Abstract
Background
High myopia (HM), characterized by a severe myopic refractive error, stands as a leading cause of visual impairment and blindness globally. HM is a multifactorial ocular disease that presents high genetic heterogeneity. Employing a genetic risk score (GRS) is useful for capturing genetic susceptibility to HM.
Methods
This study assesses the effectiveness of these strategies via incorporating rare variations into the GRS assessment. This study enrolled two independent cohorts: 12,600 unrelated individuals of Han Chinese ancestry from Myopia Associated Genetics and Intervention Consortium (MAGIC) and 8682 individuals of European ancestry from UK Biobank (UKB).
Results
Here, we first estimate the heritability of HM resulting in 0.53 (standard error, 0.06) in the MAGIC cohort and 0.21 (standard error, 0.10) in the UKB cohort by using whole-exome sequencing (WES) data. We generate, optimize, and validate an exome-wide genetic risk score (ExGRS) for HM prediction by combining rare risk genotypes with common variant GRS (cvGRS). ExGRS improved the AUC from 0.819 (cvGRS) to 0.856 for 1219 Han Chinese individuals of an independent testing dataset. Individuals with a top 5% ExGRS confer a 15.57-times (95% CI, 5.70–59.48) higher risk for developing HM compared to the remaining 95% of individuals in MAGIC cohort.
Conclusions
Our study suggests that rare variants are a major source of the missing heritability of HM and that ExGRS provides enhanced accuracy for HM prediction in Han Chinese ancestry, shedding new light on research and clinical practice.
Subject terms: Eye diseases, Population genetics
Plain language summary
High Myopia (HM) is a disease of the eyes frequently caused by one’s inherited genes. Mathematical equations can be used to predict disease risk based on a person’s genetic make-up (profile). This calculation, called a genetic risk score (GRS), doesn’t include rare genetic changes and it is challenging to consider these in the calculations. Here, we test whether combining rare genetic changes can help to predict HM risk. Our calculations not only outperformed existing methods used for HM risk, they also allow us to estimate an individual’s risk of HM, showing how important including rare genetic changes are in accurately predicting risk of this disorder.
Yuan and Qiu et al. estimate heritability for High Myopia (HM) using whole-exome sequencing data in Han Chinese ancestry and European ancestry. By combining rare risk genotypes with exome-wide association studies of HM, the authors develop an exome-wide genetic risk score for HM prediction.
Introduction
High myopia (HM) is generally defined for individuals with a spherical equivalent (SE) of -6.00 diopters (D) or lower1. HM affects 2.8% of the general population and is a risk factor for developing pathologic myopia (PM) and its complications, most notably retinal degeneration or even detachment, which can cause severe visual acuity (VA) loss and even blindness2,3. HM is more common among Asian schoolchildren (6.8–21.6%)4,5 than in non-Asians (2.0–2.3%)6.
HM is a multifactorial eye disease with a high genetic susceptibility. Twin and family studies have demonstrated that HM has a high heritability7,8. Over the past decades, numerous genome-wide association studies (GWAS) of refractive error or myopia have revealed hundreds of candidate genetic factors across different ethnic populations9–11. However, the common variant uncovered by GWAS has a small effect size independently; even the additive effects can only explain a limited fraction of myopia heritability (estimated heritability: 5.3% in Asians and 21.4% in Europeans)11,12. Whole-exome sequencing (WES) studies of HM trios or families have identified several novel mutations and genes in the Asian populations, i.e., SCO213, BSG14, CCDC102B15, and LRPAP116. Moreover, our recent WES study has also identified several HM-associated genes, including rare coding variants, which were found to have larger effect sizes17. Hence, rare variants indeed contribute to the genetic architecture of HM, although the extent to which they accounted for its heritability remains unclear, leaving ample room for further investigation.
Polygenic risk scores (PRS) summarize the cumulative genetic effects of numerous disease-associated variants, providing an overall measure of genetic susceptibility to a particular disease for an individual18,19. Blood or saliva samples can be used to predict a wide range of conditions, providing the complementary need for any additional examinations or tests for diagnosis20. In European populations, several large-scale studies have demonstrated the effectiveness of utilizing the PRS to stratify myopia risk10,11,21–24. Currently, the best-performing PRSs for refractive error explain about 19% of the variance in the trait in individuals of European ancestry and about 6% in those of East Asian ancestry23. The best area under the receiver operating characteristic curve (AUROC) for HM is 0.783 and 0.672 in European and East Asian populations, respectively23. With most large-scale myopia GWASs primarily performed among European populations, it remains unclear if these findings are generalizable to diverse populations of non-European ancestry.
Thus, in this study, we estimated heritability explained by SNP-based genetic variance and the gene-wise burden of rare alleles for HM using WES data in a large sample of 12,600 unrelated Chinese from the Myopia Associated Genetics and Intervention Consortium (MAGIC) and 8682 Europeans from the UK Biobank (UKB) program. We constructed common-variant-based genetic risk scores (cvGRS) and rare-variant-based genetic risk scores (rvGRS) models and evaluated the performance of the two models for genetic risk prediction in a subset of MAGIC. We proposed a method, exome-wide genetic risk score (ExGRS), which combined cvGRS and rvGRS, and observed further improvement in genetic risk prediction for HM. We demonstrated the creation of the ExGRS, which exhibits distinct advantages over cvGRS, by incorporating rare variants identified in HM-associated genes via burden tests, while also evaluating its portability across ancestry in the UKB European populations.
Methods
Overview of the high myopia sequencing consortium cohort
The Myopia Associated Genetics and Intervention Consortium (MAGIC) is a large-scale genomic consortium integrating myopia cohorts and sequencing data from many investigators. Over the past several years, MAGIC has been able to collected samples at the Eye Hospital of Wenzhou Medical University (Zhejiang Eye Hospital) through the Institute of Biomedical Big Data4. We recruited approximately ten thousand Chinese schoolchildren with high myopia aged from 6 to 18 years from MAGIC. In the study, high myopia is defined as a spherical equivalent refraction (SER, sphere + [cylinder/2]) of single eye -6.00 diopters(D) or less. The analysis presented here is based on 21,227 unrelated human samples collected from epidemiological studies of myopia. After removing samples showing poor sequencing quality or ambiguous sex status, population outliers identified by principal component analysis (PCA), we random selected approximately 70% (12,600) of participants as training samples, whereas the remaining 30% (5400) were assigned as validation samples.
The present study was approved by the Ethics Committee of the Wenzhou Medical University Affiliated Eye Hospital (approval numbers Wmu191204 and Wmu191205). Written informed consent conforming to the tenets of the Declaration of Helsinki and following the Guidance of Sample Collection of Human Genetic Diseases (2021SQCJ5721) by the Ministry of Public Health of China was obtained from all participating individuals or their legal guardians before the study. All procedures were carried out strictly following the guidelines of ‘Management of Human Genetic Resources’, as stipulated by the Ministry of Science and Technology of China (no. BF2022060511307 and no. BF2022060611309, effective from November 8, 2021).
