Abstract
Recently, it has been recognized that natural extracellular matrix (ECM) and tissues are viscoelastic, while only elastic property has been investigated in the past. How the viscoelastic matrix regulates stem cell patterning is critical for cell-ECM mechano-transduction. Here, this study fabricated different methacrylated hyaluronic acid (HA) hydrogels using covalent crosslinking, consisting of two gels with similar elasticity (stiffness) but different viscoelasticity, and two gels with similar viscoelasticity but different elasticity (stiffness). Meanwhile, a second set of dual network hydrogels were fabricated containing both covalent and coordinated crosslinks. Human spinal cord organoid (hSCO) patterning in HA hydrogels and co-culture with isogenic human blood vessel organoids (hBVOs) were investigated. The viscoelastic hydrogels promote regional hSCO patterning compared to the elastic hydrogels. More viscoelastic hydrogels can promote dorsal marker expression, while softer hydrogels resulted in higher interneuron marker expression. The effects of viscoelastic properties of the hydrogels become more dominant than the stiffness effects in the coculture of hSCOs and hBVOs. In addition, more viscoelastic hydrogels could lead to more Yes-associated protein nuclear translocation, revealing mechanism of cell-ECM mechano-transduction. This research provides insights on viscoelastic behaviors of the hydrogels during human organoid patterning with ECM-mimicking in vitro microenvironment for applications in regenerative medicine.
Keywords: Viscoelasticity, Hyaluronic Acid Hydrogels, Human Pluripotent Stem Cells, Spinal Cord Organoid Patterning, Vascularization
A short summary:
This study fabricated hyaluronic acid hydrogels using covalent crosslinking, consisting of two gels with similar elasticity (stiffness) but different viscoelasticity, and two gels with similar viscoelasticity but different elasticity (stiffness). Meanwhile, a second set of dual network hydrogels were fabricated containing both covalent and coordinated crosslinks. Human spinal cord organoid patterning in hydrogels and co-culture with isogenic human blood vessel organoids were investigated.
1. Introduction
The spinal cord is part of the central nervous system and provides a connection between the brain and lower back, which delivers nerve signals from the brain to the body to control locomotion and feeling sensations [1]. Human induced pluripotent stem cells (hiPSCs) can be directly induced into different types of region-specific brain organoids, including spinal cord organoids, for studying neurodevelopment and neurodegeneration [2]. These three-dimensional (3D) organoids are usually generated in suspension. To generate different subtypes of neuronal cells in vitro such as motor neurons, hiPSCs can be induced by small molecules to become functional neural cells with a high conversion rate in a 2D culture [3], and these cells can be assembled into 3D neural structure [4]. Currently, there are still many limitations for developing more complex systems in 3D organoids. For example, the lack of specific mature pattern structure, such as rostro-caudal patterning, decreased disease modeling accuracy and reduced model effectiveness [5]. Furthermore, small molecules may not be sufficient to provide spatial cues for specific cells arranged in 3D structure, which are essential for functional neuronal and synapse maturation. Additionally, environmental stimulations, such as chemical and mechanical cues, could be less effective in 3D organoids compared to in vivo environment due to the missing signaling in vitro, which may lead to the lack of function [6, 7]. Therefore, novel methods with more in vivo-like microenvironment are needed to pattern hiPSC-derived spinal cord organoids, and to provide new insights into the principles of tissue patterning during spinal neurogenesis [8].
The 3D ventral spinal cord organoids have been generated using cell cycle inhibitor and recapitulated spinal neurogenesis as well as rostro-caudal patterns for modeling motor neuron disease [9]. To promote spinal cord patterning, using novel extracellular matrix (ECM) or scaffolds may provide a 3D signaling network to better pattern spinal cord organoids [10]. Additionally, inclusion of vital structure such as blood spinal cord barrier (BSCB) in the organoid is important for studying the dysfunction of spinal cord [11]. The BSCB serves as an interface responsible for facilitating the transport of nutrients between the bloodstream and the spinal cord [12]. Due to the analogous structure to the blood brain barrier, the endothelial cells are the most important components for spinal cord vascularization. Additionally, isogenic human blood vessel organoids (hBVOs) possess the capability to generate vascular structures and can be used to co-culture with human spinal cord organoids (hSCOs) to include BSCB structure in the organoids, through spheroid fusion and assembly as shown in our previous studies [13, 14].
3D ECMs have a variety of effects on cellular process due to different characteristics. Elasticity or stiffness, nanotopography, and chemical functionalities of ECMs all have an influence on cell spreading, proliferation, migration, differentiation, and organoid formation [15–17]. Well-engineered ECMs can provide a proper microenvironment to regulate cellular behaviors including tissue regeneration due to specific biochemical and biophysical cues [18, 19]. In particular, the patterning of tissues or organoids can be tailored by 3D ECMs. 3D scaffold biomaterials especially hydrogels can be fabricated to mimic static mechanical properties of biological tissues and ECMs in the human body [20, 21]. Besides spatial mechanical properties, the viscoelasticity, or temporal (time-dependent) properties of hydrogels provides in-time cues for tissues/organoids to sense [22, 23] and dynamic stimulation to respond. The viscoelasticity of ECMs is a temporal parameter of the materials which can apply dynamic stimulation to the cells surrounded by ECMs. By regulating viscoelasticity in addition to mechanical properties such as the Young’s modulus, ECMs provide both spatial and temporal factors for neural tissue morphogenesis [24]. Recently, the viscoelasticity of biomaterials (e.g., alginate) has been assessed to regulate cell proliferation, migration, and spreading [25, 26]. In addition, the effects of ECM viscoelasticity on the generation of embryoid body-like structure from hiPSCs were revealed [22]. Using alginate hydrogels with arginine–glycine–aspartate (RGD) ligands, the hiPSC morphogenesis in 3D culture showed that RGD density and stress relaxation time influenced cell viability, proliferation, apicobasal polarization, and lumen formation [22]. Nevertheless, the influence of hydrogel viscoelasticity on the cell behaviors is at the nascent stage, and the effect on the spinal cord organoid patterning has not yet been investigated.
In the human body, the major components of ECMs in the central nervous system are hyaluronans [27, 28]. Hyaluronic acid (HA) in the tissue fluid helps the tissue resist osmotic compression and absorb compressive force [29, 30]. Additionally, the network of HA is assembled by the existence of proteoglycans. The brain and spinal cord ECMs lack the fibrous components, such as collagens [31, 32]. In the brain, the entanglement of HA network is stabilized by specific connection between tenascins and proteoglycans [33]. Furthermore, HA can be used for wound healing, tissue maintenance, and inflammation [34–36]. The specific molecular weight of HA in different body parts could promote tissue remodeling and homeostasis [37, 38]. For example, HA has a remarkable hydration capacity, and lack of HA causes reduced extracellular space volume in the brain [39]. In the brain, the entangled network of HA needs to be stabilized through linkage with proteins and chondroitin sulfate proteoglycans [33, 40]. Therefore, HA-based ECMs can be designed with various modifications and compositions to provide specific biochemical and biomechanical properties [41–44].
Hence, this study fabricated different HA-based hydrogels for the generation and recapitulation of the patterning of spinal cord organoids. The static properties such as the stiffness of the hydrogels are important for regulating the behaviors of hiPSCs. However, dynamic properties, or time-dependent feature of the polymer also have important effects on the morphogenesis and lineage-specific differentiation of hiPSCs. Therefore, HA hydrogels with different stiffness and viscoelasticity were fabricated and characterized, based on covalent bond crosslinked methacrylated HA (HAMA). Then, hiPSCs were seeded into different hydrogels and induced for hSCO differentiation and patterning. hBVOs and hSCOs from different hydrogels were cocultured and characterized for vascularization of the organoids, which may lead to the generation of blood spinal cord barrier. Furthermore, dopamine modified HA (HA-Cat) with Fe3+ coordinated crosslinked hydrogels were mixed with HAMA hydrogels to make dual network penetration (i.e., HAMA@HA-Cat) hydrogels. The dual network penetrated hydrogels also regulated hSCO patterning. Together, this study has significant implications on the role of viscoelastic properties of hydrogels in establishing human organoid model systems for disease modeling and drug screening.
2. Materials and methods
2.1. Materials and reagents
The vendors and catalog numbers of key materials and reagents are provided: Sodium hyaluronate (HA-100k, HA-200k, HA-1M, Lifecore Biomedical, Inc), Dopamine hydrochloride (Sigma, H8502), methacrylic anhydride (Sigma, 276685), sodium hydroxide (Sigma, 221465), 1-ethyl-3-(-3-dimethylaminopropyl) carbodiimide hydrochloride (EDC, D1601), N-hydroxysuccinimide (Thermo Scientific Chemicals, 157270250), poly ethylene glycol (PEG)-dithiol (Creative PEGWorks, PLS-612), 2-Hydroxy-4’-(2-hydroxyethoxy)-2-methylpropiophenone (NHS) (Sigma, 410896), Rho-associated protein kinase (ROCK) inhibitor Y27632 (Sigma, Y0503), mTeSR Plus (STEMCELL Technologies Inc., 100–0276), LDN193189 hydrochloride (Sigma, SML0559), DMEM/F-12 (Gibco™, 12400024), B-27™ Supplement (50X) (Gibco™, 17504044), CHIR99021 (a Wnt signaling activator, Sigma, SML1046), retinoic acid (RA, Sigma), purmorphamine (a sonic hedgehog signaling activator, Sigma, SML0868), Recombinant human fibroblast growth factor (FGF)-basic (bFGF, Peprotech, 100–18C), N-2 Supplement (100X) (Gibco™, 17502048), Neurobasal™ Medium (Gibco™, 21103049), Human Endothelial Serum-free Medium (hESFM) (Gibco™, 11111044), β-mercaptoethanol (Gibco™, 21985023), MEM Non-Essential Amino Acids (NEAA) Solution (100X) (Gibco™, 11140050), GlutaMAX™ Supplement (Gibco™, 35050061), Proteinase K (Research Products International Corp, P502200.1), LIVE/DEAD™ Viability/Cytotoxicity Kit for mammalian cells (Invitrogen™, L3224), N2B27 media: 50% of DMEM/F12 mix with 50% Neurobasal Medium supplemented with 0.5% N2, 2% B27, 0.5% NEAA, 1% Penicillin/ Streptomycin (P/S), 0.1% β-mercaptoethanol, and 1% GlutaMAX, Brain-derived neurotrophic factor (BDNF, Peprotech, 450–02), and Growth factor-reduced Matrigel (Corning, 354230).
2.2. Synthesis and characterization of HAMA and HA-Cat
For HAMA synthesis, methacrylation of HA was performed by adding dropwise 1.1 mL of MA at 1% (v/v) to 100 mL of 1% (w/v) HA solution in phosphate buffered saline (PBS), pH 7.4, at 4°C, under magnetic stirring for 24 h. The pH of the solution was kept between 8 and 10 with the addition of 5 N NaOH, until no further pH changes were detected, which indicated that the reaction was complete. The solution was dialyzed for 4 days with a 12–14 kDa membrane in deionized water at 4°C. Then, HAMA was frozen and lyophilized. The obtained powder material was stored at −20°C until further use.
For catechol functionalization of HAMA, i.e., HA-Cat synthesis, HAMA was dissolved in 2-(N-Morpholino) ethanesulfonic acid (MES) buffer (pH=4.5). Next, 0.03 mol/L NHS, 0.03 mol/L EDC and 0.05 mg/mL dopamine were added to a bottle and stirred overnight to fully react. After synthesis, the derivatives of HA underwent dialysis in de-ionized water for three days to purify. Then the solutions were frozen and lyophilized. After synthesis, H1-NMR (Bruker spectrometers B600, FSU-NMR Facility) was performed to characterize modification after synthesis.
2.3. HAMA and HAMA@HA-Cat hydrogel fabrication and characterization
2.3.1. Hydrogel fabrication
To obtain the covalently crosslinked HAMA gels, the HAMAs were photo-crosslinked with dithiol-PEG. A total of nine gels were synthesized. HAMA (three groups with molecular weight at 100k, 200k, and 1,000k) was dissolved at 1%, 0.5%, and 0.25% (w/w) in PBS, respectively. The mixed polymer precursor in PBS was incubated at 37°C with 0.1% (w/v) of NHS and 0.5% thiolated PEG and then cured with Dymax light shields model 5000 EC flood (intensity: 225 mW/cm2) for 30 seconds.
For fabrication of the HAMA@HA-Cat hydrogels (@ means that the hydrogel is a dual penetration network), 1% wt HAMA (three groups: 100k, 200k, and 1,000k of molecular weight) and 1% wt HA-Cat (1,000k) were mixed at a ratio of 1:1. Then, the mixed polymer precursor in PBS was incubated at 37°C with 0.1% (w/v) of NHS and 0.5% thiolated PEG and then cured with Dymax light shields model 5000 EC flood (intensity: 225 mW/cm2) for 30 seconds. After the crosslinking, 200 μL of 40 mM FeCl3 aqueous solution was added to the hydrogels. The bulk hydrogels were cut into granular hydrogels for better reaction with FeCl3 solution during HA-Cat crosslinking.
