Abstract
Increasing reports of chloroquine resistance (CQR) in Plasmodium vivax endemic regions have led to several countries, including Indonesia, to adopt dihydroarteminsin-piperaquine instead. However, the molecular drivers of CQR remain unclear. Using a genome-wide approach, we perform a genomic analysis of 1534 P. vivax isolates across 29 endemic countries, detailing population structure, patterns of relatedness, selection, and resistance profiling, providing insights into potential drivers of CQR. Selective sweeps in a locus proximal to pvmdr1, a putative marker for CQR, along with transcriptional regulation genes, distinguish isolates from Indonesia from those in regions where chloroquine remains highly effective. In 106 isolates from Indonesian Papua, the epicentre of CQR, we observe an increasing prevalence of novel SNPs in the candidate resistance gene pvmrp1 since the introduction of dihydroartemisinin-piperaquine. Overall, we provide novel markers for resistance surveillance, supported by evidence of regions under recent directional selection and temporal analysis in this continually evolving parasite.
Subject terms: Malaria, Genomics
The molecular determinants of chloroquine resistance in Plasmodium vivax are unclear. Here, Ngwana-Joseph et al. identify genomic loci under putative selection in a chloroquine-resistant population, with findings including an association with the ABC transporter family member, pvmrp1.
Introduction
Plasmodium vivax is the most geographically widespread human malaria parasite and the leading cause of malaria outside of sub-Saharan Africa. In 2022, there were 6.9 million cases across 49 endemic countries across Central and South America, East Africa, Asia, and Oceania1. Intensive efforts to combat the deadlier Plasmodium falciparum, particularly in areas that are co-endemic with P. vivax, has distributed resources away from P. vivax control programs, leading to its emergence as the dominant species, particularly in the Greater Mekong Subregion1. The absence of a long term continuous in vitro culture system2 has meant that our understanding of the parasite’s life cycle, transmission, and biology has been limited.
Chloroquine, in combination with primaquine, is the front-line treatment for the radical cure of P. vivax malaria in most endemic countries. First documented in the late 1980s, chloroquine resistance (CQR) in P. vivax, also known as chloroquine treatment failure, is characterised by the World Health Organization (WHO) as the persistence of parasitaemia on day 28 following treatment, despite a blood concentration of chloroquine-desethylchloroquine at or above 100 ng/mL. CQR has consequently led to the adoption of artemisinin-based combination therapies to replace chloroquine in several countries, including dihydroartemisinin-piperaquine in Indonesia, artesunate-mefloquine in Cambodia, and artemether-lumefantrine in Papua New Guinea (PNG)3,4. Indonesian Papua and PNG have been the epicentre of high-grade, or category 1 CQR, defined as >10% recurrences by day 28 of treatment5. Over the past two decades, increasing reports of CQR in P. vivax beyond these countries have been characterised by parasite persistence 28 days post-treatment, spurring research into the molecular determinants of CQR5–9.
Evidence regarding the molecular drivers of CQR in P. vivax is both weak and conflicting. The major candidate gene, pvmdr1, was initially posited as a mediator of CQR due to its high sequence homology with the orthologous pfmdr1, which is involved in CQR in P. falciparum10. However, subsequent studies have produced contrasting reports, showing no consistent correlation between pvmdr1 and CQR11. Profiling of polymorphisms within the pvmdr1 gene led to the association of the Y976F mutation with CQR due to increased chloroquine IC50 in samples from Indonesian Papua and Thailand12. This mutation has since become a recognised marker for CQR in studies across P. vivax endemic regions9,13,14. At least 50 pvmdr1 SNPs have been documented globally, yet none have emerged as a definitive CQR marker, questioning the extent and relevance of pvmdr1 in modulating CQR. Although profiling the prevalence of pvmdr1 polymorphisms has increased our understanding of its evolution across different drug pressure backgrounds15,16, most studies focus solely on the pvmdr1 gene itself, biasing our understanding of the acquisition of CQR, which is hypothesised to be a multifaceted process involving numerous loci.
Extensive use of antimalarial drugs over several decades has resulted in high-resolution characterisations of recent selection events associated with drug resistance in Plasmodium spp. using genome-wide sequence data. Genome-wide analyses have shown evidence for recent positive selection in P. falciparum endemic regions with artemisinin resistance17–19 and revealed selective sweeps around the pfcrt, pfmdr1, and pfaat1 genes, which are explicitly linked to CQR20,21. Similar selection metrics applied to P. vivax genomes have revealed that the orthologous loci associated with antifolate resistance in P. falciparum have been subject to selective sweeps15,16,22. More recent work has found evidence of selective sweeps around pvmrp1 in East Asian isolates, a gene associated with chloroquine and mefloquine resistance in the orthologous P. falciparum pfmrp1, necessitating further investigation16.
Understanding the population genetics and dynamics of P. vivax malaria, particularly in the context of drug resistance, is essential for successful control and elimination planning. With the expanding repertoire of whole genome sequence data for P. vivax, we can now investigate the temporal dynamics of populations pre- and post-chloroquine contraindication, to provide increased insight into the markers of CQR. Here, we leverage publicly available whole genome sequences to present a large-scale population genomics study of P. vivax. We provide an expanded insight into population structure, global ancestry, relatedness, and genomic diversity. Using a genome-wide approach, we perform intra- and inter-population analyses between isolates from regions with different degrees of reported CQR to make inferences about both previously described and novel loci that could be mediating CQR and the evolutionary forces that shape P. vivax populations.
Results
P. vivax whole genome sequence data and clonality
A total of 499,206 high-quality bi-allelic SNPs were identified in the non-hypervariable regions of the P. vivax genome after filtering, comprising 1534 isolates from 29 countries. In keeping with previous work16, we divided these countries into 7 sub-regional populations based on the degree of geographic and genetic proximity: East Africa (N = 173), West Africa (1), South America (364), South Asia (156), South East Asia (SEA) (550), Maritime SEA (66), and Oceania (224) (Supplementary Data 1). Similarly, we ascribed each site and each country an overall CQR status adapted from the categories created in a prior meta-analysis of global chloroquine efficacy5 after pooling all publicly available day 28 recurrence data (Supplementary Data 2 and 3). Here, we describe three categories: chloroquine sensitivity (<5% day 28 recurrences), low-grade CQR (5–10% day 28 recurrences), and high-grade CQR (>10% day 28 recurrences) (Supplementary Data 2 and 3, Fig. S1).
Within-sample diversity, as a metric of multiclonality, was measured using the FWS fixation index, where an FWS ≥ 0.95 indicates an infection predominated by a single genotype. Here, a large proportion of isolates (71.7%) were monoclonal (FWS ≥ 0.95). Regionally, mean FWS was greatest in Maritime SEA (0.96), compared with South Asia (0.92), South America (0.92), East Africa (0.91), SEA (0.91), and Oceania (0.86) (Fig. S2). These observations likely reflect trends in transmission intensity, where higher transmission intensity is often marked by higher clone multiplicity.
Global P. vivax isolates form distinct sub-populations to sub-continent level
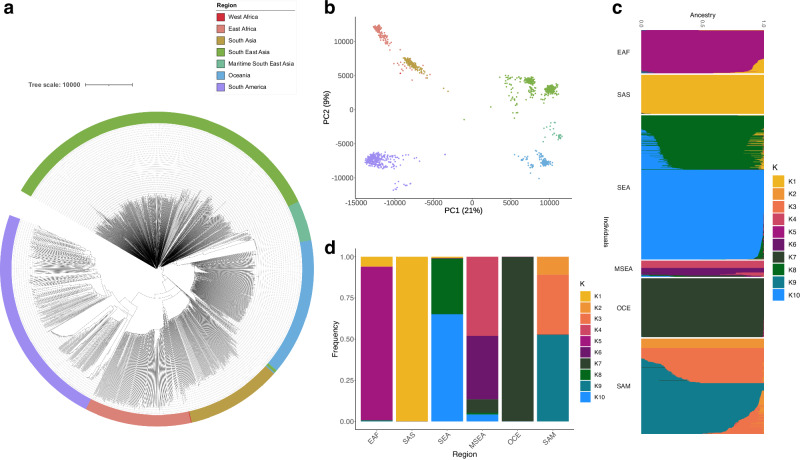
An analysis of population structure using a SNP-based neighbour-joining (NJ) tree (Fig. 1a) and principal component analysis (PCA) approach (Fig. 1b) applied to 499,206 bi-allelic SNPs revealed P. vivax isolates form distinct and independent sub-populations, reflective of their continental or sub-continental origins. In the PCA, the first principal component separated East Asian and Oceanian populations from South American, South Asian, and African populations, producing three major geographical population centres, with South American populations being the most distinct. The number of highly differentiating SNPs (FST ≥ 0.99) was positively correlated with geographic distance (Spearman’s Coefficient = 0.64) (Supplementary Data 4). These findings suggest gene flow between neighbouring territories is contributing to this phylogeographic distribution and is supported by known waves of human migration23.
Fig. 1. Population structure and ancestry in 1534 global P. vivax isolates.
P. vivax isolates form distinct populations to sub-continental level. a Neighbour-Joining tree for 1534 isolates, constructed using a distance matrix based on 499,206 high-quality bi-allelic SNPs, and coloured based on sub-regional grouping. b Principal Component Analysis (PCA) plot of the 1534 isolates, with colours based on sub-regional groupings in (a). c ADMIXTURE inference of 10 ancestral populations (K = 10) in the global dataset, visualised by bar plot, coloured by K population grouping, and summarised as frequencies in (d). EAF East Africa, MSEA Maritime South-East Asia, OCE Oceania, SAM South America, SAS South Asia, SEA South-East Asia.