UK Biobank (UKB) is a large-scale biomedical database and research resource, containing genetic and health records from half a million individuals aged 40–69 years in the United Kingdom25. There were 488,000 participants were genotyped for 805,426 markers on the UK BiLEVE Axiom array and UK Biobank Axiom array. UKB measured refractive error of 130,494 participants by non-cycloplegic autorefraction using a TomeyRC-5000 AutoRefractor Keratometer. We excluded unreliable refractometry results and calculated the spherical equivalent (SE) as spherical refractive error plus half the cylindrical error. In addition, samples identified as outliers in heterozygosity and missing rates, participants with sex discrepancy, and individuals of non-Caucasian ancestry were removed in our study according to the sample QC provided by UKB. We estimated relatedness in each cohort by PLINK and only kept one of any pair of individuals with relatedness (πˆ) >0.2. Finally, we identified 2096 HM cases (participants with SE of single eye ≤ –6.0D) and 6,586 controls (participants with SE of single eye > –0.25D). UK Biobank data has approval from the North West Multi-center Research Ethics Committee (MREC) (REC reference: 16/NW/0274). All participants gave informed consent for participation in UK Biobank. Permission to access and analyse UK Biobank data was approved under UK Biobank project 45270.
Quality control
Sample quality control (QC) and variant QC for MAGIC and UKB cohorts in our previous study are used in this study. We first selected the samples with phenotypes available and retained only the high-quality variants that passed a GATK Variant Quality Score Recalibration (VQSR) approach, and those located outside of low-complexity regions were remained. Genotypes with a genotype depth (DP) < 10 and genotype quality (GQ) < 20 and heterozygous genotype calls with an allele balance >0.8 or <0.2 were set as missing. We then excluded variants with genotype missingness rate >0.05, Hardy-Weinberg equilibrium (HWE) test P value < 10–6 or a MAC < 3 using PLINK v.1.926. Only retained individuals of East Asian (EAS) and European ancestry were retained, which were classified by a random forest algorithm with 1000 Genomes data. At the end of all the QC steps, we retained 12,600 unrelated individuals of Han Chinese and 8,682 European.
Variant annotation
The annotation of variants was performed with Ensembl’s Variant Effect Predictor (VEP v.99) for human genome assembly GRCh37. We used the VEP27 to generate additional bioinformatic predictions of variant deleteriousness (Supplementary Table 1–2). Protein-coding variants were annotated into the following four classes: (1) synonymous; (2) benign missense; (3) damaging missense; and (4) protein-truncating variants (PTVs). In detail, using VEP annotations (v.99), missense variants were classified as “inframe_deletion”, “inframe_ insertion”, “missense_variant” or “stop_lost” variants. Among the missense variants, one type of benign missense variant was predicted as “tolerated” and “benign” by PolyPhen-2 and SIFT, respectively, and another type of benign mutation showed a combined annotation dependent depletion (CADD) score <15. Furthermore, damaging missense variants were predicted as “probably damaging” and “deleterious” by PolyPhen-2 and SIFT and CADD > 15. Finally, PTVs were classified as “frameshift_variant”, “splice_acceptor_variant”, “splice_donor_ variant”, “stop_gained”, or “start_lost” variants.
Association test
We conducted a single-variant association analyses by using MLMA-LOCO28. The test statistics obtained via linear regression were inflated because of the population differentiation caused by genetic drift. Post hoc correction approaches, such as “Genomic Control”, were used to correct the inflation29. For the exome-wide association study, we first tested each variant, regardless of allele frequency, for HM associations; we applied a significance level of P < 4.3 × 10–7 for all variants30. To determine whether a single gene was enriched in or depleted of rare protein-coding variants in HM cases, we performed four gene-level association tests including Fisher’s exact test, burden, SKAT and SKAT-O, with previously defined covariates (sample sex, PC1-PC10).
Heritability estimation
In each WES dataset, we stratified SNPs into 4 MAF bins (0.0001 < MAF < 0. 0010, 0.001 < MAF < 0.010, 0.01 < MAF < 0.10 and 0.1 < MAF < 0.5). For each of the 22 autosomes, we calculated the linkage disequilibrium (LD) score of each variant with the others on a sliding window of 10 Mb using GCTA software28. Each of the four MAF bins was divided into two more bins, one for variants with LD scores above the median value of the variants in the bin (high-LD bin) and one for variants with LD score below the median (low-LD bin) (Supplementary Table 3). We then used GCTA to perform a GREML-LDMS analysis on HM in each dataset with either 20 PCs calculated from HM3 SNPs or 160 PCs (20 PCs computed from each of the 8 MAF/LD bins) fitted as fixed covariates.
Using variant annotations and the LD and MAF bins defined from the GREML-LDMS analysis on the WES data mentioned above, we further separated the low-LD and high-LD variants in the 0.0001 < MAF < 0.01 into four bins according to their predicted variant effects: PTV, D-mis, B-mis and Synonymous. We then ran a GREML-LDMS analysis with 8 Genome-wide Relationship Matrices (GRMs), fitting the 160 PCs shown to capture the effect of population stratification as well as fixed covariates in MAGIC and UKB. To compute the variance explained per SNP, we divided the estimate of variance explained for each bin by the number of variants in the bin. The s.e. was obtained by dividing the s.e. of the estimated variance explained for the bin by the number of variants in the bin. We estimated burden heritability for rare variant by using BHR (v.0.1.0), which is implemented in R, and its source code is publicly available at GitHub (https://github.com/ajaynadig/bhr). To compute the effect-size variance explained per gene, we divided the estimate of burden heritability for each bin by the number of variants in the bin.
GRS design
We derived cvGRS, rvGRS and ExGRS in the 12,600 unrelated individuals of Han Chinese ancestry from MAGIC. For cvGRS derivation, we first generated 20 pruning and thresholding (P + T) scores over a range of P value (1.0, 0.5, 0.05, 5 × 10–4 and 5 × 10-6) and r2 (0.2, 0.4, 0.6, and 0.8) thresholds. We also computed 7 candidate cvGRS using the LDPred2 algorithm31 across the following range of rho (fraction of casual variants): 1.00, 1.00 × 10–1, 1.00 × 10–2, 1.00 × 10–3, 3.00 × 10–1, 3.00 × 10–2 and 3.00 × 10-3. Additionally, the lassosum2 computational algorithm32 was used to generate a candidate GRS for HM. Each of the scores derived above was subsequently assessed for discrimination of HM cases from controls in the MAGIC validation dataset (2697 cases and 2703 controls) after adjustment for age, sex and 160 PCs of ancestry. The score with the best performance was defined by the maximal area under the receiver operator curve (AUC) and the largest fraction of variance explained. AUC confidence intervals were calculated using the ‘pROC’ package within R.
We aimed to assess if adding the rvGRS enhanced HM risk prediction. We constructed the rvGRS from the results of the rare variant burden tests. These were conducted per gene, and each gene had separate thresholds for associated to HM (P value) and pathogenicity (PTV, D-mis, B-mis and Synoymous variant) established in the training group. rvGRS models were constructed by fitting logistics regression models to HM on the rare variants (AF < 1%) in significantly associated genes. A unified PRS model, ExGRS, was also constructed, which summed the rare- and common-variant GRS models per individual. We tested the ExGRS for association with HM in this dataset.
