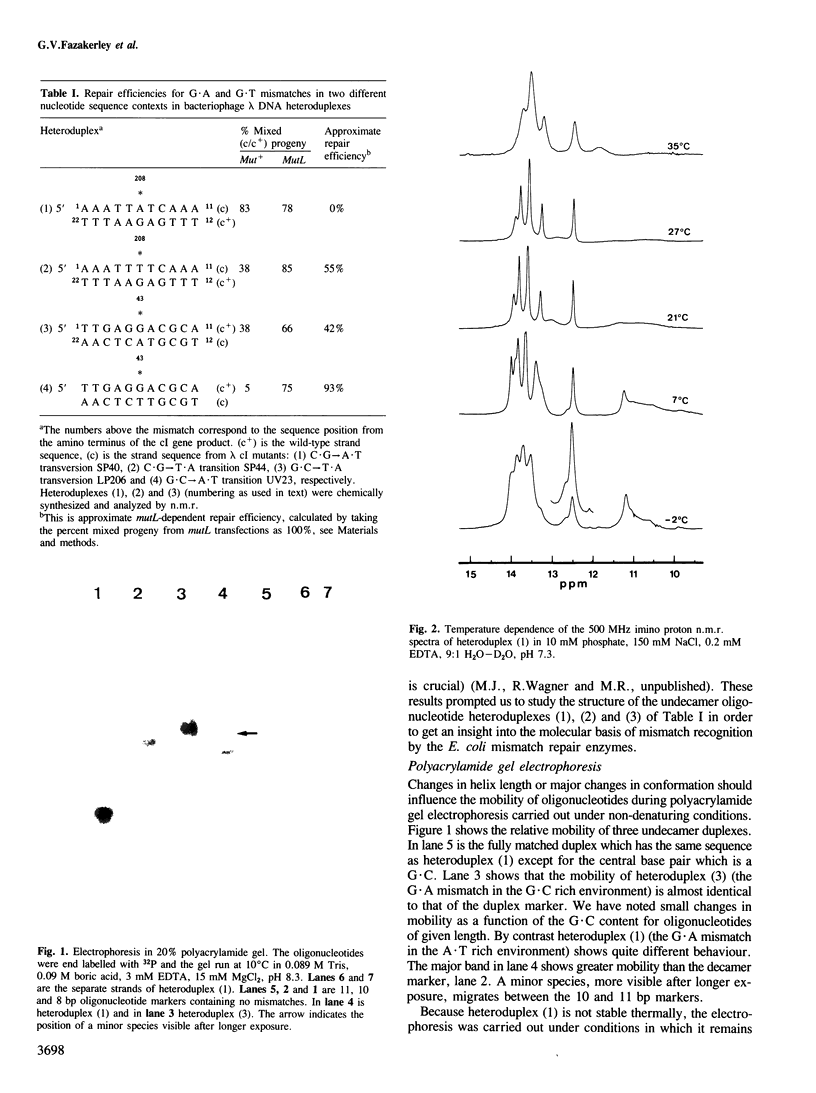
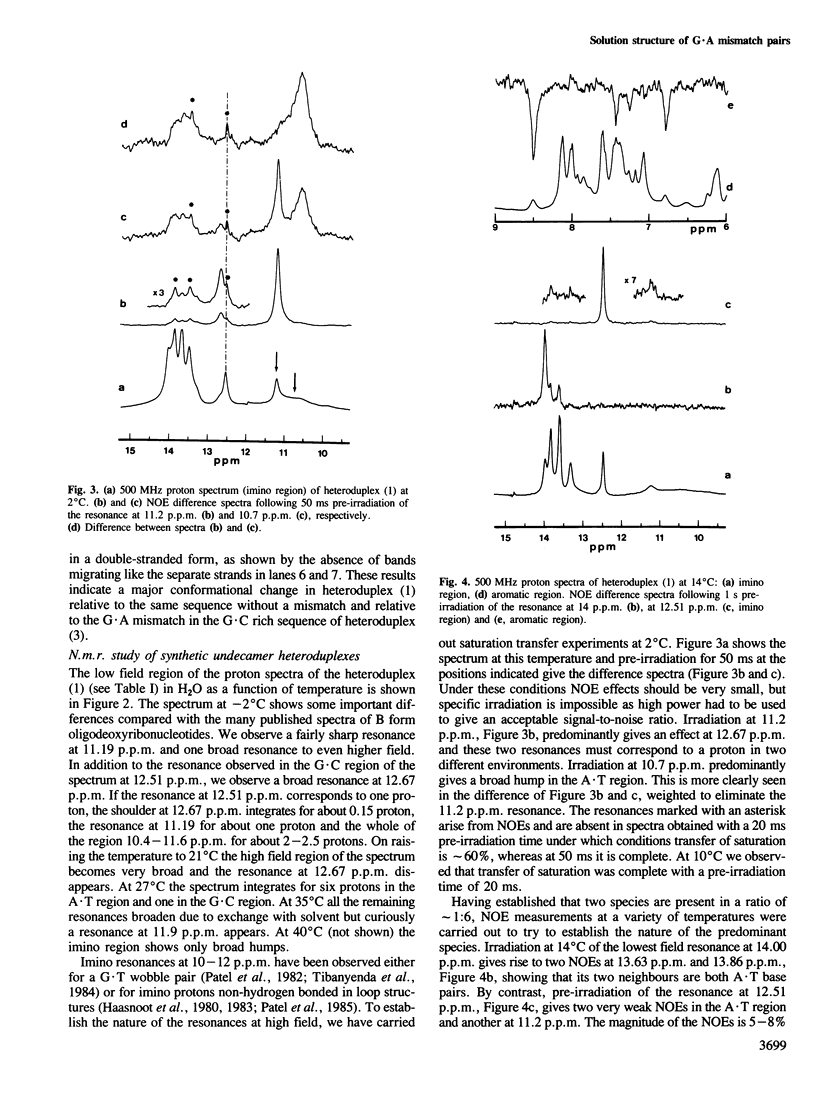
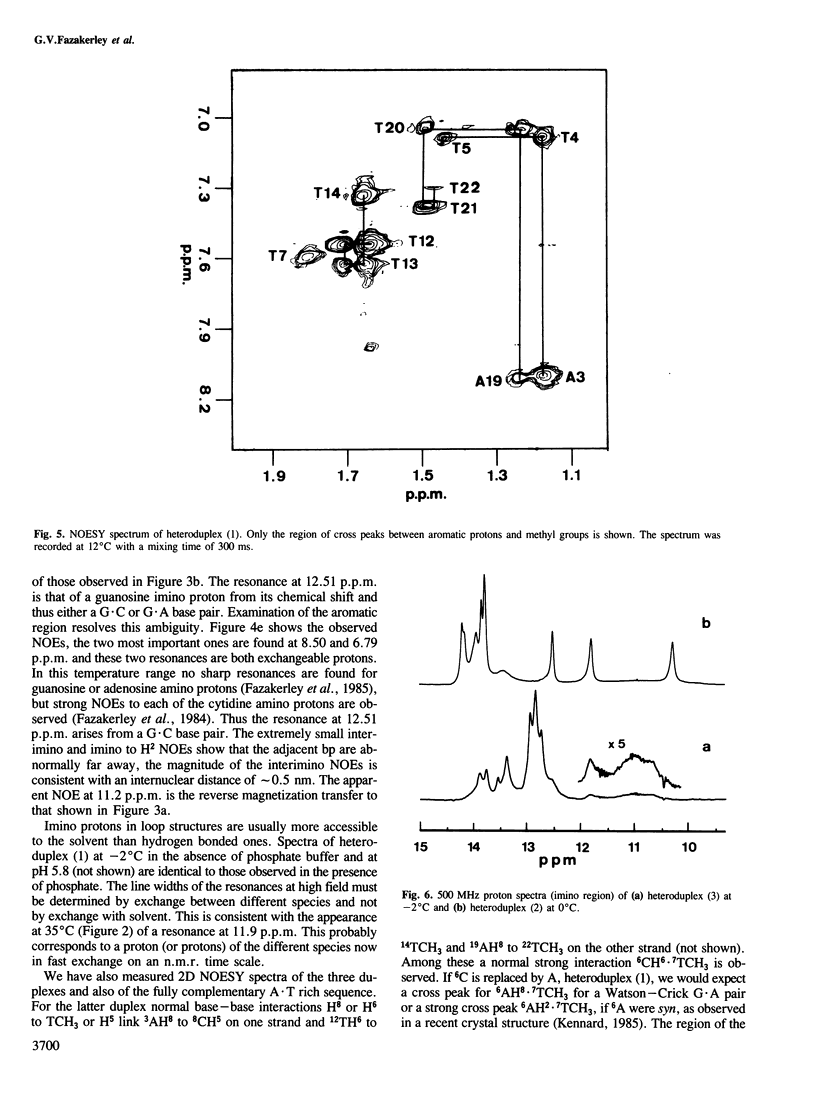
Abstract
The Escherichia coli mismatch repair system does not recognize and/or repair all mismatched base pairs with equal efficiency: whereas transition mismatches (G X T and A X C) are well repaired, the repair of some transversion mismatches (e.g. A X G or C X T) appears to depend on their position in heteroduplex DNA of phage lambda. Undecamers were synthesized and annealed to form heteroduplexes with a single base-pair mismatch in the centre and with the five base pairs flanking each side corresponding to either repaired or unrepaired heteroduplexes of lambda DNA. Nuclear magnetic resonance (n.m.r.) studies show that a G X A mismatch gives rise to an equilibrium between fully helical and a looped-out structure. In the unrepaired G X A mismatch duplex the latter predominates, while the helical structure is predominant in the case of repaired G X A and G X T mismatches. It appears that the E. coli mismatch repair enzymes recognize and repair intrahelical mismatched bases, but not the extrahelical bases in the looped-out structures.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dohet C., Wagner R., Radman M. Methyl-directed repair of frameshift mutations in heteroduplex DNA. Proc Natl Acad Sci U S A. 1986 May;83(10):3395–3397. doi: 10.1073/pnas.83.10.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohet C., Wagner R., Radman M. Repair of defined single base-pair mismatches in Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jan;82(2):503–505. doi: 10.1073/pnas.82.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazakerley G. V., Téoule R., Guy A., Fritzsche H., Guschlbauer W. NMR studies on oligodeoxyribonucleotides containing the dam methylation site GATC. Comparison between d(GGATCC) and d(GGm6ATCC). Biochemistry. 1985 Aug 13;24(17):4540–4548. doi: 10.1021/bi00338a009. [DOI] [PubMed] [Google Scholar]
