Abstract
HDAC3 has been demonstrated to play a crucial role in the progression of various tumors and the differentiation and development of T cells. However, its impact on peripheral T cells in the development of murine lung cancer remains unclear. In this experiment, a subcutaneous lung tumor model was established in C57BL/6 mice, and tumor-bearing mice were treated with the specific inhibitor of HDAC3, RGFP966, at different doses to observe changes in tumor size. Additionally, a lung tumor model was established using hdac3fl/flcd4cre+/+ mice to investigate its mechanism. Mice injected with 10 mg/kg RGFP966 had the smallest tumor volume, while those injected with 30 mg/kg RGFP966 had the largest tumors. Flow cytometry analysis revealed that the expression of HDAC3 in splenic T cells was reduced in all groups of mice, while IFN-γ and IL-17 A were increased. Moreover, the expression of granzyme B and perforin in splenic CD8+ T cells was increased in all groups of mice. Compared to the use of 30 mg/kg RGFP966 alone, the combination with anti-IL-17 A mAb reduced the infiltration of Neutrophils and exhausted T cells in mouse tumors, thereby impeding tumor development. These findings demonstrate that the use of RGFP966 or T cell-specific loss of hdac3 promotes the expression of IL-17 A in splenic T cells, leading to tumor resistance and providing insights for clinical treatment.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-83447-8.
Subject terms: Tumour immunology, Tumour immunology
Introduction
HDAC3 is a type I HDAC, homologous to the yeast RPD3 protein, and is located in the nucleus where it mainly functions to suppress gene transcription1. Previous studies have shown that high expression of HDAC3 is often associated with poor prognosis in various cancers: HDAC3 plays a crucial role in the development and therapeutic resistance of Kras-mutant non-small cell lung cancer2; in breast cancer patients, NCOR2 regulates HDAC3 to inhibit interferon regulatory factor 1 (IRF-1) dependent gene expression and interferon (IFN) signaling, thereby suppressing anti-tumor therapy3. HDAC3 expression can also inhibit tumor development: HDAC3 deficiency promotes liver cancer through a defect in H3K9ac/H3K9me3 transition4.
The fundamental principle behind HDAC inhibitors (HDACi) as anticancer drugs is based on their ability to induce apoptosis and cell cycle arrest in cancer cells5. Although HDAC inhibitors have been approved by the US Food and Drug Administration (FDA) as anticancer drugs due to their remarkable effectiveness6, their therapeutic results for solid tumors have been disappointing5. Compared to general HDAC inhibitors, HDAC3-specific inhibitors tend to have fewer side effects and are often targeted for developing cancer drugs7. RGFP966 is a specific HDAC3 inhibitor that promotes glioblastoma stem cell differentiation by blocking SMAD7-mediated TGF-β signaling, thereby combating glioblastoma8. The HDAC3-selective inhibitor 4i shows strong cytotoxicity against triple-negative breast cancer cells, promoting the expression of apoptosis-related proteins, and holds promise as a potential candidate for targeted breast cancer therapy9. HDAC3 inhibitors can also be used in combination with PD1/PDL1 monoclonal antibodies, cisplatin, and other drugs to alleviate drug resistance10. However, reports on the resistance to HDAC3 inhibitors themselves are scarce.
Previous studies have found that HDAC3 is crucial for the development of T cell subsets11. During the CD4/CD8 double-positive stage, HDAC3 is an essential factor for downregulating RORγt12. Deacetylation of CD8 lineage-associated gene loci helps maintain the potential of double-positive (DP) thymocytes to mature into CD4+ T or CD8+ T cell lineages13. T cells from the spleen and lymph nodes can migrate into the tumor microenvironment via chemotaxis, influencing tumor progression. Tumor cells with HDAC3 deficiency express high levels of these chemotactic factors, which recruit CXCR3+ T cells into the tumor microenvironment (TME) and inhibit tumor growth in immunocompetent mice14. Therefore, the application of HDAC3 inhibitors in mouse models of lung cancer can impact the differentiation of peripheral T cells, recruit these T cells to the tumor microenvironment, and mediate tumor-killing effects.
In this experiment, we used C57BL/6 mice to establish a transplanted lung cancer model and treated the mice with different doses of the HDAC3-specific inhibitor RGFP966. We found that when the drug dose was increased to 10 mg/kg, the lung cancer progression in the mice was the slowest. However, further increasing the dose of the inhibitor resulted in faster development of lung cancer in the mice. Additionally, we discovered that in T cell-specific knockout mice and tumor-bearing mice treated with 30 mg/kg RGFP966, splenic T cells differentiated into Th17 and Tc17 cells and produced IL-17 A. This resulted in the recruitment of neutrophils into the tumor microenvironment and the exhaustion of tumor-infiltrating T cells, which contributed to the accelerated development of lung cancer. Our research provides new insights for clinical cancer treatment with HDAC inhibitors.
