Abstract
The hammerhead ribozyme (HHRz) is an autocatalytic RNA motif found in subviral plant pathogens and transcripts of repetitive DNA sequences in animals. Here, we report the discovery and characterization of unique HHRzs encoded in a plant genome. Two novel sequences were identified on chromosome IV of Arabidopsis thaliana in a database search, which took into account recently defined structural requirements. The HHRzs are expressed in several tissues and coexist in vivo as both cleaved and noncleaved species. In vitro, both sequences cleave efficiently at physiological Mg2+ concentrations, indicative of functional loop–loop interactions. Kinetic analysis of loop nucleotide variants was used to determine a three-dimensional model of these tertiary interactions. Based on these results, on the lack of infectivity of hammerhead-carrying viroids in Arabidopsis, and on extensive sequence comparisons, we propose that the ribozyme sequences did not invade this plant by horizontal transfer but have evolved independently to perform a specific, yet unidentified, biological function.
INTRODUCTION
The hammerhead ribozyme (HHRz) is a small catalytic RNA motif found in a variety of subviral plant pathogens, including virus satellite RNAs and some viroids, and in transcripts from satellite DNAs in various amphibians, schistosomes, and cricket (Prody et al., 1986; Forster and Symons, 1987; Zhang and Epstein, 1996; Ferbeyre et al., 1998; Rojas et al., 2000). In the subviral pathogens, the HHRz participates in the rolling circle replication of the circular RNA. Mobile elements are likely to be the origin of the HHRz-coding satellite DNAs. The biological function of these transcripts, however, is unknown. HHRz motifs have not been identified to date in the genome of plants, with the exception of a genomically incorporated form of a retroviroid-like element in carnation (Dianthus caryophyllus) that occurs as tandem repeats (Daros and Flores, 1995; Hegedus et al., 2004).
HHRzs perform site-specific cleavage of the phosphodiester backbone by means of a transesterification reaction (summarized in Hammann and Lilley, 2002). By comparison of naturally occurring sequences, a minimal consensus motif was derived (Uhlenbeck, 1987; Ruffner et al., 1990) that consists of a strictly conserved core of 11 nucleotides from which three helices radiate (Figure 1). Such minimalist HHRzs have been studied extensively, and the Mg2+ concentration required for both folding and cleavage is ∼10 mM. Recent results from two groups (De la Peña et al., 2003; Khvorova et al., 2003) indicated that the presence of loop sequences outside of the catalytic core render the ribozyme active at considerably lower Mg2+ concentrations in the micromolar range. This is achieved by tertiary interactions of nucleotides in loops L1 and L2, which also lead to an altered metal ion binding of the HHRz core, compared with the minimalist version (Kisseleva et al., 2005). Natural HHRzs with respective loop sequences in place (Figure 1) also fold under these conditions (Penedo et al., 2004) and show fast cleavage kinetics (Canny et al., 2004). HHRzs are classified with respect to the arrangement of helices that surround the catalytic core. The motif shown in Figure 1 is of type III, in which helix III is not closed by a loop structure. Accordingly, HHRzs of types I and II have open helices I and II, respectively. To allow for productive tertiary interactions between loops L1 and L2, HHRz motifs of types I and II would need to accommodate internal bulges in helices I and II, respectively. Although HHRz motifs of type I are found in nature, for example, in transcripts of satellite DNA in amphibians (Zhang and Epstein, 1996), type II HHRzs have not been identified to date. Based on structural requirements for loop–loop interactions as described (De la Peña et al., 2003; Khvorova et al., 2003), we performed a database search for novel examples of natural HHRzs of type III.
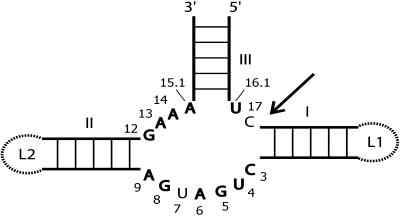
Figure 1.
The HHRz.
Scheme of the catalytic core of the HHRz with stems I, II, and III indicated. The 11 strictly conserved nucleotides are shown in boldface, with conventional numbering (Hertel et al., 1992). The cleavage site is indicated by an arrow. Ribozymes missing the loops at the ends of stems I and II (dashed lines) require Mg2+ concentrations in the 10 mM range for folding and activity (summarized in Hammann and Lilley, 2002). In the full natural type III HHRzs, as shown, the loop sequences can interact, leading to folding and accelerated cleavage in the micromolar Mg2+ range (De la Peña et al., 2003; Khvorova et al., 2003; Canny et al., 2004; Penedo et al., 2004).
RESULTS
Database Search for HHRz Motifs
Using conserved sequences of the catalytic core and structural features necessary for efficient cis hammerhead cleavage as restriction parameters, we searched the EMBL database for HHRz motifs. To this end, we used a pattern description language (D'Souza et al., 1997), as in a recent search for double-stranded RNA in Dictyostelium discoideum (Gräf et al., 2004). For type III HHRzs (Figure 1), the result of this search was 302 sequences, the majority of which (180) corresponded to annotated HHRzs from a variety of satellite RNAs and some viroids. Another 94 sequences stemmed from synthetic or patented constructs. Among the remaining sequences, we found two that originated from the genome of Arabidopsis thaliana, which were the only new examples encoded in a plant genome. This singular occurrence in only one plant genome presumably reflects the limited availability of sequence data, and we anticipate that additional examples are encoded in other plants, the genomes of which have not yet been analyzed. A summary of this database search is included in Supplemental Table 1 online. The probability of a type III HHRz (as defined by our search pattern; see Methods) to occur once in a random sequence is only 1.7 × 10−10, whereas the Arabidopsis genome has a size of 1.2 × 108 bp only, indicating that the presence of the two sequences should not occur by chance.