- Fazakerley G. V., van der Marel G. A., van Boom J. H., Guschlbauer W. Helix opening in deoxyribonucleic acid from a proton nuclear magnetic resonance study of imino and amino protons in d(CG)3. Nucleic Acids Res. 1984 Nov 12;12(21):8269–8279. doi: 10.1093/nar/12.21.8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigon J., Denny W. A., Leupin W., Kearns D. R. Proton nuclear magnetic resonance investigation of the conformation and dynamics in the synthetic deoxyribonucleic acid decamers d(ATATCGATAT) and d(ATATGCATAT). Biochemistry. 1983 Dec 6;22(25):5930–5942. doi: 10.1021/bi00294a037. [DOI] [PubMed] [Google Scholar]
- Glickman B. W., Radman M. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc Natl Acad Sci U S A. 1980 Feb;77(2):1063–1067. doi: 10.1073/pnas.77.2.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot C. A., de Bruin S. H., Berendsen R. G., Janssen H. G., Binnendijk T. J., Hilbers C. W., van der Marel G. A., van Boom J. H. Structure, kinetics and thermodynamics of DNA hairpin fragments in solution. J Biomol Struct Dyn. 1983 Oct;1(1):115–129. doi: 10.1080/07391102.1983.10507429. [DOI] [PubMed] [Google Scholar]
- Haasnoot C. A., den Hartog J. H., de Rooij J. F., van Boom J. H., Altona C. Loopstructures in synthetic oligodeoxynucleotides. Nucleic Acids Res. 1980 Jan 11;8(1):169–181. doi: 10.1093/nar/8.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan L. S., Chandrasegaran S., Pulford S. M., Miller P. S. Detection of a guanine X adenine base pair in a decadeoxyribonucleotide by proton magnetic resonance spectroscopy. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4263–4265. doi: 10.1073/pnas.80.14.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennard O. Structural studies of DNA fragments: the G.T wobble base pair in A, B and Z DNA; the G.A base pair in B-DNA. J Biomol Struct Dyn. 1985 Oct;3(2):205–226. doi: 10.1080/07391102.1985.10508412. [DOI] [PubMed] [Google Scholar]
