post‐transfection with RALA and FLR‐delivered miR combinations. N = 5.
Figure S4. Cell viability of rat caudal NP tissue following 21 days of ex vivo organ culture. (A) Representative images of maximum projection of z‐stacks of Calcein AM (green, live) and ethidium homodimer (red, dead) staining. (B) Viability assessment of live and dead cells. Healthy discs were non‐degenerated. Chondroitinase ABC (cABC) NT and FLR‐miRNA‐149 + 221‐in groups were mildly degenerated with cABC for 7 days prior to Sham or miRNA‐pair treatment. Scale bar is 150 μm. N = 4.
Figure S5. Median and worst representative histological images of rat motion segments 14 days following transfection with miRs A. 140‐5p + 141‐3p inhibitor, B. miR‐140‐5p mimic + miR‐149‐5p mimic, C. miR‐140‐5p mimic + miR‐221‐3p inhibitor, D. miRs‐141‐3p inhibitor + 149‐5p mimic, E. miR‐141‐3p inhibitor + miR‐221‐3p inhibitor, and F. miR‐149‐5p + miR‐221‐3p inhibitor following 14 days in culture post‐transfection. Alcian blue for glycosaminoglycan staining, Picrosirius red for collagen staining. Main image scale bar 500 μm, inset 150 μm. N = 3.
Table S1. Gene name abbreviations in Figure 7.
Table S2. Gene name abbreviations in Figure 8.
ACKNOWLEDGMENTS
This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (grant agreement ERC‐2019‐CoG‐864104; INTEGRATE). Open access funding provided by IReL.
Ní Néill T, Barcellona MN, Wilson N, et al. In vitro and ex vivo screening of microRNA combinations with enhanced cell penetrating peptides to stimulate intervertebral disc regeneration. JOR Spine. 2024;7(4):e1366. doi: 10.1002/jsp2.1366
REFERENCES
- 1. Dieleman JL, Baral R, Birger M, et al. US spending on personal health care and public health, 1996–2013. JAMA. 2016;316:2627‐2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kibble MJ, Domingos M, Hoyland JA, Richardson SM. Importance of matrix cues on intervertebral disc development, degeneration, and regeneration. Int J Mol Sci. 2022;23:6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gruber HE, Norton HJ, Ingram JA, Hanley EN Jr. The SOX9 transcription factor in the human disc: decreased immunolocalization with age and disc degeneration. Spine (Phila Pa 1976). 2005;30:625‐630. [DOI] [PubMed] [Google Scholar]
- 4. Buckwalter JA. Aging and degeneration of the human intervertebral disc. Spine (Phila Pa 1976). 1995;20:1307‐1314. [DOI] [PubMed] [Google Scholar]
- 5. Yaltirik CK, Timirci‐Kahraman Ö, Gulec‐Yilmaz S, Ozdogan S, Atalay B, Isbir T. The evaluation of proteoglycan levels and the possible role of ACAN gene (c.6423T>C) variant in patients with lumbar disc degeneration disease. In Vivo. 2019;33:413‐417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pockert AJ, Richardson SM, Le Maitre CL, et al. Modified expression of the ADAMTS enzymes and tissue inhibitor of metalloproteinases 3 during human intervertebral disc degeneration. Arthritis Rheum. 2009;60:482‐491. [DOI] [PubMed] [Google Scholar]
- 7. Vo NV, Hartman RA, Yurube T, Jacobs LJ, Sowa GA, Kang JD. Expression and regulation of metalloproteinases and their inhibitors in intervertebral disc aging and degeneration. Spine J. 2013;13:331‐341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gendron C, Kashiwagi M, Lim NH, et al. Proteolytic activities of human ADAMTS‐5: comparative studies with ADAMTS‐4. J Biol Chem. 2007;282:18294‐18306. [DOI] [PubMed] [Google Scholar]
- 9. Simon J, McAuliffe M, Shamim F, Vuong N, Tahaei A. Discogenic low back pain. Phys Med Rehabil Clin N Am. 2014;25:305‐317. [DOI] [PubMed] [Google Scholar]
- 10. Sherman J, Cauthen J, Schoenberg D, Burns M, Reaven NL, Griffith SL. Economic impact of improving outcomes of lumbar discectomy. Spine J. 2010;10:108‐116. [DOI] [PubMed] [Google Scholar]
