Abstract
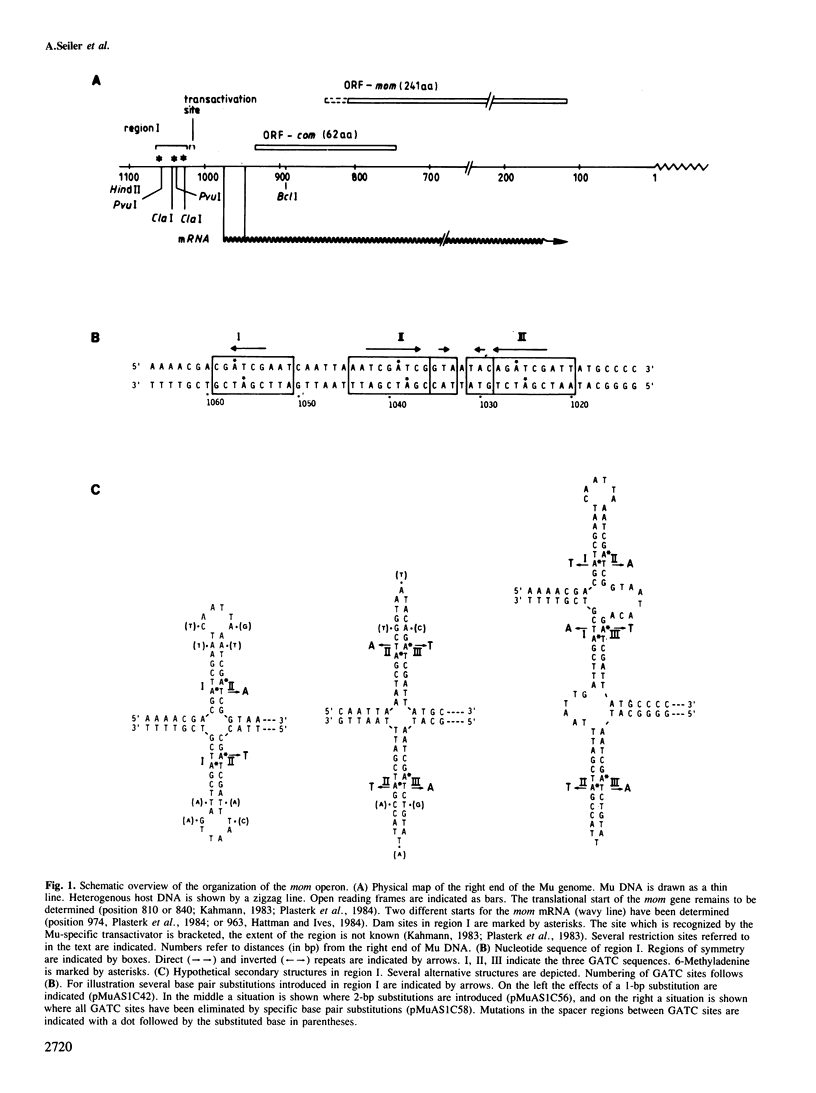
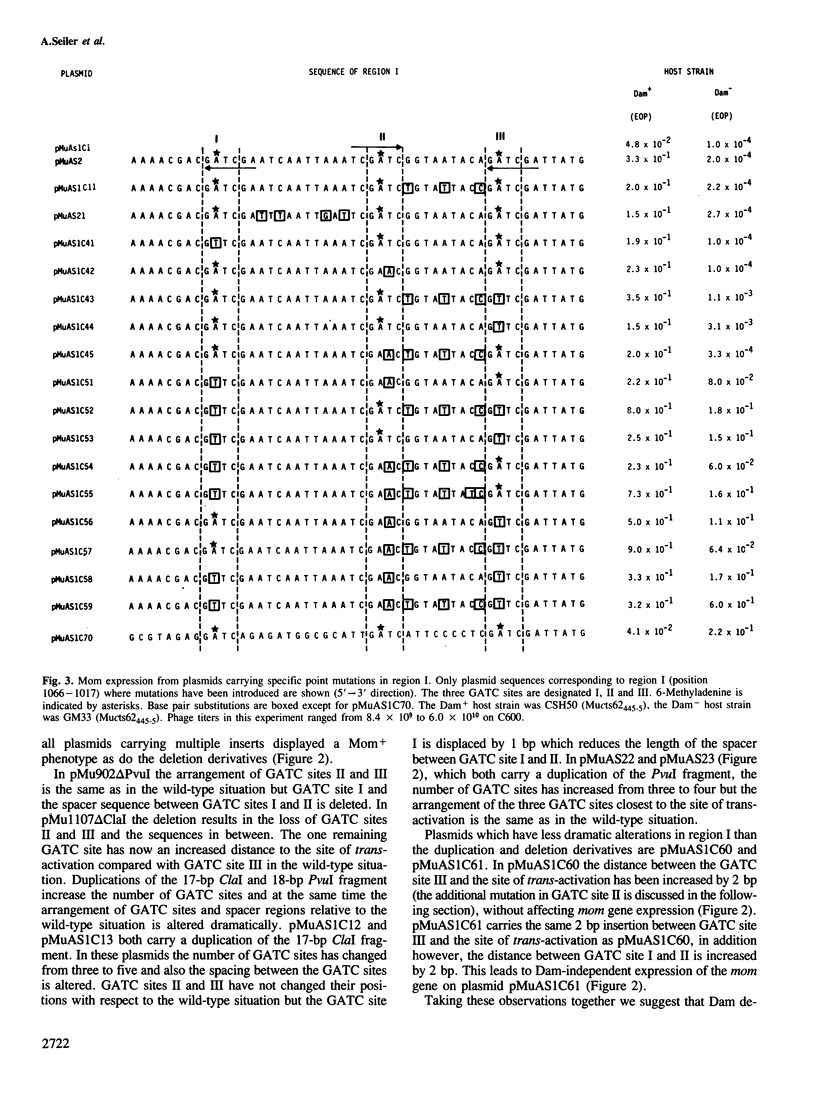
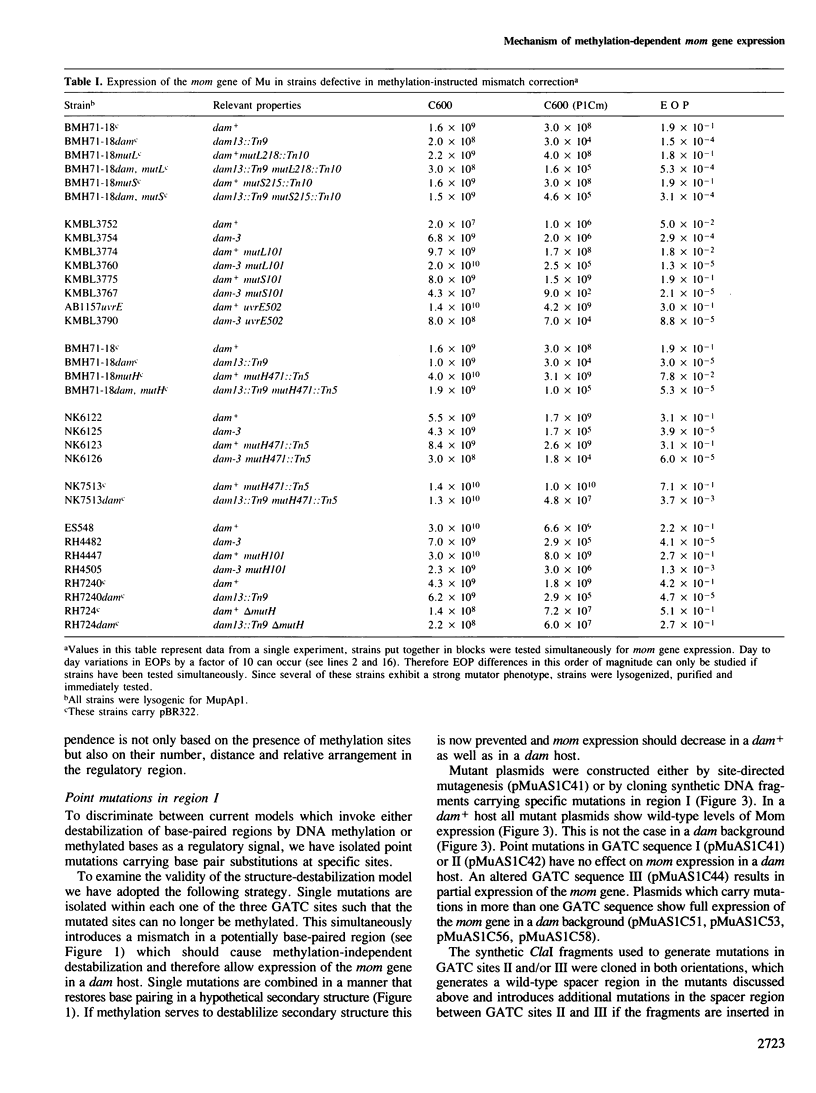
Transcription of the DNA modification gene (mom) of bacteriophage Mu requires methylation of three GATC sites upstream of the mom promoter by the Escherichia coli deoxyadenosine methylation function (Dam). The three sites map within a 40-bp segment termed region I. Small deletions, inversions, duplications and specific point mutations have been introduced in region I. Their effect on mom expression has been studied in dam+ and dam strains. Dam-dependent expression of the mom gene requires a specific arrangement of the three GATC sites and the presence of the methylated base in at least two of the three sites. We show that mom specific modification is regulated by a host protein. The Mom function is expressed in dam strains if they are defective in one component of the methylation-instructed mismatch correction system, mutH. We suggest that the product of mutH functions as a transcriptional repressor by binding to region I.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abdel-Meguid S. S., Grindley N. D., Templeton N. S., Steitz T. A. Cleavage of the site-specific recombination protein gamma delta resolvase: the smaller of two fragments binds DNA specifically. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2001–2005. doi: 10.1073/pnas.81.7.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allet B., Bukhari A. I. Analysis of bacteriophage mu and lambda-mu hybrid DNAs by specific endonucleases. J Mol Biol. 1975 Mar 15;92(4):529–540. doi: 10.1016/0022-2836(75)90307-1. [DOI] [PubMed] [Google Scholar]
- Appleyard R K. Segregation of Lambda Lysogenicity during Bacterial Recombination in Escherichia Coli K12. Genetics. 1954 Jul;39(4):429–439. doi: 10.1093/genetics/39.4.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique Eco RI sites for selection of Eco RI generated recombinant DNA molecules. Gene. 1978 Oct;4(2):121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Caillet-Fauquet P., Maenhaut-Michel G., Radman M. SOS mutator effect in E. coli mutants deficient in mismatch correction. EMBO J. 1984 Apr;3(4):707–712. doi: 10.1002/j.1460-2075.1984.tb01873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow L. T., Kahmann R., Kamp D. Electron microscopic characterization of DNAs of non-defective deletion mutants of bacteriophage Mu. J Mol Biol. 1977 Jul 15;113(4):591–609. doi: 10.1016/0022-2836(77)90224-8. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Weeks J. R., Travers A. A. Negative supercoiling induces spontaneous unwinding of a bacterial promoter. EMBO J. 1985 Apr;4(4):1025–1032. doi: 10.1002/j.1460-2075.1985.tb03734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank R., Heikens W., Heisterberg-Moutsis G., Blöcker H. A new general approach for the simultaneous chemical synthesis of large numbers of oligonucleotides: segmental solid supports. Nucleic Acids Res. 1983 Jul 11;11(13):4365–4377. doi: 10.1093/nar/11.13.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman B. W., Radman M. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc Natl Acad Sci U S A. 1980 Feb;77(2):1063–1067. doi: 10.1073/pnas.77.2.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman B., van den Elsen P., Radman M. Induced mutagenesis in dam- mutants of Escherichia coli: a role for 6-methyladenine residues in mutation avoidance. Mol Gen Genet. 1978 Jul 25;163(3):307–312. doi: 10.1007/BF00271960. [DOI] [PubMed] [Google Scholar]
- Hattman S. DNA methyltransferase-dependent transcription of the phage Mu mom gene. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5518–5521. doi: 10.1073/pnas.79.18.5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S., Goradia M., Monaghan C., Bukhari A. I. Regulation of the DNA-modification function of bacteriophage Mu. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):647–653. doi: 10.1101/sqb.1983.047.01.076. [DOI] [PubMed] [Google Scholar]
- Hattman S., Ives J., Margolin W., Howe M. M. Regulation and expression of the bacteriophage mu mom gene: mapping of the transactivation (dad) function to the C region. Gene. 1985;39(1):71–76. doi: 10.1016/0378-1119(85)90109-x. [DOI] [PubMed] [Google Scholar]
- Hattman S., Ives J. S1 nuclease mapping of the phage Mu mom gene promoter: a model for the regulation of mom expression. Gene. 1984 Jul-Aug;29(1-2):185–198. doi: 10.1016/0378-1119(84)90179-3. [DOI] [PubMed] [Google Scholar]
