Abstract
Hodgkin Reed-Sternberg (HRS) cells of classic Hodgkin lymphoma (cHL), like many solid tumors, elicit ineffective immune responses. However, patients with cHL are highly responsive to PD-1 blockade, which largely depends on HRS cell-specific retention of MHC class II and implicates CD4+ T cells and additional MHC class I-independent immune effectors. Here, we utilize single-cell RNA sequencing and spatial analysis to define shared circulating and microenvironmental features of the immune response to PD-1 blockade in cHL. Compared with non-responders, responding patients have more circulating CD4+ naïve and central memory T cells and B cells, as well as more diverse CD4+ T cell and B cell receptor repertoires. Importantly, a population of circulating and tumor-infiltrating IL1β+ monocytes/macrophages is detectable in patients with cHL but not healthy donors, and a proinflammatory, tumor-promoting signature of these circulating IL1β+ monocytes is associated with resistance to PD-1 blockade in cHL. Altogether, our findings reveal extensive immune rewiring and complementary roles of CD4+ T cells, B cells and IL1β+ monocytes in the response to PD-1 blockade and suggest that these features can be captured with a peripheral blood test.
Subject terms: Hodgkin lymphoma, Tumour immunology, Cancer microenvironment, Systems analysis
Patients with classic Hodgkin lymphoma (cHL) respond well to PD-1 blockade, but the underlying cellular insights are still lacking. Here, the authors use single-cell transcriptome and spatial analyses to identify distinct circulating and tumor-infiltrating CD4+ T cell, B cell and IL1β+ monocyte/macrophage features associated with response to PD-1 blockade in cHL.
Introduction
Classic Hodgkin lymphomas (cHLs) are composed of rare malignant Hodgkin Reed-Sternberg (HRS) cells embedded within an extensive T cell and macrophage-rich inflammatory/immune cell infiltrate1. HRS cells exhibit near-universal copy gains of chromosome 9p24.1/CD274 (PD-L1)/PDCD1LG2 (PD-L2) and copy number-dependent increased expression of these PD-1 ligands2–4. This genetic basis for enhanced PD-1 signaling prompted clinical evaluation of PD-1 blockade in patients with relapsed/refractory (R/R) cHL.
Over 70% of patients with R/R cHL obtained clinical responses to single-agent PD-1 blockade and a subset of these remissions were durable5,6. In this setting, patients with the highest level of chromosome 9p24.1 copy gain and associated PD-L1 expression had the most favorable responses to therapy7. More recently, PD-1 blockade has been added to combination regimens at relapse and initial diagnosis8–14. In a randomized trial of nivolumab (anti PD-1 antibody)/doxorubicin, vinblastine and dacarbazine (AVD) versus brentuximab/AVD in patients with newly diagnosed advanced-stage cHL, progression-free survival was significantly higher in the PD-1 blockade-containing treatment arm15.
In addition to chromosome 9p24 copy gain, cHLs frequently exhibit inactivating mutations of B2M or copy loss of B2M or MHC class I genes which limit or preclude MHC class I-mediated antigen presentation to CD8+ T cells4,7,16,17. However, malignant HRS cells often retain expression of MHC class II, likely reflecting their derivation from MHC class II+ germinal center B cells and associated ability to present antigens to CD4+ T cells7,17. In patients with R/R cHL, HRS cell expression of MHC class II, but not MHC class I, was associated with a more favorable response to PD-1 blockade7.
The cHL tumor immune microenvironment (TIME) is CD4+ T cell rich with concomitant expansion of Th1-polarized regulatory T cells and PD-1+ T effector cells18. In intact cHLs, the malignant HRS cells are in close proximity to PD-1+CD4+ T cells and PD-L1+ macrophages which form a localized immunoprotective niche19.
Despite the abundance and likely importance of tumor-infiltrating macrophages in cHL19,20, these cells were under-represented and largely uncharacterized in recent single-cell analyses of primary tumor cell suspensions18,21. However, circulating monocyte/macrophage progenitors are readily detectable and amenable to characterization at a single-cell level22.
Our previous analysis of the circulating immune signature in patients with cHL revealed the importance of recently expanded, clonally diverse CD4+ T cells and incompletely characterized innate effectors, including monocyte subsets, in the response to PD-1 blockade22.
Herein, we utilize single-cell RNA sequencing (scRNA-seq), T cell receptor (TCR) and B cell receptor (BCR) clonotype assessments and selected spatial analyses of the intact cHL TIME to define shared circulating and microenvironmental features in patients with cHL and analyze the interplay between previously unidentified CD4+ T cell, B cell and innate cell subsets in response to PD-1 blockade. These studies highlight the value of characterizing the circulating immune compartment and perturbed T cell, B cell and proinflammatory innate effectors in cHL.
Results
Analysis of the circulating immune signature in patients with cHL and control healthy donors
We performed scRNA-seq of CD3+ and CD3− peripheral blood mononuclear cells (PBMCs) from 20 patients with R/R cHL who were treated with single-agent PD-1 blockade (nivolumab)5 at two timepoints, baseline (cycle 1 day 1 [C1D1]) and on therapy (cycle 4 day 1 [C4D1]). These samples were annotated for the patients’ best overall response to PD-1 blockade: complete response (CR), 9 patients; partial response (PR), 5 patients; and progressive disease (PD), 6 patients. PBMCs from 11 patients with newly diagnosed, previously untreated cHL and 13 control healthy donors were similarly sequenced (Supplementary Data 1a).
Characterization of circulating CD3+CD8− T cell subsets, diversity and clonality
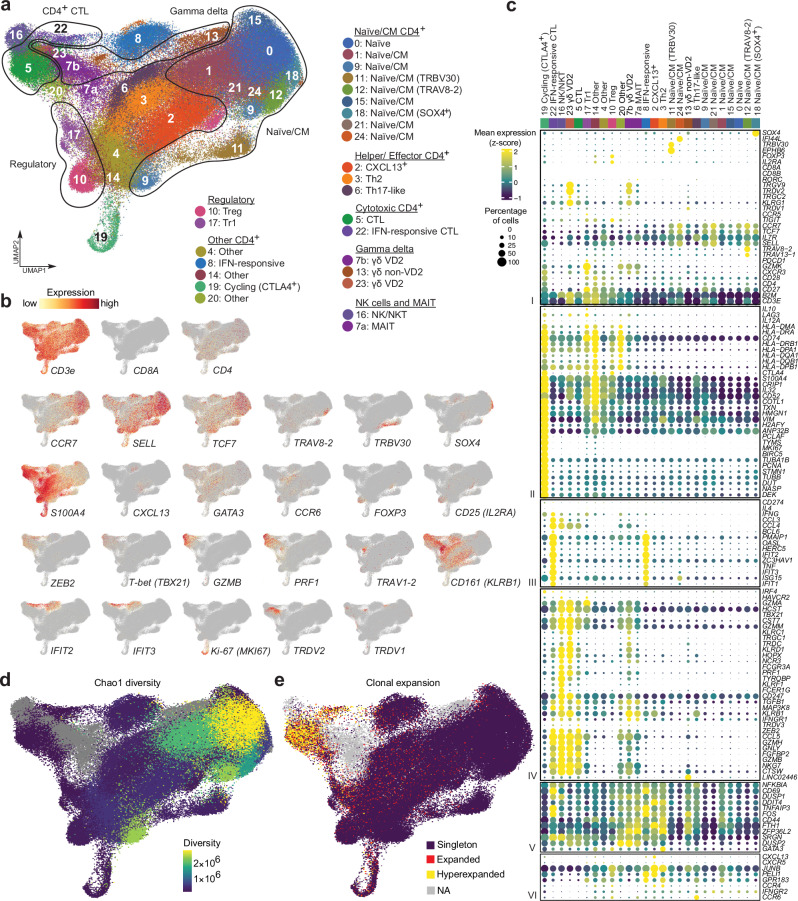
We initially focused on CD4+ T cells given their demonstrated importance in the response to PD-1 blockade in cHL7,22. First, we computationally removed all CD8A- and CD8B-expressing cells from our CD3+ scRNA-seq data and processed the CD4+ T cell enriched samples using the standard Seurat workflow (see Methods). Clusters that were present in at least 10% of the specimens at a level equal to or greater than 1% of the mean number of sequenced cells per sample were included in the analysis (Supplementary Data 2). We then visualized the CD3+CD8−, CD4+−enriched, T cell space on a Uniform Manifold Approximation and Projection (UMAP) (Fig. 1a and Supplementary Fig. 1). The 172,274 cells in 26 clusters (Fig. 1a) were annotated using a knowledge-guided manual approach based on the differential expression of canonical genes, highlighted in heatmaps and dot plots (Fig. 1b-c and Supplementary Fig. 2, 3).
Fig. 1. Analysis of circulating CD3+CD8- (CD4+ enriched) immune cell populations in healthy donors (HD), patients with newly diagnosed (ND) and relapsed/refractory (R/R) cHL.
a Annotated Uniform Manifold Approximation and Projection (UMAP) of CD3+CD8- single-cell expression profiles (n = 172,274 cells). Each cluster is labeled with a distinct color and unique number. b Feature plots of selected cell lineage and differentiation markers. c Dot plot with relative expression of selected genes in CD3+CD8- clusters following bidirectional clustering. Displayed genes were curated from the top differentially expressed genes that defined clusters using a two-sided Wilcoxon rank sum test, adjusted for multiple comparisons (adj p < 0.05, fold change > 1.75) and supplemented with relevant markers based on a priori knowledge. The size of the dot indicates the percentage of marker-expressing cells in each cluster and the z-score reflects mean marker expression across the clusters. d TCR repertoire diversity, as determined by Chao1 diversity index, obtained from single-cell TCR-seq data (n = 126,159 cells) for each T cell cluster and overlaid onto the UMAP. e TCR clonotypes expansion levels overlaid onto the UMAP (n = 126,159 cells), “Singleton” 1 clone, “Expanded” 2–5 clones, “Hyperexpanded” >5 clones. d, e Gamma delta T cell clusters (Cluster 7b, 13 and 23), MAIT (Cluster 7a) and NKT cells (Cluster 16) were not analyzed and are shown in gray.
The CD3+CD8− space was organized into specific stages of T cell differentiation, including naïve/central memory (CM), effector, cytotoxic and regulatory T cells (Fig. 1a)23. Clusters 0, 1, 9, 11, 12, 15, 18, 21 and 24 were defined by the highest expression of early differentiation markers including CCR7, SELL and TCF7 and were designated naïve/CM cells (Fig. 1b–c)24–26.
Certain naïve/CM T cells clustered together because of their expression of specific TCR variable (V) genes including TRBV30 (Cluster 11) and TRAV8-2 or TRAV13-1 (Cluster 12) or their selective expression of SOX4 (Cluster 18). The SOX4+ naïve/CM subset is of interest because this transcription factor is a downstream target of TGFβ that negatively regulates Th2 differentiation in peripheral CD4+ T cells27.
Following the T cell differentiation trajectory, we identified several helper and effector T cell populations, including Clusters 2, 3 and 6. Cluster 2 cells selectively expressed CXCL13 and shared certain features with T follicular helper (TFH) cells28 but lacked additional markers of classic TFH cells (e.g., BCL6), possibly due to incomplete polarization in the peripheral blood. In this space, we also identified circulating Th2 T cells (Cluster 3, GATA3+ and CCR4+) and Th17-like T cells (Cluster 6, CCR6+ and RORC +) (Fig. 1b, c)24,29.
Given the relative absence of MHC class I on HRS cells4,7,16,17, potential MHC class I-independent mechanisms of tumor cell killing merit particular attention. We identified several circulating CD3+CD8− T cell populations with potential cytotoxic function including CD4+ cytotoxic T lymphocytes (CTLs) (Clusters 5 and 22, GZMB+ and PRF1+)30, NKT cells (Cluster 16, GZMB+, NKG7+ and CCL5+)31, VD2 γ/δ T cells (Clusters 7b and 23, GZMB+, TRDC+, TRGC2+ and TRDV2+)32 and MAITs (Cluster 7a, CD161 [KLRB1]high and TRAV1-2+)33,34 (Fig. 1a–c). Although CD4+ T cells from Clusters 5 and 22 had similar cytotoxic signatures, the latter cluster had transcriptional features of prior IFN stimulation (OASL+, IFIT1+, IFIT2+ and IFIT3+)35,36 (Fig. 1c). An additional non-cytotoxic CD4+ T cell subset (Cluster 8) also exhibited prior IFN stimulation and activation (CD69+)37 (Fig. 1a–c).
We also identified two subsets of regulatory T cells – canonical Tregs (Cluster 10, FOXP3 + and CD25 [IL2RA] +)38 and Type 1 regulatory (Tr1) T cells (Cluster 17, IL10+ and LAG3+)39 – and additional proliferating (MKI67+) circulating CD4+ T cells that expressed the highest levels of CTLA4 and LAG3 in this space (Cluster 19) (Fig. 1a-c). Several additional subsets were classified as Other due to insufficient polarization in peripheral blood (Clusters 4, 14, 20).
