Abstract
The clinical manifestations of SARS-CoV-2 infection may range from asymptomatic or minor conditions to severe and life-threatening outcomes. The respiratory system is a principal target of the virus and in the majority of cases of severe disease, an acute form of pneumonia develops. Despite concerted global efforts to elucidate the pathogenic mechanisms of COVID-19, the progression of the infection leading to pulmonary damage remains poorly understood. The present study aimed to analyse the immunological profile of subjects with a previous SARS-CoV-2 infection and predisposition to lung injury. The results showed a strong correlation between IgG anti-membrane antibodies and lung injury.
Keywords: COVID-19, Lung injury, SARS-CoV-2 membrane protein
Subject terms: Molecular biology, Immunology
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus was first identified in China in December 20191 and rapidly spread across the globe, causing one of the most significant pandemics in the history of humanity2. Following the declaration of a global public health emergency by WHO in March 2020, the management of the pandemic, designated coronavirus disease 2019 (COVID-19), immediately proved challenging due to the wide range of clinical manifestations, the ability of the virus to spread by aerosol, and its ability to evolve into “variants” capable of evading the immune system even in previously infected or vaccinated hosts3. The virus is primarily adsorbed through the cellular receptor angiotensin 2 (ACE), which is widely expressed in the lung epithelium4. Consequently, in the severe form of COVID-19, pulmonary damage occurs in a significant proportion of subjects5.
The pathogenesis of the COVID-19 appears to exhibit a multifactorial nature. While individuals with compromised immunity, advanced age, and concomitant diseases are most susceptible to severe complications6, a subset of healthy younger individuals may also exhibit severe disease manifestations7,8. Furthermore, the incidence of disease9 and the distribution of viral strains exhibit considerable variation globally10,11, suggesting a specific host-virus interaction and then a role of the human leukocyte antigen (HLA) system12.
SARS-CoV-2 is a single-stranded, positive-sense RNA virus belonging to the Beta-coronavirus genus within the Coronaviridae family13,14. The genome of SARS-CoV-2 is approximately 30 kilobases in length and is flanked by 5ʹ and 3ʹ untranslated regions. It contains two large open reading frames (ORF1a and ORF1b), which encode 16 non-structural proteins (NSP-1–16), four structural proteins, namely spike (S), envelope (E), membrane (M), and nucleocapsid (N), and at least 9 accessory proteins15. The non-structural proteins comprise the viral replication-transcription complex16,17, whereas the structural proteins are responsible for the formation of the virion particle18. A crucial function in the virus’s capability to infect host cells is carried out by the S proteins, which specifically bind to the ACE2 receptor on the surface of human cells19. Given its pivotal role in viral entry, the S protein20 represents a primary target for vaccine development and therapeutic interventions against COVID-1921. While extensive studies have been conducted on S and N proteins22,23, comparatively less is known about the M protein. It is, however, the most abundant structural protein and could play an important role in the host immunological response24.
In this study, the immunological profile of subjects with a previous SARS-CoV-2 infection was analysed through the detection of antibodies IgG against S, N and M proteins. The results revealed a significant statistical association between IgG anti-M antibodies and pulmonary damage. Furthermore, an increasing amount of the antibodies was associated with a greater likelihood of developing lung injury.
Results
Production of recombinant proteins
Supplementary Figure S1, Panel A, depicts the three recombinant proteins purified using Ni–NTA affinity chromatography, separated by polyacrylamide gel electrophoresis. Panel B shows a Western blot analysis of the same proteins, employing a monoclonal antibody against the hexa-histidine (His) epitope.
Sample investigated
The study population consisted of 63 adults (50 male and 13 female) ranging from 18 to 81 years of age (mean = 47; median = 48). The interval between the date of molecular test positivity and serum collection exhibited a considerable range, spanning from a minimum of 21 days to a maximum of 328 days, with an average of 105 days. Image analysis revealed that 30 of 63 subjects had evidence of lung injury, of which 9 required oxygen therapy. A summary of the data is presented in Supplementary Table S1.
ELISA results and statistical analysis
Recombinant S and N proteins were found to detect IgG antibodies in 48 of 63 samples tested. Of the 15 samples that yielded negative results, 9 samples exhibited concordant results in both tests. The recombinant M protein was able to detect the presence of IgG antibodies in 40 out of 64 samples.
The χ2 association test was performed on the ELISA results of the three recombinant proteins stratified by lung injury. No association was found between the anti-S and anti-N IgG results and lung injury, conversely, a significant association with the anti-M IgG result was found (χ2 = 4.29; df = 1; p < 0.05) (Table 1).
Table 1.
