Abstract
Global health is affected by viral, bacterial, and fungal infections that cause chronic and often fatal diseases. Identifying novel antimicrobials through innovative methods that are active against human pathogens will create a new, necessary pipeline for chemical discovery and therapeutic development. Our goal was to determine whether algal production systems represent fertile ground for discovery of antibiotics and antifungals. To this end, we collected high-biomass algal-bacterial samples from outdoor mass cultivation systems, 18-L outdoor algal open cultures mesocosms, and non-axenic laboratory samples. We also cultivated 33 marine bacterial isolates for chemical extraction. Ultimately, we filtered, concentrated, extracted, and screened 77 chemically-complex mixtures using a conventional agar-based microbial growth inhibition assay against three microbes: Escherichia coli, Bacillus subtilis, and Candida albicans. We discovered that 23 of our chemical extracts (almost one-third of the chemical samples tested) exhibited some degree of growth inhibition toward B. subtilis and/or C. albicans. Our work here demonstrates the feasibility and potential of isolating bioactive natural products from high-biomass algal-bacterial samples from algal mass cultivation systems.
Keywords: Antimicrobial, Antifungal, Algal production systems, Algal mass cultivation systems, Microbial consortia, Natural products
Subject terms: Microbiology techniques, Natural products
Introduction
Emerging infectious disease continues to reveal how vulnerable humanity, our national security, and the global economy are to a single pathogen. Incidence of infection with pneumonia, fungal, and various multi-drug resistant (MDR) pathogens that have coincided with COVID-19 diagnosis has exacerbated the over-use of society’s most precious antimicrobial therapeutics and resulted in accelerated antimicrobial resistance1. Thus, demand for new antimicrobial therapeutics (e.g., antibiotics, antivirals, and antifungals) to prepare for natural and bio-engineered pathogens (bacterial, viral, and fungal) is imminent.
Over the last 75 years since the discovery of penicillins2, we have exploited, over-prescribed, and over-used the transformative antimicrobial therapeutics discovered in the 20th century. We now face a future of untreatable, incurable infections from multi-drug resistant (MDR) pathogens, leading to higher mortality rates3. By 2050, common medical procedures—childbirth, a tooth removal, chemotherapy, or an appendectomy—will be life-threatening procedures without available, effective medications for bacterial infections that frequently occur in health care facilities today4. The COVID-19 pandemic further demonstrates how inadequate arsenals of antimicrobials make humanity vulnerable to a single emergent pathogen (a virus, bacterium, or fungus). However, lack of funding5, lack of interest and monetary incentive by pharmaceutical companies6, and lack of innovative discovery platforms and pipelines7have stymied antimicrobial discovery and development for over 30 years4,8. Pharmaceutical companies, such as Novartis9, are turning away from antibiotic discovery largely due to excessive regulatory burden, lack of return on large investments10,11, and low profit margins for antimicrobials compared to designer-drugs12. Despite decades of synthesis, screening biological isolates, and high-throughput chemical-library screening efforts by pharmaceutical companies, the natural world has been, and continues to be, our best source for discovery of novel antimicrobials4.
Microorganisms (e.g., bacteria and fungi) are natural, prolific producers of chemicals—often referred to as “natural products”—with antimicrobial properties to compete against other microorganisms in their environment. These microbial natural products remain the best sources for antimicrobial therapeutics4. Notably, penicillin was discovered from a fungal strain and remains in use today – almost 100 years later. Since the 1960s, traditional antimicrobial discovery platforms have involved screening soil-derived culturable microorganisms for antimicrobial activity13. However, scientists have only focused on the 1% of bacteria that are easily grown in a laboratory14, such as Actinomycetes spp., and are yielding diminishing returns – the same antimicrobials are now commonly ‘re-discovered’4. By exclusively studying bacteria that are easily grown in a laboratory, researchers are missing out on the rich, untapped chemical potential of unknown and uncultured bacteria present in complex ecological assemblages.
Algal mass cultivation systems, used for production of biofuel and other high value products, have high biomass and high bacterial diversity15, so the competition between microbes is likely fierce. Although many microalgal strains have been surveyed for their ability to generate antimicrobial compounds16–19, the bacteria present in these natural assemblages remain unexplored for their antimicrobial potential. To survive, these algal-pond bacteria engage in ‘chemical warfare’ by expelling chemicals into their surroundings to ‘thwart’ microbial competitors. These bacterially-generated ‘chemical warfare agents’ have value as antimicrobials. Past work characterizing the bacteria in algal ponds, found that Gammaproteobacteria are among the many bacterial taxa present in mass algal cultivation systems15,20. The Gammaproteobacteria class contains many known MDR pathogens as well as several identified by the CDC as ‘Serious Threats’21, including the family Enterobacteriaceae and the genera Actinobacter, Salmonella, Shigella, Escherichia, and Pseudomonas22. Additionally, a wide variety of bacterial taxa are present in algal production ponds, including many under studied marine organisms15,20,23. These organisms survive through constant, intense microbial competition with each other. Thus, we hypothesized that there are ‘natural products’ with antimicrobial properties generated within these algal production systems, targeted not only against bacterial strains that may be related to human pathogens, including Gammaproteobacteria, but also against members of other microbial taxa and fungal pathogens. Screening for chemicals with antimicrobial properties from these mass cultivation systems could lead to the eventual discovery of novel therapeutics that could be used to treat MDR microbial infections.