- 11. Zheng L, Chen Y, Ye L, et al. miRNA‐584‐3p inhibits gastric cancer progression by repressing Yin Yang 1‐facilitated MMP‐14 expression. Sci Rep. 2017;7:8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hoang DH, Zhao D, Branciamore S, et al. MicroRNA networks in FLT3‐ITD acute myeloid leukemia. Proc Natl Acad Sci. 2022;119:e2112482119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eulalio A, Mano M, Ferro MD, et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376‐381. [DOI] [PubMed] [Google Scholar]
- 14. Gabisonia K, Prosdocimo G, Aquaro GD, et al. MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature. 2019;569:418‐422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kozomara A, Griffiths‐Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68‐D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92‐105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eichhorn SW, Guo H, McGeary SE, et al. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol Cell. 2014;56:104‐115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99‐110. [DOI] [PubMed] [Google Scholar]
- 19. Vasudevan S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip Rev RNA. 2012;3:311‐330. [DOI] [PubMed] [Google Scholar]
- 20. Miyaki S, Nakasa T, Otsuki S, et al. MicroRNA‐140 is expressed in differentiated human articular chondrocytes and modulates interleukin‐1 responses. Arthritis Rheum. 2009;60:2723‐2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang J, Qin S, Yi C, et al. MiR‐140 is co‐expressed with Wwp2‐C transcript and activated by Sox9 to target Sp1 in maintaining the chondrocyte proliferation. FEBS Lett. 2011;585:2992‐2997. [DOI] [PubMed] [Google Scholar]
- 22. Miyaki S, Sato T, Inoue A, et al. MicroRNA‐140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010;24:1173‐1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Si HB, Zeng Y, Liu SY, et al. Intra‐articular injection of microRNA‐140 (miRNA‐140) alleviates osteoarthritis (OA) progression by modulating extracellular matrix (ECM) homeostasis in rats. Osteoarthr Cartil. 2017;25:1698‐1707. [DOI] [PubMed] [Google Scholar]
- 24. Ji ML, Jiang H, Wu F, et al. Precise targeting of miR‐141/200c cluster in chondrocytes attenuates osteoarthritis development. Ann Rheum Dis. 2021;80:356‐366. [DOI] [PubMed] [Google Scholar]
- 25. Ji M‐l, Jiang H, Zhang X‐j, et al. Preclinical development of a microRNA‐based therapy for intervertebral disc degeneration. Nat Commun. 2018;9:5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu Q, Xing H, Wu J, Chen W, Zhang N. miRNA‐141 induced pyroptosis in intervertebral disk degeneration by targeting ROS generation and activating TXNIP/NLRP3 signaling in nucleus pulpous cells. Front Cell Dev Biol. 2020;8:871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Qin C, Lv Y, Zhao H, Yang B, Zhang P. MicroRNA‐149 suppresses inflammation in nucleus pulposus cells of intervertebral discs by regulating MyD88. Med Sci Monit. 2019;25:4892‐4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang Z, Hu J, Pan Y, et al. miR‐140‐5p/miR‐149 affects chondrocyte proliferation, apoptosis, and autophagy by targeting FUT1 in osteoarthritis. Inflammation. 2018;41:959‐971. [DOI] [PubMed] [Google Scholar]
- 29. Penolazzi L, Lambertini E, Bergamin LS, et al. MicroRNA‐221 silencing attenuates the degenerated phenotype of intervertebral disc cells. Aging (Albany NY). 2018;10:2001‐2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Penolazzi L, Lambertini E, Scussel Bergamin L, et al. Reciprocal regulation of TRPS1 and miR‐221 in intervertebral disc cells. Cells. 2019;8:1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shirley JL, de Jong YP, Terhorst C, Herzog RW. Immune responses to viral gene therapy vectors. Mol Ther. 2020;28:709‐722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Verdera HC, Kuranda K, Mingozzi F. AAV vector immunogenicity in humans: a long journey to successful gene transfer. Mol Ther. 2020;28:723‐746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goswami R, Subramanian G, Silayeva L, et al. Gene therapy leaves a vicious cycle. Front Oncol. 2019;9:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McCarthy HO, McCaffrey J, McCrudden CM, et al. Development and characterization of self‐assembling nanoparticles using a bio‐inspired amphipathic peptide for gene delivery. J Control Release. 2014;189:141‐149. [DOI] [PubMed] [Google Scholar]