2.3.2. Characterization of hydrogels
The static elastic properties of the hydrogels were measured via compression tests performed on an ARES-G2 Rheometer using a parallel plate geometry (d=25 mm) (TA Instruments, New Castle, DE, USA) and strain rate of 0.0000667 s−1. Each gel composition was characterized with three specimens for at least three independent measurements.
Rheological characterization was also performed with an ARES-G2 Rheometer using the parallel plate geometry (d= 25 mm, gap 0.5 mm). Oscillatory rheometery was conducted to measure the elastic and viscous modulus of the hydrogels. At first, parallel discs of 25 mm in diameter were placed on the rheometer and a 25 mm flat plate geometry was used to measure the samples across a strain sweep to find linear viscoelastic region (LVR) of the HAMA hydrogels with the parameter at 6.28 rad/s, 37 °C and within the range of strain at 0.1– 100 %. Then, the 0.5% strain was chosen for the frequency sweep to get the rheological properties of HAMA hydrogels. At least 3 samples (0.5 mm thick) for each gel composition were characterized.
The stress relaxation test was then performed in hydrogels. All samples were put between parallel discs of 25 mm in diameter and a gap of 1 mm. Next, the stress-relaxation behavior was quantified at 10% strain, with all tests lasting from 500 s to 3000 s for the samples to reach the plateau for the hydrogels. Then, the relaxation time data were regressed by Maxwell model to get the relaxation time (τ).
The morphology of the hydrogels was examined using scanning electron microscopy (SEM). The HAMA hydrogels were freeze-dried in a lyophilizer (Labconco Corporation, Kansas City, MO, USA) for 2 days. Then, the samples were taken out carefully, fixed onto a SEM stage with carbon tape, and coated with a 10 nm gold layer to better reveal the hydrogel morphology. Observations were made using a FEI Helios G4 UC multi-technique dual beam (electron and Ga ion) Field Emission Scanning Electron Microscope (FESEM) (Thermo Fisher Scientific, Pittsburgh, PA) under low‐vacuum conditions.
2.4. hiPSC 2D and 3D cultures for biocompatibility evaluation
Undifferentiated human iPSK3 cells (human foreskin fibroblasts reprogrammed with plasmid DNA encoding reprogramming factors OCT4, NANOG, SOX2, and LIN28) were maintained on Growth Factor-reduced Matrigel-coated surface in mTeSR serum-free medium as described in our previous publications [13, 14]. Prior to hiPSC seeding, the sterile HAMA precursor solutions were added into the wells of tissue culture plates and then the solutions were cured under UV for 30s to form HAMA hydrogels. For 2D culture, the hiPSC suspension (~1×105 cells) was added at 100 μL into the wells of 96-well tissue culture plates coated with Matrigel (to ensure undifferentiated hiPSC attachment), on top of which was layered with different types of hydrogels. The cells were allowed to settle down into the hydrogels for 15 min. Then additional 100 μL of media were added to each well of a 96-well plate. For 3D culture, the hydrogels were fabricated in the wells of ultralow attachment (ULA) 96-well plates (to prevent cells from attaching to the surface of the culture plates). The dissociated hiPSCs were seeded into the hydrogels by placing two concentrated droplets (50 μL each) of cells into the hydrogels, for a final density of 1×105 cells per gel. After 5 min, additional 100 μL media were added to each well. The cultures were maintained for 7 days and the cells were characterized by DNA assay for proliferation and Live/Dead assay for viability [45, 46].
2.5. Human spinal cord organoid differentiation in hydrogels
Two hSCO differentiation protocols were evaluated before the experiments using hydrogels (Supplementary Method 1) [47, 48]. After comparison, the ventral hSCO differentiation protocol was chosen for this study. Briefly, undifferentiated hiPSCs were dissociated by Accutase for 5–7 min. At day 0, the single cells were seeded in 100 μL of DMEM/F12 plus N2B27 medium with 10 μM Y-27632 in each well of a U-bottom low attachment 96-well plate at a density of 15,000 cells/well for hiPSC self-aggregation. At day 1, the cells were fed with N2B27 medium containing 10 μM Y-27632, 4 μM CHIR99021, and 0.5 μM LDN193189. At day 3, the neural induction medium containing 1μM RA was added for generating ventral patterning. The medium was changed every other day. At day 10, the spheroids were embedded into 15 μL concentrated Matrigel (1:3 dilution with neural induction medium) and incubated for one hour. Then, N2B27 media supplemented with 1 μM RA and 1 μM Purmorphamine were added to each well for neural patterning without disturbing Matrigel droplets. At day 14, the Matrigel-embedded organoids were transferred to the rocker [49, 50] or PBS Vertical Wheel bioreactor (PBS Biotech Inc., CA, USA) [51, 52] for further expansion and maturation. On day 18 and onwards, the medium was changed to N2B27 media supplemented with 10 ng/mL BDNF. To evaluate the influence of different types of hydrogels, single hiPSCs were seeded at a density at 15,000 cell/well into ULA 96-well plate. At day 3, the self-assembled spheroids were transferred into hydrogels, which were layered on top and beneath the spheroids, for further hSCO differentiation or co-culture with hBVOs.
2.6. Human blood vessel organoid differentiation
The hBVO generation was modified from previous publications [53, 54]. hiPSCs were seeded in the wells of ULA 96 well plate at a density at 10,000 cells/well in mTeSR plus supplemented with 10 μM ROCK inhibitor Y-27632. To initiate differentiation at day 0, cells were treated with 6 μM CHIR99021 (Selleckchem) in BVO1 medium: DMEM/F12 supplemented with 2% B27, 0.5% NEAA, 1% P/S, 0.1% β-mercaptoethanol, and 1% GlutaMAX. The medium was changed every other day until day 6. At day 6, the medium was switched to BVO2 medium: hESFM supplemented with 20 ng/mL bFGF, 10 μM RA, and 2% B27. At day 9, the organoids were replated to tissue culture plates or continued to grow in hESFM with 2% B27 for long-term culture.
2.7. hSCO co-culture with hBVOs
Spheroid or organoid fusion methods were evaluated for hSCO vascularization by co-culturing with hBVOs. One 9-day hBVO and one 25-day hSCO were added to the same well of ULA 96-well plate and the organoid fusion occurred spontaneously. After a two-day fusion, the assembled organoid was embedded into Matrigel. Then the organoids were transferred to a low attachment 6-well plate on the rocker. For cell tracker labeling, culture media were removed, and CellTracker™ Red (1:1000 dilution, ThermoFisher Scientific, Waltham, MA, USA) solution was added. hBVOs were incubated with CellTracker™ Red solution at 37°C for one hour. Then the staining solution was removed followed with washing. One CellTracker™ Red labeled hBVO and one hSCO were put next to each other in the same well of 96-well plate for 2 days. Finally, the assembly of the two organoids were imaged. All hSCOs from different HAMA hydrogels were extracted from hydrogels using blunt pipette tips, then they were co-cultured and assembled with hBVOs. The assembloids were characterized for the marker expression of both hSCOs and hBVOs.
2.8. Characterization of cell proliferation and biocompatibility in hydrogels
Cell proliferation was determined by DNA quantitation using Picogreen. The cells were harvested and lysed with 0.1 mg/mL proteinase K (Fisher Scientific, Pittsburgh, PA) at 50°C overnight. The lysates (100 μL) were mixed with 100 μL of 0.5% Picogreen (Molecular Probes, Eugene, OR) in a 96-well plate. The plate was incubated for 5 min in the dark and then read on a fluorescent plate reader with 485ex/528em (BioRad Laboratories, Hercules, CA). The biocompatibility of the hiPSCs in different hydrogels were evaluated using LIVE/DEAD™ Viability/Cytotoxicity Kit (Invitrogen™, Waltham, MA). The organoids were harvested and then dissociated to single cells by Accutase for 20–40 min. Then, a cell suspension at 1×106 cells/mL was prepared. Next, 6 μL of 50 μM Calcein AM and 2 μL of 2 mM ethidium homodimer-1 (EthD-1) were added to each mL of cell suspension. The mixture of dye and cells was incubated at room temperature (RT) for 15 min. The stained cells were acquired with BD FACSCanto™ II flow cytometer (Becton Dickinson) and analyzed by FlowJo software. The cell only, live only, dead only, and live and dead samples were prepared for two-color flow cytometry compensation.
2.9. Histology sectioning and immunohistochemistry
The hSCOs were harvested and placed into 1.5 mL centrifuge tubes and fixed with 10% neutral buffered formalin for 24 hours. Then, the samples were dehydrated by series of ethanol solutions. Briefly, hSCOs were sequentially transferred to 70%, 75%, 80%, and 90% ethanol for 15 min each. Next, the samples were put into 95% ethanol for 60 min twice. Lastly, samples were submerged in 100% ethanol for 60 min twice. After dehydration, hSCOs were transferred into xylene for two 30–60 min intervals. Samples were incubated with 60°C paraffin for 60 min twice and embedded with paraffin at ideal position during overnight cooling. After embedding in paraffin, the samples were sectioned by microtome at 6 μm for each slice. The slice was transferred to warm water and then dried on glass slides. Then, the sections were deparaffinized by immersing in Xylene for 3 min twice. Next, the slides were immersed into 100% ethanol for 3 min twice, 95% ethanol for 3 min, 70 % ethanol for 3 min and then put under running cold tap water to rinse. The wet sections were transferred into 95°C Sodium Citrate Buffer (10 mM Sodium Citrate, 0.05% Tween 20, pH 6.0) for 30 min and then washed under running cold tap water for 10 min. Immunocytochemistry analysis of hSCO markers were performed on the sections. Yes-associated protein (YAP) staining was also performed on the sections using a similar procedure to immunocytochemistry.
2.10. Immunocytochemistry of organoids
The hSCOs were directly replated to Matrigel (1:50) coated tissue culture plate. hBVOs were first dissociated by Accutase for 40 min, and then replated to Matrigel (1:50) coated tissue culture plate. Then, after a 3-day growth, both samples were fixed with 4% paraformaldehyde and permeabilized with cold methanol for staining intracellular markers. The samples were then blocked with 5% fetal bovine serum (FBS) and incubated with various mouse or rabbit primary antibodies (Supplementary Table S1). Next, secondary antibodies were added in staining buffer (2% FBS in PBS). The cells were washed three times each for 5 min. The samples were then stained using Hoechst 33342 (ThermoFisher, 1:2,000) to label cell nuclei and afterwards washed with PBS overnight. Images of stained organoids were captured using a fluorescent microscope (Zeiss Axio Observer) or a Zeiss LSM 880 confocal microscope.
2.11. Image analysis of organoid morphology and YAP localization
To measure spheroid or organoid circularity and area during the experiments, phase-contrast images of hSCOs were taken with a microscope using a 4× and 10× objective every day up to day 18. These images were quantified with Image J software from National Institutes of Health (NIH). Briefly, the perimeter of each individual spheroid/organoid was drawn manually, and the enclosed area and circularity was measured. For YAP localization, the sections of stained organoids were imaged using a Zeiss LSM 880 confocal microscopy. YAP localization (nuclear or cytoplasmic) was analyzed using a quantification method through ImageJ as reported in our previous study [46].
2.12. Flow cytometry analysis for phenotypic marker expression
Briefly, the hSCOs and hBVOs were dissociated into single cells using Accutase and pipetting for 40 min. Then, 1×106 cells per sample were fixed with 4% paraformaldehyde and washed with staining buffer (2% FBS in PBS). The dissociated cells were permeabilized with 100% cold methanol for intracellular markers, blocked, and then incubated with primary antibodies against Chx10, LHX3, SOX2, NKX6.1, Nkx2.2, HNF3β, OLIG2, HB9, PAX7 followed by the corresponding secondary antibody (Supplementary Table S1). The cells were acquired with BD FACSCanto™ II flow cytometer (Becton Dickinson) and analyzed against isotype controls using FlowJo software.
2.13. Reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was isolated from different cell samples using the RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. The isolated RNA samples were further treated with DNA-Free RNA Kit (Zymo, Irvine, CA, USA) to remove genomic DNA contamination [55]. Reverse transcription was carried out according to the manufacturer’s instructions using 2 ng of total mRNA, anchored oligo-dT primers (Operon, Huntsville, AL), and Superscript III (Invitrogen, Carlsbad, CA, USA). The software Oligo Explorer 1.2Primers (Genelink, Hawthorne, NY, USA) was used to design the real-time PCR primers specific for target genes (Supplementary Table S2). For normalization of expression levels, β-actin was used as an endogenous control. Using SYBR1 Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA), real-time PCR reactions were performed on an ABI7500 instrument (Applied Biosystems). The amplification reactions were performed as follows: 2 min at 50°C, 10 min at 95°C, and 40 cycles of 95°C for 15 sec and 55°C for 30 sec, and 68°C for 30 sec following with a melt curve analysis. The Ct values of the target genes were first normalized to the Ct values of the endogenous control β-actin. The corrected Ct values were then compared to the experimental control. Fold changes in gene expression were calculated using the comparative Ct method: to obtain the relative expression levels.
