Simple Summary
This study investigates the inhibitory effect of sex pheromone components of the plum fruit moth (PFM), Grapholita funebrana, loaded with different doses of (Z)-8-tetradecenyl acetate (Z8-14:Ac) or (Z)-10-tetradecenyl acetate (Z10-14:Ac) on the trapping of the closely related species, the male oriental fruit moth (OFM), G. molesta. Field tests showed that adding 5–30% of Z8-14:Ac to a mixture of Z8-12:Ac and E8-12:Ac did not significantly affect the trapping of PFM males but reduced OFM male trapping by more than 86%. In contrast, adding over 10% of Z10-14:Ac to a mixture of Z8-12:Ac and E8-12:Ac inhibited the trapping of both OFM and PFM males. GmolPBP2 from OFMs exhibited the strongest binding affinity for Z8-14:Ac compared to the other GmolPBP/GmolGOBP and was considered the most likely OBP for recognizing and transporting Z8-14:Ac. Mutating the Phe12 residue of GmolPBP2 to Ala12 resulted in a loss of affinity for Z8-14:Ac, suggesting that Phe12 was the key amino acid and that π–alkyl was the primary weak interaction maintaining the binding affinity for GmolPBP2. These findings enhance our understanding of the molecular mechanisms through which insects recognize the sex pheromones of closely related species and thereby contribute to the development of species-specific sex attractants for PFMs using secondary sex pheromones.
Keywords: Grapholita funebrana, Grapholita molesta, sex pheromone, inhibition, fluorescence competitive binding assay
Abstract
The plum fruit moth (PFM), Grapholita funebrana, and the oriental fruit moth (OFM), G. molesta, are closely related fruit moth species that severely damage fruit trees in Rosaceae. Both species share common primary sex pheromone components Z8-12:Ac and E8-12:Ac. The secondary sex pheromone components of PFMs consist of Z8-12:OH, Z8-14:Ac, and Z10-14:Ac, while those of OFMs include Z8-12:OH and 12:OH. Previous researchers have proved that the inclusion of Z8-14:Ac and Z10-14:Ac did not augment PFM catches but inhibited OFM catches in orchards in Europe, thereby maintaining the species-specificity of the PFM sex attractant. However, which of these components, Z8-14:Ac or Z10-14:Ac, plays the major role in inhibiting OFM attraction remains unclear. In the current study, electroantennogram (EAG) assays indicated that both OFM and PFM males exhibited a moderate EAG response to Z8-14:Ac and Z10-14:Ac. Rubber septa loaded with varying ratios of Z8-14:Ac (1% to 30%) or Z10-14:Ac (5% to 110%) combined with a constant dose of Z8-12:Ac and E8-12:Ac produced diverse trapping effects. Sex attractants containing Z8-14:Ac did not significantly affect the trapping of PFM males but drastically reduced the capture of OFM males, with the reduction reaching up to 96.54%. Attractants containing more than 10% of Z10-14:Ac simultaneously reduced the number of OFM and PFM males captured. Z8-14:Ac was indispensable for maintaining the specificity of sex pheromones. Fluorescence competitive binding assays of recombinant GmolPBP2 showed the lowest Ki value (0.66 ± 0.02 μM) among the PBPs/GOBPs from OFMs, suggesting that it is the most likely target for Z8-14:Ac. Molecular dynamic simulation and site-directed mutagenesis assays confirmed that the Phe12 residue, which forms a π–alkyl interaction with Z8-14:Ac, was crucial for GmolPBP2 binding to Z8-14:Ac. In conclusion, Z8-14:Ac is vital to the specificity of PFM sex pheromones inhibiting OFM attractants when added to Z8-12:Ac and E8-12:Ac. This could be potentially used to develop species-specific sex attractants for the PFM.
1. Introduction
The oriental fruit moth (OFM), Grapholita molesta, and the plum fruit moth (PFM), G. funebrana (Lepidoptera: Tortricidae), are important fruit-boring pests worldwide, causing serious economic losses to stone and pome fruits [1,2]. Previous studies assumed that OFM larvae primarily feed on peaches, pears, and apples [3,4,5], whereas PFM larvae mainly consume plums and apricots [6,7]. However, over the past two decades, with the expansion of apple, peach, and pear cultivation in Northwest China, the PFM population in orchards has rapidly increased, becoming the dominant species in regions such as Xinjiang, Shaanxi, and Gansu Provinces of China [8,9,10]. The sympatric distribution of PFMs and OFMs, along with their similar external morphologies, makes it difficult to distinguish them [11]. Moreover, the cross-attraction between the commercial sex attractants for PFMs and OFMs affects the accuracy of estimating moth populations in peak occurrences [12,13], leading to misjudgment in monitoring their occurrence periods and population sizes.
The attractiveness of a sex attractant primarily relies on the integrity of the female pheromones and the ratio of each pheromone component [14,15]. Each moth species employs a unique blend of pheromones to attract the conspecific males while inhibiting males of the closely related heterospecific species. Secondary components play a crucial role in enhancing the species specificity of sex pheromones, thereby maintaining reproductive isolation [16,17]. The sex pheromones of PFMs consist of (Z)-8-dodecenyl acetate (Z8-12:Ac), (E)-8-dodecenyl acetate (E8-12:Ac), (Z)-8-dodecen-1-ol (Z8-12:OH), (Z)-8-tetradecenyl acetate (Z8-14:Ac), and (Z)-10-tetradecenyl acetate (Z10-14:Ac) at a 100:1:2:30:5 ratio (m/m) [18]. The sex pheromones of OFMs include Z8-12:Ac, E8-12:Ac, Z8-12:OH, and 1-dodecanol (12:OH) in ratios of 100:4.2-7.2:1.1-19.1:5.4-12.0 [19,20,21,22]. Currently, commercial sex attractants for PFMs contain a 100:4 blend of Z8-12:Ac and E8-12:Ac, while those for the OFM are loaded with a 100:6:1-2 blend of Z8-12:Ac, E8-12:Ac, and Z8-12:OH [18,23,24]. The cross-attraction and the lack of species-specificity can be attributed to the shared major components and the absence of secondary components in their sex attractants [25]. An excellent study by Guerin et al. (1986) discovered that the inclusion of Z8-14:Ac and Z10-14:Ac did not augment PFM catches but inhibited OFM catches in plum orchards in Switzerland and in peach orchards bordered by plums in Hungary [18]. However, following studies did not determine how Z8-14:Ac and Z10-14:Ac from PFM females inhibit OFM male attractants.
It is widely recognized that odorant binding proteins (OBPs) are essential in detecting and transporting hydrophobic volatile semiochemicals from antennae pores to olfactory receptors [26,27]. The OBP family proteins were originally categorized into pheromone-binding proteins (PBPs), general odorant-binding proteins (GOBPs), and antennal-binding proteins (ABPs) based on their differences in binding odorant molecules [28]. PBPs show biased expression in male antennae and are distributed in long sensilla trichodea, where they function in recognizing and transporting sex pheromones and their analogs. Apart from Crambidae species, which usually possess four or more PBPs, the PBP subfamily typically consists of three members, namely, PBP1, PBP2, and PBP3 [29,30,31,32,33]. GOBPs, mainly localized at s. basiconca and short s. trichodea are equivalently expressed in both sexes in adult antennae, which are primarily involved in detecting and transporting host plant-derived volatiles and sex pheromones [33,34,35]. With the rapid expansion in genome and transcriptome sequencing techniques, an increasing number of studies have shown that GOBPs and PBPs are often clustered on the same chromosome in close proximity, suggesting that they might have derived from a single ancestral gene and then diverged through gene duplication events and various environmental selection pressures to function in detecting and recognizing sex pheromones, host plant volatiles, and even insecticides [36,37,38,39,40,41]. In our prior research, we identified three PBPs (GmolPBP1, GmolPBP2, and GmolPBP3) and two GOBPs (GmolGOBP1 and GmolGOBP2) in the antennae of the OFM using RNA-seq and RT-PCR [42]. Fluorescence competitive binding assays revealed that recombinant GmolPBPs and GmolGOBPs have different affinities for binding to sex pheromones [43,44,45]. Although Z8-14:Ac and Z10-14:Ac are not components of OFM sex pheromones, the roles of PBPs and GOBPs in OFMs when Z8-14:Ac and Z10-14:Ac are added to OFM lures remain unclear.
In the present study, we employed an electroantennogram (EAG) system to evaluate OFM and PFM males responding to individual sex pheromone components of OFMs and PFMs, including Z8-12:Ac, E8-12:Ac, Z8-12:OH, Z8-14:Ac, and Z10-14:Ac, at different doses. Then, we conducted field trapping trials to assess how OFMs respond when different doses of Z8-14:Ac or Z10-14:Ac are added to a mixture of Z8-12:Ac and E8-12:Ac. A high dose of Z10-14:Ac was found to inhibit both OFM and PFM male attraction when added to a mixture of Z8-12:Ac and E8-12:Ac (500:20) in pear orchards. In addition, GmolPBP2 was found to be the primary OBP responsible for recognizing and transporting Z8-14:Ac using fluorescence competition binding assays. Finally, the key amino acid residues and weak interactions involved in GmolPBP2 binding to Z8-14:Ac were identified via homology modeling, molecular dynamics simulations, and site-directed mutagenesis. This study advances our understanding of how moths recognize interspecific sex pheromones at the molecular level and provides data for developing species-specific PFM sex attractants using secondary sex pheromones.
2. Materials and Methods
2.1. Colony Maintenance
All individuals utilized in the experiment originated from a laboratory colony at Yan’an University in Yan’an, Shaanxi Province, China. The larvae were raised on an artificial diet [46] and kept in an artificial climate chamber maintained at 25 ± 1 °C and a relative humidity of 70 ± 5%, with a photoperiod (light/dark) of 15 h:9 h. Pupae of different sexes were separated, and newly emerged virgin males were placed in disposable plastic cups (4.0 cm bottom diameter, 6.5 cm top diameter, and 6.6 cm height) covered with a plastic film containing air vents. Adults were fed a 5% (v/v) honey solution until they were utilized for testing. Mature PFM larvae were gathered from the fruits of the flowering plum Amygdalus triloba surrounding the apple orchards in Yan’an in June 2023. After the larvae left their fruits, they were moved to moist sandy soil for pupation and adult emergence. The feeding conditions and methods used for PFM adults were the same as those used for OFMs.
2.2. Chemicals
The synthetic pheromones Z8-12:Ac, E8-12:Ac, and Z8-12:OH were sourced from Pherobio Technology Co., Ltd. (Yangling, China), while Z8-14:Ac and Z10-14:Ac were synthesized by Shenyang Beixinjingyi Trade Co., Ltd. (Shengyang, China). Chemical purity ranging from 95% to 98% and isomeric purity exceeding 98% were determined through gas chromatography (GC) (Agilent, Santa Clara, CA, USA) analysis. Additionally, the fluorescence probe N-Phenyl-1-naphthylamine (1-NPN) with a purity of 98% was obtained from Sigma-Aldrich (Milwaukee, WI, USA).