We performed single-cell TCR-sequencing for the same samples and assessed TCR diversity, measured by Chao1 scores40,41, and relative levels of clonal TCR expansion in CD3+CD8− cells (Fig. 1d, e, Supplementary Fig. 4a and Supplementary Data 3, 4). As expected, the least differentiated naïve/CM clusters had the highest Chao1 diversity scores, whereas the more mature populations of helper and effector T cells had lower Chao1 scores (Fig. 1d). Conversely, the putative cytotoxic CD4+ T cell clusters (Clusters 5 and 22) exhibited the highest levels of TCR clonal expansion (Fig. 1e).
CD3+CD8− T cell subsets in patients with cHL and healthy donors – response-related TCR diversity and naïve/CM T cell abundance
After identifying the circulating CD3+CD8− clusters in the full dataset (Fig. 1), we assessed quantitative and qualitative differences between these cells in the clinically annotated cohorts of patients with cHL and control healthy donors (Fig. 2). Patients with R/R cHL had markedly lower baseline TCR diversity than healthy donors across the entire CD4 naïve/CM T cell space (Fig. 2a and Supplementary Fig. 4b). Of note, the identified differences in TCR diversity were not age-related (Supplementary Fig. 4c and Supplementary Data 1c).
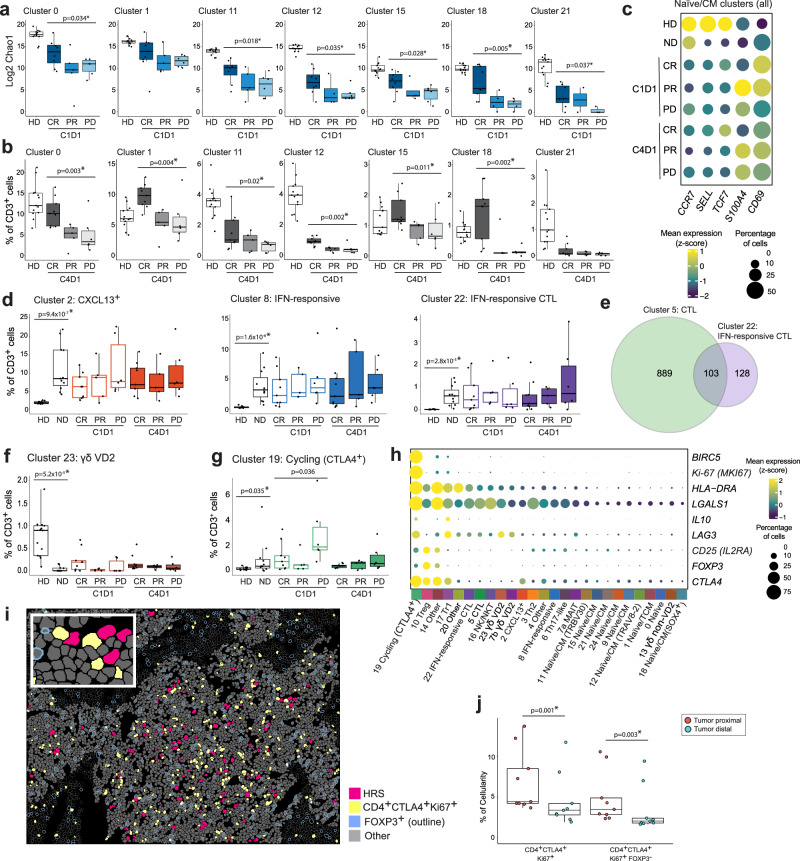
Fig. 2. Comparative analyses of CD3+CD8- (CD4+ enriched) clusters in healthy donors (HD), patients with newly diagnosed (ND) and relapsed/ refractory (R/R) cHL.
a TCR repertoire diversity, as determined by Chao1 diversity index, in CD4+ naïve/CM T cell clusters from HD (n = 13) and patients with R/R cHL at baseline [C1D1] stratified by the best overall response (BOR) to PD-1 blockade. CR Complete response, PR partial response, PD progressive disease. Number of patients with Chao1 diversity data per cluster: Cluster 0, HD (n = 13), CR (n = 8), PR (n = 5), PD (n = 6); Cluster 1, HD (n = 13), CR (n = 9), PR (n = 5), PD (n = 6); Cluster 11, HD (n = 13), CR (n = 8), PR (n = 5), PD (n = 6); Cluster 12, HD (n = 13), CR (n = 8), PR (n = 4), PD (n = 6); Cluster 15, HD (n = 13), CR (n = 8), PR (n = 4), PD (n = 6); Cluster 18, HD (n = 13), CR (n = 9), PR (n = 4), PD (n = 4); Cluster 21, HD (n = 13), CR (n = 7), PR (n = 2), PD (n = 3). b Relative abundance of CD4+ naïve/CM T cell clusters in HD (n = 13) and patients with R/R cHL on treatment [C4D1] (n = 20) stratified by BOR to PD-1 blockade, CR (n = 9), PR (n = 5), PD (n = 6). c Dot plot with relative expression of selected genes associated with T cell naïveté and activation in all CD4+ naïve/CM clusters (Cluster 0, 1, 9, 11, 12, 15, 18, 21, 24) in HD (n = 13), patients with ND cHL (n = 11) and patients with R/R cHL at baseline [C1D1] (n = 20) and C4D1 (n = 20), stratified by BOR. The size of the dot indicates the percentage of marker-expressing cells and the z-score reflects mean marker expression across cohorts. d Relative abundance of cHL-specific T cell populations. e Venn diagram illustrating the shared clonotypes between CD4+ CTLs (Cluster 5, n = 992 clonotypes) and IFN-responsive CD4+ CTLs (Cluster 22, n = 231 clonotypes) in patients with cHL. f, g Relative abundance of gamma delta VD2 T cells (Cluster 23) and cycling CTLA4+ T cells (Cluster 19). d, f, g HD (n = 13), ND (n = 11), C1D1 and C4D1: CR (n = 9), PR (n = 5), PD (n = 6). h Dot plot illustrating the mean expression (color) and percentage of cells (dot size) expressing genes that positively identified cycling CTLA4+ T cells (Cluster 19) in comparison to the additional CD3+CD8− clusters. i Cell segmentation and phenotype map for a representative multiplex IF image of ND cHL (magenta, HRS cells; yellow, CD4+CTLA4+Ki67+ cells; blue outline, FOXP3+ cells; gray-fill, Other cells less than 75 μm from HRS cells; gray-outline, Other cells greater than or equal to 75 μm from HRS cells). The experiment was performed in 9 ND cHL samples. j Relative abundance of CD4+CTLA4+Ki67+ and CD4+CTLA4+Ki67+FOXP3− cells in relation to HRS cells (red dots, less than 75 μm from HRS cells; blue dots, greater than or equal to 75 μm from the HRS cells) in patients with ND cHL (n = 9). The indicated nominal p values were calculated by using a two-sided paired T-test and adjusted for multiple comparisons using the Benjamini-Hochberg method; p values that remained significant are noted (*). a, b, d, f, g Differences between HDs and patients with ND cHL were assessed by a two-sided Wilcoxon rank-sum test. The one-sided Cuzick trend test was used to compare patients with R/R cHL by BOR (CRs, PRs and PDs). Nominal p values that were significant (p < 0.05) are listed and those that remained significant after Benjamini–Hochberg correction FDR < 0.1 are noted (*). a, b, d, f, g, j All box plots generated in R display the 25th and 75th percentiles (lower and upper hinges), median values, and whiskers. The whiskers extend from the hinges to the largest/smallest values within 1.5 times the interquartile range (IQR) from the hinge. Data points beyond the end of the whiskers are plotted individually. Source data are provided as a Source Data file.
Patients with the most favorable responses to PD-1 blockade (CRs) had significantly higher CD4+ naïve/CM TCR diversity at baseline (C1D1) and more abundant naïve/CM subsets on treatment (C4D1) (Fig. 2a, b and Supplementary Fig. 5). Taken together, these data suggest that responding patients have an increased and sustained ability to mount CD4+ T cell responses to new tumor neoantigens22,42–46.
Patients with cHL (newly diagnosed and R/R) and healthy donors had additional qualitative differences in their circulating CD4+ naïve/CM subsets. In patients with cHL, CD4+ naïve/CM cells had relatively lower levels of CCR7, SELL, TCF7 and higher levels of S100A4 and CD69 expression (Fig. 2c), potentially reflecting increased antigen exposure and activation37,47. Additionally, in patients with cHL, the relative paucity of SELL expression in circulating naïve/CM CD4+ T cells might limit their recruitment into secondary lymphoid organs48,49.
We further assessed qualitative response-related differences in circulating naïve/CM CD4+ T cells in patients who achieved CRs or progressed following PD-1 blockade. Pathway enrichment analyses revealed that naïve/CM CD4+ T cell subsets (Cluster 0, 1, 9, 11, 12, 15, 18, 21 and 24) from complete responders had increased expression of TCF7, LEF1 and LRNN3 and a more naïve cell phenotype, potentially reflecting a greater capacity for self renewal (Supplementary Fig. 6 and Supplementary Data 5)50,51.
More abundant CD3+CD8− T cell effector subsets in patients with cHL
We identified several circulating CD4+ T cell populations that were significantly more abundant in patients with newly diagnosed cHL and R/R cHL than in healthy donors: CXCL13 + T cells (Cluster 2); IFN-stimulated effectors (Cluster 8) and CTLs (Cluster 22); and cycling CTLA4+ T cells (Cluster 19) (Fig. 2d and g). The cHL-specific CXCL13 + T cells are noteworthy given the recent identification of CXCL13 + TFH-like cells that form rosettes around HRS cells and interact with CXCR5 + B cells52. This CXCL13/CXCR5 interaction reportedly attracts circulating B cells and limits their germinal center entry, activation and maturation53,54.
Qualitative and quantitative differences in circulating CD4+ CTLs in patients with cHL and healthy donors
We identified 2 subsets of circulating CD4+ CTLs, a major population in healthy donors and patients with cHL (Cluster 5) and a smaller subset primarily in patients with cHL (IFN-responsive CTLs, Cluster 22) (Figs. 1, 2d and Supplementary Fig. 5). There were response-related differences in Cluster 5 CD4+ CTLs from patients who achieved CRs or had progressive disease (PD) following treatment (C4D1) (Supplementary Data 5). These included increased expression of the inhibitory Killer cell lectin-like receptor G1 (KLRG1) in patients with progressive disease (Supplementary Fig. 6c and Supplementary Data 5). In recent preclinical models, KLRG1 expression on CD4+ T-effector cells was associated with tumor progression and lack of response to PD-1 blockade55.
To delineate the relationship between the CD4+ Cluster 5 CTLs and Cluster 22 IFN-stimulated CTLs, we characterized their TCR clonotype distributions (Supplementary Data 3, 4). In patients with cHL, Clusters 5 and 22 shared 103 clonotypes, including 10.4% (103/992) of the detected clonotypes in Cluster 5 and 44.6% (103/231) of the identified clonotypes in Cluster 22 (Fig. 2e). These results reveal the common origins of Clusters 5 and 22 CTLs and the likely additional inflammatory modulation in cHL-predominant Cluster 22 CTLs. It is also noteworthy that both CTL subsets in patients with cHL, Cluster 5 and 22, express lower levels of cytotoxic effectors such as perforin, in comparison to CTLs (Cluster 5) from healthy donors (Supplementary Fig. 3). These findings and the selective identification of IFN-stimulated cytotoxic and non-cytotoxic CD4+ T cells (Clusters 22 and 8) in patients with cHL may also reflect a broader cancer-related inflammatory process56,57. In this regard, the IFN-stimulated genes in Cluster 22 and 8 (OASL, IFIT1, IFIT2, IFIT3) are known to be induced by Type I IFN (IFNα and β) treatment of T cells58. These findings are of interest given the dual role of Type I IFNs in inflammation and feedback immunosuppression57.
Response-related proliferating Tr1-type cells in the periphery and the intact TIME
The circulating cycling CTLA4 +CD4 + T cells (Cluster 19) were also significantly more abundant in patients with cHL than in healthy donors (Fig. 2g). These proliferating cells (MKI67+) with enhanced survival (BIRC5+) shared certain features with Tr1 (Cluster 17) cells, including high-level expression of CTLA4 and LAG3 and additional IL10, and decreased expression of CD25 (IL2RA) and FOXP3 (Fig. 2h). Of note, patients who did not respond to PD-1 blockade (PD) had significantly higher numbers of circulating Cluster 19 cells at baseline (C1D1) (Fig. 2g). Using a multiplexed immunofluorescence panel, we identified a similar population of CD4+CTLA4+Ki-67+FOXP3− cells in the intact TIME of an additional cohort of patients with newly diagnosed cHL (Fig. 2i and Supplementary Fig. 7). These cycling CD4+CTLA4+ T cells were primarily observed in close proximity to HRS cells (less than 75 μm from the tumor cells) (Fig. 2j). Given their immunosuppressive features and proximity to malignant HRS cells, these newly identified Cluster 19-like cycling CD4+CTLA4+ cells may limit the response to PD-1 blockade (Fig. 2g-j).
