Abstract
The increased incidence of inflammatory diseases, infectious diseases, autoimmune disorders, and tumors in elderly individuals is closely associated with several well-established features of immunosenescence, including reduced B cell genesis and dampened immune responses. Recent studies have highlighted the critical role of dual receptor lymphocytes in tumors and autoimmune diseases. This study utilized shared data generated through scRNA-seq + scBCR-seq technology to investigate the presence of dual receptor-expressing B cells in the peritoneum of mouse and peripheral blood of healthy volunteers, and whether there are age-related differences in dual receptor B cell populations. In the peritoneum of mice, a high proportion of B cells expressing dual receptors, predominantly dual κ chains, was observed. Notably, there was an increase in dual BCR B cells in elderly mice. Subsequent analysis revealed that the elevated dual BCR B cells in elderly mice primarily originated from B1 cells.Consistent with the results we observed in healthy volunteers of different ages. Furthermore, these cells exhibited differential expressed genes compared to single BCR B cells, including Vim, Ucp2, and Zcwpw1.These findings support a hypothesis that age-related immune changes encompass not only alterations in B cell numbers but also qualitative changes in BCR diversity. Further exploration of the elevated dual BCR B cells in the elderly population can elucidate their function and their association with immune tolerance, revealing their potential role in maintaining immune surveillance and responding to age-related immune challenges.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12979-024-00493-6.
Keywords: Immunosenescence, Dual BCR B cell, Aging, scRNA-seq, scBCR-seq
Introduction
Immunosenescence refers to the gradual deterioration of the immune system that occurs with aging, characterized by changes in both innate and adaptive immunity [1]. These changes include alterations in the composition and function of immune cells, decreased responsiveness to antigens, and impaired immune surveillance against infections and tumors [2, 3]. As individuals age, the diversity of the T and B cell receptor (TCR and BCR) repertoire diminishes, which can impair the immune response to new infections and reduce the efficacy of vaccinations [4, 5]. Understanding how aging impacts T and B cell receptor diversity and function is crucial for developing strategies to enhance immune function and manage age-associated immune disorders.
Clonal selection theory forms the foundation of T and B cell development, tolerance selection, and specific immune responses by proposing that each lymphocyte expresses a receptor (TCR or BCR) derived from a single allele, ensuring recognition of unique antigens [6]. However, recent studies have identified T and B cells expressing multiple receptors, known as dual TCR T cells and dual BCR B cells. These dual receptor cells have been primarily associated with tumor and autoimmune diseases such as type 1 diabetes [7], rheumatoid arthritis [8], systemic lupus erythematosus [9], and plasmacytoma [10]. Given that these diseases have a higher prevalence in the elderly, investigating dual receptor lymphocytes in this age group is crucial for understanding their potential role in age-related immune dysregulation. Our analysis of thymic and peripheral lymphoid organs in humans and mice has revealed elevated proportions of dual TCR T cells, and we observed a significant increase in the proportion of dual TCR T cells in elderly populations [11, 12]. These findings highlight the potential role of dual receptor cells in modifying immune function during aging. However, it remains unclear whether dual receptor B cells undergo similar changes with age, warranting further investigation into their dynamics over the aging process.
Since scRNA-seq primarily focuses on gene expression levels and does not provide detailed sequence information on BCR clonotypes and complementarity-determining regions (CDRs), which are key to understanding the specificity and function of B cells.However, the revolutionary techniques of scRNA-seq combined with scBCR-seq have greatly facilitated the analysis of the homogeneity and heterogeneity of B cells across time and space [13, 14]. These methods enable the simultaneous acquisition of a large repertoire of single B cell BCRs, specifically the paired CDR3 regions of the heavy (H) and light (L) chains. Utilizing this technology, several research teams have explored age-related B cell subpopulations and their clonality in various tissues, including peritoneal B cells [15], peripheral blood B cells [16], meningeal B cells [17], and B cells in the lungs and lung-associated lymph nodes [18].
In this study, we conducted a comprehensive analysis of data from both human and mouse. For the mouse data, we utilized the shared scRNA + BCR-seq dataset from the European Bioinformatics Institute (EBI) ArrayExpress(accession number: E-MTAB-10081), which includes samples from newborn mice (day 6), young adult mice (week 10), and aged mice (month 15). For the human data, we leveraged two shared scRNA + BCR-seq datasets, with samples including six healthy pediatric samples (accession number: GSE166489) and eight healthy adult samples (accession number: GSE165080). Supplementary Tables 1–4 provide detailed information for each human and mouse sample, including accession numbers and basic sample information, as well as BCR, dual BCR, and non-paired counts and proportions for individual B1 and B2 cell subgroups.