An ADMIXTURE ancestral analysis inferred that global P. vivax isolates descend from ten ancestral populations (K1-K10), comprising two in East Africa (K1 and K5), one in South Asia (K1), three in South America (K2, K3, K9), three in SEA (K1, K8, K10), two in Maritime SEA (K4 and K6), and one in Oceania (K7) (Fig. 1c, d). There was some concordance between ADMIXTURE populations and country of origin, where several ancestral populations were made up of predominantly (>95%) one country, including K2 (Panama), K3 (Brazil), and K4 and K6 (Malaysia) (Supplementary Data 1, Fig. S3). Except for K1, which was characterised by African, South Asian, and SEA isolates, all ancestral populations comprised isolates from the same sub-regional grouping. This population structure was supported by a PCA plot coloured by dominant ancestry (Fig. S4).
Pairwise relatedness supports model of isolation by distance
To further dissect the global P. vivax population structure, pairwise relatedness of isolates was inferred by calculating identity-by-descent (IBD), which describes shared evolutionary history. Countries which presented the highest median pairwise IBD include Panama (0.97), Mexico (0.23), and Malaysia (0.19), suggesting reduced outcrossing within these populations (Supplementary Data 5 and 6). While the fractional IBD values of Malaysian and Mexican populations approach the expected value (0.25) of half-siblings in an outbred population, the value for Panama is inflated due to the persistence of a single clone in the region for a decade24. Within the remaining subpopulations, IBD sharing was low (<6%), revealing that samples were predominantly weakly related, with a minority of very highly related samples (Fig. S5). Regionally, median pairwise IBD was highest in Maritime SEA (0.15) and lowest in SEA (0.01). In agreement with FST observations, there was a moderate correlation between inter-regional median pairwise IBD and geographic distance (Spearman’s Coefficient = −0.39) (Supplementary Data 4), consistent with the model of isolation by distance25 which posits that populations in closer geographic proximity have increased genetic similarity.
IBD sharing reveals putative selective sweeps at P. vivax drug resistance loci
To investigate patterns of shared ancestry intrachromosomally, we analysed genome-wide IBD fractions calculated across 10 kb sliding windows, investigating specifically genomic regions falling in the top 1% of fractions (Fig. S6). Due to the high genetic relatedness of isolates from Malaysia, Mexico, and Panama (Fig. 1a, b) and their inflated pairwise IBD values (Fig. S7, Supplementary Data 5), we excluded them from further analysis. Overall, regions with the highest IBD fractions spanned antigenic loci (pvmsp1, PVP01_0728900; pvmsp5, PVP01_0418400; pvdbp, PVP01_0623800), genes involved in life-cycle specific processes (pvlisp2, PVP01_0304700), and candidate drug resistance loci (pvmrp1, PVP01_0203000; pvdhfr, PVP01_0526600; pvmdr1, PVP01_1010900; pvdhps, PVP01_1429500) (Supplementary Data 7).
The strongest signals (0.35) were observed in Indonesia on chromosome 10 between 320 and 330 kb (Supplementary Data 7), a region encompassing two conserved Plasmodium proteins of unknown function (PVP01_1007200 and PVP01_1007250), the former being a pseudogene. This region is of particular interest as it is ~140 kb downstream of pvmdr1, the gene putatively responsible for CQR in P. vivax. This peak of IBD sharing at 320–330 kb is also found within a large region of high IBD values spanning 200–500 kb on chromosome 10 (Figs. S8 and S9, Supplementary Data 8). Although peaking at 320–330 kb, the median fractional IBD value of the region was 0.12, and the IBD value at pvmdr1 was 0.11. Interestingly, we found high proportional IBD sharing at this downstream pvmdr1 locus (320–330 kb; PVP01_1007200, PVP01_1007250) in isolates from PNG (0.19), Sudan (0.13), and Brazil (0.11) (Fig. S10). As with Indonesian isolates, in the Sudanese population, this was found within a region spanning 280–500 kb, with a median IBD value of 0.17. At pvmdr1 itself, countries with high fractional IBD values were Sudan (0.26), Peru (0.19), Ethiopia (0.19), and Brazil (0.13).
The pvdhps and pvdhfr genes, located on chromosomes 14 and 5 respectively, are associated with resistance to the antimalarial combination therapy sulfadoxine-pyrimethamine (SP). While SP is not routinely used to treat P. vivax malaria, mutations in pvdhps and pvdhfr structurally align with resistance-conferring mutations observed in their orthologous genes, pfdhps and pfdhfr, in P. falciparum. We observed a trend of differential IBD values surrounding the genes, where isolates had high fractional IBD for one of the SP-resistance genes, but not the other. Isolates from Eritrea, Indonesia, and Myanmar had much greater fractional IBD values at pvdhps (0.25, 0.22, and 0.21, respectively) than at pvdhfr (0.11, 0.09, <0.01, respectively) (Supplementary Data 7).
Finally, we observed high fractional IBD values in pvmrp1, a gene on chromosome 2 associated with resistance to chloroquine, primaquine, and mefloquine in the orthologous P. falciparum pfmrp126 in Vietnamese (0.22), Thai (0.21), Cambodian (0.18), Burmese (0.16), and Colombian (0.14) isolates. We found no evidence of high proportional IBD sharing in any population at the resistance candidates pvcrt-o (PVP01_0109300), pvpm4 (PVP01_1340900), pvk13 (PVP01_121100), or in the sister genes to pvmrp1 and pvmdr1, pvmrp2 (PVP01_1447300) and pvmdr2 (PVP01_1259100). However, there was a signal from pvaat1 (PVP01_1120000), the orthologue of a gene newly implicated in CQR in P. falciparum21, in isolates from PNG (0.06).
Evidence of selective sweeps in candidate drug resistance loci
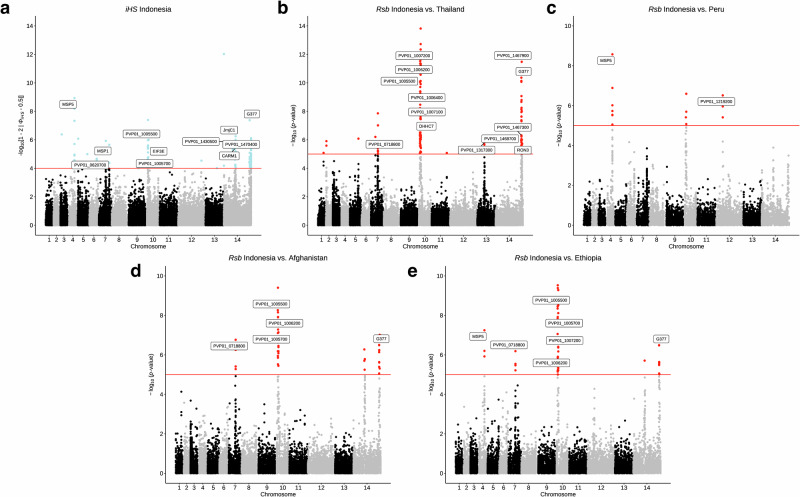
A genome-wide scan for the top 1% of genes under recent positive selection in monoclonal isolates was performed using the iHS metric (Supplementary Data 9). Multiple strong signals (N = 125) were found in the genes encoding the surface proteins pvmsp1 and pvmsp5 (P < 1 × 10−24), which are under selection pressure due to their interactions with the host immune response. The greatest values were in Afghanistan (P < 1 × 10−23), Thailand (P < 1 × 10−14), Cambodia (P < 1 × 10−11), and Vietnam (P < 1 × 10−10). Similarly, other hotspots of selection pressure were found in the cytoadherence linked asexual protein (pvclag, Ethiopia (P < 1 × 10−12), Thailand (P < 1 × 10−10)), the liver-specific protein 2 (pvlisp2, Afghanistan (P < 1 × 10−14), Indonesia (P < 1 × 10−6), Cambodia (P < 1 × 10−6)), ApiAP2 transcription factors (PVP01_1418100, Cambodia (P < 1 × 10−7); PVP01_1440600, Cambodia and Vietnam (both P < 1 × 10−5)), and the invasion proteins pvrbp1a (Cambodia, Indonesia, Pakistan; P < 1 × 10−10) and pvrbp2b (Afghanistan and India; P < 1 × 10−5). In agreement with our IBD findings, a series of 18 SNPs (P < 1 × 10−8) were under selection in Indonesian isolates downstream of pvmdr1 on chromosome 10 (243480-319423), with peaks at a SNP upstream a translation initiation factor (PVP01_1005600, P < 1 × 10−8) and a SNP in the PVP01_1007200 gene (P < 1 × 10−7). Isolates from Thailand and Pakistan also had SNPs under selection in PVP01_1007200 (P < 1 × 10−5).
To identify signals of differential selection, the cross-population metric, Rsb, was used at both a country and regional level, specifically comparing Indonesia with other countries due to known differences in CQR status. Although PNG is also a region of high-grade CQR, due to limited sample size, we excluded it from cross-population selection analysis. The most common SNPs under differential selection encompassed the cluster of SNPs with high iHS scores in the Indonesian population downstream of pvmdr1 (N = 248), differentiating Indonesian isolates from those in SEA (P < 1 × 10−13), East Africa (Eritrea and Ethiopia; P < 1 × 10−7), South Asia (Afghanistan, India, Pakistan; P < 1 × 10−10) and South America (Peru; P < 1 × 10−6) (Fig. 2, Supplementary Data 10–12). Beyond SNPs in merozoite surface antigens, we found evidence of selective sweeps via extended haplotype homozygosity at pvmrp1 in isolates from SEA (Cambodia, China, Vietnam, Thailand) when compared to isolates from South Asia (Afghanistan and India, P < 1 × 10−6). Most of these SNPs were in the promoter region, except for L1361F and 1232I. We also found SNPs within pvmrp2 under differential selection between isolates from Vietnam and China, Cambodia, and India (P < 1 × 10−6). Similarly, we observed differential signals in pvdhps in SEA isolates (Cambodia, Thailand, Vietnam; P < 1 × 10−11), and in pvdhfr in South Asian isolates (Afghanistan, India; P < 1 × 10−7). The SNPs S513R and L845F in MDR1 were under differential selection between Thai isolates compared with Vietnamese (P < 1 × 10−9) and Cambodian isolates (P < 1 × 10−6).