2.14. Whole-patch clamping for electrophysiology
Whole-cell patch clamp was used to record mature spinal cord spheroids cultured on small petri dish. The vessels were washed three times with extracellular recording solution containing 136 mM NaCl, 4 mM KCl, 2 mM MgCl, 10 mM HEPES, and 1mM EGTA (312 mOsm, pH 7.39) and then were incubated in this solution at RT during recording. Glass electrodes (resistance 1–5 MΩ) were filled with intracellular solution containing 130 mM KCl, 10 mM HEPES, and 5 mM EGTA (292 mOsm, pH 7.20). Cells were visualized under phase contrast with a Nikon Eclipse Ti-U inverted microscope with an attached DS-Qi1 monochrome digital camera. Recordings were made with an Axopatch 200B amplifier (Molecular Devices) and digitized with a Digidata 1440A system (Molecular Devices). Ionic currents were recorded under a voltage clamp protocol (-60 mV to 135 mV in 15 mV steps, 250 ms in duration).
2.15. Statistical Analysis
The differences were analyzed by independent t-test or one-way ANOVA followed by Tukey’s multiple comparisons post hoc tests. The difference was considered statistically significant at p≤0.05 and all quantitative data are presented as mean ± standard deviation.
3. Results
3.1. HA Hydrogel fabrication and characterizations
In Figure 1A, the schematic illustrations demonstrate the fabrication process of HAMA and HAMA@HA-Cat hydrogels. The modification of HA with methylate group, catechol groups, and both groups were verified from the H1NMR results (Supplementary Figure S1A). The double bond peaks introduced by the modification of MA appeared at 5.60 ppm and 6.04 ppm and the benzene ring peak introduced by the modification of dopamine appeared at 6.72 ppm, 7.10 ppm, and 7.13 ppm. Then the degrees of modification of 100k, 200k and 1,000k HAMA are 50.4%, 50.0%, 45.4%, respectively.
Figure 1. HAMA synthesis and characterization.
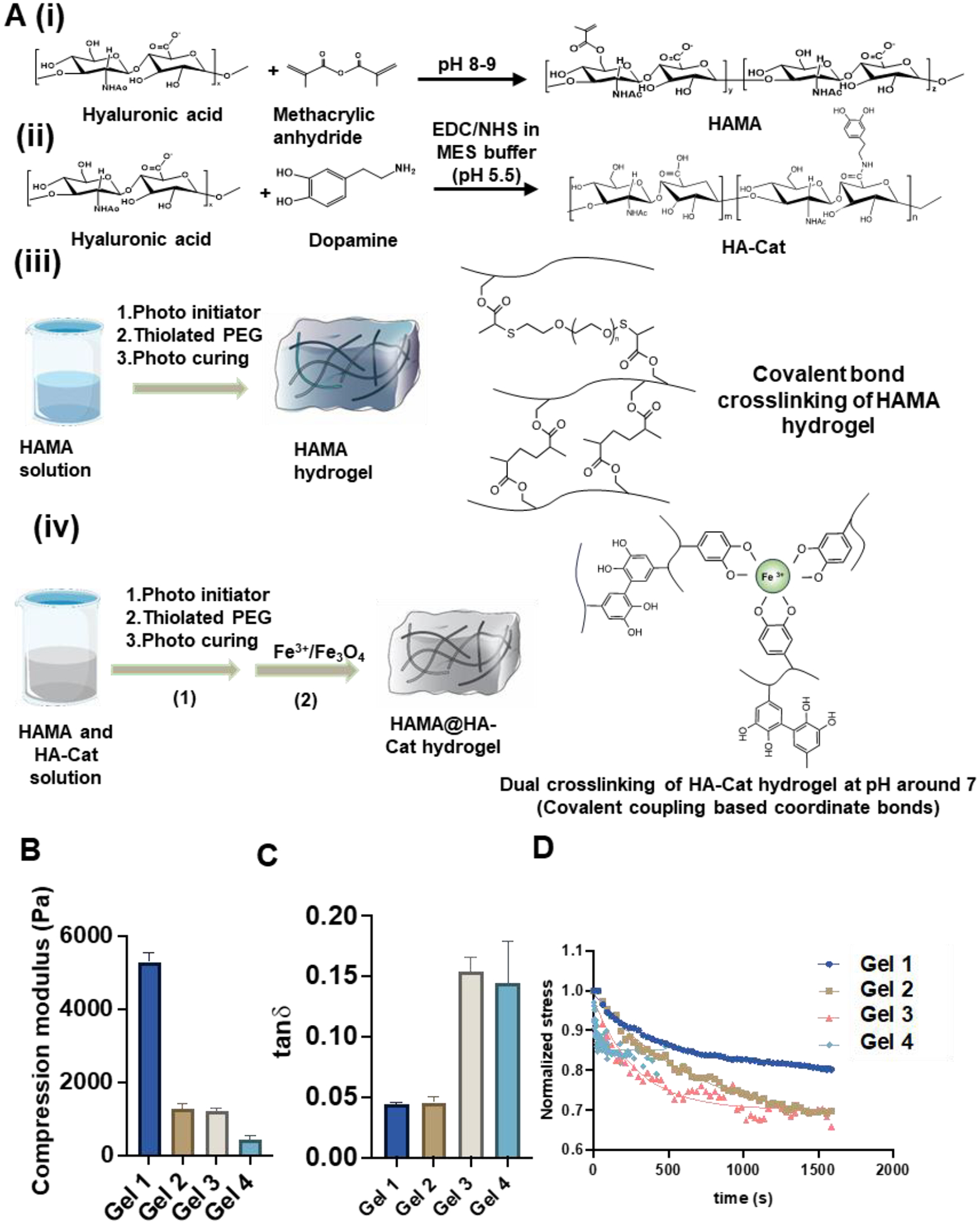
(A) Schematic illustration of methods of HAMA and HA-Cat synthesis. (i-iii) Schematic process of fabrication and synthesis of (i) HAMA, (ii) HAMA@HA-Cat, and (iii) hydrogels. (iv) Schematic illustration of the process of Fe3+ curing HA-Cat hydrogels. (B) Quantification of the compression modulus, and (C) tanδ of the hydrogels by compression and rheological test. (n > 3 measurements per gel). (D) Stress relaxation test was applied to the 4 selected HAMA hydrogels and regression was performed by a modified Maxwell model to get stress relaxation time.
HAMA hydrogels were fabricated with 3 different molecular weights and each sample was dissolved in PBS at 1%, 0.5%, and 0.25% (w/w) concentration (Supplementary Figure S1B). Different concentrations of HAMA could change the degree of crosslinking during gelation which leads to different mechanical properties [23]. Then, the mechanical and rheological properties were tested using a TA Ares-G2 (Figure 1B, 1C and Supplementary Figure S1C, S1D, S2, S3). The storage modulus of the hydrogels decreased, and viscoelasticity increased with decrease in the mass fraction. The tanδ of the gels was between 0.044 and 0.154 for the selected groups (Gel 1–4). Then a group of compression modulus (E) was derived from compression test and was found to be in the range of 400 and 7,000 Pa. In Table 1, four types of hydrogels were selected from the 9 synthesized hydrogels, which can provide stiff-elastic (Gel 1), soft-elastic (Gel 2), stiff-viscoelastic (Gel 3), and soft-viscoelastic (Gel 4) hydrogel conditions. Usually, stress relaxation is used to evaluate the viscoelasticity of the polymer materials. Therefore, the stress relaxation test was also performed for the four HAMA hydrogels and the data were regressed with updated maxwell model (Figure 1D) [56].
Table 1.
Properties of HAMA hydrogels with different molecular weights and concentrations.
| Parameters | 100k 1% | 100k 0.5% | 100k 0.25% | 200k 1% | 200k 0.5% | 200k 0.25% | 1,000k 1% | 1,000k 0.5% | 1,000k 0.25% |
|---|---|---|---|---|---|---|---|---|---|
| Tan δ | 0.037 | 0.038 | 0.079 | 0.066 | 0.113 | 0.149 | 0.041 | 0.048 | 0.158 |
| E(Pa) | 5283±265 | 1265±143 | 605±97 | 5050±348 | 2624±112 | 1208±90 | 7456±539 | 2213±384 | 419±127 |
| Stress relaxation time(s) | 420.1 | 666.7 | 283.4 | 19.7 | |||||
| Category | stiff-elastic | soft-elastic | stiff-viscoelastic | soft-viscoelastic | |||||
| ID | Gel 1 | Gel 2 | Gel 3 | Gel 4 |
Where σ stands for stress (Pa), stands for initial stress (Pa), t stands for time (s), τ stands for relaxation time (s) and C is a constant relating to the crosslink of the polymer. The four stress relaxation times are 420 s, 660 s, 266 s, and 19 s, respectively. All relaxation times (τ) of the four samples are relatively short but significantly different. The degree of crosslinking of the hydrogels may be high with less fluid or dynamic part. The morphology of HAMA hydrogels is shown in SEM images, with visible porous structure (Supplementary Figure S4). These results indicate that the hydrogels with different viscoelastic properties, i.e., stiff-elastic (Gel 1), soft-elastic (Gel 2), stiff-viscoelastic (Gel 3), and soft-viscoelastic (Gel 4), can be fabricated.
3.2. Evaluation of hSCO derivation from hiPSCs
The schematic illustration of hSCO differentiation from hiPSCs reveals that differentiation was induced using LDN193189 (inhibition of bone morphogenetic protein signaling), CHIR (Wnt activation), RA (retinoid activation), and Purmorphamine (Sonic Hedgehog signaling activation) (Figure 2A) [47]. The ventral spinal cord organoids were generated and characterized for spinal cord markers of different regions, including dorsal, interneuron, and ventral markers (Supplementary Table S3). The marker expression was compared with undifferentiated hiPSC aggregates (Supplementary Figure S5A). After 23 days of differentiation, the gene expression of ventral markers (SOX2, LHX3, NKX2.2, OLIG2) for the hSCO group was much higher than the hiPSC group, indicating the effective induction of hSCO lineage. PAX6 (a progenitor marker) had no difference between the hSCOs and the hiPSCs while ISL1 (a progenitor marker) and Nanog (a pluripotent gene) were higher for the hiPSC group. The current differentiation protocol (referred to as hSCOA) was furthered compared with a caudal hSCO differentiation protocol (referred to as hSCOB, in Supplementary Method 1), which used SB431542 (inhibition of transforming growth factor signaling), CHIR, RA, and bFGF (Supplementary Figure S5B). hSCOA conditions showed higher LHX3, NKX2.2, OLIG2 expression, while PAX6 was comparable among the three groups. Based on these results, the hSCOA protocol was selected for the subsequent experiments. The hSCOs can be replated onto Matrigel-coated surface. Extended axons from the replated organoids were observed and the edges remained intact until day 44 (Supplementary Figure S6). The hSCOs were maintained in the Vertical-wheel bioreactor for long-term culture until day 80, which showed larger organoids (~ 2 mm) with the defined organoid edges.
Figure 2. Ventral hSCO differentiation and characterization.
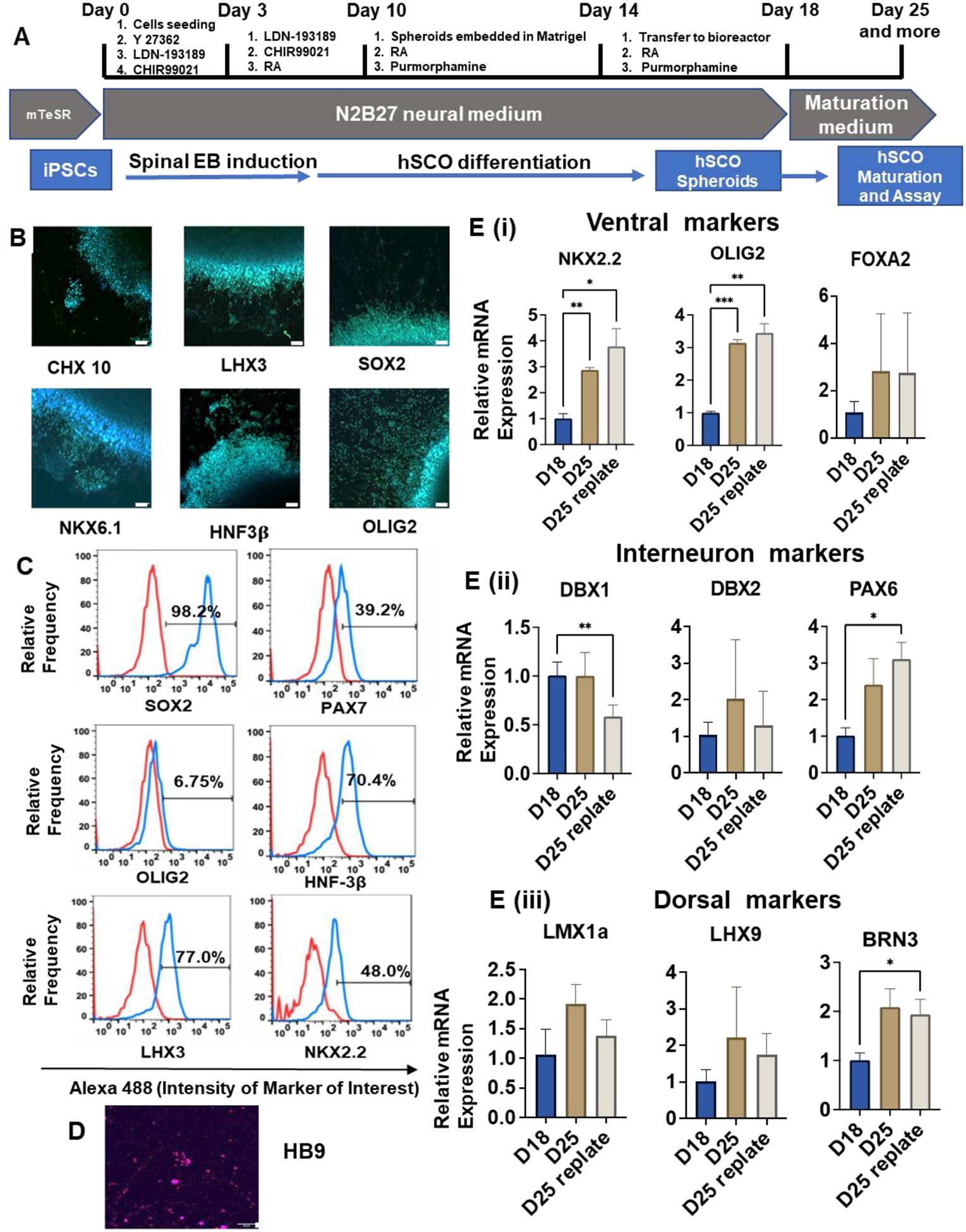
(A) Schematic illustration of ventral hSCO differentiation protocol. (B, C, D) Immunostaining and flow cytometry analysis for marker expression of hSCO differentiation. (B) and (D) were taken using confocal microscopy. Scale bar = 50 μm. (E) Quantitative RT-PCR for relative mRNA expression of various spinal cord markers after biochemical induction (n=3). (i) Ventral markers; (ii) Interneuron markers; (iii) Dorsal markers. * indicates p≤0.05, **: p≤0.01, ***: p≤0.001.