HHRzs in the Genome of Arabidopsis
The two HHRz sequences of Arabidopsis, termed Ara1 and Ara2, are located within 5 kb on chromosome IV at positions 15,027,890 to 15,027,834 and 15,032,903 to 15,032,847 (Figure 2A). They both are oriented in the antisense direction, either at the 3′ end of an open reading frame or between two open reading frames. These open reading frames are annotated as At4g30860 and At4g30870 (Arabidopsis Genome Initiative, 2000; Rhee et al., 2003). The sequences of both hits contain, as required by our database search, the 11 strictly conserved nucleotides of the catalytic core of natural HHRzs (Figure 1). In both sequences, stem I is 6 bp long and closed by an eight-nucleotide loop, whereas stem II is 4 bp long and closed by a six-nucleotide loop. However, not only these structural features but also the sequence of the HHRz domains are identical, with the exception of two nucleotides, one located in loop L1 and one in loop L2 (Figure 2B). These variable positions are occupied in Ara1 by a C in loop L1 and by a U in loop L2, whereas these nucleotides are reversed in Ara2. Also, the context of the two HHRz domains is surprisingly well conserved, as sequences 219 nucleotides upstream and 25 nucleotides downstream of Ara1 and Ara2 are 95% identical. An alignment of the two sequences is shown in Supplemental Figure 1 online. We termed this stretch of ∼300 nucleotides the HHRz-containing consensus sequence.
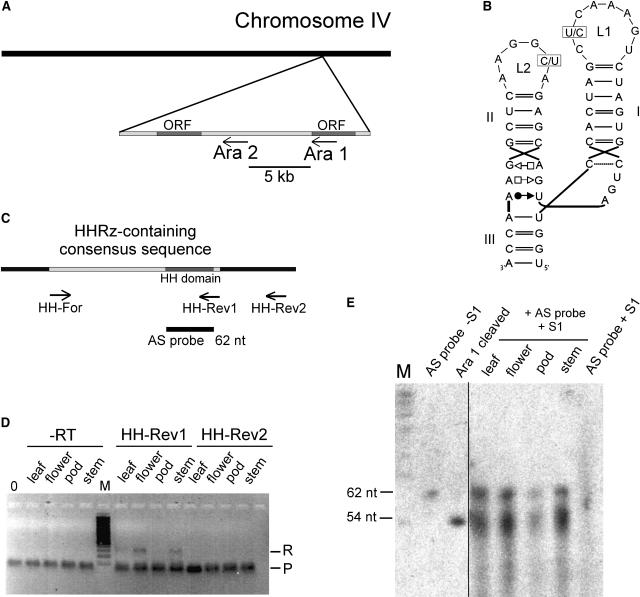
Figure 2.
HHRzs in Arabidopsis.
(A) Chromosomal localization of the two motifs in the genome of Arabidopsis. The HHRz sequences Ara1 and Ara2 are found within 5 kb in the antisense orientation to open reading frames (ORFs) on chromosome IV.
(B) Secondary structures of the HHRz domains. Variable positions in loops L1 and L2 are indicated. In Ara1, the variable positions are occupied by a C in loop L1 and a U in loop L2, whereas in Ara2, these nucleotides are replaced by U (loop L1) and C (loop L2).
(C) Design of primers and antisense RNA used in RT-PCR and the S1 nuclease assay. The HHRz-containing 300-nucleotide consensus sequence is shown in light gray, and the 66-nucleotide hammerhead motif within is shown in dark gray. The positions of primers HH-Rev1, HH-Rev2, and HH-For for RT-PCR are indicated, as is the position of the 62-nucleotide (nt) antisense (AS) probe used in the S1 nuclease assay.
(D) RT-PCR of RNA isolated from the indicated organs. For first-strand cDNA synthesis, the specific primers HH-Rev1 and HH-Rev2 were used. PCR was performed using the specific primers HH-Rev1 and HH-For that amplify the uncleaved 300-nucleotide HHRz-containing consensus sequence. R denotes the signal of the RT-PCR product, and P indicates a band generated by the primers. In the control reactions (−RT), PCR was performed on RNA that had not been subjected to first-strand cDNA synthesis, and 0 refers to a template-free control PCR. M denotes a 100-bp marker. Bands were visualized by ethidium bromide staining.
(E) Nuclease S1 protection assay. Total RNA (0.5 μg) of the indicated tissues was hybridized to a 62-nucleotide antisense RNA of the hammerhead domain. S1 nuclease was used to digest single-stranded RNA. As size markers, a non-S1–treated antisense probe and an in vitro transcript of the 66-nucleotide Ara1 sequence, which self-cleaves in the course of the transcription, were included, resulting in fragments of 54 and 12 nucleotides (the latter not visible on the gel). As a negative control, the antisense RNA probe was digested with nuclease S1 (AS probe + S1). RNA was visualized by exposure to x-ray films.