Result
The influence of RGFP966 on tumor development in mice
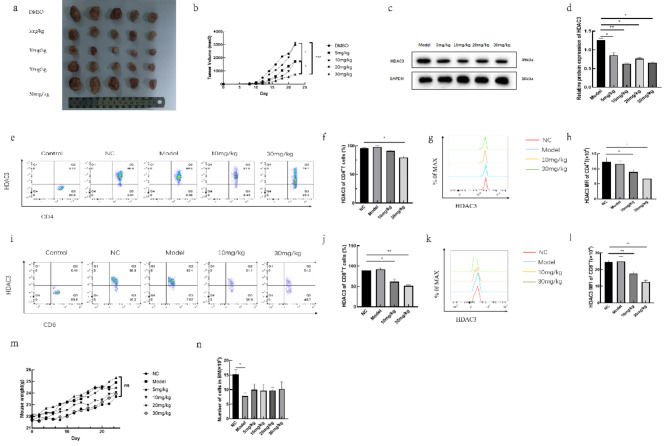
In the C57BL/6 mouse tumor models treated with different doses of RGFP966, the 10 mg/kg group exhibited the smallest lung tumor volume, while the 5 mg/kg and 20 mg/kg groups had intermediate tumor volumes. The 30 mg/kg group and the Model group had the largest tumor volumes (Fig. 1a,b). Western blot analysis of HDAC3 expression in tumor tissues showed that RGFP966 at various concentrations reduced HDAC3 levels, but the extent of inhibition did not increase with higher doses of RGFP966 (Fig. 1c,d). Flow cytometry analysis revealed a significant reduction in HDAC3 expression in CD4+ T and CD8+ T cells in the spleens of treated mice (Fig. 1e–l). Drug toxicity was assessed by monitoring changes in bone marrow cell counts and body weight. Microscopic counting of bone marrow cells and body weight measurements showed that RGFP966 treatment did not result in reduced bone marrow cell counts or body weight (Fig. 1m,n).
Fig. 1.
Injection of RGFP966 reduces the development of lung cancer in mice. The lung cancer tissues of mice in each group (a) and the volume statistics (b). Western blot images (c) and statistical graphs (d) of HDAC3 protein expression in lung cancer tissues of each group; the samples derive from the same experiment and that blots were processed in parallel. The proportion of HDAC3 expressing in CD4+T cells in the spleen (e) and the statistical graph (f), the mean fluorescence intensity (MFI) of HDAC3 expressing in CD4+ T cells in the spleen (g) and the statistical graph (h). The proportion of HDAC3 expressing in CD8+ T cells in the spleen (i) and the statistical graph (j), the MFI of HDAC3 expressing in CD8+ T cells in the spleen (k), and the statistical graph (l). Changes in body weight of mice in each group (m) and changes in bone marrow cell counts (n). *p < 0.05, **p < 0.01, ***p < 0.001, ns not significant.
The differentiation and function of T cell subsets are affected by HDAC3
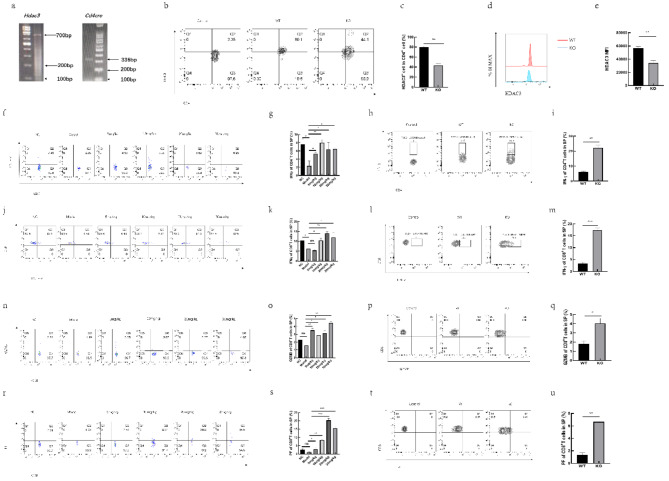
To explore how T cell-expressed HDAC3 inhibits lung cancer development by influencing T cell differentiation and function, we used CD4+ T cell-specific HDAC3 knockout mice with a C57BL/6 background to investigate the underlying mechanisms. The PCR analysis confirmed the correct genotype of the mice (Fig. 2a). Flow cytometry was employed to measure the proportion and mean fluorescence intensity (MFI) of HDAC3 expression in CD4+ T cells from the spleens of knockout mice. These results showed a reduction in HDAC3 expression in these cells compared to WT mice (Fig. 2b–e). Further analysis revealed that compared to WT mice, KO mice showed a decrease in both the proportion and number of T cells in the spleen, lymph nodes, and blood (Supplementary Fig. S1). Due to the loss of certain immune functions, the tumor development in KO tumor-bearing mice was faster than in WT mice (Supplementary Fig. S4). Further flow cytometry analysis assessed IFN-γ expression in splenic T cells from RGFP966-treated tumor mice, as well as in KO and WT mice with established lung cancer models. RGFP966 treatment led to increased IFN-γ expression in splenic CD4+ T cells (Fig. 2f,g) and CD8+ T cells (Fig. 2j,k). KO mice exhibited higher levels of IFN-γ in peripheral CD4+ T cells (Fig. 2h,i) and CD8+ T cells (Fig. 2l,m) compared to WT tumor mice. Additionally, we evaluated Gzmb and Pf expression in CD8+ T cells from the spleens of RGFP966-treated tumor mice, and in KO and WT mice with lung cancer models. RGFP966 treatment resulted in elevated Gzmb (Fig. 2n,o) and Pf (Fig. 2r,s) expression in splenic CD8+ T cells. KO mice also showed increased Gzmb (Fig. 2p,q) and Pf (Fig. 2t,u) levels in peripheral CD8+ T cells compared to WT tumor mice.