To verify the hSCO marker expression at the protein level, the day 18 organoids were replated for immunostaining (Figure 2B and Supplementary Figure S7). Seven hSCO patterning markers were evaluated, and the expression of CHX10, LHX3, NKX6.1, HNF3β, and OLIG2 was observed. In addition, neuroepithelial marker SOX2 had a high expression, indicating hSCO induction. Flow cytometry analysis was performed on day 25 samples to quantify the marker expression. Most ventral markers (except OLIG2) showed high expression (98.2% SOX2, 70.4% HNF3β, and 48.0% NKX2.2). Dorsal markers PAX7 (39.2%) and LHX3 (77.0%) were also expressed (Figure 2C). Of note, HB9, one of the motor neuron markers, was detected (Figure 2D). To evaluate hSCO patterning, RT-PCR was performed to characterize gene expression of different functional regions of the spinal cord (Figure 2E). For the ventral markers, NKX2.2 and OLIG2 had increased expression (3–4 fold) after one week of culture (day 25 vs. day 18, replated or not), when growth factors were withdrawn for maturation. FOXA2 showed no statistical difference. For interneuron markers, the expression of PAX6 was increased (~ 3-fold), but not DBX1 and DBX2. The increased PAX6 expression may be due to the maturation of specific neural cells. For dorsal markers, BRN3 was expressed more (~ 2-fold) after one week maturation, while the increase was not statistically significant for LMX1a and LHX9. These results indicate the effective hSCO derivation from hiPSCs for the investigation of hSCO patterning in the hydrogels and the extended differentiation time promotes hSCO maturation.
3.3. hSCO patterning in hydrogels with different stiffness and viscoelasticity
Next, for further hSCO differentiation within the hydrogels, the biocompatibility of the HAMA hydrogels was firstly investigated in 2D undifferentiated hiPSC culture and 3D undifferentiated hiPSC spheroids. The hiPSCs grew well when culturing with HAMA hydrogels during the 7-day period (Figure 3A). By adding Matrigel, the adhesion of hiPSCs on 2D surface was improved. For 3D culture, the morphology (e.g., size) of hiPSC spheroids was similar with or without the addition of Matrigel. Then, hiPSCs were cultured in four different gels with a cell-only control. DNA assay was performed to evaluate cell growth and Live/Dead assay was performed to measure cell viability. The normalized DNA concentration shows that the proliferation of hiPSCs cultured with different HAMA hydrogels was comparable (Figure 3B). The proliferation rates were lower than the cell-only condition which was expected because some of the hiPSCs were embedded into gels and did not proliferate much. For the Live/Dead assay, the five groups showed similar results with about 90% of live cells (Figure 3C), which indicates that all the hydrogels have good biocompatibility for hiPSCs.
Figure 3. Biocompatibility of the HAMA hydrogels and morphogenesis of the organoids.
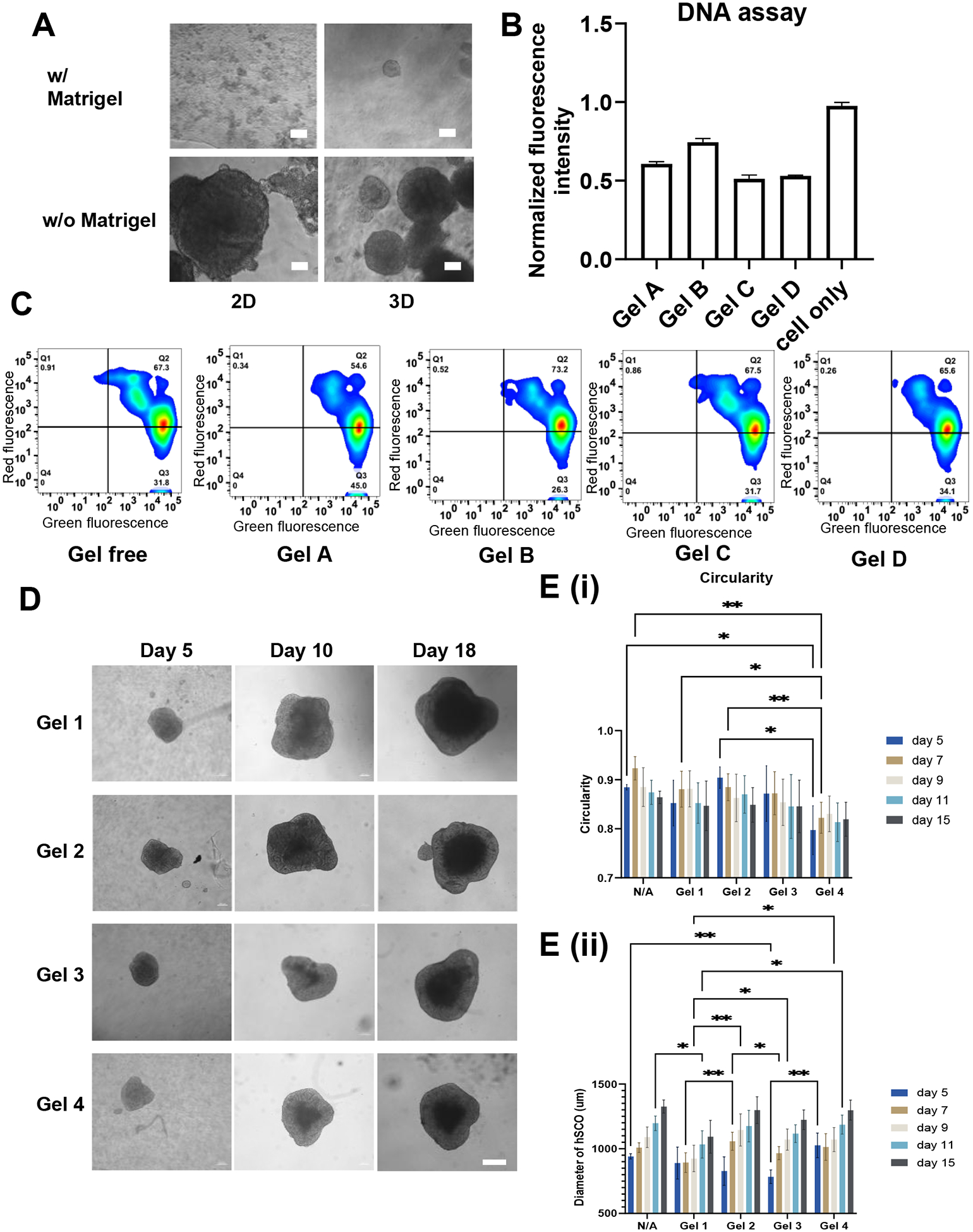
(A) hiPSC culture with HAMA and HAMA/Matrigel mixture for 7 days. Scale bar = 50 μm. (B) DNA assay and (C) Live/Dead flow cytometry analysis for determining proliferation rate and survival rate of hiPSCs cultured with different HAMA hydrogels, respectively. (D) Images of morphology of the organoids with different hydrogels over the time. Scale bar = 200 μm. (E) Quantification of diameter and circularity of hSCOs cultured in different HAMA hydrogels for morphogenesis. * indicates p≤0.05, **: p≤0.01, ***: p≤0.001.
After the biocompatibility test, the spinal cord organoids (hSCOs) derived from hiPSCs were patterned in different HAMA hydrogels. Images of the formed spheroids in the four different hydrogels were taken over 18 days of differentiation (Figure 3D and Supplementary Figure S8). The size of the spheroids increased significantly from day 5 (about 500–800 μm) to day 18 (about 1.5–1.8 mm). Image analysis was performed based on spheroid morphology to reveal if different HAMA hydrogels affect the spheroid size and shape. The quantitative summary of the diameter and circularity is shown in Figure 3E. The diameters of all spheroids were similar (~1.4 mm) for different hydrogel groups at day 15, however, they were different on the days prior to day 15, showing different growth kinetics of the spheroids. For example, the Gel 3 group started with the smaller spheroids but the spheroid size quickly increased to a size similar to the other groups. In addition, all spheroids in the HAMA hydrogels can freely grow without constriction from the hydrogels during the culture, which contributes to the size increase during the differentiation. The circularities of the spheroids all decreased over the course of differentiation (the closer to 1 the more circular). Only the spheroids in Gel 4 were less circular than the other conditions in the initial few days. These results indicate that the four types of HAMA hydrogels all support hSCO patterning.
After day 25, flow cytometry was performed to quantify hSCO marker expression at the protein level among different culture conditions (Figure 4 and Supplementary Figure S9). The ventral markers of hSCOs were evaluated, and similar expression levels among different hydrogel groups were observed. The LHX3 (70–90%) and HNF3β (60–80%) had high expression while Nkx2.2 was expressed at 17–28%, OLIG2 was 10–16%, and CHX10 was 8–12%. Nkx6.1 expression was low around 2–7%. PAX7 showed large variations of 12–67%. The data from three different runs were normalized to the cell-only group and then combined together to make comparisons (Figure 4A, 4B, and Supplementary Figure S8). There were large variations among three different runs and no statistical difference was observed, which may be attributed to organoid-to-organoid variations [57].
Figure 4. Characterization for differentiation of hSCOs in different HAMA hydrogels.