HHRz-Containing Sequences Are Expressed in Various Tissues of Arabidopsis
To assess whether the sequences are present merely at the DNA level or are transcribed in vivo, we used two independent experimental approaches, RT-PCR and S1 nuclease assay. For RT-PCR, we isolated RNA from leaves, flowers, pods, and stems. First-strand cDNA synthesis was performed using two specific primers, one located 212 nucleotides downstream of the consensus sequence (HH-Rev2) and one directed against the 3′ end of the 300-nucleotide consensus sequence (HH-Rev1) (Figure 2C). After removal of template RNA by alkaline treatment, PCR was performed using primers specific for the consensus sequence that would give a signal for an RNA in which HHRz cleavage has not occurred. No RT-PCR product was observed for the cDNAs prepared from HH-Rev2 (Figure 2D). However, RT-PCR using the HH-Rev1–primed cDNAs resulted in bands of the expected size on RNA isolated from leaves, flowers, and stems but not from pods. RT-PCR products were cloned and their identities confirmed by sequencing. An identical PCR procedure was performed on RNA that had not been subjected to an RT reaction; the negative results (−RT in Figure 2D) exclude the possibility that the observed signals for the HH-Rev1–primed cDNA were caused by contamination with genomic DNA. The observation that HH-Rev1–primed cDNAs gave rise to a PCR product, whereas HH-Rev2–primed cDNAs did not, indicates that the end of the primary transcript is located between these two primers. Our search for transcriptional motifs common to both sequences gave no result. Furthermore, we found no appropriate annotations in databases. This is not surprising, however, given the largely unknown nature of promoters for RNA genes compared with those for protein genes.
From the RT-PCR experiments, it was not possible to assess whether HHRz cleavage occurred in vivo. To address this question, we could not use RT-PCR because the first-strand cDNA primer HH-Rev1 binds very close to the HHRz cleavage site. Instead, we used a nuclease S1 protection assay. An in vitro transcribed 62-nucleotide radiolabeled antisense RNA of the HHRz motif (Figure 2C) was hybridized to 0.5 μg of RNA isolated from the aforementioned tissues and subsequently treated with DNase I. Under the experimental conditions applied here, nuclease S1 digests single-stranded RNA but not double-stranded RNA. If HHRz cleavage occurred in vivo, nuclease S1 treatment would lead to a truncated antisense RNA of the size of the 5′ cleavage product (54 nucleotides), whereas the size of the probe should remain unchanged for sequences that did not undergo HHRz cleavage. In a first exposure of the gel, when the size markers for cleaved HHRz and antisense RNA in the control lanes were already visible (Figure 2E, left), we detected no signal in the S1-treated samples (data not shown). However, upon reexposure for a longer period, we observed specific signals of the size of both cleaved and noncleaved HHRz sequences in S1 nuclease–treated RNA from leaves, flowers, and stems and a considerably weaker signal for RNA from pods (Figure 2E, right). This finding is consistent with the lack of a signal by RT-PCR on RNA from pods.
These results show that HHRz-containing transcripts are expressed in various organs in Arabidopsis and that they exist in vivo both as unprocessed and as cleaved species.
In Vitro Cleavage Activity of the Arabidopsis HHRzs
Recent results indicated that an interaction between loops in natural HHRz sequences is essential for efficient intracellular cleavage (De la Peña et al., 2003; Khvorova et al., 2003). Because we had observed HHRz cleavage in total RNA prepared from different organs, we then analyzed the cleavage behavior of the Arabidopsis sequences in vitro.
Initially, we investigated the cleavage of the two wild-type sequences Ara1 and Ara2. Full-length RNA was gel-purified from transcription reactions performed in the presence of antisense DNA oligonucleotides, the presence of which was necessary to prevent RNA self-cleavage in the course of the transcription reaction. For cleavage reactions, RNA was incubated at 25°C in 0.6 mM MgCl2, and the reaction was stopped by quenching with EDTA. Reaction products were separated by PAGE on 20% gels containing 7 M urea and visualized by phosphor imaging (Figure 3A). Data were fitted to a single exponential, resulting in a cleavage rate constant (kobs) value of 2.2 min−1 for both wild-type sequences (Figures 3C and 3D). Although cleavage at these near-physiological Mg2+ concentrations was incomplete, similar to observations made by others (Khvorova et al., 2003), the fast cleavage rates were indicative of functional loop–loop interactions (De la Peña et al., 2003; Khvorova et al., 2003; Canny et al., 2004; Penedo et al., 2004).
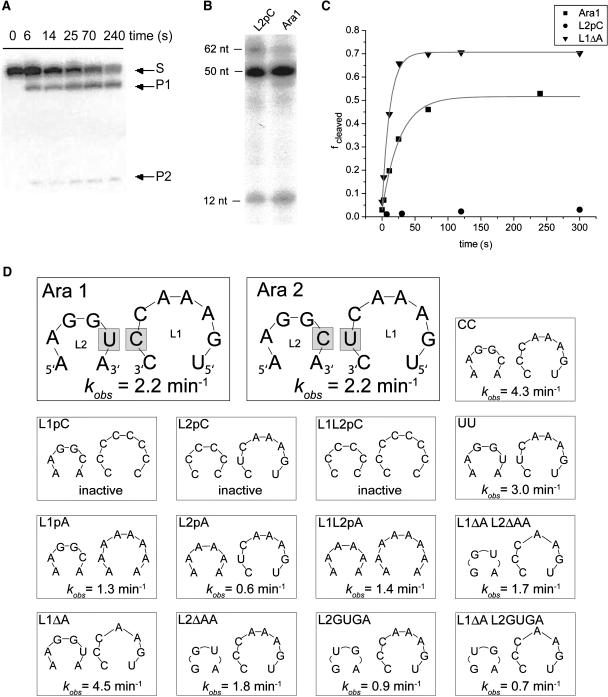
Figure 3.
