Abstract
Lipooligosaccharide (LOS), a major outer membrane component of Moraxella catarrhalis, is a possible virulence factor in the pathogenesis of human infections caused by the organism. However, information about the roles of the oligosaccharide chain from LOS in bacterial infection remains limited. Here, a kdtA gene encoding 3-deoxy-d-manno-2-octulosonic acid (Kdo) transferase, which is responsible for adding Kdo residues to the lipid A portion of the LOS, was identified by transposon mutagenesis and construction of an isogenic kdtA mutant in strain O35E. The resulting O35EkdtA mutant produced only lipid A without any core oligosaccharide, and it was viable. Physicochemical and biological analysis revealed that the mutant was susceptible to hydrophobic reagents and a hydrophilic glycopeptide and was sensitive to bactericidal activity of normal human serum. Importantly, the mutant showed decreased toxicity by the Limulus amebocyte lysate assay, reduced adherence to human epithelial cells, and enhanced clearance in lungs and nasopharynx in a mouse aerosol challenge model. These data suggest that the oligosaccharide moiety of the LOS is important for the biological activity of the LOS and the virulence capability of the bacteria in vitro and in vivo. This study may bring new insights into novel vaccines or therapeutic interventions against M. catarrhalis infections.
Moraxella catarrhalis, a gram-negative diplococcus, is now considered to be an important human respiratory tract pathogen in children and adults (6, 29). In children, it causes otitis media, the most common childhood infection and the leading cause of conductive hearing loss, while in adults it exacerbates chronic obstructive pulmonary diseases, the fourth leading cause of death in the United States. Sporadic cases of conjunctivitis, meningitis, endocarditis, ophthalmia neonatorum, keratitis, urethritis, peritonitis, and septicemia have also been reported in individuals with reduced immune defense (36). In addition, the number of antibiotic-resistant strains of M. catarrhalis has significantly increased over the past decades (3, 23). Currently, the molecular pathogenesis of M. catarrhalis infection is not fully understood. Based on limited information about this organism and other respiratory tract pathogens, it is generally believed that it has several virulence factors involved in colonization, adherence, or complement resistance that enable the organism to evade the normal host defense and to establish the infection (47).
Previous studies suggested that complement resistance involving multifactorial processes might be an important mechanism of M. catarrhalis virulence. Several M. catarrhalis isogenic mutants of UspA2 (1, 30), Fur (15), CopB (22), and outer membrane protein (OMP) E (37) showed their susceptibility to the bactericidal activity of normal human serum when compared with their parental strains. This indicates that these bacterial components may play critical roles for the bacterial resistance to killing caused by normal human serum. In addition, many studies also revealed that bacterial adherence or colonization could be another important mechanism of M. catarrhalis virulence since surface outer membrane components such as UspA1, UspA2H (30), OMP CD (24), McaP (42), MID (14), and Hag (25) have been reported as adhesion molecules that bind to host cells.
Lipooligosaccharide (LOS), a major outer membrane component of M. catarrhalis, is also considered to be a possible virulence factor in the pathogenesis of the infections (9, 13). Unlike enteric bacterial lipopolysaccharide (LPS), M. catarrhalis LOS consists of an oligosaccharide core and lipid A without an O-antigen polysaccharide side chain (10). Structural studies have revealed that the oligosaccharides from three major serotypes have branched structures, with a common inner core composed of glucosyl residues and 3-deoxy-d-manno-2-octulosonic acid (Kdo) (26). The lipid A portion is similar to that of other gram-negative bacteria but with seven shorter fatty acyl chains (C10 to C12) (10, 33).
Despite these structural data, information regarding the biosynthesis of M. catarrhalis LOS is limited. This information is critical for elucidating the virulence functions of the LOS molecule. Previously, Zaleski et al. identified a UDP-glucose-4-epimerase and showed that a corresponding knockout mutant resulted in a truncated LOS structure lacking two terminal galactosyl residues (50). Recently, Luke et al. identified a kdsA gene coding for Kdo-8-phosphate synthase and found a kdsA-deficient mutant consisting only of lipid A in its LOS structure (32). Both mutant strains showed reduced resistance to complement killing caused by normal human serum. However, the interactive roles between the LOS and the host epithelial cells in vitro or in animal challenge models remain unstudied.
In this study, we identified kdtA gene in M. catarrhalis and constructed an isogenic kdtA mutant. Based on studies from Escherichia coli and other enteric bacteria, the kdtA gene encodes Kdo transferase, which catalyzes the sequential addition of two Kdo sugars onto a lipid IVA moiety, a key precursor of lipid A. Prior to its incorporation, Kdo is synthesized and activated to CMP-Kdo by the enzymes encoded by kdsA and kdsB genes (40). Analysis of the physicochemical and biological functions of the kdtA mutant was performed to study functions of kdtA gene and its resulting LOS in vitro and in vivo.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
Bacterial strains, plasmids, and primers are described in Table 1. M. catarrhalis strains were cultured on chocolate agar plates (Remel, Lenexa, KS) or brain heart infusion (BHI) (Difco, Detroit, MI) agar plates at 37°C in 5% CO2. Mutant strains were selected on BHI agar supplemented with kanamycin at 20 μg/ml. Growth rates of wild type and mutant were measured as follows. An overnight culture was inoculated in 10 ml of BHI media (adjusted optical density at 600 nm [OD600] = 0.05) and shaken at 37°C with 250 rpm. The bacterial cultures were monitored spectrophotometrically at OD600. E. coli was grown on Luria-Bertani agar plates or broth with antibiotic supplementation. The antibiotic concentrations used for E. coli were as follows: kanamycin, 30 μg/ml; ampicillin, 50 μg/ml.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli TOP10 | Cloning strain | Invitrogen |
| M. catarrhalis O35E | Wild-type strain | 46 |
| M. catarrhalis O35E705 | Strain with EZ::TN transposon insertion in kdtA gene | This study |
| M. catarrhalis O35EkdtA | kdtA knockout mutant strain | This study |
| Plasmids | ||
| pCR2.1 | TOPO TA cloning vector | Invitrogen |
| pBluescript II SK(+) | Cloning vector | Fermentas |
| pUC4k | Kanamycin resistance cassette | Amersham |
| pCRA | kdtA cloned into pCR2.1 | This study |
| pSA | EcoRI-SalI kdtA fragment cloned into SK(+) | This study |
| pSAK | EcoRI-blunted kanamycin resistance cassette inserted into blunted HindIII site of pSA | This study |
| Primers | ||
| 31 | 5′-GAA ACC TGA GAG GTA ACT GCA GCC AGT T-3′ | This study |
| 32 | 5′-GTA TCA TTG CTC AAG ATG CCA GCT CTG C-3′ | This study |
| 33 | 5′-ATG GAC GGG CAA ATC AGG TGC TGT ATT G-3′ | This study |
| 34 | 5′-AGC CAT GAA GAT TTG GTG-3′ | This study |
| 35 | 5′-CTC GTC GAC TTG TCG CAC AAG CAA CGC-3′ (SalI site underlined) | This study |
| 36 | 5′-CTC GAA TTC CGT GAC AGT AAT GGT GAA-3′ (EcoRI site underlined) | This study |
General DNA methods.