- Hattman S. Unusual modification of bacteriophage Mu DNA. J Virol. 1979 Nov;32(2):468–475. doi: 10.1128/jvi.32.2.468-475.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisig P., Kahmann R. The sequence and mom-transactivation function of the C gene of bacteriophage Mu. Gene. 1986;43(1-2):59–67. doi: 10.1016/0378-1119(86)90008-9. [DOI] [PubMed] [Google Scholar]
- Hochschild A., Irwin N., Ptashne M. Repressor structure and the mechanism of positive control. Cell. 1983 Feb;32(2):319–325. doi: 10.1016/0092-8674(83)90451-8. [DOI] [PubMed] [Google Scholar]
- Hoess R. H., Fan D. P. Further characterization of a non-essential mutator gene in Escherichia coli K-12. J Bacteriol. 1975 Nov;124(2):650–660. doi: 10.1128/jb.124.2.650-660.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe M. M. Prophage deletion mapping of bacteriophage Mu-1. Virology. 1973 Jul;54(1):93–101. doi: 10.1016/0042-6822(73)90118-9. [DOI] [PubMed] [Google Scholar]
- Kahmann R. Methylation regulates the expression of a DNA-modification function encoded by bacteriophage Mu. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):639–646. doi: 10.1101/sqb.1983.047.01.075. [DOI] [PubMed] [Google Scholar]
- Kahmann R., Seiler A., Wulczyn F. G., Pfaff E. The mom gene of bacteriophage mu: a unique regulatory scheme to control a lethal function. Gene. 1985;39(1):61–70. doi: 10.1016/0378-1119(85)90108-8. [DOI] [PubMed] [Google Scholar]
- Kahmann R. The mom gene of bacteriophage Mu. Curr Top Microbiol Immunol. 1984;108:29–47. doi: 10.1007/978-3-642-69370-0_4. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Morisato D., Roberts D., Bender J. Mechanism and regulation of Tn10 transposition. Cold Spring Harb Symp Quant Biol. 1984;49:235–244. doi: 10.1101/sqb.1984.049.01.027. [DOI] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Fritz H. J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984 Oct;38(3):879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- Lacks S., Greenberg B. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J Mol Biol. 1977 Jul;114(1):153–168. doi: 10.1016/0022-2836(77)90289-3. [DOI] [PubMed] [Google Scholar]
- Leach D., Symonds N. The isolation and characterisation of a plaque-forming derivative of bacteriophage Mu carrying a fragment of Tn3 conferring ampicillin resistance. Mol Gen Genet. 1979 May 4;172(2):179–184. doi: 10.1007/BF00268280. [DOI] [PubMed] [Google Scholar]