To test this hypothesis, we conducted a proof-of-principal study (herein) to screen algae production ponds for antimicrobial activity against a broad-spectrum panel of human pathogen surrogates (Escherichia coli, Bacillus subtilis, and Candida albicans). We decided to test these risk-group 1 (RG1) organisms as they represent a gram-negative bacterial strain of interest for novel therapeutics (E. coli) and closely related to strains with observed drug resistance21, a gram-positive bacterial strain of interest with biofilm-forming potential (B. subtilis) with close relatives to pathogenic strains (e.g., Bacillus anthracis), and a fungal strain of rising concern (C. albicans) that is within the same genus as the drug-resistant Candida auris21. We collected pond samples from 77 high-biomass algal-bacterial co-cultures from production systems of various scales (1 L to 100,000 L) as well as unexplored marine bacterial isolates derived from algal culture systems. Samples were processed and then applied to filter disks for use in a broad-spectrum screening for antimicrobial activity. Of the 77 pond extracts screened, 23 exhibited antimicrobial activity against the gram-positive bacteria (B. subtilis) and/or the yeast (C. albicans). These results demonstrate the abundance of potential antimicrobials waiting to be discovered from marine algal production ponds.
Results and discussion
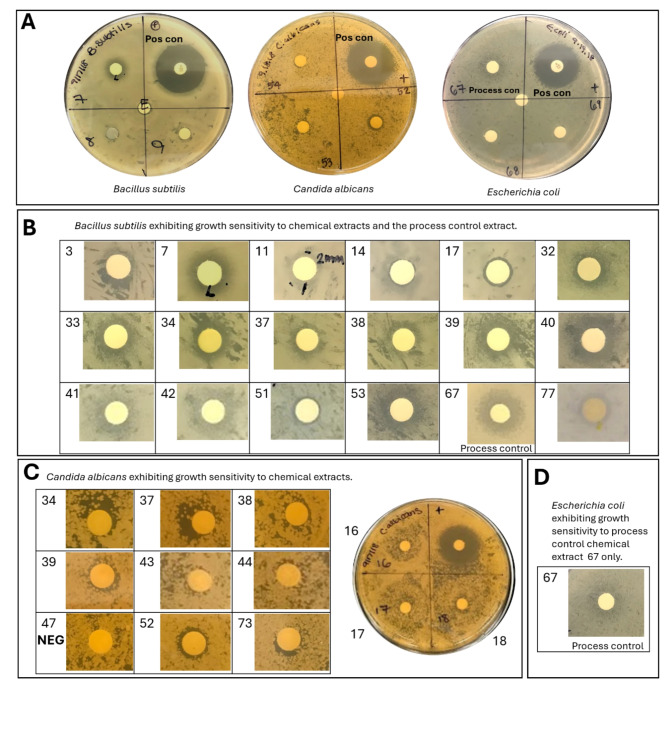
In total, 77 chemical extracts from various laboratory and outdoor cultivation systems (Tables 1 and 2) were sampled, filtered, concentrated, extracted, and then screened for bioactivity across Escherichia coli, Bacillus subtilis, and Candida albicans to assess growth sensitivity. We used a conventional Kirby-Baurer plate assay24 to characterize the antimicrobial activity of the 77 chemical extracts by screening for growth inhibition of these microorganisms. Representative plates for B. subtilis, C. albicans, and E. coli are shown with corresponding positive and negative control disks in Fig. 1A. Of the chemical extracts tested, 23 exhibited inhibitory activity against the growth of B. subtilis and/or C. albicans (Fig. 1B and C). Tables 1 and 2 catalog the chemical extracts (enumerated 1–77) tested, the biological sample type (e.g. marine isolate or algal-bacterial cultivation system) the chemical extracts were derived from and observed levels of growth inhibition. Several chemical extracts exhibited partial growth inhibition, with clearly viable microcolonies within the zone of inhibition (e.g. extracts 16–18 against C. albicans; Fig. 1C). For the purposes of this initial study and based on the likely low concentration of the inhibiting compound in each sample, even such partial growth inhibition is considered to be indicative of putative antimicrobial activity. Differences in the apparent color of the microbial growth and zones of inhibition, particularly with Bacillus subtilis, are an artifact of the image collection process and are of no biological relevance. Images were collected on several different days and the camera, illumination, and other parameters varied.
Table 1.