Decreased numbers of VD2 γδ T cells in patients with cHL
Although patients with cHL had more abundant IFN-stimulated effectors (Clusters 8 and 22) and proliferating CTLA4+LAG3+ regulatory cells (Cluster 19), they had significantly reduced numbers of cytotoxic VD2 γδ T cells (Clusters 23 and 7b) (Fig. 2f and Supplementary Fig. 5). As γδ T cells recognize antigens in the absence of MHC molecules, the paucity of these cells may be an additional mechanism of immune evasion in largely MHC class I-negative cHLs59–61.
Characterization of circulating CD3− immune cell subsets – NK cells, B cells and myeloid cells
We applied the same inclusion criteria to the CD3− space and identified 229,670 cells in 24 clusters (Fig. 3a, Supplementary Fig. 8 and Supplementary Data 2). Cell clusters were annotated using a knowledge-guided manual approach based on differentially expressed genes and prototypical lineage markers (Supplementary Fig. 9). We identified three broad categories of CD3− cells defined by their classic markers: NK cells (NKG7+, CD16 [FCGR3A]+, GZMB+ and PRF1+)62; B cells (CD19+, CD20 [MS4A1]+ and PAX5+)63; and myeloid cells, including monocytes (CD33+, CD68+ and CD14+)64, conventional dendritic cells (cDCs) (CD33+ and CLEC10A+)65 and plasmacytoid dendritic cells (pDCs) (CD123 [IL3RA]+)65 (Fig. 3a, b and Supplementary Fig. 10).
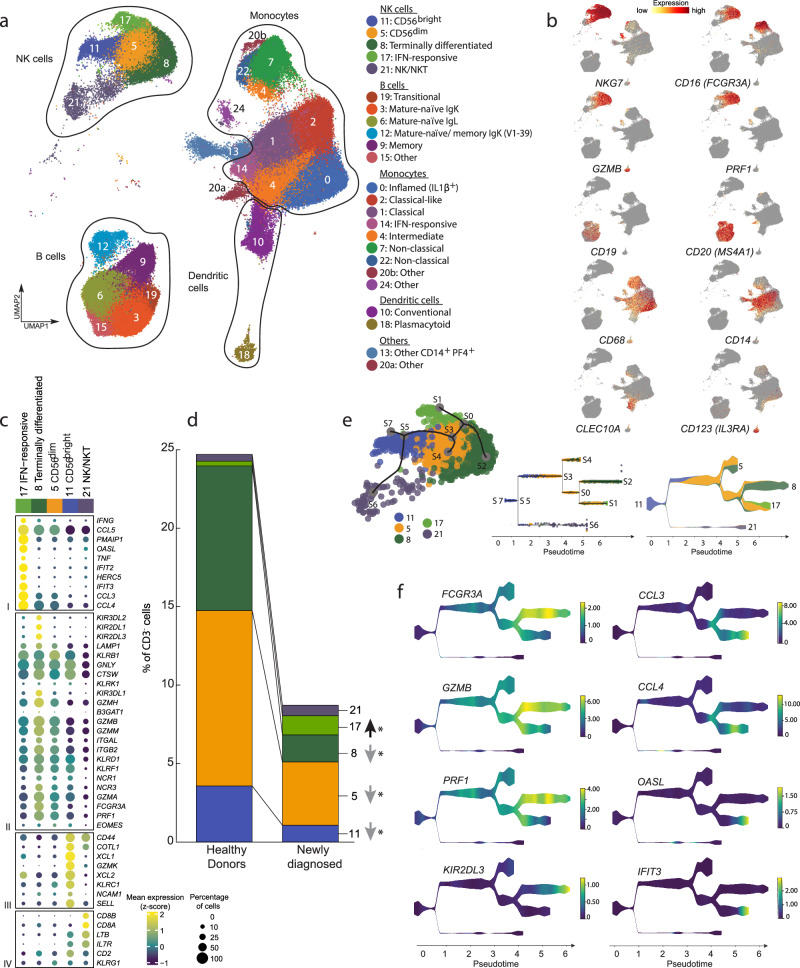
Fig. 3. Analysis of circulating CD3− immune cell populations in healthy donors (HD), patients with newly diagnosed (ND) and relapsed/refractory (R/R) cHL and detailed analysis of NK cell subsets.
a Annotated UMAP of CD3− single-cell expression profiles (n = 229,670 cells) by cell type. Each cluster is labeled with a distinct color and unique number, beginning with the most abundant cluster (Cluster 0). b Feature plots of selected cell lineage and differentiation markers. c Dot plot with relative expression of selected genes in NK cell clusters. Displayed genes were curated from the top differentially expressed genes that defined clusters using a two-sided Wilcoxon rank sum test, adjusted for multiple comparisons (adj p < 0.05, fold change > 1.75) and supplemented with relevant markers based on a priori knowledge. The size of the dot indicates the percentage of marker-expressing cells in each cluster and the z-score reflects mean marker expression across the clusters. d Relative abundance of NK cell clusters in HD (n = 13) and patients with ND cHL (n = 11). Differences between HD and patients with ND cHL were assessed by a two-sided Wilcoxon rank sum test. P values that remained significant after Benjamini–Hochberg correction FDR < 0.1 are noted (*). e Lineage trajectory analysis of NK cell populations (n = 40,844 cells) by STREAM overlaid on UMAP (left panel); Subway map plot (middle panel) showing all individual NK cells; Stream plot (right panel) visualization of cell density along different trajectories. At a given pseudotime, the width of each branch is proportional to the total number of cells. f Stream plots visualization of the selected marker genes, representing stages of NK cell maturation and/ or function. Source data are provided as a Source Data file.
Qualitative and quantitative differences in NK cell subsets in patients with cHL and healthy donors
The circulating NK cells included immature CD56 (NCAM1)bright (Cluster 11), more mature CD56dim (Cluster 5) and terminally differentiated NK cells (Cluster 8) and an additional less well-defined population that includes NKT cells (Cluster 21, CD3E+) (Fig. 3a–c)62,66. We identified an additional subset of NK cells with a signature of IFN stimulation (Cluster 17, OASL+, IFIT2+ and IFIT3+) (Fig. 3a–c and Supplementary Fig. 11a–c) that was more abundant in patients with cHL (Fig. 3d). A similar population of IFN-stimulated NK cells, termed Type I IFN-responding cells62, was described in the context of chronic viral infection62,66,67. Of note, Cluster 17 was the only circulating NK cell subset that was more abundant in patients with cHL; all other NK cell clusters were significantly decreased in comparison to healthy donors (Fig. 3d and Supplementary Fig. 11d).
To further delineate the relationship between IFN-stimulated NK cells (Cluster 17) and the additional NK subsets, we performed STREAM analysis (Single-cell Trajectories Reconstruction, Exploration and Mapping)68. As anticipated, STREAM reflected the differentiation of CD56bright (Cluster 11) into CD56dim (Cluster 5) NK cells and their further maturation into terminally differentiated NK cells (Cluster 8) (Fig. 3e). Although Cluster 17 cells were derived from CD56dim NK cells (Cluster 5), these IFN-stimulated effectors had a distinct differentiation trajectory (Fig. 3e). Additionally, Cluster 17 NK cells expressed lower levels of GZMB and PRF1 and higher levels of CCL3 and CCL4 than canonical terminally differentiated NK cells (Cluster 8) suggesting perturbed effector function (Fig. 3f). Therefore, patients with cHL had significantly reduced numbers of classically defined circulating NK cells and an expanded population of IFN-stimulated NK cells with likely decreased cytotoxic potential (Fig. 3d, f).
Response-related differences in B cell abundance and BCR diversity
The circulating B cell clusters reflected maturation stages defined by the expression of canonical marker genes and immunoglobulin heavy chains (Fig. 4a–c). A small population of transitional B cells (Cluster 19) expressed IGHD, IGHM, PLD4, CD38, MZB1 and IGLL5 63,69 and two subsets of mature naïve B cells (IGHD +, IGHM+, TCL1A+ and CD27 −)63,69 differed only in their respective light chain expression (kappa+, Cluster 3 and lambda+, Cluster 6) (Fig. 4a–c and Supplementary Fig. 12a–c). An additional memory B cell subset (CD27 + and TNFRSF13B+)63,69 included both unswitched (IGHM +) and class-switched (IGHG+ and IGHA+) populations (Cluster 9) (Fig. 4a–c). Another cluster contained mature naïve (IGHD+, IGHM +, TCL1A+ and CD27 −) and memory B cells (IGHM/IGHG + and CD27 +) that selectively expressed a specific immunoglobulin kappa variable gene, IGKV1-39 (Fig. 4a–c).
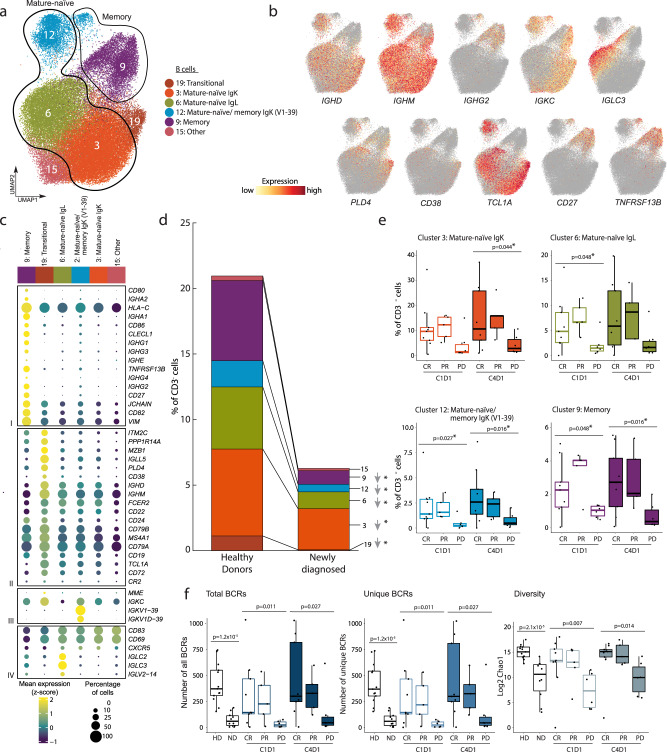
Fig. 4. Analysis of the circulating B cell populations in healthy donors (HD), patients with newly diagnosed (ND) and relapsed/refractory (R/R) cHL.
a Annotated UMAP of B cell clusters (n = 55,598 cells). Each cluster is labeled with a distinct color and unique number. b Feature plots of selected cell lineage and differentiation markers. c Dot plot with relative expression of selected genes in B cell clusters. Displayed genes were curated from the top differentially expressed genes that defined clusters using a two-sided Wilcoxon rank sum test, adjusted for multiple comparisons (adj p < 0.05, fold change >1.75) and supplemented with relevant markers based on a priori knowledge. The size of the dot indicates the percentage of marker-expressing cells in each cluster and the z-score reflects mean marker expression across the clusters. d Relative abundance of B cell clusters in HD (n = 13) and patients with ND cHL (n = 11). e Relative abundance of B cell clusters in patients with R/R cHL at baseline [C1D1] (n = 20) and C4D1 (n = 20), stratified by BOR to PD-1 blockade, CR (n = 9), PR (n = 5), PD (n = 6). f Number of total BCRs (left panel), number of unique BCRs (middle panel) and BCR repertoire diversity, as determined by Chao1 diversity index (right panel) in all B cells from HD (n = 13) and patients with ND cHL (n = 10) and R/R cHL at baseline [C1D1] (n = 20) and C4D1 (n = 20) stratified by BOR to PD-1 blockade. CR Complete response (n = 9), PR=partial response (n = 5), PD progressive disease (n = 6). d–f Differences between HD and patients with ND cHL were assessed by the two-sided Wilcoxon rank-sum test. The one-sided Cuzick trend test was used to compare patients with R/R cHL by response (CRs, PRs and PDs). d, e Nominal p values that were significant (p < 0.05) are listed and those that remained significant after Benjamini–Hochberg correction FDR < 0.1 are noted (*). In f p values are nominal with no Benjamini–Hochberg correction. e, f All box plots generated in R display the 25th and 75th percentiles (lower and upper hinges), median values, and whiskers. The whiskers extend from the hinges to the largest/smallest values within 1.5 times the interquartile range (IQR) from the hinge. Data points beyond the end of the whiskers are plotted individually. Source data are provided as a Source Data file.