Statistical analysis within the testing dataset
For HM, the ExGRS with the best discriminative capacity in the testing dataset was calculated in the testing dataset of 1219 participants in MAGIC and 8682 participants in UKB. Due to the limited availability of rare variants shared across the Han Chinese and European cohorts, we used variants with concordant direction-of-effect between MAGIC and UKB to improve the trans-ethnic performance of the score. The proportion of the population of HM individuals with a given magnitude of increased risk was determined by comparing progressively more extreme tails of the distribution with the remainder of the population. Logistic regression models were used for predicting case-control status with adjustment for age, sex, and PCs of ancestry using the glm function in R. We used the pROC R package to calculate the AUC. We also expressed the effect of the standardized risk score as ORs (with 95% CIs) per s.d. unit of the control standard-normalized risk score distribution in each of the testing cohorts. We examined the risk score discrimination at tail cutoffs corresponding to the top 20, 10, 5, 2 and 1% of the GRS distribution by deriving the ORs of disease for each tail of the distribution compared to all other individuals in each cohort.
Statistics and reproducibility
Statistical analyses were conducted using R version 4.2.1 software (The R Foundation). A two-sided P value < 0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Results
Polygenic architecture of rare to common coding variants
We used a dataset of 12,600 exomes of Han Chinese ancestry in the MAGIC project and 8682 exomes of European ancestry in the UK Biobank (Fig. 1 and Supplementary Fig. 1). We analyzed variants observed at least three times in our dataset, which corresponds to a minor allele frequency (MAF) threshold of 0.01%. After quality control (QC), 2.6 and 2.2 million variants were included in the further analysis in MAGIC and UKB cohorts, respectively. First, based on common SNPs, the estimated heritability () of HM was calculated using the residual maximum likelihood analysis (GREML) approach implemented in the software package GCTA28. This analysis utilized a selected set of 43,367 and 66,091 HapMap 3 (HM3) SNPs from the MAGIC and UKB cohorts, respectively. After correcting for the first 20 principal components (PCs) computed from HM3 SNPs, we estimated an of 0.31 (standard error, s.e. = 0.01) and 0.14 (s.e. = 0.02) for HM in MAGIC and UKB cohorts, respectively (Supplementary Fig. 2). We then applied variants with MAF greater than 0.01% to estimate and partition additive genetic variances. We grouped variants according to MAF and LD (Supplementary Fig. 3 and Supplementary Table 3), using the GREML-LDMS partitioning method with a median-based LD grouping strategy33. Corrected for the first 20 PCs from HM3 SNPs, we found the estimated heritability based on WES data () was 1.76 (s.e. = 0.03) and 0.20 (s.e. = 0.10) for HM in the MAGIC and UKB cohorts (Supplementary Fig. 2), which suggested in the MAGIC cohort may have been inflated by confounding factors such as population structure.
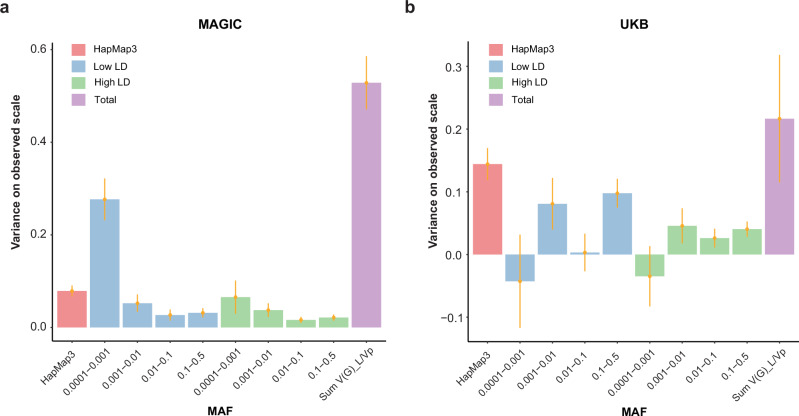
Fig. 1. GREML-LDMS estimates from WES data stratified in 8 bins, including 2 linkage disequilibrium (LD) bins for each of the 4 minor allele frequency (MAF) bins, correcting for 160 PCs (20 * 8 bins) for MAGIC and UKB.
a Estimate for HM with at 0.53 (s.e. = 0.06) in MAGIC (n = 12,600). b Estimate for HM with at 0.21 (s.e. = 0.10) in UKB (n = 8682). We stratified SNPs into 4 MAF bins (0.0001 < MAF < 0. 0010, 0.001 < MAF < 0.010, 0.01 < MAF < 0.10 and 0.1 < MAF < 0.5). Each of the 4 MAF bins was divided into two more bins, one for variants with LD scores above the median value of the variants in the bin (high-LD bin) and one for variants with LD score below the median (low-LD bin) (Supplementary Table 3). Error bars indicate standard errors (SE).
To determine the contribution of uncaptured population stratification, we utilized a linear model adjusted for PCs to assess the association of rare variants (Supplementary Fig. 4) in both cohorts. We then used 160 PCs (20 PCs computed from each of the 8 MAF/LD bins) computed from independent variants in the GREML-LDMS analyses, which decreased from 1.76 (s.e. = 0.03) to 0.53 (s.e. = 0.06) in the MAGIC cohort and increased from 0.20 (s.e. = 0.10) to 0.21 (s.e. = 0.10) in UKB (Fig. 1 and Supplementary Fig. 2). This suggests the presence of population stratification effects not captured by the 20 common variant PCs used in MAGIC. We also found that the difference of for HM between MAGIC cohort and UKB cohort is predominantly explained by rare variants, in particular those in low LD with nearby variants. For variants with an MAF > 0.01, 0.10 and 0.17 of the phenotypic variance was accounted in the MAGIC cohort and UKB cohort, respectively. For variants with an MAF < 0.01, 0.33 of the phenotypic variance in the MAGIC cohort was accounted for by variants in the low-LD group, while only 0.10 of the variance was accounted for by variants in the high-LD group. However, in the UKB cohort, only 0.04 of the phenotypic variance is accounted for by variants in the low-LD group and 0.01 by those in the high-LD group, suggesting that rare variants explain less genetic heritability than common variants in UKB’s European populations (Fig. 1). When we replaced all the called SNPs in the MAGIC cohort with overlapped variants found in both the MAGIC and the UKB WES datasets, the estimated heritability decreased from 0.53 to 0.06 (Supplementary Fig. 5), with most differences arising from variants with 0.0001 < MAF < 0.01, which are almost EAS-specific (Supplementary Fig. 6). To further estimate the association between SNP effect and MAF, we demonstrated this relationship by plotting cumulative genetic variances explained by h2 against MAF. Under an evolutionarily neutral model, h2 is linearly proportional to MAF34. We found that the curves of cumulative genetic variances in the MAGIC and UKB cohorts deviated from the neutral model, suggesting that HM is under negative selection (Supplementary Fig. 7).