Sample origin and Sanger sequencing results for tested chemical extracts (1–39) and reported sensitivity for B. subtilis, C. albicans, and E. coli.
| # | origin of sample | Sanger sequencing (top hit) or ATCC information | OD 595 | µg added to disk | Bs | Ec | Ca |
|---|---|---|---|---|---|---|---|
| 1 | O-Medium | 150 | |||||
| 2 | O-Medium | 300 | |||||
| 3 | MBI | Uncultured | 0.474 | 300 | xx | ||
| 4 | MBI | Uncultured | 0.368 | 300 | |||
| 5 | I-Lab | 150 | |||||
| 6 | I-Lab | 1000 | |||||
| 7 | O-Medium | 839 | xxx | ||||
| 8 | MBI | Pseudoalteromonas luteoviolacea, ATCC (purple isolate) | n.d. | 552 | |||
| 9 | MBI | Pseudoalteromonas luteoviolacea, ATCC (white isolate) | n.d. | 755 | |||
| 10 | MBI | Pseudoalteromonas luteoviolacea, ATCC (P.Lu 81) | n.d. | 340 | |||
| 11 | O-Mass | 150 | x | ||||
| 12 | MBI | Erythrobacter | 0.431 | 1255 | |||
| 13 | MBI | Pseudoalteromonas luteoviolacea, ATCC (P.Lu 80) | n.d. | 895 | |||
| 14 | MBI | Uncultured | 0.573 | 542 | x | ||
| 15 | MBI | Reugeria | 0.38 | 330 | |||
| 16 | MBI | Erythrobacter | 0.663 | 535 | xx | ||
| 17 | MBI | Salinimicrobium | 0.305 | 771 | xx | xx | |
| 18 | MBI | Uncultured | 0.453 | 1018 | xx | ||
| 19 | O-Mass | 5762 | |||||
| 20 | MBI | Pseudoalteromonas rubra, ATCC (P.Rub69) | 0.768 | 500 | |||
| 21 | MBI | Erythrobacter | 0.667 | 1475 | |||
| 22 | I-Lab | 785 | |||||
| 23 | I-Lab | 803 | |||||
| 24 | MBI | Pseudomonas | 0.667 | 630 | |||
| 25 | O-Mass | 545 | |||||
| 26 | O-Medium | 562 | |||||
| 27 | MBI | Rheinheimeria | 0.545 | 55 | |||
| 28 | MBI | Ruegeria | 0.584 | 300 | |||
| 29 | O-Medium | 1720 | |||||
| 30 | MBI | Uncultured | 0.465 | 325 | |||
| 31 | MBI | Uncultured | 0.092 | 782 | |||
| 32 | MBI | Erythrobacter | 0.924 | 815 | xx | ||
| 33 | O-Mass | 970 | xx | ||||
| 34 | O-Mass | 590 | xxx | xx | |||
| 35 | O-Medium | 300 | |||||
| 36 | O-Mass | 1180 | |||||
| 37 | O-Mass | 990 | xx | xx | |||
| 38 | O-Mass | 750 | xx | x | |||
| 39 | O-Mass | 300 | xx | X | |||
| Outdoor algal mass cultivation system = O-Mass; 18-L outdoor medium algal cultures = O-Med; marine bacterial isolates = MBI; indoor lab cultures = I-Lab. Growth inhibition scale: “x” = borderline, “xx” = clear inhibition, “xxx” = strong inhibition. Also “n.d.” = OD595 not determined. | |||||||
Table 2.
Sample origin and Sanger sequencing results for tested chemical extracts (40–78) and reported sensitivity for B. subtilis, C. albicans, and E. coli.