After defining the circulating B cell clusters, we assessed quantitative differences in these subsets in patients with cHL and healthy donors. Patients with newly diagnosed and R/R cHL had significantly lower numbers of almost all of the circulating B cell subsets in comparison to healthy donors (Fig. 4d and Supplementary Fig. 12d). Additionally, patients with R/R cHL who did not respond to PD-1 blockade (progressive disease, PD) had significantly fewer circulating naïve and memory B cells than responding patients (Fig. 4e). In the subset of patients with current scRNA-seq and prior CyTOF analyses, there was an excellent correlation between circulating B cell numbers (Supplementary Fig. 10c).
We also evaluated B cell abundance and proximity to HRS cells in available diagnostic biopsies from 7 of the 11 patients with newly diagnosed cHL and scRNA-seq analyses of circulating B cells (Fig. 4d and Supplementary Fig. 13). As all of the circulating B cell subsets were significantly less abundant in patients with newly diagnosed cHL than in healthy donors (Fig. 4d), we utilized a pan B cell marker, PAX5, and PD-L1 to identify small PAX5bright normal infiltrating B cells and PAX5dim/PD-L1+ HRS cells by dual immunohistochemistry and digital imaging. Normal B cells were significantly less abundant in all evaluated newly diagnosed cHL biopsies than in control lymphoid tissue (Supplementary Fig. 13c). Additionally, normal B cells were relatively excluded from the immediate HRS cell (PAX5dim/PD-L1+) niche, defined as within 25 μm of the tumor cells (Supplementary Fig. 13d).
We next utilized the circulating B cell scRNA-seq data to reconstruct individual BCR sequences with the TRUST4 algorithm70 and assess circulating BCR diversity (Supplementary Data 6). Of interest, the numbers of total and unique BCRs, as well as Chao1 diversity, were significantly lower in patients with newly diagnosed cHL in comparison to healthy donors (Fig. 4f). Among patients with R/R cHL, those who did not respond to PD-1 blockade (PD) had significantly lower numbers of unique and total BCRs and decreased BCR diversity in comparison to responding patients (Fig. 4f). The identified differences in BCR diversity were not age-related (Supplementary Fig. 12e and Supplementary Data 1c). These data highlight the likely importance of B cells in the response to PD-1 blockade in cHL via antibody production to tumor neoantigens and/or Fc receptor binding of innate effectors. Additionally, these findings build upon the recently described importance of tumor tertiary germinal centers in the response to checkpoint blockade71–74 and extend these observations to circulating B cells.
Characterization of circulating monocyte and dendritic cell subsets
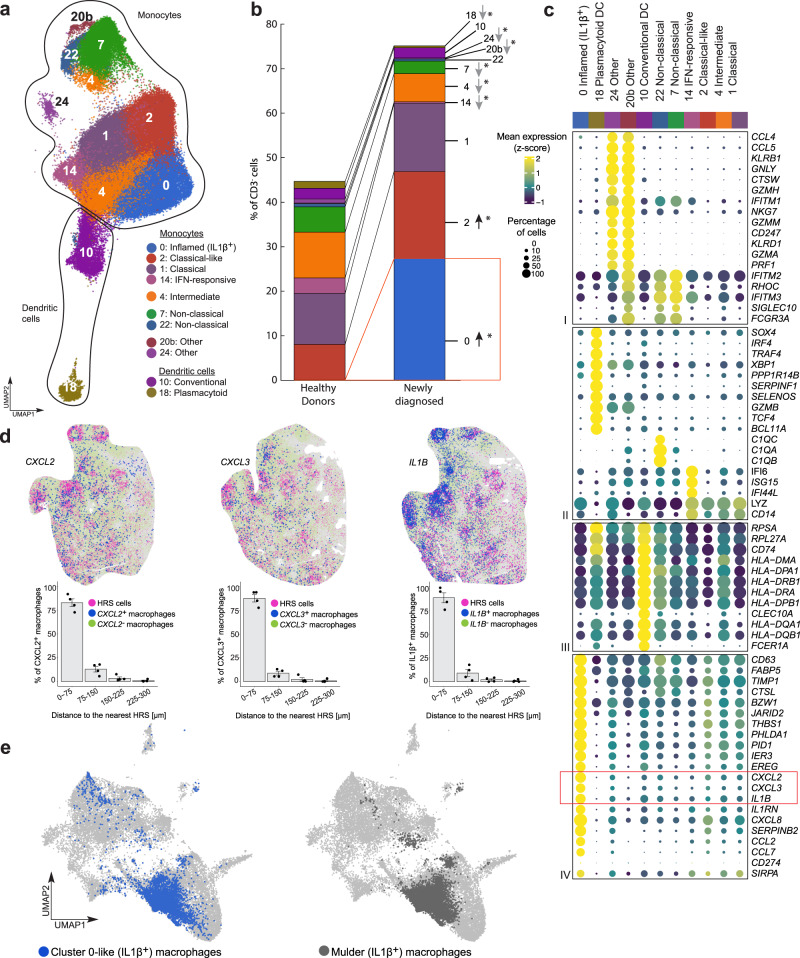
We also characterized circulating classical, intermediate and non-classical monocytes using the canonical markers, CD14 and CD16, and additional subtype-specific transcripts: classical, CD14+, CD16− (Clusters 1 and 2); intermediate, CD14+, CD16+ (Cluster 4); and non-classical, CD14−, CD16+, RHOC+, C1Q+ and SIGLEC10+ (Clusters 7 and 22) (Fig. 5a–c)64,75. Circulating monocytes with features of prior IFN stimulation (Cluster 14), Type-2 conventional dendritic cells (cDCs) (Cluster 10, CD33+, CLEC10A+, CD1c+ and MHC class II [HLA-DMA, HLA-DPA1 and HLA-DRA]high) and plasmacytoid dendritic cells (pDCs) (Cluster 18, CLEC4C+, CD123+, NRP1+ and CD11c [ITGAX] −) were also detected76 (Fig. 5a–c, Supplementary Fig. 14 and 15a, b).
Fig. 5. Analysis of the circulating monocyte and dendritic cell populations in healthy donors (HD), patients with newly diagnosed (ND) and relapsed/refractory (R/R) cHL.
a Annotated UMAP of myeloid clusters (n = 126,707 cells). Each cluster is labeled with a distinct color and unique number. b Relative abundance of myeloid clusters between HD (n = 13) and patients with ND cHL (n = 11). Differences between HD and patients with ND cHL were assessed by the two-sided Wilcoxon rank-sum test. Nominal p values that remained significant after Benjamini–Hochberg correction with an FDR < 0.1 are noted (*). c Dot plot with relative expression of selected genes in monocyte and dendritic clusters. Displayed genes were curated from the top differentially expressed genes that defined clusters using a two-sided Wilcoxon rank sum test, adjusted for multiple comparisons (adj p < 0.05, fold change >1.75) and supplemented with relevant markers based on a priori knowledge. The size of the dot indicates the percentage of marker-expressing cells in each cluster and the z-score reflects mean marker expression across the clusters. d (Top panel) Cellular phenotype maps of the representative RNAscope images from patients with ND cHL (magenta, HRS cells; blue, Cluster 0-like macrophages CXCL2+ (left), CXCL3+ (middle) IL1B+(right); green, other macrophages CXCL2-/CXCL3-/IL1B-) (all images). The experiment was performed in 4 ND cHL samples. (Bottom panel) Proximity plots for the indicated CXCL2+, CXCL3+ and IL1+ Cluster 0-like macrophages and HRS cells. Data are presented as mean values +/− SEM. e UMAP showing Cluster 0-like (IL1β+) monocytes/macrophages (our data) (left panel, blue dots, n = 5783 cells) after cell label transfer to Mulder et al. (Immunity 54, 1883-1900 e1885 (2021)) data set (light grey dots, n = 60,691 cells), overlaid on the same UMAP. The right panel represents the IL1β+ monocytes and macrophages cluster from Mulder et al. (dark grey dots, n = 7985 cells). Source data are provided as a Source Data file.
Identification of an abundant cancer-associated IL1β+ monocyte/macrophage population in the periphery and intact TIME
The most abundant subset of circulating monocytes (Cluster 0) was primarily detected in patients with cHL and virtually absent in healthy donors (Fig. 5b and Supplementary Fig. 15c). These inflamed IL1β+ monocytes (Cluster 0) resembled classical monocytes with markedly increased expression of multiple likely immunosuppressive and tumor-promoting cytokines/chemokines, including IL1Β, CXCL8, CXCL2, CXCL3 and CCL2, and more abundant PD-L1 (CD274) and SIRPA (Fig. 5c).
We postulated that the circulating inflamed IL1β+ monocytes (Cluster 0) in patients with cHL shared features with recently described “inflammatory monocytes” including differentiation into tissue-infiltrating macrophages77–80. For this reason, we asked whether tumor-associated macrophages (TAMs) in intact biopsies of newly diagnosed cHLs expressed Cluster 0 markers using RNAscope81,82. With this 4-plex in situ hybridization approach, primary HRS cells were defined by their expression of CD30 (TNFRSF8) and PD-L1 and CD68 + TAMs were assessed for their expression of the individual Cluster 0 markers, CXCL2, CXCL3 and IL1B (Fig. 5c and Supplementary Fig. 16). We identified tumor-infiltrating CD68 + monocytes/macrophages with Cluster 0 features (CXCL2, CXCL3 and IL1B expression) in the intact cHL TIME and the immediate proximity of CD30+PD-L1+ HRS cells (Fig. 5d). Proximity analysis revealed that the majority of Cluster 0 – like macrophages (83% of CD68+CXCL2+, 88% of CD68+CXCL3+ and 88% of CD68+IL1B+ cells) were localized within 75 μm of the CD30+PD-L1+ HRS cells (Fig. 5d).
To further validate our findings, we performed multiplex immunofluorescence (mIF) with a 4-plex panel (CD68, IL1β, PAX5 and PD-L1) on the same cHL cases and confirmed the presence of IL1β+ CD68+ monocytes/macrophages in the intact TIME at the protein level. IL1β+ monocytes/macrophages were primarily detected in HRS-rich regions of the intact TIME whereas monocytes/macrophages in HRS-poor regions were largely IL1β− (Supplementary Fig. 17). These mIF analyses reinforce our RNAscope results (Fig. 5d) and highlight the likely significance of tumor-infiltrating IL1β+ monocytes/macrophages in cHL.
After identifying Cluster 0-like IL1β+ monocytes/macrophages in the cHL TIME, we asked whether similar cells were detectable in other cancers. We interrogated a single-cell compendium of monocytes and macrophages from multiple solid tumors (Fig. 5e)83 and performed a cell type label transfer84 using our CD3− scRNA-seq dataset as a reference. In this publicly available compendium of tumor-associated monocytes and macrophages (60,691 total cells), we identified 5783 cells with a transcriptional profile similar to our Cluster 0 inflamed IL1β+ monocytes (Fig. 5e, left panel, blue dots). Of interest, 81% (4682/5783) of the cells with a Cluster 0 transcriptional signature were also previously identified as “IL1β+ monocytes” in the compendium (Fig. 5e, right panel, grey dots and Supplementary Data 7). There was a highly significant enrichment of Cluster 0-like cells in the previously designated IL1β+ cluster (4682 Cluster 0-like cells/7985 IL1β cells) compared to the rest of the dataset (1102 Cluster 0-like cells/52,706 Other cells, χ² = 25707.926 p < 0.0001, Chi-squared test with Yates correction). The identification of Cluster 0/IL1β+ tumor-infiltrating monocytes/macrophages in this solid tumor compendium indicates that similarly programmed inflammatory monocytes/macrophages are detectable in multiple cancers.