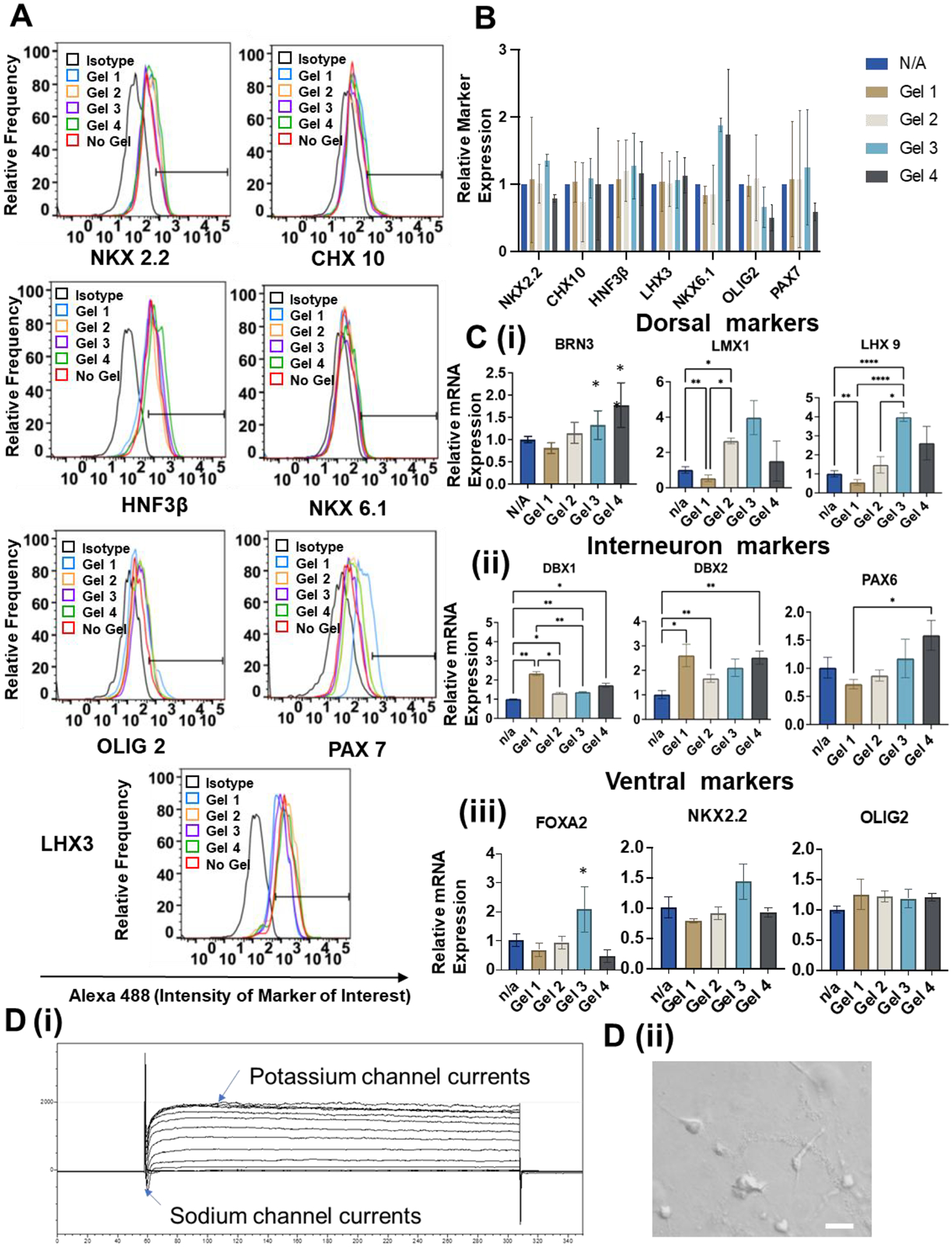
(A) Flow cytometry analysis of expression of different ventral markers when generating hSCOs in different hydrogels. (B) Summary of 3 runs of flow cytometry analysis for identification ventral hSCO marker expression. (C) RT-PCR analysis of relative mRNA expression for different region-specific patterning markers during generation of hSCOs at day 35 (n=3). * indicates p≤0.05, **: p≤0.01, ***: p≤0.001. (D) (i) Electrophysiology to show sodium and potassium currents for the replated hSCOs at day 40. (ii) Morphology of outgrowth cells of the replated hSCOs for electrophysiology. Scale bar = 20 μm.
Furthermore, RT-PCR was performed to evaluate patterning markers at the molecular level (Figure 4C). For dorsal markers, Gel 1 reduced the expression of LMX1 and LHX9 while the highest expression was observed for the Gel 3 group. BRN3 expression was higher for the Gel 3 and Gel 4 groups in comparison to the Gel 1 group. For the interneuron marker expression, the presence of the hydrogels increased the expression of interneuron markers DBX1 and DBX2. Comparing with other gels, Gel 1 promoted higher expression of DBX1. PAX6 expression was higher for the Gel 3 and Gel 4 group in comparison to the Gel 1 group. For the ventral marker expression, Gel 3 promoted higher expression of FOXA2 and NKX2.2 in comparison to other conditions. There was no statistical difference among different conditions for OLIG2. Taken together, Gel 3 (stiff-viscoelastic) promoted dorsal and ventral marker expression and Gel 1 (stiff-elastic) promoted interneuron marker expression during hSCO patterning. These results indicate that the stiffer hydrogels are preferred for hSCO differentiation and the viscoelastic hydrogels promote regional hSCO patterning compared to the elastic hydrogels.
Electrophysiology was performed to show the functional properties of hSCOs (Figure 4D). The electrophysiological properties of the outgrowth cells of the derived organoids were examined via patch clamping. The replated organoids displayed fast inward currents and long-lasting outward currents during voltage-clamp recording, suggesting the presence of functional voltage-gated Na+ and K+ channels, respectively.
3.4. Coculture of hSCOs from different hydrogels with hBVOs
The protocol of hBVO differentiation from hiPSCs was firstly evaluated for the marker expression of the endothelial cells (CD31, VWF) and tight junction (CDH5, CLDN1, ZO-1, OCLN, SELP, and GFAP) of the BSCB at different seeding densities (10,000, 20,000, and 30,000 cells/well in ULA 96-well plate) and replating (re) conditions (Supplementary Figure S10A). The vascular differentiation (vsc) from hiPSCs was also compared (Supplementary Method 2) [46]. Only CDH5 showed different expression levels among different conditions. The 30k, 30k re, and 30k vsc showed higher CDH5 expression than other densities. For the rest of the markers, the 10k conditions had higher expression in general. In addition, the replated organoids did not show higher marker expression in comparison to organoids in suspension. Based on these RT-PCR results, the suspension culture and the seeding density of 10k cells/well were selected for the generation of hBVOs. Then, the BMP4 and VEGF alone or in combination were tested as additional growth factors for hBVO generation (Supplementary Figure S10B). BMP4 significantly decreased the expression of tight junction and BSCB markers. Adding VEGF (with or without BMP4) did not significantly increase the marker expression in general. Therefore, the hBVO differentiation protocol without additional BMP4 and VEGF was used for subsequent experiments.
Afterwards, the assembly of hBVOs and hSCOs in the presence of HAMA hydrogels was performed (Figure 5A). The hBVOs were labeled with CellTracker™ Red and transferred to a well containing one hSCO. The fusion of the hBVO with the hSCO was indicated by the red color inside the hSCO. With VEGF, the fusion rate was much faster than the other two conditions of no growth factor or with BMP4. After coculture for 3 weeks, the merged organoids (i.e., hSCO-hBVO) from five different conditions (i.e., Gel 1–4 and Gel-free) were harvested for RT-PCR analysis for the expression of spinal cord markers (Figure 5B) or BSCB markers (Figure 5C) [12]. For the spinal cord markers, DBX1 and LMX1a were higher for the Gel 2 group than the Gel 1 and Gel 3 groups, but comparable to the Gel 4 group. ISL1 had no difference among different hydrogel groups. The expression of OLIG2 of hSCOs derived from the Gel 4 group was higher than the Gel 1 group. NKX2.2 expression was the highest for the Gel 2 condition compared to the other groups. Taken together, the presence of hBVOs altered the influence of different hydrogels on spinal cord organoid patterning. Gel 2 (soft-elastic) promoted dorsal and interneuron markers as well as NKX2.2, while Gel 3 and 4 (viscoelastic) promoted ventral marker OLIG2 expression. These results indicate that the effects of viscoelastic properties of the hydrogels become more dominant than the stiffness effects.
Figure 5. hSCO and hBVO coculturing for Blood-Spinal Cord Barrier (BSCB) generation.
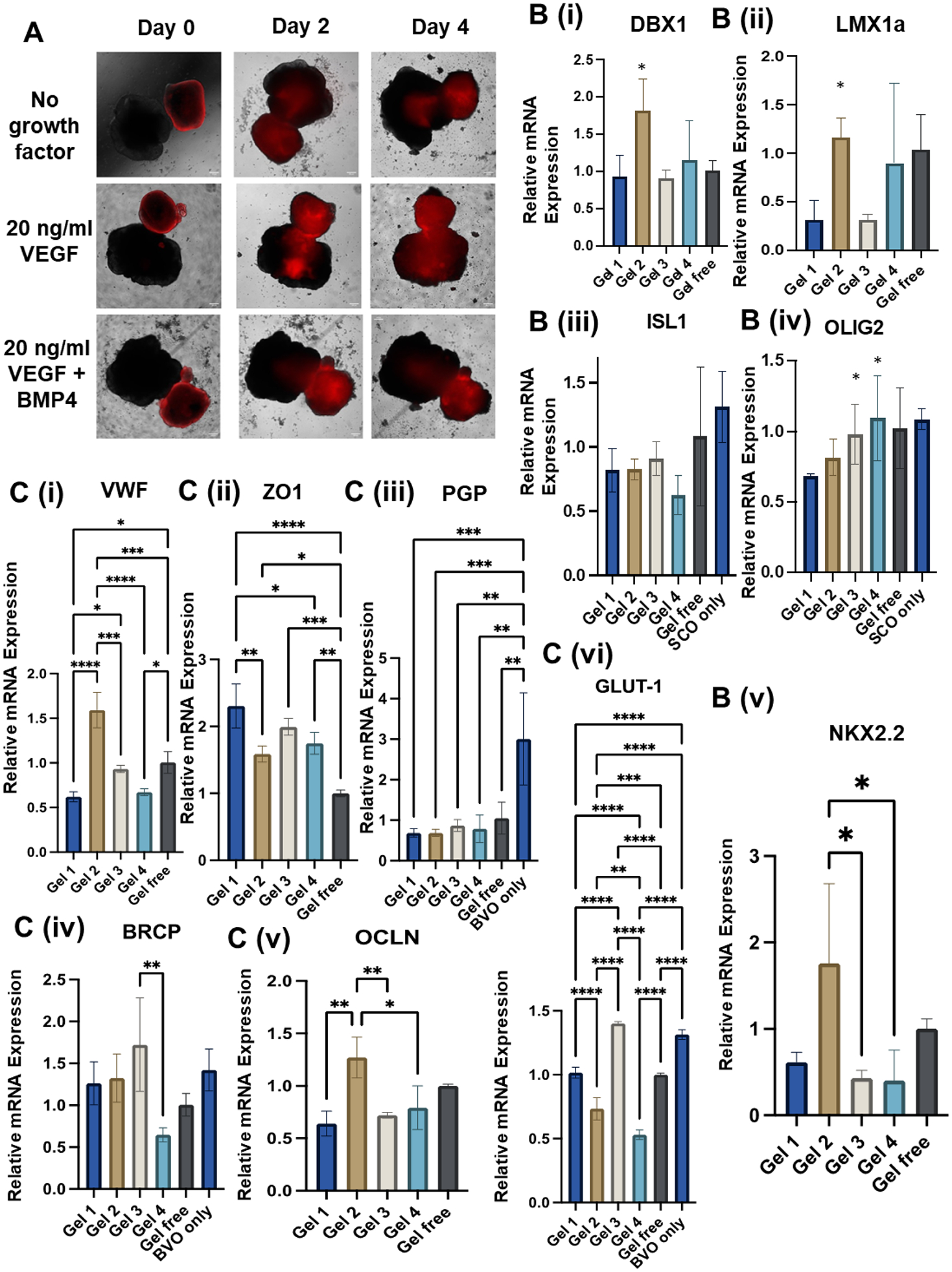
(A) Morphology of the merging process of two types of organoids indicated by cell-tracker (red) hBVOs. (B, C) RT-PCR analysis for relative mRNA expression of ventral spinal cord genes, endothelial cells (EC), and blood-brain barrier (BBB) genes during hBVO and different hSCO coculturing. n=3, ns: p>0.05, * indicates p≤0.05, **: p≤0.01, ***: p≤0.001.
For the BSCB markers in the fused hSCO-hBVO, the Gel 2 condition had the highest VWF (i) and OCLN (v) expression in comparison to the other groups (Figure 5C). These markers are important for identifying the tight junction during coculture [58]. The tight junction protein ZO-1 (ii) was expressed higher in Gel 1 and Gel 3 groups when compared to the other conditions. The expression of glucose transporter 1 (GLUT-1) and efflux transporters, BCRP and PGP, was also determined. For PGP (iii), all the hSCO-hBVO conditions showed lower expression than the hBVO only group, due to the presence of hSCO cells. BRCP (iv) expression was comparable for all the conditions except for the Gel 4 group, which had lower expression. For GLUT-1 (vi), the Gel 3 group had the highest expression while Gel 4 had the lowest expression. Taken together, the BSCB markers were differentially affected by the viscoelastic properties of the HAMA hydrogels. Gel 2 (soft-elastic) promotes the tight junction and Gel 3 (stiff-viscoelastic) promotes the expression of glucose and efflux transporters.
3.5. HAMA-Cat (Fe3+) dynamic hydrogels for continuous hydrogel improvement
In addition to single covalent hydrogels that were investigated so far, the viscoelastic dual hydrogels with dynamic crosslinking bonds may also affect hSCO patterning. HAMA was crosslinked with covalent bonds and the entanglement of the chains provides the dynamic part that contributes to the viscoelastic behaviors of the hydrogels. In addition to modifying the covalent crosslinked HAMA hydrogels with dynamic crosslinked properties, dopamine, which has a catechol group, was grafted on the HA to synthesize HA-Cat (Figure 6). The catechol group can react with ferric ions (Fe3+) with coordination. The HAMA and HA-Cat polymers were mixed with the same concentration as the four HAMA hydrogels (100 k, 1%, 100 k 0.5%, 200 k 0.25%, 1000 k 0.25%), and the HAMA@HA-Cat hydrogels were fabricated which are referred as Gel 5, Gel 6, Gel 7, and Gel 8, respectively.
Figure 6. Fabrication and characterization of HAMA@HA-Cat hydrogels.