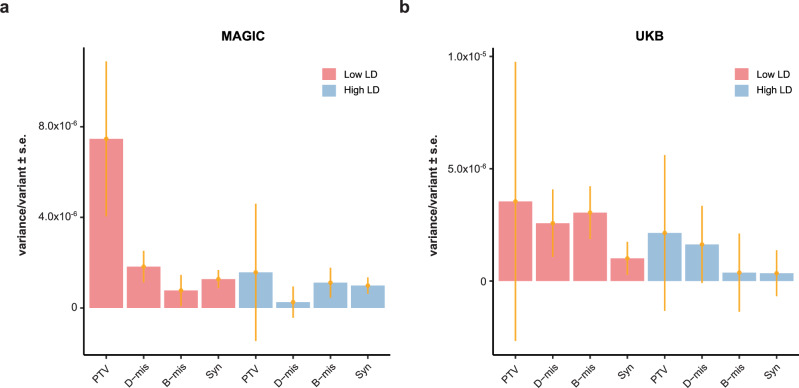
To investigate the contribution of low-LD variants with MAF < 0.01 to heritability, we partitioned these variants into bins according to the putative effects of protein-coding variants, as annotated by VEP27. Protein-coding variants are categorized into four annotations: (1) synonymous (Syn); (2) benign missense (B-mis); (3) damaging missense (D-mis); and (4) protein-truncating variants (PTVs) (Supplementary Table 4). The proportion of deleterious protein-altering variants, including PTVs and D-mis, varied across the LD and MAF groups, showing an increasing trend from low- to high-MAF bins (Supplementary Fig. 8), consistent with purifying selection on this class of variants. Interestingly, the average variance explained per variant was greater for bins with PTVs (low-LD) than for bins with other protein-altering or non-protein-altering variants (low-LD) and high-LD variants (Fig. 2). To further validate the robustness of the partitioned estimates by functional genomic annotations, we quantified the heritability explained by the gene-wise burden of rare coding variants35. We found that HM in the MAGIC cohort and UKB cohort exhibits PTVs burden heritability of 0.7% (s.e. = 0.15%) and 0.32% (s.e. = 0.25%), respectively (Supplementary Fig. 9). Burden heritability is concentrated among variants with the most severe predicted functional consequences: PTVs variants account for the majority of burden heritability, followed by D-mis, B-mis, and Syn variants, consistent with the GREML-LDMS assessment.
Fig. 2. Variance explained per variant (the estimate of genetic variance divided by the number of variants in each bin) from GREML-LDMS with rare variants partitioned into four categories according to the variant annotation.
a Variance explained per variant for Han Chinese individuals from MAGIC (n = 12,600). b Variance explained per variant for European individuals from UKB (n = 8682). Error bars indicate standard errors (SE).
Derive genetic risk scores of common coding variants for HM
The genetic risk score (GRS) serves as a reliable measure of an individual’s overall genetic susceptibility to disease, an integral part of precision medicine36. For HM in the MAGIC cohort, we created several candidate cvGRS based on summary statistics from ExWAS in 12,600 participants (6300 cases and 6300 controls) of Chinese Han ancestry (Fig. 3). Specifically, we derived 20 predictors based on a pruning and thresholding method, seven additional predictors using the LDPred2 algorithm31, and one predictor using Lassosum232. These scores were validated within the MGAIC cohort. We used a validation dataset of 5400 participants in the MAGIC cohort to select the cvGRSs with the best performance, defined as the maximum area under the receiver-operator curve (AUC). The predictors had AUCs ranging from 0.598 to 0.895 in the validation set (Supplementary Table 5; Fig. 4a). The best model, based on the P value thresholding (P + T) method, involved 40,491 variants with nonzero weights selected based on r2 = 0.2 and P = 1.0 (Supplementary Fig. 10). In the validation dataset, the polygenic component of the score explained 4.9% of the variance (R2), with one standard deviation (s.d.) of the score increasing HM risk by sevenfold (odds ratio [OR] = 6.99, 95% confidence interval [CI] = 6.34–7.75, P < 1.00 × 10–300) after controlling for age, sex and genetic ancestry.
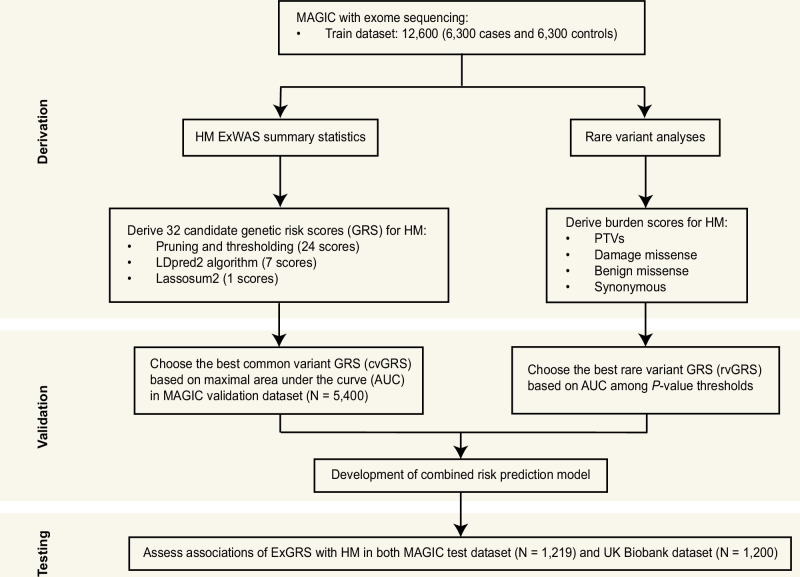
Fig. 3. Overview of the study design.
The HM GRS was designed based on the MAGIC. Validation and optimization were performed in two stages using common variant GRS (optimization 1) and rare variant GRS (optimization 2). The optimal GRS for HM was chosen based on the AUC in the MAGIC validation dataset (n = 5400 Han Chinese). ExGRS performance validation was conducted in two additional independent testing cohorts of diverse ancestries.
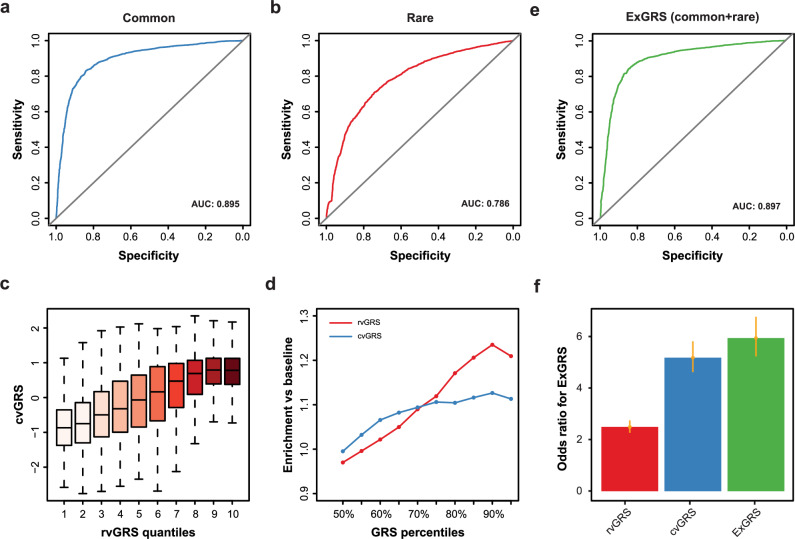
Fig. 4. Compare the GRS of common and rare variants and combine them into a unified ExGRS model.
a Receiver operating characteristic (ROC) curves for cvGRS model to detect HM in the schoolchildren from the MAGIC cohort (n = 5400). The solid black line represents chance-level prediction accuracy. b ROC curves for rvGRS model to detect HM. c cvGRS for each individual according to 10 groups of the validation dataset binned according to the quantiles of the rvGRS. d Enrichment of outlier GRS scores in individuals who are extreme HM prediction risk. GRS ordered from the 50% to the 100% percentile (x axis), and the y axis depicts the enrichment of HM for each of the percentile-defined subgroups in reference to the baseline population. e ROC curves for ExGRS model to detect HM. f Odds ratios for cvGRS, rvGRS and unified ExGRS model by comparing those in the high-risk group with the remainder of the population. Error bars based on confidence intervals (C.I.).