| # | origin of sample | Sanger sequencing (top hit) or ATCC information | OD 595 | µg added to disk | Bs | Ec | Ca |
|---|---|---|---|---|---|---|---|
| 40 | O-Mass | 300 | xxx | ||||
| 41 | O-Mass | 300 | xx | ||||
| 42 | O-Mass | 300 | xx | ||||
| 43 | MBI | Pseudomonas | n.d. | 680 | x | ||
| 44 | MBI | Pseudomonas | n.d. | 1690 | x | ||
| 45 | MBI | Pseudomonas | n.d. | 1695 | |||
| 46 | MBI | Pseudomonas | n.d. | 3490 | |||
| 47 | I-Lab | 775 | |||||
| 48 | I-Lab | 1530 | |||||
| 49 | I-Lab | 855 | |||||
| 50 | MBI | Uncultured | 0.776 | 1605 | |||
| 51 | I-Lab | 1060 | x | ||||
| 52 | I-Lab | 1055 | xxx | ||||
| 53 | I-Lab | 885 | xxx | ||||
| 54 | MBI | Vibrio | n.d. | 825 | |||
| 55 | MBI | Flavobacterium | n.d. | 663 | |||
| 56 | O-Medium | 285 | |||||
| 57 | O-Medium | 608 | |||||
| 58 | I-Lab | 832 | |||||
| 59 | I-Lab | 1090 | |||||
| 60 | MBI | Roseobacter | 0.009 | 657 | |||
| 61 | I-Lab | 1440 | |||||
| 62 | MBI | Vibrio | 0.006 | 970 | |||
| 63 | I-Lab | 1092 | |||||
| 64 | MBI | Flavobacterium | 0.702 | 1402 | |||
| 65 | O-Medium | 300 | |||||
| 66 | O-Medium | 970 | |||||
| 67 | Process control | 790 | xx | xx | |||
| 68 | O-Mass | 3987 | |||||
| 69 | MBI | Cobetia | 0.432 | 2125 | |||
| 70 | MBI | Uncultured | 0.76 | 1565 | |||
| 71 | MBI | Mariacaulis | 0.725 | 2933 | |||
| 72 | I-Lab | 885 | |||||
| 73 | O-Medium | 301 | xx | ||||
| 74 | O-Medium | 4792 | |||||
| 75 | O-Mass | 1155 | |||||
| 76 | O-Mass | 300 | |||||
| 77 | O-Mass | 300 | xx | ||||
| 78 | O-Medium | 100 |
Outdoor algal mass cultivation system = O-Mass; 18-L outdoor medium algal cultures = O-Med; marine bacterial isolates = MBI; indoor lab cultures = I-Lab. Growth inhibition scale: “x” = borderline, “xx” = clear inhibition, “xxx” = strong inhibition. Also “n.d.” = OD595 not determined.
Fig. 1.
Growth sensitivity exhibited by B. subtilis, C. albicans, and E. coli to tested chemical extracts. (A) Representative agar plates for each organism with positive control in upper right quadrant labeled “+” and “Pos con”, empty disk located in the plate center, and three tested extracts per plate with corresponding number labels. Chemical extracts resulting in distinguishable growth sensitivity to each microorganism are shown in (B), (C), and (D). A negative disk, labeled “NEG”, as an example of no exhibited growth sensitivity is shown in (C) for extract 47. Process control (chemical extract 67) results for B. subtilis and E. coli are labeled and shown in (A), (B), and (D).
Bacillus subtilis exhibited varying degrees of growth sensitivity to 17 chemical extracts tested in this study: 3, 7, 11, 14, 17, 32–34, 37–42, 51, 53, and 77 (Tables 1 and 2; Fig. 1B). Of these chemical extracts, ten were filtrates of conditioned media from outdoor mass cultivation systems (O-Mass), four were derived from cultures of marine bacterial isolates (MBI), two were from indoor laboratory cultures (I-Lab), and one was from an outdoor medium-sized culture (O-Medium). The chemical extracts exhibiting the highest discernable growth sensitivity (denoted as “xxx” in Tables 1 and 2) were extracts 7, 34, 40, and 53, all of which were derived from complex algal-bacterial mixtures (O-Medium, O-Mass, and I-Lab). Thus, it is not possible to correlate what bacteria might be responsible for the chemicals produced that elicited the sensitive growth response. In addition, the Sanger sequencing results of the marine isolates did not indicate any specific taxonomic trends correlated with antimicrobial activity; chemical extracts 3 and 4 were derived from an uncultured MBI, 17 was derived from Salinimicrobium spp., and 32 was derived from Erythrobacter spp. Not all taxa with the same Sanger sequencing result elicited the same growth response, however. For example, chemical extracts 12, 16, 21, and 32 were all classified to be within the Erythrobacter genus (Table 1), but only extract 32 showed antimicrobial activity against B. subtilis (Fig. 1B). Based on these observed differences in the antimicrobial assay, it is plausible that the Erythrobacter genera identified by the Sanger sequencing results are different species and/or strains. Alternatively, since these genera were grown to different ODs (Table 1), this could also potentially explain the differences in biological activity. Chemical extract 32, exhibiting growth sensitivity, was derived from the Erythrobacter spp. MBI with the highest OD 595 value of 0.924, possibly suggesting that a later stage or denser bacterial culture is required for extraction for antimicrobial activity. Further studies pursuing the impact of the bacterial density and the stages of growth on putative antimicrobial production are warranted. Lastly, we wanted to determine if the chemical extraction method, including the Sterlitech ultra-filtration and liquid-liquid extraction, would result in a sample with retained antibacterial activity. We generated a process control chemical extract by adding pM-µM levels of known antibiotics (chloramphenicol, carbenicillin, neomycin sulfate, erythromycin, ampicillin, tetracycline, spectinomycin, and kanamycin) to 6 L of ESAW media before the chemical extraction process. We observed that both B. subtilis and E. coli exhibited growth inhibition in response to the process control sample, chemical extract 67 (Table 1; Fig. 1B and D) confirming that the extraction methods retained antimicrobial activity of these chemicals.