DNA restriction endonucleases, T4 DNA ligase, E. coli DNA polymerase I Klenow fragment, and Taq DNA polymerase were purchased from Fermentas (Hanover, MD). Preparation of plasmid and purification of PCR products and DNA fragments were performed using kits manufactured by QIAGEN (Santa Clarita, CA). Bacterial chromosomal DNA was isolated using a genomic DNA purification kit (Promega, Madison, WI). DNA nucleotide sequences were obtained via 3070xl DNA analyzer (Applied Biosystems, Foster City, CA) and analyzed with DNASTAR software (DNASTAR Inc., Madison, WI).
Transposon mutagenesis and identification of kdtA gene.
A 20-ml portion of culture (OD600 = 0.5) of strain O35E was harvested by centrifugation and washed three times with 10% (vol/vol) glycerol in distilled water. The final cell pellet was suspended in 150 μl glycerol solution. A 20-μl portion of this cell suspension was mixed with 1 μl of EZ::TN <KAN-2> Tnp Transposome (Epicentre, Madison, WI), transferred into a microelectroporation chamber, and electroporated using a field strength of 2.2 kilovolts over a 1-mm distance for 4 ms (Micropulser, Bio-Rad, Hercules, CA). After the electroporation, the cell suspension was added to 1 ml of BHI broth, shaken at 250 rpm at 37°C for 3 h, and plated on BHI agar plates containing kanamycin. After 24 h incubation, the resulting kanamycin-resistant colonies were screened by colony blot assay and further identified by whole cell enzyme-linked immunosorbent assay (28) for loss of binding reactivity to a rabbit anti-LOS antibody. The transformants without anti-LOS antibody binding activity were subsequently evaluated by examining the LOS profiles from proteinase-K-treated whole-cell lysates (45) by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining analysis (43). The clones with no detectable LOS were selected for isolation of chromosomal DNA and nucleotide sequence analysis. A first pair of sequence primers was provided by the EZ::TN <KAN-2> Tnp Transposome kit; other primers were designed according to the results of nucleotide sequence analysis (Table 1; Fig. 1, primers 31, 32, and 33). The kdtA homologue from strain O35E was identified by BLAST searches at GenBank of the National Center for Biotechnology Information.
FIG. 1.
Genetic organization of kdtA locus in M. catarrhalis O35E genome. pepN, aminopeptidase N; HP, hypothetical protein. Large arrows represent the direction of transcription; the site of the transposon insertion identified in O35E705 is denoted as EN:TN, and the location of deletion replaced by the kanamycin resistance gene (kanR) is between two HindIII cleavage sites. The sites of primers used are indicated as small arrows (primers 31, 32, 33, 34, 35, and 36 are described in Table 1).
Cloning of kdtA homologue and construction of the knockout kdtA mutant.
The putative kdtA homologue was amplified from chromosomal DNA of strain O35E using primers 35 and 36 (Table 1; Fig. 1). This PCR product was cloned into pCR2.1 using a TOPO TA cloning kit (Invitrogen, Carlsbad, CA) to obtain pCRA. The insertion was released by EcoRI-SalI digestion and then subcloned into an EcoRI-SalI site of SK(+) to form pSA. A kanamycin resistance cassette (1,282 bp) obtained from pUC4k with EcoRI digestion was subsequently inserted into the kdtA gene using the HindIII site to form pSAK.
After verification by sequence analysis, the mutagenic construct (containing an insertion of the kanamycin resistance cassette within a deletion of the kdtA coding region) was amplified by PCR using primers 35 and 36. The PCR-generated DNA product was purified and used for electroporation. After 24 h incubation, the resulting kanamycin-resistant colonies were selected for PCR analysis of chromosomal DNA using primers 34 and 36. The inactivated kdtA mutant was verified by sequence analysis and named as O35EkdtA. OMPs of both mutant and wild-type strains were prepared by the Zwittergent extraction method (5) and examined on SDS-PAGE gels with 7.5% separating gel.
Kdo assay.
Outer membrane vesicles (OMV) were prepared by the EDTA-heat-induced method (38). Kdo was determined as described previously (48) by using 80 μg OMV preparation of wild-type or mutant strain as samples and Kdo ammonium salt (Sigma, St. Louis, MO) as a standard.
LOS determination.
A crude LOS extraction was performed from both wild-type strain O35E and the mutant O35EkdtA using the proteinase-K-treated whole-cell lysate method (45). The resulting extracts from each bacterial suspension (1.9 μg of protein concentration) were resolved by 15% SDS-PAGE and visualized by silver staining (43). The OMV preparation was also applied since there was no detectable LOS in O35EkdtA whole-cell lysates. Western blot using a rabbit anti-LOS antibody was performed (28). For the composition analysis, 30 to 35 g of wet cells from O35E and O35EkdtA were prepared for LOS purification by phenol-water extraction (21) and then the phenol-chloroform-petroleum ether (PCP) method (17), due to the lack of LOS extracts from O35EkdtA. The glycosyl compositions of O35E LOS purified by phenol-water extraction and O35EkdtA LOS purified by PCP were determined by gas chromatography-mass spectrometry (GC-MS) analysis of trimethylsilyl methylglycosides. Also fatty acid methyl esters (49) were performed on an HP-5890 GC interfaced to a mass selective detector 5970 MSD using a Supelco DB1 fused silica capillary column (30 by 0.25 mm inside diameter; J &W Scientific, Folsom, California). The fatty acyl components of samples were detected by MS analysis. The sample was dissolved in 3:1 chloroform-methanol mixture at 1 μg/μl concentration and mixed with equal volume of 0.5 M 2,5-dihydroxy benzoic acid (matrix) and spotted with 1 μl on a 100-well stainless steel matrix-associated laser desorption ionization (MALDI) plate. The mass spectra were collected on a MALDI-time of flight (TOF) instrument (Applied Biosystems) in reflectron mode using 337-nm N2 laser in negative mode.
Negative staining of bacteria.
Bacteria were suspended on a slide in a small amount of water (28). Formvar-coated 200 mesh grids (Electron Microscopy Sciences, Fort Washington, PA) were floated on the bacterial suspension for 1 min, blotted dry, and then floated on a mixture of equal volumes of 2% ammonium acetate (Mallinckrodt Chemical, Inc., Paris, KY) and 2% ammonium molybdate (Sigma). The grids were viewed with a JEOL-1010 transmission electron microscope (JEOL Ltd., Tokyo, Japan).
Limulus amebocyte lysate assay.
The chromogenic Limulus amebocyte lysate assay for endotoxin activity was performed using the QCL-1000 kit (Bio-Whittaker Inc., Walkersville, MD). Overnight cultures from chocolate agar plates were suspended in BHI broth to OD600 of 0.1, and serial dilutions of these stocks were used as samples.
