Abstract
Background
Risk prediction models can identify individuals at high risk of chronic liver disease (CLD), but there is limited evidence on the performance of various models in diverse populations. We aimed to systematically review CLD prediction models, meta-analyze their performance, and externally validate them in 0.5 million Chinese adults in the China Kadoorie Biobank (CKB).
Methods
Models were identified through a systematic review and categorized by the target population and outcomes (hepatocellular carcinoma [HCC] and CLD). The performance of models to predict 10-year risk of CLD was assessed by discrimination (C-index) and calibration (observed vs predicted probabilies).
Results
The systematic review identified 57 articles and 114 models (28.4% undergone external validation), including 13 eligible for validation in CKB. Models with high discrimination (C-index ≥ 0.70) in CKB were as follows: (1) general population: Li-2018 and Wen 1–2012 for HCC, CLivD score (non-lab and lab) and dAAR for CLD; (2) hepatitis B virus (HBV) infected individuals: Cao-2021 for HCC and CAP-B for CLD. In CKB, all models tended to overestimate the risk (O:E ratio 0.55–0.94). In meta-analysis, we further identified models with high discrimination: (1) general population (C-index ≥ 0.70): Sinn-2020, Wen 2–2012, and Wen 3–2012 for HCC, and FIB-4 and Forns for CLD; (2) HBV infected individuals (C-index ≥ 0.80): RWS-HCC and REACH-B IIa for HCC and GAG-HCC for HCC and CLD.
Conclusions
Several models showed good discrimination and calibration in external validation, indicating their potential feasibility for risk stratification in population-based screening programs for CLD in Chinese adults.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12916-024-03754-9.
Keywords: Risk prediction, Chronic liver disease, Hepatocellular carcinoma, Chinese, Systematic review, External validation
Background
Chronic liver disease (CLD), encompassing mainly liver cancer and cirrhosis, affected 1.70 billion people globally in 2021 [1]. According to the Global Burden of Disease Study (GBD), the disease burden of CLD is high in China, with 0.39 billion cases and 0.33 million deaths in 2021. With the universal coverage of hepatitis B virus (HBV) vaccination and Westernized lifestyles, the leading cause of CLD in China shifted from HBV to non-alcoholic fatty liver disease (NAFLD), which accounted for ~ 40% prevalent cases in 1990 and ~ 70% in 2019. Due to the asymptomatic progression, CLD is often diagnosed in advanced stages and has a poor prognosis [2].
Screening strategies for CLD are imperative because disease surveillance contributes to early diagnosis and overall survival [3, 4]. Screening strategies can be divided into individual-based (i.e., centering on the high-risk individuals) and population-wide approaches. The American Association for the Study of Liver Diseases (AASLD) [5] and European Association for the Study of the Liver (EASL) [6] recommend ultrasound and alpha-fetoprotein (AFP) tests to high-risk populations including patients with cirrhosis and non-cirrhotic chronic hepatitis B (CHB) as screening strategies for liver cancer. China adopts a similar approach [7]. However, because of the suboptimal sensitivity, dependency on operator skills, poor adherence, and limited accessibility of imaging modalities [8], non-invasive biomarkers and risk prediction models have emerged as promising risk stratification tools for population screening, which optimize resources and have the potential to improve the detection and prognosis of CLD [9].
Currently, international consensuses and expert views have recommended non-patented blood tests (e.g., FIB-4 or NAFLD fibrosis score) as screening tests for patients with CLD risk factors [10–12]. However, despite the satisfactory C-index (between 0.53 and 0.80) [13], the inaccessibility in the community setting hinders their generalizability as first-line tests [14, 15]. Recent studies have focused on developing models incorporating more accessible non-laboratory parameters, such as lifestyle factors and family history. Despite the development of numerous prediction models for CLD, significant gaps remain in model implementation, including inadequate validation across diverse regions and populations [10]. Furthermore, there is a lack of comprehensive comparison of CLD models across populations with different CLD etiologies, particularly in China, where the etiology differs importantly from Western countries.
Therefore, our objective was to systematically review published CLD prediction models and externally validate them using the China Kadoorie Biobank (CKB), one of the largest and geographically diverse prospective cohort studies in China. We also conducted a meta-analysis to compare model performance across populations in both published cohort studies and CKB.
Methods
Systematic literature search and identification of published models
A systematic search was conducted in the PubMed and Embase databases up to October 14, 2022. The search terms included MeSH terms in PubMed and Emtree terms in Embase as well as free-text terms. The following terms were used as index terms or free-text words: “hepatocellular carcinoma,” “liver cancer,” “chronic liver disease,” “severe liver disease,” and “risk prediction,” among other related terms. We included original research articles, systematic reviews, and conference abstracts. The references of systematic reviews were manually reviewed to identify potentially missing studies. The complete search strategy for the databases is provided in Additional file 1: Method S1.
The articles were included based on the following criteria: (1) focused on the development, update, or validation of prediction models, or the comparison of existing models; (2) included predictors involving but not limited to lifestyle and clinical risk factors; (3) designed as prospective cohorts, retrospective cohorts, case-cohort studies, or nested case–control studies; (4) included outcomes related to CLD, such as liver cancer, cirrhosis, and other liver diseases caused by various etiologies (e.g., NAFLD, alcoholic liver disease [ALD], and viral hepatitis); and (5) reported performance measures of predictive ability, including but not limited to the area under the ROC curve (AUC) or C-statistic.
We excluded ecological studies (i.e., conducted at the population level), studies comprising hospital patients who underwent CLD-related procedures (e.g., hepatectomy, liver transplantation) or received specific antiviral therapies (e.g., entecavir, ribavirin), studies focused solely on prognostic models, studies not published in English, and studies categorized as narrative reviews, letters, editorials, or commentaries.
After removing duplicate articles, two independent reviewers (XC and YC) conducted separate title and abstract screening, retrieving the full text when necessary. Discrepancies were resolved through discussion, and a third reviewer (YP) was consulted. After the initial screening, three reviewers (XC, KS, and CM) independently conducted full-text screening and data extraction to determine the final inclusion of eligible articles. The reasons for exclusion were recorded for each article during the full-text screening. To ensure the accuracy of the database, the three reviewers re-checked any discrepancies in data extraction and reviewed the reference lists of all eligible articles to identify any missed studies. The research protocol has been registered and approved in the PROSPERO international prospective register (ID: CRD42022374724). The risk of bias was assessed according to PROBAST (Prediction model Risk Of Bias Assessment Tool) [16].
Data extraction
Data extraction was based on the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement [17]. Information extracted included citation details (e.g., authors, publication date, region), study design and methods, study population, sample size, model name, model formula, included variables, measures of predictive ability, recruitment years of participants, duration of follow-up, measurement outcomes, and their corresponding International Classification of Diseases (ICD) codes, as well as outcome measurement methods. The information was extracted from each eligible risk prediction model to perform external validation. When multiple validation studies existed, a meta-analysis was performed to summarize the evidence to support and compare prediction models in a particular field according to TRIPOD-SRMA [18].
If published models had been updated regarding the predictors or coefficients, the updated data were extracted. For models providing absolute risk, the following data were also extracted to evaluate the model’s calibration: age-specific incidence rates, age-specific mortality rates, attributable risk, survival functions, mean values of risk factors in the cohort, and risk scores estimated at the mean values of all predictors.
Validation cohorts
The models were externally validated according to the TRIPOD (Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis) guidelines [19]. Details of the CKB study and methods for external validation are reported in Additional file 1: Method S2 [20, 21].
Model predictors and outcomes
During the external validation stage, we first attempted to match the predictors of the original model with the available variables in CKB. When a direct match could not be achieved, a proxy variable was defined as closely as possible to the original predictor. The measurement and definition of CKB variables are described in detail in Additional file 1: Method S3. If none of the above situations could be achieved, the model was excluded.
All eligible models were classified according to the target population. General population included population-based cohorts with no specific restrictions on CLD risk factors. HBV infected individuals included patients with CHB and individuals with positive serum hepatitis B surface antigen (HBsAg). HCV infected individuals included patients with chronic hepatitis C (CHC), individuals with positive serum anti-hepatitis C virus antibody (Anti-HCV Ab), and individuals with positive serum HCV RNA. Patients with NAFLD, diabetes, and individuals with high CLD risk (including diabetes, obesity, and high alcohol consumption) were included.