Candida albicans also exhibited apparent growth inhibition in response to 11 chemical extracts tested: 16–18, 34, 37–39, 43–44, 52, and 73 (Tables 1 and 2; Fig. 1C). Of these 11 extracts, 5 were derived from MBI, 4 were derived from O-Mass systems, 1 was from a I-Lab culture, and 1 was from an O-Medium system. Despite the asymmetric clearance zones for extracts 34 and 37 (Fig. 1C), they were still classified as “clear growth inhibition” due to the comparison to other disks with no clear growth inhibition, such as extract 47 (Fig. 1C). Chemical extracts 16, 17, and 18 in Fig. 1 all have a very large circular “halo” of sensitivity, although resistant yeast colonies were present in this region. The concentration of the chemicals in the samples is potentially very close to the minimum inhibitory concentration (MIC) for one or more chemicals. Chemical extract 52 was derived from a laboratory algal-bacterial co-culture and showed a more pronounced growth inhibition against C. albicans as denoted by “xxx” in Table 2. Lastly, C. albicans did not show any growth sensitivity to the process control chemical extract 67, as expected since only antibiotics were included in this process control sample and no antifungals.
Escherichia coli was not sensitive to any of the 77 chemical extracts tested. It is possible that this is because the dose of each chemical mixture tested was not high enough to inhibit E. coli or possibly because the strain of E. coli used was a protein production strain specifically selected for its lack of sensitivity for a variety of growth conditions. Similarly to the B. subtilis strain tested, the process control chemical extract 67 did show antimicrobial activity against E. coli (Fig. 1D; Table 2) as well. This confirms that the methods used for filtering, concentrating, and extracting the conditioned media samples from the various algal cultures used in this study were sufficient to collect organic molecules without complete destruction or loss of antimicrobial activity.
Generally, we observed that the chemical extracts with the strongest apparent antimicrobial activity were derived from high density cultivation systems rather than specific microbial isolates. These include samples 7, 34, 40, and 53 for B. subtilis specific antimicrobial activity and sample 52 for antifungal activity against C. albicans (Tables 1 and 2). In addition, five extracts (17, 34, 37–39) showed both antibacterial and antifungal activity when tested against B. subtilis and C. albicans (Fig. 1B and C; Tables 1 and 2). Of the five extracts, four were derived from high density cultivation systems and one was derived from a marine bacterial isolate from an algal culture. These results argue for the presence of multiple antimicrobial-producing strains within these samples rather than the specific growth conditions and microbial competition present in high density algal culture leading to a greater production of antimicrobial compounds. Due to the unknown chemical complexity of these samples, it is plausible that within the extract there are multiple chemicals with varied or synergistic antimicrobial activity. Further microbial analysis and chemical fractionation studies will be required to discriminate between these scenarios.
Through this work, we identified 23 chemical mixtures derived from algal-bacterial systems that elicited a growth sensitivity or inhibition response on a plate-based assay. As an innovative approach to antimicrobial discovery, we analyzed complex chemical extracts derived from high biomass algal-bacterial cultures without using the traditional laborious and biased method of isolation and culturing of microbes from an assemblage before chemical analysis. Direct isolation and characterization of marine bacteria from ocean water samples has only recently been demonstrated as an innovative approach to antimicrobial discovery25. The low biomass concentration of ocean water samples has limited in-depth exploration of marine microbial communities because ocean samples are dilute and often hard to grow and culture in laboratory settings. High density algal cultivation systems are distinct from the natural marine and aquatic systems that the algal species are derived from in that the nutrient and resulting biomass concentrations are far higher in the cultivation systems. If the concentration of chemicals is directly linked to biomass concentration then the concentration of antimicrobials should also be higher in the production system. Thus, higher biomass concentrations allow for the analysis of far smaller samples. This radically different approach from the current state-of-the-art will enable discovery of antimicrobials from all active microbes present. Despite not knowing the chemical makeup, concentration, or contamination-level from salts, or other inorganic ions, of these chemical mixtures, we were able to detect antimicrobial bioactivity by loading blanks disks with high levels (up to 6 mg) of these chemical mixtures. The complex chemical extracts very likely contain residual salt, thus the amounts listed in Tables 1 and 2 do not represent of the concentration of any growth inhibitory compounds much less any minimal inhibitory concentration. Despite the likely chemical complexity and the potentially low abundance of individual chemicals, growth inhibition was still observed in the plate assay. With these promising initial results, additional effort is justified to analyze these extracts for the chemicals responsible for the antimicrobial activity.
Conclusion
Finding new antimicrobials also means investigating novel sources and developing innovative methods for extraction. In this initial study we were able to show that approximately 30% of samples tested demonstrated antimicrobial activity. These samples have been obtained from a poorly characterized ecological niche that in nonetheless distinct from the traditional sources that have been “mined” for antimicrobials, including soil samples and to a lesser degree natural marine samples. It has been shown that the soil microbial community is distinct from marine and aquatic systems so it seems likely that the antimicrobials produced may be distinct from those derived from soil microbes. In addition, algal mass cultivation systems are a ready source conditioned media as a constant “waste stream”, eliminating the sample-limited problem and the need for going to the field to collect unique ocean samples.