Fig. 2.
HDAC3 affects the differentiation and function of T cell subsets. PCR analysis for genotype identification of hdac3fl/flcd4cre+/+ mice (a). The proportion of HDAC3 expressing T cells in KO and WT mice (b) and statistical graph (c); MFI of HDAC3 (d) and statistical graph (e). The proportion of IFN-γ expressing in CD4+ T cells in each group of mice injected with RGFP966 (f) and statistical graph (g). The proportion of IFN-γ expressing in CD4+ T cells in KO and WT mice (h) and statistical graph (i). The proportion of IFN-γ expressing in CD8+ T cells in each group of mice injected with RGFP966 (j) and statistical graph (k). The proportion of IFN-γ expressing in CD8+ T cells in KO and WT mice (l) and statistical graph (m). The proportion of Gzmb expressing in CD8+ T cells in each group of mice injected with RGFP966 (n) and statistical graph (o). The proportion of Gzmb expressing in CD8+ T cells in KO and WT mice (p) and statistical graph (q). The proportion of Pf expressing CD8+ T cells in each group of mice injected with RGFP966 (r) and statistical graph (s). Proportion of Pf expressing in CD8+ T cells in KO and WT mice (t) and statistical graph (u). *p < 0.05, **p < 0.01, ***p < 0.001, ns not significant.
The expression of PD-1 on tumor-infiltrating T cells is affected by HDAC3
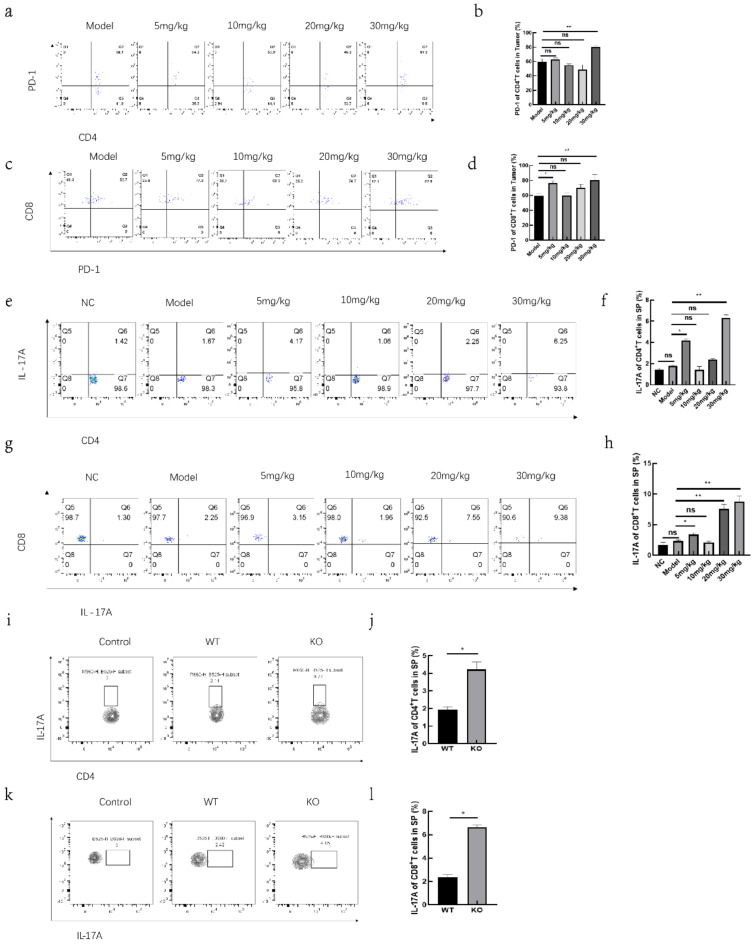
Studies have shown that splenic T cells can migrate into tumor tissues through chemotaxis to perform their immune functions. Among these tumor-infiltrating T cells, some become exhausted and express the exhaustion marker PD-1 during the anti-tumor response. Flow cytometry was used to analyze PD-1 expression on tumor-infiltrating T cells. In mice treated with various doses of the inhibitor, the 30 mg/kg group exhibited increased PD-1 expression in both CD4+ T and CD8+ T cells compared to the Model group (Fig. 3a–d), which may contribute to the faster progression of lung tumors in this group. Research also indicates that IL-17 A can increase PD-1 expression in tumor-infiltrating T cells. Flow cytometry was employed to measure IL-17 A expression in splenic T cells across different inhibitor treatment groups. The results showed that, with the exception of the 10 mg/kg group, IL-17 A expression in CD4+ T and CD8+ T cells in the spleen was higher in the other treatment groups compared to the Model group (Fig. 3e–h), and the levels of IL-17 A in the serum were also increased (Supplementary Fig. S2a). Additionally, IL-17 A expression in CD4+ and CD8+ T cells was increased in KO tumor-bearing mice compared to WT tumor-bearing mice (Fig. 3i–l), and the levels of IL-17 A in the serum were also increased (Supplementary Fig. S2b).