Susceptibility determination.
The sensitivity of strains to a panel of hydrophobic agents or a hydrophilic glycopeptide was performed using standard disk diffusion assays (39). Bacteria were cultured in BHI to an OD600 of 0.2, and 100-μl portions of the bacterial suspension were spread onto chocolate agar plates. Antibiotic disks or sterile blank paper disks (6 mm; Becton Dickinson, Cockeysville, MD) saturated with the various agents were plated on the lawn in triplicate at 37°C for 18 h. Sensitivity was assessed by measuring the diameter of the zone of growth inhibition in two axes, and the mean value was calculated.
Bactericidal assay.
A complement-sufficient normal human serum was prepared and pooled from 8 healthy adult donors. A 200-μl bactericidal assay was performed in a 96-well plate (32). Normal human serum was diluted to 0.5, 2.5, 5.0, 12.5, and 25% in pH 7.4 Dulbecco's phosphate-buffered saline containing 0.05% (wt/vol) gelatin (DPBSG). Bacteria (10 μl of 106 CFU) were inoculated into 190-μl reaction wells containing the diluted normal human serum, 25% of heat-inactivated normal human serum, or DPBSG alone and incubated at 37°C for 30 min. Serial dilutions (1:10) of each well were plated onto chocolate agar plates. The resulting colonies were counted after 24 h of incubation.
Adherence assay.
Chang (conjunctival; CCL20.2), HeLa (cervix; CCL-2), and A549 (lung; CCL-185) human epithelial lines were cultured in Eagle's minimal essential medium (American Type Culture Collection, Manassas, VA) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum in an atmosphere of 5% CO2 at 37°C. A quantitative adherence assay was performed on a 24-well tissue culture plate (Corning Incorporated, Corning, NY) (1). Adherence was expressed as the percentage of bacteria attached to the human cells relative to the original bacteria added to the well. The data represent the average of 3 independent assays.
Pulmonary and nasopharyngeal clearance patterns in animal model.
Female BALB/c mice (6 to 8 weeks of age) were obtained from Taconic Farms Inc. (Germantown, NY). The mice were housed in an animal facility in accordance with National Institutes of Health guidelines under animal study protocol 1158-04. Bacterial aerosol challenges were carried out in mice using the same OD540 value of wild-type strain O35E (1.24 × 109 CFU/ml) or mutant O35EkdtA (7.8 × 108 CFU/ml) in 10 ml DPBSG (27). The number of bacteria present in the lungs and nasal washes was measured at various time points postchallenge. The minimum detectable numbers of viable bacteria were 100 CFU per lung and 4 CFU per nasal washing. Clearance of M. catarrhalis was expressed as the percentage of bacterial CFU at each time point compared with the number deposited at time zero. Due to the lower inoculum of the mutant strain, the challenge was repeated with similar doses of 6.2 × 108 or 6.0 × 108 (CFU/ml) for O35E or O35EkdtA to confirm if the clearance rate is strain dependent rather than inoculum dependent.
Statistical analysis.
The adherence and clearance percentage was analyzed by chi-square test.
Nucleotide sequence accession number.
The nucleotide sequence of kdtA gene in M. catarrhalis strain O35E has been deposited at GenBank under accession number AY854633.
RESULTS
Identification and cloning of M. catarrhalis O35E kdtA.
M. catarrhalis mutants were constructed by transposon mutagenesis. The resulting kanamycin-resistant colonies were screened for loss of reactivity to a specific anti-LOS antibody. One clone (O35E705) with no detectable LOS band in the silver staining gel after SDS-PAGE was selected. The chromosomal DNA of O35E705 was purified and subjected to nucleotide sequence analysis which showed that the EZ:TN transposon was inserted in a 279-bp position of a DNA fragment within a single open reading frame of 1,308 bp (Fig. 1). BLAST searches at GenBank with the deduced polypeptide sequence revealed 35% or 34% identity and 52% or 51% similarity when compared with the kdtA amino acid sequence of E. coli or Neisseria meningitidis. Unlike the Kdo biosynthetic operon (pyrG-kdsA-eno) in M. catarrhalis (32), though the predicted pepN coding for an aminopeptidase N is in close proximity to the kdtA (Fig. 1), it is not related to LOS biosynthesis.
Construction of knockout kdtA mutant.
To confirm the function of the predicted kdtA gene, a kdtA knockout mutant was constructed by allelic exchange with an insertion of a kanamycin resistance cassette into the HindIII site of the kdtA gene with a 956-bp deletion (Fig. 1). Primers 34 and 36 were used to amplify the kdtA gene from kanamycin-resistant colonies. The sequence analysis of PCR products confirmed that the kanamycin resistance cassette was inserted into kdtA of O35E chromosomal DNA at the predicted position. The clone was named as O35EkdtA.
Morphology and growth rate of kdtA mutant.
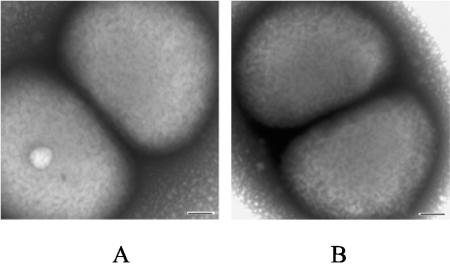
O35EkdtA formed smaller colonies on chocolate agar plates when compared with the wild-type strain. The growth rate of O35EkdtA in BHI broth was significantly slower than that of wild-type strain at logarithmic phase (Fig. 2A). Comparative analysis of OMP by SDS-PAGE indicated that both wild-type and mutant strains showed similar OMP profiles (Fig. 2B). Electron microscopy further revealed that the shape and size of the O35EkdtA mutant (Fig. 3A) is similar to those of the wild-type strain (Fig. 3B)
FIG. 2.
(A) Growth curves of M. catarrhalis wild-type strain O35E (♦) and mutant O35EkdtA (□) in BHI broth at 37°C. (B) The corresponding OMP profile of M. catarrhalis wild-type O35E (left lane) and mutant O35EkdtA (right lane) were shown by SDS-PAGE and Coomassie brilliant blue staining. Molecular size standards are shown in kilodaltons.
FIG. 3.
Electron photomicrographs of negative staining of M. catarrhalis mutant O35EkdtA (A) and wild-type strain O35E (B). Scale bars, 100 nm.
Determination of Kdo and LOS contents in kdtA mutant.
The OMV preparations were evaluated for the presence of Kdo, a specific sugar component of LOS. The concentration of Kdo in the OMV from the wild-type strain was 73 ng/80 μg of OMV, while that of the mutant was below detectable levels (<5 ng/80 μg of OMV). To detect any LOS-derived oligosaccharide component in the kdtA mutant, the LOS content of both proteinase-K-treated cell lysates and OMVs from O35E and O35EkdtA was examined by SDS-PAGE analysis. Silver staining for carbohydrate (sensitivity up to 10 ng for LOS) after SDS-PAGE revealed that LOS was detected in O35E but not in O35EkdtA (Fig. 4A). The LOS band was confirmed by Western blot analysis; here a specific anti-LOS antibody detected LOS in the O35E but not in the O35EkdtA (Fig. 4B).