GRS optimization by combining with rare variants
The second step to optimize the GRS model is to test the independent contributions of rare variants (Fig. 3). To identify genes underlying HM, we performed rare-variant burden tests for 12,600 individuals in the MAGIC cohort using four methods, namely, Fisher’s exact test [FET], Burden, SKAT, and SKAT-O. Using a MAF threshold of 0.1%, we detected 651 gene-phenotype associations with PTV variants and 1481 associations with D-mis, using a GC-corrected FET’s P value of 0.1. We observed a positive correlation between variant pathogenicity and ORs of risk genes for HM under various cut-off P values (Supplementary Fig. 11). Given the higher heritability and strong effect size of rare deleterious variants in the MAGIC cohort, we reasoned that a cvGRS combining rare variants may effectively identify individuals at high risk for HM. Here, we proposed a complementary rvGRS based on a weighted sum of rare deleterious variants from HM-associated genes. To construct the model, we initially fitted a logistic regression model to HM using the rare PTVs and D-mis in associated genes across 12,600 training subsets. Furthermore, we evaluated the predictive power of the rvGRS on 5400 MAGIC cohort individuals previously withheld for validation. We observed the best performance of rvGRS for PTVs (AUC = 0.698) and D-mis (AUC = 0.772) based on HM-associated genes selected using a FET’s P value of 0.1 (Supplementary Fig. 12-14). Then, we compared the rare-variant association study (RVAS) and rvGRS between PTVs and D-mis. For matched significance thresholds, we uncovered that only 4.3% of HM-associated genes identified by RVAS overlapped between PTV and D-mis (Supplementary Fig. 15). We further stratified the population according to rvGRS deciles in PTVs and discovered a striking gradient with respect to rvGRS in D-mis (Supplementary Fig. 15). Therefore, we derived an rvGRS to predict HM by integrating HM-associated genes carrying PTV and D-mis variants (AUC = 0.786) (Fig. 4b).
We assessed the predictive power of the rvGRS and the corresponding cvGRS, as well as a combination of the two methods, on the 5,400-participant MAGIC validation dataset. A higher cvGRS was observed in the top decile of the rvGRS (Fig. 4c). Although rvGRS underperformed for average phenotype predictions, it may outperform cvGRS for identifying individuals at risk extremes (Fig. 4d). Therefore, we combined the rare- and common-variant GRS models into a unified model (exome-wide genetic risk score, ExGRS), achieving a significant improvement in genetic risk prediction for HM. The unified ExGRS performed best, achieving a prediction AUC of 0.897, compared to 0.786 and 0.895 for the independent rare-variant and common-variant GRSs, respectively (Fig. 4e). Consistent with the AUC results, the inclusion of rvGRS enhanced HM risk prediction and improved case-control discrimination: the risk of HM for predicted cases was 5.73-times higher than for the predicted controls, surpassing the cvGRS (4.99-times) and rvGRS (2.40-times) (Fig. 4f).
Portability of ExGRSs and validation in both independent cohorts
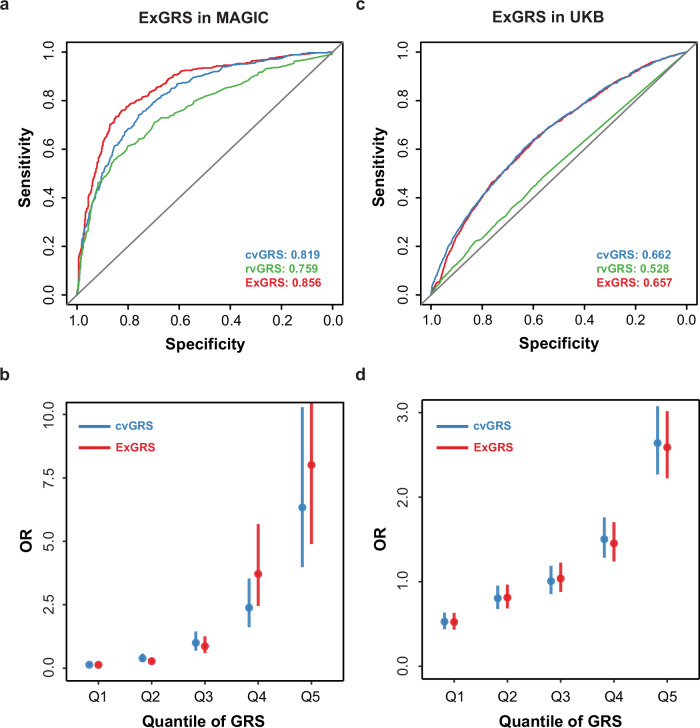
Having derived and validated a new polygenic predictor that considerably outperformed earlier scores, we explored the predictive power of the ExGRS on HM in 1219 Han Chinese individuals of an independent testing dataset. We found the ExGRS exhibited highly reproducible performance, with an AUC of 0.856 and an OR of 3.51 (95% CI: 3.05–4.07, P < 1.31 × 10–65) (Fig. 5a and Table 1). The inclusion of the rvGRS risk genotype considerably enhanced HM risk prediction in MAGIC cohorts, substantially improving tail cutoff discrimination. Compared to the remaining 95% of individuals, the risk for HM among the top 5% of individuals was approximately 9.95-fold higher in the model without rvGRS and 15.57-fold higher in the model with rvGRS (Table 1). The effects of the GRS stratified by with or without rvGRS in MAGIC cohorts are depicted in Fig. 5b.
Fig. 5. Effects of the ExGRS for HM in testing cohorts.
a ROC curves and corresponding areas under the ROC curve (AUCs) were used to assess the ability of the ExGRS to distinguish HM in MAGIC testing subgroup (n = 1219). b The x axis depicts each quantile of the ExGRS ordered from the first (Q1) to the last (Q5) quantile. The y axis depicts the ORs of HM for each of the quantile-defined subgroups in reference to the middle quantile (Q3) of cvGRS. The effect estimates (dots) and 95% CIs (vertical bars) were derived based on a fixed-effects in MAGIC testing cohorts. c ROC Curves for detecting HM using ExGRS in UKB testing subgroup of European ancestry (n = 1200). d The effects of the ExGRS in UKB. Error bars based on confidence intervals (C.I.).