Outcomes of the prediction models were categorized into hepatocellular carcinoma (HCC) and CLD. HCC was defined by the ICD-10 code C22.0 and C22.9 excluding other subtypes of liver cancer (i.e., intrahepatic bile duct carcinoma, hepatoblastoma). Sensitivity analysis was conducted using different definitions of HCC (ICD-10 code: C22.0 or C22). CLD included advanced liver disease and liver-related mortality, involving liver cirrhosis, NAFLD, and liver fibrosis (ICD-10 code: K70, K72, and K74 alongside other complications of CLD, Additional file 1: Method S4). We selected these ICD-10 codes to define CLD for the following reasons: (1) these are the standard definitions used in large-scale population-based studies including the UK Biobank and CKB and in well-established risk prediction models for CLD [22–24]; (2) several previous models included in the systematic review did not report the detailed ICD-10 codes so standard definitions need to be applied to improve the generalizability of these models. Based on the prediction time frame, models were further classified into five time intervals: < 5 years, 5 years, 5–10 years, 10 years, and > 10 years.
Statistical analysis
Ten articles with 13 models were included for external validation in CKB, and all were developed using the Cox proportional hazards model. Because 10 of the 13 models did not report enough information, we fitted Cox regression using predictors as defined in the original studies and updated the model by re-estimating the predictor coefficients in CKB (i.e., “refit”). This approach aimed to evaluate the predictive performance and improve the calibration of the models in the relatively large sample of the CKB, particularly when the provided information was limited [25]. Harrell’s C-index was used to assess discrimination, while the calibration plot and O:E ratio were used to assess calibration. Formulas for each model can be found in Additional file 1: Method S5.
For prediction models that were examined in ≥ 2 independent datasets, we did a random-effects meta-analysis to calculate a summary estimate for model performance and calibration [26]. Discrimination was assessed by comparing discriminative ability using the C-index and AUC [27]. Of the 34 studies included in the meta-analysis, 26 reported AUC, 5 reported C-index, and 3 studies reported both. Therefore, we included the parameter as reported in the original studies and referred to it as “C-index” in the meta-analysis. Where a study reported both parameters, we included C-index in the meta-analysis. Effect sizes and their 95% confidence intervals were combined for the same model across studies to obtain pooled effect estimates using the “metamisc” package [26]. All statistical analyses were performed using R version 4.2.1.
Results
Characteristics of the included models
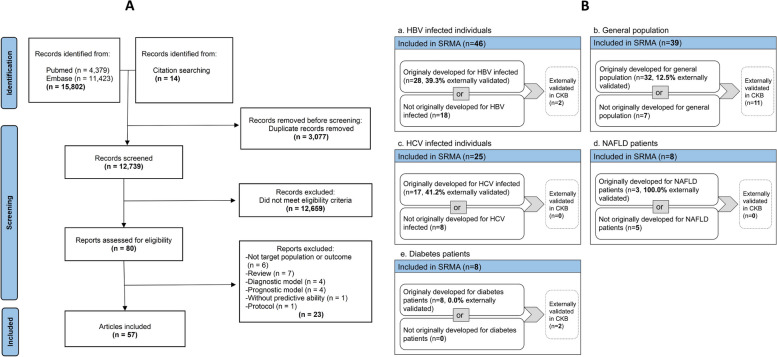
A total of 12,725 articles were initially screened and 80 relevant articles were included for full-text screening, of which 57 articles were eligible for inclusion (Fig. 1A) [22, 28–83]. The systematic review identified 114 models (39 for general, 46 for HBV, 25 for HCV, 8 for NAFLD, and 8 for diabetes, including 7 models for ≥ 2 populations) (see Additional file 2). After excluding models that were only externally validated but not originally developed for the study population (e.g., BARD initially for NAFLD, later validated in the general population), there were 32, 28, 17, 3, and 8 models developed specifically for general population, HBV infected individuals, HCV infected individuals, NAFLD patients, and diabetes patients, respectively. Among these models, 12.5% (4/32), 39.3% (11/28), 41.2% (7/17), 100.0% (3/3), and 0% (0/8), had been externally validated in the respective populations (Fig. 1B). Overall, only 28.4% of the models underwent external validation. Although there were also prediction models in alcoholic fatty liver (AFLD) patients, ALD patients, drinkers, and obese individuals, the number of models specifically developed in these sub-populations was limited. In terms of time horizon and outcomes, the commonest combinations were 5-year and 10-year HCC. Detailed information on study information and bias assessment was reported in Additional file 1: Table S1.
Fig. 1.
Flow chart. A Flow chart for screening eligible publications. B Number of models included in the systematic review and external validation of CKB. The larger box corresponds to the aggregate number of models included in the systematic review, including (1) models originally developed for the target population and (2) those previously validated within the target population (not originally developed for the target populations). The smaller box represents models externally validated in CKB, including 11 models validated in the general population and 2 models validated in HBV infected individuals. Abbreviations: CKB, China Kadoorie Biobank; HBV, hepatitis B virus; HCV, hepatitis C virus; NAFLD, non-alcoholic fatty liver disease; SRMA, systematic review and meta-analysis
Three categories of predictors were considered: non-laboratory parameters, HBV/HCV laboratory parameters, and non-HBV/HCV laboratory parameters. Additional file 1: Tables S2–4 show the number of models for these categories in the general population, HBV infected individuals, and other populations. The commonest predictors were age and gender. In prediction models developed for the general population, frequently used predictors also included (descending order by frequency): diabetes (17/32), alcohol (15/32), smoking (13/32), physical activity (12/32), alanine aminotransferase (ALT) (15/32), aspartate aminotransferase (AST) (11/32), and gamma-glutamyl transferase (GGT) (9/32). In contrast, virological parameters were common for HBV infected individuals (HBV DNA 16/28 and hepatitis B e antigen [HBeAg] 14/28).
CKB external validation
Due to the availability of predictors in the CKB study, 10 articles with 13 models were included for external validation, involving the general population, HBV infected individuals, and type 2 diabetes (T2D) patients. Among the 13 models for external validation, 3 models were based on non-lab predictors (Wen 1–2012, HLI, and CLivD score (non-lab)), while the other 10 models included blood-based biomarkers (Table 1).
Table 1.
Predictors in 13 CLD risk prediction models validated in CKB
| Model | Demographic | Lifestyle | Personal and family history | Blood-based biomarkers | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age | Sex | Alcohol | BMI | Diet | PA | Smoking | WHR | Cirrhosis | Diabetes | Hepatitis | ALT | AST | GGT | TG | |
| BARD | • | • | • | • | |||||||||||
| Cao-2021a | • | • | • | • | •b | ||||||||||
| CAP-Bc | • | • | • | • | • | •d | • | • | |||||||
| CLivD (lab) | • | • | • | • | • | • | • | ||||||||
| CLivD (non-lab) | • | • | • | • | • | • | |||||||||
| dAAR | • | • | • | ||||||||||||
| DM-HCC | • | • | • | ||||||||||||
| HLI | • | • | • | • | • | • | • | ||||||||
| Li-2018e | • | • | • | • | • | • | |||||||||
| Sinn-2020 | • | • | • | • | • | • | |||||||||
| Wen 1-2012 | • | • | • | • | • | • | |||||||||
| Wen 2-2012 | • | • | • | • | |||||||||||
| Wen 3-2012 | • | • | • | • | • | • | • | • | |||||||
Abbreviations: ALT Alanine aminotransferase, AST Aspartate aminotransferase, BMI Body mass index, GGT Gamma-glutamyl transferase, HBV Hepatitis B virus, HCV Hepatitis C virus, PA Physical activity, TG Triglycerides, WHR Waist-to-hip ratio
aPsychological trauma was included as an additional predictor in Cao-2021
bCirrhosis was defined as medical history of liver diseases in mothers
cAdditional predictors included income, statin exposure, and antiplatelet
dHepatitis was defined as hepatitis C
eAdditional predictors included HbA1c, antidiabetes medication, antihyperlipidemia medication, and THR, which was included as an additional predictor in Li-2018
Of the 8 models for HCC, 5 models were developed for the general populations, 2 models for diabetes patients (DM-HCC and Li-2018), and 1 for HBV infected individuals (Cao-2021) (Table 2). DM-HCC and Li-2018 were also validated in the general population in CKB. Among the general population, only the HLI model showed higher discrimination in CKB than in the original development study (0.68 vs 0.64). The Wen 1–2012 and Li-2018 models exhibited favorable discrimination with C-index ≥ 0.70 for both 5-year and 10-year risk. Among patients with T2D, both the DM-HCC and Li-2018 models exhibited high discrimination, but this was probably because of the small number of cases. Among HBV infected individuals, the Cao-2021 model had a C-index of 0.73 for 10-year prediction. Sensitivity analyses with different definitions of HCC (ICD-10: C22.0 and C22) showed similar results (Additional file 1: Table S5).