2.3. Electroantennogram Analysis
Electroantennogram (EAG) equipment (Syntech, Hilversum, The Netherlands) was used to record the antennal responses of the OFM and PFM adults to Z8-12:Ac, E8-12:Ac, Z8-12:OH, Z8-14:Ac, and Z10-14:Ac. These sex pheromone compounds were diluted with liquid paraffin to prepare a stimulus solution of 100 μg/μL, which was subsequently diluted to 10 and 1 μg/μL. Antenna from 3-day-old OFM and 2- to 4-day-old PFM virgin males were excised at its base, with the distal part of the terminal segment being removed. Then, the treated antenna was attached to electrode holders with SpectraR360 conducting gel. A stimulus controller (model CS-55, Syntegon Technology, Waiblingen, Germany) continuously supplied a humidified air stream at a rate of 500 mL/min. Then, 20 μL of each stimulus solution was adsorbed onto a twice-folded filter paper strip (0.5 cm × 4 cm) placed on the wide section of a l mL pipette tip, which served as an odorant cartridge. Vapor stimuli were delivered by an air compressor into the constant air stream described above, flowing in a copper delivery tube (i.d. 8 mm) with the outlet positioned approximately 1 cm from the antenna. The EAG amplitude tests of the OFM and PFM males’ responses to sex pheromones started with solvent control stimuli (20 μL of liquid paraffin), followed by stimulation with 20, 200, and 2000 μg doses (20 μL of 1.0, 10.0, and 100.0 μg/μL of liquid paraffin solutions, respectively) of stimulus in sequence. The solvent control was performed at the end of the run. To control for variations among antennae, each sample was tested on 10 antennae of different males. The EAG signals and data were analyzed using EAG 2000 software (Syntech, Hilversum, The Netherlands). The corrected EAG amplitude value = absolute EAG amplitude value − (EAGck1 + EAGck2)/2, where EAGck1 and EAGck2 represent the absolute EAG amplitude values of the solvent control before and after measuring each stimulus, respectively. The relative EAG response of the OFM and PFM males to the same doses of sex pheromones were subjected to an analysis of independent sample t-test (p = 0.05) using SPSS version 23.0 (IBM).
2.4. Determination of the Inhibitory Effects of Z8-14:Ac and Z10-14:Ac on OFM Males
To determine the inhibitory effects on OFM male adults and the impact on the trapping effects of PFM male adults, varying amounts of Z8-14:Ac (0, 5, 10, 25, 50, 75, 100, 125, 150 μg) or Z10-14:Ac (0, 25, 50, 100, 150, 250, 350, 450, and 550 μg) were added to rubber septa loaded with a constant dosage of the main pheromone (500 μg of Z8-12:Ac and 20 μg of E8-12:Ac) of the PFM. Trapping experiments of Z8-14:Ac were carried out in peach and pear orchards in Yan’an and Xinjiang (China), respectively. Trapping tests of Z10-14:Ac were performed in pear orchards in Xinjiang (China). Each orchard had nine plots, with three plots in one field. Each plot consisted of a linear arrangement of traps loaded with a constant dosage of the main pheromones and different percentages of the secondary pheromone Z8-14:Ac or Z10-14:Ac of the PFM. Plots in the same field were placed 30 m apart, and fields were ≥1 km apart. The traps were hung 1.4 m from the ground on a suspension bracket between two rows of peach or pear trees from 10 June to 25 July 2021 in Xinjiang and from 15 June to 30 July 2021 in Yan’an. The positions of the traps within the plot were randomly arranged. Every 5 days, counts were taken, and the traps were rotated to the adjacent position to avoid any potential position effects. The septa loaded with sex pheromone components were replaced with new ones after every 15 days of suspension. The data were analyzed using a one-way ANOVA, and the differences in the average trapping of OFM and PFM males using sex attractants containing different amounts of Z8-14:Ac or Z10-14:Ac were further analyzed using Tukey’s HSD test, both of which were performed using SPSS 23.0.
2.5. Tissue Expression of GmolPBPs and GmolGOBPs in OFM Adults
Tissue samples of antennae, head (without antennae), thorax, abdomen, legs, and wings were collected from 3-day-old virgin female and male adults. We utilized 200, 30, 20, 6, 100, and 100 females or males for antenna, head (without antennae), thorax, abdomen, leg, and wing samples for RNA isolation, respectively. Each sample had three independent biological replicates. The total RNA from all samples was isolated using an AG RNAex Pro reagent (AG, Changsha, China), and then the first-strand cDNA of each sample was synthesized from 2.0 μg of total RNA with EasyScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). Seven pairs of specific primers, including for five target genes GmolPBP1, GmolPBP2, GmolPBP3, GmolGOBP1, GmolGOBP2, and two reference genes β-actin (GenBank No. KF022227.1) and Elongation factor 1-α (EF1-α) (GenBank No. KT363835.1) were designed and synthesized (Table S1). The cycle threshold (Ct) and amplification efficiency (E) values of the target and reference genes in different tissues of the OFM adults were detected on a StepOnePlus Real-Time PCR Instrument (ABI, Carlsbad, CA, USA) using PerfectStart Green qPCR SuperMix (TransGen Biotech, Beijing, China). Their expression levels in each sample were calculated using Equation 1 from the study [47]. Finally, the normalized expression levels of GmolPBP1–3 and GmolGOBP1–2 in different tissues were calculated with the geometric mean of the expression of the two reference genes using Equation (2) from the study [47]. Due to the inhomogeneity of data variance, we transformed the data using the formula log10(x + ), where x represents the normalized expression level of target genes, following the method described by Kelmansky et al. [48]. Differences in expression levels in different tissues were analyzed using ANOVA followed by Tukey’s HSD test in SPSS (SPSS v23.0, IBM). Significant differences in expression levels between the sexes in the same tissue of adults were tested using an independent t-test.
2.6. Preparation of rGmolPBPs/rGmolGOBPs Proteins and Fluorescence Competitive Binding Assays
Previous researchers have successfully expressed rGmolPBP1, rGmolPBP2, rGmolPBP3, rGmolGOBP1, and rGmolGOBP2 proteins using prokaryotic expression systems [43,44,45]. In this article, the DNA sequences of these five GmolPBPs and GmolGOBPs, with the signal peptides removed and restriction sites on both ends, were inserted into the expression vector pET28a(+) linearized with the same restriction enzyme. These vectors were then transformed into Escherichia coli BL21(DE3) and induced by isopropyl-β-d-thiogalactoside (IPTG). Since all five recombinant proteins were expressed in inclusion bodies, they were denatured using 8 M urea and then renatured using a cysteine/cystine REDOX system [49]. The reduced recombinant proteins were purified using an Ni-NTA His Bind Resin column (7Sea Pharmatech Co., Ltd., Shanghai, China). Non-target proteins were washed off with washing buffer (20 mM Tris-HCl (pH 7.4), 20 mM imidazole, 1 mM PMSF, and 250 mM NaCl), and then the target proteins were eluted with elution buffer (20 mM Tris-HCl (pH 7.4), 1 mM PMSF, and 250 mM NaCl) with an increasing concentration of imidazole (50, 100, and 200 mM). Five milliliters of eluents with high target protein content and low levels of contaminants were collected and placed in dialysis tubing (Coolaber, Beijing, China) with a molecular weight cut-off of 3.5 kDa to remove NaCl and imidazole. The purified soluble proteins were stored at −80 °C before use.
The binding affinities of three rGmolPBPs and two rGmolGOBPs for Z8-14:Ac were determined using an F-2700 fluorescence spectrophotometer (Hitachi, Tokyo, Japan). First, we measured the dissociation constants (Kd) of the rGmolPBPs (or rGmolGOBPs) binding to 1−NPN. Each protein was diluted to 2 μM with 20 mM Tris-HCl buffer (pH 7.4); then, 2 mL of the protein solution was transferred to a quartz cuvette and titrated with 1 mM 1−NPN to a final concentration of 1–18 μM. As the doses of 1−NPN were increased, the increasing amplitude of the fluorescence intensity decreased, and finally, the binding of the protein to 1−NPN reached saturation. The Kd values were calculated using the nonlinear regression method and the Scatchard equation. Second, we measured the inhibitory constant (Ki) of Z8-14:Ac’s competitive binding with each rGmolPBP (or rGmolGOBP) from the protein/1−NPN complex. The initial fluorescence intensity values were measured after a 2 min reaction in 2 mL solution of 20 mM Tris-HCl (pH 7.4) containing 2 µM rGmolPBP (or rGmolGOBP) protein and 2 µM 1−NPN. Then, the solution was titrated with aliquots of 1 mM Z8-14:Ac to obtain final concentrations in the range of 1–14 µM, and the fluorescence intensity value of each reaction was recorded. The Ki values were calculated using the following equation:
| Ki = [IC50]/(1 + [1 − NPN]/Kd) |
where IC50 is the concentration of Z8-14:Ac when half of the initial fluorescence intensity value of the rGmolPBP (or GmolGOBP)/1−NPN complex is replaced, [1 − NPN] represents the concentration of free 1−NPN, and Kd is the dissociation constant of the rGmolPBP (or GmolGOBP)/1−NPN complex.
2.7. Homology Modeling and Molecular Docking
The fluorescence competitive binding assays revealed that GmolPBP2 was the most likely target for recognizing and transporting Z8-14:Ac; therefore, a three-dimensional (3D) model of GmolPBP2 was constructed and molecular dynamics simulations were used to predict the important amino acid residues and interaction forces between GmolPBP2 and Z8-14:Ac using the predicted GmolPBP2 interaction with the major sex pheromone Z8-12:Ac model for comparison. The crystal structure templates of GmolPBP2 were screened in the RCSB PDB (https://www.rcsb.org/, accessed on 1 April 2023). On the basis of the identity and coverage between the GmolPBP2 sequence and template sequences, the X-ray diffraction crystal structure of Amyelois transitella AtraPBP1 complexed with the sex pheromone component Z11, Z13-16:Ald (4INW) was utilized as a template to construct a homology model of GmolPBP2. EasyModeller 4.0 was employed to generate nine coarse homologous models of GmolPBP2. The discrete optimized protein energy (DOPE) method was used to evaluate the energy of each model. The model with the lowest energy was selected as the optimized model for GmolPBP2. The amino acid residues that form the binding pocket of GmolPBP2 were predicted using the PROTEINS PLUS online program (https://proteins.plus/, accessed on 15 April 2023). UCLA-DOE LAB—SAVES v6.0 online server (https://saves.mbi.ucla.edu/, accessed on 10 April 2023) was used to evaluate the quality of the optimized model of GmolPBP2. The 3D structure files of Z8-14:Ac (Compound CID: 5363518) and Z8-12:Ac (Compound CID: 5363377) were downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/, accessed on 15 April 2023). Molecular docking of the GmolPBP2 interaction with Z8-14:Ac and Z8-12:Ac was performed using AutoDockTools-1.5.6 software with the default parameters. The top-ranked binding mode (with the lowest binding energy score) was chosen and visualized using PyMOL (v. 2.5.2). The 2D structure of predicted amino acid residues, as well as the interaction forces between GmolPBP2 and Z8-14:Ac and Z8-12:Ac, were visualized using the Discovery Studio Visualizer software 2016.
2.8. Molecular Dynamic Simulation and Calculation of Binding Free Energy of Each Amino Acid
The molecular dynamic (MD) simulations of GmolPBP2–Z8-14:Ac and GmolPBP2–Z8-12:Ac complexes were performed for 200 nanoseconds (ns), employing GROMACS (Version 2022.2) and AMBER99SB protein forcefield. The simulations started with the solvation of complexes to neutralize the system, followed by energy with the steepest descent algorithm with a step length of 0.01 nm. The systems were equilibrated under NVT (canonical ensemble) and NPT (isobaric–isothermal ensemble) for 2 ns. After that, a 200 ns simulation with a time step of 20 fs was performed. Analyses of the root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), and radius of gyration (Rg) were executed using GROMACS (v. 2022.2) built-in tools [50] and visualized using the ggplot2 package (v. 3.4.3) in R (v. 4.3.2) [51]. Finally, the binding free energy of each amino acid of GmolPBP2 to Z8-14:Ac and Z8-12:Ac was calculated using the gmx_Molecular Mechanic and Poisson–Boltzmann Surface Area (gmx-MMPBSA) tool [52].