Table 1.
The performance metrics of the GRS in the testing cohorts
| Models | OR per s.d. (95% CI), P | AUC | PRS threshold | OR (95% CI), P | Prevalence of HM |
|---|---|---|---|---|---|
| cvGRS | 3.74 (3.19–4.44), 3.27 × 10–55 | 0.819 | Top 20% versus other 80% | 9.58 (6.43–14.68), 5.70 × 10–41 | 0.86 |
| Top 10% versus other 90% | 10.93 (5.92–22.05), 7.11 × 10–23 | 0.90 | |||
| Top 5% versus other 95% | 9.95 (4.24–28.50), 1.94 × 10–11 | 0.90 | |||
| Top 2% versus other 98% | 7.19 (2.13–37.86), 2.4 × 10−4 | 0.87 | |||
| Top 1% versus other 99% | 5.05 (1.07–47.61), 0.037 | 0.83 | |||
| rvGRS | 2.24 (1.99–2.54), 4.92 × 10–38 | 0.759 | Top 20% versus other 80% | 9.21 (6.21–14.04), 5.33 × 10–40 | 0.86 |
| Top 10% versus other 90% | 9.96 (5.50–19.54), 7.25 × 10–22 | 0.89 | |||
| Top 5% versus other 95% | 9.95 (4.24–28.50), 1.94 × 10–11 | 0.90 | |||
| Top 2% versus other 980% | 7.19 (2.13–37.86), 2.4 × 10–4 | 0.87 | |||
| Top 1% versus other 99% | 11.15 (1.61–480.32), 0.006 | 0.91 | |||
| ExGRS | 3.51 (3.05–4.07), 1.31 × 10−65 | 0.856 | Top 20% versus other 80% | 12.45 (8.07–19.86), 3.58 × 10-47 | 0.89 |
| Top 10% versus other 90% | 15.13 (7.59–34.31), 3.74 × 10−26 | 0.92 | |||
| Top 5% versus other 95% | 15.57 (5.70–59.48), 1.47 × 10–13 | 0.93 | |||
| Top 2% versus other 980% | 7.19 (2.13–37.86), 2.4 × 10–4 | 0.87 | |||
| Top 1% versus other 99% | 11.15 (1.61–480.32), 0.006 | 0.91 |
Next, we evaluated the robustness of the ExGRS in 8682 UKB European-ancestry individuals. Although there was significant between-population correlation of allelic effects (i.e., logOR) for variants clumped with different cut-off P-values (Supplementary Fig. 16), we detected significant differences in the ExGRS across ancestries (Wilcoxon rank sum test, P < 2.20 × 10–16). We then tested the final ExGRS in the UKB European cohort. Predictive models based on the MAGIC and UKB overlapped SNPs and HM-associated genes, fitted with age, sex, and population structure, were predictive of HM (versus all non-HM controls) with AUC values of 0.657, similar to 0.662 with cvGRS only (Fig. 5c). The rvGRS provides limited improvement over cvGRS in the prediction of HM risk. The combined ExGRS model resulted in an OR per s.d. of 1.46, 95% CI = 1.41–1.52 and P = 2.35 × 10–82, which is lower than cvGRS model (OR per s.d. = 1.78, 95% CI = 1.69–1.88, P = 2.14 × 10–105) (Supplementary Table 6). Contrary to MAGIC Han Chinese ancestry cohorts, the inclusion of rvGRS in the ExGRS decreased risk prediction in UKB European cohorts (Fig. 5d). Therefore, the modeled risk in individuals of European ancestry was entirely attributable to the cvGRS.
Discussion
In our study, we estimated the heritability of HM captured by both rare and common variants in unrelated individuals from two distinct ancestry cohorts. We identified additional variance attributed to rare variants, particularly rare protein-altering variants in low LD with other genomic variants, beyond what was captured by common HapMap3 variants. Our estimations largely, though not entirely, recovered the heritability estimated from pedigree data, particularly for the Han Chinese ancestry cohort, but less so for the European ancestry cohort. The remaining gap could be due to a combination of sampling variance and remaining causal variants that are not captured by the WES data. Based on the high heritability of HM, we described a systematic approach to derive and validate the ExGRS, incorporating information from rare to common genetic variants, to predict polygenic susceptibility to HM. Our studies demonstrated that the extreme tails of the risk ExGRS distribution (top 5%) conferred an approximately 15-fold increased risk for HM in the Han Chinese population. Additionally, we tested the ExGRS in participants across two ancestries and found that the top 5% risk ExGRS distribution conferred an approximately twofold increased risk for HM in European ancestry, which is lower than the 3.67-fold for cvGRS.
Beyond enhanced disease screening of asymptomatic individuals, other potential applications of the ExGRS may include improved risk stratification for schoolchildren at risk or enhanced assessment of early-onset myopia. Our results underscore the urgent need to test the individual ExGRS in this setting to better assess its impact on the risk of pathological myopia and other HM complications. The ability to quantify inborn susceptibility using ExGRS is likely generalizable across a broad range of complex diseases, contingent upon the availability of large-scale discovery WES, independent validation and testing datasets, and the extent of heritability of a given disease explained by rare and common variants. Predictive power is expected to further improve in the coming years due to larger-scale WES and WGS discovery studies and advancements in computational algorithms that integrate functional genomics annotations, variant-variant interactions, and rare large-effect variants into the predictive model.
We note that the extremes of both the cvGRS and rvGRS distributions (top 5%) identically predispose individuals to a 10.0-fold greater risk compared to the remainder of the population. Consistent with the higher heritability of rare variants, a higher risk was observed in the rvGRS model (OR = 11.15, P = 0.006) compared to cvGRS (OR = 5.05, P = 0.037) for the top 1% versus the bottom 99%. Although the combined ExGRS model substantially improved prediction performance, it demonstrated incomplete penetrance, as not all carriers manifest HM. This observation aligns with recent PGS studies that combine common and rare variants across a broad range of complex diseases, including coronary artery disease, atrial fibrillation, type 2 diabetes, inflammatory bowel disease, kidney disease, and breast cancer20,37–40. Additional studies of large unascertained populations are needed to determine whether a larger effect size for rvGRS can be found among adults, and the extent to which a favorable polygenic background can explain the absence of HM noted among many mutation carriers. Because our score is based on an ExWAS and RVAS for HM in Han Chinese ancestry, the allelic effect estimates are heavily biased by Han Chinese participants. We used variants with concordant direction-of-effect between MAGIC and UKB to improve the trans-ethnic performance of the score and further enhanced the model by including the rvGRS model. We demonstrated that rvGRS has an additive effect with cvGRS and significantly improves case-control discrimination in the Han Chinese cohort. However, because allele frequencies, linkage disequilibrium patterns, and effect sizes of polymorphisms vary by ancestry, this specific ExGRS will not have optimal predictive power for European ethnic groups.