- Kime M. J., Moore P. B. Nuclear Overhauser experiments at 500 MHz on the downfield proton spectrum of a ribonuclease-resistant fragment of 5S ribonucleic acid. Biochemistry. 1983 May 24;22(11):2615–2622. doi: 10.1021/bi00280a004. [DOI] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Fritz H. J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984 Oct;38(3):879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- Lu A. L., Clark S., Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. L., Welsh K., Clark S., Su S. S., Modrich P. Repair of DNA base-pair mismatches in extracts of Escherichia coli. Cold Spring Harb Symp Quant Biol. 1984;49:589–596. doi: 10.1101/sqb.1984.049.01.066. [DOI] [PubMed] [Google Scholar]
- Lyons S. M., Schendel P. F. Kinetics of methylation in Escherichia coli K-12. J Bacteriol. 1984 Jul;159(1):421–423. doi: 10.1128/jb.159.1.421-423.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J Bacteriol. 1973 Jun;114(3):1143–1150. doi: 10.1128/jb.114.3.1143-1150.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Hare D. R., Reid B., Ikuta S., Lander N., Itakura K. Conformation, dynamics, and structural transitions of the TATA box region of self-complementary d[(C-G)n-T-A-T-A-(C-G)n] duplexes in solution. Biochemistry. 1985 Feb 12;24(4):926–935. doi: 10.1021/bi00325a018. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Ikuta S., Itakura K. Deoxyguanosine-deoxyadenosine pairing in the d(C-G-A-G-A-A-T-T-C-G-C-G) duplex: conformation and dynamics at and adjacent to the dG X dA mismatch site. Biochemistry. 1984 Jul 3;23(14):3207–3217. doi: 10.1021/bi00309a015. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Rice J. A., Broka C., Dallas J., Itakura K., Breslauer K. J. Structure, dynamics, and energetics of deoxyguanosine . thymidine wobble base pair formation in the self-complementary d(CGTGAATTCGCG) duplex in solution. Biochemistry. 1982 Feb 2;21(3):437–444. doi: 10.1021/bi00532a003. [DOI] [PubMed] [Google Scholar]