Correlation between IgG anti-M and lung injury.
| Contingency table | |||
|---|---|---|---|
| ELISA | Lung injury | ||
| membrane_results | 0 | 1 | Total |
| Neg | 16 | 7 | 23 |
| Pos | 17 | 23 | 40 |
| Total | 33 | 30 | 63 |
| χ2 Test | |||
|---|---|---|---|
| Value | df | p | |
| χ2 | 4.29 | 1 | 0.03837 |
| N | 63 | ||
| Effect size | |||
|---|---|---|---|
| Value | |||
| Cramer’s V | 0.275 | ||
The ELISA results for the recombinant membrane protein stratified for lung injury are shown (lung injury: 0 = absence; 1 = presence) together with the result of the χ2 association test and the effect size.
A T-test analysis (Mann–Whitney U) was employed to conduct a comparison between the antibody levels of individuals with and without lung injury. For all recombinant proteins, higher levels of antibodies were found in subjects reporting lung injury, with a statistically significant difference. The largest effect size was observed for the IgG anti-M. The data are shown in Table 2.
Table 2.
Antibody titer comparison.
| (A) Independent samples T-test | ||||||
|---|---|---|---|---|---|---|
| Statistic | p | Effect size | ||||
| SPIke_OD/cutoff | Mann–Whitney U | 253 | 0.00069 | Rank biserial correlation | 0.489 | |
| Nucl_OD/cutoff | Mann–Whitney U | 288 | 0.00439 | Rank biserial correlation | 0.419 | |
| Membrane OD/cutoff | Mann–Whitney U | 235 | 0.00025 | Rank biserial correlation | 0.525 | |
| (B) Descriptive | ||||||
|---|---|---|---|---|---|---|
| Lung injury | N | Mean | Median | SD | SE | |
| SPIke_OD/cutoff | 0 | 33 | 2.03 | 1.84 | 1.345 | 0.2341 |
| 1 | 30 | 3.47 | 3.89 | 1.69 | 0.309 | |
| Nucl_OD/cutoff | 0 | 33 | 1.9 | 1.59 | 1.337 | 0.2328 |
| 1 | 30 | 3.14 | 3.24 | 1.73 | 0.316 | |
| Membrane OD/cutoff | 0 | 33 | 1.19 | 1.12 | 0.392 | 0.0683 |
| 1 | 30 | 2.13 | 1.78 | 1.38 | 0.252 | |
The non-parametric T-test (Mann–Whitney U, panel A) demonstrated a statistically significant increase in antibodies targeting the recombinant proteins under investigation in subjects presenting with lung injury. In panel B a description of the antibodies titers are shown.
A binomial regression logistic analysis was performed to assess the relationship between the antibody titers of IgG anti-S, anti-N, and anti-M and lung injury. The optical absorption, obtained in ELISA with the three antigens, was used as a continuous variable, while the presence or absence of lung injury served as the binomial variable.
The three models demonstrated statistical significance (Table 3).
Table 3.
Relationship between antibody titer and lung injury.
| (A) Model fit measures | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Model | Deviance | AIC | BIC | R2McF | R2CS | R2N | Overall model test | ||
| χ2 | df | p | |||||||
| 1 | 69.1 | 73.1 | 77.3 | 0.208 | 0.25 | 0.334 | 18.1 | 1 | 0.00002 |
| Model coefficients—lung injury | |||||||
|---|---|---|---|---|---|---|---|
| Predictor | Estimate | SE | Z | p | Odds ratio | 95% confidence interval | |
| Lower | Upper | ||||||
| Intercept | − 2.92 | 0.891 | − 3.28 | 0.00106 | 0.054 | 0.00942 | 0.31 |
| Membrane | 1.92 | 0.611 | 3.14 | 0.0017 | 6.8071 | 2.0548 | 22.551 |
| (B) Model fit measures | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Model | Deviance | AIC | BIC | R2McF | R2CS | R2N | Overall model test | ||
| χ2 | df | p | |||||||
| 1 | 74.6 | 78.6 | 82.9 | 0.144 | 0.181 | 0.241 | 12.6 | 1 | 0.00039 |
| Model coefficients—lung injury | |||||||
|---|---|---|---|---|---|---|---|
| Predictor | Estimate | SE | Z | p | Odds ratio | 95% confidence interval | |
| Lower | Upper | ||||||
| Intercept | − 1.706 | 0.568 | − 3 | 0.00267 | 0.182 | 0.0596 | 0.553 |
| Spike | 0.595 | 0.185 | 3.21 | 0.00133 | 1.813 | 1.2606 | 2.608 |
| (C) Model fit measures | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Model | Deviance | AIC | BIC | R2McF | R2CS | R2N | Overall model test | ||
| χ2 | df | p | |||||||
| 1 | 77.7 | 81.7 | 86 | 0.109 | 0.14 | 0.187 | 9.49 | 1 | 0.00207 |
| Model coefficients—lung injury | |||||||
|---|---|---|---|---|---|---|---|
| Predictor | Estimate | SE | Z | p | Odds ratio | 95% confidence interval | |
| Lower | Upper | ||||||
| Intercept | − 1.375 | 0.519 | − 2.65 | 0.00811 | 0.253 | 0.0914 | 0.7 |
| Nucleocapsid | 0.515 | 0.181 | 2.85 | 0.00437 | 1.67 | 1.1747 | 2.387 |
The three tables show the results of the logit analysis carried out to assess the relationship between IgG antibodies detected by the three recombinant proteins and lung injury. All antibodies studied were found to correlate with lung disease. However, the most significant correlation was shown by anti-M IgG (odds ratio = 6.8; Panel A) in comparison to anti-spike IgG (odds ratio = 1.81; Panel B) and anti-N IgG (odds ratio = 1.67; Panel C).