Materials and methods
Single-cell sequencing datasets of B cells
The scRNA + BCR-seq data for B cells from humans and mice were obtained from the article [15, 19, 20].This study utilized three scRNA-seq and scBCR-seq datasets from the Gene Expression Omnibus (GEO) repository and the ArrayExpress database, encompassing peritoneal B cells from C57BL/6 wild-type mice across three different age groups (E-MTAB-10081), peripheral blood B cells from healthy children (GSE166489), and peripheral blood B cells from healthy adults (GSE165080).The peritoneal B cell samples from mice (three samples in total) were collected from peritoneal tissues at Day 6, Week 10, and Month 15. The healthy children (six samples in total) had an average age of 8.4 ± 10 years, while the healthy adults (eight samples in total) had an average age of 34 ± 15 years.The accession number, basic sample information and data composition information are shown in Supplementary Tables 1–4.
Screening of B cells
The analysis steps for the shared raw scBCR-seq data table for each sample are as follows:1.Removal of non-functional sequences: Sequences with “is_cell” marked as “FALSE”, Sequences with “high_confidence” marked as “FALSE”, Sequences where “chain” is not “IGH, IGK, IGL", Sequences in “productive” that are marked as “None, FALSE”. 2.Statistics of single BCR B cells in a single B cell: (1)H + K; (2)H + L; 3.Statistics of dual BCR B cells in a single B cell: (1)H + L1 + L2; (2)H + K1 + K2; (3)H + K + L; (4)others: H1 + H2 + K; H1 + H2 + K + L;H1 + H2 + L, H1 + H2 + K1 + K2 etc.
Data analysis process and quality control
We first used R (version 4.2.3) and Seurat package (version 5.1.0) to analyze the quality control of the scRNA-seq data for each sample:
Filter the low-quality cells: cells from mice with higher than 13% of mitochondrial contents and cells with fewer than 500 UMIs or fewer than 200 features; cells from children with less than 5% of mitochondrial contents and cells of more than 1000 but fewer than 20,000 UMIs or fewer than 500 features; cells from adults with less than 5% of mitochondrial contents and cells of more than 500 UMIs or fewer than 500 features.
The DoubletFinder software package (version 2.0.4) was used to find and eliminate possible doublets, and filtering was not included in the analysis.
Based on the barcode of each cell within each sample, B cells that meet the quality control criteria for both scRNA-seq and scBCR-seq are selected. This ensures that only B cells with complete data for both RNA sequencing and B cell receptor sequencing are included in the subsequent analysis.
Cluster and single and dual BCR B1 and B2 cell transcription factor expression analysis of single-cell transcriptome data
The single-cell transcriptomic data after quality control were analyzed using R language (version 4.2.3) and the Seurat software package (version 4.4.0). Principal component analysis (PCA) was performed using the JackStraw algorithm and the PCEIbow plotting function. Subsequently, the selected principal components were subjected to Uniform Manifold Approximation and Projection (UMAP) for dimensionality reduction and clustering analysis. The UMAP method is used to map high-dimensional data onto a two-dimensional or three-dimensional space, allowing for the visualization and differentiation of cellular clusters.
Using the markers provided in the raw data, the pre-B, B1, and B2 subgroups of cells from mice on Day 6, Week 10, and Month 15 were annotated as follows: (1) Pre-B cell markers include genes such as Vpreb3, Sdc4, Sys1, etc.; (2) B1 cell markers include genes such as Bhlhe41 (also known as Plzf), Cd9, Zbtb32, etc.; (3) B2 cell markers include genes such as Fcer2a, Ighd, etc. However, due to the extremely low proportion of B1 cells in human peripheral blood, we were unable to cluster a B1 cell subgroup for analysis.For the data of children and adult individuals, they are divided based on the expression of different differential genes: (1)naive B cell makers: IL4R, (2)memory B cell makers: IGHA1,CRIP2,ITGB1;(3)marginal B cell makers: FCRL5,FGR. The cell clusters and gene expression profiles of humans and mice are shown in Supplementary Fig. 1.
To visualize B cell subgroup information using ggplot, the following steps can be taken: 1.Map the different B cell subgroups identified by their BCR pairings to the cluster: (1)H + K; (2)H + L3;H + L1 + L2;(4)H + K1 + K2; (5)H + K + L;(6) others; (7) non-pairing; 2.Map the types of BCR B cells to the cluster:(1)single BCR (2)dual BCR (3)non-pairing BCR;
For the additional analysis comparing the transcription factors between single and dual BCR B cells across three age groups in mice.