Response-related circulating IL1β+ monocyte transcriptional signature in patients with cHL
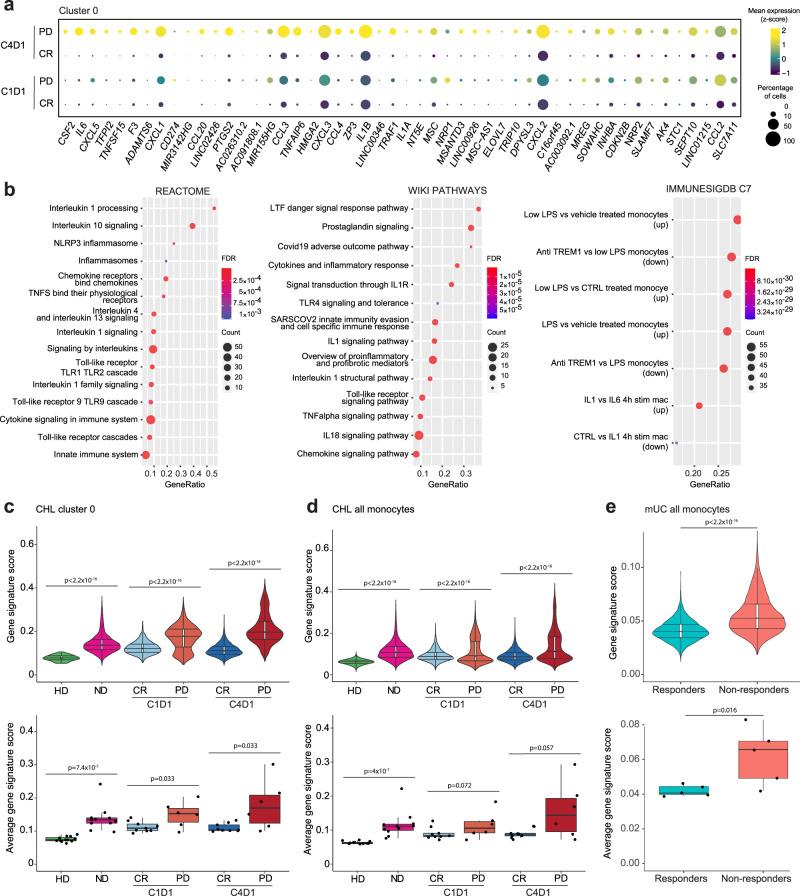
We next returned to the periphery to assess the clinical significance of circulating inflamed IL1β+ monocytes (Cluster 0) in patients with R/R cHL who were treated with PD-1 blockade. As a first step, we performed a differential analysis of response-related Cluster 0 monocyte transcripts in patients who achieved a CR or progressed (PD) on PD-1 blockade (Fig. 6a and Supplementary Table 1). In comparison with complete responders, those who progressed on nivolumab had significant upregulation of an extended Cluster 0 transcriptional signature at C4D1 (Fig. 6a). Gene set enrichment analysis (GSEA) of the response-related Cluster 0 signature revealed highly significant upregulation of pathways associated with response to prostaglandin, TNFα, Toll-like receptor (TLR) and IL-1 signaling, inflammasome activation and modulation by Triggering Receptor Expressed on Myeloid cells 1 (TREM1), an Ig-like immunoregulatory receptor and amplifier of innate immune responses (Fig. 6b and Supplementary Fig. 18)85–90. Together, these findings highlighted the likely role of prostaglandin and TNFα stimulation, TLR engagement by damage-associated molecular patterns (DAMPs), inflammasome activation and IL1β-dependent downstream signaling in Cluster 0 monocytes/macrophages.
Fig. 6. Identification of IL1β+ monocyte transcriptional signature associated with lack of response to PD-1 blockade.
a Differential expression analysis of response-related Cluster 0 transcripts in patients with relapsed/refractory (R/R) cHL who achieved a complete response (CR) or progressed on PD-1 blockade (PD). A two-sided permutation test, adjusted for multiple comparisons (adj p < 0.1, fold change > 1.5) was used to filter the differentially expressed Cluster 0 genes in patients who achieved a CR or progressed on therapy at C4D1. The top 50 of 368 differentially expressed genes are shown for patients by response at C4D1 and baseline C1D1. b Pathway enrichment analysis of response-related differentially expressed genes in Cluster 0 using Molecular Signatures Database (MSigDB) gene sets: CP:REACTOME: Reactome gene sets; CP:WIKIPATHWAYS: WikiPathways gene sets; and C7: Immunologic gene sets. Gene Ratio (x-axis) represents the number of genes in the AUCell signature over the total number of genes in a given gene set; FDR significant < 0.05; Count represents the number of genes in the overlap between the AUCell gene signature and given gene set. c AUCell signature score in circulating Cluster 0 monocytes from healthy donors (HD) and patients with newly diagnosed cHL (ND) or R/R cHL at C1D1 and C4D1 at the single-cell (upper panel: HD n = 73 cells; ND n = 8266 cells; CR C1D1 n = 3942 cells; PD C1D1 n = 2390 cells; CR C4D1 n = 5403 cells; PD C4D1 n = 4335 cells) and per-patient (lower panel: HD n = 12 samples; ND n = 11 samples; CR C1D1 n = 9 samples; PD C1D1 n = 6 samples; CR C4D1 n = 9 samples; PD C4D1 n = 5 samples) levels. d AUCell signature score in all circulating monocytes from HDs and patients with cHL at the single-cell (upper panel: HD n = 28,001 cells; ND n = 20,633 cells; CR C1D1 n = 16,851 cells; PD C1D1 n = 7464 cells; CR C4D1 n = 17,343 cells; PD C4D1 n = 10,999 cells) and per-patient (lower panel: HD n = 13 samples; ND n = 11 samples; CR C1D1 n = 9 samples; PD C1D1 n = 6 samples; CR C4D1 n = 9 samples; PD C4D1 n = 5 samples) levels. e AUCell signature score in all circulating monocytes from patients with metastatic urothelial carcinoma (mUC) (Yuen et al. Nat Med 26, 693-698 (2020)) at baseline (C1D1), stratified by the subsequent response to PD-L1 blockade, at the single-cell (upper panel: Responders n = 3718 cells, Non-responders n = 4134 cells) and per-patient (lower panel: Responders n = 5 samples, Non-responders n = 5 samples) levels. c–e A one-sided Wilcoxon rank-sum test was used to assess the distributions of AUCell gene signature scores between HD and ND, as well as between CR and PD at C1D1 and C4D1, at both the single-cell and per-patient levels. c–e (upper panel) All violin plots generated in R illustrate the data distribution with kernel density estimation. Plots display the 25th and 75th percentiles (lower and upper hinges) and median values. The width of the violin at different points reflects the density of the data. (upper panels) All box plots display the 25th and 75th percentiles (lower and upper hinges), median values, and whiskers. The whiskers extend from the hinges to the largest/smallest values within 1.5 times the interquartile range (IQR) from the hinge. Data points beyond the end of the whiskers are plotted individually. Source data are provided as a Source Data file.
We then used the response-related Cluster 0 genes (Fig. 6a and Supplementary Table 1) to develop a transcriptional (AUCell) signature score91 associated with lack of response to PD-1 blockade at C4D1 and predictive for unfavorable outcome at baseline (C1D1) (Fig. 6c and Supplementary Fig. 19a, b). On both a single-cell and per-patient level, the Cluster 0 AUCell score was significantly higher in patients with cHL who did not respond to PD-1 blockade than in complete responders (Fig. 6c, upper and lower panels). Response-related differences in the AUCell score were also detected when all circulating monocytes, rather than pre-selected Cluster 0 cells, were analyzed (Fig. 6d).
Response-related circulating IL1β+ monocyte transcriptional signature in patients with another solid tumor
After defining the response-related Cluster 0 AUCell signature in patients with R/R cHL (Fig. 6c, d), we assessed the performance of the signature in an additional series of patients with a solid tumor (metastatic urothelial carcinoma [mUC])92 who were treated with PD-L1 blockade and had publicly available PBMC scRNA-seq data (Supplementary Fig. 19c, e). In this mUC series, circulating monocytes from patients who did not respond to PD-L1 blockade had significantly higher baseline AUCell signature scores than those who responded to this treatment (Fig. 6e). Therefore, circulating Cluster 0-like monocytes were also detectable and associated with inferior responses to blockade of the PD-1 pathway in an additional solid tumor.
Discussion
In patients with cHL, scRNA-seq of the circulating immune signature revealed extensive reprogramming and/or modulation of specific CD4+ T cell, B cell and innate effectors in the setting of cancer-associated inflammation. In this largely MHC class I-negative tumor, quantitative differences in circulating CD4+ naive/CM T cell abundance and TCR diversity were associated with response to PD-1 blockade, highlighting the importance of a continued capacity to respond to new tumor neoantigens. In comparison to healthy donors, patients with cHL also exhibited qualitative differences in these essential CD4+ naive/CM T cells, with decreased expression of TCF7, CCR7 and SELL. Recent studies highlight the importance of TCF7 + T cell progenitors in the response to PD-1 blockade93. We now extend these findings to the circulating CD4+ T cell compartment.
In patients with cHL, there were also qualitative and quantitative differences in circulating MHC class I-independent cytotoxic effectors including: CD4+ CTLs with decreased perforin expression and increased expression of IFN-stimulated genes; reduced numbers of γ/δ VD2 cells; and perturbed NK cell differentiation with fewer mature perforin- and granzyme-positive cells and an expanded population of IFN-stimulated cells resembling those in chronic viral infections62,67.
Additionally, patients with cHL had significantly reduced numbers of circulating B cells across all stages of differentiation. Both the abundance of peripheral blood B cells and their BCR diversity were positively associated with response to PD-1 blockade. These findings underscore the importance of a continued capacity to mount B cell, as well as T cell, responses to new tumor neoantigens and extend prior observations regarding the prognostic significance of tumor-infiltrating B cells in cHL and additional solid tumors71–74,94,95.
Our scRNA-seq analysis also identified a major population of circulating IL1β+ monocytes with a proinflammatory, tumor-promoting cytokine/chemokine signature that was primarily detected in patients with cHL, but not in healthy donors, and associated with lack of response to PD-1/PD-L1 blockade. Monocytes/macrophages with a similar cytokine/chemokine signature were localized in proximity to HRS cells in intact cHLs and detected in multiple solid tumors. Additionally, circulating monocytes with the same transcriptional profile were associated with a lack of response to PD-L1 blockade in another solid tumor (mUC), underscoring the broad relevance of these findings.
IL1β+ TAMs with a similar transcriptional signature were also identified recently in pancreatic ductal adenocarcinoma (PDAC)80. In PDAC, these proinflammatory IL1β+ TAMs were induced by prostaglandin E2 (PGE2) and TNF, associated with disease progression and modulated by PGE2 or IL1β+ blockade80. Circulating monocytes with certain shared features were also described recently in biliary cancer96. These recent findings further highlight the importance and potential targetability of proinflammatory IL1β+ monocytes/macrophages in multiple tumors. Additionally, the data suggest that IL1β+ proinflammatory monocytes/macrophages negatively impact outcome in settings beyond PD-1 blockade.
The coordinated upregulation of cytokine/chemokines, including IL1B, CXCL2, CXCL3, CXCL8, and CCL2, and the additional response-related factors, IL-6, CXCL1 and CCL3, in Cluster 0 monocytes/macrophages identifies candidate targets for combination therapies with PD-1 blockade and suggests associated treatment strategies77,87,97–106. In previous studies, inhibition of IL-1 receptor signaling abrogated the recruitment of inflammatory monocytes into the TIME, reduced immune suppression and tumor progression, and synergized with PD-1 blockade in murine models of renal cell carcinoma (RCCa) and breast cancer77,98,99. Additionally, neutralizing CXCL8 (IL8) antibodies and CXCR2 antagonists, which block multiple cytokines/chemokines including IL-8, CXCL1, CXCL2 and CXCL3, suppress inflammasome activation, myeloid cell recruitment, tumor growth and angiogenesis, and enhance the efficacy of PD-1 blockade in certain solid tumors87,100–102,107,108. Furthermore, CCL2 blockade decreases macrophage infiltration, tumor proliferation and metastasis in inflammatory breast cancer, in part via downstream activation of CCL3103,104. Given the coordinated upregulation of multiple proinflammatory tumor-promoting cytokines/ chemokines in Cluster 0 monocyte/macrophages, it may be more effective to target the cells themselves, their polarizing program or their shared receptors rather than individual soluble factors.
Despite their abundance in the intact TIME, inflammatory IL1β+ monocyte/macrophages were poorly captured in prior single-cell analyses of disaggregated cHL tumor cell suspensions21,22, highlighting the importance and potential practical value of assessing the circulating immune compartment. More broadly, cancer-associated inflammation and perturbed CD4+ T cells, B cells and innate effectors, including newly identified Cluster 0/proinflammatory IL1β+ monocytes, can be captured and serially monitored with peripheral blood assays providing a framework for more targeted treatment.
Methods
Patient samples
Cryopreserved peripheral blood mononuclear cells (PBMCs) were obtained from: normal healthy donors; patients with newly diagnosed, previously untreated cHL; and patients with relapsed/refractory cHL who received single-agent nivolumab in the CheckMate 205 phase II trial (ClinicalTrials.gov identifier: NCT02181738)5 with Dana-Farber Cancer Institute review board (IRB) approval. Healthy donor PBMC specimens were utilized under a DFCI IRB-approved Secondary Use Protocol with a waiver of informed consent. Demographics including age and sex of patient and donor cohorts are included in Supplementary Data 1a and c. Baseline (cycle 1 day 1) and on-treatment (cycle 4 day 1) PBMCs were obtained from CheckMate 205 trial patients who received nivolumab 3 mg/kg every 2 weeks until disease progression or unacceptable toxicity. An independent review committee (IRC) used the 2007 International Working Group response criteria109 to assess best overall response (BOR) to therapy – complete response (CR), partial response (PR) or progressive disease (PD).
Formalin-fixed, paraffin-embedded (FFPE) tumor biopsies from patients with newly diagnosed cHL were obtained from the archives of Brigham & Women’s Hospital (BWH) with BWH IRB approval (Supplementary Data 1b).