FIG. 4.
LOS patterns of SDS-PAGE followed by silver staining (A) or Western blot (B) of M. catarrhalis wild-type strain O35E (lanes 1 and 3) and mutant O35E kdtA (lanes 2 and 4). Lanes 1 and 2 represent extracts from proteinase K-treated whole-cell lysates from 1.9 μg of each bacterial suspension, and lanes 3 and 4 represent 0.8 μg of each OMV. A rabbit anti-LOS antibody was used at 1:50 dilution (B).
Structural analysis.
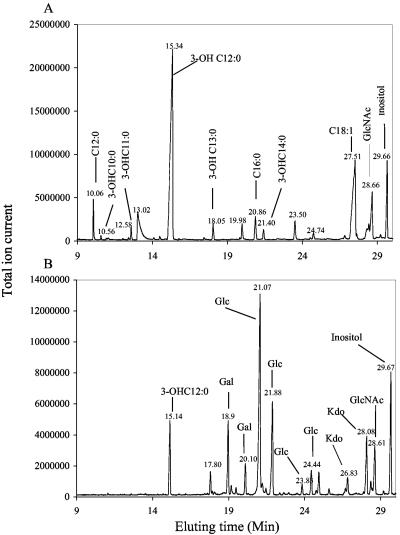
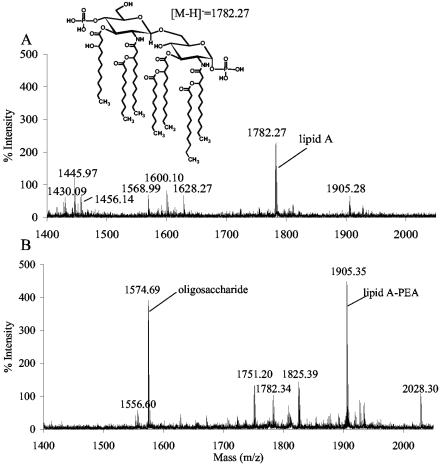
GC-MS composition analysis of the PCP-extracted “LOS” preparation from the O35EkdtA mutant showed that the only glycosyl residue present was N-acetylglucosamine, together with fatty acyl residues that are typical of the lipid A (Fig. 5A). The presence of N-acetylglucosamine is presumably due to the diglucosamine backbone of the lipid A. In contrast, LOS preparation from the wild-type strain showed a number of glycosyl components typical of the core oligosaccharide, including Kdo (Fig. 5B). The MALDI-TOF MS analysis of the O35kdtA PCP extract confirmed that the extract contained a variety of lipid A structures reflecting variations in molecular species. These are due to differences in phosphate and phosphoethanolamine (PEA) groups as well as some heterogeneity due to variations in fatty acid constituents (Fig. 6A; Table 2). A prominent species of the O35EkdtA extract was the bis-phosphorylated lipid A, which is devoid of PEA groups (m/z 1,782). However, MALDI-TOF MS analysis of the parental lipid A isolated from the LOS by mild acid hydrolysis showed a predominant lipid A species with an additional PEA group (m/z 1,905 = m/z 1,782 + 123) (Fig. 6B; Table 2). These data indicate that the LOS from O35EkdtA consists of intact lipid A structures similar to O35E but with no Kdo or other oligosaccharide components.
FIG. 5.
Total ion current spectrum from GC-MS analysis of glycosyl compositions with 100 μg of PCP extract material from M. catarrhalis mutant O35EkdtA (A) or phenol-water extract from wild-type strain O35E (B). The components of the O35EkdtA (A) were identified as indicated and show the presence of lipid A fatty acids (3-OHC12:0, 3-OHC10:0, etc.) and N-acetylglucosamine (GlcNAc), as well as the fatty acids C16:0 and C18:1, due to the presence of phospholipids (A). The components of the O35E (B) showed the sugar constituents (Kdo, Gal [galactose], Glc [glucose], and GlcNAc) and lipid A fatty acids (3-OHC12:0). Inositol was added as internal standard.
FIG. 6.
MALDI-TOF mass spectral analysis of lipid A compositions with 1 μg of PCP extract material from M. catarrhalis mutant O35EkdtA (A) or phenol-water extract from wild-type strain O35E (B). This analysis was done in the negative mode, and all ions are represented as deprotonated [M-H]− ions. The proposed lipid A composition for each ion is given in Table 2, and major ions with m/z values of 1,574.6 and 1,556.6 are due to the LOS oligosaccharide and its anhydro form, respectively. The upper portion of the figure shows the structure of the major species at 1,782.27.
TABLE 2.
Predicted lipid A compositions of LOS preparation from O35E and O35EkdtA mutant based on MALDI-TOF MS analysis
| Source | Ion m/z
|
Predicted lipid A compositiona | Relative intensityb (%) | |
|---|---|---|---|---|
| Observed | Calculated | |||
| O35E | 2,028.30 | 2,029.19 | P2-PEA2-GlcNAc2-3-OHC12:04-C10:02-C12:01 | 24 |
| 1,905.35 | 1,906.18 | P2-PEA1-GlcNAc2-3-OHC12:04-C10:02-C12:01 | 100 | |
| 1,825.39 | 1,826.18 | P1-PEA1-GlcNAc2-3-OHC12:04-C10:02-C12:01 | 32 | |
| 1,782.34 | 1,783.17 | P2-GlcNAc2-3-OHC12:04-C10:02-C12:01 | 23 | |
| 1,751.20 | 1,752.04 | P2-PEA1-GlcNAc2-3-OHC12:04-C10:01-C12:01 | 29 | |
| O35EkdtA | 1,905.28 | 1,906.18 | P2-PEA1-GlcNAc2-3-OHC12:04-C10:02-C12:01 | 28 |
| 1,782.27 | 1,783.17 | P2-GlcNAc2-3-OHC12:04-C10:02-C12:01 | 100 | |
| 1,628.27 | 1,629.03 | P2-GlcNAc2-3-OHC12:04-C10:01-C12:01 | 29 | |
| 1,600.10 | 1,601.00 | P2-GlcNAc2-3-OHC12:04-C10:02 | 35 | |
| 1,568.99 | 1,569.87 | P2-PEA1-GlcNAc2-3-OHC12:04-C10:01 | 29 | |
| 1,456.14 | 1,456.89 | P2-GlcNAc2-3-OHC12:04-C12:01 (minus H2O) | 30 | |
| 1,445.97 | 1,446.86 | P2-GlcNAc2-3-OHC12:04-C10:01 | 52 | |
| 1,430.09 | 1,430.87 | P2-GlcNAc2-3-OHC12:03-C10:01-C12:01 | 27 | |
P, phosphate moieties; GlcNAc, N-acetylglucosamine; 3-OHC12:0, 3-hydroxydodecanoyl acyl chain; C12:0, dodecanoyl acyl chain; C10:0, decanoyl acyl chain.