2.9. Site-Directed Mutagenesis and the Binding Affinities of GmolPBP2 Mutants to Z8-14:Ac
Five amino acids—Phe12, Leu68, Ile94, Arg109, and Ile113—were identified as having lower binding free energies for GmolPBP2 interaction with Z8-14:Ac. To assess their role in binding, these residues were individually mutated to alanine using site-directed mutagenesis. Primers for mutagenesis were designed following the protocols recommended by the Fast Mutagenesis System (TransGen Biotech, Beijing, China) (Table S1). These mutations were introduced into the pET-28a(+)–GmolPBP2 plasmid using a Fast Mutagenesis System kit (Vazyme, Nanjing, China) and site-directed mutagenesis primers generating the mutants F12A, L68A, I94A, R109A, and I113A, respectively. Each plasmid was verified by DNA sequencing to confirm the presence of the desired mutation. Expression and purification of the mutated proteins followed the same protocols used for the wild-type GmolPBP2. The binding affinities of the five mutants for Z8-14:Ac were determined via fluorescence competitive binding assays. An increase in Ki values for the mutants relative to the wild-type GmolPBP2 would indicate that the corresponding amino acid residues may be crucial for binding to Z8-14:Ac.
3. Results
3.1. EAG Response of OFM and PFM Males to Different Doses of Sex Pheromones
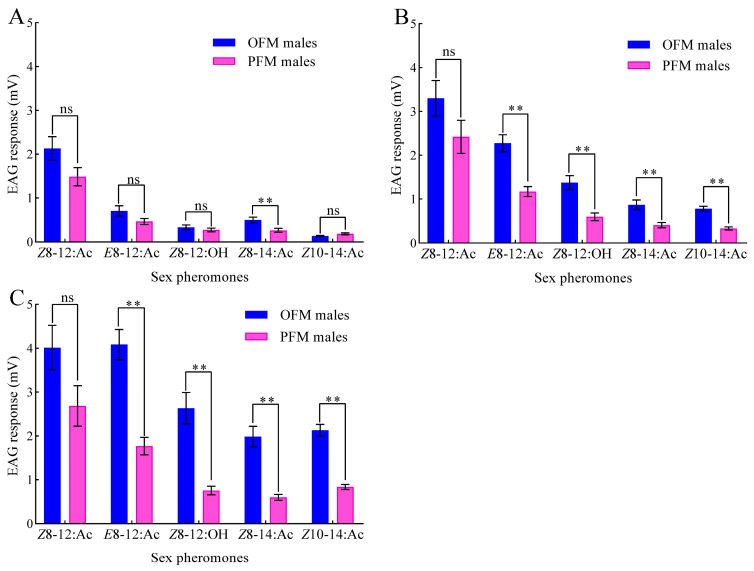
There is a dose-dependent relationship between the EAG response values of OFM and PFM males to five sex pheromones: Z8-12:Ac, E8-12:Ac, Z8-12:OH, Z8-14:Ac, and Z10-14:Ac (Figure 1). Both OFM and PFM males exhibit a higher sensitivity to the primary components Z8-12:Ac and E8-12:Ac at the same dose compared to their response to the secondary components Z8-14:Ac and Z10-14:Ac (Figure 1). Surprisingly, the EAG values of the OFM males to different doses of Z8-14:Ac stimulation were significantly higher than those of the PFM males (20 μg: t = 2.959, df = 18, p = 0.008; 200 μg: t = 3.784, df = 18, p = 0.001; 2000 μg: t = 5.708, df = 18, p < 0.001). Except for the absence of significant difference in EAG response to 20 μg of Z10-14:Ac stimulation between OFM and PFM males (t = −1.865, df = 18, p = 0.079), the EAG values of OFM males to 200 and 2000 μg of Z10-14:Ac was significantly greater than that of PFM males (200 μg: t = 6.892, df = 18, p < 0.001; 200 μg: t = 8.809, df = 18, p < 0.001).
Figure 1.
EAG dose–response of OFM and PFM males to different sex pheromone components: (A) 20 ng; (B) 200 ng; (C) 2000 ng. Data are presented as mean ± SEM (n = 10). Double asterisks and “ns” indicate extremely significant differences (p < 0.01) and no significant differences (p > 0.05), respectively, in the EAG response values between OFM and PFM males to the same sex pheromone (independent sample t-test).
3.2. Effects of Z8-14:Ac and Z10-14:Ac on Captured OFM and PFM Males
The field tests using delta traps baited with the two primary PFM pheromones and different doses of the secondary sex pheromone Z8-14:Ac showed that adding Z8-14:Ac to a mixture of Z8-12:Ac and E8-12:Ac inhibited OFM males but not PFM males in both the peach and pear orchard trials (Table 1 and Table 2). Adding 1–30% of Z8-14:Ac to the PFM sex attractants neither significantly increased nor inhibited the number of PFM males in the peach (F = 1.893, df = 8,26, p = 0.124) and pear orchards (F = 1.072, df = 8,26, p = 0.424). Compared to the mixture of Z8-12:Ac and E8-12:Ac (100:4), adding 1–2% of Z8-14:Ac significantly reduced the capture of OFM males by 58.81–63.45% in the peach orchards (1% Z8-14:Ac: t = 9.358, df = 4, p = 0.001; 2% Z8-14:Ac: t = 10.650, df = 4, p < 0.001) (Table 1) and by 63.82–65.04% in the pear orchards (1% Z8-14:Ac: t = 7.806, df = 4, p = 0.001; 2% Z8-14:Ac: t = 8.873, df = 4, p = 0.001) (Table 2). In both the peach and pear orchard trials, the number of OFM males caught by adding 5–30% of Z8-14:Ac drastically reduced (by more than 86%) and were not significantly different among the sex attractants that contained varying amounts of Z8-14:Ac (peach orchard trails/pear orchard trails: F = 2.546, df = 5,17, p = 0.094; F = 1.639, df = 5,17, p = 0.224). In contrast to Z8-14:Ac, adding more than 10% of Z10-14:Ac to the mixture of Z8-12:Ac and E8-12:Ac (100:4) inhibited the capture of both OFM and PFM males, with reductions of up to 87.77% and 71.07%, respectively (Table 3). Therefore, Z8-14:Ac combined with a mixture of Z8-12:Ac and E8-12:Ac, but not Z10-14:Ac, can serve as a specific component for PFM sex attractants to inhibit OFM male adults.
Table 1.
Captures of PFM and OFM males in a peach orchard using delta traps with varying amounts of Z8-14:Ac added to a mixture of Z8-12:Ac and E8-12:Ac.
| Pheromone Loadings (μg) | Mass Ratio of Z8-12:Ac, E8-12:Ac, and Z8-14:Ac | Total PFM Males/Trap | Total OFM Males/Trap | Inhibition Rate of OFM Males (%) | ||
|---|---|---|---|---|---|---|
| Z8-12:Ac | E8-12:Ac | Z8-14:Ac | ||||
| 500 | 20 | 0 | 100:4:0 | 302.20 ± 51.88 a | 111.87±6.24 a | 0.00 |
| 500 | 20 | 5 | 100:4:1 | 387.73 ± 18.31 a | 46.08±3.25 b | 58.81 |
| 500 | 20 | 10 | 100:4:2 | 287.22 ± 7.96 a | 40.89±2.35 b | 63.45 |
| 500 | 20 | 25 | 100:4:5 | 314.53 ± 26.14 a | 14.82±1.47 c | 86.75 |
| 500 | 20 | 50 | 100:4:10 | 331.58 ± 17.46 a | 9.04±1.08 c | 91.92 |
| 500 | 20 | 75 | 100:4:15 | 297.81 ± 4.75 a | 9.55±1.09 c | 91.46 |
| 500 | 20 | 100 | 100:4:20 | 285.62 ± 6.55 a | 9.67±1.52 c | 91.36 |
| 500 | 20 | 125 | 100:4:25 | 352.89 ± 32.42 a | 6.86±0.24 c | 93.87 |
| 500 | 20 | 150 | 100:4:30 | 348.30 ± 22.77 a | 7.96±1.19 c | 92.88 |
Note: Data in the table are presented as mean ± SEM (n = 3). The mass ratios of E8-12:Ac and Z8-14:Ac were calculated based on a Z8-12:Ac content of 100%. Different lowercase letters in each column represent significant differences using one-way ANOVA followed by Tukey’s HSD test (α = 0.05).
Table 2.
Captures of PFM and OFM males in a pear orchard using delta traps with varying amounts of Z8-14:Ac added to a mixture of Z8-12:Ac and E8-12:Ac.
| Pheromone Loadings (μg) | Mass Ratio of Z8-12:Ac, E8-12:Ac, and Z8-14:Ac | Total PFM Males/Trap | Total OFM Males/Trap | Inhibition Rate of OFM Males (%) | ||
|---|---|---|---|---|---|---|
| Z8-12:Ac | E8-12:Ac | Z8-14:Ac | ||||
| 500 | 20 | 0 | 100:4:0 | 56.49 ± 3.80 a | 169.47 ± 9.83 a | 0.00 |
| 500 | 20 | 5 | 100:4:1 | 64.07 ± 6.89 a | 59.24 ± 10.14 b | 65.04 |
| 500 | 20 | 10 | 100:4:2 | 45.47 ± 6.64 a | 61.31 ± 7.21 b | 63.82 |
| 500 | 20 | 25 | 100:4:5 | 43.06 ± 0.34 a | 21.36 ± 6.22 c | 87.40 |
| 500 | 20 | 50 | 100:4:10 | 53.73 ± 7.04 a | 12.05 ± 0.91 c | 92.89 |
| 500 | 20 | 75 | 100:4:15 | 48.57 ± 6.31 a | 8.95 ± 2.69 c | 94.72 |
| 500 | 20 | 100 | 100:4:20 | 42.36 ± 8.46 a | 12.74 ± 6.57 c | 92.48 |
| 500 | 20 | 125 | 100:4:25 | 57.52 ± 21.06 a | 5.86 ± 1.50 c | 96.54 |
| 500 | 20 | 150 | 100:4:30 | 72.67 ± 12.44 a | 10.33 ± 2.98 c | 93.90 |
Note: Data in the table are presented as mean ± SEM (n = 3). The mass ratios of E8-12:Ac and Z8-14:Ac were calculated based on a Z8-12:Ac content of 100%. Different lowercase letters in each column represent significant differences using one-way ANOVA followed by Tukey’s HSD test (α = 0.05).
Table 3.
Captures of PFM and OFM males in a pear orchard using delta traps with varying amounts of Z10-14:Ac added to a mixture of Z8-12:Ac and E8-12:Ac.
| Pheromone Loadings (μg) | Mass Ratio of Z8-12:Ac, E8-12:Ac, and Z10-14:Ac | Total PFM Males/Trap | Total OFM Males/Trap | Inhibition Rate of OFM Males (%) | ||
|---|---|---|---|---|---|---|
| Z8-12:Ac | E8-12:Ac | Z10-14:Ac | ||||
| 500 | 20 | 0 | 100:4:0 | 142.55 ± 11.16 a | 193.28 ± 60.19 a | 0.00 |
| 500 | 20 | 25 | 100:4:5 | 121.32 ± 12.10 ab | 125.55 ± 14.47 ab | 35.04 |
| 500 | 20 | 50 | 100:4:10 | 101.39 ± 7.72 b | 97.92 ± 10.31 bc | 49.34 |
| 500 | 20 | 100 | 100:4:20 | 102.73 ± 10.21 b | 83.33 ± 9.82 cd | 56.89 |
| 500 | 20 | 150 | 100:4:30 | 66.08 ± 8.27 c | 33.56 ± 2.24 e | 82.64 |
| 500 | 20 | 250 | 100:4:50 | 62.46 ± 5.63 c | 31.28 ± 3.22 e | 83.82 |
| 500 | 20 | 350 | 100:4:70 | 51.35 ± 7.21cd | 37.49 ± 3.45 e | 80.70 |
| 500 | 20 | 450 | 100:4:90 | 41.24 ± 5.10 d | 23.64 ± 1.23 e | 87.77 |
| 500 | 20 | 550 | 100:4:110 | 42.33 ± 3.95 d | 29.69 ± 2.18 e | 84.64 |
Note: Data in the table are presented as mean ± SEM (n = 3). The mass ratios of E8-12:Ac and Z10-14:Ac were calculated based on a Z8-12:Ac content of 100%. Different lowercase letters in each column represent significant differences using one-way ANOVA followed by Tukey’s HSD test (α = 0.05).