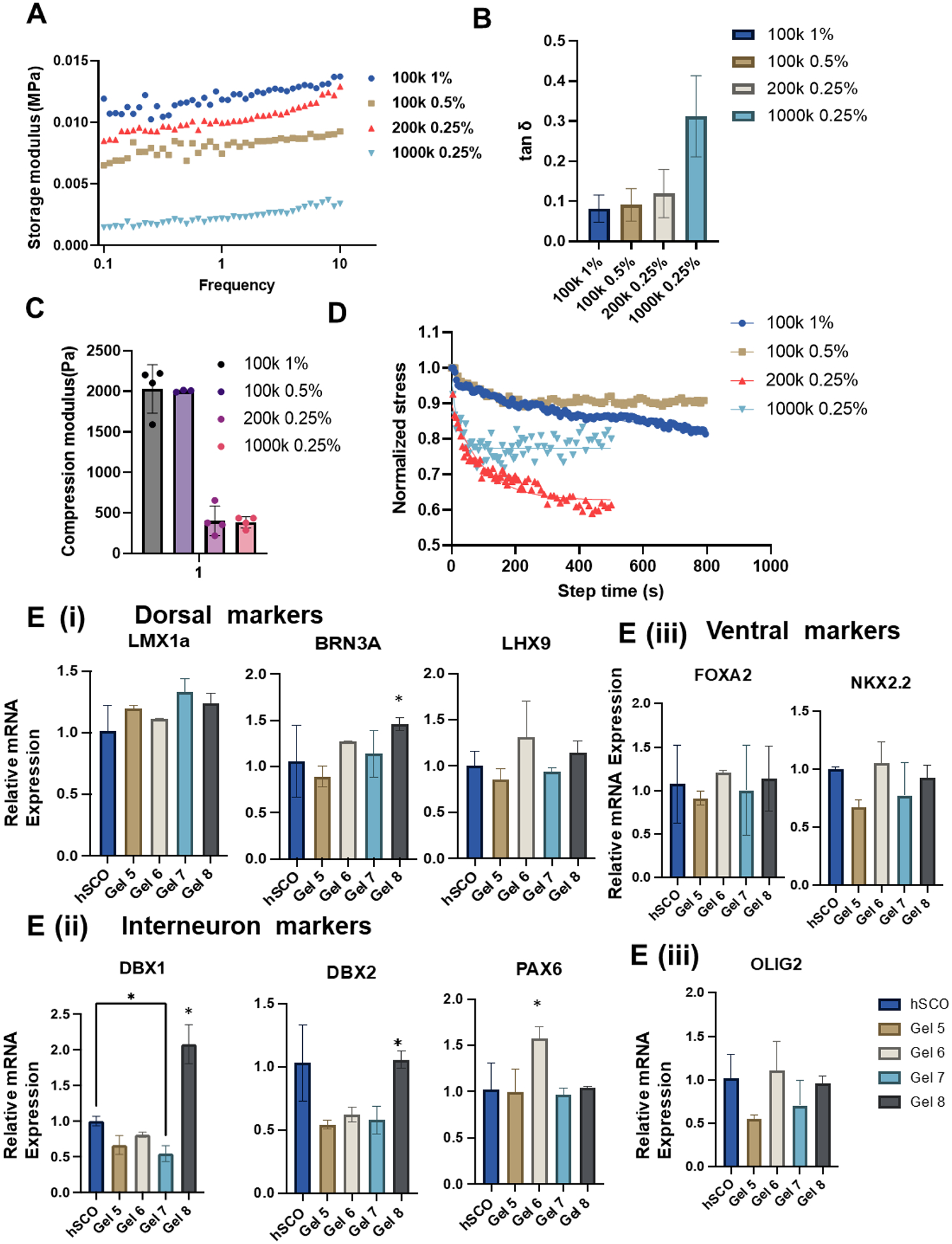
The dynamic hydrogels were fabricated to enhance the hydrogel properties and potential ability to regulate hSCO derivation. (A, B, C) Rheological test and compression test were performed to determine mechanical properties for the four new hydrogels (Gel 5–8 in sequence). (A) Storage modulus; (B) tanδ; (C) Compression modulus; (D) The viscoelasticity of the hydrogels was further determined by stress relaxation test. (E) RT-PCR analysis of relative mRNA expression for different region-specific patterning markers of hSCOs at day 35. n=3, * indicates p≤0.05.
The mechanical properties of the four HAMA@HA-Cat hydrogels (Gel 5–8) were characterized. Using rheological tests, the shear modulus (Figure 6A) and tanδ (Figure 6B) of the four gels were measured. The modulus of Gel 8 was much lower than the other three groups and it had the highest tanδ of 0.3 in this study. Meanwhile, the compression modulus was determined, where Gel 5 and Gel 6 had similar compression moduli of ~2,000 Pa and Gel 7 and Gel 8 had similar compression moduli of ~400 Pa (Figure 6C). Furthermore, the stress relaxation of the four new hydrogels was evaluated, and the Maxwell model’s regression was used to get the stress relaxation times of 373.0 s, 67.5 s, 94.6 s, and 19.4 s, respectively (Figure 6D), all of which are less than those of the HAMA hydrogels. The hSCO patterning in the four HAMA@HA-Cat hydrogels was investigated (Figure 6E). Gel 8 (the most viscoelastic HAMA@HA-Cat hydrogel) promoted the expression of dorsal marker BRN3A and interneuron markers DBX1 and DBX2 compared to other hydrogel conditions. Gel 6 promoted PAX6 expression. The ventral marker expression was not affected by different hydrogel properties. These results indicate that the viscoelasticity of dynamic hydrogels promotes hSCO patterning.
3.6. Mechanism of hydrogel effects on hSCO patterning
Considering the possible interplay between Hippo pathway and viscoelasticity of ECM, this study investigated the mechano-transduction mechanism by comparing the localization of YAP expression within the nuclei and cytoplasm (Figure 7) [46, 55, 59]. For this purpose, the hSCOs derived from different HAMA hydrogels were embedded in paraffin and sectioned into slices (6 μm) for better imaging of 3D structure. The YAP and Hoechst staining were observed in the hSCOs from different hydrogel groups (Figure 7A). In addition, the localization of YAP in the nuclei and cytoplasm was compared through image analysis by ImageJ (Figure 7B). The hSCOs from the more elastic hydrogel groups (Gel 1 and Gel 2) had lower YAP nuclear localization when comparing to the hSCOs from viscoelastic hydrogels (Gel 3 and 4). Of note, the hydrogels with similar tanδ but different modulus had no difference in YAP nuclear localization between each other, such as Gel 1and 2, Gel 3 and 4. These results indicate that more viscoelastic hydrogels could lead to more nuclear YAP localization.
Figure 7. Histological sections for YAP localization to reveal the mechanism of hydrogel effects on hSCO patterning.
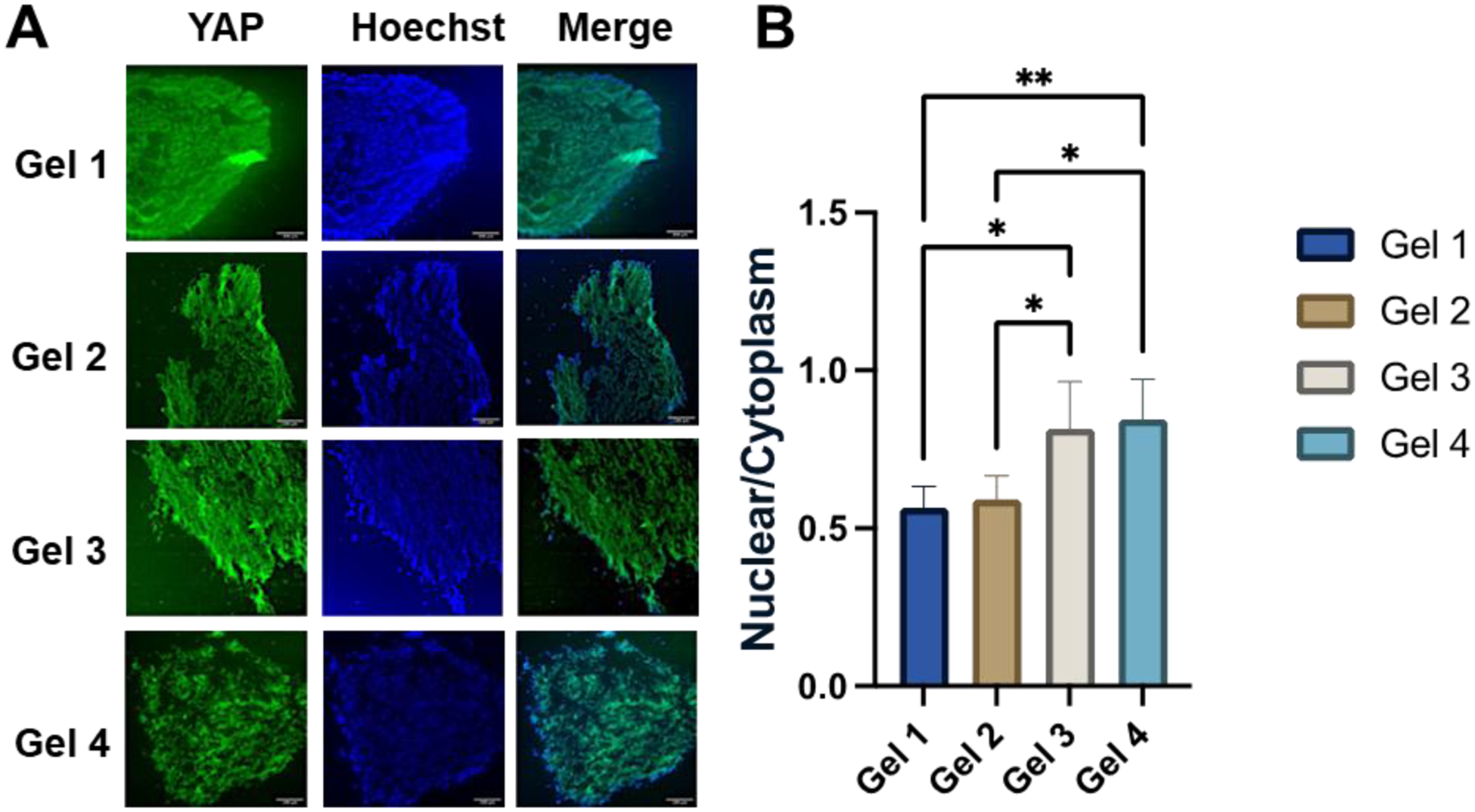
(A) Images of YAP localization. Scale bar: 50 μm. (B) The quantitative measurements of nuclear to cytoplasmic YAP localization for different hydrogel conditions. * indicates p≤0.05, **: p≤0.01.
4. Discussion
In this study, our findings provide a series of conclusions for 3D HAMA hydrogel microenvironments that influence the morphogenesis of hSCO and hSCO patterning under different viscoelasticity and stiffness of the static hydrogels as well as dynamic hydrogels. Using different concentrations and molecular weights of HA for crosslinking, the library of HAMA hydrogels with different mechanical property was established. For example, by using HA of different molecular weight with the same PEG-SH, the prorates can be manipulated. In addition, the ionic crosslinking mechanism and catechol chemistry were applied for hydrogel fabrication. Then, two groups of hydrogels with the similar modulus but different tanδ (Gel 2 and Gel 3) were selected for mimicking different ECM properties. The effects of stress relaxation were tested to reveal the influence of the viscoelasticity of HAMA hydrogels on spinal cord organoid patterning. Another two groups of hydrogels were included to show the effects of hydrogels with the similar tanδ but different modulus (Gel 1 vs. Gel 2, Gel 3 vs. Gel4). At the early stage of lineage-specific hSCO differentiation, the culture kinetics of the size and circularity of organoids were affected by different hydrogels, e.g., Gel 3 group showed initial small size but later became similar to other conditions. For the differentiation and patterning of ventral hSCOs, the stiffness and viscoelasticity of the hydrogels had a greater influence on dorsal and interneuron marker expression but less on ventral markers.
It was only recently that the viscoelastic, or time-dependent properties of the extracellular environment, has been shown to have significant influences on cell and tissue behaviors [23]. To date, only a few reports focused on the influence of tunable viscoelastic property of the 3D matrices on the interactions between stem cells and the microenvironment [60]. Conventionally, the reports of culturing mesenchymal stem cells with 2D or 3D matrices are abundant, while the investigations of hiPSC-derived organoid generation in 3D viscoelastic matrices are still limited [61]. The matrix viscoelasticity has just recently been recognized as a key component for regulating stem cell organoid morphogenesis for tumor and intestine tissue modeling [22, 23]. Tunable stress relaxation (viscoelasticity), stiffness, and RGD ligands were shown to have significant effects on hiPSC apicobasal polarization and lumen formation [22]. Furthermore, the viscoelastic properties of HA hydrogels has been observed to promote human neural progenitor cell maturation in 2D culture, with faster stress-relaxation increasing neurite extension and decreasing metabolic activity [62]. Here, the HAMA and HAMA@HACat hydrogels were fabricated by simple synthetic methods that provided a range of different biophysical properties. Four categories of hydrogels were selected for patterning hiPSC-derived spinal cord organoids which provide specific microenvironments for hSCO differentiation. Furthermore, our study can maintain the similar stiffness and the same polymer composition during the process of hydrogel fabrication while still allowing for the generation of hydrogels with different viscoelastic properties. Therefore, the effects of the viscoelasticity of hydrogels on the hSCO morphogenesis and differentiation can be isolated from the stiffness effects, serving as the main variable of the biophysical cues. The influence of the porosity of the hydrogels was thought to be small and the difference in different hydrogels was small.