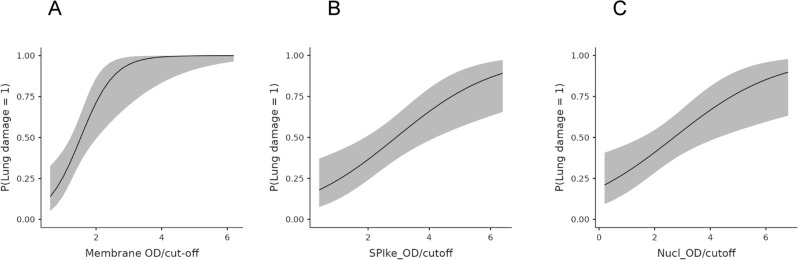
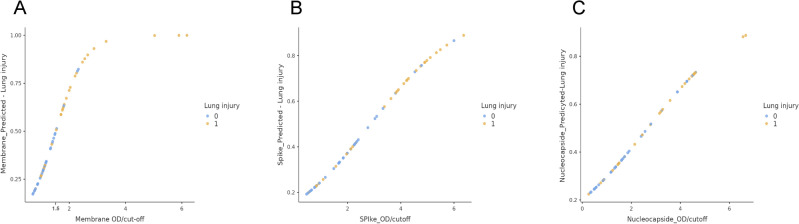
The data indicated, however, that IgG anti-M antibodies were the higher risk factor for the development of lung injury, with an odds ratio of 6.8 (95% CI, lower 2.05; upper 22.5) (Table 3, panel A). In contrast, the odds ratio for IgG anti-S and anti-N were 1.8 and 1.67, respectively (Table 3, panel B, C). The results are presented in graphical form in Fig. 1, which plots the optical density (OD) values resulting from the ELISA analysis of the three recombinant proteins against the estimated probability of developing lung injury. The IgG anti-M protein exhibited optimal logistic behaviour, reaching the plateau (probability = 1) at approximately 4 OD. Conversely, the IgG anti-S and anti-N proteins exhibited linear-like behaviour without reaching the maximum risk probability. The potential interactions between the antibodies were also investigated. Nevertheless, the various statistical mixed logit models that were evaluated yielded unsatisfactory outcomes, resulting in a reduction in the odds ratio of IgG anti-M antibodies and the loss of statistical significance for IgG anti-S and anti-N antibodies (Supplementary Tables S2, S3). Figure 2 depicts the scatterplot resulting from the logit analysis of the three antibodies above described, with the analysed samples labelled according to the presence or absence of lung injury. Upon observation of the scatterplot in relation to IgG anti-M (Fig. 2, panel A), it can be reasonably deduced that the cutoff value utilized in the ELISA test (cutoff = 1.1), which serves to designate the test result as positive, was not appropriate for differentiating between individuals at low or elevated risk of developing lung injury. A new ELISA cutoff was established by employing the optimal Youden index (OD = 1.5)25 and the resulting data were then subjected to a novel χ2 association test. The results (Table 4) indicated a statistically significant association (p < 0.00001) and a larger effect size (Cramer’s V = 0.568) than in the first contingency table. The ELISA with the new cutoff exhibited a specificity of 91% and a sensitivity of 63% in the detection of lung injury.
Fig. 1.
Graphical representation of the three logit functions. The panels illustrate the functions resulting from the estimation of the likelihood of lung injury (ordinate) plotted against the optical density (OD) resulting from the ELISA. As shown in panel A, only the membrane protein resembles a similar “S” shape and achieves the maximum probability (p = 1), while the other two proteins (spike, panel B; nucleocapsid, panel C) display a linear-like behavior and do not reach the maximum probability.
Fig. 2.
Assessment of the optimal ELISA cutoff in lung injury detection. The figure depicts the graphical representation of the logit function obtained for the M, S, and N proteins, with samples labeled according to the presence or absence of lung injury (blue = absence; yellow = presence). The OD value (cutoff) for the ELISA to differentiate individuals at high risk of lung injury (OD = 1.5), as determined by Youden’s index, is shown.