Statistical analyses
Data are presented as mean ± SD.Statistical analysis was performed using IBM SPSS Statistics 26 software.For the comparison of continuous variables between two groups, an independent samples T-test was used. For the comparison of categorical variables, a chi-square test was employed. A p-value of less than 0.05 was considered to indicate statistical significance. Data visualization was performed using R Studio (version 3.3.3), Origin (version 2022), and GraphPad Prism (version 5) software.
Results
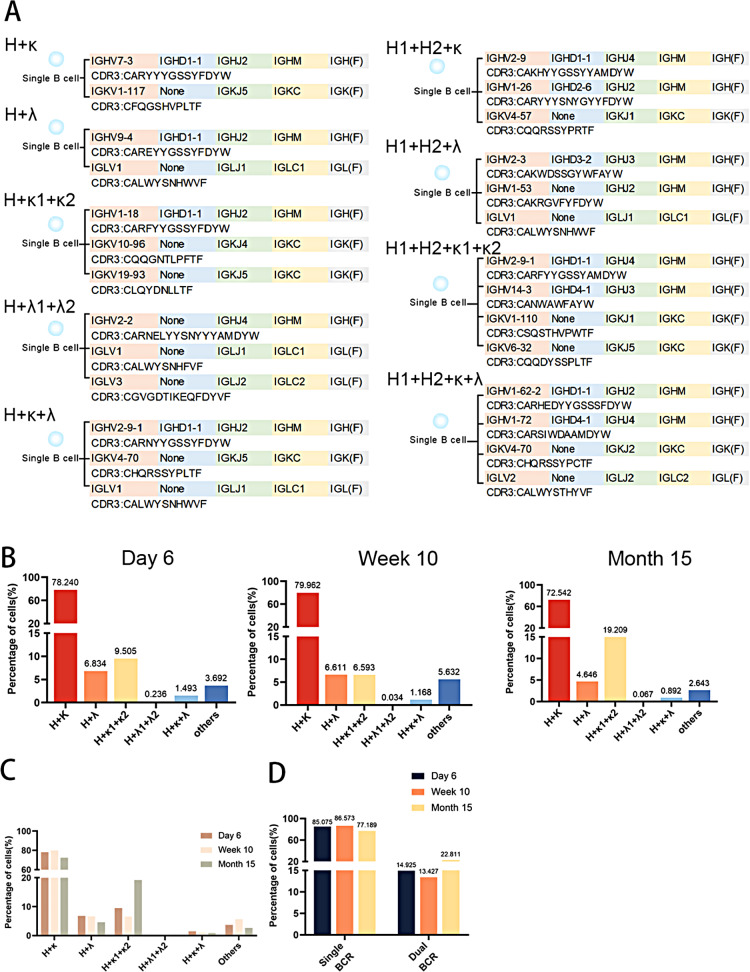
By analyzing the expression of heavy and light chains in single B cells, B cells expressing multiple heavy (H) and light (L, including κ and λ) chains were identified (Fig. 1A). Based on the pairing of BCR heavy and light chains, peritoneal total B cells were categorized into six types, detectable in mice aged 6 days, 10 weeks, and 15 months (Fig. 1B). All mice predominantly exhibited the H + κ type, with dual light chain expressions mainly comprising the H + κ1 + κ2 type. Interestingly, the numbers of H + κ and H + λ B cells decreased in elderly mice, while B cells expressing dual κ chains increased markedly (Fig. 1C). The classification of peritoneal B cells into single BCR B cells (H + κ and H + λ types) and dual BCR B cells (remaining types) revealed an increase in the number of dual BCR B cells in elderly mice compared to those at 6 days and 10 weeks of age (Fig. 1D). The accumulation of dual BCR B cells in older individuals could reflect an adaptive response to maintain immune surveillance, albeit with a potentially broader and less specific repertoire.
Fig. 1.