HA was chosen due to its abundance in human central nervous system and its ability to generate unique matrices to compare with other natural polymers, such as alginate, gelatin, etc. HA and its derivatives have been currently used as 3D matrices for cell/tissue culture, especially in 3D printing and as granular hydrogels [63–65]. The synthesis of HAMA hydrogels was based on a classical and simple method that can provide a series of hydrogels. As a result of the limited extent of grafting on the HA chains, the low degree of crosslinking leads to the entanglement of free HAMA chains and results in the viscoelastic properties of the HAMA hydrogels to some extent. Biochemical and biophysical properties are both important for hiPSC-derived organoids morphogenesis and patterning. Usually, the two factors have different effects on the regulation of fate decision of hiPSCs that are intrinsically sensitive to their biophysical and biochemical environment [66–68]. In addition, the spinal cord injury repair can be realized using the synthetic scaffolds with various biochemical and biophysical cues [69]. Once embedded within HA hydrogels, hiPSCs sense the signals from the matrix during embryoid body formation, differentiation induction, expansion, and hSCO patterning, which leads to different morphogenesis results. Furthermore, the Matrigel free condition provides low matrix affinity microenvironments for hiPSC spheroid formation and inhibits hiPSC expansion or attachment. Therefore, these HA hydrogels without any cell-attachment factors can provide biophysical signaling for organoid patterning in suspension with minimal influence of biochemical signaling. Furthermore, the analysis of YAP localization provides another angle to understand mechano-transduction mechanism of HAMA hydrogels with different modulus and viscoelasticity. Our study found that the nuclear translocation of YAP increases for the hydrogels with faster stress relaxation for both values of elastic moduli (Gel 3 and Gel 4). These results are consistent with previous study using 2D substrate culture [70, 71].
Biophysical cues such as stiffness, nanotopography, and mechanical strain can regulate the fate of the hiPSCs, such as maintaining the pluripotency or inducing differentiation. For example, substrate stiffness can influence neural induction and subtype specification of hiPSCs [72]. In addition, the topographic properties of the substrates can promote hiPSC differentiation into specific neural lineage [73]. In addition to stimulation by biophysical stimuli, directional growth and lineage-specific development of hiPSCs can be facilitated by biochemical factors. Therefore, the competition and synergistic effects between these two types of factors need to be investigated. Based on our findings, viscoelastic (i.e., high tanδ or shorter relaxation time) microenvironments promote dorsal or interneuron marker expression of hSCOs. Specifically, the stiffer hydrogels are preferred for hSCO differentiation than the softer hydrogels, and the viscoelastic hydrogels promote regional hSCO patterning compared to the elastic hydrogels. The growth factors that were added to the cultures were primarily for ventral organoid differentiation. The sonic hedgehog activator and RA are the two key factors for ventral patterning of spinal cord organoids. Comparing the ventral markers of different hSCOs from different hydrogels showed no significant difference, showing that the ventral markers are mainly affected by the differentiation factors, not the biophysical properties of hydrogels.
Vascularization is essential to the growth, maturity, and function of organoids, as a crucial component in organoid development. The ability to remove waste materials and supply nutrients and oxygen to the cells inside the organoids depends on proper vascularization. Several techniques are used to promote vascularization in organoids, including: co-culture with ECs [74], embedding in Matrigel [75], microfluidic systems [76, 77], decellularized tissue scaffolds [78], and in vivo maturation [79]. In our study, co-culturing hSCOs with hBVOh for organoid fusion was used. The presence of hBVOs altered the influence of different hydrogels on spinal cord organoid patterning. The effects of viscoelastic properties of the hydrogels become more dominant than the stiffness effects. The presence of hSCOs also had effects on the expression of EC, tight junction, and BSCB markers in the presence of hydrogels. For example, soft-elastic hydrogels promoted the tight junction and stiff-viscoelastic hydrogels appeared to promote the expression of glucose and efflux transporters. However, the hBVOs in this study were not mature and the main assessment was based on the gene expression. The vascularization structure was not assessed, which may need the mature hBVOs and dynamic perfusion culture environment.
5. Conclusion
This study fabricated HAMA hydrogels with different modulus and viscoelasticity to regulate hSCO patterning and co-culture with hBVOs. The four hydrogels are mainly separated into 2 groups, the elastic and viscoelastic groups. After testing hSCO differentiation and biocompatibility of the four hydrogels, the morphogenesis of hSCOs were observed. The viscoelasticity of the hydrogels influenced the size and circularity. Then, by comparing the gene and protein expression of hSCOs with different hydrogels, the results reveal that the stiffer hydrogels are preferred for hSCO differentiation and the viscoelastic hydrogels promote regional hSCO patterning compared to the elastic hydrogels. By coculturing hSCOs and hBVOs, this study was able to create a fusion of the two organoids. In the presence of hBVOs, the effects of viscoelastic properties became more dominant than the stiffness effects. Soft-elastic hydrogels promoted the tight junction and stiff-viscoelastic hydrogels appeared to promote the expression of glucose and efflux transporters. The viscoelasticity of dynamic hydrogels was also found to promote hSCO patterning. Furthermore, by analysis of the localization of YAP, this study found that the nuclei localization increased in the faster relaxation hydrogel groups.
Supplementary Material
Acknowledgments
The authors would thank for the support by FSU Flow Cytometry core facility, FSU High Performance Materials Institute for hydrogel characterization, and Dr. Brian K. Washburn at FSU Department of Biological Sciences for his help with RT-PCR analysis. The authors would also thank for the lyophilizers provided by Dr. Danial Hallinan and Dr. Ho Yong Chung, and access to SEM at National High Magnetic Field Laboratory. This work is supported by National Science Foundation (USA), CBET-1917618 (to YL and CZ) and CMMI-2100987 (to YL). Research reported in this publication was also partially supported by the National Institutes of Health (USA) under Award Number R01NS125016 (to YL) and R21EB033495 (to XW and YL). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing interests:
Authors declare that they have no competing interests.
Data and materials availability:
The datasets generated during and/or analyzed during the current study are available from the corresponding authors on reasonable request.
References
- [1].D’Mello R, Dickenson AH, Spinal cord mechanisms of pain, British journal of anaesthesia 101(1) (2008) 8–16. [DOI] [PubMed] [Google Scholar]
- [2].Revah O, Gore F, Kelley KW, Andersen J, Sakai N, Chen X, Li MY, Birey F, Yang X, Saw NL, Baker SW, Amin ND, Kulkarni S, Mudipalli R, Cui B, Nishino S, Grant GA, Knowles JK, Shamloo M, Huguenard JR, Deisseroth K, Pasca SP, Maturation and circuit integration of transplanted human cortical organoids, Nature 610(7931) (2022) 319–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Du Z-W, Chen H, Liu H, Lu J, Qian K, Huang C-L, Zhong X, Fan F, Zhang S-C, Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells, Nature communications 6(1) (2015) 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Yan Y, Li X, Gao Y, Mathivanan S, Kong L, Tao Y, Dong Y, Li X, Bhattacharyya A, Zhao X, Zhang SC, 3D bioprinting of human neural tissues with functional connectivity, Cell Stem Cell 31(2) (2024) 260–274 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ogura T, Sakaguchi H, Miyamoto S, Takahashi J, Three-dimensional induction of dorsal, intermediate and ventral spinal cord tissues from human pluripotent stem cells, Development 145(16) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kaitsuka T, Hakim F, Response of pluripotent stem cells to environmental stress and its application for directed differentiation, Biology 10(2) (2021) 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yamamoto T, Arita M, Kuroda H, Suzuki T, Kawamata S, Improving the differentiation potential of pluripotent stem cells by optimizing culture conditions, Scientific Reports 12(1) (2022) 14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gribaudo S, Robert R, van Sambeek B, Mirdass C, Lyubimova A, Bouhali K, Ferent J, Morin X, van Oudenaarden A, Nedelec S, Self-organizing models of human trunk organogenesis recapitulate spinal cord and spine co-morphogenesis, Nat Biotechnol (2023). [DOI] [PubMed] [Google Scholar]
- [9].Hor JH, Soh ES-Y, Tan LY, Lim VJW, Santosa MM, Ho BX, Fan Y, Soh B-S, Ng S-Y, Cell cycle inhibitors protect motor neurons in an organoid model of Spinal Muscular Atrophy, Cell death & disease 9(11) (2018) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Xue W, Li B, Liu H, Xiao Y, Li B, Ren L, Li H, Shao Z, Generation of dorsoventral human spinal cord organoids via functionalizing composite scaffold for drug testing, iScience 26(1) (2023) 105898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhou G, Pang S, Li Y, Gao J, Progress in the generation of spinal cord organoids over the past decade and future perspectives, Neural Regen Res 19(5) (2024) 1013–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jin L-Y, Li J, Wang K-F, Xia W-W, Zhu Z-Q, Wang C-R, Li X-F, Liu H-Y, Blood–spinal cord barrier in spinal cord injury: a review, Journal of neurotrauma 38(9) (2021) 1203–1224. [DOI] [PubMed] [Google Scholar]
- [13].Song L, Yuan X, Jones Z, Vied C, Miao Y, Marzano M, Hua T, Sang Q, Guan J, Ma T, Functionalization of brain region-specific spheroids with isogenic microglia-like cells. Sci Rep 9: 11055, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Song L, Yuan X, Jones Z, Griffin K, Zhou Y, Ma T, Li Y, Assembly of human stem cell-derived cortical spheroids and vascular spheroids to model 3-D brain-like tissues, Scientific Reports 9(1) (2019) 5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Murphy WL, McDevitt TC, Engler AJ, Materials as stem cell regulators, Nature materials 13(6) (2014) 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Simian M, Bissell MJ, Organoids: a historical perspective of thinking in three dimensions, Journal of Cell Biology 216(1) (2017) 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lancaster MA, Knoblich JA, Organogenesis in a dish: modeling development and disease using organoid technologies, Science 345(6194) (2014) 1247125. [DOI] [PubMed] [Google Scholar]
- [18].Lutolf MP, Hubbell J, Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering, Nature biotechnology 23(1) (2005) 47–55. [DOI] [PubMed] [Google Scholar]
- [19].Roth JG, Huang MS, Li TL, Feig VR, Jiang Y, Cui B, Greely HT, Bao Z, Paşca SP, Heilshorn SC, Advancing models of neural development with biomaterials, Nature Reviews Neuroscience 22(10) (2021) 593–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chaudhuri O, Cooper-White J, Janmey PA, Mooney DJ, Shenoy VB, Effects of extracellular matrix viscoelasticity on cellular behaviour, Nature 584(7822) (2020) 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Elosegui-Artola A, Gupta A, Najibi AJ, Seo BR, Garry R, Tringides CM, de Lazaro I, Darnell M, Gu W, Zhou Q, Weitz DA, Mahadevan L, Mooney DJ, Matrix viscoelasticity controls spatiotemporal tissue organization, Nat Mater 22 (2023) 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Indana D, Agarwal P, Bhutani N, Chaudhuri O, Viscoelasticity and adhesion signaling in biomaterials control human pluripotent stem cell morphogenesis in 3D culture, Advanced Materials 33(43) (2021) 2101966. [DOI] [PubMed] [Google Scholar]
- [23].Elosegui-Artola A, Gupta A, Najibi AJ, Seo BR, Garry R, Tringides CM, de Lázaro I, Darnell M, Gu W, Zhou Q, Matrix viscoelasticity controls spatiotemporal tissue organization, Nature Materials 22(1) (2023) 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Elosegui-Artola A, The extracellular matrix viscoelasticity as a regulator of cell and tissue dynamics, Curr Opin Cell Biol 72 (2021) 10–18. [DOI] [PubMed] [Google Scholar]
- [25].Wu DT, Jeffreys N, Diba M, Mooney DJ, Viscoelastic biomaterials for tissue regeneration, Tissue Engineering Part C: Methods 28(7) (2022) 289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tang S, Richardson BM, Anseth KS, Dynamic covalent hydrogels as biomaterials to mimic the viscoelasticity of soft tissues, Progress in Materials Science 120 (2021) 100738. [Google Scholar]