Table 2.
Discrimination of 10-year CLD model in the published literature and CKB
| Model | Population | Development cohort | Published validation cohort | CKB | ||||
|---|---|---|---|---|---|---|---|---|
| Area | Events/total | C-index (95% CI) | Events/total | C-index (95% CI) | Events/total | C-index (95% CI) | ||
| HCC | ||||||||
| DM-HCC (all)a | General | EAS | – | – | – | – | 26/15,818 | 0.66 (0.59–0.73) |
| HLI | General | EUR | 712/477,206 | 0.64 (0.57–0.70) | – | – | 1709/478,930 | 0.68 (0.67–0.70) |
| Li-2018 (all)a | General | EAS | – | – | – | – | 72/17,227 | 0.74 (0.68–0.80) |
| Sinn-2020 | General | EAS | 236/467,206 | 0.83 (0.77–0.88) | 35/91,357 | 0.92 (0.89–0.95) | 72/17,227 | 0.66 (0.60–0.72) |
| Wen 1-2012 | General | EAS | 1252/298,051 | 0.81 (0.80–0.83) | – | – | 1709/478,930 | 0.72 (0.70–0.73) |
| Wen 2-2012 | General | EAS | 1252/298,051 | 0.90 (0.90–0.92) | – | – | 72/17,227 | 0.67 (0.61–0.74) |
| Wen 3-2012 | General | EAS | 1252/298,051 | 0.91 (0.89–0.93) | – | – | 72/17,227 | 0.68 (0.61–0.74) |
| Cao-2021 | HBV | EAS | – | – | – | – | 532/13,723 | 0.73 (0.71–0.75) |
| DM-HCC | T2D | EAS | 36/2364b | 0.86 (0.85–0.88)b | – | – | 6/1348 | 0.78 (0.53–0.99) |
| Li-2018 | T2D | EAS | 493/21,149 | 0.77 (0.75–0.79) | – | – | 7/1540 | 0.96 (0.93–0.99) |
| CLD | ||||||||
| BARDc | General | EUR | – | – | 232/75,303 | 0.53 (0.50–0.57) | 150/17,227 | 0.55 (0.52–0.58) |
| CLivD score (lab) | General | EUR | 273/25,760d | 0.84 (0.75–0.93)d | 64/3049d | 0.78 (0.71–0.83)d | 141/15,945 | 0.74 (0.70–0.78) |
| CLivD score (non-lab) | General | EUR | 273/25,760d | 0.82 (0.74–0.91)d | 118/8107d | 0.70 (0.14–0.97)d | 3207/478,930 | 0.71 (0.70–0.72) |
| dAAR | General | EUR | 89/18,067e | 0.80 (0.74–0.85)e | 717/126,941 | 0.72 (0.70–0.74) | 150/17,227 | 0.72 (0.68–0.76) |
| CAP-B | HBV | EAS | 16,492/401,745 | 0.78 (0.78–0.78) | – | – | 28/394 | 0.77 (0.68–0.86) |
Abbreviations: CKB China Kadoorie Biobank, EAS East Asia, EUR Europe, HCC, Hepatocellular carcinoma, CLD Chronic liver disease
aDue to the small number of HCC cases among diabetes patients in CKB, the predictive ability of the DM-HCC and Li-2018 models was also evaluated in the general population
bThe values reported are based on the 5-year HCC outcome, given the absence of the 10-year HCC outcome in the original development cohort
cThe model was first developed for NAFLD patients in the US (lacking specific development data), but was later validated in the general population
dThe values reported are based on 15-year risk of CLD, given the absence of the 10-year CLD outcome in the development or external validation cohort
eThe values reported are based on 8.2-year risk of CLD, given the absence of the 5- or 10-year CLD outcome in the original development cohort
Of the 5 models for CLD, 4 models were developed for the general population and 1 for HBV infected individuals. In the general population, all models showed lower discrimination compared with the development cohort. BARD and CLivD score (non-lab) for 10-year prediction slightly better discrimination compared with previous external validation studies. Among all models, CLivD score (non-lab and lab) and dAAR showed C-index higher than 0.70 for 10-year CLD. For HBV infected individuals, the CAP-B model showed a C-index of 0.77 for predicting 10-year CLD (Table 2). Similar model performance was shown for 5-year HCC and 5-year CLD prediction (Additional file 1: Tables S6–7).
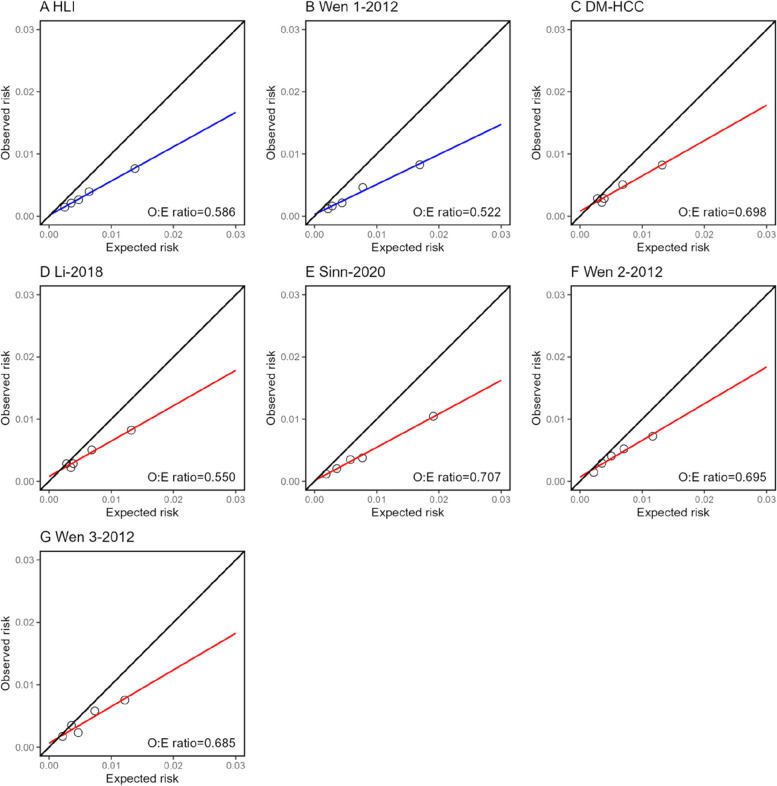
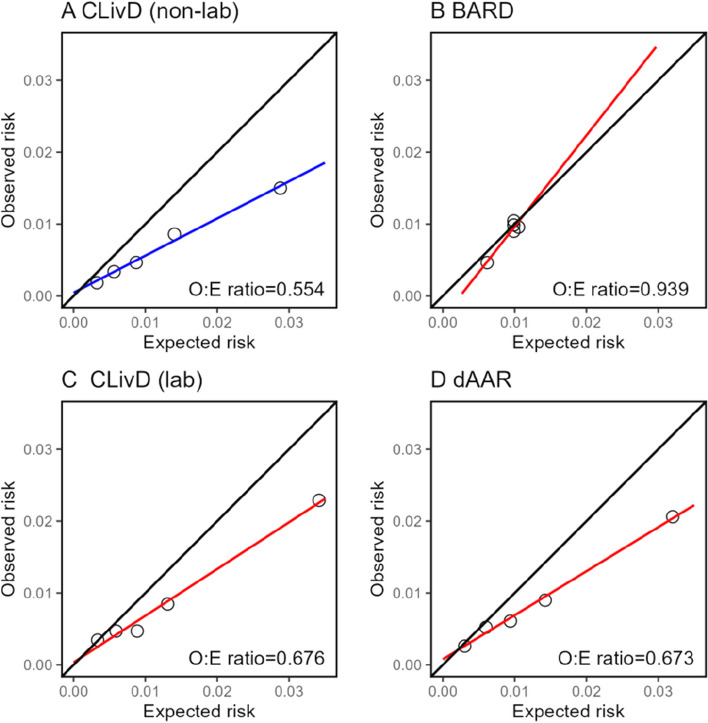
Model calibration in CKB was shown for HCC and CLD, separately. For 10-year risk of HCC, calibration across all models showed overestimation (Fig. 2). Similar patterns were observed for 5-year calibration (Additional file 1: Fig. S1). For 10-year risk of CLD, the CLivD score (non-lab and lab) and dAAR overestimated the risk, while BARD overestimated the risk at lower levels of observed risk (Fig. 3). The calibration of the BARD and dAAR score was slightly better for 5-year risk of CLD (Additional file 1: Fig. S2).