Analysis of Heavy and Light Chain Expression in Single B Cells Reveals Diversity of Peritoneal B Cells. (A) Identification of B cells expressing multiple heavy (H) and light (κ and λ) chains by single-cell analysis. (B) Classification of peritoneal B cells into six types based on BCR heavy and light chain pairing in mice aged 6 days, 10 weeks, and 15 months. (C) Decrease in H + κ and H + λ B cells and increase in dual κ chain-expressing B cells in elderly mice. (D) Increase in dual BCR B cells in elderly mice compared to 6-day-old and 10-week-old mice
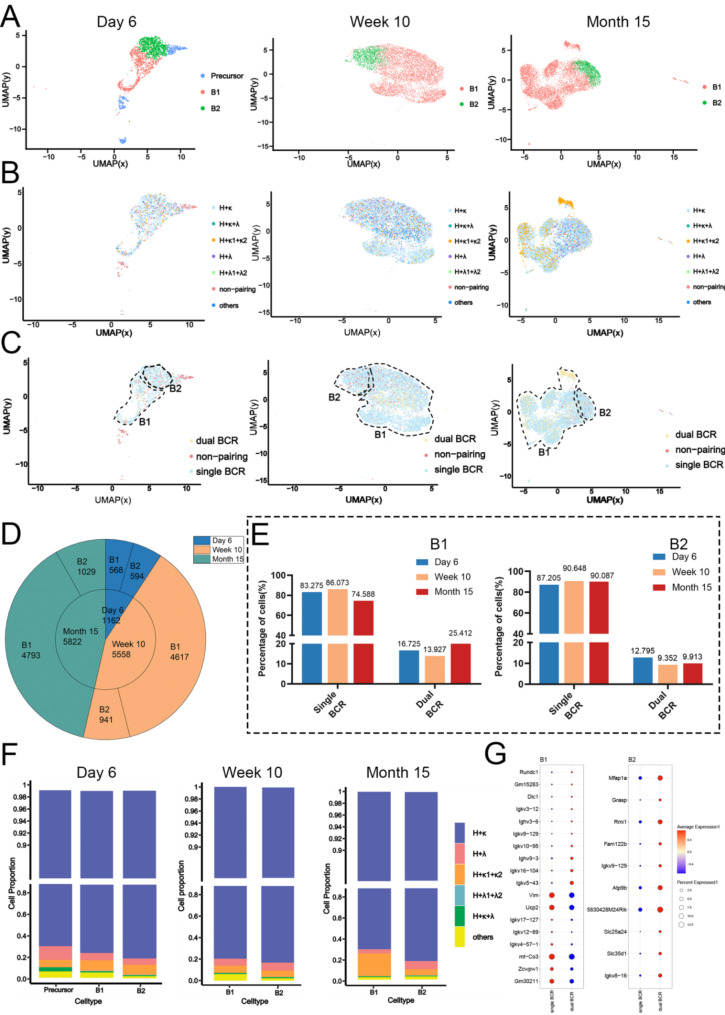
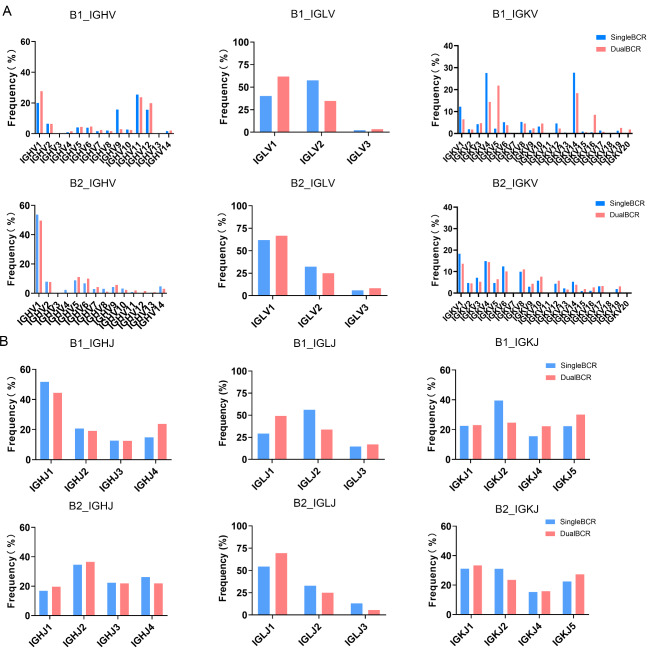
Peritoneal B cells include both B1 and B2 cells. We first strictly perform dimensionality reduction and clustering on the scRNA-seq data from mice of three different age groups.Based on the expression of marker genes, we identified B1 and B2 cell subgroups in all three groups (Fig. 2A) and mapped the various BCR combinations and single/dual BCR statuses to the B1 and B2 subgroups. Each subgroup in the three groups contains both single BCR and dual BCR B cells (Fig. 2B). Subsequently, to more intuitively observe the distribution of single, dual BCR, and unpaired cells, we categorized the BCR pairing into single BCR, dual BCR, and non-pairing (Fig. 2C). The number of B1 and B2 cells showed little difference in neonatal mice, but at 10 weeks and 15 months, B1 cells outnumbered B2 cells in the peritoneum (Fig. 2D). Notably, the proportion of dual BCR B1 cells at 15 months was much higher than in the other groups, while the proportion of dual BCR B2 cells was lower compared to 6-day and 10-week-old mice (Fig. 2E), indicating that the increase in dual BCR B cells in the peritoneum of 15-month-old mice (Fig. 1D) is primarily derived from the B1 cell population. Additionally, analysis of the BCR chain expression in peritoneal B1 cells revealed a substantial increase in H + κ1 + κ2 B1 cells in 15-month-old mice (Fig. 2F). These findings suggest that the disruption of B cell receptor composition in the peritoneum of aged mice mainly occurs within the B1 cell population. Differential gene expression analysis showed a significant decrease in Vim, Ucp2, and Zcwpw1, which are associated with cell migration capacity and metabolic regulation, in dual BCR B1 cells compared to single BCR B1 cells (Fig. 2G).Additionally, since many differential genes are germline genes, it may indicate differences in the usage of V genes.We have conducted a comparative analysis of the differences in VJ usage between single BCR B and dual BCR B cells (Fig. 3).We found that in the usage of V genes, B1 cells frequently utilize the IGHV1, 11, 12 families, while B2 cells only frequently utilize IGHV1 families. In the usage of IGKV genes, B1 cells frequently utilize the IGKV4, 14 families, whereas B2 cells show no significant preference. Regarding the usage of J genes, B1 cells frequently utilize IGHJ1 and IGLJ2 compared to B2 cells.
Fig. 2.