- 35. Bennie LA, Feng J, Emmerson C, et al. Formulating RALA/Au nanocomplexes to enhance nanoparticle internalisation efficiency, sensitising prostate tumour models to radiation treatment. J Nanobiotechnology. 2021;19:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McCrudden CM, McBride JW, McCaffrey J, et al. Gene therapy with RALA/iNOS composite nanoparticles significantly enhances survival in a model of metastatic prostate cancer. Cancer Nanotechnol. 2018;9:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Blokpoel Ferreras LA, Chan SY, Vazquez Reina S, Dixon JE. Rapidly transducing and spatially localized magnetofection using peptide‐mediated non‐viral gene delivery based on iron oxide nanoparticles. ACS Appl Nano Mater. 2021;4:167‐181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Abu‐Awwad HAM, Thiagarajan L, Dixon JE. Controlled release of GAG‐binding enhanced transduction (GET) peptides for sustained and highly efficient intracellular delivery. Acta Biomater. 2017;57:225‐237. [DOI] [PubMed] [Google Scholar]
- 39. Lai A, Gansau J, Gullbrand SE, et al. Development of a standardized histopathology scoring system for intervertebral disc degeneration in rat models: an initiative of the ORS spine section. JOR Spine. 2021;4:e1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sadowska A, Kameda T, Krupkova O, Wuertz‐Kozak K. Osmosensing, osmosignalling and inflammation: how intervertebral disc cells respond to altered osmolarity. Eur Cell Mater. 2018;36:231‐250. [DOI] [PubMed] [Google Scholar]
- 41. Jalal AR, Dixon JE. Efficient delivery of transducing polymer nanoparticles for gene‐mediated induction of osteogenesis for bone regeneration. Front Bioeng Biotechnol. 2020;8:849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Udhayakumar VK, De Beuckelaer A, McCaffrey J, et al. Arginine‐rich peptide‐based mRNA nanocomplexes efficiently instigate cytotoxic T cell immunity dependent on the amphipathic organization of the peptide. Adv Healthc Mater. 2017;6:1601412. [DOI] [PubMed] [Google Scholar]
- 43. Bennett R, Yakkundi A, McKeen HD, et al. RALA‐mediated delivery of FKBPL nucleic acid therapeutics. Nanomedicine (Lond). 2015;10:2989‐3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang J, Tian Y, Phillips KL, et al. Tumor necrosis factor α‐ and interleukin‐1β‐dependent induction of CCL3 expression by nucleus pulposus cells promotes macrophage migration through CCR1. Arthritis Rheum. 2013;65:832‐842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang J, Markova D, Anderson DG, Zheng Z, Shapiro IM, Risbud MV. TNF‐α and IL‐1β promote a disintegrin‐like and metalloprotease with thrombospondin type I motif‐5‐mediated aggrecan degradation through syndecan‐4 in intervertebral disc. J Biol Chem. 2011;286:39738‐39749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ignat'eva NY, Danilov NA, Averkiev SV, et al. Determination of hydroxyproline in tissues and the evaluation of the collagen content of the tissues. J Anal Chem. 2007;62:51‐57. [Google Scholar]