- Lu A. L., Clark S., Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G., Carraway M., Frey A. Z., Brown L., Arraj J. A. Insertion mutations in the dam gene of Escherichia coli K-12. Mol Gen Genet. 1983;192(1-2):288–289. doi: 10.1007/BF00327681. [DOI] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J Bacteriol. 1973 Jun;114(3):1143–1150. doi: 10.1128/jb.114.3.1143-1150.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Pleiotropic effects of a DNA adenine methylation mutation (dam-3) in Escherichia coli K12. Mutat Res. 1975 Apr;28(1):15–26. doi: 10.1016/0027-5107(75)90309-7. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messer W., Bellekes U., Lother H. Effect of dam methylation on the activity of the E. coli replication origin, oriC. EMBO J. 1985 May;4(5):1327–1332. doi: 10.1002/j.1460-2075.1985.tb03780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Norris K., Norris F., Christiansen L., Fiil N. Efficient site-directed mutagenesis by simultaneous use of two primers. Nucleic Acids Res. 1983 Aug 11;11(15):5103–5112. doi: 10.1093/nar/11.15.5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A., Sugimoto K., Takanami M., Hirota Y. Replication origin of the Escherichia coli K-12 chromosome: the size and structure of the minimum DNA segment carrying the information for autonomous replication. Mol Gen Genet. 1980 Apr;178(1):9–20. doi: 10.1007/BF00267207. [DOI] [PubMed] [Google Scholar]
- Plasterk R. H., Vollering M., Brinkman A., Van de Putte P. Analysis of the methylation-regulated Mu mom transcript. Cell. 1984 Jan;36(1):189–196. doi: 10.1016/0092-8674(84)90088-6. [DOI] [PubMed] [Google Scholar]
- Plasterk R. H., Vrieling H., Van de Putte P. Transcription initiation of Mu mom depends on methylation of the promoter region and a phage-coded transactivator. Nature. 1983 Jan 27;301(5898):344–347. doi: 10.1038/301344a0. [DOI] [PubMed] [Google Scholar]
- Ptashne M., Jeffrey A., Johnson A. D., Maurer R., Meyer B. J., Pabo C. O., Roberts T. M., Sauer R. T. How the lambda repressor and cro work. Cell. 1980 Jan;19(1):1–11. doi: 10.1016/0092-8674(80)90383-9. [DOI] [PubMed] [Google Scholar]