Fig. 2. Evidence of selective sweeps on chromosome 10 at downstream pvmdr1 locus.
Selection at this locus is driving differentiation of Indonesian Papua isolates from P. vivax isolates across South-East Asia, South Asia, South America, and East Africa. Manhattan plots showing integrated haplotype homozygosity scores (iHS) for SNPs in a Indonesian isolates (N = 106) and the cross-population test, Rsb, showing SNPs under differential selection between Indonesian and b Thai (N = 119), c Peruvian (N = 82) d Afghan (N = 50) and e Ethiopian (N = 102) isolates. Loci in critical regions, defined here as SNPs with an iHS score of P < 1 × 10−4 or Rsb score of P < 1 × 10−5 (two-sided tests), are highlighted in blue (iHS) and red (Rsb).
Sub-regional differences in the frequency of putative resistance mutations in P. vivax populations
Identifying mutations in putative drug resistance loci can reflect the epidemiology of transmission in both local and global contexts. We therefore evaluated the prevalence of non-synonymous mutations in all isolates in genes with a putative association to antimalarial resistance in P. vivax: pvmdr1 (33), pvmrp1 (53), pvdhfr (17), and pvdhps (20) (Table 1) and several additional genes of interest (Supplementary Data 13). Multiple pvmdr1 mutations have been previously associated with CQR or reduced chloroquine susceptibility10,13,27, including S513R, G698S, M908L, Y976F, and F1076L, and all, except S513R, are present in the P. vivax PvP01 reference. We observed the frequencies of the putative resistance alleles at 70.9% (698S), 97.9% (908L), 100% (976F), and 73.4% (1076L). The F1076L mutation was observed in all isolates from East Africa and Oceania and had lowest frequencies in South American (81.6%) and SEA (19.4%) isolates. Similarly, the G698S mutation was fixed in isolates from Oceania and Maritime SEA and had lowest frequency in South American (14.0%) isolates. The S513R mutation was found exclusively in isolates from SEA (14.0%), South Asia (37.2%), and East Africa (74.0%). There were lower frequency mutations with regional specificity, including D500N (South America, 24.2%), V221L (South America, 21.1%), and L845F (Across Asia and Oceania, 9.5%). Only one isolate from Indonesia had a non-synonymous SNP in pvmdr1 (L845F).
Table 1.
Prevalence of non-synonymous SNPs in genes that have been associated with drug resistance in P. vivax
| Gene | Chra | Positiona | Amino Acid Change | East Africa (N = 173) | South Asia (N = 156) | SEA (N = 550) | Maritime SEA (N = 66) | Oceania (N = 224) | South America (N = 364) |
|---|---|---|---|---|---|---|---|---|---|
| pvmrp1 | 2 | 154107/8 | A1606V | 5.2 | |||||
| pvmrp1 | 2 | 154168 | H1586Y | 5.1 | 2.6 | 19.8 | |||
| pvmrp1 | 2 | 154215/6 | D1570F | 14.7 | |||||
| pvmrp1 | 2 | 154992 | I1478V | 24.3 | 17.9 | 0.2 | 9.9 | ||
| pvmrp1 | 2 | 154668 | G1419A | 13.4 | 16.7 | 24.7 | |||
| pvmrp1 | 2 | 154831 | L1365F | 17.9 | |||||
| pvmrp1 | 2 | 155305 | L1207I | 81.5 | |||||
| pvmrp1 | 2 | 156208 | E906Q | ||||||
| pvmrp1 | 2 | 156563 | E787D | 0.6 | 3.8 | 3.5 | 1.5 | 1.8 | |
| pvmrp1 | 2 | 158148 | R259I | 35.3 | 27.6 | 0.6 | 11.3 | ||
| pvmrp1 | 2 | 158223 | T234M | 78.4 | |||||
| pvmrp1 | 2 | 158272 | Y218D | 82.7 | 64.1 | 92.4 | 98.5 | 87.5 | 60.4 |
| pvmrp1 | 2 | 158545 | V127I | 82.7 | 60.3 | 92.4 | 98.5 | 87.9 | 65.4 |
| pvdhfr | 5 | 1077534 | R58K | 0.5 | 5.5 | ||||
| pvdhfr | 5 | 1077535 | R58S | 37.6 | 62.8 | 2.55 | 59.1 | 6.3 | 61.3 |
| pvdhfr | 5 | 1077711 | N117T | 6.9 | 50.0 | 30.7 | 90.9 | 69.2 | 17.0 |
| pvdhfr | 5 | 1078180 | N273K | 1.7 | 10.3 | 0.2 | |||
| pvdhfr | 5 | 1077878 | I173L | 56.1 | 8.52 | ||||
| pvdhfr | 5 | 1077530 | F57I | 0.64 | 78.0 | 89.4 | 71.0 | 0.5 | |
| pvdhfr | 5 | 1077543 | T61M | 31.8 | 41.5 | 27.5 | |||
| pvmdr1 | 10 | 479908 | L1076F* | 1.9 | 19.5 | 1.5 | 81.6 | ||
| pvmdr1 | 10 | 480412 | L908M* | 0.58 | 8.52 | ||||
| pvmdr1 | 10 | 480552 | A861E | 5.8 | 3.6 | 0.82 | |||
| pvmdr1 | 10 | 480601 | L845F | 14.7 | 12.0 | 7.6 | 0.4 | ||
| pvmdr1 | 10 | 481042 | S698G | 27.2 | 51.9 | 86.0 | |||
| pvmdr1 | 10 | 481595 | S513R | 74.0 | 37.2 | 14.0 | |||
| pvmdr1 | 10 | 481636 | D500N | 23.6 | |||||
| pvmdr1 | 10 | 482473 | V221L | 21.2 | |||||
| pvdhps | 14 | 1270119 | A647V | 24.9 | 4.5 | 0.2 | |||
| pvdhps | 14 | 1270401 | A553G | 10.3 | 29.8 | 86.3 | 10.7 | ||
| pvdhps | 14 | 1270911 | G383A* | 71.1 | 83.3 | 9.6 | 4.5 | 16.1 | 39.8 |
| pvdhps | 14 | 1270914 | S382C | 0.4 | 12.9 | ||||
| pvdhps | 14 | 1270915 | S382A | 10.9 | |||||
| pvdhps | 14 | 1271444 | M205I | 71.7 | 0.6 | 96.2 | 6.1 | 1.8 | 58.5 |
| pvdhps | 14 | 1271634 | E142G | 64.7 | 59.1 | 10.7 |
Allele frequencies were calculated in isolates if, at a country level, their frequency was ≥10%. Allele frequency is bolded if ≥50%. East Africa (Eritrea, Ethiopia, Sudan); South Asia (Afghanistan, India, Pakistan); South East Asia (SEA; Cambodia, China, Myanmar, Thailand, Vietnam); Maritime SEA (Malaysia); Oceania (Indonesia, Papua New Guinea); South America (Brazil, Colombia, Mexico, Panama, Peru). Asterisked SNPs are those where the reference has the putative resistance allele. SNP prevalence was calculated solely for isolates with homozygous alternate calls.
Chr chromosome
aBased on the PvP01_v1 reference.
Mutations in the pvmrp1 orthologue, pfmrp1, decrease susceptibility to chloroquine26 (H191Y, I876V, T1007M, F1390I), piperaquine28 (H785N, I876V, T1007M), artemisinins29 (I876V, F1390I), and antifolates (K1466R). Although there were several pvmrp1 mutations approaching fixation globally (V127I; 81.2%, Y218N, 80.4%), most mutations showed regional specificity. South American isolates had the greatest number of unique pvmrp1 mutations including A1606V (5.2%), T1525I (4.4%), and C1018Y (3.3%). Other mutations with regional specificity of varying frequencies included T234M (SEA, 78.4%), L1365F (Oceania, 17.9%), F560I (East Africa (8.1%) and South Asia (1.9%)). We observed a previously undescribed mutation, D1570F, exclusive to Oceanian isolates (Indonesia (16.5%), PNG (10.0%)) (Table 1). Predictions of the domain structure of PvMRP1 revealed two AAA+ ATPase domains, with nucleotide binding domains at residues 647–774 and 1475–1642. Notably, the Oceanian-specific D1570F mutation falls within the latter domain. In silico modelling of PvMRP1, aligned with its orthologous PfMRP1, indicated that several mutations, including PvMRP1 L1365F, are located near known resistance-conferring mutations in PfMRP1, such as the PfMRP1 F1390I mutation associated with artemisinin and chloroquine resistance (Fig. S11). Based on primary structure predictions of PvMRP1, the L1365F mutation is the only Oceania-specific mutation that resides within a transmembrane helix (TMH), specifically TMH11 (Fig. S12). Outside of Oceanian populations, the only other PvMRP1 mutation found within a TMH is the L1361F mutation, a low-frequency variant present in three South Asian isolates (Afghanistan (1, 1.6%), India (2, 4.2%)) and three SEA isolates (Myanmar (1, 3.6%), Thailand (2, 1.1%)).