- [27].Fraser JRE, Laurent TC, Laurent U, Hyaluronan: its nature, distribution, functions and turnover, Journal of internal medicine 242(1) (1997) 27–33. [DOI] [PubMed] [Google Scholar]
- [28].Dijkgraaf LC, de Bont LG, Boering G, Liem RS, Normal cartilage structure, biochemistry, and metabolism: a review of the literature, Journal of oral and maxillofacial surgery 53(8) (1995) 924–929. [DOI] [PubMed] [Google Scholar]
- [29].Horkay F, Basser PJ, Hecht A-M, Geissler E, Gel-like behavior in aggrecan assemblies, The Journal of chemical physics 128(13) (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhu W, Mow VC, Rosenberg LC, Tang L-H, Determination of kinetic changes of aggrecan-hyaluronan interactions in solution from its rheological properties, Journal of biomechanics 27(5) (1994) 571–579. [DOI] [PubMed] [Google Scholar]
- [31].Fox AS, Bedi A, Rodeo ja SA, The basic science of articular cartilage: Structure, composition, and function Sports Health 1 (2009) 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zimmermann DR, Dours-Zimmermann MT, Extracellular matrix of the central nervous system: from neglect to challenge, Histochemistry and cell biology 130 (2008) 635–653. [DOI] [PubMed] [Google Scholar]
- [33].Rauch U, Brain matrix: structure, turnover and necessity, Biochemical Society Transactions 35(Pt 4) (2007) 656–660. [DOI] [PubMed] [Google Scholar]
- [34].Dicker KT, Gurski LA, Pradhan-Bhatt S, Witt RL, Farach-Carson MC, Jia X, Hyaluronan: a simple polysaccharide with diverse biological functions, Acta biomaterialia 10(4) (2014) 1558–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chanmee T, Ontong P, Itano N, Hyaluronan: A modulator of the tumor microenvironment, Cancer Letters 375(1) (2016) 20–30. [DOI] [PubMed] [Google Scholar]
- [36].Khaing ZZ, Seidlits SK, Hyaluronic acid and neural stem cells: implications for biomaterial design, Journal of Materials Chemistry B 3(40) (2015) 7850–7866. [DOI] [PubMed] [Google Scholar]
- [37].Stern R, Hyaluronan catabolism: a new metabolic pathway, European journal of cell biology 83(7) (2004) 317–325. [DOI] [PubMed] [Google Scholar]
- [38].Stern R, Asari AA, Sugahara KN, Hyaluronan fragments: an information-rich system, European journal of cell biology 85(8) (2006) 699–715. [DOI] [PubMed] [Google Scholar]
- [39].Perkins KL, Arranz AM, Yamaguchi Y, Hrabetova S, Brain extracellular space, hyaluronan, and the prevention of epileptic seizures, Reviews in the Neurosciences 28(8) (2017) 869–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lundell A, Olin AI, Mörgelin M, Al-Karadaghi S, Aspberg A, Logan DT, Structural basis for interactions between tenascins and lectican C-type lectin domains: evidence for a crosslinking role for tenascins, Structure 12(8) (2004) 1495–1506. [DOI] [PubMed] [Google Scholar]
- [41].Zhu Z, Wang Y-M, Yang J, Luo X-S, Hyaluronic acid: a versatile biomaterial in tissue engineering, Plastic and Aesthetic Research 4 (2017) 219–227. [Google Scholar]
- [42].Pedron S, Hanselman JS, Schroeder MA, Sarkaria JN, Harley BA, Extracellular hyaluronic acid influences the efficacy of EGFR tyrosine kinase inhibitors in a biomaterial model of glioblastoma, Advanced healthcare materials 6(21) (2017) 1700529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kwon MY, Wang C, Galarraga JH, Puré E, Han L, Burdick JA, Influence of hyaluronic acid modification on CD44 binding towards the design of hydrogel biomaterials, Biomaterials 222 (2019) 119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gokila S, Gomathi T, Vijayalakshmi K, Sukumaran A, Sudha P, Development of 3D scaffolds using nanochitosan/silk-fibroin/hyaluronic acid biomaterials for tissue engineering applications, International journal of biological macromolecules 120 (2018) 876–885. [DOI] [PubMed] [Google Scholar]
- [45].Cao Y, Chen X, Matarasso A, Wang Z, Yang S, Wu G, Zhang X, Sun H, Wang X, Bruchas MR, Li Y, Zhang Y, Covalently Attached Slippery Surface Coatings to Reduce Protein Adsorptions on Poly(dimethylsiloxane) Planar Surfaces and 3D Microfluidic Channels., ACS Applied Materials and Interfaces 15 (2023) 9987–9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Chen X, Liu C, Wadsworth M, Zeng EZ, Driscoll TP, Zeng C, Li Y, Surface engineering of auxetic scaffolds for neural and vascular differentiation from human pluripotent stem cells, Adv Healthc Mater 12(6) (2023) e2202511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hor J-H, Ng S-Y, Generating ventral spinal organoids from human induced pluripotent stem cells, Methods in Cell Biology, Elsevier; 2020, pp. 257–277. [DOI] [PubMed] [Google Scholar]
- [48].Lee J-H, Shin H, Shaker MR, Kim HJ, Park S-H, Kim JH, Lee N, Kang M, Cho S, Kwak TH, Production of human spinal-cord organoids recapitulating neural-tube morphogenesis, Nature Biomedical Engineering 6(4) (2022) 435–448. [DOI] [PubMed] [Google Scholar]
- [49].Jeske R, Chen X, Mulderrig L, Liu C, Cheng W, Zeng OZ, Zeng C, Guan J, Hallinan D, Yuan X, Li Y, Engineering human mesenchymal bodies in a novel 3-D printed microchannel bioreactor for studying extracellular vesicle biogenesis, Bioengineering (Basel) 9(12) (2022) 795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Yuan X, Sun L, Jeske R, Nkosi D, York S, Liu Y, Grant SC, Meckes DGJ, Li Y, Engineering Extracellular Vesicles by Three-dimensional Dynamic Culture of Human Mesenchymal Stem Cells, Journal of Extracellular Vesicles 11(6) (2022) e12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Muok L, Sun L, Esmonde C, Worden H, Vied C, Duke L, Ma S, Zeng OZ, Driscoll TP, Jung S, Li Y, Extracellular Vesicle Biogenesis of Three-dimensional Human Pluripotent Stem Cells in a Novel Vertical-Wheel Bioreactor, Journal of Extracellular Biology 3 (2024) e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Jeske R, Liu C, Duke L, Canonicco Castro ML, Muok L, Arthur P, Singh M, Sung L, Sun L, Li Y, Upscaling Human Mesenchymal Stem Cell Production in a Novel Vertical Wheel Bioreactor Enhances Extracellular Vesicle Secretion and Cargo Profile, Bioactive Materials 25 (2023) 732–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Qian T, Maguire SE, Canfield SG, Bao X, Olson WR, Shusta EV, Palecek SP, Directed differentiation of human pluripotent stem cells to blood-brain barrier endothelial cells, Science advances 3(11) (2017) e1701679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wimmer RA, Leopoldi A, Aichinger M, Wick N, Hantusch B, Novatchkova M, Taubenschmid J, Hammerle M, Esk C, Bagley JA, Lindenhofer D, Chen G, Boehm M, Agu CA, Yang F, Fu B, Zuber J, Knoblich JA, Kerjaschki D, Penninger JM, Human blood vessel organoids as a model of diabetic vasculopathy, Nature 565 (2019) 505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Song L, Wang K, Li Y, Yang Y, Nanotopography promoted neuronal differentiation of human induced pluripotent stem cells, Colloids and Surfaces B: Biointerfaces 148 (2016) 49–58. [DOI] [PubMed] [Google Scholar]
- [56].Chaudhuri O, Viscoelastic hydrogels for 3D cell culture, Biomaterials science 5(8) (2017) 1480–1490. [DOI] [PubMed] [Google Scholar]
- [57].Tan HY, Cho H, Lee LP, Human mini-brain models, Nat Biomed Eng 5(1) (2021) 11–25. [DOI] [PubMed] [Google Scholar]
- [58].Chen X, Liu C, Muok L, Zeng C, Li Y, Dynamic 3D on-chip BBB Model Design, Development, and Applications in Neurological Diseases, Cells 10 (2021) 3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bejoy J, Song L, Zhou Y, Li Y, Wnt-Yes associated protein interactions during neural tissue patterning of human induced pluripotent stem cells, Tissue Engineering Part A 24(7–8) (2018) 546–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Gattazzo F, Urciuolo A, Bonaldo P, Extracellular matrix: a dynamic microenvironment for stem cell niche, Biochimica et Biophysica Acta (BBA)-General Subjects 1840(8) (2014) 2506–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Bissoli I, D’Adamo S, Pignatti C, Agnetti G, Flamigni F, Cetrullo S, Induced pluripotent stem cell-based models: Are we ready for that heart in a dish?, Frontiers in Cell and Developmental Biology 11 (2023) 1129263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Roth JG, Huang MS, Navarro RS, Akram JT, LeSavage BL, Heilshorn SC, Tunable hydrogel viscoelasticity modulates human neural maturation, Science Advances 9(42) (2023) eadh8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hauptstein J, Böck T, Bartolf‐Kopp M, Forster L, Stahlhut P, Nadernezhad A, Blahetek G, Zernecke‐Madsen A, Detsch R, Jüngst T, Hyaluronic acid‐based bioink composition enabling 3D bioprinting and improving quality of deposited cartilaginous extracellular matrix, Advanced Healthcare Materials 9(15) (2020) 2000737. [DOI] [PubMed] [Google Scholar]
- [64].Asadikorayem M, Surman F, Weber P, Weber D, Zenobi‐Wong M, Zwitterionic Granular Hydrogel for Cartilage Tissue Engineering, Advanced Healthcare Materials (2023) 2301831. [DOI] [PubMed] [Google Scholar]
- [65].Muir VG, Qazi TH, Weintraub S, Torres Maldonado BO, Arratia PE, Burdick JA, Sticking Together: Injectable Granular Hydrogels with Increased Functionality via Dynamic Covalent Inter‐Particle Crosslinking, Small 18(36) (2022) 2201115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Ding S, Kingshott P, Thissen H, Pera M, Wang PY, Modulation of human mesenchymal and pluripotent stem cell behavior using biophysical and biochemical cues: A review, Biotechnology and bioengineering 114(2) (2017) 260–280. [DOI] [PubMed] [Google Scholar]
- [67].Wang P-Y, Thissen H, Kingshott P, Modulation of human multipotent and pluripotent stem cells using surface nanotopographies and surface-immobilised bioactive signals: A review, Acta biomaterialia 45 (2016) 31–59. [DOI] [PubMed] [Google Scholar]
- [68].Ireland RG, Simmons CA, Human pluripotent stem cell mechanobiology: manipulating the biophysical microenvironment for regenerative medicine and tissue engineering applications, Stem Cells 33(11) (2015) 3187–3196. [DOI] [PubMed] [Google Scholar]
- [69].Li Y, Dong T, Li Z, Ni S, Zhou F, Alimi OA, Chen S, Duan B, Kuss M, Wu S, Review of advances in electrospinning-based strategies for spinal cord regeneration, Materials Today Chemistry 24 (2022) 100944. [Google Scholar]
- [70].Chaudhuri O, Gu L, Klumpers D, Darnell M, Bencherif SA, Weaver JC, Huebsch N, Lee H.-p., Lippens E, Duda GN, Hydrogels with tunable stress relaxation regulate stem cell fate and activity, Nature materials 15(3) (2016) 326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Cheng B, Li M, Wan W, Guo H, Genin GM, Lin M, Xu F, Predicting YAP/TAZ nuclear translocation in response to ECM mechanosensing, Biophysical journal 122(1) (2023) 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Sun Y, Yong KMA, Villa-Diaz LG, Zhang X, Chen W, Philson R, Weng S, Xu H, Krebsbach PH, Fu J, Hippo/YAP-mediated rigidity-dependent motor neuron differentiation of human pluripotent stem cells, Nature materials 13(6) (2014) 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Pan F, Zhang M, Wu G, Lai Y, Greber B, Schöler HR, Chi L, Topographic effect on human induced pluripotent stem cells differentiation towards neuronal lineage, Biomaterials 34(33) (2013) 8131–8139. [DOI] [PubMed] [Google Scholar]
- [74].Pettinato G, Lehoux S, Ramanathan R, Salem MM, He L-X, Muse O, Flaumenhaft R, Thompson MT, Rouse EA, Cummings RD, Generation of fully functional hepatocyte-like organoids from human induced pluripotent stem cells mixed with Endothelial Cells, Scientific reports 9(1) (2019) 8920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Khanna A, Zamani M, Huang NF, Extracellular matrix-based biomaterials for cardiovascular tissue engineering, Journal of cardiovascular development and disease 8(11) (2021) 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Homan KA, Gupta N, Kroll KT, Kolesky DB, Skylar-Scott M, Miyoshi T, Mau D, Valerius MT, Ferrante T, Bonventre JV, Flow-enhanced vascularization and maturation of kidney organoids in vitro, Nature methods 16(3) (2019) 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Zhang S, Wan Z, Kamm RD, Vascularized organoids on a chip: strategies for engineering organoids with functional vasculature, Lab on a Chip 21(3) (2021) 473–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Lupon E, Lellouch AG, Acun A, Andrews AR, Oganesyan R, Goutard M, Taveau CB, Lantieri LA, Cetrulo CL, Uygun BE, Engineering vascularized composite allografts using natural scaffolds: a systematic review, Tissue Engineering Part B: Reviews 28(3) (2022) 677–693. [DOI] [PubMed] [Google Scholar]
- [79].Mansour AA, Gonçalves JT, Bloyd CW, Li H, Fernandes S, Quang D, Johnston S, Parylak SL, Jin X, Gage FH, An in vivo model of functional and vascularized human brain organoids, Nature biotechnology 36(5) (2018) 432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding authors on reasonable request.