Isolation of CD3+ and CD3− peripheral blood mononuclear cells
Unmanipulated CD3+ and CD3− cells were purified from cryopreserved bulk PBMCs by negative selection with Miltenyi Biotec separation kits. CD3+ cells were isolated using the Pan T cell isolation kit, which included CD14, CD15, CD16, CD19, CD34, CD36, CD56, CD123, and CD235a biotin-conjugated monoclonal antibodies (Miltenyi #130-096-535). CD3− cells were selected by positively depleting CD3+ cells with CD3-conjugated MicroBeads (Miltenyi #130-050-101).
Single-cell RNA (scRNA-seq) and Single-cell T cell Receptor (scTCR-seq) sequencing
Viable CD3+ and CD3− cells were resuspended in PBS (Gibco #10010023) with 0.04% UltraPure BSA (Invitrogen #AM2616) at a concentration of 1000 cells/µL and 17,000 cells were loaded onto a 10× Genomics ChromiumTM instrument according to the manufacturer’s instructions. The scRNA-seq libraries were processed using ChromiumTM single cell 5’ library & gel bead kit (10× Genomics #PN-1000165). Quality controls for amplified cDNA libraries and final sequencing libraries were performed using Bioanalyzer High Sensitivity DNA Kit (Agilent #5067-4626). For CD3+ samples, 2 µL of post ctDNA amplification material was also used to prepare scTCR-seq libraries. The sequencing libraries for scRNA-seq and scTCR-seq were normalized to 4 nM concentration, pooled using a volume ratio of 4:1 and sequenced on Illumina NovaSeq S4 300 cycle platform.
Upstream scRNA-seq analytical pipeline
The sequencing data were demultiplexed and raw single-cell RNAseq reads were aligned to GRCh38-3.0.0 using Cell Ranger version 3.1.0 pipeline (10× Genomics). The count matrix for each sample was read using Seurat V3.084 and made into a Seurat object. All of the Seurat objects were merged and batch corrected with Harmony110 for CD3+ cells and CD3− cells. The Seurat objects were processed by the standard Seurat workflow. Briefly, cells that expressed fewer than 200 genes, or greater than 5000 genes, or had >20% mitochondrial reads were removed. The count matrix was log normalized and the top 2000 most variable genes were calculated by the vst method in Seurat. The UMAP was generated using the first 50 dimensions of the Harmony coordinates. To find clusters, the shared nearest neighbor (SNN) graph was built on the first 50 dimensions of the Harmony coordinates followed by the Louvain community detection algorithm with a resolution of 1.5. Cells expressing either CD8A or CD8B were computationally removed from the CD3+ Seurat object to generate an additional CD3+CD8− (CD4+ enriched) Seurat object.
QC cluster inclusion/exclusion criteria and annotation
Based on the Seurat clustering at resolution 1.5, the mean number of sequenced cells per sample was calculated. Clusters that were present in at least 10% of the specimens at a level equal to or greater than 1% of the mean number of sequenced cells per sample were included in the downstream analyses. BBrowser v3 and BBrowserX (BioTuring) software were used to analyze the data and annotate the clusters that met the inclusion criteria. Two additional clusters were excluded from the downstream analysis because their features (canonical marker genes) were those of contaminating red blood cells or platelets.
Derivation and display of cluster marker features
Marker genes for each cluster were determined using the function wilcoxauc in the presto (https://github.com/immunogenomics/presto) R package. P adj <=0.05 and fold change >=1.75 (Wilcox rank sum test, two side, adjusted for multiple comparisons) were used to filter the top marker genes for each cluster and those genes were used to generate a heatmap using ComplexHeatmap111. Dot plot visualization was used to show the differences in the expression of selected top marker genes across clusters.
Differential expression analysis
A Bioconductor package called Distinct 112 was used to perform comparative multi-sample, multi-group single-cell RNAseq differential expression analyses. The Seurat objects were converted to SingleCellExperiment objects for analysis with Distinct.
Gene set enrichment analysis (GSEA)
Pathway enrichment analyses were carried out using the following Molecular Signatures Database (MSigDB) gene sets: CP:REACTOME: Reactome gene sets; CP:WIKIPATHWAYS: WikiPathways gene sets; and C7: immunologic gene sets113,114.
Development of the AUCell signature
AUCell was used to quantify the enrichment of a specific input gene signature in the cellular subsets91. Specifically, AUCell_buildRankings were used to rank genes in each cell by expression level, and AUCell_calcAUC was used to calculate a gene set enrichment score per cell using the most highly expressed 5% of the genes in that cell.
Single-cell trajectories reconstruction, exploration and mapping (STREAM) analysis
Single-cell trajectories reconstruction was performed using STREAM v1.068. Epg_alpha = 0.02, epg_mu = 0.05, epg_lambda = 0.05 were set in the elastic_principal_graph function and STREAM plots were plotted using the plot_stream function.
TCR and B cell receptor (BCR) analyses
The results from the TCR single-cell V(D)J sequencing were read into Immunarch115 to calculate TCR diversity. TCR diversity for each cluster was calculated using the Chao1 function in the tcr R package and super-imposed on the corresponding UMAP. TCR clonal expansion levels were binned as singleton (n = 1), expanded (2 < n ≤ 5) or hyper-expanded (n > 5), and overlaid on the corresponding UMAP.
The individual BCR sequences were reconstructed by TRUST470 from the single-cell RNAseq BAM files. The TRUST4 output for the reconstructed BCRs was also imported to Immunarch to calculate BCR Chao1 diversity.
RNAscopeTM multiplex fluorescent assay
FFPE tissue sections (4 µm) from patients with newly diagnosed cHL (demographics in Supplementary Data 1b) were selected for hybridization and simultaneous visualization of four RNA targets using the RNAscopeTM LS Multiplex Fluorescent Assay (Advanced Cell Diagnostics (ACD) #322800) and automated Leica Biosystems’ BOND RX System. RNA-specific probes were hybridized to target transcripts (set I: CD68-channel 1 (ACD #560598), IL1B-channel 2 (ACD #310368-C2), PD-L1-channel 3 (ACD #600868-C3), CD30-channel 4 (ACD #310838-C4); set II: CD68-channel 1 (ACD #560598), CXCL2-channel 2 (ACD #425258-C2), PD-L1-channel 3 (ACD #600868-C3), CD30-channel 4 (ACD #310838-C4); set 3: CD68-channel 1 (ACD #560598), CXCL3-channel 2 (ACD #1002158-C2), PD-L1-channel 3 (ACD #600868-C3), CD30-channel 4 (ACD #310838-C4)) followed by staining with nuclear counterstain/4′,6-diamidino-2-phenylindole (DAPI). Signals were amplified and detected using the OpalTM dyes (Akoya Biosciences #OP-001001, #OP-001003, #OP-001004 and #OP-001006) specific for each channel (channel 1 – Opal 520, channel 2- Opal 570, channel 3 – Opal 620, channel 4 – Opal 690) (Supplementary Table 2a). Slides were mounted with Prolong Gold Anti-fade reagent (Invitrogen #P36934) mounting medium and whole slide tissue scans were imaged and digitized using the PhenoImager HT multispectral imaging platform (Akoya Biosciences).
Digitized whole slide scans were analyzed with HALO and HALO AI software (Indica Labs HALO Image Analysis Platform version 3.5.3577 and HALO AI version 3.5.3577). Tissue scans were first evaluated for RNA quality and regions without RNA signals were excluded from the analysis. The AI-custom nuclear segmentation algorithm was then trained and developed for cell segmentation. Cell phenotyping was subsequently performed based on the combination of marker genes expression and HALO spatial plots were generated. Proximity analyses were utilized to identify specific cell subsets in the immediate vicinity of malignant HRS cells (< 75 um from an HRS cell) and illustrated with proximity histograms.
Immunohistochemistry
Double staining of PD-L1 (G.J.F.19 clone 405.9A11) and PAX5 (BD Biosciences #610862) was performed using the BOND RX fully automated stainer (Leica Biosystems)116. 4 μm thick FFPE tissue sections from patients with newly diagnosed cHL (demographics in Supplementary Data 1a) were baked for 1 h at 60 °C and subsequently dewaxed and rehydrated. Epitope retrieval was performed using Epitope Retrieval Solution 2 (pH 8) for 30 min (Leica Biosystems #AR9961). The PD-L1 primary antibody was incubated for 2 h, twice. Thereafter, a post-primary blocking reagent was applied for 8 min, horseradish peroxidase-labeled polymer was added for 12 min, and peroxidase blocking was performed for 5 min, followed by 15 min of DAB development. PAX5 immunostaining was subsequently performed with two 2-h application of the primary antibody, a 20-min incubation with a post-primary AP-blocking reagent, and a 15-min application of an AP-labeled polymer followed by 10 min of Red substrate development. Slides were subsequently counterstained with hematoxylin for 10 min, dehydrated, and cover-slipped (Supplementary Table 2b).
Images were acquired using the PhenoImager HT multispectral imaging platform (Akoya Biosciences). Slides underwent whole-slide imaging at 20× resolution and images were viewed with Phenochart viewing software (Akoya Biosciences) to select 3–10 regions of interest (ROIs) under the guidance of an expert pathologist (SJR). The images were spectrally separated and analyzed using inForm 2.6.0 (Akoya Biosciences). The ROIs were segmented, and each marker was quantified using the inForm analysis tools. Data was exported from inForm and examined through a custom data extraction pipeline to obtain cell population densities and perform cell spatial analysis.
Spatial quantitative analysis was performed by establishing cellular labels for each cell being proximal (within 25 µm) or distal (>25 um) to a cellular feature of interest, in this case the HRS cells. This was accomplished using our analysis software (https://github.com/dfci/pythologist) and these features enabled the measurement of percent cellularity of cell-types in regions proximal-to and distal-from HRS cells. The same software was used to generate heat-map visualizations of the local densities of cell-types of interest.
Multiplex immunofluorescence
Multiplex Immunofluorescence staining was performed using the BOND RX fully automated stainer (Leica Biosystems)117. 5 μm thick FFPE tissue sections from patients with newly diagnosed cHL (demographics in Supplementary Data 1b) were initially baked at 60 °C for 3 h, then deparaffinated (Leica Biosystems #AR9590) and rehydrated with a series of graded ethanol solutions to deionized water. Antigen retrieval was conducted at 98 °C using Epitope Retrieval Solution 1 (pH 6) or 2 (pH 9) (Leica Biosystems #AR9961 and #AR9640). Primary antibodies were applied for 40 min. Subsequently, secondary antibodies, including anti-mouse and anti-rabbit Opal Polymer Horseradish Peroxidase (Akoya Biosciences #ARH1001EA), along with Post Primary Block (Leica Biosystems #RE7159) and Polymer (Leica Biosystems #RE7161), were applied as detailed in Supplementary Table 2c. The antibody complex was then incubated for 5 min with the corresponding Opal Fluorophore Reagents (Akoya Biosciences #NEL871001KT) to visualize the signal. Following the final fluorophore application, samples were removed from the BOND autostainer and incubated with NucBlue Fixed Cell Stain ReadyProbes reagent (Invitrogen #R37606) for nuclei detection. Samples were air-dried and mounted with Prolong Diamond Anti-fade mounting medium (Life Technologies #P36965). Details on target antigens, antibody clones, marker dilutions, diluents, secondary applications, and antigen retrieval are provided in Supplementary Table 2c.
Images were acquired using PhenoImager HT multispectral imaging platform (Akoya Biosciences). Slides underwent whole-slide imaging at 20× resolution and images were viewed with Phenochart viewing software (Akoya Biosciences) to select 4–6 regions of interest (ROIs) under the guidance of an expert pathologist (SJR). The images were spectrally separated and analyzed using inForm 2.6.0 (Akoya Biosciences). The ROIs were segmented, and each marker was quantified using the inForm analysis tools. Data was exported from inForm and examined through a custom data extraction pipeline to obtain cell population densities (number of cells per mm2) and perform cell spatial analysis.
Spatial regions were annotated based on their proximity, <75 um or ≥75 um, to HRS cells. In these respective regions, the percent cellularity of prioritized cell types (CD4+CTLA4+Ki67+ and CD4+CTLA4+Ki67+FOXP3−) was measured in each sample. Within each sample, the cumulative regions across all ROIs were treated as a singular aggregated entity for <75 um or ≥75 um from HRS cells. The differences in percent cellularity of the indicated cell types <75 um or ≥75 um from HRS cells were evaluated using a two-sided paired T-test and P-values that were adjusted for multiple comparisons using the Benjamini-Hochberg method.
External scRNA-seq datasets
An external PBMC scRNA-seq dataset from patients with metastatic urothelial carcinoma (mUC) (EGAD00001005481, EGAS00001004008 and GSE145281) who were treated with PD-L1 blockade92 was also used to evaluate the Cluster 0 monocyte/macrophage AUCell signature. The count matrix for this study was obtained from GEO, converted into Seurat objects and normalized using NormalizeData in Seurat and batch corrected with Harmony. A UMAP was generated using the first 20 dimensions of the Harmony coordinates. Cells were clustered using an SNN graph and Louvain algorithm at a resolution of 0.5 based on the batch-corrected reduction. Clusters were manually annotated and inspected in the BioTuring browser v2.