The highest value of each strain represents 100% and others compared with the highest one.
Susceptibility of kdtA mutant.
To determine if inactivation of the kdtA gene had an effect on the permeability of the outer membrane of the mutant strain, a broad range of hydrophobic agents and a hydrophilic glycopeptide were used to test the mutant's susceptibility. As shown in Table 3, O35EkdtA was more susceptible to most hydrophobic antibiotics, reagents, and a hydrophilic glycopeptide, vancomycin, than the wild-type strain.
TABLE 3.
Susceptibilities of M. catarrhalis wild-type strain O35E and mutant O35EkdtA to a panel of hydrophobic agents or a hydrophilic glycopeptide
| Compound (amt or concn) | Zone of growth inhibition for strains (mm)a
|
|
|---|---|---|
| O35E | O35EkdtA | |
| Azithromycin (15 μg) | 29.0 ± 0.5 | 35.4 ± 0.5 |
| Fusidic acid (10 mg/ml) | 29.0 ± 0.9 | 32.5 ± 0.5 |
| Novobiocin (5 μg) | 13.8 ± 0.3 | 16.1 ± 0.3 |
| Polymycin B (300 iu) | 11.5 ± 0.5 | 15.5 ± 0.0 |
| Rifampin (5 μg) | 22.1 ± 0.3 | 29.0 ± 0.5 |
| Vancomycin (5 μg) | <6.0b | 8.2 ± 0.3 |
| Deoxycholate (100 mg/ml) | 22.0 ± 0.9 | 26.8 ± 0.3 |
| Triton X-100 (5% [wt/vol]) | 15.5 ± 0.5 | 24.5 ± 0.5 |
| Tween 20 (5% [vol/vol]) | 11.5 ± 0.5 | 14.2 ± 0.3 |
Sensitivity was assessed by measuring the diameter of the zone of growth inhibition in two axes, and the mean value was calculated. Values are the means ± standard deviations. The data represent the averages of three separate experiments.
Stands for no inhibition for bacterial growth.
Biological activity of kdtA mutant.
To investigate the effect on bacterial toxicity or virulence due to loss of functional kdtA gene, the kdtA mutant was tested for LOS-associated biological activity. In a Limulus amebocyte lysate assay, whole-cell suspensions (OD600 = 0.1) gave 3.7 × 103 endotoxin units (EU)/ml for O35E and 6 × 102 EU/ml for the O35EkdtA mutant, a sixfold reduction. In a bactericidal assay with normal human serum, strain O35E survived at 25% normal human serum, which was the highest concentration used. However, 50% of the mutant cells died at 2.5% normal human serum and 95% of the mutant cells died at 25% normal human serum (Fig. 7). The results indicated that the mutant strain showed reduced resistance to the complement killing activity of the normal human serum.
FIG. 7.
Bactericidal activity of normal human serum against M. catarrhalis wild-type strain O35E (black bar) and mutant O35E kdtA (gray bar). “HI” represents the group of 25% heat-inactivated normal human serum. The data represent the averages of three independent assays.
To test the adherence of the kdtA mutant to human epithelial cells, Chang, HeLa, and A549 cell lines were used (Table 4). Lack of expression of the kdtA gene in the mutant caused an approximately 50% reduction in adherence to Chang or HeLa epithelial cells, and no reduction for A549 epithelial cells.
TABLE 4.
Adherence of M. catarrhalis wild-type strain O35E and mutant O35EkdtA to human epithelia
| Strain | Adherencea (%)
|
||
|---|---|---|---|
| Chang cells | HeLa cells | A549 cells | |
| O35E | 44.0 ± 6.6 | 42.6 ± 7.6 | 25.9 ± 11.2 |
| O35EkdtA | 20.6 ± 6.4* | 19.0 ± 6.3* | 25.1 ± 2.6 |
Adherence of M. catarrhalis organisms to human epithelia is expressed as mean (± standard deviation) percentage of bacteria attached to the human cells, based on the number of original bacteria that were added to the well. *, compared with O35E (P < 0.01, using chi-square test).
To investigate the effect on survival of the kdtA mutant in a murine respiratory tract clearance model, mice were challenged with wild-type or mutant strain by aerosolization (Table 5). Mutant O35EkdtA present in the lungs showed a significantly accelerated clearance rate compared to the wild type at 3 h (86.0% versus 68.2%, P < 0.01) or 6 h (97.2% versus 84.0%, P < 0.01). Mutant O35EkdtA present in the nasopharynx showed a similar pattern at 3 h (92.3% versus 70.2%, P < 0.01) or 6 h (99.3% versus 90.1%, P < 0.05).
TABLE 5.
Time course of bacterial recovery in mouse respiratory tract after an aerosol challenge with M. catarrhalis wild-type strain O35E and mutant O35EkdtA
| Groupa (sample source) | Bacterial recoveryb (CFU/mouse) (SD range) and reductionc (%) at:
|
||||||
|---|---|---|---|---|---|---|---|
| 0 h | 3 h
|
6 h
|
24 h
|
||||
| Recovery | Recovery | Reduction | Recovery | Reduction | Recovery | Reduction | |
| O35E (lung) | 112,489 (77,097-164,129) | 35,782 (23,721-53,977) | 68.2* | 18,480 (11,764-29,031) | 84.0* | 102 (50-211) | NDd |
| O35EkdtA (lung) | 63,400 (54,963-73,130) | 8,979 (7,861-10,256) | 86.0** | 1,771 (787-3,986) | 97.2** | <100 | ND |
| O35E (nasal washes) | 15,661 (12,596-19,470) | 4,662 (2,776-7,829) | 70.2* | 1,541 (1,104-2,149) | 90.1*** | 26 (11-61) | ND |
| O35EkdtA (nasal washes) | 14,614 (9,976-21,410) | 1,123 (659-1,912) | 92.3** | 101 (56-181) | 99.3**** | 4 (1-12) | ND |
Mice were challenged by aerosol with 10 ml of 1.24 × 109 CFU/ml O35E or 7.8 × 108 CFU/ml O35EkdtA.
Lung and nasal washes were collected for bacterial counts, and each time point represents a geometric mean bacterial CFU of six mice.
Compared with time point zero. * versus **, significant difference in bacterial reduction between O35E and O35EkdtA at P < 0.01. *** versus ****, significant difference at P < 0.05. The chi-square test was used for all comparisons.
ND, bacterial recoveries in some mice were under detectable level.
Due to the lower inoculum of the mutant strain, the challenge was repeated with similar doses of 6.2 × 108 or 6.0 × 108 (CFU/ml) for O35E or O35EkdtA. The numbers of O35E and O35EkdtA present in lungs at 0 h were 32,854 (26,099 to 41,356) and 22,238 (16,276 to 30,386), respectively. Similar to the first challenge, the mutant O35EkdtA present in the lungs showed an accelerated clearance rate compared to the wild type at 3 h (85.6% versus 67.9%, P < 0.01) or at 6 h (94.5% versus 83.8%, P < 0.05) (data not shown). Meanwhile, the numbers of O35E and O35EkdtA present in nasopharynx at 0 h were 9,646 (6,865 to 13,556) and 7,916 (5,598 to 11,193), respectively. The mutant O35EkdtA present in the nasopharynx also showed an accelerated clearance rate compared to the wild type at 3 h (85.6% versus 61.4%, P < 0.01) (data not shown).
