Abstract
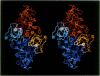
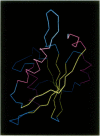
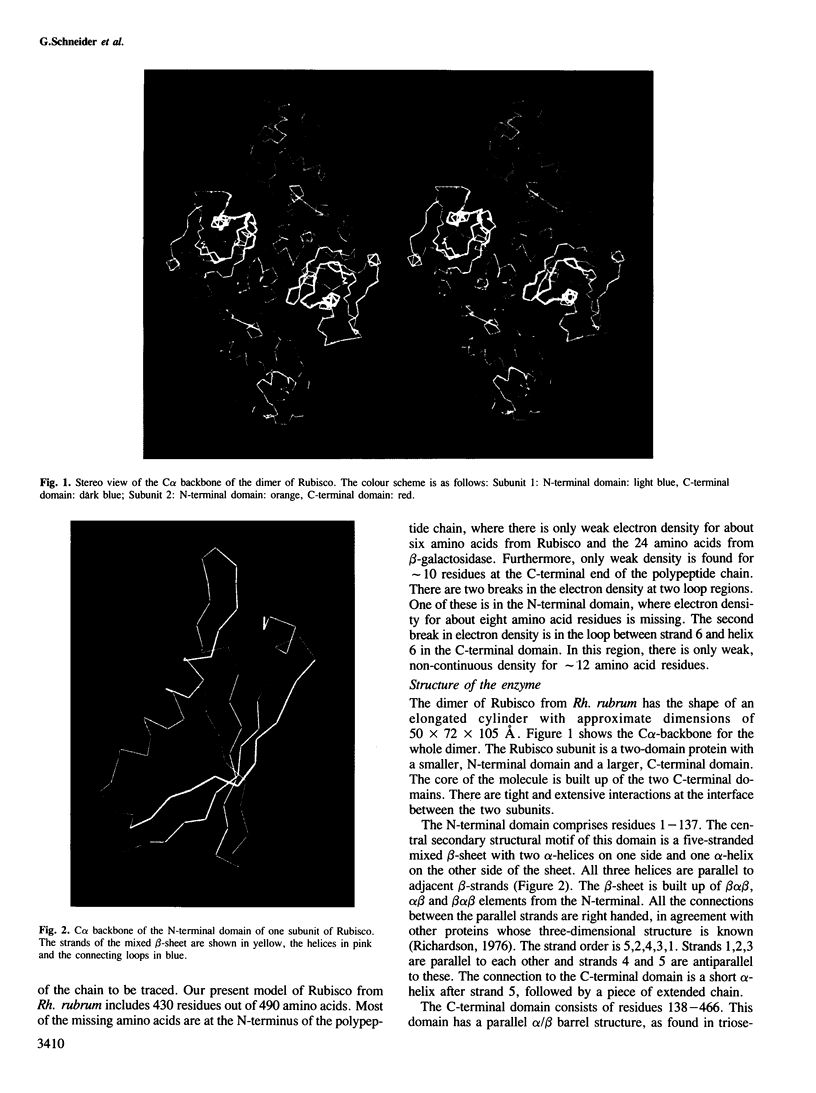
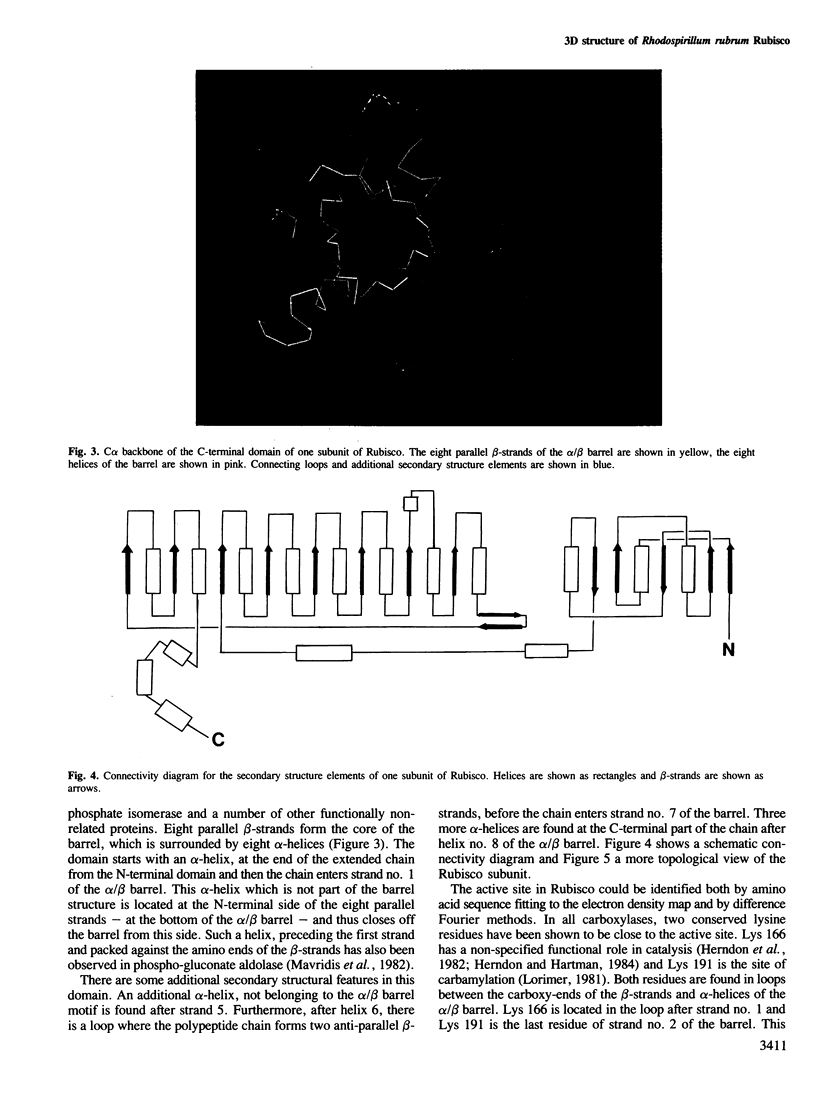
The three-dimensional structure of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Rhodospirillum rubrum has been determined at 2.9 Å resolution by X-ray crystallographic methods. The MIR-electron density map was substantially improved by two-fold non-crystallographic symmetry averaging. The polypeptide chains in the dimer were traced using a graphics display system with the help of the BONES option in FRODO. The dimer has approximate dimensions of 50 x 72 x 105 Å. The enzyme subunit is a typical two-domain protein. The smaller, N-terminal domain consists of 137 amino acid residues and forms a central, mixed five-stranded β-sheet with α-helices on both sides of the sheet. The larger C-terminal domain consists of 329 amino acid residues. This domain has an eight-stranded parallel α/β barrel structure as found in triosephosphate isomerase and a number of other functionally non-related proteins. The active site in Rubisco determined by difference Fourier techniques and fitting of active site residues to the electron density map, is located at the carboxy-end of the β-strands in the α/β barrel of the C-terminal domain. There are few domain–domain interactions within the subunit. The interactions at the interface between the two subunits of the dimer are tight and extensive. There are tight contacts between the two C-terminal domains, which build up the core of the molecule. There are also interactions between the N-terminal domain of one subunit and the C-terminal domain of the second subunit, close to the active site.
Keywords: Rubisco, protein crystallography, photosynthesis
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bowien B., Gottschalk E. M. Influence of the activation state on the sedimentation properties of ribulose bisphosphate carboxylase from Alcaligenes eutrophus. J Biol Chem. 1982 Oct 25;257(20):11845–11847. [PubMed] [Google Scholar]
- Carrell H. L., Rubin B. H., Hurley T. J., Glusker J. P. X-ray crystal structure of D-xylose isomerase at 4-A resolution. J Biol Chem. 1984 Mar 10;259(5):3230–3236. [PubMed] [Google Scholar]