Changes in Single and Dual BCR B1 and B2 Cells in the Peritoneum. (A) Categorization of peritoneal B cells into B1 and B2 subsets. (B) Mapping of various BCR combinations within B1 and B2 subsets. (C) Distribution of single and dual BCR statuses in B1 and B2 cells. (D) Comparison of B1 and B2 cell numbers in neonatal, 10-week-old, and 15-month-old mice. (E) Proportion of dual BCR B1 and B2 cells at different ages. (F) Increase in H + κ1 + κ2 B1 cells in 15-month-old mice. (G) Differential gene expression analysis in dual BCR B cells compared to single BCR B cells
Fig. 3.
V and J Gene Usage in B1 and B2 Cells. (A) Distribution of V gene usage in B1 and B2 cells. (B) Distribution of J gene usage in B1 and B2 cells
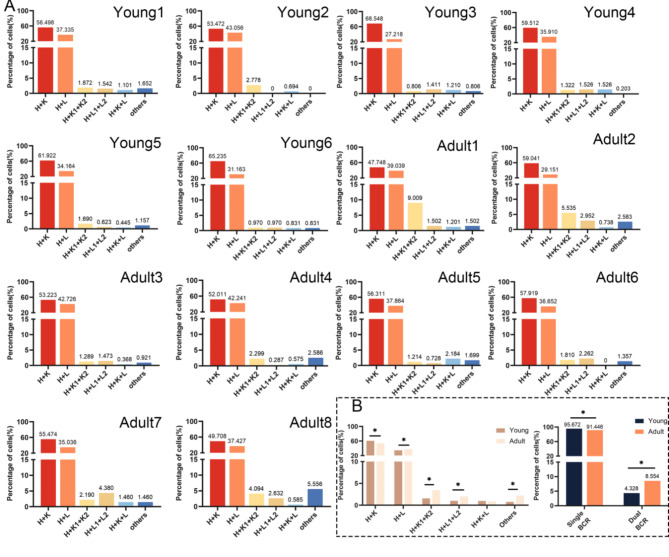
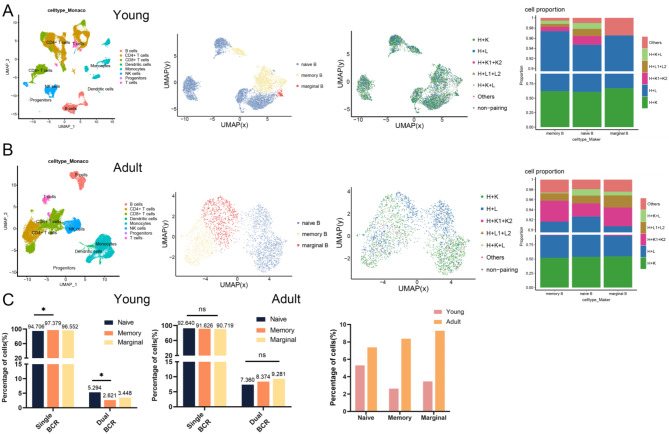
To further investigate the changes in single and dual BCR B cells in different ages, we analyzed scRNA + scBCR-seq data from two groups of healthy children and adult individuals. Figure 4A shows the proportion of BCR types in samples of different ages. The results indicate that the primary BCR pairing types in both children and adult individuals are H + κ, with dual BCR predominantly being H + κ1 + κ2, consistent with previous mouse analyses. Notably, in adult samples, there is an increase in cells with the H + κ1 + κ2 and Other pairing types, which is significantly different from the children’s group (Fig. 4B). Similarly, when we statistically analyzed the differences between single BCR and dual BCR B cells in total B cells, we found significant differences between the two age groups. However, due to the extremely low proportion of B1 cells in human peripheral blood, we were unable to cluster a B1 cell subgroup for analysis.We then divided B cells from both groups into three subgroups based on the expression of different differential genes: naive, memory, and marginal B cells, to compare the changes in single and dual BCR B cells within each subgroup (Fig. 5A and B). The results show that the increase in dual BCR B cells in children is mainly derived from naive B cells, but no significant difference is observed in the adult group. To compare the changes in dual BCR B cells across the three cell subgroups, we statistically analyzed the dual BCR B cells in children and adult groups and found a significant increase in the proportion of dual BCR B cells in the adult group (Fig. 5C).
Fig. 4.
Proportions of single and dual BCR B cells from children and adult. (A) Proportions and types of single BCR B cells and dual BCR B cells in health children and adult.(B) Statistical analysis of 6 types of dual BCR B cells in each group and single/dual BCR B cells among two groups
Fig. 5.
Distribution and proportions of single and dual BCR B cells across different subgroups. (A) The UMAP plot illustrates the distribution of 3 B cell subgroups of children; The figure depicts the distribution and proportion of 7 types of BCR B cells of children (B) The UMAP plot illustrates the distribution of 3 B cell subgroups of adult; The figure depicts the distribution and proportion of 7 types of BCR B cells of adult. (C) Statistical analysis reveals the proportions of total single and total dual BCR B cells of children and adult. *P < 0.05