In general, this study demonstrates the feasibility and potential of isolating bioactive compounds from high-biomass algal-bacterial co-culture samples. Here, we concentrated and extracted conditioned-media samples, derived from high biomass systems, with organic solvent to generate complex chemical extracts, of which 23 showed bioactivity against the growth of Bacillus subtilis and/or Candida albicans. Based on these results, we feel confident that algal-bacterial cultivation systems represent a unique source of unexplored chemical diversity that should be further analyzed for novel chemicals with therapeutic value. We aim to continue exploring algal cultivation systems, and other natural microbial assemblages, for antimicrobial discovery.
Methods
Acquisition of algal-bacterial co-cultures
Tables 1 and 2 summarize acquisition information for 77 samples obtained from outdoor algal mass cultivation systems (O-Mass), 18-L outdoor medium algal cultures (O-Med), marine bacterial isolates (MBI) from algal cultures, or from non-axenic indoor laboratory cultures (I-Lab), and one process control (“Process con”). Several xenic and axenic cultures of Microchloropsis salina CCMP 1776 (Bigelow Labs, ME, USA) were grown at laboratory scale (~ 1 L) as described previously20. Additionally, outdoor medium scale (~ 18 L) cultures of Microchloropsis salina CCMP 1776 were grown with and without the addition of probiotic bacterial consortia; previously described20. After 7–14 days of cultivation, biomass from 8-L samples from each culture were harvested by sequential filtration using 0.8-µm cellulose nitrate Nalgene filters (ThermoFisher Scientific, MA, USA) and 0.2-µm filters (VWR, PA, USA) and the final 0.2-µm filtrate was concentrated through ultrafiltration using a Sterlitech skid Sepa Cell with NFW membranes (Sterlitech, WA, USA) that have a low retention for NaCl and MgSO4and a molecular weight cut off (MWCO) of 300. Filtrate from selected samples (16–24 L volumes) were concentrated by ultrafiltration resulting in 1 L samples. Lastly, marine bacterial isolates (MBI), originally derived from algal-bacterial co-cultures and identified by Sanger sequencing (Genewiz, NJ, USA) (Tables 1 and 2), were grown in 300 mL of ESAW + NPM medium (ESAW medium26,27containing an organic stock solution28) for 48 h in 30 °C at 160 RPM to the ODs listed in Tables 1 and 2. Additionally, a process control sample (Process con) was generated by adding 1 µM chloramphenicol, 1 µM carbenicillin, 1 nM neomycin sulfate, 1 pM erythromycin, 1 pM ampicillin, 1 nM tetracycline, 1 pM spectinomycin, and 1 pM kanamycin (all final concentrations) to 6 L ESAW media (all antibiotics purchased from ThermoFisher Scientific, MA, USA or Millipore Sigma, MA, USA).
Conditioned media concentration and chemical extraction
Biomass was removed through centrifugation at 4,000 x g for 10 min and filtration using 0.2 μm pore size filters depending on the concentration of the biomass and the size of the sample (between 300 and 1000 mL). The resulting conditioned media samples (ranging from ~ 300 to 1000 mL in volume) were extracted three times with 300–600 mL of ethyl acetate (Fisher Optima; ThermoFisher Scientific, MA, USA) each time in separatory funnels. The organic layer was then concentrated to 1–4 mL using roto-evaporation. Resulting samples were moved to pre-weighed 4 mL glass vials and completely dried down to generate approximate quantities of organic material, even with visible salt observed. Samples were resuspended in methanol (Fisher Optima; ThermoFisher Scientific, MA, USA), with the salt largely in suspension.
Bacterial and fungal cultures for microbial growth inhibition assay
For chemical extract testing by Kirby Bauer assay24, Escherichia coli C3000 was cultivated at 37 °C and Bacillus subtilis 168 was cultivated at 30 °C in LB broth. Candida albicans was cultivated in YEPD broth (Teknova, Hollister CA) at 30 °C. Cultures were grown in their respective media and temperature with shaking at 240 RPM. Mid-logarithmic phase cultures were used as indicator organisms in the plate assays. 100 µL of E. coli and B. subtilis at an OD 600 of 0.2 (1 × 108 cells/mL) were each spread on Mueller-Hinton (MH) agar plates (Teknova, Hollister CA). For antifungal testing, 100 µL C. albicans at an O.D. 600 of 0.05 (1 × 106 cells/mL) was spread on YEPD agar plates. After spreading, plates were allowed to dry for at least 20 min before addition of disks containing chemical extracts and controls.