Fig. 2.
Calibration plots of 10-year HCC risk prediction models in CKB. Non-lab models and lab models are shown using different colors (blue for non-lab models and red for lab models). Observed to expected (O:E) ratio are shown in lower-right corner of each panel
Fig. 3.
Calibration plots of 10-year CLD risk prediction models in CKB. Convention as in Fig. 2
Meta-analysis of the model performance
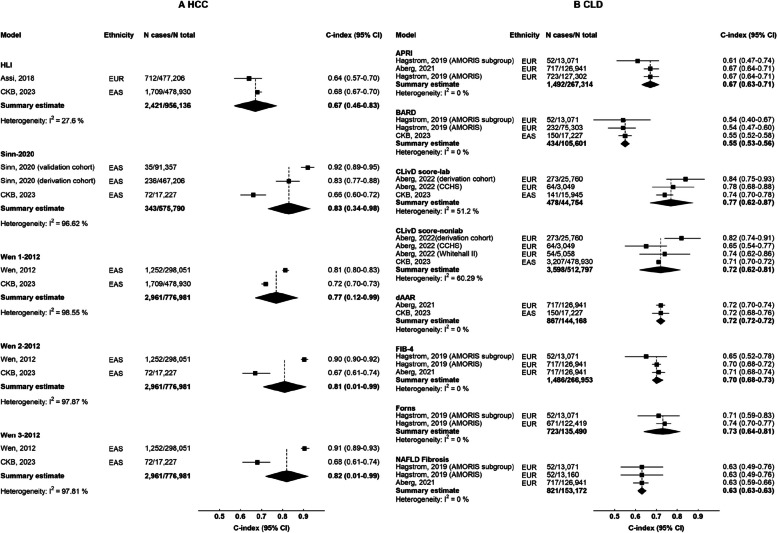
In the general population, there were 5 models for HCC and 8 models for CLD eligible for meta-analysis. For 10-year risk of HCC (Fig. 4A), Sinn-2020, Wen-1 2012, Wen-2 2021, and Wen-3 2012 showed good performance (C-index ≥ 0.70), albeit large heterogeneity between studies. For 10-year risk of CLD (Fig. 4B), CLivD score (lab and non-lab), dAAR, FIB-4, and Forns showed good performance (C-index ≥ 0.70). Model performance for 5-year and > 10-year are shown in Additional file 1: Fig. S3. Detailed results for all models are reported in Additional file 1: Fig. S4.
Fig. 4.
C-index of risk prediction models for HCC and CLD in meta-analysis of CKB and published studies. Boxes represent the C-index for predicting 10-year A HCC or B CLD in the general population. Diamonds represent summary C-index for each model, with the size of the diamond showing 95% confidence interval. For each model, published studies are sorted according to number of participants. Estimates and 95% CI of the summary C-index are shown in bold. The CIs of the summary estimates for HCC models were truncated because of the relatively low SE calculated using the “metamisc” package
In HBV infected individuals, there were 13 models for HCC and 2 models for CLD eligible for meta-analysis. The majority of models had C-index above 0.70 in previous external validation. The RWS-HCC, GAG-HCC, and REACH-B IIa models exhibited C-index ≥ 0.80 for 10-year HCC prediction, followed by mREACH-B model (C-index: 0.79–0.83 for < 10-year HCC prediction). FIB-4, Ishak fibrosis, PAGE-B, and PAGE-B + Ishak showed favorable discrimination (C-index ≥ 0.85), but they were only externally validated in one study (Additional file 1: Fig. S5). GAG-HCC, PAGE-B, and PAGE-B + Ishak also showed good discrimination (C-index ≥ 0.80) for predicting 10-year risk of CLD (Additional file 1: Fig. S6).
In HCV infected individuals, there were 5 models for HCC and 3 models for CLD eligible for meta-analysis, with FIB-4 showing the highest discrimination for HCC and CLD across all time frames (C-index 0.76–0.85 for < 5, 5, 5–10 years) (Additional file 1: Fig. S7). In NAFLD patients, there were 3 models for HCC and 1 model for CLD eligible for meta-analysis. LS-Based Model 2 and dAAR showed the highest discrimination for HCC and CLD (C-index 0.78 and 0.84), respectively (Additional file 1: Fig. S8).
For calibration, there was considerable variability in reporting across different studies, including (1) calibration plots (n = 13); (2) correlation coefficients between observed and predicted risks (n = 2); (3) the O:E ratio (n = 1, Kurosaki-2012 [32]); and (4) Brier scores (n = 1, An-2021 [70]). The majority of models (78/92) did not report calibration in external validation, and meta-analysis was therefore not feasible.
Discussion
We conducted a systematic review of CLD prediction models developed for different populations, meta-analyzed their performance, and independently validated selected models in the CKB cohort. Our findings showed substantial variation in model predictors and predictive performance. Lifestyle factors were the commonest predictors for the general population, while virological and biochemical markers were the commonest predictors for individuals with HBV, HCV, and NAFLD. A total of 13 models were included for external validation in the CKB cohort, showing distinct differences in discrimination, with C-index ranging from 0.55 to 0.96. Of all 11 models validated in the general population, Li-2018 and Wen 1–2012 had good discrimination for 10-year HCC, while CLivD score (non-lab and lab) and dAAR had the highest discrimination for 10-year CLD in CKB.
We summarized the best-performing models for 10-year risk of CLD by study populations according to the results of the meta-analysis and external validation in CKB. For the general population, Li-2018, Wen 1–2012, Wen 2–2012, Wen 3–2012, and Sinn-2020 models showed good discrimination for HCC, while CLivD score (non-lab and lab), dAAR, FIB-4, and Forns showed good discrimination for CLD. Additionally, our meta-analysis showed good discrimination of the Forns score for CLD, with higher C-index than other non-invasive scores for fibrosis (i.e., BARD, FIB-4, and APRI); however, validation in CKB was hindered by the lack of timely on-site testing of platelet count in large-scale cohort studies [84]. For individuals with diabetes, both the Li-2018 model and DM-HCC model showed good performance for HCC in the original development and CKB; however, the high C-index may be due to the limited number of CLD cases among diabetes patients in CKB. For HBV infected individuals, our meta-analysis highlighted the favorable performance of the RWS-HCC, REACH-B IIa, and GAG-HCC models. Only Cao-2021 and CAP-B models were externally validated in CKB, both showing good discrimination for CLD. For HCV infected individuals, NAFLD patients, and high-risk populations, there was a lack of adequate studies for model development and validation. Specifically, prediction models for HCV infected individuals lacked long-term prediction (> 10 years). FIB-4 showed the best discrimination for short- to medium-term risks of HCC and CLD. Prediction models for NAFLD patients relied heavily on transient elastography technology and had limited generalizability.
Although 9 out of 13 models showed slightly lower predictive performance in the CKB cohort compared to their previous development cohorts, our study findings supported the transportability of CLD models in Chinese adults by showing generally good performance. Indeed, the discrepancy in model performance between CKB and previous studies may be attributed to differences in the etiology of CLD and risk factor profiles, particularly between East Asians and Caucasians. The GBD 2019 study showed that the leading causes of CLD prevalence were NAFLD (82%) and hepatitis B (9%) in high Socio-Demographic Index (SDI) countries (primarily European and North American countries) and NAFLD (69%) and hepatitis B (26%) in China (Additional file 1: Table S8). Although previous studies in CKB reported lifestyle and metabolic risk factors for CLD similar to those in Western populations (Additional file 1: Table S9) [24, 85–88], the magnitude of associations differed for several risk factors (e.g., adiposity and physical activity). Although the predictive performance in CKB was relatively lower, we showed the transportability of risk prediction models developed in Western populations to the Chinese population. Of note, this favorable performance of CLD models was comparable to risk prediction models for cardiovascular disease (CVD) [89] and colorectal cancer (CRC) [90] in CKB. The WHO risk chart with non-laboratory data to predict 10-year CVD risk achieved a C-statistic of 0.75, where well-established models for CRC involving non-laboratory parameters had C-statistics between 0.65 and 0.70. Despite the different etiologies of CLD and magnitude of associations between primary risk factors and CLD, HLI, Li-2018, and BARD models performed better in the CKB cohort compared to its original development or validation cohorts. Risk prediction models incorporating combined risk factors for CLD, analogous to the Framingham risk score or PCE score used for CVD, would be promising to risk-stratify individuals before clinical onset of advanced liver disease.