Cleavage Activity of the Arabidopsis HHRzs and Sequence Variants.
(A) Representative time course of the cleavage reaction of Ara2. The reaction was performed at 25°C in 10 mM Tris-HCl, pH 7.6, 0.1 mM EDTA, and 10 mM NaCl and started by the addition of 0.6 mM MgCl2. The reaction was stopped at different times by mixing aliquots with EDTA, products were separated by PAGE on a 20% gel containing 7 M urea, and the RNA was visualized by phosphor imager analysis.
(B) Representative cleavage reactions of Ara1 and the L2pC variant in the course of an in vitro transcription reaction at 20 mM MgCl2, in which HHRz cleavage was not inhibited by a DNA oligonucleotide. The sizes of the full-length transcripts (62 nucleotides [nt]) used in this experiment and of the cleavage products (50 and 12 nucleotides, respectively) are indicated. The RNA was visualized by phosphor imager analysis.
(C) Representative kinetic analysis of the cleavage reactions of Ara1 and the loop variants L1ΔA and L2pC at 0.6 mM MgCl2. The cleaved fraction (fcleaved) was plotted against time, and data were fit to a single exponential. Cleavage reactions were performed as described for (A).
(D) Summary of loop sequences in the two natural variants (Ara1 and Ara2) and several artificial variants used in this study. Each panel shows loop L1 (5′ to 3′ direction) at right and loop L2 (3′ to 5′ direction) at left. The cleavage rate constants (kobs), obtained from cleavage reactions as described for (A), are indicated at the bottom of each panel.
To study the importance of loop nucleotides for the cleavage reaction, we created a series of sequence variants (Figure 3D). As HHRz sequences without any loop–loop interactions, we designed variants L1pC and L2pC, in which nucleotides in loops L1 and L2, respectively, were replaced by a consecutive stretch of cytidines, as well as the combination of both variants (L1L2pC; Figure 3D). Independent of productive loop interactions, HHRzs do cleave under increased, nonphysiological MgCl2 concentrations, at which they behave as minimalist ribozymes. Therefore, it was not surprising that complete cleavage of these variants, similar to the wild-type Ara1 sequence, was observed in the course of an uninhibited transcription reaction that contained 20 mM MgCl2 (Figure 3B; data not shown). In stark contrast to this, the L1pC, L2pC, and L1L2pC variants were essentially inactive when tested for cleavage activity at 0.6 mM MgCl2 (Figures 3C and 3D). These results show that the cleavage activity of the Arabidopsis sequences depends under near-physiological Mg2+ concentrations on productive loop–loop interaction, as observed for other natural HHRzs (De la Peña et al., 2003; Khvorova et al., 2003; Canny et al., 2004; Penedo et al., 2004).
To analyze loop–loop interactions further, we changed the variable loop position CU (Ara1) and UC (Ara2; Figure 2B) to CC and UU (Figure 3D). Observed cleavage rates for these variants were near those of the wild type, showing that these changes did not affect interactions adversely. When loop sequences were changed to consecutive stretches of adenosines (L1pA, L2pA, and L1L2pA; Figure 3D), we observed HHRz cleavage, although at slightly reduced rates. These findings indicate that the structures of the loops and their resulting interactions are suboptimal with all-purine loops. When one of the three adenosines was removed from loop L1 (variant L1ΔA; Figure 3D), cleavage was faster than for the wild-type sequences. Conversely, shortening loop L2 by the removal of the two 5′ adenosines (variant L2ΔAA; Figure 3D) or exchanging its sequence with the loop L2 sequence GUGA of the satellite RNA of Tobacco ringspot virus (sTRSV; Buzayan et al., 1990) (variant L2GUGA; Figure 3D) caused a decrease of kobs. This effect could not be rescued for either variant by also changing loop L1 to the otherwise advantageous variant L1ΔA (variants L1ΔA L2ΔAA and L1ΔA L2GUGA, respectively; Figure 3D).