Although the average refractive error has increased substantially across multiple populations, the variability within a given population has also increased, suggesting that an increasingly myopiagenic environment may have led to a preferential “unmasking” of inherited susceptibility in those with the highest genetic risk12,41,42. For example, prior studies suggest that the effects of education, metabolism, near work, and time outdoors on refractive error are most pronounced in individuals with a genetic predisposition43–46. The ability to identify high-risk individuals from birth may facilitate targeted strategies for HM prevention with increased effectiveness or cost-efficiency. The ExGRS permits the identification of individuals from birth who inherit high susceptibility, even before clinical disease manifests itself. Careful study of individuals at the extremes of an ExGRS distribution might uncover new causal risk factors or underlying disease pathways. Similarly, clinical and multi-omic profiling of individuals at the extremes of an ExGRS distribution for HM may reveal the contributions and molecular correlates of pathways related to ocular development47, neurotransmission48, and scleral remodeling49, and might enable the identification of clinically relevant subtypes of severe myopia that most benefit from a given pharmacologic or behavioral intervention.
Several important limitations of this work need to be discussed. First, our study is significantly limited by the lack of large-scale WES for HM across multiethnic populations, as well as the small size of existing cohorts that could optimize performance in European and Asian groups. The assumption of fixed allelic effects across different ancestry groups is likely inaccurate, as many disease-related lifestyle factors and environmental exposures associated with ancestry can modify allelic effects. Accordingly, the overall tail discrimination of the score was lower in European cohorts than in Han Chinese cohorts, with notably lower sensitivity at the top 5% GPS cutoff. Although overcoming this limitation is not possible in the present study, our ExGRS approach could be refined by incorporating larger WES studies for HM when they become available in the future. Second, performance comparisons between different ancestral groups might be biased due to differences in genotyping platforms and ascertainment methods employed by various biobanks. For example, the UKB represents a population-based cohort that recruits European participants aged 40-60, while the MAGIC case-control cohorts target schoolchildren. The inclusion of older participants in UKB cohorts might lead to some cases being misclassified due to age-related refractive error decline, resulting in underestimated risk in cohorts with older participants. Finally, it is crucial to consider other factors that can affect PRS transferability. Environmental and lifestyle factors, such as near-work activities, outdoor exposure, and educational attainment, play significant roles in the development and progression of high myopia. In light of these complexities, a comprehensive assessment of an individual’s genetic susceptibility, considering specific environmental factors, should be developed for the future.
In summary, this study highlighted the importance of rare variants in addressing the current gap in heritability of various traits or diseases, using WES data. In this study, we derived, optimized, and validated a new tool, ExGRS, for HM prediction across ancestries. The variants uncovered by cvGRS and rvGRS had additive effects on HM, resulting in a nearly 15-fold increased risk for HM among individuals in the highest 5% of the risk score distribution. This result underscores the significance of genetic risk scores that combine rare variants, which may provide higher prediction accuracy for many polygenic diseases. The potential implications of the ExGRS include its ability to identify at-risk individuals before the disease or trait manifests. With the cost of WES no longer being prohibitive, a population-based genetic screening approach for common eye diseases may prove to be a cost-effective public health strategy. While our study marks an initial step in this direction, prospective studies are warranted to evaluate the performance of this approach in clinical practice and to analyze its cost-effectiveness.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work was supported by the National Natural Science Foundation of China (U20A20364 and 81830027) and the Zhejiang Provincial Key Research and Development Program Grant (2021C03102) to J. Qu; the National Natural Science Foundation of China (82172882, 81930068) to J. Su.
Author contributions
The study was conceived, designed and supervised by J.S., J.Q. and L.Q. Analysis of data was performed by J.Y., R.Q., Y.W., H.S., W.D., Y.Y. and K.L. Patient sample recruitment was conducted by R.Z. and S.X. with member of Myopia Associated Genetics and Intervention Consortium. DNA extraction and sequencing were carried out by X.Y. The manuscript was written by J.Y. and Z.C. with contributions from all other authors.
Peer review
Peer review information
Communications Medicine thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
The raw genetic sequencing data for patients and control individuals generated in this study have been deposited in the Genome Sequence Archive (GSA, https://ngdc.cncb.ac.cn/gsa-human/) under accession numbers HRA007816 in BIG Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences. All raw sequencing data deposited in GSA are under restricted access, and only academic use will be approved via email to Jianzhong Su (sujz@wmu.edu.cn). A response would be expected within a week. Source data underlying Fig. 1 and Fig. 2 is supplied as Supplementary Data 1, as well as in the public repository figshare with the identifier (10.6084/m9.figshare.27896016)50. All other data are available from the corresponding author on reasonable request. The data are not publicly available due to them containing information that could compromise research participant privacy/consent. Researchers who would like to obtain the data related to this study will be presented with a Data Use Agreement which requires that participants will not be reidentified and no data will be shared between individuals, or third parties, or uploaded onto public domains. Upon reasonable request, a data sharing agreement can be initiated between the interested parties and the clinical institution following institution-specific guidelines.
Code availability
All the code used is publicly available at https://github.com/sulab-wmu/MAGIC-PIPELINE51.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jian Yuan, Ruowen Qiu.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Liya Qiao, Email: dr13621035879@gmail.com.
Jia Qu, Email: jia.qu@eye.ac.cn.
Jianzhong Su, Email: sujz@wmu.edu.cn.
Myopia Associated Genetics and Intervention Consortium:
Yinghao Yao, Ran Zhuo, Jianzhong Su, Liangde Xu, Fan Lyu, Hong Wang, Jian Yuan, Zhen Ji Chen, Yunlong Ma, Zhengbo Xue, Hui Liu, Wei Dai, Riyan Zhang, Xiaoguang Yu, and Jia Qu
Supplementary information
The online version contains supplementary material available at 10.1038/s43856-024-00718-1.