Table 4.
A new contingency table using optimized ELISA data.
| Contingency table | |||
|---|---|---|---|
| ELISA membrane results | Lung injury | ||
| (cutoff, OD = 1.5) | 0 | 1 | Total |
| Neg | 30 | 11 | 41 |
| Pos | 3 | 19 | 22 |
| Total | 33 | 30 | 63 |
| χ2 test | |||
|---|---|---|---|
| Value | df | p | |
| χ2 | 20.3 | 1 | < 0.00001 |
| N | 63 | ||
| Effect size | |||
|---|---|---|---|
| Value | |||
| Cramer’s V | 0.568 |
The new ELISA cutoff was obtained by means of Youden’s index, and the results demonstrate superior performance, primarily due to a high effect size and increased specificity (93%) in predicting lung injury.
Discussion
Despite global effort, the multifactorial nature of COVID-19 has thus far prevented elucidation of the pathogenetic mechanism leading to progressive lung damage26,27. The immunological system appears to play an important role, given the unbalanced immune response often observed in critical patients28,29. The role of the humoral response remains a matter of contention, with several studies reporting conflicting results regarding the correlation between virus-specific antibody titers and the progression to severe disease. The interpretation of the data is challenging because of the differing kinetics of the antibody response observed in patients following infection. A number of studies agree on a correlation between higher IgG antibody titers and more severe disease, with antibody levels sustained during the first 6 months, followed by a decline between six and twelve months. Conversely, in patients with mild or asymptomatic infection, IgG titers are lower, and antibody decay is faster in the first months after infection30–34. Noteworthy, these studies have primarily focused on antibodies against the spike and nucleocapsid proteins. The contribution of the two other structural proteins, the membrane and the envelope, remains substantially unknown.
The objective of the present study was to examine the immunoglobulin response of subjects with a previous SARS-CoV-2 infection and lung injury. The immunological profile of the sample under investigation was evaluated by the detection of antibodies using an in-house ELISA which employed specific portions of the spike, nucleocapsid, and membrane proteins.
A total of 63 subjects were analysed, of whom 30 exhibited evidence of lung injury (Supplementary Table 1). The preliminary analyses (Table 1) indicated a significant association (p < 0.05) between positivity to IgG anti-M and lung injury. However, the strength of this association was low (V = 0.275). The data were subsequently subjected to a binary logistic regression analysis. The results of the logistic regression analysis were consistent with those of the previous analysis and demonstrated a strong correlation between the risk of developing pulmonary lesions and the growing amount of IgG anti-M, with an odds ratio of 6.8 (Table 3A).
Similarly, the logistic regression analysis of the IgG anti-S and anti-N antibodies demonstrated a correlation with lung injury (Table 3B,C), with an odds ratio of 1.83 and 1.67, respectively. Analysis of the potential interactions of these antibodies with IgG anti-M did not, however, indicate any synergistic effects on the pulmonary damage. The results of the logit analysis of IgG anti-M in conjunction with the other two proteins and pulmonary damage produced a loss of statistical significance for IgG anti-S and anti-N. Contextually, the odds ratio for the IgG anti-M decreased from 6.8 to lower values (Supplementary Tables 2, 3). These results may suggest that the elevated levels of antibodies against S and N observed in subjects with lung injury (Table 2) are the consequence of a physiological, protective immune response masked by a concomitant underlying factor, rather than an indicator of pathological processes. Furthermore, as illustrated in Fig. 2 (panels B and C), there is no discernible pattern in the distribution of samples exhibiting or lacking lung injury within a broad range of optical density with respect to the S and N proteins. Conversely, the M protein (Fig. 2, panel A) showed a cluster of samples, primarily from subjects without lung lesions, within a narrow range of optical density (OD) values. The ELISA results with the M protein elaborated through the choice of a more appropriate ELISA cutoff point (OD = 1.5) in relation to lung injury (Table 4), revealed marked statistical significance (p < 0.00001) and an increased effect size (V = 0.568). The assay demonstrated a 91% specificity and a 63% sensitivity, indicating a very low risk of developing lung disease in the absence of antibodies IgG anti-M protein.
The data are also consistent with disease severity. As shown in Fig. 3, by stratifying the subjects on an ordinal scale in relation to lung injury as revealed by image analysis (low: grade 1; moderate: grade 2; high: grade 3), logistic analysis identified 6 out of 9 subjects who required oxygen therapy as having a high predicted probability of developing lung injury: specifically, 4 subjects in grade 3 (p = 0.9–1) and 2 subjects in grade 2 (p = 0.7–0.9).
Fig. 3.