Modeling Loop–Loop Interactions in the Arabidopsis HHRz
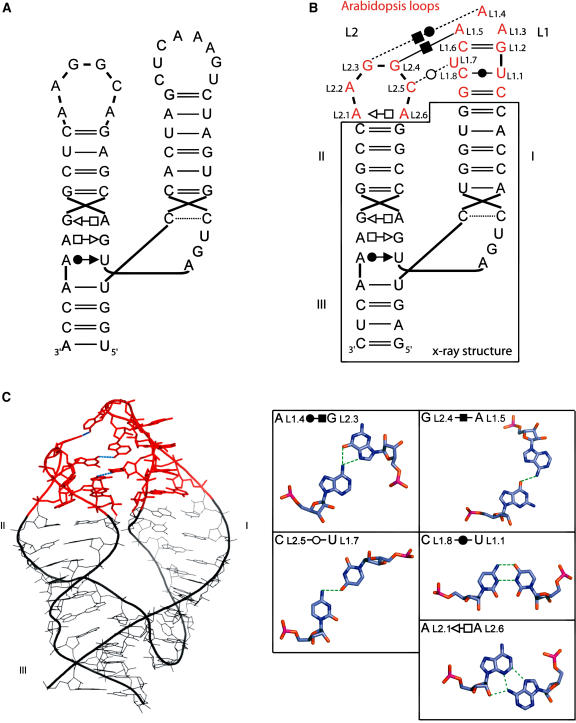
The structure of the HHRz core has been determined by crystallography (Pley et al., 1994; Scott et al., 1995). However, this minimal ribozyme is 100-fold less efficient than the entire ribozyme in which helices I and II interact through their apical loops. The specific tertiary interactions between these loops in various natural HHRzs have been modeled (Khvorova et al., 2003). In the case of the Arabidopsis HHRz, we proceeded as previously by keeping intact the core structure and modeling the loop–loop interactions. Throughout this section, we use the conventional numbering system (Hertel et al., 1992). As in the previous model (Khvorova et al., 2003), the first and last nucleotides of loop L1 (L1.1 and L1.8, respectively; Figure 4B) form a noncanonical cis Watson-Crick-Watson-Crick CU base pair adjacent to a bulged pyrimidine, L1.7 (Leontis and Westhof, 2001). The remaining five bases (L1.2 to L1.6) can form a triloop closed by a standard base pair GC, as seen for positions 326 to 330 in the Haloarcula marismortui 23S rRNA structure (Ban et al., 2000; Lee et al., 2003). To build loop L2, the hexaloop closing helix 68 in H. marismortui 23S rRNA was used. In the x-ray structure, the closing pair of this loop is a trans Hoogsteen-Sugar edge, and nucleotides at positions 4 and 5 are implicated in tertiary contacts. In our model (Figure 4C), the corresponding nucleotides at positions L2.4 and L2.5, as well as that at position L2.3, make interactions with loop L1. In this model, the Watson-Crick faces of L1.7 and L2.5 form a pseudo-trans Watson-Crick-Watson-Crick base pair, L1.5 and L2.4 form a cis Hoogsteen-Hoogsteen base pair, and L1.4 and L2.3 form a pseudo-cis Watson-Crick-Hoogsteen base pair. This model is in good agreement with the kinetic data from the loop variants (Figure 3D). The observed rate constants are near wild type when bases at variable positions L1.7 and L2.5 are replaced by CC or UU (the trans Watson-Crick-Watson-Crick base pair is more stable in these two cases than with CU or UC). The catalytic efficiency is improved by the removal of one A in loop L1, which confirms the absence of a stable GNRA tetraloop in spite of the GAAA sequence. The reduction of kobs caused by the replacement of loop L2 with the shorter loops GUGA or GGCA might seem surprising, given the involvement of such a stable GUGA tetraloop in equivalent interactions in the sTRSV RNA. However, in sTRSV RNA, loop L1 has a different sequence, allowing for other types of contacts that could not be formed with the native Arabidopsis L1 loop (Khvorova et al., 2003).
Figure 4.
Modeling of the Arabidopsis HHRz Ara2.
(A) Secondary structure.
(B) Secondary structure of the same motif with the proposed tertiary contacts of the model. Loop nucleotides are numbered according to the conventional numbering system (Hertel et al., 1992). The secondary structure contained within the black box is derived from the crystallographic structure reported previously (Scott et al., 1995), and the modeled regions are shown in red. The tertiary interactions and other noncanonical base pairs are indicated using specific symbols (Leontis and Westhof, 2001).
(C) Proposed three-dimensional model. The color code corresponds to that used for the secondary structure. The non–Watson-Crick base pairs within and between the loops are shown at right.
DISCUSSION
Research on the HHRz has received a new direction from the ground-breaking data produced by the groups of Khvorova and Flores (De la Peña et al., 2003; Khvorova et al., 2003), who realized that interacting loops at the ends of stems I and II of natural sequences are required for efficient cleavage of HHRzs under near-physiological Mg2+ concentrations, whereas minimal versions are virtually inactive (Figure 1). For our database search, we used a descriptive pattern that included structural requirements for these tertiary interactions, which led to the discovery of new HHRzs in Arabidopsis. The in vitro cleavage behavior of these ribozymes is similar to that of other natural sequences with intact loops in place (De la Peña et al., 2003; Khvorova et al., 2003; Lilley, 2003; Penedo et al., 2004), as they are active at Mg2+ concentrations in the submillimolar range with cleavage being incomplete (Figure 3A). In agreement with this, cleaved and noncleaved RNA species coexist in vivo (Figure 2E). To investigate the tertiary interactions between the apical loops responsible for this behavior, we created a series of sequence variants (Figure 3D). Based on their kinetic properties and on previous crystallographic data, a three-dimensional model with a complex array of interactions within and between bases in loops L1 and L2 is proposed (Figure 4).