Antifolate resistance is attributed to mutations in DHFR and DHPS. In pvdhfr and pvdhps, numerous SNPs structurally correspond to resistance conferring mutations in P. falciparum. The pvdhfr S58R and S117T mutations and the pvdhps A383G mutation were present in the PvP01 reference, so we report the frequencies of the wild-type (reference) allele (Table 1). The triple pvdhfr mutants F57L-S58R-S117N (LRN) coupled with the pvdhps double mutant A383G-A553G (GG) are associated with clinical SP failure30. We found 3 instances of the pvdhfr LRN haplotype in isolates from Myanmar, PNG, and Peru (Supplementary Data 14). The pvdhps GG haplotype was globally more prevalent, found in India (33.3%), Indonesia (1.5%), Malaysia (3.2%), and Thailand (1.9%). Another pvdhfr haplotype of concern is the pvdhfr L/IRMT(F57L/I-S58R-T61M-S117N/T) quadruple mutant, found at moderate prevalence in East Asian and Oceanian isolates: China (16.7%), Malaysia (30.6%), Thailand (65.4%), Myanmar (28.6%), Indonesia (64.9%), and PNG (10.0%). Although we did not observe the pvdhps SAKAV mutant (S382A-A383G-K512E-A553G-V585A)31, we observed the alternate variant, MSAAK (M205I-S382A-A383G-A553G-K512M), exclusive to Thai isolates (4.5%). Finally, although not structurally linked to sulfadoxine resistance, we observed the Q142G mutation in various haplotypes specific to isolates from East Africa (Ethiopia, Eritrea, and Sudan), and the S382C-M205I double mutant specific to isolates from South America (Brazil, Guyana, Panama, and Peru).
A moderate degree of sequence conservation in global mdr1 haplotypes
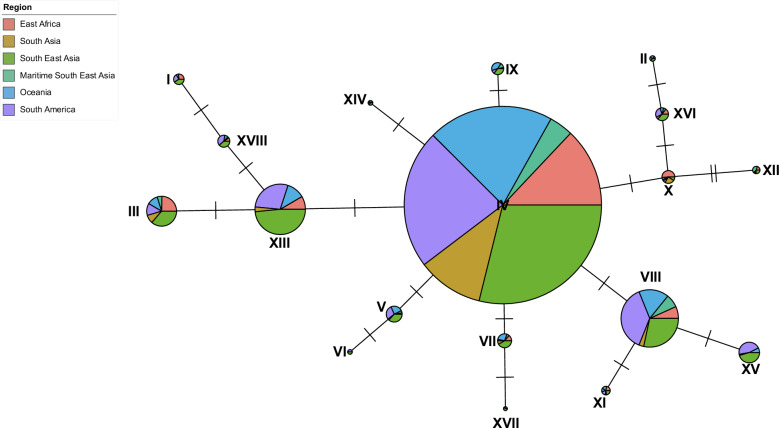
Haplotype networks were constructed to visualise the global diversity of pvmdr1 and explore haplotypes in regions with high-grade CQR, such as Indonesian Papua, against those with high clinical efficacy of chloroquine (Supplementary Data 2). We observed 169 distinct haplotypes, of which 93 (55.1%) were singletons and 151 (89.3%) had less than 10 observations. A median-joining haplotype network was estimated for the remaining 18 (10.7%) haplotypes identified across 1238 samples (Fig. 3). Haplotype IV, representing the 598P synonymous mutation, was the most prevalent, present in all geographic subregions (41.1%). Most Oceanian isolates (80.8%) were represented by this major haplotype, indicating that isolates from low and high-grade CQR regions have high pvmdr1 sequence similarity. Globally, 44.0% of isolates had pvmdr1 haplotypes comprised solely of synonymous mutations, suggesting a substantial degree of conservation at pvmdr1. Intra-region estimates of haplotype diversities (h) ranged from 0.33 to 0.93, with Oceanian isolates having the lowest genetic diversity (h = 0.33), and South Asian isolates with the highest (h = 0.93) (Supplementary Data 15).
Fig. 3. Median joining haplotype network constructed using pvmdr1 gene sequences from 1238 global isolates.
Each node represents a unique haplotype. Segments within nodes represent isolates from the 6 different subregional groupings and coloured accordingly. Node size is in proportion to the number of samples represented by that haplotype. The number of ticks between nodes represents the number of SNP differences between the two haplotypes.
Temporal trends in markers under selection in Indonesian isolates
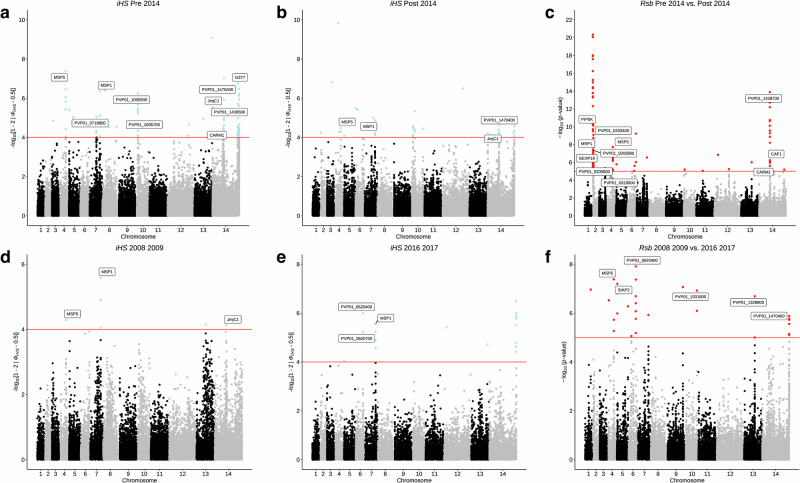
An ex vivo susceptibility study of P. vivax isolates from Indonesian Papua revealed the persistence of CQR between 2005–2018, despite dihydroartemisinin-piperaquine replacing chloroquine in 200432. With conflicting evidence of pvmdr1’s role in mediating CQR and maintained phenotypic CQR, we sought to investigate temporal trends in genome-wide signals of differential selection within the Indonesian population. We divided all monoclonal isolates into two sub-populations: pre-2014 (N = 75) and post-2014 (N = 29), with 2014 being 10 years post contraindication of chloroquine in Indonesia, and a timeframe we perceive adequate for genotypic changes to occur. Comparing iHS data revealed that the hotspot of SNPs downstream pvmdr1 was present solely in the pre-2014 population (Fig. 4a, b). A scan for the top 1% of genes under differential selection (Rsb) between the pre- and post-2014 populations revealed the Y218D and V127I pvmrp1 mutations had high hits (P < 1 × 10−8) (Fig. 4a–c). Contrastingly, pairwise FST revealed that the D1570F pvmrp1 mutation was the most highly differentiating between the two populations (FST = 0.34; Median FST = 0.027) (Supplementary Data 16, Fig. S13). This SNP first occurred in 2 isolates from 2011, and was present in 58.6% of post-2014 samples, compared with only 20.0% of pre-2014 isolates.
Fig. 4. Recent directional selection in Indonesian Papua (N = 104) isolates.
Manhattan plots showing integrated haplotype homozygosity scores (iHS) for SNPs in a pre-2014 isolates (N = 75), b post-2014 isolates (N = 29), d 2008–2009 isolates (N = 18), e 2016–2017 isolates (N = 10), and the cross-population test, Rsb, showing SNPs under differential selection between c pre-2014 vs. post-2014 isolates and f 2008–2009 vs. 2016–2017 isolates. Loci in critical regions, defined here as SNPs with an iHS score of P < 1 × 10−4 or Rsb score of P < 1 × 10−5 (two-sided tests), are highlighted in blue (iHS) and red (Rsb).
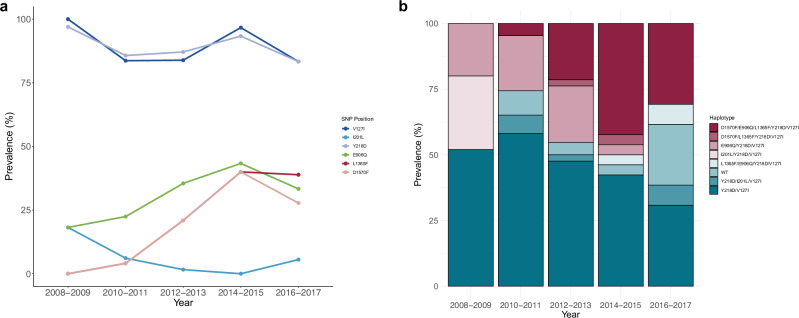
Taking this further, we divided all Indonesian isolates into 5 groups based on year of sample collection (2008–2009 (N = 33), 2010–2011 (N = 49), 2012–2013 (N = 62), 2014–2015 (N = 30), and 2016–2017 (N = 18)) and applied the same intra- and inter-population selection metrics. Across all groups, hotspots of selection were observed on chromosomes 4 and 7, corresponding to pvmsp5 and pvsmp1, and on chromosome 14, corresponding to proteins expressed in gametocytes (G377, PVP01_1467200), proteins for red blood cell adherence (CLAG, PVP01_1401400), and histone methylation machinery (CARM1, PVP01_142800; JmJC1, PVP01_1430400) (Fig. 4d–f, Supplementary Data 17). High iHS scores at the downstream pvmdr1 locus were observed only in populations 2012–2013 and 2014–2015 (all P < 1 × 10−6), indicative of transient directional selection at this locus. Differential selection metrics applied between the divergent 2008–2009 and 2016–2017 groups (4 years vs. 12 years post contraindication of chloroquine) revealed SNPs in the sporozoite-invasion associated protein 2 (pvsiap2, P < 1 × 10−8) and the cysteine-rich protective antigen (pvcyrpa, P < 1 × 10−7) under differential selection (Supplementary Data 18 and 19). Contrastingly, the FST metric revealed the pvmrp1 SNP L1365F was the most highly differentiating (FST = 0.54) between the two populations, with prevalence of the SNP increasing from 0% in 2008–2009 to 38.5% in 2016–2017 (Fig. 5).