DISCUSSION
A kdtA gene from M. catarrhalis was identified by using random transposon mutagenesis and further confirmed by the construction and characterization of a kdtA knockout mutant. As expected, the mutant O35EkdtA did not produce a detectable LOS band by silver staining or Western blot from whole-cell lysates or OMVs. No Kdo component was detected in OMV prepared from O35EkdtA. These observations were supported by GC-MS and MS analysis, which showed that the LOS extracted from the O35EkdtA consisted of only the bis-phosphorylated lipid A with seven fatty acyl chains. These data indicate that inactivation of the kdtA gene in M. catarrhalis results in viable bacteria with only an intact lipid A structure, similar to a known kdtA mutant in N. meningitidis (45) and a kdsA mutant in M. catarrhalis (32).
In the lipid A biosynthesis pathway of E. coli and other enteric bacteria, the late acyltransferases require the presence of the Kdo disaccharide in their substrate for activity since lipid IVA is the major component that accumulates in all mutants with defects in Kdo (4, 7, 8). In our study, the lipid A precursor of M. catarrhalis is completely acylated prior to addition of Kdo. This phenomenon has been only observed in Pseudomonas aeruginosa (19, 34) and N. meningitidis (45). In addition, our data suggest that late acyltransferases of M. catarrhalis incorporate one or two secondary fatty acyl chains to each glucosamine unit with an asymmetrical pattern, while the acyltransferases of P. aeruginosa or N. meningitidis add one secondary fatty acyl chain in a symmetrical pattern.
The M. catarrhalis kdtA mutant showed complete or incomplete septum separation in binary cells with similar shapes and sizes as the wild type, as determined by electron microscopy. However the growth rate of the kdtA mutant was dramatically reduced in comparison to the wild type. In N. meningitidis, the kdtA mutant also showed a reduced growth rate, but with incomplete septum separation in tetrads. This is believed due to a defect in murein hydrolase activity or other cell division enzymatic activities (45). In addition, our kdtA mutant was susceptible to hydrophobic compounds as well as a hydrophilic glycopeptide that is normally excluded by the intact bacterial outer membrane, and also sensitive to the bactericidal activity of human normal serum. It is not clear if these changes are caused by a direct loss of the oligosaccharide moiety of the LOS on the outer membrane, or an indirect effect on surface display of the membrane molecules, such as proteins. It has been shown that UspA2 expression is involved in serum resistance by M. catarrhalis (1, 30) and the UspA2 tightly associated with the LOS (11). The lack of oligosaccharide moiety of the LOS may affect surface display of UspA2 and cause sensitive phenotype of the kdtA mutant. These data suggest that the oligosaccharide moiety of the LOS molecule is an important component in maintaining complement resistance of the bacteria to human normal serum and the integrity of the bacterial outer membrane.
Although lipid A has long been recognized as the active moiety for endotoxic activity (16), variations in the saccharide content of LOS/LPS have been reported as structural determinants of macrophage activation by different LOS/LPS. For example, the polysaccharide portion covalently bound to lipid A played a principal role in Salmonella LPS-induced activation (35). Kdo2 linked to meningococcal lipid A was structurally required for maximal activation of the macrophages (51). In the Limulus amebocyte lysate assay, our kdtA mutant showed a sixfold reduction of endotoxin activity compared to that of the wild type. The data imply that oligosaccharides in M. catarrhalis LOS may play an important role in preserving its biological activity.
Bacterial adherence to the surface of epithelial cells plays a critical role in colonization and is believed to be the first step in the pathogenesis of microbial infections. It has been reported that several LOSs from respiratory tract bacteria are associated with bacterial adherence (2, 18, 20, 41, 44). Fitzgerald et al. showed that the bacterial adherence of M. catarrhalis might be mediated by carbohydrate moieties (12). Our previous study revealed that a specific anti-LOS monoclonal antibody significantly inhibits the adherence of M. catarrhalis to human epithelial cells (28). In this report, attachment to human epithelial cells by the kdtA mutant showed more than a 50% reduction in attachment to Chang or cervix epithelial cells while there was no reduction in attachment to lung epithelial cells. These data imply that the mutant has variable affinity to different types of epithelial cells and that LOS in M. catarrhalis is involved in adherence to certain epithelial cells, which may be associated with M. catarrhalis pathogenesis in specific organs. Besides the LOS, several OMPs have shown as adhesion molecules. Among them, UspA1 appeared spontaneous phase variation in vitro resulting in significant changes in binding to host cells (31). To rule out a possibility that the reduced adherence of the kdtA mutant was caused by such phase variation, the expression of uspA1 was examined. It was reported that expression of the UspA1 was correlated with both adherence ability in vitro and the number of guanine (G) residues in a homopolymeric [poly (G)] tract located upstream of the uspA1 open reading frame (31). High expression of UspA1 (10 Gs) would have higher binding capability while low expression of UspA1 (9 Gs) resulted in lower binding capability. When nucleotide sequence analysis of the uspA1 was performed, both mutant and wild type strains showed 9 G residues in their uspA1 poly(G) tracts (data not shown), suggesting that both strains retained a lower expression level of UspA1 protein, and the expression of UspA1 in kdtA mutant did not undergo phase variation spontaneously. In addition, the kdtA mutant showed enhanced clearance from the mouse nasopharynx and lung after an aerosol challenge. The involvement of LOS in the ability of M. catarrhalis to survive in vivo raises the possibility that LOS might be essential for the virulence of the organism in human respiratory tract.
In summary, we identified a M. catarrhalis kdtA homologue by a transposon mutagenesis approach and constructed an isogenic kdtA mutant that produces truncated LOS with only intact lipid A structure. The resulting mutant was sensitive to the bactericidal activity of human normal serum, and presented reduced endotoxin activity, reduced bacterial attachment in vitro, and/or increased bacterial clearance in vivo. This indicates that LOS is an important virulence factor in the pathogenesis of M. catarrhalis infections. The finding may lead to implications for the development of vaccines and to designs of therapeutic intervention against M. catarrhalis infections.
Acknowledgments
We thank Eric J. Hansen for providing strain O35E, Wenzhou Hong for assisting in the animal challenge, and Robert Morell and Yandan Yang for helping in DNA sequencing.
The analytical work was supported by a grant from the Department of Energy (DE-FG09-93ER20097) to the Complex Carbohydrate Research Center.