HHRz sequences in satellite DNA from amphibians, cave cricket, and schistosomes (Forster and Symons, 1987; Zhang and Epstein, 1996; Ferbeyre et al., 1998; Rojas et al., 2000) are likely to have originated from genomic incorporation of mobile genetic elements. In the plant kingdom, there is also an example of genomic incorporation of a HHRz-containing sequence, that of the Carnation small viroid–like RNA (CarSV RNA). The DNA form of this retroviroid-like element exists in carnation as tandem repeats and is also found fused to the DNA of a carnation caulimovirus (Daros and Flores, 1995; Hegedus et al., 2004). For the sequences from Arabidopsis, such an incorporation event seems less likely for a variety of reasons. Despite some structural similarity, no sequence similarities were found between the 300-nucleotide conserved stretch (Figure 2C) containing the HHRzs in Arabidopsis and known viroids, particularly those of the HHRz-bearing family Avsunviroidae (Flores et al., 2004; Tabler and Tsagris, 2004). Also, in viroids of this family, HHRz motifs are found on strands of both polarities, presumably for processing of multimeric viroid RNAs in chloroplasts. In the antisense strand of our sequences, we detected no hammerhead or hairpin ribozyme motif, nor was the antisense RNA of the consensus sequence catalytically active (data not shown). Furthermore, Arabidopsis is naturally not infected by viroid species carrying HHRzs, and recent data using this plant as an artificial host indicate that a viroid of the family Avsunviroidae is poorly processed in planta (Daros and Flores, 2004; Matousek et al., 2004). This argues against an evolutionary origin of the HHRzs in Arabidopsis by incorporation of the cDNA of a hammerhead-like viroid RNA. However, it does not preclude the possibility that these sequences could stem from viral satellite RNAs, in which the HHRz was found originally (Prody et al., 1986), because HHRzs in some of these pathogenic RNAs are found exclusively on the plus polarity strand. Yet, the lack of sequence similarity to viral satellite RNAs argues against such a possibility, and the presence of only two HHRz sequences in the genome of Arabidopsis is also not consistent with this idea, because a much greater frequency of occurrence would be expected. Given the high conservation of the sequences that surround the two HHRz motifs in Arabidopsis, as well as their close chromosomal location, it is likely that they are derived from one another by duplication and translocation events. This fact suggests that these sequences originated from a single, independent event, in agreement with the proposed multiple origins for HHRzs inferred from in vitro selection experiments (Salehi-Ashtiani and Szostak, 2001). An independent evolution of the HHRzs in Arabidopsis is supported by the lack of any apparent sequence similarity to any other plant entry in the databases, both for the catalytic motif (Figure 2B) and for the 300-nucleotide conserved stretches that contain them (Figure 2C). The sequence conservation, expression in various organs, and in vivo activity of the novel HHRzs suggest a still unidentified biological function in the plant. Because of their unique features, also compared with the DNA form of CarSV RNA, we conclude that the Arabidopsis sequences are unique examples of HHRzs encoded in a plant genome and, in a more general context, the only HHRz motifs genomically encoded but not forming part of a satellite DNA.
METHODS
Database Search
As a pattern for a hammerhead type III ribozyme, we used the conserved sequence features (Uhlenbeck, 1987) and the structure descriptions (De la Peña et al., 2003; Khvorova et al., 2003) that were proposed to be critical for activity in low Mg2+ concentrations: that is, a stem III of 5 to 6 bp beginning with an AU (positions 15.1:16.1 in standard nomenclature), a stem I of 5 to 7 bp with a 3- to 13-nucleotide hairpin loop, and a stem II of 4 to 5 bp beginning with a GC (positions 10.1:11.1) and a 3- to 6-nucleotide hairpin loop. We used PatScan (D'Souza et al., 1997) with this pattern, allowing for one mismatch in stem I, for a search in the EMBL library (release 78). Details of this pattern and the outcome of the search are presented in Supplemental Table 1 online. After the elimination of redundant hits, they were sorted according to the thermodynamic stability of their secondary structures, as predicted by RNAfold (Hofacker, 2003). The hits to Arabidopsis thaliana chromosome IV sequence ranked highest on the final list, with a free energy ΔG0 = −25.7 kcal/mol. An alignment of the two conserved sequences, which contain the HHRzs from Arabidopsis, was performed using Clustal (Jeanmougin et al., 1998) and is shown in Supplemental Figure 1 online.
For estimation of the number of expected hits, we calculated the probability of occurrence of our HHRz pattern. For the probability of nucleotides, we used frequencies according to the base composition of the Arabidopsis genome, and for helical regions, we took into account the base pairing rules including GU wobble pairs, then added the probabilities according to all different alternatives attributable to the allowed mismatch in stem I and the stem length alternatives.
RNA Isolation
RNA was isolated from Arabidopsis ecotype Columbia (Col-0, N1092). Fresh tissue (100 mg of flowers, stems, leaves, or pods) was shock-frozen in liquid nitrogen and ground in a tissue mill precooled at −80°C. One milliliter of lysis buffer (50% guanidinium thiocyanate, 0.5% sarkosyl, and 25 mM sodium citrate, pH 7.0) was added, and the mixture was phenolized immediately. RNA was precipitated with ethanol and resuspended, with its concentration being spectrophotometrically adjusted and its integrity checked on 1.6% agarose gels containing 20 mM guanidinium thiocyanate.