Our study findings may have public health implications. Currently, EASL and AASLD recommend ultrasound screening for liver cancer among individuals diagnosed with cirrhosis, CHB, or CHC. In China, the screening criteria also encompass individuals with a family history of liver cancer, and serum AFP is used as an additional screening measure (Additional file 1: Table S10) [5–7]. However, these screening strategies are limited by the high number needed to screen and the reliance on secondary or tertiary healthcare centers. The international community of hepatologists has recommended screening for high-risk individuals by non-invasive tests that are widely available in primary healthcare settings, followed by second-line confirmatory tests (e.g., abdominal ultrasound). In this context, we showed feasibility of combining non-laboratory parameters and routinely measured liver biomarkers in risk stratification of CLD. Two models (Wen 1–2012 and CLivD score (non-lab)) were based on non-laboratory parameters and achieved C-index ≥ 0.70 in the general Chinese population, while CLivD score (lab) incorporated non-laboratory parameters plus GGT also had good performance. Such predictive models can enable early case-finding and individualized follow-up for adults at risk of liver disease in primary care and non-liver healthcare settings. They may also help prevent disease progression by facilitating timely interventions, such as weight loss or alcohol rehabilitation, thereby reducing the risk of severe liver conditions and associated mortality. Furthermore, these models provide valuable data on CLD risk to local policymakers and health authorities, helping the development of public health strategies.
Study limitations included uncertainty in risk estimates when dealing with relatively rare outcomes, especially among patients with diabetes. This may lead to overly optimistic estimation of discrimination. In addition, the eligibility criteria varied across the original development cohorts. Of note, the Sinn-2020 model for 10-year HCC prediction showed the highest discrimination in the meta-analysis but moderate performance in the CKB cohort. This discrepancy could be attributed to the fact that the Sinn-2020 model originated from a health screening cohort where volunteer selection bias might exist, potentially limiting the generalizability. Moreover, only 3 out of 13 models provided complete information to assess model performance, including regression coefficients and baseline survival rates. For this reason, a refitting approach was employed to evaluate calibration, which might lead to underestimation or overestimation because of the inherent limitations or biases in the study design and predictors of the original model. Lastly, the CKB involved 5 urban and 5 rural areas in China and was not nationally representative. However, the large sample size, the diversity of regions covered, the heterogeneity in exposures, and consistent findings from subgroup analyses suggest that our model’s performance results are largely generalizable to the broader Chinese population.
Conclusions
In conclusion, our meta-analysis and external validation in Chinese adults showed that several models had good discrimination and calibration with potential to identify high-risk populations for CLD, who would be referred to liver clinics for further assessment and be the target population of lifestyle modifications. Future studies are warranted to validate the performance of CLD prediction models in diverse populations and to assess the cost-effectiveness of screening strategies for CLD.
Supplementary Information
Additional file 1: Method S1 Search strategy. Method S2 China Kadoorie Biobank (CKB) information. Method S3 The measurement and definition of CKB predictor variables. Method S4 ICD-10 of CLD events in CKB. Method S5 Predictors and equations for included models. Table S1 Bias assessment. Table S2 Predictors of prediction models for general population. Table S3 Predictors of prediction models for HBV infected individuals. Table S4 Predictors of prediction models for other populations. Table S5 Sensitivity analysis for PLC and HCC. Table S6 CLD risk model for 5-year HCC discrimination in the published literature and CKB. Table S7 CLD risk model for 5-year CLD discrimination in the published literature and CKB. Table S8 CLD diseases and causes in high-income countries and China in 2019. Table S9 Risk factors (HR (95% CI)) for CLD in CKB and Western populations. Table S10 Summary of liver cancer screening strategies. Fig. S1 Calibration plots of 5-year HCC risk prediction models in the CKB. Fig. S2 Calibration plots of 5-year CLD risk prediction models in the CKB. Fig. S3 Discrimination of CLD risk prediction models in the published literature and CKB. Fig. S4 Discrimination of HCC risk prediction models in the general population. Fig. S5 Discrimination of HCC risk prediction models in the HBV infected individuals. Fig. S6 Discrimination of CLD risk prediction models in the HBV infected individuals. Fig. S7 Discrimination of CLD risk prediction models in the HCV infected individuals. Fig. S8 Discrimination of CLD risk prediction models in the NAFLD patients.
Acknowledgements
The chief acknowledgment is to the participants, the project staff, and the China National Centre for Disease Control and Prevention (CDC) and its regional offices for access to death and disease registries. The Chinese National Health Insurance scheme provided electronic linkage to all hospital admission data.
Abbreviations
- AASLD
American Association for the Study of Liver Diseases
- AFLD
Alcoholic fatty liver
- AFP
Alpha-fetoprotein
- ALD
Alcoholic liver disease
- ALT
Alanine aminotransferase
- Anti-HCV Ab
Anti-hepatitis C virus antibody
- AST
Aspartate aminotransferase
- AUC
Area under the ROC curve
- BMI
Body mass index
- CHB
Chronic hepatitis B
- CHC
Chronic hepatitis C
- CKB
China Kadoorie Biobank
- CLD
Chronic liver disease
- CRC
Colorectal cancer
- CVD
Cardiovascular disease
- EAS
East Asia
- EASL
European Association for the Study of the Liver
- EUR
Europe
- GBD
Global Burden of Disease Study
- GGT
Gamma-glutamyl transferase
- HBsAg
Hepatitis B surface antigen
- HBeAg
Hepatitis B e antigen
- HBV
Hepatitis B virus
- HCV
Hepatitis C virus
- HCC
Hepatocellular carcinoma
- ICD
International Classification of Diseases
- NAFLD
Non-alcoholic fatty liver disease
- PA
Physical activity
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
- PROBAST
Prediction model Risk Of Bias Assessment Tool
- ROC
Receiver operating characteristic
- SDI
Socio-Demographic Index
- SLD
Severe liver disease
- SRMA
Systematic review and meta-analysis
- T2D
Type 2 diabetes
- TG
Triglycerides
- TRIPOD
Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis
- WHR
Waist-to-hip ratio
Authors’ contributions
XC conducted the literature review, performed formal analysis, interpreted data, and wrote the original draft. SS and LY reviewed and edited the manuscript. KS and CM were responsible for data extraction, while YC1 handled literature screening. JL, CY, DS, PP, LY, and YC2 contributed to conceptualization, data curation, and reviewed and edited the manuscript. SW collected data and performed data curation. XY and RS contributed to conceptualization and reviewed and edited the manuscript. LL and ZC had full access to the data. YP, CK, LL, and ZC conducted data analysis and are responsible for accuracy of the results and the decision to submit for publication. All authors were involved in study design, conduct, long-term follow-up, review and coding of disease events, interpretation of the results, or writing the report. All authors have read and approved the final version of the manuscript.
Funding
This work was supported by the National Key R&D Program of China (2023YFC3606300) and the National Natural Science Foundation of China (82304223, 82192904, 82192901, 82192900). The CKB baseline survey and the first re-survey were supported by a grant from the Kadoorie Charitable Foundation in Hong Kong. The long-term follow-up is supported by grants from the UK Wellcome Trust (212946/Z/18/Z, 202922/Z/16/Z, 104085/Z/14/Z, 088158/Z/09/Z), grants (2016YFC0900500) from the National Key R&D Program of China, National Natural Science Foundation of China (81390540, 91846303, 81941018), and Chinese Ministry of Science and Technology (2011BAI09B01). The funders had no role in the study design, data collection, data analysis and interpretation, writing of the report, or the decision to submit the article for publication.
Data availability
The materials are available upon request; some restrictions will apply.
Declarations
Ethics approval and consent to participate
Central ethical approvals were obtained from Oxford University and the China National CDC. Approvals were also obtained from institutional research boards at the local CDCs in the ten areas: Qingdao, Qingdao CDC; Heilongjiang, Provincial CDC; Hainan, Provincial CDC; Jiangsu, Provincial CDC; Guangxi, Provincial CDC; Sichuan, Provincial CDC; Gansu, Provincial CDC; Henan, Provincial CDC; Zhejiang, Provincial CDC; and Hunan, Provincial CDC. The study protocol of the resurvey was approved by the Peking University Institutional Review Board (No. IRB00001052-20040), and separate written informed consent was obtained from all participants before accelerometer data collection.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Christiana Kartsonaki and Yuanjie Pang contributed equally to this work.
Contributor Information
Christiana Kartsonaki, Email: christiana.kartsonaki@dph.ox.ac.uk.
Yuanjie Pang, Email: yuanjie.p@gmail.com.