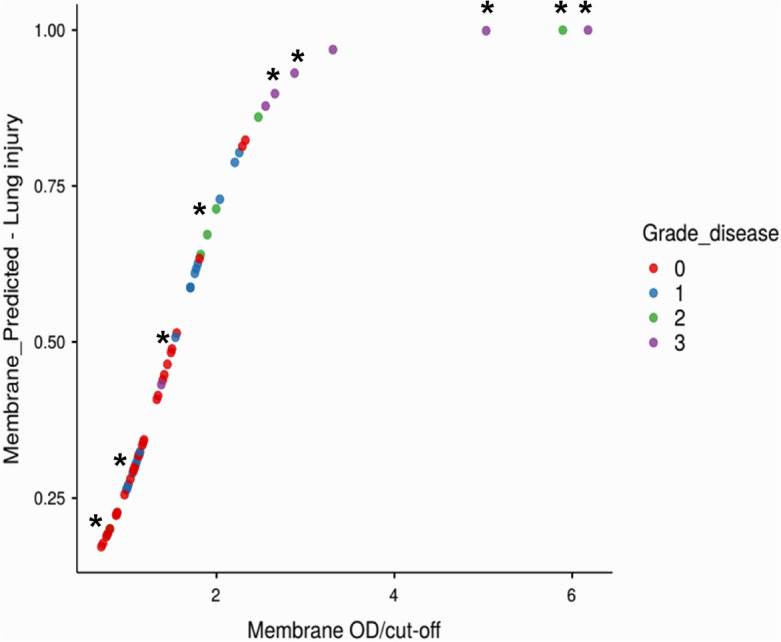
Relationship between disease likelihood and oxygen therapy. The figure shows the logit function obtained for M protein, with samples labelled according to the degree of lung injury as revealed by image analysis. The grading system employed was as follows: G0 (red) = absence of injury; G1 (blue) = low-grade injury; G2 (green) = medium-grade injury; G3 (violet) = high-grade injury. The asterisk indicates subjects who required oxygen therapy.
Regarding the low sensitivity of the ELISA employing the M antigen, it is important to note that the assay was developed using a portion of the protein, which was produced in a prokaryotic system, purified under denaturing conditions and then used with an in-house assay.
Furthermore, given the multifactorial nature of the disease, it is very likely that IgG anti-M is not the only factor involved12,35,36.
A risk factor for disease progression has been identified in the AB0 system. Despite the lack of complete agreement on which blood groups are protective and which are not, the majority of studies have revealed a protective role for the 0 group and a risk factor for the A group37.
The present study included 44 samples with available data on the AB0 system. The results of the association study with lung injury showed a significant relationship between the AB0 system and lung injury, but in contrast to the findings of previous studies. The A group was identified as a protective factor, while the 0 group was identified as a risk factor (χ2 = 10.7; df = 3; Fisher’s exact test, p = 0.0113; Cramer’s V = 0.494) (Supplementary Table 5). No synergistic effects with the ELISA results were observed.
Finally, subjects below 30 years old did not show lung injury (χ2 = 6.93; df = 1; Fisher’ exact test, p = 0.012; Cramer’s V = 0.334) (Supplementary Table 6).
This study has some limitations. As far as the authors know, this is the first description of a relationship between IgG anti-M and the risk of lung injury in subjects infected with SARS-CoV-2. Further studies are required to confirm these findings.
It is well established that levels of anti-S and anti-N antibodies decline over time. In this study, the sample collection period ranged from 15 to 328 days, suggesting the potential for underestimating antibody levels in response to the S and N proteins. Conversely, antibodies against the M protein have been reported to remain stable for at least 1 year38.
The samples were collected during the initial phase of the epidemic, and the recombinant proteins were produced using the circulating strain at the time (Wuhan-Hu-1, Spike_D614G). The subsequent viral variants’ behaviour remains unconfirmed, despite the fact that the viral M gene is well conserved among the different strains24,39.
The potential utility of IgG anti-M as a prognostic marker is contingent upon the temporal dynamics of antibody appearance. As illustrated in Table 1, the interval between molecular test positivity and serum collection ranges from 21 to 328 days, with only five serum samples collected within three weeks following the molecular diagnosis of infection.
It is also not possible to ascertain whether the risk of damage is due to the cross reactivity of IgG anti-M or to the antigen itself. The M protein represents the most abundant structural protein of SARS-CoV-2. It has been demonstrated that the antigen modulates the host immune response by reducing interferon β and λ production. Furthermore, it has been shown that the M protein promotes apoptosis and enhances the permeability of lung cells39–42.
It has recently been reported, however, that IgG anti-M can induce antibody-dependent cell-mediated cytotoxicity (ADCC) in effector cells of the immune system through the interaction of the Fc region with the Fcγ receptor. In addition, it was found that IgG anti-M antibodies from patients with severe COVID-19 were more potent stimulators of cytotoxicity than anti-M antibodies from patients with mild COVID-1943.