3.3. Tissue Expression of GmolPBPs and GmolGOBPs
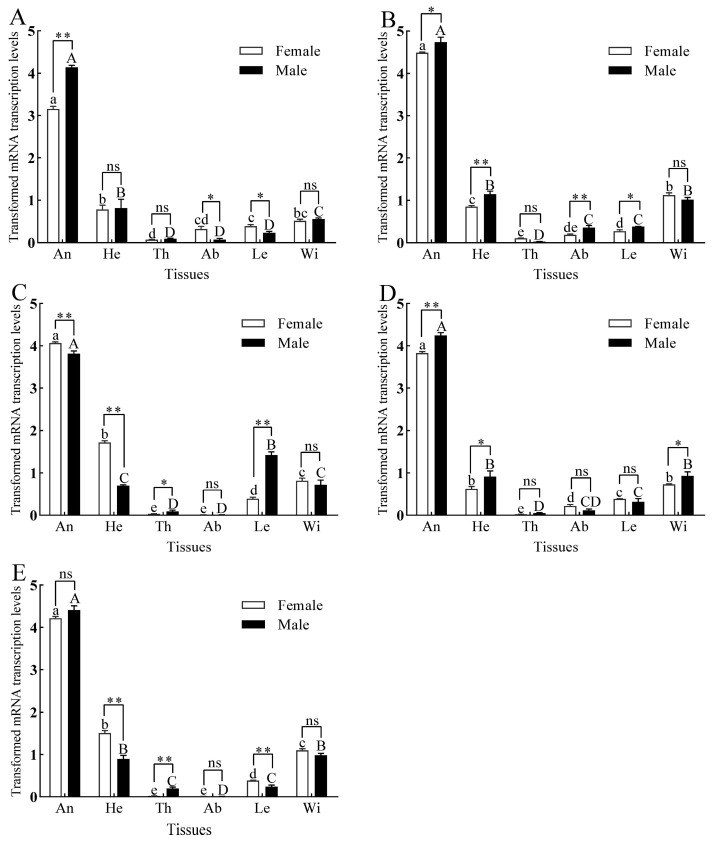
All three GmolPBPs and two GmolGOBPs were predominantly expressed in the antennae of both male and female adults, with slight expression in the heads, wings, and legs and minimal expression in the thoraxes and abdomens (Figure 2). The expression levels of GmolPBP1, GmolPBP2, and GmolGOBP1 were significantly higher in the male antennae than in the female antennae (Figure 2A,B,D), whereas the expression of GmolPBP3 was notably higher in the female antennae compared to the male antennae (Figure 2C). No significant difference in expression was observed for GmolGOBP2 between male and female antennae (Figure 2E). Beyond the antennae, sex-specific differences in expression were also observed in other tissues. For instance, in the head, GmolPBP3 and GmolGOBP2 expression levels were significantly higher in the females (Figure 2C,E), whereas GmolPBP2 and GmolGOBP1 levels were significantly higher in males (Figure 2B,D). In the legs, the expression of GmolPBP1 and GmolGOBP2 were higher in females (Figure 2A,E), while the expression of GmolPBP2 and GmolPBP3 were higher in males (Figure 2B,C). Notably, the expression of GmolGOBP1 in male wings was significantly elevated compared to female wings (Figure 2D), while no significant sex differences were observed for the other four GmolPBPs/GmolGOBPs in wings (Figure 2A–C,E).
Figure 2.
Tissue expression patterns of three GmolPBPs and two GmolGOBPs. (A) GmolPBP1. (B) GmolPBP2. (C) GmolPBP3. (D) GmolGOBP1. (E) GmolGOBP2. Different lowercase and capital letters above each bar indicate significant differences in relative transcript levels of male and female adults across various tissues, as determined by one-way ANOVA followed by Tukey’s HSD test (p < 0.05). Double asterisks, single asterisks, and “ns” indicated extremely significant differences (p < 0.01), significant differences (p < 0.05), and no significant difference (p > 0.05), respectively, in expression levels between males and females within the same tissue (independent sample t-test).
3.4. Binding Affinities of rGmolPBPs and rGmolGOBPs for Z8-14:Ac
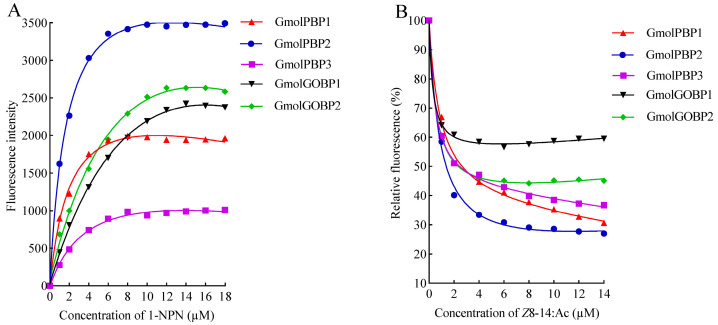
We successfully expressed and purified rGmolPBP1, rGmolPBP2, rGmolPBP3, rGmolGOBP1, and rGmolGOBP2 proteins using an E. coli BL21(DE3) expression system (Figure S1). Fluorescence saturation was observed in these five proteins when titrated with increasing concentrations of the fluorescent probe 1−NPN, indicating the suitability of 1−NPN for measuring their binding affinities for odorant ligands (Figure 3A). The calculated Kd values of 1−NPN interacting with rGmolPBP1, rGmolPBP2, rGmolPBP3, rGmolGOBP1, and rGmolGOBP2 were 1.29 ± 0.02, 1.09 ± 0.02, 11.24 ± 0.08, 5.76 ± 0.01, and 4.32 ± 0.02 μM, respectively (Table 4). The Ki value of rGmolGOBP1 for Z8-14:Ac could not be calculated because 50% inhibition was not reached, and an IC50 value could not be extrapolated. The other four proteins showed strong binding affinities for Z8-14:Ac, with Ki values of 1.66 ± 0.04, 0.66 ± 0.02, 2.56 ± 0.27, and 2.86 ± 0.18 μM, respectively (Figure 3B). The lowest Ki value for rGmolPBP2 binding to Z8-14:Ac suggests that it is the most likely target for Z8-14:Ac, indicating a better fit in the GmolPBP2 binding pocket, resulting in stronger interactions with the protein.
Figure 3.
Curves of rGmolPBPs and rGmolGOBPs binding to 1−NPN probe (A) and sex pheromone Z8-14:Ac (B). In (A), each rGmolPBP and rGmolGOBP protein was diluted to a concentration of 2 µM with 20 mM Tris-HCl buffer (pH 7.4), and then aliquots of 1−NPN were added to reach a final concentration of 1 to 18 µM. The calculated Kd values of rGmolPBP1, rGmolPBP2, rGmolPBP3, rGfunGOBP1, and rGmolGOBP2 were 1.29 ± 0.02, 1.09 ± 0.02, 11.24 ± 0.08, 5.76 ± 0.01, and 4.32 ± 0.02 µM, respectively. In (B), the fluorescence intensity after Z8-14:Ac displaced the 1−NPN probe bound to rGmolPBPs and rGmolGOBPs is displayed as a percentage of the initial fluorescence intensity. The calculated Ki values of the rGmolPBPs and rGmolGOBPs with Z8-14:Ac are listed in Table 4.
Table 4.
Binding affinities of rGmolPBPs and rGmolGOBPs for Z8-14:Ac.
| Protein Name | Kd (μM) | IC50 (μM) | Ki (μM) |
|---|---|---|---|
| GmolPBP1 | 1.29 ± 0.02 | 2.93 ± 0.07 | 1.66 ± 0.04 c |
| GmolPBP2 | 1.09 ± 0.02 | 1.26 ± 0.04 | 0.66 ± 0.02 d |
| GmolPBP3 | 11.24 ± 0.08 | 2.78 ± 0.32 | 2.56 ± 0.27 b |
| GmolGOBP1 | 5.76 ± 0.01 | >14 | >14 a |
| GmolGOBP2 | 4.32 ± 0.02 | 3.53 ± 0.22 | 2.86 ± 0.18 b |
Note: IC50: the concentration of Z8-14:Ac when replacing 1−NPN to reduce the initial fluorescence intensity of the rGmolPBP(rGmolGOBP)/1−NPN complex to 50%; Kd: the dissociation constants of each rGmolPBP (rGmolGOBP) binding to 1−NPN; Ki: the inhibition constants of Z8-14:Ac competitive binding with each GmolPBP (GmolGOBP) from the protein/1−NPN complex. Data in the table are presented as mean ± SE (n = 3). Different lowercase letters after the data in the same column indicate significant differences in the affinities of different recombinant proteins for binding to Z8-14:Ac (p < 0.05, one-way ANOVA followed by Tukey’s HSD test).
3.5. Homology Modeling and Molecular Dynamics Simulation of GmolPBP2
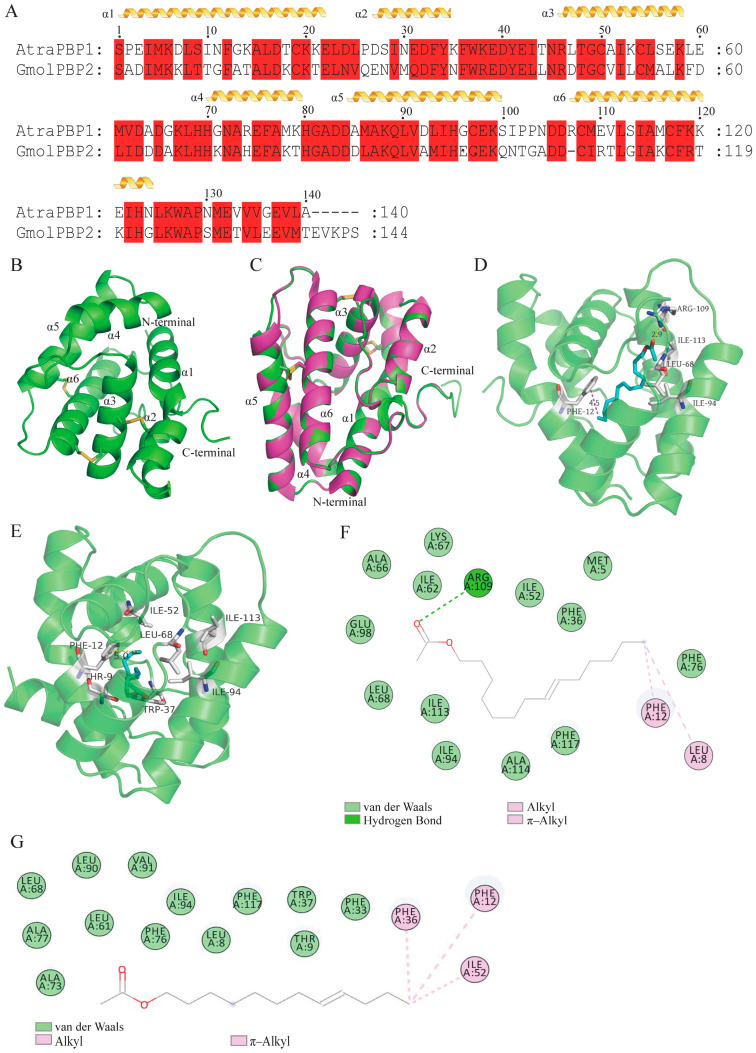
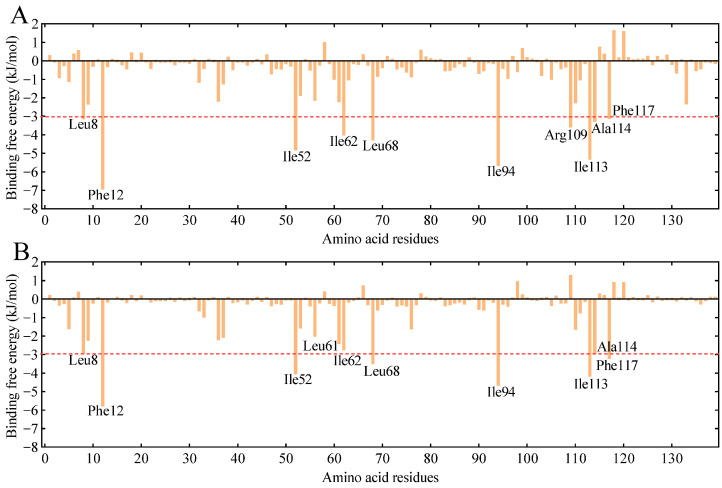
The 3D structure of GmolPBP2 was constructed using the X-ray diffraction crystal structure of AtraPBP1 from the A. transitella complex (4INW) as a template (Figure 4B,C). A BLASTP search revealed a 53.96% amino acid similarity between GmolPBP2 and AtraPBP1, with a sequence coverage of 97% (Figure 4A). Ramachandran plots indicated that 94.0% of the residues in GmolPBP2 were situated in the favorable region, while 6.0% were in the allowed region, indicating a high quality of this model (Figure S2). The predicted 3D structure of GmolPBP2 featured six typical α-helices located between residues Ser1-Glu22 (α1), Glu27-Tyr34 (α2), Arg46-Lys58 (α3), His70-Thr79 (α4), Asp84-Gln100 (α5), and Asp106-G123 (α6). Three pairs of disulfide bridges connected Cys19 in α1 and Cys54 in α3, Cys50 in α3 and Cys107 in α6, and Cys97 in α5 and Cys116 in α6 (Figure 4B). It was found that 33 amino acids, including Met5, Leu8, Thr9, Phe12, Phe33, Phe36, Trp37, Ile52, Leu53, Met55, Ala56, Leu61, Ile62, Ala66, Lys67, Leu68, Ala73, His74, Phe76, Ala77, Leu86, Ala87, Leu90, Ala91, Ile94, Glu98, Arg109, Thr110, Ile113, Ala114, Phe117, Arg118, and Val133, were involved in the formation of the hydrophobic binding pockets of GmolPBP. The proportion of hydrophobic amino acids was 78.79% (Table S2).