Application of chemical extracts to disks for antimicrobial bioassay
Chemical extracts were stored at 4 °C until use in a Kirby-Bauer assay (described above). Prior to use in the bioassay, the chemical extracts (at different concentrations, quantities of which are summarized in Tables 1 and 2) were brought to room temperature (~ 22–25 °C) and 10 µL of each chemical extract was applied to one side of the empty Oxoid disk. Oxoid disks containing 30 ug of kanamycin (Oxoid, ThermoFisher Scientific, MA, USA) were used as positive control disks for E. coli and B. subtilis. Empty Oxoid disks were used as a negative control. Fluconazole (Millipore-Sigma, MA, USA) was resuspended at 33 mg/mL in DMSO (Millipore-Sigma, MA, USA) and then diluted 1:10 in methanol to 3.3 µg/µL, 10 µL was added to an empty disk as a positive control for C. albicans. All disks with added chemicals were allowed to dry for at least 20–30 min before application to assay plates. No more than five disks were added to each plate, with each plate containing a positive control with 10 µL of methanol added, a blank disk control, and then 2–3 chemical extract test samples. C. albicans and B. subtilis plates were incubated at 30 °C for 16–20 h and E. coli plates were incubated at 37 °C for 14–18 h before plates were assessed for sensitivity.
Acknowledgements
The authors would like to thank several colleagues and collaborators for their heroic support for this project: Matthew Hirakawa for fungal culture training, Dave Brekke for installing monkey bars in the chemical fume hood for expediting chemical extraction procedures and ensuring the project could be finished on the expedited timeline, George Buffleben for help setting up the ultrafiltration system, Sam Schrauth for ensuring that the physical and mental support of the postdoctoral researcher on this project, the Büchi Rotovap for being the ultimate work horse of this entire project, and the algal outdoor cultivation samples generously sent from our collaborators at Global Algal Innovations, California Polytechnic State University, Arizona State University and the Algal Testbed Public-Private Partnership (ATP3). C.L.F. acknowledges Sandia National Laboratories’ support from Lab Directed Research and Development (LDRD) project for the experimental work performed and Lawrence Livermore National Laboratory’s support from LDRD project (23-LW-029) for the writing, editing, and publishing of the results of this work. This work was performed at Sandia National Laboratories, a multi-mission laboratory managed and operated by National Technology and Engineering Solutions of Sandia, LLC., a wholly owned subsidiary of Honeywell International, Inc., for the U.S. Department of Energy’s National Nuclear Security Administration under contract DE-NA0003525. This paper describes objective technical results and analysis. Any subjective views or opinions that might be expressed in the paper do not necessarily represent the views of the U.S. Department of Energy or the United States Government. This manuscript has been authored by Lawrence Livermore National Security, LLC under Contract No. DE-AC52-07NA2 7344 with the US. Department of Energy (LLNL-JRNL-868492). The United States Government retains, and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes.
Author contributions
C.L.F., P.D.L, and T.W.L. conceived the experiments; C.L.F., H.L.W., P.D.L., and C.M. conducted the experiments; all authors analyzed the results; C.L.F., H.L.W., and T.W.L. drafted the manuscript; all authors reviewed the manuscript.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper. Should any raw data files be needed in another format they are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Ethics declarations
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lai, C. C., Chen, S. Y., Ko, W. C. & Hsueh, P. R. Increased antimicrobial resistance during the COVID-19 pandemic. Int. J. Antimicrob. Agents. 57 (4), 106324. 10.1016/j.ijantimicag.2021.106324 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sir Alexander Fleming’s speech at the Nobel Banquet in Stockholm. December 10, (1945). https://www.nobelprize.org/uploads/2018/06/fleming-lecture.pdf
- 3.Boucher, H. W. et al. Bad Bugs, No Drugs: No ESKAPE! An Update from the Infectious Diseases Society of America, Clinical Infectious Diseases, 48(1), pg 1–12. DOI: (2009). 10.1086/595011 [DOI] [PubMed]
- 4.Lewis, K. The Science of Antibiotic Discovery. Cell181 (1), 29–45. 10.1016/j.cell.2020.02.056 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Simpkin, V. L., Renwick, M. J., Kelly, R. & Mossialos, E. Incentivising innovation in antibiotic drug discovery and development: progress, challenges and next steps. J. Antibiot.70, 1087–1096. 10.1038/ja.2017.124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ventola, C. L. The Antibiotic Resistance Crisis. Pharm Therap, 2015; 40(4): 277–283. PMID: 25859123; PMCID: PMC4378521. (2015). [PMC free article] [PubMed]
- 7.Genilloud Natural products discovery and potential for new antibiotics, Current Opinion in Microbiology 2019, 51:81–87. DOI: (2019). 10.1016/j.mib.2019.10.012 [DOI] [PubMed]
- 8.Lewis, K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 12, 371–387. 10.1038/nrd3975 (2013). [DOI] [PubMed] [Google Scholar]
- 9.LeMieux, J. As Novartis Exists, Who Will Make New Antibiotics? Genetic Engineering & Biotechnology News, (2018). https://www.genengnews.com/insights/as-novartis-exits-who-will-make-new-antibiotics/ (accessed Mar 14, 2022).