Discussion
B1 cells primarily participate in innate immune responses, quickly recognizing and responding to pathogens. The peritoneum, as a major reservoir of B1 cells, provides a unique microenvironment where these cells can promptly detect and react to intraperitoneal pathogens. Due to the proportion of dual BCR B cells reported by transgenic mouse models and FCM technology in physiological and pathological states being very different, ranging from 1 to 30% [21].In order to analyze the universality of dual BCR B cells, utilizing scRNA + scBCR-seq technology to analyze peritoneal B cells from three groups of mice, we found that the proportion of dual BCR B1 cells (20%) in the mouse peritoneum is higher than that of dual BCR B2 cells (10%), and this proportion tends to increase with age. Additionally, in our previous work, we have observed that the proportion of dual BCR B cells in various tissue samples from mice, including bone marrow and peripheral blood, is approximately 20%. Notably, the proportion of dual BCR B cells in humans is significantly lower than in mice.This trend is consistent with our previous findings in human and mouse [11, 12, 22], suggesting that receptor rearrangement disruptions occur in both T and B cells during early life and old age. This dysregulation may be linked to the decreased efficiency of central tolerance mechanisms in elderly individuals, which typically eliminate self-reactive B cells during early development [23].Given these findings, it is evident that the changing characteristics and significance of the proportion of dual BCR B cells with increasing age in humans and mice warrant further investigation. Such studies should be conducted with larger sample sizes and across a broader range of tissues to better understand the implications of these changes. Additionally, we found that all mouse expressed dual light chains as H + κ1 + κ2, and the predominance of dual κ chains suggests that allelic exclusion may not be stringent during BCR light chain rearrangement, and that the rearrangement of κ chains could be favored over λ chains, which helps in more effectively recognizing a broader range of antigens [24]. At the same time, we found significant differences in gene expression in dual BCR B1 cells from aged mice, including the downregulation of genes such as Vim, Ucp2, and Zcwpw1. These differentially expressed genes may have a substantial impact on B cell function, affecting processes such as cell migration, metabolism, and signal transduction. For instance, the downregulation of the Vim gene may diminish the migratory capacity of the cells [25], while changes in the expression of the Ucp2 gene could be associated with the metabolic regulation of the cells [26]. These alterations may be related to the decline in B cell function during the process of immunosenescence and might also be linked to the increased susceptibility of the elderly to infections and the elevated risk of developing autoimmune diseases.