- 47. Huang H‐Y, Lin Y‐C‐D, Cui S, et al. miRTarBase update 2022: an informative resource for experimentally validated miRNA–target interactions. Nucleic Acids Res. 2021;50:D222‐D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Licursi V, Conte F, Fiscon G, Paci P. MIENTURNET: an interactive web tool for microRNA‐target enrichment and network‐based analysis. BMC Bioinformatics. 2019;20:545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498‐2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web‐server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28:3442‐3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Szklarczyk D, Kirsch R, Koutrouli M, et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638‐D646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jiang H, Moro A, Wang J, Meng D, Zhan X, Wei Q. MicroRNA‐338‐3p as a novel therapeutic target for intervertebral disc degeneration. Exp Mol Med. 2021;53:1356‐1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang W, Yang C, Wang X, et al. MicroRNA‐129 and ‐335 promote diabetic wound healing by inhibiting Sp1‐mediated MMP‐9 expression. Diabetes. 2018;67:1627‐1638. [DOI] [PubMed] [Google Scholar]
- 54. Kasinski AL, Kelnar K, Stahlhut C, et al. A combinatorial microRNA therapeutics approach to suppressing non‐small cell lung cancer. Oncogene. 2015;34:3547‐3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schoolmeesters A, Eklund T, Leake D, et al. Functional profiling reveals critical role for miRNA in differentiation of human mesenchymal stem cells. PLoS One. 2009;4:e5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mariner PD, Johannesen E, Anseth KS. Manipulation of miRNA activity accelerates osteogenic differentiation of hMSCs in engineered 3D scaffolds. J Tissue Eng Regen Med. 2012;6:314‐324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Castaño IM, Raftery RM, Chen G, et al. Dual scaffold delivery of miR‐210 mimic and miR‐16 inhibitor enhances angiogenesis and osteogenesis to accelerate bone healing. Acta Biomater. 2023;172:480‐493. [DOI] [PubMed] [Google Scholar]
- 58. Dixon JE, Osman G, Morris GE, et al. Highly efficient delivery of functional cargoes by the synergistic effect of GAG binding motifs and cell‐penetrating peptides. Proc Natl Acad Sci USA. 2016;113:E291‐E299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jensen C, Teng Y. Is it time to start transitioning from 2D to 3D cell culture? Front Mol Biosci. 2020;7:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ndikung J, Storm D, Violet N, et al. Restoring circadian synchrony in vitro facilitates physiological responses to environmental chemicals. Environ Int. 2020;134:105265. [DOI] [PubMed] [Google Scholar]
- 61. Kaeffer B, Pardini L. Clock genes of mammalian cells: practical implications in tissue culture. In Vitro Cell Dev Biol Anim. 2005;41:311‐320. [DOI] [PubMed] [Google Scholar]
- 62. Yang M, Zhang L, Gibson GJ. Chondrocyte miRNAs 221 and 483‐5p respond to loss of matrix interaction by modulating proliferation and matrix synthesis. Connect Tissue Res. 2015;56:236‐243. [DOI] [PubMed] [Google Scholar]
- 63. Lolli A, Narcisi R, Lambertini E, et al. Silencing of antichondrogenic microRNA‐221 in human mesenchymal stem cells promotes cartilage repair in vivo. Stem Cells. 2016;34:1801‐1811. [DOI] [PubMed] [Google Scholar]
- 64. Lambertini E, Penolazzi L, Notarangelo MP, et al. Pro‐differentiating compounds for human intervertebral disc cells are present in Violina pumpkin leaf extracts. Int J Mol Med. 2023;51:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chen Q, Wu S, Wu Y, Chen L, Pang Q. MiR‐149 suppresses the inflammatory response of chondrocytes in osteoarthritis by down‐regulating the activation of TAK1/NF‐κB. Biomed Pharmacother. 2018;101:763‐768. [DOI] [PubMed] [Google Scholar]
- 66. Santini P, Politi L, Vedova PD, Scandurra R, Scotto d'Abusco A. The inflammatory circuitry of miR‐149 as a pathological mechanism in osteoarthritis. Rheumatol Int. 2014;34:711‐716. [DOI] [PubMed] [Google Scholar]