- 10.Planckett, B. Why big pharma has abandoned antibiotics. Nat. Retrieved 03/14/2022: (2020). https://www.nature.com/articles/d41586-020-02884-3
- 11.Bartlett, J. G., Gilbert, D. N. & Spellberg, B. Seven ways to preserve the miracle of antibiotics, Clin Infect Dis. 2013; 56(10):1445-50. DOI: (2013). 10.1093/cid/cit070 [DOI] [PubMed]
- 12.BioSpace : Topical antibiotics market: growing prevalence of skin infections Worldwide to Aid Market Growth, Biospace, Retrieved 03/14/2022: (2020). https://www.biospace.com/article/topical-antibiotics-market-growing-prevalence-of-skin-infections-worldwide-to-aid-market-growth
- 13.Lewis, K. New approaches to antimicrobial discovery. Biochem. Pharmy. 134, 87–98. 10.1016/j.bcp.2016.11.002 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Schloss, P. D. & Handelsman, J. Status of the microbial census. Microbiol. Mol. Biol. Rev.68 (4), 686–691. 10.1128/MMBR.68.4.686-691.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geng, H., Sale, K. L., Tran-Gyamfi, M. B., Lane, T. W. & Yu, E. Longitudinal analysis of Microbiota in Microalga Nannochloropsis salina cultures. Microb. Ecol.72, 14–24. 10.1007/s00248-016-0746-4 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Shaima, A. F. et al. Unveiling antimicrobial activity of microalgae Chlorella sorokiniana (UKM2), Chlorella sp. (UKM8) and Scenedesmus sp. (UKM9). Saudi J. Biol. Sci.29 (2), 1043–1052. 10.1016/j.sjbs.2021.09.069 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alsenani, F. et al. Evaluation of microalgae and cyanobacteria as potential sources of antimicrobial compounds. Saudi Pharma J.28 (12), 1834–1841. 10.1016/j.jsps.2020.11.010 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falaise, C. et al. Antimicrobial compounds from eukaryotic microalgae against human pathogens and diseases in aquaculture. Mar. Drugs. 14, 159. 10.3390/md14090159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jusidin, M. R. et al. In vitro antibacterial activity of marine microalgae extract against Vibrio harveyi. Appl. Sci.12, 1148. 10.3390/app12031148 (2022). [Google Scholar]
- 20.Fisher, C. L. et al. Bacterial communities protect the alga Microchloropsis salina from grazing by the rotifer Brachionus plicatilis. Algal Res.40, 101500. 10.1016/j.algal.2019.101500 (2019). [Google Scholar]
- 21.Antibiotic Resistance Threats in the United States. US Centers for Disease Control and Prevention. (2019). www.cdc.gov/DrugResistance/Biggest-Threats.html
- 22.Berman, J. J. Chapter 7 – Gamma Proteobacteria, Taxonomic Guide to Infectious diseases. Elsevier Inc. 37–47. 10.1016/B978-0-12-415895-5.00007-6 (2012).
- 23.Fulbright, S. P. et al. Bacterial community changes in an industrial algae production system. Algal Res.31, 147–156. 10.1016/j.algal.2017.09.010 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hudzicki, J. Kirby-Bauer disk Diffusion Susceptibility Test Protocol. Am Soc Microbiol. (2009). https://asm.org/getattachment/2594ce26-bd44-47f6-8287-0657aa9185ad/Kirby-Bauer-Disk-Diffusion-Susceptibility-Test-Protocol-pdf.pdf
- 25.Chbel, A. et al. Molecular identification and antibacterial potential of Marine Bacteria from Deep Atlantic Ocean of Morocco. Avicenna J. Med. Biotechnol.14 (3), 206–215. 10.18502/ajmb.v14i3.9827 (2022 Jul-Sep). PMID: 36061126; PMCID: PMC9376996. [DOI] [PMC free article] [PubMed]
- 26.Berges, J. A., Franklin, D. J. & Harrison, P. J. Evolution of an artificial seawater medium: improvements in enriched seawater, artificial water over the past two decades. J. Phycol.37, 1138–1145 (2001). [Google Scholar]
- 27.Harrison, P. J., Waters, R. E. & Taylor, F. J. R. A broad spectrum artificial seawater medium for coastal and open ocean phytoplankton. J. Phycol.16, 28–35 (1980). [Google Scholar]
- 28.Guillard, R. R. L. A mutant of Chlamydomonas moewusii lacking contractile vacuoles. J. Protozool. 7, 262–268 (1960). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper. Should any raw data files be needed in another format they are available from the corresponding author upon reasonable request.