A recently published integrated dataset of mononuclear phagocytes from 13 tissues across 41 single-cell RNAseq datasets83 (GSE178209 and https://gustaveroussy.github.io/FG-Lab/) was used to generate a compendium of conserved monocyte/macrophage gene programs. We used a cell type label transfer method and implemented it in Seurat84 to assess the presence of the cHL Cluster 0 monocytes transcriptional program in specific subsets of cancer-associated monocytes in other solid tumors. Briefly, cell type label transfer was performed by projecting the PCA structure of our reference dataset onto the query dataset. First, common anchors were identified between our dataset and the “MoMac-VERSE” dataset83 using FindTransferAnchors, and then TransferData was used to classify the cells in the MoMac-VERSE dataset based on the cell type labels of our dataset. Principal component analysis was used as the dimensionality reduction method, and the first 50 principal components were used for this analysis. Statistical significance was assessed by performing a Chi-square test for enrichment of Cluster 0-like cells in the IL1ß+ monocyte cluster versus all other cell types in the Mulder et al. dataset.
Statistical analyses
Two-group comparisons including comparisons between cell abundances, TCR or BCR diversity and AUCell gene signature score distributions were measured by Wilcoxon rank-sum test. Three-group (CR, PR, PD) comparisons were assessed with Cuzick trend test (PMCMRplus package, Pohlert T (2014). The Pairwise Multiple Comparison of Mean Ranks Package (PMCMR). R package, https://CRAN.R-project.org/package=PMCMR). For both types of analyses, nominal one-sided p-values were reported (p-values < 0.05) and adjusted for multiple comparisons using the Benjamini-Hochberg method (FDR < 0.1). Separate Benjamini-Hochberg corrections were performed in the following immune cell subsets: CD3+CD8− naïve/CM, non-naïve and gamma delta T cell clusters; CD3− NK cell, B cell and myeloid clusters. Separate Benjamini–Hochberg corrections were also performed to assess TCR diversity in CD3+CD8− naïve/CM and non-naïve T cells.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We gratefully acknowledge support from a Blood Cancer Discovery grant co-funded by the Leukemia & Lymphoma Society, Paul G. Allen Foundation and Mark Foundation (8019-20, MAS and SJR), Leukemia & Lymphoma Society (6671-24, MAS; 5650-24, JP), Lymphoma Research Foundation (816323, EM), an AIRC-FGB fellowship Gianni Bonadonna (28076, EC), Bristol-Myers Squibb (MAS and SJR), and the Miller Family Foundation (MAS). JP is a Fellow of The Leukemia & Lymphoma Society. We thank Lindy Boyne for editorial assistance.
Author contributions
J.P., M.T., K.T.W., S.J.R., X.S.L. and M.A.S. conceived and led the project and analyzed the data. J.P., M.T., K.T.W., L.S., K.L., V.S., E.L.W., J.L.W., N.B., H.O., B.A.P., F.Z.C., E.M., J.O., G.B. and L.N.L. performed experiments and analyzed the data. K.L.P., R.A.R., E.C. and P.A. contributed to the analysis and scientific discussions. J.P., M.T., X.S.L. and M.A.S. wrote the paper. All authors reviewed and approved the manuscript.
Peer review
Peer review information
Nature Communications thanks Xianwen Ren and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The scRNA-seq and scTCR-seq data generated in this study have been deposited in the European Genome-phenome Archive (EGA) under the following accession numbers: EGAS00001007569 and EGAD00001011360. Additional data are included in the Supplementary Information. Two external datasets that were used as validation in this study are available under the following accession numbers: EGAD00001005481, EGAS00001004008, GSE145281, and GSE178209 and https://gustaveroussy.github.io/FG-Lab/. The raw numbers for charts and graphs are available in the Source Data file. Source data are provided with this paper.
Code availability
The code is available at https://github.com/crazyhottommy/hodgkin_lymphoma_publication_scRNAseq_analysis and https://github.com/dfci/pythologist.
Competing interests
M.T. is a full-time employee of Astra Zeneca. K.L. is a full-time employee of PathAI. F.Z.C. is a full-time employee of AstraZeneca. J.O. is a full-time employee of Bristol Myers Squibb. P.A. consults for Merck, Bristol Myers Squibb, A.D.C. Therapeutics, GenMab, Enterome, Genentech/Roche, ATB Therapeutics, Foresight Diagnostics, AstraZeneca, MSD, Tessa Therapeutics, Regeneron, Xencor, and receives institutional research funding from Merck, Bristol Myers Squibb, Genentech/Roche, KITE/Gilead, Adaptive Biotechnologies, IGM, AstraZeneca, MSD, Aamed Therapeutics and honoraria from Merck. S.J.R. receives institutional research funding from Bristol Myers Squibb, KITE/Gilead and is an advisory board member of Immunitas Therapeutics. X.S.L. is the founder and CEO of GV20 Therapeutics, LLC. M.A.S. has received research funding from Bristol Myers Squibb, AstraZeneca, Bayer Abbvie, Genentech and Novartis and has served on advisory boards for Bristol Myers Squibb and AstraZeneca. The remaining authors declare no competing financial interests. No authors have non-financial competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Julia Paczkowska, Ming Tang, Kyle T. Wright.
Supplementary information
The online version contains Supplementary Material available at 10.1038/s41467-024-54512-7.
References
- 1.Weniger, M. A. & Kuppers, R. Molecular biology of Hodgkin lymphoma. Leukemia35, 968–981 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green, M. R. et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B cell lymphoma. Blood116, 3268–3277 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roemer, M. G. et al. PD-L1 and PD-L2 genetic alterations define classical Hodgkin lymphoma and predict outcome. J. Clin. Oncol.34, 2690–2697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wienand, K. et al. Genomic analyses of flow-sorted Hodgkin Reed-Sternberg cells reveal complementary mechanisms of immune evasion. Blood Adv.3, 4065–4080 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ansell, S. M. et al. Nivolumab for relapsed/refractory classical Hodgkin lymphoma: 5-year survival from the pivotal phase 2 CheckMate 205 study. Blood Adv. 2023;7(20):6266-6274. Blood Adv.8, 829–831 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armand, P. et al. Five-year follow-up of KEYNOTE-087: pembrolizumab monotherapy for relapsed/refractory classical Hodgkin lymphoma. Blood142, 878–886 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roemer, M. G. M. et al. Major histocompatibility complex Class II and programmed death Ligand 1 expression predict outcome after programmed death 1 blockade in classic Hodgkin Lymphoma. J. Clin. Oncol.36, 942–950 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armand, P. et al. PD-1 blockade with pembrolizumab for classical Hodgkin lymphoma after autologous stem cell transplantation. Blood134, 22–29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Advani, R. H. et al. Brentuximab vedotin in combination with nivolumab in relapsed or refractory Hodgkin lymphoma: 3-year study results. Blood138, 427–438 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Moskowitz, A. J. et al. Phase II trial of pembrolizumab plus gemcitabine, vinorelbine, and liposomal doxorubicin as second-line therapy for relapsed or refractory classical Hodgkin Lymphoma. J. Clin. Oncol.39, 3109–3117 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramchandren, R. et al. Nivolumab for newly diagnosed advanced-stage classic Hodgkin Lymphoma: safety and efficacy in the phase II CheckMate 205 Study. J. Clin. Oncol.37, 1997–2007 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen, P. B. et al. Pembrolizumab followed by AVD in untreated early unfavorable and advanced-stage classical Hodgkin lymphoma. Blood137, 1318–1326 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brockelmann, P. J. et al. Efficacy of nivolumab and AVD in early-stage unfavorable classic hodgkin lymphoma: the randomized phase 2 german hodgkin study group NIVAHL trial. JAMA Oncol.6, 872–880 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrera, A. F. et al. Nivoumab (N)-AVD improves progression-free survival compared to brentuximab vedotin (BV)-AVD in advanced stage (AS) classic Hodgkin lymphoma (HL): Results of SWOG S1826. Hematolog. Oncol.41, 33–35 (2023). [Google Scholar]
- 15.Herrera, A. F. et al. Nivolumab+AVD in advanced-stage classic Hodgkin’s Lymphoma. N. Engl. J. Med.391, 1379–1389 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reichel, J. et al. Flow sorting and exome sequencing reveal the oncogenome of primary Hodgkin and Reed-Sternberg cells. Blood125, 1061–1072 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Roemer, M. G. et al. Classical Hodgkin Lymphoma with reduced beta2M/MHC Class I expression is associated with inferior outcome independent of 9p24.1 Status. Cancer Immunol. Res.4, 910–916 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cader, F. Z. et al. Mass cytometry of Hodgkin lymphoma reveals a CD4(+) regulatory T cell-rich and exhausted T-effector microenvironment. Blood132, 825–836 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carey, C. D. et al. Topological analysis reveals a PD-L1-associated microenvironmental niche for Reed-Sternberg cells in Hodgkin lymphoma. Blood130, 2420–2430 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steidl, C. et al. Tumor-associated macrophages and survival in classic Hodgkin lymphoma. N. Engl. J. Med.362, 875–885 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aoki, T. et al. Single-cell transcriptome analysis reveals disease-defining T cell subsets in the tumor microenvironment of classic Hodgkin Lymphoma. Cancer Discov.10, 406–421 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Cader, F. Z. et al. A peripheral immune signature of responsiveness to PD-1 blockade in patients with classical Hodgkin lymphoma. Nat. Med26, 1468–1479 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mousset, C. M. et al. Comprehensive phenotyping of T cells using flow cytometry. Cytom. A95, 647–654 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Szabo, P. A. et al. Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat. Commun.10, 4706 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyu, M. et al. Dissecting the landscape of activated CMV-stimulated CD4+ T cells in humans by linking single-cell RNA-Seq with T cell receptor sequencing. Front Immunol.12, 779961 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den Broek, T., Borghans, J. A. M. & van Wijk, F. The full spectrum of human naive T cells. Nat. Rev. Immunol.18, 363–373 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Kuwahara, M. et al. The transcription factor Sox4 is a downstream target of signaling by the cytokine TGF-beta and suppresses T(H)2 differentiation. Nat. Immunol.13, 778–786 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crotty, S. T follicular helper cell differentiation, function, and roles in disease. Immunity41, 529–542 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang, P. et al. Immune cell subset differentiation and tissue inflammation. J. Hematol. Oncol.11, 97 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cenerenti, M., Saillard, M., Romero, P. & Jandus, C. The Era of Cytotoxic CD4 T cells. Front Immunol.13, 867189 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou, L. et al. Single-Cell RNA-Seq analysis uncovers distinct functional human NKT cell sub-populations in peripheral blood. Front Cell Dev. Biol.8, 384 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pizzolato, G. et al. Single-cell RNA sequencing unveils the shared and the distinct cytotoxic hallmarks of human TCRVdelta1 and TCRVdelta2 gammadelta T lymphocytes. Proc. Natl Acad. Sci. USA116, 11906–11915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Godfrey, D. I., Koay, H. F., McCluskey, J. & Gherardin, N. A. The biology and functional importance of MAIT cells. Nat. Immunol.20, 1110–1128 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Garner, L. C. et al. Single-cell analysis of human MAIT cell transcriptional, functional and clonal diversity. Nat. Immunol.24, 1565–1578 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, S. Y., Sanchez, D. J., Aliyari, R., Lu, S. & Cheng, G. Systematic identification of Type I and type II interferon-induced antiviral factors. Proc. Natl Acad. Sci. USA109, 4239–4244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lukhele, S., Boukhaled, G. M. & Brooks, D. G. Type I interferon signaling, regulation and gene stimulation in chronic virus infection. Semin. Immunol.43, 101277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cibrian, D. & Sanchez-Madrid, F. CD69: from activation marker to metabolic gatekeeper. Eur. J. Immunol.47, 946–953 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shevyrev, D. & Tereshchenko, V. Treg heterogeneity, function, and homeostasis. Front Immunol.10, 3100 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roncarolo, M. G., Gregori, S., Bacchetta, R., Battaglia, M. & Gagliani, N. The biology of T regulatory Type 1 cells and their therapeutic application in immune-mediated diseases. Immunity49, 1004–1019 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Chao, A. Nonparametric estimation of the number of classes in a population. Scand. J. Stat.11, 265–270 (1984). [Google Scholar]
- 41.Chao, A., Ma, M. & Yang, M. Stopping rules and estimation for recapture debugging with unequal failure rates. Biometrics43, 783–791 (1993). [Google Scholar]