Figure 4.
Construction of a 3D model of GmolPBP2 and analysis of the amino acid residues and interaction forces in GmolPBP2–Z8-14:Ac and GmolPBP2–Z8-12:Ac. (A) Sequence alignment of GmolPBP2 and AtraPBP1. α-helices are displayed as squiggles, while residues that are strictly identical are highlighted with a red background. (B) Predicted 3D model of GmolPBP2. (C) Structure alignment of AtraPBP1 (4INW) and GmolPBP2. (D,E) represent the predicted amino acid residues and interaction forces of GmolPBP2 binding to Z8-14:Ac and Z8-12:Ac, respectively (3D structure). (F,G) represent the predicted amino acid residues and interaction forces of GmolPBP2 binding to Z8-14:Ac and Z8-12:Ac, respectively (2D structure).
GmolPBP2–Z8-14:Ac and GmolPBP2–Z8-12:Ac complexes were obtained by docking the Z8-14:Ac and Z8-12:Ac molecules into the binding pocket of GmolPBP2. Then, 200 nm MD simulations of the complexes were performed, followed by calculation of the theoretical binding free energy for each amino acid residue in the GmolPBP2–Z8-14:Ac and GmolPBP2–Z8-12:Ac complexes. A lower binding free energy indicated a stronger binding between the residue and the ligands. Ten residues exhibited binding free energies to Z8-14:Ac lower than −3.0 kJ/mol, including Phe12 (−6.960 kJmol), Ile94 (−5.674 kJ/mol), Ile113 (−5.350 kJ/mol), Ile52 (−4.841 kJ/mol), Leu68 (−4.305 kJ/mol), Ile62 (−4.033 kJ/mol), Arg109 (−3.596 kJ/mol), Ala114 (−3.299 kJ/mol), Leu8 (−3.150 kJ/mol), and Phe117 (−3.140 kJ/mol), suggesting their importance in Z8-14:Ac binding (Figure 5A). Notably, the NH2 atom in Arg109 formed a hydrogen bond (H-bond) with the carbonyl oxygen atom in the Z8-14:Ac acetyl group, with a bond distance of 2.9 Å (Figure 4D). Meanwhile, the phenyl side of Phe12 engaged in a π–alkyl interaction with the C14 atom of Z8-14:Ac, with a π–C14 distance of 4.5 Å (Figure 4D). Van der Waals forces were observed between the residues Leu68, Ile52, Ile94, and Ile113 and Z8-14:Ac (Figure 4F). The ten lowest binding free energies for GmolPBP2–Z8-12:Ac were found between Phe12 (−5.800 kJ/mol), Ile94 (−4.692 kJ/mol), Ile113 (−4.189 kJ/mol), Ile52 (−4.053 kJ/mol), Leu68 (−3.526 kJ/mol), 117Phe (−3.233), 114Ala (−2.968), 8Leu (−2.965), 62Ile (−2.758), 61Leu (−2.427) and Z8-12:Ac (Figure 5B). Like the GmolPBP2–Z8-14:Ac complex, van der Waals forces were the primary contributor to the binding of Z8-12:Ac (Figure 4G). However, differences arose from the hydrogen bonds: hydrogen bonds were formed between the side chain of Arg109 in GmolPBP2 and the acetyl of Z8-14:Ac, whereas no hydrogen bond was formed between GmolPBP2 and Z8-12:Ac (Figure 4E).
Figure 5.
Total binding free energy contributions of each residue of GmolPBP2 to Z8-14:Ac (A) and Z8-12:Ac (B). The top ten residues with the lowest binding free energies are labeled. The red line signifies that the binding free energy value at this point is 3.0 kJ/mol.
3.6. Validation of Key Amino Acid Residues for GmolPBP2 Binding to Z8-14:Ac
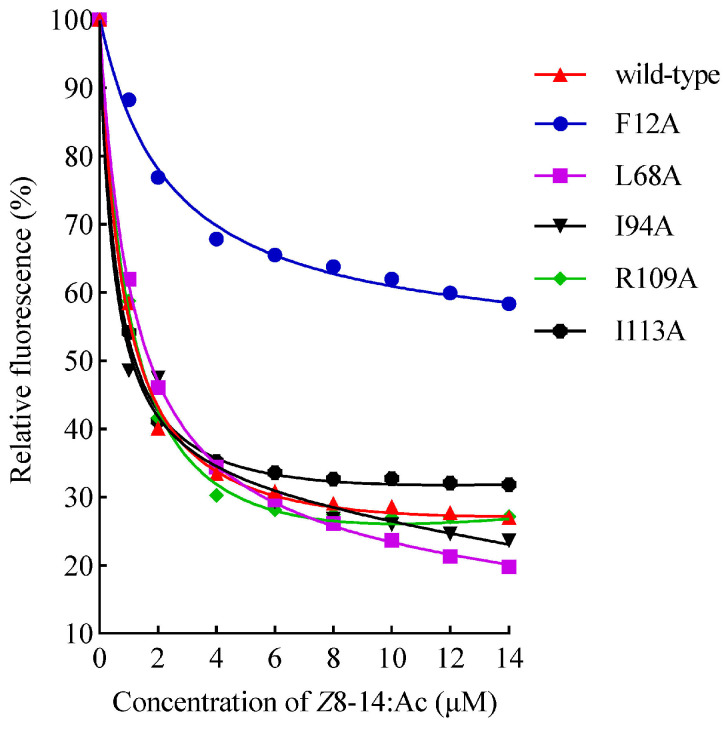
Based on the results of the aforementioned GmolPBP2–Z8-14:Ac docking analysis, five potential key residues (Phe12, Leu68, Ile94, Arg109, and Ile113) were individually substituted with Ala to generate the mutant proteins named F12A, L68A, I94A, R109A, and I113A, respectively. The results of SDS-PAGE showed that these mutants and wild-type rGmolPBP2 were all presented as inclusion bodies (Figure S3). The F12A mutant lost its affinity for binding Z8-14:Ac, as shown by the lack of IC50 and Ki values for Z8-14:Ac when replacing the fluorescence probe, 1–NPN, in the F12A/1–NPN complex, suggesting that the π–alkyl interaction between Phe12 and the C14 atom of Z8-14:Ac was key to the binding affinity of GmolBPB2 for Z8-14:Ac (Figure 6). In contrast, the R109A mutant, which disrupted the hydrogen bond between Arg109 and the carbonyl oxygen of Z8-14:Ac, did not significantly affect the binding affinity. The L68A, I94A, and I113A mutants displayed binding affinities comparable to the wild-type GmolPBP2, suggesting that Leu68, Ile94, and Ile113 were not key amino acid residues that bind to Z8-14:Ac (Table 5).
Figure 6.
Curves of the binding of rGmolPBP2 (wild-type) and its five mutants to Z8-14:Ac. The fluorescence intensity after Z8-14:Ac displaced the 1–NPN probe bound to rGmolPBP2 is displayed as a percentage of the initial fluorescence intensity. The calculated Ki values of rGmolPBP2 (wild-type) and mutants with Z8-14:Ac are listed in Table 5.
Table 5.
Binding affinities of wild-type and mutants of GmolPBP2 to Z8-14:Ac.
| Protein Name | IC50 (μM) | Ki (μM) |
|---|---|---|
| wild-type | 1.260 ± 0.036 | 0.660 ± 0.019 |
| F12A | >14 | >14 |
| L68A | 1.129 ± 0.026 | 0.589 ± 0.007 |
| I94A | 1.082 ± 0.008 | 0.564 ± 0.004 |
| R109A | 1.257 ± 0.029 | 0.655 ± 0.015 |
| I113A | 0.980 ± 0.038 | 0.520 ± 0.020 |
Note: IC50: the concentration of Z8-14:Ac when replacing 1–NPN to reduce the initial fluorescence intensity of rGmolPBP2 (and its mutants)/1–NPN complex to 50%; Ki: the inhibition constants of Z8-14:Ac competitive binding with wild-type GmolPBP2 and each mutant from the protein/1–NPN complex. Data in the table are presented as mean ± SE (n = 3).
4. Discussion
4.1. Z8-14:Ac Exhibits an Inhibitory Effect on OFM Males
Typically, species that share major sex pheromone components often use unique secondary components to generate species-specific signals [17,53,54]. Some secondary components serve as part of an intraspecific attractive message, while others may function as antagonists to closely related sympatric species [16,55,56]. Z8-14:Ac is a secondary sex pheromone component of female PFMs, and EAG tests indicated that Z8-14:Ac elicited EAG responses in both intraspecific PFM males and interspecific OFM males. Adding 1–30% of Z8-14:Ac to the PFM sex attractants (Z8-12:Ac/E8-14:Ac = 100:4, m/m) neither significantly increased nor inhibited the number of PFM males captured, but significantly reduced OFM males captured. Tòth et al. (1991) found that adding Z6-12:Ac (a sex pheromone analog) to OFM sex attractants caused a reduction in the capture of both OFM and PFM males, with a more pronounced effect on PFMs [57]. Z6-12:Ac acts as an attractive inhibitor for male PFMs, making OFM sex attractants more species-specific. Heliotis peltigera, H. virescens, and H. assulta are closely related sympatric species, and (Z)-9-tetradecenal (Z9-14:Al) is a sex pheromone component of female adult H. peltigera and H. virescens. Adding Z9-14:Al to the sex attractant of H. assulta notably decreased the capture of male H. assulta [57,58]. Guern et al. discovered that Z8-14:Ac and Z10-14:Ac were secondary sex pheromone components of female PFMs that did not increase G. funebrana catches but inhibited G. molesta [18]. Since that wonderful discovery, there have been few field trials specifically examining the individual roles of Z8-14:Ac and Z10-14:Ac in OFM inhibition, nor have the molecular mechanisms behind this inhibition been explored. At present, commercial PFM sex attractants contain only Z8-12:Ac and E8-12:Ac, indicating that Z8-14:Ac and Z10-14:Ac were not considered to have a synergistic effect during the development of these attractants. In the current study, field trials revealed that adding various amounts of Z8-14:Ac to a mix of sex attractants neither significantly increased nor inhibited the trapping of PFM males, but reduced the trap capture of OFM males by up to 96.54%. Additionally, as the concentration of Z10-14:Ac in the sex attractant increased, it exerted an inhibitory effect on both PFMs and OFMs. Therefore, it is hypothesized that the Z8-14:Ac of the PFM primarily functions to inhibit males of closely related sympatric species when added to a mixture of Z8-12:Ac and E8-12:Ac, thereby ensuring the species specificity of the sex pheromone.