References
- 1.Global incidence, prevalence, years lived with disability (YLDs), disability-adjusted life-years (DALYs), and healthy life expectancy (HALE) for 371 diseases and injuries in 204 countries and territories and 811 subnational locations, 1990–2021: a systematic analysis for the Global Burden of Disease Study 2021. Lancet (London, England). 2024;403(10440):2133–61. https://pubmed.ncbi.nlm.nih.gov/38642570/. [DOI] [PMC free article] [PubMed]
- 2.Wang MM, Wang GS, Shen F, Chen GY, Pan Q, Fan JG. Hepatic steatosis is highly prevalent in hepatitis B patients and negatively associated with virological factors. Dig Dis Sci. 2014;59(10):2571–9. [DOI] [PubMed] [Google Scholar]
- 3.Singal AG, Zhang E, Narasimman M, Rich NE, Waljee AK, Hoshida Y, et al. HCC surveillance improves early detection, curative treatment receipt, and survival in patients with cirrhosis: a meta-analysis. J Hepatol. 2022;77(1):128–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chalmers J, Wilkes E, Harris R, Kent L, Kinra S, Aithal GP, et al. The development and implementation of a commissioned pathway for the identification and stratification of liver disease in the community. Frontline Gastroenterol. 2020;11(2):86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singal AG, Llovet JM, Yarchoan M, Mehta N, Heimbach JK, Dawson LA, et al. AASLD practice guidance on prevention, diagnosis, and treatment of hepatocellular carcinoma. Hepatology. 2023;78:1922–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.EASL clinical practice guidelines. management of hepatocellular carcinoma. J Hepatol. 2018;69(1):182–236. [DOI] [PubMed] [Google Scholar]
- 7.[Guideline for stratified screening and surveillance of primary liver cancer (2020 edition)]. Zhonghua Gan Zang Bing Za Zhi. 2021;29(1):25–40. https://pubmed.ncbi.nlm.nih.gov/33541021/. [DOI] [PubMed]
- 8.Parikh ND, Tayob N, Singal AG. Blood-based biomarkers for hepatocellular carcinoma screening: approaching the end of the ultrasound era? J Hepatol. 2023;78(1):207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tzartzeva K, Obi J, Rich NE, Parikh ND, Marrero JA, Yopp A, et al. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: a meta-analysis. Gastroenterology. 2018;154(6):1706-18.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anstee QM, Castera L, Loomba R. Impact of non-invasive biomarkers on hepatology practice: past, present and future. J Hepatol. 2022;76(6):1362–78. [DOI] [PubMed] [Google Scholar]
- 11.Sterling RK, Lissen E, Clumeck N, Sola R, Correa MC, Montaner J, et al. Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology. 2006;43(6):1317–25. [DOI] [PubMed] [Google Scholar]
- 12.Wai CT, Greenson JK, Fontana RJ, Kalbfleisch JD, Marrero JA, Conjeevaram HS, et al. A simple noninvasive index can predict both significant fibrosis and cirrhosis in patients with chronic hepatitis C. Hepatology. 2003;38(2):518–26. [DOI] [PubMed] [Google Scholar]
- 13.Innes H, Morling JR, Buch S, Hamill V, Stickel F, Guha IN. Performance of routine risk scores for predicting cirrhosis-related morbidity in the community. J Hepatol. 2022;77(2):365–76. [DOI] [PubMed] [Google Scholar]
- 14.Armstrong MJ, Schmidt-Martin D, Rowe IA, Newsome PN. Caution in using non-invasive scoring systems in NAFLD beyond highly selected study populations. Am J Gastroenterol. 2017;112(4):653–4. [DOI] [PubMed] [Google Scholar]
- 15.Caballería L, Pera G, Arteaga I, Rodríguez L, Alumà A, Morillas RM, et al. High prevalence of liver fibrosis among European adults with unknown liver disease: a population-based study. Clin Gastroenterol Hepatol. 2018;16(7):1138-45.e5. [DOI] [PubMed] [Google Scholar]
- 16.Moons KGM, Wolff RF, Riley RD, Whiting PF, Westwood M, Collins GS, et al. PROBAST: a tool to assess risk of bias and applicability of prediction model studies: explanation and elaboration. Ann Intern Med. 2019;170(1):W1-w33. [DOI] [PubMed] [Google Scholar]
- 17.Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JP, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339:b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snell KIE, Levis B, Damen JAA, Dhiman P, Debray TPA, Hooft L, et al. Transparent reporting of multivariable prediction models for individual prognosis or diagnosis: checklist for systematic reviews and meta-analyses (TRIPOD-SRMA). BMJ. 2023;381: e073538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ. 2015;350:g7594. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z, Chen J, Collins R, Guo Y, Peto R, Wu F, Li L. China Kadoorie Biobank of 0.5 million people: survey methods, baseline characteristics and long-term follow-up. Int J Epidemiol. 2011;40(6):1652–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang G, Rao C, Ma J, et al. Validation of verbal autopsy procedures for adult deaths in China. Int J Epidemiol. 2006;35(3):741–8. [DOI] [PubMed] [Google Scholar]
- 22.Åberg F, Luukkonen PK, But A, Salomaa V, Britton A, Petersen KM, et al. Development and validation of a model to predict incident chronic liver disease in the general population: the CLivD score. J Hepatol. 2022;77(2):302–11. [DOI] [PubMed] [Google Scholar]
- 23.Emdin CA, Haas M, Ajmera V, Simon TG, Homburger J, Neben C, et al. Association of genetic variation with cirrhosis: a multi-trait genome-wide association and gene-environment interaction study. Gastroenterology. 2021;160(5):1620-33.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pang Y, Kartsonaki C, Turnbull I, Guo Y, Clarke R, Chen Y, et al. Diabetes, plasma glucose, and incidence of fatty liver, cirrhosis, and liver cancer: a prospective study of 0.5 million people. Hepatology. 2018;68(4):1308–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Calster B, McLernon DJ, van Smeden M, Wynants L, Steyerberg EW. Calibration: the Achilles heel of predictive analytics. BMC Med. 2019;17(1):230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Debray TP, Damen JA, Riley RD, Snell K, Reitsma JB, Hooft L, et al. A framework for meta-analysis of prediction model studies with binary and time-to-event outcomes. Stat Methods Med Res. 2019;28(9):2768–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerds TA, Cai T, Schumacher M. The performance of risk prediction models. Biom J. 2008;50(4):457–79. [DOI] [PubMed] [Google Scholar]
- 28.Yuen MF, Tanaka Y, Fong DY, Fung J, Wong DK, Yuen JC, et al. Independent risk factors and predictive score for the development of hepatocellular carcinoma in chronic hepatitis B. J Hepatol. 2009;50(1):80–8. [DOI] [PubMed] [Google Scholar]
- 29.Wong VW, Chan SL, Mo F, Chan TC, Loong HH, Wong GL, et al. Clinical scoring system to predict hepatocellular carcinoma in chronic hepatitis B carriers. J Clin Oncol. 2010;28(10):1660–5. [DOI] [PubMed] [Google Scholar]
- 30.Yang HI, Sherman M, Su J, Chen PJ, Liaw YF, Iloeje UH, et al. Nomograms for risk of hepatocellular carcinoma in patients with chronic hepatitis B virus infection. J Clin Oncol. 2010;28(14):2437–44. [DOI] [PubMed] [Google Scholar]
- 31.Yang HI, Yuen MF, Chan HL, Han KH, Chen PJ, Kim DY, et al. Risk estimation for hepatocellular carcinoma in chronic hepatitis B (REACH-B): development and validation of a predictive score. Lancet Oncol. 2011;12(6):568–74. [DOI] [PubMed] [Google Scholar]
- 32.Kurosaki M, Hiramatsu N, Sakamoto M, Suzuki Y, Iwasaki M, Tamori A, et al. Data mining model using simple and readily available factors could identify patients at high risk for hepatocellular carcinoma in chronic hepatitis C. J Hepatol. 2012;56(3):602–8. [DOI] [PubMed] [Google Scholar]
- 33.Michikawa T, Inoue M, Sawada N, Iwasaki M, Tanaka Y, Shimazu T, et al. Development of a prediction model for 10-year risk of hepatocellular carcinoma in middle-aged Japanese: the Japan Public Health Center-based Prospective Study Cohort II. Prev Med. 2012;55(2):137–43. [DOI] [PubMed] [Google Scholar]