- 67. Wang Y, Che M, Xin J, Zheng Z, Li J, Zhang S. The role of IL‐1β and TNF‐α in intervertebral disc degeneration. Biomed Pharmacother. 2020;131:110660. [DOI] [PubMed] [Google Scholar]
- 68. Du J, Pfannkuche JJ, Lang G, et al. Proinflammatory intervertebral disc cell and organ culture models induced by tumor necrosis factor alpha. JOR Spine. 2020;3:e1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li H, Wang X, Pan H, et al. The mechanisms and functions of IL‐1β in intervertebral disc degeneration. Exp Gerontol. 2023;177:112181. [DOI] [PubMed] [Google Scholar]
- 70. Johnson ZI, Schoepflin ZR, Choi H, Shapiro IM, Risbud MV. Disc in flames: roles of TNF‐α and IL‐1β in intervertebral disc degeneration. Eur Cell Mater. 2015;30:104‐116. discussion 116–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Deng X, Zhao F, Kang B, Zhang X. Elevated interleukin‐6 expression levels are associated with intervertebral disc degeneration. Exp Ther Med. 2016;11:1425‐1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Weber KT, Alipui DO, Sison CP, et al. Serum levels of the proinflammatory cytokine interleukin‐6 vary based on diagnoses in individuals with lumbar intervertebral disc diseases. Arthritis Res Ther. 2016;18:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Haddadi K, Abediankenari S, Alipour A, et al. Association between serum levels of interleukin‐6 on pain and disability in lumbar disc herniation surgery. Asian J Neurosurg. 2020;15:494‐498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Huang D, Xiao J, Deng X, et al. Association between Fas/FasL gene polymorphism and musculoskeletal degenerative diseases: a meta‐analysis. BMC Musculoskelet Disord. 2018;19:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Han D, Ding Y, Liu S‐L, et al. Double role of Fas ligand in the apoptosis of intervertebral disc cells in vitro. Acta Biochim Biophys Sin. 2009;41:938‐947. [DOI] [PubMed] [Google Scholar]
- 76. Yamamoto J, Maeno K, Takada T, et al. Fas ligand plays an important role for the production of pro‐inflammatory cytokines in intervertebral disc nucleus pulposus cells. J Orthop Res. 2013;31:608‐615. [DOI] [PubMed] [Google Scholar]
- 77. Qin C, Zhang B, Zhang L, et al. MyD88‐dependent toll‐like receptor 4 signal pathway in intervertebral disc degeneration. Exp Ther Med. 2016;12:611‐618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bayer AL, Alcaide P. MyD88: at the heart of inflammatory signaling and cardiovascular disease. J Mol Cell Cardiol. 2021;161:75‐85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zheng C, Chen J, Chu F, Zhu J, Jin T. Inflammatory role of TLR‐MyD88 signaling in multiple sclerosis. Front Mol Neurosci. 2020;12:314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wang WJ, Yu XH, Wang C, et al. MMPs and ADAMTSs in intervertebral disc degeneration. Clin Chim Acta. 2015;448:238‐246. [DOI] [PubMed] [Google Scholar]
- 81. Rastogi A, Kim H, Twomey JD, Hsieh AH. MMP‐2 mediates local degradation and remodeling of collagen by annulus fibrosus cells of the intervertebral disc. Arthritis Res Ther. 2013;15:R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhang Y, Gu Z, Qiu G. Association of the polymorphism of MMP2 with the risk and severity of lumbar disc degeneration in the Chinese Han population. Eur Rev Med Pharmacol Sci. 2013;17:1830‐1834. [PubMed] [Google Scholar]
- 83. Fan D, Kassiri Z. Biology of tissue inhibitor of metalloproteinase 3 (TIMP3), and its therapeutic implications in cardiovascular pathology. Front Physiol. 2020;11:661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tortorella MD, Burn TC, Pratta MA, et al. Purification and cloning of aggrecanase‐1: a member of the ADAMTS family of proteins. Science. 1999;284:1664‐1666. [DOI] [PubMed] [Google Scholar]