Fig. 3.
HDAC3 affects the expression of PD-1 in tumor-infiltrating T cells. In tumor-bearing mice injected with RGFP966, expression of PD-1 in CD4+ T cells (a) and statistical graph (b). In tumor-bearing mice injected with RGFP966, expression of PD-1 in CD8+ T cells (c) and statistical graph (d). In mice injected with RGFP966, expression of IL-17 A in CD4+ T cells (e) and statistical graph (f). In mice injected with RGFP966, expression of IL-17 A in CD8+ T cells (g) and statistical graph (h). Expression of IL-17 A in CD4+ T cells in WT and KO tumor-bearing mice (i) and statistical graph (j). Expression of IL-17 A in CD8+ T cells in WT and KO tumor-bearing mice (k) and statistical graph (l). *p < 0.05, **p < 0.01, ns not significant.
The influence of HDAC3 on the differentiation of Th17 and Tc17 cells
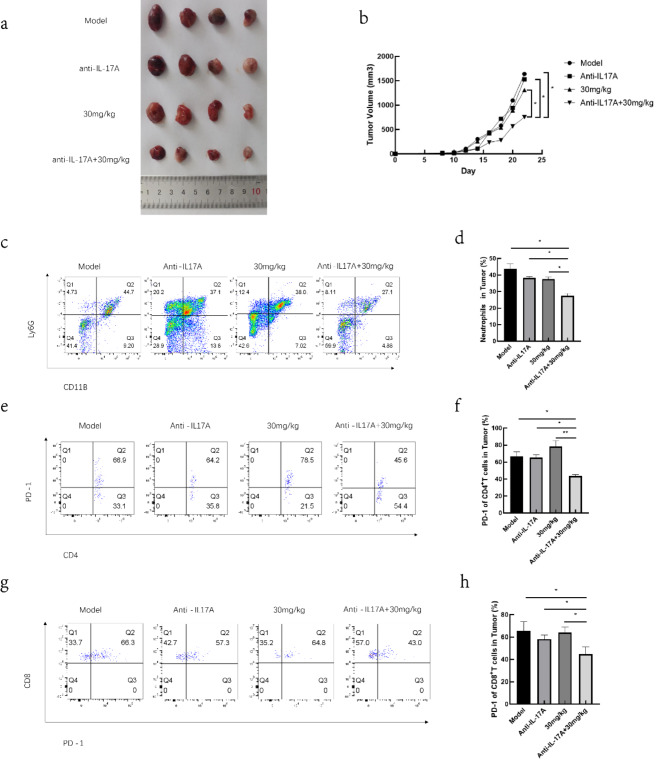
To further explore the impact of IL-17 A on the exhaustion of tumor-infiltrating T cells and its effect on lung cancer progression, we combined anti-IL-17 A with 30 mg/kg RGFP966. The results showed that this combination treatment significantly slowed lung cancer progression compared to RGFP966 alone, anti-IL-17 A alone, and the Model control group (Fig. 4a,b). Additionally, the combination treatment markedly reduced the number of tumor-infiltrating neutrophils (Fig. 4c,d) and decreased the proportion of PD-1 expressing in CD4+ T and CD8+ T cells in the tumor tissue compared to the other groups (Fig. 4e–h).
Fig. 4.
HDAC3 affects the differentiation of Th17 and Tc17 cells. Tumors in each group of mice (a) and their volumes (b). Neutrophil infiltration in tumor tissues of each group of mice (c) and statistical graph (d). PD-1 expression in CD4+ T cells infiltrating tumor tissues of each group of mice (e) and statistical graph (f). PD-1 expression in CD8+ T cells infiltrating tumor tissues of each group of mice (g) and statistical graph (h). *p < 0.05, **p < 0.01.