- 34.Wen CP, Lin J, Yang YC, Tsai MK, Tsao CK, Etzel C, et al. Hepatocellular carcinoma risk prediction model for the general population: the predictive power of transaminases. J Natl Cancer Inst. 2012;104(20):1599–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gavilán JC, Ojeda G, Arnedo R, Puerta S. Predictive factors of risk of hepatocellular carcinoma in chronic hepatitis C. Eur J Intern Med. 2013;24(8):846–51. [DOI] [PubMed] [Google Scholar]
- 36.Kim DY, Song KJ, Kim SU, Yoo EJ, Park JY, Ahn SH, et al. Transient elastography-based risk estimation of hepatitis B virus-related occurrence of hepatocellular carcinoma: development and validation of a predictive model. Onco Targets Ther. 2013;6:1463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee MH, Yang HI, Liu J, Batrla-Utermann R, Jen CL, Iloeje UH, et al. Prediction models of long-term cirrhosis and hepatocellular carcinoma risk in chronic hepatitis B patients: risk scores integrating host and virus profiles. Hepatology. 2013;58(2):546–54. [DOI] [PubMed] [Google Scholar]
- 38.Lin YJ, Lee MH, Yang HI, Jen CL, You SL, Wang LY, et al. Predictability of liver-related seromarkers for the risk of hepatocellular carcinoma in chronic hepatitis B patients. PLoS ONE. 2013;8(4):e61448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee MH, Lu SN, Yuan Y, Yang HI, Jen CL, You SL, et al. Development and validation of a clinical scoring system for predicting risk of HCC in asymptomatic individuals seropositive for anti-HCV antibodies. PLoS One. 2014;9(5):e94760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong GL, Chan HL, Wong CK, Leung C, Chan CY, Ho PP, et al. Liver stiffness-based optimization of hepatocellular carcinoma risk score in patients with chronic hepatitis B. J Hepatol. 2014;60(2):339–45. [DOI] [PubMed] [Google Scholar]
- 41.Jung KS, Kim SU, Song K, Park JY, Kim DY, Ahn SH, et al. Validation of hepatitis B virus-related hepatocellular carcinoma prediction models in the era of antiviral therapy. Hepatology. 2015;62(6):1757–66. [DOI] [PubMed] [Google Scholar]
- 42.Shin SH, Kim SU, Park JY, Kim DY, Ahn SH, Han KH, et al. Liver stiffness-based model for prediction of hepatocellular carcinoma in chronic hepatitis B virus infection: comparison with histological fibrosis. Liver Int. 2015;35(3):1054–62. [DOI] [PubMed] [Google Scholar]
- 43.Suh B, Park S, Shin DW, Yun JM, Yang HK, Yu SJ, et al. High liver fibrosis index FIB-4 is highly predictive of hepatocellular carcinoma in chronic hepatitis B carriers. Hepatology. 2015;61(4):1261–8. [DOI] [PubMed] [Google Scholar]
- 44.Abu-Amara M, Cerocchi O, Malhi G, Sharma S, Yim C, Shah H, et al. The applicability of hepatocellular carcinoma risk prediction scores in a North American patient population with chronic hepatitis B infection. Gut. 2016;65(8):1347–58. [DOI] [PubMed] [Google Scholar]
- 45.Duarte-Salles T, Misra S, Stepien M, Plymoth A, Muller D, Overvad K, et al. Circulating osteopontin and prediction of hepatocellular carcinoma development in a large European population. Cancer Prev Res (Phila). 2016;9(9):758–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poh Z, Shen L, Yang HI, Seto WK, Wong VW, Lin CY, et al. Real-world risk score for hepatocellular carcinoma (RWS-HCC): a clinically practical risk predictor for HCC in chronic hepatitis B. Gut. 2016;65(5):887–8. [DOI] [PubMed] [Google Scholar]
- 47.Rau HH, Hsu CY, Lin YA, Atique S, Fuad A, Wei LM, et al. Development of a web-based liver cancer prediction model for type II diabetes patients by using an artificial neural network. Comput Methods Programs Biomed. 2016;125:58–65. [DOI] [PubMed] [Google Scholar]
- 48.Si WK, Chung JW, Cho J, Baeg JY, Jang ES, Yoon H, et al. Predictors of increased risk of hepatocellular carcinoma in patients with type 2 diabetes. PLoS One. 2016;11(6):e0158066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang HI, Tseng TC, Liu J, Lee MH, Liu CJ, Su TH, et al. Incorporating serum level of hepatitis B surface antigen or omitting level of hepatitis B virus DNA does not affect calculation of risk for hepatocellular carcinoma in patients without cirrhosis. Clin Gastroenterol Hepatol. 2016;14(3):461-8.e2. [DOI] [PubMed] [Google Scholar]
- 50.Brouwer WP, van der Meer AJP, Boonstra A, Plompen EPC, Pas SD, de Knegt RJ, et al. Prediction of long-term clinical outcome in a diverse chronic hepatitis B population: role of the PAGE-B score. J Viral Hepat. 2017;24(11):1023–31. [DOI] [PubMed] [Google Scholar]
- 51.Butt AA, Ren Y, Lo Re V 3rd, Taddei TH, Kaplan DE. Comparing Child-Pugh, MELD, and FIB-4 to predict clinical outcomes in hepatitis C virus-infected persons: results from ERCHIVES. Clin Infect Dis. 2017;65(1):64–72. [DOI] [PubMed] [Google Scholar]
- 52.Daheim M, Lang S, Goeser T, Steffen HM, Demir M. Real-world risk score for hepatocellular carcinoma risk prediction in CHBV: a validation outside of Asia. Gut. 2017;66(7):1346–7. [DOI] [PubMed] [Google Scholar]
- 53.Kao WY, Yang SH, Liu WJ, Yeh MY, Lin CL, Liu CJ, et al. Genome-wide identification of blood DNA methylation patterns associated with early-onset hepatocellular carcinoma development in hepatitis B carriers. Mol Carcinog. 2017;56(2):425–35. [DOI] [PubMed] [Google Scholar]
- 54.Konerman MA, Lu D, Zhang Y, Thomson M, Zhu J, Verma A, et al. Assessing risk of fibrosis progression and liver-related clinical outcomes among patients with both early stage and advanced chronic hepatitis C. PLoS One. 2017;12(11):e0187344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seo YS, Jang BK, Um SH, Hwang JS, Han KH, Kim SG, et al. Validation of risk prediction models for the development of HBV-related HCC: a retrospective multi-center 10-year follow-up cohort study. Oncotarget. 2017;8(68):113213–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sinn DH, Lee JH, Kim K, Ahn JH, Lee JH, Kim JH, et al. A novel model for predicting hepatocellular carcinoma development in patients with chronic hepatitis B and normal alanine aminotransferase levels. Gut Liver. 2017;11(4):528–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tseng TC, Liu CJ, Su TH, Yang WT, Chen CL, Yang HC, et al. Fibrosis-4 index helps identify HBV carriers with the lowest risk of hepatocellular carcinoma. Am J Gastroenterol. 2017;112(10):1564–74. [DOI] [PubMed] [Google Scholar]
- 58.Assi N, Gunter MJ, Thomas DC, Leitzmann M, Stepien M, Chajès V, et al. Metabolic signature of healthy lifestyle and its relation with risk of hepatocellular carcinoma in a large European cohort. Am J Clin Nutr. 2018;108(1):117–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jeon MY, Lee HW, Kim SU, Kim BK, Park JY, Kim DY, et al. Feasibility of dynamic risk prediction for hepatocellular carcinoma development in patients with chronic hepatitis B. Liver Int. 2018;38(4):676–86. [DOI] [PubMed] [Google Scholar]
- 60.Li TC, Li CI, Liu CS, Lin WY, Lin CH, Yang SY, et al. Risk score system for the prediction of hepatocellular carcinoma in patients with type 2 diabetes: Taiwan Diabetes Study. Semin Oncol. 2018;45(5–6):264–74. [DOI] [PubMed] [Google Scholar]
- 61.Paik N, Sinn DH, Lee JH, Oh IS, Kim JH, Kang W, et al. Non-invasive tests for liver disease severity and the hepatocellular carcinoma risk in chronic hepatitis B patients with low-level viremia. Liver Int. 2018;38(1):68–75. [DOI] [PubMed] [Google Scholar]
- 62.Fan C, Li M, Gan Y, Chen T, Sun Y, Lu J, et al. A simple AGED score for risk classification of primary liver cancer: development and validation with long-term prospective HBsAg-positive cohorts in Qidong. China Gut. 2019;68(5):948–9. [DOI] [PubMed] [Google Scholar]
- 63.Hagström H, Nasr P, Ekstedt M, Stål P, Hultcrantz R, Kechagias S. Accuracy of noninvasive scoring systems in assessing risk of death and liver-related endpoints in patients with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2019;17(6):1148-56.e4. [DOI] [PubMed] [Google Scholar]