- 85. He M, Pang J, Sun H, Zheng G, Lin Y, Ge W. Overexpression of TIMP3 inhibits discogenic pain by suppressing angiogenesis and the expression of substance P in nucleus pulposus. Mol Med Rep. 2020;21:1163‐1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Er ZJ, Yin CF, Wang WJ, Chen XJ. Serum CXCL12/SDF‐1 level is positively related with lumbar intervertebral disc degeneration and clinical severity. Innate Immun. 2020;26:341‐350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhang H, Zhu T, Zhang L, Wu Q. Stromal cell‐derived factor‐1 induces matrix metalloproteinase expression in human endplate chondrocytes, cartilage endplate degradation in explant culture, and the amelioration of nucleus pulposus degeneration in vivo. Int J Mol Med. 2018;41:969‐976. [DOI] [PubMed] [Google Scholar]
- 88. Liu Z, Ma C, Shen J, Wang D, Hao J, Hu Z. SDF‐1/CXCR4 axis induces apoptosis of human degenerative nucleus pulposus cells via the NF‐κB pathway. Mol Med Rep. 2016;14:783‐789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Liu Z‐c, Wang Z‐l, Huang C‐y, et al. Duhuo Jisheng decoction inhibits SDF‐1‐induced inflammation and matrix degradation in human degenerative nucleus pulposus cells in vitro through the CXCR4/NF‐κB pathway. Acta Pharmacol Sin. 2018;39:912‐922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zhong H, Zhou Z, Guo L, et al. The miR‐623/CXCL12 axis inhibits LPS‐induced nucleus pulposus cell apoptosis and senescence. Mech Ageing Dev. 2021;194:111417. [DOI] [PubMed] [Google Scholar]
- 91. Kluba T, Niemeyer T, Gaissmaier C, Gründer T. Human anulus fibrosis and nucleus pulposus cells of the intervertebral disc: effect of degeneration and culture system on cell phenotype. Spine (Phila Pa 1976). 2005;30:2743‐2748. [DOI] [PubMed] [Google Scholar]
- 92. Gantenbein B, Calandriello E, Wuertz‐Kozak K, Benneker LM, Keel MJB, Chan SCW. Activation of intervertebral disc cells by co‐culture with notochordal cells, conditioned medium and hypoxia. BMC Musculoskelet Disord. 2014;15:422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Liu Z, Zheng Z, Qi J, et al. CD24 identifies nucleus pulposus progenitors/notochordal cells for disc regeneration. J Biol Eng. 2018;12:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Aguiar DJ, Johnson SL, Oegema TR. Notochordal cells interact with nucleus pulposus cells: regulation of proteoglycan synthesis. Exp Cell Res. 1999;246:129‐137. [DOI] [PubMed] [Google Scholar]
- 95. Gantenbein‐Ritter B, Chan SC. The evolutionary importance of cell ratio between notochordal and nucleus pulposus cells: an experimental 3‐D co‐culture study. Eur Spine J. 2012;21(suppl 6):S819‐S825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Rijal G, Li W. Native‐mimicking in vitro microenvironment: an elusive and seductive future for tumor modeling and tissue engineering. J Biol Eng. 2018;12:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. McDonnell EE, Buckley CT. Investigating the physiological relevance of ex vivo disc organ culture nutrient microenvironments using in silico modeling and experimental validation. JOR Spine. 2021;4:e1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Chan SC, Ferguson SJ, Gantenbein‐Ritter B. The effects of dynamic loading on the intervertebral disc. Eur Spine J. 2011;20:1796‐1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Dou Y, Sun X, Ma X, Zhao X, Yang Q. Intervertebral disk degeneration: the microenvironment and tissue engineering strategies. Front Bioeng Biotechnol. 2021;9:592118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Frapin L, Clouet J, Delplace V, Fusellier M, Guicheux J, le Visage C. Lessons learned from intervertebral disc pathophysiology to guide rational design of sequential delivery systems for therapeutic biological factors. Adv Drug Deliv Rev. 2019;149–150:49‐71. [DOI] [PubMed] [Google Scholar]