- Eklund H., Nordström B., Zeppezauer E., Söderlund G., Ohlsson I., Boiwe T., Söderberg B. O., Tapia O., Brändén C. I., Akeson A. Three-dimensional structure of horse liver alcohol dehydrogenase at 2-4 A resolution. J Mol Biol. 1976 Mar 25;102(1):27–59. doi: 10.1016/0022-2836(76)90072-3. [DOI] [PubMed] [Google Scholar]
- Estelle M., Hanks J., McIntosh L., Somerville C. Site-specific mutagenesis of ribulose-1,5-bisphosphate carboxylase/oxygenase. Evidence that carbamate formation at Lys 191 is required for catalytic activity. J Biol Chem. 1985 Aug 15;260(17):9523–9526. [PubMed] [Google Scholar]
- Fraij B., Hartman F. C. 2-Bromoacetylaminopentitol 1,5-bisphosphate as an affinity label for ribulose bisphosphate carboxylase/oxygenase from Rhodospirillum rubrum. J Biol Chem. 1982 Apr 10;257(7):3501–3505. [PubMed] [Google Scholar]
- Gutteridge S., Sigal I., Thomas B., Arentzen R., Cordova A., Lorimer G. A site-specific mutation within the active site of ribulose-1,5-bisphosphate carboxylase of Rhodospirillum rubrum. EMBO J. 1984 Dec 1;3(12):2737–2743. doi: 10.1002/j.1460-2075.1984.tb02204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman F. C., Stringer C. D., Lee E. H. Complete primary structure of ribulosebisphosphate carboxylase/oxygenase from Rhodospirillum rubrum. Arch Biochem Biophys. 1984 Jul;232(1):280–295. doi: 10.1016/0003-9861(84)90544-7. [DOI] [PubMed] [Google Scholar]
- Hartman F. C., Stringer C. D., Omnaas J., Donnelly M. I., Fraij B. Purification and sequencing of cyanogen bromide fragments from ribulosebisphosphate carboxylase/oxygenase from Rhodospirillum rubrum. Arch Biochem Biophys. 1982 Dec;219(2):422–437. doi: 10.1016/0003-9861(82)90174-6. [DOI] [PubMed] [Google Scholar]
- Herndon C. S., Hartman F. C. 2-(4-Bromoacetamido)anilino-2-deoxypentitol 1,5-bisphosphate, a new affinity label for ribulose bisphosphate carboxylase/oxygenase from Rhodospirillum rubrum. Determination of reaction parameters and characterization of an active site peptide. J Biol Chem. 1984 Mar 10;259(5):3102–3110. [PubMed] [Google Scholar]