- 42.Linnemann, C. et al. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nat. Med21, 81–85 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Zuazo, M. et al. Functional systemic CD4 immunity is required for clinical responses to PD-L1/PD-1 blockade therapy. EMBO Mol. Med11, e10293 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagasaki, J. et al. The critical role of CD4+ T cells in PD-1 blockade against MHC-II-expressing tumors such as classic Hodgkin lymphoma. Blood Adv.4, 4069–4082 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuazo, M. et al. Systemic CD4 immunity as a Key Contributor to PD-L1/PD-1 Blockade Immunotherapy Efficacy. Front Immunol.11, 586907 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kagamu, H. et al. CD4(+) T cell immunity in the peripheral blood correlates with response to Anti-PD-1 therapy. Cancer Immunol. Res.8, 334–344 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Weatherly, K. et al. Functional profile of S100A4-deficient T cells. Immun. Inflamm. Dis.3, 431–444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Masopust, D. & Schenkel, J. M. The integration of T cell migration, differentiation and function. Nat. Rev. Immunol.13, 309–320 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Sallusto, F., Lenig, D., Forster, R., Lipp, M. & Lanzavecchia, A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature401, 708–712 (1999). [DOI] [PubMed] [Google Scholar]
- 50.Cano-Gamez, E. et al. Single-cell transcriptomics identifies an effectorness gradient shaping the response of CD4(+) T cells to cytokines. Nat. Commun.11, 1801 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, F., Cheng, F. & Zheng, F. Stem cell like memory T cells: a new paradigm in cancer immunotherapy. Clin. Immunol.241, 109078 (2022). [DOI] [PubMed] [Google Scholar]
- 52.Aoki, T. et al. Single-cell profiling reveals the importance of CXCL13/CXCR5 axis biology in lymphocyte-rich classic Hodgkin lymphoma. Proc. Natl. Acad. Sci. USA118, e2105822118 (2021). [DOI] [PMC free article] [PubMed]
- 53.Kazanietz, M. G., Durando, M. & Cooke, M. CXCL13 and its receptor CXCR5 in cancer: inflammation, immune response, and beyond. Front Endocrinol. (Lausanne)10, 471 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saez de Guinoa, J., Barrio, L., Mellado, M. & Carrasco, Y. R. CXCL13/CXCR5 signaling enhances BCR-triggered B cell activation by shaping cell dynamics. Blood118, 1560–1569 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Ager, C. R. et al. KLRG1 marks tumor-infiltrating CD4 T cell subsets associated with tumor progression and immunotherapy response. J. Immunother. Cancer11, e006782 (2023). [DOI] [PMC free article] [PubMed]
- 56.Greten, F. R. & Grivennikov, S. I. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity51, 27–41 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Snell, L. M., McGaha, T. L. & Brooks, D. G. Type I interferon in chronic virus infection and cancer. Trends Immunol.38, 542–557 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rusinova, I. et al. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res.41, D1040–D1046 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mensurado, S., Blanco-Dominguez, R. & Silva-Santos, B. The emerging roles of gammadelta T cells in cancer immunotherapy. Nat. Rev. Clin. Oncol.20, 178–191 (2023). [DOI] [PubMed] [Google Scholar]
- 60.Sebestyen, Z., Prinz, I., Dechanet-Merville, J., Silva-Santos, B. & Kuball, J. Translating gammadelta (gammadelta) T cells and their receptors into cancer cell therapies. Nat. Rev. Drug Discov.19, 169–184 (2020). [DOI] [PubMed] [Google Scholar]
- 61.de Vries, N. L. et al. gammadelta T cells are effectors of immunotherapy in cancers with HLA class I defects. Nature613, 743–750 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith, S. L. et al. Diversity of peripheral blood human NK cells identified by single-cell RNA sequencing. Blood Adv.4, 1388–1406 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morgan, D. & Tergaonkar, V. Unraveling B cell trajectories at single cell resolution. Trends Immunol.43, 210–229 (2022). [DOI] [PubMed] [Google Scholar]
- 64.Yang, J., Zhang, L., Yu, C., Yang, X. F. & Wang, H. Monocyte and macrophage differentiation: circulation inflammatory monocyte as biomarker for inflammatory diseases. Biomark Res.2, (2014). [DOI] [PMC free article] [PubMed]
- 65.Villani, A. C. et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science356, eaah4573 (2017). [DOI] [PMC free article] [PubMed]
- 66.Yang, C. et al. Heterogeneity of human bone marrow and blood natural killer cells defined by single-cell transcriptome. Nat. Commun.10, 3931 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kramer, B. et al. Early IFN-alpha signatures and persistent dysfunction are distinguishing features of NK cells in severe COVID-19. Immunity54, 2650–2669.e2614 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen, H. et al. Single-cell trajectories reconstruction, exploration and mapping of omics data with STREAM. Nat. Commun.10, 1903 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carsetti, R. et al. Comprehensive phenotyping of human peripheral blood B lymphocytes in healthy conditions. Cytom. A101, 131–139 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Song, L. et al. TRUST4: immune repertoire reconstruction from bulk and single-cell RNA-seq data. Nat. Methods18, 627–630 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim, S. S. et al. Role of B cells in responses to checkpoint blockade immunotherapy and overall survival of cancer patients. Clin. Cancer Res.27, 6075–6082 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Helmink, B. A. et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature577, 549–555 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cabrita, R. et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature577, 561–565 (2020). [DOI] [PubMed] [Google Scholar]
- 74.Petitprez, F. et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature577, 556–560 (2020). [DOI] [PubMed] [Google Scholar]
- 75.Ravenhill, B. J., Soday, L., Houghton, J., Antrobus, R. & Weekes, M. P. Comprehensive cell surface proteomics defines markers of classical, intermediate and non-classical monocytes. Sci. Rep.10, 4560 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Collin, M. & Bigley, V. Human dendritic cell subsets: an update. Immunology154, 3–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chittezhath, M. et al. Molecular profiling reveals a tumor-promoting phenotype of monocytes and macrophages in human cancer progression. Immunity41, 815–829 (2014). [DOI] [PubMed] [Google Scholar]
- 78.Sanin, D. E. et al. A common framework of monocyte-derived macrophage activation. Sci. Immunol.7, eabl7482 (2022). [DOI] [PubMed] [Google Scholar]
- 79.Loke, P. & Lin, J. D. Redefining inflammatory macrophage phenotypes across stages and tissues by single-cell transcriptomics. Sci. Immunol.7, eabo4652 (2022). [DOI] [PubMed] [Google Scholar]
- 80.Caronni, N. et al. IL-1beta(+) macrophages fuel pathogenic inflammation in pancreatic cancer. Nature623, 415–422 (2023). [DOI] [PubMed]
- 81.Wang, F. et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn.14, 22–29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Millar, M. Mixed multiplex staining: automated RNAscope and OPAL for multiple targets. Methods Mol. Biol.2148, 277–298 (2020). [DOI] [PubMed] [Google Scholar]
- 83.Mulder, K. et al. Cross-tissue single-cell landscape of human monocytes and macrophages in health and disease. Immunity54, 1883–1900.e1885 (2021). [DOI] [PubMed] [Google Scholar]
- 84.Stuart, T. et al. Comprehensive integration of single-cell data. Cell177, 1888–1902.e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Raggi, F. & Bosco, M. C. Targeting mononuclear phagocyte receptors in cancer immunotherapy: new perspectives of the triggering receptor expressed on myeloid cells (TREM-1). Cancers (Basel)12, 1337 (2020). [DOI] [PMC free article] [PubMed]
- 86.Wu, Q. et al. Blocking Triggering receptor expressed on myeloid cells-1-positive tumor-associated macrophages induced by hypoxia reverses immunosuppression and anti-programmed cell death ligand 1 resistance in liver cancer. Hepatology70, 198–214 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boro, M. & Balaji, K. N. CXCL1 and CXCL2 regulate NLRP3 inflammasome activation via G-protein-coupled receptor CXCR2. J. Immunol.199, 1660–1671 (2017). [DOI] [PubMed] [Google Scholar]
- 88.Mantovani, A., Dinarello, C. A., Molgora, M. & Garlanda, C. Interleukin-1 and related cytokines in the regulation of inflammation and immunity. Immunity50, 778–795 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jang, J. H., Kim, D. H. & Surh, Y. J. Dynamic roles of inflammasomes in inflammatory tumor microenvironment. NPJ Precis Oncol.5, 18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guo, B., Fu, S., Zhang, J., Liu, B. & Li, Z. Targeting inflammasome/IL-1 pathways for cancer immunotherapy. Sci. Rep.6, 36107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods14, 1083–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yuen, K. C. et al. High systemic and tumor-associated IL-8 correlates with reduced clinical benefit of PD-L1 blockade. Nat. Med.26, 693–698 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kurtulus, S. et al. Checkpoint Blockade Immunotherapy Induces Dynamic Changes in PD-1(−)CD8(+) Tumor-Infiltrating T Cells. Immunity50, 181–194.e186 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Grund, J. et al. Low B cell content is associated with a CD73-low tumour microenvironment and unfavourable prognosis in classic Hodgkin lymphoma. Br. J. Haematol.201, 1097–1102 (2023). [DOI] [PubMed] [Google Scholar]
- 95.Tudor, C. S. et al. B cells in classical Hodgkin lymphoma are important actors rather than bystanders in the local immune reaction. Hum. Pathol.44, 2475–2486 (2013). [DOI] [PubMed] [Google Scholar]
- 96.Keenan, B. P. et al. Circulating monocytes associated with anti-PD-1 resistance in human biliary cancer induce T cell paralysis. Cell Rep.40, 111384 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hou, J., Karin, M. & Sun, B. Targeting cancer-promoting inflammation - have anti-inflammatory therapies come of age? Nat. Rev. Clin. Oncol.18, 261–279 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Aggen, D. H. et al. Blocking IL1 beta promotes tumor regression and remodeling of the myeloid compartment in a renal cell carcinoma model: multidimensional analyses. Clin. Cancer Res.27, 608–621 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kaplanov, I. et al. Blocking IL-1beta reverses the immunosuppression in mouse breast cancer and synergizes with anti-PD-1 for tumor abrogation. Proc. Natl Acad. Sci. USA116, 1361–1369 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Highfill, S. L. et al. Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Sci. Transl. Med.6, 237ra267 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Steele, C. W. et al. CXCR2 inhibition profoundly suppresses metastases and augments immunotherapy in pancreatic ductal adenocarcinoma. Cancer Cell29, 832–845 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ghallab, A. M., Eissa, R. A. & El Tayebi, H. M. CXCR2 small-molecule antagonist combats chemoresistance and enhances immunotherapy in triple-negative breast cancer. Front Pharm.13, 862125 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rogic, A. et al. High endogenous CCL2 expression promotes the aggressive phenotype of human inflammatory breast cancer. Nat. Commun.12, 6889 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kitamura, T. et al. CCL2-induced chemokine cascade promotes breast cancer metastasis by enhancing retention of metastasis-associated macrophages. J. Exp. Med.212, 1043–1059 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Qian, B. Z. et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature475, 222–225 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pittet, M. J., Michielin, O. & Migliorini, D. Clinical relevance of tumour-associated macrophages. Nat. Rev. Clin. Oncol.19, 402–421 (2022). [DOI] [PubMed] [Google Scholar]
- 107.Di Mitri, D. et al. Re-education of tumor-associated macrophages by CXCR2 blockade drives senescence and tumor inhibition in advanced prostate cancer. Cell Rep.28, 2156–2168.e2155 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu, H. et al. Neutralizing IL-8 potentiates immune checkpoint blockade efficacy for glioma. Cancer Cell41, 693–710.e698 (2023). [DOI] [PubMed] [Google Scholar]
- 109.Cheson, B. D. et al. Revised response criteria for malignant lymphoma. J. Clin. Oncol.25, 579–586 (2007). [DOI] [PubMed] [Google Scholar]
- 110.Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 112.Tiberi, S., Crowell, H. L., Samartsidis, P., Weber, L. M. & Robinson, M. D. Distinct: a novel approach to differential distribution analyses. Ann. Appl. Stat.17, 1681–1700, 1620 (2023). [Google Scholar]
- 113.Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci.102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liberzon, A. et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst.1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Popov, A. et al. Immunomind/immunarch: Immunarch 0.9.1 (0.9.1). Zenodo. 10.5281/zenodo.3367200 (2024).
- 116.Ansell, S. M. et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N. Engl. J. Med372, 311–319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Alessi, J. V. et al. Low peripheral blood derived neutrophil-to-lymphocyte ratio (dNLR) is associated with increased tumor T cell infiltration and favorable outcomes to first-line pembrolizumab in non-small cell lung cancer. J. Immunother. Cancer9, e003536 (2021). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The scRNA-seq and scTCR-seq data generated in this study have been deposited in the European Genome-phenome Archive (EGA) under the following accession numbers: EGAS00001007569 and EGAD00001011360. Additional data are included in the Supplementary Information. Two external datasets that were used as validation in this study are available under the following accession numbers: EGAD00001005481, EGAS00001004008, GSE145281, and GSE178209 and https://gustaveroussy.github.io/FG-Lab/. The raw numbers for charts and graphs are available in the Source Data file. Source data are provided with this paper.
The code is available at https://github.com/crazyhottommy/hodgkin_lymphoma_publication_scRNAseq_analysis and https://github.com/dfci/pythologist.