Fig. 5. Temporal trends in pvmrp1 haplotypes in Indonesian Papua (N = 104) isolates.
a Change in frequency of non-synonymous mutations in pvmrp1 across five time periods (2008–2009, 2010–2011, 2012–2013, and 2016–2017). b Proportion of major pvmrp1 haplotypes from the same time periods.
Discussion
Understanding the epidemiological and evolutionary dynamics of malaria using genome-wide data can provide essential biological insights that can be harnessed in the global malaria control and elimination agenda. This is of particular importance in the context of P. vivax malaria, where the biological determinants of resistance to the front-line treatment, chloroquine, are still unknown. Here, using genome-wide SNPs, we were able to show distinct subpopulations of vivax isolates to sub-continent level, with classic signs of population differentiation by geographic separation. By profiling SNPs at candidate resistance genes and across the P. vivax genome, we have identified new loci exhibiting population differentiation between isolates from Indonesian Papua—a region with a longstanding history of high-grade CQR—and those from chloroquine-sensitive regions. We also describe the temporal dynamics of SNPs near pvmdr1, a gene proposed as a determinant of CQR, in Indonesian Papua isolates, and offer hypotheses for the absence of selection pressure within the gene itself over a decade after chloroquine contraindication. Additionally, our selection and temporal SNP dynamics analyses suggest that pvmrp1 is a promising molecular research and surveillance target. Further investigation is required to fully understand its contribution to antimalarial resistance in P. vivax.
Exploration of the population structure of global P. vivax isolates revealed the genetic structure largely reflects geographic structure, with significantly higher FST values between more spatially disparate populations, consistent with previous findings15,16,33–35. This observation also agrees with a model of isolation by distance, which indicates increased gene flow and genetic similarity between isolates that are more geographically proximal. At a local scale, we observed that intra-subcontinental populations were less structurally defined, except for South American and, to a lesser extent, SEA populations. These observations are not only a reflection of the complex geographic and ecological niches within these subcontinents, but in a South American context, could be reflective of independent introductions of P. vivax in the region, as seen in P. falciparum36. ADMIXTURE analysis of nuclear SNP data revealed moderate shared South Asian ancestry in East African isolates from Eritrea, Uganda, Sudan, and Madagascar, which has been previously described16,37, and coincides with human migration of South Asians to East African regions outside the Horn of Africa38. However, this did not reveal an Ethiopian-specific ancestral population, which agrees with prior observations of Ethiopian populations being less structured and more diversified35. In the SEA subgrouping, Cambodian and Vietnamese isolates were genetically and ancestrally distinct from isolates from Thailand, Myanmar, and China, which has recently been documented39. This is likely a result of the intimate historical links between the two countries throughout the 20th century.
Genome-wide IBD analyses provided helpful insights into the genomic architecture of global P. vivax populations, especially in the East African context. We observed, across all three major populations, high fractional IBD at pvdhps (Eritrea) and pvdhfr (Ethiopia, Sudan). This is most likely a relic of the intensive use of SP as a first-line drug in the treatment of P. falciparum malaria, which is co-endemic with P. vivax in the region, and due to inadvertent SP drug pressure on P. vivax isolates by seasonal malaria chemoprevention regimes40. When we paired these data with our SNP profiling results, we observed low frequencies of single and double mutants in pvdhfr (S58R, S117N/T) that structurally corresponded with residues linked to pyrimethamine resistance in P. falciparum, directly contrasting prior findings of pvdhfr double mutants approaching fixation in Ethiopian vivax populations41. Similarly, in pvdhps, we observed an abundance of the wildtype alleles (553A/383G) or other pvdhps mutants that have not yet been associated with sulfadoxine resistance (M205I/E142G). The apparent heterogeneity in pvdhfr/pvdhps mutants, coupled with clear evidence of P. falciparum treatment strategies shaping the vivax resistance landscape necessitates further investigation in the East African context, especially due to the moderate rise in P. vivax cases in the region between 2019–20221.
While in vitro susceptibility studies have shown a role of the Y976F mutation of PvMDR1 in modulating CQR in isolates from Indonesian Papua12, we found this mutation fixed in global populations (100%). Our observation agrees with no significant change found in chloroquine sensitivity in P. cynomolgi introduced with the Y976F mutation42. Similarly, we observed the M908L mutation fixed globally (97.9%). With ex vivo measurements associating M908L with reduced susceptibility to chloroquine, dihydroartemisinin, and mefloquine43, if M908L and Y976F do mediate CQR, this implies that CQR is intrinsic in vivax populations. The F1076L mutation, another potential CQR marker, is approaching fixation in P. vivax isolates from Sabah, Malaysia44,45 and Indonesian Papua10, yet again, as with the Y976F and M908L mutations, we found it at fixation in several countries with high clinical efficacy of chloroquine, such as Afghanistan46. The presence of geographically diverse pvmdr1 haplotypes with representation of isolates from regions with both low and high clinical efficacy of chloroquine supports the finding of multiple independent local diversification events at pvmdr147, and strengthens the evidence against these mutations being reliable markers for CQR.
Given the trend of declining chloroquine sensitivity in regions such as Sabah, Malaysia44, Vietnam48, and the China-Myanmar border43, we hypothesised that if pvmdr1 is mediating CQR, there should be detectable signals of selection in pvmdr1 indicative of this resistance. We found no evidence of selective sweeps encompassing the pvmdr1 gene itself within or between any population, despite our genome-wide IBD analyses inferring putative selection events at pvmdr1 in Sudanese, Ethiopian and Brazilian isolates. Moreover, differential selection of pvmdr1 SNPs was within the SEA population only. One of the strongest selective sweeps in Indonesian Papua isolates was at a locus proximal to pvmdr1, encompassing 18 SNPs in a ~76 kb region. Strong directional selection of this region specific to the Indonesian population at genes PVP01_1007200-250 has been identified previously49, however, investigations into its role, if any, in mediating CQR have been limited. From IBD data, we could also infer putative selection events at this locus in isolates from PNG and Sudan but limited further investigations due to limited sample size. Here, our study of the temporal dynamics of this cluster of SNPs in the Indonesian context revealed that the selective sweep takes place between 2012 and 2015. Given the temporally transient nature of this genomic signature, several hypotheses can be generated. Firstly, one could hypothesise that these SNPs have no impact on the parasite’s fitness or ability to withstand chloroquine pressure. These alleles are therefore neutral and not linked to CQR. Conversely, one could hypothesise that these SNPs reflect changing local dynamics, considering that chloroquine is still readily available in the private sector in Indonesian Papua50. In this scenario, the temporal signature could be a result of indirect selection, as the SNPs may be beneficial in periods of increasing chloroquine use, but non-beneficial in periods of increasing dihydroartemisinin-piperaquine use. It is unclear of the role these SNPs play in PNG and Sudan.
Strong signals of positive selection at the pvmrp1 gene have led to its emergence as a candidate drug-resistance gene16,51,52. We found the previously described L1207I SNP under selection in Thai isolates and the pvmrp1 gene with high fractional IBD in Vietnamese, Thai, and Cambodian isolates. Although not under selection, we observed the mutation D1570F in Indonesian isolates. Investigating the temporal dynamics of this mutation revealed an intriguing pattern. The first appearance of the corresponding SNP was in an isolate from 2011, and it had strong population differentiation effects between isolates pre- and post-2014. Further investigation of pvmrp1 between isolates specifically from 2008–2009 to 2016–2017 groupings revealed that along with the D1570F mutation, the L1365F mutation was the most differentiating SNP between the two populations. Dihydroartemisinin-piperaquine has replaced chloroquine as the frontline antimalarial against P. vivax in Indonesia, with primaquine still remaining in the treatment strategy to target hypnozoite stages. We observed that, since its introduction, increasing dihydroartemisinin-piperaquine pressure on Indonesian Papuan isolates over time is correlated with a rising prevalence of pvmrp1 SNPs. Between 2008 and 2018, ex vivo susceptibility of P. vivax to piperaquine in the Indonesian Papua population declined32. Although clinical efficacy of dihydroartemisinin-piperaquine is reportedly high in Indonesian Papua, therapeutic efficacy studies have shown day 42 recurrence rates ranging from 1.2%53 to 11.3%54.
In P. falciparum, knockout of the orthologous pfmrp1 gene resulted in increased susceptibility to piperaquine26. Moreover, in a P. falciparum genetic cross, progeny with mutant pfmrp1 were only found in parasites harbouring SNPs in kelch13 that confer artemisinin resistance55. Allele frequency trajectories are not solely driven by selection pressures exerted by drug regimens; changes in the human or vector genomic backgrounds and climate change may also play a role. The appearance of novel pvmrp1 haplotypes after the introduction of dihydroartemisinin-piperaquine in Indonesia could reflect changing local dynamics, could mitigate the cost of previously fixed alleles, such as those in pvmdr1, or could in fact be resistance-conferring. The presence of the PvMRP1 L1365F mutation in TMH11, near the PfMRP1 F1390I mutation associated with chloroquine and dihydroartemisinin resistance, is noteworthy. If PvMRP1 TMHs play an integral role in interacting with antimalarials, mutations at these sites can impact drug influx and efflux, enabling the parasite to evade drug action. Leveraging the P. cynomolgi or P. knowlesi56 models to introduce the PvMRP1 D1570F and L1365F mutations and investigating the in vitro efficacy of chloroquine, dihydroartemisinin, and piperaquine will be essential in determining whether drug pressure is the ultimate driver in the appearance of these mutations, and if pvmrp1 plays a role in mediating resistance.