Overall, the data indicate a clear correlation between IgG anti-M antibodies and lung injuries, providing compelling evidence that this is an area worthy of further investigation.
Material and methods
Study and samples
The study adopted an observational approach. Samples were collected during the initial phase of the pandemic, from April 2020 to April 2021. They included 63 subjects diagnosed as positive for SARS-CoV-2 infection by molecular analysis of an oro-pharyngeal swab. None of the individuals had been vaccinated. Lung injury was evaluated by imaging techniques, including computed tomography (CT) scans and X-rays. All image analyses were conducted on subjects with respiratory distress or dyspnoea after the positive result of the molecular test. A specialized radiologist made the diagnosis of lung injury. Serum samples were collected and stored at a temperature of − 80 °C. A quantitative immunological profile was established for each participant through the detection of antibodies directed against specific portions of the viral proteins S, N and M, using enzyme-linked immunosorbent assay (ELISA).
Sera from healthy donors collected prior to the onset of the SARS-CoV-2 pandemic were used to set up the ELISA and served as negative controls.
Ethics declarations
The study was conducted in accordance with the Declaration of Helsinki.
All the data in this study were anonymized by deleting all sensitive information. All study participants provided informed consent, as required by Article 40 of Decree-Law No. 23 dated April 8, 2020, in Italy.
Ethical Committee of National Institute of Infectious Diseases (INMI) Lazzaro Spallanzani—IRCCS approved the study (approval number 2/2020).
Production of recombinant proteins
The recombinant viral proteins produced were as follows: Spike S1 (RBD), AA 292–573 (YP_009724390); Nucleocapsid, AA 1–209 (YP_009724397); Membrane: AA 109–222 (YP_009724393). The PCR fragments encoding the three recombinant proteins were retrotranscribed and amplified using the strain SARS-CoV-2 isolated in Italy during the initial epidemic wave (GSAID code: Italy/CDG1/2020|EPI ISL 412973|2020-02-20) as a template. The three RT-PCRs were assembled using the same protocol and amplification program. The reactions were set up in a final volume of 50 μl using the SuperScript™ III One-Step RT-PCR System kit (Thermo Fisher Scientific) with 0.3 μM of each primer and 1 μl of genomic RNA. The amplification programme consisted of a reverse transcription at 50 °C for 30 min, followed by 95 °C for 2 min and then 12 cycles of: 95 °C × 10 s, 58 °C × 30 s (ΔT − 0.5 °C each cycle), 68 °C × 40 s. A further 30 cycles were carried out with the following temperature profile: 95 °C × 10 s, 60 °C × 30 s and 68 °C × 1 min. The primers used are listed in Supplementary Table 4. All oligonucleotides were synthesised by Eurofins Genomics (https://eurofinsgenomics.eu/).
The three fragments were purified and digested with the restriction endonucleases listed in Supplementary Table 4, cloned into the previously opened pRSETC vector by the T4 DNA ligase, and subsequently propagated into the One Shot™ TOP10 chemically competent E. coli cells (Thermofisher). Purified vectors underwent sequencing analysis using the automated DNA sequencer CEQ 8000 (Beckman, Brea, CA, USA) prior to transformation with BL21-AI™ One Shot® Chemically Competent E. coli. Transformed cells were induced for 3 h and His-tagged recombinant proteins were purified by Ni–NTA affinity chromatography kit according to manufacturer’s instructions (QIAGEN, Hilden, Germany). The recombinant proteins were analysed and quantified by polyacrylamide gel electrophoresis (PAGE) and 2100 Bioanalyzer Instrument (Agilent, www.agilent.com). The specificity of the three recombinant proteins was also evaluated by western blot analysis using a monoclonal antibody targeting the poly-His tag.
Enzyme-linked immunosorbent assay (ELISA)
ELISAs were carried out using Costar 96-well plates (Corning, USA) coated with 100 ng/well recombinant protein in carbonate-bicarbonate buffer pH 9.6 for 18 h at 4 °C. The plates were blocked using 300 µl of 5% low-fat milk (www.genespin.com) in wash buffer (WB, 1× PBS buffer, 0.1% Tween20) at room temperature (RT) for a minimum of three hours. The sera samples were diluted 1/100 in 2% milk WB and incubated for 1 h at RT. All samples were run in duplicate. Four wash cycles were performed with 300 µl of WB. Subsequently, the samples were incubated with 100 µl of secondary antibody (peroxidase-conjugated anti-human IgG) diluted at 1:10,000 in WB for 1 h at room temperature. Following four cycles of washing, 100 µl of the substrate (OPD, SIGMA) was added, incubated, and the reaction stopped according to the manufacturer’s instructions. The absorbance was quantified at 490 nm using the VICTOR X4 instrument (PerkinElmer 2023 Multilabel Reader). The cut-off value was calculated by summing the mean of three or more negative sera and adding three times the standard deviation.