- 101. Guerrero J, Häckel S, Croft AS, Hoppe S, Albers CE, Gantenbein B. The nucleus pulposus microenvironment in the intervertebral disc: the fountain of youth? Eur Cell Mater. 2021;41:707‐738. [DOI] [PubMed] [Google Scholar]
- 102. Altun I. Cytokine profile in degenerated painful intervertebral disc: variability with respect to duration of symptoms and type of disease. Spine J. 2016;16:857‐861. [DOI] [PubMed] [Google Scholar]
- 103. Aripaka SS, Bech‐Azeddine R, Jørgensen LM, Mikkelsen JD. The expression of metalloproteinases in the lumbar disc correlates strongly with Pfirrmann MRI grades in lumbar spinal fusion patients. Brain Spine. 2022;2:100872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Bachmeier BE, Nerlich A, Mittermaier N, et al. Matrix metalloproteinase expression levels suggest distinct enzyme roles during lumbar disc herniation and degeneration. Eur Spine J. 2009;18:1573‐1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Silagi ES, Shapiro IM, Risbud MV. Glycosaminoglycan synthesis in the nucleus pulposus: dysregulation and the pathogenesis of disc degeneration. Matrix Biol. 2018;71–72:368‐379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Raftery RM, Walsh DP, Blokpoel Ferreras L, et al. Highly versatile cell‐penetrating peptide loaded scaffold for efficient and localised gene delivery to multiple cell types: from development to application in tissue engineering. Biomaterials. 2019;216:119277. [DOI] [PubMed] [Google Scholar]
- 107. Lee S, Millecamps M, Foster DZ, Stone LS. Long‐term histological analysis of innervation and macrophage infiltration in a mouse model of intervertebral disc injury‐induced low back pain. J Orthop Res. 2020;38:1238‐1247. [DOI] [PubMed] [Google Scholar]
- 108. Wuertz K, Haglund L. Inflammatory mediators in intervertebral disk degeneration and discogenic pain. Global Spine J. 2013;3:175‐184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supporting information.
Figure S1. Transfection of scramble negative control miR did not significantly alter the production of DNA, sulfated glycosaminoglycans (sGAGs), or collagen in monolayer. Delivery of a negative control miR showed no significant changes to biochemical quantification 14 days post‐transfection. N = 5; ns, nonsignificant.
Figure S2. Biochemical analysis of rat nucleus pulposus monolayer cultures following vector‐miR pairing delivery demonstrated some positive increases 14 days post‐transfection. (A) Sulfated glycosaminoglycans (sGAG) and (B) collagen (COL). Data is displayed as sGAG or COL normalized to DNA content and the non‐transfected (NT) control. N = 5.
Figure S3. Biochemical quantification of DNA, sulfated glycosaminoglycan (sGAG), and collagen (COL) levels 14 days post‐transfection with RALA and FLR‐delivered miR combinations. N = 5.
Figure S4. Cell viability of rat caudal NP tissue following 21 days of ex vivo organ culture. (A) Representative images of maximum projection of z‐stacks of Calcein AM (green, live) and ethidium homodimer (red, dead) staining. (B) Viability assessment of live and dead cells. Healthy discs were non‐degenerated. Chondroitinase ABC (cABC) NT and FLR‐miRNA‐149 + 221‐in groups were mildly degenerated with cABC for 7 days prior to Sham or miRNA‐pair treatment. Scale bar is 150 μm. N = 4.
Figure S5. Median and worst representative histological images of rat motion segments 14 days following transfection with miRs A. 140‐5p + 141‐3p inhibitor, B. miR‐140‐5p mimic + miR‐149‐5p mimic, C. miR‐140‐5p mimic + miR‐221‐3p inhibitor, D. miRs‐141‐3p inhibitor + 149‐5p mimic, E. miR‐141‐3p inhibitor + miR‐221‐3p inhibitor, and F. miR‐149‐5p + miR‐221‐3p inhibitor following 14 days in culture post‐transfection. Alcian blue for glycosaminoglycan staining, Picrosirius red for collagen staining. Main image scale bar 500 μm, inset 150 μm. N = 3.
Table S1. Gene name abbreviations in Figure 7.
Table S2. Gene name abbreviations in Figure 8.