References
- 1.Yu, X. et al. Whole-exome sequencing among school-aged children with high myopia. JAMA Netw. Open6, e2345821–e2345821 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morgan, I. G., Ohno-Matsui, K. & Saw, S.-M. Myopia. Lancet379, 1739–1748 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Saw, S. M., Gazzard, G., Shih‐Yen, E. C. & Chua, W. H. Myopia and associated pathological complications. Ophthalmic Physiol. Opt.25, 381–391 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Xu, L. et al. COVID-19 quarantine reveals that behavioral changes have an effect on myopia progression. Ophthalmology128, 1652–1654 (2021). [DOI] [PMC free article] [PubMed]
- 5.You, Q. S. et al. Prevalence of myopia in school children in greater Beijing: The Beijing Childhood Eye Study. Acta Ophthalmol.92, e398–e406 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Wong, Y.-L. & Saw, S.-M. Epidemiology of pathologic myopia in Asia and worldwide. Asia-Pac. J. Ophthalmol.5, 394–402 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Lopes, M. C., Andrew, T., Carbonaro, F., Spector, T. D. & Hammond, C. J. Estimating heritability and shared environmental effects for refractive error in twin and family studies. Investig. Ophthalmol. Vis. Sci.50, 126–131 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Guggenheim, J. A., Kirov, G. & Hodson, S. A. The heritability of high myopia: A reanalysis of Goldschmidt’s data. J. Med. Genet.37, 227–231 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verhoeven, V. J. et al. Genome-wide meta-analyses of multiancestry cohorts identify multiple new susceptibility loci for refractive error and myopia. Nat. Genet.45, 314–318 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tedja, M. et al. Genome-wide association meta-analysis highlights light-induced signaling as a driver for refractive error. Nat. Genet.50, 834–848 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hysi, P. et al. Meta-analysis of 542,934 subjects of European ancestry identifies new genes and mechanisms predisposing to refractive error and myopia. Nat. Genet.52, 401–407 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan, I. G. et al. IMI risk factors for myopia. Investig. Ophthalmol. Vis. Sci.62, 3–3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tran-Viet, K. et al. Mutations in SCO2 are associated with autosomal-dominant high-grade myopia. Am. J. Hum. Genet.92, 820–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin, Z. et al. Trio-based exome sequencing arrests de novo mutations in early-onset high myopia. Proc. Natl. Acad. Sci. USA114, 4219–4224 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosoda, Y. et al. CCDC102B confers risk of low vision and blindness in high myopia. Nat. Commun.9, 1782 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aldahmesh, M. et al. Mutations in LRPAP1 are associated with severe myopia in humans. Am. J. Hum. Genet.93, 313–320 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su, J. et al. Sequencing of 19,219 exomes identifies a low-frequency variant in FKBP5 promoter predisposing to high myopia in a Han Chinese population. Cell Rep.42, 112510 (2023). [DOI] [PubMed]
- 18.Hao, L. et al. Development of a clinical polygenic risk score assay and reporting workflow. Nat. Med.28, 1006–1013 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wray, N. R. et al. From basic science to clinical application of polygenic risk scores: A primer. JAMA Psychiatry78, 101–109 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Khera, A. V. et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet.50, 1219–1224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mojarrad, N. G., Plotnikov, D., Williams, C., & Guggenheim, JA. Association between polygenic risk score and risk of myopia. JAMA Ophthalmol.138, 7–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tideman, J. et al. Evaluation of shared genetic susceptibility to high and low myopia and hyperopia. JAMA Ophthalmol.139, 601–609 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark, R. et al. A new polygenic score for refractive error improves detection of children at risk of high myopia but not the prediction of those at risk of myopic macular degeneration. EBioMedicine91, 104551 (2023). [DOI] [PMC free article] [PubMed]
- 24.Kassam, I. et al. The potential of current polygenic risk scores to predict high myopia and myopic macular degeneration in multi-ethnic Singapore adults. Ophthalmology129, 890–902 (2022). [DOI] [PubMed]
- 25.Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet.81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLaren, W. et al. The ensembl variant effect predictor. Genome Biol.17, 1–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet.88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devlin, B. & Roeder, K. Genomic control for association studies. Biometrics55, 997–1004 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Sveinbjornsson, G. et al. Weighting sequence variants based on their annotation increases power of whole-genome association studies. Nat. Genet.48, 314–317 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Privé, F., Arbel, J. & Vilhjálmsson, B. J. LDpred2: better, faster, stronger. Bioinformatics36, 5424–5431 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mak, T. S. H., Porsch, R. M., Choi, S. W., Zhou, X. & Sham, P. C. Polygenic scores via penalized regression on summary statistics. Genet. Epidemiol.41, 469–480 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Wainschtein, P. et al. Assessing the contribution of rare variants to complex trait heritability from whole-genome sequence data. Nat. Genet.54, 263–273 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng, J. et al. Signatures of negative selection in the genetic architecture of human complex traits. Nat. Genet.50, 746–753 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Weiner, D. J. et al. Polygenic architecture of rare coding variation across 394,783 exomes. Nature614, 492–499 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torkamani, A., Wineinger, N. E. & Topol, E. J. The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet.19, 581–590 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Fiziev, P. P. et al. Rare penetrant mutations confer severe risk of common diseases. Science380, eabo1131 (2023). [DOI] [PubMed] [Google Scholar]
- 38.Khan, A. et al. Genome-wide polygenic score to predict chronic kidney disease across ancestries. Nat. Med.28, 1412–1420 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dornbos, P. et al. A combined polygenic score of 21,293 rare and 22 common variants improves diabetes diagnosis based on hemoglobin A1C levels. Nat. Genet.54, 1609–1614 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khera, A. V. et al. Polygenic prediction of weight and obesity trajectories from birth to adulthood. Cell177, 587–596.e589 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morgan, I. & Rose, K. How genetic is school myopia? Prog. Retinal Eye Res.24, 1–38 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Morgan, I. G. et al. The epidemics of myopia: Aetiology and prevention. Prog. Retinal Eye Res.62, 134–149 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Fan, Q. et al. Childhood gene-environment interactions and age-dependent effects of genetic variants associated with refractive error and myopia: The CREAM Consortium. Sci. Rep.6, 25853 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Enthoven, C. A. et al. Interaction between lifestyle and genetic susceptibility in myopia: The Generation R study. Eur. J. Epidemiol.34, 777–784 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wojciechowski, R., Yee, S. S., Simpson, C. L., Bailey-Wilson, J. E. & Stambolian, D. Matrix metalloproteinases and educational attainment in refractive error: Evidence of gene–environment interactions in the Age-Related Eye Disease Study. Ophthalmology120, 298–305 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fan, Q. et al. Meta-analysis of gene–environment-wide association scans accounting for education level identifies additional loci for refractive error. Nat. Commun.7, 11008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wallman, J. & Winawer, J. Homeostasis of eye growth and the question of myopia. Neuron43, 447–468 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Troilo, D. et al. IMI–Report on experimental models of emmetropization and myopia. Investig. Ophthalmol. Vis. Sci.60, M31–M88 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, H. et al. Scleral hypoxia is a target for myopia control. Proc. Natl. Acad. Sci.115, E7091–E7100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jian Y. Exome-wide genetic risk score (ExGRS) to predict high myopia across multi-ancestry populations. figshare, 10.6084/m9figshare27896016 (2024).
- 51.Jian Y. MAGIC-PIPELINE. Source Code, https://githubcom/sulab-wmu/MAGIC-PIPELINE (2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The raw genetic sequencing data for patients and control individuals generated in this study have been deposited in the Genome Sequence Archive (GSA, https://ngdc.cncb.ac.cn/gsa-human/) under accession numbers HRA007816 in BIG Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences. All raw sequencing data deposited in GSA are under restricted access, and only academic use will be approved via email to Jianzhong Su (sujz@wmu.edu.cn). A response would be expected within a week. Source data underlying Fig. 1 and Fig. 2 is supplied as Supplementary Data 1, as well as in the public repository figshare with the identifier (10.6084/m9.figshare.27896016)50. All other data are available from the corresponding author on reasonable request. The data are not publicly available due to them containing information that could compromise research participant privacy/consent. Researchers who would like to obtain the data related to this study will be presented with a Data Use Agreement which requires that participants will not be reidentified and no data will be shared between individuals, or third parties, or uploaded onto public domains. Upon reasonable request, a data sharing agreement can be initiated between the interested parties and the clinical institution following institution-specific guidelines.
All the code used is publicly available at https://github.com/sulab-wmu/MAGIC-PIPELINE51.