- Pukkila P. J., Peterson J., Herman G., Modrich P., Meselson M. Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics. 1983 Aug;104(4):571–582. doi: 10.1093/genetics/104.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. W., Garland A. M., Herman G., Enns R. E., Baker T. A., Zyskind J. W. Importance of state of methylation of oriC GATC sites in initiation of DNA replication in Escherichia coli. EMBO J. 1985 May;4(5):1319–1326. doi: 10.1002/j.1460-2075.1985.tb03779.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N. Evidence that adenine methylation influences DNA-protein interactions in Escherichia coli. J Bacteriol. 1985 Oct;164(1):490–493. doi: 10.1128/jb.164.1.490-493.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinton D., Hattman S., Crain P. F., Cheng C. S., Smith D. L., McCloskey J. A. Purification and characterization of the unusual deoxynucleoside, alpha-N-(9-beta-D-2'-deoxyribofuranosylpurin-6-yl)glycinamide, specified by the phage Mu modification function. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7400–7404. doi: 10.1073/pnas.80.24.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint A. DNA modification of bacteriophage Mu-1 requires both host and bacteriophage functions. J Virol. 1977 Sep;23(3):825–826. doi: 10.1128/jvi.23.3.825-826.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint A., Desmet L., Faelen M. Mapping of the modification function of temperate phage Mu-1. Mol Gen Genet. 1980 Jan;177(2):351–353. doi: 10.1007/BF00267450. [DOI] [PubMed] [Google Scholar]
- Toussaint A. The DNA modification function of temperate phage Mu-1. Virology. 1976 Mar;70(1):17–27. doi: 10.1016/0042-6822(76)90232-4. [DOI] [PubMed] [Google Scholar]
- Vaccaro K. K., Siegel E. C. The frameshift mutability of polA1 and recA1 derivatives of mutator strains of Escherichia coli. Mutat Res. 1977 Mar;42(3):443–446. doi: 10.1016/s0027-5107(77)80048-1. [DOI] [PubMed] [Google Scholar]
- Wilson G. G., Young K. Y., Edlin G. J., Konigsberg W. High-frequency generalised transduction by bacteriophage T4. Nature. 1979 Jul 5;280(5717):80–82. doi: 10.1038/280080a0. [DOI] [PubMed] [Google Scholar]
- Youderian P., Vershon A., Bouvier S., Sauer R. T., Susskind M. M. Changing the DNA-binding specificity of a repressor. Cell. 1983 Dec;35(3 Pt 2):777–783. doi: 10.1016/0092-8674(83)90110-1. [DOI] [PubMed] [Google Scholar]