Statistical methods
The ELISA results were subjected to a t-test for independent samples (Mann–Whitney U-test), χ2-tests for associations, and binary logistic regressions (logit) using the Jamovi software (https://www.jamovi.org).
Supplementary Information
Acknowledgements
The study was supported by the Italian Ministry of Defense. We would like to thank Colonel Fabrizio Frisoni for his valuable logistical support.
Author contributions
GF and FL designed the study and drafted the manuscript. GF and FM performed statistical analysis and analyzed the data. GF, FM, RDS, EL carried out the experiments. PG performed the diagnosis of lung injury. AM, FM, GP, DM, DC collected and provided the samples. OR critically reviewed the draft and the English language. All authors critically reviewed and approved the final draft.
Data availability
Data are available from the corresponding authors upon reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-78381-8.
References
- 1.Zhu, N. et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med.20, 727–733. 10.1056/NEJMoa2001017 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu, B., Guo, H., Zhou, P. & Shi, Z. L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol.19, 141–154. 10.1038/s41579-020-00459-7 (2021). Erratum in: Nat. Rev. Microbiol.20, 315. 10.1038/s41579-022-00711-2 (2022). [DOI] [PMC free article] [PubMed]
- 3.Koelle, K., Martin, M. A., Antia, R., Lopman, B. & Dean, N. E. The changing epidemiology of SARS-CoV-2. Science11, 1116–1121. 10.1126/science.abm4915 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li, W. et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature27, 450–454. 10.1038/nature02145 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu, Z. & McGoogan, J. M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA7, 1239–1242. 10.1001/jama.2020.2648 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Zhang, J. J., Dong, X., Liu, G. H. & Gao, Y. D. Risk and protective factors for COVID-19 morbidity, severity, and mortality. Clin. Rev. Allergy Immunol.64, 90–107. 10.1007/s12016-022-08921-5 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou, X. et al. Clinical features of 162 fatal cases of COVID-19: A multi-center retrospective study. Emerg. Crit. Care Med.2, 109–115. 10.1097/EC9.0000000000000026 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Owusu, M. et al. Epidemiological profile of SARS-CoV-2 among selected regions in Ghana: A cross-sectional retrospective study. PLoS ONE15, e0243711. 10.1371/journal.pone (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports.
- 10.Faggioni, G. et al. Identification and characterization of SARS-CoV-2 clusters in the EU/EEA in the first pandemic wave: Additional elements to trace the route of the virus. Infect. Genet. Evol.96, 105108. 10.1016/j.meegid.2021.105108 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Islam, M. R. et al. Author Correction: Genome-wide analysis of SARS-CoV-2 virus strains circulating worldwide implicates heterogeneity. Sci. Rep.11, 20568. 10.1038/s41598-021-00133-9 (2021). Erratum for: Sci. Rep.10, 14004 (2020). [DOI] [PMC free article] [PubMed]
- 12.Tavasolian, F. et al. HLA, immune response, and susceptibility to COVID-19. Front. Immunol.11, 601886. 10.3389/fimmu.2020 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu, R. et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet395, 565–574. 10.1016/S0140-6736(20)30251-8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhama, K. et al. Coronavirus disease 2019-COVID-19. Clin. Microbiol. Rev.33, e00028. 10.1128/CMR.00028-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naqvi, A. A. T. et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta Mol. Basis Dis.1866, 165878. 10.1016/j.bbadis.2020.165878 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romano, M., Ruggiero, A., Squeglia, F., Maga, G. & Berisio, R. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells9, 1267. 10.3390/cells9051267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, D. et al. The architecture of SARS-CoV-2 transcriptome. Cell181, 914–921. 10.1016/j.cell.2020.04.011 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardenbrook, N. J. & Zhang, P. A structural view of the SARS-CoV-2 virus and its assembly. Curr. Opin. Virol.52, 123–134. 10.1016/j.coviro.2021.11.011 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann, M. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell181, 271–280. 10.1016/j.cell.2020.02.052 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heald-Sargent, T. & Gallagher, T. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses4, 557–580. 10.3390/v4040557 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kyriakidis, N. C., López-Cortés, A., González, E. V., Grimaldos, A. B. & Prado, E. O. SARS-CoV-2 vaccines strategies: A comprehensive review of phase 3 candidates. NPJ Vaccines6, 28. 10.1038/s41541-021-00292-w (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu, W., Cheng, Y., Zhou, H., Sun, C. & Zhang, S. The SARS-CoV-2 nucleocapsid protein: Its role in the viral life cycle, structure and functions, and use as a potential target in the development of vaccines and diagnostics. Virol. J.20, 6. 10.1186/s12985-023-01968-6 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walls, A. C. et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell181, 281–292. 10.1016/j.cell.2020.02.058 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marques-Pereira, C. et al. SARS-CoV-2 membrane protein: From genomic data to structural new insights. Int. J. Mol. Sci.23, 2986. 10.3390/ijms23062986 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Youden, W. J. Index for rating diagnostic tests. Cancer3, 32–35. 10.1002/1097-0142(1950)3:1%3c32::aid-cncr2820030106%3e3.0.co;2-3 (1950). [DOI] [PubMed] [Google Scholar]