Discussion
HDAC3 has emerged as a promising target for cancer therapy development. RGFP966, a specific HDAC3 inhibitor, induces apoptosis in tumor cells by causing DNA damage and disrupting the S phase of the cell cycle15. However, the effects of RGFP966 on the differentiation and function of peripheral immune cells in lung cancer mice are not fully understood. In this study, lung cancer mice were treated with RGFP966 at four different doses. The results indicated that the 10 mg/kg dose group exhibited the smallest tumor volume, while the 5 mg/kg and 20 mg/kg dose groups showed intermediate tumor volumes. The largest tumor volumes were observed in the Model group and the 30 mg/kg dose group. Treatment with RGFP966 at all doses did not cause weight loss or reduce bone marrow cell counts in the mice. This suggests that an appropriate dose of RGFP966 can inhibit lung cancer progression without causing significant drug-related toxicity. RGFP966 effectively reduced HDAC3 levels in peripheral T cells, with the reduction proportional to the drug dose, confirming the successful inhibition of HDAC3. Although RGFP966 decreased HDAC3 levels in lung tumor tissues, the extent of inhibition did not increase with higher doses. This suggests that the tumor barrier limits drug penetration into the tumor microenvironment. Increasing the dose of RGFP966 does not enhance its direct tumor-killing effect but instead affects lung cancer progression through modulation of the immune system.
The clinical therapeutic effect of RGFP966 was still unclear, but it had been shown to inhibit various types of tumors. RGFP966 induces acetylation of WISP2 at lysine K6 and prevents its degradation, leading to reduced cell proliferation and increased apoptosis in AML cells16. It also inhibits the proliferation and migration of HCC cells17, and suppresses the growth of PTEN-deficient and SPOP-mutant prostate cancer cells in mouse cultures, patient-derived organoids, and xenografts18. Based on these previous findings and the results of the current study, we believed RGFP966 held potential as a clinical cancer treatment.
PD-1 is a marker of exhausted T cells that infiltrate tumors19. Within these tumor-infiltrating T cells, a higher proportion expressing PD-1 is associated with a weaker ability to suppress the tumor20. In this research, in the 30 mg/kg group, lung cancer tissues exhibited the highest levels of PD-1 expression on infiltrating T cells, indicating the greatest degree of T cell exhaustion. This group had the largest tumor volume and the highest proportion of T cell infiltration, leading to the most exhausted T cells and the strongest immune suppression. Therefore, the 30 mg/kg group showed the most extensive PD-1 expression on both peripheral and tumor-infiltrating T cells, contributing to its highest level of drug resistance.
Peripheral T cells can migrate into the tumor microenvironment and perform their functions through chemotaxis14. This study explored how HDAC3 affects the differentiation and function of peripheral T cells and the reasons for the increased PD-1 expression on tumor-infiltrating T cells in lung cancer. The results demonstrated that RGFP966 at various doses enhanced the expression of IFN-γ, granzyme B, and perforin in peripheral T cells. The KO mice exhibit the same changes as the WT mice. Due to the tumor-killing abilities of Tc1, Th1, and CTL cells21–23, reducing the expression of HDAC3 in spleen T cells inhibits the development of lung cancer by promoting the differentiation of T cells into Th1, Tc1, and CTL. However, except the 10 mg/kg dose, other doses of RGFP966 also promoted the differentiation of peripheral Th17 and Tc17 cells.
IL-17 A can recruit neutrophils24 and, by activating the VEGFR1-PI3K-AKT pathway, promotes neutrophil polarization, which induces CD8+ T cell exhaustion and creates an immunosuppressive environment25. Tumor-infiltrating neutrophils further suppress T cell function in the tumor microenvironment by producing NO, GM-CSF, AHRR, and IL-626–28. These neutrophils also express high levels of PD-L1 via the GM-CSF/JAK/STAT3 pathway29. The interaction between elevated PD-L1 on neutrophils and PD-1 on T cells helps tumors evade immune-induced apoptosis and facilitates tumor progression30. When tumor-infiltrating neutrophils express higher levels of PD-L1, surrounding T cells may receive stronger inhibitory signals. In response, T cells may upregulate PD-1 to better respond to PD-L1 signals31. Thus, we hypothesize that Th17 and Tc17 cells, through IL-17 A secretion, recruit neutrophils and lead to increased PD-1 expression on tumor-infiltrating T cells in lung cancer. Our study tested this hypothesis using an anti-IL-17 A antibody. Compared to the Model group, anti-IL-17 A group, and 30 mg/kg RGFP966 group, the combination treatment significantly reduced the number of tumor-infiltrating neutrophils and decreased T cell exhaustion, thereby slowing lung cancer progression. This supports our hypothesis. In the 10 mg/kg RGFP966 group, T cells secreted the least IL-17 A, and the lung tumors were the smallest. Therefore, 10 mg/kg appears to be the optimal dose of RGFP966 for inhibiting lung cancer progression. This study provides insights into how RGFP966 affects lung cancer development and resistance by modulating peripheral T cell differentiation, offering valuable implications for cancer treatment in clinical settings.