- 64.Konerman MA, Beste LA, Van T, Liu B, Zhang X, Zhu J, et al. Machine learning models to predict disease progression among veterans with hepatitis C virus. PLoS One. 2019;14(1):e0208141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poynard T, Peta V, Deckmyn O, Munteanu M, Moussalli J, Ngo Y, et al. LCR1 and LCR2, two multi-analyte blood tests to assess liver cancer risk in patients without or with cirrhosis. Aliment Pharmacol Ther. 2019;49(3):308–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sinn DH, Kim SE, Kim BK, Kim JH, Choi MS. The risk of hepatocellular carcinoma among chronic hepatitis B virus-infected patients outside current treatment criteria. J Viral Hepat. 2019;26(12):1465–72. [DOI] [PubMed] [Google Scholar]
- 67.Hagström H, Talbäck M, Andreasson A, Walldius G, Hammar N. Ability of noninvasive scoring systems to identify individuals in the population at risk for severe liver disease. Gastroenterology. 2020;158(1):200–14. [DOI] [PubMed] [Google Scholar]
- 68.Sinn DH, Kang D, Cho SJ, Paik SW, Guallar E, Cho J, et al. Risk of hepatocellular carcinoma in individuals without traditional risk factors: development and validation of a novel risk score. Int J Epidemiol. 2020;49(5):1562–71. [DOI] [PubMed] [Google Scholar]
- 69.Åberg F, Danford CJ, Thiele M, Talbäck M, Rasmussen DN, Jiang ZG, et al. A dynamic aspartate-to-alanine aminotransferase ratio provides valid predictions of incident severe liver disease. Hepatol Commun. 2021;5(6):1021–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.An C, Choi JW, Lee HS, Lim H, Ryu SJ, Chang JH, et al. Prediction of the risk of developing hepatocellular carcinoma in health screening examinees: a Korean cohort study. BMC Cancer. 2021;21(1):755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cao M, Li H, Sun D, He S, Xia C, Lei L, et al. A primary screening method for liver cancer in chronic hepatitis B carriers: a prospective community-based cohort study. Front Oncol. 2021;11:762662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kamalapirat T, Yingcharoen K, Ungtrakul T, Soonklang K, Dechma J, Chunnuan P, et al. Assessing risk scores for predicting hepatocellular carcinoma in Thai patients with chronic hepatitis B. J Viral Hepat. 2021;28(7):1034–41. [DOI] [PubMed] [Google Scholar]
- 73.Kang N, Chung JW, Jang ES, Jeong SH, Kim JW. Computed tomography-measured liver volume predicts the risk of hepatocellular carcinoma development in chronic hepatitis C patients. Dig Dis Sci. 2021;66(12):4536–44. [DOI] [PubMed] [Google Scholar]
- 74.Le AK, Yang HI, Yeh ML, Jin M, Trinh HN, Henry L, et al. Development and validation of a risk score for liver cirrhosis prediction in untreated and treated chronic hepatitis B. J Infect Dis. 2021;223(1):139–46. [DOI] [PubMed] [Google Scholar]
- 75.Lee JS, Sinn DH, Park SY, Shin HJ, Lee HW, Kim BK, et al. Liver stiffness-based risk prediction model for hepatocellular carcinoma in patients with nonalcoholic fatty liver disease. Cancers (Basel). 2021;13(18):4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liang LY, Lee HW, Wong VW, Yip TC, Tse YK, Hui VW, et al. Serum fibrosis index-based risk score predicts hepatocellular carcinoma in untreated patients with chronic hepatitis B. Clin Mol Hepatol. 2021;27(3):499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Poynard T, Lacombe JM, Deckmyn O, Peta V, Akhavan S, de Ledinghen V, et al. External validation of LCR1-LCR2, a multivariable HCC risk calculator, in patients with chronic HCV. JHEP Rep. 2021;3(4):100298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Y, Wang M, Li H, Chen K, Zeng H, Bi X, et al. A male-ABCD algorithm for hepatocellular carcinoma risk prediction in HBsAg carriers. Chin J Cancer Res. 2021;33(3):352–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu C, Song C, Lv J, Zhu M, Yu C, Guo Y, et al. Prediction and clinical utility of a liver cancer risk model in Chinese adults: a prospective cohort study of 0.5 million people. Int J Cancer. 2021;148(12):2924–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Costa APM, da Silva M, Castro RS, Sampaio ALO, Alencar Júnior AM, da Silva MC, et al. PAGE-B and REACH-B predicts the risk of developing hepatocellular carcinoma in chronic hepatitis B patients from Northeast, Brazil. Viruses. 2022;14(4):732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jo AJ, Choi WM, Kim HJ, Choi SH, Han S, Ko MJ, et al. A risk scoring system to predict clinical events in chronic hepatitis B virus infection: a nationwide cohort study. J Viral Hepat. 2022;29(2):115–23. [DOI] [PubMed] [Google Scholar]
- 82.Johnson PJ, Innes H, Hughes DM, Kalyuzhnyy A, Kumada T, Toyoda H. Evaluation of the aMAP score for hepatocellular carcinoma surveillance: a realistic opportunity to risk stratify. Br J Cancer. 2022;127(7):1263–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thomas J, Liao LM, Sinha R, Patel T, Antwi SO. Hepatocellular carcinoma risk prediction in the NIH-AARP diet and health study cohort: a machine learning approach. J Hepatocell Carcinoma. 2022;9:69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rory C. UK Biobank: protocol for a large-scale prospective epidemiological resource. United Kingdom: UK Biobank; 2007. [Google Scholar]
- 85.Im PK, Wright N, Yang L, Chan KH, Chen Y, Guo Y, et al. Alcohol consumption and risks of more than 200 diseases in Chinese men. Nat Med. 2023;29(6):1476–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Im PK, Millwood IY, Kartsonaki C, Guo Y, Chen Y, Turnbull I, et al. Alcohol drinking and risks of liver cancer and non-neoplastic chronic liver diseases in China: a 10-year prospective study of 0.5 million adults. BMC Med. 2021;19(1):216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pang Y, Kartsonaki C, Guo Y, Chen Y, Yang L, Bian Z, et al. Central adiposity in relation to risk of liver cancer in Chinese adults: a prospective study of 0.5 million people. Int J Cancer. 2019;145(5):1245–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chan KH, Wright N, Xiao D, Guo Y, Chen Y, Du H, et al. Tobacco smoking and risks of more than 470 diseases in China: a prospective cohort study. Lancet Public Health. 2022;7(12):e1014–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang S, Han Y, Yu C, Guo Y, Pang Y, Sun D, et al. Development of a model to predict 10-year risk of ischemic and hemorrhagic stroke and ischemic heart disease using the China Kadoorie Biobank. Neurology. 2022;98(23):e2307–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Abhari RE, Thomson B, Yang L, Millwood I, Guo Y, Yang X, et al. External validation of models for predicting risk of colorectal cancer using the China Kadoorie Biobank. BMC Med. 2022;20(1):302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Method S1 Search strategy. Method S2 China Kadoorie Biobank (CKB) information. Method S3 The measurement and definition of CKB predictor variables. Method S4 ICD-10 of CLD events in CKB. Method S5 Predictors and equations for included models. Table S1 Bias assessment. Table S2 Predictors of prediction models for general population. Table S3 Predictors of prediction models for HBV infected individuals. Table S4 Predictors of prediction models for other populations. Table S5 Sensitivity analysis for PLC and HCC. Table S6 CLD risk model for 5-year HCC discrimination in the published literature and CKB. Table S7 CLD risk model for 5-year CLD discrimination in the published literature and CKB. Table S8 CLD diseases and causes in high-income countries and China in 2019. Table S9 Risk factors (HR (95% CI)) for CLD in CKB and Western populations. Table S10 Summary of liver cancer screening strategies. Fig. S1 Calibration plots of 5-year HCC risk prediction models in the CKB. Fig. S2 Calibration plots of 5-year CLD risk prediction models in the CKB. Fig. S3 Discrimination of CLD risk prediction models in the published literature and CKB. Fig. S4 Discrimination of HCC risk prediction models in the general population. Fig. S5 Discrimination of HCC risk prediction models in the HBV infected individuals. Fig. S6 Discrimination of CLD risk prediction models in the HBV infected individuals. Fig. S7 Discrimination of CLD risk prediction models in the HCV infected individuals. Fig. S8 Discrimination of CLD risk prediction models in the NAFLD patients.
Data Availability Statement
The materials are available upon request; some restrictions will apply.