- Petruska J., Goodman M. F. Influence of neighboring bases on DNA polymerase insertion and proofreading fidelity. J Biol Chem. 1985 Jun 25;260(12):7533–7539. [PubMed] [Google Scholar]
- Pukkila P. J., Peterson J., Herman G., Modrich P., Meselson M. Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics. 1983 Aug;104(4):571–582. doi: 10.1093/genetics/104.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman M., Wagner R. Effects of DNA methylation on mismatch repair, mutagenesis, and recombination in Escherichia coli. Curr Top Microbiol Immunol. 1984;108:23–28. doi: 10.1007/978-3-642-69370-0_3. [DOI] [PubMed] [Google Scholar]
- Tibanyenda N., De Bruin S. H., Haasnoot C. A., van der Marel G. A., van Boom J. H., Hilbers C. W. The effect of single base-pair mismatches on the duplex stability of d(T-A-T-T-A-A-T-A-T-C-A-A-G-T-T-G) . d(C-A-A-C-T-T-G-A-T-A-T-T-A-A-T-A). Eur J Biochem. 1984 Feb 15;139(1):19–27. doi: 10.1111/j.1432-1033.1984.tb07970.x. [DOI] [PubMed] [Google Scholar]
- Topal M. D., Fresco J. R. Complementary base pairing and the origin of substitution mutations. Nature. 1976 Sep 23;263(5575):285–289. doi: 10.1038/263285a0. [DOI] [PubMed] [Google Scholar]
- Wagner R., Dohet C., Jones M., Doutriaux M. P., Hutchinson F., Radman M. Involvement of Escherichia coli mismatch repair in DNA replication and recombination. Cold Spring Harb Symp Quant Biol. 1984;49:611–615. doi: 10.1101/sqb.1984.049.01.069. [DOI] [PubMed] [Google Scholar]
- Wagner R., Jr, Meselson M. Repair tracts in mismatched DNA heteroduplexes. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4135–4139. doi: 10.1073/pnas.73.11.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]