Conclusion
In lung cancer mice with peripheral T cells lacking HDAC3 or those treated with RGFP966, there is enhanced differentiation of Th1 and Tc1 cells and increased CTL cytotoxicity, which helps suppress lung cancer progression. However, at a dose of 30 mg/kg, RGFP966 promotes the differentiation of Th17 and Tc17 cells, leading to increased PD-1 expression on tumor-infiltrating T cells and contributing to drug resistance in lung cancer.
Methods
Mice
C57BL/6 and hdac3fl/flcd4cre+/+ mice(KO mice) were used for experiments. Mice carrying a conditional floxed allele of hdac3 (hdac3flox, Stock No: 024119) were backcrossed onto the C57BL/6 background for 5 generations and then mated to C57BL/6 mice carrying the cd4 enhancer/ promoter/silence cre allele (obtained from The Jackson Laboratory, Stock No:022071) to generate the hdac3 conditional knockout mice, designated as hdac3fl/flcd4cre+/+. Mice were housed in a specific pathogen-free barrier unit. This experimental plan has been approved by the Animal Care and Use Committee at North China University of Science and Technology (No. LX201834). The handling of mice and experimental procedures were conducted in accordance with the relevant guidelines and regulations established by the committee. Mice were anesthetized and euthanized by intraperitoneal injection of sodium pentobarbital.
Cell line
The Lewis lung carcinoma cell line (LLC) and the mouse lymphoma cell line (EL4) were both purchased from Wuhan Pricella Biotechnology. Cells were cultured at 37 °C in a 5% CO2 environment. LLC cells were cultured in a complete medium composed of 10% fetal bovine serum(Pricella), 1% penicillin-streptomycin (Gibco), and 89% high-glucose DMEM basal medium (Hyclone). EL4 cells were cultured in a complete medium composed of 10% fetal bovine serum, 1% penicillin-streptomycin (Gibco), and 89% 1640 basal medium (Hyclone).
Genotyping
Offsprings were genotyped using the following PCR primer pairs: for cd4cre mice, Wild type Reverse: 5’-TATGCTCTAAGGACAAGAATTGACA-3’; Mutant Reverse: 5’-CTTTGCAGAGGGCTAACAGC-3’; Common: 5’-GTTCTTTGTATATATTGAATGTTAGCC-3’. For hdac3 mice, Wild type Forward: 5’-TGGTGGTGAATGGCTTTAATC-3’, reverse: 5’-TAACGGGAGCAGAACTCGAA-3’. The hdac3fl/fl, cd4cre+/+ and cd4cre+/− mice produce DNA fragments of 700 bp; 336 bp; 336 bp, and 262 bp respectively after PCR amplification.
Mouse modeling
Sex-matched C57BL/6 mice were subcutaneously injected with PBS containing 1 × 106 LLC cells. A week later, the presence of a palpable tumor mass at the injection site indicated successful modeling. hdac3fl/flcd4cre+/+ mice were modeled in the same way. Fifty successfully modeled C57BL/6 mice were randomly divided into five groups: DMSO control group, 5 mg/kg RGFP966 group, 10 mg/kg RGFP966 group, 20 mg/kg RGFP966 group, and 30 mg/kg RGFP966 group. The treatment was administered every two days for two weeks. Additionally, 10 untreated C57BL/6 mice were randomly selected as the normal control group. Four hdac3fl/flcd4cre+/+ and hdac3fl/flcd4cre−/− genotype mice were used to establish subcutaneous lung tumor models. Forty successfully modeled C57BL/6 mice were randomly divided into four groups: solvent control group, 30 mg/kg RGFP966 group, 0.1 mg anti-IL-17 antibody (17F3, BioXCell) group, and 30 mg/kg RGFP966 plus anti-IL-17 antibody group. RGFP966 and anti-IL-17 antibody were administered via intraperitoneal injection, every two days for two weeks. The tumor size and body weight of the mice were measured. After the injections were completed, the mice were anesthetized and euthanized, and the number of bone marrow cells was counted under a microscope.
Flow cytometry analysis
Anesthetize and euthanize the mice, then isolate the spleen or tumor tissues to prepare single-cell suspensions. Use NH4Cl to lyse red blood cells, and filter the suspension through a 100-micron mesh. Incubate the cell suspension at 4 °C with Fc Block reagent (clone 2.4G2) for 10 min to block nonspecific binding. Add the following antibodies and perform cell surface staining at 4 °C for 30 min: anti-CD4-APC(Biolegend), anti-CD8-PerCP-Cy5.5(BD Biosciences), anti-CD11b-APC (BD Biosciences), Anti-Ly6G-PerCP-Cy5.5(eBioscience), anti-PD-1-PE(BD Biosciences). The cell was washed with PBS buffer and then analyzed with a Beckman flow cytometer. The data were analyzed by FlowJo software.