In the analysis of dual BCR B cells in peripheral blood samples from healthy volunteers of different ages, we found a consistent trend similar to that observed in mouse, namely that the proportion of dual BCR B cells in human increases with age. The increase in H + κ1 + κ2 dual BCR B cells in human peripheral blood samples aligns with the trends observed in mouse, suggesting that this trend may be conserved across different species. However, due to the extremely low proportion of B1 cells in human peripheral blood, we were unable to cluster a B1 cell subgroup for analysis. In this study, we compared the proportions and characteristics of dual BCR B cells in naïve B cells, memory B cells, and marginal B cells in the peripheral blood of healthy children and adult individuals. We found that the proportion of dual BCR B cells in naïve B cells is the highest. With increasing age, the proportion of dual BCR B cells increases, suggesting a potential correlation with immune senescence. This insight may provide new perspectives and research directions for studying the relationship between B cells and aging.
In summary, our study highlights the changes in the B cell repertoire with age, particularly the emergence of dual BCR B cells. Further research is needed to understand the functional significance of these cells and their potential role in immune regulation associated with aging. By elucidating the mechanisms underlying the development of dual BCR B cells, we can gain valuable insights into the complex interplay between aging and immune function.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Luo Y, Ma J, Anjali Ramaswamy and colleagues for sharing their single-cell RNA sequencing and single-cell BCR sequencing data, which were instrumental in our research.
Author contributions
HW, JL, and YX contributed equally to this work and are considered co-first authors. HW and YX involved in the design of the study, data collection, and data analysis. JL carried out the gene expression analyses and contributed to the writing of the manuscript. XY supervised the project, provided critical feedback on the experimental design and data interpretation, and was responsible for the final approval of the manuscript. All authors contributed to reviewing and editing the final manuscript.
Funding information
This study was supported by the National Natural Science Foundation of China (82160279) and the Guizhou Provincial Hundred level Talent Fund [No (2018)5637].
Data availability
The data analyzed in this research are provided in the supplementary materials accompanying this manuscript. Accession numbers and details of the scRNA + BCR-seq data of mice can also be accessed through the public database ArrayExpress (https://www.ebi.ac.uk/arrayexpress/E-MTAB-10081).. The original and processed data related to healthy children in this article can be obtained from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE166489. The original and processed data related to healthy adult samples can be obtained from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE165080.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Huifang Wang, Jun Li and Yuanyuan Xu are joint first authors.
References
- 1.Zhou D, Borsa M, Simon AK. Hallmarks and detection techniques of cellular senescence and cellular ageing in immune cells. Aging Cell. 2021;20:e13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Z, Liang Q, Ren Y, Guo C, Ge X, Wang L, et al. Immunosenescence: molecular mechanisms and diseases. Signal Transduct Target Ther. 2023;8:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikolich-Žugich J. The twilight of immunity: emerging concepts in aging of the immune system. Nat Immunol. 2018;19:10–9. [DOI] [PubMed] [Google Scholar]
- 4.Amoriello R, Mariottini A, Ballerini C. Immunosenescence and Autoimmunity: exploiting the T-Cell receptor repertoire to investigate the impact of aging on multiple sclerosis. Front Immunol. 2021;12:799380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunn-Walters DK, Ademokun AA. B cell repertoire and ageing. Curr Opin Immunol. 2010;22:514–20. [DOI] [PubMed] [Google Scholar]
- 6.Burnet FM. A modification of Jerne’s theory of antibody production using the concept of clonal selection. CA Cancer J Clin. 1976;26:119–21. [DOI] [PubMed] [Google Scholar]