- Herndon C. S., Norton I. L., Hartman F. C. Reexamination of the binding site for pyridoxal 5'-phosphate in ribulosebisphosphate carboxylase/oxygenase from Rhodospirillum rubrum. Biochemistry. 1982 Mar 16;21(6):1380–1385. doi: 10.1021/bi00535a043. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Thirup S. Using known substructures in protein model building and crystallography. EMBO J. 1986 Apr;5(4):819–822. doi: 10.1002/j.1460-2075.1986.tb04287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimer F. W., Machanoff R., Hartman F. C. A reconstruction of the gene for ribulose bisphosphate carboxylase from Rhodospirillum rubrum that expresses the authentic enzyme in Escherichia coli. Gene. 1986;41(1):113–120. doi: 10.1016/0378-1119(86)90273-8. [DOI] [PubMed] [Google Scholar]
- Lindqvist Y., Brändén C. I. Structure of glycolate oxidase from spinach. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6855–6859. doi: 10.1073/pnas.82.20.6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorimer G. H., Miziorko H. M. Carbamate formation on the epsilon-amino group of a lysyl residue as the basis for the activation of ribulosebisphosphate carboxylase by CO2 and Mg2+. Biochemistry. 1980 Nov 11;19(23):5321–5328. doi: 10.1021/bi00564a027. [DOI] [PubMed] [Google Scholar]
- Lorimer G. H. Ribulosebisphosphate carboxylase: amino acid sequence of a peptide bearing the activator carbon dioxide. Biochemistry. 1981 Mar 3;20(5):1236–1240. doi: 10.1021/bi00508a028. [DOI] [PubMed] [Google Scholar]
- Matsuura Y., Kusunoki M., Harada W., Tanaka N., Iga Y., Yasuoka N., Toda H., Narita K., Kakudo M. Molecular structure of taka-amylase A. I. Backbone chain folding at 3 A resolution. J Biochem. 1980 May;87(5):1555–1558. doi: 10.1093/oxfordjournals.jbchem.a132896. [DOI] [PubMed] [Google Scholar]
- Mavridis I. M., Hatada M. H., Tulinsky A., Lebioda L. Structure of 2-keto-3-deoxy-6-phosphogluconate aldolase at 2 . 8 A resolution. J Mol Biol. 1982 Dec 5;162(2):419–444. doi: 10.1016/0022-2836(82)90536-8. [DOI] [PubMed] [Google Scholar]
- Miziorko H. M., Lorimer G. H. Ribulose-1,5-bisphosphate carboxylase-oxygenase. Annu Rev Biochem. 1983;52:507–535. doi: 10.1146/annurev.bi.52.070183.002451. [DOI] [PubMed] [Google Scholar]
- Niyogi S. K., Foote R. S., Mural R. J., Larimer F. W., Mitra S., Soper T. S., Machanoff R., Hartman F. C. Nonessentiality of histidine 291 of Rhodospirillum rubrum ribulose-bisphosphate carboxylase/oxygenase as determined by site-directed mutagenesis. J Biol Chem. 1986 Aug 5;261(22):10087–10092. [PubMed] [Google Scholar]