Intracellular cytokine staining
Splenic cells from the WT-control, the KO-control, the WT-RA, and the KO-RA mice were suspended in RPMI1640 cell culture medium in the presence of 2 µg Activation Cocktail(550583, BD Biosciences), incubated 4 h at 37 °C. The cells were collected and first performed by cell surface staining. Then, the cells were fixed and permeated with a Cytofix/ Cytoperm kit (554722, BD Biosciences). After washing with PBS, the cells were intracellularly stained with anti-mouse-IFN-γ-PE(clone XMG1.2, Biolegend), anti-mouse-IL-4-APC(clone 11B11, Biolegend, ) and anti-mouse-IL-17 A-FITC (clone TC11-18H10.1, Biolegend) for 30 min at 4 °C, washed with PBS buffer, and then analyzed with a Beckman flow cytometer. The data were analyzed by Flow Jo software.
Western blot
Isolate mouse tumor tissues and prepare a single-cell suspension. Add 1 ml of RIPA buffer and 10 µl of PMSF to extract the proteins, then measure the protein concentration using a BCA assay kit (Solarbio). Load 10 µg of the protein sample onto a 10% SDS-PAGE gel for electrophoresis (80 V, 30 min and then 120 V, 90 min), transferred to PVDF membrane (1 h), sealed with 5% skimmed milk powder (1 h), and then incubated overnight with anti-mouse HDAC3 antibody (1:1000, Abcam), rabbit anti-mouse GAPDH antibody (1:1000, Abcolonal) in 4 °C refrigerator. The next day, they were incubated with goat anti-rabbit antibodies (1:5000, Proteintech) for 1 h and observed protein bands with ECL kit.
ELISA
Collected mouse whole blood and left it at room temperature for 1 h. After the blood coagulated, centrifuged at 1000 g, 4 °C for 5 min to obtain the serum, and then performed detection using the Mouse IL-17 A ELISA Kit (ab199081, abcam).
Statistical analysis
SPSS17 software was employed for the data processing and statistical analysis. One-way ANOVA and Student’s t-test were used to analyze the statistical difference of multiple or two groups of samples respectively. The data were presented as the mean ± SD, and p < 0.05 indicates that the difference is statistically significant.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Author contributions
H.C. and Q.Z. conceived and supervised study. F.X., A.Q., and Q.Z. designed the study and contributed to the literature search. H.C., A.Q., F.X., S.G., and G.Z. performed the experiments and contributed to data collection and data interpretation. A.Z., W.L., F.T., and H.C. contributed to data analysis. H.C. and Q.Z. wrote the paper. All authors discussed the results and commented on the manuscript.
Funding
The National Natural Science Foundation of China (No. 8137311) and the Hebei Provincial Health Commission (No. 20190105) provided fundings for this research.
Data availability
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Accordance
We confirmed that all experiments in this study were performed in accordance with the relevant guidelines and regulations.
Arrive
All the procedure of the study is followed by the ARRIVE guidelines.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Emiliani, S., Fischle, W., Van Lint, C., Al-Abed, Y. & Verdin, E. Characterization of a human RPD3 ortholog, HDAC3. Proc. Natl. Acad. Sci. USA95, 2795–2800. 10.1073/pnas.95.6.2795 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eichner, L. J. et al. HDAC3 is critical in tumor development and therapeutic resistance in Kras-mutant non-small cell lung cancer. Sci. Adv.9, eadd3243. 10.1126/sciadv.add3243 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsai, K. K. et al. Screening of organoids derived from patients with breast cancer implicates the repressor NCOR2 in cytotoxic stress response and antitumor immunity. Nat. Cancer3, 734–752. 10.1038/s43018-022-00375-0 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ji, H. et al. HDAC3 deficiency promotes liver cancer through a defect in H3K9ac/H3K9me3 transition. Cancer Res.79, 3676–3688. 10.1158/0008-5472.can-18-3767 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vancurova, I., Uddin, M. M., Zou, Y. & Vancura, A. Combination therapies targeting HDAC and IKK in solid tumors. Trends Pharmacol. Sci.39, 295–306. 10.1016/j.tips.2017.11.008 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang, Z. et al. Tucidinostat plus exemestane for postmenopausal patients with advanced, hormone receptor-positive breast cancer (ACE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol.20, 806–815. 10.1016/s1470-2045(19)30164-0 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Sarkar, R., Banerjee, S., Amin, S. A., Adhikari, N. & Jha, T. Histone deacetylase 3 (HDAC3) inhibitors as anticancer agents: A review. Eur. J. Med. Chem.192, 112171. 10.1016/j.ejmech.2020.112171 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Liang, H. et al. RGFP966, a histone deacetylase 3 inhibitor, promotes glioma stem cell differentiation by blocking TGF-beta signaling via SMAD7. Biochem. Pharmacol.180, 114118. 10.1016/j.bcp.2020.114118 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Pulya, S. et al. Selective HDAC3 inhibitors with potent in vivo antitumor efficacy against triple-negative breast cancer. J. Med. Chem.66, 12033–12058. 10.1021/acs.jmedchem.3c00614 (2023). [DOI] [PubMed] [Google Scholar]