4.2. GmolPBPs and GmolGOBPs Predominantly Expressed in OFM Adult Antennae
The expression patterns of insect OBPs are closely correlated with their physiological functions [59]. Three GmolPBPs and two GmolGOBPs were predominantly expressed in the antennae, suggesting their roles in chemoreception. There were variations in the expression levels of these five GmolOBPs in female and male adult antennae. GmolPBP1, GmolPBP2, and GmolGOBP1 exhibited significantly higher expression levels in male antennae compared to female antennae. Conversely, GmolPBP3 displayed significantly higher expression levels in female antennae than in male antennae. However, there was no significant difference in the expression level of GmolGOBP2 between male and female antennae. Based on their expression patterns in the antennae, GmolPBP1, GmolPBP2, and GmolGOBP1 are more likely to be involved in binding and transporting sex pheromones. Both PBPs and GOBPs in lepidopteron insects were highly expressed in the antennae of adults. However, the different expression levels in the antennae of both sexes depended on the species, such as in Carposina sasakii [60], G. funebrana [61], Peridroma saucia [62], and Chilo suppressalis [63,64]. What is more, the expression patterns of insect OBPs in different tissues seemed to be related to age after eclosion, mating status, and circadian rhythm [43,65,66]. In the present study, the expression levels of GmolPBPs and GmolGOPBs were only quantified in different tissues. Age-dependent, mating-dependent, and circadian rhythm-dependent expression patterns should be further illuminated.
4.3. GmolPBP2 Was the Most Likely Target for Binding Z8-14:Ac
Z8-14:Ac is a secondary PFM sex pheromone, as well as one of the sex pheromone components of Ctenopseutis herana, C. obliquana, Planotortrix octo, P. excessana, Pandamis cerasana, Spilonota oceana, and S. laricana [67,68,69]. In the current study, the ligand-binding assays revealed that rGmolPBP1, rGmolPBP2, rGmolPBP3, and rGmoGOBP2 all possess binding capabilities for Z8-14:Ac, indicating their overlapping functions in binding this component. Among these four rGmolPBPs/rGmolGOBPs, rGmolPBP2 exhibited the strongest binding affinity for Z8-14:Ac, potentially functioning as the primary GmolOBP responsible for perceiving and transporting Z8-14:Ac. Four PBPs (GfunPBP1.1, GfunPBP1.2, GfunPBP2, and GfunPBP3) and three GOBPs (GfunGOBP1, GfunGOBP2, and GfunGOBP3) were identified in PFMs antennae. rGfunPBP1.1, rGfunPBP1.2, and rGfunGOBP3 exhibited significantly stronger binding affinities for Z8-14:Ac than rGfunPBP2, rGfunPBP3, rGfunGOBP1, and rGfunGOBP2, indicating that the PBPs/GOBPs of PFMs and OFMs differ in their binding to Z8-14:Ac [2,61]. The sex pheromones released by OFM females consist of Z8-12:Ac, E8-12:Ac, Z8-12:OH, and 12:OH, with GmolPBP2 showing a binding preference for Z8-12:Ac and E8-12:Ac [43], GmolGOBP2 exhibiting the strongest binding affinity for 12:OH [44], and GmolPBP1 demonstrating a preference for binding to Z8-12:OH [45]. The strong binding affinity of GmolPBP2 for Z8-14:Ac may be related to Z8-14:Ac sharing a similar chemical structure and functional group with Z8-12:Ac. However, direct evidence of GmolPBP2 binding to Z8-14:Ac needs further analysis using protein crystallography to fully elucidate the interactions of the GmolPBP2–Z8-14:Ac complex. In addition to chemoreception, insect PBPs/GOBPs are involved in binding and transporting “non-semiochemical” ligands, such as BmorPBP1 and BmorGOBP2 in Bombyx mori, which bind vitamins, insecticides, and juvenile hormones [65,70]. Athetis lepigone AlepPBP2, AlepPBP3, and AlepGOBP2 have high binding affinities for insecticide phoxim and play an important role in phoxim adaptation [71]. Glyphodes pyloalis GpylPBP1 exhibits a strong binding affinity for chlorpyrifos and phoxim, with the Phe12, Ile52, and Phe118 residues being crucial binding sites for both insecticides [40]. Further research is needed to explore the functions of GmolPBPs/GmolGOBPs beyond chemoreception.
4.4. Key Amino Acid Residues and Interaction Forces of GmolPBP2 Binding to Z8-14:Ac
Classical OBPs possess a binding pocket formed by six typical α-helices. OBPs possess the capability to bind sex pheromones and other volatile semiochemicals as the molecules of these compounds enter the binding pocket and form specific binding interactions with the residues within the binding cavities [72,73,74]. The interactions between OBPs and odorant ligands encompass hydrogen bonds, electrostatic forces, van der Waals, π-π interactions, and π–alkyl interactions [75,76,77]. Hydrogen bonds frequently serve as the primary interaction force between insect PBPs and sex pheromone molecules, as observed in various examples such as Bombyx mori BmorPBP1 and bombykol [72], Epiphyas Postvittana EposPBP3 and E11-14:OH [78], and Apis mellifera ASP1 and 9-keto-2(E)-decenoic acid (9-ODA) [79]. Tian et al. analyzed the binding interactions of the GmolPBP2–Z8-12:Ac complex [80]. In our study, the GmolPBP2 sequence differs by one additional amino acid on the N-terminus due to the use of a different version of SignalP software (version 5.0). Additionally, Tian et al. used AMMBER (v.12) and we used GROMACS (v. 2023.3) for molecular dynamics simulations of GmolPBP2–Z8-12:Ac [80]. As a result, the calculated binding free energies in this study of GmolPBP2–Z8-12:Ac and hydrogen bonds were based on the GROMACS results. The five lowest energies of residues for GmolPBP2–Z8-12:Ac were found at Phe12 (−5.800 kJ/mol), Ile94 (−4.692 kJ/mol), Ile113 (−4.189 kJ/mol), Ile52 (−4.053 kJ/mol), and Leu68 (−3.526 kJ/mol). Likewise, the five lowest energies for GmolPBP2–Z8-14:Ac were observed at Phe12 (−7.352 kJ/mol), Ile94 (−5.863 kJ/mol), Ile113 (−4.103 kJ/mol), Ala114 (−4.071 kJ/mol), and Leu68 (−3.897 kJ/mol). Van der Waals forces were the primary contributors to the binding between GmolPBP2 and both sex pheromone components. However, differences arose in the hydrogen bonds: no hydrogen bond was found in GmolPBP2–Z8-12:Ac, while a hydrogen bond was formed between the side chain of Arg109 in GmolPBP2 and the carbonyl oxygen of Z8-14:Ac. According to the site-mutagenesis results, only the F12A mutant lost affinity for Z8-14:Ac, suggesting that Phe12 was the key amino acid residue for GmolPBP2 binding to Z8-14:Ac. The π–alkyl interaction, formed between the phenyl sidechain of Phe12 and the C14 atom of Z8-14:Ac, was identified as the primary interaction force in GmolPBP2’s binding to Z8-14:Ac, whereas the H-bond between the NH2 atom of Arg109 and the carbonyl oxygen atom in Z8-14:Ac was not. Li et al. discovered that GfunPBP1.1 of the PFM preferentially bound to Z8-14:Ac, whereas a hydrogen bond was observed between the OA atom of Ser56 and the O atom from the acetoxy group of Z8-14:Ac [2]. Moreover, a π–alkyl interaction formed between the phenyl of Trp37 and the C14 atom of Z8-14:Ac. These results imply that, even among closely related species, there are variations in the residues and binding forces of PBPs when binding to the same ligand.
Acknowledgments
We are grateful for the assistance of Bingqiang Xu at the Institute of Plant Protection, Xinjiang Academy of Agricultural Sciences, Urumchi 830091, China.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/insects15120918/s1, Figure S1: SDS-PAGE results showing the expressed products and purified rGmolPBP1 (A), rGmolPBP2 (B), rGmolPBP3 (C), rGmolGOBP1 (D), and rGmolGOBP2 (E) of OFMs. Figure S2: The Ramachandran plots of simulated GmolPBP2 display the residues in the most favored regions, additional allowed regions, and disallowed regions. Figure S3: SDS-PAGE of the rGmolPBP2 (wild-type) (A) and the mutants F12A (B), L68A (C), I94A (D), R109A (E), and I113A (F). Table S1: Specific primers used for gene expression level detection, prokaryotic expression, and site-directed mutagenesis. Table S2: The 33 amino acids that constitute the binding pocket of GmolPBP2.