- 26.Gilyazova, I. et al. COVID-19: Mechanisms, risk factors, genetics, non-coding RNAs and neurologic impairments. Noncoding RNA Res.8, 240–254. 10.1016/j.ncrna.2023.02.007 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun, Y., Luo, B., Liu, Y., Wu, Y. & Chen, Y. Immune damage mechanisms of COVID-19 and novel strategies in prevention and control of epidemic. Front. Immunol.14, 1130398. 10.3389/fimmu.2023.1130398 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sette, A. & Crotty, S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell184, 861–880. 10.1016/j.cell.2021.01.007 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zelek, W. M. & Harrison, R. A. Complement and COVID-19: Three years on, what we know, what we don’t know, and what we ought to know. Immunobiology228, 152393. 10.1016/j.imbio.2023.152393 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazzoni, A. et al. Cell-mediated and humoral adaptive immune responses to SARS-CoV-2 are lower in asymptomatic than symptomatic COVID-19 patients. Eur. J. Immunol.50, 2013–2024. 10.1002/eji.202048915 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Rijkers, G. et al. Differences in antibody kinetics and functionality between severe and mild severe acute respiratory syndrome coronavirus 2 infections. J. Infect. Dis.222, 1265–1269. 10.1093/infdis/jiaa463 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucas, C. et al. Delayed production of neutralizing antibodies correlates with fatal COVID-19. Nat. Med.27, 1178–1186. 10.1038/s41591-021-01355-0 (2021). Erratum in: Nat. Med.27, 1309. 10.1038/s41591-021-01416-4 (2021). [DOI] [PMC free article] [PubMed]
- 33.Legros, V. et al. A longitudinal study of SARS-CoV-2-infected patients reveals a high correlation between neutralizing antibodies and COVID-19 severity. Cell Mol. Immunol.18, 318–327. 10.1038/s41423-020-00588-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abril, A. G. et al. Author Correction: Titers of IgG and IgA against SARS-CoV-2 proteins and their association with symptoms in mild COVID-19 infection. Sci. Rep.14, 15918. 10.1038/s41598-024-66711-9 (2024). Erratum for: Sci. Rep.14, 12725. 10.1038/s41598-024-59634-y (2024). [DOI] [PMC free article] [PubMed]
- 35.COVID-19 Host Genetics Initiative. Mapping the human genetic architecture of COVID-19. Nature600, 472–477. 10.1038/s41586-021-03767-x (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schultze, J. L. & Aschenbrenner, A. C. COVID-19 and the human innate immune system. Cell184, 1671–1692. 10.1016/j.cell.2021.02.029 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pereira, E. et al. ABO blood group and link to COVID-19: A comprehensive review of the reported associations and their possible underlying mechanisms. Microb. Pathog.169, 105658. 10.1016/j.micpath.2022.105658 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amjadi, M. F. et al. Anti-membrane antibodies persist at least one year and discriminate between past coronavirus disease 2019 infection and vaccination. J. Infect. Dis.28, 1897–1902. 10.1093/infdis/jiac263 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bianchi, M. et al. Sars-CoV-2 envelope and membrane proteins: Structural differences linked to virus characteristics?. Biomed. Res. Int.2020, 4389089. 10.1155/2020/4389089 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong, N. A. & Saier, M. H. The SARS-coronavirus infection cycle: A survey of viral membrane proteins, their functional interactions and pathogenesis. Int. J. Mol. Sci.22, 1308. 10.3390/ijms22031308 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang, Y. et al. SARS-CoV-2 membrane protein causes the mitochondrial apoptosis and pulmonary edema via targeting BOK. Cell Death Differ.29, 1395–1408. 10.1038/s41418-022-00928-x (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng, G., Qiu, G., Qian, H., Shu, Q. & Xu, J. Multifaceted role of SARS-CoV-2 structural proteins in lung injury. Front. Immunol.15, 1332440. 10.3389/fimmu.2024.1332440 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernández-Soto, D. et al. SARS-CoV-2 membrane protein-specific antibodies from critically ill SARS-CoV-2-infected individuals interact with Fc receptor-expressing cells but do not neutralize the virus. J. Leukoc. Biol.115, 985–991. 10.1093/jleuko/qiae017 (2024). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available from the corresponding authors upon reasonable request.