Overall, the work described here of global P. vivax isolates provides insights on population structure, admixture, markers of IBD, differentiation, and selection signatures in the context of drug resistance. Although the markers of CQR remain elusive, our findings of directional selection and selective sweeps in the candidate resistance gene pvmrp1 highlights the need for broader genotypic and phenotypic surveillance of P. vivax to complement elimination efforts.
Methods
Sequence data and raw reads processing
Illumina whole genome sequencing data were obtained from the publicly available Pv4 dataset generated by the MalariaGEN Community Project57 for P. vivax (N = 1895) and newly sequenced isolates sent to the UKHSA Malaria Reference Laboratory from imported UK cases (N = 47), totalling 1942 isolates available for analysis. After quality control, the final dataset included 1534 (79.0%) isolates from 29 countries: East Africa (N = 173, Eritrea (13), Ethiopia (138), Madagascar (4), Sudan (13), Uganda (5)), South America (N = 364, Brazil (139), Colombia (58), Guyana (3), Mexico (20), Panama (47), Peru (97)), South Asia (N = 156, Afghanistan (63), Bangladesh (1), India (48), Iran (5), Pakistan (37), Sri Lanka (2)), South East Asia (SEA) (N = 550, Cambodia (218), China (12), Laos (2), Myanmar (28), North Korea (1), Thailand (179), Vietnam (110)), Maritime SEA (N = 66, Malaysia (62), The Philippines (4)), Oceania (n = 224, Indonesia (194), PNG (30)), and West Africa (N = 1, Mauritania (1)).
Variant calling and quality control
All raw Illumina sequencing reads were first trimmed using trimmomatic58 (v0.39, parameters PE -phred33, LEADING:3, TRAILING:3, SLIDINGWINDOW:4:20 MINLEN:36). Trimmed reads were aligned to the reference genome, PvP01_v159 using bwa-mem (v0.7.17, default parameters)60. The resultant BAM files were processed with samtools functions fixmate and markdup (v1.9, default parameters)61. A training-set of high-quality P. vivax single nucleotide polymorphisms (SNPs) from previously published literature was used to calibrate variant calling16. Using this set, the GATK BaseRecalibrator and ApplyBQSR functions62 were run to produce improved BAM files for all isolates15,16. SNPs and indels were identified using the GATK HaplotypeCaller function (v.4.4.0.0, parameters: -ERC GVCF) to produce individual sample variant call format (VCF) files. The resultant VCFs were merged to create a multi-sample VCF using the GATK CombineGVCF function (default parameters). A total of 3,671,958 unfiltered SNPs were identified across 1942 isolates. The merged VCF was filtered iteratively (described in full here16) to produce the final dataset for subsequent analyses. Briefly, variants identified in the hypervariable subtelomeric regions were removed by mapping the 14 chromosomal sequences against the P. vivax PvP01_v1 reference, leaving variants only within the core P. vivax genome. Variants with a Variant Quality Score Log-Odds (VQSLOD) score <0, representing variants that are most likely false, were excluded. Variants where >40% of SNPs had missing genotype data were excluded. Monomorphic SNPs, heterozygous SNPs, and indels were also excluded. A set of 175 isolates from Indonesia that passed quality control, were taken from the original Pv4 release VCF (ftp://ngs.sanger.ac.uk/production/malaria/Resource/30) and merged into the VCF created within the present study. The final dataset encompassed 499,206 high-quality bi-allelic SNPs across 1534 samples. SNPs were annotated with predictions of their downstream coding effects using the SnpEFF (v5.1) software63.
Chloroquine resistance status designation
To ascribe a chloroquine resistance status to each site and each country in this study, we extended the work within a systematic review of Price et al.5 and used the Worldwide Antimalarial Resistance Network’s (WWARN) Vivax Surveyor (http://www.wwarn.org/vivax/surveyor/#0). The Vivax Surveyor houses a repository of all P. vivax clinical trial data, including studies from Price et al.’s systematic review and meta-analysis on global chloroquine resistance. We downloaded all P. vivax clinical trial data from 1950 to 2019 (N = 260) from the WWARN Vivax Surveyor. First, we filtered out studies without a chloroquine arm, resulting in 215 studies. We then excluded studies not conducted in countries with publicly available sequence data in our dataset, reducing the number to 191. Finally, we excluded studies from countries within our dataset with fewer than 10 publicly available P. vivax genome sequences or those certified malaria-free by the WHO, leaving a total of 169 studies for our analysis.
To extend the work of Price et al.5 and include studies published post-2019, we applied an identical research methodology. We systematically searched in the PubMed, Embase, Web of Science, and the Cochrane Database of Systematic Reviews databases, using the same search terms for prospective studies of chloroquine treatment of vivax malaria published in English, filtering for “vivax” in the title or abstract. This time, we screened for studies published between January 1, 2019, and June 28, 2024. Estimates of chloroquine efficacy for each site in each study, defined as the proportion of patients with recurrent P. vivax parasitaemia at day 28, were extracted, and 95% confidence intervals were calculated using the Wilson score interval procedure. Together with the existing pre-2019 data, a pooled 95% confidence interval was calculated for each site and each country. Based on these pooled day 28 recurrence rates and 95% confidence intervals, we ascribed one of three chloroquine resistance (CQR) statuses to each country, based on and adapted from the a priori categories created by Price et al.5: (i) High-Grade CQR: greater than 10% recurrences by day 28 (with a lower 95% CI of >5%), corresponding to Price et al.’s5 Category 1; (ii) Low-Grade CQR: between 5 and 10% recurrences by day 28, corresponding to a combination of Price et al.’s5 Categories 2 and 3; (iii) Chloroquine Sensitive (CQS): less than 5% recurrences by day 28 with chloroquine monotherapy (no primaquine given before day 28) in trials of at least 10 patients, corresponding to Price et al.’s5 Category 4.
Population genetics analyses
A binary matrix of pairwise genetic distances was constructed from the filtered biallelic VCF in PLINK (v1.9, default parameters)64. Using the distance matrix, population structure was assessed by conducting a Principal Component Analysis (PCA) and constructing a neighbour-joining (NJ) tree using the R package ape (v5.7). The biallelic VCF of 499,206 SNPs was filtered using bcftools65 (v1.17) view function and used to select SNPs with a minor allele frequency (MAF) > 1%, producing a multi-sample VCF of 115,022 high-quality bi-allelic SNPs. The NJ tree was visualised in iTOL66. To further investigate population structure, we used the ADMIXTURE software (v1.3.0)67, a tool used to estimate individual ancestries and population allele frequencies in SNP genotype datasets. PLINK (v1.9)64 was used to convert the MAF-filtered multi-sample VCF to a BED file. Using ADMIXTURE, the most likely number of subpopulations (K) was obtained using cross-validation error of 1–10 dimensions of eigenvalue decay. The output was visualised in R (v4.2.3). The population differentiation metric, fixation index (FST), was calculated pairwise to assess SNPs driving allele frequency differences between populations at a country and regional level using the vcftools (v0.1.17)68 function -weir-fst-pop.
Within-infection genomic diversity or multiplicity of infection, expressed as the variant of inbreeding coefficient FWS, was calculated at an individual level using the R package moimix (v0.0.2.9001, https://github.com/bahlolab/moimix). The FWS metric expresses the probability of the heterozygosity of parasites within an individual against the heterozygosity within a parasite population. Samples with an FWS score of ≥0.95 are highly considered to be monoclonal, whereas samples with an FWS < 0.95 suggests mixed strain infections, and therefore, a poorly defined population sub-structure. In all selection analyses, only monoclonal isolates (FWS ≥ 0.95) were considered.
IBD and directional selection analyses
Relatedness between all samples was explored in a pairwise manner through identity by descent (IBD) analysis, conducted by the hmmIBD69 package (v2.0.4, default parameters). hmmIBD implements a hidden Markov model for inference of pairwise IBD between haploid genotypes, enabling detection of DNA segments with shared ancestry. Only populations of monoclonal samples (FWS ≥ 0.95) at both a country and regional level with >10 isolates and biallelic SNPs with a MAF > 1% were used for analysis. An additional filtering step to the binary SNP matrix replaced all missing calls to a reference call and all mixed calls to alternative calls. Using sliding windows of 10 kb, IBD was cumulatively calculated and plotted by chromosomal location in R (v4.2.3).
To detect signals of recent positive selection, all samples were screened in a pairwise manner at both a country and regional level using the R package rehh (v.3.2.2)70 on SNPs with MAF > 1%. We calculated the summary statistics integrated haplotype homozygosity score (iHS)71 for within-population selection, and the cross-population ratio of extended haplotype homozygosity (EHH) expressed as Rsb and XP-EHH for differential selection between populations72. For iHS analysis, we describe the ancestral and derived alleles as the reference and non-reference alleles respectively. A positive iHS score suggests that the reference allele has undergone selection, whereas a negative iHS score suggests selection of a non-reference allele. Critical loci were identified using 10 kb sliding windows, which included at least 5 SNPs with a p value < 1 × 10−4 for iHS and <1 × 10−5 for Rsb and XP-EHH. These cutoffs were calculated using a Gaussian approximation method. XP-EHH detects selective sweeps in which a selected allele has approached or achieved fixation in one population while remaining polymorphic in the other population by comparing the lengths of the haplotypes associated with the selected allele in both populations73. A positive XP-EHH indicates selection occurring in population 1, whereas a negative XP-EHH indicates selection occurring in population 2. The Rsb metric is the ratio of EHH between two populations, normalised to 1. In-house scripts for analysis are available on GitHub (https://github.com/LSHTMPathogenSeqLab/malaria-hub).