Author Contributions
Conceptualization, G.L. and B.L.; methodology, Y.L. and X.C.; software, B.L. and K.L.; validation, G.L. and S.X.; formal analysis, Y.L. and G.L.; investigation, Y.L., X.C. and S.X.; data curation, Y.L. and B.L.; writing—original draft preparation, Y.L. and K.L.; writing—review and editing, G.L. and K.L.; supervision, G.L.; project administration, G.L.; funding acquisition, X.C. and G.L. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
Data are available upon request from Guangwei Li.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of this study, in the collection, analysis, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Funding Statement
This research was funded by the National Natural Science Foundation of China (32160636, 32260669), the Natural Science Foundation Research Project of Shaanxi province of China (2021JQ-621), Open Special Project of Institute of Bio (Chemical) Informational Materials from Pherobio Technology Co., Ltd. (PHROBIO2023ZJSF04), Shaanxi Province “Two Chains” Integration Key Project (2023LLRH-01), and The Youth Innovation Team of Shaanxi Universities (2022).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Myers C.T., Hull L.A., Krawczyk G. Effects of orchard host plants (apple and peach) on development of oriental fruit moth (Lepidoptera: Tortricidae) J. Econ. Entomol. 2007;100:421–430. doi: 10.1093/jee/100.2.421. [DOI] [PubMed] [Google Scholar]
- 2.Li L.L., Xu B.Q., Li C.Q., Li B.L., Luo K., Li G.W., Chen X.L. Functional disparity of four pheromone-binding proteins from the plum fruit moth Grapholita funebrana Treitscheke in detection of sex pheromone components. Int. J. Biol. Macromol. 2023;225:1267–1279. doi: 10.1016/j.ijbiomac.2022.11.186. [DOI] [PubMed] [Google Scholar]
- 3.Najar-Rodriguez A., Orschel B., Dorn S. Season-long volatile emissions from peach and pear trees in situ, overlapping profiles, and olfactory attraction of an oligophagous fruit moth in the laboratory. J. Chem. Ecol. 2013;39:418–429. doi: 10.1007/s10886-013-0262-7. [DOI] [PubMed] [Google Scholar]
- 4.Yang X.F., Fan F., Ma C.S., Wang C., Wei G.S. Effect of host plants on the development, survivorship and reproduction of Grapholita molesta (Lepidoptera: Tortricidae) under laboratory conditions. Austral Entomol. 2016;55:433–438. doi: 10.1111/aen.12204. [DOI] [Google Scholar]
- 5.Sarker S., Lim U.T. Olfactory responses of Grapholita molesta to different developmental stages of immature fruit of peach, plum, and apple. J. Asia-Pac. Entomol. 2023;26:102035. doi: 10.1016/j.aspen.2022.102035. [DOI] [Google Scholar]
- 6.Rizzo R., Farina V., Saiano F., Lombardo A., Ragusa E., Verde G.L. Do Grapholita funebrana infestation rely on specific plum fruit features? Insects. 2019;10:444. doi: 10.3390/insects10120444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stefanova D., Vasilev P., Kutinkova H., Andreev R., Palagacheva N., Tityanov M. Possibility for control of plum fruit moth Grapholita funebrana Tr. by pheromone dispensers. J. Biopestic. 2019;12:153–156. [Google Scholar]
- 8.Zheng Y., Wu R.X., Dorn S., Chen M.H. Diversity of Tortricid moths in apple orchards: Evidence for a cryptic species of Grapholita (Lepidoptera: Tortricidae) from China. Bull. Entomol. Res. 2017;107:268–280. doi: 10.1017/S0007485316000973. [DOI] [PubMed] [Google Scholar]
- 9.Yang L., Sun H.M., Lin H.Z., Chen L.S., Wang S.S. Species and population dynamics of fruit borer in Shihezi reclamation zone. North Horticult. 2017;12:132–135. [Google Scholar]
- 10.Chen X.L., Chen Y.X., Bao L.J., Kang L., Sun Y., Li G.W. Species and population dynamic of fruit-borer in apple orchard of Yan’an area. Plant Protect. 2021;47:219–225. [Google Scholar]
- 11.Zheng Y., Qiao X.F., Wang K., Dorn S., Chen M.H. Population genetics affected by pest management using fruitbagging: A case study with Grapholita molesta (Lepidoptera: Tortricidae) in China. Entomol. Exp. Appl. 2015;156:117–127. doi: 10.1111/eea.12316. [DOI] [Google Scholar]
- 12.Sziràki G. Specificity of sexual attractant traps for signalization of oriental fruit moth, Grapholitha molesta Busck. Acta Phytopathol. Acad. Sci. Hung. 1978;13:205–213. [Google Scholar]
- 13.Tòth M., Sziràki G., Szöcs G., Sàringer E. A pheromone inhibitor for male Grapholitha funebrana Tr., and its use for increasing the specifcity of the lure for G. molesta Busck (Lepidoptera: Tortricidae) Agric. Ecosyst. Environ. 1991;35:65–72. [Google Scholar]
- 14.Renou M., Anton S. Insect olfactory communication in a complex and changing world. Curr. Opin. Insect Sci. 2020;42:1–7. doi: 10.1016/j.cois.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Deng J.Y., Shen Z.J., Wang F.M., Liu T., Hong W.Y., Fang M.H., Wo L.F., Chu S.J. Enhancement of attraction to sex pheromone of Grapholita molesta (Busck) (Lepidoptera: Tortricidae) by structurally unrelated sex pheromone compounds of Conogethes punctiferalis (Guenée) (Lepidoptera: Crambidae) J. Asia-Pac. Entomol. 2022;25:101859. doi: 10.1016/j.aspen.2021.101859. [DOI] [Google Scholar]
- 16.Meier L., Zou Y.F., Millar J., Mongold-Diers J., Hanks L. Synergism between enantiomers creates species-specific pheromone blends and minimizes cross-attraction for two species of Cerambycid beetles. J. Chem. Ecol. 2016;42:1181–1192. doi: 10.1007/s10886-016-0782-z. [DOI] [PubMed] [Google Scholar]
- 17.Silva W.D., Millar J.G., Hanks L.M., Costa C.M., Leite M.O.G., Tonelli M., Bento J.M.S. Interspecific cross-attraction between the South American Cerambycid beetles Cotyclytus curvatus and Megacyllene acuta is averted by minor pheromone components. J. Chem. Ecol. 2018;44:268–275. doi: 10.1007/s10886-018-0933-5. [DOI] [PubMed] [Google Scholar]
- 18.Guerin P.M., Arn H., Buser H.R., Charmillot P., Tth M., Szirki G. Sex pheromone of Grapholita funebrana. Occurrence of Z-8- and Z-10-tetradecenyl acetate as secondary components. J. Chem. Ecol. 1986;12:1361–1368. doi: 10.1007/BF01012355. [DOI] [PubMed] [Google Scholar]
- 19.Lacey M.J., Sanders C.J. Chemical composition of sex pheromone of oriental fruit moth and rates of release by individual female moths. J. Chem. Ecol. 1992;18:1421–1435. doi: 10.1007/BF00994366. [DOI] [PubMed] [Google Scholar]
- 20.Cardé A.M., Baker T.C., Cardé R.T. Identification of a four component sex pheromone of the female oriental fruit moth, Grapholita molesta (Lepidoptera: Tortricidae) J. Chem. Ecol. 1979;5:423–427. doi: 10.1007/BF00987927. [DOI] [Google Scholar]
- 21.Han K.S., Jung J.K., Choi K.H., Lee S.W., Boo K.S. Sex pheromone composition and male trapping of oriental fruit moth, Grapholita molesta (Lepidoptera: Tortricidae) in Korea. J. Asia-Pac. Entomol. 2001;4:31–35. doi: 10.1016/S1226-8615(08)60099-0. [DOI] [Google Scholar]
- 22.Yang C.Y., Jung J.K., Han K.S., Boo K.S., Yiem M.S. Sex pheromone composition and monitoring of the oriental fruit moth Grapholitha molesta (Lepidoptera: Tortricidae) in Naju pear orchards. J. Asia-Pac. Entomol. 2002;5:201–207. doi: 10.1016/S1226-8615(08)60153-3. [DOI] [Google Scholar]
- 23.Stelinski L.L., Miller J.R., Ledebuhr R., Siegert P., Gut L.J. Season-long mating disruption of Grapholita molesta (Lepidoptera: Tortricidae) by one machine application of pheromone in wax drops. J. Pest Sci. 2007;80:109–117. doi: 10.1007/s10340-007-0162-0. [DOI] [Google Scholar]
- 24.Rosu-Mares S.D., Feneșan M.P., Ciotlaus I., Balea A., Andreica A. Pheromone monitoring of harmful Lepidoptera present in Bistrita area in apple and plum orchards. Rom. J. Hortic. 2021;2:99–106. [Google Scholar]
- 25.Puskás J., Nowinszky L., Barczikay G., Kúti Z.S. The pheromone trap catch of harmful moths in connection with solar activity featured by q-index. Appl. Ecol. Env. Res. 2010;8:261–266. [Google Scholar]
- 26.Pelosi P., Maida R. Odorant-binding proteins in insects. Comp. Biochem. Phys. B. 1995;111:503–514. doi: 10.1016/0305-0491(95)00019-5. [DOI] [PubMed] [Google Scholar]
- 27.Brito N.F., Moreira M.F., Melo A.C.A. A look inside odorant-binding proteins in insect chemoreception. J. Insect Physiol. 2016;95:51–65. doi: 10.1016/j.jinsphys.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 28.Zhou J.J., Robertson G., He X.L., Dufour S., Hooper A.M., Pickett J.A., Keep N.H., Field L.M. Characterisation of Bombyx mori odorant-binding proteins reveals that a general odorant-binding protein discriminates between sex pheromone components. J. Mol. Biol. 2009;389:529–545. doi: 10.1016/j.jmb.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 29.Forstner M., Gohl T., Breer H., Krieger J. Candidate pheromone binding proteins of the silk moth Bombyx mori. Invertebr. Neurosci. 2006;6:177–187. doi: 10.1007/s10158-006-0032-0. [DOI] [PubMed] [Google Scholar]
- 30.Zhou J.J. Odorant-binding proteins in insects. Vitam. Horm. 2010;83:241–272. doi: 10.1016/S0083-6729(10)83010-9. [DOI] [PubMed] [Google Scholar]
- 31.Cao D.P., Liu Y., Wei J.J., Liao X.Y., Walker W.B., Li J.H., Wang G.R. Identifcation of candidate olfactory genes in Chilo suppressalis by antennal transcriptome analysis. Int. J. Biol. Sci. 2014;10:846–860. doi: 10.7150/ijbs.9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao W., Yang L., Xu Z.F., He L. Transcriptomics and identifcation of candidate chemosensory genes in antennae of Conogethes punctiferalis (Lepidoptera: Crambidae) J. Asia-Pac. Entomol. 2016;19:911–920. doi: 10.1016/j.aspen.2016.08.007. [DOI] [Google Scholar]
- 33.Jing D.P., Zhang T.T., Prabu S., Bai S.X., He K.L., Wang Z.Y. Molecular characterization and volatile binding properties of pheromone binding proteins and general odorant binding proteins in Conogethes pinicolalis (Lepidoptera: Crambidae) Int. J. Biol. Macromol. 2020;146:263–272. doi: 10.1016/j.ijbiomac.2019.12.248. [DOI] [PubMed] [Google Scholar]
- 34.Huang G.Z., Liu J.T., Zhou J.J., Wang Q., Dong J.Z., Zhang Y.J., Li X.C., Li J., Gu S.H. Expressional and functional comparisons of two general odorant binding proteins in Agrotis ipsilon. Insect Biochem. Mol. Biol. 2018;98:34–47. doi: 10.1016/j.ibmb.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 35.Yang H.H., Li S.P., Yi M.Z., Zhu X.Y., Li J.B., Zhang Y.N., Li X.M. Functional differentiation of two general odorant-binding proteins to sex pheromones in Spodoptera frugiperda. Pestic. Biochem. Physiol. 2023;191:105348. doi: 10.1016/j.pestbp.2023.105348. [DOI] [PubMed] [Google Scholar]
- 36.Xia Q., Zhou Z., Lu C., Cheng D., Dai F., Li B., Zhao P., Zha X., Cheng T., Chai C., et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori) Science. 2004;306:1937–1940. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- 37.Zhan S., Merlin C., Boore J.L., Reppert S.M. The monarch butterfly genome yields insights into long-distance migration. Cell. 2011;147:1171–1185. doi: 10.1016/j.cell.2011.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dasmahapatra K.K., Walters J.R., Briscoe A.D., Davey J.W., Whibley A., Nadeau N.J., Zimin A.V., Hughes D.S.T., Ferguson L.C., Martin S.H., et al. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487:94–98. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.You M., Yue Z., He W., Yang X., Yang G., Xie M., Zhan D., Baxter S.W., Vasseur L., Gurr G.M., et al. A heterozygous moth genome provides insights into herbivory and detoxification. Nat. Genet. 2013;45:220–225. doi: 10.1038/ng.2524. [DOI] [PubMed] [Google Scholar]