RT-PCR
Before the RT reaction, the RNA preparation was digested with DNase I to prevent contamination by genomic DNA. This RNA (0.5 μg) was hybridized to 5.4 pmol of the first-strand primers HH-Rev1 and HH-Rev2. Synthesis of cDNA was performed using 200 units of Moloney murine leukemia virus reverse transcriptase (Fermentas, St. Leon-Rot, Germany) for 50 min at 42°C, according to the manufacturer's instructions. After alkaline treatment for 20 min, 30 cycles of PCR were performed using primers HH-For and HH-Rev1. For the −RT negative control, RNA that had not been subjected to reverse transcription but that otherwise had been treated identically was used as template in the PCR. RT-PCR products were cloned into pGEM-Teasy (Promega, Mannheim, Germany) and their identities confirmed by sequencing.
S1 Nuclease Protection Assay
For this purpose, a 62-nucleotide radiolabeled antisense RNA, which corresponds to the hammerhead domain, was prepared by in vitro transcription (see below). DNase I–treated total RNA (0.5 μg) was hybridized overnight to this antisense probe at 62°C in a total volume of 30 μL of hybridization buffer (75% formamide, 40 mM Pipes, pH 6.4, 1 mM EDTA, and 400 mM NaCl). Nuclease S1 (150 units) was added in 300 μL of S1 buffer (Fermentas), and the mixture was incubated for 30 min at 37°C. After proteinase K treatment, the mixture was phenolized and the RNA was precipitated with ethanol, resuspended, and resolved by PAGE on 16% gels containing 7 M urea. Bands were visualized by exposure to x-ray films.
Transcription of RNA
Templates for the transcription of antisense RNA and ribozymes were made by recursive PCR from synthetic DNA oligonucleotides. To ensure efficient transcription, either GGG or GCG was inserted after the T7 promoter sequence. RNA was transcribed using T7 RNA polymerase (Milligan et al., 1987) in 40 mM Tris-HCl, pH 8.0, 20 mM MgCl2, 2 mM spermidine, 0.01% Triton X-100, with 0.5 mM rATP, rGTP, or rCTP or 0.1 mM rUTP and in the presence of [α-32P]UTP. For the transcription of ribozyme sequences, DNA antisense oligonucleotides covering the catalytic core were included. Full-length RNA was purified by PAGE on 20% gels containing 7 M urea and eluted from excised gel slices with 0.7% SDS/1× TE (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA) overnight. Eluted RNA was phenolized, recovered by ethanol precipitation, and dissolved in water. RNA was snap-cooled in 10 mM Tris-HCl, pH 7.6, 0.1 mM EDTA, and 10 mM NaCl, and cleavage reactions at 25°C were started by the addition of MgCl2 to a final concentration of 0.6 or 20 mM. Reactions were stopped by adding 3 volumes of loading solution (95% formamide and 50 mM EDTA). Cleavage products were resolved by PAGE on 16 or 20% gels containing 7 M urea and quantified by phosphor imager analysis. Values for kobs were obtained by fitting data to the equation F(t) = F0 + F∞(1 − e−kt) (Stage-Zimmermann and Uhlenbeck, 1998). Cleavage reactions were performed at least in duplicate, and errors in kobs were not >10%.
Structural Modeling
Structural homology modeling was performed using the MANIP package (Massire and Westhof, 1998).
Supplementary Material
Acknowledgments
We thank Michael Krieg and Bertram Daum for help with cleavage analyses, Branimira E. Borisova for sequencing, Markus Kuhlmann for regularly providing us with plants and instructions on their maintenance (water), and Michael Schmitz for stimulating discussions about the probability calculations. This work was supported by a grant from the Zentrale Forschungförderung of the University of Kassel to C.H. and by a grant from the Deutsche Forschungsgemeinschaft to G.S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Christian Hammann (c.hammann@uni-kassel.de).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.032730.
References
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. (2000). The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289, 905–920. [DOI] [PubMed] [Google Scholar]
- Buzayan, J.M., van Tol, H., Feldstein, P.A., and Bruening, G. (1990). Identification of a non-junction phosphodiester that influences an autocatalytic processing reaction of RNA. Nucleic Acids Res. 18, 4447–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canny, M.D., Jucker, F.M., Kellogg, E., Khvorova, A., Jayasena, S.D., and Pardi, A. (2004). Fast cleavage kinetics of a natural hammerhead ribozyme. J. Am. Chem. Soc. 126, 10848–10849. [DOI] [PubMed] [Google Scholar]
- Daros, J.A., and Flores, R. (1995). Identification of a retroviroid-like element from plants. Proc. Natl. Acad. Sci. USA 92, 6856–6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daros, J.A., and Flores, R. (2004). Arabidopsis thaliana has the enzymatic machinery for replicating representative viroid species of the family Pospiviroidae. Proc. Natl. Acad. Sci. USA 101, 6792–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Peña, M., Gago, S., and Flores, R. (2003). Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 22, 5561–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza, M., Larsen, N., and Overbeek, R. (1997). Searching for patterns in genomic data. Trends Genet. 13, 497–498. [DOI] [PubMed] [Google Scholar]