Editor: J. D. Clements
REFERENCES
- 1.Aebi, C., E. R. Lafontaine, L. D. Cope, L. Latimer, S. L. Lumbley, G. H. McCracken, Jr., and E. J. Hansen. 1998. Phenotype effect of isogenic uspA1 and uspA2 mutations on Moraxella catarrhalis 035E. Infect. Immun. 66:3113-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albiger, B., L. Johansson, and A. B. Jonsson. 2003. Lipooligosaccharide-deficient Neisseria meningitidis shows altered pilus-associated characteristics. Infect. Immun. 71:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bandak, S. I., M. R. Turnak, B. S. Allen, L. D. Bolzon, D. A. Preston, S. K. Bouchillon, and D. J. Hoban. 2001. Antibiotic susceptibilities among recent clinical isolates of Haemophilus influenzae and Moraxella catarrhalis from fifteen countries. Eur. J. Clin. Microbiol. Infect. Dis. 20:55-60. [DOI] [PubMed] [Google Scholar]
- 4.Brozek, K. A., and C. R. Raetz. 1990. Biosynthesis of lipid A in Escherichia coli. Acyl carrier protein-dependent incorporation of laurate and myristate. J. Biol. Chem. 265:15410-15417. [PubMed] [Google Scholar]
- 5.Campagnari, A. A., T. F. Ducey, and C. A. Rebmann. 1996. Outer membrane protein B1, an iron-repressible protein conserved in the outer membrane of Moraxella (Branhamella) catarrhalis, binds human transferrin. Infect. Immun. 64:3920-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Catlin, B. W. 1990. Branhamella catarrhalis: an organism gaining respect as a pathogen. Clin. Microbiol. Rev. 3:293-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clementz, T., J. J. Bednarski, and C. R. Raetz. 1996. Function of the htrB high temperature requirement gene of Escherchia coli in the acylation of lipid A: HtrB catalyzed incorporation of laurate. J. Biol. Chem. 271:12095-12102. [DOI] [PubMed] [Google Scholar]
- 8.Clementz, T., Z. Zhou, and C. R. Raetz. 1997. Function of the Escherichia coli msbB gene, a multicopy suppressor of htrB knockouts, in the acylation of lipid A. Acylation by MsbB follows laurate incorporation by HtrB. J. Biol. Chem. 272:10353-10360. [DOI] [PubMed] [Google Scholar]
- 9.Doyle, W. J. 1989. Animal model of otitis media: other pathogens. Pediatr. Infect. Dis. J. 81(Suppl.):S45-S47. [PubMed] [Google Scholar]
- 10.Edebrink, P., P. E. Jansson, M. M. Rahman, G. Widmalm, T. Holome, M. Rahman, and A. Weintraub. 1994. Structural studies of the O-polysaccharide from the lipopolysaccharide of Moraxella (Branhamella) catarrhalis serotype A (strain ATCC 25238). Carbohydr. Res. 257:269-284. [DOI] [PubMed] [Google Scholar]
- 11.Fiske, M. J., R. A. Fredenburg, K. R. VanDerMeid, J. C. McMichael, and R. Arumugham. 2001. Method for reducing endotoxin in Moraxella catarrhalis UspA2 protein preparations. J. Chromatogr. B. Biomed. Sci. Appl. 753:269-278. [DOI] [PubMed] [Google Scholar]
- 12.Fitzgerald, M., S. Murphy, R. Mulcahy, C. Keane, D. Coakley, and T. Scott. 1999. Tissue culture adherence and haemagglutination characteristics of Moraxella (Branhamella) catarrhalis. FEMS Immunol. Med. Microbiol. 24:105-114. [DOI] [PubMed] [Google Scholar]
- 13.Fomsgaard, J. S., A. Fomsgaard, N. Hoiby, B. Bruun, and C. Galanos. 1991. Comparative immunochemistry of lipopolysaccharides from Branhamella catarrhalis strains. Infect. Immun. 59:3346-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forsgren, A., M. Brant, M. Karamehmedovic, and K. Riesbeck. 2003. The immunoglobulin D-binding protein MID from Moraxella catarrhalis is also an adhesin. Infect Immun. 71:3302-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furano, K., and A. A. Campagnari. 2003. Inactivation of the Moraxella catarrhalis 7169 ferric uptake regulator increases susceptibility to the bactericidal activity of normal human sera. Infect. Immun. 71:1843-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galanos, C., O. Luderitz, E. T. Rietschel, O. Westphal, H. Brade, L. Brade, M. Freudenberg, U. Schade, M. Imoto, H. Yoshimura, et al. 1985. Synthetic and natural Escherichia coli free lipid A express identical endotoxic activities. Eur. J. Biochem. 148:1-5. [DOI] [PubMed] [Google Scholar]
- 17.Galanos, C., O. Luderitz, and O. Westphal. 1969. A new method for the extraction of R lipopolysaccharides. Eur. J. Biochem. 9:245-249. [DOI] [PubMed] [Google Scholar]
- 18.Gibson, B. W., A. A. Campagnari, W. Melaugh, N. J. Phillips, M. A. Apicella, S. Grass, J. Wang, K. L. Palmer, and R. S. Munson, Jr. 1997. Characterization of a transposon Tn916-generated mutant of Haemophilus ducreyi 35000 defective in lipooligosaccharide biosynthesis. J. Bacteriol. 179:5062-5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldman, R. C., C. C. Doran, S. K. Kadam, and J. O. Capobianco. 1988. Lipid A precursor from Pseudomonas aeruginosa is completely acylated prior to addition of 3-deoxy-D-manno-octulosonate. J. Biol. Chem. 263:5217-5223. [PubMed] [Google Scholar]
- 20.Gorter, A. D., J. Oostrik, P. van der Ley, P. S. Hiemstra, J. Dankert, and L. van Alphen. 2003. Involvement of lipooligosaccharides of Haemophilus influenzae and Neisseria meningitidis in defensin-enhanced bacterial adherence to epithelial cells. Microb. Pathog. 34:121-130. [DOI] [PubMed] [Google Scholar]
- 21.Gu, X. X., J. Chen, S. J. Barekamp, J. B. Robbins, C. M. Tsai, D. J. Lim, and J. Battey. 1998. Synthesis and characterization of lipooligosaccharide-based conjugates as vaccine candidates for Moraxella (Branhamella) catarrhalis. Infect. Immun. 66:1891-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helminen, M. E., I. Maciver, M. Paris, J. L., Latimer, S. L. Lumbley, L. D. Cope, G. H. McCracken, Jr., and E. J. Hansen. 1993. A mutation affecting expression of a major outer membrane protein of Moraxella catarrhalis alters serum resistance and survival in vivo. J. Infect. Dis. 168:1194-1201. [DOI] [PubMed] [Google Scholar]
- 23.Hoban, D. J., G. V. Doern, A. C. Fluit, M. Roussel-Delvallez, and R. N. Jones. 2001. Worldwide prevalence of antimicrobial resistance in Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis in the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin. Infect. Dis. 32:S81-S93. [DOI] [PubMed] [Google Scholar]