- 40.Li Y.J., Gu F.M., Chen H.C., Liu Z.X., Song W.M., Wu F.A., Sheng S., Wang J. Binding characteristics of pheromone-binding protein 1 in Glyphodes pyloalis to organophosphorus insecticides: Insights from computational and experimental approaches. Int. J. Biol. Macromol. 2024;260:129339. doi: 10.1016/j.ijbiomac.2024.129339. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X.Q., Yan Q., Li L.L., Xu J.W., Mang D., Wang X.L., Hoh H.H., Ye J., Ju Q., Ma Y., et al. Different binding properties of two general odorant binding proteins in Athetis lepigone with sex pheromones, host plant volatiles and insecticides. Pestic. Biochem. Physiol. 2020;164:173–182. doi: 10.1016/j.pestbp.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 42.Li G.W., Du J., Li Y.P., Wu J.X. Identification of putative olfactory genes from the oriental fruit moth Grapholita molesta via an antennal transcriptome analysis. PLoS ONE. 2015;10:e0142193. doi: 10.1371/journal.pone.0142193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song Y.Q., Dong J.F., Qiao H.L., Wu J.X. Molecular characterization, expression patterns and binding properties of two pheromone-binding proteins from the oriental fruit moth, Grapholita molesta (Busck) J. Integr. Agr. 2014;13:2709–2720. doi: 10.1016/S2095-3119(13)60686-3. [DOI] [Google Scholar]
- 44.Li G.W., Chen X.L., Li B.L., Zhang G.H., Li Y.P., Wu J.X. Binding properties of general odorant binding proteins from the oriental fruit moth, Grapholita molesta (Busck) (Lepidoptera: Tortricidae) PLoS ONE. 2016;11:e0155096. doi: 10.1371/journal.pone.0155096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang G.H., Chen J., Yu H.L., Tian X.L., Wu J.X. Molecular and functional characterization of pheromone binding protein 1 from the oriental fruit moth, Grapholita molesta (Busck) Sci. Rep. 2018;8:2276. doi: 10.1038/s41598-018-20719-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du J., Wang Y.R., Wu J.X. Effect of four different artificial diets on development and reproduction of Grapholitha molesta (Lepidoptera: Tortricidae) J. Shanxi Agric. Univ. Nat. Sci. Ed. 2010;30:228–231. [Google Scholar]
- 47.Vandesompele J., Preter K.D., Pattyn F., Poppe B., Roy N.V., Paepe A.D., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by genometric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034.1. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kelmansky D.M., Martínez E.J., Leiva V. A new variance stabilizing transformation for gene expression data analysis. Stat. Appl. Genet. Mol. Biol. 2013;12:653–666. doi: 10.1515/sagmb-2012-0030. [DOI] [PubMed] [Google Scholar]
- 49.Zhang T.T., Mei X.D., Feng J.N., Berg B.G., Zhang Y.J., Guo Y.Y. Characterization of three pheromone-binding proteins (PBPs) of Helicoverpa armigera (Hübner) and their binding properties. J. Insect Physiol. 2012;58:941–948. doi: 10.1016/j.jinsphys.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 50.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 51.Wickham H. Ggplot2: Elegant Graphics for Data Analysis. Springer; New York, NY, USA: 2016. [Google Scholar]
- 52.Valdés-Tresanco M.S., Valdés-Tresanco M.E., Valiente P.A., Moreno E. gmx_MMPBSA: A new tool to perform end-state free energy calculations with GROMACS. J. Chem. Theory Comput. 2021;17:6281–6291. doi: 10.1021/acs.jctc.1c00645. [DOI] [PubMed] [Google Scholar]
- 53.Grant A.J., O’Connell R.J., Hammond A.M. A comparative study of pheromone perception in two species of Noctuid moths. J. Insect Behav. 1988;1:75–96. doi: 10.1007/BF01052505. [DOI] [Google Scholar]
- 54.Mozūraitis R., Liblikas I., Noreika R. Sex pheromone communication of tentiform leaf-miners Phyllonorycter insignitella and Phyllonorycter nigrescentella from two related species groups. Chemoecology. 2009;18:171–176. doi: 10.1007/s00049-008-0400-9. [DOI] [Google Scholar]
- 55.Greenberg L., Johnson C.A., Trager J.C., McElfresh J.S., Rodstein J., Millar J.G. Sex attractant pheromones of virgin queens of sympatric slave-making ant species in the genus Polyergus, and their possible roles in reproductive isolation. J. Chem. Ecol. 2018;44:547–555. doi: 10.1007/s10886-018-0966-9. [DOI] [PubMed] [Google Scholar]
- 56.Xu T., Hansen L., Cha D.H., Hao D., Zhang L., Teale S.A. Identification of a female-produced pheromone in a destructive invasive species: Asian longhorn beetle, Anoplophora glabripennis. J. Pest Sci. 2020;93:1321–1332. doi: 10.1007/s10340-020-01229-3. [DOI] [Google Scholar]
- 57.Klun J.A., Bierl-Leonhardt B.A., Plimmer J.R., Sparks A.N., Primiani M., Chapman O.L., Lepone G., Lee G.H. Sex pheromone chemistry of the female tobacco budworm moth, Heliothis virescens. J. Chem. Ecol. 1980;6:177–183. doi: 10.1007/BF00987536. [DOI] [Google Scholar]
- 58.Dunkelblum E., Kehat M. Female sex pheromone components of Heliothis peltigera (Lepidoptera: Noctuidae). Chemical identification from gland extracts and male response. J. Chem. Ecol. 1989;15:2233–2245. doi: 10.1007/BF01014112. [DOI] [PubMed] [Google Scholar]
- 59.Chang X.Q., Nie X.P., Zhang Z., Zeng F.F., Lv L. De novo analysis of the oriental armyworm Mythimna separata antennal transcriptome and expression patterns of odorant-binding proteins. Comp. Biochem. Phys. D. 2017;22:120–130. doi: 10.1016/j.cbd.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 60.Tian Z.Q., Sun L., Li Y.Y., Quan L.F., Zhang H.J., Yan W.T., Yue Q., Qiu G.S. Antennal transcriptome analysis of the chemosensory gene families in Carposina sasakii (Lepidoptera: Carposinidae) BMC Genom. 2018;19:544. doi: 10.1186/s12864-018-4900-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li L.L., Xu B.Q., Li C.Q., Li B.L., Chen X.L., Li G.W. Different binding affinities of three general odorant-binding proteins in Grapholita funebrana (Treitscheke) (Lepidoptera: Tortricidae) to sex pheromones, host plant volatiles, and insecticides. J. Econ. Entomol. 2022;115:1129–1145. doi: 10.1093/jee/toac063. [DOI] [PubMed] [Google Scholar]
- 62.Dong J., Wang K., Sun Y.L., Tian C.H., Wang S.L. Antennal transcriptome analysis of odorant-binding proteins and characterization of GOBP2 in the variegated cutworm Peridroma saucia. Front. Physiol. 2023;14:1241324. doi: 10.3389/fphys.2023.1241324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chang H., Liu Y., Yang T., Pelosi P., Dong S., Wang G. Pheromone binding proteins enhance the sensitivity of olfactory receptors to sex pheromones in Chilo suppressalis. Sci. Rep. 2015;5:13093. doi: 10.1038/srep13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khuhro S.A., Liao H., Dong X.T., Yu Q., Yan Q., Dong S.L. Two general odorant binding proteins display high bindings to both host plant volatiles and sex pheromones in a pyralid moth Chilo suppressalis (Lepidoptera: Pyralidae) J. Asia-Pac. Entomol. 2017;20:521–528. doi: 10.1016/j.aspen.2017.02.015. [DOI] [Google Scholar]
- 65.Guo X., Xuan N., Liu G., Xie H., Lou Q., Arnaud P., Offmann B., Picimbon J.-F. An expanded survey of the moth PBP/GOBP clade in Bombyx mori: New insight into expression and functional roles. Front. Physiol. 2021;12:712593. doi: 10.3389/fphys.2021.712593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jain R., Brockmann A. Sex-specific molecular specialization and activity rhythm-dependent gene expression in honey bee antennae. J. Exp. Biol. 2020;223:jeb217406. doi: 10.1242/jeb.217406. [DOI] [PubMed] [Google Scholar]
- 67.Tonini C., Cassani G., Piccardi P., Maini S., Castellari P.L., Pasqualini E. Sex pheromone components of the leafroller moth Pandemis cerasana. J. Insect Physiol. 1982;28:443–446. doi: 10.1016/0022-1910(82)90072-5. [DOI] [Google Scholar]
- 68.Priesner E., Buser H.R., Bengtsson M., Chambon P.J., Wildbolz T., Arn H., Witzgall P. Sex pheromones of Spilonota ocellana and Spilonota laricana. Entomol. Exp. Appl. 1991;60:219–223. [Google Scholar]
- 69.Alessandro G., Thrimawithana A.H., Bernd S., Newcomb R.D. Differential gene expression in the evolution of sex pheromone communication in New Zealand’s endemic leafroller moths of the genera Ctenopseustis and Planotortrix. BMC Genom. 2018;19:94. doi: 10.1186/s12864-018-4451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pelosi P., Iovinella I., Zhu J., Wang G.R., Dani F.R. Beyond chemoreception: Diverse tasks of soluble olfactory proteins in insects. Biol. Rev. 2018;93:184–200. doi: 10.1111/brv.12339. [DOI] [PubMed] [Google Scholar]
- 71.Abendroth J.A., Moural T.W., Wei H.S., Zhu F. Roles of insect odorant binding proteins in communication and xenobiotic adaptation. Front. Insect Sci. 2023;3:1274197. doi: 10.3389/finsc.2023.1274197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sandler B.H., Nikonova L., Leal W.S., Clardy J. Sexual attraction in the silkworm moth: Structure of the pheromone-binding-protein-bombykol complex. Chem. Biol. 2000;7:143–151. doi: 10.1016/S1074-5521(00)00078-8. [DOI] [PubMed] [Google Scholar]
- 73.Tian Z., Liu J.Y., Zhang Y.L. Key residues involved in the interaction between Cydia pomonella pheromone binding protein 1 (CpomPBP1) and codlemone. J. Agric. Food Chem. 2016;64:7994–8001. doi: 10.1021/acs.jafc.6b02843. [DOI] [PubMed] [Google Scholar]
- 74.Falchetto M., Ciossani G., Scolari F., Di Cosimo A., Nenci S., Field L.M., Mattevi A., Zhou J.J., Gasperi G., Forneris F. Structural and biochemical evaluation of Ceratitis capitata odorant-binding protein 22 affinity for odorants involved in intersex communication. Insect Mol. Biol. 2019;28:431–443. doi: 10.1111/imb.12559. [DOI] [PubMed] [Google Scholar]
- 75.Lescop E., Briand L., Pernollet J.C., Guittet E. Structural basis of the broad specificity of a general odorant-binding protein from Honeybee. Biochemistry. 2009;48:2431–2441. doi: 10.1021/bi802300k. [DOI] [PubMed] [Google Scholar]
- 76.Tsitsanou K.E., Thireou T., Drakou C.E., Koussis K., Keramioti M.V., Leonidas D.D., Eliopoulos E., Iatrou K., Zographos S.E. Anopheles gambiae odorant binding protein crystal complex with the synthetic repellent DEET: Implications for structure-based design of novel mosquito repellents. Cell. Mol. Life Sci. 2012;69:283–297. doi: 10.1007/s00018-011-0745-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y.N., Zhang X.Q., Zhang X.C., Xu J.W., Li L.L., Zhu X.Y., Wang J.J., Wei J.Y., Mang D.Z., Zhang F., et al. Key amino acid residues influencing binding affinities of pheromone-binding protein from Athetis lepigone to two sex pheromones. J. Agric. Food Chem. 2020;68:6092–6103. doi: 10.1021/acs.jafc.0c01572. [DOI] [PubMed] [Google Scholar]
- 78.Hamiaux C., Carraher C., Löfstedt C., Corcoran J.A. Crystal structure of Epiphyas postvittana pheromone binding protein 3. Sci. Rep. 2020;10:16366. doi: 10.1038/s41598-020-73294-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pesenti M.E., Spinelli S., Bezirard V., Briand L., Pernollet J.C., Campanacci V., Tegoni M., Cambillau C. Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release. J. Mol. Biol. 2009;390:981–990. doi: 10.1016/j.jmb.2009.05.067. [DOI] [PubMed] [Google Scholar]
- 80.Tian Z., Li Y., Xing Y.J., Li R.C., Liu J.Y. Structural insights into two representative conformations of the complex formed by Grapholita molesta (Busck) pheromone binding protein 2 and Z-8-dodecenyl acetate. J. Agric. Food Chem. 2019;67:4425–4434. doi: 10.1021/acs.jafc.9b00052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon request from Guangwei Li.