- Ferbeyre, G., Smith, J.M., and Cedergren, R. (1998). Schistosome satellite DNA encodes active hammerhead ribozymes. Mol. Cell. Biol. 18, 3880–3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores, R., Delgado, S., Gas, M.E., Carbonell, A., Molina, D., Gago, S., and De la Peña, M. (2004). Viroids: The minimal non-coding RNAs with autonomous replication. FEBS Lett. 567, 42–48. [DOI] [PubMed] [Google Scholar]
- Forster, A.C., and Symons, R.H. (1987). Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell 49, 211–220. [DOI] [PubMed] [Google Scholar]
- Gräf, S., Borisova, B.E., Nellen, W., Steger, G., and Hammann, C. (2004). A database search for double-strand containing RNAs in Dictyostelium discoideum. Biol. Chem. 385, 961–965. [DOI] [PubMed] [Google Scholar]
- Hammann, C., and Lilley, D.M. (2002). Folding and activity of the hammerhead ribozyme. ChemBioChem 3, 690–700. [DOI] [PubMed] [Google Scholar]
- Hegedus, K., Dallmann, G., and Balazs, E. (2004). The DNA form of a retroviroid-like element is involved in recombination events with itself and with the plant genome. Virology 325, 277–286. [DOI] [PubMed] [Google Scholar]
- Hertel, K.J., Pardi, A., Uhlenbeck, O.C., Koizumi, M., Ohtsuka, E., Uesugi, S., Cedergren, R., Eckstein, F., Gerlach, W.L., Hodgson, R., and Symons, R.H. (1992). Numbering system for the hammerhead. Nucleic Acids Res. 20, 3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker, I.L. (2003). Vienna RNA secondary structure server. Nucleic Acids Res. 31, 3429–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanmougin, F., Thompson, J.D., Gouy, M., Higgins, D.G., and Gibson, T.J. (1998). Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23, 403–405. [DOI] [PubMed] [Google Scholar]
- Khvorova, A., Lescoute, A., Westhof, E., and Jayasena, S.D. (2003). Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 10, 708–712. [DOI] [PubMed] [Google Scholar]
- Kisseleva, N., Khvorova, A., Westhof, E., and Schiemann, O. (2005). Binding of manganese(II) to a tertiary stabilized hammerhead ribozyme as studied by electron paramagnetic resonance spectroscopy. RNA 11, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.C., Cannone, J.J., and Gutell, R.R. (2003). The lonepair triloop: A new motif in RNA structure. J. Mol. Biol. 325, 65–83. [DOI] [PubMed] [Google Scholar]
- Leontis, N.B., and Westhof, E. (2001). Geometric nomenclature and classification of RNA base pairs. RNA 7, 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley, D.M. (2003). Ribozymes: A snip too far? Nat. Struct. Biol. 10, 672–673. [DOI] [PubMed] [Google Scholar]
- Massire, C., and Westhof, E. (1998). MANIP: An interactive tool for modelling RNA. J. Mol. Graph. Model. 16, 197–205, 255–257. [DOI] [PubMed] [Google Scholar]
- Matousek, J., Orctova, L., Steger, G., Skopek, J., Moors, M., Dedic, P., and Riesner, D. (2004). Analysis of thermal stress-mediated PSTVd variation and biolistic inoculation of progeny of viroid “thermomutants” to tomato and Brassica species. Virology 323, 9–23. [DOI] [PubMed] [Google Scholar]
- Milligan, J.F., Groebe, D.R., Witherall, G.W., and Uhlenbeck, O.C. (1987). Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penedo, J.C., Wilson, T.J., Jayasena, S.D., Khvorova, A., and Lilley, D.M. (2004). Folding of the natural hammerhead ribozyme is enhanced by interaction of auxiliary elements. RNA 10, 880–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pley, H.W., Flaherty, K.M., and McKay, D.B. (1994). Three-dimensional structure of a hammerhead ribozyme. Nature 372, 68–74. [DOI] [PubMed] [Google Scholar]
- Prody, G.A., Bakos, J.T., Buzayan, J.M., Schneider, I.R., and Bruening, G. (1986). Autolytic processing of dimeric plant virus satellite RNA. Science 231, 1577–1580. [DOI] [PubMed] [Google Scholar]
- Rhee, S.Y., et al. (2003). The Arabidopsis Information Resource (TAIR): A model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 31, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas, A.A., Vazquez-Tello, A., Ferbeyre, G., Venanzetti, F., Bachmann, L., Paquin, B., Sbordoni, V., and Cedergren, R. (2000). Hammerhead-mediated processing of satellite pDo500 family transcripts from Dolichopoda cave crickets. Nucleic Acids Res. 28, 4037–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffner, D.E., Stormo, G.D., and Uhlenbeck, O.C. (1990). Sequence requirements of the hammerhead RNA self-cleavage reaction. Biochemistry 29, 10695–10702. [DOI] [PubMed] [Google Scholar]
- Salehi-Ashtiani, K., and Szostak, J.W. (2001). In vitro evolution suggests multiple origins for the hammerhead ribozyme. Nature 414, 82–84. [DOI] [PubMed] [Google Scholar]
- Scott, W.G., Finch, J.T., and Klug, A. (1995). The crystal structure of an all-RNA hammerhead ribozyme: A proposed mechanism for RNA catalytic cleavage. Cell 81, 991–1002. [DOI] [PubMed] [Google Scholar]
- Stage-Zimmermann, T.K., and Uhlenbeck, O.C. (1998). Hammerhead ribozyme kinetics. RNA 4, 875–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabler, M., and Tsagris, M. (2004). Viroids: Petite RNA pathogens with distinguished talents. Trends Plant Sci. 9, 339–348. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck, O.C. (1987). A small catalytic oligoribonucleotide. Nature 328, 596–600. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., and Epstein, L.M. (1996). Cloning and characterization of extended hammerheads from a diverse set of caudate amphibians. Gene 172, 183–190. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.