- 10.Wang, H. et al. Enhanced histone H3 acetylation of the PD-L1 promoter via the COP1/c-Jun/HDAC3 axis is required for PD-L1 expression in drug-resistant cancer cells. J. Exp. Clin. Cancer Res.39, 29. 10.1186/s13046-020-1536-x (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stengel, K. R. et al. Histone deacetylase 3 is required for efficient T cell development. Mol. Cell. Biol.35, 3854–3865. 10.1128/MCB.00706-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Philips, R. L. et al. HDAC3 is required for the downregulation of RORgammat during thymocyte positive selection. J. Immunol.197, 541–554. 10.4049/jimmunol.1502529 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Philips, R. L. et al. HDAC3 restrains CD8-lineage genes to maintain a bi-potential state in CD4(+)CD8(+) thymocytes for CD4-lineage commitment. Elife8, e43821. 10.7554/elife.43821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, L. et al. HDAC3 inhibition promotes antitumor immunity by enhancing CXCL10-mediated chemotaxis and recruiting of immune cells. Cancer Immunol. Res.11, 657–673. 10.1158/2326-6066.cir-22-0317 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wells, C. E. et al. Inhibition of histone deacetylase 3 causes replication stress in cutaneous T cell lymphoma. PLoS One8, e68915. 10.1371/journal.pone.0068915 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang, H. et al. Acetylation stabilizes the signaling protein WISP2 by preventing its degradation to suppress the progression of acute myeloid leukemia. J. Biol. Chem.299, 102971. 10.1016/j.jbc.2023.102971 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu, X. et al. RGFP966 suppresses tumor growth and migration through inhibition of EGFR expression in hepatocellular carcinoma cells in vitro. Drug Des. Dev. Ther.14, 121–128. 10.2147/DDDT.S234871 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan, Y. et al. Dual inhibition of AKT-mTOR and AR signaling by targeting HDAC3 in PTEN- or SPOP-mutated prostate cancer. EMBO Mol. Med.10, e8478. 10.15252/emmm.201708478 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wherry, E. J. T cell exhaustion. Nat. Immunol.12, 492–499. 10.1038/ni.2035 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Chow, A., Perica, K., Klebanoff, C. A. & Wolchok, J. D. Clinical implications of T cell exhaustion for cancer immunotherapy. Nat. Rev. Clin. Oncol.19, 775–790. 10.1038/s41571-022-00689-z (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basu, A. et al. Differentiation and regulation of T(H) cells: a balancing act for cancer immunotherapy. Front. Immunol.12, 669474. 10.3389/fimmu.2021.669474 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang, H., Fu, D., Bidgoli, A. & Paczesny, S. T. Cell subsets in graft versus host disease and graft versus tumor. Front. Immunol.12, 761448. 10.3389/fimmu.2021.761448 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nava, L. C. et al. Linoleic acid potentiates CD8(+) T cell metabolic fitness and antitumor immunity. Cell Metab.35, 633–650. 10.1016/j.cmet.2023.02.013 (2023). [DOI] [PubMed] [Google Scholar]
- 24.Zhang, Y. et al. Interleukin-17-induced neutrophil extracellular traps mediate resistance to checkpoint blockade in pancreatic cancer. J. Exp. Med.217, e20190354. 10.1084/jem.20190354 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, Z. et al. γδT Cell-IL17A-neutrophil axis drives immunosuppression and confers breast cancer resistance to high-dose anti-VEGFR2 therapy. Front. Immunol.12, 699478. 10.3389/fimmu.2021.699478 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogel, C. F. et al. Transgenic overexpression of aryl hydrocarbon receptor repressor (AhRR) and AhR-mediated induction of CYP1A1, cytokines, and acute toxicity. Environ. Health Perspect.124, 1071–1083. 10.1289/ehp.1510194 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koyama, S. et al. STK11/LKB1 deficiency promotes neutrophil recruitment and proinflammatory cytokine production to suppress T-cell activity in the lung tumor microenvironment. Cancer Res.76, 999–1008. 10.1158/0008-5472.can-15-1439 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao, Y. et al. Neutrophils resist ferroptosis and promote breast cancer metastasis through aconitate decarboxylase 1. Cell Metab.35, 1688–1703. 10.1016/j.cmet.2023.09.004 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, T. T. et al. Tumour-activated neutrophils in gastric cancer foster immune suppression and disease progression through GM-CSF-PD-L1 pathway. Gut66, 1900–1911. 10.1136/gutjnl-2016-313075 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lei, Q., Wang, D., Sun, K., Wang, L. & Zhang, Y. Resistance mechanisms of anti-PD1/PDL1 therapy in solid tumors. Front. Cell Dev. Biol.8, 672. 10.3389/fcell.2020.00672 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan, O. et al. TOX transcriptionally and epigenetically programs CD8(+) T cell exhaustion. Nature571, 211–218. 10.1038/s41586-019-1325-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.