- Phillips D. C., Sternberg M. J., Thornton J. M., Wilson I. A. An analysis of the structure of triose phosphate isomerase and its comparison with lactate dehydrogenase. J Mol Biol. 1978 Feb 25;119(2):329–351. doi: 10.1016/0022-2836(78)90440-0. [DOI] [PubMed] [Google Scholar]
- Pierce J., Andrews T. J., Lorimer G. H. Reaction intermediate partitioning by ribulose-bisphosphate carboxylases with differing substrate specificities. J Biol Chem. 1986 Aug 5;261(22):10248–10256. [PubMed] [Google Scholar]
- Pierce J., Reddy G. S. The sites for catalysis and activation of ribulosebisphosphate carboxylase share a common domain. Arch Biochem Biophys. 1986 Mar;245(2):483–493. doi: 10.1016/0003-9861(86)90241-9. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. Handedness of crossover connections in beta sheets. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2619–2623. doi: 10.1073/pnas.73.8.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss J. V., Phares E. F., Long M. V., Norton I. L., Stringer C. D., Hartman F. C. Isolation, characterization, and crystallization of ribulosebisphosphate carboxylase from autotrophically grown Rhodospirillum rubrum. J Bacteriol. 1979 Jan;137(1):490–501. doi: 10.1128/jb.137.1.490-501.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
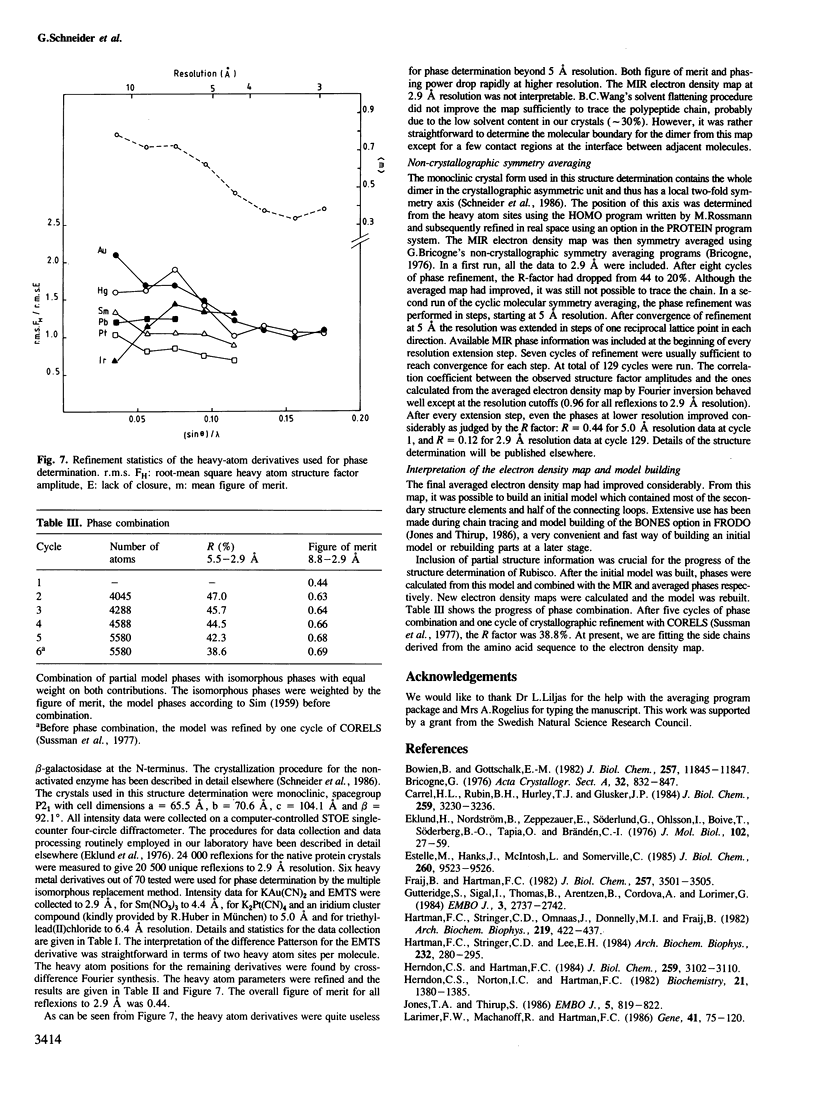
- Schneider G., Brändén C. I., Lorimer G. New crystal forms of ribulose-1,5-bisphosphate carboxylase/oxygenase from Rhodospirillum rubrum. J Mol Biol. 1986 Jan 5;187(1):141–143. doi: 10.1016/0022-2836(86)90415-8. [DOI] [PubMed] [Google Scholar]
- Stuart D. I., Levine M., Muirhead H., Stammers D. K. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 A. J Mol Biol. 1979 Oct 15;134(1):109–142. doi: 10.1016/0022-2836(79)90416-9. [DOI] [PubMed] [Google Scholar]
- Terzaghi B. E., Laing W. A., Christeller J. T., Petersen G. B., Hill D. F. Ribulose 1,5-bisphosphate carboxylase. Effect on the catalytic properties of changing methionine-330 to leucine in the Rhodospirillum rubrum enzyme. Biochem J. 1986 May 1;235(3):839–846. doi: 10.1042/bj2350839. [DOI] [PMC free article] [PubMed] [Google Scholar]