- 7.Ahmed R, Omidian Z, Giwa A, Cornwell B, Majety N, Bell DR, et al. A public BCR Present in a unique dual-receptor-expressing lymphocyte from type 1 diabetes patients encodes a potent T cell Autoantigen. Cell. 2019;177:1583–e159916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradley CP, Teng F, Felix KM, Sano T, Naskar D, Block KE, et al. Segmented filamentous Bacteria provoke lung autoimmunity by Inducing Gut-Lung Axis Th17 cells expressing dual TCRs. Cell Host Microbe. 2017;22:697–e7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sang A, Danhorn T, Peterson JN, Rankin AL, O’Connor BP, Leach SM, et al. Innate and adaptive signals enhance differentiation and expansion of dual-antibody autoreactive B cells in lupus. Nat Commun. 2018;9:3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuoka R, Sakamoto N, Kato T, Chiba S, Noguchi M. A case of solitary plasmacytoma of bone showing co-expression of both immunoglobulin light chains. Eur J Med Res. 2021;26:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jun L, Lanwei Z, Jiayi W, Xinsheng Y. A new immunological index for the elderly: high proportion of multiple TCR T cells based on scRNA-seq. Aging Dis. 2023. 10.14336/AD.2023.0509. [DOI] [PubMed] [Google Scholar]
- 12.Zhu L, Peng Q, Li J, Wu Y, Wang J, Zhou D, et al. scRNA-seq revealed the special TCR β & α V(D)J allelic inclusion rearrangement and the high proportion dual (or more) TCR-expressing cells. Cell Death Dis. 2023;14:487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. 2018;18:35–45. [DOI] [PubMed] [Google Scholar]
- 14.He J, Shen J, Luo W, Han Z, Xie F, Pang T, et al. Research progress on application of single-cell TCR/BCR sequencing technology to the tumor immune microenvironment, autoimmune diseases, and infectious diseases. Front Immunol. 2022;13:969808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo Y, Wang J, Li K, Li M, Xu S, Liu X, et al. Single-cell genomics identifies distinct B1 cell developmental pathways and reveals aging-related changes in the B-cell receptor repertoire. Cell Biosci. 2022;12:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu M-Z, Wang C, Wu Z, Zhao Q, Zhao Z, Huang C-Y, et al. Dynamic single-cell mapping unveils Epstein–Barr virus-imprinted T-cell exhaustion and on-treatment response. Signal Transduct Target Ther. 2023;8:370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brioschi S, Wang W-L, Peng V, Wang M, Shchukina I, Greenberg ZJ, et al. Heterogeneity of meningeal B cells reveals a lymphopoietic niche at the CNS borders. Science. 2021;373:eabf9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.M R, G J, K, R, O H, L L, F L, et al. Induction of bronchus-associated lymphoid tissue is an early life adaptation for promoting human B cell immunity. Nat Immunol. 2023. 10.1038/s41590-023-01557-3. [DOI] [PMC free article] [PubMed]
- 19.Ramaswamy A, Brodsky NN, Sumida TS, Comi M, Asashima H, Hoehn KB, et al. Immune dysregulation and autoreactivity correlate with disease severity in SARS-CoV-2-associated multisystem inflammatory syndrome in children. Immunity. 2021;54:1083–e10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma J, Bai H, Gong T, Mao W, Nie Y, Zhang X, et al. Novel skewed usage of B-cell receptors in COVID-19 patients with various clinical presentations. Immunol Lett. 2022;249:23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pelanda R. Dual immunoglobulin light chain B cells: Trojan horses of autoimmunity? Curr Opin Immunol. 2014;27:53–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu L, Peng Q, Wu Y, Yao X. scBCR-seq revealed a special and novel IG H&L V(D)J allelic inclusion rearrangement and the high proportion dual BCR expressing B cells. Cell Mol Life Sci. 2023;80:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fong S, Chen PP, Vaughan JH, Carson DA. Origin and age-associated changes in the expression of a physiologic autoantibody. Gerontology. 1985;31:236–50. [DOI] [PubMed] [Google Scholar]
- 24.Mostoslavsky R, Alt FW, Rajewsky K. The lingering enigma of the allelic exclusion mechanism. Cell. 2004;118:539–44. [DOI] [PubMed] [Google Scholar]
- 25.Eckes B, Colucci-Guyon E, Smola H, Nodder S, Babinet C, Krieg T, et al. Impaired wound healing in embryonic and adult mice lacking vimentin. J Cell Sci. 2000;113(Pt 13):2455–62. [DOI] [PubMed] [Google Scholar]
- 26.Brand MD, Esteves TC. Physiological functions of the mitochondrial uncoupling proteins UCP2 and UCP3. Cell Metab. 2005;2:85–93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data analyzed in this research are provided in the supplementary materials accompanying this manuscript. Accession numbers and details of the scRNA + BCR-seq data of mice can also be accessed through the public database ArrayExpress (https://www.ebi.ac.uk/arrayexpress/E-MTAB-10081).. The original and processed data related to healthy children in this article can be obtained from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE166489. The original and processed data related to healthy adult samples can be obtained from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE165080.