- 24.Holm, M. M., S. L. Vanlerberg, I. M. Foley, D. D. Sledjeski, and E. R. Lafontaine. 2004. The Moraxella catarrhalis porin-like outer membrane protein CD is an adhesin for human lung cells. Infect. Immun. 72:1906-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holm, M. M., S. L. Vanlerberg, D. D. Sledjeski, and E. R. Lafontaine. 2003. The Hag protein of Moraxella catarrhalis strain O35E is associated with adherence to human lung and middle ear cells. Infect. Immun. 71:4977-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holme, T., M. Rahman, P. E. Jansson, and G. Widmalm. 1999. The lipopolysaccharide of Moraxella catarrhalis structural relationships and antigenic properties. Eur. J. Biochem. 265:524-529. [DOI] [PubMed] [Google Scholar]
- 27.Hu, W. G., J. Chen, F. M. Collins, and X. X. Gu. 2000. An aerosol challenge mouse model for Moraxella catarrhalis. Vaccine 18:799-804. [DOI] [PubMed] [Google Scholar]
- 28.Hu, W. G., J. Chen, J. C. McMichael, and X. X. Gu. 2001. Functional characteristics of a protective monoclonal antibody against serotype A and C lipooligosaccharides from Moraxella catarrhalis. Infect. Immun. 69:1358-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karalus, R., and A. Campagnari. 2000. Moraxella catarrhalis: a review of an important human mucosal pathogen. Microbes Infect. 2:547-559. [DOI] [PubMed] [Google Scholar]
- 30.Lafontaine, E. R., L. D. Cope, C. Aebi, J. L. Latimer, G. H. McCracken, Jr., and E. J. Hansen. 2000. The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol. 182:1364-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lafontaine, E. R., N. J. Wagner, and E. J. Hansen. 2001. Expression of the Moraxella catarrhalis UspA1 protein undergoes phase variation and is regulated at the transcriptional level. J. Bacteriol. 183:1540-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luke, N. R., S. Allen, B. W. Gibson, and A. A. Campagnari. 2003. Identification of a 3-deoxy-d-manno-octulosonic acid biosynthetic operon in Moraxella catarrhalis and analysis of a KdsA-deficient isogenic mutant. Infect. Immun. 71:6426-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masoud, H., M. B. Perry, and J. C. Richards. 1994. Characterization of the lipopolysaccharide of Moraxella catarrhalis. Structural analysis of the lipid A from M. catarrhalis serotype A lipopolysaccharide. Eur. J. Biochem. 220:209-216. [DOI] [PubMed] [Google Scholar]
- 34.Mohan, S., and C. R. Raetz. 1994. Endotoxin biosynthesis in Pseudomonas aeruginosa: enzymatic incorporation of laurate before 3-deoxy-d-manno-octulosonate. J. Bacteriol. 176:6944-6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muroi, M., and K. Tanamoto. 2002. The polysaccharide portion plays an indispensable role in Salmonella lipopolysaccharide-induced activation of NF-κB through human toll-like receptor 4. Infect. Immun. 70:6043-6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy, T. F. 1996. Branhamella catarrhalis: epidemiology, surface antigenic structure, and immune response. Microbiol. Rev. 60:267-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy, T. F., A. L. Brauer, N. Yuskiw, and T. J. Hiltke. 2000. Antigenic structure of outer membrane protein E of Moraxella catarrhalis and construction and characterization of mutants. Infect. Immun. 68:6250-6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy, T. F., and M. R. Loeb. 1989. Isolation of the outer membrane of Branhamella catarrhalis. Microb. Pathog. 6:159-174. [DOI] [PubMed] [Google Scholar]
- 39.Ochsner, U. A., A. I. Vasil, Z. Johnson, and M. L. Vasil. 1999. Pseudomonas aeruginosa fur overlaps with a gene encoding a novel outer membrane lipoprotein, OmlA. J. Bacteriol. 181:1099-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swords, W. E., B. A. Buscher, K. Ver Steeg Ii, A. Preston, W. A. Nichols, J. N. Weiser, B. W. Gibson, and M. A. Apicella. 2000. Non-typeable Haemophilus influenzae adhere to and invade human bronchial epithelial cells via an interaction of lipooligosaccharide with the PAF receptor. Mol. Microbiol. 37:13-27. [DOI] [PubMed] [Google Scholar]
- 42.Timpe, J. M., M. M. Holm, S. L. Vanlerberg, V. Basrur, and E. R. Lafontaine. 2003. Identification of a Moraxella catarrhalis outer membrane protein exhibiting both adhesin and lipolytic activities. Infect. Immun. 71:4341-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 44.Tullius, M. V., N. J. Phillips, N. K. Scheffler, N. M. Samuels, R. S. Munson, Jr., E. J. Hansen, M. Stevens-Riley, A. A. Campagnari, and B. W. Gibson. 2002. The lbgAB gene cluster of Haemophilus ducreyi encodes a β-1,4-galactosyltransferase and an α-1,6-dd-heptosyltransferase involved in lipooligosaccharide biosynthesis. Infect. Immun. 70:2853-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tzeng, Y. L., A. Datta, V. K. Kolli, R. W. Carlson, and D. S. Stephens. 2002. Endotoxin of Neisseria meningitidis composed only of intact lipid A: inactivation of the meningococcal 3-deoxy-d-manno-octulosonic acid transferase. J. Bacteriol. 184:2379-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Unhanand, M., I. Maciver, O. Ramilo, O. Arencibia-Mireles, J. C. Argyle, G. H. McCracken, Jr., and E. J. Hansen. 1992. Pulmonary clearance of Moraxella catarrhalis in an animal model. J. Infect. Dis. 165:644-650. [DOI] [PubMed] [Google Scholar]
- 47.Verduin, C. M., C. Hol, A. Fleer, H. van Dijk, and A. van Belkum. 2002. Moraxella catarrhalis: from emerging to established pathogen. Clin. Microbiol. Rev. 15:125-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weissbach, A., and J. Hurwitz. 1959. The formation of 2-keto-3-deoxyheptonic acid in extracts of Escherichia coli B. I. Identification. J. Biol. Chem. 234:705-709. [PubMed] [Google Scholar]
- 49.York, W. S., A. G. Darvill, M. McNeil, T. T. Stevenson, and P. Albersheim. 1985. Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 118:3-40. [Google Scholar]
- 50.Zaleski, A., N. K. Scheffler, P. Densen, F. K. Lee, A. A. Campagnari, B. W. Gibson, and M. A. Apicella. 2000. Lipooligosaccharide Pk (Galα1-4 Galβ1-4Glc) epitope of Moraxella catarrhalis is a factor in resistance to bactericidal activity mediated by normal human serum. Infect. Immun. 68:5261-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zughaier, S. M., Y. L. Tzeng, S. M. Zimmer, A. Datta, R. W. Carlson, and D. S. Stephens. 2004. Neisseria meningitidis lipooligosaccharide structure-dependent activation of the macrophage CD14/Toll-like receptor 4 pathway. Infect. Immun. 72:371-380. [DOI] [PMC free article] [PubMed] [Google Scholar]