Haplotype network estimation
The aligned FASTA files for pvmdr1 and its downstream locus were used to estimate haplotype networks using the pegas (v0.11)74 package in R, along with nucleotide and haplotype diversity statistics.
In silico protein structural prediction of PvMRP1 and PfMRP1
MRP1 amino acid sequences from the P. vivax P01 (PvMRP1; PVP01_0203000) and P. falciparum 3D7 (PfMRP1; PF3D7_0112200) reference genomes were obtained from PlasmoDB75, and aligned using Clustal Omega76. The resultant alignment was visualised in JalView (v2.11.3.3)77. The PvMRP1 and PfMRP1 tertiary protein structures were predicted using AlphaFold3 (https://alphafoldserver.com)78, and aligned and visualised with UCSF ChimeraX (v1.17.3)79. The primary structure of PvMRP1 was predicted using DeepTMHMM80 (https://biolib.com/DTU/DeepTMHMM/), and domain structure predicted using InterPro81.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
G.C.N-J is funded by a BBSRC LIDo PhD studentship (BB/T008709/1). T.G.C. and S.C. are funded by UKRI MRC (IAA2129, MR/R026297/1, and MR/X005895/1) and EPSRC (EP/Y018842/1), and Wellcome iTPA Translational Accelerator Award (214227/Z/18/Z) grants. C.R.F.M was supported by the São Paulo Research Foundation-FAPESP (2020/06747-4 and 2022/13150-0) and the National Council for Scientific and Technological Development-CNPq (302917/2019-5). J.G.D was supported by fellowships from FAPESP (2019/12068-5 and 2022/02771-3). The SMRU is part of the Mahidol Oxford Research Unit supported by the Wellcome Trust of Great Britain. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
T.G.C. and S.C. conceived and designed the study. G.C.N.-J. performed all bioinformatic analyses and interpreted results under the supervision of T.G.C. and S.C. J.E.P. and E.M. contributed bioinformatic tools and insights. J.G.D., M.S.-M., S.S., G.V.-Z., A.T.C., R.L.D.M., C.R.F.M., D.N., F.N., and C.J.S. contributed samples and sequence data. S.C. sequenced samples. G.C.N.-J. wrote the first draft of the manuscript with inputs from T.G.C. and S.C. All authors provided comments on the versions of the manuscript, approving of the final manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The data used in this study are available at the European Nucleotide Archive (https://www.ebi.ac.uk/ena). For samples from imported P. vivax cases diagnosed at the UKHSA Malaria Reference Laboratory, data are under accession codes PRJEB44419 and PRJEB56411. For samples from the MalariaGEN P. vivax Genome Variation Project57, data are under accession codes PRJEB2136, PRJEB2140, PRJEB4409, PRJEB4410, PRJEB10888, PRJNA65119, PRJNA67065, PRJNA67237, PRJNA67239, PRJNA175266, PRJNA240366, PRJNA240531, PRJNA271480, PRJNA284437, PRJNA295233, PRJNA420510, PRJNA432819, PRJNA603279, PRJNA643698, and PRJNA655141. The raw data for 175 Indonesian Papua isolates from the MalariaGEN P. vivax Genome Variation Project are available as an unfiltered VCF at ftp://ngs.sanger.ac.uk/production/malaria/Resource/30. All accession codes and sample provenance are detailed in Supplementary Data 1.
Code availability
For analysis scripts, please see the GitHub repository available at https://github.com/LSHTMPathogenSeqLab/malaria-hub. All scripts use open-source software (see “Methods”). For population genetics analyses, SnpEFF63 (version 5.1), PLINK64 (v1.9), ape (v5.7), bcftools65 (v.1.17), R (v.4.2.3), vcftools68 (v.0.1.17), moimix (v.0.0.2.9001, https://github.com/bahlolab/moimix), ADMIXTURE67 (v.1.3.0), hmmIBD69 (v2.04), rehh70 (v3.2.2), pegas74 (v0.11). For protein structural analysis we used Clustal Omega (https://www.ebi.ac.uk/jdispatcher/msa/clustalo), AlphaFold378 (https://alphafoldserver.com), DeepTMHMM80 (https://dtu.biolib.com/DeepTMHMM), InterPro81 (https://www.ebi.ac.uk/interpro/), JalView77 (v2.11.3.3) and UCSF ChimeraX79 (v1.17.3).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Susana Campino, Email: Susana.Campino@lshtm.ac.uk.
Taane G. Clark, Email: Taane.Clark@lshtm.ac.uk
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54964-x.
References
- 1.World Health Organisation. World Malaria Report 2023.
- 2.Bermúdez, M., Moreno-Pérez, D. A., Arévalo-Pinzón, G., Curtidor, H. & Patarroyo, M. A. Plasmodium vivax in vitro continuous culture: the spoke in the wheel. Malar. J.17, 301 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu, C. S. & White, N. J. Management of relapsing Plasmodium vivax malaria. Expert Rev. Anti Infect. Ther.14, 885–900 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Price, R. N., Commons, R. J., Battle, K. E., Thriemer, K. & Mendis, K. Plasmodium vivax in the Era of the Shrinking P. falciparum Map. Trends Parasitol.36, 560–570 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Price, R. N. et al. Global extent of chloroquine-resistant Plasmodium vivax: a systematic review and meta-analysis. Lancet Infect. Dis.14, 982–991 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nyunt, M. H. et al. Clinical and molecular surveillance of drug resistant vivax malaria in Myanmar (2009–2016). Malar. J.16, 117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soe, M. T. et al. Therapeutic efficacy of chloroquine for uncomplicated Plasmodium vivax malaria in southeastern and western border areas of Myanmar. Infection50, 681–688 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ketema, T., Getahun, K. & Bacha, K. Therapeutic efficacy of chloroquine for treatment of Plasmodium vivax malaria cases in Halaba district, South Ethiopia. Parasites Vectors4, 46 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnadas, C. et al. Plasmodium vivax resistance to chloroquine in Madagascar: clinical efficacy and polymorphisms in pvmdr1 and pvcrt-o genes. Antimicrob. Agents Chemother.52, 4233–4240 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brega, S. et al. Identification of the Plasmodium vivax mdr-like gene (pvmdr1) and analysis of single-nucleotide polymorphisms among isolates from different areas of endemicity. J. Infect. Dis.191, 272–277 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Sá, J. M. et al. Plasmodium vivax: allele variants of the mdr1 gene do not associate with chloroquine resistance among isolates from Brazil, Papua, and monkey-adapted strains. Exp. Parasitol.109, 256–259 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Suwanarusk, R. et al. Chloroquine resistant Plasmodium vivax: in vitro characterisation and association with molecular polymorphisms. PLoS ONE2, e1089 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orjuela-Sánchez, P. et al. Analysis of single-nucleotide polymorphisms in the crt-o and mdr1 genes of Plasmodium vivax among chloroquine-resistant isolates from the Brazilian Amazon region. Antimicrob. Agents Chemother.53, 3561–3564 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mekonnen, S. K. et al. Return of chloroquine-sensitive Plasmodium falciparum parasites and emergence of chloroquine-resistant Plasmodium vivax in Ethiopia. Malar. J.13, 244 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benavente, E. D. et al. Genomic variation in Plasmodium vivax malaria reveals regions under selective pressure. PLoS ONE12, e0177134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benavente, E. D. et al. Distinctive genetic structure and selection patterns in Plasmodium vivax from South Asia and East Africa. Nat. Commun.12, 3160 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheeseman, I. H. et al. A major genome region underlying artemisinin resistance in malaria. Science336, 79–82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miotto, O. et al. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat. Genet45, 648–655 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takala-Harrison, S. et al. Genetic loci associated with delayed clearance of Plasmodium falciparum following artemisinin treatment in Southeast Asia. Proc. Natl Acad. Sci. USA110, 240–245 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wootton, J. C. et al. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature418, 320–323 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Amambua-Ngwa, A. et al. Chloroquine resistance evolution in Plasmodium falciparum is mediated by the putative amino acid transporter AAT1. Nat. Microbiol8, 1213–1226 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winter, D. J. et al. Whole genome sequencing of field isolates reveals extensive genetic diversity in Plasmodium vivax from Colombia. PLoS Negl. Trop. Dis.9, e0004252 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodrigues, P. T. et al. Human migration and the spread of malaria parasites to the New World. Sci. Rep.8, 1993 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buyon, L. E. et al. Population genomics of Plasmodium vivax in Panama to assess the risk of case importation on malaria elimination. PLoS Negl. Trop. Dis.14, e0008962 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slatkin, M. Isolation by distance in equilibrium and non‐equilibrium populations. Evolution47, 264–279 (1993). [DOI] [PubMed] [Google Scholar]
- 26.Raj, D. K. et al. Disruption of a Plasmodium falciparum multidrug resistance-associated protein (PfMRP) alters its fitness and transport of antimalarial drugs and glutathione. J. Biol. Chem.284, 7687–7696 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spotin, A. et al. Global assessment of genetic paradigms of Pvmdr1 mutations in chloroquine-resistant Plasmodium vivax isolates. Trans. R. Soc. Trop. Med Hyg.114, 339–345 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Bai, Y. et al. Longitudinal surveillance of drug resistance in Plasmodium falciparum isolates from the China-Myanmar border reveals persistent circulation of multidrug resistant parasites. Int. J. Parasitol. Drugs Drug Resist.8, 320–328 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veiga, M. I. et al. Antimalarial exposure delays plasmodium falciparum intra-erythrocytic cycle and drives drug transporter genes expression. PLoS ONE5, e12408 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Desai, M. et al. Impact of sulfadoxine-pyrimethamine resistance on effectiveness of intermittent preventive therapy for malaria in pregnancy at clearing infections and preventing low birth weight. Clin. Infect. Dis.62, 323–333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korsinczky, M. et al. Sulfadoxine resistance in Plasmodium vivax is associated with a specific amino acid in dihydropteroate synthase at the putative sulfadoxine-binding site. Antimicrob. Agents Chemother.48, 2214–2222 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marfurt, J. et al. Longitudinal ex vivo and molecular trends of chloroquine and piperaquine activity against Plasmodium falciparum and P. vivax before and after introduction of artemisinin-based combination therapy in Papua, Indonesia. Int. J. Parasitol. Drugs Drug Resist.17, 46–56 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koepfli, C. et al. Plasmodium vivax diversity and population structure across four continents. PLoS Negl. Trop. Dis.9, e0003872 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hupalo, D. N. et al. Population genomics studies identify signatures of global dispersal and drug resistance in Plasmodium vivax. Nat. Genet48, 953–958 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rougeron, V. et al. Human Plasmodium vivax diversity, population structure and evolutionary origin. PLoS Neglected Trop. Dis.14, e0008072 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yalcindag, E. et al. Multiple independent introductions of Plasmodium falciparum in South America. Proc. Natl Acad. Sci.109, 511–516 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gartner, V. et al. Genomic insights into Plasmodium vivax population structure and diversity in central Africa. Malar. J.23, 27 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Culleton, R. et al. The origins of African Plasmodium vivax; insights from mitochondrial genome sequencing. PLoS ONE6, e29137 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, Y. et al. Retrospective analysis of Plasmodium vivax genomes from a pre-elimination China inland population in the 2010s. Front. Microbiol.14, 1071689 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flegg, J. A. et al. Spatiotemporal spread of Plasmodium falciparum mutations for resistance to sulfadoxine-pyrimethamine across Africa, 1990–2020. PLoS Comput Biol.18, e1010317 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kebede, A. M. et al. Genomic analysis of Plasmodium vivax describes patterns of connectivity and putative drivers of adaptation in Ethiopia. Sci. Rep.13, 20788 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward, K. E. et al. Integrative genetic manipulation of Plasmodium cynomolgi reveals multidrug resistance-1 Y976F associated with increased in vitro susceptibility to Mefloquine. J. Infect. Dis.227, 1121–1126 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li, J. et al. Ex vivo susceptibilities of Plasmodium vivax isolates from the China-Myanmar border to antimalarial drugs and association with polymorphisms in Pvmdr1 and Pvcrt-o genes. PLoS Negl. Trop. Dis.14, e0008255 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Auburn, S. et al. Genomic analysis of a pre-elimination Malaysian Plasmodium vivax population reveals selective pressures and changing transmission dynamics. Nat. Commun.9, 2585 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheong, F.-W., Dzul, S., Fong, M.-Y., Lau, Y.-L. & Ponnampalavanar, S. Plasmodium vivax drug resistance markers: Genetic polymorphisms and mutation patterns in isolates from Malaysia. Acta Tropica206, 105454 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Awab, G. R. et al. Dihydroartemisinin-piperaquine versus chloroquine to treat vivax malaria in Afghanistan: an open randomized, non-inferiority, trial. Malar. J.9, 105 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schousboe, M. L. et al. Multiple origins of mutations in the mdr1 Gene—a putative marker of chloroquine resistance in P. vivax. PLoS Negl. Trop. Dis.9, e0004196 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thanh, P. V. et al. Confirmed Plasmodium vivax resistance to chloroquine in Central Vietnam. Antimicrob. Agents Chemother.59, 7411–7419 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pearson, R. D. et al. Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nat. Genet48, 959–964 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kenangalem, E. et al. Malaria morbidity and mortality following introduction of a universal policy of artemisinin-based treatment for malaria in Papua, Indonesia: a longitudinal surveillance study. PLoS Med.16, e1002815 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dharia, N. V. et al. Whole-genome sequencing and microarray analysis of ex vivo Plasmodium vivax reveal selective pressure on putative drug resistance genes. Proc. Natl Acad. Sci. USA107, 20045–20050 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Flannery, E. L. et al. Next-generation sequencing of plasmodium vivax patient samples shows evidence of direct evolution in drug-resistance genes. ACS Infect. Dis.1, 367–379 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poespoprodjo, J. R. et al. Therapeutic response to dihydroartemisinin–piperaquine for P. falciparum and P. vivax nine years after its introduction in Southern Papua, Indonesia. Am. J. Trop. Med Hyg.98, 677–682 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Asih, P. B. S. et al. Efficacy and safety of dihydroartemisinin–piperaquine for the treatment of uncomplicated Plasmodium falciparum and Plasmodium vivax malaria in Papua and Sumatra, Indonesia. Malar. J.21, 95 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mok, S. et al. Mapping the genomic landscape of multidrug resistance in Plasmodium falciparum and its impact on parasite fitness. Sci. Adv.9, eadi2364 (2023). [DOI] [PMC free article] [PubMed]
- 56.Mohring, F. et al. Rapid and iterative genome editing in the malaria parasite Plasmodium knowlesi provides new tools for P. vivax research. eLife8, e45829 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.MalariaGEN. et al. An open dataset of Plasmodium vivax genome variation in 1,895 worldwide samples. Wellcome Open Res.7, 136 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Auburn, S. et al. A new Plasmodium vivax reference sequence with improved assembly of the subtelomeres reveals an abundance of pir genes. Wellcome Open Res.1, 4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics27, 2987–2993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res.20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet.81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res.49, W293–W296 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alexander, D. H. & Lange, K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinform.12, 246 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Danecek, P. et al. The variant call format and VCFtools. Bioinformatics27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schaffner, S. F., Taylor, A. R., Wong, W., Wirth, D. F. & Neafsey, D. E. hmmIBD: software to infer pairwise identity by descent between haploid genotypes. Malar. J.17, 196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gautier, M. & Vitalis, R. rehh: an R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics28, 1176–1177 (2012). [DOI] [PubMed] [Google Scholar]
- 71.Voight, B. F., Kudaravalli, S., Wen, X. & Pritchard, J. K. A map of recent positive selection in the human genome. PLoS Biol.4, e72 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tang, K., Thornton, K. R. & Stoneking, M. A new approach for using genome scans to detect recent positive selection in the human genome. PLoS Biol.5, e171 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sabeti, P. C. et al. Genome-wide detection and characterization of positive selection in human populations. Nature449, 913–918 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paradis, E. pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics26, 419–420 (2010). [DOI] [PubMed] [Google Scholar]
- 75.Aurrecoechea, C. et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res.37, D539–D543 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sievers, F. & Higgins, D. G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci.27, 135–145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Procter, J. B. et al. Alignment of biological sequences with jalview. in Multiple Sequence Alignment: Methods and Protocols (ed Katoh, K.) 203–224 (Springer US, New York, NY, 2021). 10.1007/978-1-0716-1036-7_13. [DOI] [PMC free article] [PubMed]
- 78.Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature630, 493–500 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci.30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hallgren, J. et al. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. Preprint at bioRxivhttps://www.biorxiv.org/content/10.1101/2022.04.08.487609v1 (2022).
- 81.Paysan-Lafosse, T. et al. InterPro in 2022. Nucleic Acids Res.51, D418–D427 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The data used in this study are available at the European Nucleotide Archive (https://www.ebi.ac.uk/ena). For samples from imported P. vivax cases diagnosed at the UKHSA Malaria Reference Laboratory, data are under accession codes PRJEB44419 and PRJEB56411. For samples from the MalariaGEN P. vivax Genome Variation Project57, data are under accession codes PRJEB2136, PRJEB2140, PRJEB4409, PRJEB4410, PRJEB10888, PRJNA65119, PRJNA67065, PRJNA67237, PRJNA67239, PRJNA175266, PRJNA240366, PRJNA240531, PRJNA271480, PRJNA284437, PRJNA295233, PRJNA420510, PRJNA432819, PRJNA603279, PRJNA643698, and PRJNA655141. The raw data for 175 Indonesian Papua isolates from the MalariaGEN P. vivax Genome Variation Project are available as an unfiltered VCF at ftp://ngs.sanger.ac.uk/production/malaria/Resource/30. All accession codes and sample provenance are detailed in Supplementary Data 1.
For analysis scripts, please see the GitHub repository available at https://github.com/LSHTMPathogenSeqLab/malaria-hub. All scripts use open-source software (see “Methods”). For population genetics analyses, SnpEFF63 (version 5.1), PLINK64 (v1.9), ape (v5.7), bcftools65 (v.1.17), R (v.4.2.3), vcftools68 (v.0.1.17), moimix (v.0.0.2.9001, https://github.com/bahlolab/moimix), ADMIXTURE67 (v.1.3.0), hmmIBD69 (v2.04), rehh70 (v3.2.2), pegas74 (v0.11). For protein structural analysis we used Clustal Omega (https://www.ebi.ac.uk/jdispatcher/msa/clustalo), AlphaFold378 (https://alphafoldserver.com), DeepTMHMM80 (https://dtu.biolib.com/DeepTMHMM), InterPro81 (https://www.ebi.ac.uk/interpro/), JalView77 (v2.11.3.3) and UCSF ChimeraX79 (v1.17.3).